Abstract
Several early genes of murine cytomegalovirus (MCMV) encode proteins that mediate immune evasion by interference with the major histocompatibility complex class I (MHC-I) pathway of antigen presentation to cytolytic T lymphocytes (CTL). Specifically, the m152 gene product gp37/40 causes retention of MHC-I molecules in the endoplasmic reticulum (ER)-Golgi intermediate compartment. Lack of MHC-I on the cell surface should activate natural killer (NK) cells recognizing the “missing self.” The retention, however, is counteracted by the m04 early gene product gp34, which binds to folded MHC-I molecules in the ER and directs the complex to the cell surface. It was thus speculated that gp34 might serve to silence NK cells and thereby complete the immune evasion of MCMV. In light of these current views, we provide here results demonstrating an in vivo role for gp34 in protective antiviral immunity. We have identified an antigenic nonapeptide derived from gp34 and presented by the MHC-I molecule Dd. Besides the immunodominant immediate-early nonapeptide consisting of IE1 amino acids 168-176 (IE1168-176), the early nonapeptide m04243-251 is the second antigenic peptide described for MCMV. The primary immune response to MCMV generates significant m04-specific CD8 T-cell memory. Upon adoptive transfer into immunodeficient recipients, an m04-specific CTL line controls MCMV infection with an efficacy comparable to that of an IE1-specific CTL line. Thus, gp34 is the first noted early protein of MCMV that escapes viral immune evasion mechanisms. These data document that MCMV is held in check by a redundance of protective CD8 T cells recognizing antigenic peptides in different phases of viral gene expression.
Cytomegalovirus (CMV) disease with severe organ manifestations is restricted to the immunocompromised or immature host (for overviews, see references 22, 36, 57, and 66). This finding implies that an intact immune system is effectual in controlling CMV infection at crucial organ sites. Accordingly, the redundant molecular strategies reported for human CMV (HCMV) and murine CMV (MCMV) to interfere with the major histocompatibility complex (MHC) class I (MHC-I) pathway of antigenic peptide presentation (for an overview, see reference 19) do not result in an immune evasion of productively infected tissue cells.
MHC-I-restricted CD8 T cells are the principal antiviral effector cells in the long-term control of HCMV (56, 58) and MCMV (42, 48, 52, 54, 61; for a review, see reference 33) infections, whereas natural killer (NK) cells form the first line of defense early in the infection (3). Specifically, in vivo depletion of the CD8 subset, but not of the CD4 subset, resulted in lethal multiple-organ MCMV disease in an experimental setting of reconstitution after bone marrow transplantation (BMT) (41). Accordingly, CD8 T cells, but not CD4 T cells, isolated from pulmonary infiltrates after BMT and MCMV infection (24) were effectual in controlling MCMV in immunodeficient recipients after adoptive cell transfer (1). The nature of the viral antigenic peptides presented to CD8 T cells via the MHC-I pathway of antigen presentation is therefore an issue central to the understanding of immunity to CMV.
In the case of MCMV infection of BALB/c mice (H-2d), which express the MHC-I molecules Kd, Dd, and Ld, we (50) have demonstrated an immunodominance of immediate-early (IE)-phase proteins in the recognition of infected target cells by CD8-expressing cytolytic T lymphocytes (CTL). Thereafter, the immunodominant antigen was mapped to exon 4 of the ie1 gene encoding the IE1 protein pp89 (10). It was identified as IE1 peptide YPHFMPTNL presented by Ld, first by systematic screening of synthetic peptides (51, 53) and finally by its identification among the peptides naturally processed in infected cells and organs (8, 16, 24). The capacity of this single peptide to induce protective immunity was documented with a recombinant vaccinia virus containing the peptide-coding 27-nucleotide sequence (9). The findings were later confirmed by the demonstration of protective immunity induced by vaccination with IE1-encoding plasmid DNA (17).
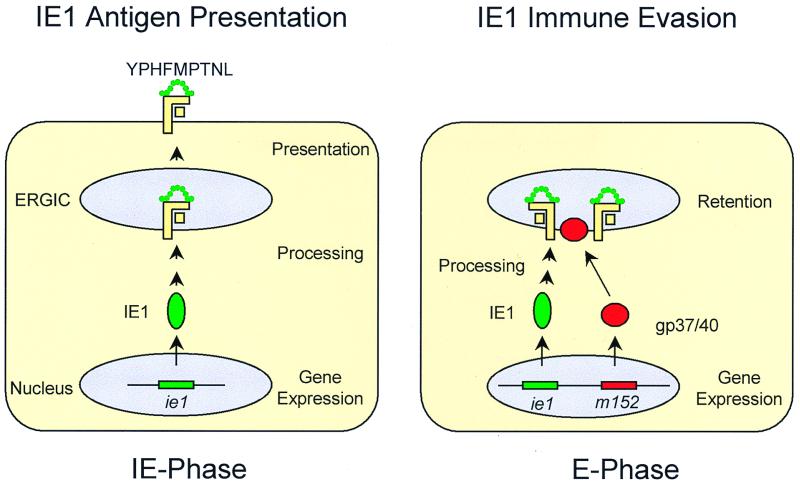
The reason for the immunodominance of the IE1 peptide remained unclear, but an attractive explanation (Fig. 1) was provided by the observation of IE1 immune evasion effective in the early (E) phase of the viral replication cycle (7, 47). Molecular analysis of the phenomenon identified redundant immune evasion mechanisms effected by E proteins (for an overview, see reference 19). Specifically, the m152 E gene product gp37/40 was identified as the viral factor responsible for the retention of peptide-loaded MHC-I complexes in the endoplasmic reticulum (ER)-Golgi intermediate compartment and cis-Golgi, thereby preventing peptide presentation to CD8 T cells (6, 63, 67); more recently, the m06 E gene product gp48 was shown to target MHC-I complexes for lysosomal degradation (55). Since these mechanisms are not peptide specific, presentation of antigenic peptides should generally be precluded in the E-phase; consequently, antigenic peptides derived from E proteins were not expected to exist. The expression of IE1 precedes expression of the E genes, a temporal advantage that might explain the immunodominance of the IE1 protein.
FIG. 1.
State-of-the-art models of IE1 presentation and evasion. (Left) IE1 presentation model, based on references 8, 10, 49, and 53. In the IE phase, and in particular under IE conditions enforced by the metabolic inhibitors cycloheximide and actinomycin D, the IE1 protein pp89 encoded by gene ie1 is expressed and processed, and the nonapeptide YPHFMPTNL is presented in the haplotype H-2d by the MHC-I molecule Ld. (Right) IE1 immune evasion model, based on references 6, 7, 47, 63, and 67. In the E phase of viral gene expression, gp37/40 encoded by gene m152 causes the retention of peptide-loaded MHC-I molecules in the ER-Golgi intermediate compartment (ERGIC) and cis-Golgi, thereby preventing the presentation of the IE1 peptide. It is proposed that lack of MHC-I surface expression renders the E-phase cells susceptible to recognition by NK cells.
However, a number of long-neglected observations argue against this explanation. (i) In the original work from 1984 describing the existence of IE-phase-specific CTL among polyclonal MCMV-primed CTL, recognition of E-phase target cells as well as of late (L)-phase target cells was already apparent (49). (ii) Under conditions of IE1 immune evasion, E-phase target cells were lysed by a CTL clone that recognized an unidentified antigen specified in EcoRI fragment F of the MCMV genome and presented by Ld (7). (iii) Antiviral CTL derived from pulmonary infiltrates after BMT preferentially lysed E-phase target cells (24). Even though the IE1 nonapeptide proved to be the only antigenic peptide detected by polyclonal pulmonary CTL among naturally processed peptides derived from MCMV-infected lungs, a comparison between IE1-specific CTL activity and overall CTL activity in pulmonary infiltrates predicted the existence of a multitude of subdominant antigenic peptides invisible as individual entities (24), many of which are expected to be E-phase specific. (iv) In mixed bone marrow chimeras resulting from the transplantation of BALB/c bone marrow cells into the Ld gene deletion mutant BALB/c-H-2dm2, a significant component of the protection against MCMV infection of the Ld-negative parenchymal tissue cells of the recipient was attributed to recipient-derived Ld-negative CD8 T cells (1). This finding clearly implied the existence of undefined antigenic peptides presented by Kd or Dd. (v) Most recently, work by Morello et al. gave evidence for protective immunity induced by the E protein specified by the M84 gene of MCMV, the positional homolog of HCMV UL84 (40). Collectively, these data indicated the existence of more than one antigenic E-phase peptide, but no such peptide has ever been identified in its amino acid sequence.
In this work we report on the identification of an antigenic peptide presented by Dd and specified by the E gene m04, known to encode gp34 (31). A CTL line (CTLL) specific for the m04 nonapeptide (m04-CTLL) protected against MCMV infection in vivo with an efficacy comparable to that of an IE1 nonapeptide-specific CTLL (IE1-CTLL). The m04 nonapeptide is the second antigenic peptide known for MCMV and the first antigenic E-phase peptide that apparently escapes the redundant immune evasion mechanisms of MCMV.
MATERIALS AND METHODS
Generation of CTLL with specificity for MCMV peptides.
Immunocompetent, 8- to 10-week-old female BALB/c mice (MHC haplotype H-2d) were sensitized by subcutaneous infection at the left hind footpad with 105 PFU of purified MCMV (35), strain Smith ATCC VR-194/1981. At >3 months after infection, at a time when productive infection is cleared in all organs, spleen cells were prepared according to standard protocols to serve as a source of MCMV-specific memory T cells. Throughout, cell culture was performed under standard conditions at 37°C, 5% CO2, and humidified atmosphere.
(i) Bulk culture for the generation of peptide-specific CTLL.
The cultivation protocol was as used in our previous work (46), with some modification. In brief, 1.5 × 107 spleen cells were seeded in a volume of 2 ml in 24-well culture plates in clone medium, minimal essential medium alpha (product no. 22561-021; Gibco BRL, Eggenstein, Germany) supplemented as described elsewhere (46) except that fetal calf serum (FCS) was reduced to 7.5% (vol/vol) and the antibiotics kanamycin and amphotericin B were not routinely added. Restimulation of peptide-specific memory T cells in the spleen cell population was achieved by synthetic peptides added in a molarity optimized for each peptide. Conditions for the generation of IE1-CTLL were reported previously (24). Usually, restimulations were performed weekly by a 1:1 split and readdition of 1 ml of clone medium containing the appropriate peptide and 200 U of recombinant human interleukin-2 (rhIL-2; 8.2 × 106 U per mg of pure protein; generously supplied by the Sandoz Research Institute, Vienna, Austria). Monospecificity of the CTLL was assessed by congruence between peptide-specific lysis and antigen-independent CD3ɛ-redirected lysis of P815 mastocytoma cells (24). Usually, but with some variance depending on the peptide, this stage was reached after three restimulations; by cytofluorometric monitoring, all lines were classified as CD8 T cells.
(ii) Microculture system for the screening of naturally processed peptides.
Peptides derived from MCMV-infected mouse embryofetal fibroblasts (MEF) were fractionated by high-performance liquid chromatography (HPLC; see below). Of each 0.8-ml fraction, 60-μl aliquots were plated in triplicate in 96-well 0.2-ml round-bottom microwell plates (CELLSTAR; product no. 650 180; Greiner, Nürtingen, Germany) and lyophilized to remove the HPLC solvent and for storage. Thus, each HPLC run gave four replicate sets of microwell plates. To each well of the first set, 2 × 106 memory spleen cells were added in 0.2 ml of clone medium. After 5 days of cultivation, cultures were supplemented with 20 U of rhIL-2. For weekly restimulation with the HPLC-fractionated peptides (HPLC-peptides), 0.1-ml aliquots of the cultures were transferred to a replicate set of microwell plates that contained the HPLC-peptides in 0.1 ml of clone medium supplemented with 20 U of rhIL-2 and 5 × 104 γ-irradiated (20 Gy) unprimed spleen cells as stimulator cells. After two restimulations (that is, after three stimulations with the HPLC-peptides), cytolytic activity was tested (see below) by using the final replicate set of peptide-containing microwell plates.
(iii) Microculture system for the screening of synthetic peptide libraries.
Synthetic peptides dissolved in clone medium were plated in microwell plates in 20-μl aliquots of 10-fold the desired final concentrations, which were 10−6, 10−8, and 10−10 M, each in triplicate. Per culture, 2 × 106 memory spleen cells were added in a volume of 180 μl. After 5 days of cultivation, cultures were supplemented with 20 U of rhIL-2, and weekly restimulations were performed by transfer to replicate plates essentially as described above for HPLC-peptides.
Cytolytic assay.
Cytolytic activity of the CTLL was tested in a standard 4-h 51Cr release assay performed with 103 target cells per 0.2-ml microwell. Data represent percentages of specific lysis and are given either for individual cultures (specifically indicated) or as the mean value of three replicate cultures. Target cells were P815 (H-2d) mastocytoma cells, except in the MHC restriction analysis that was performed with L fibroblasts (H-2k; parental line L-tk−) transfected with the MHC-I genes Kd, Dd, and Ld, referred to as L-Kd (5), L-Dd (kindly provided by M. Cochet and J. P. Abastado, Institute Pasteur, Paris, France) and L-Ld cells (10), respectively.
(i) Cytolytic assay for bulk culture-derived CTLL. The CTL were harvested, and the assay was performed with 0.1 ml of effector cells and 0.1 ml of target cells at defined effector-to-target (E/T) ratios. The target cells were preincubated with synthetic peptides at the indicated peptide molarities.
(ii) Cytolytic assay for microculture-derived CTLL. After the third stimulation with HPLC-fractionated or synthetic peptides, 0.1 ml of the CTL suspension of each well was transferred to its corresponding well in the fourth replicate microwell plate, containing the peptide in 0.1 ml of target cell suspension. For the MHC restriction analysis with L-tk− cells and the three transfectants as targets, four additional replicate sets of microwell plates were prepared with HPLC-peptides derived from a second HPLC run performed with another aliquot of the same cell extract that had been used for generation of the CTLL.
Peptides. (i) Isolation of endogenously processed peptides from MCMV-infected cells.
Peptides were acid extracted from infected cells by an adaptation of a previously published protocol (59). Specifically, MEF were plated in 10-cm petri dishes (usually 50 to 100 dishes per extraction) and were infected with purified MCMV in the third cell culture passage at a multiplicity of infection of 2, which corresponds to 0.1 PFU per cell under conditions of centrifugal enhancement of infectivity (25, 35). Cells were harvested by trypsinization in the L phase at 22 h postinfection, washed three times with ice-cold phosphate-buffered saline (PBS) in the absence of FCS, counted, and sedimented. Acidification to a pH of 2 was achieved by subsequent addition of equal volumes of 0.1 and 1% (vol/vol) trifluoroacetic acid (TFA) under permanent stirring. The acidified suspension was homogenized with a Dounce homogenizer, sonicated (Cup ultrasonicator; Branson Ultrasonics, Danbury, Conn.), and kept on ice for 30 min. Particulate components were removed from the extract by ultracentrifugation at 4°C for 45 min at 100,000 × g, and the supernatant was passed through an 0.2-μm-pore-size filter. Low-molecular-weight fractions obtained by size exclusion chromatography (Sephadex G-25 column; Pharmacia) were concentrated to a final volume of 2 ml by solid-phase extraction (SepPak C18 reversed-phase unit; Waters, Eschborn, Germany) followed by vacuum centrifugation. Separation of peptides was finally performed by HPLC with a SuperPac Sephasil C18 5-μm reversed-phase column (Pharmacia). In detail, 1 ml of the concentrated SepPak eluate, which corresponded to the extract derived from 0.5 × 108 to 2 × 108 infected MEF, was loaded on the HPLC column, and peptides were eluted at a flow rate of 0.8 ml per min on a linear acetonitrile gradient (solution A, 0.1% [vol/vol] TFA; solution B, 80% [vol/vol] acetonitrile–0.09% [vol/vol] TFA). The gradient was generated as follows: min 0 to 6, solution A; min 6 to 46, linear increase to 100% solution B; min 46 to 50, solution B; min 50 to 54, linear decrease to 0% solution B; and min 54 to 58, solution A. Aliquots of the 0.8-ml fractions were distributed into wells of microwell plates and lyophilized for storage until used for the generation of CTLL and for pulsing of target cells in the cytolytic assays.
(ii) Synthetic peptides.
Custom peptide synthesis in ca. 1-mg scale and a purity of >75% was performed by JERINI Bio Tools GmbH (Berlin, Germany). Peptides were dissolved in 30% (vol/vol) acetonitrile in PBS at a concentration of 10−3 M. Further dilutions were performed in medium.
IFN-γ-based ELISPOT assay.
Peptide-specific CTL derived from the spleen during acute MCMV infection (2 weeks after infection) or after the establishment of viral latency and immunological memory (>3 months after infection) were quantitated by an enzyme-linked immunospot (ELISPOT) assay detecting single CD8 T cells that secrete gamma interferon (IFN-γ) upon resensitization with peptide (for the principle of the assay, see reference 21). A modification of the established protocol (39, 62) published previously (15) was used, with some further modification. The assay was performed in nylon membrane-backed 96-well microwell plates (Biodyne B nylon 66 membrane product no. 256154; Nunc, Wiesbaden, Germany). The membrane bottom of the wells was coated with rat anti-mouse IFN-γ monoclonal antibody (MAb) (clone RMMG-1; product no. AMC 4834; Biosource Europe, Ratingen, Germany) contained in bicarbonate coating buffer (pH 9.6) at a concentration of 10 μg per ml. Surplus antibody was washed out, and the membrane was saturated with clone medium (with no IL-2) for 1 h at 37°C. Peptide-presenting cells (PPC) were added to the wells of the assay plate in a concentration of 105 cells per 50 μl of clone medium (with no IL-2). PPC were P815 mastocytoma cells stably transfected with human B7-1 (CD80) cDNA (2), referred to as P815-B7 cells (used with the kind permission of L. L. Lanier, DNAX, Palo Alto, Calif.), pulsed with the appropriate peptide at a final concentration of 10−8 M. As observed by one of us (G. Geginat, unpublished data), the use of P815-B7 instead of P815 significantly improves the specificity of the assay by enhancing the signal-to-background ratio. Excess peptide was washed out before use of the PPC in the ELISPOT assay. As IFN-γ-secreting effector cells to be quantitated, graded numbers of spleen cells (here 105 and 106 cells) were added in 50 μl of clone medium (with no IL-2). After incubation for ca. 16 h at 37°C, cells were removed by extensive washing with PBS containing 0.25% (vol/vol) Tween 20 (PBS-Tween). Bound IFN-γ was labeled for 2 h at ca. 20°C with biotinylated rat anti-mouse IFN-γ MAb (50 μl of 1 μg/ml; clone XMG1.2, product no. 18112D; Pharmingen, Hamburg, Germany), washed with PBS-Tween, and then incubated for 2 h at ca. 20°C with horseradish peroxidase-streptavidin conjugate (50 μl of 4 μg/ml; Dianova, Hamburg, Germany). After further washing, 50 μl of aminoethylcarbazole solution (15) was added as the substrate, yielding a brown precipitate. After 5 to 10 min, the membranes were washed with water and then air dried for storage. Spots, representing individual peptide-specific cells, were counted under a stereomicroscope.
Cytofluorometric detection of intracellular IFN-γ.
The functional capacity of CTLL to produce IFN-γ upon sensitization with the specific peptide was tested by the cytofluorometric staining of intracellular IFN-γ according to an established protocol (43; for the principle of the assay, see reference 21), with some modification. Peptide-specific CTLL were seeded in 96-well round-bottom microwell plates at a concentration of 106 cells in 0.2 ml of RPMI 1640 medium (product no. 31870-025; Gibco-BRL) supplemented with 5% (vol/vol) FCS, 50 μM 2-mercaptoethanol, 10 mM HEPES, 50 U of rhIL-2 per ml, stimulating peptide or control peptide at a concentration of 10−6 M, and 10 μg of brefeldin A (product no. B7651; Sigma) per ml. Brefeldin A inhibits the secretory pathway, thus retaining newly synthesized IFN-γ in the ER. The cells were harvested for cytofluorometric analysis after 5 h of incubation. Note that subsequent steps were performed in the presence of 10 μg of brefeldin A per ml, until the cells were fixed. Harvested cells were pooled, and aliquots of 4 × 106 cells were washed twice with 500 μl of PBS supplemented with 1% (wt/vol) bovine serum albumin and 0.1% (wt/vol) NaN3. For the blocking of unspecific binding sites, cells were resuspended in 50 μl of PBS supplemented as specified above, and 0.4 μg of MAb anti-FcγRII/III (clone 2.4G2; product no. 01241D; Pharmingen, San Diego, Calif.) was added per sample. Surface staining was performed with phycoerythrin (PE)-conjugated MAb anti-CD8a (clone 53.6.7; product no. 01045A, Pharmingen). The cells were washed twice with PBS to remove excess protein and were resuspended in 100 μl of PBS. Cell fixation was performed for 20 min at ca. 20°C by addition of 100 μl of PBS containing 2% (vol/vol) paraformaldehyde. After washing twice with PBS, cells were permeabilized for 10 min at ca. 20°C by resuspension in permeabilization solution, which is PBS containing 1% (wt/vol) bovine serum albumin, 0.1% (wt/vol) NaN3, and 2% (wt/vol) saponin (product no. 9622.1; Roth, Karlsruhe, Germany). For intracellular IFN-γ staining, half of the aliquot (i.e., 2 × 106 cells) was labeled with 0.05 μg of fluorescein-conjugated MAb rat anti-mouse IFN-γ (clone XMG1.2, product no. 18114A; Pharmingen), and the remaining half of the aliquot was incubated with 0.05 μg of the appropriate fluorescein-conjugated isotype control MAb (rat immunoglobulin G1; clone R3-34, product no. 20614A; Pharmingen). In either case, labeling was performed in 50 μl of permeabilization solution for 30 min on ice. Finally, cells were washed twice in PBS, resuspended in 250 μl of PBS, and analyzed immediately with a FACSort cytofluorometer (Becton Dickinson, San Jose, Calif.). Data were processed by using CellQuest software (Becton Dickinson).
Adoptive CTL transfer.
Recipients of adoptive transfer were 8-week-old, female BALB/c mice that were immunosuppressed by γ-irradiation with a dose of 6.5 Gy and were infected in the left hind footpad with 105 PFU of purified MCMV. Under these conditions, all mice die of multiple organ histopathology (54) and bone marrow aplasia (37) between days 10 and 18 after infection unless they receive protective T cells. CTL were transferred intravenously 2 h before infection, and antiviral function was assessed on day 12 after infection.
Quantitation of virus infection and T-cell infiltration in tissues. (i) Determination of virus titers.
Infectious virus was quantitated in organ homogenates by a plaque assay performed on subconfluent second-passage MEF monolayers under conditions of centrifugal enhancement of infectivity in 48-well culture plates as described previously (54). The virus titers represent the amount of infectious virus per organ and are expressed as PFU* to indicate the ca. 20-fold enhancement of infectivity (25, 35) achieved by the centrifugally enforced virus penetration.
(ii) Quantitative two-color immunohistology.
Livers were fixed with PBS (pH 7.4) containing 4% (vol/vol) formalin. The tissue was then processed for paraffin embedding. Sections of 2 μm were dewaxed with xylene and subjected to two-color immunohistology with hematoxylin counterstaining precisely as described previously (1, 24). In essence, liver-infiltrating T cells were visualized by membrane staining with a rat MAb directed against murine CD3ɛ, biotinylated anti-rat antibody, and the avidin-biotin-peroxidase complex with diaminobenzidine tetrahydrochloride as the substrate. The staining was enhanced with ammonium nickel sulfate hexahydrate, yielding a black precipitate. Infected hepatocytes were visualized by detection of intranuclear viral IE1 protein with MAb CROMA 101 (kindly provided by S. Jonjic, Medical Faculty of the University of Rijeka, Rijeka, Croatia), goat anti-mouse immunoglobulin antibody (no. M5899; Sigma), and the alkaline phosphatase–anti-alkaline phosphatase complex with new fuchsin as the substrate, yielding a brilliant red precipitate. Liver-infiltrating T cells and infected hepatocytes were counted for representative 10-mm2 areas of tissue sections.
Analysis of viral gene expression.
The kinetics of viral gene expression in infected MEF monolayer cultures (10-cm petri dishes with ca. 2 × 106 cells per dish) was assessed by reverse transcriptase (RT)-mediated PCR (RT-PCR). Specifically, MEF were infected with purified MCMV in the third cell culture passage at a multiplicity of infection of 4, which corresponds to 0.2 PFU per cell under conditions of centrifugal enhancement of infectivity. Under these conditions, >90% of the cells were infected as assessed by immunofluorescence specific for the intranuclear IE1 protein pp89 (30, 60). The extraction buffer of a QuickPrep-Micro mRNA purification kit (Pharmacia Biotech) was added to the MEF monolayers, and detachment of cells was facilitated by using a cell scraper. Poly(A)+ RNA was purified on the basis of oligo(dT)-cellulose affinity.
(i) DNase treatment of poly(A)+ RNA prior to RT-PCR.
For genes with an exon-intron structure, such as MCMV ie1 (29), primers were placed in neighboring exons, yielding different-sized amplificates from mRNA and contaminating DNA. In the case of all other viral genes, contaminating DNA may result in false-positive signals for gene expression and must be digested before RT-PCR. DNase treatment of poly(A)+ RNA samples was performed with RNase-free DNase (product no. M6101; Promega, Madison, Wis.). The reaction was performed for 30 min at 37°C in a volume of 10 μl with 1 μg of poly(A)+ RNA adjusted to 8 μl with RNase-free water (Braun, Melsungen, Germany), 1 μl of a 10-fold-concentrated reaction buffer (400 mM Tris-HCl [pH 8.0], 100 mM MgSO4, 10 mM CaCl2), and 1 μl containing 1 U of the RNase-free DNase. The digestion was stopped by inactivating the DNase with 1 μl of stop solution (20 mM EGTA, pH 8.0) and incubation for 10 min at 65°C. The clean poly(A)+ RNA was diluted 1:50 in RNase-free water to reduce the Mg2+ concentration that would otherwise affect the RT reaction. Aliquots of 10 μl were subjected to RT-PCR.
(ii) Design of primers and probes for RT-PCR.
Throughout, oligo(dT) priming was used for the RT reactions. Forward and reverse primers for the detection of hprt (hypoxanthine phosphoribosyltransferase) cDNA (32) were 5′-GTTGGATACAGGCCAGACTTTGTTG-3′ and 5′-GCCTAAGATGAGCGCAAGTTGAATC-3′, respectively. The 163-bp hprt amplificate was specifically detected with the internal oligonucleotide probe 5′-GTTGTTGGATATGCCCTTGAC-3′. Positions of primers and probes for viral gene expression refer to the genomic sequence of the Smith strain of MCMV (45; GenBank accession no. MCU68299 [complete genome]). For detection of ie1 cDNA, forward primer 5′-181750-181726-3′ (5′-CCAGTTGCAACATGATCATGATCGC-3′) and reverse primer 5′-181374-181398-3′ (5′-GCAGCATGCTTGATGGCCATGTCGC-3′) were used. The 280-bp ie1 amplificate was detected by probe 5′-181504-181531-3′ (5′-GACCTGACTCTGGAGGACATGTTGGGA-3′) directed against the exon 3-exon 4 splicing junction. m04 cDNA was detected with forward primer 5′-3423-3446-3′ (5′-GGCATCAACCCGACTAAGGTCCCG-3′) and reverse primer 5′-3746-3723-3′ (5′-AGCGCTGCCTGTGGTGGCGTTCGG-3′). The 324-bp m04 amplificate was detected by probe 5′-3616-3640-3′ (5′-CACTCCCATGCACGGATTACGTAC-3′). Forward and reverse primers for detection of m152 cDNA were 5′-210423-210446-3′ (5′-GCTTGAGACCGTGTCCCCTGTGCC-3′) and 5′-210928-210904-3′ (5′-CTCGGGTCTCCGCCGTACTCTCCC-3′), respectively. The 506-bp m152 amplificate was detected by probe 5′-210674-210697-3′ (5′-TTCGCGAGACTGATGTTGTACGCC-3′).
(iii) RT-PCR.
Reactions were carried out by using an automated thermal cycler (GeneAmp PCR System 9700; Perkin-Elmer Applied Biosystems, Norwalk, Conn.). The RT reaction was performed with 10 μl of poly(A)+ RNA (ca. 10 ng, representing the yield of ca. 2 × 104 infected MEF) adjusted with RT reaction buffer to a total volume of 20 μl [final concentrations: 60 mM KCl, 15 mM Tris-HCl (pH 8.4), 3 mM MgCl2, 10 mM dithiothreitol, 20% (vol/vol) glycerol, 1 mM each deoxynucleoside triphosphate, 12.5 pmol of oligo(dT) primer, 10 U of RNasin, and 100 U of Moloney murine leukemia virus RT (Gibco BRL)] for 10 min at 28°C followed by 30 min at 42°C. The reaction was stopped by heating to 95°C for 5 min, and the cDNA samples were kept on ice until PCR. For the amplification of cDNA sequences by PCR, 30 μl of PCR mix (60 mM KCl, 15 mM Tris-HCl [pH 8.4], 3 mM MgCl2, 20% [vol/vol] glycerol, 25 pmol of each primer, 1.5 U of Taq DNA polymerase [Eurobio, Raunheim, Germany]) was added. The PCR time-temperature profile for cycles 2 to 29 was as follows: denaturation for 30 s at 96°C, annealing for 1 min at the primer-dependent temperature (58°C for hprt and ie1 and 65°C for m04 and m152), and elongation for 1 min at 72°C. In the first cycle, denaturation was performed for 3 min at 95°C. In the last cycle (cycle 30), the elongation time was extended to 5 min. Amplification products (15 μl) were visualized by standard procedures of 2% (wt/vol) agarose gel electrophoresis, Southern blot, hybridization with the appropriate γ-32P-end-labeled oligonucleotide probe, and autoradiography.
RESULTS
Search for novel MCMV-specific antigenic peptides.
In a previous report on the detection of MCMV-specific MHC-I-presented peptides by pulmonary CTL of BALB/c mice (24), the long-known immunodominant, Ld-presented IE1 peptide YPHFMPTNL (53) was the only peptide detectable in HPLC fractions derived from an extract of infected lungs. However, infected target cells were recognized in the E phase of the viral replication cycle (24), even though the IE1 peptide is not presented in the E phase (7, 47). Furthermore, compared with the overall cytolytic activity assessed by CD3ɛ-redirected lysis, IE1-specific CTL apparently accounted for an only minor part of the pulmonary CTL response to MCMV (24). This finding implied the existence of further antigenic peptides that remained invisible as individual entities for polyclonal pulmonary ex vivo CTL but collectively constituted a strong immune response. It was therefore necessary to develop a strategy for making the postulated “invisible” peptides visible.
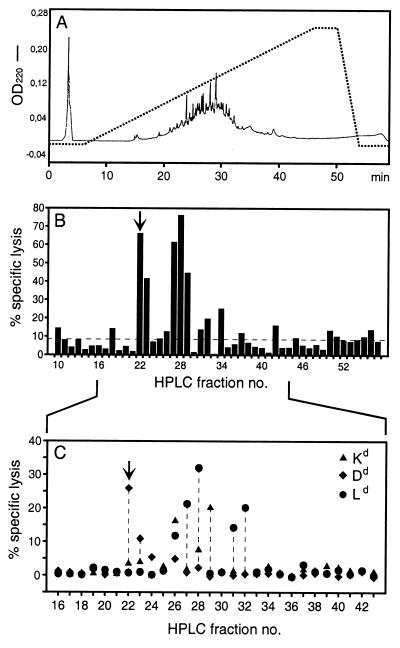
Antigenic peptides, which are involved in the CD8 T-cell response in vivo, leave their imprint in the specificity repertoire of memory T-cell populations. If an antigenic peptide that was naturally processed in MCMV-infected cells is present in an HPLC fraction of the cell extract, and if memory T cells with the specific T-cell receptor (TCR) are present in the spleen cell population of MCMV-sensitized mice, repeated restimulations with that particular HPLC fraction should lead to the selection of an HPLC fraction-specific CTLL. Fractions of an HPLC control run performed with an extract from uninfected MEF did not select any CTLL from memory spleen cells (not shown). By contrast, restimulation of memory spleen cells with HPLC fractions derived from productively infected MEF identified several fractions with antigenic activity (Fig. 2). Most peptides elute from the HPLC column between fractions 20 and 40 (Fig. 2A). Antigenic peptides were found to be contained most prominently in fractions 22, 23, 27, 28, and 29, and there were minor candidates as well (Fig. 2B).
FIG. 2.
Antigenic activities in HPLC fractions derived from productively infected MEF. (A) Elution profile of the HPLC. The dotted line describes the acetonitrile gradient from 0 to 80%. OD220, optical density at 220 nm. (B) Detection of naturally processed antigenic peptides in HPLC fractions by short-term microculture CTLL generated from MCMV-specific memory spleen cells by repeated restimulation with the corresponding HPLC fractions. The cytolytic assay was performed with P815 target cells. Data represent the mean of triplicates. Fractions 27 and 28 contain the already known IE1 peptide. The arrow points to fraction 22, which represents the most consistently observed novel antigenic activity. The dashed line represents the baseline of the assay, as defined with no HPLC fraction added to the cultures. (C) MHC-I restriction analysis for the antigenic activities in HPLC fractions, performed with MHC-I transfectants L-Kd, L-Dd, and L-Ld as target cells. Data represent the mean of triplicates. The arrow marks the Dd-restricted activity in fraction 22.
When the HPLC fractions containing naturally processed MCMV peptides were used to pulse target cells for a cytolytic assay performed with IE1-CTLL as effector cells, fractions 27 and 28 were positive. Likewise, IE1-CTL detected antigenic activity in fractions 27 and 28 of an HPLC separation of extract from uninfected MEF supplemented with the synthetic IE1 nonapeptide YPHFMPTNL. Thus, fractions 27 and 28 contained the already known IE1 peptide (not shown).
In conclusion, the strategy of repeated restimulation of memory T cells did not reveal numerous antigenic HPLC fractions, but fractions 22, 23, and 29 each contained at least one antigenic peptide distinct from the known IE1 peptide. Actually, this experiment was the first to clearly indicate the existence of MCMV-specific antigenic peptides in addition to the immunodominant IE1 peptide.
The MHC restriction of the thus selected HPLC fraction-specific CTLL was tested with target cells expressing any of the three MHC-I genes of the H-2d haplotype selectively, namely, the L-cell transfectants L-Ld, L-Dd, and L-Kd (Fig. 2C). It should be noted that no cytolytic activity was detected with the parental L-tk− cells, which are of H-2k haplotype (not shown). As predicted from the known Ld presentation of the IE1 peptide, Ld restriction was seen for CTLL raised with HPLC fractions 27 and 28. Fractions 22 and 23 contained at least one peptide presented by Dd. Fraction 29 contained at least one peptide presented by Kd. Fractions 31 and 32, although less prominent in the assay shown in Fig. 2B, were found to contain peptides presented by Ld. Fraction 26 revealed a promiscuous restriction, which might indicate a mixture of different peptides. The activity in fractions 34 and 42 was not confirmed by the MHC restriction analysis. It should be noted that the pattern can vary with different memory spleen cell populations, and thus a negative result in a particular experiment does not exclude the presence of an antigenic peptide in a negative fraction. The activity in fraction 22 was most consistently observed in our experiments, and therefore we focused first on peptides in that fraction. In summary, MCMV specifies a strong and novel antigenic peptide presented by Dd, and further candidates exist.
Dd motif peptide library of the full-length MCMV Smith genome.
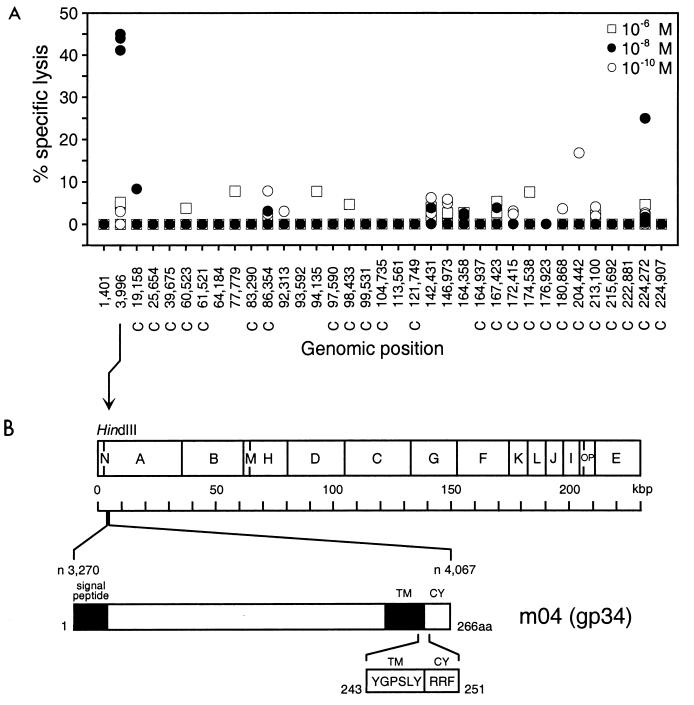
An approach to identify the peptide in fraction 22 by mass spectrometry failed due to the heterogeneity and low amount of peptides present in the HPLC-derived fraction (T. Ruppert, personal communication). Given the existence of at least one Dd-presented peptide (see above), we performed a screening of synthetic peptides (Fig. 3) based on Rammensee's Dd binding motif (12; for an extensive overview and motif listing, see reference 44). Specifically, the nonameric Dd motif is xGPxxxxx[L, I, F] with MHC anchor residues glycine and proline at positions 2 and 3, respectively, and leucine, isoleucine, or phenylalanine at C-terminal position 9. The search for the motif was made for all open reading frames of the entire genome of MCMV, strain Smith, based on the complete genomic sequence published by Rawlinson et al. (45). The search revealed 35 potentially antigenic peptides proposed to be presentable by Dd. The respective synthetic peptides were each used in three different molarities for the restimulation of MCMV-sensitized memory spleen cells. Screening for a wide range of peptide concentrations proved to be essential, because the affinity of an antigenic peptide for MHC binding is not predictable from its sequence and because suboptimal as well as supraoptimal peptide concentrations in the restimulation assay can give false-negative results (4).
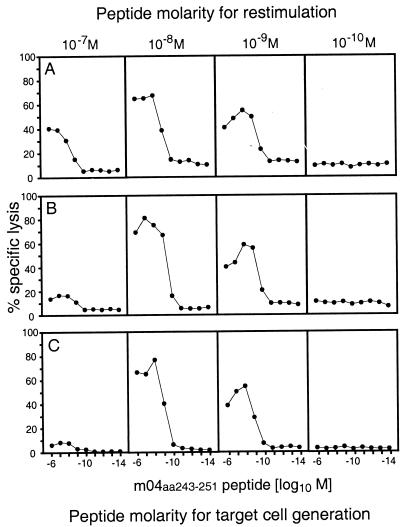
FIG. 3.
Identification of an antigenic peptide by screening of a Dd motif peptide library of MCMV. (A) A search for the Dd binding motif xGPxxxxx[L, I, F] (44) was performed for all open reading frames of the full-length genomic sequence of MCMV (45), and corresponding synthetic nonapeptides were used at the indicated concentrations for the generation of short-term microculture CTLL by repeated restimulation of MCMV-specific memory spleen cells. The cytolytic assay was performed with P815 target cells. Data represent the cytolytic activity in individual microcultures. The genomic positions of the nonapeptide-coding sequences are given by the positions of the first nucleotides according to the listing by Rawlinson et al. (45). C, complementary strand. (B) Map of the location of the prominent antigenic peptide (nucleotide [n] positions 3996 to 4022), representing aa 243 to 251 of gp34 encoded by gene m04 (31). TM, transmembrane region; CY, cytoplasmic tail. The peptide sequence is given in one-letter code.
With some peptides, CTL were generated only in one culture of the triplicate. This finding is likely to reflect a very low frequency of memory T cells specific for those peptides. A prominent Dd-presented peptide, seen in triplicate cultures after restimulation with a peptide concentration of 10−8 M, mapped to positions 3996 to 4022 of the MCMV genome (Fig. 3A). This sequence is located in HindIII fragment A (11) and within gene m04 encoding gp34 (31). The corresponding peptide with the sequence YGPSLYRRF represents amino acids (aa) 243 to 251 located at the junction between transmembrane region and cytoplasmic tail of gp34 (Fig. 3B).
It was an obvious question to ask in retrospect whether this Dd-presented synthetic peptide was indeed identical with the Dd-presented naturally processed peptide that had eluted in the experiment of Fig. 2 in HPLC fraction 22. Therefore, an HPLC run was performed with synthetic peptide YGPSLYRRF. Surprisingly, the peptide did not elute in fraction 22 but proved to be contained in fraction 24, which was seen before (Fig. 2C) to contain a minor Dd-presented candidate peptide. Work is in progress to identify the fraction 22 peptide. Although fraction 24 peptide was thus discovered by serendipity, its derivation from the well-defined and particularly interesting viral NK decoy glycoprotein gp34 (31) led us to continue with working on it.
H-2d class I motif peptide library for gene m04 and frequencies of m04-specific CD8 T cells.
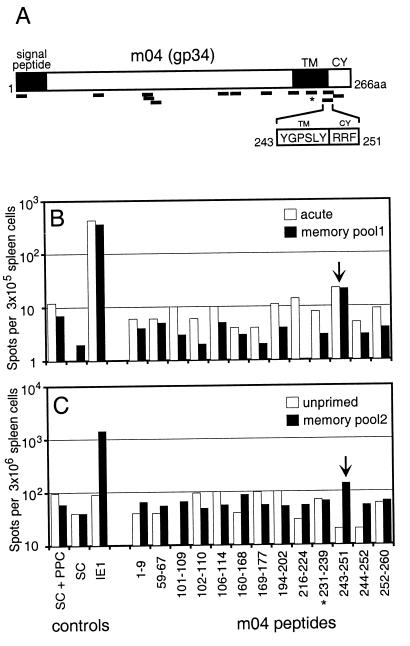
Notably, m04 contains only a single Dd motif. To identify further antigenic peptides processed from gp34 and presented by MHC-I molecules, memory spleen cells were repeatedly restimulated with synthetic peptides representing Kd and Ld motifs specified by the m04 gene. Specifically, the nonameric Kd motif is x[Y, F]xxxxxx[I, L, V], and the nonameric Ld motif is x[P, S]xxxxxx[F, L, M] (44). The search revealed a single Kd motif and 11 Ld motifs within the m04 open reading frame, but none of the corresponding synthetic peptides could be identified as an antigenic peptide generating short-term microculture CTLL (not shown). It is worth noting that the group of nonantigenic motifs includes an Ld motif (aa 1 to 9), which is located in the signal sequence, and were thus potentially independent of proteasomal processing (18, 64).
As a second approach for the identification of relevant antigenic peptides, the 13 synthetic peptides of the H-2d class I motif library of m04 (Fig. 4A) were used to stimulate acutely primed and memory spleen cells in an ELISPOT assay based on IFN-γ secretion by activated CD8 T cells (Fig. 4B and C). In accordance with the results of the CTLL microcultures, the Dd-presented peptide consisting of aa 243 to 251 (peptide 243-251) proved to be the only m04-encoded peptide that gave a reproducible and significant signal with acutely primed spleen cells (Fig. 4B) as well as with two independent memory spleen cell pools (Fig. 4B and C). Unprimed spleen cells gave no signal above the assay background (Fig. 4C) that was defined with all components except peptide. f-m04, the frequency of pool 1 memory cells (Fig. 4B) specific for the m04 peptide 243-251 (hereafter referred to as m04 peptide), was 1 per 14,000 spleen cells. As a positive reference, the ELISPOT assays were all also performed with the IE1 peptide 168-176, i.e., peptide YPHFMPTNL presented by Ld (53). The frequency of pool 1 memory cells specific for the IE1 peptide (f-IE1) was found to be 1 per 700 spleen cells. Since the proportion of CD8 T cells in the spleen is ca. 10% (varying between 8 and 12% in memory spleen cell populations), corrected frequencies f-m04 and f-IE1 are 1 per 1,400 and 1 per 70 CD8 T cells, respectively. Altogether, these assays have impressively reconfirmed the previously noted relative immunodominance of the IE1 peptide (24, 50), but the m04 peptide was clearly identified as a second antigenic peptide of quantitative significance in the in vivo CD8 T-cell response and generation of immunological memory to MCMV.
FIG. 4.
Screening of an MHC-I motif peptide library of gene m04 by ELISPOT assays. (A) Searches for the Kd motif x[Y, F]xxxxxx[I, L, V] and the Ld motif x[P, S]xxxxxx[F, L, M] (44) were performed for the m04 open reading frame. Besides the single Dd motif (Fig. 3), a single Kd motif and 11 Ld motifs were found. The locations of the motifs within the gp34 sequence are indicated by bars. The asterisk marks the single Kd motif. For abbreviations, see the legend to Fig. 3. (B) Corresponding synthetic peptides were tested in an IFN-γ-based ELISPOT assay performed with spleen cells derived from acutely infected BALB/c mice at 2 weeks after infection (open bars) or from memory spleen cells (pool 1) derived from three latently infected BALB/c mice at 4 months after infection (filled bars). (C) IFN-γ-based ELISPOT assay performed with spleen cells from adult, unprimed BALB/c mice (open bars) or from memory spleen cells (independent pool 2) derived from another three latently infected BALB/c mice at 4 months after infection (filled bars). The arrow points to results obtained for peptide 243-251. The asterisk marks the peptide that corresponds to the Kd motif. Controls from left to right: (i) spleen cells (SC) and P815-B7 as PPC, but no peptide added; (ii) SC only; (iii) SC, PPC, and, as a positive reference, the IE1 nonapeptide YPHFMPTNL. Throughout, peptides were used at a concentration of 10−8 M.
Expression of the immune evasion gene m152 precedes the expression of m04.
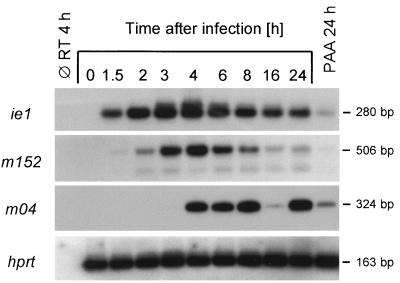
The m152 E-phase gene product gp37/40 prevents presentation of antigenic peptides by retention of peptide-loaded MHC-I complexes in the ER-Golgi intermediate compartment and cis-Golgi (67). Expression of the IE-phase gene ie1 precedes expression of m152, and therefore presentation of the IE1 peptide may prime CD8 T cells before evasion mechanisms become effectual. This plausible explanation for antigenicity might apply also to other peptides presented before the expression of gp37/40. We therefore studied the kinetics of MCMV gene expression in infected MEF by RT-PCR (Fig. 5). Consistent with previous findings (28), ie1 gene expression was apparent at 1.5 h postinfection and the IE1-specific poly(A)+ RNA remained detectable throughout the viral replication cycle, with a phosphonoacetic acid (PAA)-sensitive reexpression in the L phase (47). In agreement with a previous study (67), expression of m152 was seen early in the E phase, with a maximum of m152-specific poly(A)+ RNA detectable at 4 h postinfection, declining thereafter. Expression of the E-phase gene m04 encoding gp34 clearly occurs later in the E phase. Most importantly, Kleijnen et al. have previously shown that expression of m152 precedes expression of m04 also on the protein level (31). We could show here PAA-sensitive reexpression of m04 in the L phase. One may therefore conclude that presentation of the m04 peptide occurs after rather than before the MHC-I retention mediated by m152-encoded gp37/40.
FIG. 5.
Kinetics of MCMV gene expression in MEF. Poly(A)+ RNA, derived from infected MEF harvested at the indicated time points after infection, was subjected to RT-PCRs specific for the indicated genes, including cellular hprt as a control. First lane, all reagents except RT; last lane, poly(A)+ RNA derived from MEF infected for 24 h in the presence of PAA (250 μg per ml). Shown are autoradiographs obtained after agarose gel electrophoresis, Southern blotting, and hybridization with the respective γ-32P-end-labeled oligonucleotide probes.
Establishment of an m04-CTLL.
An analysis of the properties of m04-specific CTL requires their large-scale propagation. CTLL were therefore generated in bulk cultures by stimulation of MCMV-sensitized memory spleen cells with the synthetic m04 peptide YGPSLYRRF (Fig. 6). We document here the development of the cultures during three rounds of restimulation (Fig. 6) to emphasize the crucial importance of the peptide concentration used for restimulation. It may be considered trivial that low concentrations of the stimulating peptide, in this case 10−10 M, can fail to provide the required stimulation. Even though not without precedent (4), it is still less well known that generation of CTLL can also fail because of supraoptimal peptide concentration. Thus, at 10−7 M m04 peptide used for restimulation, CTL activity vanished after three restimulations. The optimal condition for the generation of an m04-CTLL proved to be restimulation with 10−8 M peptide, and plateau lysis in the cytolytic assay was reached when target cells were pulsed with 10−8 to 10−7 M peptide.
FIG. 6.
Generation of m04-CTLL. Memory spleen cells derived from BALB/c mice at 4 months after infection with MCMV were restimulated under bulk culture conditions with synthetic m04 peptide YGPSLYRRF at the molar concentrations indicated. Panels A to C show cytolytic activity of CTLL after one to three rounds of restimulation, respectively. The cytolytic assay was performed at an E/T ratio of 15:1. Target cells were P815 mastocytoma cells pulsed with the m04 peptide at the molar concentrations indicated.
Functional activity in vitro: comparison between IE1-CTLL and m04-CTLL.
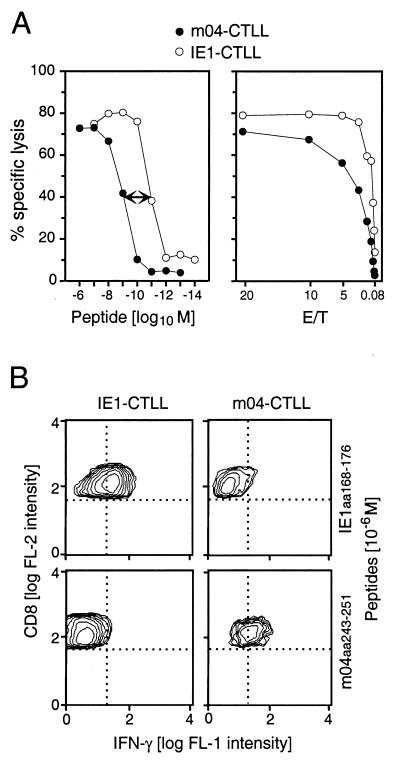
If we wish to compare antiviral functions of CTLL with different peptide specificities, it is of interest to know the requirements for triggering an effector function. The affinities of the interactions between peptide and presenting MHC molecule and between TCR and MHC-peptide complex are determinants of the amount of peptide that needs to be processed and presented in infected cells in order to elicit antiviral effector function. An IE1-CTLL, generated from memory spleen cells by restimulation with 10−9 M IE1 peptide YPHFMPTNL, required 100-fold less peptide for cytolytic activity (Fig. 7A, left). Specifically, half-maximal lysis of peptide-pulsed target cells occurred at 10−11 M and plateau lysis occurred at 10−9 M, compared with 10−9 and 10−7 M, respectively, required with the m04-CTLL. Furthermore, even at the optimal conditions for target cell formation, the IE1-CTL were superior in terms of cytolytic potential (Fig. 7A, right).
FIG. 7.
Properties of m04-CTLL and IE1-CTLL in vitro. (A) Comparison of cytolytic effector function. (Left) Peptide dose dependence of target cell recognition. Target cells were P815 mastocytoma cells pulsed with the indicated molar concentrations of the appropriate synthetic peptide. The assays were performed at an E/T cell ratio of 15:1. The 100-fold peptide molarity difference between the two CTLL is highlighted by a two-headed arrow. (Right) Cytolytic potential of the two CTLL, compared by E/T titration. P815 mastocytoma target cells were pulsed with optimal molar concentrations of synthetic peptide, 10−7 and 10−8 M for m04-CTLL and IE1-CTLL, respectively. (B) Detection of intracellular IFN-γ by two-color cytofluorometric analysis. Production of IFN-γ was stimulated in IE1-CTLL and m04-CTLL by sensitization with 10−6 M synthetic peptide, with the heterologous peptides serving as negative controls. CTL were stained for the expression of IFN-γ (fluorescein fluorescence [FL-1]; abscissa) and CD8 (PE fluorescence, FL-2; ordinate). Quadrants (dotted lines) were defined by omission of PE-conjugated specific antibody in the case of CD8 and by a fluorescein-conjugated isotype control antibody in the case of IFN-γ. All cells of both CTLL expressed CD8. Data obtained with 25,000 and 12,500 cells for IE1-CTLL and m04-CTLL, respectively, are shown as contour plots in a 50% log mode.
Expression and secretion of IFN-γ upon sensitization with MHC-presented peptide is another important effector function of CD8 T cells. Both CTLL were capable of expressing IFN-γ after stimulation with a saturating concentration of their corresponding peptides (Fig. 7B). Neither of the two CTLL expressed IFN-γ spontaneously, and the induction of IFN-γ required recognition of the specific MHC-peptide complex.
In conclusion, both CTLL exert effector functions when their specific peptide is adequately presented, but the IE1-CTLL requires less peptide for its effector function.
Control of MCMV infection in vivo: comparison between IE1-CTLL and m04-CTLL.
Gene m04 is expressed in the E phase (31) and reexpressed in the L phase of the viral replication cycle (Fig. 5). It was therefore obvious to examine whether infected target cells are lysed by m04-CTL in the E phase and/or in the L phase. The assay was performed, as usual in this field, with MEF infected in vitro. To our surprise, the result was negative (not shown). Admittedly, this was at first a disappointment. We therefore addressed the possibility that the m04 peptide might result from gp34 processing after exogenous loading of the MHC-I pathway following virus penetration (for a review, see reference 26) rather than in the course of viral gene expression. Such a mechanism was described previously by Reddehase et al. for structural proteins of MCMV (49, 50) and appears to apply also to the processing of the HCMV tegument protein pp65 (38, 65). However, even at a multiplicity of infection of 100 carried out under inhibition of viral gene expression (49), the m04 peptide was not presented (not shown). We therefore concluded that the antigenicity of gp34 does not result from exogenous loading of the MHC-I pathway of antigen processing and presentation.
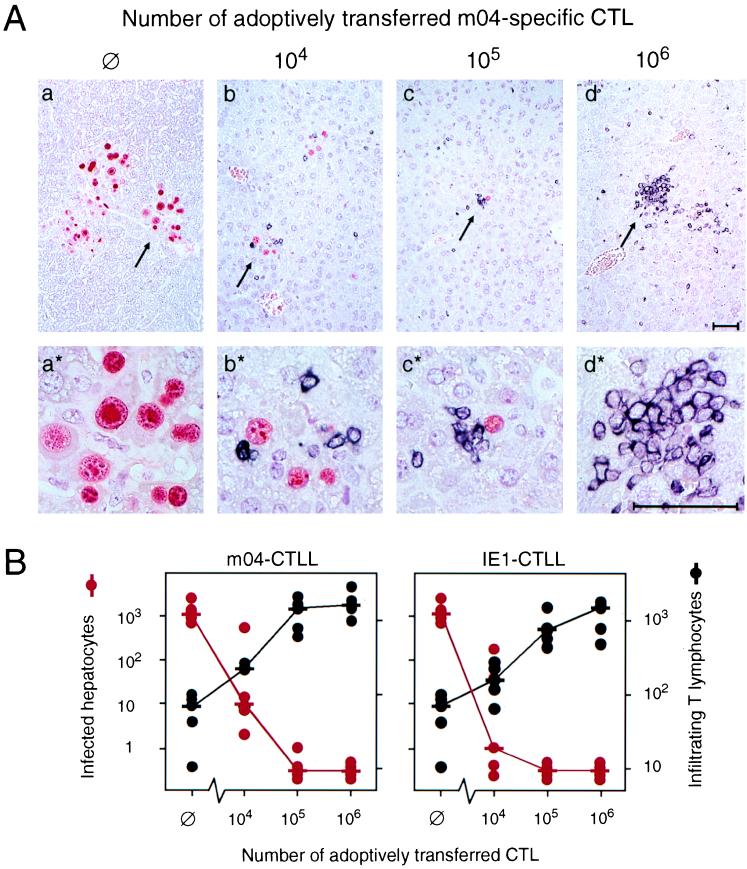
Moreover, the m04 peptide was not detected with m04-CTLL among naturally processed peptides isolated from MEF during the E phase (not shown). These in vitro data clearly contrast with the fact that m04/gp34-specific memory cells were generated during in vivo infection at a frequency of ca. 1 in 1,400 CD8 T cells and that CTLL with that specificity were so easily retrieved from memory spleen cell populations. Apparently, infected tissue cells in vivo must have adequately processed and presented this peptide, and results obtained with MEF in vitro are thus prone to mislead most likely because MEF are not representative of the cell types relevant to infection in vivo. To make the point clear, we tested the in vivo antiviral potential of the two CTLL in comparison by adoptive transfer into immunosuppressed, lethally infected recipients. Virus infection and T-cell infiltration of the liver were assessed by quantitative two-color immunohistology, simultaneously visualizing infected hepatocytes by red-stained intranuclear IE1 protein and infiltrating CTL by black-stained CD3ɛ surface molecules (Fig. 8). In the absence of CTL transfer, only few residual liver T cells were present (Fig. 8B) and failed to control the infection, resulting in plaque-like, necrotic foci of infection in liver parenchyma with no signs of inflammation (Fig. 8A, panel a for overview and panel a* for detail). Adoptively transferred m04-CTL controlled the infection in a dose-dependent manner (Fig. 8A, panels b to d for overview and b* to d* for detail). Notably, the infiltration of infected tissue is not random, but the transferred CTL are specifically attracted by infected cells, thus forming inflammatory foci (best seen in Fig. 8A, c and c*). After transfer of 106 m04-CTL, the infection was resolved, and CTL were found in clusters (Fig. 8A, d and d*). Transfer of IE1-CTL gave essentially the same histological images (therefore not shown). Quantitation of the data indicates a somewhat higher efficacy of IE1-CTL, in particular at low cell numbers (Fig. 8B).
FIG. 8.
Control of hepatic MCMV infection by liver-infiltrating CTL. (A) Two-color immunohistological analysis visualizing infected hepatocytes (red intranuclear staining) and liver-infiltrating CD3ɛ-expressing T cells (black membrane staining). The indicated cell numbers of m04-specific CTL (from the same CTLL as characterized in Fig. 7) were adoptively transferred into immunocompromised, infected BALB/c recipients. ∅, control recipients with no cell transfer. The analysis was performed on day 12 after infection and cell transfer. (a to d) Overviews. The arrows point to sites of interest shown enlarged in corresponding panels (a* to d*). Bar markers represent 50 μm. (B) Quantitative two-color immunohistology after adoptive transfer of m04-CTLL (left, corresponding to panel A) or of IE1-CTLL (right). The numbers of infected hepatocytes (red dots) and of infiltrating T lymphocytes (black dots) refer to representative 10-mm2 areas of liver tissue sections. Each dot represents an individual transfer recipient. Median values are marked by a horizontal bar.
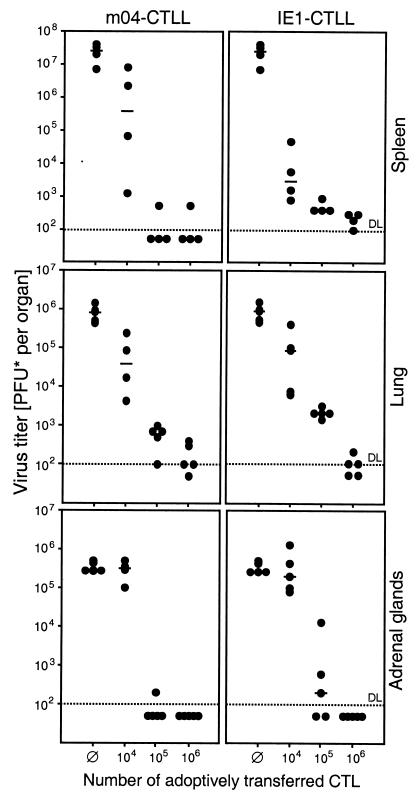
The antiviral function of the two CTLL was not restricted to liver tissue. Both controlled the infection, as measured by virus titers, also in other organs relevant to CMV disease, such as the spleen, lungs, and adrenal glands (Fig. 9). The minor advantage of IE1-CTL over m04-CTL, as discussed above for the liver, holds true also for the spleen, whereas m04-CTL were slightly more effectual in the lungs and adrenal glands.
FIG. 9.
Control of MCMV replication in further organs relevant to CMV disease. Infectious virus in the indicated organs of immunocompromised BALB/c recipients (same experiment as in Fig. 8) was quantitated by an in vitro plaque assay performed on day 12 after adoptive transfer of the indicated cell numbers of m04-CTL (left) or of IE1-CTL (right). ∅, control recipients with no cell transfer. Virus titers are given as PFU* determined under conditions of centrifugal enhancement of infectivity. Each dot represents an individual transfer recipient. Median values are marked by a horizontal bar. DL and dotted line, detection limit of the plaque assay.
Our general conclusion from this set of experiments is that both CTLL were more or less equally efficient in controlling MCMV infection in a variety of organs. Altogether, MCMV appears to be held in check by a redundance of protective CD8 T cells differing in their peptide specificities. Notably, in spite of immune evasion mechanisms operative in the E phase of the viral replication cycle, a peptide derived from the E-phase protein gp34 mediates antiviral protection.
DISCUSSION
Recent research on CMV immunology has focused on strategies developed by HCMV and MCMV to evade recognition of infected cells by CD8 T cells and NK cells (for a review, see reference 19). While the precise molecular mechanisms of immune evasion differ somewhat between these two viruses, they conform to the general concept that genes expressed in the E phase code for proteins that interfere at various steps with the MHC-I pathway of antigen processing and presentation. Redundance in this function indicates a biological relevance from the viewpoint of the virus but also implies a leakiness inherent to each individual molecular mechanism involved.
In the specific case of MCMV, the IE-phase gene ie1 codes for an antigenic peptide that is immunodominant in the H-2d haplotype (24, 50). Since its molecular identification in 1989 (53), this IE1 nonapeptide had remained the only antigenic peptide known for MCMV. The reason for its immunodominance may lie in the fact that it is processed and presented before immune evasion mechanisms become effectual (Fig. 1, left). The E-phase gene m152 encodes gp37/40, which mediates retention of peptide-loaded MHC-I complexes in the ER-Golgi intermediate compartment and cis-Golgi (6, 63, 67) (Fig. 1, right). Another E-phase gene, m06, codes for gp48, which targets MHC-I complexes for lysosomal degradation (55). As a consequence, both mechanisms in concert should prevent the presentation of antigenic peptides to CD8 T cells and simultaneously should reduce the expression of MHC-I on the cell surface.
In light of these findings, one may see no reason to search for antigenic peptides generated and presented in the E phase or L phase. However, there are a number of observations (listed in the introduction) that suggest the existence of further antigenic peptides of MCMV. Previous work has shown a prevention of IE1 peptide presentation in the E phase (7, 47), whereas the same E-phase target cells were lysed by a CTL clone, then named clone E1, which recognized an antigen presented by Ld and specified by EcoRI fragment F (7). This early finding was set aside when the later discovery of MHC-I retention predicted a more general inhibition of presentation in the E phase (6). It is worth noting that the E1 clone, originally isolated by Del Val et al. (7), still exists and was recently revived in our lab. We could reproduce the earlier results showing that clone E1 recognizes E- and L-phase target cells but not IE-phase target cells (not shown). However, despite all indirect evidence, no antigenic E-phase peptide was ever identified. It was therefore high time to replace evidence by proof. We have here identified the first antigenic peptide of MCMV that is derived from an E-phase protein.
Our search for novel antigenic peptides of MCMV was free from any supposition regarding candidate proteins, including gp34. It was therefore more than a surprise when a gp34-derived peptide was identified. Owing to the careful work by Kleijnen et al. (31), gp34 is well characterized. It is encoded by gene m04 and is expressed late in the E phase, clearly after the expression of the MHC-I retention protein gp37/40 (31, 67). Particular interest in gp34 arose from the finding that it is complexed with MHC-I molecules at the cell surface. Notably, in the H-2d haplotype, gp34 was found to coprecipitate with Dd and Ld but not with Kd. It was concluded that gp34 binds to newly synthesized MHC-I in the ER, transports the complex to the cell surface, and thereby restores MHC-I surface expression in cells in which the presentation of antigenic peptides is prevented by retention of peptide-loaded MHC-I (Fig. 10, left). Since cells in the E phase are not recognized by Ld-restricted IE1-specific CTL (7, 47), it is apparent that the MHC-I surface transport mediated by gp34 does not rescue already retained IE1-peptide–MHC-I complexes. Kleijnen et al. (31) favored the interpretation that gp34 serves to prevent the attack by NK cells, which would otherwise recognize the lack of MHC-I surface expression as missing self (23, 27). Thus, similar to the MHC-I homolog of MCMV that is encoded by gene m144 (13), gp34 is proposed to contribute to immune evasion of MCMV through interference with NK cell-mediated clearance.
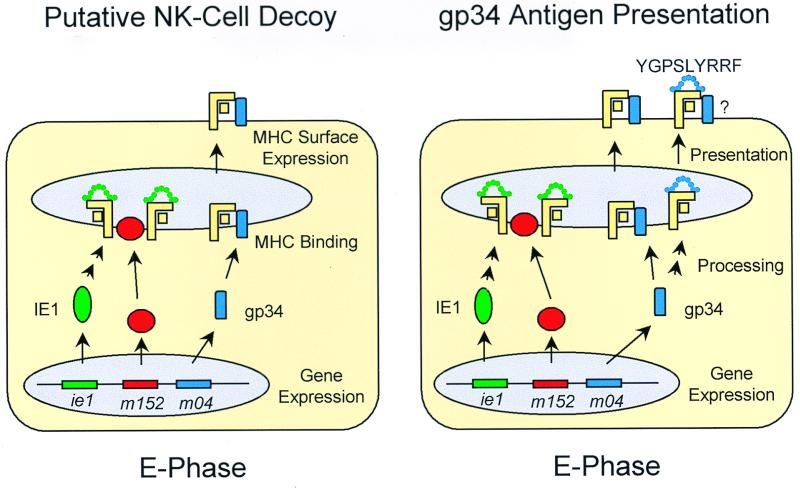
FIG. 10.
State-of-the-art models of the role of gp34 in the E phase. (Left) NK cell decoy model, based on reference 31. The m04 E-gene product gp34 binds to MHC-I molecules in the ER and directs the complex to the cell surface. It is proposed that restoration of MHC-I surface expression silences the NK cells. (Right) Model of gp34 antigen presentation in the E phase, based on this report. The E-phase protein gp34 is by itself subject of antigen processing, and peptide YGPSLYRRF is presented in the haplotype H-2d by the MHC-I molecule Dd. Although not documented for cultured fibroblasts, the priming of memory T cells and the antiviral function of m04-CTL imply peptide presentation in infected tissue cells in vivo. It remains open to question whether the gp34 peptide-presenting MHC-I molecules are also complexed with native gp34 protein.
We wish to point out that our data do not argue against a putative role for gp34 in silencing NK cells. However, our data do add an unexpected new aspect to the role of gp34 (Fig. 10, right) by demonstrating that gp34 is by itself subject of antigen processing and accounts for an antigenic peptide, m04243-251, with the sequence YGPSLYRRF presented by the MHC-I molecule Dd. Kleijnen et al. suggested that the binding of gp34 to a peptide-presenting MHC-I molecule may interfere with recognition of this complex by specific CD8 T cells (31). However, there was no experimental evidence to support this idea. With the same logic, one may also speculate that gp34, while it cannot rescue already retained peptide–MHC-I complexes, can guide peptide-loaded, newly synthesized MHC-I complexes to the cell surface to prevent NK cell recognition and simultaneously restore the control by CD8 T cells. Promising work is in progress to identify further antigenic peptides specified in the E phase. We therefore do not wish to convey the impression that there is any special link between native gp34 and the presentation of a gp34-derived peptide. If native gp34 does indeed aid the presentation of a peptide derived from its own processing, this is most likely just a fancy of nature. At the moment, it is not known whether the MHC-I molecules that present the gp34 peptide in the E phase are biochemically complexed with native gp34. Whether native gp34 is needed for antigen presentation in the E phase as a transporter for peptide-loaded MHC-I molecules could theoretically be tested with an MCMV m04 gene deletion mutant. However, since deletion of m04 also eliminates the gp34-derived peptide, performance of this attractive experiment must await the identification of other E-phase peptides.
Alternatively, one can also envisage the possibility that presentation in the late E phase does not require assistance by gp34. A possible reason for an escape of peptide–MHC-I complexes in the late E phase may lie in the fact that expression of the m152 retention gene wanes in the late E phase, while m04 is still expressed. An exhaustion of available retention protein gp37/40 might also offer an explanation for the occasionally observed recognition of L-phase target cells by IE1-specific CTL (7, 47), because the ie1 gene was found to be reexpressed in the L-phase (47).
A problem to be considered is the possibility that a structural viral protein may be processed after virion penetration by exogenous loading of the MHC-I pathway (reviewed in reference 26). Thus, presentation of antigenic peptide may precede the immune evasion mechanisms, even though normal expression of the protein occurs in the E or L phase of the viral replication cycle. Such a mechanism has been described for MCMV (49), and CTLL that recognized structural antigens of MCMV processed and presented in absence of viral gene expression were then established (46). The question of whether gp34 is a virion protein of MCMV was not specifically addressed in the work by Kleijnen et al. (31). Even if it is not a structural protein, virion preparations used for infection may include membranes of infected cells carrying gp34–MHC-I complexes. However, when we used extremely high doses of virions to intentionally load the alternative MHC-I pathway, the cells were not recognized by the gp34-specific CTL (not shown). We therefore see no reason to assume that gp34 is processed along this pathway.
To our surprise, in vitro-infected MEF were not lysed by gp34-specific CTL in either the E phase or the L phase (not shown). At present, we can only speculate that the sensitivity of the gp34-specific CTLL was too low to detect presented peptide, most likely because of the low affinity of the Dd-m04 peptide-TCR interaction, which proved to be 100-fold lower than the affinity of the Ld-IE1 peptide-TCR interaction observed for the IE1-CTLL (Fig. 7A). Based on these negative in vitro findings, one would not propose a role for an m04 peptide in protective immunity to MCMV. The in vivo data, therefore, have taught us the valuable lesson that assessing peptide presentation in cultured fibroblasts is inappropriate for predicting the in vivo relevance of an antigen. Specifically, as documented here, the in vivo infection with MCMV had generated significant m04 peptide-specific T-cell memory, and m04 peptide-specific CTL controlled the infection in all tissues analyzed. This implies that unlike the fibroblasts in culture, the various infected cell types in tissues must have presented the m04 peptide in an amount adequate for eliciting T-cell effector function. Enhancement of antigenic peptide presentation by IFN-γ during inflammation in vivo may contribute to the antiviral function of CTL (16, 20). That the in vivo protective function of virus-specific T cells does not always correlate with the hierarchy of CTL responses to naturally processed viral peptides is a finding reported previously for another virus infection (14).
We have here identified the first antigenic peptide of MCMV that is derived from an E-phase protein. Is this the end of the road? Certainly not! One can predict the existence of further antigenic peptides assigned to the E phase. Specifically, Del Val et al.'s proposed peptide still awaits identification. This peptide is clearly distinct from the m04 peptide (located in EcoRI-G within HindIII-A), as it maps to a distant region of the MCMV genome, namely, to EcoRI-F within HindIII-E (7, 11), and as it is presented by Ld. Unlike the m04 peptide, this undefined peptide is recognized by the E1-CTLL during the E phase in MEF (7). In addition, a second E-phase-specific CTLL isolated by Del Val et al., clone E2, is proposed to recognize a peptide that does not map to EcoRI-F. Furthermore, for MCMV strain K181, recent work by Morello et al. (40) predicts an antigenic peptide encoded by gene M84 located within HindIII-C. Finally, data shown in the present report (Fig. 2) suggest the existence of a more prominent Dd-presented peptide in HPLC fraction 22 and of a significant Kd-presented peptide in fraction 29. The latter candidate is of particular interest because, as discussed above, Kd is not complexed with gp34 in the E phase (31).
Conclusion.
We have shown here that MCMV is controlled in vivo by a redundance of CD8 T cells with specificities assigned to different phases of viral gene expression. Even though recent work by Krmpotic et al. has demonstrated an in vivo modulatory function of the immune evasion gene m152 of MCMV (34), it is apparent from the course of CMV infections that all of the sophisticated viral strategies of molecular immune evasion do not eventually prevent effectual immune control. We have identified an antigenic peptide derived from gp34, a viral E-phase glycoprotein intimately involved in the viral manipulation of the MHC-I pathway in that it assists the traffic of MHC-I molecules to the cell surface. We predict that our work will stimulate an era of CMV immunology beyond immune evasion.
ACKNOWLEDGMENTS
We thank Ronda Cardin (at the time of this study at St. Jude Children's Research Hospital, Memphis, Tenn.) and Norbert Palm (Institute for Immunology, Mainz, Germany) for advice and practical help regarding the intracellular staining of IFN-γ. We appreciate the expert technical assistance by Doris Dreis.
Support was provided by grants to M.J.R. from the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 311 (Immunopathogenesis), project A18, and individual projects RE 712/3-2 and RE 712/4-1.
REFERENCES
- 1.Alterio de Goss M, Holtappels R, Steffens H-P, Podlech J, Angele P, Dreher L, Thomas D, Reddehase M J. Control of cytomegalovirus in bone marrow transplantation chimeras lacking the prevailing antigen-presenting molecule in recipient tissues rests primarily on recipient-derived CD8 T cells. J Virol. 1998;72:7733–7744. doi: 10.1128/jvi.72.10.7733-7744.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azuma M, Cayabyab M, Buck D, Philipps J H, Lanier L L. CD28 interaction with B7 costimulates primary allogeneic proliferative responses and cytotoxicity mediated by small, resting T lymphocytes. J Exp Med. 1992;175:353–360. doi: 10.1084/jem.175.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bukowski J F, Woda B A, Welsh R M. Pathogenesis of murine cytomegalovirus infection in natural killer cell-depleted mice. J Virol. 1984;52:119–128. doi: 10.1128/jvi.52.1.119-128.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Busch D H, Pamer E G. MHC class I/peptide stability: implications for immunodominance, in vitro proliferation, and diversity of responding CTL. J Immunol. 1998;160:4441–4448. [PubMed] [Google Scholar]
- 5.Cochet M, Kast W M, Kummer A-M, Transy C, Melief C J M, Kourilsky P. Alternative splicing in the mouse H-2Kd gene is not necessary for the classical Kd antigen function. Immunogenetics. 1986;24:267–274. doi: 10.1007/BF00364531. [DOI] [PubMed] [Google Scholar]
- 6.Del Val M, Hengel H, Häcker H, Hartlaub U, Ruppert T, Lucin P, Koszinowski U H. Cytomegalovirus prevents antigen presentation by blocking the transport of peptide-loaded major histocompatibility complex class I molecules into the medial-Golgi compartment. J Exp Med. 1992;176:729–738. doi: 10.1084/jem.176.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Del Val M, Münch K, Reddehase M J, Koszinowski U H. Presentation of CMV immediate-early antigen to cytolytic T lymphocytes is selectively prevented by viral genes expressed in the early phase. Cell. 1989;58:305–315. doi: 10.1016/0092-8674(89)90845-3. [DOI] [PubMed] [Google Scholar]
- 8.Del Val M, Schlicht H-J, Ruppert T, Reddehase M J, Koszinowski U H. Efficient processing of an antigenic sequence for presentation by MHC class I molecules depends on its neighboring residues in the protein. Cell. 1991;66:1145–1153. doi: 10.1016/0092-8674(91)90037-y. [DOI] [PubMed] [Google Scholar]
- 9.Del Val M, Schlicht H-J, Volkmer H, Messerle M, Reddehase M J, Koszinowski U H. Protection against lethal cytomegalovirus infection by a recombinant vaccine containing a single nonameric T-cell epitope. J Virol. 1991;65:3641–3646. doi: 10.1128/jvi.65.7.3641-3646.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Val M, Volkmer H, Rothbard J B, Jonjic S, Messerle M, Schickedanz J, Reddehase M J, Koszinowski U H. Molecular basis for cytolytic T-lymphocyte recognition of the murine cytomegalovirus immediate-early protein pp89. J Virol. 1988;62:3965–3972. doi: 10.1128/jvi.62.11.3965-3972.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebeling A, Keil G M, Knust E, Koszinowski U H. Molecular cloning and physical mapping of murine cytomegalovirus DNA. J Virol. 1983;47:421–433. doi: 10.1128/jvi.47.3.421-433.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falk K, Rötzschke O, Stevanovic S, Jung G, Rammensee H-G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 13.Farrell H E, Vally H, Lynch D M, Fleming P, Shellam G R, Scalzo A A, Davis-Poynter N J. Inhibition of natural killer cells by a cytomegalovirus MHC class I homologue in vivo. Nature. 1997;386:510–514. doi: 10.1038/386510a0. [DOI] [PubMed] [Google Scholar]
- 14.Gallimore A, Dumrese T, Hengartner H, Zinkernagel R M, Rammensee H-G. Protective immunity does not correlate with the hierarchy of virus-specific cytotoxic T cell responses to naturally processed peptides. J Exp Med. 1998;187:1647–1657. doi: 10.1084/jem.187.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geginat G, Nichterlein T, Kretschmar M, Schenk S, Hof H, Lalic-Mülthaler M, Goebel W, Bubert A. Enhancement of the Listeria monocytogenes p60-specific CD4 and CD8 T cell memory by nonpathogenic Listeria innocua. J Immunol. 1999;162:4781–4789. [PubMed] [Google Scholar]
- 16.Geginat G, Ruppert T, Hengel H, Holtappels R, Koszinowski U H. IFN-γ is a prerequisite for optimal antigen processing of viral peptides in vivo. J Immunol. 1997;158:3303–3310. [PubMed] [Google Scholar]
- 17.Gonzales Armas J C, Morello C S, Cranmer L D, Spector D H. DNA immunization confers protection against murine cytomegalovirus infection. J Virol. 1996;70:7921–7928. doi: 10.1128/jvi.70.11.7921-7928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson R A, Michel H, Sakaguchi K, Shabanowitz J, Appella E, Hunt D F, Engelhard V H. HLA-A2.1-associated peptides from a mutant cell line: a second pathway of antigen presentation. Science. 1992;255:1264–1266. doi: 10.1126/science.1546329. [DOI] [PubMed] [Google Scholar]
- 19.Hengel H, Brune W, Koszinowski U H. Immune evasion by cytomegalovirus—survival strategies of a highly adapted opportunist. Trends Microbiol. 1998;6:190–197. doi: 10.1016/s0966-842x(98)01255-4. [DOI] [PubMed] [Google Scholar]
- 20.Hengel H, Lucin P, Jonjic S, Ruppert T, Koszinowski U H. Restoration of cytomegalovirus antigen presentation by gamma interferon combats viral escape. J Virol. 1994;68:289–297. doi: 10.1128/jvi.68.1.289-297.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hickling, J. K. 13 October 1998, posting date. Measuring human T-lymphocyte function. Expert Rev. Mol. Med. [Online.] http://www-ermm.cbcu.cam.ac.uk/jhc/txt001jhc.htm. [DOI] [PubMed]
- 22.Ho M. Cytomegaloviruses. In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. New York, N.Y: Churchill Livingstone; 1995. pp. 1351–1364. [Google Scholar]
- 23.Höglund P, Sundbäck J, Olsson-Alheim M Y, Johansson M, Salcedo M, Öhlen C, Ljunggren H-G, Sentman C L, Kärre K. Host MHC class I gene control of NK-cell specificity in the mouse. Immunol Rev. 1997;155:11–28. doi: 10.1111/j.1600-065x.1997.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 24.Holtappels R, Podlech J, Geginat G, Steffens H-P, Thomas D, Reddehase M J. Control of murine cytomegalovirus in the lungs: relative but not absolute immunodominance of the immediate-early 1 nonapeptide during the antiviral cytolytic T-lymphocyte response in pulmonary infiltrates. J Virol. 1998;72:7201–7212. doi: 10.1128/jvi.72.9.7201-7212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hudson J B, Misra V, Mosmann T R. Cytomegalovirus infectivity: analysis of the phenomenon of centrifugal enhancement of infectivity. Virology. 1976;72:235–243. doi: 10.1016/0042-6822(76)90326-3. [DOI] [PubMed] [Google Scholar]
- 26.Jondal M, Schirmbeck R, Reimann J. MHC class-I-restricted CTL responses to exogenous antigens. Immunity. 1996;5:295–302. doi: 10.1016/s1074-7613(00)80255-1. [DOI] [PubMed] [Google Scholar]
- 27.Kärre K. Express yourself or die: peptides, MHC molecules, and NK cells. Science. 1995;267:978–979. doi: 10.1126/science.7863341. [DOI] [PubMed] [Google Scholar]
- 28.Keil G M, Ebeling-Keil A, Koszinowski U H. Temporal regulation of murine cytomegalovirus transcription and mapping of viral RNA synthesized at immediate-early times after infection. J Virol. 1984;50:784–795. doi: 10.1128/jvi.50.3.784-795.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keil G M, Ebeling-Keil A, Koszinowski U H. Sequence and structural organization of murine cytomegalovirus immediate-early gene 1. J Virol. 1987;61:1901–1908. doi: 10.1128/jvi.61.6.1901-1908.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keil G M, Fibi M R, Koszinowski U H. Characterization of the major immediate-early polypeptides encoded by murine cytomegalovirus. J Virol. 1985;54:422–428. doi: 10.1128/jvi.54.2.422-428.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleijnen M F, Huppa J B, Lucin P, Mukherjee S, Farrell H, Campbell A E, Koszinowski U H, Hill A B, Ploegh H L. A mouse cytomegalovirus glycoprotein, gp34, forms a complex with folded class I MHC molecules in the ER which is not retained but is transported to the cell surface. EMBO J. 1997;16:685–694. doi: 10.1093/emboj/16.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konecki D S, Brennand J, Fuscoe J C, Caskey C T, Chinault A C. Hypoxanthine-guanine phosphoribosyltransferase genes of mouse and Chinese hamster: construction and sequence analysis of cDNA recombinants. Nucleic Acids Res. 1982;10:6763–6775. doi: 10.1093/nar/10.21.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koszinowski U H, Reddehase M J, Jonjic S. The role of T-lymphocyte subsets in the control of cytomegalovirus infection. In: Thomas D B, editor. Viruses and the cellular immune response. New York, N.Y: Marcel Dekker; 1993. pp. 429–445. [Google Scholar]
- 34.Krmpotic A, Messerle M, Crnkovic-Mertens I, Polic B, Jonjic S, Koszinowski U H. The immunoevasive function encoded by the mouse cytomegalovirus gene m152 protects the virus against T cell control in vivo. J Exp Med. 1999;190:1–11. doi: 10.1084/jem.190.9.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurz S, Steffens H-P, Mayer A, Harris J R, Reddehase M J. Latency versus persistence or intermittent recurrences: evidence for a latent state of murine cytomegalovirus in the lungs. J Virol. 1997;71:2980–2987. doi: 10.1128/jvi.71.4.2980-2987.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ljungman P, Einsele H. Cytomegalovirus infection. Curr Opin Hematol. 1994;1:418–422. [PubMed] [Google Scholar]
- 37.Mayer A, Podlech J, Kurz S, Steffens H-P, Maiberger S, Thalmeier K, Angele P, Dreher L, Reddehase M J. Bone marrow failure by cytomegalovirus is associated with an in vivo deficiency in the expression of essential stromal hemopoietin genes. J Virol. 1997;71:4589–4598. doi: 10.1128/jvi.71.6.4589-4598.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLaughlin-Taylor E, Pande H, Forman S J, Tanamachi B, Li C R, Zaia J A, Greenberg P D, Riddell S R. Identification of the major late human cytomegalovirus matrix protein pp65 as a target antigen for CD8+ virus-specific cytotoxic T lymphocytes. J Med Virol. 1994;43:103–110. doi: 10.1002/jmv.1890430119. [DOI] [PubMed] [Google Scholar]
- 39.Miyahira Y, Murata K, Rodriguez D, Rodriguez J R, Esteban M, Rodrigues M M, Zavala F. Quantification of antigen-specific CD8+ T cells using an ELISPOT assay. J Immunol Methods. 1995;181:45–54. doi: 10.1016/0022-1759(94)00327-s. [DOI] [PubMed] [Google Scholar]
- 40.Morello C S, Cranmer L D, Spector D H. In vivo replication, latency, and immunogenicity of murine cytomegalovirus mutants with deletions in the M83 and M84 genes, the putative homologs of human cytomegalovirus pp65 (UL83) J Virol. 1999;73:7678–7693. doi: 10.1128/jvi.73.9.7678-7693.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Podlech J, Holtappels R, Wirtz N, Steffens H-P, Reddehase M J. Reconstitution of CD8 T cells is essential for the prevention of multiple-organ cytomegalovirus histopathology after bone marrow transplantation. J Gen Virol. 1998;79:2099–2104. doi: 10.1099/0022-1317-79-9-2099. [DOI] [PubMed] [Google Scholar]
- 42.Polic B, Hengel H, Krmpotic A, Trgovcich J, Pavic I, Lucin P, Jonjic S, Koszinowski U H. Hierarchical and redundant lymphocyte subset control precludes cytomegalovirus replication during latent infection. J Exp Med. 1998;188:1047–1054. doi: 10.1084/jem.188.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prussin C, Metcalfe D D. Detection of intracytoplasmic cytokine using flow cytometry and directly conjugated anti-cytokine antibodies. J Immunol Methods. 1995;188:117–128. doi: 10.1016/0022-1759(95)00209-x. [DOI] [PubMed] [Google Scholar]
- 44.Rammensee H-G, Bachmann J, Stevanovic S. MHC ligands and peptide motifs. Austin, Tex: Molecular Biology Intelligence Unit, Landes Bioscience; 1997. [Google Scholar]
- 45.Rawlinson W D, Farrell H E, Barrell B G. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reddehase M J, Bühring H-J, Koszinowski U H. Cloned long-term cytolytic T-lymphocyte line with specificity for an immediate-early membrane antigen of murine cytomegalovirus. J Virol. 1986;57:408–412. doi: 10.1128/jvi.57.1.408-412.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reddehase M J, Fibi M R, Keil G M, Koszinowski U H. Late-phase expression of a murine cytomegalovirus immediate-early antigen recognized by cytolytic T lymphocytes. J Virol. 1986;60:1125–1129. doi: 10.1128/jvi.60.3.1125-1129.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reddehase M J, Jonjic S, Weiland F, Mutter W, Koszinowski U H. Adoptive immunotherapy of murine cytomegalovirus adrenalitis in the immunocompromised host: CD4-helper-independent antiviral function of CD8-positive memory T lymphocytes derived from latently infected donors. J Virol. 1988;62:1061–1065. doi: 10.1128/jvi.62.3.1061-1065.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reddehase M J, Keil G M, Koszinowski U H. The cytolytic T lymphocyte response to the murine cytomegalovirus. II. Detection of virus replication stage-specific antigens by separate populations of in vivo active cytolytic T lymphocyte precursors. Eur J Immunol. 1984;14:56–61. doi: 10.1002/eji.1830140111. [DOI] [PubMed] [Google Scholar]
- 50.Reddehase M J, Koszinowski U H. Significance of herpesvirus immediate early gene expression in cellular immunity to cytomegalovirus infection. Nature. 1984;312:369–371. doi: 10.1038/312369a0. [DOI] [PubMed] [Google Scholar]
- 51.Reddehase M J, Koszinowski U H. Redistribution of critical major histocompatibility complex and T cell receptor-binding functions of residues in an antigenic sequence after biterminal substitution. Eur J Immunol. 1991;21:1697–1701. doi: 10.1002/eji.1830210717. [DOI] [PubMed] [Google Scholar]
- 52.Reddehase M J, Mutter W, Münch K, Bühring H-J, Koszinowski U H. CD8-positive T lymphocytes specific for murine cytomegalovirus immediate-early antigens mediate protective immunity. J Virol. 1987;61:3102–3108. doi: 10.1128/jvi.61.10.3102-3108.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reddehase M J, Rothbard J B, Koszinowski U H. A pentapeptide as minimal antigenic determinant for MHC class I-restricted T lymphocytes. Nature. 1989;337:651–653. doi: 10.1038/337651a0. [DOI] [PubMed] [Google Scholar]
- 54.Reddehase M J, Weiland F, Münch K, Jonjic S, Lüske A, Koszinowski U H. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J Virol. 1985;55:264–273. doi: 10.1128/jvi.55.2.264-273.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reusch U, Muranyi W, Lucin P, Burgert H G, Hengel H, Koszinowski U H. A cytomegalovirus glycoprotein re-routes MHC class I complexes to lysosomes for degradation. EMBO J. 1999;18:1081–1091. doi: 10.1093/emboj/18.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reusser P, Riddell S R, Meyers J D, Greenberg P D. Cytotoxic T-lymphocyte response to cytomegalovirus after human allogeneic bone marrow transplantation: pattern of recovery and correlation with cytomegalovirus infection and disease. Blood. 1991;78:1373–1380. [PubMed] [Google Scholar]
- 57.Riddell S R. Pathogenesis of cytomegalovirus pneumonia in immunocompromised hosts. Semin Respir Infect. 1995;10:199–208. [PubMed] [Google Scholar]
- 58.Riddell S R, Watanabe K S, Goodrich J M, Li C R, Agha M E, Greenberg P D. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science. 1992;257:238–241. doi: 10.1126/science.1352912. [DOI] [PubMed] [Google Scholar]
- 59.Rötzschke O, Falk K, Deres K, Schild H, Norda M, Metzger J, Jung G, Rammensee H-G. Isolation and analysis of naturally processed viral peptides as recognized by cytotoxic T cells. Nature. 1990;348:252–254. doi: 10.1038/348252a0. [DOI] [PubMed] [Google Scholar]
- 60.Schickedanz J, Philipson L, Ansorge W, Pepperkok R, Klein R, Koszinowski U H. The 89,000-Mr murine cytomegalovirus immediate-early protein stimulates c-fos expression and cellular DNA synthesis. J Virol. 1988;62:3341–3347. doi: 10.1128/jvi.62.9.3341-3347.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steffens H-P, Kurz S, Holtappels R, Reddehase M J. Preemptive CD8 T-cell immunotherapy of acute cytomegalovirus infection prevents lethal disease, limits the burden of latent viral genome, and reduces the risk of virus recurrence. J Virol. 1998;72:1797–1804. doi: 10.1128/jvi.72.3.1797-1804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taguchi T, McGhee J R, Coffman R L, Beagley K W, Eldridge J H, Takatsu K, Kyono H. Detection of individual mouse splenic T cells producing IFN-γ and IL-5 using the enzyme-linked immunospot (ELISPOT) assay. J Immunol Methods. 1990;128:65–73. doi: 10.1016/0022-1759(90)90464-7. [DOI] [PubMed] [Google Scholar]
- 63.Thäle R, Szepan U, Hengel H, Geginat G, Lucin P, Koszinowski U H. Identification of the mouse cytomegalovirus genomic region affecting major histocompatibility complex class I molecule transport. J Virol. 1995;69:6098–6105. doi: 10.1128/jvi.69.10.6098-6105.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei M L, Cresswell P. HLA-A2 molecules in an antigen-processing mutant cell contain signal sequence-derived peptides. Nature. 1992;256:443–446. doi: 10.1038/356443a0. [DOI] [PubMed] [Google Scholar]
- 65.Wills M R, Carmichael A J, Mynard K, Jin X, Weekes M P, Plachter B, Sissons J G P. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J Virol. 1996;70:7569–7579. doi: 10.1128/jvi.70.11.7569-7579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winston D J, Ho W G, Champlin R E. Cytomegalovirus infections after bone marrow transplantation. Rev Infect Dis. 1990;12:S776–S792. doi: 10.1093/clinids/12.supplement_7.s776. [DOI] [PubMed] [Google Scholar]
- 67.Ziegler H, Thäle R, Lucin P, Muranyi W, Flohr T, Hengel H, Farrell H, Rawlinson W, Koszinowski U H. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity. 1997;6:57–66. doi: 10.1016/s1074-7613(00)80242-3. [DOI] [PubMed] [Google Scholar]