Abstract
A cytopathic variant of feline immunodeficiency virus (FIV) strain PPR emerged after passage of wild-type virus on an interleukin-2-independent cell line. The virus, termed FIV-PPRglial, displayed a phenotype markedly different from the parental virus, including the ability to productively infect previously refractory cell lines, induction of large syncytia, and accelerated kinetic properties. A chimeric molecular clone, FIV-PPRchim42, containing the FIV-PPRglial envelope within the backbone of FIV-PPR, exhibited all the characteristics of the FIV-PPRglial phenotype, demonstrating that the viral envelope was responsible for the acquired traits. Subsequent molecular characterization revealed that the FIV-PPRglial envelope contained five amino acid substitutions relative to wild-type FIV-PPR. Mutagenic analyses further demonstrated that the acquired phenotype was minimally attributable to a combination of three mutations, specifically, a glutamine-to-proline change within the second constant domain of the surface protein (SU); a threonine-to-proline change within the V4 loop, also in the SU; and a premature stop codon in the cytoplasmic tail of the transmembrane protein. All three changes were required to produce the FIV-PPRglial phenotype. Cotransfection studies with mutant viruses in combination with each other and with FIV-PPR indicated that the truncated cytoplasmic tail was responsible for the induction of syncytium formation. Receptor usage analyses were pursued, and distinctions were observed between FIV-PPR and FIV-PPRglial. In vitro infections with FIV-PPR, FIV-PPRglial, and FIV-34TF10 on two adherent cell lines were ablated in the presence of SDF1α, the natural ligand for CXCR4. In contrast, viral infection of T cells was not limited to CXCR4 usage, and inhibition studies indicate the potential involvement of a CC chemokine receptor.
The feline immunodeficiency virus (FIV) is a lentivirus of the cat and the etiologic agent of feline AIDS (40). The virus has a broad host cell range both in vivo and in vitro that includes CD4+ cells, CD8+ cells, monocytes/macrophages, and a subset of immunoglobulin G-bearing B cells, (4, 5, 11, 20) yet it remains partial to cells of the CD4+ lineage (1, 4, 60). While CD4 is the primary receptor of the primate lentiviruses, it does not mediate the entry of FIV into target cells (30, 35, 59). The primary receptor for FIV has yet to be determined, although earlier studies suggested a role for the tetraspannin, CD9 (30). Subsequent studies demonstrated that the inhibition by anti-feline CD9 monoclonal antibody involved the prevention of virion release from the cell rather than a block in binding and entry (12). Recently, studies have shown that the alpha 7TMG protein-coupled receptor, CXCR4, the coreceptor for syncytium-inducing, T-cell tropic, and dually tropic isolates of human immunodeficiency virus (HIV) (14), and some laboratory-adapted strains of simian immunodeficiency virus (SIV) (34), is also able to mediate fusion of FIV with Crandell feline kidney cells and additionally with human and murine cells ectopically expressing human CXCR4 (43, 61; B. J. Willett, M. J. Hosie, J. C. Neil, J. Turner, and J. A. Hoxie, Letter, Nature 385:587, 1997). It has yet to be determined whether CXCR4 acts as a primary or a secondary receptor for FIV, although certain isolates of the primate lentiviruses are able to utilize CXCR4 and CCR5 independently of CD4 (17, 19, 28, 31, 44, 45).
Sequence variations and conformational changes within the viral envelope proteins are responsible for determining receptor usage. Recently, chemokine receptor tropism has been linked to sequence variations in the highly charged V3 loop of the envelope surface (SU) protein (10). However, other domains of SU distinct from V3, including the V1/V2 (38) and V4 loops (10, 36) and the constant domains (16), are also important determinants and/or codeterminants of cytotropism in the primate lentiviruses. A study with FIV also reported the importance of the V3 loop in cytotropism, wherein a singular point mutation, conferring a greater charge to the domain, was responsible for the acquired CrFK tropism of this virus (58).
The multifunctional transmembrane protein (TM) also plays a significant role in the determination of host cell range and of the fusogenic and cytopathogenic potential of lentiviruses. It was reported that a single residue alteration within the ectodomain of the TM expanded the host cell range of a Dutch isolate of FIV (56), and cats that were experimentally infected with FIV after immunization with a peptide derived from the membrane-proximal ectodomain experienced a delay in the infection (48). In the primate lentiviruses, alterations made within various regions of the ectodomain including the N-terminal fusion peptide, the leucine zipper, the dicysteine loop, membrane-spanning domains, and the regions which span between these functional domains dramatically alter syncytium formation and/or fusogenicity (3, 8, 15, 23, 24, 27, 37), infectivity (8, 9, 15, 22), cytopathogenicity (8), and envelope subunit association (6).
In an interesting adaptation, the lentiviruses have dealt genotypically with ex vivo culture by truncating the cytoplasmic domain of the TM by the introduction of a premature stop codon. Such truncations to the cytoplasmic tails of the primate lentiviruses, SIV (7, 32, 50, 53) and both HIV-1 and HIV-2 (51, 52), and to equine infectious anemia virus (47) cause an expansion of the viral host cell range and increases in growth kinetics, envelope fusogenicity, and cytopathogenicity. Truncations to the cytoplasmic domain also occur in oncoretroviruses, such as murine leukemia virus (MuLV); they are mediated by the cleavage of the carboxyl-terminal 16-residue R peptide by the viral protease prior to virus maturation (25). The MuLV R peptide is a potent inhibitor of fusogenicity, and after its cleavage from the TM, the fusogenic potential of the virus increases dramatically (62). Thus, removal of the carboxyl-terminal portion of the cytoplasmic tail from diverse retroviruses results in altered syncytium-forming properties.
Here we have detailed the molecular nature, receptor usage patterns, and in vitro characteristics of a phenotypically divergent clone of FIV strain PPR (FIV-PPR), FIV-PPRglial. This isolate arose after passage of FIV-PPR through an interleukin-2- (IL-2)-independent cell line. The virus possesses an expanded host cell range and markedly increased fusogenic properties. The results show that the phenotype is dictated by an interesting combination of altered amino acid changes in both SU and TM.
MATERIALS AND METHODS
Cell lines and propagation.
The feline T-cell lines MCH5-4 and 104-C1 (33) were kindly provided by Chris Grant (Custom Monoclonals, Sacramento, Calif.). The two continuous IL-2-independent cell lines, MCH5-4DL and 104-C1DL, were isolated from their parental lines, MCH5-4 and 104-C1, respectively, in our laboratory after undergoing crisis in the absence of IL-2 and concanavalin A (33). Crandell feline kidney cells (CrFK) were obtained from the American Type Culture Collection ATCC (21), and G355-5, a feline glial cell line, was kindly provided by Don Blair (National Institutes of Health, Bethesda, Md.). The T-cell lines, MCH5-4 and 104-C1, were cultured in RPMI 1640 (Mediatech) supplemented with 10% heat-inactivated fetal bovine serum (Gemini-BioProducts), 1× MEM-vitamins (Sigma, St. Louis, Mo.), 1× nonessential amino acids (Sigma), 1× sodium pyruvate (Sigma), 200 μM l-glutamine (Sigma), 5.5 × 10−5 M β-mercaptoethanol (Gibco-BRL), 50 μg of gentamicin (Gemini-BioProducts) per ml, 50 U of recombinant human IL-2 (rhIL-2) (a generous gift of Hoffmann-La Roche), and 2.5 μg of concanavalin A (Boehringer Mannheim) per ml. The IL-2-independent T-cell lines, as well as the CrFK and G355-5 cells, were cultured as above without the addition of IL-2 or concanavalin A.
Viruses.
The viral isolates used in these studies included FIV-PPR, a molecular clone derived from a cat in San Diego, Calif. (42); FIV-34TF10, a molecular clone derived from the Petaluma isolate (55); and FIV-PPRglial, an isolate which emerged after ex vivo passage of FIV-PPR through the IL-2-independent cell line 104-C1DL (33).
Viral chimeras.
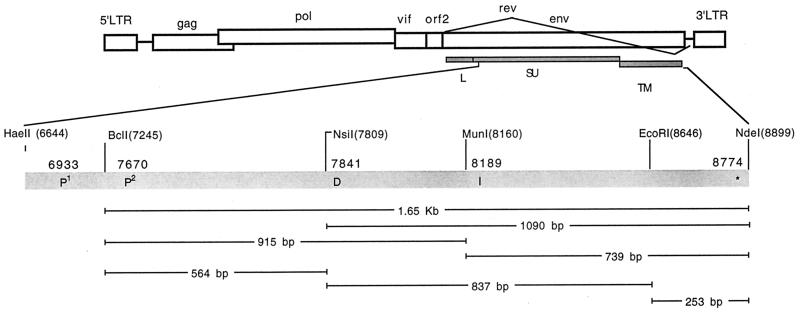
The envelope of FIV-PPRglial was amplified by reverse transcriptase PCR (RT-PCR) (Stratagene) with the following primers: 5′-CCCATTTAGAGTACCTGCAG-3′ (6382 to 6401) and 5′-CGAATAGTTTTCTGAAGCGGTC TTCTAAATCTGTCAT CAT-3′ (9001 to 9040). A 94°C 5-min presoak was followed by 35 cycles of 94°C for 30 s, 58°C for 1 min, and 72°C for 3 min, ending with a 10-min 72°C extension. We used 1× Advantage cDNA polymerase mix (Clontech, Palo Alto, Calif.) with 1× Advantage cDNA polymerase buffer, 200 μM each deoxynucleoside triphosphate (Promega, Madison, Wis.), and 700 ng of each primer were used per amplification reaction. The 2,658-bp product was digested with HaeII (which cuts at position 6644) and NdeI (which cuts at position 8899), yielding a fragment of 2,255 bp, which was reserved for later use. FIV-PPR was digested with EcoRV (which cuts at 4218) and NdeI (which cuts at 8899). The digest produced two fragments of the virus/pUC119 vector, of 4,681 and 7,953 bp. The 4,681-bp fragment was further digested with HaeII. Two similar-sized fragments resulted: 2,426 and 2,255 bp. A ligation using high-concentration T4 DNA polymerase (Gibco-BRL) was set up with the reserved 2,255-bp HaeII-NdeI fragment from FIV-PPRglial, the 2,426-bp EcoRV-HaeII fragment from FIV-PPR, and the EcoRV-NdeI 7,953-bp backbone from FIV-PPR. This resulted in a chimera of PPR/PPRglial which contained the distal 150 bp of the leader sequence and the entire SU/TM of FIV-PPRglial ligated into the remainder of the viral genome of FIV-PPR.
Mutant constructs.
Most of the mutant constructs were made by using the natural restriction sites within the 1.65-kb BclI-NdeI fragment of FIV-PPR (see Fig. 1) and ligating combinations of the resulting smaller fragments into the 11-kb backbone of either FIV-PPR or FIV-PPRchim42 with the pUC112 vector. Two of the constructs (P2 and ∗) were made using PCR and mutagenic primers. The single-mutant constructs were assembled as follows. The P1 construct was made by ligating the 1.65-kb fragment of FIV-PPR into the 11-kb backbone of FIV-PPRchim42. The D mutant was made by preligating the 915-bp fragment from FIV-PPRchim 42 with the 739-bp fragment of FIV-PPR and then ligating the whole into the FIV-PPR backbone. The I construct was made by preligating the 739-bp piece of FIV-PPRchim 42 to the 915-bp piece of FIV-PPR and then ligating the whole into the FIV-PPR backbone. The P2 construct was generated with primers 5′-TTGTCATATTGAAATGCATAATAAGGTCATCTACCTTC ATAGTAAACCCGTTTTGTAAGGAACAGTTATACATTTTATTTGCATA CATGGTACAATCTACAGGATTGGCACCTGAAACATTTAGATTTTTTC CACATGTATCAATGAGTGAGGGATTATC-3′, which ran through the NsiI site (italicized) and contained a mutation to change the natural threonine to a proline (underlined), and 5′-CAGACCCATTACAAATCCCACTGATCAATTATA-3′, which ran through the BclI site (italicized). The 100-μl reaction mixture contained 200 μM each deoxynucleoside triphosphate, 1× Advantage cDNA PCR reaction buffer with 1× Advantage cDNA polymerase mix, and 700 ng of each primer. The parameters were 35 cycles of 94°C for 15 s, 58°C for 15 s, and 72°C for 30 s. This generated a 599-bp product that overlapped the BclI-NsiI 564-bp fragment. The product was digested with BclI and NsiI and then ligated to the 1,090-bp NsiI-NdeI PPR product before being ligated into the PPR backbone. The stop (∗) construct was made using the 3′ megaprimer, 5′-AAAATG GATTCATATGACATATCTTCCTCAAAGGGAAGAAATCAGCTCAGCA CCTCTCCTGCATTAAGCAGTTCTGAGGCATCTCATCATTCCTCCTCTT TTTCAGACATGCCACATTGCCTGCCATTTTTCCTCAATTACATTTG T-3′ with the NdeI site italicized and the nucleotide that alters the natural glutamic acid to the premature stop codon underlined. The same 5′ oligonucleotide that was used in the P2 construct was used to generate a fragment that spanned the BclI and NdeI sites. The PCR parameters were 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 2 min. The reaction conditions were the same as previously described except for the addition of 5% dimethyl sulfoxide.
FIG. 1.
Schematic representation of mutant construction strategy. The figure illustrates the restriction enzyme sites used in the generation of the mutant viruses and the location of the nucleotide mutations found with the envelope of FIV-PPRglial and FIV-PPRchim42. LTR, long terminal repeat.
The DI mutant was made by preligating the 837-bp fragment of FIV-PPRchim42 to the 564- and 253-bp fragments of FIV-PPR, which in turn were then ligated into the backbone of FIV-PPR. The P1* mutant was made by ligating the 1.65-kb fragment of the stop construct into the backbone of FIV-PPRchim42. The P2* construct was made by preligating the 564-bp fragment of the P2 construct with the 1,090-bp fragment of the stop construct and then ligating the whole into the FIV-PPR backbone. The P1P2 mutant was made by ligating the 1.65-kb fragment of the P2 construct into the backbone of FIV-PPRchim 42. The P1P2* mutant was made by ligating the 1.65-kb fragment of the P2* construct into the FIV-PPRchim 42 backbone. The P2DI* mutant was made by ligating the 1.65-kb fragment of FIV-PPRchim 42 in to the backbone of FIV-PPR. The P1P2DI* mutant (regeneration of FIV-PPRchim 42) was made by ligating the 1.65-kb fragment of the P2DI* mutant into the FIV-PPRchim 42 backbone.
All ligated products were electroporated into electrocompetent SURE cells (Stratagene, La Jolla, Calif.) in 0.1-cm Gene Pulser cuvettes (Bio-Rad, Hercules, Calif.) at 8 V in a Gene Pulser. Clones were sequenced with Sequenase 2.0 (United States Biochemical) to verify fidelity.
Transfection and infection studies.
One day prior to transfection, 106 G355-5 cells were plated into 100-mm tissue culture plates (Falcon). A 10-μg portion of plasmid DNA (Qiagen Maxi Preps) was transfected into cells with Superfect reagent (Qiagen) as specified by the manufacturer. The cells were incubated with serum-containing medium for 4 h at 37°C, after which the inoculate was aspirated and 15 ml of fresh medium was added per plate. Four days later, the supernatants were harvested and assayed for RT activity.
MCH5-4 and MCH5-4DL cells were added 24 h posttransfection and cocultivated overnight with the G355-5 cells. The following day, the cells were washed and then cultivated separately for 4 days, at which point the supernatant was harvested and assayed for RT activity.
RT assay.
Assessment of viral infection was determined by using virion-associated, pelletable RT as previously described (33). The only exception was that the supernatant volume was reduced to 500 μl and pelleted at 60,000 rpm for 30 min in a TLA 120.1 rotor (Beckman). Accordingly, the entire pellet was used in the assay in a total volume of 25 μl.
Immunocytochemical analyses.
Determination of virus-specific syncytium formation was performed as described previously (39) with some modifications. CrFK cells (2.5 × 105 per well) were plated out per well in six-well plates (Costar). Plasmid DNA (2 μg) was transfected with Superfect reagent for 4 h at 37°C. The cells were washed, and fresh medium was added. The following day, each well was trypsinized and brought up to a volume of 4 ml. Then 0.5 ml was transferred into each well of a four-well slide chamber (Nunc) and incubated for 2 days at 37°C. The cells were fixed with 2% formaldehyde and then permeabilized with 0.2× Triton X-100 for 15 min. Slides were blocked with 1% bovine serum albumin (Sigma) for 1 h and then incubated with the anti-CA monoclonal antibody, PAK3-2C (kindly provided by C. Grant), at 10 μg/ml for an additional hour. Subsequent incubations and development were as previously described (39).
Cotransfection studies.
The cotransfection assay was set up essentially as described above. FIV-PPRchim42 DNA (1 μg) was transfected into CrFK cells together with 1 μg of plasmid DNA from either FIV-PPR, FIV-PPRchim42, P1 (containing the native threonine in V4 and full-length TM), P2 (containing the native glutamine in C2 and full-length TM), or stop constructs (containing both the native threonine and glutamine and a truncated TM) in combination with 1 μg of pRSV–β-gal DNA as an internal control. A standard β-galactosidase (β-gal) assay was performed to assess transfection efficiency (26). The cells were lifted after 1 day, and equal volumes were seeded into slide chambers for the fusion assay or 12-well plates for the β-gal assay. Multiple fields of FIV-positive cells were scored, and the results tabulated.
Chemokine studies.
CrFK cells (5 × 104) or MCH5-4 cells (5 × 105) were plated into each well of a 12-well plate and allowed to incubate at 37°C overnight. The cells were then pelleted to remove the supernatant (MCH5-4), or the supernatant was aspirated (CrFK). The synthetic chemokines, human stromal cell-derived factor 1-alpha (SDF1α), human regulated upon activation, normal T cells, expressed and secreted (RANTES), and human macrophage inflammatory protein 1-β (MIP1-β), each kind gifts of Gryphon Pharmaceuticals, were added in a volume of 500 μl at a concentration of 1 μg/ml and incubated for 1 h at 37°C. Then 250 μl of viral supernatant stocks with an RT activity of 106/ml was added per well and further incubated for 1 h. The cells were then washed twice, and 2 ml of medium containing 1 μg of chemokine per ml was added per well. After 5 days, supernatants were recovered for RT assays. The cells (CrFK) were fixed with 2% formaldehyde, and immunocytochemistry was performed as described above.
RESULTS
Generation of FIV-PPRglial and its molecular clone, FIV-PPRchim42.
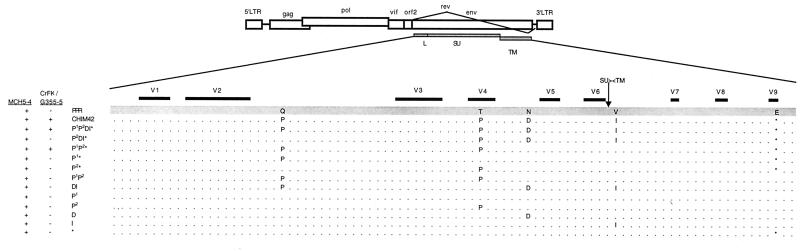
A previous study described the generation of an uncharacterized cytopathic pool of FIV-PPR, termed FIV-PPRglial, possessing an altered cytotropism concomitant with a vigorous increase in viral kinetics and the ability to form syncytia upon adherent cell lines (33). To generate a molecular clone, CrFK cells were acutely infected with FIV-PPRglial. The envelope was amplified by RT-PCR and subsequently cloned into the HaeII (position 6644) and NdeI (position 8899) sites of the parental virus, FIV-PPR (Fig. 1). This mutagenesis resulted in several chimeric clones of FIV-PPR containing the C-terminal portion of the envelope leader and the entire SU and TM of FIV-PPRglial. One chimeric clone, FIV-PPRchim42, was chosen for further analysis based on its ability to replicate the altered phenotype. The envelope of FIV-PPRchim42 was sequenced, revealing five amino acid substitutions relative to wild-type FIV-PPR (Fig. 1 and 2). Specifically, the substitutions consisted of a glutamine-to-proline change at residue 224 in C2, a threonine-to-proline change at residue 470 in V4, an asparagine-to-aspartic-acid change at residue 527 in C4, a valine-to-isoleucine change at residue 643 between the polar domain and the leucine zipper of TM, and the introduction of a premature stop codon in the cytoplasmic tail, abbreviating the TM protein by 17 residues.
FIG. 2.
Schematic representation of FIV-PPR mutant profiles with cytotropism. A total of five mutations were noted between FIV-PPR and FIV-PPRchim42. The minimal requirement for growth on adherent cells was a specific triad of mutations, specifically, two within SU at positions 224 and 470 and the incorporation of a premature stop codon in TM at position 838. All of the mutants grew productively on the MCH5-4 T-cell line. LTR, long terminal repeat.
Twelve clones were constructed on the parental FIV-PPR background, containing each of the five mutations singly and in combination, by using restriction enzyme sites and mutagenic oligonucleotides in combination with PCR (Fig. 1). Plasmid DNA was transfected into G355-5 cells, which were in turn cocultivated with MCH5-4 T cells. Infections were quantitated by measuring the RT activity of pelleted culture supernatants.
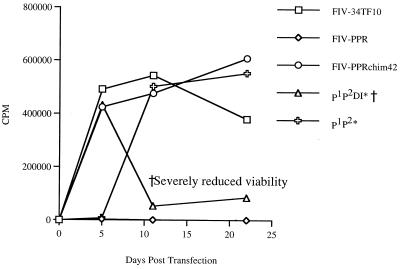
Each of the construct viruses retained the ability to productively replicate upon the MCH5-4 T-cell line (Fig. 2). However, only viruses containing a specific triad of mutations were able to reproduce the characteristics of FIV-PPRglial upon adherent cells (Fig. 2 and 3). The phenotype was minimally attributable to the synergistic combination of the proline in the second constant domain of SU at residue 224 with the proline in the fourth hypervariable loop of SU at residue 470, and the premature truncation of the cytoplasmic tail of gp41 residue 838. Single or double mutants in any combination failed to mimic the characteristics acquired by FIV-PPR after passage through the IL-2-independent cells (data not shown).
FIG. 3.
Graphic representation of transfection studies with the mutant constructs. G355-5 cells were transfected with FIV-PPR, FIV-PPRchim42, FIV-34TF10, and 11 mutant FIVs possessing combinations of Env mutations that distinguish FIV-PPR and FIV-PPRchim42. Release of pelletable RT activity into the culture supernatant was then monitored over a 3-week period. The results indicated that only viral constructs containing a specific combination of three mutations were able to replicate on G355-5 cells and mimic the phenotype of FIV-PPRglial. Specifically, these were a glutamine-to-proline change in C2, a threonine-to-proline change in V4, and the premature truncation of the cytoplasmic tail by the incorporation of a stop codon. All of the mutants were replication competent to wild-type levels on MCH5-4 T cells (data not shown); therefore, the inability to grow on the adherent cells was not due to replication-deficient constructs. All single and double mutants (listed in Fig. 2) were negative of productive growth in G355-5 or CrFK cells but were fully infectious on MCH5-4 T cells (data not shown).
Inhibition of syncytium formation and viral spread by cotransfection of FIV-PPRchim42 with viral mutants.
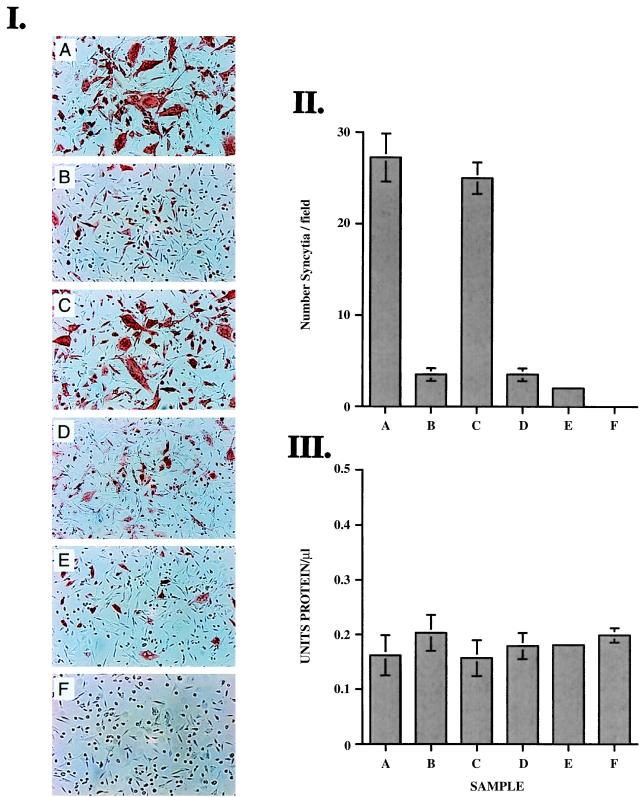
In an effort to further define the role of the mutations upon syncytium formation, cotransfection studies were initiated. CrFK cells were transfected with FIV-PPRchim42 plasmid DNA and concurrently cotransfected with one of several mutant virus plasmids in combination with a pRSV–β-gal plasmid. The infections were quantitated by immunocytochemical means with the Gag-specific monoclonal antibody, PAK32C (Fig. 4I), and a standard β-gal assay was used to evaluate transfection efficiencies (Fig. 4III). FIV-positive cells were scored in multiple-field analyses (Fig. 4II). It was observed that when plasmids expressing the full-length cytoplasmic tail of the TM (nonpermissive for replication on CrFK) were cotransfected with FIV-PPRchim42 (permissive replication on CrFK), the infection was severely inhibited, in terms of both viral spread and syncytium formation, presumably due to the oligomeric glycoprotein complex formation between the envelopes of the full-length and truncated variants. Cotransfections with viruses containing the truncated TM did not interfere with syncytium formation (Fig. 4I, panels A and C). Likewise, mutants containing the wild-type glutamine at residue 224 in C2 (Fig. 4I, panel E) or the native threonine at position 470 in V4 of SU (Fig. 4I, pandl D) did not inhibit the infection or fusogenicity of FIV-PPRchim42 upon CrFK cells. These results suggest that the mutations within SU are dispensable for fusion events and that the abbreviated cytoplasmic tail of TM is the crucial factor in induction of syncytium formation, as has been observed with SIV (50).
FIG. 4.
Results of cotransfection studies with FIV-PPRchim42 and mutant viruses. (I) Photomicrography on CrFK cells cotransfected with FIV-PPRchim42 and FIV-PPRchim42 (+ control) (A), FIV-PPRchim42 and FIV-PPR (B), FIV-PPRchim42 and the stop construct (which contains the premature truncation of the TM) (C), FIV-PPRchim42 and P1 (which contains the native threonine in V4) (D), FIV-PPRchim42 and P2 (containing the native glutamine in C2) (E), and no-virus β-gal control (F). Note the giant syncytia and viral spread present when constructs containing the truncated TM were used as input virus. This phenotype is disrupted when viruses containing the full-length TM are cotransfected with FIV-PPRchim42. (II) Graphic quantitation of syncytia (more than three nuclei per cell) from multiple fields from each cotransfection. There was a log-fold reduction in the number of syncytia when constructs containing a full-length TM were cotransfected with FIV-PPRchim42. (III) β-Gal assay results, demonstrating that the transfection efficiencies were consistent.
Inhibition of FIV-PPR, FIV-34TF10, and FIV-PPRchim42 by chemokines.
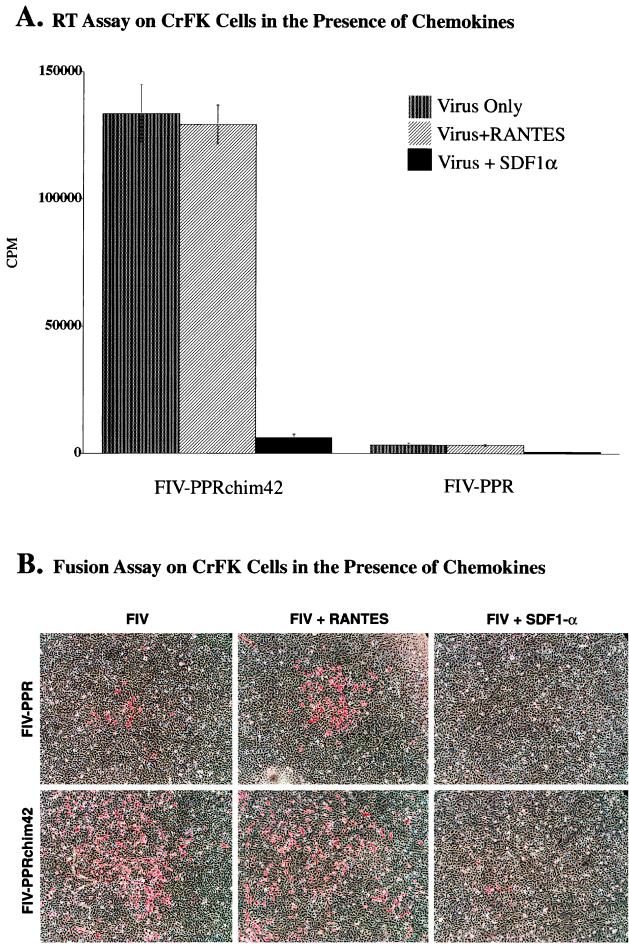
Cells were pre-incubated in the presence of the synthetically derived chemokines SDF1α, MIP1-β (data not shown), and RANTES, the natural ligands for the α-chemokine receptor CXCR4 and the β-chemokine receptors, such as CCR1, CCR3, and CCR5, respectively. The chemokines were added prior to the addition of virus and then maintained throughout the culture period. Following several days in culture, the virus was quantitated by both RT activity measurement (Fig. 5A and 6) and immunocytochemical methods (Fig. 5B).
FIG. 5.
Results from chemokine inhibition assays on CrFK cells infected with FIV-PPR and FIV-PPRchim42. (A) Graphic representation showing the low-level infection of FIV-PPR infection upon adherent cells and the contrasting highly productive infection of CrFK by FIV-PPRchim42. The panel also shows the nearly complete inhibition of FIV-PPRchim42 and FIV-PPR in the presence of SDF1α, indicating that these viruses utilize CXCR4 for entry. RANTES had no effect on the infections. (B) Immunocytochemical analyses of FIV-PPR and FIV-PPRchim42 infections on CrFK cells in the presence of chemokines. FIV-PPR infection is readily detected by this method, whereas the infection is below the level of sensitivity of the RT assay, as observed in panel A. Also, the foci of infection were enlarged in the FIV-PPR infections in the presence of RANTES and the foci were sparse compared with those observed with FIV-PPRchim42, which were not limited to focal points. Importantly, it was observed that SDF1α was able to severely inhibit the infections correlating with the RT data presented in panel A.
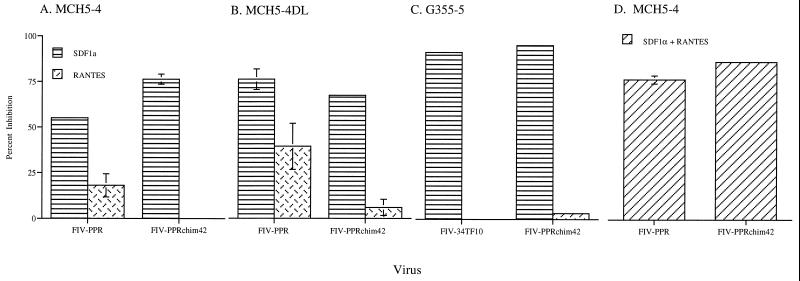
FIG. 6.
Graphic representation of infections of IL-2-dependent cells (MCH5-4), IL-2-independent cells (MCH5-4DL), and an adherent glial cell line (G355-5) in the presence of SDF1α and RANTES. A 55% inhibition was observed when SDF1α was added to cultures of FIV-PPR-infected MCH5-4 cells. The inhibition by SDF1α was increased to 80% in cultures of FIV-PPR-infected MCH5-4DL cells. FIV-PPRchim42 infections were inhibited to a greater extent on MCH5-4 cells and to near equal levels to FIV-PPR inhibition on the MCH5-4DL cells in the presence of SDF1α. SDF1α inhibited FIV-34TF10 and FIV-PPRchim42 infections on G355-5 cells to nearly 100%. RANTES inhibited FIV-PPR infection by 30%, whereas there was no effect on FIV-PPRchim42 infection of MCH5-4 cells. RANTES was able to minimally inhibit FIV-PPRchim42 infection of the IL-2-independent MCH5-4DL cells; however, the most marked effect with RANTES was observed for the FIV-PPR infection of these cells, although the graph represents a percent inhibition of a low-level infection wherein the RT values were less than 25,000 cpm. RANTES had a very minimal to no effect on infections of G355-5 cells.
The addition of either MIP1-β or RANTES had no observable effect upon FIV-34TF10 on either G355-5 cells (Fig. 6C, MIP1-β data not shown) or CrFK cells (data not shown). Likewise, FIV-PPRchim42 infection of CrFK and G355-5 cells was not blocked by RANTES or MIP1-β (data not shown) on CrFK cells, as discerned by RT activity measurement (Fig. 5A and 6C, respectively) or by cell-staining patterns (Fig. 5B). However, the presence of RANTES appeared to enlarge the foci of infection in FIV-PPR (Fig. 5B). This increase in focal size was undetectable by the RT assay, and the presence of FIV-PPR infection of CrFK cells was demonstrated only by immunocytochemical means. It is important to mention that the foci of infection with FIV-PPR were sporadic and not distributed evenly over the entire culture area, as seen in FIV-34TF10-infected (data not shown) and FIV-PPRchim42-infected cultures (Fig. 5B). In direct contrast to the RANTES result, SDF1α significantly reduced the number of FIV-positive cells in the FIV-PPRchim42 infection (Fig. 5B) and the FIV-34TF10 infection (not shown) and completely ablated the low-level infection of CrFK by FIV-PPR (Fig. 5B). These results are consistent with the results of recent studies that indicated that the Petaluma-derived FIV-34TF10 isolate can utilize CXCR4 for entry (29). The studies also yielded the unexpected finding that FIV-PPR utilizes CXCR4 in the infection of CrFK cells, although the level of FIV-PPR infection upon the adherent cells was minimal (Fig. 5A). SDF1α also inhibited the FIV-PPRchim42 infection of G355-5 cells by 95% (Fig. 6C). The results are consistent with the notion that all three FIV isolates utilize CXCR4 for entry into CrFK and G355-5 cells.
In contrast, distinct differences were noted between FIV-PPR and FIV-PPRchim42 receptor usage on T cells (Fig. 6A and B). FIV-34TF10 was excluded from these studies, since it is unable to productively infect MCH5-4 cells due to an interruption of the orf2 gene (33). An 85% reduction in the FIV-PPRchim42 infection of T cells was observed in the presence of SDF1α, whereas the FIV-PPR infection was inhibited by only 55%. These data indicate that FIV-PPR and FIV-PPRchim42 both utilize CXCR4 for entry into CrFK cells and to different degrees for entry into T cells. However, the results indicate that alternative receptors are present on T cells. MIP1-β was also used in inhibition studies with MCH5-4 cells (data not shown). The results were variable and inconsistent, perhaps indicating that while RANTES and MIP1-β share certain receptors, the affinities may vary and MIP1-β may not interact with the viral receptor as avidly as RANTES does.
In contrast to the results on infection of CrFK or G355-5 cells, RANTES was able to curtail FIV-PPR infection of MCH5-4 T cells by 25% (Fig. 6A), and with a combination of SDF1α and RANTES, the infection was inhibited by 77% (Fig. 6D); that is, the effect was additive. In contrast, RANTES was unable to inhibit the FIV-PPRchim42 infection of MCH5-4 cells, and there was no observable augmentation of inhibition when RANTES was used in combination with SDF1α (Fig. 6A and D).
Additionally, when either SDF1α or RANTES was added to FIV-PPR-infected MCH5-4DL cells, which are IL-2 independent (33), the inhibition was greater than that observed with the IL-2-dependent cell line, MCH5-4. However, it must be noted that this was a percent inhibition of a low-level infection whose RT activities were less than 25,000 cpm. This greater inhibition may be due to differential receptor expression between the two cell lines, although the MCH5-4DL cells originated from the MCH5-4 cell stock. In a previous study, we demonstrated that the two cell lines differed with respect to certain topically expressed and secreted proteins, including CCR5 (33), and thus it is highly plausible that other receptors may also be differentially expressed.
DISCUSSION
In this study we have characterized a chimeric molecular clone of FIV containing the envelope from a phenotypically divergent pool of FIV-PPR (FIV-PPRglial) which arose after ex vivo passage in IL-2-independent T cells. The chimeric clone, FIV-PPRchim42, was able to faithfully replicate the phenotype of FIV-PPRglial, which included an enhanced host cell range, increased cytopathogenicity, and enhanced fusogenic properties. It was further determined through sequence and mutational analyses that of the five amino acid alterations present within the envelope of FIV-PPRglial, three were minimally sufficient to generate the phenotype. Additional experimentation revealed that one of the mutations, a stop codon in the cytoplasmic domain of the TM, was responsible for increased syncytium formation. Subsequent chemokine inhibition studies were pursued in which both the alpha chemokine SDF1α and the beta chemokine RANTES were used as potential viral inhibitors. We observed that SDF1α was able to ablate FIV-34TF10 infection of G355-5 cells and CrFK cells (data not shown) and FIV-PPR and FIV-PPRchim42 infections of CrFK cells. Neither RANTES nor MIP1-β had any effect on infection of CrFK or G355-5 cells by any of the FIV strains. SDF1α inhibited FIV-PPR and FIV-PPRchim42 infection of T cells by 55 and 85%, respectively. Addition of the CC chemokine RANTES in combination with SDF1α increased the inhibition of FIV-PPR by 25% but had no additive effect on FIV-PPRchim42 infection of T cells. Taken together, the findings suggest that CXCR4 is the sole FIV receptor on CrFK and G355-5 cells. However, the results are consistent with the involvement of one or more CC chemokine receptors in FIV infection of T cells.
Initial sequence analyses revealed that the envelope proteins of FIV-PPRglial contained five amino acid substitutions, three in the SU protein and two in the TM protein. None of these mutations occurred within the highly charged third hypervariable loop (V3) of SU, suggesting an alternative mechanism to those previously reported (57, 58), by which FIV may become CrFK tropic. The mutations within SU were found in the second and fourth constant domains (C2 and C4, respectively) and in the fourth hypervariable loop (V4). Both the mutations in C2 and V4 involved the substitution of proline, which apparently induced conformational changes in protein structure. Neither of the residues were found in domains predicted to have alpha-helical structures as judged by computer profiling using MacVector (Oxford Molecular) (data not shown). However, the prolines most probably confer an altered conformation to SU and could be an important factor in determining receptor preference and/or avidity of receptor binding. The mutation in C4 introduced an aspartic acid in place of the native asparagine and was found to be dispensable to the phenotype.
There were two mutations in the TM; the first was in the ectodomain at the single position that separates the polar domain from the leucine zipper and substituted an isoleucine for the native valine, and the second introduced a premature stop codon within the cytoplasmic domain that shortened the tail by 17 residues. The isoleucine-to-valine mutation within the ectodomain was dispensable. Mutational analyses confirmed that the phenotype of FIV-PPRglial was minimally attributable to three synergistic mutations, the insertion of the two prolines within the SU protein and the truncation of the cytoplasmic tail of the TM.
In an effort to further delineate the residue(s) responsible for certain aspects of the phenotype, studies were initiated in which the mutant viruses were concurrently cotransfected with one another and with the parental FIV-PPR in the presence of a reporter plasmid. It was predicted that coexpression of the different viruses would result in mixed envelope trimers with potential changes in phenotype, similar to results reported for SIV (50) and proposed for MuLV (46). When FIV-PPRchim42 was cotransfected with mutants containing the full-length cytoplasmic tail, including FIV-PPR, syncytium formation was severely curbed, as was viral spread, presumably, due to oligomeric glycoprotein complexes formed between the full-length and truncated proteins. The syncytium-forming, cytopathic properties of FIV-PPRchim42 were not disrupted by cotransfection with any construct containing a truncated TM, independent of any of the other changes. Conversely, all constructs encoding full-length TM caused a marked reduction in syncytium formation by FIV-PPRchim42. It is unlikely that the truncated tail is solely responsible for the enhanced cytotropic nature of FIV-PPRglial, as has been observed with SIV, since only viruses containing the truncated TM in combination with the two changes in SU were able to replicate in either adherent cell line.
The cytoplasmic domain is multifunctional, and a recent report determined that the cytoplasmic tails of HIV and SIV are posttranslationally modified by palmitic acid linkage to cysteine residues (63). Whether FIV TM is palmitoylated has yet to be determined. Further studies to determine whether palmitoylation, or loss thereof, of the cytoplasmic tails of FIV-PPR and FIV-PPRchim42 is an important phenotypic determinant are underway.
The ex vivo adaptation to growth in particular cell types has been studied extensively in the primate and equine lentiviruses. In particular, truncations of TM have been observed in HIV-1 and HIV-2 (51, 52), SIV (7, 32, 50, 53), and equine infectious anemia virus (47). The mechanism(s) by which this truncation induces cytopathogenicity, accelerated kinetics, and expanded host cell ranges is not entirely clear. What is clear is that syncytium formation and increased fusogenicity are common themes. Precisely what causes this to occur is speculative. Some studies have reasoned that it is due to an increase in envelope expression on the cell and viral surfaces (53), while others have concluded that there is an enhanced budding profile and hence more viral accessibility (2). We used capture assays in which both cell-associated and shed SU and p24 Gag expression were measured. These studies determined that the ratios of SU to p24 expression were identical in cultures of FIV-PPR and FIV-PPRchim42 (data not shown). Thus, the increase in syncytium formation and fusogenicity in FIV-PPRchim42 is not due to an increase in envelope expression. However, another earlier study proposed that the truncated cytoplasmic tail was able to induce a conformational change in the ectodomain of the protein (54), and it is possible that the same is true of FIV-PPRchim42. Similar assays were not performed in this study but are planned. Finally, with respect to the truncated TM, oncoretroviruses lose a 16-residue peptide, termed the R peptide, from the carboxyl terminus of the cytoplasmic tail prior to viral maturation (25). The removal of the R peptide increases the fusogenicity of the virus, and exogenous addition of the peptide was recently shown to severely reduce the fusogenic nature of MuLV (62). The premature truncation of the lentivirus cytoplasmic tail and the cleavage of the R peptide from the same domain by the viral protease in the murine retrovirus MuLV yield viruses much more fusogenic than their precursors containing the full-length proteins.
As with the primate lentiviruses, seven-transmembrane G-protein-coupled receptors have recently been reported to play a role in the FIV life cycle. Specifically, the alpha chemokine receptor CXCR4 was able to mediate the fusion of CrFK-tropic isolates of FIV in feline cells (29) and to mediate fusion with human (29, 43, 61; Willett et al., letter) and murine (43) cells bearing the human CXCR4 homolog. Additionally, two very recent reports showed that the bicyclam AMD-3100, a small molecule that interacts with CXCR4 and that had previously been shown to inhibit CXCR4-tropic isolates of the primate lentiviruses (13), also inhibits FIV (18, 49). It remains unclear whether CXCR4 acts as a primary or secondary receptor for FIV. Certain isolates of HIV and SIV are able to use CXCR4 or CCR5, respectively, for entry in the absence of their primary receptor, CD4, implying that chemokine receptors can facilitate viral entry independently, at least, of other known receptors.
In these studies, we have also found that the CrFK-tropic isolate FIV-34TF10 utilizes CXCR4 for entry into both CrFK (data not shown) and G355-5 cells. We had surmised that FIV-PPR, which does not productively infect CrFK or G355-5 cells (33, 41), failed to do so because of the lack of a specific receptor for this isolate in the adherent cells. However, the results are consistent with the notion that FIV-PPR utilizes CXCR4 for entry into these cells. Whether the limited spread of FIV-PPR in CrFK cells is a consequence of the lowered receptor affinity or the need for coutilization of another receptor remains to be determined.
A distinct picture emerged from chemokine inhibition studies of entry of FIV-PPR and FIV-PPRchim42 into T cells. Inhibition of infection with SDF1α was incomplete with both viruses but was more efficacious for FIV-PPRchim42 (85%) than for FIV-PPR (55%). Additionally, the CC chemokine RANTES blocked FIV-PPR infection of T cells by 20 to 40% but had negligible effect on FIV-PPRchim42. Taken together, the results suggest that the broader host cell range of the latter FIV strain coincides with an increased reliance on CXCR4 for entry into host cells and/or a decreased reliance on a secondary interaction with a CC chemokine. Conversely, the poor infection of FIV-PPR on CrFK cells may be due to the absence of such an interaction on these cells and the successful infection of this virus on T cells may be facilitated by such an interaction. Which CC chemokine receptors are involved remains to be determined, since RANTES interacts with several receptors, including CCR5, CCR1, CCR3, and DARC Duffy antigen. Thus, any of these receptors, and possibly still others, might be utilized by certain FIV strains. Work is in progress to further define receptor interactions by these FIV strains.
ACKNOWLEDGMENTS
We thank the following for their generous contributions to this work. Specifically, we thank Chris Grant for the monoclonal antibody PAK3-2C and for the T-cell line MCH5-4; Bruce Torbett, Steve Kent, and Gryphon Pharmaceuticals for SDF1α, RANTES, and MIP1-β; Robert Turner for his expertise in photomicography, Hoffmann-La Roche for rhIL-2; Pamela Foye for oligonucleotide synthesis; Udayan Chatterji, Aymeric de Parseval, and Ying-Chuan Lin for manuscript review; and C. Kat Kiser for administrative assistance.
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (AI25825) and the National Institute of Mental Health (GM47680) of the National Institutes of Health.
REFERENCES
- 1.Ackley C D, Hoover E A, Cooper M D. Identification of a CD4 homologue in the cat. Tissue Antigens. 1990;35:92–98. doi: 10.1111/j.1399-0039.1990.tb01762.x. [DOI] [PubMed] [Google Scholar]
- 2.Ball J M, Mulligan M J, Compans R W. Basolateral sorting of the HIV type 2 and SIV envelope glycoproteins in polarized epithelial cells: role of the cytoplasmic domain. AIDS Res Hum Retroviruses. 1997;13:665–675. doi: 10.1089/aid.1997.13.665. [DOI] [PubMed] [Google Scholar]
- 3.Bergeron L, Sullivan N, Sodroski J. Target cell-specific determinants of membrane fusion within the human immunodeficiency virus type 1 gp120 third variable region and gp41 amino terminus. J Virol. 1992;66:2389–2397. doi: 10.1128/jvi.66.4.2389-2397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown W C, Bissey L, Logan K S, Pedersen N C, Elder J H, Collisson E W. Feline immunodeficiency virus infects both CD4+ and CD8+ T lymphocytes. J Virol. 1991;65:3359–3364. doi: 10.1128/jvi.65.6.3359-3364.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunner D, Pedersen N C. Infection of peritoneal macrophages in vitro and in vivo with feline immunodeficiency virus. J Virol. 1989;63:5483–5488. doi: 10.1128/jvi.63.12.5483-5488.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao J, Bergeron L, Helseth E, Thali M, Repke H, Sodroski J. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J Virol. 1993;67:2747–2755. doi: 10.1128/jvi.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakrabarti L, Emerman M, Tiollais P, Sonigo P. The cytoplasmic domain of simian immunodeficiency virus transmembrane protein modulates infectivity. J Virol. 1989;63:4395–4403. doi: 10.1128/jvi.63.10.4395-4403.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen S S. Functional role of the zipper motif region of human immunodeficiency virus type 1 transmembrane protein gp41. J Virol. 1994;68:2002–2010. doi: 10.1128/jvi.68.3.2002-2010.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S S, Lee S F, Hao H J, Chuang C K. Mutations in the leucine zipper-like heptad repeat sequence of human immunodeficiency virus type 1 gp41 dominantly interfere with wild-type virus infectivity. J Virol. 1998;72:4765–4774. doi: 10.1128/jvi.72.6.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho M W, Lee M K, Carney M C, Berson J F, Doms R W, Martin M A. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean G A, Reubel G H, Moore P F, Pedersen N C. Proviral burden and infection kinetics of feline immunodeficiency virus in lymphocyte subsets of blood and lymph node. J Virol. 1996;70:5165–5169. doi: 10.1128/jvi.70.8.5165-5169.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Parseval A, Lerner D L, Borrow P, Willett B J, Elder J H. Blocking of feline immunodeficiency virus infection by a monoclonal antibody to CD9 is via inhibition of virus release rather than interference with receptor binding. J Virol. 1997;71:5742–5749. doi: 10.1128/jvi.71.8.5742-5749.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donzella G A, Schols D, Lin S W, Este J A, Nagashima K A, Maddon P J, Allaway G P, Sakmar T P, Henson G, De Clercq E, Moore J P. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4:72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 14.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 15.Dubay J W, Roberts S J, Brody B, Hunter E. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J Virol. 1992;66:4748–4756. doi: 10.1128/jvi.66.8.4748-4756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumonceaux J, Nisole S, Chanel C, Quivet L, Amara A, Baleux F, Briand P, Hazan U. Spontaneous mutations in the env gene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent entry phenotype. J Virol. 1998;72:512–519. doi: 10.1128/jvi.72.1.512-519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edinger A L, Mankowski J L, Doranz B J, Margulies B J, Lee B, Rucker J, Sharron M, Hoffman T L, Berson J F, Zink M C, Hirsch V M, Clements J E, Doms R W. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent simian immunodeficiency virus strain. Proc Natl Acad Sci USA. 1997;94:14742–14747. doi: 10.1073/pnas.94.26.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egberink H F, De Clercq E, Van Vliet A L, Balzarini J, Bridger G J, Henson G, Horzinek M C, Schols D. Bicyclams, selective antagonists of the human chemokine receptor CXCR4, potently inhibit feline immunodeficiency virus replication. J Virol. 1999;73:6346–6352. doi: 10.1128/jvi.73.8.6346-6352.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 20.English R V, Johson C M, Douglas D H, Tompkins M B. In vivo lymphocyte tropism of feline immunodeficiency virus. J Virol. 1993;67:5175–5186. doi: 10.1128/jvi.67.9.5175-5186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischinger P, Peebles P, Nomura S, Haapala D. Isolation of a RD114-like oncornavirus from a cat cell line. J Virol. 1973;11:978–985. doi: 10.1128/jvi.11.6.978-985.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freed E O, Delwart E L, Buchschacher G L, Jr, Panganiban A T. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc Natl Acad Sci USA. 1992;89:70–74. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freed E O, Myers D J. Identification and characterization of fusion and processing domains of the human immunodeficiency virus type 2 envelope glycoprotein. J Virol. 1992;66:5472–5478. doi: 10.1128/jvi.66.9.5472-5478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freed E O, Myers D J, Risser R. Characterization of the fusion domain of the human immunodeficiency virus type 1 envelope glycoprotein gp41. Proc Natl Acad Sci USA. 1990;87:4650–4654. doi: 10.1073/pnas.87.12.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green N, Shinnick T M, Witte O, Ponticelli A, Sutcliffe J G, Lerner R A. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc Natl Acad Sci USA. 1981;78:6023–6027. doi: 10.1073/pnas.78.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall C V, Jacob P E, Ringold G M, Lee F. Expression and regulation of Escherichia coli lacZ gene fusions in mammalian cells. J Mol Appl Genet. 1983;2:101–109. [PubMed] [Google Scholar]
- 27.Helseth E, Olshevsky U, Gabuzda D, Ardman B, Haseltine W, Sodroski J. Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J Virol. 1990;64:6314–6318. doi: 10.1128/jvi.64.12.6314-6318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hesselgesser J, Halks-Miller M, DelVecchio V, Peiper S C, Hoxie J, Kolson D L, Taub D, Horuk R. CD4-independent association between HIV-1 gp120 and CXCR4: functional chemokine receptors are expressed in human neurons. Curr Biol. 1997;7:112–121. doi: 10.1016/s0960-9822(06)00055-8. [DOI] [PubMed] [Google Scholar]
- 29.Hosie M J, Broere N, Hesselgesser J, Turner J D, Hoxie J A, Neil J C, Willett B J. Modulation of feline immunodeficiency virus infection by stromal cell-derived factor. J Virol. 1998;72:2097–2104. doi: 10.1128/jvi.72.3.2097-2104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hosie M J, Willett B J, Dunsford T H, Jarrett O, Neil J C. A monoclonal antibody which blocks infection with feline immunodeficiency virus identifies a possible non-CD4 receptor. J Virol. 1993;67:1667–1671. doi: 10.1128/jvi.67.3.1667-1671.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoxie J A, LaBranche C C, Endres M J, Turner J D, Berson J F, Doms R W, Matthews T J. CD4-independent utilization of the CXCR4 chemokine receptor by HIV-1 and HIV-2. J Reprod Immunol. 1998;41:197–211. doi: 10.1016/s0165-0378(98)00059-x. [DOI] [PubMed] [Google Scholar]
- 32.Johnston P B, Dubay J W, Hunter E. Truncations of the simian immunodeficiency virus transmembrane protein confer expanded virus host range by removing a block to virus entry into cells. J Virol. 1993;67:3077–3086. doi: 10.1128/jvi.67.6.3077-3086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lerner D, Grant C, de Parseval A, Elder J. FIV infection of IL-2-dependent and -independent feline lymphocyte lines: host cells range distinctions and specific cytokine upregulation. Vet Immunol Immunopathol. 1998;65:277–297. doi: 10.1016/S0165-2427(98)00162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marx P A, Chen Z. The function of simian chemokine receptors in the replication of SIV. Semin Immunol. 1998;10:215–223. doi: 10.1006/smim.1998.0135. [DOI] [PubMed] [Google Scholar]
- 35.Norimine J, Miyazawa T, Kawaguchi Y, Tomonaga K, Shin Y S, Toyosaki T, Kohmoto M, Niikura M, Tohya Y, Mikami T. Feline CD4 molecules expressed on feline non-lymphoid cell lines are not enough for productive infection of highly lymphotropic feline immunodeficiency virus isolates. Arch Virol. 1993;130:171–178. doi: 10.1007/BF01319005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Overbaugh J, Rudensey L M, Papenhausen M D, Benveniste R E, Morton W R. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J Virol. 1991;65:7025–7031. doi: 10.1128/jvi.65.12.7025-7031.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owens R J, Burke C, Rose J K. Mutations in the membrane-spanning domain of the human immunodeficiency virus envelope glycoprotein that affect fusion activity. J Virol. 1994;68:570–574. doi: 10.1128/jvi.68.1.570-574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmer C, Balfe P, Fox D, May J C, Frederiksson R, Fenyo E M, McKeating J A. Functional characterization of the V1V2 region of human immunodeficiency virus type 1. Virology. 1996;220:436–449. doi: 10.1006/viro.1996.0331. [DOI] [PubMed] [Google Scholar]
- 39.Pancino G, Castelot S, Sonigo P. Differences in feline immunodeficiency virus host cell range correlate with envelope fusogenic properties. Virology. 1995;206:796–806. doi: 10.1006/viro.1995.1002. [DOI] [PubMed] [Google Scholar]
- 40.Pedersen N C, Ho E H, Brown M L, Yamamoto J K. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science. 1987;235:790–793. doi: 10.1126/science.3643650. [DOI] [PubMed] [Google Scholar]
- 41.Phillips T R, Talbott R L, Lamont C, Muir S, Lovelace K, Elder H. Comparison of two host cell range variants of feline immunodeficiency virus. J Virol. 1990;64:4605–4613. doi: 10.1128/jvi.64.10.4605-4613.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phillips T R, Talbott R L, Lamont C, Muir S, Lovelace K, Elder J H. Comparison of two host cell range variants of feline immunodeficiency virus. J Virol. 1990;64:4605–4613. doi: 10.1128/jvi.64.10.4605-4613.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poeschla E M, Looney D J. CXCR4 is required by a nonprimate lentivirus: heterologous expression of feline immunodeficiency virus in human, rodent, and feline cells. J Virol. 1998;72:6858–6866. doi: 10.1128/jvi.72.8.6858-6866.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reeves J D, McKnight A, Potempa S, Simmons G, Gray P W, Power C A, Wells T, Weiss R A, Talbot S J. CD4-independent infection by HIV-2 (ROD/B): use of the 7-transmembrane receptors CXCR-4, CCR-3, and V28 for entry. Virology. 1997;231:130–134. doi: 10.1006/viro.1997.8508. [DOI] [PubMed] [Google Scholar]
- 45.Reeves J D, Schulz T F. The CD4-independent tropism of human immunodeficiency virus type 2 involves several regions of the envelope protein and correlates with a reduced activation threshold for envelope-mediated fusion. J Virol. 1997;71:1453–1465. doi: 10.1128/jvi.71.2.1453-1465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rein A, Yang C, Haynes J A, Mirro J, Compans R W. Evidence for cooperation between murine leukemia virus Env molecules in mixed oligomers. J Virol. 1998;72:3432–3435. doi: 10.1128/jvi.72.4.3432-3435.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rice N R, Henderson L E, Sowder R C, Copeland T D, Oroszlan S, Edwards J F. Synthesis and processing of the transmembrane envelope protein of equine infectious anemia virus. J Virol. 1990;64:3770–3778. doi: 10.1128/jvi.64.8.3770-3778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richardson J, Moraillon A, Crespeau F, Baud S, Sonigo P, Pancino G. Delayed infection after immunization with a peptide from the transmembrane glycoprotein of the feline immunodeficiency virus. J Virol. 1998;72:2406–2415. doi: 10.1128/jvi.72.3.2406-2415.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richardson J, Pancino G, Merat R, Leste-Lasserre T, Moraillon A, Schneider-Mergener J, Alizon M, Sonigo P, Heveker N. Shared usage of the chemokine receptor CXCR4 by primary and laboratory-adapted strains of feline immunodeficiency virus. J Virol. 1999;73:3661–3671. doi: 10.1128/jvi.73.5.3661-3671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ritter G D, Jr, Mulligan M J, Lydy S L, Compans R W. Cell fusion activity of the simian immunodeficiency virus envelope protein is modulated by the intracytoplasmic domain. Virology. 1993;197:255–264. doi: 10.1006/viro.1993.1586. [DOI] [PubMed] [Google Scholar]
- 51.Shimizu H, Hasebe F, Tsuchie H, Morikawa S, Ushijima H, Kitamura T. Analysis of a human immunodeficiency virus type 1 isolate carrying a truncated transmembrane glycoprotein. Virology. 1992;189:534–546. doi: 10.1016/0042-6822(92)90577-c. [DOI] [PubMed] [Google Scholar]
- 52.Shimizu H, Morikawa S, Yamaguchi K, Tsuchie H, Hachimori K, Ushijima H, Kitamura T. Shorter size of transmembrane glycoprotein of an HIV-1 isolate. AIDS. 1990;4:575–576. doi: 10.1097/00002030-199006000-00013. [DOI] [PubMed] [Google Scholar]
- 53.Spies C P, Compans R W. Effects of cytoplasmic domain length on cell surface expression and syncytium-forming capacity of the simian immunodeficiency virus envelope glycoprotein. Virology. 1994;203:8–19. doi: 10.1006/viro.1994.1449. [DOI] [PubMed] [Google Scholar]
- 54.Spies C P, Ritter G D, Jr, Mulligan M J, Compans R W. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters the conformation of the external domain. J Virol. 1994;68:585–591. doi: 10.1128/jvi.68.2.585-591.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Talbott R L, Sparger E E, Lovelace K M, Fitch W M, Pedersen N C, Luciw P A, Elder J H. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc Natl Acad Sci USA. 1989;86:5743–5747. doi: 10.1073/pnas.86.15.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vahlenkamp T W, Verschoor E J, Schuurman N N, van Vliet A L, Horzinek M C, Egberink H F, de Ronde A. A single amino acid substitution in the transmembrane envelope glycoprotein of feline immunodeficiency virus alters cellular tropism. J Virol. 1997;71:7132–7135. doi: 10.1128/jvi.71.9.7132-7135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.VandeWoude S, O'Brien S J, Langelier K, Hardy W D, Slattery J P, Zuckerman E E, Hoover E A. Growth of lion and puma lentiviruses in domestic cat cells and comparisons with FIV. Virology. 1997;233:185–192. doi: 10.1006/viro.1997.8587. [DOI] [PubMed] [Google Scholar]
- 58.Verschoor E J, Boven L A, Blaak H, van Vliet A L, Horzinek M C, de Ronde A. A single mutation within the V3 envelope neutralization domain of feline immunodeficiency virus determines its tropism for CRFK cells. J Virol. 1995;69:4752–4757. doi: 10.1128/jvi.69.8.4752-4757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Willett B J, de Parseval A, Peri E, Rocchi M, Hosie M J, Randall R, Klatzmann D, Neil J C, Jarrett O. The generation of monoclonal antibodies recognising novel epitopes by immunisation with solid matrix antigen-antibody complexes reveals a polymorphic determinant on feline CD4. J Immunol Methods. 1994;176:213–220. doi: 10.1016/0022-1759(94)90315-8. [DOI] [PubMed] [Google Scholar]
- 60.Willett B J, Hosie M J, Dunsford T H, Neil J C, Jarrett O. Productive infection of T-helper lymphocytes with feline immunodeficiency virus is accompanied by reduced expression of CD4. AIDS. 1991;5:1469–1475. doi: 10.1097/00002030-199112000-00009. [DOI] [PubMed] [Google Scholar]
- 61.Willett B J, Picard L, Hosie M J, Turner J D, Adema K, Clapham P R. Shared usage of the chemokine receptor CXCR4 by the feline and human immunodeficiency viruses. J Virol. 1997;71:6407–6415. doi: 10.1128/jvi.71.9.6407-6415.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang C, Compans R W. Analysis of the murine leukemia virus R peptide: delineation of the molecular determinants which are important for its fusion inhibition activity. J Virol. 1997;71:8490–8496. doi: 10.1128/jvi.71.11.8490-8496.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang C, Spies C P, Compans R W. The human and simian immunodeficiency virus envelope glycoprotein transmembrane subunits are palmitoylated. Proc Natl Acad Sci USA. 1995;92:9871–9875. doi: 10.1073/pnas.92.21.9871. [DOI] [PMC free article] [PubMed] [Google Scholar]