Summary
Approximately 20% of breast cancer cases are attributed to increased family risk, yet variation in BRCA1/2 can only explain 20%–25% of cases. Historically, only single gene or single variant testing were common in at-risk family members, and further sequencing studies were rarely offered after negative results. In this study, we applied an efficient and inexpensive targeted sequencing approach to provide molecular diagnoses in 245 human samples representing 134 BRCA mutation-negative (BRCAX) hereditary breast and ovarian cancer (HBOC) families recruited from 1973 to 2019 by Dr. Henry Lynch. Sequencing identified 391 variants, which were functionally annotated and ranked based on their predicted clinical impact. Known pathogenic CHEK2 breast cancer variants were identified in five BRCAX families in this study. While BRCAX was an inclusion criterion for this study, we still identified a pathogenic BRCA2 variant (p.Met192ValfsTer13) in one family. A portion of BRCAX families could be explained by other hereditary cancer syndromes that increase HBOC risk: Li-Fraumeni syndrome (gene: TP53) and Lynch syndrome (gene: MSH6). Interestingly, many families carried additional variants of undetermined significance (VOUSs) that may further modify phenotypes of syndromic family members. Ten families carried more than one potential VOUS, suggesting the presence of complex multi-variant families. Overall, nine BRCAX HBOC families in our study may be explained by known likely pathogenic/pathogenic variants, and six families carried potential VOUSs, which require further functional testing. To address this, we developed a functional assay where we successfully re-classified one family’s PMS2 VOUS as benign.
Keywords: Cancer, Hereditary cancer, Lynch Syndrome, Hereditary breast and ovarian cancer (HBOC) syndrome, Targeted sequencing, Variants of undetermined significance
This study utilizes multi-gene targeted sequencing to identify cancer risk variants in BRCA1/2-negative families. It highlights the value of family-based biobanks to better understand hereditary cancer genetics. The results emphasize that expanded clinical sequencing must be used in conjunction with functional validation of variants to clarify cancer risk.
Introduction
Breast cancer remains the most common cancer diagnosed among women and is a leading cause of cancer-related death across all races and ethnicities,1 representing a critical public health challenge. Having a relative with breast cancer raises an individual’s risk for developing the disease by as high as 5.7-fold over the course of a lifetime.2,3 Approximately 20% of breast and ovarian cancers are familial, signifying the presence of underlying genetic risk factors; however, only 20%–25% of hereditary breast and ovarian cancer (HBOC) cases have inherited mutations in BRCA1 [MIM: 113705] or BRCA2 [MIM: 600185].4,5,6,7,8 While BRCA1/2 are the most common targets for clinical sequencing in suspected HBOC cases, these are not the only known genetic risk factors.9,10 A current leading cause of breast cancer diagnoses is CHEK2 [MIM: 604373] mutations.11,12 Of note, CHEK2 mutations contribute an additional 20%–40% increased risk for breast cancer during one’s lifetime. On a molecular level, CHEK2 is interconnected with both TP53 [MIM: 191170] and ATM [MIM: 607585], creating the ATM-CHEK2-p53 axis. This axis is required for key cellular mechanisms, such as responses to DNA damage.11 Mutations in both ATM13 and TP5314 also carry increased risks for breast cancer, as do mutations in additional genes involved in other DNA damage repair responses, like mismatch repair (MMR).15 Many of these genes define well-described hereditary cancer conditions like Lynch syndrome (LS) [MIM: 120435, 609310, 613244, 614337, 614350]16 and Li-Fraumeni syndrome (LFS) [MIM: 151623].17 Yet, the genetic architecture of HBOC is still not well understood in many HBOC cases.18,19,20 Early screening and prophylactic measures, such as a bilateral mastectomy, have proven to be successful in significantly decreasing (≤90%) the incidence of breast cancer diagnoses in known BRCA1/2 mutation carriers.21 Although several other hereditary cancer susceptibility genes have been identified, protocols have not been well established for interpreting risk in BRCA mutation-negative (BRCAX) individuals and families related to secondary cancers, family planning, early surveillance, and prophylactic measures.
In this study, we performed targeted sequencing of 28 high-risk cancer genes using a custom molecular inversion probe (MIP) gene panel in 134 HBOC families from the Lynch Memorial Biobank. While whole-genome sequencing is becoming more accessible over time, MIPs provide an inexpensive, accurate, and scalable approach for the identification of high-risk variants in potential HBOC families and have been used across a variety of disease contexts.22,23,24,25,26 Combined with the deep clinical power of our biobank, we (1) describe previously undefined HBOC risk, (2) identify novel HBOC risk variants, and (3) resolve the functional impact and penetrance of VOUSs. Eight BRCAX families could be resolved by known pathogenic mutations in other known HBOC genes. Through segregation analysis, we identified a BRIP1 VOUS in one family that is likely pathogenic. Several families carried one or more VOUSs. To examine VOUSs further, we developed an assay for testing the direct functional impact of a PMS2 [MIM: 600259] VOUS present in one family with significant clinical evidence of LS. Our data led to the surprising re-classification of this variant as benign, in agreement with our pedigree cosegregation analysis. In the future, this assay may be more broadly applied to test additional VOUSs. Still, many of the families in this study remain undefined by variants in known HBOC risk genes, suggesting that there is still much work to be done in hereditary cancer genetic risk discovery.
Materials and methods
Research participants
Human samples were obtained from the Lynch Memorial Biobank under Creighton IRB protocol #1185786-1 (approved 6 November 2018). These individuals were drawn from suspected HBOC families recruited for research studies and biobanked from 1973 to 2019. HBOC risk was previously determined by Dr. Henry Lynch through deep pedigree analysis. For this study, we included only families that (1) had two or more individuals diagnosed with breast or ovarian cancer and (2) had previously tested negative for BRCA variants under a research or clinical protocol. Stored DNA extracted from either blood or saliva samples, cancer diagnoses, and family pedigrees were provided de-identified to the research team.
Gene selection and MIP design
MIP probes were designed to span all RefSeq exons plus 10 base pairs of flanking introns for 31 candidate HBOC risk genes (Table 1) using the MIPGEN design tool27 and the hg19 reference genome (e.g., BRCA2 shown in Figure S1). In total, 1,855 MIP probes were synthesized by Integrated DNA Technologies (IDT) (Table S1) for targeted capture.
Table 1.
Target genes
| Risk gene | Associated cancer(s) |
|---|---|
| BRCA1, BRCA2, RAD51C, RAD51D, CHEK2, PALB2, ATM, NBN, CDH1, BRIP1, and RAD50 | breast and ovarian |
| MLH1, MSH2, MLH3a, MSH6, PMS1a, PMS2, EPCAM, and RINT1 | endometrial, colorectal, ovarian, stomach, small intestinal, pancreatic, kidney, brain, and bile duct |
| BARD1, ABRAXAS1, FANCC, and XRCC2 | breast |
| TP53 | osteosarcoma, soft-tissue sarcoma, leukemia, brain (CNS), adrenal cortex, and breast |
| PTEN | breast, thyroid, and endometrial |
| APC | brain, colorectal, desmoid, gastric, hepatoblastoma, thyroid, and breast |
| STK11a | breast, skin, pancreatic, and testicular |
| MRE11 | breast, ovarian, bladder, colorectal, and rectal |
| NF1 | peripheral nerve sheath, CNS, breast, pheochromocytoma, gastrointestinal, fibrous histiocytoma, and thyroid |
| CDKN2A | melanoma, pancreatic, and breast |
| PIK3CA | breast, lung, ovarian, stomach, brain, colorectal, and rectal |
Genes were not included in the final capture design due to poor performance during optimization.
MIP capture, amplification, and sequencing
The MIP protocol was performed as previously published.28 In brief, column-synthesized MIPs were initially pooled at equal ratios (1×) with a probe:DNA ratio of 800:1. DNA capture was performed over 22 h, followed by PCR amplification to add individual barcode tags (Table S2) to each sample. Protocol optimization was performed on an Illumina MiSeq instrument at Creighton University using a 150 base-pair paired-end protocol (300 cycles; V2 chemistry). Empirical rebalancing of the MIPs was used to further improve capture uniformity (Table S3). All experimental samples captured using this final MIP pool were sequenced (N = 384 per run) at the University of Nebraska Medical Center Genomics Core facility on an Illumina NextSeq 500 MidOutput 300 cycle V2 flowcell.
MIP data analysis
MIP data were analyzed using the MIPGEN27 data analysis tools and mapped to the hg19 human reference genome. Variant calling was performed using Freebayes v1.1.0-3-g961e5f3 on mapped bam files. High-quality variants (depth >8×; quality score >20) were annotated using the Ensembl Variant Effect Predictor tool (GRCh37 release 110).29 Variants were compared with both gnomAD v2.1.1 (https://gnomad.broadinstitute.org/) and the COSMIC databases (http://cancer.sanger.ac.uk/cosmic). Additional manual annotation of variants was performed using ClinVar30 and potential pathogenic risk was ranked using these criteria.
Validation of variants
Potential risk variants were PCR amplified using variant-spanning primers (IDT) and 2× PCR Master Mix (Roche). Reactions were either (1) treated with ExoSAP-IT (Thermo Fisher Scientific) and Sanger validated at the University of Nebraska Medical Center Genomics Core facility or (2) cleaned up with 1.8× Ampure XP beads (Beckman Coulter), eluted in nuclease free water, and Sanger sequenced at Genewiz/Azenta.
PMS2 functional assay
The human HAP1 haploid cell line (Horizon Discovery) was cultured in IMDM (Gibco) supplemented with 10% fetal bovine serum (Gibco) in a 37°C incubator with 5% CO2 and used to generate a PMS2 knockout (KO) clonal cell line using CRISPR-Cas9 technology. A CRISPR guide RNA was designed to exon 7 of the predominant PMS2 transcript (NM_000535.7), an exon shared across all annotated PMS2 transcripts. The AltR gRNA/tracRNA/Cas9 complex (IDT) was electroporated (Neon, Invitrogen) into the wild-type (WT) or parental HAP1 cell line to introduce insertions and/or deletions at the target site. Limiting dilution was used to isolate an isogenic HAP1 cell line carrying a protein-disruptive variant (frameshifting or nonsense) confirmed by Sanger sequencing (Genewiz/Azenta). Absence of PMS2 protein expression in the KO line was further validated by western blot.
The PMS2 NM_000535.7 coding sequence was cloned into a Gateway entry plasmid (Invitrogen) and recombined into the pCW57.1 lentiviral vector (Addgene) using a Gateway LR reaction (Invitrogen). Q5 site-directed mutagenesis (NEB) was performed on the PMS2-pCW57.1 plasmid to generate PMS2 variants of interest. PMS2-pCW57.1 constructs were packed into lentiviral particles produced by transient transfection of 293T cells using PolyJet transfection reagent as directed by the manufacturer (SignaGen Laboratories) using a 2.5:2.5:1 ratio of pCW57.1 plasmid, psPAX2 packaging vector, and pMD2.G VSVg envelope expression vector (Addgene). After transfection, each supernatant was collected twice at 24-h intervals (48 h total) and combined. After the second collection, the viral supernatant was clarified by centrifugation at 4,500×g for 5 min, aliquoted, and stored at −80°C. These were used for a MMR functional assay as previously described31 with the following revisions. Briefly, 750 μL viral supernatant (with 8 μg/mL polybrene) was added to approximately 5 × 106 cells seeded into two tissue-culture treated 100-mm dishes (Falcon). After 48 h, the media was replaced with media containing 1 μg/mL G418 (Roche) for 2 weeks. To turn on PMS2 expression (from the integrated PMS2-pCW57.1 cassette), doxycycline (1 μg/mL; Sigma) was added to cells seeded at 3 × 105 in T25 flasks; media with doxycycline was replaced every 24 h. After 72 h, cells were re-seeded in media with either 6-thioguanine (6-TG; 5 μM) or with a vehicle (media only) control at a cell density of 3 × 104 into 12-well plates for MMR selection. After 72 h, cell supernatants were collected, adherent cells were trypsinized, and cell solutions were combined to seed triplicate wells of each treatment condition into white, opaque 96-well half well plates (PerkinElmer). An equal volume,15 μL, of cell solution and CellTiter-Glo buffer was added to each well to measure cell viability. Cell viability data were normalized first to the 0 h time point to control for variability in cell growth and then for doxycycline treatment. Finally, 6-TG treatment data were compared with the 6-TG vehicle control. Data were analyzed using a one-way ANOVA followed by post hoc Tukey’s multiple comparisons tests with GraphPad Prism 9. Data are presented as means of at least three independent experiments (each measured in technical triplicate ± SEM).
MMR functional assay of Epstein-Barr virus-transformed lymphocyte lines
Epstein-Barr virus (EBV)-transformed lymphocyte cell lines were retrieved from the Lynch Memorial Biobank. These samples were previously generated from blood samples collected from research participants. Lymphocyte cell lines were cultured in RPMI 1640 medium (Gibco) supplemented with 10%–15% fetal bovine serum (Gibco) and 1× penicillin-streptomycin (Gibco) in a 37°C incubator with 5% CO2. Three slow-growing individual lines were additionally supplemented with 1× insulin-transferrin-selenium (Gibco). Cell lines were tested routinely for mycoplasma, and all tests were negative.
The same PMS2 functional assay described above was used with the lymphocyte lines with the following modifications. Cells were seeded at 5 × 105 in 12-well plates in media with either 6-TG (15 μM) or vehicle (media only) for MMR selection. After 72 h, cells were re-suspended and re-seeded into triplicate wells of white, opaque 96-well half-well plates (PerkinElmer). An equal volume of cell solution and CellTiter-Glo buffer (15 μL) was added to each well for cell viability testing. Cell viability data were normalized first to the 0 h time point and then to the vehicle control. Data were analyzed using an unpaired t test with GraphPad Prism 9. Data are presented as means of at least three independent experiments (each measured in technical triplicate ± SEM).
Western blotting
Approximately 5 × 106 cells were collected and lysed using RIPA lysis buffer (Thermo Scientific) with Halt protease inhibitor cocktail (Thermo Fisher Scientific). Lysates were quantified using the Pierce Rapid Gold BCA Protein Assay (Thermo Fisher Scientific). Five micrograms of denatured lysates were separated on 4%–20% MP TGX stain-free gels (BioRad) at 200 V for 30 min. Gels were transferred to mini polyvinylidene fluorise membranes using the Trans-Blot Turbo Transfer System (BioRad) and blocked with EveryBlot Blocking buffer (BioRad). Blots were probed with a 1:1,000 dilution of mouse anti-PMS2 (BD Biosciences, A16-4) antibody or a 1:20,000 dilution of rabbit anti-GAPDH (Abcam, ab181602) antibody followed by a 1:20,000 dilution of goat anti-mouse IgG H&L HRP (Abcam, ab205719) or a 1:10,000 dilution of goat anti-rabbit IgG H&L HRP (Abcam, ab205718) secondary antibody, respectively. Blots were visualized using Clarity Western ECL substrate (BioRad) on a ChemiDoc Touch (BioRad).
Results
Multi-gene-targeted sequencing identifies high-risk variations in BRCAX families
Under the hypothesis that a portion of BRCAX HBOC cases can be explained by inherited coding mutations involving high-risk cancer genes other than BRCA1/2, we designed a targeted sequencing strategy to simultaneous and inexpensively sequence all the coding regions of 31 high-risk breast and ovarian cancer genes (Table 1) using MIPs. While all probes were initially tested at equimolar concentrations (1×), increasing the concentration of all moderate- and poor-performing MIPs (n = 382) to 30× improved overall capture uniformity (Figure S2). Poor MIP performance across MLH3 [MIM: 604395], PMS1 [MIM: 600258], and STK11 [MIM: 602216] prompted removal of these genes from the final capture design, resulting in a total of 28 target genes. In total, we captured 245 participant samples representing both cancer positive (n = 173) and negative (n = 72) individuals from 134 BRCAX families (Tables 2 and S4).
Table 2.
Individual samples tested from BRCAX families
| Aff with Br/Ov | Aff with other | Unaffa | |
|---|---|---|---|
| BRCAX family members (n = 134 families) | 149 | 24 | 72 |
Aff, Cancer diagnosis; Unaff, unaffected; Br/Ov, breast or ovarian cancer; Other, any other cancer diagnosis (not Br/Ov).
No cancer diagnosis at time of last contact.
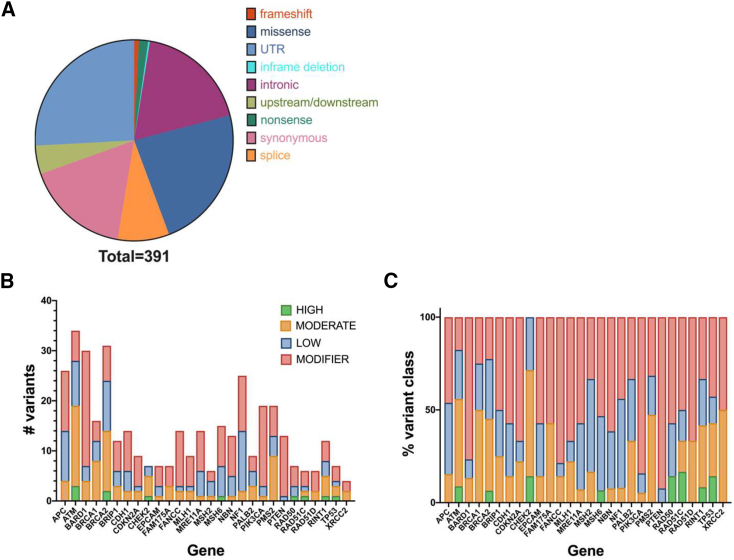
Sequencing identified 1,144 variants that were reduced to 391 when limited by threshold depth and quality (depth > 8; quality > 20) across all samples. Filtered out variants were more likely attributed to low sequencing depth (Figure S3, and Table S4) across many samples rather than issues of sequencing quality. Of the 391 variants, 235 survived further filtering criteria at the level of at least one individual sample. These were considered the highest quality variants (evidence = high) (Table S5); however, lower quality variants were retained (evidence = low; n = 156) (Table S5) for validations where warranted. All 391 variants were annotated using the Ensembl Variant Effect Predictor tool.29 As expected, given the exon-targeted capture design, many variants were predicted to be protein coding (Figures 1A–1C, and Table S5). Coding variants (nonsense, frameshifting, missense, and splice) were further binned based on their disease-causing potential to identify potentially pathogenic variants. This was based on a combination of prediction algorithms and ClinVar data.
Figure 1.
Variant summary
(A) Identified variants limited by depth and quality filtering.
(B and C) High-quality variants (n = 235/391) binned by predicted functional impact. Data shown as the total number of variants (B) and variant type by percentage (C) by gene.
Known pathogenic variants in TP53 [MIM: 191170], CHEK2, BRCA2, RAD50 [MIM: 604040], RAD51C [MIM: 179617], and MSH6 [MIM: 600678] were identified among this collection (Table S6). Predicted high-risk coding variants (nonsense, frameshifting, or splice) were also identified in ATM [MIM: 607585], BRCA2, and RINT1 [MIM: 610089]. Only one of these variants—a nonsense variant in BRCA2 (ENST00000544455.1:c.9976A>T; ENSP00000439902.1:p.Lys3326Ter)—has been extensively studied (rs11571833). While identification of this variant in non-cancer populations prompted an initial likely benign classification, more recent work suggests that this variant may indeed be associated with some level of breast cancer risk.32 For the remaining variants, we manually inspected the sequencing quality of each using the Integrated Genomics Viewer (Broad Institute) and mined the literature for data supporting potential pathogenicity (Table S6). Twenty-four variants had data supporting a potential VOUS clinical classification. These variants were in the following genes: ATM, BRIP1 [MIM: 605882], CDH1 [MIM: 192090], CHEK2, FAM175A [MIM: 611143], FANCC [MIM: 613899], MLH1 [MIM: 120436], MRE11A [MIM: 600814], NF1 [MIM: 613113], PMS2, RINT1, TP53, and XRCC2 [MIM: 600375]. All suspected pathogenic and VOUS variants were Sanger validated (Table S7). For each family with a validated variant, all other available family members were also tested by Sanger sequencing (Table S8).
Known breast cancer variants and hereditary cancer syndromes resolve a portion of BRCAX families
CHEK2
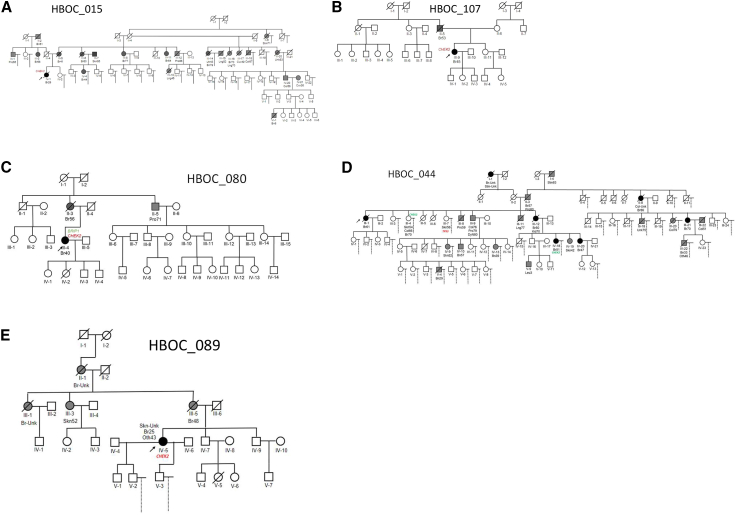
Five families in this study—families 015, 044, 080, 084, and 107—carried the same pathogenic CHEK2 frameshifting variant (ENST00000382580.2:c.1229del; ENSP00000372023.2 p.Thr410MetfsTer15). Families 015 (Figure 2A) and 107 (Figure 2B) carried only this variant. Among white women of northern European descent, this variant (also known as CHEK2∗1100delC) has been estimated as present in 1.5% of familial breast cancer cases and is associated with a 24% increased lifetime risk for developing breast cancer.33 Of the CHEK2∗c.1100delC families, three carried additional pathogenic or VOUS variants (084, 044, and 080) that may have contributed to additional cancer risk. The breast cancer affected proband (III-4; diagnosed at age 40) in family 080 (Figure 2C) carried both the frameshifting variant in CHEK2 (ENST00000382580.2:c.1229del) and a private 3′ UTR variant in BRIP1 (NC_000017.10:g.59756617A>T). This BRIP1 variant has been reported on ClinVar twice: once with a likely benign and once with a VOUS classification (VCV000891958.7). However, we were unable to assess their cosegregation with cancer in this family, as only the proband sample was available for testing. It is interesting that CHEK2 and BRIP1 converge on BRCA1 suggesting the combination of these two intermediate penetrance variants could compound HBOC risk.34 Given the breast cancer diagnosis at age 56 in the proband's mother (II-3), the HBOC genetic risk is suspected to have derived from that lineage. In family 044 (Figure 2D), the CHEK2 variant was identified in an affected sister (III-4; diagnoses of skin cancer at 54, colorectal cancer at 66, and breast cancer at 70) of the affected proband (III-1) and one of their shared affected nieces (IV-18; diagnosed with breast cancer at 51). Another sister of the proband (III-7; diagnosed with skin cancer at 56) carried a 3′ UTR variant in TP53 (NC_000017.10:g.7571752T>G) that is predicted to be likely benign on ClinVar (VCV000035555.22).30 Importantly, no one in this family carried both variants. The CHEK2 variant cosegregates with the HBOC phenotype with expected moderate penetrance, whereas the TP53 variant may have been introduced from a different founder and may contribute to risk. Family 084 will be discussed further later.
Figure 2.
Families identified with CHEK2 pathogenic mutations
(A–D) Family pedigrees carrying the CHEK2∗c.1100delC variant (E) Family pedigree carrying a CHEK2 c.1412C>T variant. Cancer types: Bn, brain; Br, breast; Col, colorectal; Cvx, cervix; Lng, lung; Oth, unknown source; Pro, prostate; Skn, skin; Stm, stomach; Utr, uterine.
A different CHEK2 variant (ENST00000382580.2:c.1412C>T; ENSP00000372023.2:p.Ser471Phe) was identified in family 089 (Figure 2E). This variant has been associated with approximately a 2-fold increased risk (odds ratio, 2.13; 95% confidence interval, 1.26–3.69; p = 0.004) of breast cancer among individuals of Ashkenazi Jewish ancestry.35 Functional studies have shown this variant results in protein loss of function.35,36 The relative increased frequency of this variant among cancer cases versus unaffected controls and its segregation among hereditary cancer families all suggest that it is a pathogenic, low-penetrance variant.35,36 Only the female proband (IV-5) had a DNA sample available for testing in this family. She was diagnosed with breast cancer at age 25, skin cancer at an unknown age, and an unknown cancer at age 43 (Figure 2E). Her mother (III-5) was diagnosed with breast cancer at age 48, as were the proband’s maternal aunt and grandmother (III-1, II-1) at unknown ages, suggesting this risk variant was likely inherited through the maternal lineage. Historic clinical records for this family confirmed evidence of Ashkenazi Jewish ancestry. Taken together, we conclude that this is likely a primary genetic risk factor for breast cancer in this family.
BRCA2
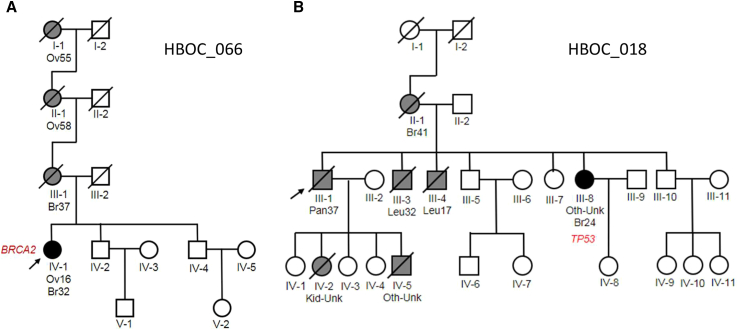
The most surprising finding was a known pathogenic BRCA2 variant (ENST00000544455.1:c.573_574del; ENSP00000439902.1:p.Met192ValfsTer13) seen in family 066 (Figure 3A). The variant was identified in the proband (IV-1), who was diagnosed at age 16 with ovarian cancer and breast cancer at 32. While no samples were available for other members of the family, the proband’s mother (III-1) was diagnosed with breast cancer at age 37. The maternal grandmother and great-grandmother both had a history of ovarian cancer (II-1, I-1). Limited pedigree information suggested an autosomal-dominant, highly penetrant variant. This is consistent with our BRCA2 finding.
Figure 3.
Families with known pathogenic variants defining hereditary cancer syndromes
(A) Family identified with BRCA2 c.573_574del variant. Pedigree supports a highly penetrant autosomal dominant pattern of inheritance.
(B) Family identified with TP53 c.586C>T variant. The variant is pathogenic and associated with LFS. Proband is an affected non-carrier. Cancer types: Br, breast; Kid, kidney; Leu, leukemia; Oth, unknown source; Ov, ovarian; Pan, pancreatic.
TP53
Cancer diagnoses in family 018 could be explained by a known pathogenic variant in TP53 (ENST00000269305.4:c.586C>T; ENSP00000269305.4:p.Arg196Ter), which defines another hereditary cancer syndrome, LFS (Figure 3B). Classic LFS is characterized by osteosarcomas, soft tissue sarcomas, acute leukemia, brain cancer, and adrenal cortical tumors. LFS breast cancer risk ranges from 54% to 85%.37 Some pedigrees also present with melanomas, Wilms' tumor (kidney cancer), gastrointestinal, and lung and germ cell cancers.38,39 Indeed, the proband (III-1) in this family presented with pancreatic cancer at age 37. His mother (II-1) was affected with breast cancer at age 41, two brothers with leukemia at 32 and 17 years of age (III-3, III-4), and a sister (III-8) with breast cancer at age 24 and an undefined cancer type at an unknown age. Two of the proband’s five biological children (IV-2, IV-5) were also affected by cancers as infants (consistent with LFS). The only family member who provided a DNA sample was the proband’s affected sister (III-8), where the pathogenic TP53 variant was identified and validated (Figure 3B).
False positives and negatives
The pathogenic variant in RAD51C (ENST00000337432.4:c.97C>T; ENSP00000336701.4:p.Gln33Ter) identified in an unaffected family member of family 075 proved to be a false positive by Sanger sequencing. This was surprising, given the MIP sequencing read support for this variant (ALT 5/14 reads), suggesting that this variant may be due to a clonal hematopoiesis of indeterminate potential or a historic sample labeling error. The CHEK2 variant (ENST00000382580.2:c.1412C>T; ENSP00000372023.2:p.Ser471Phe) identified in family 025 was also a false positive; however, this was a low evidence call, with poor read support (1/8 reads for the ALT allele) (Table S6), suggesting this was due to PCR or sequencing error. The pathogenic RAD50 insertion/deletion variant (ENST00000265335.6:c.2165del; ENSP00000265335.6:p.Lys722ArgfsTer14) identified in 34 individuals in the collection also proved to be a false positive by Sanger sequencing. This variant falls within a poly-A tract that caused poor sequencing and mapping quality, likely contributing to the false-positive signal. These results highlight the critical importance of orthogonal Sanger sequencing validation. Our validation rate is consistent with previous MIP studies.23,28
Highly variable DNA quality and preparations among these historic samples (aged anywhere from 15 to 40 years) likely contributed to low sequencing depth (Figure S3) and false negatives in this study. These challenges also precluded copy number variant analyses. Unfortunately, our false-negative rate could not be formally evaluated, as none of the families in the study carried a confirmed research or clinical variant in any of the target genes. Additional sequencing in families void of variants in this study is warranted.
VOUS variants may describe a portion of families
RINT1
Three families carried a single variant in RINT1. A nonsense VOUS in RINT1 (ENST00000257700.2:c.2361G>A; ENSP00000257700.2:p.Trp787Ter) was identified in the proband of family 076 (Figure S4A). This VOUS was predicted to have a high functional impact with a Combined Annotation Dependent Depletion (CADD) score of 44. Three submissions on ClinVar (SCV003807679.1, SCV002735756.2, SCV000940575.6) have all reported this variant as a VOUS. The female proband (IV-1) was diagnosed with breast cancer at age 44 and her daughter (V-1) with thyroid cancer at age 22. The only DNA samples available for testing in this family were the proband, her affected daughter, and the biological father of this offspring. The RINT1 variant was not present in the cancer-affected child of the proband nor the child’s father (IV-2). The proband’s sister was also diagnosed with breast cancer at age 31 (IV-5). Additional pedigree analysis revealed the proband’s maternal grandmother (II-3) was also diagnosed with breast cancer at age 59, as were the grandmother’s sister and niece (ages of diagnoses unknown), which suggested the risk allele was likely derived from this lineage. However, absence of the RINT1 variant in the cancer-affected child of the proband suggest additional risk variants may be present in this family.
Two independent missense RINT1 variants were also identified in families 028 (ENST00000257700.2:c.376C>T; ENSP00000257700.2:p.His126Tyr; Figure S4B) and 021 (ENST00000257700.2:c.41C>T; ENSP00000257700.2:p.Pro14Leu; Figure S4C), respectively. The p.His126Tyr variant has been observed in 0.09% of cancer cases and 0.07% of controls across two previous studies.40 There are only two submissions on ClinVar for this variant, one likely benign (SCV002623211.1) and one VOUS (SCV000551653.7). For this study, DNA was available from two affected and two unaffected members of family 028. The variant was validated in the unaffected proband (III-2) (Figure S4B) and her unaffected father (II-2), but was not present in her breast cancer-affected mother (II-1), maternal aunt (II-3), or maternal grandmother (I-1) who was diagnosed with cervical (age 47) and lung (age 77) cancers (sample unavailable for I-1). Additional pedigree analysis showed a significant family history of cancers in the proband’s paternal lineage, including endometrial and both female and male breast cancers, consistent with RINT1-associated cancer risks.40,41 While it is possible that this is a modifier variant, the inheritance pattern of the variant in this family supports a likely benign variant classification. For the p.Pro14Leu variant, this variant was observed in 0.05% of breast cancer cases and 0.1% of controls in a previous study.40 Reports on ClinVar for this variant include one likely benign (SCV002626673.1) and two VOUS (SCV002011404.3, SCV000551662.7) classifications. This variant was not present in the proband (II-1) (diagnosed with breast cancer at age 61) of family 021 (Figure S4C), but was validated in his breast cancer-affected younger sister (II-8) (diagnosed at age 46) and their mother (I-1), who was diagnosed with skin cancer at an unknown age. Given the lack of cancer-unaffected DNA samples for this family, we cannot rule out this VOUS as a risk factor. However, we can confidently say that it did not contribute to the cancer diagnosis of the proband.
ATM
We identified three potentially interesting missense variants in ATM in this study. ATM is a major factor in the signaling pathway required for the DNA double-strand break (DDSB) damage response.42 The first ATM variant (ENST00000278616.4:c.8734A>G; ENSP00000278616.4:p.Arg2912Gly) was identified in family 132 (Figure S5A). This variant has been reported on ClinVar 16 times in the context of cancer (RCV000131723.27, RCV001196874.15, RCV001355184.9, RCV003492518.1). The most recent of these reports from 2023 all report this as a VOUS (SCV000292188, SCV004206344, SCV004239605). The proband (III-1) was the only member of this family with a DNA sample available and was diagnosed with ovarian cancer at age 46 with clinical evidence of historic endometriosis. Her father (II-2) was diagnosed with skin cancer at age 36 and then again several decades later; however, no other family history was available for the paternal lineage. The p.Arg2912Gly variant has been observed among individuals with cancer.43,44,45,46,47,48,49 Human lymphocytes carrying this variant showed an altered ability to phosphorylate specific downstream targets, suggesting at least a partial loss in kinase activity.50 Based on this evidence, we cannot rule out this variant’s impact on this family’s risk of cancer.
The second ATM missense variant (ENST00000278616.4:c.6293T>C; ENSP00000278616.4:p.Leu2098Pro) was the only variant identified in family 065 (Figure S5B). This variant was predicted to be probably damaging by PolyPhen and had a CADD score of 26.5. All eight reports on ClinVar classify this variant as a VOUS (VCV000135766.33). However, there are conflicting interpretations as to whether this variant has an impact on protein structure and function. This variant was observed in an individual with breast cancer in another hereditary gene panel study.51 Only two DNA samples were available for this family, the proband (II-1) (diagnosed with breast cancer at age 64 and ovarian cancer at 66) and the proband’s oldest daughter (III-1) (diagnosed with breast cancer at age 40). The ATM variant was validated in the daughter, but was not present in the proband. While several of the proband’s siblings (II-3, II-9, II-5, II-14) were affected by pancreatic, lung, and brain cancers, the ATM variant was not likely derived from this lineage and, therefore, does not explain the extended family’s cancer risk.
The third ATM missense variant (ENST00000278616.4:c.1703G>T; ENSP00000278616.4:p.Arg568Ile) was confirmed in family 041. In the context of cancer, this variant has been reported six times on ClinVar (RCV000129176.18, RCV000515396.8, RCV001357003.5). While three reports in the context of hereditary cancer-predisposing syndrome predict this variant to be likely benign (RCV000129176.18), two reports in the context of ataxia-telangiectasia syndrome familial cancer of breast predict this to be a VOUS (RCV000515396.8). For this family, additional variants were identified and are described in further detail later.
TP53
Family 073 (Figure S6A) carried a 3′ UTR variant in TP53 (ENST00000269305.4:c.∗1175A>C, rs78378222); this same mutation was also identified in families 011 and 051. This variant has been implicated in several studies as a low-penetrance risk factor in LFS and cancer.52,53,54 In ClinVar, this variant has been reported in the context of LFS twice (RCV000412103.10) and in the context of hereditary cancer-predisposing syndrome twice (RCV000492363.7). Under both diseases, there is both a benign/likely benign and a VOUS classification. While the proband (III-1) of family 073 (diagnosed with breast cancer at age 43) was not available for genetic testing, this variant was found in two of her three siblings (III-3, III-5) (one sister diagnosed with lobular breast cancer at age 44 and another sister that was unaffected). The third sister (III-7), who was also unaffected, did not carry this variant. The father (II-2) of these four females was diagnosed with an undefined cancer at age 72 and the mother (II-3) with lobular breast cancer at age 57. There were no other cancers of note in the extended family pedigree. While it is possible that this variant contributes to the development of breast cancer, its segregation among unaffected and affected individuals in this pedigree support a low penetrance or even benign impact, further supporting the ClinVar likely benign designations.
FANCC
Family 048 carried a 3′ UTR variant (ENST00000289081.3:c.∗1483T>C) in FANCC (Figure S6B). This variant has not been reported on ClinVar in the context of cancer (VCV000367585.5). The proband (III-6), who had a breast cancer diagnosis at age 40, was the only DNA sample available for this family. However, her family history revealed a wide variety of other cancers likely derived from a paternally inherited risk factor. The proband’s youngest sister (III-9) was diagnosed and succumbed to leukemia at age 21 and their father (II-5) had a history of bladder, kidney, and skin cancers (at ages 62, 62, and 71, respectively). His brother (II-1) was also diagnosed with skin cancer at age 75 and his sister (II-3) with ovarian cancer at age 55. Nieces from both of those lineages were diagnosed with gynecological cancers between the ages of 39 and 42 (III-1, III-4). We noted several other extended relatives in this pedigree with gastrointestinal, blood, and gynecological cancers. This variant has not been previously reported in ClinVar to be associated with any cancer cases (only as a VOUS for Fanconi anemia complementation group C) [MIM: 227645]. Rates of this variant in gnomAD suggest population frequencies of 0.05%–0.5% with no observed homozygotes. Further, this variant lies in a non-conserved region of the genome collectively supporting a likely benign classification.
NF1
The proband in family 133 (Figure S6C) carried a missense variant in NF1 (ENST00000358273.4:c.7396A>G; ENSP00000351015.4:p.Ile2466Val). While this variant was predicted to be tolerated by SIFT and PolyPhen, it was private in our study. This base is weakly conserved across species and has been reported four times on ClinVar in the context of cancer and/or neurofibromatosis (RCV000575889.4, RCV000200710.10, RCV003390940.4, RCV000764116.2) as a VOUS. The proband (II-1) was diagnosed with breast cancer at age 64 and was the only member of this family to have a DNA sample available. Pedigree analysis revealed a brother (II-3) who died of pancreatic cancer at age 55. Their mother (I-1) had a history of both breast and endometrial cancers, while their father (I-2) was affected by lung cancer (ages of diagnoses unknown). Of note, the proband’s niece (III-17) was also diagnosed with breast cancer at age 48 and two males, the proband’s first cousin (not shown) and great nephew (IV-1), in the extended pedigree were diagnosed with leukemia at ages 24 and 16, respectively. While we cannot rule this variant out as a risk factor in this family, other factors likely contributed to the extensive cancer diagnoses.
MRE11A
One affected individual from family 058 (Figure S6D) carried a missense variant in MRE11A (ENST00000323929.3:c.1475C>A). We only had one available DNA sample from this family, the proband’s daughter (IV-3) who was affected with breast cancer at age 35. The MRE11A variant was confirmed in this individual. There are conflicting reports and predictions regarding the pathogenicity of this variant. While PolyPhen predicted a disruptive effect, SIFT did not. The variant has a CADD score of 22.8. Recent ClinVar reports show an increased likelihood that this variant is likely benign or benign in the context of cancer (VCV000127031.58).30 While we cannot rule this variant out as a risk factor in this family, other factors likely contributed to cancer diagnoses.
CHEK2
The proband in family 121 (Figure S6E) carried a missense variant in CHEK2 (ENST00000382580.2:c.1465A>G; ENSP00000372023.2:p.Asn489Asp). SIFT and PolyPhen characterized this mutation as likely benign, and the variant had a CADD score of 18.55. This variant has been reported in the context of cancer nine times on ClinVar (VCV000126909.50) including three likely benign and six VOUS classifications. The proband’s mother (II-2) was diagnosed with colorectal cancer at age 56, their sister (III-5) with ovarian cancer at age 46, another sister (III-7) with breast cancer at age 32, and their daughter (IV-3) with uterine cancer at age 26. However, only the proband (III-1), an affected carrier, had a DNA sample available for sequencing. Functional studies of CHEK2 variation have shown this amino acid change does not likely affect protein function.55,56 While we cannot rule out this variant as a contributing factor to cancer risk in this family, these data support a likely benign classification.
A portion of families carry multiple VOUS variants
Two potential VOUSs were identified in family 041 (Figure S7A), a missense variant in ATM (ENST00000278616.4:c.1703G>T; described above) and a 5′ UTR PMS2 (ENST00000265849.7:c.-7T>C) variant. SIFT and PolyPhen scores characterized the ATM variant as likely benign, and it had a CADD score of 17.28. ClinVar contains both likely benign and VOUS predictions for this variant in the context of cancer (VCV000140916.55). This variant was confirmed in the proband’s unaffected father (III-3) and sister (IV-3) affected with breast cancer (diagnosed at age 53). The PMS2 variant identified was in the Kozak sequence. While the MIP sequencing data were convincing (Figure S7B), the high GC content of this region prevented Sanger validation. However, an abundance of clinical reports, and our own recent study,57 suggested that this PMS2 variant was benign or likely benign, including 8 of 10 ClinVar submissions (VCV000140831.20). We conclude that neither of these variants are the primary genetic risk factor for cancer diagnoses in this family.
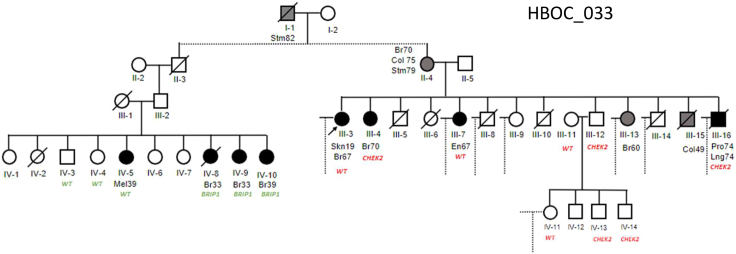
Family 033 carried both a missense VOUS in BRIP1 (ENST00000259008.2:c.2087C>T; ENSP00000259008.2:p.Pro696Leu) and a missense VOUS in CHEK2 (ENST00000382580.2:c.1182G>T; ENSP00000372023.2:p.Glu394Asp). This was a very large family pedigree with numerous cases of breast cancer. We limited Figure 4 to the two branches of this tree with available DNA for Sanger validations. The BRIP1 variant is predicted to be damaging by SIFT and PolyPhen and had a CADD score of 29.8. Currently, all 16 ClinVar submissions report a VOUS classification for this variant (VCV000128167.43). Clinical reports have observed this variant in multiple individual cases of breast cancer. As shown in this family pedigree, this variant was isolated to a single branch and generation of this family (second cousins of the proband), including nine sisters and one brother. In this branch, three sisters (IV-8, IV-9, IV-10) carried the BRIP1 mutation and were diagnosed with breast cancer under the age of 40. One sister (IV-4) and the brother (IV-3) were not carriers and were unaffected by cancer. One sister (IV-5) was not a carrier, but was affected by melanoma at age 39. Only one other individual in this 495-member pedigree was diagnosed with melanoma; moreover, melanoma risk has a known strong environmental contribution (e.g., UV exposure) suggesting that it may fall outside of this family’s risk profile. The CHEK2 variant was also isolated to a single, multi-generational branch of this pedigree that included the proband. This variant has been classified in ClinVar 15 times (VCV000142151.32), 14 times as a VOUS, and once as a likely pathogenic variant (SCV004221712.1). Importantly, no individual in this pedigree carried both variants. While the proband (III-3) was affected with skin cancer at age 19 and breast cancer at age 67, they were not a carrier of either variant. Within this branch, the CHEK2 variant was found in the proband’s sister (III-4; diagnosed with breast cancer at age 70) and one of their shared nieces (diagnosed with breast cancer at age 41; not shown). None of the niece’s children with an available DNA sample (n = 5 daughters) were carriers and were not affected by cancer at last contact. The proband’s brother (III-16) who was affected with prostate and lung cancers (diagnosed at age 74) and nephew diagnosed with lymphoma at age 46 (not shown) were also carriers. At the time of testing, no other immediate relatives of the proband were carriers or were affected with cancer. We identified 18 family members who were carriers of this variant but were unaffected with cancer (Table S8). We conclude that the BRIP1 VOUS is likely pathogenic and a highly penetrant risk factor for the early development of breast cancer. Further, this variant was most likely inherited from the married-in mother of the affected carriers, which reflects the isolation of this variant to the single branch. Further, the segregation of the CHEK2 variant in this family supports a likely benign classification.
Figure 4.
Multiple VOUSs identified in family 033
Family 033 carried a BRIP1 c.2087C>T variant isolated to a single branch of the pedigree. This variant is likely pathogenic and highly penetrant. The family also carried a CHEK2 c.1182G>T variant, which is also isolated to a separate branch of the pedigree. This variant is likely benign due to numerous unaffected carriers in this family. Cancer types: Br, breast; Col, colorectal; En, endometrial; Lng, lung; Mel, melanoma; Pro, prostate; Skn, skin; Stm, stomach. If more than one variant was present, WT (wild-type sequence carrier) was labeled in the corresponding gene color.
LS may explain a portion of BRCAX families
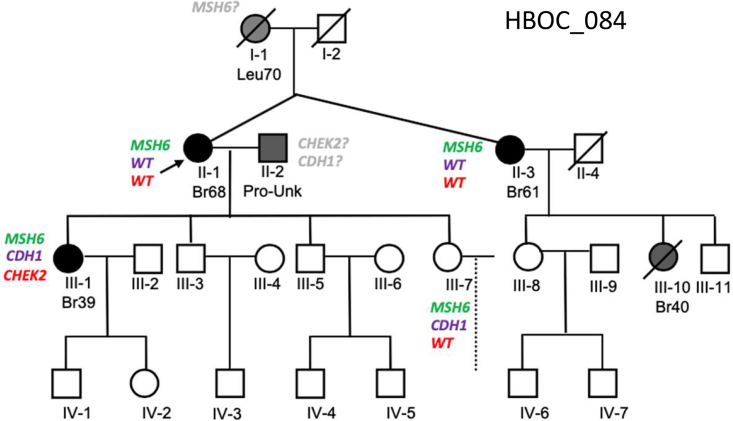
LS is the most common hereditary cancer and can include breast and ovarian cancer diagnoses.58 We found two prominent MMR gene mutations, in MSH6 and PMS2, in two families, which we conclude are atypical LS presentations. The first, family 084 (Figure 5) carried two pathogenic variants, a nonsense variant in MSH6 (ENST00000234420.5:c.892C>T; ENSP00000234420.4:p.Arg298Ter) and the frameshifting variant in CHEK2 (ENST00000382580.2:c.1229del) identified in other families in this study, as well as a missense variant in CDH1 (ENST00000261769.5:c.808T>G; ENSP00000261769.4:p.Ser270Ala). Of the ClinVar reports for the CDH1 variant, 12 of the 14 support a likely benign or benign classification (VCV000142011.30). The MSH6 and CDH1 variants were initially identified in unaffected individual (III-7) and the CHEK2 variant in III-1 (Table S6). Sanger sequencing confirmed that individual III-1, in fact, carried all three variants (MSH6, CHEK2, and CDH1), and that the MSH6 variant was inherited from the mother/proband (II-1) who was diagnosed with breast cancer at age 68. The proband’s identical twin sister (II-3) carries the same MSH6 variant and was diagnosed with breast cancer at age 61. This variant is predicted to have contributed to breast cancer in the proband’s niece (III-10), who was diagnosed at age 40 (DNA not available for testing). The proband passed on this variant to a second unaffected daughter (III-7), who also carried the CDH1 variant but not the CHEK2 variant. A review of this pedigree did not reveal an obvious upstream lineage source for this MSH6 variant. In fact, the paucity of cancers in extended family members on both sides of the proband’s pedigree was surprising, given the known pathogenicity of the MSH6 variant. This could suggest that the variant arose de novo or non-paternity. The CDH1 variant was likely introduced from the married-in spouse of the proband (II-2); however, no extended family history was available for this individual. The CHEK2 variant was only identified in individual III-1 of the samples tested (which did not include her father, II-2). The variant arising de novo is highly unlikely given its frequency in the human population. It is most likely that this variant was also inherited from the paternal lineage. Most interesting and alarming was the confirmed MSH6 pathogenic variant found in individual III-7. This individual was diagnosed with endometriosis and had a hysterectomy at age 44 which would have significantly reduced her risk of developing endometrial and ovarian cancers.59 It is reasonable that this may have contributed to her being unaffected with cancer at the time of DNA collection.
Figure 5.
Family identified with multiple pathogenic VOUS(s) likely explained by LS
Family 084 carried a pathogenic MSH6 variant c.892C>T, a pathogenic CHEK2 c.1229del variant, and a missense CDH1 c.808T>G variant. While the MSH6 variant was likely transmitted through the proband’s maternal lineage, the CHEK2 and CDH1 variants were likely transmitted to individual III-1 from their married-in father. If more than one variant was identified, WT was written in the corresponding gene color. Cancer types: Br, breast; Leu, leukemia; Pro, prostate; Unk, unknown age.
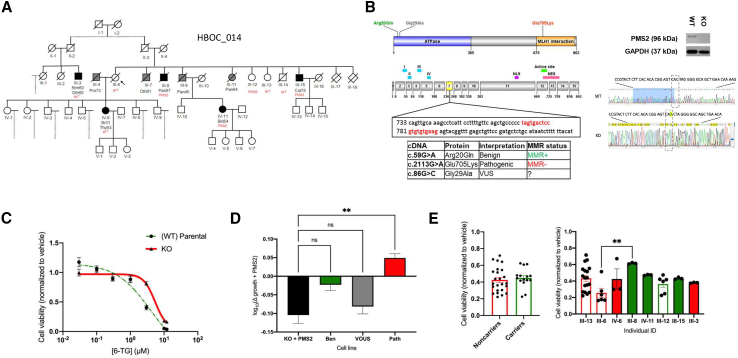
A PMS2 missense variant (ENST00000265849.7:c.86G>C; ENSP00000265849.7:p.Gly29Ala) was identified in family 014 (Figure 6A). High rates of LS-linked cancers in this pedigree suggested that this variant could in fact contribute to LS risk. This variant was predicted by both SIFT and PolyPhen to be protein damaging. However, conflicting interpretations of this variant’s (p.Gly29Ala; MAF: 0.000440) pathogenicity have been reported in ClinVar (VCV000041721.54): 11 likely benign or benign and 4 VOUS classifications.30 The proband of this family (III-6) was not a PMS2 variant carrier nor affected by cancer. There were numerous cases of cancer in this family, with an abundance of pancreatic cancer. After reviewing the proband’s medical history, we discovered she was taking an experimental drug due to her risk of pancreatic cancer at the time of collection. We do not know if this drug influenced the proband’s risk of developing cancer. The proband had 10 siblings: 3 with pancreatic cancer (III-8, III-9, III-11), 1 with colorectal (III-15), 1 with an unknown cancer type (III-7), 4 with no history of cancer (III-12, III-13, III-14), and 2 who died in their first year of life supposedly unrelated to cancer (III-17, III-18). As shown in the pedigree, the daughter of the proband (IV-6) was diagnosed with breast cancer at age 31 and thyroid cancer at age 53. Of these family members, four carried the PMS2 variant (III-8, IV-11, III-12, III-15), three affected and one unaffected. An analysis of the extended pedigree revealed a first cousin of the proband with stomach and an unknown cancer at age 60 (III-3), a cousin with breast cancer at age 64 (not shown on pedigree), and a niece with bladder cancer at age 54 (IV-11). Segregation analysis indicated that this variant had either moderate penetrance or other variants were contributing to cancer risk.
Figure 6.
Re-classification of a LS VOUS through family history analysis and a functional in vitro assay
(A) Family 014 carried a PMS2 VOUS c.86G>C. The proband is an unaffected carrier. The variant can be seen in unaffected and affected carriers. Cancer types: Bld, bladder; Br, breast; Col, colorectal; Oth, unknown source; Pan, pancreas; Stm, stomach; Thy, thyroid. WT, wild-type sequence carrier.
(B) Shown are the PMS2 protein map (top) and the corresponding exon structure (below) including 15 exons and two functional domains (ATPase binding [blue] and MLH1 interaction [orange]). The locations of variants of interest in the model are notated by their clinical interpretation: benign (green), pathogenic (red), and VOUS (gray). The CRISPR-Cas9 cut site is located in exon 7 (red on the exon map). The induced CRISPR-Cas9 frameshifting mutation in PMS2 (insA) was confirmed by Sanger sequencing and western blot analysis.
(C) The HAP1 PMS2 KO line was tested for MMR sensitivity (i.e., 6-TG sensitivity) in vitro alongside the HAP1 PMS2 parental line. We selected logarithmic doses of 6-TG (0.03, 0.1, 0.3, 1, 3, 10, and 12 μM) and analyzed the IC50 of 6-TG over 72 h as a functional surrogate for MMR function. Shown are cell viability data (mean ± SEM) from three independent replicate experiments (N = 3) with triplicate wells per condition.
(D) Bar graph shows cell viability after MMR selection of PMS2 variant lines. The PMS2 KO + full-length PMS2 (rescue) cell line is in black, PMS2 benign line is green, PMS2 VOUS line is gray, and PMS2 pathogenic line is red. Shown are the mean ± SEM cell viability data from three independent replicate experiments (N = 3) with triplicate wells per condition. Triplicates were first averaged, averages were normalized to 0 h, normalized to vehicle, and then log transformed to normalize for the shared PMS2 KO line background. ∗∗p < 0.01; ns, not significant.
(E) Shown are the mean ± SEM cell viability data from EBV-transformed lymphocyte lines of this LS family. PMS2 VOUS noncarriers and carriers were analyzed (left). Cell viability data (mean ± SEM) for each individual line is shown on the right. ∗∗p < 0.01.
Generation of a functional assay for LS variant re-classification
To address the penetrance of the PMS2 p.Gly29Ala variant, we developed a functional assay to determine whether this variant has altered MMR activity in vitro. CRISPR-Cas9 technology was used to generate a PMS2 KO human HAP1 haploid cell line (Figure 6B). MMR function was quantified by measuring cell viability after 72 h of 6-TG treatment; loss of PMS2 activity/MMR function was expected to increase survival based on previous studies.31 As expected, our PMS2 KO line was significantly more resistant to 6-TG than the MMR-proficient parental line (WT) (p < 0.0001), indicating reduced MMR activity in the KO state (Figure 6C).
Full-length WT human PMS2 and specific variants were then re-introduced into the KO line using a lentiviral vector to test for functional MMR rescue. Variants tested included a well-annotated benign variant (c.59G>A, p.Arg20Gln), a pathogenic variant (c.2113G>A, p.Glu705Lys), and the VOUS (c.86G>C, p.Gly29Ala; Figure 6D). Importantly, our introduced PMS2 constructs were regulated by an inducible promoter such that we could regulate when the introduced PMS2 variant was expressed. As expected, induced WT PMS2 expression significantly increased 6-TG sensitivity in the PMS2 KO line (p < 0.0001) and showed that MMR activity could be rescued (Figure 6D). Induction of the known pathogenic variant significantly increased cell growth after 6-TG treatment, indicating 6-TG resistance and MMR deficiency (∗∗p = 0.0015), whereas the benign variant had decreased cell viability that was not statistically different from the KO + WT PMS2 condition (Figure 6D), indicating MMR proficiency (not significant; p = 0.0825). Our VOUS had decreased cell growth under 6-TG treatment that was not statistically different from the KO + WT PMS2 condition (not significant; p = 0.8599). These results indicate that the PMS2 VOUS p.Gly29Ala is MMR proficient and equivalent to WT PMS2.
Finally, we tested in vitro MMR activity using immortalized EBV-transformed lymphocyte lines derived from eight individuals in family 014. We found no statistically significant difference in cell viability after 6-TG treatment between VOUS carriers (n = 4) and noncarriers (n = 4; not significant; p = 0.2153) (Figure 6E). These data collectively support a benign classification for the PMS2 p.Gly29Ala variant.
Discussion
Herein, we present the results of targeted sequencing for potential hereditary cancer risk variants in one of the oldest and most comprehensive hereditary cancer biobanks. This biobank was founded by Dr. Henry Lynch and was used to steadily collect and study suspected HBOC families from 1973 to 2019. While this biobank has contributed families to many of the seminal hereditary cancer studies of its time,60,61 it has never been systematically sequenced. The effect of loss-of-function mutations on cancer risk in genes such as BRCA1/2, MSH2/6, and others are generally well understood; yet, family-based biobanks still offer unique opportunities to better understand VOUSs, modifiers, and multi-genic variant conditions that may increase cancer risk over time. Indeed, clinical sequencing identifies far more VOUSs than known pathogenic variants annually,62 highlighting the desperate need for understanding these variants.
Through multi-gene targeted sequencing, we identified several potential hereditary cancer risk variants other than BRCA1/2 in BRCAX families. Importantly, we included BRCA1 and BRCA2 in our panel to confirm that all families were in fact BRCAX, as our biobanked families have been evaluated by many different research and clinical protocols over the course of more than 40 years such that previous full-gene BRCA coverage could not be confirmed across all samples. A summary of our most interesting VOUSs, likely pathogenic, and pathogenic variants can be found in Table 3. Known breast cancer risk variants and hereditary cancer syndromes resolved eight BRCAX families with potential resolution of a ninth family by a BRIP1 VOUS that is likely pathogenic. While a combination of sequencing data, pedigree analysis, and ClinVar data helped us to re-classify eight potential VOUSs as likely benign, several VOUSs remain that may contribute to cancer risk. These are enriched for variants in the ATM-CHEK2-p53 pathway. The ATM-CHEK2-p53 axis is known to be required in the damage response to DDSBs. In response to DDSBs, ATM phosphorylates CHEK2, allowing CHEK2 to phosphorylate p53, directly contributing to G1 cell-cycle arrest. Accordingly, this axis is critical to DNA fidelity, with mutations predisposing individuals to cancer. Other known hereditary cancer genes also interact with this pathway, including BRCA1, which CHEK2 can also phosphorylate. Of the genes in this pathway, CHEK2 is the most susceptible to mutations11 and had the largest number of pathogenic mutations in this study. Breast cancer in carriers of pathogenic germline CHEK2 mutations often have clinicopathological characteristics, such as bilateral breast cancer as seen in family 015 (Figure 2A). CHEK2 variation has also been associated with an increased risk for sarcomas and stomach, kidney, prostate,63 and skin64 cancers. However, variants in CHEK2 are considered to have moderate penetrance, increasing risk by 2- to 3-fold. Recent literature has shown that tumors with low levels of ATM but normal TP53 and CHEK2 expression are predicted to be chemo-resistant and, further, that loss of function of the ATM-CHEK2-p53 cascade is strongly associated with resistance to anthracycline/mitomycin-containing chemotherapy in breast cancer.65 This is important, because individuals who carry these mutations may require additional or different treatment plans.
Table 3.
Summary of VOUS and likely pathogenic/pathogenic variants identified in BRCAX families
| Gene | Variant pathogenicity | Family # | Diagnoses |
|---|---|---|---|
| ATM | VOUS VOUS |
HBOC_065 HBOC_132 |
Br, Ov, Pan, Lng, Bn, Skn Ov, Skn |
| BRCA2 | pathogenic | HBOC_066 | Br, Ov |
| BRIP1 | likely pathogenic | HBOC_033 | Br, Mel, Col, Stm, En, Pro, Lng, Lym, Skn |
| CHEK2 | pathogenic pathogenic pathogenic pathogenic pathogenic pathogenic |
HBOC_015 HBOC_044 HBOC_080 HBOC_084 HBOC_089 HBOC_107 |
Br, Bn, Pro, Skn, Lng, Utr, Cvx, Col, Stm Br, Leu, Col, Skn Br Br Br, Skn, Unk Br |
| MSH6 | pathogenic | HBOC_084 | Br |
| NF1 | VOUS | HBOC_133 | Br, En, Pan, Lng, Leu |
| PMS2 | VOUS | HBOC_014 | Br, Stm, Pan, Col, Thy, Bld, Pro, Unk |
| RINT1 | VOUS VOUS |
HBOC_021 HBOC_076 |
Br, Skn, Lym Br, Thy |
| TP53 | pathogenic | HBOC_018 | Br, Pan, Leu, Kid, Other |
Bld, bladder; Bn, brain; Br, breast; Col, colorectal; Cvx, cervical; En, endometrial; Kid, kidney; Leu, leukemia; Lng, lung; Lym, lymphoma; Mel, melanoma; Ov, ovarian; Pan, pancreatic; Pro, prostate; Skn, skin; Stm, stomach; Thy, thyroid; Unk/Other, unknown/other; Utr , uterine.
The relationship between genotype and phenotype is further complicated by genetic penetrance. Given the increasing acquisition of somatic mutations over time in hereditary cancer syndromes, penetrance depends on (1) which somatic mutations occur and (2) age. However, our study shows that we cannot discount the variable of clinical intervention in estimates of penetrance. For example, in family 084 (Figure 5) individual III-7 carried a known pathogenic MSH6 variant (c.892C>T, p.Arg298Ter) that likely contributed to cancer diagnoses in other members of her family; however, she seems to be unaffected. While this could be interpreted as reduced penetrance, the average age of diagnosis for extracolonic cancers in LS is 49.1 years.66 In a study of only MSH6 mutation carriers, overall cancer risk was increased 22% by age 50 and 73% by age 70.67 At the time of testing, this individual was only 43 years of age, and she had already had a hysterectomy to treat severe endometriosis. Colorectal and endometrial cancer diagnoses are most common among LS families.67,68 The fact that this individual had one of these tissues removed while she was quite young is predicted to be protective against the pathogenic MSH6 variant that she carries.
Further, it is possible, and in fact likely, that a portion of the genetic penetrance in hereditary cancer families can be explained by additional genetic burden. Indeed, we identified several potential multi-hit families in this study, suggesting that they may be more common than we appreciate. Again, family 084 provides an excellent example of two moderate penetrance breast cancer variants—CHEK2 (c.1229del) and the MSH6 (c.892C>T, p.Arg298Ter) LS variant—that likely synergized to cause an earlier breast cancer onset in individual III-1 (age 39). In fact, many families in this study presented with earlier diagnoses across successive generations. While reasonable, the genetic anticipation hypothesis in LS remains controversial and is likely confounded by multigenic events (such as in family 084) and improved surveillance measures. While updated guidelines recommending conservative gene panel testing are available to some, only whole genome sequencing for all will allow us to calculate the prevalence of multi-hit families, detect non-traditional genotype-phenotype patterns, and formally test genetic anticipation in an unbiased way. Even our study design, which targeted only exons with a 10 basepair flank into introns, was designed to maximize pathogenic or likely pathogenic variant yield, yet surely missed known pathogenic non-coding variants that have been reported previously in many of our target genes.69,70 Whole genome coverage will be required to (1) surmount the lack of diversity in control databases to better understand how variant frequencies differ across populations, (2) see how different haplotypes (i.e., combinations of variants), and polygenic risk may interact to increase disease risk,71,72 and (3) understand the full genetic contribution of each gene (non-coding and coding variants) to hereditary cancer risk.
Expanded clinical sequencing must be developed in tandem with high-throughput functional validation of VOUSs. VOUSs are not clinically actionable, but must be functionally re-classified to apply deeper clinical value to sequencing. As a proof of concept in this study, we developed a mid-throughput assay for functionally validating the MMR activity of the PMS2 VOUS (p.Gly29Ala) identified in family 014 (Figure 6). Combined pedigree analysis and functional data all suggest that this variant is benign. The continued development of multiplexed assays of variant effects73 are an obvious match for this significant clinical need. Functional re-classification of variants will require dedicated partnerships between basic scientists and the clinical genetics community. In addition, protocols will need to be standardized to establish (1) how large genomics data will be stored, (2) how often data are re-analyzed for novel variants and re-classifications, (3) who pays for re-analysis, (4) exactly which variants are actionable, and (5) how to handle incidental findings. In addition to these prospective challenges, there will also be an obligation for clinicians to offer updated sequencing to individuals that have previously participated in genetic testing with the risk of proving previous findings false. As an example, from this study, we identified a known pathogenic BRCA2 variant (p.Met192ValfsTer13) in family 066 (Figure 3). This family was tested under research protocols in 1999 and 2004 for a limited number of specific BRCA1/2 mutations and was found to be negative. At the time of testing, clinical sequencing for hereditary cancer risk was limited or not available, and clinical decision-making was often done based on research findings such as these. Genetic counselors are likely to be an invaluable resource in this near-future scenario to help navigate the interface between the evolving field of clinical genetics and patients.
Data and code availability
The datasets generated during this study are available at the National Library of Medicine Sequence Read Archive (SRA) [SUB14326985/PRJNA1090944/https://www.ncbi.nlm.nih.gov/bioproject/1090944].
Acknowledgments
We sincerely thank the families who have contributed samples and data to the Lynch Memorial Biobank. We would like thank Dr. Henry T. Lynch for being at the forefront of hereditary cancer research and pioneering the investigation and biobanking of these families. Also, we would like to thank C. L. Snyder, M. Christensen, and R. S. Aziz-Seible for technical assistance with the project and the University of Nebraska Medical Center Genomics Core facility for sequencing services. Funding for this project was provided by the State of Nebraska Department of Health and Human Services Cancer and Smoking Disease Research Program LB506 and LB595 grants to H.A.F.S. Plasmid pCW57.1 was a gift from David Root (Addgene plasmid # 41393; http://n2t.net/addgene:41393; RRID:Addgene_41393). Plasmids psPAX2 (Addgene plasmid # 12260; http://n2t.net/addgene:12260; RRID:Addgene_12260) and pMD2.G (Addgene plasmid # 12259; http://n2t.net/addgene:12259; RRID:Addgene_12259) were gifts from Didier Trono.
Author contributions
H.A.F.S. designed the study. H.A.F.S., J.N.P., E.J.M., C.J.W., and S.P.A. performed the experiments. H.A.F.S., E.J.M., C.J.W., and Y.F. analyzed the sequencing data. J.N.P., E.J.M., L.V.U., S.B.D., B.A.S., and M.L.S. performed pedigree analyses and contributed to participant chart review. M.A.B. provided some cell line and lentivirus resources for this study. E.E.B. and C.D.H. provided intellectual input regarding data interpretation. H.A.F.S., J.N.P., and E.J.M. wrote the manuscript with input from all authors.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2024.100306.
Web resources
GnomAD: https://gnomad.broadinstitute.org/
OMIM: https://www.omim.org/
Supplemental information
References
- 1.Siegel R.L., Miller K.D., Wagle N.S., Jemal A. Cancer statistics, 2023. CA. Cancer J. Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Collaborative Group on Hormonal Factors in Breast Cancer Familial breast cancer: collaborative reanalysis of individual data from 52 epidemiological studies including 58,209 women with breast cancer and 101,986 women without the disease. Lancet. 2001;358:1389–1399. doi: 10.1016/S0140-6736(01)06524-2. [DOI] [PubMed] [Google Scholar]
- 3.Easton D.F. Familial risks of breast cancer. Breast Cancer Res. 2002;4:179–181. doi: 10.1186/bcr448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraus C., Hoyer J., Vasileiou G., Wunderle M., Lux M.P., Fasching P.A., Krumbiegel M., Uebe S., Reuter M., Beckmann M.W., Reis A. Gene panel sequencing in familial breast/ovarian cancer patients identifies multiple novel mutations also in genes others than BRCA1/2. Int. J. Cancer. 2017;140:95–102. doi: 10.1002/ijc.30428. [DOI] [PubMed] [Google Scholar]
- 5.Hall J.M., Lee M.K., Newman B., Morrow J.E., Anderson L.A., Huey B., King M.C. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250:1684–1689. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 6.Narod S.A., Feunteun J., Lynch H.T., Watson P., Conway T., Lynch J., Lenoir G.M. Familial breast-ovarian cancer locus on chromosome 17q12-q23. Lancet. 1991;338:82–83. doi: 10.1016/0140-6736(91)90076-2. [DOI] [PubMed] [Google Scholar]
- 7.Miki Y., Swensen J., Shattuck-Eidens D., Futreal P.A., Harshman K., Tavtigian S., Liu Q., Cochran C., Bennett L.M., Ding W., et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 8.Wooster R., Neuhausen S.L., Mangion J., Quirk Y., Ford D., Collins N., Nguyen K., Seal S., Tran T., Averill D., et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994;265:2088–2090. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- 9.Hu C., Hart S.N., Gnanaolivu R., Huang H., Lee K.Y., Na J., Gao C., Lilyquist J., Yadav S., Boddicker N.J., et al. A Population-Based Study of Genes Previously Implicated in Breast Cancer. N. Engl. J. Med. 2021;384:440–451. doi: 10.1056/NEJMoa2005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breast Cancer Association Consortium. Dorling L., Carvalho S., Allen J., González-Neira A., Luccarini C., Wahlström C., Pooley K.A., Parsons M.T., Fortuno C., et al. Breast Cancer Risk Genes - Association Analysis in More than 113,000 Women. N. Engl. J. Med. 2021;384:428–439. doi: 10.1056/NEJMoa1913948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stolarova L., Kleiblova P., Janatova M., Soukupova J., Zemankova P., Macurek L., Kleibl Z. CHEK2 Germline Variants in Cancer Predisposition: Stalemate Rather than Checkmate. Cells. 2020;9:2675. doi: 10.3390/cells9122675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bell D.W., Varley J.M., Szydlo T.E., Kang D.H., Wahrer D.C., Shannon K.E., Lubratovich M., Verselis S.J., Isselbacher K.J., Fraumeni J.F., et al. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science. 1999;286:2528–2531. doi: 10.1126/science.286.5449.2528. [DOI] [PubMed] [Google Scholar]
- 13.Stucci L.S., Interno V., Tucci M., Perrone M., Mannavola F., Palmirotta R., Porta C. The ATM Gene in Breast Cancer: Its Relevance in Clinical Practice. Genes. 2021;12:727. doi: 10.3390/genes12050727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schon K., Tischkowitz M. Clinical implications of germline mutations in breast cancer: TP53. Breast Cancer Res. Treat. 2018;167:417–423. doi: 10.1007/s10549-017-4531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts M.E., Jackson S.A., Susswein L.R., Zeinomar N., Ma X., Marshall M.L., Stettner A.R., Milewski B., Xu Z., Solomon B.D., et al. MSH6 and PMS2 germ-line pathogenic variants implicated in Lynch syndrome are associated with breast cancer. Genet. Med. 2018;20:1167–1174. doi: 10.1038/gim.2017.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch H.T., Lynch P.M., Lanspa S.J., Snyder C.L., Lynch J.F., Boland C.R. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin. Genet. 2009;76:1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hisada M., Garber J.E., Fung C.Y., Fraumeni J.F., Jr., Li F.P. Multiple primary cancers in families with Li-Fraumeni syndrome. J. Natl. Cancer Inst. 1998;90:606–611. doi: 10.1093/jnci/90.8.606. [DOI] [PubMed] [Google Scholar]
- 18.Lynch H., Wen H., Kim Y.C., Snyder C., Kinarsky Y., Chen P.X., Xiao F., Goldgar D., Cowan K.H., Wang S.M. Can unknown predisposition in familial breast cancer be family-specific? Breast J. 2013;19:520–528. doi: 10.1111/tbj.12145. [DOI] [PubMed] [Google Scholar]
- 19.Stafford J.L., Dyson G., Levin N.K., Chaudhry S., Rosati R., Kalpage H., Wernette C., Petrucelli N., Simon M.S., Tainsky M.A. Reanalysis of BRCA1/2 negative high risk ovarian cancer patients reveals novel germline risk loci and insights into missing heritability. PLoS One. 2017;12 doi: 10.1371/journal.pone.0178450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sinilnikova O.M., Dondon M.G., Eon-Marchais S., Damiola F., Barjhoux L., Marcou M., Verny-Pierre C., Sornin V., Toulemonde L., Beauvallet J., et al. GENESIS: a French national resource to study the missing heritability of breast cancer. BMC Cancer. 2016;16:13. doi: 10.1186/s12885-015-2028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rebbeck T.R., Friebel T., Lynch H.T., Neuhausen S.L., van 't Veer L., Garber J.E., Evans G.R., Narod S.A., Isaacs C., Matloff E., et al. Bilateral prophylactic mastectomy reduces breast cancer risk in BRCA1 and BRCA2 mutation carriers: the PROSE Study Group. J. Clin. Oncol. 2004;22:1055–1062. doi: 10.1200/JCO.2004.04.188. [DOI] [PubMed] [Google Scholar]
- 22.O'Roak B.J., Vives L., Fu W., Egertson J.D., Stanaway I.B., Phelps I.G., Carvill G., Kumar A., Lee C., Ankenman K., et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stessman H.A.F., Xiong B., Coe B.P., Wang T., Hoekzema K., Fenckova M., Kvarnung M., Gerdts J., Trinh S., Cosemans N., et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat. Genet. 2017;49:515–526. doi: 10.1038/ng.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panneman D.M., Hitti-Malin R.J., Holtes L.K., de Bruijn S.E., Reurink J., Boonen E.G.M., Khan M.I., Ali M., Andréasson S., De Baere E., et al. Cost-effective sequence analysis of 113 genes in 1,192 probands with retinitis pigmentosa and Leber congenital amaurosis. Front. Cell Dev. Biol. 2023;11 doi: 10.3389/fcell.2023.1112270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hany U., Watson C.M., Liu L., Smith C.E.L., Harfoush A., Poulter J.A., Nikolopoulos G., Balmer R., Brown C.J., Patel A., et al. Heterozygous COL17A1 variants are a frequent cause of amelogenesis imperfecta. J. Med. Genet. 2024;61:347–355. doi: 10.1136/jmg-2023-109510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernandez-Neuta I., Magoulopoulou A., Pineiro F., Lisby J.G., Gulberg M., Nilsson M. Highly multiplexed targeted sequencing strategy for infectious disease surveillance. BMC Biotechnol. 2023;23:31. doi: 10.1186/s12896-023-00804-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyle E.A., O'Roak B.J., Martin B.K., Kumar A., Shendure J. MIPgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics. 2014;30:2670–2672. doi: 10.1093/bioinformatics/btu353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Roak B.J., Stessman H.A., Boyle E.A., Witherspoon K.T., Martin B., Lee C., Vives L., Baker C., Hiatt J.B., Nickerson D.A., et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat. Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W., et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia X., Burugula B.B., Chen V., Lemons R.M., Jayakody S., Maksutova M., Kitzman J.O. Massively parallel functional testing of MSH2 missense variants conferring Lynch syndrome risk. Am. J. Hum. Genet. 2021;108:163–175. doi: 10.1016/j.ajhg.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson E.R., Gorringe K.L., Rowley S.M., Li N., McInerny S., Wong-Brown M.W., Devereux L., Li J., Lifepool Investigators. Trainer A.H., et al. Reevaluation of the BRCA2 truncating allele c.9976A > T (p.Lys3326Ter) in a familial breast cancer context. Sci. Rep. 2015;5 doi: 10.1038/srep14800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang S., Phelan C.M., Zhang P., Rousseau F., Ghadirian P., Robidoux A., Foulkes W., Hamel N., McCready D., Trudeau M., et al. Frequency of the CHEK2 1100delC mutation among women with breast cancer: an international study. Cancer Res. 2008;68:2154–2157. doi: 10.1158/0008-5472.CAN-07-5187. [DOI] [PubMed] [Google Scholar]
- 34.Torabi Dalivandan S., Plummer J., Gayther S.A. Risks and Function of Breast Cancer Susceptibility Alleles. Cancers. 2021;13 doi: 10.3390/cancers13163953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaag A., Walsh T., Renbaum P., Kirchhoff T., Nafa K., Shiovitz S., Mandell J.B., Welcsh P., Lee M.K., Ellis N., et al. Functional and genomic approaches reveal an ancient CHEK2 allele associated with breast cancer in the Ashkenazi Jewish population. Hum. Mol. Genet. 2005;14:555–563. doi: 10.1093/hmg/ddi052. [DOI] [PubMed] [Google Scholar]
- 36.Roeb W., Higgins J., King M.C. Response to DNA damage of CHEK2 missense mutations in familial breast cancer. Hum. Mol. Genet. 2012;21:2738–2744. doi: 10.1093/hmg/dds101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mai P.L., Best A.F., Peters J.A., DeCastro R.M., Khincha P.P., Loud J.T., Bremer R.C., Rosenberg P.S., Savage S.A. Risks of first and subsequent cancers among TP53 mutation carriers in the National Cancer Institute Li-Fraumeni syndrome cohort. Cancer. 2016;122:3673–3681. doi: 10.1002/cncr.30248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bougeard G., Renaux-Petel M., Flaman J.M., Charbonnier C., Fermey P., Belotti M., Gauthier-Villars M., Stoppa-Lyonnet D., Consolino E., Brugières L., et al. Revisiting Li-Fraumeni Syndrome From TP53 Mutation Carriers. J. Clin. Oncol. 2015;33:2345–2352. doi: 10.1200/JCO.2014.59.5728. [DOI] [PubMed] [Google Scholar]
- 39.Schneider K., Zelley K., Nichols K.E., Garber J. In: GeneReviews((R)) Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K., Amemiya A., editors. 1993. Li-Fraumeni Syndrome. [Google Scholar]
- 40.Li N., Thompson E.R., Rowley S.M., McInerny S., Devereux L., Goode D., LifePool Investigators. Wong-Brown M.W., Scott R.J., Trainer A.H., et al. Reevaluation of RINT1 as a breast cancer predisposition gene. Breast Cancer Res. Treat. 2016;159:385–392. doi: 10.1007/s10549-016-3944-3. [DOI] [PubMed] [Google Scholar]
- 41.Park D.J., Tao K., Le Calvez-Kelm F., Nguyen-Dumont T., Robinot N., Hammet F., Odefrey F., Tsimiklis H., Teo Z.L., Thingholm L.B., et al. Rare mutations in RINT1 predispose carriers to breast and Lynch syndrome-spectrum cancers. Cancer Discov. 2014;4:804–815. doi: 10.1158/2159-8290.CD-14-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stracker T.H., Roig I., Knobel P.A., Marjanović M. The ATM signaling network in development and disease. Front. Genet. 2013;4:37. doi: 10.3389/fgene.2013.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thorstenson Y.R., Roxas A., Kroiss R., Jenkins M.A., Yu K.M., Bachrich T., Muhr D., Wayne T.L., Chu G., Davis R.W., et al. Contributions of ATM mutations to familial breast and ovarian cancer. Cancer Res. 2003;63:3325–3333. [PubMed] [Google Scholar]
- 44.Pylkas K., Tommiska J., Syrjakoski K., Kere J., Gatei M., Waddell N., Allinen M., Karppinen S.M., Rapakko K., Kaariainen H., et al. Evaluation of the role of Finnish ataxia-telangiectasia mutations in hereditary predisposition to breast cancer. Carcinogenesis. 2007;28:1040–1045. doi: 10.1093/carcin/bgl237. [DOI] [PubMed] [Google Scholar]
- 45.Goldgar D.E., Healey S., Dowty J.G., Da Silva L., Chen X., Spurdle A.B., Terry M.B., Daly M.J., Buys S.M., Southey M.C., et al. Rare variants in the ATM gene and risk of breast cancer. Breast Cancer Res. 2011;13:R73. doi: 10.1186/bcr2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ring K.L., Bruegl A.S., Allen B.A., Elkin E.P., Singh N., Hartman A.R., Daniels M.S., Broaddus R.R. Germline multi-gene hereditary cancer panel testing in an unselected endometrial cancer cohort. Mod. Pathol. 2016;29:1381–1389. doi: 10.1038/modpathol.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villacis R.A.R., Basso T.R., Canto L.M., Pinheiro M., Santiago K.M., Giacomazzi J., de Paula C.A.A., Carraro D.M., Ashton-Prolla P., Achatz M.I., Rogatto S.R. Rare germline alterations in cancer-related genes associated with the risk of multiple primary tumor development. J. Mol. Med. 2017;95:523–533. doi: 10.1007/s00109-017-1507-7. [DOI] [PubMed] [Google Scholar]
- 48.Zidan J., Zhou A.Y., van den Akker J., Laitman Y., Schayek H., Schnaider J., Friedman E. Inherited predisposition to breast and ovarian cancer in non-Jewish populations in Israel. Breast Cancer Res. Treat. 2017;166:881–885. doi: 10.1007/s10549-017-4474-3. [DOI] [PubMed] [Google Scholar]
- 49.Randon G., Fucà G., Rossini D., Raimondi A., Pagani F., Perrone F., Tamborini E., Busico A., Peverelli G., Morano F., et al. Prognostic impact of ATM mutations in patients with metastatic colorectal cancer. Sci. Rep. 2019;9:2858. doi: 10.1038/s41598-019-39525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall M.J., Bernhisel R., Hughes E., Larson K., Rosenthal E.T., Singh N.A., Lancaster J.M., Kurian A.W. Germline Pathogenic Variants in the Ataxia Telangiectasia Mutated (ATM) Gene are Associated with High and Moderate Risks for Multiple Cancers. Cancer Prev. Res. 2021;14:433–440. doi: 10.1158/1940-6207.CAPR-20-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fanale D., Incorvaia L., Filorizzo C., Bono M., Fiorino A., Calo V., Brando C., Corsini L.R., Barraco N., Badalamenti G., et al. Detection of Germline Mutations in a Cohort of 139 Patients with Bilateral Breast Cancer by Multi-Gene Panel Testing: Impact of Pathogenic Variants in Other Genes beyond BRCA1/2. Cancers. 2020;12:2415. doi: 10.3390/cancers12092415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macedo G.S., Araujo Vieira I., Brandalize A.P., Giacomazzi J., Inez Palmero E., Volc S., Rodrigues Paixão-Côrtes V., Caleffi M., Silva Alves M., Achatz M.I., et al. Rare germline variant (rs78378222) in the TP53 3' UTR: Evidence for a new mechanism of cancer predisposition in Li-Fraumeni syndrome. Cancer Genet. 2016;209:97–106. doi: 10.1016/j.cancergen.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y., Wu X.S., He J., Ma T., Lei W., Shen Z.Y. A novel TP53 variant (rs78378222 A > C) in the polyadenylation signal is associated with increased cancer susceptibility: evidence from a meta-analysis. Oncotarget. 2016;7:32854–32865. doi: 10.18632/oncotarget.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voropaeva E.N., Orlov Y.L., Pospelova T.I., Gurageva A.A., Voevoda M.I., Maksimov V.N., Seregina O.B., Churkina M.I. The rs78378222 prevalence and the copy loss of the protective allele A in the tumor tissue of diffuse large B-cell lymphoma. PeerJ. 2020;8 doi: 10.7717/peerj.10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delimitsou A., Fostira F., Kalfakakou D., Apostolou P., Konstantopoulou I., Kroupis C., Papavassiliou A.G., Kleibl Z., Stratikos E., Voutsinas G.E., Yannoukakos D. Functional characterization of CHEK2 variants in a Saccharomyces cerevisiae system. Hum. Mutat. 2019;40:631–648. doi: 10.1002/humu.23728. [DOI] [PubMed] [Google Scholar]
- 56.Boonen R.A.C.M., Wiegant W.W., Celosse N., Vroling B., Heijl S., Kote-Jarai Z., Mijuskovic M., Cristea S., Solleveld-Westerink N., van Wezel T., et al. Functional Analysis Identifies Damaging CHEK2 Missense Variants Associated with Increased Cancer Risk. Cancer Res. 2022;82:615–631. doi: 10.1158/0008-5472.CAN-21-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matoy E.J., Plowman J.N., Watson C.J., Belshan M.A., Blue E.E., Huff C.D., Stessman H.A.F. In vitro data suggest a role for PMS2 Kozak sequence mutations in Lynch syndrome risk. HGG Adv. 2024;5:100298. doi: 10.1016/j.xhgg.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jarvinen H.J., Renkonen-Sinisalo L., Aktan-Collan K., Peltomaki P., Aaltonen L.A., Mecklin J.P. Ten years after mutation testing for Lynch syndrome: cancer incidence and outcome in mutation-positive and mutation-negative family members. J. Clin. Oncol. 2009;27:4793–4797. doi: 10.1200/JCO.2009.23.7784. [DOI] [PubMed] [Google Scholar]
- 59.Adachi M., Banno K., Yanokura M., Iida M., Nakamura K., Nogami Y., Umene K., Masuda K., Kisu I., Ueki A., et al. Risk-reducing surgery in hereditary gynecological cancer: Clinical applications in Lynch syndrome and hereditary breast and ovarian cancer. Mol. Clin. Oncol. 2015;3:267–273. doi: 10.3892/mco.2014.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cantor D. The frustrations of families: Henry Lynch, heredity, and cancer control, 1962-1975. Med. Hist. 2006;50:279–302. [PMC free article] [PubMed] [Google Scholar]
- 61.Sinilnikova O.M., Mazoyer S., Bonnardel C., Lynch H.T., Narod S.A., Lenoir G.M. BRCA1 and BRCA2 mutations in breast and ovarian cancer syndrome: reflection on the Creighton University historical series of high risk families. Fam. Cancer. 2006;5:15–20. doi: 10.1007/s10689-005-2571-7. [DOI] [PubMed] [Google Scholar]
- 62.Adam F., Fluri M., Scherz A., Rabaglio M. Occurrence of variants of unknown clinical significance in genetic testing for hereditary breast and ovarian cancer syndrome and Lynch syndrome: a literature review and analytical observational retrospective cohort study. BMC Med. Genom. 2023;16:7. doi: 10.1186/s12920-023-01437-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Naslund-Koch C., Nordestgaard B.G., Bojesen S.E. Increased Risk for Other Cancers in Addition to Breast Cancer for CHEK2∗1100delC Heterozygotes Estimated From the Copenhagen General Population Study. J. Clin. Oncol. 2016;34:1208–1216. doi: 10.1200/JCO.2015.63.3594. [DOI] [PubMed] [Google Scholar]
- 64.Bui A.N., LeBoeuf N.R., Nambudiri V.E. Skin cancer risk in CHEK2 mutation carriers. J. Eur. Acad. Dermatol. Venereol. 2021;35:353–359. doi: 10.1111/jdv.16729. [DOI] [PubMed] [Google Scholar]
- 65.Knappskog S., Chrisanthar R., Løkkevik E., Anker G., Østenstad B., Lundgren S., Risberg T., Mjaaland I., Leirvaag B., Miletic H., Lønning P.E. Low expression levels of ATM may substitute for CHEK2/TP53 mutations predicting resistance towards anthracycline and mitomycin chemotherapy in breast cancer. Breast Cancer Res. 2012;14:R47. doi: 10.1186/bcr3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pande M., Wei C., Chen J., Amos C.I., Lynch P.M., Lu K.H., Lucio L.A., Boyd-Rogers S.G., Bannon S.A., Mork M.E., Frazier M.L. Cancer spectrum in DNA mismatch repair gene mutation carriers: results from a hospital based Lynch syndrome registry. Fam. Cancer. 2012;11:441–447. doi: 10.1007/s10689-012-9534-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hendriks Y.M.C., Wagner A., Morreau H., Menko F., Stormorken A., Quehenberger F., Sandkuijl L., Møller P., Genuardi M., Van Houwelingen H., et al. Cancer risk in hereditary nonpolyposis colorectal cancer due to MSH6 mutations: impact on counseling and surveillance. Gastroenterology. 2004;127:17–25. doi: 10.1053/j.gastro.2004.03.068. [DOI] [PubMed] [Google Scholar]
- 68.Bonadona V., Bonaïti B., Olschwang S., Grandjouan S., Huiart L., Longy M., Guimbaud R., Buecher B., Bignon Y.J., Caron O., et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA. 2011;305:2304–2310. doi: 10.1001/jama.2011.743. [DOI] [PubMed] [Google Scholar]
- 69.Santana Dos Santos E., Lallemand F., Burke L., Stoppa-Lyonnet D., Brown M., Caputo S.M., Rouleau E. Non-Coding Variants in BRCA1 and BRCA2 Genes: Potential Impact on Breast and Ovarian Cancer Predisposition. Cancers. 2018;10 doi: 10.3390/cancers10110453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoberg-Vetti H., Ognedal E., Buisson A., Vamre T.B.A., Ariansen S., Hoover J.M., Eide G.E., Houge G., Fiskerstrand T., Haukanes B.I., et al. The intronic BRCA1 c.5407-25T>A variant causing partly skipping of exon 23-a likely pathogenic variant with reduced penetrance? Eur. J. Hum. Genet. 2020;28:1078–1086. doi: 10.1038/s41431-020-0612-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Manrai A.K., Funke B.H., Rehm H.L., Olesen M.S., Maron B.A., Szolovits P., Margulies D.M., Loscalzo J., Kohane I.S. Genetic Misdiagnoses and the Potential for Health Disparities. N. Engl. J. Med. 2016;375:655–665. doi: 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ahearn T.U., Zhang H., Michailidou K., Milne R.L., Bolla M.K., Dennis J., Dunning A.M., Lush M., Wang Q., Andrulis I.L., et al. Common variants in breast cancer risk loci predispose to distinct tumor subtypes. Breast Cancer Res. 2022;24:2. doi: 10.1186/s13058-021-01484-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gasperini M., Starita L., Shendure J. The power of multiplexed functional analysis of genetic variants. Nat. Protoc. 2016;11:1782–1787. doi: 10.1038/nprot.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during this study are available at the National Library of Medicine Sequence Read Archive (SRA) [SUB14326985/PRJNA1090944/https://www.ncbi.nlm.nih.gov/bioproject/1090944].