Abstract
Human polyomavirus JC (JCV) can encode the three capsid proteins VP1, VP2, and VP3, downstream of the agnoprotein in the late region. JCV virions are identified in the nucleus of infected cells. In this study, we have elucidated unique features of JCV capsid formation by using a eukaryotic expression system. Structures of JCV polycistronic late RNAs (M1 to M4 and possibly M5 and M6) generated by alternative splicing were determined. VP1 would be synthesized from M2 RNA, and VP2 and VP3 would be synthesized from M1 RNA. The presence of the open reading frame of the agnoprotein or the leader sequence (nucleotides 275 to 409) can decrease the expression level of VP1. VP1 was efficiently transported to the nucleus in the presence of VP2 and VP3 but distributed both in the cytoplasm and in the nucleus in their absence. Mutation analysis indicated that inefficiency in nuclear transport of VP1 is due to the unique structure in the N-terminal sequence, KRKGERK. Within the nucleus, VP1 was localized discretely and identified as speckles in the presence of VP2 and VP3 but distributed diffusely in their absence. These results suggest that VP1 was efficiently transported to the nucleus and localized in the discrete subnuclear regions, possibly with VP2 and VP3. By electron microscopy, recombinant virus particles were identified in the nucleus, and their intranuclear distribution was consistent with distribution of speckles. This system provides a useful model with which to understand JCV capsid formation and the structures and functions of the JCV capsid proteins.
Human polyomavirus JC (JCV) persists asymptomatically in healthy individuals of most of the human population. However, in immunocompromised individuals, JCV can cause progressive multifocal leukoencephalopathy (PML), a fatal demyelinating disorder of the central nervous system. JCV has a genome of a double-stranded circular DNA composed of the early region and the late region. Genome organization of JCV is closely related to that of simian virus 40 (SV40), which shares 70% identity in nucleotide sequence. The JCV replication cycle is divided into the early stage and the late stage. During the late stage, the capsid proteins are synthesized in the cytoplasm and then transported to the nucleus to be assembled into progeny virions. By electron microscopy, JCV virions are identified as round particles or filamentous forms in the nuclei of infected oligodendrocytes of PML brains (41, 44, 69).
JCV Tokyo-1 strain was isolated from the brain tissue of a Japanese PML patient (43, 44), and the viral genomic DNA was cloned (40). The virus particles of Tokyo-1 were purified, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and stained with Coomassie brilliant blue. The detected protein components indicated the presence of the major capsid protein VP1 (43 kDa), the minor capsid proteins VP2 (39 kDa) and VP3 (27 kDa), and histones (16, 15, and 14 kDa) incorporated into the viral capsids (2).
Based on the crystal structures of other polyomaviruses (26, 35, 62, 63), the JCV capsid is likely composed of 360 molecules of VP1 and approximately 1/10 molecules of VP2 and VP3. The coding sequences of VP1, VP2, and VP3 are encoded in an overlapping manner downstream of the coding sequence of the agnoprotein in the late region (19). It is not known how these capsid proteins are expressed, transported to the nucleus, and assembled into viral capsids. The capsid proteins may be translated from different JCV late RNAs generated by alternative splicing, and both expression and nuclear transport of the capsid proteins may be regulated to allow assembly of the capsid proteins in an appropriate ratio. Therefore, this study was initiated to investigate JCV capsid formation, in particular, splicing of late RNAs, translation and nuclear transport of the major capsid protein VP1, and capsid assembly.
SV40 has two classes of late RNAs, 16S and 19S, which are generated by alternative splicing from common pre-mRNAs (24). Each class of late RNAs contains several species which are heterogeneous in the leader sequence. In the leader sequence, 80% of the 16S RNAs (64% of the total late RNAs) and 5% of the 19S RNAs (1% of the total late RNAs) encode the agnoprotein (30). The coding sequence of the agnoprotein has been reported to regulate transcription (3, 27) and splicing (60) of the late RNAs. The presence of the upstream AUG start codon of the agnoprotein downregulates translation of VP1 on 16S RNAs (25, 52) and translation of VP2 and VP3 on 19S RNAs (23, 52). After translation, SV40 VP1 is transported to the nucleus by a bipartite nuclear localization signal (NLS) (28). It has been suggested that SV40 VP1 is associated with VP2 and VP3 in the cytoplasm and is transported to the nucleus as a complex (29). The SV40 agnoprotein has also been reported to affect nuclear transport of VP1 (8, 52), capsid assembly, and virion maturation (5, 38, 46).
The slow and inefficient replication of JCV has been a major barrier in studying the late events of the JCV replication cycle. JCV late RNAs are not detected until 10 days after infection of primary human fetal glial cells (15), and the VP1 protein is not detected until 2 weeks after infection of IMR-32 cells (2). To obtain sufficient virus titers, a long culture period of 3 to 4 weeks is required (2, 36, 37, 48, 57), and viral yield is generally low. To overcome this difficulty, we established an expression system for JCV capsid proteins (58) and studied JCV capsid formation. The complete sequence of JCV Tokyo-1 strain was determined and the genomic fragments of Tokyo-1 were inserted downstream of the powerful SRα promoter in eukaryotic expression vector pcDL-SRα296 (64). Structures of JCV late RNAs were determined by reverse transcription-PCR (RT-PCR) and sequencing, using a biopsy specimen of a PML brain and vector-transfected COS-7 cells (22). Potential roles of the agnoprotein and its coding sequence were analyzed in cells transfected with the vector encoding both the agnoprotein and the capsid proteins (AVP231-SRα [“A” for “agnoprotein”]) or the vector encoding only the capsid proteins (VP231-SRα). Nuclear transport of VP1 was studied in cells transfected with the vector encoding only VP1 (VP1-SRα) or the vectors encoding the three capsid proteins with or without the agnoprotein (AVP231-SRα or VP231-SRα). Nuclear transport of VP1 was further investigated by mutating its N-terminal sequence, which corresponds to the NLS of SV40 VP1. Intranuclear localization of VP1 was analyzed by confocal microscopy, and assembly of JCV recombinant particles in transfected cells was analyzed by electron microscopy. These results were compared with previous reports on SV40 to better understand unique features of JCV capsid formation.
MATERIALS AND METHODS
Virus genome.
JCV Tokyo-1 strain was isolated from the brain tissue of a Japanese PML patient (44). The plasmid clones were obtained from the PML brain tissue (the pJCT-Br clone) and from primary human fetal glial cells after viral passage (the pJCT-TC clone) (40). The complete DNA sequence of pJCT-TC was determined from both strands with a T7 Sequenase sequencing kit version 2.0 (Amersham) (1).
Plasmids, cells, and transfection.
The eukaryotic expression vector pcDL-SRα296 is an SV40-based plasmid vector with the SRα promoter (64). Expression vectors VP231-SRα, AVP231-SRα (previously called LP-SRα), and VP1-SRα were constructed by inserting fragments of pJCT-TC into pcDL-SRα296 as previously described (58). COS-7 cells (ATCC CRL 1651) (22) were incubated at 37°C and 5% CO2 in Dulbecco's modified Eagle's minimum essential medium supplemented with 10% fetal bovine serum. COS-7 cells were transfected by Lipofectamine (GIBCO BRL) according to the manufacturer's procedure.
RT-PCR analysis and sequencing of RT-PCR products.
Total RNA was extracted from a biopsy specimen of a PML brain and from COS-7 cells 72 h posttransfection (hpt) by using an RNeasy total RNA kit (Qiagen). The extracts were treated with RQ1 RNase-free DNase (Promega). The RNA was analyzed by RT-PCR using the Access RT-PCR system (Promega). The RT-PCR products were electrophoresed on agarose gels and extracted by a QIAquick gel extraction kit (Qiagen). Splice junctions were determined by sequencing RT-PCR products with the Thermo Sequenase radiolabeled terminator cycle sequencing kit (Amersham). Nucleotide sequences and positions of PCR primers are as follows. The 5′ primers are RR15 (5′-GAGCTGTTTTGGCTTGTCAC-3′, nucleotides [nt] 246 to 265), V1 (5′-AGAAACACAGTGGTTTGACT-3′, nt 429 to 448), V2 (5′-AGTACCTCTGAGGCTATAGC-3′, nt 680 to 699), and V13 (5′-GTTCATGGGTGCCGCACTTG-3′, nt 520 to 539). The 3′ primers are VAS3 (5′-CAACATTCAACAGGATATGC-3′, nt 2068 to 2049), VAS5 (5′-TCTACCTCTGTAATTGAGTC-3′, nt 1588 to 1569), VAS8 (5′-CCAACTGAGCAATAGCACTA-3′, nt 815 to 796), VAS9 (5′-GCAGCCTCAGAAACAGTAGC-3′, nt 579 to 560), and VAS12 (5′-TTCAGGCAAAGCACTGTATG-3′, nt 475 to 456). The 3′ primer Vect4 (5′-TTCTTTCCGCCTCAGAAGGT-3′) is located in the vector sequence of pcDLSRα-296, immediately downstream of the insert-vector junction. As an internal control for COS-7 cells, SV40 T antigen RNA was analyzed with primers SVT1 (5′-TGCAGCTAATGGACCTTCTA-3′, SV40 nt 5132 to 5113) and SVTAS1 (5′-GAGTAGAATGTTGAGAGTCA-3′, SV40 nt 4445 to 4464). These primers were designed from the SV40 nucleotide sequence assigned GenBank accession no. J02400.
Antibody.
VP1 was detected with rabbit polyclonal antibodies, which are anti-JCV antibody and the anti-VP1BC antibody. The anti-JCV antibody was prepared by inoculating purified JCV (43). This antibody recognizes VP1 but not the agnoprotein, VP2, or VP3 (58). The anti-VP1BC antibody was prepared against a synthetic peptide of VP1, RGFSKSISISDTFESD.
Radioimmunoprecipitation.
COS-7 cells were cultured 60 hpt for 4 h in methionine- and cysteine-free medium and then labeled for 20 h with [35S] Protein Labeling Mix (150 μCi/ml; NEN). Cells were dispersed in radioimmunoprecipitation assay buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, 0.1% sodium deoxycholate); then the chromosomal DNA was sheared with 26-gauge needles. Cell lysates were incubated with the anti-JCV antibody followed by incubation with protein A-Sepharose beads (Sigma). The immune complex was washed six times with radioimmunoprecipitation buffer. The samples were treated at 97°C for 5 min in SDS-PAGE loading buffer (100 mM Tris-HCl [pH 6.8], 10% 2-mercaptoethanol, 4% SDS, 0.2% bromophenol blue, 20% glycerol). After electrophoresis, the radiolabeled proteins were detected by autoradiography.
Mutagenesis.
The 5′ region of the VP1 coding sequence was mutated by PCR. The sequence of the first VP1 mutant, Mut-1, was generated by using the 5′ primer 5′-CTGCTGCAGCCAAGATGGCCCCAACAAAAAGAAAAGGAGAATGTCCAGGGGCAGCTCCCAGGAAGGACCCCGTGCAAG-3′. The sequence of the second mutant, Mut-2, was generated by using the 5′ primer 5′-CTGCTGCAGCCAAGATGGCCCCAACAAAAAGAAAAGGAGAATGTCCAGGGGCAGCTCCCAAAAAACCAAAGGACCCCGTGCAAGTT-3′. In these primer sequences, CTGCAG is a PstI recognition site and underlined sequences are nucleotides inserted into the VP1 coding sequence. Using each of these 5′ primers, we first synthesized single-stranded DNA. VP1-SRα, the plasmid DNA carrying the sequence of wild-type VP1, was used as a template. The single-stranded DNA was purified and then amplified by PCR using each of the two 5′ primers and the 3′ primer, VAS13 (5′-ATATTTCCACAGGTTAGATCCTCATTTAGA-3′, nt 1765 to 1736). PCR products were digested by PstI and EcoRI and then replaced with a PstI-EcoRI fragment in VP1-SRα. The sequences of the VP1 mutant vectors were confirmed by sequencing reactions.
Immunofluorescence microscopy.
COS-7 cells were grown on tissue culture glass slides (Falcon) and transfected with the expression vectors. Cells were fixed 72 hpt for 15 min in 2% paraformaldehyde, then incubated with the anti-VP1BC antibody, and finally incubated with a fluorescein isothiocyanate-conjugated goat anti-rabbit antibody (Biomeda). The slides were mounted and examined with an Olympus FV500 confocal microscope. The confocal image and the differential interference contrast image were sequentially acquired and superposed.
Electron microscopy.
Cells transfected with the expression vectors were harvested 72 hpt, washed with phosphate-buffered saline, fixed with 2.5% glutaraldehyde and 4.0% paraformaldehyde in 0.1 M phosphate buffer (pH 7.3), and embedded in epoxy resin. Ultrathin sections were prepared, stained with uranyl acetate and lead citrate, and examined with an H-800 electron microscope (Hitachi).
Nucleotide sequence accession number.
The GenBank accession number for JCV Tokyo-1 strain is AF030085.
RESULTS
Genomic sequences of JCV Tokyo-1 strain in the regulatory and late regions.
The complete sequence of JCV Tokyo-1 strain (5,128 bp) was determined by using the plasmid clone of the viral genomic DNA named pJCT-TC (40). Tokyo-1 shares 70% identity with SV40 (65), 76% identity with BK virus (BKV) Dunlop strain (55), and 97.1% identity with JCV Mad1 strain (19). The sequences of the regulatory and late regions of Tokyo-1 are presented in Fig. 1. Numbering begins with the presumed origin of DNA replication (19), and the first G in GCCTCG is nt 1.
FIG. 1.
Nucleotide sequence of JCV Tokyo-1 strain (5,128 bp) and amino acid sequences of the late proteins of Tokyo-1. The nucleotide sequences of the regulatory region (nt 5012 to 274) and the late region (nt 275 to 2600) are presented. In the regulatory region, the sequences identical to the 25- and 18-bp blocks of the archetype and the partial sequences corresponding to the 55- and 66-bp blocks of the archetype are marked [25], [18], [55], and [66], respectively. the 75-bp repeats (nt 38 to 112 and 134 to 208) and the 20-bp repeats (nt 113 to 132 and 273 to 292) are underlined with solid lines and dotted lines with the notations (75) and (20), respectively. The 75-bp repeats consist of the partial sequences of the 55-, 66-, and 18-bp blocks of the archetypal sequence. The 20-bp repeats, which are unique to Tokyo-1, contain the duplicated ATG start codons of the agnoprotein. The sequences identical to the surrogate TATA signal, TACCTA, are indicated in boldface at nt 64 to 69 and nt 160 to 165. The putative polyadenylation signal, AATAAA, of the late RNAs is present at nt 2566 to 2571. The 5′ splice sites (nt 490, 730, and 1583) and the 3′ splice sites (nt 1425 and 1952), determined in this study, are indicated by pairs of back-to-back brackets and labeled 5′ sp and 3′ sp, respectively. The nucleotides identical to the consensus sequences defined for splice sites are in boldface. Amino acid sequences of the four late proteins are deduced based on the sequence similarity to SV40 and BKV: the agnoprotein (71 amino acids, nt 275 to 490), VP2 (344 amino acids, nt 524 to 1558), VP3 (225 amino acids, nt 881 to 1558), and VP1 (354 amino acids, nt 1467 to 2531). Though there are two potential translation initiation sites for VP1 at nt 1461 and 1467, the downstream ATG (nt 1467 to 1469) is tentatively assigned as a translation initiation signal. If translation begins at nt 1461, VP1 has additional two amino acids (M K) at the N terminus. The favorable nucleotides, R−3 and G+4 (see text) for translation initiation signals are indicated by boldface based on the scanning model (33).
The regulatory region of Tokyo-1 spans nt 5012 to 274 (391 bp) between the ATG codons of large T antigen and the agnoprotein. In the regulatory region, sequences on the late side of the presumed DNA replication origin are highly divergent among JCV isolates from brain tissues of PML patients. It has been proposed that divergent regulatory sequences have evolved, by deletion and duplication, from the sequence of the archetypal JCV, which was originally found in urine of nonimmunosuppressed patients and healthy individuals (68). The regulatory sequence of Tokyo-1 was compared with the archetypal sequence in the variable region (Fig. 1). The regulatory sequence of the archetype is divided into blocks of 25, 23, 55, 66, and 18 bp (68). Tokyo-1 has the sequence identical to the 25-bp block of the archetype at nt 12 to 36. The sequence of the 23-bp block of the archetype is deleted in the pJCT-TC clone (40). Tokyo-1 has the 75-bp repeats (nt 38 to 112 and 134 to 208), which are composed of the partial sequences of the 55- and 66-bp blocks and the entire sequences of the 18-bp block of the archetype. These observations indicate that the regulatory sequence of Tokyo-1 has evolved from the archetypal sequence by deletion and duplication. In addition to the 75-bp repeats, Tokyo-1 has the specific 20-bp repeats (nt 113 to 132 and 273 to 292), in which the sequence surrounding the ATG start codon for the agnoprotein is duplicated. The regulatory sequences of Tokyo-1 and Mad4 (39) are similar in that they have only one 25-bp block and lack the 23-bp block of the archetype. The regulatory sequence of Tokyo-1 is substantially different from that of Mad1, which has two 25-bp blocks of the archetype within the 98-bp tandem repeats (19).
Transcription of the late RNAs of Tokyo-1 can be initiated from heterogeneous start sites around nt 5112 to 240, based on the analysis of the RNA start sites determined in Mad1 (15, 31) and Mad4 (15). Some of the RNA start sites in Mad1 and Mad4 were mapped downstream of putative surrogate TATA signals, encoded in the sequence corresponding to the 55-bp block of the archetype within the repeat structures (15, 31). By sequence comparison, two putative surrogate TATA signals, TACCTA in Tokyo-1, are identified at nt 64 to 69 and nt 160 to 165 within the 75-bp repeats. One of the candidates for the late RNA start sites in Tokyo-1 is nt 196 to 201, approximately 30 bp downstream of the putative surrogate TATA signal at nt 160 to 165. A potential poly(A) signal, AATAAA, is present at nt 2566 to 2571. JCV has multiple species of late RNAs generated by alternative splicing, with the 5′ splice sites (nt 490, 1583, and possibly 730) and the 3′ splice sites (nt 1425 and 1952), which will be described later. In this report, positions of 5′ or 3′ splice sites will be indicated by the last or first nucleotides of exons, respectively.
Amino acid sequences of the four late proteins of Tokyo-1 were deduced, as in Mad1 (19), based on the sequence similarities to SV40 and BKV: the agnoprotein (71 amino acids, nt 275 to 490), VP2 (344 amino acids, nt 524 to 1558), VP3 (225 amino acids, nt 881 to 1558), and VP1 (354 amino acids, nt 1467 to 2531). The putative coding sequence of the agnoprotein is located downstream of the divergent sequence of the regulatory region, followed by the coding sequences of VP2, VP3, and VP1. The coding sequences of the two minor capsid proteins VP2 and VP3 and that of the major capsid protein VP1 are encoded in an overlapping manner. The entire coding sequence of VP3 overlaps the C-terminal coding sequence of VP2 in the same reading frames. The C-terminal coding sequences of VP2 and VP3 partially overlap the N-terminal coding sequence of VP1 in different reading frames.
Expression vectors carrying the late region of the JCV genomic DNA.
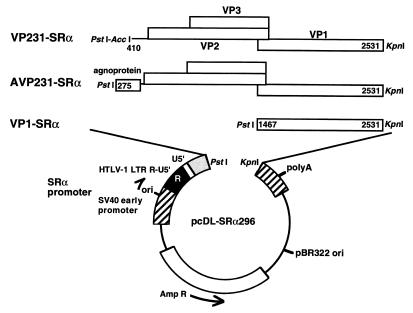
The slow and inefficient replication of JCV in culture systems has been a major barrier in studying the late events of the viral replication cycle. Reduced lytic activities of JCV are due in part to weak promoter-enhancer signals. Therefore, we developed highly efficient eukaryotic expression vectors to study the JCV late region. Fragments of the late region from the genomic DNA of Tokyo-1 were inserted between PstI and KpnI sites of the plasmid vector, pcDL-SRα296 (50, 64) (Fig. 2). Expression of the capsid proteins was under the control of the SRα promoter. This promoter is composed of the SV40 promoter-enhancer unit and R-U5′, the R segment and a part of the U5 sequence from the long terminal repeat of human T-cell leukemia virus type 1 (HTLV-1) (64). The SRα promoter is 50 to 100 times more efficient than the original SV40 promoter (34). The expression vectors VP231-SRα, AVP231-SRα, and VP1-SRα were constructed. VP231-SRα contains the fragment (nt 410 to 2531) which includes the coding sequences of VP2, VP3, and VP1. In VP231-SRα, the 5′ termini of the agnoprotein coding sequence (nt 275 to 409, 135 bp) was deleted to study roles of the agnoprotein and its coding sequence. In contrast, AVP231-SRα contains the fragment (nt 275 to 2531) which includes the coding sequences of the agnoprotein, VP2, VP3, and VP1. VP1-SRα, which contains only the coding sequence of VP1 (nt 1467 to 2531), was constructed to study VP1 expression in the absence of the agnoprotein, VP2, and VP3. Transcription of the inserted JCV late genes initiates at the SV40 RNA start sites and utilizes the SV40 poly(A) signal of pcDL-SRα296 (64). Since pcDL-SRα296 contains the SV40 replication origin, the vector can replicate to high copy number in the presence of SV40 T antigen. High efficiency of protein expression is expected in COS-7 cells, which stably express SV40 T antigen (22).
FIG. 2.
Structures of the three expression vectors VP231-SRα, AVP231-SRα, and VP1-SRα. The eukaryotic expression vector pcDL-SRα296 has the powerful SRα promoter, composed of the SV40 promoter-enhancer unit and a partial long terminal repeat of HTLV-1 (R-U5′). The fragments of the late region of Tokyo-1 were cloned between PstI and KpnI sites downstream of the SRα promoter of pcDL-SRα296. VP231-SRα contains the fragment from JCV Tokyo-1, nt 410 to 2531, encoding VP2, VP3, and VP1. AVP231-SRα contains the fragment from nt 275 to 2531, encoding the agnoprotein, VP2, VP3, and VP1. VP1-SRα contains the fragment from nt 1467 to 2531, the coding sequence of VP1. Transcription of the inserted JCV late genes is initiated from the SV40 RNA start sites within the SRα promoter and is processed by the poly(A) signal of pcDL-SRα296. Since the vectors contain the SV40 replication origin and replicate to high copy number in the presence of SV40 T antigen, highly efficient expression is expected in COS-7 cells, which stably express SV40 T antigen. Abbreviations: ori, SV40 replication origin; polyA, SV40 poly(A) signal; Amp R, ampicillin resistance gene.
The leader sequence of the JCV late RNAs.
Structures of JCV late RNAs have not been determined. In SV40, late mRNAs are highly heterogeneous in the leader sequence upstream of the capsid proteins. The open reading frame (ORF) of the agnoprotein is present in the leader sequences of approximately two-thirds of the SV40 late RNAs. On other SV40 late RNAs, the ORF of the agnoprotein is disrupted due to splicing within the leader sequence (24, 59) (see Fig. 10). Therefore, to analyze the presence of the ORF of the JCV agnoprotein, JCV late RNAs were examined to determine whether they are spliced in the leader sequence.
FIG. 10.
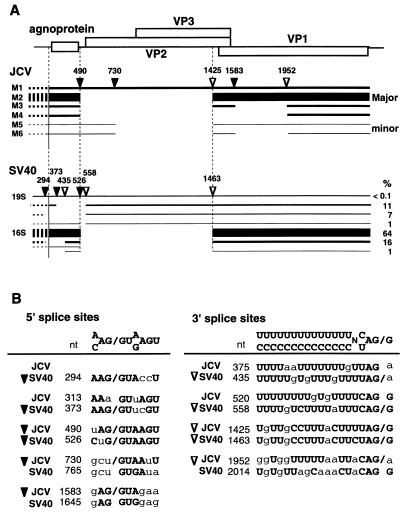
Structures and splice sites of late RNAs of JCV and SV40. JCV and SV40 share 70% nucleotide identity and have similarities in organization of the genomic DNA. Structures and splice sites of the late RNAs of the two viruses show similarity to some extent but are substantially different from each other. (A) Structures of the late RNAs of JCV and SV40 illustrated under the genome organization. The 5′ and 3′ splice sites are indicated by closed and open triangles, respectively. In addition to the corresponding splice sites between the two viruses, JCV has splice sites which disrupt the ORF of VP1 and possibly VP2, while SV40 has splice sites which disrupt the ORF of the agnoprotein. The levels of the different species of the SV40 late RNAs are from Good et al. (24) and Somasekhar and Mertz (61). In the JCV late RNAs, dotted lines indicate the 5′ ends of the late RNAs, which have not been determined experimentally in Tokyo-1. In the SV40 late RNAs, dotted lines indicate the heterogeneous 5′ ends of the late RNAs resulting from the multiple RNA start sites. (B) Alignment of nucleotide sequences of the splice sites and the corresponding sequences between JCV and SV40. Nucleotides identical to the consensus sequences defined for splice sites are indicated by boldfaces uppercase letters. Exon-intron boundaries are indicated by slashes. The 5′ and the 3′ splice sites are marked with closed and open triangles, respectively.
Potential splice sites of JCV Tokyo-1 were analyzed by using the computer program HSPL of BCM Genefinder (59). In the leader sequence of the late RNAs, 5′ splice sites including nt 296, 313, 317, 398, 490, and 730 and 3′ splice sites including nt 269, 375, 399, 520, and 575 were predicted (Fig. 3A). In this report, splicing reactions and splice junctions will be indicated by positions of 5′ and 3′ splice sites separated by a slash. For example, nt 313/520 indicates the splicing reaction or the splice junction using the 5′ splice site at nt 313 and the 3′ splice site at nt 520. The pairs of JCV potential splice sites, nt 313/520 and nt 490/520, correspond to the pairs of SV40 splice sites, nt 373/558 and nt 526/558, respectively (Fig. 3A; see also Fig. 10).
FIG. 3.
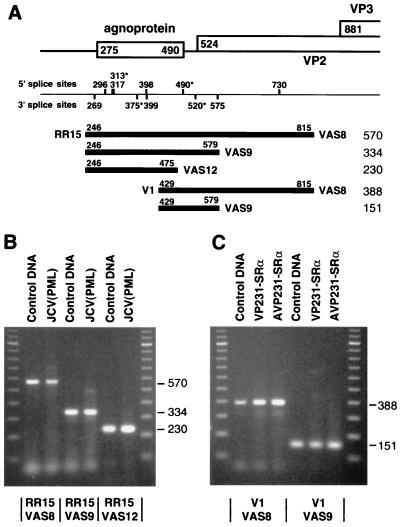
Leader sequence of JCV late RNAs. The structure of the leader sequence of JCV late RNAs was analyzed by RT-PCR using total RNA extracted from a biopsy specimen of a PML brain [JCV(PML)] and from COS-7 cells transfected with VP231-SRα or AVP231-SRα. Most of JCV late RNAs are not spliced in the leader sequence and encode the ORF of the agnoprotein. (A) Experimental designs of RT-PCR and summary of results. Below the genome structure, potential splice sites in the leader sequence are indicated based on the prediction by the computer program BCM Genefinder (59). The potential JCV splice sites marked with asterisks (nt 313, 375, 490, and 520) correspond to the splice sites used in SV40 late RNAs. Below the potential splice sites, the fragments amplified by RT-PCR with five pairs of primers are illustrated. The 5′ primers (and positions) are RR15 (nt 246 to 265) and V1 (nt 429 to 448); and the 3′ primers are VAS8 (nt 815 to 796), VAS9 (nt 579 to 560), and VAS12 (nt 475 to 456). The length of each product (in nucleotides) is shown on the right. (B) RT-PCR products amplified from the JCV(PML) RNA with primer pairs RR15-VAS8, RR15-VAS9, and RR15-VAS12. (C) RT-PCR products amplified from the VP231-SRα- or AVP231-SRα-transfected cell RNA with primer pairs V1-VAS8 and V1-VAS9. The control DNA is the cloned JCV genomic DNA, pJCT-TC of Tokyo-1 (40). PCR products and 100-bp markers were electrophoresed on 2% agarose gels in Tris-acetate-EDTA buffer.
JCV RNAs from a biopsy specimen of a PML brain were analyzed by RT-PCR with three pairs of primers: RR15-VAS8, RR15-VAS9, and RR15-VAS12. The locations of these primers are illustrated in Fig. 3A. JCV potential splice sites nt 313/520 and nt 490/520 were spanned with primer pairs RR15-VAS8 and RR15-VAS9. The RT-PCR products of JCV RNAs were the same length as the PCR products of JCV genomic DNA (control DNA), which were 570, 334, and 230 bp in length, with primer pairs RR15-VAS8, RR15-VAS9, and RR15-VAS12, respectively (Fig. 3B). JCV RNAs spliced at either nt 313/520 or nt 490/520 were not detected. This result indicates that most of the JCV late RNAs are not spliced in the leader sequence.
RNAs from COS-7 cells transfected with VP231-SRα or AVP231-SRα were examined to further analyze the presence of late RNAs that are spliced at nt 490/520 with primer pairs V1-VAS8 and V1-VAS9 (Fig. 3A). The 5′ primer V1 was used since these expression vectors do not contain the annealing site of RR15. The RT-PCR products of the vector-derived RNAs were the same length as the PCR products of JCV genomic DNA (control DNA), and RNAs spliced at nt 490/520 were not detected (Fig. 3C). These results consistently indicate that most of the JCV late RNAs are not spliced in the leader sequence. Therefore, the ORF of the JCV agnoprotein is present in the leader sequence upstream of those of the capsid proteins.
Structures of the M1 to M6 late RNAs.
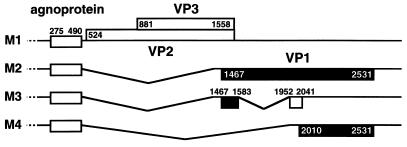
Next, structures of the late RNAs encoding the ORFs of VP1, VP2, and VP3 were determined. As summarized in Fig. 4A, multiple species of JCV late RNAs, designated M1 to M6 RNAs, are generated by alternative splicing. M1 is an unspliced RNA. M2 to M4 RNAs are alternatively spliced by using the common 5′ splice site at nt 490, and potential M5 and M6 RNAs are spliced by using the common 5′ splice site at nt 730. Splice sites were identified in M2 at nt 490/1425 (934-nt intron), in M3 at nt 490/1425 (934-nt intron) and nt 1583/1952 (368-nt introns), in M4 at nt 490/1952 (1,461-nt intron), in M5 at nt 730/1425 (694-nt intron), and in M6 at nt 730/1425 (694-nt intron) and nt 1583/1952 (368-nt intron). By RT-PCR, long RNA fragments containing the entire ORFs of VP1, VP2, and VP3 were hardly detected from a biopsy specimen of a PML brain. Since PML is a degenerative disorder accompanied with cell death, RNA from a PML brain tissue is substantially degraded. Therefore, we analyzed RNA from cells transfected with the expression vectors by using primers which span long fragments containing the ORFs of the three capsid proteins. Then the results were confirmed in RNA from the biopsy specimen by using primers spanning shorter fragments.
FIG. 4.
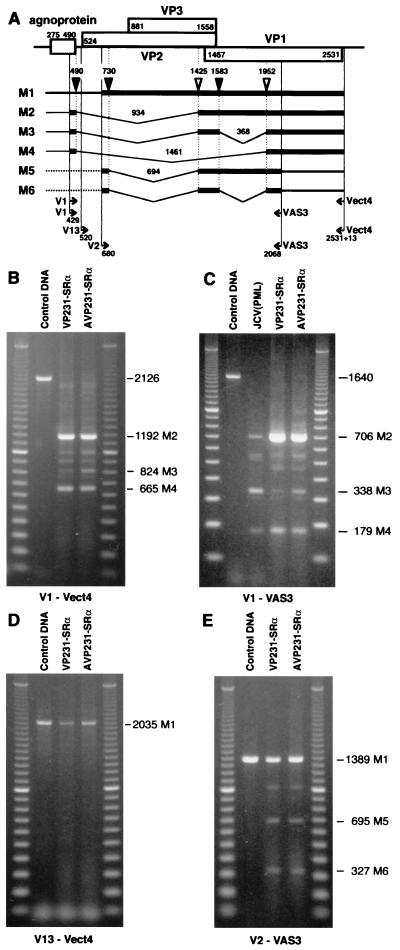
Structures of M1 to M6 RNAs. JCV late RNAs, which span the ORFs of VP1, VP2, and VP3, were analyzed by RT-PCR using total RNA extracted from the biopsy specimen of a PML brain and those from COS-7 cells transfected with VP231-SRα or AVP231-SRα. Six species of RNAs (M1 to M6) were detected, and splice sites were determined by sequencing the RT-PCR products. (A) Structures of M1 to M6 late RNAs schematically illustrated below the organization of viral genomic DNA. Three 5′ splice sites (filled triangles) were identified at nt 490, 730, and 1583; two 3′ splice sites (open triangles) were identified at nt 1425 and 1952. Bold lines indicate the regions amplified by RT-PCR; dotted lines indicate regions which are not determined experimentally. Positions of the 5′ termini of the primers are indicated below the structures of the late RNAs. The 5′ primers (and positions) are V1 (nt 429 to 448), V13 (nt 520 to 539), and V2 (nt 680 to 699); the 3′ primer is VAS3 (nt 2068 to 2049). The 5′ terminus of the 3′ primer Vect4 is located 23 nt downstream of the stop codon of VP1 (2531+23). (B) RT-PCR products amplified with the primer pair V1- Vect4 by using RNAs from cells transfected with VP231-SRα or AVP231-SRα. The products of 1,192, 824, and 665 bp correspond to M2, M3, and M4 RNAs, respectively. (C) RT-PCR products amplified with the primer pair V1-VAS3 by using RNAs from the PML brain RNA [JCV(PML)] and those from COS-7 cells transfected with VP231-SRα or AVP231-SRα. The products of 706, 338, and 179 bp correspond to M2, M3, and M4 RNAs, respectively. (D) RT-PCR products amplified with the primer pair V13-Vect4 by using RNAs from COS-7 cells transfected with VP231-SRα or AVP231-SRα. The product of 2,035 bp corresponds to M1 RNA. (E) RT-PCR products amplified with the primer pair V2-VAS3 from COS-7 cells transfected with VP231-SRα or AVP231-SRα. The products of 1,389, 695, and 327 bp correspond to M1, M5, and M6 RNAs, respectively. The control DNA is VP231-SRα, which contains the genomic fragment of Tokyo-1. PCR products and 100-bp markers were electrophoresed on 0.8% agarose gels (B, D, and E) or 2% agarose gels (C) in Tris-acetate-EDTA buffer.
First, the late RNAs were analyzed in COS-7 cells transfected with VP231-SRα or AVP231-SRα with the primer pair V1-Vect4. As illustrated in Fig. 4A, the 5′ primer V1 was located upstream of the 5′ splice site at nt 490, and the 3′ primer Vect4 was located immediately downstream of the 3′ end of VP1, within the vector sequence of pcDL-SRα296. With the primer pair V1-Vect4, fragments corresponding to M2 (1,192 bp), M3 (824 bp), and M4 (665 bp) were amplified (Fig. 4B). Splice sites used in these RNAs were identified by sequencing the RT-PCR products. The sequences of the 5′ splice sites, nt 490 (uAG/GUAAGU) and nt 1583 (gAG/GUAgaa) (the nucleotides identical to the consensus sequences are indicated in uppercase) conformed to the consensus sequence for 5′ splice sites, (A or C)AG/GU (A or G)AGU. The sequences of the 3′ splice sites, nt 1425 (UgUUgCCUUUaCUUUUAG/G) and nt 1952 (ggUggUUUUUaaUUaCAG/a) conformed to the consensus sequence for 3′ splice sites, (C or U) nN (C or U)AG/G (56).
The presence of M2 to M4 RNAs was examined in the PML brain tissue with the primer pair V1-VAS3, which spans a short region including the splice sites identified in the vector-derived RNAs (Fig. 4A). With the primer pair V1-VAS3, fragments corresponding to M2 (706 bp), M3 (338 bp), and M4 (179 bp) were amplified both from the PML brain tissue and from the transfected COS-7 cells (Fig. 4C). The splice sites of M2-M4 RNAs were identical both in the PML brain tissue and in the transfected cells by sequencing the RT-PCR products.
Next, M1 RNA was analyzed in transfected cells with the primer pair V13-Vect4. To detect M1 but not M2 to M4, the 5′ primer V13 (nt 520 to 539) was located downstream of the 5′ splice site at nt 490 within the introns of M2 to M4 (Fig. 4A). The primer pair V13-Vect4 spans the ORF of VP2, VP3, and VP1. The RT-PCR products corresponding to M1 (2,035 bp) were of the same size as the PCR product of JCV genomic DNA (control DNA). The presence of M1 was also confirmed in the PML brain tissue by using several pairs of primers, including V13-VAS5 (VAS5, nt 1588 to 1569), which spans the ORF of VP2 and VP3 (data not shown).
Finally, two minor species, M5 and M6 RNAs, were detected in transfected COS-7 cells but not in the PML brain tissue (Fig. 4E). The presence or absence of rare M5 and M6 RNAs in the PML brain could not be determined. The sequence of the 5′ splice site at 730 used in M5 and M6, gcu/GUAAuU, is weakly homologous to the consensus sequence for 5′ splice sites. In addition to M1 to M6 RNAs, several other fragments were amplified from the PML brain tissue and transfected cells, although their sequences remain unresolved. These are the fragments about 2,000, 1,000, and 900 bp in length with V1-Vect4 (Fig. 4B), that of about 540 bp with V1-VAS3 (Fig. 4C), and that of about 1,010 bp with V2-VAS3 (Fig. 4E). Some of them were consistently detected with different pairs of primers, which may suggest that JCV has spliced RNAs other than M1 to M6.
JCV late RNAs are polycistronic RNAs.
On JCV M1 to M4 RNAs, potential ORFs are deduced as presented in Fig. 5. According to the scanning model (32, 33), translation begins efficiently at the first AUG codon in the 5′ proximity, although translation also begins at a downstream AUG codon in some cases. Within the most efficient sequence for translation initiation, GCC ACC AUG G, counting A of the AUG codon as +1, a purine, preferably A at the −3 position (R−3 = A−3 or G−3) and G at the +4 position (G+4) are particularly significant (33). Nucleotides which are significant for translation initiation are underlined.
FIG. 5.
ORFs encoded on M1 to M4 RNAs, deduced based on the scanning model (32, 33). M1 RNA encodes the agnoprotein (71 amino acids, nt 275 to 490), VP2 (344 amino acids, nt 524 to 1558), and VP3 (225 amino acids, nt 881 to 1558). M2 RNA encodes the agnoprotein and VP1 (354 amino acids, nt 1467 to 2531). M3 RNA can encode the agnoprotein and a new potential ORF (68 amino acids, nt 1467 to 1583 and 1952 to 2041). M4 RNA can encode the agnoprotein and another new potential ORF (173 amino acids, nt 2010 to 2531).
M1 encodes the ORFs of the agnoprotein, VP2, VP3, and VP1. However, it would be only the agnoprotein, VP2, and VP3, but not VP1, that are translated from M1 RNA. The AUG start codon for the agnoprotein is surrounded by the sequence cug GCC AUG G (nt 269 to 278), which has both the favorable nucleotides, G−3 and G+4 (nucleotides identical to the consensus sequence are indicated in uppercase). The AUG start codon for VP2 surrounded by the sequence of agg uuC AUG G (nt 518 to 527) has G+4 but lacks R−3, while the AUG start codon for VP3 surrounded by the sequence of cCa GCu AUG G (nt 875 to 884) has both favorable nucleotides, G−3 and G+4. This observation suggests that the translation initiation signal for VP3 may be more efficient than the initiation signal for VP2. The ORF of VP1 is also present on M1 RNA; however, it is unlikely that VP1 is translated from M1 RNA due to the presence of at least 13 AUG triplets upstream of the AUG start codon for VP1. Indeed, in the case of SV40, VP1 is not translated from the 19S RNAs, although the ORF of VP1 is present on 19S RNAs (23).
M2 encodes the agnoprotein and VP1. Downstream of the AUG start codon for the agnoprotein, the two AUG codons (underlined) are located in frame at nt 1461 and 1467, CAU AUG AAG AUG GCC (nt 1458 to 1472). These two AUG codons are the potential AUG start codons for VP1. The second AUG codon, surrounded by the sequence, aug Aag AUG G, has the favorable nucleotides, A−3 and G+4; in contrast, the first AUG codon has neither R−3 nor G+4. Even though it is not determined which AUG is used for translation of VP1, the second AUG (nt 1467 to 1469) is tentatively assigned as the VP1 translation initiation signal as previously suggested for JCV Mad1 strain (19).
M3 and M4 encode the agnoprotein and potentially new ORFs. On M3 RNA, the ORF of VP1 is disrupted by splicing at nt 1583/1952, and a new putative ORF (68 amino acids, nt 1467 to 1583 and 1952 to 2041) is generated. The N-terminal region of this new ORF is identical to the N-terminal region of VP1, but the frame is shifted in the C-terminal region downstream of the splice junction. On M4, the intron containing the AUG start codons for VP2, VP3, and VP1 is excised by splicing at nt 490/1952. Following the AUG start codon for the agnoprotein, the next three AUG codons are located within the sequences aCC cag AUG G (nt 1960 to 1969), caa Aga AUG c (nt 1981 to 1990), and caa Gua AUG a (nt 2004 to 2013). Each of the AUG codons has either R−3 or G+4. If translation is initiated from these AUG codons, the lengths of the translated peptides will be 14, 7, and 173 amino acids, respectively. The ORF (173 amino acids, nt 2010 to 2531) which is identical to the C-terminal region of VP1 is tentatively presented as a potential new ORF (Fig. 5).
The presence of the ORF of the agnoprotein or the leader sequence (nt 275 to 409) decreases the expression level of VP1.
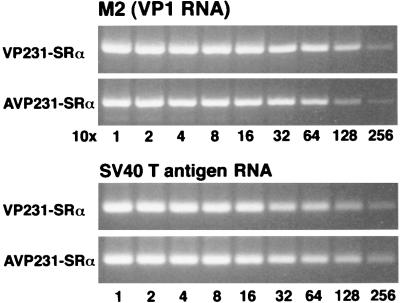
JCV M1 to M4 RNAs are polycistronic RNAs and encode the ORF of the agnoprotein in the leader sequence. It is not known whether the leader sequence encoding the agnoprotein influences expression of the downstream ORFs. To investigate its potential effect on expression of major capsid protein VP1, we transfected COS-7 cells with VP231-SRα or AVP231-SRα and then compared the expression levels of M2 (VP1 mRNA) and VP1 protein in cells transfected with each of the expression vectors. Transfection and replication efficiencies of the two vectors were identical, based on analysis of plasmid DNAs extracted from transfected cells.
First, to study the expression of M2 (VP1 mRNA), total RNA was extracted from cells transfected with each of the vectors. The harvested RNA was first diluted 1:10 and then serially diluted twofold (1:10 to 1:2,560). The serially diluted RNA was analyzed by RT-PCR with the primer pair V1-VAS3. The products corresponding to the M2 fragment (706 bp) were semiquantitatively compared. M2 expression from VP231-SRα was slightly higher than that from AVP231-SRα (Fig. 6). This small difference in M2 expression levels was within twofold. RNA recovery from transfected cells was found to be nearly identical by analyzing SV40 large T antigen RNA as an internal control for COS-7 cells (Fig. 6).
FIG. 6.
M2 (VP1 RNA) expression levels in cells transfected with VP231-SRα or AVP231-SRα. To test potential roles of the agnoprotein and its coding sequence in expression of M2, COS-7 cells were transfected with VP231-SRα or AVP231-SRα, and total RNA was extracted from the transfected cells. Total RNA was first diluted 1:10 and then serially diluted twofold (1:10 to 1:2,560). The RNA was analyzed by RT-PCR with the primer pair V1-VAS3; then the products from M2 (706 bp) were semiquantitatively compared. M2 expression from VP231-SRα-transfected cells was slightly higher than that from AVP231-SRα-transfected cells. This small difference in M2 expression levels was roughly within twofold. The same RNA recovery was demonstrated by analyzing SV40 large T antigen RNA as an internal control for COS-7 cells.
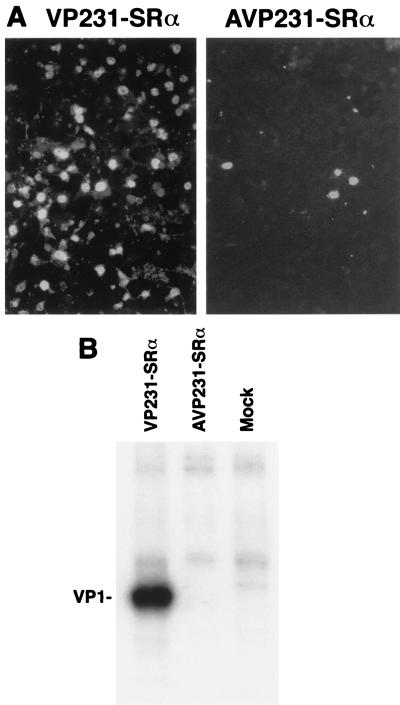
Next, VP1 protein expression was examined in cells transfected with VP231-SRα or AVP231-SRα by immunocytochemistry and radioimmunoprecipitation (Fig. 7). By immunocytochemistry, when cells were transfected with VP231-SRα, 60 to 70% of cells were VP1 positive, while when cells were transfected with AVP231-SRα, less than 5% of cells were VP1 positive (Fig. 7A). By radioimmunoprecipitation, VP1 was detected as a strikingly dominant band in cells transfected with VP231-SRα but was undetectable in those transfected with AVP231-SRα. There was no evidence of degraded VP1 molecules in cells transfected with AVP231-SRα (Fig. 7B).
FIG. 7.
Enhanced expression of VP1 by deleting the 5′ termini of the agnoprotein coding sequence (nt 275 to 409). To test potential roles of the agnoprotein and its coding sequence in expression of VP1, COS-7 cells were transfected with VP231-SRα or AVP231-SRα. Expression of VP1 was analyzed by immunocytochemistry and radioimmunoprecipitation using the anti-JCV antibody, which recognizes VP1. VP1 expression by VP231-SRα was strikingly enhanced by deleting the 5′-terminal region of the agnoprotein coding sequence (nt 275 to 409, 135 bp). The increase in VP1 protein expression level was markedly greater than the increase in M2 (VP1 RNA) RNA expression level, suggesting enhanced translation efficiency. (A) Immunocytochemistry of cells transfected with VP231-SRα (left) and AVP231-SRα (right). When cells were transfected with VP231-Srα, 60 to 70% of cells were VP1 positive; when cells were transfected with AVP231-SRα, less than 5% of cells were VP1 positive. (B) Immunoprecipitation of 35S-labeled VP1 expressed by VP231-SRα and by AVP231-SRα. VP1 expression by VP231-SRα was detected as a strikingly dominant band, while VP1 expression by AVP231-SRα was undetectable.
Therefore, the expression level of the VP1 protein was markedly higher in cells transfected with VP231-SRα, in which the sequence of nt 275 to 409, the 5′ terminus of the agnoprotein, is deleted. This observation suggests that the presence or absence of the ORF of the agnoprotein or the leader sequence at nt 275 to 409 could regulate the expression level of VP1. Since the difference in M2 (VP1 mRNA) expression levels between the two vectors was much smaller, the presence of the ORF of the agnoprotein or the leader sequence at nt 275 to 409 may decrease the expression level of VP1 mainly by suppressing VP1 translation efficiency.
Inefficiency in nuclear transport of JCV VP1 is due to the unique structure in the N-terminal sequence, KRKGERK.
The major capsid protein VP1 and minor capsid proteins VP2 and VP3 are translated in the cytoplasm; then the three capsid proteins are transported to the nucleus and assembled into the virus particles. To study how VP1 is transported to the nucleus, COS-7 cells were transfected with each of the three expression vectors, AVP231-SRα, VP231-SRα, and VP1-SRα, and then subcellular distribution of VP1 was examined by fluorescence immunocytochemistry using a confocal microscope. When cells were transfected with AVP231-SRα or VP231-SRα, VP1 was efficiently transported to the nucleus (Fig. 8B and C). In contrast, when cells were transfected with VP1-SRα, VP1 was present both in the cytoplasm and in the nucleus (Fig. 8D). In some cells transfected with VP1-SRα, VP1 was distributed predominantly in the cytoplasm and at lower levels in the nucleus. These results indicate that JCV VP1 is efficiently transported to the nucleus in the presence of VP2 and VP3 but is distributed both in the cytoplasm and in the nucleus in their absence.
FIG. 8.
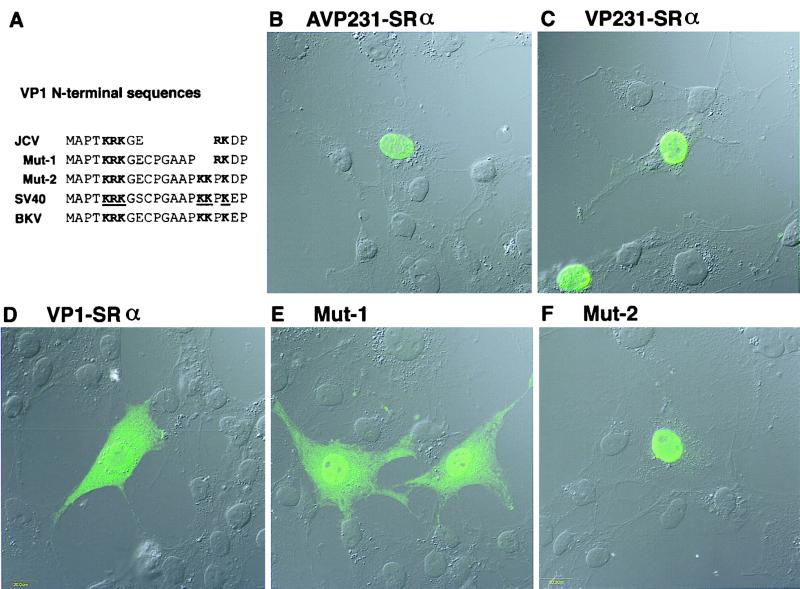
Distribution of VP1 in cells transfected with AVP231-SRα, VP231-SRα, or VP1-SRα and distribution of the two VP1 mutants, Mut-1 and Mut-2. To study nuclear transport of VP1, COS-7 cells were transfected with each of the three expression vectors, and distribution of VP1 or the VP1 mutants was investigated by immunocytochemistry using a confocal microscope. (A) The N-terminal sequences of JCV VP1, JCV VP1 mutants Mut-1 and Mut-2, SV40 VP1, and BKV VP1 are aligned. In the N-terminal sequence of JCV VP1, the basic amino acids KRK and RK (in boldface) are encoded in a monopartite structure. In contrast, in the N-terminal sequence of SV40 VP1, the two clusters of basic amino acids KRK and KKPK are encoded in a bipartite structure and identified as the NLS, as indicated with boldface and underlining (28). Mut-1 encodes the basic amino acids KRK and RK in a bipartite structure, and Mut-2 encodes KRK and KKPK in a bipartite structure. In each row of amino acid sequence, basic amino acids which are likely responsible for nuclear transport are indicated in boldface. (B and C) When cells were transfected with AVP231-SRα or VP231-SRα, VP1 was efficiently transported to the nucleus and identified as numerous speckles, indicating that VP1 is accumulated in discrete subnuclear regions, possibly with VP2 and VP3. (D) When cells were transfected with VP1-SRα, VP1 was distributed both in the cytoplasm and in the nucleus. VP1 was distributed more diffusely in the nucleus. (E) Mut-1 was transported to the nucleus more efficiently and detected more prominently in the nucleus than wild-type VP1. However, Mut-1 was still distributed both in the cytoplasm and in the nucleus. In the nucleus, Mut-1 was distributed diffusely except for nucleoli, and no speckle was identified. (F) Mut-2 was efficiently transported to the nucleus and distributed diffusely except for nucleoli. No speckle was identified in the nucleus.
In contrast to JCV VP1, SV40 VP1 has been shown to be efficiently transported to the nucleus in the absence of VP2 and VP3. The NLS of SV40 VP1 has been mapped to the N-terminal region, K5R6K7 and K16K17, K19 (superscripts indicate positions of residues in the amino acid sequence; boldfaced residues are basic) (Fig. 8A) (28). This NLS of SV40 VP1 is encoded in the sequence, MAPTKRKGSCPGAAPKKPKE, in which two clusters of basic amino acids, KRK and KKPK, are separated with eight residues, GSCPGAAP. Therefore, the NLS of SV40 VP1 is in a bipartite structure. A nearly identical sequence is present in the N-terminal region of BKV VP1. However, in the N-terminal region of JCV VP1, the basic amino acids K5R6K7and R10K11 are encoded in the sequence MAPTKRKGERKD, in which KRK and RK are separated with only two amino acids, GE, and the residues corresponding to the six SV40 amino acids CPGAAP are missing. Thus, unlike the NLS of SV40 VP1, the basic amino acids of JCV VP1, KRK and RK, are encoded in a monopartite structure. A recent study has reported that karyopherin α, which recognizes NLS, harbors two NLS binding sites, suggesting that NLS in a bipartite structure is more efficient than NLS in a monopartite structure (13, 47). Therefore, we hypothesized that the basic amino acids of JCV VP1, KRK and RK, in a monopartite structure may not be as efficient in nuclear transport as the NLS of SV40 VP1. To examine this hypothesis, we mutated the JCV VP1 sequence by inserting the SV40 six amino acids CPGAAP between KRK and RK. This VP1 mutant, designated Mut-1, has the sequence MAPTKRKGECPGAAPRKD, in which KRK and RK are encoded in a bipartite structure. When Mut-1 was expressed in COS-7 cells, Mut-1 was transported to the nucleus more efficiently and detected more prominently in the nucleus than wild-type VP1. However, Mut-1 was still distributed both in the cytoplasm and in the nucleus (Fig. 8E). This observation indicates that inefficiency in nuclear transport of JCV VP1, compared with SV40 VP1, is not due only to a monopartite structure of KRK and RK. Next, we thought that Mut-1 is not transported to the nucleus efficiently, possibly because the basic amino acids RK in JCV VP1 are not as potent in nuclear transport as KKPK in SV40 VP1. Therefore, we replaced the basic amino acids RK in Mut-1 with KKPK to create Mut-2. The N-terminal sequence of Mut-2, MAPTKRKGE9CPGAAPKKPKD20 is identical to the N-terminal sequence of SV40 VP1 except for underlined residues. When Mut-2 was expressed in COS-7 cells, Mut-2 was efficiently transported to the nucleus (Fig. 8F). This result indicates that inefficiency in nuclear transport of JCV VP1 is, also in part, due to the basic amino acids of RK, which is not as potent in nuclear transport as KKPK in SV40 VP1. Taken together, these results suggest that inefficiency in nuclear transport of JCV VP1 is due to the unique N-terminal sequence KRKGERK, a monopartite structure of basic amino acids of KRK, RK, and low potency of the RK residues in nuclear transport.
Discrete intranuclear localization of VP1 in cells transfected with AVP231-SRα and in those transfected with VP231-SRα.
Intranuclear distributions of VP1 and VP1 mutants were further observed with a confocal microscope. When cells were transfected with AVP231-SRα, VP1 was identified as speckles in the nucleus, suggesting that VP1 is localized to discrete subnuclear regions (Fig. 8B). When cells were transfected with VP231-SRα, VP1 was expressed at higher levels and distributed discretely as speckles in the nucleus (Fig. 8C). Distribution of speckles in the nucleus was variable, depending on levels of expression. When VP1 was expressed at a low level, well-isolated speckles were identified mostly near the nuclear membrane. When VP1 was expressed at a high level, speckles were distributed in the entire area of the nucleus, with higher density near the nuclear membrane. These features suggest that the speckles are formed near the nuclear membrane and then spread to the center of the nucleus when more speckles are synthesized. When cells were transfected with VP231-SRα, VP1 was localized both in the nucleus and in the cytoplasm (Fig. 8D). In the nucleus, VP1 was distributed at higher density near the nuclear membrane and at lower density near the center. However, VP1 was distributed more diffusely in the nucleus. The two VP1 mutants, Mut-1 and Mut-2, were transported to the nucleus more efficiently than wild-type VP1 (Fig. 8E and F). In the nucleus, both Mut-1 and Mut-2 were diffusely distributed in the nucleus except for nucleoli, and speckles were not identified. Taken together, these results suggest that in the presence of VP2 and VP3, VP1 proteins are discretely localized within the nucleus and identified as speckles. Although the nature of speckles is not known, the discrete intranuclear localization of VP1 suggests the presence of distinct nuclear regions in which VP1 proteins, possibly with VP2 and VP3, accumulated to a high density.
Assembly of JCV recombinant particles in nuclei of COS-7 cells transfected with VP231-SRα.
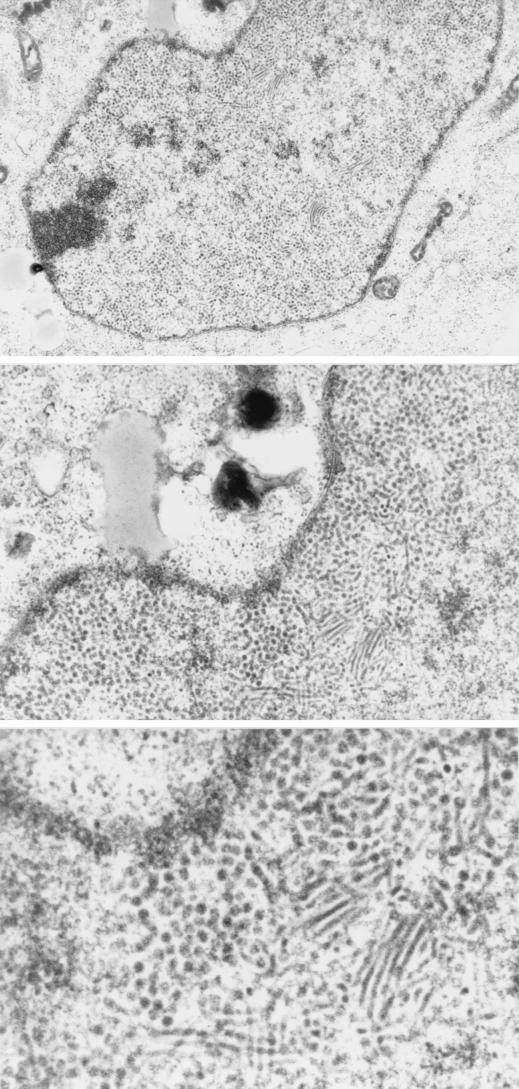
Finally we investigated whether the recombinant capsid proteins synthesized from the expression vectors could be assembled into virus particles in transfected cells (Fig. 9). COS-7 cells were transfected with VP1-SRα or VP231-SRα and analyzed by electron microscopy. When cells were transfected with VP1-SRα, no virus particles were identified either in the nucleus or in the cytoplasm. In contrast, when cells were transfected with VP231-SRα, a large number of round particles (40 nm in diameter) and filamentous forms (30 nm in diameter) of the recombinant particles were identified in the nucleus but not were found in the cytoplasm. The morphological features of these particles were quite similar to those of JCV Tokyo-1 identified in oligodendrocytes in the brain of a Japanese PML patient (43, 44). The proportion of round particles and filamentous forms was variable in each COS-7 cell. Within the nucleus, both round particles and filamentous forms were observed at higher density near the nuclear membrane and at lower density near the center. In most of cells, round particles were present closer to the nuclear membrane compared with the filamentous forms. In PML brains, the crystalloid arrangements of virions were detected in the nucleus and clusters of virions surrounded by membranes were observed in the cytoplasm by electron microscopy (41, 43, 44). However, these structures were not identified in cells transfected with VP231-SRα.
FIG. 9.
Assembly of JCV recombinant particles in the nucleus of COS-7 cells transfected with VP231-SRα. A large number of round particles (40 nm in diameter) and filamentous forms (30 nm in diameter) of the recombinant capsids were detected in the nucleus. The morphological features of these particles were identical to those of JCV Tokyo-1 strain in oligodendrocytes of the brain of a Japanese PML patient (43, 44). Magnification: top, ×10,000; center, ×20,000; bottom, ×40,000.
DISCUSSION
Structures of JCV late RNAs.
JCV and SV40 are similar with respect to genome organization in the late region encoding the agnoprotein, VP2, VP3, and VP1. However, the structures of JCV late RNAs are substantially different from those of SV40 late RNAs due to different splicing patterns (Fig. 10A). In JCV, splice sites are absent within the ORF of the agnoprotein but present within the ORF of VP1. In SV40, however, splice sites are present within the ORF of the agnoprotein but absent within the ORF of VP1. To better understand the differences between the two viruses, sequences of the 5′ and 3′ splice sites in JCV and SV40 are compared with those of the corresponding region as shown in Fig. 10B.
JCV M1 corresponds to the SV40 19S RNAs. In JCV, RNAs spliced in the leader sequence were not detected. In contrast, in SV40 19S RNAs, the leader sequence of most RNAs is spliced at either nt 294/558, nt 373/558, or nt 526/558. This difference can be explained by the absence of 5′ splice sites in the JCV leader sequence. The JCV sequence does not have a 5′ splice site corresponding to the SV40 5′ splice site at nt 294 due to the sequence divergence. The JCV sequence of AAa313GUuAGU (JCV nt 311 to 319) corresponds to the SV40 5′ splice site at nt 373 in the sequence of AAG373/GUucGU (SV40 nt 371 to 379). However, in JCV, the a at nt 313 differs from the consensus sequence for the 5′ splice sites, while in SV40 the G at nt 373 conforms to the consensus sequence. At nt 490 and 520, the JCV sequences well correspond to the SV40 5′ splice sites at 526 and 3′ splice site 558, respectively. The JCV sequences at these positions also well conform to the consensus sequences for 5′ or 3′ splice sites. Although JCV splicing at nt 490/520 was not detectable in this study, it is still possible that JCV late RNAs are spliced at nt 490/520 at a low level, since SV40 RNA spliced at 526/558 represents only 1% of the total late RNAs (24, 61).
JCV M2 corresponds to the SV40 16S RNAs. The sequences of the 5′ splice sites (JCV nt 490; SV40 nt 526) and the 3′ splice sites (JCV nt 1425; SV40 1463) used in these RNAs are highly conserved between the two viruses and nearly identical to the consensus sequences for splice sites. About one-fifth of the SV40 16S RNAs (16% of the total late RNAs) is spliced in the leader sequence at nt 294/435. However, in JCV the sequence corresponding to the SV40 5′ splice site of nt 294 is not present due to the sequence divergence.
JCV M3 and M4, and potentially M5 and M6, may be RNAs unique to JCV. SV40 late RNAs which correspond to JCV M3 to M6 have not been reported. JCV M3, M4, and M6 RNAs use the 3′ splice site at nt 1952, ggUggUUUUUaaUUaCAG/a (JCV 1934 to 1952). The corresponding SV40 sequence UgUgUUagCaaaCUaCAG G (SV40 1996 to 2014) does not have a pyrimidine stretch as in JCV, and so this position in SV40 may not be used as a 3′ splice site. M5 and M6 are minor species of JCV late RNAs. The low amounts of M5 and M6 can be explained by the sequence of the 5′ splice site at nt 730, gcu/GUAAuU (nt 728 to 736), which is weakly homologous to the consensus sequence.
Translation.
Our data suggested that the presence of the ORF of the agnoprotein or the leader sequence at nt 275 to 409 can decrease the expression level of JCV VP1 (Fig. 7). In SV40, the presence of the AUG start codon for the agnoprotein decreases VP1 translation efficiency on the major 16S RNA (64% of the total late RNAs) (Fig. 10A) (25, 52). However, the SV40 16S RNAs are heterogeneous in the leader sequence, in one of which the AUG start codon for the agnoprotein is removed by splicing at nt 294/435 (16% of the total late RNAs). VP1 is translated more efficiently from this RNA. It has been also suggested that this 16S RNA is more frequently used for VP1 translation despite the lower level of RNA expression than the major 16S RNA (4). In the same manner, SV40 VP2 and VP3 are efficiently translated from the 19S RNAs that are spliced in the leader sequence and the AUG start codon of the agnoprotein is deleted (23, 52).
In JCV, however, splicing is not evident in the leader sequence of the late RNAs. The AUG start codon for the agnoprotein is present upstream of the AUG start codon for VP1 on M2 RNA and also upstream of those for VP2 and VP3 on M1 RNA. Therefore, in JCV, translation of the capsid proteins may not be as efficient as in SV40. As shown in Fig. 7, the presence of the ORF of the agnoprotein or the leader sequence at nt 275 to 409 can decrease the expression level of VP1. Although a low, steady expression level of VP1 may be due to reduced VP1 synthesis or rapid VP1 degradation, it is likely that in JCV, as in SV40, the presence of the AUG start codon for the agnoprotein would decrease the translation efficiency of VP1 and possibly VP2 and VP3.
Nuclear transport.
JCV VP1 is unique in that it is distributed both in the cytoplasm and in the nucleus in the absence of VP2 and VP3 (Fig. 8). In contrast, SV40 VP1 (28) and mouse polyomavirus VP1 (11, 42) are autonomously transported to the nucleus. Our mutation analysis indicated that inefficient nuclear transport of JCV VP1 is due to the unique structure in the N-terminal sequence, KRKGERK (Fig. 8). Although in insect cells JCV VP1 has been reported to be autonomously transported to the nucleus (9), this discrepancy can be explained, in part, by distinct mechanisms of nuclear transport in mammalian cell lines and in insect cells. A discrepancy has been also reported for mouse polyomavirus VP2, which was localized in the nucleus in COS-7 cells (10) but found primarily in the cytoplasm and nuclear periphery in insect cells (17).
JCV VP1 is efficiently transported to the nucleus in the presence of VP2 and VP3 (Fig. 8). Cooperation of JCV capsid proteins in nuclear transport was further examined by cotransfection experiments. The vector VP23-SRα was constructed by inserting the fragment of JCV Tokyo-1 (nt 410 to 1725) encoding the coding sequences of VP2 and VP3. When cells were cotransfected with both VP1-SRα and VP23-SRα, VP1 was efficiently transported to the nucleus (data not shown). Similarly, in SV40 and mouse polyomavirus, cotransfection experiments have demonstrated cooperative nuclear transport of the capsid proteins (7, 29). When the NLS of VP1 in SV40 or mouse polyomavirus was deleted or mutated, VP1 was localized in the cytoplasm, but it was transported to the nucleus in the presence of VP2 or VP3. It has been interpreted that the NLS-defective VP1 is associated with VP2 or VP3 in the cytoplasm and then cotransported to the nucleus via the NLS of VP2 and VP3. Direct interaction of VP1 with VP2 and VP3 has also been demonstrated by coimmunoprecipitation (6, 7, 16, 18, 20). The NLSs of SV40 VP2 and VP3 (GPNKKKRKL) and of mouse polyomavirus VP2 and VP3 (EEDGPQKKKRRL) have been mapped to their C-terminal regions (10, 21, 67). In the C-terminal region of JCV VP2 and VP3, we find the amino acid sequence GPNKKKRRK, which is nearly identical to the NLS of SV40 VP2 and VP3 and related to the NLS of mouse polyomavirus VP2 and VP3 (Fig. 11). Therefore, in JCV, it is also likely that VP1 is associated with VP2 or VP3 in the cytoplasm and then cotransported to the nucleus via the NLS of VP2 or VP3.
FIG. 11.
Comparison of the C-terminal sequence of VP2/VP3 between JCV Tokyo-1, Mad1, BKV, SV40, and mouse polyomavirus (PyV). The NLS of SV40 VP2/VP3 mapped by Clever et al. (12) and that of PyV VP2/VP3 was mapped by Chang et al. (10) are indicated with boldface underlined letters. By sequence comparison, the putative NLS of JCV VP2/VP3 (GPNKKKRRK) as well as that of BKV VP2/VP3 are indicated in boldface.
In JCV, interaction of VP1 with VP2 or VP3 in the cytoplasm may be a critical step for efficient nuclear transport, since VP1 is distributed both in the cytoplasm and in the nucleus in the absence of VP2 and VP3. In contrast, in SV40 and mouse polyomavirus, each capsid protein has its own NLS, and so the capsid proteins have the potential to be transported to the nucleus individually (10–12, 18, 21, 28, 42, 67). Our findings for JCV support previous interpretations regarding in SV40 and mouse polyomavirus (6, 7, 16–18, 20, 29) that the capsid proteins interact in the cytoplasm and then cooperatively are transported to the nucleus. We are now preparing antibodies for VP2 and VP3 to further study interactions of VP1, VP2, and VP3 for nuclear transport.
Capsid assembly in the nucleus.
We have shown that JCV recombinant particles are assembled in the nucleus of cells transfected with VP231-SRα encoding VP1, VP2, and VP3 (Fig. 9). The agnoprotein was not essential for capsid assembly in our system. Both round particles and filamentous forms were identified, similarly to oligodendrocytes of the brains of PML patients.
When cells were transfected with AVP231-SRα or VP231-SRα, VP1 was identified as speckles in the nucleus with a confocal microscope. This observation indicates that VP1 is localized to discrete subnuclear regions and not distributed uniformly in the nucleus. In contrast, when cells were transfected with VP1-SRα, VP1 was distributed more diffusely. By electron microscopy, recombinant virus particles were identified as clusters of round and filamentous forms in the nucleus of cells transfected with VP231-SRα. Distribution of clusters of virus particles detected by electron microscopy was consistent with distribution of speckles detected by confocal microscopy. The nature of speckles remains unresolved; however, observation of these speckles indicates that VP1 molecules are accumulated in discrete regions within the nucleus, possibly with VP2 and VP3. It is in these subnuclear regions that the three capsid proteins can be assembled into virus particles, or the virus particles assembled elsewhere can gather each other and organize clusters. Detection of nuclear speckles with a confocal microscope is particularly important, since it suggests the future application of this system in studying dynamic interaction of the three capsid proteins during capsid assembly in analyses using living cells.
The structures of SV40 and mouse polyomavirus have been determined by crystallography (26, 35, 62, 63). The outer surface of these virus particles is composed of 72 pentamers of VP1. At the inner surface, VP2 and/or VP3 has been reported to be extended from the axial cavity of the 72 VP1 pentamers to the core of the virus particles (26). A similar viral structure is expected for JCV. Slow and inefficient replication has been a major barrier in studying the late life cycle of JCV. By using a eukaryotic expression system, we have overcome this difficulty and elucidated unique features of the JCV late life cycle, especially in splicing, translation, nuclear transport, and capsid assembly. Our system provides a useful model with which to understand the late events of the JCV replication cycle and the structures and functions of the three capsid proteins, VP1, VP2, and VP3.
ACKNOWLEDGMENTS
We thank Alison Deckhut for critical review of the manuscript, Caroline Ryschkewitsch for sequencing, and Mami Sato for electron microscopy.
This work was supported in part by Special Coordination Funds (SPSBS) from the Science and Technology Agency of the Japanese Government, a grant for Research Project on Slow Virus Infection from the Japanese Ministry of Health and Welfare, and a grant for special viral research from the Tokyo Metropolitan Government. Y.S.-H. was a recipient of fellowship sponsored by Japan Society Promotion of Science.
REFERENCES
- 1.Agostini H T, Shishido-Hara Y, Baumhefner R W, Singer E J, Ryschkewitsch C F, Stoner G L. JC virus type 2: definition of subtypes based on DNA sequence analysis of ten complete genomes. J Gen Virol. 1998;79:1143–1151. doi: 10.1099/0022-1317-79-5-1143. [DOI] [PubMed] [Google Scholar]
- 2.Akatani K, Imai M, Kimura M, Nagashima K, Ikegami N. Propagation of JC virus in human neuroblastoma cell line IMR-32. J Med Virol. 1994;43:13–19. doi: 10.1002/jmv.1890430104. [DOI] [PubMed] [Google Scholar]
- 3.Aloni Y, Hay N. Attenuation and modulation of mRNA secondary structure in a feedback control system regulating SV40 gene expression. Mol Biol Rep. 1983;9:91–100. doi: 10.1007/BF00777479. [DOI] [PubMed] [Google Scholar]
- 4.Barkan A, Mertz J E. The number of ribosomes on simian virus 40 late 16S mRNA is determined in part by the nucleotide sequence of its leader. Mol Cell Biol. 1984;4:813–816. doi: 10.1128/mcb.4.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barkan A, Welch R C, Mertz J E. Missense mutations in the VP1 gene of simian virus 40 that compensate for defects caused by deletions in the viral agnogene. J Virol. 1987;61:3190–3198. doi: 10.1128/jvi.61.10.3190-3198.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barouch D H, Harrison S C. Interactions among the major and minor coat proteins of polyomavirus. J Virol. 1994;68:3982–3989. doi: 10.1128/jvi.68.6.3982-3989.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai X, Chang D, Rottinghaus S, Consigli R A. Expression and purification of recombinant polyomavirus VP2 protein and its interactions with polyomavirus proteins. J Virol. 1994;68:7609–7613. doi: 10.1128/jvi.68.11.7609-7613.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carswell S, Alwine J C. Simian virus 40 agnoprotein facilitates perinuclear-nuclear localization of VP1, the major capsid protein. J Virol. 1986;60:1055–1061. doi: 10.1128/jvi.60.3.1055-1061.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang D, Fung C Y, Ou W C, Chao P C, Li S Y, Wang M, Huang Y L, Tzeng T Y, Tsai R T. Self-assembly of the JC virus major capsid protein, VP1, expressed in insect cells. J Gen Virol. 1997;78:1435–1439. doi: 10.1099/0022-1317-78-6-1435. [DOI] [PubMed] [Google Scholar]
- 10.Chang D, Haynes II J I, Brady J N, Consigli R A. Identification of a nuclear localization sequence in the polyomavirus capsid protein VP2. Virology. 1992;191:978–983. doi: 10.1016/0042-6822(92)90276-u. [DOI] [PubMed] [Google Scholar]
- 11.Chang D, Haynes II J I, Brady J N, Consigli R A. The use of additive and subtractive approaches to examine the nuclear localization sequence of the polyomavirus major capsid protein VP1. Virology. 1992;189:821–827. doi: 10.1016/0042-6822(92)90615-v. [DOI] [PubMed] [Google Scholar]
- 12.Clever J, Kasamatsu H. Simian virus 40 Vp2/3 small structural proteins harbor their own nuclear transport signal. Virology. 1991;181:78–90. doi: 10.1016/0042-6822(91)90472-n. [DOI] [PubMed] [Google Scholar]
- 13.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 14.Dabrowski C, Alwine J C. Translational control of synthesis of simian virus 40 late proteins from polycistronic 19S late mRNA. J Virol. 1988;62:3182–3192. doi: 10.1128/jvi.62.9.3182-3192.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniel A M, Frisque R J. Transcription initiation sites of prototype and variant JC virus early and late messenger RNAs. Virology. 1993;194:97–109. doi: 10.1006/viro.1993.1239. [DOI] [PubMed] [Google Scholar]
- 16.Delos S E, Cripe T P, Leavitt A D, Greisman H, Garcea R L. Expression of the polyomavirus minor capsid proteins VP2 and VP3 in Escherichia coli: in vitro interactions with recombinant VP1 capsomeres. J Virol. 1995;69:7734–7742. doi: 10.1128/jvi.69.12.7734-7742.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delos S E, Montross L, Moreland R B, Garcea R L. Expression of the polyomavirus VP2 and VP3 proteins in insect cells: coexpression with the major capsid protein VP1 alters VP2/VP3 subcellular localization. Virology. 1993;194:393–398. doi: 10.1006/viro.1993.1274. [DOI] [PubMed] [Google Scholar]
- 18.Forstova J, Krauzewicz N, Wallace S, Street A J, Dilworth S M, Beard S, Griffin B E. Cooperation of structural proteins during late events in the life cycle of polyomavirus. J Virol. 1993;67:1405–1413. doi: 10.1128/jvi.67.3.1405-1413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frisque R J, Bream G L, Cannella M T. Human polyomavirus JC virus genome. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gharakhanian E, Takahashi J, Clever J, Kasamatsu H. In vitro assay for protein-protein interaction: carboxyl-terminal 40 residues of simian virus 40 structural protein VP3 contain a determinant for interaction with VP1. Proc Natl Acad Sci USA. 1988;85:6607–6611. doi: 10.1073/pnas.85.18.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gharakhanian E, Takahashi J, Kasamatsu H. The carboxyl 35 amino acids of SV40 Vp3 are essential for its nuclear accumulation. Virology. 1987;157:440–448. doi: 10.1016/0042-6822(87)90286-8. [DOI] [PubMed] [Google Scholar]
- 22.Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 23.Good P J, Welch R C, Barkan A, Somasekhar M B, Mertz J E. Both VP2 and VP3 are synthesized from each of the alternative spliced late 19S RNA species of simian virus 40. J Virol. 1988;62:944–953. doi: 10.1128/jvi.62.3.944-953.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Good P J, Welch R C, Ryu W S, Mertz J E. The late spliced 19S and 16S RNAs of simian virus 40 can be synthesized from a common pool of transcripts. J Virol. 1988;62:563–571. doi: 10.1128/jvi.62.2.563-571.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grass D S, Manley J L. Selective translation initiation on bicistronic simian virus 40 late mRNA. J Virol. 1987;61:2331–2335. doi: 10.1128/jvi.61.7.2331-2335.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffith J P, Griffith D L, Rayment I, Murakami W T, Caspar D L. Inside polyomavirus at 25-A resolution. Nature. 1992;355:652–654. doi: 10.1038/355652a0. [DOI] [PubMed] [Google Scholar]
- 27.Hay N, Aloni Y. Attenuation of late simian virus 40 mRNA synthesis is enhanced by the agnoprotein and is temporally regulated in isolated nuclear systems. Mol Cell Biol. 1985;5:1327–1334. doi: 10.1128/mcb.5.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishii N, Minami N, Chen E Y, Medina A L, Chico M M, Kasamatsu H. Analysis of a nuclear localization signal of simian virus 40 major capsid protein Vp1. J Virol. 1996;70:1317–1322. doi: 10.1128/jvi.70.2.1317-1322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishii N, Nakanishi A, Yamada M, Macalalad M H, Kasamatsu H. Functional complementation of nuclear targeting-defective mutants of simian virus 40 structural proteins. J Virol. 1994;68:8209–8216. doi: 10.1128/jvi.68.12.8209-8216.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jay G, Nomura S, Anderson C W, Khoury G. Identification of the SV40 agnogene product: a DNA binding protein. Nature. 1981;291:346–349. doi: 10.1038/291346a0. [DOI] [PubMed] [Google Scholar]
- 31.Kenney S, Natarajan V, Salzman N P. Mapping 5′ termini of JC virus late RNA. J Virol. 1986;58:216–219. doi: 10.1128/jvi.58.1.216-219.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 33.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 34.Kriegler M. Gene transfer and expression: a laboratory manual. New York, N.Y: Stockton Press; 1990. [Google Scholar]
- 35.Liddington R C, Yan Y, Moulai J, Sahli R, Benjamin T L, Harrison S C. Structure of simian virus 40 at 3.8-A resolution. Nature. 1991;354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- 36.Major E O, Miller A E, Mourrain P, Traub R G, de Widt E, Sever J. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc Natl Acad Sci USA. 1985;82:1257–1261. doi: 10.1073/pnas.82.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandl C W, Frisque R J. Characterization of cells transformed by the human polyomavirus JC virus. J Gen Virol. 1986;67:1733–1739. doi: 10.1099/0022-1317-67-8-1733. [DOI] [PubMed] [Google Scholar]
- 38.Margolskee R F, Nathans D. Suppression of a VP1 mutant of simian virus 40 by missense mutations in serine codons of the viral agnogene. J Virol. 1983;48:405–409. doi: 10.1128/jvi.48.2.405-409.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin J D, King D M, Slauch J M, Frisque R J. Differences in regulatory sequences of naturally occurring JC virus variants. J Virol. 1985;53:306–311. doi: 10.1128/jvi.53.1.306-311.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuda M, Jona M, Yasui K, Nagashima K. Genetic characterization of JC virus Tokyo-1 strain, a variant oncogenic in rodents. Virus Res. 1987;7:159–168. doi: 10.1016/0168-1702(87)90077-3. [DOI] [PubMed] [Google Scholar]
- 41.Mazlo M, Tariska I. Morphological demonstration of the first phase of polyomavirus replication in oligodendroglia cells of human brain in progressive multifocal leukoencephalopathy (PML) Acta Neuropathol. 1980;49:133–143. doi: 10.1007/BF00690753. [DOI] [PubMed] [Google Scholar]
- 42.Moreland R B, Garcea R L. Characterization of a nuclear localization sequence in the polyomavirus capsid protein VP1. Virology. 1991;185:513–518. doi: 10.1016/0042-6822(91)90811-o. [DOI] [PubMed] [Google Scholar]
- 43.Nagashima K, Yamaguchi K, Nakase H, Miyazaki J. Progressive multifocal leukoencephalopathy. A case report and review of the literature. Acta Pathol Jpn. 1982;32:333–343. [PubMed] [Google Scholar]
- 44.Nagashima K, Yamaguchi K, Yasui K, Ogiwara H. Progressive multifocal leukoencephalopathy. Neuropathology and virus isolation. Acta Pathol Jpn. 1981;31:953–961. [PubMed] [Google Scholar]
- 45.Nagashima K, Yasui K, Kimura J, Washizu M, Yamaguchi K, Mori W. Induction of brain tumors by a newly isolated JC virus (Tokyo-1 strain) Am J Pathol. 1984;116:455–463. [PMC free article] [PubMed] [Google Scholar]
- 46.Ng S C, Mertz J E, Sanden-Will S, Bina M. Simian virus 40 maturation in cells harboring mutants deleted in the agnogene. J Biol Chem. 1985;260:1127–1132. [PubMed] [Google Scholar]
- 47.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 48.Nukuzuma S, Yogo Y, Guo J, Nukuzuma C, Itoh S, Shinohara T, Nagashima K. Establishment and characterization of a carrier cell culture producing high titres of polyoma JC virus. J Med Virol. 1995;47:370–377. doi: 10.1002/jmv.1890470413. [DOI] [PubMed] [Google Scholar]
- 49.Ohsumi S, Ikehara I, Motoi M, Ogawa K, Nagashima K, Yasui K. Induction of undifferentiated brain tumors in rats by a human polyomavirus (JC virus) Jpn J Cancer Res. 1985;76:429–431. [PubMed] [Google Scholar]
- 50.Okayama H, Berg P. A cDNA cloning vector that permits expression of cDNA inserts in mammalian cells. Mol Cell Biol. 1983;3:280–289. doi: 10.1128/mcb.3.2.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Padgett B L, Walker D L, ZuRhein G M, Varakis J N. Differential neurooncogenicity of strains of JC virus, a human polyoma virus, in newborn Syrian hamsters. Cancer Res. 1977;37:718–720. [PubMed] [Google Scholar]
- 52.Resnick J, Shenk T. Simian virus 40 agnoprotein facilitates normal nuclear location of the major capsid polypeptide and cell-to-cell spread of virus. J Virol. 1986;60:1098–1106. doi: 10.1128/jvi.60.3.1098-1106.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rinaldo C H, Traavik T, Hey A. The agnogene of the human polyomavirus BK is expressed. J Virol. 1998;72:6233–6236. doi: 10.1128/jvi.72.7.6233-6236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sedman S A, Good P J, Mertz J E. Leader-encoded open reading frames modulate both the absolute and relative rates of synthesis of the virion proteins of simian virus 40. J Virol. 1989;63:3884–3893. doi: 10.1128/jvi.63.9.3884-3893.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seif I, Khoury G, Dhar R. The genome of human papovavirus BKV. Cell. 1979;18:963–977. doi: 10.1016/0092-8674(79)90209-5. [DOI] [PubMed] [Google Scholar]
- 56.Sharp P A, Burge C B. Classification of introns: U2-type or U12-type. Cell. 1997;91:875–879. doi: 10.1016/s0092-8674(00)80479-1. [DOI] [PubMed] [Google Scholar]
- 57.Shinohara T, Nagashima K, Major E O. Propagation of the human polyomavirus, JCV, in human neuroblastoma cell lines. Virology. 1997;228:269–277. doi: 10.1006/viro.1996.8409. [DOI] [PubMed] [Google Scholar]
- 58.Shishido Y, Nukuzuma S, Mukaigawa J, Morikawa S, Yasui K, Nagashima K. Assembly of JC virus-like particles in COS7 cells. J Med Virol. 1997;51:265–272. [PubMed] [Google Scholar]
- 59.Solovyev V V, Salamov A A, Lawrence C B. Predicting internal exons by oligonucleotide composition and discriminant analysis of spliceable open reading frames. Nucleic Acids Res. 1994;22:5156–5163. doi: 10.1093/nar/22.24.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Somasekhar M B, Mertz J E. Exon mutations that affect the choice of splice sites used in processing the SV40 late transcripts. Nucleic Acids Res. 1985;13:5591–5609. doi: 10.1093/nar/13.15.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Somasekhar M B, Mertz J E. Sequences involved in determining the locations of the 5′ ends of the late RNAs of simian virus 40. J Virol. 1985;56:1002–1013. doi: 10.1128/jvi.56.3.1002-1013.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stehle T, Gamblin S J, Yan Y, Harrison S C. The structure of simian virus 40 refined at 3.1 A resolution. Structure. 1996;4:165–182. doi: 10.1016/s0969-2126(96)00020-2. [DOI] [PubMed] [Google Scholar]
- 63.Stehle T, Yan Y, Benjamin T L, Harrison S C. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature. 1994;369:160–163. doi: 10.1038/369160a0. [DOI] [PubMed] [Google Scholar]
- 64.Takebe Y, Seiki M, Fujisawa J, Hoy P, Yokota K, Arai K, Yoshida M, Arai N. SR alpha promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tooze J. DNA tumor viruses. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 66.Walker D L, Padgett B L, ZuRhein G M, Albert A E, Marsh R F. Human papovavirus (JC): induction of brain tumors in hamsters. Science. 1973;181:674–676. doi: 10.1126/science.181.4100.674. [DOI] [PubMed] [Google Scholar]
- 67.Wychowski C, Benichou D, Girard M. The intranuclear location of simian virus 40 polypeptides VP2 and VP3 depends on a specific amino acid sequence. J Virol. 1987;61:3862–3869. doi: 10.1128/jvi.61.12.3862-3869.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yogo Y, Kitamura T, Sugimoto C, Ueki T, Aso Y, Hara K, Taguchi F. Isolation of a possible archetypal JC virus DNA sequence from nonimmunocompromised individuals. J Virol. 1990;64:3139–3143. doi: 10.1128/jvi.64.6.3139-3143.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zu Rhein G M, Chou S-M. Particles resembling Papova viruses in human cerebral demyelinating disease. Science. 1965;11:1477–1479. doi: 10.1126/science.148.3676.1477. [DOI] [PubMed] [Google Scholar]
- 70.Zu Rhein G M, Varakis J N. Perinatal induction of medulloblastomas in Syrian golden hamsters by a human polyoma virus (JC) Natl Cancer Inst Monogr. 1979;1979:205–208. [PubMed] [Google Scholar]