Abstract
Helicase/nucleoside triphosphatase (NTPase) motifs have been identified in many RNA virus genomes. Similarly, all the members of the Flaviviridae family contain conserved helicase/NTPase motifs in their homologous NS3 proteins. Although this suggests that this activity plays a critical role in the viral life cycle, the precise role of the helicase/NTPase in virus replication or whether it is essential for virus replication is still unknown. To determine the role of the NS3 helicase/NTPase in the viral life cycle, deletion and point mutations in the helicase/NTPase motifs of the bovine viral diarrhea virus (BVDV) (NADL strain) NS3 protein designed to abolish either helicase activity alone (motif II, DEYH to DEYA) or both NTPase and helicase activity (motif I, GKT to GAT and deletion of motif VI) were generated. The C-terminal domain of NS3 (BVDV amino acids 1854 to 2362) of these mutants and wild type was expressed in bacteria, purified, and assayed for RNA helicase and ATPase activity. These mutations behaved as predicted with respect to RNA helicase and NTPase activities in vitro. When engineered back into an infectious cDNA for BVDV (NADL strain), point mutations in either the GKT or DEYH motif or deletion of motif VI yielded RNA transcripts that no longer produced infectious virus upon transfection of EBTr cells. Further analysis indicated that these mutants did not synthesize minus-strand RNA. These findings represent the first report unequivocably demonstrating that helicase activity is essential for minus-strand synthesis.
The Flaviviridae family is comprised of three genera, Flavivirus (such as Yellow fever virus and Dengue virus types 1 to 4), Hepacivirus (such as Hepatitis C virus [HCV]), and Pestivirus (such as Bovine viral diarrhea virus [BVDV]) (28). BVDV infection represents an economically important disease of cattle, and BVDV has been identified as the causative agent of viral diarrhea-mucosal disease (reviewed in references 1, 12, 25, and 37). Like the other members of the Flaviviridae, BVDV is an enveloped, plus-stranded RNA virus whose genome consists of a nonsegmented single-stranded RNA molecule. BVDV genomic RNA is approximately 12.5 kb and encodes a single open reading frame of approximately 3,900 amino acids (7–9, 24). The polyprotein translated from the open reading frame is subsequently processed by virally encoded and cellular proteases into 12 individual proteins (13, 30, 31, 35, 44). These individual proteins function either as structural components of the virion or presumably, at least in part, as components of the viral RNA replicase complex as described for other Flaviviridae family members (2, 6, 17). RNA replicons derived from defective interfering particles have shown that the 5′ and 3′ nontranslated regions (NTRs) along with the nonstructural proteins NS3, NS4A, NS4B, NS5A, and NS5B can support RNA replication (4, 45). However, the essentiality of the individual nonstructural proteins has not yet been tested.
The NS5B proteins of both BVDV (46) and the related HCV (3, 23) display RNA-dependent RNA polymerase (RdRp) activities. In addition to the RdRp, the NS3 proteins of several other members of the Flaviviridae family have been shown to possess nucleoside triphosphatase (NTPase) activity (10, 18–20, 32–34, 40, 42, 43). The BVDV NS3 protein (p80) has been shown previously to encode both NTPase (32, 34) and RNA helicase activities (41). These activities localize to the carboxy terminus of NS3, which contains canonical amino acid motifs present in all superfamily II RNA helicases (14, 16).
Although many plus-strand RNA viruses encode proteins either postulated or demonstrated to have RNA helicase activity (reviewed in reference 16), the precise function of these helicases in viral RNA replication remains unclear. Possibly, the RNA helicase could act during the initiation of minus-strand template synthesis by unwinding the secondary structures, such as those present in the 3′ NTR of the viral genomic RNA (11, 45), thus allowing initiation by the RdRp. The RNA helicase could also unwind secondary structures within the genomic RNA, facilitating RdRp processivity during both minus- and plus-strand synthesis. Alternatively, the RNA helicase could aid in the release of nascent genomic plus strands from the minus-strand template, thereby allowing their packaging into the progeny virions. Finally, the RNA helicase could somehow assist in the formation of progeny virions by unwinding, and thus exposing, key RNA elements necessary for recognition by the viral RNA packaging complex.
To begin to address the biological role of the NS3 NTPase/RNA helicase activity in the BVDV life cycle, we cloned and expressed the C terminus of BVDV NS3 in bacteria. This protein contains both RNA helicase and NTPase activity. We then made a series of mutations in the conserved helicase motifs designed to eliminate either the helicase activity alone or the NTPase/RNA helicase activities. These mutant proteins were expressed, purified, and assayed for ATPase and RNA helicase activities. These same mutations were then individually transferred back into the full-length genomic cDNA clone of a cytopathic BVDV strain (NADL strain) that yields infectious RNA transcripts (38), and their effects upon virus production were examined.
MATERIALS AND METHODS
Cells and viruses.
BVDV-free MDBK cells (CCL 22) and EBTr (embryonic bovine tracheal) cells (CCL-44) were obtained from the American Type Culture Collection and propagated in Dulbecco's modified minimal essential medium supplemented with penicillin (500 U/ml), streptomycin (500 U/ml), and 10% horse serum. Cells were maintained in a humidified incubator at 37°C with 5% CO2.
BVDV (NADL strain) was obtained from the American Type Culture Collection, plaque purified, and amplified in MDBK cells. For infections, virus inoculum was added in complete medium and adsorbed for 1 h at 37°C, and the inoculum was removed and replaced with fresh medium. Cultures were then incubated at 37°C for 48 h or until cytopathic effects (CPE) were observed. Virus stocks were prepared by freeze-thawing the infected cells and culture supernatant three times followed by centrifugation at 1,000 × g for 5 min. Stock titers were determined, and stocks were aliquoted and stored at −80°C.
BVDV plaque assays.
MDBK cells were seeded into six-well plates at a density of 2.5 × 105 cells per well. Twenty-four hours later, the cells were infected with 10-fold dilutions of virus. After adsorption for 1 h at 37°C, the inoculum was removed, and cells were overlaid with medium containing 1.5% SeaPlaque GTG agarose (FMC Bioproducts) and incubated at 37°C for 3 days or until plaques were visible. Agarose plugs were removed, and plaques were visualized by staining with crystal violet in 70% methanol for 15 min.
Plasmid constructs and site-directed mutagenesis.
Plasmid pVVNADL was used as the parental plasmid for all cloning and was generously provided by Ruben Donis (University of Nebraska). pVVNADL contains a full-length genomic cDNA clone of BVDV (NADL strain) capable of generating RNA transcripts which are infectious in vivo (38). Nucleotides encoding the helicase/NTPase domain of NS3 (amino acids 1853 to 2362) were amplified using PCR with primers which added BglII and HindIII sites to the 5′ and 3′ ends, respectively. The BglII-to-HindIII PCR fragment was then cloned into the BamHI and HindIII sites of pET-21b(+) (Novagen) to create pET-21bHB.
pACCX, a derivative of pACYC177, was constructed by adding a polylinker sequence containing cleavage sites for the restriction enzymes ClaI, SalI, XmaI, ApaLI, and XbaI into AatII- and StuI-digested pACYC177 (5a). The BVDV complete genomic cDNA from pVVNADL was cloned into pACCX in three steps. First, the 5.6-kb ClaI-SalI fragment of the 5′ ends of the BVDV genome was cloned into the ClaI- and SalI-digested pACCX to generate pACCX5.6. Then the SalI-XbaI fragment containing the 3′ end of the genome was cloned into pACCX5.6 to create pACCX5.6+2.7. Finally, the BVDV internal 4.3-kb SalI fragment was ligated to SalI-digested pACCX5.6+2.7 to generate pACCX-BVDV. The polylinker in pACCX is positioned such that the BVDV genomic cDNA was oriented in an opposite direction from that of transcription of the ampicillin resistance gene. The 4.3-kb SalI fragment of pVVNADL was also cloned into the SalI site of pUC19 to generate pSal. For mutagenesis, the BglII-to-Tth111I fragment of BVDV containing the NS3 region from pSal was subcloned into pUC19 to generate pD2.2. PCR-based mutagenesis was performed using pD2.2 as a template and the QuickChange site-directed mutagenesis kit (Stratagene) as per the manufacturer's instructions. To create the GKT-to-GAT mutation, the following oligonucleotides were used: 5′-CTTTGGCAACAGGGGCAGGCGCCACCACAGAACTCCCAAA-3′ and 5′-TTTGGGAGTTCTGTGGTGGCGCCTGCCCCTGTTGCCAAAG-3′. This mutation introduced a novel KasI site (underlined). Likewise, the oligonucleotides 5′-CATATTCTTAGATGAATACGCGTGTGCCACTCCTGAACAA-3′ and 5′-TTGTTCAGAGTGGCACACGCGTATTCATCTAAGAATATG-3′ were used to make the DEYH-to-DEYA mutation and to introduce a novel MluI site. To make a deletion mutation in motif VI, two oligonucleotides (5′-CCTTAAGAGGATG-3′ and 5′-CCGGCATCCTCTTAAGG-3′) were annealed and used to replace the StuI-XmaI fragment in pD2.2, resulting in a net loss of 54 bp. The sequence of all mutations was confirmed by restriction digest analysis and DNA sequencing. The mutations in NS3 were then transferred to plasmid pSal by replacing the wild-type BglII-to-Tth111I fragment in pSal with the BglI-to-Tth111I fragments from pD2.2 containing the various mutations. The mutations were transferred to the complete BVDV genomic cDNA by excising the SalI fragments from the NS3 mutations in pSal and ligating them to SalI-digested pACCX5.6+2.7 in parallel with the wild-type SalI fragment. A mutation in which the GDD catalytic motif of the NS5B region was mutated to GAA was also made. All clones were confirmed using DNA sequencing and restriction digest and PCR analysis. For riboprobes, the NS4A region of BVDV was amplified by PCR using primers which added EcoRI and XbaI sites to the 5′ and 3′ ends. The resulting fragment was then cloned into the EcoRI and XbaI sites of pGEM-4Z (Promega) to generate plasmid pGEM0.2.
Expression and purification of BVDV NS3 helicase domain.
To express the wild-type and mutant forms of the BVDV helicase/NTPase domain, Escherichia coli strain BL21 (DE3) was transformed with either pET-21bHB or pET-21bHB encoding the mutant forms of the BVDV helicase domain. A single transformant was used to inoculate Luria-Bertani broth. Mid-log-phase cultures were induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and grown at 37°C for 3 h. Cells were pelleted, resuspended, and sonicated in buffer containing 20% glycerol, 20 mM Tris-HCl (pH 7.9), 500 mM NaCl, 0.1% Triton X-100, 50 μg of lysozyme per ml, and a protease inhibitor cocktail (Complete tablets without EDTA; Roche Molecular Biochemicals) at 4°C, followed by centrifugation. The BVDV helicase was purified from the clarified supernatant by using Ni-nitrilotriacetic acid affinity chromatography on a Talon column (Clontech). The eluted protein was dialyzed against buffer containing 50 mM HEPES (pH 7.5), 20% glycerol, 0.1 mM dithiothreitol, and 1 mM MgCl2. Total protein concentration was determined using the Bio-Rad protein assay.
Antisera, sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis, and Western blotting.
A synthetic peptide corresponding to amino acids 2180 to 2197 of the BVDV (NADL strain) polyprotein was coupled to keyhole limpet hemocyanin and used to immunize New Zealand White rabbits. This antiserum (512) immunoreacts with an 80-kDa protein present in BVDV-infected MDBK cells but absent in uninfected MDBK cells (data not shown). Antibody to the hexahistidine tag (H-15) was obtained from Santa Cruz Biotechnology or Clontech.
RNA helicase and ATPase assays.
ATPase assays were performed using a malachite green colorimetric assay as previously described (21). Briefly, dilutions of enzyme were added to buffer containing 10 mM Tris (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, and 1 mM ATP in a final volume of 50 μl. After incubation for 1 h at 37°C, the reactions were terminated by the addition of 75 μl of a solution comprised of three parts malachite green (0.45 g/liter; Sigma) and one part ammonium molybdate (42 g/liter in 4 M HCl; Sigma) which had been previously mixed for 30 min at room temperature. The reaction was allowed to develop for 5 min, and then the optical density was read at a wavelength of 670 nm.
RNA helicase assays were performed as previously described (41) with the following modifications. A 28-mer oligoribonucleotide, 5′-GGGAGACCGGCCUCGAGCAGCUGAAGCU-3′, was synthesized and 5′- end labeled using [γ-32P]ATP and T4 polynucleotide kinase. This labeled 28-mer was annealed to a 41-mer oligoribonucleotide (5′-UCGAAGAGAAGCUGCUCGAGGCCGGUCUCCCAGAGAGAG-3′) to create the substrate. Helicase reactions were performed in 10 μl of buffer having a final constitution of 50 mM HEPES-KOH (pH 6.5), 6 mM ATP, 3 mM MgCl2, and 10% glycerol, with 0.2 nM 28-mer RNA annealed to 0.4 nM 41-mer RNA, and 0.1 μg of BVDV helicase. Reaction mixtures were incubated for 30 min at 37°C, and reactions were terminated by the addition of 2.5 μl of 5× RNA sample buffer (100 mM Tris-HCl [pH 7.5]), 50 mM EDTA, 0.1% Triton X-100, 0.5% SDS, 50% glycerol, 0.1% bromophenol blue). Mixtures were electrophoresed on 6% polyacrylamide (1× Tris-borate-EDTA) gels at 100 V. Gels were dried and autoradiographed.
In vitro transcription reactions.
Plasmids pACCX-BVDV and the NS3 mutant constructs, pCLK, pCLM21, and pCLΔ21, were digested to completion with SacII, extracted with phenol-chloroform and precipitated with ethanol, and used as templates for in vitro transcription by T7 RNA polymerase. In vitro transcription was performed with 1 μg of linearized DNA template in 20 μl by using a T7-MEGAscript kit (Ambion). To determine the RNA quality, parallel reactions were performed with the addition of traces of [α-32P]UTP. After incubation of the reaction at 37°C for 5 h, the DNA template was digested with RNase-free DNase I (2 U) for 30 min at 37°C. The reaction mixture was then extracted twice with acid phenol (Ambion), and the RNA was precipitated with ethanol. Formaldehyde denaturing agarose gel analysis was performed on RNA that had been labeled with [α-32P]UTP to monitor the quality of transcription.
Transfection of bovine cells.
MDBK or EBTr cells were electroporated as previously described (38) with minor modifications. Briefly, trypsinized monolayers were washed twice with serum-free medium, resuspended in serum-free medium at a concentration of 5 × 106 cells/ml, and stored on ice. Two hundred microliters of the cell suspension was added to a chilled 0.2-cm-gap cuvette containing 5 μg of RNA transcripts and electroporated with a double pulse charge using a Bio-Rad electroporation unit on a setting of 280 V and 125 μF. Cuvettes were placed on ice for 1 min, 5 ml of complete medium was added, and the cells were transferred to the incubator in a T25 flask and monitored for the appearance of CPE (3 to 4 days). Supernatants were removed on day 4, and titers were determined by plaquing on MDBK cells.
RNase protection assays.
RNase protection assays were performed as previously described (4, 45) with slight modifications. Plus-strand RNA probes were generated by in vitro transcription of pGEM0.2 DNA which had been linearized with XbaI using SP6 RNA polymerase followed by digestion with RNase-free DNase. Minus-strand RNA probes were generated by in vitro transcription of pGEM0.2 DNA which had been linearized with EcoRI using T7 RNA polymerase followed by digestion with RNase-free DNase. Cytoplasmic RNA or total RNA was prepared at different times after transfection by using the TRIzol Reagent according to the protocol supplied by the manufacturer (Gibco BRL). For RNase protection assay, an RNase protection assay kit (RPA-II; Ambion) was used. Briefly, 2 μg of total RNA was mixed with 4.5 × 104 cpm of labeled probe in 20 μl of hybridization buffer (80% formamide, 100 mM sodium citrate [pH 6.4], 300 mM sodium acetate [pH 6.4], 1 mM EDTA), denatured at 95°C for 5 min, and allowed to hybridize overnight at 42°C. Unhybridized RNAs were degraded by adding 0.5 U of RNase A and 20 U of RNase T1 to the hybridization mixture and incubating the mixture at 37°C for 30 min. Protected RNA was then precipitated, resuspended in 10 μl of loading buffer, and electrophoresed on a 1× Tris-borate-EDTA–6% urea sequencing gel. The gel was dried onto 3M paper and exposed to film.
RESULTS
Expression of BVDV NS3 helicase domain.
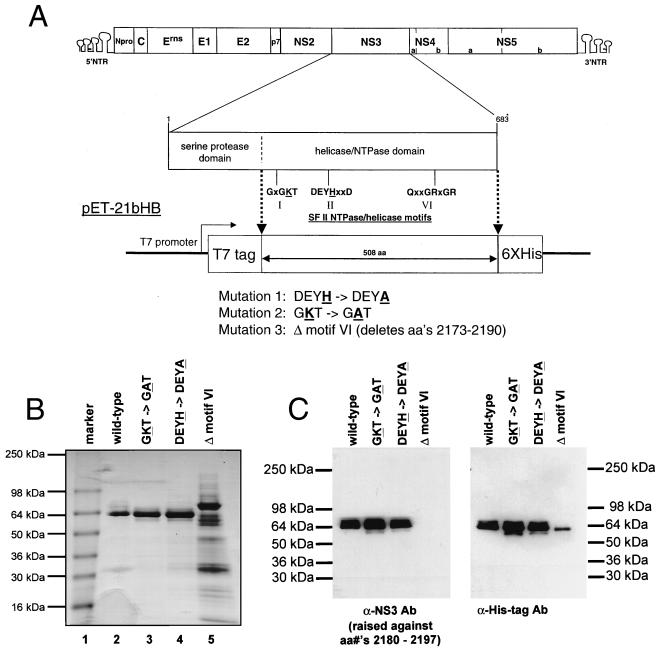
To examine the effects of mutations on the biochemical properties of the BVDV NS3 protein, the helicase domain of the NS3 region of the BVDV genomic cDNA (corresponding to amino acids 1854 to 2362 of the BVDV [NADL strain] polyprotein) was amplified using PCR and cloned into a bacterial expression vector. This protein was engineered such that the T7 epitope tag was added to the amino terminus and a hexahistidine tag was added to the carboxy terminus (Fig. 1A). Point and deletion mutations were generated in the helicase domain in three motifs known to be critical for helicase/NTPase activity. The lysine residue in the Walker motif A (GKT in BVDV) which binds the terminal phosphate groups of the NTP cofactor (39) was changed to an alanine (GAT). The histidine residue in the Walker motif B (DEYH in BVDV) responsible for coordinating the Mg2+ of the Mg-NTP complex (39) was changed to an alanine (DEYA). A similar change was shown to dissociate helicase and NTPase activities in the HCV NS3 helicase (15). A deletion of 18 residues (residues 2174 to 2190 of the polyprotein) encompassing motif VI (QRRGRVGR) was also constructed. All proteins were expressed in bacteria, purified using Ni-nitrilotriacetic acid affinity chromatography, and analyzed on SDS-containing polyacrylamide gels by Coomassie blue staining (Fig. 1B). Proteins were approximately 85 to 90% pure except for the deletion mutant. Difficulty was encountered in purifying this protein to similar levels of purity. As shown in Fig. 1C, all proteins were recognized by the monoclonal antibody against the hexahistidine tag. Additionally, the identity of the proteins as NS3 was confirmed using a BVDV NS3 antipeptide-specific antiserum raised against residues 2180 to 2196 of the BVDV polyprotein (Fig. 1C). This antipeptide antiserum recognizes all the recombinant proteins except the deletion mutant which lacks the antiserum epitope (Fig. 1C).
FIG. 1.
(A) Schematic drawing of the BVDV genome and NS3 helicase domain expression constructs. The individual proteins and 5′ and 3′ NTRs of the BVDV genome (NADL strain) are shown. The amino acid residues, serine protease and helicase/NTPase domains, and helicase motifs within the NS3 region are indicated below the genome. The BVDV NS3 helicase domain bacterial expression vector pET-21bHB is shown. The T7 promoter, N-terminal T7 tag, and C-terminal hexahistidine tags are indicated. The mutations made in the helicase/NTPase motifs in this study are indicated. aa, amino acid(s); SF II, superfamily II. (B) A Coomassie blue-stained SDS-polyacrylamide gel of the purified BVDV NS3 helicase domain proteins expressed in bacteria. Molecular size markers (kilodaltons) are indicated in lane 1. The wild-type BVDV NS3 helicase domain (lane 2), the GKT mutant (lane 3), the DEYH mutant (lane 4), and the deletion mutation (lane 5) are shown. (C) Western blot analysis of the purified BVDV NS3 helicase domain proteins expressed in bacteria. Purified proteins were analyzed by SDS-polyacrylamide gel electrophoresis followed by Western blotting with either a rabbit polyclonal antipeptide antiserum (α-NS3 Ab) raised against BVDV residues 2180 to 2197 or a monoclonal antibody against the hexahistidine affinity tag (Clontech) (α-His-tag Ab). Positions of molecular mass markers are indicated. aa, amino acid.
Separation of the ATPase and RNA helicase activities of the BVDV NS3 helicase domain.
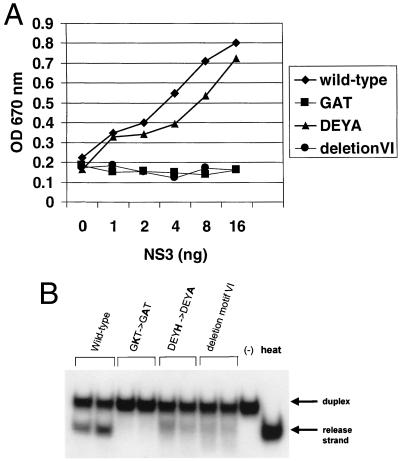
The ATPase activities of the wild-type and mutant BVDV NS3 helicase domain proteins were tested using a colorimetric assay (21). Equal amounts of protein as determined by Coomassie blue staining and Western blotting were titrated (between 1 and 16 ng of NS3 protein), and the resulting ATPase activities are shown (Fig. 2A). As expected, mutation of the Walker motif A and deletion of motif VI abolished ATPase activity. Moreover, mutation of Walker motif B had no major effect upon ATPase activity compared to wild type. This finding is in agreement with results obtained with the HCV NS3 protein (15).
FIG. 2.
(A) ATPase activity of the wild-type and mutant BVDV NS3 helicase domains. ATPase activity is measured using a colorimetric assay and by monitoring an increase in absorbance (optical density [OD] at 670 nm). A titration of NS3 protein was used. (B) RNA-unwinding activity of the wild-type and mutant BVDV NS3 helicase domains. Equal amounts of wild-type (lanes 1 and 2) or mutant (lanes 3 to 8) NS3 proteins were analyzed in duplicate for RNA helicase activity as described in Materials and Methods. Native duplex substrate alone (“−” lane) or heat-denatured duplex (heat lane) is indicated. Positions of duplex and release strand are shown.
The RNA helicase activity of the wild-type and mutant NS3 proteins was tested using a gel-based RNA duplex unwinding assay. Again, equal amounts of protein were used, and the resulting RNA helicase activities are shown in Fig. 2B. As expected, mutation of the Walker motif A abolished RNA helicase activity (Fig. 2B, lanes 3 and 4) compared to the wild-type protein. Mutation of Walker motif B or deletion of motif VI greatly reduced helicase activity (Fig. 2B, lanes 5 to 8). These results confirm that ATPase activity is essential for RNA helicase activity and notably demonstrate that the NTPase and RNA helicase activities of the BVDV NS3 protein can be functionally uncoupled by a mutation in the Walker motif B.
The BVDV NS3 helicase and NTPase activities are essential for BVDV replication.
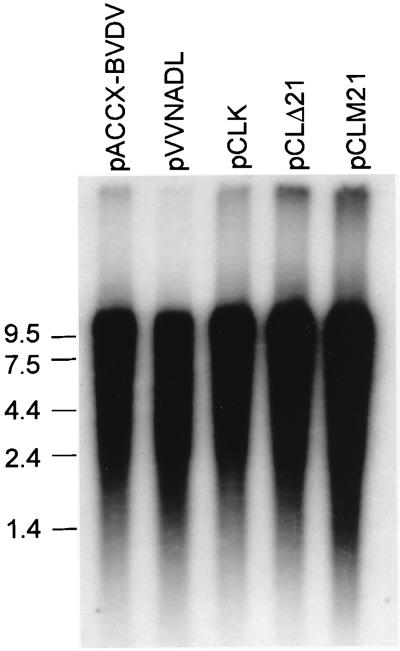
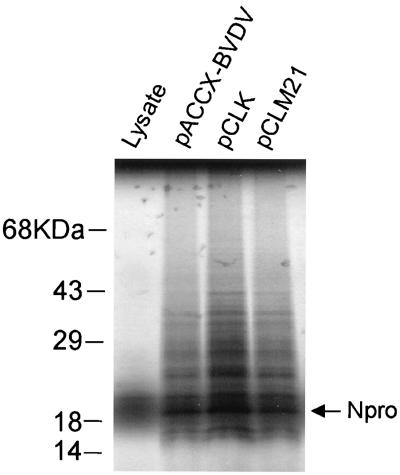
To test the essentiality of the BVDV NS3 NTPase/RNA helicase activities for virus replication, these point and deletion mutations were transferred into an infectious cDNA for BVDV (NADL strain) (38). To overcome plasmid stability issues and the difficulties in introducing the mutations in pVVNADL, we transferred the viral genome to a low-copy-number vector, pACCX. The orientation of the BVDV genome was such that transcription is in the opposite direction from that of the ampicillin resistance gene. Repeated attempts to make a construct containing the BVDV genomic cDNA in the reverse orientation failed. To minimize the possibility of mutations introduced by PCR mistakes, we used a small plasmid, pD2.2, as the template for mutagenesis, after which the insert region was sequenced in its entirety. The mutagenized fragment was then transferred back into the full-length genomic cDNA as described in Materials and Methods. Plasmid pCLK contains the BVDV genomic cDNA with the GKT-to-GAT mutation in Walker motif I of NS3. Plasmid pCLM21 contains the BVDV genomic cDNA with the DEYH-to-DEYA mutation in Walker motif II of NS3. Plasmid pCLΔ21 contains the BVDV genomic cDNA with the deletion in Walker motif VI of NS3. Full-length genomic RNA transcripts were generated by in vitro transcription of SacII-linearized DNA from either wild-type or mutant DNAs using T7 RNA polymerase. To monitor the production of full-length transcripts during the transcription reaction, a portion of the RNA transcripts from the reaction were labeled with traces of [α-32P]UTP and analyzed on denaturing agarose gels (Fig. 3). The mutant genomes and the wild-type genome were transcribed equally well. These transcripts were then transfected into either EBTr or MDBK cells. Cells were monitored for the production of infectious virus as evidenced by the development of CPE. Transfection with wild-type BVDV genomic RNA transcripts, derived from either pACCX-BVDV or parental plasmid pVVNADL, consistently produced approximately 2 × 106 PFU/ml of culture supernatant. However, repeated transfection with the NS3 mutant BVDV genomic RNA transcripts failed to yield infectious virus (Table 1). Even when the cultures were passaged for up to 2 weeks after transfection, CPE was never observed, strongly suggesting that the NTPase/RNA helicase activity of NS3 is required for viral replication. To rule out the possibility that the NS3 mutations had somehow affected either translation or polyprotein processing, the RNA transcripts from either wild type (Fig. 4, lane pACCX-BVDV) or NS3 mutants (Fig. 4, lanes pCLK and pCLM21) were used to program a rabbit reticulocyte lysate extract in the presence of [35S]methionine. All RNA transcripts produced similar levels of a 20-kDa protein corresponding to Npro, indicating that the translational efficiency of the genome was not affected by the NS3 mutations.
FIG. 3.
In vitro transcription of BVDV wild-type and mutant genomes. Plasmids containing wild-type (pACCX-BVDV or pVVNADL) or mutant (pCLK, pCLΔ21, or pCLM21) NS3 in the BVDV genomic cDNA linearized by SacII were transcribed in vitro by T7 RNA polymerase. A portion of the reaction mixture was labeled with traces of [α-32P]UTP and analyzed on denaturing agarose gels. The positions of the RNA size markers (kilobases) are indicated.
TABLE 1.
Production of virus by full-length BVDV genomic RNA transcripts containing either wild type or mutations in NS3a
| Construct | Virus titer (PFU/ml) |
|---|---|
| pACCX-BVDV | 1.8 × 106 |
| pVVNADL | 1.8 × 106 |
| pCLK (GKT→GAT) | 0 |
| pCLM21 (DEYH→DEYA) | 0 |
| pCLΔ21 (delete motif VI) | 0 |
Yields of virus are from supernatants derived from cells 4 days posttransfection with genomic RNA transcripts from either wild type (pACCX-BVDV or pVVNADL) or NS3 mutants (pCLK, pCLΔ21, or pCLM21).
FIG. 4.
In vitro translation efficiency of BVDV RNAs. Rabbit reticulocyte lysates were programmed with BVDV genomic RNA transcripts derived by in vitro transcription of plasmids encoding either wild-type (pACCX-BVDV) or mutant (pCLK or pCLM21) NS3 proteins in the presence of [35S]methionine, and reactions were resolved by electrophoresis on SDS–10% polyacrylamide gels. Labeled proteins were visualized by autoradiography of the dried gel. Positions of molecular mass markers are indicated.
The BVDV NS3 helicase and NTPase activities are required for efficient minus-strand synthesis.
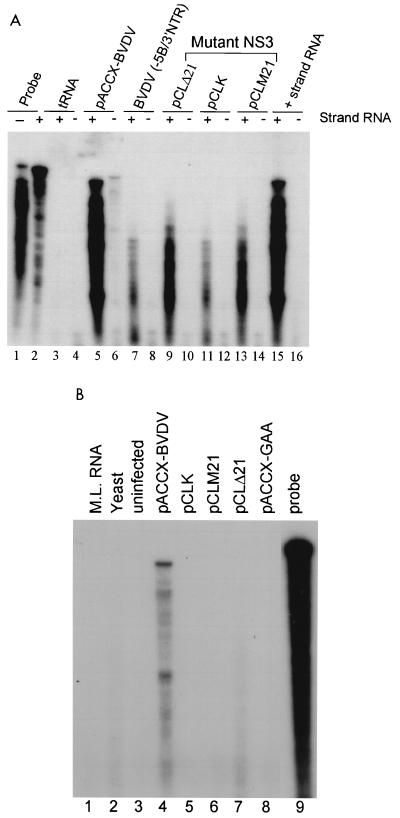
Since BVDV can also exist as a noncytopathic form, it was critical to determine if the mutations in NS3 made the virus noncytopathic. RNase protection assays using plus- or minus-strand-specific probes were used to examine the level of RNA replication. As an additional negative control, the wild-type BVDV genomic cDNA was truncated near the amino terminus of the NS5B region, thereby deleting most of NS5B and the 3′ NTR. To test the specificity of the plus- and minus-strand probes, in vitro-transcribed wild-type BVDV genomic plus-strand RNA was used as a positive control (Fig. 5A, lanes 15 and 16), and yeast tRNA was used as a negative control (Fig. 5A, lanes 3 and 4). Total RNA was harvested 72 h posttransfection. Unlike transfections with the wild-type BVDV genomic RNA transcripts (Fig. 5A, lane 5), no fully protected RNA band was observed with any of the NS3 mutants (Fig. 5A, lanes 9 to 14) or the NS5B/3′ NTR deletion (Fig. 5A, lane 7), indicating that the input RNA from the NS3 mutants was degrading with time. In contrast to the wild type, the level of detectable input plus-strand RNA of the NS3 mutants decreased with time, indicating a lack of sustainable RNA replication (data not shown). Furthermore, only with wild-type BVDV was a band corresponding to the minus strand readily observed (Fig. 5A, lane 6).
FIG. 5.
(A) RNase protection analysis of transfected BVDV RNAs 72 h posttransfection. Total RNA was harvested 72 h posttransfection, hybridized with 32P-labeled plus- and minus-strand-specific riboprobes from the BVDV NS4A region, and analyzed by RNase protection. 32P-labeled plus (+)- and minus (−)-strand probes used for protection are shown in lanes 1 and 2. Yeast tRNA (tRNA, lanes 3 and 4) and in vitro-transcribed plus-strand BVDV genomic RNA (+ strand RNA, lanes 15 and 16) were used as negative and positive controls for probe specificity, respectively. Samples derived from transfection with RNA transcripts encoding wild-type NS3 (pACCX-BVDV, lanes 5 and 6) or a transcript containing a deletion of NS5B and the 3′ NTR [BVDV(−5B/3′NTR), lanes 7 and 8] were used as positive and negative controls, respectively. Samples derived from transfection with RNA transcripts encoding mutant NS3 proteins (pCLΔ21, pCLK, and pCLM21, lanes 9 to 14) are shown and indicated above lanes. (B) RNase protection analysis of BVDV RNA from cytoplasmic RNA 24 h posttransfection. Cytoplasmic RNA was harvested 24 h posttransfection and subjected to a single round of RNase digestion followed by hybridization with a minus-strand probe and RNase protection as described in Materials and Methods. Mouse liver RNA (lane 1, M.L. RNA), yeast tRNA (lane 2, yeast), and uninfected MDBK cell RNA (lane 3, uninfected) were used as negative controls for probe specificity. Protected RNAs from cells transfected with wild-type BVDV genomic RNA transcripts (lane 4, pACCX-BVDV), NS5B mutant (lane 8, pACCX-GAA), or NS3 mutants (lanes 5 to 7, pCLK, pCLM21, and pCLΔ21, respectively) are shown. The 32P-labeled minus-strand probe used for protection is shown in lane 9 (probe).
To more closely examine the level of minus-strand RNA synthesis, a more sensitive modified RNase protection assay was used. As an additional negative control, a double point mutation was generated in the conserved catalytic GDD motif of the NS5B RNA polymerase (GDD changed to GAA). MDBK cells were transfected with RNA transcripts derived from either the wild-type, NS5B double point mutant, or NS3 mutant template DNAs. Twenty-four hours posttransfection, cytoplasmic RNAs were harvested and subjected to a cycle of hybridization and RNase treatment without an external probe as previously described (4, 26). This was followed by hybridization with a minus-strand-specific probe and RNase treatment. This procedure has been shown to increase the sensitivity of minus-strand RNA detection by reducing the background signal of excess plus-strand RNA derived from the transfected input plus-strand or disproportionate plus-strand synthesis. Using this modified procedure, we could easily detect minus-strand synthesis by the wild-type BVDV transcript (Fig. 5B, lane 4) 24 h posttransfection. However, we could not detect any level of minus-strand RNA synthesis by either the NS5B double point mutant (Fig. 5B, lane 8) or any of the NS3 mutants (Fig. 5B, lanes 5 to 7). The specificity of the minus-strand probe was shown by its lack of hybridization with RNA derived from either uninfected MDBK cells (Fig. 5B, lane 3), yeast tRNA (Fig. 5B, lane 2), or mouse liver (Fig. 5B, lane 1). Taken together, these results indicate that the NTPase/helicase activity of the BVDV NS3 protein is essential for the synthesis of minus-strand RNA.
We have attempted to rescue our NS3 mutants using a variety of methods (cotransfection with NS5B mutant RNA transcripts, cotransfection with NS3 expression plasmids, etc.) to supply NS3 in trans; however, we have never been successful using any method (data not shown).
DISCUSSION
Although most plus-strand RNA viruses either have proteins which contain helicase/NTPase motifs or have been shown to encode helicase/NTPase activity, the function of these activities in the life cycle of the virus is unknown. The data presented here show that, for BVDV, these functions encoded by the NS3 protein are absolutely essential for the synthesis of minus-strand RNA, and thus the production of infectious virus particles. While our results demonstrate the essentiality of the NS3 NTPase/helicase function of NS3 for minus-strand RNA synthesis, they do not reveal anything about the mechanism or role these activities play in minus-strand synthesis. Possibly, the helicase activity is needed to unwind the secondary structures present in the 3′ NTR to allow initiation by NS5B or to facilitate the processivity of NS5B during elongation by unwinding secondary structures within the coding region. Furthermore, since the NTPase activity is absolutely required for helicase activity, we cannot rule out the possibility that NTP hydrolysis independent of nucleic acid unwinding may play some role in the viral life cycle. Indeed, although the NTPase activity of the HCV NS3 helicase domain is greatly stimulated by single-stranded nucleic acid, the protein still possesses a rather high intrinsic NTPase activity in the absence of nucleic acid (27), unlike other helicases such as bacterial Rep protein. The function of the unstimulated NTPase activity is unknown, although it alone is not sufficient to support minus-strand synthesis.
It is possible that the NS3 NTPase/helicase activity may play additional roles in steps subsequent to minus-strand synthesis, such as release of newly synthesized plus-strand RNA from the minus strand or RNA packaging. However, since minus-strand synthesis is the first step in the synthesis of RNA for the production of progeny virions, we were not able to examine any of these possibilities with the NS3 knockout mutants. Temperature shift experiments with a temperature-sensitive mutation in NS3 would be very useful for investigating these possibilities.
We have used mutational analysis to demonstrate that the NS3 NTPase/helicase activities of the BVDV NS3 protein can be uncoupled and that both of these activities are essential for the synthesis of minus-strand RNA and subsequently the production of infectious virus.
Although the NTPase/helicase motifs represent some of the most highly conserved motifs throughout all the plus-stranded RNA viruses, little is known about their function in the life cycle of these viruses. The 2C protein of poliovirus has been shown to possess NTPase activity (29). Furthermore, certain 2C mutations are lethal due to a lack of viral RNA synthesis (5, 22, 36); however, 2C has not yet been shown to encode helicase activity. This is the first demonstration that the RNA helicase activity of any plus-strand RNA viral protein is essential for minus-strand synthesis and virus growth.
ACKNOWLEDGMENTS
We thank Ruben Donis and Ventzislav Vassilev (University of Nebraska) for their most generous gift of the infectious BVDV cDNA clone, pVVNADL. We thank Sven-Erik Behrens (Justus-Liebig-Universität, Giessen, Germany) for helpful discussion and communicating results prior to publication and Susan Dillon, Klaus Esser, and Robert Sarisky for critical reading of the manuscript.
ADDENDUM IN PROOF
After submission of the manuscript, similar findings were reported by Grassmann et al. (C. W. Grassmann, O. Isken, and S.-E. Behrens, J. Virol. 73:9196–9205, 1999).
REFERENCES
- 1.Baker J C. Bovine viral diarrhea virus: a review. J Am Vet Med Assoc. 1987;190:1449–1458. [PubMed] [Google Scholar]
- 2.Bartholomeusz A I, Wright P J. Synthesis of dengue virus RNA in vitro: initiation and the involvement of proteins NS3 and NS5. Arch Virol. 1993;128:111–121. doi: 10.1007/BF01309792. [DOI] [PubMed] [Google Scholar]
- 3.Behrens S-E, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens S-E, Grassmann C W, Thiel H-J, Meyers G, Tautz N. Characterization of an autonomous subgenomic pestivirus RNA replicon. J Virol. 1998;72:2364–2372. doi: 10.1128/jvi.72.3.2364-2372.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein H D, Sarnow P, Baltimore D. Genetic complementation among poliovirus mutants derived from an infectious cDNA clone. J Virol. 1986;60:1040–1049. doi: 10.1128/jvi.60.3.1040-1049.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C-J, Kuo M-D, Chien L-J, Hsu S-L, Wang Y-M, Lin J-H. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J Virol. 1997;71:3466–3473. doi: 10.1128/jvi.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collett M S, Larson R, Gold C, Strick D, Anderson D K, Purchio A F. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology. 1988;165:191–199. doi: 10.1016/0042-6822(88)90672-1. [DOI] [PubMed] [Google Scholar]
- 8.Collett M S, Larson R, Belzer S, Retzel E. Proteins encoded by bovine viral diarrhea virus: the genome organization of a pestivirus. Virology. 1988;165:200–208. doi: 10.1016/0042-6822(88)90673-3. [DOI] [PubMed] [Google Scholar]
- 9.Collett M S. Molecular genetics of pestiviruses. Comp Immunol Microbiol Infect Dis. 1992;15:145–154. doi: 10.1016/0147-9571(92)90087-8. [DOI] [PubMed] [Google Scholar]
- 10.Cui T, Sugrue R J, Xu Q, Lee A K W, Chan Y-C, Fu J. Recombinant dengue virus type 1 NS3 protein exhibits specific viral RNA binding and NTPase activity regulated by the NS5 protein. Virology. 1998;246:409–417. doi: 10.1006/viro.1998.9213. [DOI] [PubMed] [Google Scholar]
- 11.Deng R, Brock K V. 5′ and 3′ untranslated regions of pestivirus genome: primary and secondary structure analysis. Nucleic Acids Res. 1993;21:1949–1957. doi: 10.1093/nar/21.8.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donis R O. Molecular biology of bovine viral diarrhea virus and its interactions with the host. Vet Clin N Am Food Anim Pract. 1995;11:393–423. doi: 10.1016/S0749-0720(15)30459-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbers K, Tautz N, Becher P, Rumenapf T, Thiel H-J. Processing in the pestivirus E2-NS2 region: identification of the nonstructural proteins p7 and E2p7. J Virol. 1996;70:4131–4135. doi: 10.1128/jvi.70.6.4131-4135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuller-Pace F V. RNA helicases: modulators of RNA structure. Trends Cell Biol. 1994;4:271–274. doi: 10.1016/0962-8924(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 15.Heilek G M, Peterson M G. A point mutation abolishes the helicase but not the nucleoside triphosphatase activity of hepatitis C virus NS3 protein. J Virol. 1997;71:6264–6266. doi: 10.1128/jvi.71.8.6264-6266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadare G, Haenni A-L. Virus-encoded RNA helicases. J Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapoor M, Zhang L, Ramachandara M, Kusukawa J, Ebner K E, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270:19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 18.Kim D W, Gwack Y, Han J H, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 19.Kim D W, Kim J, Gwack Y, Han J H, Choe J. Mutational analysis of the hepatitis C virus RNA helicase. J Virol. 1997;71:9400–9409. doi: 10.1128/jvi.71.12.9400-9409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuo M-D, Chin C, Hsu S-L, Shiao J-Y, Wang T-M, Lin J-H. Characterization of the NTPase activity of Japanese encephalitis virus NS3 protein. J Gen Virol. 1996;77:2077–2084. doi: 10.1099/0022-1317-77-9-2077. [DOI] [PubMed] [Google Scholar]
- 21.Lanzetta P A, Alvarez L J, Reinach P S, Candia O A. An improved assay for nanomole amounts of inorganic phosphate. Anal Chem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 22.Li J P, Baltimore D. Isolation of poliovirus 2C mutants defective in viral RNA synthesis. J Virol. 1988;62:4016–4021. doi: 10.1128/jvi.62.11.4016-4021.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohmann V, Korner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motif essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyers G, Thiel H-J. Molecular characterization of pestiviruses. Adv Virus Res. 1996;47:53–118. doi: 10.1016/s0065-3527(08)60734-4. [DOI] [PubMed] [Google Scholar]
- 25.Moennig V, Plagemann P G. The pestiviruses. Adv Virus Res. 1992;41:53–98. doi: 10.1016/s0065-3527(08)60035-4. [DOI] [PubMed] [Google Scholar]
- 26.Novak J E, Kirkegaard K. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J Virol. 1991;65:3384–3387. doi: 10.1128/jvi.65.6.3384-3387.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preugschat F, Averett D R, Clarke B E, Porter D J T. A steady-state and pre-steady-state kinetic analysis of the NTPase activity associated with the hepatitis C virus NS3 helicase domain. J Biol Chem. 1996;271:24449–24457. doi: 10.1074/jbc.271.40.24449. [DOI] [PubMed] [Google Scholar]
- 28.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields' virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Press; 1996. pp. 931–959. [Google Scholar]
- 29.Rodriguez P L, Carrasco L. Poliovirus protein 2C has ATPase and GTPase activities. J Biol Chem. 1993;268:8105–8110. [PubMed] [Google Scholar]
- 30.Rumenapf T, Unger G, Strauss J H, Thiel H-J. Processing of the envelope glycoproteins of pestiviruses. J Virol. 1993;67:3288–3295. doi: 10.1128/jvi.67.6.3288-3294.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stark R, Meyers G, Rumenapf T, Thiel H-J. Processing of pestivirus polyprotein: cleavage site between autoprotease and nucleocapsid protein of classical swine fever virus. J Virol. 1993;67:7088–7095. doi: 10.1128/jvi.67.12.7088-7095.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzich J A, Tamura J K, Palmer-Hill F, Warrener P, Grakoui A, Rice C M, Feinstone S M, Collett M S. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J Virol. 1993;67:6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takegami T, Sakamuro D D, Furukawa T. Japanese encephalitis virus nonstructural protein NS3 has RNA binding and ATPase activities. Virus Genes. 1995;9:105–112. doi: 10.1007/BF01702653. [DOI] [PubMed] [Google Scholar]
- 34.Tamura J K, Warrener P, Collett M S. RNA-stimulated NTPase activity associated with the p80 protein of the pestivirus bovine viral diarrhea virus. Virology. 1993;193:1–10. doi: 10.1006/viro.1993.1097. [DOI] [PubMed] [Google Scholar]
- 35.Tautz N, Elbers K, Stoll D, Meyers G, Thiel H-J. Serine protease of pestiviruses: determination of cleavage sites. J Virol. 1997;71:5415–5422. doi: 10.1128/jvi.71.7.5415-5422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teterina N L, Kean K M, Gorbalenya A E, Agol V I, Girard M. Analysis of the functional significance of amino acid residues in the putative NTP-binding pattern of the poliovirus 2C protein. J Gen Virol. 1992;73:1977–1986. doi: 10.1099/0022-1317-73-8-1977. [DOI] [PubMed] [Google Scholar]
- 37.Thiel H J, Plagemann G W, Moennig V. Pestiviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields' virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Press; 1996. pp. 1059–1073. [Google Scholar]
- 38.Vassilev V B, Collett M S, Donis R O. Authentic and chimeric full-length genomic cDNA clones of bovine viral diarrhea virus that yield infectious transcripts. J Virol. 1997;71:471–478. doi: 10.1128/jvi.71.1.471-478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthetase, myosin, kinases, and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warrener P, Tamura J K, Collett M S. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J Virol. 1993;67:989–996. doi: 10.1128/jvi.67.2.989-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warrener P, Collett M S. Pestivirus NS3 (p80) protein possesses helicase activity. J Virol. 1995;69:1720–1726. doi: 10.1128/jvi.69.3.1720-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wengler G, Wengler G. The carboxy-terminal part of the NS3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology. 1991;184:707–715. doi: 10.1016/0042-6822(91)90440-m. [DOI] [PubMed] [Google Scholar]
- 43.Wengler G, Wengler G. The NS3 nonstructural protein of flaviviruses contains an RNA triphosphatase activity. Virology. 1993;197:265–273. doi: 10.1006/viro.1993.1587. [DOI] [PubMed] [Google Scholar]
- 44.Xu J, Mendez E, Caron P R, Lin C, Murcko M A, Collett M C, Rice C M. Bovine viral diarrhea virus: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J Virol. 1997;71:5312–5322. doi: 10.1128/jvi.71.7.5312-5322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu H, Grassmann C W, Behrens S-E. Sequence and structural elements at the 3′ terminus of bovine viral diarrhea virus genomic RNA: functional role during RNA replication. J Virol. 1999;73:3638–3648. doi: 10.1128/jvi.73.5.3638-3648.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong W, Gutshall L L, Del Vecchio A M. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J Virol. 1998;72:9365–9369. doi: 10.1128/jvi.72.11.9365-9369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]