Abstract
For determining cellular functions of the interferon-inducible human cytoplasmic protein P56, we undertook a Saccharomyces cerevisiae two-hybrid screen that identified Int6 as a P56-interacting protein. That the interaction also occurs in human cells was confirmed by coimmunoprecipitation and the observed cytoplasmic displacement of nuclear Int6 upon coexpression of P56. Because Int6 has been claimed to be both a cytoplasmic and a nuclear protein, we investigated the structural basis of this discrepancy. By mutational analyses, we showed that the Int6 protein contains a bipartite nuclear localization signal and a nuclear export signal at the far end of the amino terminus. The 20 amino-terminal residues of Int6, when they were attached to a different nuclear protein, were sufficient to translocate that protein to the cytoplasm. Within this region, replacement of any of the three leucine residues with alanine destroyed the function of the export signal. The specific domain of P56 that is required for its interaction with Int6 was mapped using the yeast two-hybrid assay and a mammalian coimmunoprecipitation assay. Both assays demonstrated that the C-terminal region of P56 containing three specific tetratricopeptide motifs is required for this interaction. In contrast, removal of an internal domain of P56 enhanced the interaction, as quantified by the two-hybrid assay.
The interferon (IFN) system is the first line of cellular defense against virus infection (24). In addition to their antiviral effects, however, IFNs have many other effects on cell physiology (25). The plethora of cellular effects of IFNs is thought to be mediated by the numerous cellular proteins whose synthesis is induced at the transcriptional level upon IFN treatment of cells (24). However, the specific biochemical and cellular functions of only a few of those proteins have as yet been determined, the most notable among them being PKR and 2-5 (A) synthetases (25). The study presented here deals with the function of another IFN-inducible protein, P56.
Gene 561, which encodes P56, is one of the first IFN-inducible genes cloned (5, 15). The transcriptional regulation of 561 mRNA has been extensively studied in our laboratory (3). The mRNA is undetectable in cells but it is rapidly induced upon IFN treatment and reaches very high levels. Indeed, in a recent gene array analysis, it scored as the most abundant IFN-induced mRNA among more than a hundred such mRNAs (6). The 561 mRNA is induced in cells not only by IFN treatment but also by double-stranded RNA (dsRNA) or virus infection (26). The transcriptional signaling used by dsRNA to induce 561 mRNA is independent and distinct from the JAK-STAT pathway used by IFNs (4). Because the cellular level of P56 is upregulated so dramatically upon treatment of cells with IFNs or dsRNA, we speculated that it has important cellular functions. Analysis of the primary structure of this protein did not reveal any functional motifs other than the eight tetratricopeptide (TPR) motifs, spaced evenly along the entire protein. TPR motifs have loose sequence identities, but they are known to mediate protein-protein interaction (12). For that reason, we searched for P56-interacting proteins using P56 as a bait to screen a HeLa cell cDNA library by a Saccharomyces cerevisiae two-hybrid transcriptional activation assay. This screen identified several proteins that may potentially interact with P56 in mammalian cells.
One potential P56-interacting protein was identified as Int6. Int6 was discovered as the product of a gene whose disruption by the integration of a mouse mammary tumor virus genome causes mammary carcinoma in mice (18). Appropriate expression of human Int6 is claimed to be affected in many human breast tumors as well (19). These observations suggest an important cell growth regulatory activity of Int6. The human Int6 protein is identical to the mouse Int6 protein, and it was shown to interact with the human T-cell leukemia virus oncoprotein Tax (7). An unexpected connection of Int6 to protein synthesis was made by the observation that the P48 subunit of the translation initiation factor eIF-3 is identical to Int6 (1). There have been conflicting reports regarding the subcellular location of the Int6 protein. It was claimed to be a nuclear protein by one group (7) and a cytoplasmic protein by others (8, 20).
Recently, we have shown that the interaction of the P56 protein with the cytoplasmic P48/Int6 subunit of eIF-3 causes inhibition of translation (13). To investigate the nature of the interaction further, in this study, we mapped the Int6-interacting domain of P56. Moreover, we provide experimental evidence to demonstrate that Int6 contains a bipartite nuclear localization signal and a nuclear export signal (NES) located at the far end of the amino terminus. Thus, Int6 can have both nuclear and cytoplasmic isoforms, and although P56 is a cytoplasmic protein, it can interact with the nuclear form of Int6 as well.
MATERIALS AND METHODS
Antibody.
The rabbit polyclonal antibody that recognizes P56 was raised by injection of a rabbit with purified bacterially expressed P56 (13a).
Construction of P56 clones.
The full-length P56 cDNA was constructed by PCR using an existing partial clone (our unpublished data) (Table 1). The cDNA sequence was inserted into pBluescript KS (II). pCMV-P56 was constructed by excising full-length P56 cDNA from pBluescript KS (II) and inserting it into pCB6+, a eukaryotic expression vector (23).
TABLE 1.
PCR primers for generating wt P56, mutant P56, Int6 constructs, and wt NES-DRBP76-Flag and mutant NES-DRBP76-Flag constructs
| Protein(s) | Terminus | Sequence |
|---|---|---|
| P56 | 5′ | 5′-GAAGGCCTCATATGAGTACAAATGGTGATGATC-3′ |
| P56, Δ2-8 | 3′ | 5′-CGAATAATGGATCCTCTGGGTGCCTAAGGACCTTGTC-3′ |
| Δ2-8 | 5′ | 5′-GAATGACTCATATGGCTGGGTATGCGATCTCTGC-3′ |
| Δ1-2 | 5′ | 5′-GAATGACTCATATGAGTACAAATGGTGATGATC-3′ |
| Δ1-2 | 3′ | 5′-CGATATATGGATCCTCTGGGTGCCTAAGGACCTTGTC-3′ |
| Δ3-5 P1, Δ6-8 | 5′ | 5′-ACAGCCCGGGGATCCGAACCATGGGCATGAGTACAAATGGTGATGATC-3′ |
| Δ3-5 P2 | 5′-GGTGATGCAGTAAGACGGATTCAGGTTTTCAGG-3′ | |
| Δ3-5 P3 | 5′-AACCCTGAATCCGTCTTACTGCATCACCAGATAG-3′ | |
| Δ3-5 P4 | 5′-ACAGGATATCGTCGACCTAAGGACCTTGTCTCACAGAG-3′ | |
| Δ6-8 | 3′ | 5′-ACAGGATATCGTCGACCTAAAATGTGGGCTTTTTTTCCACTGC-3′ |
| Int6NES | 5′ | 5′-GGCCGATATCGGTACCAAGATGGCGGAGTACGACTTGAC-3′ |
| Int6NES | 3′ | 5′-GCGAATTCCCCGGAAAGACTAGATGCCGATC-3′ |
| L6AInt6NES | 5′ | 5′-ATGGTACCAAGATGGCGGAGTACGACGCAACTACTCGCATC-3′ |
| L14AInt6NES | 3′ | 5′-GCGAATTCCCCGGAAAGACTAGATGCCGATCTGCAAAGTGC-3′ |
| L18AInt6NES | 3′ | 5′-GCGAATTCCCCGGAAAGACTGCATGCCGATC-3′ |
| Int6B | 5′ | 5′-GTCAGGATCCGCGAATTCCCACCATGGACTACAAGG-3′ |
| Int6B | 3′ | 5′-GTCAGATATCGGTTCTTCATGTTATGACTGCTGTAGTC-3′ |
| Int6D | 5′ | 5′-ATGAATTCATGGCGGAGTACGACTTGACTACTCGCATCGCGCACTTTTTGG-3′ |
| Int6D, Int6E | 3′ | 5′-TCGGATCCTCACTTGTCATCGTCGTCCTTGTAGTCGTAGAAGCCAGAATC-3′ |
| Int6E | 5′ | 5′-ATGAATTCGAGATATACCATGATCGCGCACTTTTTGGAT-3′ |
Transfection.
HT1080 cells were seeded into a 100-mm-diameter plate. The next day cells were ∼60% confluent and transfection was performed using the Fugene 6 transfection method (Boehringer Mannheim). For transfection in a 100-mm-diameter plate, 16 μg of a single plasmid or 8 μg of each of two plasmids was used. Twenty-four hours after transfection, cells were harvested and whole-cell extracts were prepared as described previously (16).
Immunoprecipitation and Western blotting.
Immunoprecipitation of Flag-tagged protein was done in Tow-concentration-salt buffer (20 mM Tris [pH 7.5], 50 mM KCl, 200 mM NaCl, 1 mM EDTA, 20% glycerol, 0.05% Triton X-100, 0.2 mM phenylmethylsulfonyl fluoride). The M2 anti-Flag Sepharose beads (Kodak Scientific Imaging System) were presoaked with 3 μg of bovine serum albumin for 15 min. Whole-cell extracts containing equal amounts of protein were mixed with 500 ml of low-salt buffer and 20 μl of preincubated anti-Flag Sepharose beads at 4°C for 2 h. The immunoprecipitates were washed four times with the low-salt buffer and subjected to denaturing polyacrylamide gel electrophoresis. Western blotting as described before (16) was done with a 1:2,000 dilution of anti-P56 antibody.
Immunofluorescence.
Immunofluorescence was performed as described previously (16) with the following modifications. HT1080 cells were plated on coverslips in six-well plates the night before transfection. At the time transfection was performed, the cells were 50 to 60% confluent. A single plasmid (1.6 μg) or two plasmids (0.8 μg each) were transfected into HT1080 cells on coverslips using the Fugene 6 transfection method (Boehringer Mannheim). Twenty-four hours after transfection, cells were fixed and incubated with a 1:2,000 dilution of anti-P56 antibody and a 1:2,000 dilution of anti-Flag M2 antibody (Kodak Scientific Imaging Systems) to detect P56 and Flag-tagged proteins, respectively. Antibody binding was detected with fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit antibody (GIBCO) and Texas Red-conjugated goat anti-mouse antibody (Molecular Probes). The coverslips were finally washed, mounted, and examined. Filters to detect FITC or Texas Red were used for labeling of different antibodies and standard optics was used for phase-contrast microscopy. If IFN treatment was needed, 12 h after transfection, cells were treated with 1,000 U of IFN-β per ml for 16 h. Then the cells were washed, fixed, and labeled the same way as described above.
Construction of Int6 clones.
Int6A was a gift of P. Jalinot. It was referred to as pSGF-Int6 by Desbois et al. (7). Int6B was generated by PCR using Int6A as a template (Table 1) and subcloned into pSG5 (Promega). Int6B lacks amino acids (aa) 264 to 445 of the Int6 sequence, which includes the bipartite nuclear localization signal. Int6C was constructed by digesting Int6A with EcoRI and BglII. The released fragment was inserted into pSG5 EcoRI and BglII sites. Int6C contains aa 1 to 276 of the Int6 sequence. Int6D contains the full-length Int6 coding sequence with a Flag tag at the C terminus and was subcloned into pCB6+. Full-length Int6-Flag was generated by PCR using Int6A as a template. The 5′ PCR primer encoded 14 residues, including residues 2 to 9 of the Int6 coding sequence, which were missing in Int6A, eliminating the Flag and the 13 extraneous residues present at the amino terminus of Int6A, and the 3′ PCR primer included the Flag tag sequence (Table 1). Int6E has the Int6 coding sequence but lacks aa 2 to 9 (8 aa) and has the Flag tag at the C terminus. Int6E was generated by PCR using Int6D as a template and subcloned into pCB6+. The 5′ PCR primer has the first methionine of the Int6 coding sequence and then connects to the 10th amino acid of the Int6 coding sequence (Table 1).
Construction of NES-DRBP76-Flag or mutant NES-DRBP76-Flag clones.
DRBP76-Flag in pcDNA3 (Invitrogen) was constructed as described previously (22). The NES of Int6 was the 1 to 20 aa of Int6 that were generated by PCR (Table 1). The PCR product of NES was fused in frame to the N terminus of DRBP76-Flag in pcDNA3 to produce NES-DRBP76-Flag. L6ANES was generated by PCR, and the mutation of leucine at position 6 to alanine (L6A) was generated in the 5′ PCR primer (Table 1). L14ANES and L18ANES were generated by PCR, and the L14A and L18A mutations were generated in the 3′ PCR primer (Table 1). Then L6ANES, L14ANES, and L18ANES PCR products were fused in frame to the N terminus of DRBP76-Flag in pcDNA3 to produce L6ANES-CRBP76-Flag, L14ANES-DRBP76-Flag, and L18ANES-DRBP76-Flag, respectively.
Construction of P56 deletion mutant proteins.
Mutant protein Δ2-8 containing residues 1 to 95 of P56 was generated by PCR (Table 1). The PCR product was subcloned in frame into pGBT9 (Clontech) to generate BD-Δ2-8. Δ1-2 containing residues 179 to 478 of P56 was generated by PCR (Table 1). The PCR product was subcloned in frame into pGBT9 to produce BD-Δ1-2. pCMV-Δ1-2 was generated by subcloning the Δ1-2 PCR product into pcDNA3. Δ3-5 has a deletion of nucleotides 534 to 855 of P56 cDNA and was produced by using oligonucleotide-directed mutagenesis as described below. Then the PCR product was subcloned into pGBT9 to generate BD-Δ3-5 or into pcDNA3 to produce pCMV-Δ3-5. Δ6-8 contains aa 1 to 339 of P56 and was generated by PCR (Table 1). The Δ6-8 PCR product was subcloned into pGBT9 to produce BD-Δ6-8 or into pcDNA3 to produce pCMV-Δ6-8.
Oligonucleotide-directed mutagenesis.
An overlap extension PCR method (17) was used to introduce the deletion of nucleotides 534 to 855 (TPRs 3 to 5) of P56 cDNA. Briefly, two separate PCRs were performed to amplify both halves of a complete gene, using four primers (Table 1). An outside-forward primer (P1) was paired with a middle-reverse primer (P2) to synthesize the first half of the gene; an outside-reverse primer (P4) was paired with a middle-forward primer (P3) to synthesize the second half. The deletion mutation was introduced by the middle two primers (P2 and P3). Both primers contained nucleotides 520 to 534 and 856 to 870 of P56 cDNA and looped out nucleotides 534 to 855. Then the two PCR products, which were overlapping, were gel purified and put into the third PCR with the two outside primers (P1 and P4) to produce Δ3-5, the full-length P56 cDNA with the deletion of nucleotides 534 to 855.
Yeast two-hybrid assay.
The yeast two-hybrid system (9) was used to assay for in vivo protein-protein interaction. The yeast strain used was Y190 (Clontech). Combinations of DNA activation domain (AD) and DNA binding domain (BD) plasmids described in the legend to Fig. 5 were cotransfected into Y190 using the lithium acetate method as described in the Clontech yeast two-hybrid system manual. All colonies which contained expression constructs were tested for respective fusion protein expression by immunoblotting as described below. In order to measure the apparent strengths of protein-protein interactions, a dual-reporter system of the histidine (His) and β-galactosidase (β-Gal) contained in yeast strain Y190 was used. The physical interaction of AD and BD plasmids caused the expression of both His and β-Gal reporter genes. Transfectants were plated onto synthetic defined (SD) medium lacking tryptophan and leucine but containing histidine (+His medium) or lacking histidine (−His medium) in the presence of 25 mM 3-aminotriazole and incubated at 30°C. Seven days after incubation, plates containing −His medium were scored for growth (data not shown). Quantitative interaction was measured by a liquid β-Gal assay. Colonies containing both indicated AD and BD plasmids were picked from plates containing +His medium and grown in 5 ml of SD +His medium at 30°C overnight. The next day, liquid culture was added to 5 ml of YPD medium (the standard medium for growing the yeast strain [20 g of Difco peptone per liter, 10 g of yeast extract per liter, 2% glucose]) at a final concentration of 0.2 optical density at 600 nm (OD600) equivalent and incubation was continued at 30°C with shaking until the OD600 reached 0.8. The yeast cells were spun down and washed once with Tris-HCl, pH 7.5. Then cells were resuspended in lysis buffer provided in a β-Gal detection kit (Tripix) and frozen and thawed three times at −80°C. Cell extracts were used for the β-Gal assay with the detection kit (Tripix) and normalized with equal cell numbers.
FIG. 5.
Interactions between P56 deletion mutant proteins and Int6 in human cells. pCMV-P56, pCMV-Δ1-2, pCMV-Δ3-5, or pCMV-Δ6-8 was cotransfected with Int6D into HT1080 cells. Twenty-four hours later, cells were harvested. (A) Western blotting (W.B.) was performed to show that P56 and all the deletion mutant proteins were expressed in cells. (B) Immunoprecipitation (IP) was done using Flag M2 antibody and followed by Western blotting with P56 antibody to show the interaction between P56 or P56 deletion mutant proteins and Int6.
Immunoblot assay to detect the AD and BD fusion proteins in yeast.
An immunoblot assay using anti-P56 antibody was performed to verify the expression of each of the BD-P56 and BD-P56 deletion mutant proteins in yeast. Yeast colonies containing each BD plasmid were grown in SD +His medium overnight. Then the overnight culture was transferred to 5 ml of YPD medium at a final OD600 of 0.2 and incubated at 30°C until the cultures reached a final OD600 of 0.8. Cells were harvested by centrifugation and washed once with water. Then cell pellets were resuspended in an equal volume of chilled 2× sodium dodecyl sulfate sample buffer and acid-washed glass beads (Sigma) in the presence of 0.2 mM phenylmethlsulfonyl fluoride. Cells were lysed by vortexing them three times for 30 s each time and chilled on ice between each burst. Equal volumes of extracts were subjected to immunoblot analysis using a 1:2,000 dilution of anti-P56 antibody.
RESULTS
Interaction of nuclear Int6 with P56.
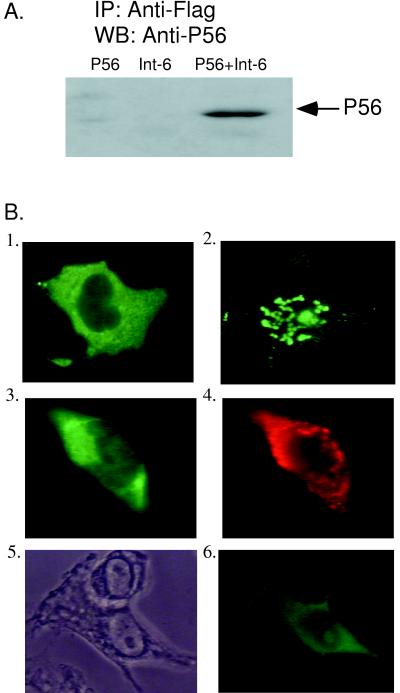
We initially identified Int6 as a P56-binding protein by a yeast two-hybrid screen of a HeLa cell cDNA library. Because the Int6 cDNA isolated by this screen encoded only a partial segment of the Int6 protein, we obtained a complete human Int6 cDNA from Desbois et al. (7). The latter clone was tagged with a Flag epitope, which helped us to monitor the protein because a high-quality antibody to the protein itself was not available. We used expression vectors for Flag Int6 and P56 and antibodies to Flag and P56 for examining interactions between the two proteins in human cells. Human HT1080 cells were transfected with the two expression vectors individually or in combination. From the cell extracts, proteins were immunoprecipitated with anti-Flag antibody, and the immunoprecipitates were subjected to Western blot analysis using P56 antiserum. As shown in Fig. 1A, when both proteins were expressed, P56 was coimmunoprecipitated with Int6. However, as expected, when they were individually expressed, no signal was obtained because of the absence of the other protein in the cell. The interaction between P56 and Int6 in human cells was confirmed further by the assays in which we monitored the translocation of nuclear Int6 to the cytoplasm by interaction with P56 using immunofluorescence as the detection method (Fig. 1B). IFN-β-induced P56 was cytoplasmic (Fig. 1B, image 1), whereas Int6 was located in the punctuated promyelocytic leukemia (PML) bodies in the nucleus (image 2) as reported by Desbois et al. (7). When the two proteins were coexpressed by transfection, both were in the cytoplasm (images 3 and 4). Instead of P56 being expressed by transfection, when endogenous P56 was induced by IFN treatment, it also translocated nuclear Int6 to the cytoplasm (images 5 and 6). Although the subcellular locations of the proteins in single cells are shown in different images of this panel, similar distributions were observed in many cells. The results shown in Fig. 1 demonstrated that P56 and nuclear Int6 interact in vivo, causing a redistribution of the Int6 protein from the nucleus to the cytoplasm.
FIG. 1.
Interaction of P56 and Int6. (A) pCMV-P56 (first lane), Int6A (second lane), or both (third lane) were transfected into HT1080 cells. Twenty-four hours posttransfection, cells were fixed and immunoprecipitation (IP) was performed using Flag M2 antibody, followed by Western blotting (WB) with P56 antibody to detect the interaction between P56 and Int6A. (B) (Image 1) HT1080 cells were treated with IFN-β for 14 h to induce endogenous P56. Cells were fixed, and immunofluorescence was performed with P56 antibody to show the localization of P56. (Image 2) Int6A was transfected into HT1080 cells. Twenty-four hours posttransfection, cells were fixed and immunofluorescence was done with a Flag probe (D-8) to detect the subcellular location of Int6A. (Images 3 and 4) HT1080 cells were transfected with pCMV-P56 and Int6A. Twenty-four hours posttransfection, cells were fixed and a double-immunofluorescence assay was performed using both P56 and Flag M2 antibody. The subcellular locations of P56 (image 3) and Int6 (image 4) in the same cell are shown. (Images 5 and 6) HT1080 cells were transfected with Int6A, and 12 h posttransfection, cells were treated with 1,000 U of IFN-β per ml for 14 h. Then cells were fixed and immunofluorescence was performed using the Flag probe (D-8) to detect the subcellular location of Int6A. Phase-contrast microscopy and (image 5) immunofluorescence (image 6) are shown.
Structural basis of the subcellular location of Int6.
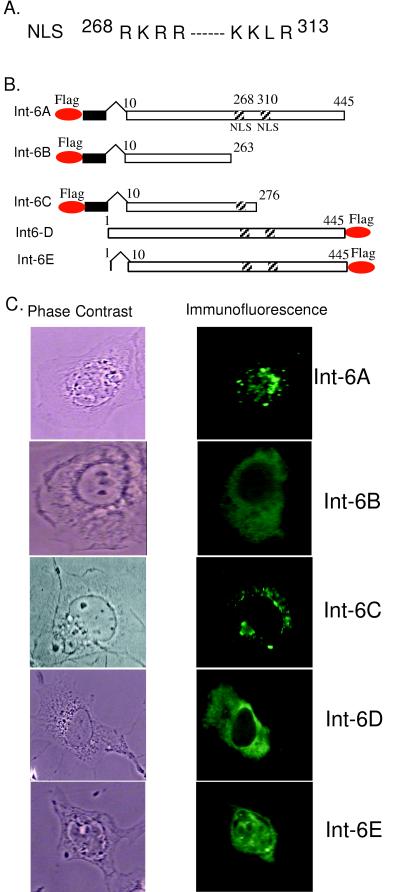
As shown in Fig. 1, the Int6 clone obtained from Desbois et al. (7) (Int6A) encoded a protein that was nuclear. Others have reported, however, that the Int6 protein is cytoplasmic. To resolve this apparent discrepancy, we analyzed the primary structure of the protein for the presence of apparent structural motifs which may determine its subcellular location. In Int6A there are two stretches of basic residues starting at positions 268 and 310 (Fig. 2A) that may constitute a bipartite nuclear localization signal. Indeed, when both motifs (Int6B) or only one of them (Int6C) was deleted, the proteins became cytoplasmic (Fig. 2B and C), indicating that we have identified a functional nuclear localization signal in the Int6 protein.
FIG. 2.
(A) Bipartite nuclear localization signal (NLS) of Int6. (B) Maps of different Int6 constructs. Oval circles represent the Flag epitope. Black rectangles represent an unrelated sequence of 13 residues. Hatched squares represent the nuclear localization signals. (C) Subcellular location of different Int6 proteins in cells. HT1080 cells was transfected with Int6A, Int6B, Int6C, Int6D, or Int6E. Twenty-four hours posttransfection, immunofluorescence was performed using the Flag probe (D-8) antibody to detect different Int6 proteins. Phase-contrast (left panel) and immunofluorescence (right panel) images are shown.
Sequencing of the Int6A cDNA revealed that the encoded Int6 protein lacked the first nine natural residues. Instead, it contained 13 extraneous residues at the N terminus preceded by the Flag tag. To determine whether any of the above-described changes at the N terminus of the protein influence its subcellular location, we constructed a new clone (Int6D) (Fig. 2B) that encoded the full-length Int6 with its natural N terminus but a Flag tag at the C terminus. To our surprise, Int6D was found to be cytoplasmic (Fig. 2C). Int6D showed that the missing nine residues of Int6A, and not the location of the Flag tag, were the determining factors. Unlike Int6D, Int6E was nuclear (Fig. 2C), demonstrating that residues 2 through 8 were required for the cytoplasmic location of Int6. Note that the nuclear staining of Int6E was much more uniform than that of Int6A, indicating that the localization of the Int6A protein to the PML bodies was probably determined by the extraneous 13 residues encoded by the vector sequences present between the Flag tag and the body of the protein.
Characterization of the NES of Int6.
The above results suggest the presence of an NES at the far end of the N terminus of the Int6 protein. For delineating this putative signal further, we compared the sequence of the first 20 residues of Int6 to those of several well-characterized NES motifs (Table 2). Although the primary sequences of these motifs are not conserved, all contain critical leucine residues that are required for NES function. Int6 has three such conserved Leu residues at positions 6, 14, and 18 (Table 2). At position 10, Ile replaces Leu in Int6. Thus, it appeared that the first 20 residues of Int6 might constitute a functional NES.
TABLE 2.
NES of Int6a
| Protein | NES sequence |
|---|---|
| Int6 | 6L T T R I A H F L D R H L V F20 |
| Rex | 82L S A Q L Y S S L S – – L D S94 |
| PKI | 37L A – – L K – – L A G – L D I46 |
| Rev | 75L P P – L E R – L T – – L D84 |
| Ahr | 64L D K – L S V – L R – – L S73 |
The NES sequence of Int6 is compared with the known NES sequences of human T-cell leukemia virus type I Rex (21), heat-stable protein kinase inhibitor (PKI) (28), human immunodeficiency virus type 1 Rev (10), and the human aryl hydrocarbon receptor (Ahr) (14). The leucines that are critical for the function of NES are underlined. The numbers at the beginning and the end show the positions of the NES in the respective protein sequences.
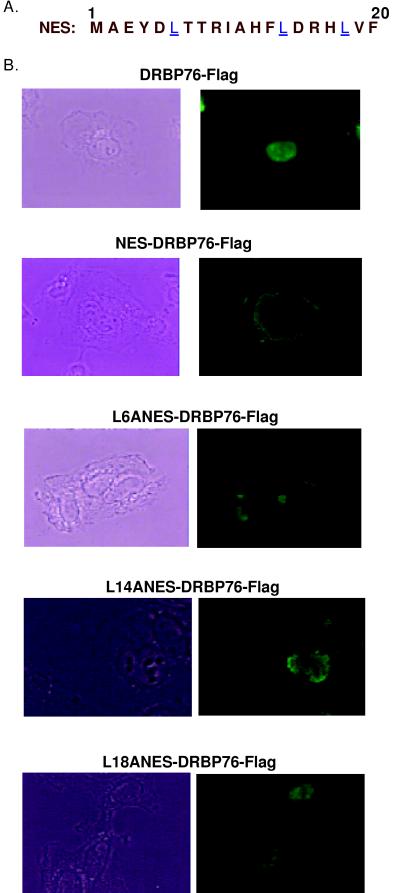
The putative NES of Int6 was functionally tested in the experiments shown in Fig. 3. We have recently cloned a human nuclear protein, DRBP76 (22). When the putative NES of Int6 (Fig. 3A) was attached to the N terminus of DRBP76, it was translocated to the cytoplasm (Fig. 3B), demonstrating that the first 20 residues of Int6 constitute a functional NES that is sufficient for the cytoplasmic export of an authentic nuclear protein. The essential residues required for the appropriate functioning of the NES were identified by mutations of the Leu residues at positions 6, 14, and 18. When any of those three Leu residues was replaced by Ala, the NES became nonfunctional and the NES-DRBP76 fusion protein was retained in the nucleus (Fig. 3B). Thus, the conserved Leu residues of Int6 NES were absolutely required for its function.
FIG. 3.
NES of Int6. (A) The NES sequence of Int6, containing amino acids 1 to 20 of Int6, is shown. The leucines which are critical for the function of the NES are underlined. (B) The wt NES of Int6 was fused in frame to the N terminus of the Flag-tagged nuclear protein DRBP76 to produce NES-DRBP76-Flag and DRBP76-Flag or NES-DRBP76-Flag was transfected into HT1080 cells. Twenty-four hours posttransfection cells were fixed and immunofluorescence was performed using Flag antibody to detect the subcellular location of DRBP76-Flag or NES-DRBP76-Flag fusion proteins. Phase-contrast (left side) and immunofluorescence (right side) images are shown. The leucine at position 6, 14, or 20 of the NES of Int6 was mutated singly to alanine, and the mutated NES was fused in frame to DRBP76-Flag to produce L6ANES-DRBP76-Flag, L14ANES-DRBP76-Flag, or L18ANES-DRBP76-Flag. Then the three mutant proteins were expressed singly in HT1080 cells in the same way as described above. Immunofluorescence was performed using Flag antibody to detect the subcellular location of different mutant proteins.
Mapping of the Int6-interacting domain of P56.
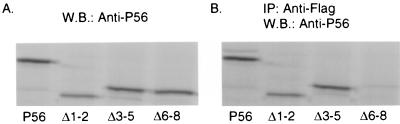
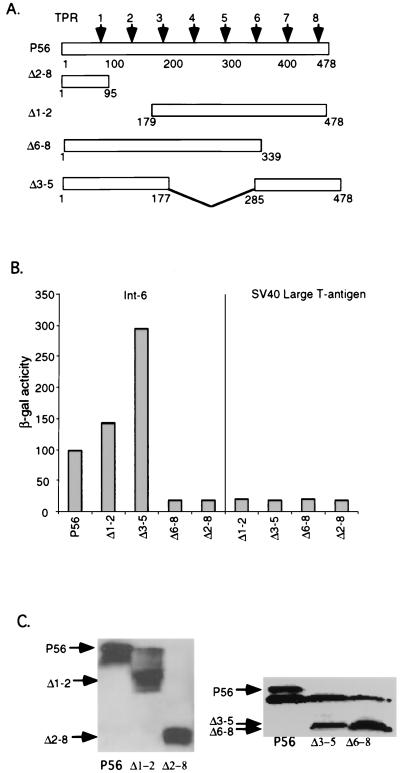
In the next series of experiments, we mapped the domain of P56 that is required for its interaction with Int6. In the experiments shown in Fig. 4, we monitored the interaction, in yeast, of wild-type (wt) P56 or deletion mutant of P56 with clone 6, which encodes a fusion protein of the Gal4 activation domain and the C-terminal region of Int6. The extent of interaction was quantitated by measuring the level of expression of the β-Gal gene, whose transcription is driven by the interacting fusion proteins. The deletion mutants were designed to delete two or more TPR motifs from the P56 protein (Fig. 4A). The wt protein and its four mutants were expressed in yeast approximately to similar levels (Fig. 4C), and none of these proteins interacted with the simian virus large-T antigen (Fig. 4B). As expected, wt P56 interacted strongly with Int6, but Δ6-8 and Δ2-8, which lack TPR motifs 6 to 8 and 2 to 8, respectively, did not interact at all. On the other hand, the Δ1-2 mutant protein lacking TPR 1 and 2 interacted slightly better than the wt protein, and the Δ3-5 mutant protein lacking TPR motifs 3 to 5 was more than three times better than the wt protein in driving β-Gal expression.
FIG. 4.
Mapping of the domain of P56 interacting with Int6. (A) Maps of full-length P56 and P56 deletion mutants. The arrows represent eight TPR motifs. (B) Interaction of P56 or its deletion mutant with Int6 in yeast. BD-P56, BD-Δ1-2, BD-Δ3-5, BD-Δ6-8, or BD-Δ2-8 was cotransfected with AD-Int6 or AD-simian virus 40 large-T antigen into yeast strain Y190. Transfectants were plated on selection plates. A liquid β-Gal assay as described in Materials and Methods was performed to quantify the interaction strength of Int6 with P56 or its deletion mutant proteins. The β-Gal activity is presented in arbitrary units, and the values represent the means of results of two independent experiments. (C) Immunoblot assays of the yeast extracts used to obtain the data in panel B were performed to detect the BD fusion proteins in yeast. The arrows on the left show the positions of different P56 proteins.
The above pattern of interaction in yeast between P56 and Int6 was confirmed in human cells. Full-length Int6 and P56 or mutants of the latter were coexpressed in HT1080 cells, and coimmunoprecipitation of the two proteins was monitored. wt P56 and three mutant P56 proteins were expressed equally well (Fig. 5A), but the Δ6-8 mutant protein, and not the other two mutant proteins, failed to interact with Int6 (Fig. 5B). The results shown in Fig. 4 and 5 demonstrated that the C-terminal region of P56, containing TPR motifs 6 to 8, is required for its interaction with Int6. Removal of the three internal TPR motifs, 3 to 5, on the other hand, enhanced the interaction between the two proteins.
DISCUSSION
Int6 was discovered to be the protein encoded by a cellular genetic locus through which mouse mammary tumor virus was integrated in several mammary tumors (18). Evidence was presented that these integrations caused disruption of the gene and the production of truncated Int6 mRNAs and proteins. The human Int6 protein, which has a sequence identical to the mouse Int6 sequence, was first identified as a protein that interacts with the Tax oncoprotein of human T-cell leukemia virus (7). In that study, the researchers observed that the human Int6 protein is nuclear and localized in the PML bodies. In contrast, Diella et al. (8) claimed that the mouse protein is cytoplasmic. This claim received strong support from the unexpected observation that Int6 is identical to the P48 subunit of the translation initiation factor elF-3 (1), which predominantly, if not exclusively, resides in the cytoplasm. The results reported here strongly indicate that the protein can exist in both compartments of the cell. Like many eukaryotic shuttle proteins, it has an authentic nuclear localization signal and an NES (Fig. 6A). Desbois et al. (7) concluded that the protein is nuclear because their cDNA clone (Int6A) was missing a few residues at the N terminus of Int6, where, fortuitously, the NES resides. Our experiments conclusively demonstrated that it is a functional NES. Although in other proteins, the NES is located internally and not at the N terminus, the Int6 NES shared their property of requiring the presence of Leu residues for its function. Our results explained why Int6A is nuclear, but they also demonstrated that its observed localization in PML bodies is probably not physiological. Without the extraneous sequence, the nuclear Int6, Int6E (Fig. 2), did not give a speckled staining pattern.
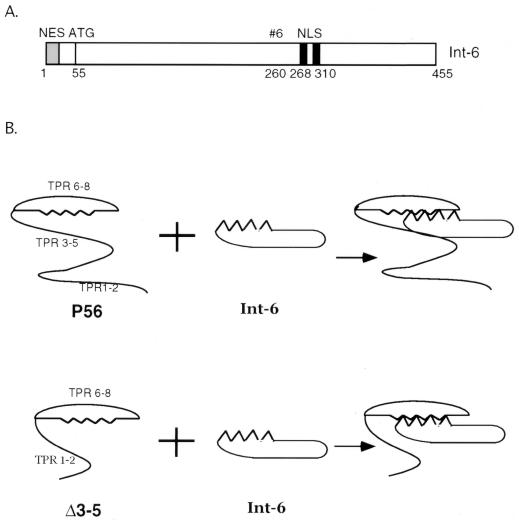
FIG. 6.
Model for interaction between P56 and Int6. (A) The full-length Int6 protein is shown. The N-terminal NES is shown by the gray box. The black boxes represent the bipartite nuclear localization signal (NLS). The internal ATG at amino acid 55 is shown. #6 represents the start point of the yeast clone which contains aa 260 to 455 of Int6 and was identified by the yeast two-hybrid assay in interactions of Int6 with P56. (B) Model for interaction between P56 and Int6. The upper panel shows that in wt P56, TPR motifs 6 to 8 form a scaffold that interacts with Int6. However, the presence of TPR motifs 3 to 5 makes it difficult for Int6 to access the scaffold. In the lower panel, deletion of TPR motifs 3 to 5 opens up the scaffold and Int6 can freely get in touch with TPR motifs 6 to 8 of P56. As a result, the interaction between Int6 and Δ3-5 is stronger than that between Int6 and wt P56.
Being a shuttle protein, the full-length Int6 is capable of distributing itself to both the nuclear and the cytoplasmic compartment of the cell, but does an exclusively nuclear isoform of Int6 exist in cells? In principle, such an isoform will arise if the Int6 protein lacks a functional NES. Inspection of the Int6 cDNA sequence revealed that the sequence beginning at position 55, an internal methionine, is in an excellent Kozak sequence (Fig. 6A). If translation starts at that methionine, the product will lack the NES and be exclusively nuclear. Although Diella et al. (8) observed such a protein as an in vitro translation product, the existence of such a truncated protein in cells awaits further investigation. Similarly, future investigation will reveal the complete cellular functions of Int6. One important function as a subunit of elF-3 has already been defined, but are there additional functions, especially of the nuclear isoform? Could it have an independent effect on cell growth and thus does its dysfunction cause tumorigenesis? The fact that the protein is designed as a shuttle protein strongly suggests that it has a nuclear function.
Our interests in Int6 arose from the observation that it interacts with P56. We have shown that P56 binds to the P48 subunit of elF-3 and inhibits translation (13). Data presented here confirmed that P56 also binds to the nuclear isoform of Int6 and, surprisingly, that as a result, the nuclear Int6 gets relocated to the cytoplasm. From this observation, one can predict that the presence of P56 would affect the cellular functions of nuclear Int6 as well. We suspected that the P56 and Int6 interactions are mediated by specific TPR motifs of P56. The TPR motifs are 34-residue-long sequences that are quite degenerate, and they often appear in tandem arrays which form scaffolds for protein-protein interactions. Our data indicate that TPR motifs 6, 7, and 8 of P56 probably form such a scaffold and that it interacts with Int6 (Fig. 6B). It also appears that the middle portion of the protein encompassing motifs 3, 4, and 5 interferes with that interaction because removal of this region from P56 enhanced its interaction with Int6. Thus, potentially, TPR motifs 3 to 5 may be able to regulate the function of TPR motifs 6 to 8 in vivo. Such putative regulations might be exerted by the binding of other cellular proteins to TPR motifs 3 to 5 because specific TPRs mediating binding to specific proteins have been documented for other proteins. For example, P58 uses an internal TPR for its interaction with the protein kinase PKR, causing inhibition of its enzyme activity (11).
P56 is a member of a family of structurally related IFN-induced proteins which include P54 (27), P60 (2), and RIG-G (29). Like P56, the other members also contain TRP motifs. It will be of considerable interest to examine whether they also interact with Int6 or whether they are designed for interaction with other cellular proteins. Because untreated cells do not contain a detectable level of P56 and the protein is induced to a high level upon IFN treatment, we believe that it has major cellular regulatory effects. Results presented here indicate that such potential effects may be mediated by its interaction with different isoforms of Int6. Similar regulations may also be achieved in virus-infected cells because the protein is strongly induced by dsRNA in an IFN-independent manner (26). Many viruses such as Sendai virus, vesicular stomatitis virus, encephalomyocarditis virus, and cytomegalovirus can also induce P56, presumably through an intracellular dsRNA product (13a). The effects of P56 on viral gene expression in an infected cell remain to be determined. Finally, P56 may be used as a tool for analyzing the mechanism of Int6-mediated mammary carcinogenesis in mice. For example, if a block in the production of the Int6 protein, because of mouse mammary tumor virus insertion in its genome, is the primary cause of tumorigenesis, the functional equivalent can be achieved by transgenic expression of P56 in mammary epithelial cells. In contrast, if the production of a truncated Int6 is required for pathogenesis, neutralization of full-length Int6 by P56 will not have the same effect (18).
ACKNOWLEDGMENTS
We thank P. Jalinot for the pSGF-Int6 clone. We thank Judith Drazba for helping us with the microscope. We also thank Deborah Vestal, Rekha C. Patel, and Michael Molstad for helpful discussion.
This work was supported in part by the National Institutes of Health grants CA-68782 and CA-62220.
REFERENCES
- 1.Asano K, Merrick W C, Hershey J W. The translation initiation factor eIF3-p48 subunit is encoded by int-6, a site of frequent integration by the mouse mammary tumor virus genome. J Biol Chem. 1997;272:23477–23480. doi: 10.1074/jbc.272.38.23477. [DOI] [PubMed] [Google Scholar]
- 2.Baker E, de Veer M J, Devenish R J, Sutherland G R, Ralph S J. Interferon- and virus-inducible gene ISG-60. Map position 10q23.3. Chromosome Res. 1997;5:572. [PubMed] [Google Scholar]
- 3.Bandyopadhyay S K, Kalvakolanu D V, Sen G C. Gene induction by interferons: functional complementation between trans-acting factors induced by alpha interferon and gamma interferon. Mol Cell Biol. 1990;10:5055–5063. doi: 10.1128/mcb.10.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandyopadhyay S K, Leonard G T, Jr, Bandyopadhyay T, Stark G R, Sen G C. Transcriptional induction by double-stranded RNA is mediated by interferon-stimulated response elements without activation of interferon-stimulated gene factor 3. J Biol Chem. 1995;270:19624–19629. doi: 10.1074/jbc.270.33.19624. [DOI] [PubMed] [Google Scholar]
- 5.Chebath J, Merlin G, Metz R, Benech P, Revel M. Interferon-induced 56,000 Mr protein and its mRNA in human cells: molecular cloning and partial sequence of the cDNA. Nucleic Acids Res. 1983;11:1213–1226. doi: 10.1093/nar/11.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Der S D, Zhou A, Williams B R, Silverman R H. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desbois C, Rousset R, Bantignies F, Jalinot P. Exclusion of Int-6 from PML nuclear bodies by binding to the HTLV-I Tax oncoprotein. Science. 1996;273:951–953. doi: 10.1126/science.273.5277.951. [DOI] [PubMed] [Google Scholar]
- 8.Diella F, Levi G, Callahan R. Characterization of the INT6 mammary tumor gene product. DNA Cell Biol. 1997;16:839–847. doi: 10.1089/dna.1997.16.839. [DOI] [PubMed] [Google Scholar]
- 9.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 10.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 11.Gale M, Jr, Tan S L, Wambach M, Katze M G. Interaction of the interferon-induced PKR protein kinase with inhibitory proteins P58IPK and vaccinia virus K3L is mediated by unique domains: implications for kinase regulation. Mol Cell Biol. 1996;16:4172–4181. doi: 10.1128/mcb.16.8.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goebl M, Yanagida M. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991;16:173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- 13.Guo J, Sen G C. The interferon-induced protein, P56, binds to the P48 subunit of the translation initiation factor eIF-3 and inhibits translation. Eur Cytokine Netw. 1998;9:325. [Google Scholar]
- 13a.Guo, J., K. Peters, and G. C. Sen. Induction of the human protein P56 by interferon, double-stranded RNA or virus infection. Virology, in press. [DOI] [PubMed]
- 14.Ikuta T, Eguchi H, Tachibana T, Yoneda Y, Kawajiri K. Nuclear localization and export signals of the human aryl hydrocarbon receptor. J Biol Chem. 1998;273:2895–2904. doi: 10.1074/jbc.273.5.2895. [DOI] [PubMed] [Google Scholar]
- 15.Kusari J, Sen G C. Regulation of synthesis and turnover of an interferon-inducible mRNA. Mol Cell Biol. 1986;6:2062–2067. doi: 10.1128/mcb.6.6.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leonard G T, Sen G C. Effects of adenovirus E1A protein on interferon-signaling. Virology. 1996;224:25–33. doi: 10.1006/viro.1996.0503. [DOI] [PubMed] [Google Scholar]
- 17.Ling M M, Robinson B H. Approaches to DNA mutagenesis: an overview. Anal Biochem. 1997;254:157–178. doi: 10.1006/abio.1997.2428. [DOI] [PubMed] [Google Scholar]
- 18.Marchetti A, Buttitta F, Miyazaki S, Gallahan D, Smith G H, Callahan R. Int-6, a highly conserved, widely expressed gene, is mutated by mouse mammary tumor virus in mammary preneoplasia. J Virol. 1995;69:1932–1938. doi: 10.1128/jvi.69.3.1932-1938.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyazaki S, Imatani A, Ballard L, Marchetti A, Buttitta F, Albertsen H, Nevanlinna H A, Gallahan D, Callahan R. The chromosome location of the human homolog of the mouse mammary tumor-associated gene INT6 and its status in human breast carcinomas. Genomics. 1997;46:155–158. doi: 10.1006/geno.1997.4996. [DOI] [PubMed] [Google Scholar]
- 20.Neuveut C, Jin D Y, Semmes O J, Diella F, Callahan R, Jeang K-T. Divergent subcellular locations of HTLV-1 Tax and Int-6: a contrast between in vitro protein-protein binding and intracellular protein colocalization. Biomed Sci. 1997;4:229–234. doi: 10.1007/BF02253422. [DOI] [PubMed] [Google Scholar]
- 21.Palmeri D, Malim M H. The human T-cell leukemia virus type 1 posttranscriptional trans-activator Rex contains a nuclear export signal. J Virol. 1996;70:6442–6445. doi: 10.1128/jvi.70.9.6442-6445.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel R C, Vestal J D, Xu Z, Bandyopadhyay S, Guo W, Erme S M, Williams B R G, Sen G C. DRBP76, a double-stranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. J Biol Chem. 1999;274:20432–20437. doi: 10.1074/jbc.274.29.20432. [DOI] [PubMed] [Google Scholar]
- 23.Patel R C, Stanton P, Sen G C. Specific mutations near the amino terminus of double-stranded RNA-dependent protein kinase (PKR) differentially affect its double-stranded RNA binding and dimerization properties. J Biol Chem. 1996;271:25657–25663. doi: 10.1074/jbc.271.41.25657. [DOI] [PubMed] [Google Scholar]
- 24.Sen G C, Ransohoff R M. Interferon-induced antiviral actions and their regulation. Adv Virus Res. 1993;42:57–102. doi: 10.1016/s0065-3527(08)60083-4. [DOI] [PubMed] [Google Scholar]
- 25.Stark G R, Kerr I M, Williams B R G, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 26.Tiwari R K, Kusari J, Kumar R, Sen G C. Gene induction by interferons and double-stranded RNA: selective inhibition by 2-aminopurine. Mol Cell Biol. 1988;8:4289–4294. doi: 10.1128/mcb.8.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wathelet M G, Clauss I M, Content J, Huez G A. The IFI-56K and IFI-54K interferon-inducible human genes belong to the same gene family. FEBS Lett. 1988;231:164–171. doi: 10.1016/0014-5793(88)80724-5. [DOI] [PubMed] [Google Scholar]
- 28.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 29.Yu M, Tong J H, Mao M, Kan L X, Liu M M, Sun Y W, Fu G, Jing Y K, Yu L, Lepaslier D, Lanotte M, Wang Z Y, Chen Z, Waxman S, Wang Y X, Tan J Z, Chen S J. Cloning of a gene (RIG-G) associated with retinoic acid-induced differentiation of acute promyelocytic leukemia cells and representing a new member of a family of interferon-stimulated genes. Proc Natl Acad Sci USA. 1997;94:7406–7411. doi: 10.1073/pnas.94.14.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]