Abstract
The human immunodeficiency virus envelope glycoproteins, gp120 and gp41, function in cell entry by binding to CD4 and a chemokine receptor on the cell surface and orchestrating the direct fusion of the viral and target cell membranes. On the virion surface, three gp120 molecules associate noncovalently with the ectodomain of the gp41 trimer to form the envelope oligomer. Although an atomic-level structure of a monomeric gp120 core has been determined, the structure of the oligomer is unknown. Here, the orientation of gp120 in the oligomer is modeled by using quantifiable criteria of carbohydrate exposure, occlusion of conserved residues, and steric considerations with regard to the binding of the neutralizing antibody 17b. Applying similar modeling techniques to influenza virus hemagglutinin suggests a rotational accuracy for the oriented gp120 of better than 10°. The model shows that CD4 binds obliquely, such that multiple CD4 molecules bound to the same oligomer have their membrane-spanning portions separated by at least 190 Å. The chemokine receptor, in contrast, binds to a sterically restricted surface close to the trimer axis. Electrostatic analyses reveal a basic region which faces away from the virus, toward the target cell membrane, and is conserved on core gp120. The electrostatic potentials of this region are strongly influenced by the overall charge, but not the precise structure, of the third variable (V3) loop. This dependence on charge and not structure may make electrostatic interactions between this basic region and the cell difficult to target therapeutically and may also provide a means of viral escape from immune system surveillance.
The human immunodeficiency viruses (types 1 [HIV-1] and 2 [HIV-2]) and related simian viruses (SIVs) cause the depletion and functional dysregulation of CD4+ lymphocytes, resulting in the development of AIDS. The HIV envelope contains two glycoproteins: gp120, the exterior receptor-binding component, and its noncovalently interacting partner, gp41, the transmembrane envelope glycoprotein. Only half of gp41 is exposed in the ectodomain; the other half, separated by a transmembrane region, is thought to anchor the envelope complex to the underlying matrix. New infections are initiated by interaction of gp120 with the N-terminal membrane-distal domain of CD4, a glycoprotein on the surface of specific cells of the immune system (12, 29). A second interaction of gp120, with a member of the chemokine receptor family, primarily CCR5 or CXCR4, is believed to trigger conformational changes in gp41, which ultimately mediates virus-cell membrane fusion (20, 38).
HIV receptor binding takes place in the context of an oligomeric viral spike. Atomic-level structures have been determined for many of the component molecules: the entire extracellular portion of CD4 (60), the complex of monomeric core gp120 (a truncated version of gp120 with deletions of the gp41-interactive region at the N and C termini as well as of the variable V1/V2 and V3 loops) with the two N-terminal domains of CD4 and the antigen binding fragment (Fab) of the neutralizing antibody 17b (32), and a final fusion-active state of the gp41 trimer (10, 52, 56). Despite extensive effort, the structure of the oligomeric spike has resisted atomic-level investigation and is only known from electron microscopy (21, 22).
Accumulating evidence suggests that the HIV viral spike is a trimer of gp120-gp41 heterodimers. The most convincing evidence comes from the structural resemblance of the fusion-active state of gp41 to other fusion-active trimeric coiled-coils, including the equivalent transmembrane envelope proteins from Moloney murine leukemia virus (19), influenza virus (6), and Ebola virus (55). Other suggestive evidence comes from the introduction of cysteines into the coiled-coil to create disulfide-stabilized trimers (16), the trimeric nature of the underlying HIV matrix which interacts with gp41 (25), the trimerization of various ectodomain constructs of gp120-gp41 (X. Yang, L. Florin, M. Farzan, P. Kolchinsky, P. D. Kwong, J. Sodroski, and R. Wyatt, submitted for publication) and the therapeutic success of a peptide which appears to work by stabilizing an intermediate trimeric state in the gp120-gp41 fusion process (11, 28, 58).
Although the conformation of core gp120 is known by antigenic studies to be similar in the gp120-gp41 complex, molecular docking of gp120 onto the trimeric gp41 is not feasible because gp41 undergoes large conformational changes (6, 10, 56). Nonetheless, by correlating the antigenic map of gp120 with its atomic structure, a preliminary model of oligomeric gp120 was defined (62). Because this model was based on antibody binding, which occurs in the context of a ∼600-Å2 epitope, it was of relatively low resolution. Still it showed that the regions of the oligomer facing the target cell after CD4 binding consisted of two components: a conserved portion of the core and a sequence-variable excursion, the V3 loop. These components have been shown by mutational analysis to interact with the chemokine receptor CCR5 (45).
Here we more precisely model the HIV-1 envelope glycoprotein oligomer, using quantifiable criteria based on carbohydrate exposure, occlusion of conserved surface residues that are solvent exposed on the gp120 protomer, and steric constraints imposed by the binding of the 17b antibody. We have applied the same modeling techniques to influenza virus hemagglutinin to estimate the modeling precision, since the structures of both the monomeric “HA top” of hemagglutinin (the gp120 core equivalent) and the HA1-HA2 heterotrimer (the gp120-gp41 complex equivalent) have been determined (4, 59). We have also investigated the electrostatic nature of the gp120 region facing the target cell, examining in particular the dependence of the potential on the structure and overall charge of the V3 loop. In a companion study (39), we tested the electrostatic predictions of our model on the binding of heparan sulfate and dextran sulfate to different variants of gp120. Finally, we examine the consequences of our results with regard to initial virus-cell attachment, viral mechanisms of immune evasion, and the feasibility of anion-based therapeutic strategies.
MATERIALS AND METHODS
Structural modeling of the exterior envelope oligomer.
Superpositions, center-of-mass calculations, model rotations and translations, radius of gyration analysis, and solvent exposure calculations were performed with the software XPLOR (5). The structure of the monomeric core gp120 complexed with the two membrane-distal domains of CD4 (D1D2) and the neutralizing antibody 17b (Protein Data Bank [PDB] accession code 1gc1) was superimposed onto the corresponding D1D2 domains (residues 1 to 178) of the structure of the four-domain CD4 (the entire extracellular region) (PDB accession code 1wio) (60). The superposition gave an RM5 deviation of 2.12 Å for all atoms in residues 1 to 178.
The coordinate system used for modeling the gp120 oligomer (Fig. 1) had the following properties: (i) the z axis was coincident with the gp120 trimer axis; (ii) all three rotational degrees of freedom were permitted; (iii) since the coordinate system was threefold symmetric, only one other translation axis, chosen here to be the x axis, was independent (translations along the y axis were thus related by a Z rotation and x axis translation); and (iv) the initial position of the gp120 protomer was oriented by the superposition described above and placed with its center of mass at x = 35 Å and y = 0. (To distinguish between translations and rotations, lowercase letters x, y, and z are used to specify both the axis and the translational position along each axis and uppercase letters X, Y, and Z are used to specify the rotations about each axis.)
FIG. 1.
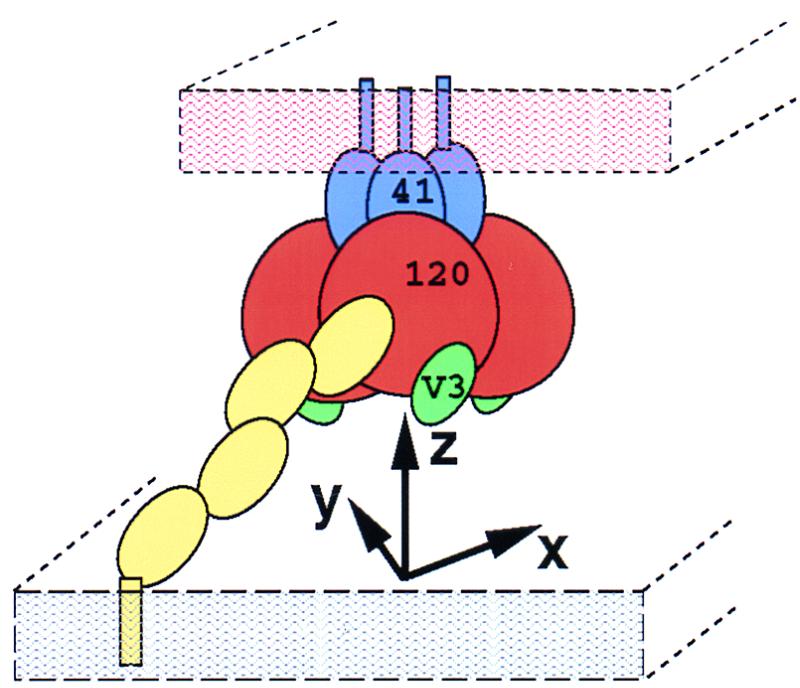
Coordinate system used to model the envelope oligomer shown in the context of the various interactions of gp120 during virus-cell attachment. CD4 (yellow) is shown reaching from the target cell membrane (gray) to bind gp120 (120; brownish red), which is arranged in a symmetrical fashion around the ectodomain of the trimeric gp41 (41; blue), shown jutting out of the viral membrane (orange). The V3 loop (green) can be seen on gp120 just above the cell membrane. The coordinate system used for modeling has the z axis coincident with the gp41 trimer axis, the x axis connecting the center of mass of a gp120 protomer with the z axis, and the y axis mutually perpendicular to the x and z axis and sharing a common origin. Thus, X and Y rotations (that is, about the x axis and y axis, respectively) specify the orientation of a gp120 protomer with respect to the membranes, and rotations about the z axis determine the orientation with respect to the envelope oligomer.
The gp120 protomer was rotated around its center of mass. The distance from the z axis of each of the various criteria chosen for quantification was calculated at 10° intervals. These calculations were performed with the program GRASP, with the normal to the z axis being determined by calculating the minimum of the distance matrix between the criterion being analyzed and a set of pseudoatoms placed at 1-Å intervals along the z axis.
Three criteria were chosen for quantification: carbohydrate exposure, conserved and exposed residues, and steric constraint of the 17b Fab fragment. For calculations of carbohydrate exposure, the structure of gp120 used was a model described previously (62), which comprised the crystallographic core (32), the modeled residues 88 and 89 and the V4 loop, and the central (N-acetylglucosamine)2(mannose)3 glycan moiety of all N-linked glycosylations. Because the positions of the mannose residues were poorly defined in the model (62), the N-acetylglucosamine residues, which are more proximal to the protein and appear less conformationally flexible, were used to represent the carbohydrate. The molecular surface of the N-acetylglucosamine residues was constructed, and the distance of this surface to the z axis was calculated as a function of gp120 orientation.
Conserved residues were conserved across all HIV-1 isolates (32). The fractional solvent accessibility for individual amino acids of core gp120 which were extracted from the 1gc1 complex was calculated as the ratio of the solvent-accessible surface area for atoms of an amino acid residue X in the protein to that area obtained after reducing the structure to a Gly-X-Gly tripeptide (47). Residues were considered exposed if they had a solvent accessibility of more than 40%. The resultant set of conserved and exposed residues on the gp120 core (33 residues) was further delineated by removal of those within van der Waals radii of either CD4 or the 17b Fab (10 residues excluded) or those that were glycosylated (2 residues excluded). Finally, a clustering analysis which excluded outliers more than 5 Å from the main cluster of conserved solvent-exposed residues (six residues excluded) was performed. The residues chosen by imposing the above criteria were 102, 103, 113, 114, 204, 208, 209, 211, 213, 214, 216, 221, 250, 439, and 491. The molecular surface of these residues was constructed, and the distance of this surface to the z axis was calculated as a function of the gp120 orientation.
To quantify steric constraint of the 17b Fab, its molecular surface was calculated as oriented by the position of the gp120 protomer, and the distance from this surface to the z axis was calculated. This criterion was less strict than the others since it was dependent on translational positioning, for which there was little constraint. To account for this, an orientation was considered sterically forbidden only if the distance from the 17b surface to the z axis was less than 3 Å for both the orientation being considered and the previous 10° rotation, effectively adding a 10° buffer zone to the sterically forbidden region.
Determination of modeling accuracy.
The accuracy of the surface criterion optimization procedure was tested with the monomeric HA top of influenza virus hemagglutinin complexed with Fab HC19 (PDB accession code 2vir) (4). The “correct” oligomeric orientation was defined by the HA1-HA2 heterotrimer (PDB accession code 5hmg) (54, 59), positioned with its trimer axis coincident with the z axis. Superposition of the HA top onto a protomer of the oriented oligomeric HA1-HA2 heterotrimer gave an RMS deviation of 1.09 Å for all atoms in residues 43 to 309.
Quantification of surface criteria described above for gp120 was performed on the HA top structure with several modifications. For the carbohydrate criterion, a BLAST search (1) of the GenBank database (release 113.0) with the HA top sequence enabled 485 HA1 sequences to be aligned. All sites of potential N-linked glycosylation in the aligned sequences were identified. These mostly nonconserved sites of glycosylation were at residues 45, 63, 81, 122, 126, 133, 144, 165, 246, 276, and 285. A model of the molecular surface of the nonbackbone portions of these residues in the HA top structure (2vir) was constructed, and the distance from this surface to the z axis was calculated as a function of the HA top orientation.
For the conserved exposed residue criterion, a threshold of 99% identity was used with the same 485 sequences. A 40% solvent exposure criterion was used, as calculated for the HA top (2vir) structure. Because the receptor (sialic acid) is small and no main cluster of conserved exposed residues could be identified, receptor distance exclusion and outlier rejection were not used. Conserved exposed residues identified were 55, 57, 104, 107, 110, 129, 165, 169, 187, 208 to 210, 212, 221, 222, 225, 238, 240, 263, 269, 271, 285, 289 to 291, 293, 304, and 308. A model of the molecular surface of the nonbackbone portions of these residues in the HA top structure was constructed, and the distance of this surface to the z axis was calculated as a function of the HA top orientation.
Finally, for the steric constraint criterion, the Fab fragment of the influenza virus-neutralizing antibody HC19 was used in place of the HIV-neutralizing antibody 17b.
V3 loop modeling.
In addition to the typical modeling criteria, such as avoiding steric clashes and maximizing hydrogen bonding, the position of the 17b antibody was used to provide additional constraints since it is known that this antibody binds to both native gp120 and V3 loop-truncated gp120. Three different models of the V3 loop were constructed by using the program O (27). Using the helix model, called alpha, the g and h helices of sperm whale myoglobin, residues 106 to 137 of 1mbo, were extracted and substituted for the GAG residues of the V3 loop in 1gc1. The myoglobin sequence was replaced by the HXBc2 sequence, and successive rounds of stereochemical optimization coupled to manual rebuilding were performed to remove steric clashes. Using the beta-strand model, called beta, the f and g strands of the immunoglobulin Bence-Jones protein, residues 80 to 107 of 1rei, were grafted onto the core gp120 in the same manner as described for the alpha model. The nmr model was derived from nuclear magnetic resonance (NMR) analyses of V3 loops from several different HIV-1 isolates (7, 8) and consisted primarily of random-coil secondary structure. Five different NMR structures were examined in relation to the core gp120. Only two, from the mn and Haiti isolates, both refined from H2O-trifluoroethanol (TFE) mixtures, could be grafted onto the core without extensive clashes. Upon completion of structure building, clashes with the carbohydrate at position 332 of the gp120 core could not be resolved with the model derived from the Haiti isolate, and so this isolate was not used in further analyses. The nmr model thus derives solely from the NMR analysis of the mn isolate in H2O-TFE (8).
Electrostatic analysis.
Electrostatic analyses were performed analytically with the program DelPhi (41) with a protein dielectric of 2.0, a solvent dielectric of 80, an ion exclusion radius of 2.0 Å, a probe radius of 1.4 Å, and an ionic strength of 0.14 M. For pictorial display, the precise Delphi potentials were read into the program GRASP (42), with the local potentials displayed at the solvent-accessible surface.
Homology modeling.
Homology modeling was carried out with the program PrISM (64), with sequence alignments and homologous models constructed based on the 1gc1 gp120 structure.
RESULTS
Modeling.
A three-dimensional model of the gp120 portion of the trimeric HIV envelope glycoprotein complex was developed subject to the conditions that the surface of gp120 that is occluded in the oligomer interface should (i) maximize carbohydrate exclusion, (ii) minimize conserved residue exposure, and (iii) be sterically compatible with binding of the 17b antibody. Since gp120 interacts with gp41, which is a symmetric trimer in its isolated form and remains trimeric when in the complex, the model was constrained to be threefold symmetric. The occluded interface in the oligomer is expected to comprise gp120-gp41 contacts, gp120-gp120 contacts, and occluded surfaces that large ligands such as antibodies cannot access. The constraining conditions for the model were chosen because, a priori, they represent reasonable expectations about the oligomer interface and because they could be reduced to quantifiable criteria.
Glycosylation sites occur almost exclusively on the exposed surfaces of protein molecules, and they occur near protein interfaces only at the periphery. Carbohydrate residues in N-linked glycans tend to be both flexible and highly hydrated; although they can be secured by protein contacts, the resultant entropic loss is large, making such interactions generally unlikely. As a consequence, we could expect the oligomeric interface to be free of glycosylation.
We would also expect exposed surface residues on gp120 to be variable, a consequence of immune pressure. Possible factors of conservation are limited primarily to occlusion at the oligomer interface, involvement with receptor binding, or constraints of folding topology. Conservation due to the last two criteria could be eliminated by examining residues for proximity to either the CD4 or the 17b Fab binding site (the 17b site here served as a surrogate for the chemokine receptor binding site [61]) and by performing a clustering analysis to remove statistical outliers (constraints on exposed residues for structural purposes should be rare).
Several ligands are known to bind to gp120 in the context of the oligomeric complex as well as to isolated gp120. The sites for such ligands must be oriented on the oligomer interface and also appropriately oriented for biological interactions. The 17b antibody is known to bind to oligomeric gp120 (53) and does not cause appreciable dissociation of the gp120 from the oligomer (44). Therefore, any valid oligomer model must be sterically compatible with 17b binding. Similarly, productive binding of HIV to CD4-positive cells is known to involve intact glycoprotein oligomers. Therefore, one expects the CD4 binding site to be both free on the oligomer surface and appropriately oriented for attachment to CD4 on the cell surface.
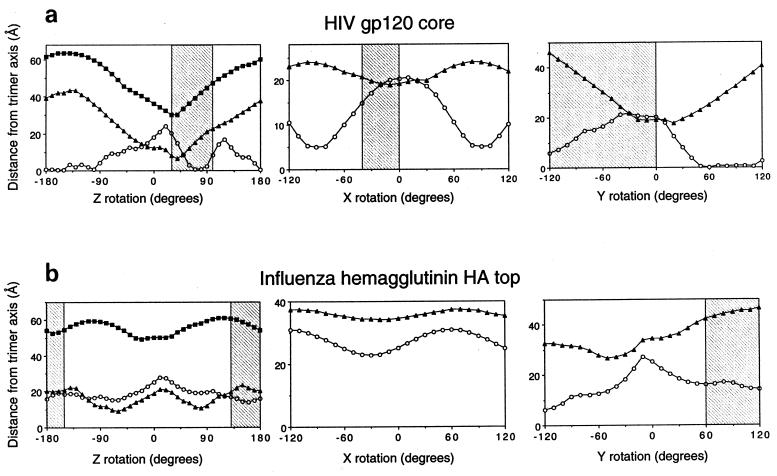
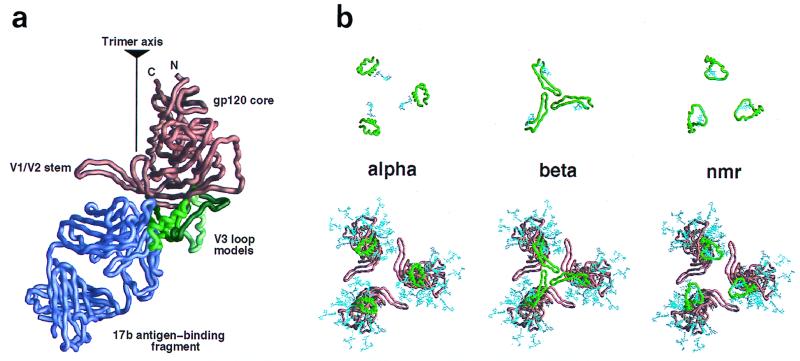
The crystal structure of core gp120 was first elaborated with a modeled completion of two N-terminal residues (one of which is glycosylated), of the V4 loop, and of the two N-acetylglucosamine residues at each site of N-linked glycosylation. This elaborated core was then oriented as a rigid body relative to a coordinate system established with a threefold-symmetry axis perpendicular to the viral surface (Fig. 1). The initial orientation of gp120 was set such that the D1D2 portion of CD4 in the core gp120-CD4 complex would be superimposed onto a promoter of D1D2 in the structure of dimeric soluble CD4, oriented with its diad axis perpendicular to the hypothetical cell surface. (The dimeric soluble CD4, which consists of the entire extracellular portion of CD4, crystallizes as a dimer in three different space groups [60]; its orientation is physiologically relevant and thus serves to position the hypothetical cell surface.) The gp120 protomer was then reoriented about its center of mass, displaced from the triad axis sufficiently to avoid collisions with other protomers. First, all rotational orientations about the z axis were tested with respect to the quantifiable criteria, and the optimal value was found to be at 30° (Fig. 2a, left panel). Then, with the Z orientation at the optimum, rotations were made successively about the X and Y rotational axes (Fig. 2a, middle and right panels). A protomer in the optimized model can be obtained from the 1gc1 PDB coordinates by the Euler rotation (Θ1 = 6.90, Θ2 = 112.34, Θ3 = 22.60) followed by the translation (tx = −25.91, ty = −71.24, tz = 30.90). This procedure of successive rotations, which was used for computational economy, does not sample all of the rotational space; nevertheless, we expect the result to be close to the optimum for the three conditions since it preserves the initial orientation relative to CD4 on the cell surface. In addition, visual inspection of the final orientation confirms that it is close to, if not at, the global optimum.
FIG. 2.
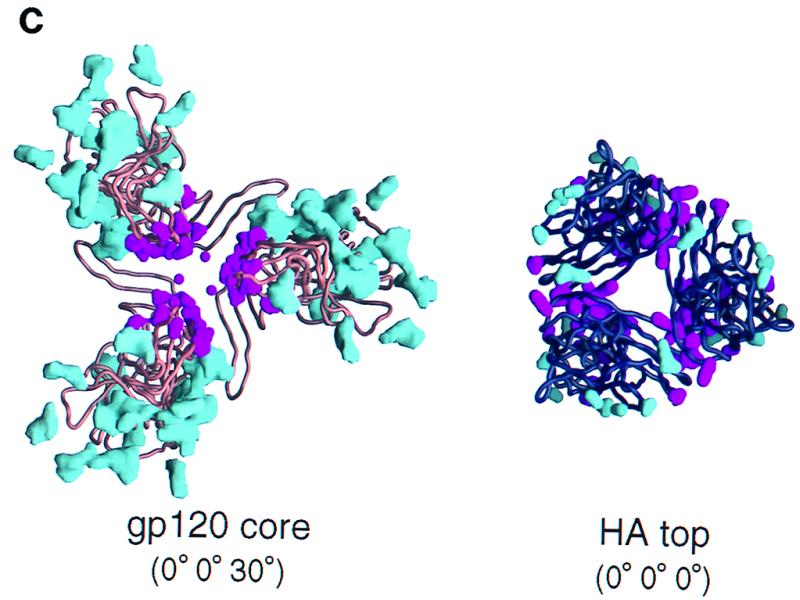
Quantitative modeling of the gp120 oligomer. Quantitative modeling employed three criteria: carbohydrate exposure (open circles), occlusion of conserved residues which are solvent exposed on a gp120 protomer (filled symbols), and steric considerations of 17b binding (forbidden regions [shaded]). These criteria have been graphed with respect to their distances from the trimer axis (depicted here on the vertical axis) as the rotational parameter (horizontal axis) was varied. The coordinate system was as defined in the legend to Fig. 1. (a) Rotational orientation of the HIV gp120 core. (Left panel) The orientations of X and Y in this panel were determined by the superposition of CD4 in the gp120-CD4 complex (32) with its orientation in the four-domain CD4 structure (60). The maximum (filled squares) and minimum (filled triangles) distances of the conserved, solvent-exposed residues from the trimer axis were determined at 10° intervals in Z; minimum values correspond to regions of greater oligomer occlusion. The distance of the carbohydrate (open circles) from the trimer axis is shown at 10° intervals; peaks correspond to carbohydrate-free regions at which the overall exposure of carbohydrate is greatest. The highest peak in this panel corresponds to a large carbohydrate-free region centered about the first helix in the inner domain of core gp120 (32). The second-highest peak corresponds to the CD4 binding site, which is also free of carbohydrate. (Middle panel). Given the optimal Z orientation, rotations were made around the x axis. Here the minimum and maximum distances for the conserved, solvent-exposed residues have been averaged (filled triangles). The 0 of the X rotation corresponds to the orientation of X as determined by the initial superposition with CD4. (Right panel) Given the optimal orientation for X and Z as determined in the previous two panels, the remaining rotational axis was varied. Again, the minimum and maximum distances for the conserved, solvent-exposed residues have been averaged (filled triangles). The extended carbohydrate-free region corresponds to the overlap between the carbohydrate-free chemokine receptor region and the presumed oligomer interface. Steric constraints from binding of the 17b antibody (forbidden regions [shaded]) helped to distinguish these regions. As with the X rotation, the 0 of the Y rotation corresponds to the orientation of Y as determined by the initial superposition with CD4, independent of the modeling criteria used here. (b) Surface criterion optimization of the influenza virus hemagglutinin HA top. The origin of each panel corresponds to the orientation of the HA top superimposed on the HA1-HA2 heterotrimer (59). The symbols used for the criteria are the same as described for panel a. (c) Surface criteria depicted from the perspective of the viral membrane at the (0° 0° 30°) orientation for the gp120 core and the (0° 0° 0°) orientation for the HA top. (Left image) the gp120 core is depicted as a copper-brown Cα backbone worm. The molecular surfaces of all of the N-acetylglucosamine residues used in the modeling are colored cyan. The molecular surfaces of the side chains of the conserved exposed residues on the monomeric gp120 core are colored magenta. The gp120 protomer has been positioned such that the distance of its center of mass from the trimer axis is proportional to that observed in hemagglutinin. (Right image) The HA top is depicted as a black Cα backbone worm. The molecular surfaces of side chains that are sites of glycosylation in at least 1% of the 485 aligned hemagglutinin sequences are shown in blue. The molecular surfaces of side chains which are conserved and exposed on the HA top monomer are colored magenta. The orientation shown here corresponds to the superposition of the HA top onto the HA1-HA2 heterotrimer.
The carbohydrate criterion and the exposed and conserved surface residue criterion were completely independent. Nonetheless, quantification of these two criteria led to maxima and minima within 20° of each other for all independent axial rotations (with the 17b steric criterion used bifunctionally to eliminate incompatible orientations) (Fig. 2). Taken together, the three criteria produced well-defined peaks with all three independent rotational parameters, thereby allowing a “best” orientation to be derived at an X rotation of 0°, a Y rotation of 0°, and a Z rotation of 30° (0° 0° 30°). This best orientation actually corresponds to two possible alignments with respect to the viral membrane, “up” and “down.” One of these alignments could be eliminated due to CD4 (or 17b) steric constraints; the binding of CD4 (or 17b) in this orientation would bury it in the viral membrane (Fig. 3), thus permitting a single unique best orientation to be derived.
FIG. 3.
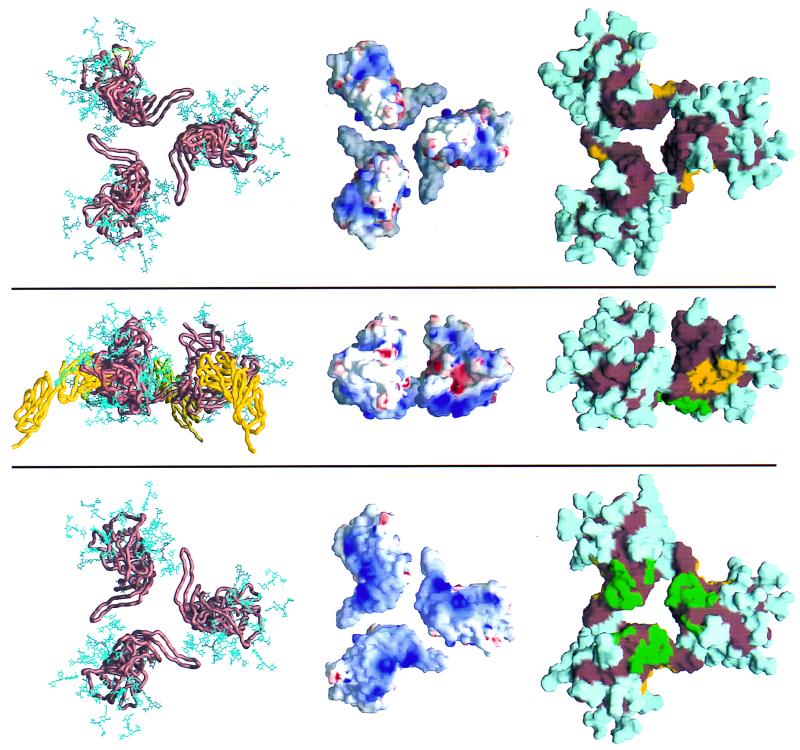
Trimeric model of gp120. Three orientations of the model are shown. The images at the top depict the view from the orientation of the viral membrane. The middle images depict the view from the side, in between the viral and target cell membranes. The images at the bottom depict the view from the target cell membrane. The left-most three images are Cα worm representations of core gp120 (copper brown) and the two membrane-distal domains of CD4 (yellow). Also shown are the gp120 carbohydrate cores (blue), the (N-acetylglucosamine)2-(mannose)3 cores shared by both high-mannose and complex N-linked glycan moieties. The carbohydrate shown here represents approximately half the carbohydrate on gp120, with the rest extending further from the gp120 surface. The middle images show the electrostatic surface of gp120 for the core. The electrostatic potential is depicted at the solvent-accessible surface, which is colored according to the local electrostatic potential, ranging from dark blue (most positive) to red (negative). The right-most images show the gp120 core with carbohydrate, with the solvent-accessible surface colored cyan for carbohydrate, yellow for the surface of gp120 less than 3 Å from CD4, green for the surface of gp120 less than 3 Å from the 17b antibody, and copper brown for the remaining surface of core gp120. The degree to which carbohydrate covers all of the solvent-accessible trimer surface is remarkable. Other than a small region at the viral proximal portion of the oligomer (where the missing N and C termini most likely reside), the only carbohydrate-free surfaces large enough to serve as an antibody epitope correspond to regions of receptor binding.
The resultant model of the core oligomer (Fig. 3) displayed several relevant features that were not included in the modeling criteria. The observed and deduced binding sites for CD4 and neutralizing antibodies were all on exposed surfaces. The variable loops (V1 to V5) were all well exposed on the oligomer. The N and C termini were clustered at the end pointing toward the viral membrane (with the C terminus proximal to the oligomer axis poised to interact with the trimeric gp41). Additionally, the region identified by substitutional mutagenesis as interacting with chemokine receptors (45) was free of carbohydrate and pointed directly at the target membrane. These features are consistent with the biology of the gp120 trimer and with the qualitative characteristics of the rough model obtained previously (62).
Modeling accuracy.
To estimate the precision of the surface criterion quantification procedure, we turned to the influenza virus hemagglutinin system. Atomic-level structures are known for both the HA1-HA2 heterotrimer (59) (equivalent to the gp120-gp41 oligomeric complex) and a monomeric proteolytic fragment, the HA top, complexed with the Fab of a neutralizing antibody (4) (equivalent to the gp120-17b complex). In addition, the fusion-activated forms of gp41 and HA2 are structurally similar, and although core gp120 and the HA top show no sequence similarity, they have almost identical radii of gyration (20.6 and 20.8 Å, respectively), making rotations about a position displaced the same distance from the trimer axis reasonable.
The HA top contains much less glycosylation than core gp120. Only three sites are present, although no carbohydrate is modeled in the 2vir coordinates (X31 isolate). In contrast, 18 sites are present on the gp120 core model (HXBc2 isolate). To increase the accuracy of this criterion for the HA top, all nonconserved sites of glycosylation were identified and then used to enhance overall surface coverage, increasing the residues used in the carbohydrate criterion to 11. Even so, this was less than two-thirds of that used for gp120 (compare the structures in Fig. 2c). In addition, because side chains, instead of glycan moieties, were used to mark the positions of the carbohydrates, criterion uncertainty increased. Nevertheless, the residues identified tended to be on the outside of the trimer, and for rotations in Y and Z this criterion produced well-defined maxima close to the known trimer orientation (Fig. 2b and c).
The hemagglutinin sequence is much less divergent than the gp120 sequence. This makes it difficult to find a reasonable set of conserved exposed residues. (If there were no divergence, for example, this criterion would be meaningless.) Using 485 aligned HA1 sequences and a minimum solvent exposure of 40%, 39 residues showed 98% identity, 28 showed 99% identity, and 12 showed 100% identity. The 100% criterion was judged too strict because sequencing errors might account for some divergence, and so a 99% criterion was used. This stringent 99% criterion selected approximately the same number of surface residues as the less-restrictive gp120 criterion (for which all single-atom substitutions [e.g., Gln to Glu] were included, as well as larger substitutions as long as they did not change the character of the amino acid [e.g., Lys to Arg or Tyr to Trp]). While these residues tended to cluster at the known oligomer interface, the minima produced by this criterion were not well-defined (Fig. 2b and c). For example, the minimum Z rotation of the furthest conserved residues appears close to the maximum for the nearest (Fig. 2b, left panel), and the X rotation showed very little change in parameter distance (Fig. 2b, middle panel). This lack of definition may be related to the packing of the hemagglutinin trimer, with conserved residues clustering at two discrete interfaces, as opposed to gp120, for which one central cluster was observed (Fig. 2c). Such clustering would account, for example, for the local maximum at the origin of the Z rotation for the minimum conserved residue distance.
Finally, because the HC19 Fab binds further from the trimer axis than 17b, it does not produce as much of a steric clash. Indeed, for rotations in X, no angles are restricted.
Judging from the poor shape of the quantification curves, the surface criteria optimization did not work as well with the HA top as with the gp120 core. Overall results of the optimization procedures are shown in Table 1. The internal agreement of the criterion optimizations, as judged by the peak orientational difference between the carbohydrate and conserved-residue extrema, was much better for gp120. Despite the poor agreement, the mean of the extrema for the HA top was within 20° of the correct orientation, suggesting that averaging these independent criteria reduced the overall error. In the case of gp120, the 17b steric criterion reduced the mean deviation even further, to one-fourth that observed for the HA top. This suggested that the error associated with the rotational parameters of the resultant gp120 oligomeric model was only 5°.
TABLE 1.
Summary of modeling results
| Criterion | Degrees of rotation of:
|
|||||
|---|---|---|---|---|---|---|
| Hemagglutinin HA top
|
gp120 corea
|
|||||
| Z | X | Y | Z | X | Y | |
| Carbohydrate maximum | 10 | 60 | −10 | −10 | 10 | −30 (0) |
| Conserved residue minimum | −30 | −20 | −50 | 10 (0) | −10 (0) | 20 |
| Peak positional difference | 40 | 80 | 40 | 20 (10) | 20 (10) | 50 (20) |
| Mean of extrema | −10 | 20 | −30 | 0 (−5) | 0 (5) | −5 (10) |
| Absolute mean deviations | ||||||
| Of peak rotational differences (X, Y, Z) | 53 | 30 (13) | ||||
| From the correct trimer orientation (of the mean of the extrema) | 20 | 11 (5)b | ||||
Numbers in parentheses are extrema if differences occur when forbidden regions determined from the Fab steric criterion are taken into account.
The mean deviation here was calculated assuming that the ratio of the mean deviation of the modeling parameters to the mean deviation from the correct trimer obtained in the hemagglutinin test case holds for gp120.
While it is conceivable that the deletion of the gp120 N and C termini (57 and 19 amino acids, respectively) could alter the modeling results, we feel that this is unlikely. With respect to the carbohydrate and 17b steric criteria, the missing termini are carbohydrate free and 17b binds to core gp120. Thus, both criteria should be unaffected. With respect to the conserved and exposed amino acid criterion, similar extrema were observed for both the furthest and closest residues (Fig. 2a, left panel). If some of the 15 exposed and conserved amino acids are covered by the termini, a subset should give similar results. In the event that all or nearly all 15 are covered, this would localize much of the missing termini in the same region as the previous exposed amino acids; since the missing termini correspond to the epitopes of virtually all of the nonneutralizing antibodies, this would again place the expected oligomer interface in a similar region. Finally, the agreement between the carbohydrate and exposed-residue criteria was so good that even if the exposed-residue criterion were omitted, the extrema would change by only 5° on average. Thus, we feel that the presence of the missing termini is unlikely to substantially alter the results obtained here.
Although the modeling precisely defined the rotational parameters of the trimer, the translational parameter was only partially determined. Steric constraints defined a minimum approach, and distance constraints between the C terminus of gp120 and the N terminus of gp41 defined a maximum, but these two criteria did not discriminate sharply. With the center of rotation for a protomer placed at 35 Å from the trimer axis, which was close to the sterically constrained minimum approach, the Cα-Cα diameter of the model was ∼110 Å; if the (mannose)3 carbohydrate extensions were included, the diameter increased to ∼150 Å. These dimensions agreed with electron microscopic observations of the viral spike, although these observations vary widely, from 100 Å (by negative staining of gp120 from SIV [22]) to 150 Å (by ultrathin section tannic acid cytochemistry and surface replica electron microscopy of HIV-1 [21]). If the distance from the trimer axis is proportional to the dimensions of the protomer in the x and y directions, using the relative proportions of hemagglutinin as a guide would place the gp120 core 30.5 Å from the trimer axis (Fig. 2c). A protomer of this hemagglutinin proportionate model can be obtained from the 1gc1 coordinates with the Euler rotation specified previously followed by the translation (tx = −30.43, ty = −71.24, tz = 30.90). At this distance, steric clashes occur between neighboring gp120 protomers with the carbohydrate at position 197, although these are easily resolved by minor movements of this flexible portion of the model. (This proportionate model is shown in Fig. 2c and 7; the rest of the figures depict the gp120 protomer with a center of mass at x = 35 Å.)
FIG. 7.
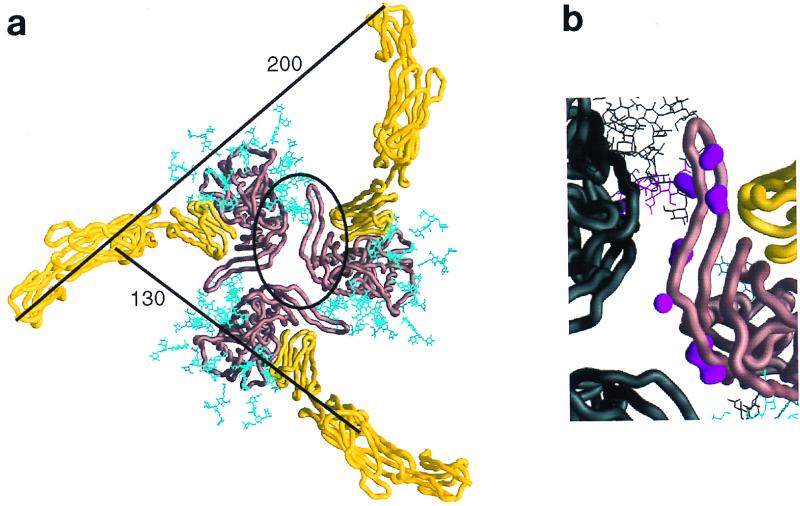
Selected features of the oligomeric gp120 model. (a) The entire extracellular portion of CD4 (yellow Cα worm) is shown binding to the oriented gp120 oligomer (copper-brown Cα worm). Carbohydrate on the gp120 is colored cyan. Distances between adjacent CD4 molecules are shown for the Cα of residue 178 at the end of the second domain and the Cα of residue 363, the last ordered residues prior to the transmembrane-spanning region. The contact region highlighted in panel b has been circled. The hemagglutinin proportionate model is used here, and thus the gp120 is close to the minimum protomer packing distance. (b) An enlargement of the contact site between gp120 core protomers in the oligomer. The coloring for one protomer is the same as in panel a; the other protomers are colored black. In addition, the molecular surfaces of side chains of conserved exposed residues in this contacting region (residues 195 to 210) are colored magenta. Starting from the top of the figure, these include residues 196, 197, 198, 201, 204, 208, and 209. Residue 197 is glycosylated, and its carbohydrate has been depicted in magenta. This region is sensitive to the binding of CD4, which is seen to the right of the figure in close proximity. The orientation of both images is shown from the perspective of the target cell membrane.
Electrostatic analysis of the oligomeric core gp120.
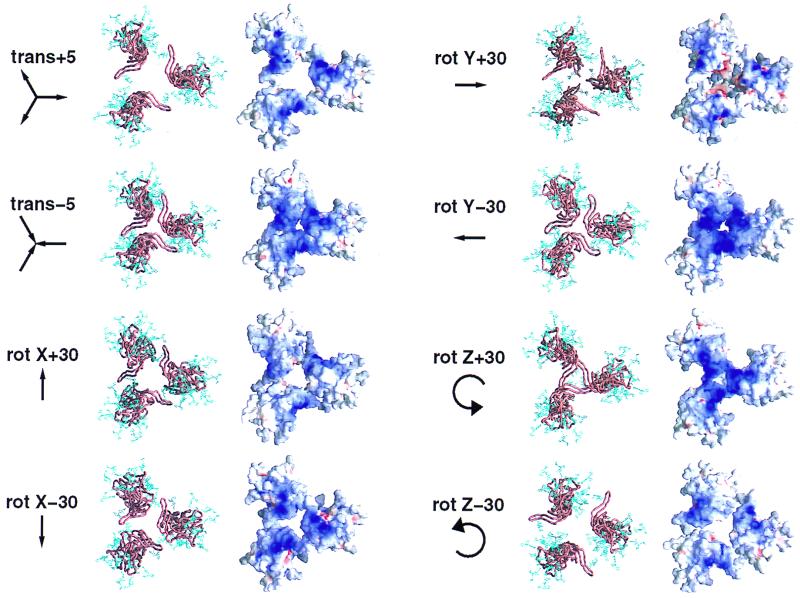
Electrostatic analysis of our model of the oligomeric core gp120 defined an electropositive surface which would face the target cell membrane. We tested the robustness of this basic region to changes in the modeling parameters. As can be seen in Fig. 4, rotations of ±30° and translations of ±5 Å maintained the electropositivity of the region.
FIG. 4.
Robustness of the basic cell-facing surface of core gp120 to variations in modeling parameters. The oligomer modeling was dependent on four independent parameters, one translational (trans) and three rotational (rot). The effect of varying these parameters on the basicity of the cell-facing surface of the proposed envelope oligomer is shown here. All of the images in this figure are depicted from the view of the target cell membrane. A Cα worm diagram of core gp120 (copper brown) with carbohydrate (blue) is shown to aid in orienting each variation in modeling parameter. The surface diagrams depict the local electrostatic potential. A dark blue (basic) surface can be seen on core gp120 (HIV-1, HXBc2 isolate) in all modeling parameter variations.
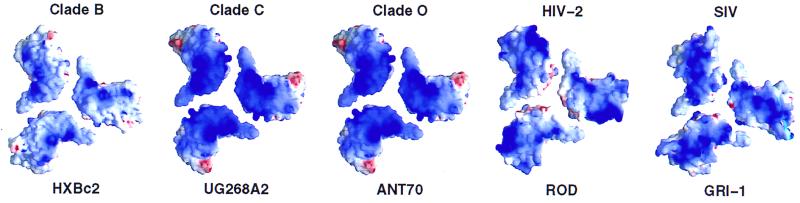
We used sequence analysis combined with homology modeling to test the conservation of this basic region throughout the primate immunodeficiency viruses. Although the charge of the region differed among viruses, its basic nature was conserved in different clades as well as with HIV-2 and SIV (Fig. 5).
FIG. 5.
Conservation of basic cell-facing surface on core gp120. Homology modeling was used to construct the corresponding structures of core gp120 for HIV-1 clades C and O as well as HIV-2 and SIV, starting with the crystal structure of HIV-1 clade B core gp120 (32) modeled as the envelope oligomer. The electrostatic potentials are depicted at the solvent-accessible surface, from the perspective of the target cell membrane. A basic (dark blue) surface can be seen in all isolates, although the precise charge distribution and the degree of overall basicity show variation.
Analysis of the V3 loop.
The V3 loop comprises roughly 30 amino acids. It is too large to be modeled correctly without experimental constraints. We used the steric constraints inherent in connecting the V3 loop to core gp120 as well as those consistent with 17b Fab binding. Nevertheless, very different V3 loop structures could be successfully built (Fig. 6a).
FIG. 6.
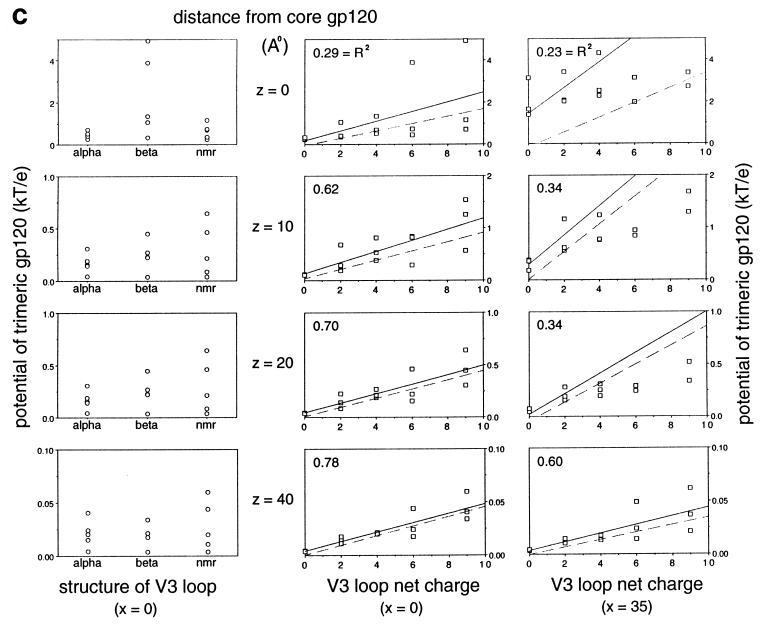
Modeling and electrostatic contribution of the V3 loop. (a) Different V3 loop models were constructed: alpha (green), beta (dark green; toward the back left of gp120 in this orientation), Haiti (forest green; depicted as the front right model in this orientation), and nmr (light green) (see text for details). These are displayed as Cα worm diagrams in the context of core gp120 (copper brown) and the 17b Fab fragment (purple violet). As can be seen, steric constraints, primarily from 17b, separate the V1/V2 stem from the V3 loop on a gp120 protomer, although quite diverse V3 loop structures can be successfully grafted onto the gp120 core. The position of the trimer axis (as determined by quantitative modeling) and the N and C termini of core gp120 are labeled for reference. (b) Cα worm diagrams for the different V3 loops (green) are depicted by themselves (top) and with core gp120 (copper brown) (bottom) for the HXBc2 strain of HIV-1 gp120 as arranged in the envelope oligomer. The orientation of each image is shown from the perspective of the target cell membrane. (c) Analysis of the electrostatic potential of the cell-facing region. Models of the trimeric gp120 core have been constructed with different V3 loops (described above for panels a and b), and the charge on the V3 loop for each of these structure has been varied between 0 and 9. For each of these 15 different models, the electrostatic potential has been calculated at various positions, either along the trimer axis x = 0 or below the center of mass of a protomer x = 35. Increasing numbers in z correspond to increasing distances away from gp120 (moving away from the virus). The position in z varies from z = 0, which corresponds to 0.5 Å below the gp120 core, to z = 40, which is 40.5 Å below the core but only approximately 25 Å below the V3 loop. The difference between core and V3 loop distances occurs because the V3 loop models extend roughly 15 Å below the core (as can be seen in panel a). (Left panels) The electrostatic potential from each model, that is, the three different V3 loop models shown in panel b, is graphed as a function of structure. No simple correspondence is seen. (Center and right panels) The electrostatic potential from each model is graphed as a function of charge. A rough linear correspondence is seen. This has been least-squares fitted (solid line), and the r2 fit, which shows how well the data correlate to the line, is depicted (an r2 of 1.0 corresponds to a perfect fit). The correlation is poor close to the gp120 core but increases as the distance from the virus increases. The dotted line corresponds to the least-squares fit if the potential is calculated for only the V3 loop. For the top three panels on the right, in order to use a reasonable scale, two outlier points, for charges of 6 and 9, are not depicted, although they have been used in the least-squares fitting.
The overall charge of the V3 loop ranges from +2 to +10, with that of a CCR5-using isolate generally in the range of +3 to +5 and that of a CXCR4-using isolate (often a T-cell-line-adapted strain) being from +7 to +10. With the HXBc2 sequence (+9 charge on the V3 loop), we analyzed the electrostatics of three very different V3 loop structures, as well as the effect of varying the overall net V3 loop charge (Fig. 6c).
Analysis of the potential near the cell-facing region of the oligomer showed a correlation between the potential and the overall V3 loop charge, especially at distances further than 20 Å from the gp120 core (Fig. 6c, middle and right panels). In contrast, the potential did not seem to correlate with the precise structure of the V3 loop (Fig. 6c, left panels).
The relatively low electrostatic potential for a zero V3 loop net charge suggested that the contribution of the core to the overall potential was small. We tested this explicitly by calculating the electrostatic potential as a function of only the V3 loop (Fig. 6c). At long distances and with a high charge (for example, CXCR4-using isolates), the V3 loop potential dominated, approximating closely the potential for the entire core with V3 loop. At short distances and with a low V3 loop charge, the core contributed significantly to the electrostatic potential. This was especially true at x = 35, y = 0, directly below the protomer, close to the above-described basic conserved region on the core.
DISCUSSION
From the earliest days of structural biology, for example, with myoglobin and hemoglobin, biologically important results have been extracted from low-resolution oligomeric models of more highly resolved protomers. The construction of such models often requires little additional information; if the protomer conformation does not change, only three angles and one distance are needed to specify a symmetric oligomer. Here, optimization of quantifiable surface criteria was used to precisely determine the orientation of gp120 in the oligomeric viral spike. The orientational precision of better than 10° corresponded to an average positional error of less than 3.5 Å for a molecule with a 20-Å radius of gyration. Because we did not have precise experimental limits on the translation component, however, our analysis of the model was limited to properties that depend primarily on rotational parameters. Still, the resultant model defined several interesting features, and here we explore their functional, immunological, and therapeutic implications.
Receptor binding.
Studies of the effect of soluble CD4 on virus entry suggest that more than one CD4 molecule must bind the envelope glycoprotein oligomer to initiate virus-cell fusion (33, 37, 50). The model derived here demonstrates that three CD4 molecules can bind without CD4:CD4 steric interference (in the hemagglutinin proportionate model, the closest CD4-to-CD4 contact is over 35 Å). CD4 binds obliquely to the sides of the oligomer, with the third and fourth domains of CD4 being almost parallel to the cell membrane. Because CD4 is an extended molecule and it binds gp120 at the N-terminal membrane-distal domain, the membrane-spanning portion of CD4 is positioned far from the oligomer axis. With the hemagglutinin proportionate model, the end of the second CD4 domain (residue 178) would be 127 Å from the same position on an adjacent CD4 and the last ordered extracellular residue (363) would be 198 Å away (Fig. 7). Even accounting for a tighter protomer packing or for the known segmental flexibility between domains 2 and 3 of CD4, membrane-spanning portions of adjacent CD4 molecules would be separated by at least 190 Å.
The chemokine receptor, in contrast, binds close to the trimer axis, at a gp120 surface which is almost 20 Å closer to the target cell membrane. This arrangement is consistent with simultaneous CD4 and chemokine receptor binding to a protomer. In terms of multiple chemokine receptors binding to the oligomer, our data show that the 17b Fab fragment, whose epitope overlaps the chemokine receptor binding site (45), lies at the edge of the sterically restricted “forbidden zone” in all three rotational parameters (Fig. 2). This suggests that multiple chemokine receptors bound to the same oligomer may be sterically strained. Since the chemokine receptor is an integral membrane protein and z translations are constrained, such strain could only be relieved by movement of the gp120 protomer away from the trimer axis. Such movement may serve to signal the binding state of gp120 at the target cell membrane to the gp41-interactive regions, roughly 50 Å distal, triggering gp41 fusion-related conformational changes at the appropriate moment. In this regard, it may be relevant that some of the substitutions that effect chemokine receptor binding map closer to the trimer axis than the 17b epitope, and many of these axis-proximal substitutions induce dissociation of gp120 from the oligomer (45).
Multivalent CD4 molecules have been constructed with the intent of binding multiple gp120 molecules to enhance avidity. Our results show that this is only possible with an extremely long and flexible linker, thus ruling out enhanced affinity with, for example, the two N-terminal domains of CD4 in an antibody format (9). Indeed, the dimensions are such that although the position of viral spikes is fixed by the underlying matrix, binding to a neighboring spike (which is roughly 380 Å away, assuming 72 viral spikes per virion and a virion diameter of 1,000 Å) would be almost equally feasible. (Such orientational steric restrictions should also apply to antibodies that bind to the CD4 binding site.)
Finally, with regard to the known segmental flexibility of CD4, the optimal orientations of the x and y axes of gp120 in the quantitative modeling were found to coincide with the dimer orientation of CD4. Given that the orientations were based on completely different criteria, this coincidence suggests that the orientation of CD4 on the cell surface may be more rotationally constrained with respect to the plane of the membrane than previously thought (60).
Oligomeric contacts.
A central mechanistic question is how CD4 activates gp120 on the virion, transforming the stable envelope into a reactive fusogen. CD4 is known to cause conformational changes in gp120, including most prominently the formation of the bridging sheet (32, 53). Interestingly, the edge of this conserved sheet can be seen as a pivotal site of contact in the oligomer; although the specific details of the contact are dependent on the ill-defined translation component, it is the well-defined rotation parameters which position the bridging sheet in its prominent location. In the hemagglutinin proportionate model, steric clashes are found with residue 197 in this region. Moreover, deletion of the outer two strands of the bridging sheet generates an oligomer to which CD4 binds without inducing shedding (63), and mutations in this region, especially of the glycosylation site at residue 197, correlate with a CD4-independent infection phenotype (30). All of these lines of evidence are consistent with the notion that activation of the trimer may involve close contact of the bridging sheet with the neighboring protomer (Fig. 7).
Another region of potential protomer:protomer contact involves the sequence-variable V1/V2 and V3 loops, which were truncated in the core gp120 structure. In the V3 loop modeling it was clear that 17b acts as a barrier between the V1/V2 stem and the V3 loop (Fig. 6a). Unless 17b binding substantially alters the positioning of the V1/V2 and/or V3 loop, this suggests that these loops probably do not contact each other within a protomer. Data from experiments in which revertants were obtained subsequent to mutation of the V3 loop, however, show that changes in the V1/V2 loop can rescue changes in the V3 loop (46). Moreover, some neutralizing antibodies in simian-human immunodeficiency virus-infected monkeys apparently recognize a discontinuous V2-V3 determinant (15), and a functional interaction of the V2 and V3 loops is able to determine coreceptor choice and neutralization resistance (50). These data suggest that, in the context of the oligomer, the V1/V2 loop and the V3 loop are near one another. In this regard, it may be relevant that the model derived here for the trimer displayed the base of the V3 loop juxtaposed to the V1/V2 stem from the neighboring protomer.
Biological implications of the basicity of the cell-facing surface of the gp120 oligomer.
Nonspecific electrostatic interactions of the basic cell-facing surface may affect entry of virus into a cell in several ways. With respect to initial attachment, these interactions may play a role in binding of virus to polyanions such as heparan sulfate. This conjecture agrees with biochemical results which show that heparan sulfate influences the binding of HIV virions to some cells (36). In addition, the model derived here allows us to analyze the electrostatic properties of a wide variety of HIV strains. In a companion study (39), we found that the observed binding of polyanions to different isolates of gp120 as well as to mutants of gp120 with and without the V3 loop correlates extremely well with model-based electrostatic predictions.
Other acidic surfaces may interact with the basic cell-facing region described here. For example, negatively charged lipid head groups may create an acidic zone in the target membrane itself. The degree of membrane acidity is dependent on the composition of the lipid head groups, which may differ in different cells as well as locally, from patch to patch, on the surface of an individual cell. The target membrane compositional variation may explain the ability of heparinase to affect initial binding in some cell types and not in others (36, 43).
Numerous studies have documented the binding of membrane-associated proteins through both a hydrophobic interaction and an electrostatic attraction to membrane head groups (34, 40). A theoretical and experimental analysis of the binding of the polylysine peptides (Lys)3, (Lys)5, and (Lys)7 to a 33% acidic membrane (a 2:1 mixture of phosphotidylcholine and phosphotidylglycerol) showed that each charge enhances the binding of the polymer to the membrane by roughly 1 order of magnitude (3). While it is difficult to correlate the polylysine results with the V3 loop/core basicity determined here, that analysis does suggest that if the cell-facing surface of oligomeric gp120 is highly basic, direct interactions with the membrane will be enhanced. Moreover, our results suggest that the wide variation in overall charge on the V3 loop will be a primary determinant of the magnitude of this electrostatic interaction. (We note, however, that the relatively poor correlations between potential and charge at shorter distances suggest that the precise structure of the V3 loop will influence short-range interactions.)
After gp120 is bound to CD4, nonspecific electrostatics may play a role in virus entry and chemokine receptor binding. CD4 itself is quite basic, with a net overall charge for the first two domains of +5, which should serve to increase the overall basicity of the CD4-gp120 complex. In addition to the membrane head group interactions detailed above, nonspecific electrostatics may effect binding to the sulfonated, acidic N terminus of the chemokine receptor (particularly CXCR4) (14, 17, 18). Relevant to this, mutagenesis of the chemokine receptor binding site on gp120 showed that substitutions of negatively charged amino acids for positively charged ones often have substantial effects on binding (45).
Implications of electrostatics for therapeutic development and immune surveillance.
Our results suggest that the biological effects of polyanions are primarily the consequence of nonspecific electrostatic interactions. If so, this may explain the dramatic differences in in vitro and in vivo effectiveness of such polyanions as dextran sulfate. Since the primary interaction appears to be dependent on charge density rather than a particular structural motif (57), it seems unlikely that the specificity essential for effective therapeutic treatment is present. This does not bode well for drug development based on polyanions such as sulfated polysaccharides (2, 24, 35), carboxylated albumins (26, 51), porphyrins (13, 49), or DNA or RNA derivatives (31). Nevertheless, the unusual basicity observed for the CXCR4-using viruses may, in this particular case, permit anion-based intervention.
Because generation of broadly neutralizing antibodies depends on detection of conserved sequences and nonspecific electrostatic interactions can be achieved through variable regions like the V3 loop, the nonspecific binding predicted here, which does not appear to be sensitive to the precise structure of the V3 loop, may serve as a mechanism of gp120 immune system evasion. Additionally, the structures of several neutralizing antibodies complexed with their V3 loop epitopes revealed that the V3 loop exists in at least two quite different conformation (48). This suggests that the V3 loop may adopt different structures, adding yet another layer of difficulty to its recognition by the immune system.
Initial nonspecific electrostatic attachment may help to conceal the conserved chemokine receptor site, which is on the cell-facing surface. Studies have shown that polyanions compete effectively with antibodies for binding to the V3 loop (23, 39). The chemokine receptor binding site, which in any case may only be exposed after a conformational change is induced by CD4 (32, 53), may thus be multiply shielded from immune system surveillance.
Last, we note that although electrostatic potentials are calculated from the exact positions of the atoms in the atomic structure, the effect of electrostatics on biological processes such as initial viral attachment and virus-cell membrane fusion depends on much-lower-resolution parameters. Since the modeling criteria used here are not well constrained with respect to protomer translations, the relative insensitivity of the electrostatics to this parameter bodes well for the analysis. Indeed, the basic surface defined here appears to be robust to relatively large changes in modeling parameters, both for the core (Fig. 4) and for the V3 loop (Fig. 6). This robustness, coupled with the low resolution of the effects, increases our confidence in the biological significance of our conclusions.
ACKNOWLEDGMENTS
We thank An-Suei Yang for assistance with the program PrISM and homology modeling, Barry Honig and Ben Hitz for help with the programs DelPhi and GRASP, Lawrence Shapiro for a thorough reading of the manuscript, and a reviewer for suggesting the use of influenza virus hemagglutinin to test the modeling procedure.
This work was supported by grants from the National Institutes of Health (AI 31783 and AI 39420); by the ANRS, INSERM, and CNRS of France; and by a Center for AIDS Research grant to the Dana-Farber Cancer Institute (AI 28691). The Dana-Farber Cancer Institute is also the recipient of a Cancer Center Grant from the National Institutes of Health (CA 06516). Columbia University is a participant in the Center for AIDS Research (AI 42848). This work was made possible by gifts from the late William McCarty-Cooper, from the G. Harold and Leila Y. Mathers Charitable Foundation, and from the Aaron Diamond Foundation as well as Douglas and Judi Krupp. R.W. was a fellow of the American Foundation for AIDS Research, and P.D.K. is a recipient of a Burroughs Wellcome Career Development award.
REFERENCES
- 1.Altschul S, Gish W, Miller W, Myer E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Baba M, Nakajima M, Schols D, Pauwels R, Balzarini J, De Clercq E. Pentosan polysulfate, a sulfated oligosaccharide, is a potent and selective anti-HIV agent in vitro. Antivir Res. 1988;9:335–343. doi: 10.1016/0166-3542(88)90035-6. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Tal N, Honig B, Peitzsch R M, Denisov G, McLaughlin S. Binding of small basic peptides to membranes containing acidic lipids: theoretical models and experimental results. Biophys J. 1996;71:561–575. doi: 10.1016/S0006-3495(96)79280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bizebard T, Benoit G, Rigolet P, Rasmussen B, Diat O, Bosecke P, Wharton S, Skehel J, Knossow M. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature. 1995;376:92–94. doi: 10.1038/376092a0. [DOI] [PubMed] [Google Scholar]
- 5.Brunger A T. X-PLOR version 3.1 manual. New Haven, Conn: Yale University; 1993. [Google Scholar]
- 6.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 7.Catasti P, Bradbury E M, Gupta G. Structure and polymorphism of HIV-1 third variable loops. J Biol Chem. 1996;271:8236–8242. doi: 10.1074/jbc.271.14.8236. [DOI] [PubMed] [Google Scholar]
- 8.Catasti P, Fontenot J D, Bradbury E M, Gupta G. Local and global structural properties of the HIV-MN V3 loop. J Biol Chem. 1995;270:2224–2232. doi: 10.1074/jbc.270.5.2224. [DOI] [PubMed] [Google Scholar]
- 9.Chamow S, Duliege A, Ammann A, Kahn J, Allen J, Eichberg J, Byrn R, Capon D, Ward R, Ashenazi A. CD4 immunoadhesins in anti-HIV therapy: new developments. Int J Cancer Suppl. 1992;7:69–72. [PubMed] [Google Scholar]
- 10.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 11.Chan D C, Kim P S. HIV entry and its inhibition. Nat Struct Biol. 1998;5:276–279. [Google Scholar]
- 12.Dalgleish A G, Beverley P C L, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 13.Debnath A K, Jiang S, Strick N, Lin K, Haberfield P, Neurath A R. Three-dimensional structure-activity analysis of a series of porphyrin derivatives with anti-HIV-1 activity targeted to the V3 loop of the gp120 envelope glycoprotein of the human immunodeficiency virus type 1. J Med Chem. 1994;37:1099–1108. doi: 10.1021/jm00034a007. [DOI] [PubMed] [Google Scholar]
- 14.Dragic T, Trkola A, Lin S W, Nagashima K A, Kajumo F, Zhao L, Olson W C, Wu L, Mackay C R, Allaway G P, Sakmar T P, Moore J P, Maddon P J. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency type 1 entry. J Virol. 1998;72:279–285. doi: 10.1128/jvi.72.1.279-285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etemad-Moghadam B, Karlsson G B, Halloran M, Sun Y, Schenten D, Fernandes M, Letvin N L, Sodroski J. Characterization of simian-human immunodeficiency virus envelope glycoprotein epitopes recognized by neutralizing antibodies from infected monkeys. J Virol. 1998;72:8437–8445. doi: 10.1128/jvi.72.10.8437-8445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farzan M, Choe H, Desjardins E, Sun Y, Kuhn J, Cao J, Archambault D, Kolchinsky P, Koch M, Wyatt R, Sodroski J. Stabilization of human immunodeficiency virus type 1 envelope glycoprotein trimers by disulfide bonds introduced into the gp41 glycoprotein ectodomain. J Virol. 1998;72:7620–7625. doi: 10.1128/jvi.72.9.7620-7625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farzan M, Choe H, Vaca L, Martin K, Sun Y, Desjardin E, Ruffing N, Wu L, Wyatt R, Gerard N, Gerard C, Sodroski J. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J Virol. 1998;72:1160–1164. doi: 10.1128/jvi.72.2.1160-1164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard N P, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 19.Fass D, Davey R A, Hamson C A, Kim P S, Cunningham J M, Berger J M. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 20.Feng F, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 21.Gelderblom H R, Hausmann E H S, Ozel M, Pauli G, Koch M A. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 22.Grief C, Hockley D J, Fromholc C E, Kitchin P A. The morphology of simian immunodeficiency virus as shown by negative staining electron microscopy. J Gen Virol. 1989;70:2215–2219. doi: 10.1099/0022-1317-70-8-2215. [DOI] [PubMed] [Google Scholar]
- 23.Harrop H A, Coombe D R, Rider C C. Heparin specifically inhibits binding of V3 loop antibodies to HIV-1 gp120, an effect potentiated by CD4 binding. AIDS. 1994;8:183–192. doi: 10.1097/00002030-199402000-00005. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi K, Hayashi T, Kojima I. A natural sulfated polysaccharide, calcium spirulan, isolated from Spirulina platensis: in vitro and ex vivo evaluation of anti-herpes simplex virus and anti-human immunodeficiency virus activities. AIDS Res Hum Retroviru. 1996;12:1463–1471. doi: 10.1089/aid.1996.12.1463. [DOI] [PubMed] [Google Scholar]
- 25.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structure of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jansen R W, Schols D, Pauwels R, De Clercq E, Meijer D K. Novel, negatively charged, human serum albumins display potent and selective in vitro anti-human immunodeficiency virus type 1 activity. Mol Pharmacol. 1993;44:1003–1007. [PubMed] [Google Scholar]
- 27.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr Sect A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 28.Kilby J M, Hopkins S, Venetta T M, DiMassimo B, Cloud G A, Lee J Y, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson M R, Nowak M A, Shaw G M, Saag M S. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 29.Klatzmann D, Champagne E, Charmaret S, Gruest J, Guetard D, Hercend T, Gluckman J C, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 30.Kolchinsky P, Mirzabekov T, Farzan M, Kiprilov E, Cayabyab M, Mooney L, Choe H, Sodroski J. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J Virol. 1999;73:8120–8126. doi: 10.1128/jvi.73.10.8120-8126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krust B, Callebaut C, Hoanessian A G. Inhibition of entry of HIV into cells by poly(A) · poly(U) AIDS Res Hum Retrovir. 1993;9:1087–1090. doi: 10.1089/aid.1993.9.1087. [DOI] [PubMed] [Google Scholar]
- 32.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Layne S, Merges M, Dembo M, Spouge J, Nara P. HIV requires multiple gp120 molecules for CD4-mediated infection. Nature. 1990;346:277–279. doi: 10.1038/346277a0. [DOI] [PubMed] [Google Scholar]
- 34.McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem Sci. 1995;20:272–276. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- 35.Mitsuya H, Looney D J, Kuno S, Ueno R, Wong-Staal F, Broder S. Dextran sulfate suppression of viruses in the HIV family: inhibition of virion binding to CD4+ cells. Science. 1988;240:646–649. doi: 10.1126/science.2452480. [DOI] [PubMed] [Google Scholar]
- 36.Mondor I, Ugolini S, Sattentau Q J. Human immunodeficiency virus type 1 attachment to HeLa cells is CD4 independent and gp120 dependent and requires cell surface heparans. J Virol. 1998;72:3623–3634. doi: 10.1128/jvi.72.5.3623-3634.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore J, McKeating J, Norton W, Sattentau Q. Direct measurement of soluble CD4 binding to human immunodeficiency virus type 1 virions: gp120 dissociation and its implication for virus-cell binding and fusion reactions and their neutralization by soluble CD4. J Virol. 1991;65:1133–1140. doi: 10.1128/jvi.65.3.1133-1140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore J P. Coreceptors: implications for HIV pathogenesis and therapy. Science. 1997;276:51–52. doi: 10.1126/science.276.5309.51. [DOI] [PubMed] [Google Scholar]
- 39.Moulard M, Lortat-Jacob H, Mondor I, Roca G, Wyatt R, Sodroski J, Maddon P J, Olson W, Kwong P D, Sattentau Q J. Selective interactions of polyanions with basic surfaces on human immunodeficiency virus type 1 gp120. J Virol. 1999;74:1948–1960. doi: 10.1128/jvi.74.4.1948-1960.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray D, Hermida-Matsumoto L, Buser C A, Tsang J, Sigal C T, Ben-Tal N, Honig B, Resh M D, McLaughlin S. Electrostatics and the membrane association of Src: theory and experiment. Biochemistry. 1998;37:2145–2159. doi: 10.1021/bi972012b. [DOI] [PubMed] [Google Scholar]
- 41.Nicholls A, Honig B. A rapid finite difference algorithm, utilizing successive over-relaxation to solve the Poisson-Boltzmann equation. J Comput Chem. 1991;12:435–445. [Google Scholar]
- 42.Nicholls A, Sharp K A, Honig B. Protein folding and association: insight from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 43.Patel M, Yanagishita M, Roderiquez G, Bou-Habib D C, Oravecz T, Hascall V C, Norcross M A. Cell-surface heparan sulfate proteoglycan mediates HIV-1 infection of T-cell lines. AIDS Res Hum Retrovir. 1993;9:167–174. doi: 10.1089/aid.1993.9.167. [DOI] [PubMed] [Google Scholar]
- 44.Poignard P, Fouts T, Naniche D, Moore J P, Sattentau Q J. Neutralizing antibodies to human immunodeficiency virus type 1 gp120 induce envelope glycoprotein subunit dissociation. J Exp Med. 1996;183:473–484. doi: 10.1084/jem.183.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rizzuto C, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. Identification of a conserved human immunodeficiency virus gp120 glycoprotein structure important for chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 46.Ross T M, Cullen B R. The ability of HIV type 1 to use CCR-3 as a coreceptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc Natl Acad Sci USA. 1998;95:7682–7286. doi: 10.1073/pnas.95.13.7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheriff S, Hendrickson W A, Stenkamp R E, Sieker L C, Jensen L H. Influence of solvent accessibility and intermolecular contacts on atomic mobilities in hemerythrins. Proc Natl Acad Sci USA. 1985;82:1104–1107. doi: 10.1073/pnas.82.4.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stanfield R L, Cabezas E, Satterthwait A C, Stura E A, Profy A T, Wilson I A. Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing Fabs. Structure. 1999;7:131–142. doi: 10.1016/s0969-2126(99)80020-3. [DOI] [PubMed] [Google Scholar]
- 49.Staudinger R, Abraham N G, Levere R D, Kappas A. Inhibition of human immunodeficiency virus-1 reverse transcriptase by heme and synthetic heme analogs. Proc Assoc Am Physicians. 1996;108:47–54. [PubMed] [Google Scholar]
- 50.Sullivan N, Sun Y, Binley J, Lee J, Barbas III C, Parren P W H I, Burton D R, Sodroski J. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J Virol. 1998;72:6332–6338. doi: 10.1128/jvi.72.8.6332-6338.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swart P J, Beljaars E, Smit C, Pasma A, Schuitemaker H, Meijer D K. Comparative pharmacokinetic, immunologic and hematologic studies on the anti-HIV-1/2 compounds aconitylated and succinylated HSA. J Drug Target. 1996;4:109–116. doi: 10.3109/10611869609046269. [DOI] [PubMed] [Google Scholar]
- 52.Tan K, Lui J, Wang J, Shen S, Lu M. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weis W, Brunger A, Skehel J, Wiley D. Refinement of the influenza virus hemagglutinin by simulated annealing. J Mol Biol. 1990;212:737–761. doi: 10.1016/0022-2836(90)90234-D. [DOI] [PubMed] [Google Scholar]
- 55.Weissenhorn W, Carfi A, Lee K-H, Skehel J J, Wiley D C. Crystal structure of the Ebola virus membrane fusion subunit, Gp2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 56.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 57.Weisz P B, Joullie M M, Hunter C M, Kumor K M, Zhang Z, Levine E, Macarak E, Weiner D, Barnathan E S. A basic compositional requirement of agents having heparin-like cell-modulating activities. Biochem Pharmacol. 1997;54:149–157. doi: 10.1016/s0006-2952(97)00157-3. [DOI] [PubMed] [Google Scholar]
- 58.Wild C T, Shugars D C, Greenwell T K, McDanal C B, Matthews T J. Peptides corresponding to a predictive α-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson I A, Skehel J J, Wiley D C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;287:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 60.Wu H, Kwong P D, Hendrickson W A. Dimeric association and segmental variability in the structure of human CD4. Nature. 1997;387:527–530. doi: 10.1038/387527a0. [DOI] [PubMed] [Google Scholar]
- 61.Wu L, Gerard N P, Wyatt R, Choe H, Parlin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 62.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J. The antigenic structure of the human immunodeficiency virus gp120 envelope glycoprotein. Nature. 1998;393:705–710. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 63.Wyatt R, Sullivan N, Thali M, Repke H, Ho D, Robinson J, Posner M, Sodroski J. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J Virol. 1993;67:4557–4565. doi: 10.1128/jvi.67.8.4557-4565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang A-S, Honig B. Sequence to structure alignment in comparative modeling using PrISM. Proteins. 1999;37(Suppl. 3):66–72. doi: 10.1002/(sici)1097-0134(1999)37:3+<66::aid-prot10>3.3.co;2-b. [DOI] [PubMed] [Google Scholar]