Abstract
Due to the growing demand for natural carotenoids, researchers have been searching for strains that are capable of efficient synthesis of these compounds. This study tested 114 red yeast strains collected from various natural environments and food specimens in Poland. The strains were isolated by their ability to produce red or yellow pigments in rich nutrient media. According to potential industrial significance of the carotenoids, both their total production and share of individual carotenoids (β-carotene, γ-carotene, torulene, and torularhodin) were analyzed. The total content of carotenoid pigments in the yeast dry matter ranged from 13.88 to 406.50 µg/g, and the percentages of individual carotenoids highly varied among the strains. Most of the yeast isolates synthesized torulene at the highest amount. Among the studied strains, isolates with a total carotenoid content in biomass greater than 200 µg/g and those containing more than 60% torularhodin were selected for identification (48 strains). The identified strains belonged to six genera: Rhodotorula, Sporidiobolus, Sporobolomyces, Buckleyzyma, Cystofilobasidium, and Erythrobasidium. The largest number of isolates belonged to Rhodotorula babjevae (18), Rhodotorula mucilaginosa (7), Sporidiobolus pararoseus (4), and Rhodotorula glutinis (4).
Keywords: Red yeast, Carotenoid pigments, Torulene, Torularhodin, Rhodotorula
Introduction
Carotenoids are a group of compounds widely used in pharmaceutical, chemical, food, and feed industries due to their ability to act as vitamin A precursors as well as their coloring, antioxidant, and potentially disease-inhibiting properties. Many species of yeasts classified under Basidiomycota, which grow as pigmented colonies and are therefore known as “red yeast,” are capable of producing carotenoids. These include genera such as Rhodotorula, Rhodosporidium, Sporobolomyces, Sporidiobolus, Xanthophyllomyces, and Cystofilobasidium. These yeasts synthesize compounds including astaxanthin, β-carotene, torulene, or torularhodin, which give their colonies a characteristic orange, salmon, pink, or red color [1].
Humans and animals cannot synthesize carotenoids; therefore, these compounds must be supplied in the diet. Because various microorganisms can synthesize several important carotenoids, they are recognized as a potential natural source of these compounds. Biotechnologically produced carotenoids are an alternative to synthetic compounds, which, according to studies, may have negative effects on health, and even increase the risk of cancer when consumed in excess [2]. Currently, the microalga Haematococcus pluvialis and the mold Blakeslea trispora are mainly used for the biotechnological production of carotenoids (astaxanthin and β-carotene, respectively) [3].
β-Carotene belongs to the group of nonoxidized carotenoids and is one of the widely studied pigments. It is composed of two β-ionone rings connected together by a polyene chain consisting of nine conjugated double bonds [4]. β-Carotene is a strong antioxidant that can protect cells against damage by light and oxygen. It exhibits the highest provitamin A activity compared to other carotenoids. One molecule of β-carotene is converted into two molecules of vitamin A, and thus, this compound is considered as an important source of vitamin A for humans. β-Carotene is a fat-soluble, red-orange pigment used not only in the food industry but also as a feed additive and in the cosmetic industry [5]. Some yeasts of the genera Rhodotorula, Sporobolomyces, and Sporidiobolus are efficient producers of this compound [3].
Torulene and torularhodin are fungal carotenoids. Torulene contains one β-ionone ring attached to a polyene chain. It also has 13 double bonds, and thus exhibits stronger antioxidant properties than β-carotene. As β-ionone ring is the backbone of vitamin A, torulene can act as a precursor of this vitamin. This orange pigment is predominantly produced by yeasts of the genera Rhodotorula, Sporidiobolus, and Sporobolomyces [6].
Torularhodin is a red carotenoid produced mainly by Rhodotorula and Sporobolomyces yeasts. Due to the presence of a carboxyl group at the end of the polyene chain, this compound is classified under the group of xanthophylls. Torularhodin is produced as a result of the oxidation of torulene. It is a potent antioxidant that helps to stabilize cell membranes under stressful conditions. Torularhodin can more effectively quench singlet oxygen than β-carotene, remove free radicals, and inhibit the formation of lipid peroxides, protecting cells, among others, against tumors. Moreover, this compound exhibits antimicrobial activity against both Gram-positive and Gram-negative bacteria [7].
Red yeasts are widespread in nature. These organisms can be found in flowers, air, fruit, and leaves, and can contaminate foods such as Greek yogurt, cream, and ground pork. Red yeasts have previously been isolated from the natural environment on all continents [8], including extreme environments such as Antarctic glaciers [9, 10].
During the study, 408 specimens were sampled from various natural environments and food products in Poland. The goals of research were to study distribution and diversity of red yeasts and, as well, to study production of carotenoids by the isolates for their possible industrial application.
Materials and Methods
Isolation of Red Yeast
Yeast isolation was carried out from February 2019 to September 2020 in Poland (Fig. 1). 408 specimens were sampled from various sources for isolation of red yeasts: air (124), flowers (78), tree leaves and bark (56), fruit (56), tree slime fluxes (31), propolis (15), cereals (13), soil (9), vegetables (8), bee pollen (6), cheese (4), meat (3), bee bread (2), the Baltic Sea water (2), and chocolate (1). For the isolation of yeast from the air, Petri dishes containing the YPD medium were left open for 15 min and then incubated at 22℃ for 96 h. Other materials were placed in sterile bags or containers for their transportation to the research laboratory. Samples that could not be immediately transported to the laboratory after collection were stored at − 18℃. The samples were transferred to 100 mL of liquid YPD medium (20 g/L glucose, 20 g/L peptone, 10 g/L yeast extract, BTL, Poland) and incubated at 22℃ for 48 h. After incubation, the proliferated material was inoculated into Sabouraud agar with chloramphenicol (BTL, Poland), and then incubated again at 22℃ for 96 h. After incubation, orange-, red-, or pink-colored colonies were observed and were reductively transferred to Sabouraud agar with chloramphenicol for the isolation of pure yeast cultures. The obtained yeast isolates were taken by loop, transferred to 80 mL of liquid YPD medium, grown on a shaker (New Brunswick™ Innova® 44R, Eppendorf) at 140 rpm for 24 or 48 h at 22℃, and then frozen with glycerol (50%, v/v) at − 85℃.
Fig. 1.
Cities in Poland where samples were taken for the isolation of red yeasts
Culture Conditions and Media
Pure yeast cultures were streaked on YPD agar medium and incubated at 22℃ for 96 h. After incubation, a yeast colony was taken using a loop and transferred to a liquid medium (glucose 30 g/L, yeast extract 10 g/L, KH2PO4 1.5 g/L, MgSO4∙7H2O 1 g/L, (NH4)2SO4 5 g/L, CaCl2 0.15 g/L, pH 5.6) in 500-mL flatbottomed flasks. The isolates were grown on a shaker (New Brunswick™ Innova® 44R, Eppendorf) at 140 rpm for 96 h at 22℃.
Biomass Yield
To determine the biomass yield, 10 mL of yeast cultures was centrifuged (Eppendorf Centrifuge 5804R) at 10,000 rpm for 10 min. The resulting supernatant was removed, and the samples were dried at 105℃ for 24 h. The results below are presented as grams of dry matter per liter of the medium (g/L).
Determination of Carotenoid Content by Spectrophotometric Method
To determine the carotenoid content, 1 mL of liquid yeast culture was centrifuged (10,000 rpm for 10 min), suspended in 2 mL of dimethyl sulfoxide, and 0.5 mm glass beads were added to disrupt yeast cells. The samples were shaken for 1 h at 70 rpm (Rotator Multi Bio RS-24, Biosan) to extract intracellular carotenoids. Then, 2 mL of acetone, petroleum ether (with 0.25% BHT), and 20% sodium chloride were added. The samples thus prepared were repeatedly shaken for 1 h and centrifuged at 3500 rpm for 5 min to separate the phases of the mixture. The colored ether phase was transferred to a cuvette, and its absorbance was measured in a spectrophotometer at a wavelength of 490 nm. The total carotenoid content (µg/gd.m.) was calculated using the formula = Amax × D × V/(E × W), where Amax is the absorbance at 490 nm, D is the sample dilution factor, V is the volume of the ether phase, E is the carotenoids extinction coefficient (0.16), and W values were calculated from the biomass yield data. If the yeast biomass remained colored after the first extraction step, the process was repeated analogously with 2 mL of DMSO [11, 12].
Analysis of the Carotenoid Profile by HPLC
To determine the percentages of β-carotene, γ-carotene, torulene, and torularhodin in the total pool of carotenoids, the samples were extracted using the same method applied for the determination of the total content of carotenoids by spectrophotometric method. Then, petroleum ether was evaporated under a nitrogen atmosphere, and 1 mL of mobile phase (a mixture of acetonitrile, isopropanol, and ethyl acetate in a 4:4:2 volume ratio) was added. The sample was filtered through a 0.45-µm filter and separated on a C18 analytical column (Bionacom, 250 mm × 4.6 mm, 5 μm). The flow rate of the mobile phase was set at 0.7 mL/min (isocratic). Detection was performed with a UV–VIS detector at a wavelength of 490 nm. Identification of β-carotene and γ-carotene was carried out based on the retention of standards (Sigma-Aldrich, Poland). Torulene and torularhodin were identified based on the retention time of the patterns separated by thin-layer liquid chromatography [13]. The percentages of β- and γ-carotene, torulene, and torularhodin in the carotenoids mixture were determined based on the area of the peaks.
DNA Isolation Using the Chloroform–Phenol Method
For the isolation of DNA from yeast, a colony was taken from the YPD agar and suspended in sterile water. The samples were thoroughly mixed and centrifuged at 14,000 rpm for 10 min (Mini Spin Plus, Eppendorf). Then, the supernatant was removed, 300 µL of lysis buffer was added to the biomass, and the samples were incubated at 500 rpm at 37℃ for 1 h (Thermomixer Comfort, Eppendorf). After incubation, 200 µL of TE buffer was added and each sample was mixed for 1 min. Next, 200 µL of phenol:chloroform:isoamyl alcohol (25:24:1) at pH 8.0 (Sigma-Aldrich, Poland) was added, and each tube was mixed for 1 min. The samples were centrifuged at 14,000 rpm for 10 min. The top layer was collected, and 600 µL of cold 96% ethanol was added and mixed for 30 s. The prepared samples were incubated for 30 min at − 18 °C, and centrifuged at 14,000 rpm for 10 min. The supernatant was removed, 500 µL of 70% ethanol was added, and the mixture was centrifuged again under the same conditions. Finally, ethanol was removed and the extracted DNA was resuspended in 25 µL of sterile nuclease-free water.
DNA Amplification
The molecular method used for yeast identification was based on amplification and sequence analysis of the D1/D2 domain of the large subunit ribosomal DNA (28 S rDNA). The LSU domain was amplified using the primers NL1 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and NL4 (5′-GGTCCGTGTTTCAAGACGG-3′). The PCR mixture contained, apart from the other primers (NL1 and NL4, 20 pmol each), the same components as for the reaction with the ITS primers. The amplification conditions were as follows: initial denaturation—91℃ for 2 min; 36 cycles—denaturation at 91℃ for 1 min, hybridization at 60℃ for 1 min, and extension at 72℃ for 1 min; final extension—72℃ for 15 min. The PCR products were sent to Genomed S.A. (Warsaw, Poland) for sequencing by enzymatic synthesis (Sanger’s dideoxy). The sequences obtained in the form of a chromatogram were analyzed using the Chromas Lite program (version 2.6.6). Further analysis of sequences from each isolate was performed using BLAST from the National Center for Biotechnology Information (NCBI) with Genbank® genetic sequence database. Strain sequences were submitted to GenBank. The strains were also deposed in the Collection of Microorganisms of the Institute of Food Sciences (CMIFS) of the Warsaw University of Life Sciences.
Morphological and Physiological Characteristics
The isolates were assessed to determine their ability to assimilate nitrates and various carbon sources (glucose, xylose, galactose, arabinose, rhamnose, fructose, maltose, sucrose, lactose, raffinose, lactic acid, tartaric acid, citric acid, glucuronic acid, methanol, ethanol, glycerol, mannitol, trehalose, xylitol, erythritol, inulin), ability to grow in the absence of vitamins, and ability to grow at 35℃. The cell shape and reproduction method were also determined. The obtained results were compared with the taxonomic keys included in “The Yeast—A Taxonomic Study” [8].
Statistical Analysis
Pearson correlation coefficient was determined for the yield of cellular biomass, the total content of carotenoids, and the percentages of individual carotenoid fractions (β-carotene, γ-carotene, torulene, and torularhodin). The significance of differences between the mean lipid content in yeast cell biomass was assessed using the one-way analysis of variance and Tukey’s test. The significance level was set at α = 0.05. All analyses were performed in R Studio (version 8.9).
Results and Discussion
Isolation of Red Yeast
More than 130 yeast strains were isolated from the sampled specimens, and the colonies of the strains were characterized by a salmon, orange, pink, or pink-red color. Unfortunately, some of them did not survive freezing (with the addition of a cryoprotective substance) at − 85℃. As a result, a total of 114 pure cultures of red yeasts were finally obtained. A majority of the strains were isolated from air (37) and flower petals (36). Moreover, red yeast was not isolated from any tested samples of propolis (15), soil (9) and water from the Baltic Sea (2).
Carotenoid Biosynthesis
For carotenoid biosynthesis, red yeast culture was carried out in a liquid medium at 22℃. The growth of the yeasts varied greatly, and the yield of cellular biomass ranged from 2.00 to 19.55 g/L (Table 1). The lowest yields (below 5 g/L) were obtained for A-007, A-022, A-024, A-040, A-042, A-051, and A-090 strains. The biomass yield of 56 strains ranged from 15.00 to 19.55 g/L, while that of 51 strains from 5.60 to 14.90 g/L. A comparison of the biomass yields obtained in this study with that of Chreptowicz et al. [14] showed that the yeast strains isolated here were characterized by high cellular biomass yields. The cited authors also investigated the growth and carotenoid biosynthesis capacity of new red yeast isolates and determined cellular biomass in the range of 4.5–9.7 g/L. A similar thematic scope was observed in the work of El-Banna et al. [15], who achieved a biomass yield of 9.5–11.5 g/L for the tested red yeast strains.
Table 1.
Characteristics of isolated red yeasts and the values of their dried biomass yield, total carotenoid content in the dried biomass, and percentage of carotenoids after 96 h of cultivation
| Strain no. | Source | Location | Biomass yield (g/L) | Carotenoid content in biomass (µg/g) | Percentages of individual fractions of carotenoids (%) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Torularhodin | Torulene | γ-Carotene | β-Carotene | Others | |||||
| 001 | Birch sap (Betula pendula) | Grudziądz (53°29′02″N, 18°45′13″E) | 17.45 ± 0.49 | 175.46 ± 3.13 | 9.29 ± 1.20 | 77.35 ± 1.49 | 2.63 ± 0.59 | 11.77 ± 0.29 | 0.00 |
| 002 | Birch sap (Betula pendula) | Wyszków (52°35′34″N, 21°27′30″E) | 15.43 ± 1.93 | 222.62 ± 6.88 | 26.73 ± 4.01 | 61.57 ± 3.63 | 2.22 ± 0.42 | 9.46 ± 0.04 | 0.00 |
| 003 | Birch bark (Betula pendula) | Skarżysko-Kamienna (51°06′47″N, 20°52′17″E) | 17.65 ± 0.21 | 106.94 ± 0.28 | 1.06 ± 0.24 | 87.51 ± 1.02 | 2.23 ± 0.62 | 9.20 ± 0.64 | 0.00 |
| 004 | Birch bark (Betula pendula) | Otwock (52°06′20″N, 21°15′40″E) | 18.23 ± 0.33 | 141.36 ± 14.84 | 9.32 ± 1.00 | 73.01 ± 1.29 | 6.22 ± 0.06 | 11.44 ± 0.35 | 0.00 |
| 005 | Birch bark (Betula pendula) | Skarżysko-Kamienna (51°06′47″N, 20°52′17″E) | 17.07 ± 0.38 | 101.84 ± 2.06 | 14.08 ± 1.68 | 81.68 ± 1.69 | 1.34 ± 0.15 | 2.89 ± 0.16 | 0.00 |
| 006 | Birch bark (Betula pendula) | Skarżysko-Kamienna (51°06′47″N, 20°52′17″E) | 17.10 ± 0.33 | 132.15 ± 8.75 | 4.68 ± 1.16 | 74.64 ± 1.96 | 2.25 ± 0.41 | 18.42 ± 1.21 | 0.00 |
| 007 | Birch slime flux (Betula pendula) | Skarżysko-Kamienna (51°06′47″N, 20°52′17″E) | 4.0 ± 0.71 | 127.70 ± 11.90 | 0.00 | 0.00 | 3.34 ± 1.39 | 46.74 ± 2.28 | 49.93 ± 2.28 |
| 008 | Birch slime flux (Betula pendula) | Skarżysko-Kamienna (51°06′47″N, 20°52′17″E) | 17.25 ± 0.49 | 180.29 ± 10.20 | 23.00 ± 1.33 | 60.30 ± 0.47 | 2.30 ± 0.17 | 12.85 ± 0.41 | 1.55 ± 0.28 |
| 009 | Oak leaf (Quercus L.) | Warszawa (52°13′44″N, 21°00′59″E) | 15.60 ± 0.85 | 223.53 ± 13.86 | 25.89 ± 1.36 | 50.06 ± 0.61 | 1.85 ± 0.41 | 22.20 ± 1.56 | 0.00 |
| 010 | Oak leaf (Quercus L.) | Żyrardów (52°02′55″N, 20°26′45″E) | 15.75 ± 0.07 | 204.75 ± 3.57 | 29.92 ± 0.87 | 16.02 ± 0.18 | 3.14 ± 0.01 | 48.43 ± 1.39 | 0.79 ± 0.22 |
| 011 | Goatsbeard leaf (Aruncus dioicus) | Skarżysko-Kamienna (51°06′47″N, 20°52′17″E) | 26.85 ± 1.20 | 139.49 ± 21.49 | 12.61 ± 1.94 | 61.90 ± 0.62 | 1.77 ± 0.75 | 23.73 ± 2.07 | 0.00 |
| 012 | Astilbe leaf (Astilbe spp.) | Rogów (51°49′03″N, 19°53′11) | 14.00 ± 0.57 | 235.22 ± 19.61 | 5.99 ± 1.39 | 59.99 ± 0.38 | 2.75 ± 0.04 | 31.28 ± 0.97 | 0.00 |
| 013 | Linden leaf (Tilia L.) | Radzyń Podlaski (51°46′59″N, 22°37′00″E) | 15.40 ± 0.71 | 206.67 ± 4.32 | 44.48 ± 3.02 | 45.75 ± 2.56 | 2.31 ± 0.10 | 7.47 ± 0.56 | 0.00 |
| 014 | Linden leaf (Tilia L.) | Nakło nad Notecią (53°08′31″N, 17°36′06″E) | 17.10 ± 0.28 | 212.90 ± 21.81 | 26.58 ± 0.95 | 58.06 ± 0.57 | 3.06 ± 0.02 | 11.01 ± 1.41 | 1.31 ± 0.14 |
| 015 | Pine needles (Pinus L.) | Kalisz (51°45′39″N, 18°05′27″E) | 16.05 ± 0.64 | 168.85 ± 7.62 | 43.29 ± 2.91 | 38.25 ± 0.99 | 3.59 ± 0.61 | 10.85 ± 0.87 | 1.90 ± 0.03 |
| 016 | Air, street | Warszawa (52°13′44″N, 21°00′59″E) | 14.05 ± 1.91 | 13.88 ± 1.26 | 0.00 | 0.00 | 19.45 ± 1.57 | 80.55 ± 1.57 | 00 |
| 017 | Air, forest | Skarżysko-Kamienna (51°06′47″N, 20°52′17″E) | 13.63 ± 0.33 | 76.77 ± 4.88 | 1.91 ± 0.78 | 22.24 ± 1.62 | 3.61 ± 1.19 | 6.43 ± 0.19 |
10.16 ± 0.47 19.30 ± 1.20 22.72 ± 0.64 13.66 ± 1.48 |
| 018 | Air, garden | Piastów (52°11′03″N, 20°50′22″E) | 13.67 ± 0.75 | 48.15 ± 0.79 | 8.71 ± 1.39 | 41.26 ± 1.87 | 19.85 ± 1.12 | 10.23 ± 0.30 | 19.97 ± 1.31 |
| 019 | Air, forest | Skarżysko-Kamienna (51°06′47″N, 20°52′17″E) | 13.15 ± 0.07 | 25.19 ± 0.81 | 0.00 | 70.35 ± 2.84 | 29.65 ± 2.84 | 0.00 | 0.00 |
| 020 | Air, garden | Kraków (50°03′41″N, 19°56′11″E) | 14.07 ± 0.28 | 228.55 ± 12.16 | 2.15 ± 0.25 | 53.19 ± 2.89 | 7.31 ± 0.01 | 37.36 ± 3.16 | 0.00 |
| 021 | Air, garden | Nakło nad Notecią (53°08′31″N, 17°36′06″E) | 9.03 ± 0.61 | 161.64 ± 19.45 | 33.38 ± 1.63 | 41.18 ± 1.34 | 4.94 ± 0.99 | 20.50 ± 1.27 | 0.00 |
| 022 | Air, forest | Nadarzyn (52°05′39″N, 20°48′27″E) | 4.93 ± 1.13 | 180.50 ± 8.19 | 14.67 ± 1.51 | 62.21 ± 1.76 | 1.29 ± 0.08 | 20.24 ± 3.72 | 1.59 ± 0.37 |
| 023 | Air, street | Warszawa (52°13′44″N, 21°00′59″E) | 17.65 ± 3.61 | 134.01 ± 22.37 | 15.84 ± 0.14 | 78.49 ± 0.08 | 0.00 | 5.68 ± 0.06 | 0.00 |
| 024 | Air, garden | Pruszków (52°10′14″N, 20°48′43″E) | 2.10 ± 0.33 | 190.18 ± 3.79 | 13.44 ± 1.54 | 30.45 ± 1.28 | 4.44 ± 1.28 | 49.74 ± 4.95 | 1.94 ± 0.85 |
| 025 | Air, Botanic Garden | Warszawa (52°13′44″N, 21°00′59″E) | 6.55 ± 1.06 | 142.46 ± 27.12 | 26.23 ± 3.48 | 46.66 ± 2.59 | 9.08 ± 0.16 | 18.03 ± 1.05 | 0.00 |
| 026 | Air, Botanic Garden | Warszawa (52°13′44″N, 21°00′59″E) | 14.77 ± 0.14 | 158.35 ± 12.29 | 65.49 ± 2.78 | 21.68 ± 0.14 | 2.88 ± 0.62 | 7.36 ± 1.72 | 2.60 ± 0.30 |
| 027 | Air, street | Wrocław (51º6′32″N, 17º1′57″E) | 11.80 ± 0.42 | 142.75 ± 14.34 | 26.79 ± 2.19 | 34.40 ± 2.90 | 7.00 ± 0.80 | 31.82 ± 1.51 | 0.00 |
| 028 | Air, street | Warszawa (52°13′44″N, 21°00′59″E) | 16.17 ± 0.42 | 200.23 ± 21.72 | 36.87 ± 1.23 | 45.45 ± 1.17 | 4.31 ± 0.19 | 12.03 ± 0.40 | 1.35 ± 0.26 |
| 029 | Air, Botanic Garden | Warszawa (52°13′44″N, 21°00′59″E) | 5.60 ± 0.42 | 115.59 ± 16.65 | 36.87 ± 1.23 | 45.45 ± 1.17 | 4.31 ± 0.19 | 12.03 ± 0.40 | 0.00 |
| 030 | Air, garden | Piastów (52°11′03″N, 20°50′22″E) | 13.37 ± 1.56 | 209.78 ± 21.33 | 30.87 ± 2.02 | 52.07 ± 1.89 | 4.29 ± 0.95 | 11.02 ± 0.86 | 1.77 ± 0.23 |
| 031 | Air, Arboretum | Rogów (51°49′03″N, 19°53′11″E) | 16.76 ± 0.04 | 284.31 ± 13.31 | 53.99 ± 0.52 | 32.38 ± 0.03 | 2.77 ± 0.09 | 8.64 ± 0.06 | 1.45 ± 0.18 |
| 032 | Air, garden | Żyrardów (52°02′55″N, 20°26′45″E) | 14.90 ± 1.27 | 14.24 ± 5.96 | 0.00 | 33.85 ± 1.97 | 3.85 ± 0.57 | 62.31 ± 2.54 | 0.00 |
| 033 | Air, street | Kalisz (51°45′39″N, 18°05′27″E) | 9.23 ± 0.90 | 197.34 ± 1.66 | 44.51 ± 5.24 | 11.65 ± 1.00 | 2.44 ± 0.45 | 41.41 ± 4.70 | 0.00 |
| 034 | Air, forest | Nadarzyn (52°05′39″N, 20°48′27″E) | 8.05 ± 0.92 | 124.41 ± 3.23 | 29.82 ± 2.53 | 39.57 ± 0.83 | 8.20 ± 0.69 | 22.42 ± 1.10 | 0.00 |
| 035 | Air, street | Sokołów Podlaski (52°24′24″N, 22°15′11″E) | 13.57 ± 0.33 | 179.87 ± 16.97 | 2.14 ± 0.45 | 38.88 ± 2.22 | 3.75 ± 0.70 | 55.24 ± 2.47 | 0.00 |
| 036 | Air, university campus | Warszawa (52°13′44″N, 21°00′59″E) | 12.18 ± 0.83 | 273.55 ± 12.93 | 0.00 | 42.39 ± 3.37 | 2.01 ± 0.17 | 55.60 ± 3.56 | 0.00 |
| 037 | Air, street | Warszawa (52°13′44″N, 21°00′59″E) | 9.15 ± 29.67 | 232.15 ± 29.67 | 14.56 ± 2.69 | 12.87 ± 1.55 | 6.29 ± 1.34 | 66.29 ± 2.90 | 0.00 |
| 038 | Air, Botanic Garden | Warszawa (52°13′44″N, 21°00′59″E) | 13.45 ± 1.34 | 248.29 ± 12.32 | 32.00 ± 3.47 | 39.85 ± 2.11 | 3.69 ± 1.01 | 24.47 ± 0.35 | 0.00 |
| 039 | Air, university campus | Warszawa (52°13′44″N, 21°00′59″E) | 7.35 ± 0.35 | 140.56 ± 10.37 | 9.84 ± 1.94 | 42.95 ± 3.38 | 3.12 ± 0.52 | 39.29 ± 1.12 | 4.81 |
| 040 | Air, street | Koszalin (54°11′39″N, 16°10′19″E) | 2.30 ± 0.42 | 131.25 ± 8.84 | 9.84 ± 1.97 | 42.95 ± 3.38 | 3.12 ± 0.52 | 39.29 ± 1.12 | 4.81 ± 0.81 |
| 041 | Air, garden | Podkowa Leśna (52°07′20″N, 20°43′35″E) | 12.50 ± 0.85 | 295.20 ± 5.90 | 73.67 ± 2.38 | 18.66 ± 2.05 | 1.38 ± 0.38 | 6.30 ± 0.71 | 0.00 |
| 042 | Air, street | Pruszków (52°10′14″N, 20°48′43″E) | 4.50 ± 0.42 | 37.20 ± 6.31 | 0.00 | 12.37 ± 0.07 | 13.84 ± 2.12 | 73.79 ± 2.05 | 0.00 |
| 043 | Air, street | Pruszków (52°10′14″N, 20°48′43″E) | 7.30 ± 0.28 | 234.21 ± 24.82 | 40.11 ± 0.85 | 26.01 ± 1.55 | 7.94 ± 1.41 | 25.95 ± 0.71 | 0.00 |
| 044 | Air, street | Sokołów Podlaski (52°24′24″N, 22°15′11″E) | 8.50 ± 0.71 | 235.59 ± 10.56 | 19.81 ± 2.02 | 31.24 ± 1.56 | 1.99 ± 0.34 | 46.97 ± 0.79 | 0.00 |
| 045 | Air, university campus | Warszawa (52°13′44″N, 21°00′59″E) | 12.00 ± 1.41 | 162.19 ± 21.32 | 19.81 ± 2.02 | 31.24 ± 1.56 | 1.99 ± 0.34 | 46.97 ± 0.79 | 0.00 |
| 046 | Air, university campus | Warszawa (52°13′44″N, 21°00′59″E) | 17.83 ± 1.46 | 255.79 ± 4.44 | 0.00 | 9.40 ± 1.23 | 1.76 ± 0.12 | 14.05 ± 1.69 | 74.80 ± 0.58 |
| 047 | Air, Botanic Garden | Warszawa (52°13′44″N, 21°00′59″E) | 10.15 ± 0.64 | 219.32 ± 3.30 | 9.07 ± 1.10 | 18.35 ± 1.43 | 5.57 ± 1.92 | 67.01 ± 0.61 | 0.00 |
| 048 | Air, Botanic Garden | Warszawa (52°13′44″N, 21°00′59″E) | 9.80 ± 0.71 | 268.12 ± 10.42 | 25.95 ± 2.26 | 60.01 ± 1.77 | 1.36 ± 0.26 | 12.69 ± 0.75 | 0.00 |
| 049 | Air, street | Olsztyn (53º46′36″N, 20º28′40″E) | 18.45 ± 0.21 | 145.76 ± 16.05 | 20.07 ± 3.97 | 53.01 ± 2.61 | 1.85 ± 0.01 | 25.09 ± 1.35 | 0.00 |
| 050 | Air, street | Warszawa (52°13′44″N, 21°00′59″E) | 14.75 ± 1.20 | 107.20 ± 10.44 | 11.03 ± 0.13 | 63.73 ± 2.18 | 1.95 ± 0.33 | 23.30 ± 1.98 | 0.00 |
| 051 | Air, street | Warszawa (52°13′44″N, 21°00′59″E) | 2.00 ± 0.85 | 224.59 ± 1.94 | 11.67 ± 1.16 | 22.04 ± 1.70 | 1.90 ± 0.62 | 41.30 ± 2.60 |
9.06 ± 1.97 14.04 ± 4.12 |
| 052 | Air, street | Warszawa (52°13′44″N, 21°00′59″E) | 10.55 ± 0.35 | 118.93 ± 8.58 | 17.57 ± 1.88 | 42.42 ± 2.01 | 5.15 ± 0.43 | 30.97 ± 0.06 | 3.90 ± 0.62 |
| 053 | Redcurrant fruit (Ribes spicatum) | Skarżysko-Kamienna (51°06′47″N, 20°52′17″E) | 19.30 ± 0.57 | 144.30 ± 12.93 | 24.45 ± 1.77 | 64.77 ± 2.27 | 0.43 ± 0.10 | 10.36 ± 0.60 | 0.00 |
| 054 | Redcurrant fruit (Ribes spicatum) | Skarżysko-Kamienna (51°06′47″N, 20°52′17″E) | 15.30 ± 0.57 | 206.67 ± 1.60 | 62.32 ± 4.35 | 15.64 ± 1.24 | 3.30 ± 1.18 | 17.18 ± 1.61 | 1.58 ± 0.33 |
| 055 | Blackcurrant fruit (Ribes nigrum) | Gdynia (54°31′08″N, 18°31′54″E) | 17.65 ± 0.35 | 155.08 ± 1.90 | 71.05 ± 3.58 | 24.35 ± 3.05 | 1.19 ± 0.21 | 3.42 ± 0.32 | 0.00 |
| 056 | Wild strawberry fruit (Fragaria spp.) | Piastów (52°11′03″N, 20°50′22″E) | 14.47 ± 0.38 | 234.72 ± 23.21 | 42.01 ± 2.45 | 22.37 ± 1.59 | 1.58 ± 0.34 | 27.95 ± 1.44 | 6.10 ± 0.92 |
| 057 | Apple fruit (Malus spp.) | Piastów (52°11′03″N, 20°50′22″E) | 17.65 ± 0.21 | 202.28 ± 13.95 | 34.72 ± 2.97 | 52.95 ± 2.74 | 0.48 ± 0.08 | 11.86 ± 0.30 | 0.00 |
| 058 | Apple fruit (Malus spp.) | Gdynia (54°31′08″N, 18°31′54″E) | 14.30 ± 0.00 | 338.72 ± 10.51 | 24.19 ± 1.66 | 41.90 ± 0.81 | 4.03 ± 0.21 | 29.89 ± 0.64 | 0.00 |
| 059 | Kamchatka berry fruit (Lonicera caerulea var. kamtschatica) | Skarżysko-Kamienna (51°06′47″N, 20°52′17″E) | 17.90 ± 1.84 | 134.24 ± 16.75 | 18.70 ± 1.27 | 70.34 ± 0.88 | 1.34 ± 0.08 | 9.63 ± 0.47 | 0.00 |
| 060 | Strawberry fruit (Fragaria × ananassa Duchesne) | Gorzów Wielkopolski (52°44′12″N, 15°13′43″E) | 18.70 ± 0.14 | 120.98 ± 1.92 | 63.27 ± 0.21 | 27.49 ± 0.78 | 0.44 ± 0.13 | 5.38 ± 0.86 | 0.00 |
| 061 | Blueberry fruit (Vaccinium corymbosum) | Ostrowiec Świętokrzyski (50°51′N, 21°23′E) | 16.25 ± 1.48 | 209.15 ± 6.60 | 27.97 ± 2.66 | 39.67 ± 1.02 | 0.57 ± 0.13 | 31.79 ± 1.51 | 0.00 |
| 062 | Pumpkin fruit (Cucurbita pepo L.) | Piastów (52°11′03″N, 20°50′22″E) | 16.70 ± 0.71 | 198.74 ± 0.48 | 41.30 ± 0.49 | 42.33 ± 0.93 | 2.36 ± 0.08 | 14.02 ± 0.36 | 0.00 |
| 063 | Grape (Vitis vinifera L.) | Skarżysko-Kamienna (51°06′47″N, 20°52′17″E) | 17.75 ± 0.64 | 185.64 ± 4.17 | 56.00 ± 3.28 | 38.85 ± 3.71 | 2.05 ± 0.30 | 3.11 ± 0.13 | 0.00 |
| 064 | Grape (Vitis vinifera L.) | Skarżysko-Kamienna (51°06′47″N, 20°52′17″E) | 16.30 ± 0.42 | 269.62 ± 10.92 | 29.36 ± 1.49 | 43.92 ± 2.04 | 0.64 ± 0.16 | 14.95 ± 0.09 |
7.22 ± 0.06 3.93 ± 0.68 |
| 065 | Homemade fermented carrot juice | Warszawa (52°13′44″N, 21°00′59″E) | 16.60 ± 1.27 | 110.14 ± 5.25 | 2.08 ± 0.18 | 91.39 ± 1.85 | 1.65 ± 0.47 | 4.89 ± 1.21 | 0.00 |
| 066 | Tomato fruit (Solanum lycopersicum) | Kraków (50°03′41″N, 19°56′11″E) | 13.55 ± 0.92 | 94.05 ± 12.25 | 29.79 ± 1.48 | 48.06 ± 3.15 | 1.83 ± 0.14 | 20.33 ± 4.48 | 0.00 |
| 067 | Rose petals (Rosa L.) | Kraków (50°03′41″N, 19°56′11″E) | 17.50 ± 1.56 | 184.86 ± 20.98 | 17.12 ± 0.28 | 66.25 ± 1.49 | 1.14 ± 0.15 | 15.51 ± 1.07 | 0.00 |
| 068 | Rose petals (Rosa L.) | Tomaszów Mazowiecki (51°31′52″N, 20°00′30″E) | 16.53 ± 1.04 | 231.24 ± 20.56 | 33.48 ± 0.75 | 56.07 ± 2.64 | 2.73 ± 0.02 | 7.73 ± 1.92 | 0.00 |
| 069 | Rose petals (Rosa L.) | Żyrardów (52°02′55″N, 20°26′45″E) | 16.20 ± 0.28 | 116.48 ± 3.42 | 13.71 ± 0.66 | 78.37 ± 1.43 | 0.66 ± 0.18 | 7.27 ± 0.59 | 0.00 |
| 070 | Lily flower (Lilium L.) | Koszalin (54°11′39″N, 16°10′19″E) | 14.65 ± 0.07 | 212.43 ± 12.25 | 21.07 ± 0.33 | 37.66 ± 0.33 | 4.32 ± 0.09 | 36.96 ± 0.09 | 0.00 |
| 071 | Lily flower (Lilium L.) | Warka (51°47′03″N, 21°11′27″E) | 17.90 ± 0.71 | 156.06 ± 17.03 | 18.89 ± 0.21 | 70.43 ± 0.16 | 1.69 ± 0.02 | 9.00 ± 0.32 | 0.00 |
| 072 | Rhododendron flowers (Rhododendron L.) | Rogów (51°49′03″N, 19°53′11) | 16.10 ± 0.14 | 172.06 ± 20.73 | 13.35 ± 0.06 | 60.12 ± 0.06 | 9.53 ± 1.54 | 17.01 ± 1.55 | 0.00 |
| 073 | Rhododendron flowers (Rhododendron L.) | Rogów (51°49′03″N, 19°53′11) | 14.75 ± 1.48 | 178.57 ± 28.76 | 30.97 ± 0.07 | 47.28 ± 1.05 | 0.67 ± 0.18 | 17.47 ± 1.70 | 3.62 ± 0.76 |
| 074 | Rhododendron flowers (Rhododendron L.) | Rogów (51°49′03″N, 19°53′11) | 16.60 ± 1.13 | 158.39 ± 7.60 | 13.60 ± 2.61 | 50.63 ± 1.32 | 9.82 ± 0.07 | 25.96 ± 1.36 | 0.00 |
| 075 | Rhododendron flowers (Rhododendron L.) | Warszawa (52°13′44″N, 21°00′59″E) | 15.65 ± 0.35 | 194.61 ± 10.61 | 26.46 ± 2.03 | 52.20 ± 1.62 | 2.91 ± 0.11 | 18.44 ± 0.52 | 0.00 |
| 076 | Rhododendron flowers (Rhododendron L.) | Tomaszów Mazowiecki (51°31′52″N, 20°00′30″E) | 14.25 ± 0.07 | 292.55 ± 2.07 | 9.97 ± 0.08 | 55.68 ± 0.35 | 2.00 ± 0.03 | 31.62 ± 0.27 | 0.74 ± 0.13 |
| 077 | Tulip flowers (Tulipa L.) | Sokołów Podlaski (52°24′24″N, 22°15′11″E) | 13.65 ± 1.91 | 217.89 ± 25.29 | 9.90 ± 1.80 | 64.40 ± 0.83 | 5.07 ± 0.49 | 20.64 ± 0.48 | 0.00 |
| 078 | Tulip flowers (Tulipa L.) | Piastów (52°11′03″N, 20°50′22″E) | 16.95 ± 0.07 | 193.22 ± 3.93 | 38.54 ± 4.79 | 38.12 ± 2.46 | 2.90 ± 0.36 | 16.51 ± 1.37 | 3.94 ± 1.32 |
| 079 | Marigold flowers (Tagetes L.) | Warszawa (52°13′44″N, 21°00′59″E) | 6.10 ± 0.28 | 204.35 ± 19.62 | 31.94 ± 0.24 | 50.59 ± 0.54 | 2.82 ± 0.42 | 13.31 ± 0.18 | 1.34 ± 0.18 |
| 080 | Marigold flowers (Tagetes L.) | Warszawa (52°13′44″N, 21°00′59″E) | 15.15 ± 0.49 | 198.10 ± 30.39 | 31.94 ± 0.24 | 50.59 ± 0.54 | 2.82 ± 0.42 | 13.31 ± 0.18 | 0.00 |
| 081 | Marigold flowers (Tagetes L.) | Gorzów Wielkopolski (52°44′12″N, 15°13′43″E) | 15.30 ± 1.70 | 176.60 ± 9.77 | 11.29 ± 1.34 | 62.70 ± 0.64 | 8.92 ± 0.76 | 17.11 ± 0.06 | 0.00 |
| 082 | Marigold flowers (Tagetes L.) | Warszawa (52°13′44″N, 21°00′59″E) | 9.20 ± 0.99 | 120.57 ± 18.74 | 12.93 ± 1.41 | 63.61 ± 0.45 | 2.02 ± 0.14 | 21.45 ± 1.10 | 0.00 |
| 083 | Cornflower flowers (Centaurea cyanus) | Opole (50°40′19″N, 17°55′31″E) | 11.15 ± 0.07 | 49.32 ± 2.86 | 23.24 ± 3.03 | 48.15 ± 2.96 | 2.59 ± 0.19 | 26.03 ± 0.11 | 0.00 |
| 084 | Cornflower flowers (Centaurea cyanus) | Tomaszów Mazowiecki (51°31′52″N, 20°00′30″E) | 16.00 ± 2.64 | 70.90 ± 19.80 | 4.35 ± 1.75 | 63.69 ± 1.14 | 2.04 ± 0.42 | 29.93 ± 1.04 | 0.00 |
| 085 | Cornflower flowers (Centaurea cyanus) | Kołobrzeg (54°10′32″N, 15°35′00″E) | 10.80 ± 0.42 | 176.62 ± 6.12 | 50.57 ± 3.34 | 39.68 ± 2.22 | 1.60 ± 0.33 | 8.16 ± 1.45 | 0.00 |
| 086 | Cornflower flowers (Centaurea cyanus) | Kołobrzeg (54°10′32″N, 15°35′00″E) | 8.90 ± 0.42 | 231.91 ± 7.08 | 70.12 ± 3.23 | 20.59 ± 1.90 | 0.79 ± 0.08 | 8.52 ± 1.41 | 0.00 |
| 087 | Common poppy flowers (Papaver rhoeas) | Opole (50°40′19″N, 17°55′31″E) | 17.30 ± 0.85 | 270.13 ± 10.69 | 17.87 ± 3.83 | 52.48 ± 1.71 | 3.33 ± 0.39 | 26.33 ± 1.73 | 0.00 |
| 088 | Rudbeckia flowers (Rudbeckia L.) | Piastów (52°11′03″N, 20°50′22″E) | 15.60 ± 1.70 | 224.70 ± 13.68 | 41.44 ± 0.47 | 48.65 ± 2.51 | 2.67 ± 0.81 | 6.23 ± 1.26 | 1.02 ± 0.03 |
| 089 | Rudbeckia flowers (Rudbeckia L.) | Nadarzyn (52°05′39″N, 20°48′27″E) | 15.87 ± 0.09 | 127.58 ± 16.33 | 5.77 ± 0.61 | 59.71 ± 2.54 | 3.95 ± 0.57 | 30.58 ± 1.36 | 0.00 |
| 090 | Rudbeckia flowers (Rudbeckia L.) | Sokołów Podlaski (52°24′24″N, 22°15′11″E)) | 3.65 ± 0.35 | 158.51 ± 20.20 | 44.83 ± 2.19 | 28.51 ± 1.49 | 5.79 ± 0.78 | 20.88 ± 2.91 | 0.00 |
| 091 | Rudbeckia flowers (Rudbeckia L.) | Sokołów Podlaski (52°24′24″N, 22°15′11″E) | 13.47 ± 0.57 | 161.28 ± 10.73 | 10.79 ± 1.56 | 48.63 ± 0.70 | 6.64 ± 1.29 | 33.95 ± 0.98 | 0.00 |
| 092 | Hydrangea flowers (Hydrangea L.) | Warszawa (52°13′44″N, 21°00′59″E) | 18.57 ± 1.18 | 109.98 ± 0.63 | 14.62 ± 1.46 | 54.06 ± 0.79 | 8.15 ± 1.36 | 23.17 ± 0.89 | 0.00 |
| 093 | Hydrangea flowers (Hydrangea L.) | Żyrardów (52°02′55″N, 20°26′45″E) | 19.55 ± 1.91 | 168.95 ± 9.72 | 20.17 ± 0.11 | 66.02 ± 0.11 | 2.05 ± 0.12 | 11.77 ± 1.32 | 0.00 |
| 094 | Flowers of the delphinium (Delphinium consolida L.) | Sokołów Podlaski (52°24′24″N, 22°15′11″E) | 14.43 ± 0.52 | 181.47 ± 22.06 | 12.94 ± 2.18 | 70.64 ± 2.48 | 3.86 ± 0.57 | 12.58 ± 0.28 | 0.00 |
| 095 | Blue phacelia flowers (Phacelia tanacetifolia) | Sokołów Podlaski (52°24′24″N, 22°15′11″E) | 16.60 ± 0.57 | 168.55 ± 14.80 | 65.74 ± 0.34 | 19.68 ± 0.45 | 3.41 ± 0.23 | 9.55 ± 0.22 | 1.64 ± 0.11 |
| 096 | Common chamomile flower (Matricaria chamomilla L.) | Nadarzyn (52°05′39″N, 20°48′27″E) | 15.00 ± 0.57 | 161.51 ± 13.75 | 49.39 ± 2.69 | 33.23 ± 1.36 | 4.77 ± 0.26 | 10.45 ± 1.52 | 2.18 ± 0.45 |
| 097 | Alfalfa flower (Medicago sativa L.) | Radzyń Podlaski (51°46′59″N, 22°37′00″E) | 7.25 ± 0.49 | 103.36 ± 2.70 | 30.20 ± 2.84 | 37.80 ± 2.04 | 8.84 ± 0.40 | 23.16 ± 0.41 | 0.00 |
| 098 | Thymus alpestris flowers | Zakopane (49°17′58″N, 19°57′07″E) | 12.57 ± 2.03 | 68.90 ± 7.36 | 3.50 ± 0.16 | 44.54 ± 0.37 | 17.68 ± 0.01 | 34.29 ± 0.54 | 0.00 |
| 099 | Goldenrod flowers (Solidago virgaurea L.) | Ryki (51°37.5444′N, 21°55.9644′E) | 17.30 ± 0.99 | 214.40 ± 5.63 | 64.06 ± 0.21 | 22.19 ± 0.48 | 3.60 ± 0.04 | 6.39 ± 0.88 | 3.77 ± 0.16 |
| 100 | Sonchus arvensis L. flowers | Ryki (51°37.5444′N, 21°55.9644′E) | 15.85 ± 0.49 | 216.68 ± 12.90 | 43.31 ± 1.56 | 41.52 ± 2.98 | 4.05 ± 1.40 | 9.94 ± 0.09 | 1.20 ± 0.11 |
| 101 | Flowers hypochaeris rough (Hypochaeris radicata L.) | Ustrzyki górne (49°06′17″N 22°39′02″E) | 17.07 ± 1.23 | 214.09 ± 7.41 | 38.29 ± 3.03 | 48.06 ± 2.89 | 1.83 ± 0.09 | 11.83 ± 0.23 | 0.00 |
| 102 | Cosmos bipinnatus flowers | Warszawa (52°13′44″N, 21°00′59″E) | 14.20 ± 0.42 | 226.46 ± 15.48 | 59.29 ± 0.89 | 32.02 ± 0.23 | 1.68 ± 0.02 | 7.02 ± 0.64 | 0.00 |
| 103 | A blade of Avena L. | Rogów (51°49′03″N, 19°53′11) | 13.63 ± 1.56 | 143.67 ± 11.21 | 67.81 ± 0.25 | 25.06 ± 1.00 | 3.35 ± 0.81 | 2.15 ± 0.06 | 1.64 ± 0.11 |
| 104 | A blade of Avena L. | Rogów (51°49′03″N, 19°53′11) | 14.30 ± 1.56 | 243.69 ± 7.14 | 19.97 ± 1.23 | 43.50 ± 0.29 | 2.16 ± 0.30 | 34.38 ± 0.64 | 0.00 |
| 105 | A blade of Avena L. | Warszawa (52°13′44″N, 21°00′59″E) | 10.05 ± 1.06 | 108.61 ± 19.38 | 8.30 ± 0.08 | 71.46 ± 2.52 | 2.09 ± 0.36 | 18.17 ± 2,81 | 0.00 |
| 106 | Feta cheese | Warszawa (52°13′44″N, 21°00′59″E) | 14.03 ± 0.42 | 194.28 ± 6.71 | 81.97 ± 2.26 | 2.73 ± 0.57 | 2.95 ± 1.09 | 9.32 ± 1.46 | 3.04 ± 1.32 |
| 107 | Minced pork | Warszawa (52°13′44″N, 21°00′59″E) | 18.05 ± 0.64 | 163.14 ± 16.77 | 18.29 ± 0.71 | 67.56 ± 1.39 | 2.30 ± 0.22 | 11.86 ± 1.87 | 0.00 |
| 108 | Bee pollen | Białystok (53°08′07″N, 23°08′44″E) | 17.80 ± 1.70 | 157.41 ± 9.55 | 9.69 ± 0.49 | 77.02 ± 1.54 | 2.20 ± 0.62 | 10.60 ± 1.42 | 0.00 |
| 109 | Bee bread | Białystok (53°08′07″N, 23°08′44″E) | 15.10 ± 0.57 | 137.63 ± 10.65 | 2.51 ± 0.47 | 86.48 ± 1.34 | 1.72 ± 0.02 | 9.30 ± 0.85 | 0.00 |
| 110 | Homemade yogurt | Warszawa (52°13′44″N, 21°00′59″E) | 14.39 ± 0.85 | 221.92 ± 13.04 | 34.13 ± 2.46 | 52.35 ± 4.45 | 1.83 ± 0.14 | 11.69 ± 0.73 | 0.00 |
| 111 | Salda mix | Warszawa (52°13′44″N, 21°00′59″E) | 16.95 ± 0.07 | 232.47 ± 18.05 | 20.66 ± 3.31 | 48.98 ± 1.99 | 5.16 ± 0.45 | 25.21 ± 0.87 | 0.00 |
| 112 | Chocolate bar | Warszawa (52°13′44″N, 21°00′59″E) | 15.70 ± 2.55 | 166.42 ± 10.09 | 18.80 ± 2.38 | 52.89 ± 2.43 | 4.44 ± 1.36 | 23.88 ± 1.32 | 0.00 |
| 113 | Laboratory contamination | Warszawa (52°13′44″N, 21°00′59″E) | 16.20 ± 2.07 | 168.52 ± 8.85 | 12.32 ± 1.34 | 55.68 ± 2.96 | 2.26 ± 0.12 | 29.75 ± 4.43 | 0.00 |
| 114 | Sour cream | Radom (51°24′09″N, 21°08′49″E | 11.33 ± 0.28 | 406.50 ± 9.61 | 50.30 ± 3.35 | 41.05 ± 2.28 | 1.74 ± 0.39 | 6.92 ± 0.69 | 0.00 |
The isolated yeast strains produced carotenoid pigments, the total content of which in the cellular biomass ranged from 13.88 to 406.50 µg/g (Table 1). In the available literature, different values are provided for the content of carotenoids synthesized by microorganisms. In order to sort them out, El-Banna et al. [15] proposed the classification of carotenoid-producing strains into low-efficiency (below 100 µg/g), medium-efficiency (between 100 and 500 µg/g), and high-efficiency (above 500 µg/g) strains. Among the investigated red yeast strains, 10 can be considered as low-efficiency producers, and the remaining 104 strains as medium-efficiency producers, of which 61 isolates synthesized carotenoids at an amount of 101.84–198.74 µg/g and 41 strains in the range of 200.23–295.20 µg/g. The other two strains synthesized significantly more carotenoids. The greatest amount of total carotenoids was produced by the A-058 strain (338.72 µg/g) isolated from apple and the A-114 strain (406.50 µg/g) which was originally a contaminant of cream stored under refrigerated conditions.
The carotenoid-biosynthesizing capacity of yeasts depends primarily on the strain and culture conditions. Of the 16 red yeast strains isolated by Chreptowicz et al. [14], Sporobolomyces roseus WUT182 synthesized as much as 1.12 mg of carotenoids per gram of cellular biomass. In another work by Allahkarami et al. [16], red yeast was isolated from Pinaceae forest ecosystems. The most efficient of the isolated strains, R. mucilaginosa, synthesized only 223.5 µg/g. In the study by Libkind and Van Broock [17], 15 strains of carotenogenic yeast were isolated from the natural environment in northwestern Patagonia. Similar to the work of Allahkarami et al. [16] the highest amount of carotenoids (301 µg/gd.m.) was synthesized by a R. mucilaginosa strain. The strains with the highest biomass yield were characterized by lower overall content of carotenoids in the cellular biomass.
The next stage of the present study was to determine the percentages of torularhodin, torulene, β-carotene, and γ-carotene. Torulene occurs in the highest amounts in yeast cells [18], which is confirmed by the results of this study. Among the isolated yeast strains, torulene represented more than 50% of the total carotenoid pool in 49 strains. Only in two strains (A-016 and A-007), this compound was not detected, while four strains (A-065, A-003, A-109, and A-005) had more than 80% of torulene in the total carotenoid pool (Table 1).
Currently, there is a demand for yeasts that can synthesize high amounts of torularhodin due to the anticancer properties of this compound [19]. In this study, seven isolated strains (A-016, A-042, A-032, A-036, A-007, A-046, and A-019) were not found to synthesize this carotenoid. Only in the case of 15 strains, torularhodin constituted more than 50% of the total carotenoid pool (Table 1). Strain A-106, which was originally isolated as a contaminant in feta cheese stored under refrigerated conditions, produced the highest amount of this compound (81.97%).
Eight yeast strains (A-016, A-042, A-047, A-037, A-032, A-040, A-035, and A-036) synthesized more than 50% of β-carotene in the carotenoid pool. β-Carotene was not detected in the biomass of only one yeast isolate (A-019). Interestingly, this strain synthesized the highest amount of γ-carotene (29.65%) which was found in the smallest amount in most of the isolated strains.
A positive correlation was found between the content of β-carotene and γ-carotene. This can be attributed to the biosynthetic pathway of these compounds. γ-Carotene is a direct precursor of β-carotene [3]. The strains that synthesized more β-carotene and γ-carotene produced a less amount of cellular biomass. The opposite tendency was observed in the case of torulene: a positive correlation was found between the yield of cellular biomass and the content of this compound (Fig. 2). A positive correlation was also observed between the total content of carotenoids and the share of torularhodin.
Fig. 2.
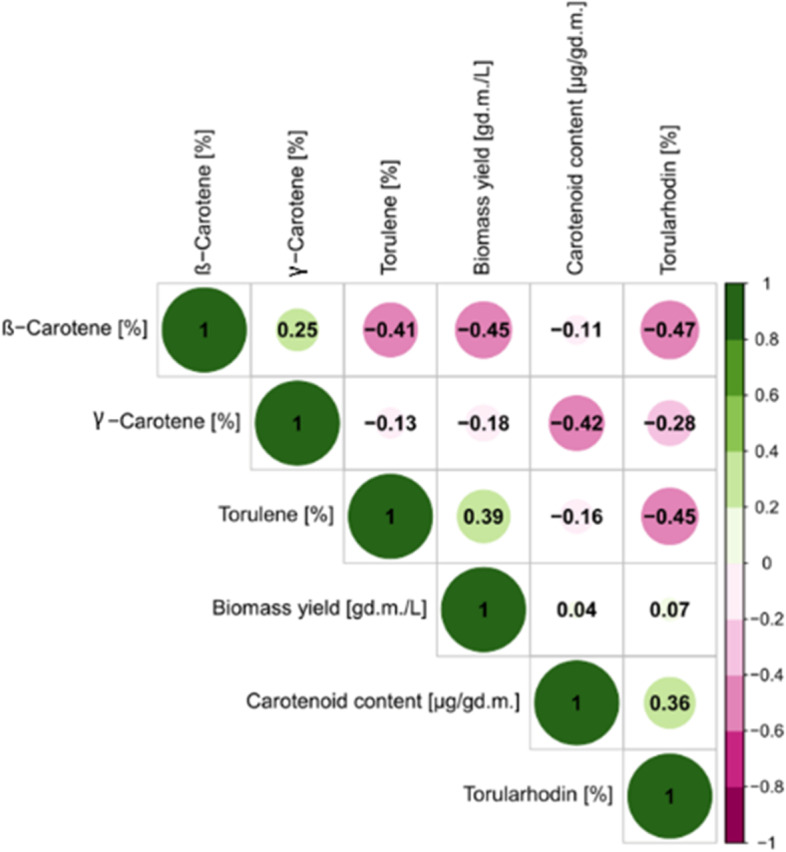
Correlations between the yield of cellular biomass, total carotenoid content and the shares of individual carotenoid fractions (β-carotene, γ-carotene, torulene and torularhodin)
Identification of Selected Yeast Strains
To identify the genus and species of the yeast strains, a selection process was carried out. Yeasts were identified both genotypically and phenotypically. Strains that synthesized more than 200 µg of carotenoids per gram of dry matter (A-002, A-009, A-010, A-012, A-013, A-014, A-020, A-028, A-030, A-031, A-036, A-037, A-038, A-041, A-043, A-044, A-046, A-047, A-04, A-051, A-054, A-056, A-057, A-058, A-061, A-064, A-068, A-070, A-076, A-077, A-079, A-086, A-087, A-088, A-099, A-100, A-101, A-102, A-104, A-110, A-111 and A-114) were selected for further analysis. Those strains that synthesized more than 60% torularhodin (A-026, A-041, A-054, A-055, A-060, A-095, A-099, A-103 and A-106) were also selected. A total of 48 strains of yeast have been identified. The identification process revealed that the selected isolates belonged to the following genera: Rhodotorula, Sporidiobolus, Sporobolomyces, Buckleyzyma, Cystofilobasidium, and Erythrobasidium (Table 2).
Table 2.
Red yeast strains identified by sequence comparison with the BLASTn match with the NCBI GenBank database
| No. | Strain no. | Top BLAST search results | Query cover (%) | Identity (%) | Proposed taxa (GenBank acc. number) | CMIFS collection number |
|---|---|---|---|---|---|---|
| 1 | A-002 | Rhodotorula mucilaginosa | 100 | 100 |
Rhodotorula mucilaginosa (OM256524) |
CMIFS 004 |
| 2 | A-009 | Rhodotorula graminis | 100 | 100 |
Rhodotorula babjevae (ON202973) |
CMIFS 020 |
| Rhodotorula babjevae | 100 | 100 | ||||
| Rhodotorula glutinis | 100 | 100 | ||||
| 3 | A-010 | Rhodotorula graminis | 100 | 100 |
Rhodotorula babjevae (OQ996825) |
CMIFS 094 |
| Rhodotorula babjevae | 100 | 100 | ||||
| Rhodotorula glutinis | 100 | 100 | ||||
| Rhodotorula diobovata | 100 | 100 | ||||
| 4 | A-012 | Sporidiobolus pararoseus | 100 | 100 |
Sporidiobolus pararoseus (ON202961) |
CMIFS 012 |
| Sporidiobolus sp. | 100 | 100 | ||||
| Rhodotorula sp. | 100 | 100 | ||||
| 5 | A-013 | Rhodotorula babjevae | 99 | 100 |
Rhodotorula babjevae (OQ996826) |
CMIFS 096 |
| Rhodotorula glutinis | 99 | 100 | ||||
| Rhodotorula diobovata | 99 | 100 | ||||
| Rhodotorula graminis | 99 | 100 | ||||
| 6 | A-014 | Rhodotorula babjevae | 100 | 100 |
Rhodotorula babjevae (OQ990346) |
CMIFS 040 |
| Rhodotorula glutinis | 99 | 99.82 | ||||
| Rhodotorula diobovata | 99 | 99.82 | ||||
| 7 | A-020 | Rhodotorula graminis | 100 | 100 |
Rhodotorula graminis (ON203044) |
CMIFS 030 |
| Rhodotorula babjevae | 100 | 100 | ||||
| Rhodotorula glutinis | 100 | 100 | ||||
| 8 | A-026 | Rhodotorula graminis | 100 | 99.81 |
Rhodotorula babjevae (ON202975) |
CMIFS 021 |
| Rhodotorula babjevae | 100 | 99.81 | ||||
| Rhodotorula glutinis | 100 | 99.81 | ||||
| Rhodotorula diobovata | 100 | 99.81 | ||||
| Ustilentyloma graminis | 100 | 99.81 | ||||
| 9 | A-028 | Rhodotorula mucilaginosa | 100 | 100 |
Rhodotorula mucilaginosa (OR100603) |
CMIFS 167 |
| Rhodotorula alborubescens | 100 | 100 | ||||
| 10 | A-030 | Rhodotorula graminis | 100 | 100 |
Rhodotorula babjevae (OR020690) |
CMIFS 123 |
| Rhodotorula babjevae | 100 | 100 | ||||
| Rhodotorula glutinis | 100 | 100 | ||||
| Ustilentyloma graminis | 100 | 100 | ||||
| Rhodotorula diobovata | 100 | 100 | ||||
| 11 | A-031 | Rhodotorula diobovata | 100 | 100 |
Rhodotorula glutinis (ON203045) |
CMIFS 032 |
| Rhodotorula glutinis | 100 | 100 | ||||
| Rhodotorula graminis | 99 | 100 | ||||
| 12 | A-036 | Rhodotorula diobovata | 100 | 99.81 |
Rhodotorula babjevae (OR032918) |
CMIFS 171 |
| Rhodotorula glutinis | 100 | 99.81 | ||||
| Rhodotorula graminis | 100 | 99.81 | ||||
| Rhodotorula babjevae | 100 | 99.81 | ||||
| 13 | A-037 | Rhodotorula diobovata | 100 | 100 |
Rhodotorula graminis (OQ996256) |
CMIFS 031 |
| Rhodotorula glutinis | 100 | 100 | ||||
| Rhodotorula graminis | 100 | 100 | ||||
| Rhodotorula babjevae | 100 | 100 | ||||
| 14 | A-038 | Sporidiobolus pararoseus | 100 | 100 |
Sporidiobolus pararoseus (ON202963) |
CMIFS 013 |
| Sporidiobolus sp. | 100 | 100 | ||||
| Sporobolomyces sp. | 100 | 100 | ||||
| 15 | A-041 | Rhodotorula babjevae | 100 | 100 |
Rhodotorula babjevae (OQ826838) |
CMIFS 022 |
| Rhodotorula diobovata | 100 | 100 | ||||
| Rhodotorula glutinis | 100 | 100 | ||||
| 16 | A-043 | Sporobolomyces ruberrimus | 100 | 100 |
Sporobolomyces ruberrimus (ON202960) |
CMIFS 010 |
| Sporidiobolus metaroseus | 100 | 100 | ||||
| 17 | A-044 | Sporidiobolus metaroseus | 100 | 100 |
Sporobolomyces roseus (ON202962) |
CMIFS 011 |
| Sporobolomyces roseus | 100 | 100 | ||||
| Sporobolomyces sp. | 100 | 100 | ||||
| 18 | A-046 | Buckleyzyma aurantiaca | 100 | 99.82 | Buckleyzyma aurantiaca (OM305030) | CMIFS 009 |
| Rhodotorula sp. | 100 | 99.82 | ||||
| 19 | A-047 | Rhodotorula mucilaginosa | 100 | 100 |
Rhodotorula mucilaginosa (OR100624) |
CMIFS 170 |
| 20 | A-048 | Rhodotorula diobovata | 100 | 100 |
Rhodotorula diobovata (OQ826837) |
CMIFS 019 |
| 21 | A-051 | Erythrobasidium hasegawianum | 100 | 100 |
Erythrobasidium hasegawianum (ON202967) |
CMIFS 018 |
| 22 | A-054 | Rhodotorula graminis | 100 | 100 |
Rhodotorula babjevae (OQ828067) |
CMIFS 023 |
| Rhodotorula babjevae | 100 | 100 | ||||
| Rhodotorula glutinis | 100 | 100 | ||||
| Rhodotorula diobovata | 100 | 100 | ||||
| Ustilentyloma graminis | 100 | 100 | ||||
| 23 | A-055 | Rhodotorula glutinis | 100 | 100 |
Rhodotorula glutinis (OQ828070) |
CMIFS 033 |
| Rhodotorula diobovata | 100 | 100 | ||||
| 24 | A-056 | Cystofilobasidium macerans | 100 | 100 |
Cystofilobasidium macerans (ON202966) |
CMIFS 017 |
| 25 | A-057 | Rhodotorula mucilaginosa | 100 | 100 |
Rhodotorula mucilaginosa (OR100518) |
CMIFS 165 |
| Rhodotorula alborubescens | 100 | 100 | ||||
| 26 | A-058 | Sporidiobolus pararoseus | 100 | 100 |
Sporidiobolus pararoseus (ON202964) |
CMIFS 014 |
| Sporidiobolus sp. | 100 | 100 | ||||
| Sporobolomyces sp. | 100 | 100 | ||||
| 27 | A-060 | Rhodotorula mucilaginosa | 100 | 100 |
Rhodotorula mucilaginosa (OQ826830) |
CMIFS 005 |
| 28 | A-061 | Rhodotorula taiwanensis | 100 | 100 |
Rhodotorula taiwanensis (OR036248) |
CMIFS 164 |
| 29 | A-064 | Rhodotorula graminis | 100 | 100 |
Rhodotorula sp. (ON203116) |
CMIFS 035 |
| Rhodotorula babjevae | 100 | 100 | ||||
| 30 | A-068 | Rhodotorula graminis | 100 | 100 |
Rhodotorula babjevae (ON340724) |
CMIFS 024 |
| Rhodotorula babjevae | 100 | 100 | ||||
| Rhodotorula glutinis | 100 | 100 | ||||
| Ustilentyloma graminis | 100 | 100 | ||||
| 31 | A-070 | Rhodotorula graminis | 100 | 100 |
Rhodotorula babjevae (OR004714) |
CMIFS 091 |
| Rhodotorula babjevae | 100 | 100 | ||||
| Rhodotorula glutinis | 100 | 100 | ||||
| Rhodotorula diobovata | 100 | 100 | ||||
| 32 | A-076 | Sporidiobolus pararoseus | 95 | 99.82 |
Sporobolomyces sp. (ON202965) |
CMIFS 016 |
| Sporobolomyces sp. | 95 | 99.82 | ||||
| 33 | A-077 | Rhodotorula mucilaginosa | 100 | 100 |
Rhodotorula mucilaginosa (OR100595) |
CMIFS 166 |
| Rhodotorula alborubescens | 100 | 100 | ||||
| 34 | A-079 | Cystofilobasidium infirmominiatum | 100 | 100 |
Cystofilobasidium infirmominiatum (OQ197480) |
CMIFS 048 |
| Cystofilobasidium sp. | 100 | 100 | ||||
| 35 | A-086 | Rhodotorula graminis | 100 | 100 |
Rhodotorula babjevae (OQ828069) |
CMIFS 025 |
| Rhodotorula babjevae | 100 | 100 | ||||
| Rhodotorula glutinis | 100 | 100 | ||||
| Rhodotorula diobovata | 100 | 100 | ||||
| Ustilentyloma graminis | 100 | 100 | ||||
| 36 | A-087 | Rhodotorula toruloides | 100 | 100 |
Rhodotorula toruloides (OQ990341) |
CMIFS 007 |
| 37 | A-088 | Rhodotorula graminis | 100 | 100 |
Rhodotorula glutinis (ON204010) |
CMIFS 034 |
| Rhodotorula babjevae | 100 | 100 | ||||
| Rhodotorula glutinis | 100 | 100 | ||||
| Rhodotorula diobovata | 100 | 100 | ||||
| Ustilentyloma graminis | 100 | 100 | ||||
| 38 | A-095 | Rhodotorula graminis | 100 | 100 |
Rhodotorula babjevae (ON203025) |
CMIFS 026 |
| Rhodotorula babjevae | 100 | 100 | ||||
| Rhodotorula glutinis | 100 | 100 | ||||
| Rhodotorula diobovata | 100 | 100 | ||||
| Ustilentyloma graminis | 100 | 100 | ||||
| 39 | A-099 | Rhodotorula graminis | 100 | 100 |
Rhodotorula babjevae (ON203027) |
CMIFS 027 |
| Rhodotorula babjevae | 100 | 100 | ||||
| Rhodotorula glutinis | 100 | 100 | ||||
| Rhodotorula diobovata | 100 | 100 | ||||
| 40 | A-100 | Rhodotorula graminis | 100 | 100 |
Rhodotorula babjevae (OR039570) |
CMIFS 169 |
| Rhodotorula babjevae | 100 | 100 | ||||
| Rhodotorula glutinis | 100 | 100 | ||||
| Rhodotorula diobovata | 100 | 100 | ||||
| 41 | A-101 | Rhodotorula glutinis | 100 | 99.81 |
Rhodotorula glutinis (OR039629) |
CMIFS 126 |
| Rhodotorula babjevae | 100 | 99.81 | ||||
| Rhodotorula diobovata | 100 | 99.81 | ||||
| 42 | A-102 | Rhodotorula graminis | 96 | 100 |
Rhodotorula babjevae (ON203028) |
CMIFS 028 |
| Rhodotorula babjevae | 95 | 100 | ||||
| 43 | A-103 | Rhodotorula graminis | 100 | 100 |
Rhodotorula babjevae (ON203029) |
CMIFS 029 |
| Rhodotorula babjevae | 100 | 100 | ||||
| Rhodotorula glutinis | 100 | 100 | ||||
| Rhodotorula diobovata | 100 | 100 | ||||
| Ustilentyloma graminis | 100 | 100 | ||||
| 44 | A-104 | Sporidiobolus pararoseus | 100 | 100 |
Sporidiobolus pararoseus (ON204008) |
CMIFS 015 |
| Sporidiobolus sp. | 100 | 100 | ||||
| Rhodotorula sp. | 100 | 100 | ||||
| 45 | A-106 | Rhodotorula graminis | 100 | 99.45 |
Rhodotorula sp. (ON203118) |
CMIFS 036 |
| Rhodotorula babjevae | 100 | 99.45 | ||||
| Rhodotorula glutinis | 100 | 99.45 | ||||
| Rhodotorula diobovata | 100 | 99.45 | ||||
| Ustilentyloma graminis | 100 | 99.45 | ||||
| 46 | A-110 | Rhodotorula babjevae | 99 | 100 |
Rhodotorula babjevae (OR041764) |
CMIFS 163 |
| Rhodotorula graminis | 99 | 100 | ||||
| 47 | A-111 | Rhodotorula toruloides | 100 | 99.81 |
Rhodotorula toruloides (OM281704) |
CMIFS 008 |
| Rhodosporidium toruloides | 100 | 99.81 | ||||
| 48 | A-114 | Rhodotorula mucilaginosa | 100 | 100 | Rhodotorula mucilaginosa (OM257175) | CMIFS 006 |
| Rhodotorula alborubescens | 100 | 100 |
Seven red yeast strains (A-002, A-028, A-047, A-057, A-060, A-077 and A-114) were identified as Rhodotorula mucilaginosa. The sequences of the LSU domain of strains A-002, A-047 and A-060 showed 100% similarity only to R. mucilaginosa, while strains A-028, A-057, A-077 and A-114 also to Rhodotorula alborubescens. This species was introduced during a phylogenetic revision of the yeast subdivision Pucciniomycotina by Wang et al. in 2015 [20]. Its basionym is Sporobolomyces alborubescens, which was first described in 1930 by Derx [21]. Rhodotorula mucilaginosa and Sporobolomyces alborubescens have an identical sequence of the LSU domain and are physiologically very similar, while the characteristic feature of the yeast S. alborubescens is the ability to use D-glucosamine as a nitrogen source [22]. Furthermore, the yeast R. alborubescens CBS 482, recognized as a typical representative of this species, does not show the ability to assimilate sorbitol and citric acid, but assimilates L-tartaric acid and Tween 80 [23]. All tested isolates did not assimilate D-glucosamine (nitrogen source) and L-tartaric acid and Tween 80 (as carbon sources), but they were able to metabolize sorbitol and citric acid as carbon sources (Table 3). The species R. mucilaginosa is cosmopolitan and has been isolated from natural environments worldwide [8]. It has wide biotechnological applications. Depending on the strain, R. mucilaginosa exhibits the ability to biosynthesize exopolysaccharides [20], patulin-degrading aldolases [24], carotenoids [25], proteases [26], and Single Cell Oil (SCO) [27], and can be used as a biological agent in biocontrol [28]. The yeast R. mucilaginosa is also characterized by detoxification mechanisms that can neutralize the toxic effects of various elements (including selenium) present in the culture medium by forming selenium nanoparticles. Furthermore, this species can transform inorganic selenium into organic and elemental forms (Se0) [29].
Table 3.
Physiological characteristics of Rhodotorula mucilaginosa and Buckleyzyma aurantiaca isolates
| Assimilation | A-002 | A-028 | A-046 | A-047 | A-057 | A-060 | A-077 | A-114 | R. mucilaginosa** | B. aurantiaca* |
|---|---|---|---|---|---|---|---|---|---|---|
| Glucose | + | + | + | + | + | + | + | + | + | + |
| D-Xylose | + | + | + | + | + | + | + | + | + | + |
| Galactose | + | + | w | + | s | + | s | + | +/s | –/w/s |
| L-Arabinose | + | + | + | + | + | + | + | + | + | + |
| L-Rhamnose | – | – | – | – | – | – | – | – | – | – |
| Fructose | + | + | – | + | + | + | + | + | n | n |
| Maltose | + | + | s | + | + | + | + | + | + | s |
| Saccharose | + | + | + | + | + | + | + | + | + | + |
| Lactose | – | – | – | – | – | – | – | – | – | – |
| Glycerol | + | + | + | + | + | + | + | + | + | + |
| D-Mannitol | + | + | + | + | s | + | + | + | v | + |
| Trehalose | + | + | – | + | + | + | + | + | + | v |
| Xylitol | – | + | – | + | + | + | + | – | + | –/w/s |
| Erythritol | – | – | – | – | – | – | – | – | – | – |
| Inulin | – | – | – | – | – | – | – | – | – | – |
| DL-Lactate | + | + | – | + | + | + | + | + | + | –/s |
| L-Tartaric acid | – | – | – | – | – | – | – | – | – | – |
| Citrate | + | + | + | + | + | + | + | + | + | + |
| Ethanol | + | + | + | + | + | + | + | + | + | +/w/s |
| Methanol | – | – | – | – | – | – | – | – | – | – |
| Raffinose | + | + | – | + | + | + | + | + | + | – |
| Nitrates | – | – | – | – | – | – | – | – | – | v |
| D-glucosamine | – | – | x | – | – | – | – | – | – | – |
| Tween 80 | – | – | x | – | – | – | – | – | – | n |
| Sorbitol | + | + | x | + | + | + | + | + | + | n |
| Vanillic acid | + | + | – | + | + | + | + | + | + | – |
| Gallic acid | – | – | – | – | – | – | – | – | – | – |
| Vitamins-free | – | – | – | – | – | – | – | – | – | – |
| Growth at 35 °C | + | + | – | - | + | – | + | + | v | – |
*Typical strains from [8]; ** Typical strains from [8] and [22, 23]; +, positive; l, latent (rapid development of a positive reaction after a lag period); s, positive but slow; w, weak; ws, weak and slow; lw, latent but weak (rapid development of a weak reaction after a lag period); v, variable; –, negative; n, no data; x - not determined
The strain A-046 was identified as Buckleyzyma aurantiaca (Table 2). Previously, this species was classified as Rhodotorula aurantiaca. Later, in 2015, a new genus Buckleyzyma was created by Wang and colleagues, and the species B. aurantiaca was considered typical of this genus. In this study, the A-046 isolate grew slowly on the YPD agar and formed small, intense orange colonies. In assimilation tests, it showed significant similarity to the strains described in “The Yeasts—A Taxonomic Study” (Table 3). A characteristic feature of the isolate was that it could not grow at temperatures above 25℃. Some strains of B. aurantiaca have been classified as psychrophiles and were isolated, for example, in Antarctica [30]. The species B. aurantiaca has been shown to play a role in the biotechnological production of γ-decalactone with a peach flavor [31].
The sequence analysis of the A-043 isolate showed 100% similarity to the sequences of Sporobolomyces ruberrimus and Sporidiobolus metaroseus strains. Therefore, classical physiological tests were carried out, which showed the significant similarity of the A-043 strain to the known strains of S. ruberrimus. The characteristic features of this strain included the inability to assimilate arabinose and lactic acid (Table 4). In addition, the strain was unable to reproduce sexually by producing ellipsoidal teliospores, which is characteristic of S. metaroseus. The species S. ruberrimus was first described by Yamasaki and Fujii in 1950[32] and found to synthesize carotenoids[33] and lipases [34].
Table 4.
Physiological characteristics of Sporobolomyces, Sporidiobolus, Cystofilobasidium, and Erythrobasidium isolates
| Assimilation | A-012 | A-038 | A-043 | A-044 | A-051 | A-056 | A-058 | A-076 | A-079 | A-104 | S. ruberrimus* | S. metaroseus* | S. pararoseus* |
S. patagonicus
** |
C. macerans
* |
C. infirmominiatum* | E. hasegawianum* |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glucose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| D-Xylose | – | + | – | – | + | + | s | – | + | + | – | v | v | – | + | s | + |
| Galactose | + | + | + | + | + | + | + | – | – | + | l | + | + | l | +/s/w | v | s |
| L-Arabinose | + | – | – | – | + | s | + | – | + | s | – | – | v | – | + | + | + |
| L-Rhamnose | – | – | – | – | – | – | – | – | + | – | – | – | – | n | – | + | – |
| Fructose | + | + | + | + | + | + | + | + | + | + | n | n | n | n | n | n | n |
| Maltose | + | + | s | + | + | + | + | + | + | + | + | + | + | v | + | + | w |
| Saccharose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Lactose | – | – | – | – | – | – | – | – | – | – | – | – | – | – | v | v | – |
| Glycerol | + | + | + | + | + | s | + | + | s | + | l | +/s | + | l | s | s | + |
| D-Mannitol | + | + | + | + | + | + | + | + | + | + | l | + | + | l | + | + | + |
| Trehalose | + | + | s | + | + | + | + | + | - | + | + | + | + | v | + | s | + |
| Xylitol | + | + | – | + | – | + | + | s | + | s | n | – | s | n | + | + | n |
| Erythritol | – | – | – | – | – | + | – | – | – | – | – | – | – | – | + | – | – |
| Inulin | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| DL-Lactate | s | s | – | + | s | s | + | – | + | – | – | + | v | – | +/s | + | s |
| L-Tartaric acid | – | – | – | – | – | – | – | – | + | – | n | – | – | n | – | + | – |
| Citrate | + | + | + | + | + | + | – | + | + | + | l | + | + | – | + | + | + |
| Ethanol | + | + | + | + | – | + | + | + | + | + | + | + | + | – | + | + | – |
| Methanol | – | – | – | – | – | – | – | – | – | – | – | – | – | n | – | – | – |
| Raffinose | + | + | + | + | – | + | + | + | + | + | + | + | + | + | + | + | – |
| Nitrates | – | – | + | + | + | + | – | + | + | – | + | + | – | + | + | + | + |
| Vanillic acid | + | + | + | + | – | – | + | + | – | + | n | + | + | n | – | – | – |
| Gallic acid | – | – | – | – | – | – | – | – | – | – | n | – | – | n | – | – | – |
| Vitamins-free | + | + | + | + | – | – | + | + | – | + | + | + | + | + | – | – | – |
| Growth at 35 °C | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
*Typical strains from [8]; +, positive; l, latent (rapid development of a positive reaction after a lag period); s, positive but slow; w. weak; ws, weak and slow; lw, latent but weak (rapid development of a weak reaction after a lag period); v, variable; –, negative; n, no data
The obtained nucleotide sequence indicated that the A-044 strain belonged to S. roseus or S. metaroseus (Table 2). These species are very closely related; therefore, in 2008, it was assumed that S. metaroseus is a teleomorphic form of S. roseus [35]. All examined physiological features of the A-044 isolate corresponded to those described in “The Yeast—A Taxonomic Study” for typical representatives of S. metaroseus (anam. S. roseus) (Table 4). The A-044 strain showed no ability to produce teliospores on corn meal agar. Due to its inability to reproduce, this strain has been classified under S. roseus which has the ability to biosynthesize carotenoids [36] and aspartic protease [37], as well as take part in the transformation of wood lignins into technical lignins [38].
The isolates A-012, A-038, A-058, and A-104 were identified as Sporidiobolus pararoseus (Table 2). A characteristic feature of this yeast that distinguishes it from S. metaroseus and a significant number of Sporobolomyces yeasts is its inability to assimilate nitrates. On a solid medium, the strains of S. pararoseus produced ballistoconidia, which resulted in a mirror image of the colony in the upper Petri dish. All physiological tests of these isolates were consistent with the identification key of [8] for typical representatives of S. pararoseus (Table 4). Some S. pararoseus strains can be used in biocontrol [39] and in the production of exopolysaccharides [40], carotenoids, lipids [41], and enzymes for the synthesis of wine aromas [42].
Sequence analysis showed that the A-076 strain belonged to S. pararoseus or Sporobolomyces sp. with a 99.85% probability and to Sporobolomyces patagonicus with a 99.25% probability (Table 2). However, the membership of this isolate to S. patagonicus was excluded based on physiological tests. The name patagonicus was first proposed by Libkind et al. in 2005 for yeast strains isolated from aquatic environments in Patagonia (Argentina) [43]. So far, only two strains of this species (CBS 9657 and 9658) have been deposited in the collection of the Westerdijk Fungal Biodiversity Institute, which proves that these microorganisms are limited to specific regions of the world. The A-076 strain did not show similarity to S. pararoseus, as it could not assimilate galactose or nitrates (Table 4).
The strain A-056 was identified as Cystofilobasidium macerans, and strain A-079 as Cystofilobasidium infirmominiatum (Table 2). The genus Cystofilobasidium was first described by Oberwinkler in 1983 [44]. Originally, two species, namely C. bisporidiis and C. capitatum, were included in this genus, and later C. infirmominiatum and C. ferigula were added [45]. In 2009, Libkind et al. proposed to include two more species, C. lacus-mascardi and C. macerans, of which the last one is the sexual form of Cryptocoocus macerans [46]. The A-056 and A-079 strains showed all the morphological and physiological characteristics that are described as typical for the representatives of this species in ‘The Yeast – A Taxonomic Study’. So far, the biotechnological use of Cystofilobasidium yeasts has not been well described in the literature. These yeasts can synthesize carotenoids and polygalacturonase [47].
The A-051 strain was isolated from air and identified as Erythrobasidium hasegawianum (Table 2). The identification of selected physiological characteristics of this strain confirmed its genus and species (Table 4). The genus Erythrobasidium was created by Hamamoto, Sugiyama, and Komagata in 1988 and included only one species, which was previously classified as Rhodotorula hasegawae [48]. In 2015, Wang et al. included two more species (E. elongatum and E. yunnanense) which were previously classified in the genus Sporobolomyces [21]. The yeast Erythrobasidium is not widely distributed in the environment and has only been isolated from some sources, including the waters of the Calderone glacier (Italy) [49]. The potential biotechnological uses of Erythrobasidium have not been described so far.
Strain A-061, isolated from blueberry fruit, was identified as Rhodotorula taiwanensis (Tables 2 and 5). A yeast strain of this species was first isolated by Huang et al. in 2011 from the stem of the Artemisia argyi plant in Taiwan [50]. Since then, this yeast has also been isolated from the leaves of Celtis bungeana Bl. plants [51], sugarcane (Saccharum officinarum Linn.) [52] and traditional starter culture of Assam [53]. A characteristic feature of Rhodotorula taiwanensis yeasts is the lack of the ability to produce ballistoconidia or sexual reproduction, as well as the assimilation of ethanol and nitrites [50]. It was found that yeast strains belonging to this species can be a source of lipids for the production of biodiesel [51], biosurfactants [54] and carotenoids, including β-Cryptoxanthin [53], and also conduct the process of aluminum detoxification [55].
Table 5.
Physiological characteristics of Rhodotorula graminis, Rhodotorula glutinis, Rhodotorula diobovata, Rhodotorula taiwanensis, Rhodotorula toruloides and Rhodotorula sp. isolates
| Assimilation | A-020 | A-031 | A-037 | A-048 | A-055 | A-061 | A-064 | A-087 | A-088 | A-101 | A-106 | A-111 |
R. graminis * |
R. glutinis * |
R. diobovata* |
R. toruloides * |
R. taiwanensis ** |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glucose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| D-Xylose | s | s | + | + | s | + | + | + | s | + | + | + | + | s | + | + | + |
| Galactose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | +/s | + |
| L-Arabinose | + | + | + | + | + | + | + | + | s | + | – | + | +/w | + | v | + | + |
| L-Rhamnose | + | – | + | – | – | – | + | – | – | – | + | – | + | – | v | – | – |
| Fructose | + | + | + | + | + | + | + | + | + | + | + | – | + | n | + | n | n |
| Maltose | – | + | – | + | + | + | + | – | + | + | + | – | – | + | + | + | + |
| Saccharose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Lactose | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Glycerol | + | + | + | + | + | + | + | + | + | + | s | + | + | + | + | + | + |
| D-Mannitol | + | + | + | w | + | + | + | + | + | + | s | + | + | + | + | + | + |
| Trehalose | s | s | + | w | + | + | + | + | s | + | + | + | + | + | + | + | + |
| Xylitol | – | + | – | + | s | + | – | + | + | + | – | – | –/s | + | + | + | + |
| Erythritol | – | – | – | – | – | – | + | – | – | – | – | – | – | – | – | – | – |
| Inulin | – | – | – | – | – | – | – | – | – | – | – | – | – | – | +/s | – | – |
| DL-Lactate | – | – | – | – | – | – | + | – | – | – | – | – | – | – | – | v | – |
| L-Tartaric acid | – | + | – | – | + | – | + | – | + | + | + | – | – | + | – | – | n |
| Citrate | + | + | + | + | + | + | s | + | + | + | + | + | + | + | + | +/s | + |
| Ethanol | + | + | + | + | + | – | + | + | + | + | s | – | + | + | + | + | – |
| Methanol | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Raffinose | + | + | + | + | + | + | + | + | + | s | + | + | + | +/s | + | + | + |
| Nitrates | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Vanillic acid | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | n |
| Gallic acid | – | – | – | + | – | – | – | + | – | – | – | + | – | – | + | + | n |
| Glucuronic acid | – | – | – | – | – | – | – | + | – | – | – | – | – | – | n | + | n |
| Vitamins-free | + | + | + | + | + | – | + | + | + | + | + | + | + | + | + | + | n |
| Growth at 35 °C | – | – | – | + | – | + | – | + | – | – | – | + | – | – | + | + | + |
The isolates A-087 and A-111 were identified as belonging to R. toruloides. The results of the biochemical tests were consistent with the identification key of Kurtzman et al. (2011). Only the A-111 strain could not grow in the medium lacking vitamins (Table 5). The species R. toruloides is classified under the family Sporidiobolaceae, the order Sporidiobolales, the class Microbotryomycetes, and the class Basidiomycota. Some edible mushrooms, rusts, and smuts are also included in this group. Originally, R. toruloides was classified as Rhodosporidium toruloides, but in 2015 Wang and colleagues proposed to include this species as well as Rhodosporidium diobovatum, Rhodosporidium babjevae, Rhodosporidium kratochvilovae, Rhodosporidium paludigenum, and Rhodosporidium sphaerocarpum in the genus Rhodotorula. The yeast R. toruloides can biosynthesize lipids and carotenoids [57–59].
Sequence analysis of the the remaining red yeast strains revealed that they possibly belonged mainly to Rhodotorula glutinis, Rhodotorula graminis, Rhodotorula babjevae, Rhodotorula diobovata or Ustilentyloma graminis (Table 2). Gadanho and Sampaio reported that R. glutinis, R. babjevae, and R. graminis show close phenetic and phylogenetic proximity and indicated various assimilation tests which allow distinguishing these species from one another. A characteristic feature of R. glutinis is its ability to assimilate L-arabinose, maltose, tartaric acid, and nitrates and inability to assimilate L-rhamnose, gallic acid, and lactates. The yeast R. graminis can metabolize both L-rhamnose and L-arabinose, and does not absorb maltose, gallic and tartaric acid, and lactate. The typical strains of R. babjevae can assimilate L-rhamnose, maltose, lactate, and tartaric acid. A characteristic feature of R. diobovata is that it can grow up to 35℃ and assimilate L-arabinose, maltose, and gallic acid [56]. Rhodotorula hordea was first included in the genus Ustilentyloma by Wang et al. in 2015 under the name Ustilentyloma graminis [21]. The characteristic features of this yeast are the yellow-cream color of the colony, inability to assimilate L-rhamnose, L-arabinose, xylose, ethanol, xylitol, vanillic acid, gallic acid, and ability to grow in a nutrient-free medium [8]. None of the yeast isolates tested in this study showed these features (Tables 5 and 6).
Table 6.
Physiological characteristics of Rhodotorula babjevae and isolates
| Assimilation | A-009 | A-010 | A-013 | A-014 | A-026 | A-030 | A-036 | A-041 | A-054 | A-068 | A-070 | A-086 | A-095 | A-099 | A-100 | A-102 | A-103 | A-110 | R. babjevae* |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glucose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| D-Xylose | + | + | + | + | + | + | + | w | + | + | + | + | + | + | + | + | + | + | + |
| Galactose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| L-Arabinose | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| L-Rhamnose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Fructose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | n |
| Maltose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Saccharose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Lactose | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Glycerol | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| D-Mannitol | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Trehalose | + | + | + | + | s | + | + | – | s | + | + | s | + | + | + | s | + | s | + |
| Xylitol | + | + | + | + | s | + | + | – | + | + | + | + | + | + | + | + | + | + | + |
| Erythritol | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Inulin | – | – | s | – | – | – | s | – | – | – | s | – | – | – | – | s | – | – | s/– |
| DL-Lactate | + | s | + | + | + | + | s | s | + | + | + | s | + | + | + | + | + | s | +/s |
| L-Tartaric acid | + | + | + | – | – | – | + | + | + | + | + | – | + | + | + | + | + | – | v |
| Citrate | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Ethanol | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Methanol | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Raffinose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Nitrates | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Vanillic acid | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Gallic acid | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Glucuronic acid | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Vitamins-free | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Growth at 35℃ | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
Only the A-048 isolate (Table 5) showed all characteristics of R. diobovata. It was the only tested strain to assimilate gallic acid. Another characteristic feature of this strain was the ability to grow up to 35℃. In 1970, Newell and Hunter isolated the first strain of the yeast Rhodosporidium diobovatum (teleomorphic form) from seawater collected in South Florida [60]. This yeast was also isolated from deep-sea sediments of the eastern Pacific Ocean [61] and cotton grown in Egypt [62]. The yeast R. diobovata may have great biotechnological potential. It is classified as oleaginous and can synthesize and accumulate mainly triacylglycerols. It also has the ability to use waste carbon and nitrogen sources, which reduces the environmental burden of industrial waste [63]. Some strains of R. diobovata can effectively remove inorganic forms of nitrogen from the environment and thus play an important role in bioremediation and wastewater treatment [64].
As reported by Gadanho and Sampaio, a characteristic feature of R. babjevae is the ability to assimilate D-glucuronate, which cannot be found in the species R. glutinis and R. graminis and thus allows easy differentiation between them [56]. In this study, the D-glucuronate-assimilating capacity was exhibited by strains A-009, A-010, A-013, A-014, A-026, A-030, A-036, A-041, A-054, A-068, A-070, A-086, A-095, A-099, A-100, A-102, A-103 and A-110 (Table 6). The analysis of other physiological features confirmed that these strains belonged to R. babjevae. Gadanho and Sampaio also reported that R. babjevae is widespread in nature and some of the strains previously identified as R. glutinis may actually belong to R. babjevae. The possibilities of using these microorganisms in various industries have been widely investigated by researchers. The most important areas of research include the production of biosurfactants [65], carotenoids [66], microbial lipids [67], and glycolipids [68].
The typical features of R. glutinis were shown by isolates A-031, A-055, A-088 and A-101. These strains were able to assimilate L-arabinose, maltose, tartaric acid, and nitrates, and failed to absorb L-rhamnose and gallic acid (Table 5). The yeast R. glutinis was first isolated from sour cream in 1850. Currently, this species is considered typical of the entire genus Rhodotorula [8]. This yeast can be used as a source of SCO [69], carotenoids [70], α-L-arabinofuranosidase [71], PAL [72], and lipases [73].
Strains A-020 and A-037 were identified as belonging to R. graminis (Table 2) because they could not assimilate maltose, tartaric acid, and lactic acid, which allowed distinguishing from R. babjevae and R. glutinis (Table 5). The species R. graminis belongs to the group of R. glutinis sensu stricto together with R. glutinis and R. babjevae; however, it is much less frequently isolated compared to these species [56]. In 1958, Margaret di Menna isolated the first strain of R. graminis from the leaf surface of pasture grasses on the North Island of New Zealand [74]. Since then, the yeasts of this species have been considered as a valuable source of lipids [75], L-phenylalanine ammonia-lyase (PAL) [76] and carotenoids [77].
Strains A-064 and A-106 were identified as belonging only to the genus Rhodotorula because the assimilation profiles of the tested compounds did not allow for their unambiguous classification into R. graminis, R. babjevae, or R. glutinis species.
Summary
Red yeasts are representatives of natural microflora found in different environments in Poland. In this study, a total of 114 red yeast strains were isolated from plant organisms (e.g., flowers, leaves) and environments (e.g., air). All isolated strains which had been pre-selected by formation of colored colonies, synthesized carotenoid pigments. The total content of carotenoids in cellular biomass ranged from 13.88 to 406.50 µg/g. Torulene is a compound that occurs in the highest amounts in yeast cells. Among the yeast isolates, torulene accounted for more than 50% of the total carotenoid pool in 49 strains. The largest number of strains belonged to the genera Rhodotorula (37), Sporidiobolus (4), Sporobolomyces (3), Cystofilobasidium (2), Buckleyzyma (1) and Erythrobasidium (1).
Acknowledgements
The authors would like to thank all employees and students of the WULS Department of Food Biotechnology and Microbiology for providing samples for yeast isolation from various regions of Poland.
Author Contributions
Conceptualization: A.K.; methodology: A.K. and M.K.; formal analysis, A.K.; investigation, A.K., W.S., M.K., K.P. and R.B.; data curation, A.K.; writing—original draft preparation, A.K.; writing—review and editing, A.K and S.B.; visualization, A.K. All authors have read and agreed to the published version of the manuscript.
Funding
Research equipment was purchased as part of the “Food and Nutrition Centre—modernisation of the WULS campus to create a Food and Nutrition Research and Development Centre (CŻiŻ)” project co-financed by the European Union from the European Regional Development Fund under the Regional Operational Programme of the Mazowieckie Voivodeship for 2014–2020 (Project No. RPMA.01.01.00-14-8276/17).
Data Availability
The data presented in this study are available on request from the corresponding author.
Declarations
Ethics Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mannazzu I, Landolfo S, da Silva TL, Buzzini P. Red yeasts and carotenoid production: Outlining a future for non-conventional yeasts of biotechnological interest. World Journal of Microbiology and Biotechnology. 2015;31:1665–1673. doi: 10.1007/s11274-015-1927-x. [DOI] [PubMed] [Google Scholar]
- 2.Frengova GI, Beshkova DM. Carotenoids from Rhodotorula and Phaffia: Yeasts of biotechnological importance. Journal of Industrial Microbiology and Biotechnology. 2009;36:163–180. doi: 10.1007/s10295-008-0492-9. [DOI] [PubMed] [Google Scholar]
- 3.Mussagy CU, Khan S, Kot AM. Current developments on the application of microbial carotenoids as an alternative to synthetic pigments. Critical Reviews in Food Science and Nutrition. 2022;62(25):6932–6946. doi: 10.1080/10408398.2021.1908222. [DOI] [PubMed] [Google Scholar]
- 4.Grune T, Lietz G, Palou A, Ross AC, Stahl W, Tang G, Thurnham S, Yin SA, Biesalski HK. β-carotene is an important vitamin a source for humans. Journal of Nutrition. 2010;140:2268S–2285S. doi: 10.3945/jn.109.119024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gul K, Tak A, Singh AK, Singh P, Yousuf B, Wani AA. Chemistry, encapsulation, and health benefits of β-carotene - A review. Cogent Food & Agriculture. 2015;1(1):1018696. doi: 10.1080/23311932.2015.1018696. [DOI] [Google Scholar]
- 6.Kot AM, Błazejak S, Gientka I, Kieliszek M, Bryś J. Torulene and torularhodin: “New” fungal carotenoids for industry? Microbial Cell Factories. 2018;17:49. doi: 10.1186/s12934-018-0893-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mussagy CU, Gonzalez-Miquel M, Santos-Ebinuma VC, Pereira JFB. Microbial torularhodin – a comprehensive review. Critical Reviews in Biotechnology. 2022;43(4):540–558. doi: 10.1080/07388551.2022.2041540. [DOI] [PubMed] [Google Scholar]
- 8.Kurtzman, C., Fell, J., & Boekhout, T. (2011). The Yeasts - A Taxonomic Study. Elsevier.
- 9.Ray MK, Shivaji S, Shyamala Rao N, Bhargava PM. Yeast strains from the Schirmacher Oasis, Antarctica. Polar Biology. 1989;9(5):305–309. doi: 10.1007/BF00287428. [DOI] [Google Scholar]
- 10.Pavlova K, Grigorova D, Hristozova T, Angelov A. Yeast strains from Livingston Island, Antarctica. Folia Microbiologica. 2001;46(5):397–401. doi: 10.1007/BF02814428. [DOI] [PubMed] [Google Scholar]
- 11.Cheng YT, Yang CF. Using strain Rhodotorula mucilaginosa to produce carotenoids using food wastes. Journal of the Taiwan Institute of Chemical Engineers. 2016;61:270–275. doi: 10.1016/j.jtice.2015.12.027. [DOI] [Google Scholar]
- 12.Kot AM, Błażejak S, Kieliszek M, Gientka I, Bryś J. Simultaneous production of lipids and carotenoids by the red yeast Rhodotorula from waste glycerol fraction and potato wastewater. Applied Biochemistry and Biotechnology. 2019;189(2):589–607. doi: 10.1007/s12010-019-03023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kot AM, Błażejak S, Kurcz A, Bryś J, Gientka I, Bzducha-Wróbel A, Maliszewska, Reczek M. Effect of initial pH of medium with potato wastewater and glycerol on protein, lipid and carotenoid biosynthesis by Rhodotorula glutinis. Electronic Journal of Biotechnology. 2017;27:25–31. doi: 10.1016/j.ejbt.2017.01.007. [DOI] [Google Scholar]
- 14.Chreptowicz K, Mierzejewska J, Tkáčová J, Młynek M, Čertik M. Carotenoid-producing yeasts: Identification and characteristics of environmental isolates with a valuable extracellular enzymatic activity. Microorganisms. 2019;7(12):653. doi: 10.3390/microorganisms7120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Banna AAER, El-Razek A, El-Mahdy AR. Isolation, udentification and screening of carotenoid-producing strains of Rhodotorula glutinis. Food and Nutrition Sciences. 2012;03(05):627–633. doi: 10.4236/fns.2012.35086. [DOI] [Google Scholar]
- 16.Allahkarami S, Sepahi AA, Hosseini H, Razavi MR. Isolation and identification of carotenoid-producing Rhodotorula sp. from Pinaceae forest ecosystems and optimization of in vitro carotenoid production. Biotechnology Reports. 2021;32:e00687. doi: 10.1016/j.btre.2021.e00687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Libkind D, Van Broock M. Biomass and carotenoid pigment production by patagonian native yeasts. World Journal of Microbiology and Biotechnology. 2006;22(7):687–692. doi: 10.1007/s11274-005-9091-3. [DOI] [Google Scholar]
- 18.Sperstad S, Lutnæs BF, Stormo SK, Liaaen-Jensen S, Landfald B. Torularhodin and torulene are the major contributors to the carotenoid pool of marine Rhodosporidium babjevae (Golubev) Journal of Industrial Microbiology & Biotechnology. 2006;33(4):269–273. doi: 10.1007/s10295-005-0065-0. [DOI] [PubMed] [Google Scholar]
- 19.Bao R, Gao N, Lv J, Ji C, Liang H, Li S, Yu C, Wang Z, Lin X. Enhancement of torularhodin production in Rhodosporidium toruloides by Agrobacterium tumefaciens-mediated transformation and culture condition optimization. Journal of Agricultural and Food Chemistry. 2019;67(4):1156–1164. doi: 10.1021/acs.jafc.8b04667. [DOI] [PubMed] [Google Scholar]
- 20.Ashraf SS, Parivar K, Hayati Roodbari N, Mashayekhan S, Amini N. Fabrication and characterization of biaxially electrospun collagen/alginate nanofibers, improved with Rhodotorula mucilaginosa sp. GUMS16 produced exopolysaccharides for wound healing applications. International Journal of Biological Macromolecules. 2022;196:194–203. doi: 10.1016/j.ijbiomac.2021.11.132. [DOI] [PubMed] [Google Scholar]
- 21.Wang QM, Yurkov AM, Göker M, Lumbsch HT, Leavitt SD, Groenewald M, Theelen B, Liu XZ, Boekhout T, Bai FY. Phylogenetic classification of yeasts and related taxa within Pucciniomycotina. Studies in Mycology. 2015;81:149–189. doi: 10.1016/j.simyco.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong SG, Chun J, Nam JS, Park YD, Bae KS. Phylogenetic analysis of genus Sporobolomyces based on partial sequences of 26S rDNA. Journal of Microbiology and Biotechnology. 2000;10(3):363–366. [Google Scholar]
- 23.Touchette D, Altshuler I, Gostinčar C, Zalar P, Raymond-Bouchard I, Zajc J, McKay CP, Gunde-Cimerman N, Whyte LG. Novel Antarctic yeast adapts to cold by switching energy metabolism and increasing small RNA synthesis. The ISME Journal. 2021;16(1):221–232. doi: 10.1038/s41396-021-01030-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li N, Cui R, Zhang F, Meng X, Liu B. A novel enzyme from Rhodotorula mucilaginosa Aldolase: Isolation, identification and degradation for patulin in apple juice. Process Biochemistry. 2022;116:148–156. doi: 10.1016/j.procbio.2022.03.001. [DOI] [Google Scholar]
- 25.Sharma R, Ghoshal G. Optimization of carotenoids production by Rhodotorula mucilaginosa (MTCC-1403) using agro-industrial waste in bioreactor: A statistical approach. Biotechnology Reports. 2020;25:e00407. doi: 10.1016/j.btre.2019.e00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lario LD, Pillaca-Pullo OS, Durães Sette L, Converti A, Casati P, Spampinato C, Pessoa A. Optimization of protease production and sequence analysis of the purified enzyme from the cold adapted yeast Rhodotorula mucilaginosa CBMAI 1528. Biotechnology Reports. 2020;28:e00546. doi: 10.1016/j.btre.2020.e00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bandhu S, Bansal N, Dasgupta D, Junghare V, Sidana A, Kalyan G, Hazra S, Ghosh D. Overproduction of single cell oil from xylose rich sugarcane bagasse hydrolysate by an engineered oleaginous yeast Rhodotorula mucilaginosa IIPL32. Fuel. 2019;254:115653. doi: 10.1016/j.fuel.2019.115653. [DOI] [Google Scholar]
- 28.Gu N, Zhang X, Gu X, Zhao L, Dhanasekaran S, Qian X, Zhang H. Proteomic analysis reveals the mechanisms involved in the enhanced biocontrol efficacy of Rhodotorula mucilaginosa induced by chitosan. Biological Control. 2020;149:104325. doi: 10.1016/j.biocontrol.2020.104325. [DOI] [Google Scholar]
- 29.Ashengroph M, Tozandehjani S. Optimized resting cell method for green synthesis of selenium nanoparticles from a new Rhodotorula mucilaginosa strain. Process Biochemistry. 2022;116:197–205. doi: 10.1016/j.procbio.2022.03.014. [DOI] [Google Scholar]
- 30.Sabri A, Jacques P, Weekers F, Baré G, Hiligsmann S, Moussaïf M, Thonart P. Effect of temperature on growth of psychrophilic and psychrotrophic members of Rhodotorula aurantiaca. Applied Biochemistry and Biotechnology. 2000;84–86:391–399. doi: 10.1385/abab:84-86:1-9:391. [DOI] [PubMed] [Google Scholar]
- 31.Alchihab M, Destain J, Aguedo M, Majad L, Ghalfi H, Wathelet JP, Thonart P. Production of γ-decalactone by a psychrophilic and a mesophilic strain of the yeast Rhodotorula aurantiaca. Applied Biochemistry and Biotechnology. 2009;158(1):41–50. doi: 10.1007/s12010-008-8297-x. [DOI] [PubMed] [Google Scholar]
- 32.Fell JW, Scorzetti G, Statzell-Tallman A, Pinel N, Yarrow D. Recognition of the basidiomycetous yeast Sporobolomyces ruberrimus sp. nov. as a distinct species based on molecular and morphological analyses. FEMS Yeast Research. 2002;1(4):265–270. doi: 10.1111/j.1567-1364.2002.tb00044.x. [DOI] [PubMed] [Google Scholar]
- 33.Cardoso LAC, Jäckel S, Karp SG, Framboisier X, Chevalot I, Marc I. Improvement of Sporobolomyces ruberrimus carotenoids production by the use of raw glycerol. Bioresource Technology. 2016;200:374–379. doi: 10.1016/j.biortech.2015.09.108. [DOI] [PubMed] [Google Scholar]
- 34.Rigoli Ferraz L, dos Santos de Oliveira D, Fernandes Silva M, Rigo E, Di Luccio M, Oliveira JV, Oliviera D, Treichel H. Production and partial characterization of multifunctional lipases by Sporobolomyces ruberrimus using soybean meal, rice meal and sugarcane bagasse as substrates. Biocatalysis and Agricultural Biotechnology. 2012;1(3):243–252. doi: 10.1016/j.bcab.2012.03.008. [DOI] [Google Scholar]
- 35.Valério E, Gadanho M, Sampaio JP. Reappraisal of the Sporobolomyces roseus species complex and description of Sporidiobolus metaroseus sp. nov. International Journal of Systematic and Evolutionary Microbiology. 2008;58(3):736–741. doi: 10.1099/ijs.0.65580-0. [DOI] [PubMed] [Google Scholar]
- 36.Davoli P, Mierau V, Weber RWS. Carotenoids and fatty acids in red yeasts Sporobolomyces roseus and Rhodotorula glutinis. Applied Biochemistry and Microbiology. 2004;40(4):392–397. doi: 10.1023/B:ABIM.0000033917.57177.f2. [DOI] [PubMed] [Google Scholar]
- 37.Krysiak J, Bialkowska A, Turkiewicz M. Aspartic protease – unique enzyme from psychrotolerant yeast Sporobolomyces roseus. New Biotechnology. 2016;33:S109. doi: 10.1016/j.nbt.2016.06.1101. [DOI] [Google Scholar]
- 38.Košíková B, Sláviková E. Biotransformation of lignin polymers derived from beech wood pulping by Sporobolomyces roseus isolated from leafy material. Biotechnology Letters. 2004;26(6):517–519. doi: 10.1023/B:BILE.0000019560.88769.f4. [DOI] [PubMed] [Google Scholar]
- 39.Han P, Ni L, Wei Y, Jiang S, Xu F, Wang H, Shao X. Optimization of the freeze-drying of marine yeast Sporidiobolus pararoseus ZMY-1 for its application in biocontrol of fungal infections. Biological Control. 2021;161:104707. doi: 10.1016/j.biocontrol.2021.104707. [DOI] [Google Scholar]
- 40.Wang H, Hu B, Liu J, Qian H, Xu J, Zhang W. Co-production of lipid, exopolysaccharide and single-cell protein by Sporidiobolus pararoseus under ammonia nitrogen-limited conditions. Bioprocess and Biosystems Engineering. 2020;43(8):1403–1414. doi: 10.1007/s00449-020-02335-3. [DOI] [PubMed] [Google Scholar]
- 41.Manowattana A, Techapun C, Watanabe M, Chaiyaso T. Bioconversion of biodiesel-derived crude glycerol into lipids and carotenoids by an oleaginous red yeast Sporidiobolus pararoseus KM281507 in an airlift bioreactor. Journal of Bioscience and Bioengineering. 2018;125(1):59–66. doi: 10.1016/j.jbiosc.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 42.Baffi MA, Martin N, Tobal TM, Ferrarezi AL, Lago JHG, Boscolo M, Gomes E, Da-Silva R. Purification and characterization of an ethanol-tolerant β-glucosidase from Sporidiobolus pararoseus and its potential for hydrolysis of wine aroma precursors. Applied Biochemistry and Biotechnology. 2013;171(7):1681–1691. doi: 10.1007/s12010-013-0471-0. [DOI] [PubMed] [Google Scholar]
- 43.Libkind D, Gadanho M, van Broock M, Sampaio JP. Sporidiobolus longiusculus sp. nov. and Sporobolomyces patagonicus sp. nov., novel yeasts of the Sporidiobolales isolated from aquatic environments in Patagonia, Argentina. International Journal of Systematic and Evolutionary Microbiology. 2005;55(1):503–509. doi: 10.1099/ijs.0.63322-0. [DOI] [PubMed] [Google Scholar]
- 44.Oberwinkler F, Bandoni R, Blanz P, Kisimova-Horovitz L. Cystofilobasidium: A new genus in the Filobasidiaceae. Systematic and Applied Microbiology. 1983;4(1):114–122. doi: 10.1016/S0723-2020(83)80039-3. [DOI] [PubMed] [Google Scholar]
- 45.Fell JW. Cystofilobasidiales, a new order of basidiomycetous yeasts. International Journal of Systematic Bacteriology. 1999;49(2):907–913. doi: 10.1099/00207713-49-2-907. [DOI] [PubMed] [Google Scholar]
- 46.Libkind D, Gadanho M, van Broock M, Sampaio JP. Cystofilobasidium lacus-mascardii sp. nov., a basidomycetous yeast species isolated from aquatic environments of the Patagonian Andes, and Cystofilobasidium macerans sp. nov., the sexual stage of Cryptococcus macerans. International Journal of Systematic and Evolutionary Microbiology. 2009;59(3):622–630. doi: 10.1099/ijs.0.004390-0. [DOI] [PubMed] [Google Scholar]
- 47.Nakagawa T, Nagaoka T, Miyaji T, Tomizuka N. Cold-active polygalacturonase from psychrophilic-basidiomycetous yeast Cystofilobasidium capitatum strain PPY-1. Bioscience Biotechnology and Biochemistry. 2005;69(2):419–421. doi: 10.1271/bbb.69.419. [DOI] [PubMed] [Google Scholar]
- 48.Hamamoto M, Sugiyama J, Komagata K. Transfer of Rhodotorula hasegawae to a new basidiomycetous genus Erythrobasidium as Erythrobasidium hasegawae comb. nov. The Journal of General and Applied Microbiology. 1988;34(3):279–287. doi: 10.2323/jgam.34.279. [DOI] [Google Scholar]
- 49.Branda E, Turchetti B, Diolaiuti G, Pecci M, Smiraglia C, Buzzini P. Yeast and yeast-like diversity in the southernmost glacier of Europe (Calderone Glacier, Apennines, Italy) FEMS Microbiology Ecology. 2010;72(3):354–369. doi: 10.1111/j.1574-6941.2010.00864.x. [DOI] [PubMed] [Google Scholar]
- 50.Huang CH, Lee FL, Tien CJ, Hsieh PW. Rhodotorula taiwanensis sp. nov., a novel yeast species from a plant in Taiwan. Antonie van Leeuwenhoek International Journal of General and Molecular Microbiology. 2011;99(2):297–302. doi: 10.1007/s10482-010-9489-2. [DOI] [PubMed] [Google Scholar]
- 51.Miao Z, Tian X, Liang W, He Y, Wang G. Bioconversion of corncob hydrolysate into microbial lipid by an oleaginous yeast Rhodotorula taiwanensis AM2352 for biodiesel production. Renewable Energy. 2020;161:91–97. doi: 10.1016/J.RENENE.2020.07.007. [DOI] [Google Scholar]
- 52.Nasanit R, Tangwong-O-Thai A, Tantirungkij M, Limtong S, Boekhout T. The assessment of epiphytic yeast diversity in sugarcane phyllosphere in Thailand by culture-independent method. Fungal Biology. 2015;119:1145–1157. doi: 10.1016/j.funbio.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 53.Parasar DP, Ramakrishnan E, Kabilan S, Kotoky J, Sarma HK. Characterization of β-Cryptoxanthin and other carotenoid derivatives from Rhodotorula taiwanensis, a novel yeast isolated from traditional starter culture of Assam. Chemistry & Biodiversity. 2020;17(12):e2000198. doi: 10.1002/CBDV.202000198. [DOI] [PubMed] [Google Scholar]
- 54.Lyman M, Rubinfeld B, Leif R, Mulcahy H, Dugan L, Souza B. Rhodotorula taiwanensis MD1149 produces hypoacetylated PEFA compounds with increased surface activity compared to Rhodotorula babjevae MD1169. PLoS One. 2018;13(1):e0190373. doi: 10.1371/JOURNAL.PONE.0190373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang C, Wang CY, Zhao XQ, Chen RF, Lan P, Shen F. Proteomic analysis of a high aluminum tolerant yeast Rhodotorula taiwanensis RS1 in response to aluminum stress. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2013;1834:1969–1975. doi: 10.1016/j.bbapap.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 56.Gadanho M, Sampaio JP. Polyphasic taxonomy of the basidiomycetous yeast genus Rhodotorula: Rh. glutinis sensu stricto and Rh. Dairenensis comb. nov. FEMS Yeast Research. 2002;2(1):47–58. doi: 10.1111/j.1567-1364.2002.tb00068.x. [DOI] [PubMed] [Google Scholar]
- 57.Liu Z, Natalizio F, Dragone G, Mussatto SI. Maximizing the simultaneous production of lipids and carotenoids by Rhodosporidium toruloides from wheat straw hydrolysate and perspectives for large-scale implementation. Bioresource Technology. 2021;340:125598. doi: 10.1016/j.biortech.2021.125598. [DOI] [PubMed] [Google Scholar]
- 58.Saran S, Mathur A, Dalal J, Saxena RK. Process optimization for cultivation and oil accumulation in an oleaginous yeast Rhodosporidium toruloides A29. Fuel. 2017;188:324–331. doi: 10.1016/j.fuel.2016.09.051. [DOI] [Google Scholar]
- 59.Bonturi N, Crucello A, Viana AJC, Miranda EA. Microbial oil production in sugarcane bagasse hemicellulosic hydrolysate without nutrient supplementation by a Rhodosporidium toruloides adapted strain. Process Biochemistry. 2017;57:16–25. doi: 10.1016/j.procbio.2017.03.007. [DOI] [Google Scholar]
- 60.Newell SY, Hunter IL. Rhodosporidium diobovatum sp. n., the perfect form of an asporogenous yeast (Rhodotorula sp) Journal of Bacteriology. 1970;104(1):503–508. doi: 10.1128/jb.104.1.503-508.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeng LP, Huang JF, Qiu GZ, Chu FY, Chen D, Tong J, Bin, Luo XG. Isolation and identification of Rhodosporidium diobovatum DS-0205 from deep-sea sediment of eastern Pacific Ocean. Journal of Central South University of Technology. 2009;16(6):942–947. doi: 10.1007/s11771-009-0157-5. [DOI] [Google Scholar]
- 62.Osman ME, Abdel-Razik AB, Zaki KI, Mamdouh N, El-Sayed H. Isolation, molecular identification of lipid-producing Rhodotorula diobovata: Optimization of lipid accumulation for biodiesel production. Journal of Genetic Engineering and Biotechnology. 2022;20:32. doi: 10.1186/s43141-022-00304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fakankun I, Spicer V, Levin DB. Proteomic analyses of the oleaginous and carotenogenic yeast Rhodotorula diobovata across growth phases under nitrogen- and oxygen-limited conditions. Journal of Biotechnology. 2021;332:11–19. doi: 10.1016/j.jbiotec.2021.03.016. [DOI] [PubMed] [Google Scholar]
- 64.Civiero E, Pintus M, Ruggeri C, Tamburini E, Sollai F, Sanjust E, Zucca P. Physiological and phylogenetic characterization of Rhodotorula diobovata DSBCA06, a nitrophilous yeast. Biology. 2018;7(3):39. doi: 10.3390/biology7030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sen S, Borah SN, Bora A, Deka S. Production, characterization, and antifungal activity of a biosurfactant produced by Rhodotorula babjevae YS3. Microbial Cell Factories. 2017;16:95. doi: 10.1186/s12934-017-0711-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim JI, Lee NK, Yeo IC, Ryu YJ, Park HS, Kim BY, Kim HK, Hahm YT. Isolation of carotenoid-producing yeast, Rhodosporidium babjevae JI-1, and evaluation of cell extract toxicity against rat hepatic cells. Journal of the Korean Society for Applied Biological Chemistry. 2012;55(1):137–140. doi: 10.1007/s13765-012-0024-1. [DOI] [Google Scholar]
- 67.Munch G, Sestric R, Sparling R, Levin DB, Cicek N. Lipid production in the under-characterized oleaginous yeasts, Rhodosporidium babjevae and Rhodosporidium diobovatum, from biodiesel-derived waste glycerol. Bioresource Technology. 2015;185:49–55. doi: 10.1016/j.biortech.2015.02.051. [DOI] [PubMed] [Google Scholar]
- 68.Guerfali M, Ayadi I, Mohamed N, Ayadi W, Belghith H, Bronze MR, Ribeiro MH, Gargouri A. Triacylglycerols accumulation and glycolipids secretion by the oleaginous yeast Rhodotorula babjevae Y-SL7: Structural identification and biotechnological applications. Bioresource Technology. 2019;273:326–334. doi: 10.1016/j.biortech.2018.11.036. [DOI] [PubMed] [Google Scholar]
- 69.Maza DD, Viñarta SC, García-Ríos E, Guillamón JM, Aybar MJ. Rhodotorula glutinis T13 as a potential source of microbial lipids for biodiesel generation. Journal of Biotechnology. 2021;331:14–18. doi: 10.1016/j.jbiotec.2021.03.002. [DOI] [PubMed] [Google Scholar]
- 70.Mussagy CU, Guimarães AAC, Rocha LVF, Winterburn J, de Santos-Ebinuma V, Pereira JF. Improvement of carotenoids production from Rhodotorula glutinis CCT-2186. Biochemical Engineering Journal. 2021;165:107827. doi: 10.1016/j.bej.2020.107827. [DOI] [Google Scholar]
- 71.Martínez C, Gertosio C, Labbe A, Pérez R, Ganga MA. Production of Rhodotorula glutinis: A yeast that secretes α-L-arabinofuranosidase. Electronic Journal of Biotechnology. 2006;9:407–413. doi: 10.2225/vol9-issue4-fulltext-8. [DOI] [Google Scholar]
- 72.Zhu L, Zhou L, Cui W, Liu Z, Zhou Z. Mechanism-based site-directed mutagenesis to shift the optimum pH of the phenylalanine ammonia-lyase from Rhodotorula glutinis JN-1. Biotechnology Reports. 2014;3:21–26. doi: 10.1016/j.btre.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khayati G, Alizadeh S. Extraction of lipase from Rhodotorula glutinis fermentation culture by aqueous two-phase partitioning. Fluid Phase Equilibria. 2013;353:132–134. doi: 10.1016/j.fluid.2013.05.037. [DOI] [Google Scholar]
- 74.Di Menna ME. Two new species of yeasts from New Zealand. Journal of General Microbiology. 1958;18(1):269–272. doi: 10.1099/00221287-18-1-269. [DOI] [PubMed] [Google Scholar]
- 75.Galafassi S, Cucchetti D, Pizza F, Franzosi G, Bianchi D, Compagno C. Lipid production for second generation biodiesel by the oleaginous yeast Rhodotorula graminis. Bioresource Technology. 2012;111:398–403. doi: 10.1016/j.biortech.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 76.Rowles I, Groenendaal B, Binay B, Malone KJ, Willies SC, Turner NJ. Engineering of phenylalanine ammonia lyase from Rhodotorula graminis for the enhanced synthesis of unnatural L-amino acids. Tetrahedron. 2016;72(46):7343–7347. doi: 10.1016/j.tet.2016.06.026. [DOI] [Google Scholar]
- 77.Buzzini P, Martini A, Gaetani M, Turchetti B, Pagnoni UM, Davoli P. Optimization of carotenoid production by Rhodotorula graminis DBVPG 7021 as a function of trace element concentration by means of response surface analysis. Enzyme and Microbial Technology. 2005;36(5–6):687–692. doi: 10.1016/j.enzmictec.2004.12.028. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.