Summary
The studies reported here focus on the impact of pre-existing CD4 T cell immunity on the first encounter with SARS-CoV-2. They leverage PBMC samples from plasma donors collected after a first SARS-CoV-2 infection, prior to vaccine availability and compared to samples collected prior to the emergence of SARS-CoV-2. Analysis of CD4 T cell specificity across the entire SARS-CoV-2 proteome revealed that the recognition of SARS-CoV-2-derived epitopes by CD4 memory cells prior to the pandemic are enriched for reactivity toward non-structural proteins conserved across endemic CoV strains. However, CD4 T cells after primary infection with SARS-CoV-2 focus on epitopes from structural proteins. We observed little evidence for preferential recall to epitopes conserved between SARS-CoV-2 and seasonal CoV, a finding confirmed through use of selectively curated conserved and SARS-unique peptides. Our data suggest that SARS-CoV-2 CD4 T cells elicited by the first infection are primarily established from the naive CD4 T cell pool.
Subject areas: Health sciences, Immunology, Immune response, Virology
Graphical abstract
Highlights
-
•
Pre-COVID memory selectively recognize peptides conserved between SARS-CoV-2 and sCoV
-
•
Post-infection responses display no biases toward conserved epitopes or proteins
-
•
The CD4 T cell response magnitude to SARS-CoV-2 is host-specific across viral proteins
Health sciences; Immunology; Immune response; Virology
Introduction
Soon after the onset of SARS-CoV-2 pandemic, studies sought to understand the nature of CD4 T cell immunity to SARS-CoV-2 infection. Distinct experimental approaches were used by different groups to detect and quantify these SARS-CoV-2 specific CD4 T cells. These approaches differed by the epitopes surveyed (see1,2,3 for reviews) and include peptides selected by predictive algorithms,4,5,6,7 peptide class II tetramers,8,9 overlapping peptide libraries representing the sequence of major structural proteins such as Spike or nucleocapsid,10,11 or a subset of proteins12 or at the far extreme, the total proteome.5 Different assays were used to quantify the frequency of the SARS-CoV-2-reactive CD4 T cells (reviewed in13). The most common assay used to detect SARS-CoV-2-reactive T cells directly ex vivo has been the “AIM” assay that detects the upregulation of cell surface molecules that occurs upon T cell receptor (TcR) engagement, allowing for the sampling of limited PBMC and detection of CD4 T cells independently of cytokine production.5,14 A more limited subset of studies has used cytokine secretion, most commonly IFN-γ EliSpot assays, to quantify SARS-CoV-2-reactive CD4 T cells.10,12 Also, studies differ with regard to whether they were performed directly ex vivo4,5 or after in vitro expansion.14,15 Each of these approaches have both strengths and limitations and are important to consider in deriving models for immunity elicited by SARS-CoV-2 infection.
The study design in this article was enabled by the availability of large samples of human PBMC collected from COVID convalescent plasma donors elicited by this first confrontation with SARS-CoV-2 prior to availability of vaccines, and a similarly large and diverse sample of PBMC banked prior to the emergence of SARS-CoV-2. These large sample sizes enabled the assessment of issues that are currently unresolved in the field. First, by tracking the reactivity to the peptide epitopes contained in the entire proteome and through analyses of selectively curated peptide pools, we sought to address whether circulating memory CD4 T cells established by seasonal CoV (“sCoV”) are preferentially recruited by SARS-CoV-2 infection. This question remains of high interest because the degree of conservation in sequence is highly disparate across the different proteins encoded by the SARS-CoV-2 genome. The experimental findings that SARS-CoV-2 derived peptides can activate CD4 T cells from seronegative subjects or those sampled prior to the emergence of SARS-CoV-216,17,18,19 has raised the following questions. First, how extensively are CD4 T cells specific for peptides conserved between SARS-CoV-2 and previously established memory cells drawn into the response to SARS-CoV-2 infection? Second, how substantially do CD4 T cells from sCoV contribute to protection from the first encounter by infection with SARS-CoV-2?
Here, we quantify responses toward all the viral classes by cytokine secretion assays, stressing the functionality of the SARS-CoV-2 reactive CD4 T cells. By obtaining PBMC samples from plasma donors collected early after the emergence of SARS-CoV-2, prior to the availability of SARS-CoV-2 vaccines, we were able to probe the primary response to infection. The sample sizes provided sufficient cell numbers to enrich CD4 T cells and exclude CD8 T cells as well as other cells that can produce cytokines. These post-infection assays were compared to CD4 T cell responses from pre-pandemic PBMC samples collected prior to autumn of 2019 that represents memory from periodic infections with different seasonal CoV (“sCoV”). Combining the large number of CD4 T cells with the availability of overlapping peptides libraries of the proteome from both cohorts enabled the evaluation of CD4 T cell reactivity toward the entire SARS-CoV-2 proteome in discreet and unbiased protein-specific patterns. A peptide-dependent direct ex vivo cytokine secretion EliSpot assay was used, allowing the detection of CD4 T cells with anti-viral function without any expansion in vitro. This approach has been used in our laboratory as a strategy to quantify human influenza-specific CD4 T cells to a wide range of antigen specificities.20 These samples were also large enough to enable the purification of CD4 T cells and detection of SARS-CoV-2 peptide-reactive cells in a protein-specific manner to the entire proteome. This strategy provided an estimation of the potential recall of sCoV reactive CD4 T cells by SARS-CoV-2-derived CD4 T cell epitopes and patterns of overall and generalizable protein specificity and immunogenicity. Together, these studies enable an assessment of whether and how extensively cross-reactive CD4 T cells were recalled and expanded by the first infection with SARS-CoV-2.
These analyses revealed striking features in CD4 T cell immunity to SARS-CoV-2 in the post-infection, convalescent subjects. First, despite the broad range in responses that were noted in response to infection, SARS-CoV-2 specific CD4 T cell abundance across different viral proteins was tightly correlated, suggesting that selective HLA-linked epitope dominance was not a factor in the magnitude of the CD4 T response among subjects. Second, when separately querying curated pools of peptides from the spike protein that were highly conserved or highly disparate with endemic sCoV, responses were not enriched for recall toward the conserved epitopes. Finally, when the patterns and preferences in responses between pre-2019 and post-infection samples were compared, they were remarkably distinct. The CD4 T cells from pre-pandemic samples exhibited high reactivity to conserved epitopes within non-structural proteins, accessory proteins, or those regions of structural proteins with stretches of amino acids shared with SARS-CoV-2, while CD4 T cells elicited by infection exhibited strong preferences toward structural proteins and perhaps most importantly, were not detectably biased toward conserved epitopes. Thus, the comprehensive studies reported here provide little evidence for the recall of memory CD4 T cells established by periodic and lifetime encounter with sCoV during the host’s first encounter with SARS-CoV-2.
Results
Human CD4 T cell responses to the SARS-CoV-2 proteome-derived epitopes after the first infection with SARS-CoV-2
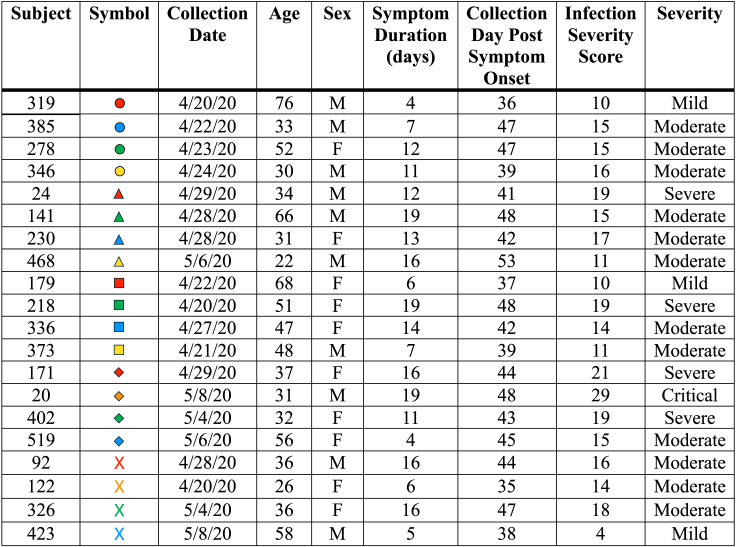
We sought to address the specificity and immunodominance patterns in human CD4 T cells after the recovery of subjects from primary SARS-CoV-2 infection. Shown in Figure 1 are the demographic features and disease parameters of the convalescent plasma donors sampled for reactivity to individual peptide pools representing each viral protein in the SARS-CoV-2 proteome. This set of PBMC was comprised of adult subjects who were plasma donors soon after SARS-CoV-2 first emerged, where plasma was used as a treatment strategy (reviewed in22,23,24). The sample dates were in April and May of 202025,26 and thus prior to the availability of SARS-CoV-2 vaccines.
Figure 1.
Demographics and infection data, including the duration of symptoms, infection severity score, and defined severity based on symptomology in SARS-CoV-2 convalescent subjects collected in April and May of 2020
Infection severity score was calculated based on 12 symptoms including fatigue, cough, shortness of breath, loss of taste and smell, and fever with a potential score of up to 18. Also included in the scoring was hospitalization (not hospitalized - 0), hospitalized without oxygen (6) or with oxygen (8) and ICU admission (10). Finally, the duration of symptoms with a value of 0 (0 days) to 8 (21+ days) was included. These values lead to cumulative scores indicated in the infection severity score column (<10 to 35) and the cumulative scores we used to rank severity (<10 mild, 11–18 moderate, 19–27 severe and 28–35 critical). The exact criteria are referenced in Guthmiller et al.21
In order to quantify the reactivity to SARS-CoV-2 post infection, we used cytokine EliSpot assays, used in conjunction with overlapping peptide libraries comprising the entire translated sequence of each of the SARS-CoV-2 proteins. These libraries represented each of the structural proteins contained in the virion, the non-structural proteins encoded by the Orf1a and Orf1b, and many of the ORF accessory proteins that were sequenced at the time that the peptide libraries were acquired (see Table S1). Cryopreserved PBMC were thawed, rested overnight, then washed and depleted of CD8 and NK cells, yielding a CD4 T cell-enriched population that contain primarily host antigen-presenting cells (APC) and CD4 T cells. Cytokine EliSpots (IFN-γ) were chosen because they serve as a straightforward and objective method to quantify the T cell recognition of antigen to viral pathogens (reviewed in13,27,28) and detect cells that respond to TcR engagement with a functional response with a cytokine that has potent anti-viral functionality. This approach also overcomes the limitations in the detection of T cells due to complexity of HLA molecules that makes predictive algorithms or HLA-derived peptide tetramers inadequate for the quantification of all potential epitopes recognized by the CD4 T cells. Our assays thus enabled an unbiased comprehensive assessment of the CD4 T cells elicited by human SARS-CoV-2 infection. This type of assay has been used by our laboratory to study human memory CD4 T cells,20 human vaccine responses,29,30 and responses to human endemic sCoV.31
Peptide pools were incubated with CD4 T cell-enriched cell populations and cultured for 36 h with the protein-specific peptide pools. The frequency of reactive cells was scored for cytokine secreting cells by an automated plate reader (see Figure S1 for representative images). The number of spots detected reflect a direct ex vivo measurement of antigen-specific CD4 T cells and thus enabled the assessment of the relative immunodominance of CD4 T cells specific for the multiple proteins expressed by SARS-CoV-2 after the first infection of humans with this virus.
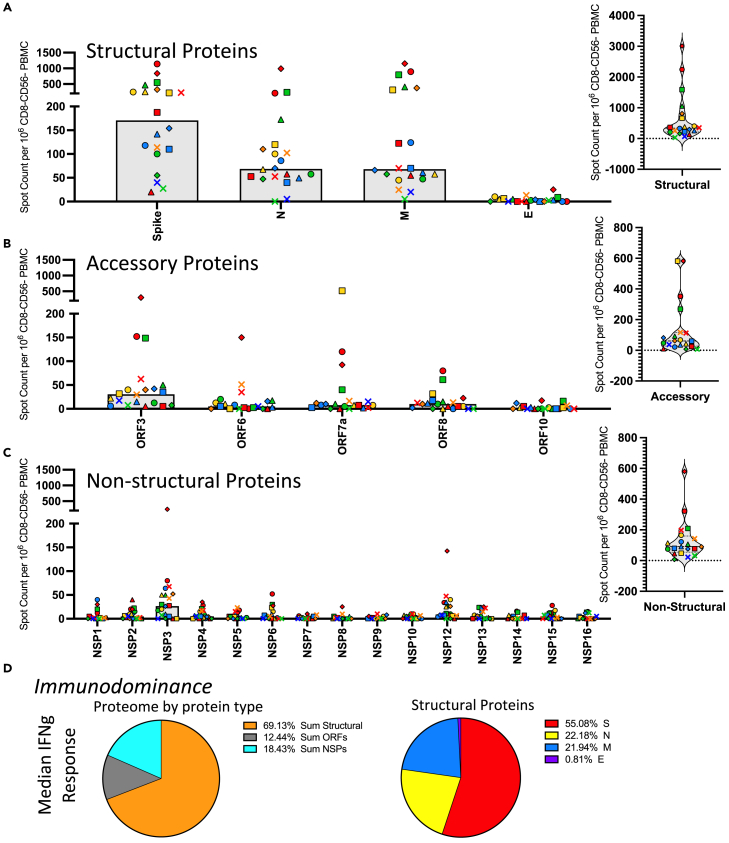
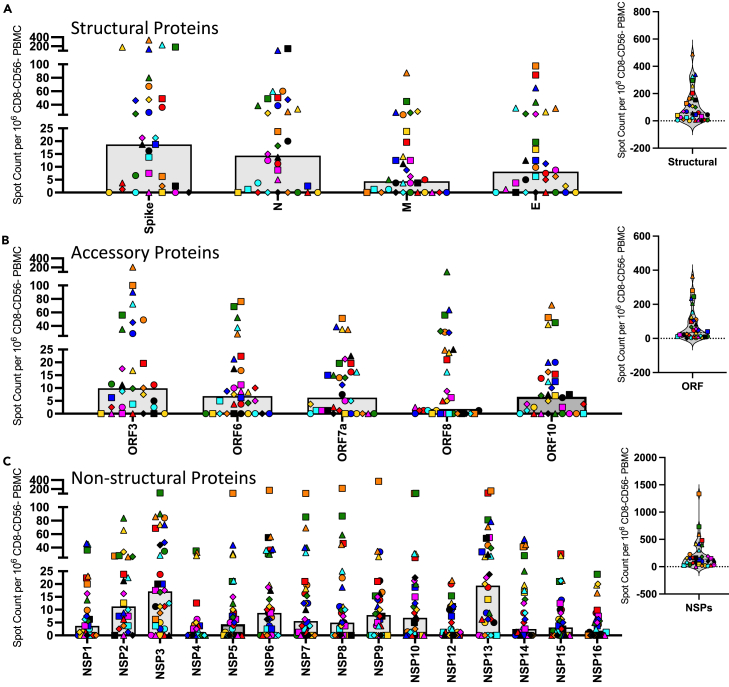
Figure 2 shows the results of these analyses, where each SARS-CoV-2 protein is indicated below the Figure panel with the number of cytokine-producing cells per million CD4 T cell enriched population shown on the Y axis. In this Figure, each subject is noted by a unique symbol and color that matched the data presented in Figure 1. The SARS-CoV-2 virus protein specificities are divided into 3 classes of proteins, represented by Figures 2A–2C, where the response to the structural proteins is shown in Panel A, the accessory proteins in Panel B, and the non-structural proteins (NSP) from ORF-1 shown in Panel C. The distribution of the total responses is shown as pie graphs in Panel D.
Figure 2.
CD4 T cell responses to the SARS-CoV-2 proteome in PCR infection confirmed convalescent subjects are dominated by reactivity to structural proteins and spike-specific responses
CD4 T cell responses to the entire SARS-CoV-2 proteome were evaluated in 20 subjects that had PCR confirmed SARS-CoV-2 infections in April or May of 2020 at a convalescent time point (D35-53 post symptom onset). Responses were measured using IFN-γ production in Enzyme-linked (EliSpot) analyses. The proteome has been divided into three classes of proteins (A) structural proteins, (B) accessory proteins and (C) non-structural proteins. Shown are the spot count per million CD8− and CD56− depleted PBMC with background subtracted. The median CD4 T cell response to each antigen is shown as a gray bar and individual subjects are indicated by unique symbols (shown in Figure 1). The right most panels of A, B and C show the total response to these classes of SARS-CoV-2 proteins. Panel D shows the average relative immunodominance as pie charts illustrating the dominant response to the structural proteins. The peptide specific CD4 T cell frequencies were summed based on the protein class for each individual subject and the median frequency for the protein classes, structural proteins (orange), accessory proteins (gray) and non-structural proteins (turquoise) was plotted as a pie chart. In the right pie chart the structural proteins that accounted for the largest proportion of the response and were further broken down into specific structural proteins spike (red), nucleocapsid (yellow), membrane (blue), and envelope (purple).
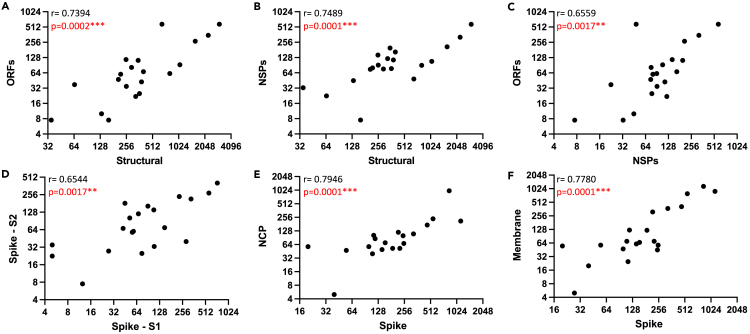
Several observations were made in this first level of analyses. It is clear, first, that there is a tremendous range in overall response magnitude, with occasional subjects exhibiting extremely modest responses, and others displaying an exceptionally high frequency of SARS-CoV-2-specific CD4 T cells across many protein specificities. These differences in response magnitude did not detectably correlate with disease severity (see Figure S2). Second, CD4 T cell responses to the SARS-CoV-2-derived epitopes from different proteins and classes of viral proteins exhibit generalizable differences in immunodominance, despite differences in responses magnitude. The structural proteins are the dominant specificity in cytokine-producing cells in post infection samples, with spike-reactive CD4 T cells being the most dominant, averaging over 150 spots per million of enriched CD4 T cells (Figure 2A). ORF3 elicits the greatest response among the accessory proteins tested, and averages approximately 30 cytokine producing cells per million (Figure 2B), while most of the non-structural proteins (NSP) elicit very few CD4 T cells after infection. The NSP that recruits the greatest number of T cells is NSP3, followed by NSP12, with the others eliciting less than 10 cytokine producing cells per million (Figure 2C). The overall dominance of the structural proteins in the CD4 T cell responses relative to the non-structural proteins and accessory proteins is even more apparent when one represents these response preferences in the CD4 T cell response by pie diagrams (Figure 2D). The average fractional CD4 T cell response to different classes of SARS-CoV-2 proteins (structural, accessory, and NSP) is shown for all subjects. Almost 70% of the CD4 T cell response is specific for epitopes in the structural proteins, while approximately 20% of the response is specific for the sum of all 15 of the NSP. The remaining 14% of the response is specific for the sum of the ORF. The relative response to each of the 4 structural proteins is shown in the right panel of Figure 2D, where Spike-derived epitopes are dominant, recruiting almost 70% of the response, followed by approximately equal responses to M and N. Only a very small fraction (<1%) of the total responses to structural proteins are specific for the small E protein. Interestingly, when the abundance of cytokine -producing CD4 cells reactive with the peptides encoded by the different structural proteins were analyzed for correlation coefficients (Figures 3A–3F), there were clear and statistically positive correlations between the relative response magnitude to one protein (e.g., spike) relative to the others (e.g., NCP (Figure 3E) or Membrane (Figure 3F)). Similarly, when reactivity to subdomains of spike (Figure 3D) was compared for correlations, the same highly statistical significance was obtained. This result is important in several respects. First, it indicates that robust reactivity to a given SARS-CoV-2 protein is not driven by dominance toward a single epitope or limited subset of epitopes, perhaps dictated by HLA-allele distribution among the subjects. Second, this concurrence of relative reactivity toward all major sources of epitopes makes efforts to assess the relationship between protein-specific antibody and CD4 T cell specificity4,5,32 difficult because subjects that display robust CD4 T cell responses to one protein will almost certainly exhibit robust responses to the others. Finally, it suggests that humans confronted with SARS-CoV-2 for the first time have substantial and inherent differences in the magnitude of their CD4 T cell response.
Figure 3.
CD4 T cell responses to SARS-CoV-2 proteins correlate
The abundance of cytokine producing CD4 cells reactive with the peptides encoded by the indicated classes of proteins as well as individual structural proteins or segments were analyzed for correlation coefficients. Using XY plots, the sum of the CD4 T cell IFN-γ response to the structural proteins (X axis) was plotted against the sum of the IFN-γ CD4 T cell response to the accessory proteins (A) and the non-structural proteins (B). In panel C the response to the nonstructural and accessory proteins are correlated. In panel D the CD4 T cell IFN-γ response to S1 was plotted against the response to S2 and in panels E and F the responses to NCP and Membrane, respectively, were evaluated against the response to spike. The r and p values shown in the left corner of each panel were calculated using the Spearman correlation.
Estimation of the immunogenicity of SARS-CoV-2 viral proteins in the elicitation of CD4 T cells after SARS-CoV-2 infection
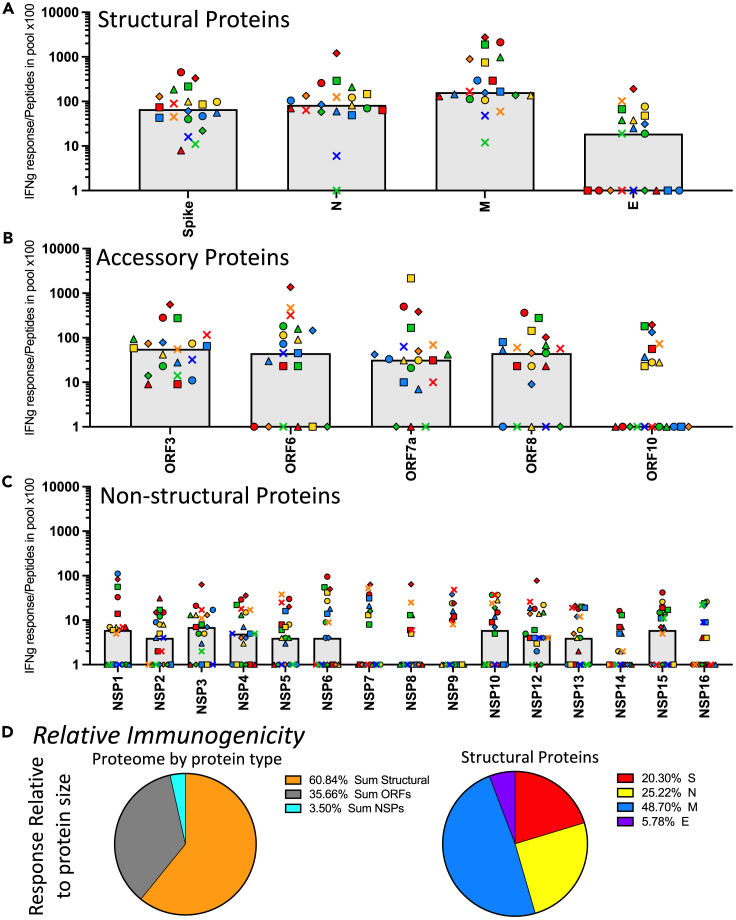
Beyond the absolute immunodominance exhibited in response preferences, it is useful to evaluate the factors that contribute to immunogenicity of different viral proteins. As a first step in this evaluation, we estimated relative “immunogenicity” of each protein and each class of proteins by taking into account the size of the proteins, and thus the potential number of CD4 T cell peptide epitopes available for recognition. The total number of peptides in each pool is shown in Table S1 and is proportional to the size and total number of amino acids in each protein. When the response magnitude (as spots per million) for each peptide pool was divided by the number of peptides in the peptide pool for that protein, the size-normalized response is obtained (Figures 4A–4C), it is clear that the structural proteins are the most immunogenic in the primary response to infection, with the membrane protein (“M”) specific cells most immunogenic and the small envelope protein (“E”) the most poorly immunogenic. Spike and nucleocapsid were similarly immunogenic. NSP 1 through 15 were discovered to be very poorly immunogenic in the CD4 T response to SARS-CoV-2 infection, even relative to most of the accessory proteins (ORF 3, 6, 7a, 8 and 10), when normalized to their variable size.
Figure 4.
Evaluation of the immunogenicity of SARS-CoV-2 proteins demonstrates dominance of membrane and poor immunogenicity of the NSP
Based on the CD4 T cell responses to the SARS-CoV-2 proteome, shown in Figure 2, the relative of immunogenicity of each protein was calculated. Responses measured using cytokine producing cells per million, shown in Figure 2, were normalized to the number of peptides contained in each pool by dividing the frequency of cytokine spots per million CD8− and CD56− depleted PBMC by the number of peptides contained in the pool (Table S1) and multiplying by 100. The immunogenicity of the SARS-CoV-2 proteome is shown in the three classes of proteins (A) structural proteins, (B) accessory proteins, and (C) non-structural proteins. Donors are indicated by unique symbols (identified in Figure 1), with the median value illustrated by a gray shaded bar. In panel D, the average immunogenicity is represented by pie diagrams. The median response of the population for the sum of the proteins from each class, relative to peptide pool size, indicating the relative immunogenicity is shown in the left pie diagram; structural proteins (orange), accessory proteins (grey), and non-structural proteins (turquoise). To the right, the relative immunogenicity of each of the structural proteins, spike (red), nucleocapsid (yellow), membrane (blue), and envelope (purple) are shown.
The differences in immunogenicity are even more evident when one constructs a pie diagram for each of the viral protein classes (Figure 4D). The left panel shows that almost 60% of the response is specific for epitopes from the structural proteins (S, M E, and N), when normalized for size, and approximately 35% of the response is dedicated to the sum of ORF3, ORF6, ORF7a, ORF8, and ORF10-specific CD4 T cells, and less than 5% of the response is dedicated to NSP, when the sum of these responses, many of these which are quite large, are normalized to their size. The right panel of Figure 4D the fractional response to the different structural proteins was calculated, where Spike recruits over half of the response relative to the other structural proteins, with the remainder approximately equally shared between M and N, while very few T cells reactive to E. Strikingly, when normalized to size, the M protein is the most immunogenic, with spike and nucleocapsid equivalent to each other. M is known to be the most abundant SARS-CoV-2 protein in the virion, while N is abundant in infected cells. Thus, viral protein abundance may be one key parameter that contributes to the overall ability to recruit CD4 T cells and the resulting immunogenicity.
The relative reactivity of CD4 T cells elicited toward sCoV-conserved vs. SARS-CoV-2-specific epitopes after the first encounter with SARS-CoV-2
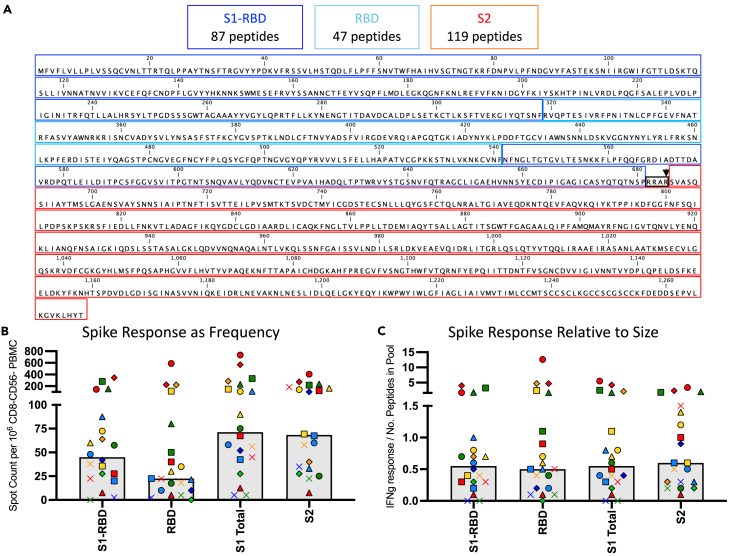
To examine the potential for recall responses from memory generated by the endemic sCoV, we then examined the CD4 T cell response distribution to different domains of Spike as well as their relative immunogenicity when normalized to size. This issue, which has been a bit contentious in the literature, is important to address to anticipate the preservation of T cell recognition of spike in the face of the extensive mutation and selection that has continued takes place the course of the pandemic.33,34,35 In order to assess the relative distribution of CD4 T cell reactivity to different segments of the Spike protein, we constructed and tested separate pools of peptides from S2, and two discreet subdomains of S1 (RBD and S1 “minus” RBD), the latter pool including all of the peptides in S1, excluding those in the RBD segment. Figure 5A illustrates the sequence and the boundaries of each segment. Figure 5B shows that in most subjects, each segment elicits readily detectable cytokine-producing cells. There are many subjects who display robust responses to each segment of spike and a few subjects who have very few detectable spike-specific cells. Interestingly, when normalized to size and thus the potential number of peptide epitopes (Figure 5C), each of the 3 subdomains S1-RBD, RBD, and S2), displayed indistinguishable immunogenicity. This result indicates that CD4 T cell reactivity to spike, established after the first SARS-CoV-2 infection, will likely persist upon subsequent vaccination or infection with drifted variants. Even when mutations to sCoV-2-derived epitopes have been noted that altered T cell recognition,36,37,38,39,40 the broad reactivity of the CD4 repertoire allowed persistent reactivity to be maintained. Our limited analyses of CD4 T cell reactivity to Wuhan vs. Omicron derived peptide pools agree with this conclusion (data not shown). Most of the experimental studies assessing this issue support this conclusion (reviewed in41,42).
Figure 5.
The CD4 T cell response to SARS-CoV-2 spike protein post infection is distributed across the spike protein
(A) The amino acid sequence of the spike protein from SARS-CoV-2 is indicated as defined by 3 different segments of the protein including, S1-RBD (blue), which is the S1 segment with the RBD portion of S1 removed, RBD (turquoise) and S2 (red). These delineations formed the basis of the construction of the separate pools of peptides used for stimulation in the CD4 T cell EliSpot assay. The number of peptides contained in each segment that were pooled and used are indicated above the Figure. The CD4 T cell response measured by IFN-γ EliSpot assay to these segments of spike is shown with the frequency in Panel B and the response relative to each segment’s size and peptide number in the pool is in Panel C. The average median response is illustrated by a gray box and individual subjects are indicated by their unique symbols.
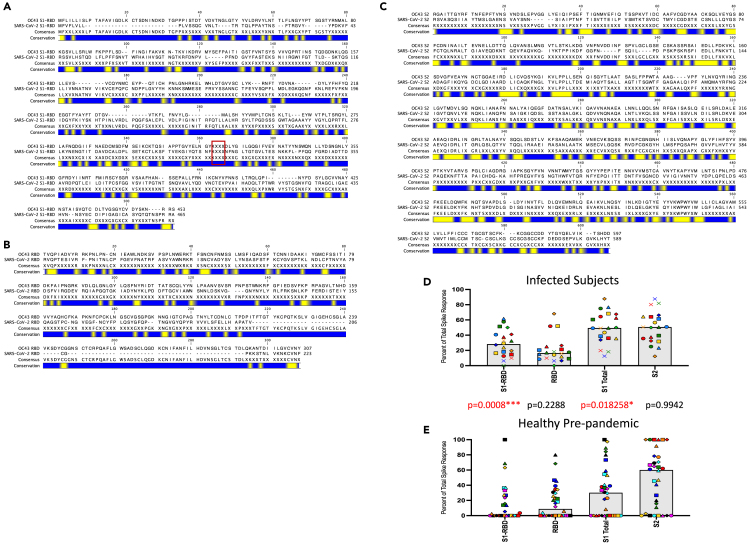
The reactivity of CD4 T cells to the S1 and S2 segments of spike is also shown as separate bar graphs, Figure 5B. The pattern of relative reactivity to these segments of spike was unexpected. The S2 domain has considerably more sequence identity with the endemic sCoV than does the S1 domain or RBD domain, as is shown in Figures 6A–6C and thus the S2 segment has the potential to share CD4 T cell epitopes. Our previous analyses31 of subjects whose PBMC were collected prior to the fall of 2019, showed that almost all of the spike-reactive CD4 T cells were specific for epitopes in the conserved S2 domain. Because of this disparity in the memory specificity to SARS-spike S1 and S2, we expected that upon the first confrontation of subjects with SARS-CoV-2, CD4 T cells recruited from memory established by lifelong exposure to endemic sCoV, CD4 T cells reactive to epitopes in the S2 domain would be selectively expanded and thus be more highly represented in post-infection samples. However, the CD4 T cell response to infection was equally balanced between S1 and S2 (Figure 6D).
Figure 6.
Distinct patterns of CD4 T cell immunodominance within the spike protein in post-convalescent subjects versus pre-pandemic samples
Amino acid sequence alignments of spike proteins for sCoV OC43 and SARS-CoV-2, broken down in the three segments, S1-RBD (A), RBD (B) and S2 (C). Blue bars represent segments of sequence variation and yellow segments indicate stretches of sequence identity. The boxed S1-RBD segment indicates the gap in sequence where the RBD would be with XXXX. Sequence files were downloaded from PubMed (accession numbers YP_009555241.1and YP_009724390.1 for OC43 and SARS-CoV-2 spike proteins, respectively). To assess the percent of the CD4 T cell response dedicated to each segment of spike protein, the number of cytokine-producing cells elicited by each pool were summed and the percent response for each segment was divided by the total number of spike specific CD4 cells qualified multiplied by 100. Panel D shows the response from the infected convalescent samples and Panel E, results shown are from a cohort of healthy adults collected between spring 2014 and spring 2019. The average median response is illustrated by a gray box and individual subjects for each cohort are indicated by unique symbols. Between panels D (convalescent) and E (healthy) are p values calculated using the Mann-Whitney test comparing each stimulation condition for the two cohorts. p values that reach statistical significance are indicated in red font.
The SARS-CoV-2-reactive human CD4 T cell memory compartment prior to the pandemic
To explore this issue more fully, we extended our studies to banked PBMC that were collected prior to the emergence of SARS-CoV-2 in 2020. These samples, by virtue of the date collected, are from subjects never exposed to SARS-CoV-2 (see Figure 7). The PBMC were enriched for CD4 T cells and tested for reactivity to SARS-CoV-2-derived peptides from Spike. This comparative study of the two cohorts was analyzed for each subject in the two cohorts as a fraction of the total response to spike specific for the three pools of spike derived peptides: RBD, S1-RBD and S2 peptides (see Figure 6D, previously infected; Figure 6E, pre-pandemic, with the statistical treatment of data comparing the cohorts). These data demonstrate T cells sampled prior to 2020 have negligible reactivity to RBD and S1-RBD and that many subjects have 100% of their response specific for S2.
Figure 7.
Healthy donor (HD) subject demographics and unique symbols
Collection times are indicated by the season (S-spring/summer, W-winter, F-fall) and the year.
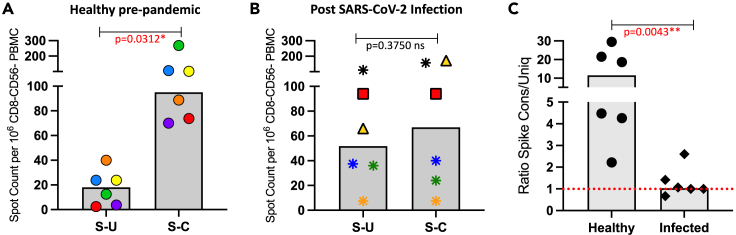
We then examined this issue more stringently by selectively curating peptides from spike that were either primarily conserved or unique, independently of their localization in the spike protein. The criteria used for “unique” were those 17mer peptides that had fewer than 4 contiguous amino acids of identity between SARS-CoV-2 spike and the endemic beta sCoV OC43. Conserved peptides were selected based on at least 6 amino acids in the 17mer that were shared between the two sources. Approximately 65 peptides were used in each pool to both normalize the overall potential for CD4 T cell recognition and to gain as robust a response as possible. These two pools of peptides (sequences shown in Table S2) were tested for their ability to stimulate the pre-2020 banked samples and the post-COVID convalescent samples (Figures 8 and S3). Panel A shows that in the samples collected from healthy donors prior to 2020, the response is restricted mainly to the conserved pool, with only very modest responses to the unique pool. These results are consistent with the view that SAR-CoV-2 Spike S2-specific responses, detected in CD4 T cells collected prior to the pandemic, are likely due to accumulated memory due to previous seasonal coronavirus infections and have true cross-reactivity rather than spurious reactivity to unrelated peptides, as has been argued in some studies43,44 (see16 for discussion). In contrast, in the CD4 T cells isolated from post-infection samples Panel 8B shows that the response is balanced between conserved and unique in all subjects but one (marked as gold triangle), which may reflect the recognition of a dominant epitope in the conserved peptide pool. Despite the overall range in responses in both cohorts, a careful examination of the responses between the two peptide pools for each of the subjects, which can be tracked by comparing the symbols in both bar graphs representing the same individual, demonstrate the disparity in the response patterns in the two cohorts. The equivalent recognition of conserved or unique spike-specific epitopes supports the view that SARS-CoV-2 infection does not preferentially recruit CD4 T cells from the circulating memory pool and instead is likely a primary response. Because there are both “high” and “low” responders in each cohort, a particularly useful way of examining these results is to calculate a ratio of CD4 T cell responses to conserved versus unique peptides for each subject. Figure 8C shows that the ratio of responses of conserved to unique peptide pools is about 13 in pre-COVID samples, whereas in infected and convalescent subjects, it is approximately 1.
Figure 8.
Reactivity of CD4 T cells to conserved peptides shared between sCoV OC43 and SARS-CoV-2 vs. peptides unique to SARS-CoV-2 suggests lack of recall of CD4 T cells from the memory pool
Curated pools of peptides representing unique and conserved regions of the spike protein were designed based on alignments between seasonal CoV OC43 and SARS-CoV-2 (see Table S2). Each pool contained an equal number of peptides. CD4 T cell responses from 6 healthy adult subjects collected prior to 2019 (Panel A) and from 6 subjects following PCR confirmed SARS-CoV-2 infection (April-May 2020) at a convalescent time point (Panel B) were quantified by IFN-γ EliSpot. Shown are the spot counts per million CD8− and CD56− depleted PBMC with background subtracted. The median CD4 T cell response to each peptide pool is shown as a gray bar and individual subjects are indicated by unique symbols. Panel C illustrates the ratio of the responses to conserved vs. unique peptide pools for each individual. The red dotted line indicating a value of 1, represents an equal response between conserved and unique. p values are indicated above each plot and have been calculated using the Wilcoxon test for panels A and B and the Mann-Whitney test for panel C. Statistically significant p values are indicated in red text.
To extend the comparison of response patterns between pre-pandemic and post-infection samples, we examined CD4 T cell reactivity to proteins in the entire proteome of SARS-CoV-2 in samples collected prior to the pandemic. Figure 9 shows the reactivity of CD4 T cells from pre-COVID subjects across the three classes of SARS-CoV-2 proteins examined earlier in the convalescent samples. While some subjects displayed little reactivity toward many of the SARS-CoV-2 proteins, a substantial number displayed readily detectable cytokine-producing CD4 T cells, with frequencies of over 200 per million enriched CD4 T cells. Among the structural proteins (Figure 9A), Spike-derived and nucleocapsid-derived epitopes elicited the greatest response although many displayed no detectable responses. The responses to the accessory proteins (Figure 9B) were readily detectable among many subjects, with the exception of ORF8, which is among the most highly genetically variable accessory protein of beta coronaviruses (reviewed in45,46). Among the NSP (Figure 9C), helicase (NSP 13) and NSP3-derived epitopes recruit the most CD4 T cells from circulating memory from endemic sCoV. These proteins are relatively highly conserved across human coronaviruses, across alpha and beta sCoV, SARS-CoV and SARS-CoV-2. Although the NSP are low in abundance relative to the structural proteins,47,48 it is possible that repeated exposure over the adults’ lifetimes ultimately allows the accumulation of CD4 T cells specific for some of the more conserved NSP.
Figure 9.
CD4 T cell responses to the entire SARS-CoV-2 proteome evaluated in 32 healthy subjects collected pre-pandemic reveals striking differences from post SARS-CoV-2 infection samples
Shown are immunodominance patterns of CD4 T cells collected prior to the onset of the SARS-CoV-2 pandemic (December 2019), using the same pools of peptides as tested in results shown in Figure 2. Responses were measured using IFN-γ production in EliSpot assays. The proteome was divided into three classes of proteins (A) structural proteins, (B) accessory proteins and (C) non-structural proteins. Shown are the spot count per million CD8− and CD56− depleted PBMC with background subtracted. The median CD4 T cell response to each antigen is shown as a gray bar and individual subjects are indicated by unique symbols (Figure 7). The right most panels of A, B and C show the total response to these classes of SARS-CoV-2 proteins.
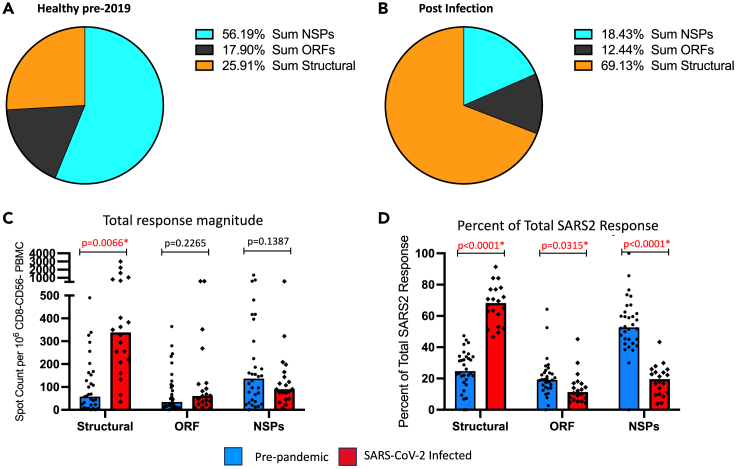
A comparison of the relative distribution of SARS-CoV-2 reactive CD4 T cells between pre-2020 and convalescent subjects is shown in Figure 10, where the sum of the CD4 T cell responses for the structural proteins, the accessory proteins and the NSP are indicated for each of the two cohorts. The differences in distribution are striking. In the samples collected prior to the emergence of SARS-CoV-2 (Figure 10A), circulating CD4 T cells displayed an average of 56% of their overall reactivity toward NSP, with the remainder were dedicated at the sum of structural proteins (25%) and the accessory proteins ORF (18%). In contrast, as is shown in Figure 10B, in the post-infection convalescent samples, an average of almost 70% of the response is dedicated to epitopes derived from the structural proteins, with 18% being specific for NSP and 12% specific for accessory ORF proteins. Because there is a large range in overall reactivity of the CD4 T cell responses in both the pre-pandemic and convalescent samples (Figure 10C), we analyzed the data in an additional way to normalize these responses. The average response distribution across the 3 classes of proteins for each individual was calculated and is displayed as bar graphs (Figure 10D). When contrasted in this way, it is clear that the two cohorts have dramatically and statistically significant differences in reactivity to the SARS-CoV-2 structural proteins, accessory proteins and NSP.
Figure 10.
Distribution of the CD4 T cell response in pre-pandemic healthy subjects and SARS-CoV-2 convalescent subjects express distinct patterns of epitope dominance
The frequencies of CD4 T cells reactive to discreet pools of peptides measured by IFNγ EliSpot assays to the entire SARS-CoV-2 proteome were evaluated in 32 healthy subjects that had been collected prior to the pandemic and 20 post-infection convalescence samples, collected in April and May of 2020, were summed for each protein group: structural, accessory and non-structural proteins. Panel A represents the median distribution of the summed SARS-CoV-2 CD4 responses in the pre-pandemic samples and panel B the distribution of sum of reactivity in the convalescent infected subjects, shown as pie charts, indicated by colors representing the response distribution. The individual subject responses are shown in panel C where blue bars indicate the median response in healthy pre-pandemic subjects and red bars depict the median response in post-infection samples. The percent of the total SARS-CoV-2 response was calculated for each protein type and illustrated in panel D as a % response distribution. Indicated above the bar graphs are the calculated p values with statistically significant values indicated in red. Statistical analysis was done using multiple unpaired t tests with Welch correction.
Discussion
In this study, we have analyzed the CD4 T cell response to SARS-CoV-2 infection from PBMC collected from recovered plasma donors early during the pandemic. We have then compared them to the accumulated CD4 T cell memory to seasonal endemic CoV from samples collected prior to the emergence of SARS-CoV-2. These studies utilized cytokine EliSpot assays and overlapping peptide libraries that represented the expressed proteome of SARS-CoV-2 in a completely unbiased way assumed into protein-specific pools. These assays thus allowed us to estimate CD4 T cell reactivity across the individual expressed proteins during SARS-CoV-2 infection in a comprehensive, functionally relevant, and unbiased way directly ex vivo.
These studies revealed several striking features of CD4 T cells elicited in the primary response to SARS-CoV-2 infection. First, among COVID recovered subjects, at a global level, there was a dramatic range in overall response magnitude, with occasional subjects exhibiting very modest to undetectable responses, and others who displayed an exceptionally high frequency of SARS-CoV-2-specifc cells. When reactivity to each protein was comparatively analyzed by examining individual patterns among the members of the cohorts or through correlation coefficients, it was notable that these extreme ends of response magnitude were quite often displayed across many different proteins in the SARS-CoV-2 proteome. In previous studies of influenza-specific memory,20,30 we observed similar variations in CD4 T cell abundance among human subjects and speculated that some of these differences might reflect the history of infection or vaccination of each host over their lifetime. Because these samples were collected soon after the onset of the pandemic, the CD4 T cells studied here likely represent the primary CD4 T cell response after a single confrontation with SARS-CoV-2. Interestingly, the overall response magnitude did not appear to correlate with disease severity or duration of symptoms by the criteria used in these studies (see Figure 1 for details). Also, in the post-COVID subjects either very high or low CD4 T cell response magnitudes did not correlate detectably with age (data not shown). The underlying factors that lead to the variation in response magnitude among the subjects studied are not understood. Because each subject will express a diverse "palette" of different HLA class II alleles, the antigen-presenting cells (APC) from each will present a distinctive and almost certainly distinct array of peptides for the recruitment of CD4 T cells.49 It is possible that there are dominant HLA class II molecules driving the response in some individuals as has been recently observed for a spike derived peptide (S167-180) presented by common HLA-DP allele HLA-DP0404 in HLA-haplotypes50 and a dominant HLA-DR15-restricted spike derived epitope (S751-767).36,51 Even in these cases, it is not clear what fraction of the pre-existing memory cells for these cross-reactive memory cells are recalled from memory relative to cells drawn from the naive repertoire. Overall, we postulate that the broad diversity of MHC class II allele distribution and complexity of HLA-class II molecules expressed in each different individual in US populations makes it unlikely that in general a single or even limited epitope specificity dominates based on the HLA-DR, DP, and DQ molecules expressed in the subjects. Also arguing strongly against the possibility of limited, HLA-linked, peptide selection dominating the response is our finding that response magnitudes correlate tightly across proteins. Thus, subjects who were robust responders to spike-derived epitopes from S1 also displayed robust responses to S2. And the overall CD4 T cell reactivity to spike correlated with reactivity to nucleocapsid and membrane-derived epitopes in the COVID elicited response. Because of these strong correlative relationships, we speculate that host factors or viral features distinct from the HLA genotype dictate the overall magnitude of the CD4 T cell response to infection. Accordingly, the high or low total CD4 T cell response magnitude may reflect other parameters in the SARS-CoV-2-infected host such as differences in viral protein abundance or viral persistence during priming. This could promote continued expansion or recruitment of a broad repertoire of elicited CD4 T cells. Alternatively, or additionally, there are likely other elements in the host such as innate activation or inherent host-dependent properties of protein-bearing dendritic cells that influence CD4 T cell priming or expansion post infection. It is also conceivable that the response magnitude detected here in peripheral blood CD4 T cells could reflect host-dependent differences in CD4 T cell memory cell distribution in vivo after infection, with asymmetries in the localization of memory cells in circulation and the respiratory tract where the viral infection dominates.
These studies also revealed strong protein-specific preferences in the proteins that elicit CD4 T cells during the response to SARS-CoV-2 infection. When the average responses among members of the cohort were analyzed, it was clear that the epitopes from the four structural proteins and to a lesser extent, the accessory proteins, were the most dominant in the CD4 T cell response to infection. In contrast, the overall responses to NSP accounted for less than 20% of the total CD4 T cell response. In a study of selected epitopes for CD8 T cells, NSP-derived specificities were also found to be lacking in dominance post infection.52 It is likely that one major factor that dictates immunogenicity of different proteins after SARS-CoV-2 viral infection is the copy number of different SARS-CoV-2 proteins that are available uptake and presentation by host dendritic cells. Proteomics studies suggest that among the total proteome, structural proteins are abundant in both the virion and in infected cells and among these, the membrane protein and nucleocapsid are the most abundant, while the small envelope protein has the lowest copy number.48,53,54 An additional element that may control protein-specific memory generation is the persistence of the antigen in vivo, either at the local site or in the lymph node.55,56,57 A final factor that may favor initial reactivity and establishment of CD4 T cell memory to structural proteins or even in selective viral proteins such as spike is the role that B cell presentation may play in CD4 T cell expansion and establishment of memory.58,59,60 Thus, proteins with many B cell epitopes such as SARS-CoV-2 spike protein might tend to have an enriched CD4 T cell memory population that is not predicted by their absolute abundance or persistence after infection.
In these studies, a very striking and perhaps the most unexpected finding was an apparent lack of selective recruitment of CD4 T cells established in the CD4 T cell memory pool, presumably accumulated by intermittent and repeated infections with endemic sCoV. The degree of amino acid conservation between SARS-CoV-2 proteins and endemic seasonal β-CoV (HKU1, OC43) varies across the genome and is enriched for some NSP proteins (e.g., NSP 15 (2 methyltransferase), and NSP 13 (helicase)) and for some segments of the structural proteins (e.g., Spike S2 domain and N). The circulating CD4 T cells isolated from PBMC samples collected prior to the emergence of SARS-CoV-2 were almost exclusively activated by peptide pools consisting of SARS-CoV-2-derived peptides with this high degree of sequence relatedness to sCoV. A recent study examining the response to the β-CoV OC43 has provided evidence that when tracked over time in young children, the abundance of circulating memory CD4 T cells diminishes with time and continues this pattern through adulthood.61 Our own studies of human CD4 T cell reactivity to 3 of the seasonal coronaviruses (two alpha CoV, 229E and NL63) and a beta CoV OC43) showed a general pattern of diminished apparent reactivity in older adults relative to younger adults.31 These studies suggest the possibility that repeated and mild infections with endemic CoV may lead to abortive expansion, more rapid contraction or change in the elicited phenotype of the elicited CD4 T cells, as the recent study indicates.61
In any case, proteins that have relative high conservation with SARS-CoV-2 were not particularly immunogenic or preferentially expanded after infection with SARS-CoV-2. For example, very few post-infection samples had detectable responses to the highly conserved NSP proteins, such as helicase, but were highly enriched for reactivity to poorly genetically conserved membrane protein, which was the most immunogenic. Even among the highly dominant spike-specific CD4 T cell responses, CD4 T cell reactivity in the post infection samples was equally balanced across “conserved” and “SARS-CoV-2 unique” peptides. Overall, our data indicate that the first infection with SARS-CoV-2 in humans drives its own CD4 T cell epitope immunodominance hierarchy, driven perhaps by robust innate activation and by the inherent immunogenicity of the various viral proteins.
Studies in animal models have demonstrated the many advantages of memory T cells relative to naive T cells, due at least in part to their higher abundance and lower thresholds of activation (reviewed in62,63,64). However, our studies here provide little evidence for this recall. We can envision at least four explanations for this lack of recall. First, it is possible that the abundant protein expression after SARS-CoV-2 infection leads to an exceptionally high peptide epitope density presented by HLA-class II molecules on APC after infection in the priming lymph node. Under these conditions, differences in sensitivity in CD4 T cell receptor signaling and activation between naive and memory cells might be irrelevant to CD4 T cell activation and expansion, because the threshold of activation is met for all potential responding CD4 T cells, including those from the naive CD4 T cell repertoire. Second, it is possible that the SARS-CoV-2 cross-reactive CD4 T cells are activated and expand but do not produce IFN-γ in the in vitro restimulation assay. We have tested for the production of IL-2 in response to the “spike-conserved” and “spike-unique” peptide pools (Figure S3) and do not find the preferential expansion of CD4 T cells specific for the conserved epitopes by this measure and have not detected alternative cytokines in SARS-CoV2 reactive cells. It is also possible that the breadth of the CD4 T cell response in the acute phase is very broad and includes the memory cells originally elicited by the endemic CoV, but that although these are recruited, they do not expand or rapidly contract or decay in the weeks following infection. It is of course possible that the cross-reactive cells do not readily produce cytokines that are easily measured after SARS-CoV-2 infection, perhaps deviating from this functionality after repeated activation after infections with endemic sCoV over a lifetime prior to the confrontation with SARS-CoV-2 in vivo. It is known that humans experience repeated infections with seasonal CoV, sometimes with the same virus in the same year.65,66,67 These findings suggest that protective T cell immunity to these viruses can be short-lived or not recruited efficiently or with a protective phenotype in the response during re-infection which is common for endemic CoV.68,69 This latter explanation could account for the lack of boosting of specificities of cross-reactive SARS-specific CD4 T cells into the infection with SARS-CoV-2. By the same logic, it is possible that the circulating cross-reactive CD4 T cells are not recruited to the lung draining lymph node after infection due to the loss or attenuation of homing and chemokine receptors that ordinarily recruit T cells to these sites after respiratory infection (reviewed in70,71,72,73), perhaps also as a consequence of repeated activation and expansion after seasonal infection. Finally, it should be noted that any limited recall of memory cells may be masked by an exceptionally robust primary response to SARS-CoV-2 infection. Future experiments focusing on the consequences of repeated infections by seasonal endemic CoV on the longevity and recruitment potential by related sCoV will be useful in understanding these complex issues. In any case, our results suggest that the primary CD4 T cell response to the first SARS-CoV-2 infection is not primarily drawn from the memory pool established by seasonal CoV and may involve largely a naive CD4 T cell population.
Limitations of the study
There are several limitations of this work. First, we have not sampled all potential sources of CD4 T cells elicited by SARS-CoV-2 in vivo within the human subjects, because these were living donors and sampling of lung-localized T cells after recovery is not feasible. Second, we have not studied the same subjects sampled before and after the first infection with SARS-COV-2, and thus we are drawing our conclusions on generalized patterns in CD4 T cell epitope specificity on the large cohorts sampled prior to and after infection. Finally, there may be phenotypic characteristics of recruited sCoV memory CD4 T cells that have diminished our ability to detect them.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Purified anti-human IFNg clone 1-D1K | MabTech | Cat# 3420-3-1000; RRID: AB_907282 |
| Biotinylated anti-human IFNg clone 7-B6-1 | MabTech | Cat# 3420-6-250; RRID: AB_907272 |
| Streptavidin conjugated alkaline phosphatase | Jackson ImmunoResearch | Cat# 016-050-084 |
| Purified anti-human IL-2 clone MT2A91/2C95 | MabTech | Cat# 3445-3-1000 |
| Biotinylated anti-human IL-2 clone MT8G10 | MabTech | Cat# 3445-6-250 |
| Biological samples | ||
| PMBC from SARS-CoV-2 convalescent subjects | P. Wilson, University of Chicago, clinical trial identifier NCT04340050 | N/A |
| PMBC from healthy adults collected prior to winter 2019 | University of Rochester Medical Center, Healthy Donor Protocol 14-0064. | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Synthetic peptide set Sin Nombre Virus, NM-H10 Glycoprotein Precursor protein | BEI Resources | NR-4764 |
| Overlapping peptide set, 17-mers with 11aa overlap, NSP1 sequence from accession number YP_009725297.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, NSP2 sequence from accession number YP_009725298.1 | Genscript | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, NSP3 sequence from accession number YP_009725299.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, NSP4 sequence from accession number YP_009725300.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, NSP5 sequence from accession number YP_009725301.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, NSP6 sequence from accession number YP_009725302.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, NSP7 sequence from accession number YP_009725303.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, NSP8 sequence from accession number YP_009725304.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, NSP9 sequence from accession number YP_009725305.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, NSP10 sequence from accession number YP_009725306.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, NSP12 sequence from accession number YP_009725307.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, NSP13 sequence from accession number YP_009725308.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, NSP14 sequence from accession number YP_009725309.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, NSP15 sequence from accession number YP_009725310.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, NSP16 sequence from accession number YP_009725311.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, ORF3 sequence from accession number QHD43417.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, ORF6 sequence from accession number QHD43420.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, ORF7a sequence from accession number QHD43421.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, ORF8 sequence from accession number QDH43422.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, ORF10 sequence from accession number QHI42199.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, Spike sequence from accession number QHD43416.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, Nucleocapsid sequence from accession number QHD43423.2 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, Membrane sequence from accession number QHD43419.1 | Mimotopes | N/A |
| Overlapping peptide set, 17-mers with 11aa overlap, Envelope sequence from accession number QHD43418.1 | Mimotopes | N/A |
| Critical commercial assays | ||
| CD8 microbeads, human | Miltenyi Biotec | 130-045-201 |
| CD56 microbeads, human | Miltenyi Biotec | 130-097-042 |
| LD columns | Miltenyi Biotec | 130-042-901 |
| Vector Blue Substrate Kit | Vector Labs | SK-5300 |
| Software and algorithms | ||
| ImmunoSpot Reader series 5.2 and ImmunoSpot software v5.1 | Cellular Technology Limited | N/A |
| GraphPad Prism 10 v10.2.1 | GraphPad | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Andrea J. Sant (andrea_sant@urmc.rochester.edu).
Materials availability
This study did not generate new unique reagents. Requests for materials should be directed to the lead contact.
Data and code availability
The full complement of data accumulated for these studies is available from the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Cohort 1
SARS-CoV-2 infected cohort. Frozen PBMC from 20 SARS-CoV-2 infected adults were received from Dr. Patrick Wilson, University of Chicago, where samples were collected following approval of the University of Chicago institutional review board IRB20-0523 and University of Chicago, University of Wisconsin-Madison, and Washington University in St. Louis institutional biosafety committees. Informed consent was obtained after the research applications and possible consequences of the studies were disclosed to study subjects. This clinical trial was registered at ClinicalTrials.gov with identifier NCT04340050. Subjects were over the age of 18 and had a PCR-confirmed SARS-CoV-2 infection. All samples were collected in April and May of 2020 at a convalescent timepoint (D35-53 post symptom onset) as described in21,25. Details regarding age, sex, disease severity, duration of symptoms and time of blood draw are indicated in Figure 1. Subjects ranged in age from 22 to 76 years and there is equal representation of males and females (10 each). No additional subject information was collected for this study. This data does not evaluate associations with sex or gender.
Cohort 2
Healthy adult cohort, following approval from DMID and the University of Rochester Research Subjects Review Boards (protocol 14-0064), blood was obtained from 32 healthy adult subjects aged 18-70 residing in the Rochester, NY area, that had provided informed consent between spring 2014 and spring 2019. Details regarding age, sex and time of blood draws are indicated in Figure 7. Subjects used in this study ranged in age from 22 to 68 years and both sexes are represented with 13 males and 19 females. These are the only demographic information collected for this study. This data does not evaluate associations with sex or gender.
Method details
Synthetic peptides
Overlapping peptide sets encompassing the translated sequence of the SARS-CoV-2 proteins indicated in the figures were obtained from Mimotopes (Mulgrave Australia) and Genscript (Piscataway, NJ). The sequence of the SARS-CoV-2 proteins used for derivation of peptide pools, their accession numbers and their nomenclature in the literature is indicated in Table S1. A peptide pool made with peptides from Sin Nombre Virus, NM-H10, Glycoprotein Precursor protein (BEIR, NR-4764) was used as a negative control. Individual lyophilized 17-mer peptides were reconstituted in DMSO and water based on solubility, with or without DTT based on cysteine containing sequences at a final concentration of 10 mM. Peptides were pooled and diluted into specific pools (Tables S1 and S2). Peptides from the SARS-CoV-2 proteome were divided into pools representing each protein, with larger proteins divided into multiple pools, such as NSP3 divided into two pools, to keep the number of peptides in any given pool below 200 peptides. The spike protein was divided into three pools representing the subdomains of the protein (S1-RBD, RBD and S2) to probe the different segments of the spike protein individually. Additionally, two curated pools of peptides from the spike protein were designed to evaluate responses to unique peptides, defined as peptide sequences that did not have more than 4 contiguous amino acids shared with sCoV OC43 spike protein, or conserved peptides, defined as peptide sequences that had at least 6 shared amino acids when aligned with the sCoV OC43 spike sequence. These pools only contained a subset of the peptides within the spike protein (Table S2). Peptides were pooled with each peptide present at a final concentration of 0.5 μM in assays.
Isolation of PBMCs from human blood
For the healthy adult cohort, following approval from DMID and the University of Rochester Research Subjects Review Boards (protocol 14-0064), blood was obtained from 32 healthy adult subjects aged 18-70 residing in the Rochester, NY area, that had provided informed consent between spring 2014 and spring 2019 (Figure 7). PBMCs were isolated from drawn blood samples and frozen in fetal calf serum containing 10% DMSO at 15 million cells/ml. For the SARS-CoV-2 infected cohort (Figure 1) cells were received from Dr. Patrick Wilson, University of Chicago where samples were collected following approval of the University of Chicago institutional review board IRB20-0523 and University of Chicago, University of Wisconsin-Madison, and Washington University in St. Louis institutional biosafety committees. Informed consent was obtained after the research applications and possible consequences of the studies were disclosed to study subjects. This clinical trial was registered at ClinicalTrials.gov with identifier NCT04340050. Subjects were over the age of 18 and had a PCR-confirmed SARS-CoV-2 infection. Peripheral blood mononuclear cells were isolated from leukoreduction filters and frozen in fetal calf serum containing 10% DMSO from subjects that had a PCR confirmed infection in April and May 2020 at a convalescent timepoint (D35-53 post symptom onset) as described in.21,25
EliSpot assay
After thawing and overnight rest in RPMI media containing 10% FBS cultured at 37°C and 5% CO, PBMC were washed and depleted of CD8 and CD56 cells using MACS microbeads per manufacturer instructions (Miltenyi Biotec). EliSpot assays were performed as previously described.31 Briefly, CD8- and CD56-depleted PBMCs (250,000-350,000 cells per well) were cultured with pools of peptides on plates coated with 10 μg/ml anti-human IFNγ or anti-human IL-2 (clone 1-D1K, or clone MT2A91/2C95, respectively, MabTech) for 36 hr at 37°C, 5% CO2. After incubation, plates were washed and incubated with biotinylated detection antibodies human IFNγ or IL-2 (2 μg/ml, clone 7-B6-1, and 1 μg/ml clone MT8G10, respectively, MabTech) in wash buffer with 10% FBS for 2hr followed by washing and incubation for 30min with streptavidin conjugated-alkaline phosphatase (1:1000 dilution in wash buffer with 10% FBS, JacksonImmuno Labs) and development using Vector Blue substrate in 100mM Tris, pH 8.2 (Vector Labs) to detect mediators as previously described.20 Quantification of cytokine-secreting cells was performed with an Immunospot reader series 5.2, using Immunospot software, v5.1. Data are presented as the frequency of mediator-producing cells per million CD8- and CD56-depleted PBMCs with background response subtracted. All responses with background subtracted are shown, including zero values, a median response that is three-fold over background (average background 5 spots) is considered a positive response. Any statistical treatment of data is described in the legend of the respective figure. All statistical analysis was completed using Graphpad Prism 10 v10.2.1.
Quantification and statistical analysis
ELISPOT assays were imaged using an Immunospot reader series 5.2, and spots were counted using Immunospot software, v5.1. Data are presented as the frequency of mediator-producing cells per million CD8- and CD56-depleted PBMCs with background response subtracted. Shown are both the median values (bars) and individual responses (symbols), unless otherwise indicated in the figure legend. All responses with background subtracted are shown, including zero values. A median response that is three-fold over background (average background 5 spots) is considered a positive response. Statistical analyses included calculations of r and p values using the Spearman correlation, The Mann-Whitney test comparing the two cohorts, the Wilcoxon test for paired analysis, and multiple unpaired t tests with Welch correction to calculate p values for different comparative analyses. Any calculated p value is indicated and those that reach significance are indicated in red font. Specific statistical treatment of data is described in the legend of the respective figure. All statistical analysis was completed using Graphpad Prism 10 v10.2.1.
Additional resources
The SARS-CoV-2 convalescent samples were collected as part of the clinical trial registered at ClinicalTrials.gov with identifier NCT04340050. https://classic.clinicaltrials.gov/ct2/show/study/NCT04340050.
Acknowledgments
We thank Maria Lucia Madariaga and Kumaran Shanmugarajah at the University of Chicago Dept. of Medicine for obtaining SARS-CoV-2 patient samples. The authors would like to acknowledge the contribution of the clinical core in enrolling study subjects, and for processing clinical samples. We also recognize our study participants, for their willingness to contribute to scientific research. Finally, we thank Samantha Conflitti for her help with the graphical abstract. This work was supported with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, U01 [AI14461602S1], the NIAID Centers of Excellence for Influenza Research and Response (CEIRR-CIDER) contract number 75N93021C00018 and NIAD Collaborative Influenza Vaccine Innovation Centers (CIVR-HRP) Contract number 75N93019C00052 sub-contracts to AJS. National Institute of Allergy and Infectious Diseases (NIAID); National Institutes of Health grant numbers 1U19AI168632 (PCW), P01AI165077 (PCW), 5U01AI144616 (PCW), and the NIAID Centers of Excellence for Influenza Research and Response (CEIRR) grant number 75N93019R00028 (PCW), and the NIAD Collaborative Influenza Vaccine Innovation Centers (CIVIC) grant number 75N93019C00051 (PCW). This work was supported with ALSAC funds at St. Jude (PT), and with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, U01 [AI144616], and contracts 75N93019C00052 (CIVR-HRP), 75N93021C00016 (SJCEIRR) to PT.
Author contributions
KAR, AJS, PW, and PGT: conceptualization of studies. PGT: funding acquisition and provision of unique reagents. KAR and AJS data analyses and visualization. KAR and AJS: writing of the article, KAR, AJS, PW, and PGT editing of article, KAR, AJS, PW, and SC: recruitment of subjects and curation of samples. All authors approved the submitted article.
Declaration of interests
P.G.T. has consulted or received travel support from JNJ, Pfizer, Illumina, 10X Genomics, Merck, and PACT Pharma and served on the SAB of Immunoscape, Shennon Bio, and Cytoagents.
No other authors have competing interests.
Published: May 17, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109992.
Supplemental information
References
- 1.Grifoni A., Sidney J., Vita R., Peters B., Crotty S., Weiskopf D., Sette A. SARS-CoV-2 human T cell epitopes: Adaptive immune response against COVID-19. Cell Host Microbe. 2022;30:1788. doi: 10.1016/j.chom.2022.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sette A., Sidney J., Crotty S. T Cell Responses to SARS-CoV-2. Annu. Rev. Immunol. 2023;41:343–373. doi: 10.1146/annurev-immunol-101721-061120. [DOI] [PubMed] [Google Scholar]
- 4.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R., Moore E., Rubiro P., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strazar M., Park J., Abelin J.G., Taylor H.B., Pedersen T.K., Plichta D.R., Brown E.M., Eraslan B., Hung Y.M., Ortiz K., et al. HLA-II immunopeptidome profiling and deep learning reveal features of antigenicity to inform antigen discovery. Immunity. 2023;56:1681–1698.e13. doi: 10.1016/j.immuni.2023.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uttamrao P.P., Sathyaseelan C., Patro L.P.P., Rathinavelan T. Revelation of Potent Epitopes Present in Unannotated ORF Antigens of SARS-CoV-2 for Epitope-Based Polyvalent Vaccine Design Using Immunoinformatics Approach. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.692937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson R.W., Chen Y., Venezia O.L., Majerus R.M., Shin D.S., MGH COVID-19 Collection & Processing Team. Carrington M.N., Yu X.G., Wesemann D.R., Moon J.J., Luster A.D. SARS-CoV-2 epitope-specific CD4(+) memory T cell responses across COVID-19 disease severity and antibody durability. Sci. Immunol. 2022;7 doi: 10.1126/sciimmunol.abl9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansson A.M., Malhotra U., Kim Y.G., Gomez R., Krist M.P., Wald A., Koelle D.M., Kwok W.W. Cross-reactive and mono-reactive SARS-CoV-2 CD4+ T cells in prepandemic and COVID-19 convalescent individuals. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1010203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogbe A., Kronsteiner B., Skelly D.T., Pace M., Brown A., Adland E., Adair K., Akhter H.D., Ali M., Ali S.E., et al. T cell assays differentiate clinical and subclinical SARS-CoV-2 infections from cross-reactive antiviral responses. Nat. Commun. 2021;12:2055. doi: 10.1038/s41467-021-21856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang X.L., Wang G.L., Zhao X.N., Yan F.H., Yao L., Kou Z.Q., Ji S.X., Zhang X.L., Li C.B., Duan L.J., et al. Lasting antibody and T cell responses to SARS-CoV-2 in COVID-19 patients three months after infection. Nat. Commun. 2021;12:897. doi: 10.1038/s41467-021-21155-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Law J.C., Watts T.H. Considerations for Choosing T Cell Assays during a Pandemic. J. Immunol. 2023;211:169–174. doi: 10.4049/jimmunol.2300129. [DOI] [PubMed] [Google Scholar]
- 14.Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Low J.S., Vaqueirinho D., Mele F., Foglierini M., Jerak J., Perotti M., Jarrossay D., Jovic S., Perez L., Cacciatore R., et al. Clonal analysis of immunodominance and cross-reactivity of the CD4 T cell response to SARS-CoV-2. Science. 2021;372:1336–1341. doi: 10.1126/science.abg8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehmann A.A., Kirchenbaum G.A., Zhang T., Reche P.A., Lehmann P.V. Deconvoluting the T Cell Response to SARS-CoV-2: Specificity Versus Chance and Cognate Cross-Reactivity. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.635942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M., Lübke M., Bauer J., Rieth J., Wacker M., et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 18.Quiros-Fernandez I., Poorebrahim M., Fakhr E., Cid-Arregui A. Immunogenic T cell epitopes of SARS-CoV-2 are recognized by circulating memory and naive CD8 T cells of unexposed individuals. EBioMedicine. 2021;72 doi: 10.1016/j.ebiom.2021.103610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 20.Richards K.A., Treanor J.J., Nayak J.L., Sant A.J. Overarching Immunodominance Patterns and Substantial Diversity in Specificity and Functionality in the Circulating Human Influenza A and B CD4 T Cell Repertoire. J. Infect. Dis. 2018;218:1169–1174. doi: 10.1093/infdis/jiy288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guthmiller J.J., Stovicek O., Wang J., Changrob S., Li L., Halfmann P., Zheng N.Y., Utset H., Stamper C.T., Dugan H.L., et al. SARS-CoV-2 Infection Severity Is Linked to Superior Humoral Immunity against the Spike. mBio. 2021;12 doi: 10.1128/mBio.02940-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ripoll J.G., van Helmond N., Senefeld J.W., Wiggins C.C., Klassen S.A., Baker S.E., Larson K.F., Murphy B.M., Andersen K.J., Ford S.K., et al. Convalescent Plasma for Infectious Diseases: Historical Framework and Use in COVID-19. Clin. Microbiol. Newsl. 2021;43:23–32. doi: 10.1016/j.clinmicnews.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloch E.M., Shoham S., Casadevall A., Sachais B.S., Shaz B., Winters J.L., van Buskirk C., Grossman B.J., Joyner M., Henderson J.P., et al. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J. Clin. Invest. 2020;130:2757–2765. doi: 10.1172/JCI138745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casadevall A., Pirofski L.A., Joyner M.J. The Principles of Antibody Therapy for Infectious Diseases with Relevance for COVID-19. mBio. 2021;12 doi: 10.1128/mBio.03372-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dugan H.L., Stamper C.T., Li L., Changrob S., Asby N.W., Halfmann P.J., Zheng N.Y., Huang M., Shaw D.G., Cobb M.S., et al. Profiling B cell immunodominance after SARS-CoV-2 infection reveals antibody evolution to non-neutralizing viral targets. Immunity. 2021;54:1290–1303.e7. doi: 10.1016/j.immuni.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Changrob S., Fu Y., Guthmiller J.J., Halfmann P.J., Li L., Stamper C.T., Dugan H.L., Accola M., Rehrauer W., Zheng N.Y., et al. Cross-Neutralization of Emerging SARS-CoV-2 Variants of Concern by Antibodies Targeting Distinct Epitopes on Spike. mBio. 2021;12 doi: 10.1128/mBio.02975-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ranieri E., Netti G.S., Gigante M. CTL ELISPOT Assay and T Cell Detection. Methods Mol. Biol. 2021;2325:65–77. doi: 10.1007/978-1-0716-1507-2_5. [DOI] [PubMed] [Google Scholar]
- 28.Yang F., Patton K., Kasprzyk T., Long B., Gupta S., Zoog S.J., Tracy K., Vettermann C. Validation of an IFN-gamma ELISpot assay to measure cellular immune responses against viral antigens in non-human primates. Gene Ther. 2022;29:41–54. doi: 10.1038/s41434-020-00214-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards K.A., Moritzky S., Shannon I., Fitzgerald T., Yang H., Branche A., Topham D.J., Treanor J.J., Nayak J., Sant A.J. Recombinant HA-based vaccine outperforms split and subunit vaccines in elicitation of influenza-specific CD4 T cells and CD4 T cell-dependent antibody responses in humans. NPJ Vaccines. 2020;5:77. doi: 10.1038/s41541-020-00227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moritzky S.A., Richards K.A., Glover M.A., Krammer F., Chaves F.A., Topham D.J., Branche A., Nayak J.L., Sant A.J. The Negative Effect of Preexisting Immunity on Influenza Vaccine Responses Transcends the Impact of Vaccine Formulation Type and Vaccination History. J. Infect. Dis. 2023;227:381–390. doi: 10.1093/infdis/jiac068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards K.A., Glover M., Crawford J.C., Thomas P.G., White C., Sant A.J. Circulating CD4 T Cells Elicited by Endemic Coronaviruses Display Vast Disparities in Abundance and Functional Potential Linked to Antigen Specificity and Age. J. Infect. Dis. 2021;223:1555–1563. doi: 10.1093/infdis/jiab076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loyal L., Braun J., Henze L., Kruse B., Dingeldey M., Reimer U., Kern F., Schwarz T., Mangold M., Unger C., et al. Cross-reactive CD4(+) T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science. 2021;374 doi: 10.1126/science.abh1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazarevic I., Pravica V., Miljanovic D., Cupic M. Immune Evasion of SARS-CoV-2 Emerging Variants: What Have We Learnt So Far? Viruses. 2021;13 doi: 10.3390/v13071192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez-Vazquez L.D., Arenas M. Molecular Evolution of SARS-CoV-2 during the COVID-19 Pandemic. Genes. 2023;14 doi: 10.3390/genes14020407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chatterjee S., Bhattacharya M., Nag S., Dhama K., Chakraborty C. A Detailed Overview of SARS-CoV-2 Omicron: Its Sub-Variants, Mutations and Pathophysiology, Clinical Characteristics, Immunological Landscape, Immune Escape, and Therapies. Viruses. 2023;15 doi: 10.3390/v15010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tye E.X.C., Jinks E., Haigh T.A., Kaul B., Patel P., Parry H.M., Newby M.L., Crispin M., Kaur N., Moss P., et al. Mutations in SARS-CoV-2 spike protein impair epitope-specific CD4(+) T cell recognition. Nat. Immunol. 2022;23:1726–1734. doi: 10.1038/s41590-022-01351-7. [DOI] [PubMed] [Google Scholar]
- 37.Mazzoni A., Vanni A., Spinicci M., Capone M., Lamacchia G., Salvati L., Coppi M., Antonelli A., Carnasciali A., Farahvachi P., et al. SARS-CoV-2 Spike-Specific CD4+ T Cell Response Is Conserved Against Variants of Concern, Including Omicron. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.801431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., Bloom N.I., Goodwin B., Phillips E., Mallal S., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185:847–859.e11. doi: 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Y., Cai C., Grifoni A., Müller T.R., Niessl J., Olofsson A., Humbert M., Hansson L., Österborg A., Bergman P., et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat. Med. 2022;28:472–476. doi: 10.1038/s41591-022-01700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarke A., Sidney J., Methot N., Yu E.D., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., Wang E., et al. Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grifoni A., Sette A. From Alpha to omicron: The response of T cells. Curr. Res. Immunol. 2022;3:146–150. doi: 10.1016/j.crimmu.2022.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kedzierska K., Thomas P.G. Count on us: T cells in SARS-CoV-2 infection and vaccination. Cell Rep. Med. 2022;3 doi: 10.1016/j.xcrm.2022.100562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pothast C.R., Dijkland R.C., Thaler M., Hagedoorn R.S., Kester M.G.D., Wouters A.K., Hiemstra P.S., van Hemert M.J., Gras S., Falkenburg J.H.F., Heemskerk M.H.M. SARS-CoV-2-specific CD4(+) and CD8(+) T cell responses can originate from cross-reactive CMV-specific T cells. Elife. 2022;11 doi: 10.7554/eLife.82050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eggenhuizen P.J., Ng B.H., Chang J., Cheong R.M.Y., Yellapragada A., Wong W.Y., Ting Y.T., Monk J.A., Gan P.Y., Holdsworth S.R., Ooi J.D. Heterologous Immunity Between SARS-CoV-2 and Pathogenic Bacteria. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.821595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vinjamuri S., Li L., Bouvier M. SARS-CoV-2 ORF8: One protein, seemingly one structure, and many functions. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1035559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arduini A., Laprise F., Liang C. SARS-CoV-2 ORF8: A Rapidly Evolving Immune and Viral Modulator in COVID-19. Viruses. 2023;15 doi: 10.3390/v15040871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Münch C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grenga L., Gallais F., Pible O., Gaillard J.C., Gouveia D., Batina H., Bazaline N., Ruat S., Culotta K., Miotello G., et al. Shotgun proteomics analysis of SARS-CoV-2-infected cells and how it can optimize whole viral particle antigen production for vaccines. Emerg. Microb. Infect. 2020;9:1712–1721. doi: 10.1080/22221751.2020.1791737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker R., Partridge T., Wormald C., Kawahara R., Stalls V., Aggelakopoulou M., Parker J., Powell Doherty R., Ariosa Morejon Y., Lee E., et al. Mapping the SARS-CoV-2 spike glycoprotein-derived peptidome presented by HLA class II on dendritic cells. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mudd P.A., Minervina A.A., Pogorelyy M.V., Turner J.S., Kim W., Kalaidina E., Petersen J., Schmitz A.J., Lei T., Haile A., et al. SARS-CoV-2 mRNA vaccination elicits a robust and persistent T follicular helper cell response in humans. Cell. 2022;185:603–613.e15. doi: 10.1016/j.cell.2021.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wragg K.M., Lee W.S., Koutsakos M., Tan H.X., Amarasena T., Reynaldi A., Gare G., Konstandopoulos P., Field K.R., Esterbauer R., et al. Establishment and recall of SARS-CoV-2 spike epitope-specific CD4(+) T cell memory. Nat. Immunol. 2022;23:768–780. doi: 10.1038/s41590-022-01175-5. [DOI] [PubMed] [Google Scholar]
- 52.Swaminathan S., Lineburg K.E., Ambalathingal G.R., Crooks P., Grant E.J., Mohan S.V., Raju J., Panikkar A., Le Texier L., Tong Z.W.M., et al. Limited Recognition of Highly Conserved Regions of SARS-CoV-2. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.02780-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duart G., García-Murria M.J., Grau B., Acosta-Cáceres J.M., Martínez-Gil L., Mingarro I. SARS-CoV-2 envelope protein topology in eukaryotic membranes. Open Biol. 2020;10 doi: 10.1098/rsob.200209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blair D.A., Turner D.L., Bose T.O., Pham Q.M., Bouchard K.R., Williams K.J., McAleer J.P., Cauley L.S., Vella A.T., Lefrançois L. Duration of antigen availability influences the expansion and memory differentiation of T cells. J. Immunol. 2011;187:2310–2321. doi: 10.4049/jimmunol.1100363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams M.A., Bevan M.J. Shortening the infectious period does not alter expansion of CD8 T cells but diminishes their capacity to differentiate into memory cells. J. Immunol. 2004;173:6694–6702. doi: 10.4049/jimmunol.173.11.6694. [DOI] [PubMed] [Google Scholar]
- 57.Swain S.L., Jones M.C., Devarajan P., Xia J., Dutton R.W., Strutt T.M., McKinstry K.K. Durable CD4 T-Cell Memory Generation Depends on Persistence of High Levels of Infection at an Effector Checkpoint that Determines Multiple Fates. Cold Spring Harbor Perspect. Biol. 2021;13 doi: 10.1101/cshperspect.a038182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Welsh R.A., Song N., Sadegh-Nasseri S. How Does B Cell Antigen Presentation Affect Memory CD4 T Cell Differentiation and Longevity? Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.677036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Linton P.J., Harbertson J., Bradley L.M. A critical role for B cells in the development of memory CD4 cells. J. Immunol. 2000;165:5558–5565. doi: 10.4049/jimmunol.165.10.5558. [DOI] [PubMed] [Google Scholar]
- 60.Barr T.A., Gray M., Gray D. B cells: programmers of CD4 T cell responses. Infect. Disord. Drug Targets. 2012;12:222–231. doi: 10.2174/187152612800564446. [DOI] [PubMed] [Google Scholar]
- 61.Humbert M., Olofsson A., Wullimann D., Niessl J., Hodcroft E.B., Cai C., Gao Y., Sohlberg E., Dyrdak R., Mikaeloff F., et al. Functional SARS-CoV-2 cross-reactive CD4(+) T cells established in early childhood decline with age. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2220320120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kunzli M., Masopust D. CD4(+) T cell memory. Nat. Immunol. 2023;24:903–914. doi: 10.1038/s41590-023-01510-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pepper M., Jenkins M.K. Origins of CD4(+) effector and central memory T cells. Nat. Immunol. 2011;12:467–471. doi: 10.1038/ni.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krueger P.D., Osum K.C., Jenkins M.K. CD4(+) Memory T-Cell Formation during Type 1 Immune Responses. Cold Spring Harbor Perspect. Biol. 2021;13 doi: 10.1101/cshperspect.a038141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edridge A.W.D., Kaczorowska J., Hoste A.C.R., Bakker M., Klein M., Loens K., Jebbink M.F., Matser A., Kinsella C.M., Rueda P., et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- 66.Galanti M., Shaman J. Direct Observation of Repeated Infections With Endemic Coronaviruses. J. Infect. Dis. 2021;223:409–415. doi: 10.1093/infdis/jiaa392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petrie J.G., Bazzi L.A., McDermott A.B., Follmann D., Esposito D., Hatcher C., Mateja A., Narpala S.R., O'Connell S.E., Martin E.T., Monto A.S. Coronavirus Occurrence in the Household Influenza Vaccine Evaluation (HIVE) Cohort of Michigan Households: Reinfection Frequency and Serologic Responses to Seasonal and Severe Acute Respiratory Syndrome Coronaviruses. J. Infect. Dis. 2021;224:49–59. doi: 10.1093/infdis/jiab161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galanti M., Shaman J. Direct Observation of Repeated Infections With Endemic Coronaviruses. J. Infect. Dis. 2021;223:409–415. doi: 10.1093/infdis/jiaa392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kiyuka P.K., Agoti C.N., Munywoki P.K., Njeru R., Bett A., Otieno J.R., Otieno G.P., Kamau E., Clark T.G., van der Hoek L., et al. Human Coronavirus NL63 Molecular Epidemiology and Evolutionary Patterns in Rural Coastal Kenya. J. Infect. Dis. 2018;217:1728–1739. doi: 10.1093/infdis/jiy098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gregor C.E., Foeng J., Comerford I., McColl S.R. Chemokine-Driven CD4(+) T Cell Homing: New Concepts and Recent Advances. Adv. Immunol. 2017;135:119–181. doi: 10.1016/bs.ai.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 71.Griffith J.W., Sokol C.L., Luster A.D. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu. Rev. Immunol. 2014;32:659–702. doi: 10.1146/annurev-immunol-032713-120145. [DOI] [PubMed] [Google Scholar]
- 72.Rahimi R.A., Luster A.D. Chemokines: Critical Regulators of Memory T Cell Development, Maintenance, and Function. Adv. Immunol. 2018;138:71–98. doi: 10.1016/bs.ai.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Groom J.R. Regulators of T-cell fate: Integration of cell migration, differentiation and function. Immunol. Rev. 2019;289:101–114. doi: 10.1111/imr.12742. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full complement of data accumulated for these studies is available from the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.