Abstract
Electron transfer is a fundamental process in life that is very often coupled to catalysis within redox enzymes through a stringent control of protein conformational movements. Mammalian nitric oxide synthase (NOS) proteins are redox flavo-hemoproteins consisting of multiple modular domains. The NOS enzyme is exquisitely regulated in vivo by its partner, the Ca2+ sensing protein calmodulin (CaM), to control production of nitric oxide (NO). The importance of functional domain motion in NOS regulation has been increasingly recognized. The significant size and flexibility of NOS is a tremendous challenge to the mechanistic studies. Herein recent applications of modern biophysical techniques to NOS problems have been critically analyzed. It is important to note that any current biophysical technique alone can only probe partial aspects of the conformational dynamics due to limitations in the technique itself and/or the sample preparations. It is necessary to combine the latest methods to comprehensively quantitate the key conformational aspects (conformational states and distribution, conformational change rates, and domain interacting interfaces) governing the electron transfer. This is to answer long-standing central questions about the NOS isoforms by defining how specific CaM-NOS interactions and regulatory elements underpin the distinct conformational behavior of the NOS isoform, which in turn determine unique electron transfer and NO synthesis properties. This review is not intended as comprehensive, but as a discussion of prospects that promise impact on important questions in the NOS enzymology field.
Keywords: Nitric Oxide Synthases, CaM-NOS, Electron transfer, Protein Conformational Movements, Calmodulin, Review
2. INTRODUCTION
2.1. Electron transfer and protein dynamics
Electron transfer (ET) is a fundamental reaction in biology and the efficient and controlled transfer of electrons is essential to living organisms. Proteins are dynamic molecules. Large-scale domain motions and conformational changes facilitate the transfer of electrons over long distance in redox enzymes. Characterizing the dynamics and large-scale movements within proteins is a frontier challenge for understanding electron transfer mechanisms. Our current knowledge of the spatial distribution and temporal exploration of the conformational transitions are limited, attributed mainly to a dearth of combining structural and biophysical tools to fully capture such information. Mammalian nitric oxide synthase (NOS) proteins are redox flavo-hemoproteins consisting of multiple modular domains, the chemistry of which is coupled to major dynamical excursions during the enzyme catalysis. The mechanism for conformational changes in the electron transfer in NOSs is the subject of several excellent reviews (1–3). Here, this review is not intended as comprehensive, but as a critical analysis of the recent advances in the field, along with discussion of prospects that promise impact on important questions in the NOS enzymology field.
2.2. NOS enzymology
NOSs are a family of proteins catalyzing biosynthesis of nitric oxide (NO) from l-arginine (l-Arg) with NADPH and O2 as co-substrates (4). They are important enzymes because of the crucial roles of NO in signaling processes. NO, a free radical with an unpaired electron, is reactive and short-lived. As such, NO signaling must be tightly controlled through regulation of NOS activation. The NOS enzyme is exquisitely regulated in vivo by its partner, the Ca2+ sensing protein calmodulin (CaM). In addition to control via Ca2+/CaM, eNOS & nNOS are regulated by posttranslational modifications including phosphorylation at several sites, and functional importance of posttranslational regulation of NOSs has been established in vivo (5, 6). Such a complex set of modulators provides stringent regulation in both time and place of NO production in response to a wide variety of stimuli such as shear stress.
Eukaryotic NOS evolved via a series of gene fusion events, resulting in a self-sufficient modular heme- and flavin-containing enzyme that produces NO by very intricately controlled redox processes. Each subunit of the NOS homo-dimer has two domains joined by a CaM-binding linker (Figure 1): a C-terminal electron-supplying reductase (NOSred) domain, which consists of smaller (sub)domains with binding sites for NADPH (the electron source), flavin adenine dinucleotide (FAD), and flavin mononucleotide (FMN), and an N-terminal heme-containing oxygenase (NOSoxy) domain; the terms “oxygenase domain” and “heme domain” are interchangeable. There are three NOS isoforms: endothelial, neuronal, and inducible NOS (eNOS, nNOS, and iNOS, respectively). The CaM-binding linker in iNOS binds CaM at a basal level of Ca2+, while in nNOS and eNOS the CaM binding requires an increase in intracellular Ca2+ concentration. Binding of CaM to NOS acts as a “molecular switch”, which activates NO production by enabling electron flow from the flavin prosthetic groups to the heme (Figure 1).
Figure 1.
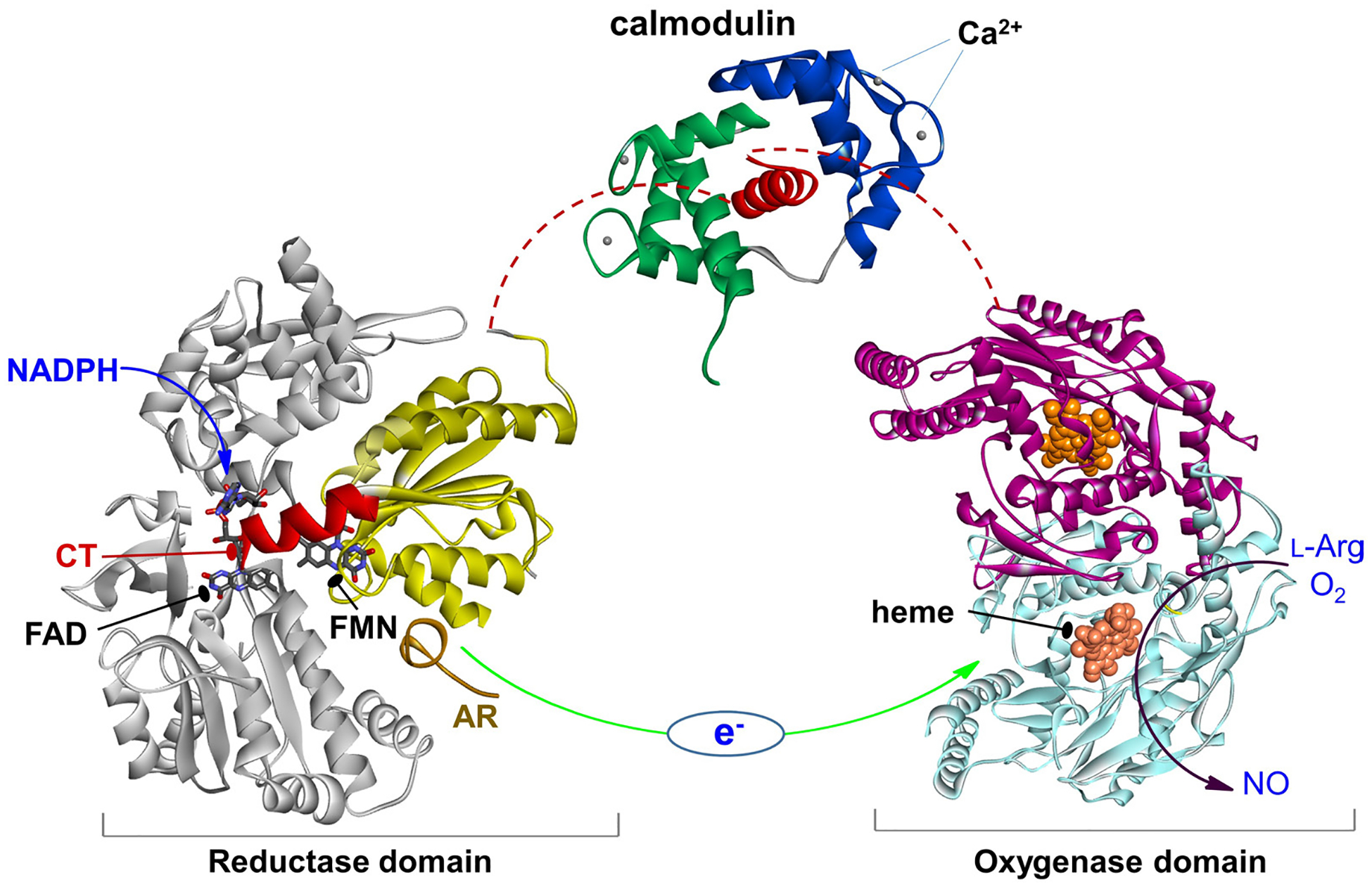
The NOS domains/modules: reductase domain with the FMN (sub)domain colored in yellow and the rest in gray, an a helical CaM-binding linker with bound CaM, and dimeric heme domain. Only the individual domain structures are available to date. CaM binding to NOS enables the electron transfer between the FMN and heme domains (green arrow). This involves a conformational change, in which the FMN domain shuttles between the FAD and heme domains to transfer the NADPH-derived electrons.
Extensive crystallographic studies of the NOSoxy domains of all three NOS isoforms have been conducted (7–10); the substrate, l-Arg, and a cofactor, (6R)-5,6,7,8 tetrahydrobiopterin (H4B), both bind near the heme center in the oxygenase domain (11). These efforts aim to guide rational development of isoform-specific NOS inhibitors targeting the heme active site (recently reviewed in (12)) and substrate access channel (10, 13). In addition to the NOSoxy structures, only a few crystal structures of partial (14) and intact (15) rat nNOS reductase domains, CaM bound human iNOS FMN domain (16), as well as CaM bound to a peptide corresponding to the CaM-binding sequence in human eNOS (17) have been reported. Crystal structures of the CaM bound peptide corresponding to the CaM-binding linker in iNOS and eNOS are also deposited in the PDB database. However, numerous attempts to crystallize the full-length NOS enzyme have not been successful because of considerable NOS domain motions. While the molecular basis for CaM binding to NOS became clearer (16, 18), the structural and dynamical details related to the interaction and assembly of NOS domains and CaM during catalysis remain largely unknown.
A combination of kinetics methods/simulations with site-directed mutagenesis have provided useful information about the roles that specific amino acid residues play in regulating NOS function (19–25). Besides the site-directed mutants, CaM (26) and NOS (27, 28) chimeras have been used to probe roles of specific domains/regions in NOS function. A general picture emerging from these extensive biochemical studies is that structural and conformational features that are common to di-flavin reductases combine with those unique to NOS enzymes (including CaM and intrinsic regulatory elements; see below) to afford distinct mechanisms of NOS regulation (28, 29).
2.3. Interdomain electron transfer processes controlled by CaM binding are key steps in NOS catalysis
Although the details of the novel NOS catalytic/chemical mechanism remain debatable (30), it has been established that interdomain electron transfer (IET) processes (following the flow of NADPH→FAD→FMN→heme) are key steps in NO synthesis through coupling reactions between domains (Figure 2) (4, 31). A tight control of the IET processes is important for the normal function because an uncoupled NOS (in which the usual catalytic electron flow within the enzyme is diverted to molecular oxygen to yield superoxide) generates reactive oxygen species, resulting in deleterious effects (32–35).
Figure 2.
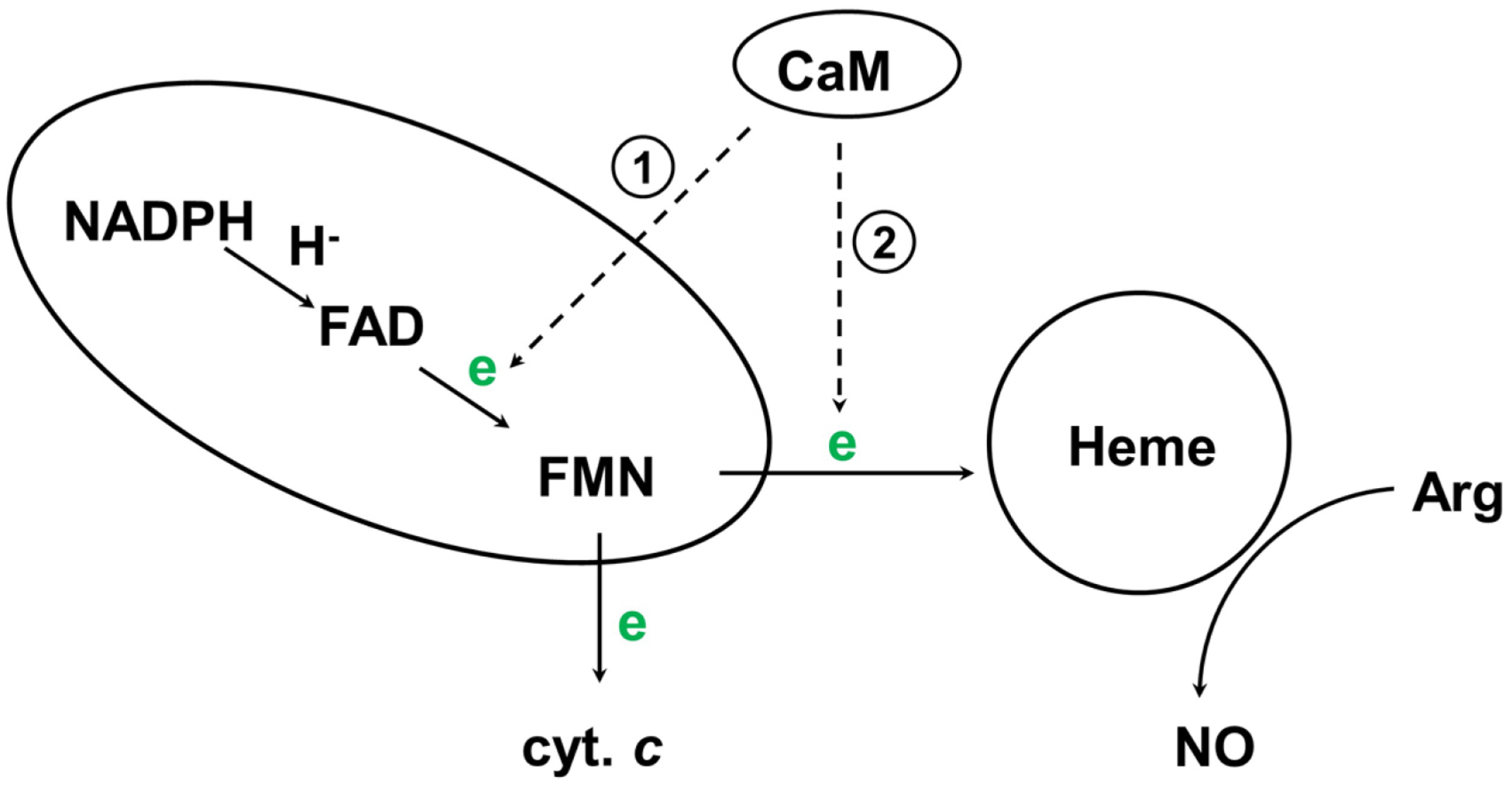
Electron flow in NOS. In eNOS/nNOS, CaM-binding facilitates IET between FAD and FMN within the reductase domain (reaction 1), and triggers the inter-subunit FMN-heme IET (reaction 2). Cytochrome c reduction is a measure of the release of the FMN domain from the reductase complex.
Unlike iNOS, eNOS and nNOS synthesize NO in a Ca2+/CaM dependent manner. Here CaM-binding facilitates the IET reaction from the FAD hydroquinone to FMN semiquinone (FMNH·) within one subunit (reaction 1) (36), and enables IET from the FMN hydroquinone to the catalytic heme iron in the oxygenase domain of another subunit (reaction 2) (37–40). The FMN hydroquinone has been recently suggested as its neutral form of FMNH2 (20), while direct spectroscopic evidence such as NMR is still needed to definitely assign its protonation state. The CaM binding has little or no effect on the thermodynamics of redox processes in NOS (41–43), indicating that the regulation by CaM is accomplished kinetically through controlling redox-linked conformational changes required for effective IET (see below).
2.4. Intrinsic control elements contribute to the CaM dependence of IET
Control of the electron transfer processes in NOS by CaM involves the CaM binding site and intrinsic control elements including an autoregulatory (AR) insert (~40 amino acids) within the FMN domain of eNOS/nNOS (44), which does not exist in iNOS, and a C-terminal tail (CT) differing in length among the three isoforms (45); AR and CT are depicted in Figure 1. Electron transfer modulation appears also to involve a much smaller insertion present in the hinge subdomain (46). Control element deletion studies indicate that the control elements suppress electron flow into and out of the NOS reductase domain in the CaM-free state ((47); also reviewed in (48)), and CaM-binding to NOS relieves the repression. A crystal structure of an intact nNOS reductase domain revealed key information about how the control elements may repress electron transfer in the CaM-free state (15). A unique AR (~ 40 amino acids) in the FMN domain of eNOS/nNOS pins the FMN domain to the reductase complex via a network of hydrogen bonds in the CaM-free state (15). The AR insert and CT elements do not exist in the homologous P450 reductase (i.e., these CaM-responsive regulatory elements are NOS-unique).
3. REGULATION OF INTERDOMAIN ELECTRON TRANSFER IN NOS BY CAM THROUGH BIASING CONFORMATIONAL DYNAMICS
Evidence from the nNOS reductase domain structure (15) and our earlier work (49) leads to the idea that domain motion plays an important role in the NOS activation by CaM (50). This motion is described by a tethered shuttle model originally proposed by Salerno (50, 51) (Figure 3): the FMN domain shuttles the NADPH-derived electrons by interacting alternatively with the FAD and heme domains. The shuttling mechanism also operates in the ancestral electron transfer systems (e.g., P450 reductase) (52), while the novel aspect of the NOS enzyme is the involvement of CaM protein (see 3.2. below). The domain shuttle model has been supported by studies of discrete electron transfer kinetics (31, 53, 54).
Figure 3.
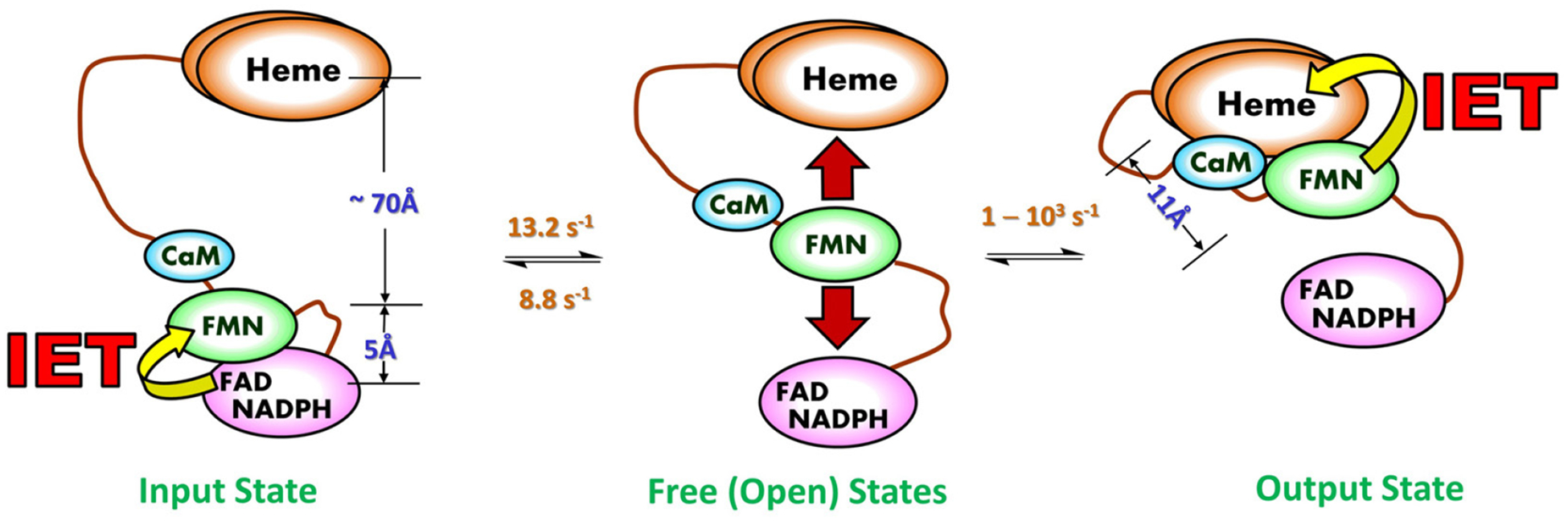
Tethered shuttle model (with the tethers corresponding to the interdomain FAD-FMN and FMN-heme linkers). This involves motion of the FMN domain between electron-accepting “input state” and electron-donating “output state”. The intermediate undocked free/open state(s) between the terminal docking positions is characterized by a continuum of conformations and exists in equilibrium with the docked conformations of the input and output states. The IET related edge-to-edge distances between redox cofactors (FAD, FMN and heme) in a rat nNOS input state model are displayed in blue, along with the edge-to-edge distance between FMN and heme in an iNOS output state model. The interchange rates between the NOS states are also displayed; the rates are of the CaM-bound eNOS isoform, which are currently available in the literature (63, 64). The conformational rates are at best semi-quantitative, and the docked/undocked rates of the interconversion between the free and output states can only be estimated within a large range (two to three orders of magnitude) (63). It is of note that the docked FMN/heme complex is transient (58), indicating that the docked and undocked rates differ significantly. As the three NOS isozymes show different turnover numbers, the isoform should have distinct conformational rates and distance.
The docked FMN/heme complex is defined as output state in the literature (50). The structure of the output state has not yet been elucidated. To favor observation of the output state of the tethered shuttle mechanism, Ghosh designed a series of two-domain NOS oxyFMN constructs in which only the oxygenase and FMN domains along with the CaM binding sequence are expressed (55). This was done to preclude FAD/FMN interactions and favor interactions between the FMN-binding domain and the oxygenase domain. The oxyFMN construct was also constructed afterward by other laboratories (56–58), and has become an established model of minimal electron transfer complex of the NOS output state for NO production (31).
It is imperative to discuss two pieces of key information obtained from various biophysical methods: (i) the distance changes between domains during the “docked” and “free/open” states; and (ii) the rate constants of conformational steps, especially those involved in the IET processes (Figure 3). Crystal structure of a CaM-free intact nNOS reductase domain reveal that FMN is in close proximity with FAD (within 5 Å), as required for electron transfer in the input state of FMN from NADPH through FAD (15). This structure is not as it exists in the output state; the FMN cofactor is ~ 70 Å apart from the heme domain in the constructed model (15). On the other hand, in the structures of the output state models (59, 60), the FMN-heme edge-to-edge distance is 11.2 – 13.1 Å. Therefore, a large-scale motion (61) and rotation (62) of the FMN domain are required in the electron transport through the NOS protein. Compared to relatively clear picture of the distance changes among the NOS states, the conformational change rates remain vague and are at best semi-quantitatively estimated at amplitude level. For example, interconversion between the conformational states in eNOS holoenzyme were estimated to take place on tens of milliseconds to seconds from analysis of the smFRET trajectories (63). The on and off rates of the FAD-FMN docking have been estimated to be 8.8 and 13.2 s−1, respectively, for the CaM-bound eNOS reductase domain, while the rates are much slower in the absence of CaM (4.2 and 0.63 s−1, respectively) (64). Overall the domain association and dissociation occur on milliseconds to seconds time scale, governing the bulk electron transfer.
3.1. Electron transfer from FMN to the surrogate acceptor cytochrome c
The catalytic action of the NOSred domain involves three primary events: hydride transfer from NADPH to FAD, electron transfer from FAD to FMN, and electron transfer from FMN to heme or surrogate electron acceptor cyt. c (Figure 2). The transfer of an electron from FMN hydroquinone to cyt. c provides an estimate of the accessibility of the FMN domain. It can be measured by the stopped-flow mixing of chemically reduced NOSred domain with stoichiometric amount of cyt. c, in the presence and absence of CaM and NADPH (65). Stuehr’s laboratory expanded the approach by simulating the stopped-flow traces with a four-state kinetic model (66) that links the electron transport through the NOSred domain to the conformational equilibria and motions of its FMN (sub) domain. Simulations of this model have revealed how the NOSred domain can support its certain rate of electron flux (i.e., the cyt. c reduction activity) based on its unique conformational equilibrium constant value and rates of the FMN domain motions (21, 29, 64). CaM binding to NOS have multiple effects: it alters the conformational equilibrium of the reductase domain, and increases the rate of the conformational transitions (64).
3.2. CaM controlled electron transfer between FMN and heme
The FMN domain motion (Figure 3) facilitates efficient IET between the FMN and heme centers in the NOS output state. The formation of the NOS output state involves two primary steps: (i) CaM-binding triggered dissociation of the FMN domain from its reductase binding site, and (ii) subsequent re-association of the FMN domain with the oxygenase domain (50). The NOS state formed at step (i) is competent to reduce cyt. c, which requires accessibility of FMN to cyt. c (as described in 3.1. above). This is a necessary but not a sufficient condition for NO production, which in addition requires additional CaM-dependent promotion of the oxygenase/FMN domain interactions. This model is strongly supported by the results of a study showing that NAPDH reduction is 3 orders of magnitude faster than NO production, and cyt. c reduction is intermediate between the two (26). Previously, CaM control of NOS electron transfer was believed to be primarily localized within the reductase portion of the enzyme (67, 68). We are the first to directly demonstrate the importance of productive interactions between the FMN domain and heme-containing oxygenase domain in the CaM-activation of formation of the NOS output state (50, 69). The FMN-heme IET within the NOS output state (reaction 2 in Figure 2) is essential in the delivery of electrons required for O2 activation in the heme domain and the subsequent NO synthesis. It is thus of current interest to decipher the mechanism through which CaM activates this electron transfer, as discussed next.
4. STATE-OF-THE-ART IN STUDYING THE NOS CONFORMATIONAL DYNAMICS
4.1. Current structural and biophysical methods applied to NOS
Functional domain dynamics poses a central challenge for studying the NOS structure-function relationship. Crystal structures of individual NOS domains represent a significant step toward understanding NOS mechanisms. Yet the mobility of the NOS domains makes crystallization of the full-length enzyme very challenging, and the structure of full-length NOS has not been obtained. Even if such crystal structures become available, the molecular mechanisms that promote function may remain elusive, since NOS is a highly dynamic protein existing in a broad range of interconverting conformations (Figure 3). The crystals can only stabilize some specific structural substates from a full range of conformers that define the functional state of the protein. Thus, X-ray crystallography alone will not solve the NOS structural problems and may only shed light on some of the static structural details without addressing the dynamics aspects. NMR spectroscopy is a powerful complement to X-ray crystallography, but the size of the NOS holoenzymes (~120–160 kDa per subunit) has prevented application of NMR to NOS proteins with methods available to date. Importantly, 13C- and 15N-labeled CaM proteins were used in the NMR studies of structure and dynamics of CaM bound to peptides corresponding to the CaM-binding linker in the NOS proteins (70–72). An excellent review in this Special issue by Guillemette summarizes the related NMR work.
Several other modern techniques including cryo-electron microscopy (EM) (61, 62, 73), single molecule fluorescence resonance energy transfer (smFRET) (74), hydrogen deuterium exchange mass spectrometry (HDX MS) (18), and pulsed electron paramagnetic resonance (EPR) (75, 76) have been lately applied to decipher the NOS conformational dynamics. Studying the flavin reduction and overall turnover activities of nNOS holoenzyme as a function of pressure, viscosity and ionic strength (77, 78) have also revealed some interesting insights. These techniques do not require crystallization, and can provide structural and dynamic information for protein samples in solution. Table 1 lists the techniques and the information one can learn from the data. Collectively, these recent studies have significantly advanced our understanding of how CaM activates NOS by controlling its conformational fluctuation dynamics. However, individual technique alone can only probe partial facets of the NOS conformational dynamics due to limitations in the current method itself and/or the sample preparations.
Table 1.
Current biophysical approaches in studying conformational control of NOSs
| Technique | Results | Information obtained from the data |
|---|---|---|
| Cryo EM | Transmission electron microscopes images | Higher-order NOS domain architecture and quaternary structure |
| smFRET | FRET signal of individual protein molecules in real time | Conformational states distribution and interconversion rates |
| Pulsed EPR | Distances between paramagnetic centers & distance distribution | Structure of the docking complex and conformational equilibrium |
| HDX MS | Monitoring perturbations in amide proton exchange rate changes | Interdomain interaction surfaces (at peptide level) induced by CaM and/or phosphorylation |
| Laser flash photolysis | Kinetics of the discrete FMN-heme IET process | A direct measure of the interdomain FMN-heme complex formation |
| Solvent perturbation stopped flow | Electron transfer rates as a function of pressure, viscosity and ionic strength | Functional significance of the rugged energy landscape |
For example, cryo-EM is a powerful method for determining structures of large proteins and protein complexes. Recently, researchers were racing to reveal high-order architecture of NOS holoenzymes using this technique. Four cryo-EM studies of the three mammalian NOS isoforms were published within one and half years in 2013–2014 (61, 62, 73, 79). This also attests an urgent need of elucidating conformational changes required for efficient electron transfer in NOS proteins. These EM studies explicitly confirmed the shuttling motion of the FMN domain, and revealed a range of conformations enabled by the flexible tethers. These conformations reveal that CaM activates NOS by constraining rotational motions and by directly docking onto the heme domain (61, 73). However, at low resolutions (23 Å (62) and 60–74 Å (61)), cryo-EM cannot provide definitive solutions to the problem of the interaction of the FMN and heme domains. Moreover, the authors used negative-stain EM methods in some reports (61), which involved fixing the protein on a carbon-coated surface and treating it with a high-contrast heavy metal stain. While this is a powerful approach for observing protein complexes, the grid surface may distort the sample and lead to a debatable asymmetry (61).
smFRET is another unique tool to detect CaM-induced NOS conformational changes in real time, but a cys-lite construct, in which reactive cysteines have been mutated to serines, has to be used for specific labelling of nNOS reductase domain by fluorescent dye molecules (74). This is a concern because the cys-lite mutant responds to CaM binding to a much less extent than the wild type protein (19). In other words, the observed CaM-induced conformational fluctuations may not accurately reflect what occurs in the native NOS protein. Moreover, while the reductase domain is more tolerant to cysteine knocked out, the NOS heme domain contains several surface cysteines that are essential for proper folding of the heme domain, and the cysteine residues thus can’t be replaced to ensure specific fluorescent labeling of the heme domain. Not surprisingly, smFRET of dye-labeled NOS holoprotein has not been reported yet.
4.2. Pulsed EPR spectroscopy in studying the functional domain dynamics in NOS proteins
Compared to the methods discussed above, pulsed EPR spectroscopy represents a promising tool for determining distance relationships in NOS: EPR can be used to study the samples in (frozen) solutions where the full structural distribution of NOS is realized and can provide information about both the fixed distances between rigid protein fragments and the distributed distances between mobile segments. By combining the data from these studies with the crystal structural data for the NOS modules, a full picture of dynamic, structurally distributed NOS protein shall emerge. The access to the distance distribution profiles provided by pulsed EPR is especially valuable for understanding of the NOS dynamics. The fixed-distance resolution in the pulsed EPR studies is relatively high: for long distances of 55–60 Å (corresponding to the dipolar oscillation period of 3–4 ms), the resolution is better than 2.4 Å (4%). For shorter distances, the absolute resolution improves even more since it scales as R4. For example, we have determined the distance between the FMNH· and Fe(III) centers in iNOS (18.8. ± 0.1. Å) (60) and the CaM-heme domain docking statistics in nNOS (75). Thus, the pulsed EPR measurements can give exclusive insight into the alterations in the interdomain alignment and docked state population caused by CaM binding and/or the mutations at the docking interface.
Pulsed electron-electron double resonance (ELDOR, also known as double electron-electron resonance, DEER) is capable of measuring the distances between paramagnetic centers from 15 to over 60 Å and resolving different conformational states existing simultaneously within the sample (80). ELDOR has been employed to study the diflavosemiquinoid form of nNOS proteins (76). Another pulsed EPR distance measurement technique is relaxation-induced dipolar modulation enhancement (RIDME) (81, 82). Compared to the conventional DEER technique, RIDME is uniquely suited for measuring the distances in pairs where one or both spins are paramagnetic metal ions such as low spin ferric heme center in NOS (82). In addition, RIDME enables quantitative measurements for the absolute population of the spin pair, which is required for obtaining statistical weight of docked and undocked states in the NOS system (75, 82), while in DEER the absolute spin pair population is determined semiquantitatively at best.
The FMN⋯Fe distance information is important for a better understanding of the FMN-heme IET mechanism, because the distance between redox centers is critical for electron transfer processes in proteins (83). We have applied the RIDME technique (81) to determine the distance between the FMN and heme centers in the (Fe(III))(FMNH·) form of human iNOS (60); some key factors in successful measurement of the distance are selection of heme iron spin state, and trapping of sufficient amount of intermediate containing the spin pair. The obtained Fe(III)⋯FMNH· distance of 18.8 ± 0.1 Å is in agreement with the IET rate (56). We have also probed the relative orientation of the two centers (84), and the FMN-heme domain docking model (60) was found to be in qualitative agreement with the combined experimental results of our pulsed EPR work (60, 84).
To obtain the structural information about the CaM/heme domain docking complex, we have used spin-labeled CaM and determined its magnetic dipole interaction with the low-spin ferric heme centers in the oxygenase domain (75). The RIDME trace for CaM-bound nNOS represents a monotonic decay without well-defined oscillations. The simulation of this trace has allowed us to obtain information about the geometry of the CaM docking and the statistical weights of the docking complex (15 ± 3 %) and the undocked open/free states (85 ± 3 %) (75). These results support the concept of CaM docking with the heme domain. The low docked state population indicates that the CaM-controlled FMN-heme docking is highly dynamic.
A common assumption used in structural studies by pulsed EPR is that the structural distribution in a frozen solution represents a “snapshot” of the dynamic equilibrium in a fluid solution. Although the validity of this assumption may be debatable for some systems, we have recently shown that the correspondence between the conformational distributions of nNOS in frozen solution and in liquid at room temperature is at least semi-quantitative (75). To validate the “snapshot” model for the NOS mutants, the statistical weights of the conformational distributions in frozen solution obtained by pulsed EPR should be compared with the room temperature distributions estimated from fluorescence lifetime measurements (85). While the assignment of fluorescence lifetimes to particular conformational states is somewhat speculative, the combined use of EPR and fluorescence will dramatically enhance the reliability of the conformational equilibrium estimates.
4.3. Direct determination of the FMN-heme electron transfer kinetics by CO photolysis
The accuracy of the stopped-flow technique previously employed for the measurement of the FMN-heme IET kinetics (86) is compromised by the low resolution of the multiple kinetic phases observed. It is therefore important to use alternative methods to more accurately determine the rates of the individual IET steps. We have developed a laser flash photolysis approach to directly measure kinetics of IET between catalytically significant redox couples of FMN and heme centers (i.e., Fe3+/Fe2+ and FMNH·/FMN hydroquinone). Briefly, we first partially reduce the NOS protein to a (Fe(II)–CO)(FMNH·) form by photo-illumination in the presence of 5-deazariboflavin, and then use 450 nm laser to flash CO off the pre-reduced Fe(II)-CO complex, triggering the IET. We can individually time-resolve consumption of FMNH· and Fe(II) (due to the IET) and CO rebinding, and thereby reliably measure the rapid FMN-heme IET followed by a much slower CO re-binding process (Figure 4). The stopped flow approach is unable to distinguish these two reactions as only formation of the Fe(II)–CO complex was observed (37, 86). Thus, our CO photolysis approach offers clear advantages since it allows us to observe both the IET and CO rebinding processes directly.
Figure 4.
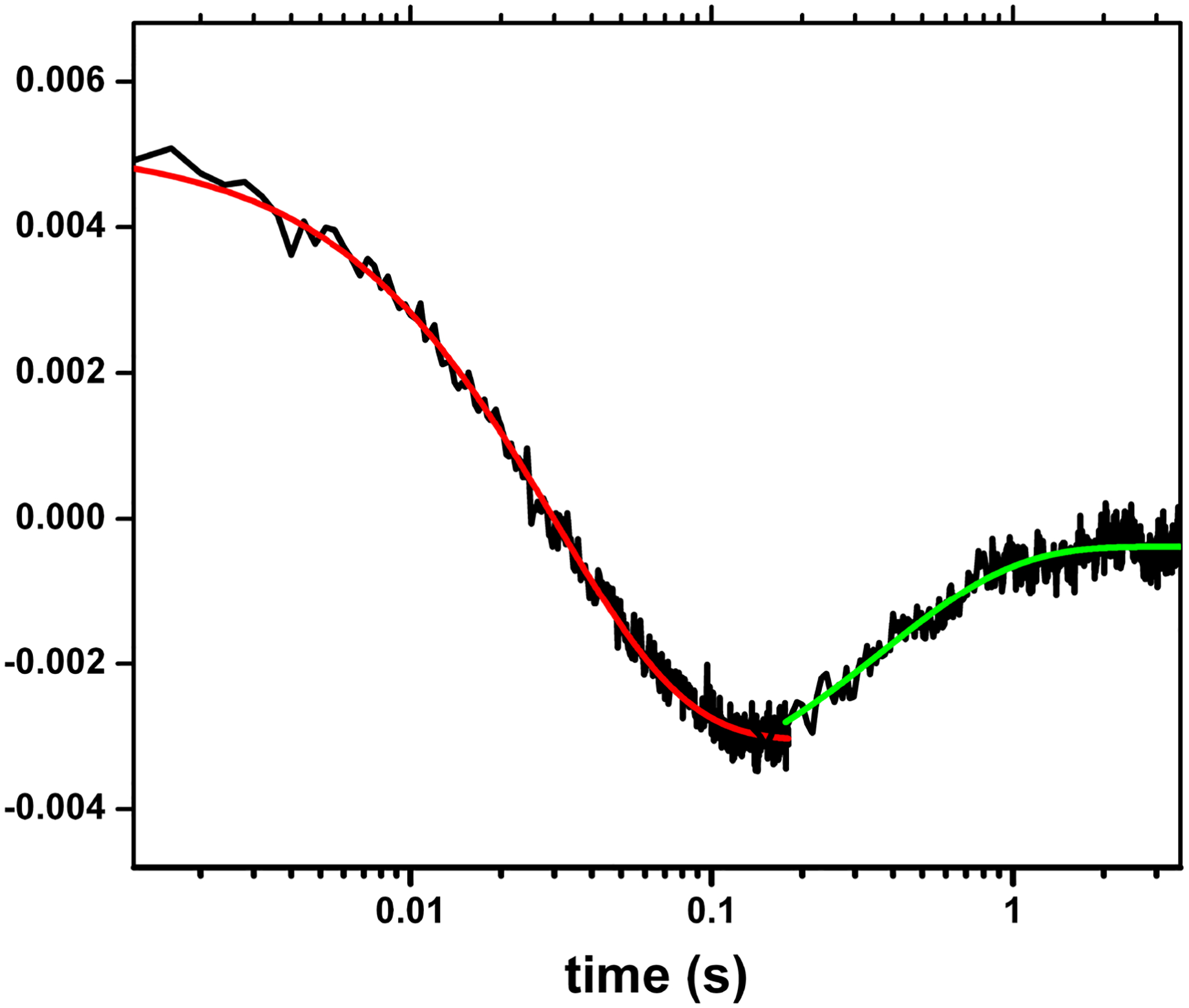
Transient traces at 460 nm at 0 – 4 s obtained for the (Fe(II)-CO)(FMNH·) form of nNOSm holoenzyme flashed by 450 nm laser. The graph is a combined plot of two individual traces at 0 – 0.2. s and 0 – 4 s using a logarithmic timescale; solid lines correspond to the best single-exponential fit to the data in different parts: the red and green lines are of the FMN-heme IET and the CO re-binding processes, respectively. This is to show the spectral “transition”, i.e., a reversal in direction of absorption changes over time in the traces. Such transition feature indicates successful observation of the FMN-heme IET, and can only be detected at 580–600 nm (that is due to FMNH· reduction), and 465 or 460 nm (that is due to heme re-oxidation in iNOS and nNOS respectively), if the IET takes place. Sample temperature was set at 21 °C. Anaerobic solution contained ~ 10 mM NOS protein, ~ 20 mM dRF and 5 mM fresh semicarbazide in pH 7.6. buffer (40 mM Bis-Tris propane, 400 mM NaCl, 2 mM l-Arg, 20 mM H4B, 1 mM Ca2+ 4 and 10 % glycerol).
Regarding the choice of wavelength to monitor the laser-induced electron transfer, we can choose either 580–600 nm (where FMNH· has a broad absorbance peak) or 460/465 nm for nNOS/iNOS, which is an isosbestic point for the process of CO rebinding to free Fe2+ in nNOS and iNOS (Figure 5), respectively. We have confirmed that the observed IET rates are independent of the selected wavelength (56). Since absorbance change at 460/465 nm (due to re-oxidation of Fe2+) is larger than that of 580 nm, one can more reliably measure the kinetics at 460/465 nm when the amplitude of the trace at 580 nm is low. For example, the IET kinetics of E546N iNOS mutant were measured at 465 nm because of weak signal at 580 nm (due to lower docked state%) (87).
Figure 5.
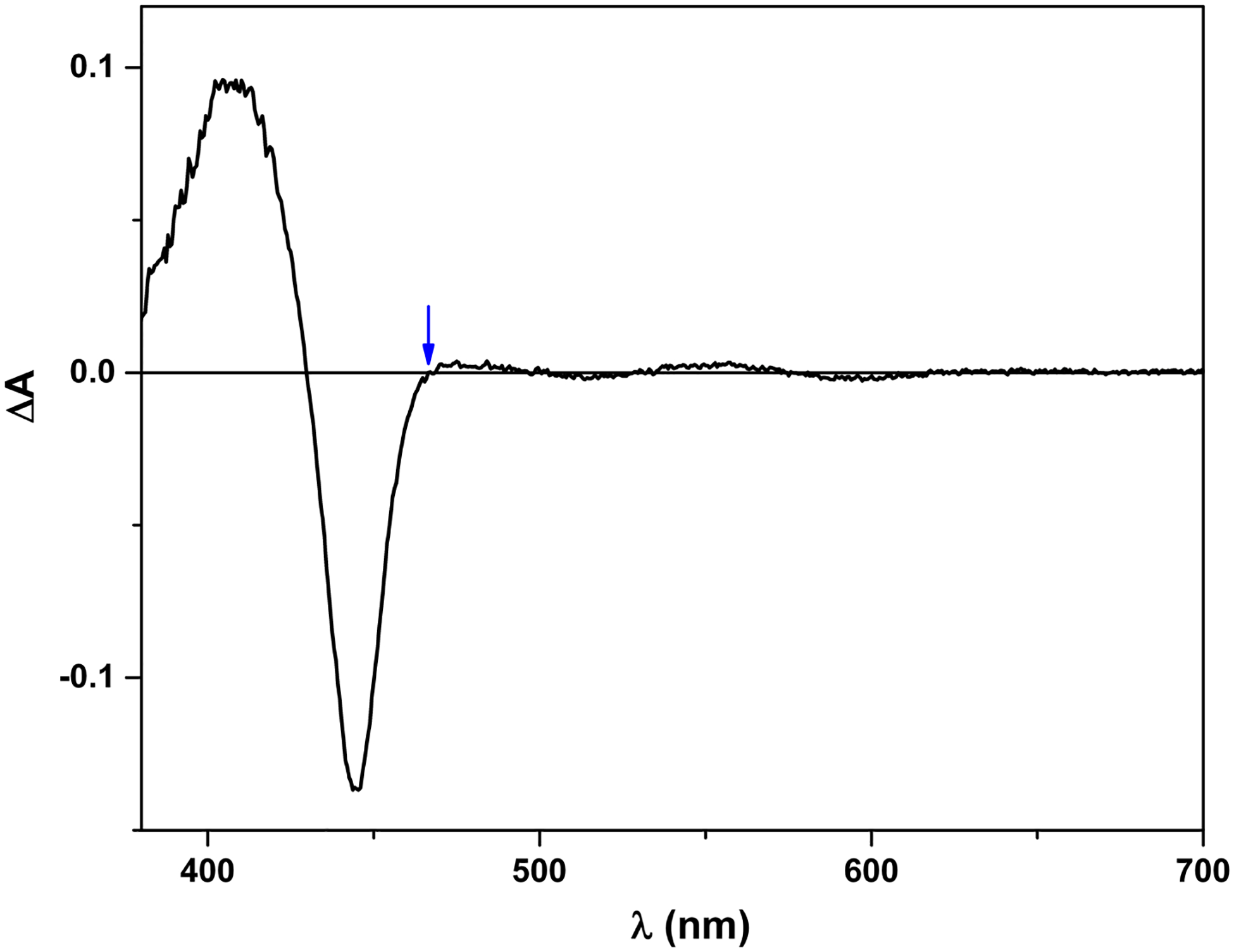
Laser-induced difference spectrum of iNOSoxy. The protein was first reduced to Fe2+-CO form, and then excited by 450 nm laser. The difference spectrum was collected on an Edinburgh L900 laser flash photolysis instrument in spectral mode using an ANDOR iCCD camera. Note that 465 nm is an isosbestic point in the spectrum (marked by an arrow). In other words, the CO rebinding process does not contribute to the absorbance change at 465 nm, which gives a (narrow) window to observe re-oxidation of Fe2+ (due to the FMN-heme IET).
The direct measurement of the IET kinetics by laser flash photolysis has allowed us to significantly advance understanding of the FMN-heme IET mechanism. Our viscosity dependence results indicated that a large conformational change is required for the FMN-heme IET, and6that the FMN/heme docking is transient (88). We have also determined the kinetics over the temperature range from 283 to 304 K in the wild type (wt) full-length (89), wt oxyFMN (89), and E546N (charge neutralization) mutant oxyFMN (87) of human iNOS. Interestingly, the order of the observed IET rate constants (wt oxyFMN > E546N oxyFMN > wt full-length) is in agreement with the order of the activation entropies (87). This result is a key finding with important mechanistic implications: it provides solid evidence for a conformational sampling mechanism for the FMN-heme IET in NOS protein, in which a sampling of a continuum of conformational states gives a range of transient donor-acceptor complexes, only a subset of which are IET-competent (58, 90, 91). The observed IET rate is thus limited by the relatively infrequent formation of the IET-competent docking complexes.
We have conducted a comparative study of nNOSμ and nNOSα proteins (92). nNOSμ contains 34 extra residues in the AR region but is otherwise identical to nNOSα, both nNOSs being splicing variants of the same gene. We have found that the order of the IET rate constants (nNOSα > nNOSμ) also correlates with the order of the activation entropies (92). The larger negative activation entropy of nNOSμ indicates that the FMN domain needs to search for the IET-active conformation among a larger number of conformations that are not IET-competent compared to the nNOSα protein, which results in a less frequent formation of the FMN/heme complex and slower subsequent IET (92). This supports the role of the AR insert in modulating probability of the output state formation, a model originally proposed by us in 2008 (93).
4.4. Combination of pulsed EPR and laser flash photolysis in studying NOS
Our recent study of the E546N mutant of human iNOS oxyFMN (87) has nicely demonstrated how one can employ our combined laser flash photolysis/pulsed EPR approach (Figure 6) to directly examine the role of a specific residue in the FMN-heme IET. Glu546 is a conserved charged residue located at surface of the human iNOS FMN domain, and was proposed to participate in the interdomain FMN/heme interactions (94). Extensive studies were conducted on the analogous residue in nNOS (95, 96), where neutralization at the equivalent residue induced the largest effect on heme reduction (decreasing the heme reduction rate by about 60 % (95)). However, the underlying mechanism for the slower electron transfer in the nNOS mutant was unclear. We found that the iNOS E546N mutation retards the IET by significantly raising the activation entropic barrier (87). Moreover, our pulsed EPR data showed that the geometry of the docked FMN/heme complex in the mutant is basically the same as in the wt, whereas the probability of formation of such a complex is about two-fold lower (87). The IET kinetics and pulsed EPR data together demonstrated conclusively that the E546N mutation retards the IET process by significantly altering the probability of the docked state formation (87).
Figure 6.
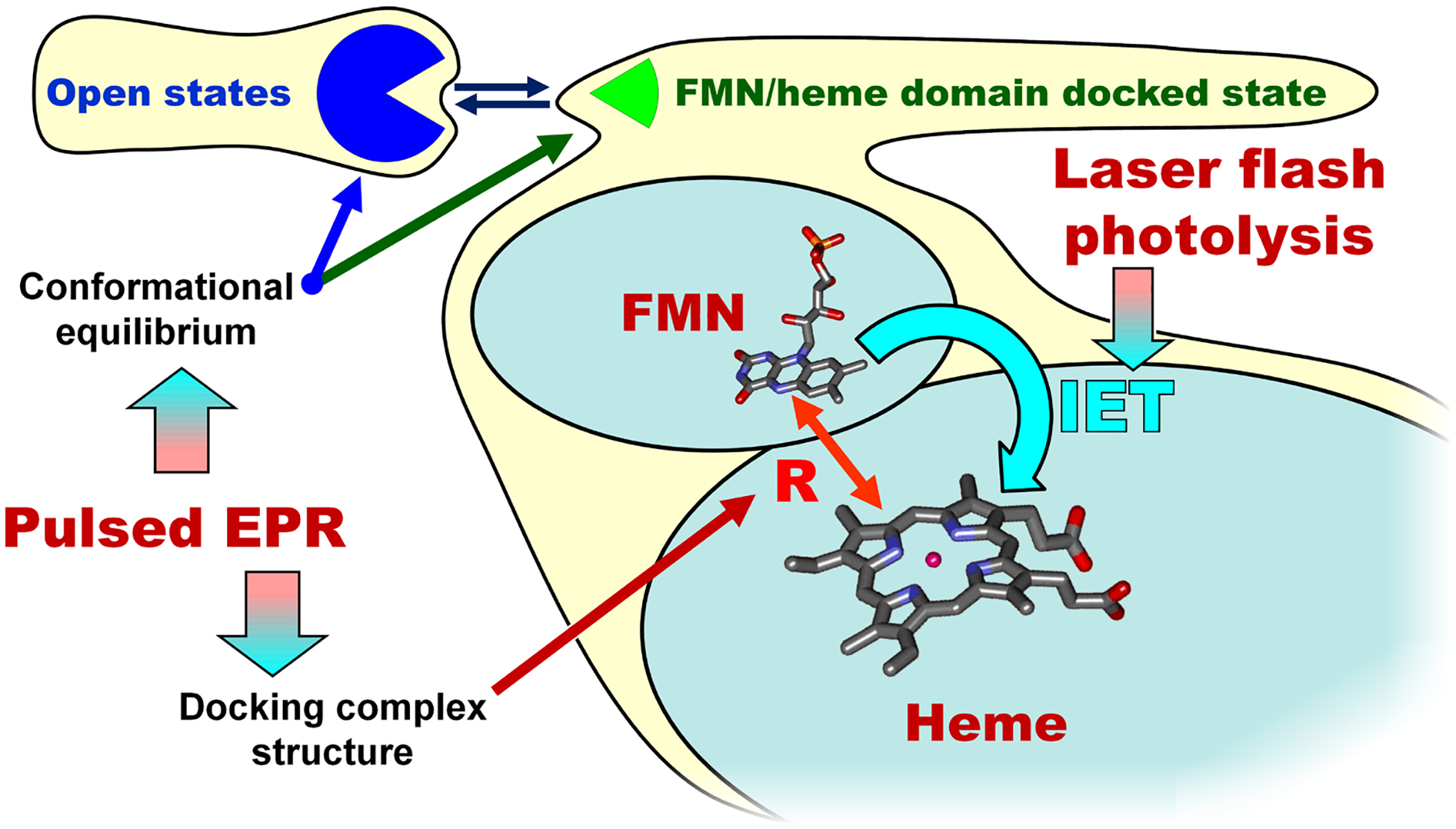
A combined laser flash photolysis and pulsed EPR approach in studying the docked FMN/heme complex.
4.5. Computational simulations of the structural rearrangements during the electron transfer
To investigate the structural rearrangements and the domain interactions before and after the FMN-heme IET, we carried out molecular dynamics (MD) simulations on iNOS models in the two redox states, (Fe(III))(FMNH−) and (Fe(II))(FMNH·) (97). The MD simulations were conducted up to 60 ns. The computational results indicate the departure of FMN domain from the heme center after the IET, i.e., the conformational change is redox-dependent. Moreover, we have envisaged specific residues of the heme, FMN and CaM domains important for optimal docking of the FMN domain to the heme domain. Predictions of some of the key interacting sites in the IET-competent conformations are well supported by the experimental data in the literature. The control of FMN domain motion by these specific interactions is important for NOS function since it facilitates directional electron transfer across the protein by appropriately modifying the conformational space available for the NOS protein. Another elegant computational work was done by Poulos’ laboratory nearly at the same time, using a 105 ns MD trajectory to provide a detailed working model of the FMN-heme IET step in iNOS (59).
It is of note that the bulk FMN-heme electron transfer takes place at a much slower milliseconds time scale, which requires a lot of computational resources for MD calculations. A modeled NOS protein complex of extremely large size will take lots of time to equilibrate, probably on microsecond range, and dissociation would occur on even longer time scale. It is thus necessary to utilize new MD simulations method to accelerate the computation. Additionally, one should confirm that the docking complex is indeed equilibrated by carefully examining the RMSD graph.
5. PERSPECTIVE
Since the discovery of the role of the FMN domain motions in NOS function, intensive efforts have been devoted to deciphering the conformational dynamics. The general picture that emerges from these work is that NOS enzymes exist in an equilibrium of conformations, comprising a rugged conformational landscape, with a key regulatory role for CaM in driving the electron transport by altering the conformational equilibrium. Although these are important developments, important questions on conformational control of NOSs by CaM remain unanswered, and certainly merit further investigation at the molecular level. These are summarized below.
1. We still lack insight into how the conformational changes correlate with the obligatory electron transfer steps in the catalytic cycle of the NOS enzymes. Because individual technique alone can only probe partial facets of the NOS conformational dynamics (see above), it is necessary to utilize complementary and synergistic techniques to study full aspects (temporal, spatial and kinetics) of the dynamically controlled electron transfer. This is a must because the bulk FMN-heme IET rate is significantly affected by the conformational dynamics that determines the formation and dissociation rates of the docking complex between the FMN and heme domains (98). Complimentary approach should be implemented to ensure that the obtained results are robust and unbiased. The NOS conformations and conformational changes can be tracked by time-resolved and single-molecule fluorescence methods at room temperature. The inter-domain interfaces at peptide level can be observed from HDX MS. Because smFRET is best suitable for detecting conformational change, but rather limited at obtaining precise distance, it should be complemented with distance and distribution measurements by pulsed EPR in frozen solution. To identify and assign the observed conformational states, selected site-directed mutants that disrupt specific subdomain interactions should be probed by fluorescence, pulsed EPR and mass spectrometry. As such, the combined use of fluorescence, HDX MS, pulsed EPR, and mutagenesis will dramatically enhance the reliability of the conformational state assignment and conformational equilibrium evaluation.
2. What are the rates of interconversions between conformational states? In optimal situations, one can obtain this information from the analysis of single-molecule trajectories by applying generalized concepts of time correlation functions (TCFs) (99). For example, analysis of TCFs of immobilized eNOS provided preliminary estimate of interconversion rates between free/open and docking states (tens of milliseconds to seconds), which are consistent with previous enzymatic kinetics data (63). In deducing the transition rates between various states, researchers rely on simple, but often arbitrary methods of extracting rates from smFRET trajectories. Although these have proven satisfactory in cases of well-separated, two- or three-state systems, they become less reliable when applied to a system of greater complexity such as NOS. Hidden Markov Model (HMM) analysis allows one to determine, based on probability alone, the most likely FRET states underlying the fluorescence trajectory and their interconversion rates while simultaneously determining the most likely time sequence of underlying states for each trajectory (100). In other words, HHM analysis is probabilistic, less user-dependent and unbiased, and thus more useful for sorting out the kinetics model in the NOS systems.
3. What induces the motions of the FMN domain? In homologous P450 reductase, it is mainly the redox state of the FMN cofactor that biases the motion (101). But it remains unclear if this also stands for the NOS proteins, and how CaM and NOS-unique intrinsic regulatory elements fit in the picture. Our MD simulations suggest that the FMN domain only docks onto the heme domain when it is needed to transfer the electron to the heme center, and dissociates from the docked state afterward (97). Iyanagi proposed an interesting model in which solvation dynamics and protein conformation near the FMN isoalloxazine ring are closely linked to the flavin redox states, which in turn can be correlated to the IET properties (102). It is interesting to experimentally test these models and probe the conformations of NOSs at various redox states. A promising approach is single molecule enzymology, which has been applied to P450 reductase reconstituted in nanodisc (103).
4. Emerging evidence demonstrates that the NOS-unique control elements AR (92) and CT (64) are involved in the FMN-heme IET. These two regions are only partially resolved in crystal structure of the CaM-free nNOS reductase (15), speaking of their high flexibility nature. A wider variety of experiments need to be proposed that would begin to provide new insights as to the underlying mechanism by which the AR region promotes interactions between the FMN and heme domains (93). For example, one can ask: what conformational space is explored by the AR region and how is this changed by CaM binding; also how is it that CaM binds, but that it does so without sterically blocking the interaction between the FMN and heme domains? These are challenging questions to address, and require more structural information than is presently available. To solve this long-standing mystery, it is necessary to obtain the distance information for the AR insert and CT, relative to CaM and the FMN and heme domains in the NOS proteins. Cryo-EM at current low resolution can’t resolve these elements in the images. Pulsed EPR studies of NOSs labeled with paramagnetic tag (such as Cu2+, Mn2+ or nitroxide spin label) at these sites have great potential in providing such critical distance information. The critical steps in making the pulsed EPR samples are site-specific labelling of cysteine-rich NOSs and selection of the paramagnetic label(s) and label-NOS linkage bond that are reduction-resistant.
5. There is a real need to develop new strategies for selective intervention of the NOS isoforms. The current NOS inhibitors targeting the heme active sites encounter major obstacles due to high degree conservation of the heme site. Moreover, the NOS heme inhibition may cause an adverse production of unwanted reactive oxygen species (e.g., superoxide) through the leakage of the electron to oxygen from the reduced heme site. An understanding of conformational dynamics (at both the spatial and temporal levels) that is linked to NOS function will provide new opportunities for inhibitor design by targeting interfaces between the domain modules. The “interfacial inhibitor” concept is becoming mainstream in drug discovery, and many of such new drugs have been recently FDA-approved for other targets (104). The inter-domain FMN/heme interacting surfaces are well understood (24, 95), but the docking site is not a promising target to selectively modulate the NOS isoforms because it is basically conserved among the NOS isoforms. Equally important, the CaM-heme(NOS) docking is required for the FMN/heme domain alignment (18). In contrast to the FMN/heme interface, the CaM/heme interface is predicted to be isoform-specific (97). It is thus important to further define the CaM/NOS docking interface and its role in NOS isoform function, and screen compounds to target the dynamic CaM-NOS docking interface.
6. CONCLUSIONS
Characterizing the dynamics and large-scale movements within proteins is a frontier challenge for understanding electron transfer mechanisms. Mammalian NOS enzymes are redox flavor-hemoproteins consisting of multiple modular domains. They are important proteins because of the crucial roles of NO in signaling processes. The importance of conformational changes in the function of complex enzymes like NOS has become increasingly recognized. Despite significant progress in general understanding the structural rearrangements during NOS catalysis, how the observed conformational changes correlate with the obligatory electron transfer steps in NOS catalysis remains unclear. A roadblock to answering this central question is that any current biophysical method alone cannot grasp full aspects of the conformational dynamics. It is necessary to combine state-of-the-art methods in pulsed EPR, fluorescence and HDX MS with rapid kinetics (laser flash photolysis and stopped flow) techniques. This is to correlate the spatial and temporal observations with functional kinetics data on electron transfer and catalysis - information that on the long term may provide new opportunities for selective intervention of the NOS isoforms. Furthermore, a combined approach established for quantitating the complex heterogenous NOS systems can be applicable to other biomolecular systems where the dynamic interactions between constituent domains or proteins determine the chemical mechanism. This is clearly a fruitful area for future studies in many years to come.
ACKNOWLEDGEMENT
The research was supported by grants to CF from the National Institutes of Health (GM081811). CF acknowledges the support of UNM College of Pharmacy pilot award.
REFERENCES
- 1.Hedison TM, Hay S and Scrutton NS: A perspective on conformational control of electron transfer in nitric oxide synthases. Nitric Oxide, 63, 61–67 (2017). DOI: 10.1016/j.niox.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leferink NGH, Hay S, Rigby SEJ and Scrutton NS: Towards the free energy landscape for catalysis in mammalian nitric oxide synthases. FEBS J, 282(16), 3016–3029 (2015). DOI: 10.1111/febs.13171 [DOI] [PubMed] [Google Scholar]
- 3.Stuehr DJ, Tejero J and Haque MM: Structural and mechanistic aspects of flavoproteins: Electron transfer through the nitric oxide synthase flavoprotein domain FEBS J, 276(15), 3959–3974 (2009). DOI: 10.1111/j.1742-4658.2009.07120.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alderton WK, Cooper CE and Knowles RG: Nitric oxide synthases: Structure, function and inhibition. Biochem J, 357, 593–615 (2001). DOI: 10.1042/bj3570593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fulton DJR: Chapter two - Transcriptional and posttranslational regulation of eNOS in the endothelium In: Advances in Pharmacology. Ed Raouf AK. Academic Press, (2016). [DOI] [PubMed] [Google Scholar]
- 6.Fulton D, Gratton JP, McCabe TJ, Fontana J, Fujio Y, Walsh K, Franke TF, Papapetropoulos A and Sessa WC: Regulation of endothelium-derived nitric oxide production by the protein kinase Akt. Nature, 399(6736), 597–601 (1999). DOI: 10.1038/21218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crane BR, Arvai AS, Ghosh DK, Wu CQ, Getzoff ED, Stuehr DJ and Tainer JA: Structure of nitric oxide synthase oxygenase dimer with pterin and substrate. Science, 279(5359), 2121–2126 (1998). DOI: 10.1126/science.279.5359.2121 [DOI] [PubMed] [Google Scholar]
- 8.Raman CS, Li HY, Martasek P, Kral V, Masters BSS and Poulos TL: Crystal structure of constitutive endothelial nitric oxide synthase: A paradigm for pterin function involving a novel metal center. Cell, 95(7), 939–950 (1998). DOI: 10.1016/S0092-8674(00)81718-3 [DOI] [PubMed] [Google Scholar]
- 9.Li HY, Igarashi J, Jamal J, Yang WP and Poulos TL: Structural studies of constitutive nitric oxide synthases with diatomic ligands bound. J Biol Inorg Chem, 11(6), 753–768 (2006). DOI: 10.1007/s00775-006-0123-8 [DOI] [PubMed] [Google Scholar]
- 10.Fedorov R, Vasan R, Ghosh DK and Schlichting I: Structures of nitric oxide synthase isoforms complexed with the inhibitor AR-R17477 suggest a rational basis for specificity and inhibitor design. Proc Natl Acad Sci USA, 101(16), 5892–5897 (2004). DOI: 10.1073/pnas.0306588101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei CC, Crane BR and Stuehr DJ: Tetrahydrobiopterin radical enzymology. Chem Rev, 103(6), 2365–2383 (2003). DOI: 10.1021/cr0204350 [DOI] [PubMed] [Google Scholar]
- 12.Poulos TL and Li H: Nitric oxide synthase and structure-based inhibitor design. Nitric Oxide, 63, 68–77 (2017). DOI: 10.1016/j.niox.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcin ED, Arvai AS, Rosenfeld RJ, Kroeger MD, Crane BR, Andersson G, Andrews G, Hamley PJ, Mallinder PR, Nicholls DJ, St-Gallay SA, Tinker AC, Gensmantel NP, Mete A, Cheshire DR, Connolly S, Stuehr DJ, Aberg A, Wallace AV, Tainer JA and Getzoff ED: Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase. Nature Chem Biol, 4(11), 700–707 (2008). DOI: 10.1038/nchembio.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Martasek P, Paschke R, Shea T, Masters BSS and Kim JJP: Crystal structure of the FAD/NADPH-binding domain of rat neuronal nitric-oxide synthase - Comparisons with NADPH-cytochrome P450 oxidoreductase. J Biol Chem, 276(40), 37506–37513 (2001). DOI: 10.1074/jbc.M105503200 [DOI] [PubMed] [Google Scholar]
- 15.Garcin ED, Bruns CM, Lloyd SJ, Hosfield DJ, Tiso M, Gachhui R, Stuehr DJ, Tainer JA and Getzoff ED: Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase. J Biol Chem, 279(36), 37918–37927 (2004). DOI: 10.1074/jbc.M406204200 [DOI] [PubMed] [Google Scholar]
- 16.Xia C, Misra I, Iyanagi T and Kim J-JP: Regulation of interdomain interactions by calmodulin in inducible nitric oxide synthase. J Biol Chem, 284(44), 30708–30717 (2009). DOI: 10.1074/jbc.M109.031682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aoyagi M, Arvai AS, Tainer JA and Getzoff ED: Structural basis for endothelial nitric oxide synthase binding to calmodulin. EMBO J, 22(4), 766–775 (2003). DOI: 10.1093/emboj/cdg078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith BC, Underbakke ES, Kulp DW, Schief WR and Marletta MA: Nitric oxide synthase domain interfaces regulate electron transfer and calmodulin activation. Proc Natl Acad Sci USA, 110(38), E3577–E3586 (2013). DOI: 10.1073/pnas.1313331110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai Y, Haque MM and Stuehr DJ: Restricting the conformational freedom of the neuronal nitric-oxide synthase flavoprotein domain reveals impact on electron transfer and catalysis. J Biol Chem, 292(16), 6753–6764 (2017). DOI: 10.1074/jbc.M117.777219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, Zheng H, Li W, Li W, Miao Y and Feng C: Role of a conserved tyrosine residue in the FMN–heme interdomain electron transfer in inducible nitric oxide synthase. J Phys Chem A, 120(39), 7610–7616 (2016). DOI: 10.1021/acs.jpca.6b08207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haque MM, Bayachou M, Fadlalla MA, Durra D and Stuehr DJ: Charge-pairing interactions control the conformational setpoint and motions of the FMN domain in neuronal nitric oxide synthase. Biochem J, 450(3), 607–617 (2013). DOI: 10.1042/BJ20121488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benson MA, Batchelor H, Chuaiphichai S, Bailey J, Zhu H, Stuehr DJ, Bhattacharya S, Channon KM and Crabtree MJ: A pivotal role for tryptophan 447 in enzymatic coupling of human endothelial nitric oxide synthase (eNOS): Effects on tetrahydrobiopterin-dependent catalysis and eNOS dimerization. J Biol Chem, 288(41), 29836–29845 (2013). DOI: 10.1074/jbc.M113.493023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tejero J, Haque MM, Durra D and Stuehr DJ: A bridging interaction allows calmodulin to activate NO synthase through a bi-modal mechanism. J Biol Chem, 285(34), 25941–25949 (2010). DOI: 10.1074/jbc.M110.126797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tejero J, Hannibal L, Mustovich A and Stuehr DJ: Surface charges and regulation of FMN to heme electron transfer in nitric-oxide synthase. J Biol Chem, 285(35), 27232–27240 (2010). DOI: 10.1074/jbc.M110.138842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan Z-W, Haque MM, Wei C-C, Garcin ED, Getzoff ED and Stuehr DJ: Lys842 in neuronal nitric-oxide synthase enables the autoinhibitory insert to antagonize calmodulin binding, increase FMN shielding, and suppress interflavin electron transfer. J Biol Chem, 285(5), 3064–3075 (2010). DOI: 10.1074/jbc.M109.000810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newman E, Spratt DE, Mosher J, Cheyne B, Montgomery HJ, Wilson DL, Weinberg JB, Smith SME, Salerno JC, Ghosh DK and Guillemette JG: Differential activation of nitric-oxide synthase isozymes by calmodulin-troponin C chimeras. J Biol Chem, 279(32), 33547–33557 (2004). DOI: 10.1074/jbc.M403892200 [DOI] [PubMed] [Google Scholar]
- 27.Wang Z-Q, Haque MM, Binder K, Sharma M, Wei C-C and Stuehr DJ: Engineering nitric oxide synthase chimeras to function as NO dioxygenases. J Inorg Biochem, 158, 122–130 (2016). DOI: 10.1016/j.jinorgbio.2016.03.002 [DOI] [PubMed] [Google Scholar]
- 28.Jachymova M, Martasek P, Panda S, Roman LJ, Panda M, Shea TM, Ishimura Y, Kim JJP and Masters BSS: Recruitment of governing elements for electron transfer in the nitric oxide synthase family. Proc Natl Acad Sci USA, 102(44), 15833–15838 (2005). DOI: 10.1073/pnas.0506522102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haque MM, Bayachou M, Tejero J, Kenney C, Pearl NM, Im S-C, Waskell L and Stuehr DJ: Distinct conformational behaviors of four mammalian dual-flavin reductases (cytochrome P450 reductase, methionine synthase reductase, neuronal nitric oxide synthase, endothelial nitric oxide synthase) determine their unique catalytic profiles. FEBS J, 281(23), 5325–5340 (2014). DOI: 10.1111/febs.13073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Y and Silverman RB: Revisiting heme mechanisms. A perspective on the mechanisms of nitric oxide synthase (NOS), heme oxygenase (HO), and cytochrome P450s (CYP450s). Biochemistry, 47(8), 2231–2243 (2008). DOI: 10.1021/bi7023817 [DOI] [PubMed] [Google Scholar]
- 31.Feng C: Mechanism of nitric oxide synthase regulation: Electron transfer and interdomain interactions. Coord Chem Rev, 256(3–4), 393–411 (2012). DOI: 10.1016/j.ccr.2011.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo S, Lei H, Qin H and Xia Y: Molecular mechanisms of endothelial NO synthase uncoupling. Curr Pharm Des, 20(22), 3548–53 (2014). DOI: 10.2174/13816128113196660746 [DOI] [PubMed] [Google Scholar]
- 33.Zweier JL, Chen C-A and Druhan LJ: S-Glutathionylation reshapes our understanding of endothelial nitric oxide synthase uncoupling and nitric oxide/reactive oxygen species-mediated signaling. Antioxidants & Redox Signaling, 14(10), 1769–1775 (2011). DOI: 10.1089/ars.2011.3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vasquez-Vivar J, Kalyanaraman B, Martasek P, Hogg N, Masters BSS, Karoui H, Tordo P and Pritchard KA: Superoxide generation by endothelial nitric oxide synthase: The influence of cofactors. Proc Natl Acad Sci USA, 95(16), 9220–9225 (1998). DOI: 10.1073/pnas.95.16.9220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen CA, Druhan LJ, Varadharaj S, Chen YR and Zweier JL: Phosphorylation of endothelial nitric-oxide synthase regulates superoxide generation from the enzyme. J Biol Chem, 283(40), 27038–27047 (2008). DOI: 10.1074/jbc.M802269200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panda K, Adak S, Konas D, Sharma M and Stuehr DJ: A conserved aspartate (Asp-1393) regulates NADPH reduction of neuronal nitric-oxide synthase - Implications for catalysis. J Biol Chem, 279(18), 18323–18333 (2004). DOI: 10.1074/jbc.M310391200 [DOI] [PubMed] [Google Scholar]
- 37.Panda K, Ghosh S and Stuehr DJ: Calmodulin activates intersubunit electron transfer in the neuronal nitric-oxide synthase dimer. J Biol Chem, 276(26), 23349–23356 (2001). DOI: 10.1074/jbc.M100687200 [DOI] [PubMed] [Google Scholar]
- 38.Abusoud HM and Stuehr DJ: Nitric-oxide synthases reveal a role for calmodulin in controlling electron transfer. Proc Natl Acad Sci USA, 90(22), 10769–10772 (1993). DOI: 10.1073/pnas.90.22.10769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abusoud HM, Yoho LL and Stuehr DJ: Calmodulin controls neuronal nitric-oxide synthase by a dual mechanism - Activation of intradomain and interdomain electron transfer. J Biol Chem, 269(51), 32047–32050 (1994). [PubMed] [Google Scholar]
- 40.Tiso M, Konas DW, Panda K, Garcin ED, Sharma M, Getzoff ED and Stuehr DJ: C-terminal tail residue Arg(1400) enables NADPH to regulate electron transfer in neuronal nitric-oxide synthase. J Biol Chem, 280(47), 39208–39219 (2005). DOI: 10.1074/jbc.M507775200 [DOI] [PubMed] [Google Scholar]
- 41.Noble MA, Munro AW, Rivers SL, Robledo L, Daff SN, Yellowlees LJ, Shimizu T, Sagami I, Guillemette JG and Chapman SK: Potentiometric analysis of the flavin cofactors of neuronal nitric oxide synthase. Biochemistry, 38(50), 16413–16418 (1999). DOI: 10.1021/bi992150w [DOI] [PubMed] [Google Scholar]
- 42.Gao YT, Smith SME, Weinberg JB, Montgomery HJ, Newman E, Guillemette JG, Ghosh DK, Roman LJ, Martasek P and Salerno JC: Thermodynamics of oxidation-reduction reactions in mammalian nitric-oxide synthase Isoforms. J Biol Chem, 279(18), 18759–18766 (2004). DOI: 10.1074/jbc.M308936200 [DOI] [PubMed] [Google Scholar]
- 43.Daff S, Noble MA, Craig DH, Rivers SL, Chapman SK, Munro AW, Fujiwara S, Rozhkova E, Sagami I and Shimizu T: Control of electron transfer in neuronal NO synthase. Biochem Soc Trans, 29, 147–152 (2001). DOI: 10.1042/bst0290147 [DOI] [PubMed] [Google Scholar]
- 44.Salerno JC, Harris DE, Irizarry K, Patel B, Morales AJ, Smith SME, Martasek P, Roman LJ, Masters BSS, Jones CL, Weissman BA, Lane P, Liu Q and Gross SS: An autoinhibitory control element defines calcium-regulated isoforms of nitric oxide synthase. J Biol Chem, 272(47), 29769–29777 (1997). DOI: 10.1074/jbc.272.47.29769 [DOI] [PubMed] [Google Scholar]
- 45.Roman LJ, Martasek P, Miller RT, Harris DE, de la Garza MA, Shea TM, Kim JJP and Masters BSS: The C termini of constitutive nitric-oxide synthases control electron flow through the flavin and heme domains and affect modulation by calmodulin. J Biol Chem, 275(38), 29225–29232 (2000). DOI: 10.1074/jbc.M004766200 [DOI] [PubMed] [Google Scholar]
- 46.Jones RJ, Smith SME, Gao YT, DeMay BS, Mann KJ, Salerno KM and Salerno JC: The function of the small insertion in the hinge subdomain in the control of constitutive mammalian nitric-oxide synthases. J Biol Chem, 279(35), 36876–36883 (2004). DOI: 10.1074/jbc.M402808200 [DOI] [PubMed] [Google Scholar]
- 47.Nishida CR and de Montellano PRO: Control of Electron Transfer in Nitric-oxide Synthases: SWAPPING OF AUTOINHIBITORY ELEMENTS AMONG NITRIC-OXIDE SYNTHASE ISOFORMS. J Biol Chem, 276(23), 20116–20124 (2001). DOI: 10.1074/jbc.M101548200 [DOI] [PubMed] [Google Scholar]
- 48.Roman LJ, Martasek P and Masters BSS: Intrinsic and extrinsic modulation of nitric oxide synthase activity. Chem Rev, 102(4), 1179–1189 (2002). DOI: 10.1021/cr000661e [DOI] [PubMed] [Google Scholar]
- 49.Feng CJ, Tollin G, Hazzard JT, Nahm NJ, Guillemette JG, Salerno JC and Ghosh DK: Direct measurement by laser flash photolysis of intraprotein electron transfer in a rat neuronal nitric oxide synthase. J Am Chem Soc, 129(17), 5621–5629 (2007). DOI: 10.1021/ja068685b [DOI] [PubMed] [Google Scholar]
- 50.Feng CJ, Tollin G, Holliday MA, Thomas C, Salerno JC, Enemark JH and Ghosh DK: Intraprotein electron transfer in a two-domain construct of neuronal nitric oxide synthase: The output state in nitric oxide formation. Biochemistry, 45(20), 6354–6362 (2006). DOI: 10.1021/bi060223n [DOI] [PubMed] [Google Scholar]
- 51.Ghosh DK and Salerno JC: Nitric oxide synthases: Domain structure and alignment in enzyme function and control. Frontiers in Bioscience, 8, D193–D209 (2003). DOI: 10.2741/959 [DOI] [PubMed] [Google Scholar]
- 52.Hedison TM, Hay S and Scrutton NS: Real-time analysis of conformational control in electron transfer reactions of human cytochrome P450 reductase with cytochrome c. FEBS J, 282(22), 4357–4375 (2015). DOI: 10.1111/febs.13501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Das A, Sibhatu H, Jamal J, Sligar SG and Poulos TL: Exploring the electron transfer properties of neuronal nitric oxide synthase by reversal of the FMN redox potential. J Biol Chem, 283(50), 34762–34772 (2008). DOI: 10.1074/jbc.M806949200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunford AJ, Rigby SEJ, Hay S, Munro AW and Scrutton NS: Conformational and thermodynamic control of electron transfer in neuronal nitric oxide synthase. Biochemistry, 46(17), 5018–5029 (2007). DOI: 10.1021/bi7001339 [DOI] [PubMed] [Google Scholar]
- 55.Ghosh DK, Holliday MA, Thomas C, Weinberg JB, Smith SME and Salerno JC: Nitric-oxide synthase output state - Design and properties of nitric-oxide synthase oxygenase/FMN domain constructs. J Biol Chem, 281(20), 14173–14183 (2006). DOI: 10.1074/jbc.M509937200 [DOI] [PubMed] [Google Scholar]
- 56.Feng CJ, Dupont A, Nahm N, Spratt D, Hazzard JT, Weinberg J, Guillemette J, Tollin G and Ghosh DK: Intraprotein electron transfer in inducible nitric oxide synthase holoenzyme. J Biol Inorg Chem, 14(1), 133–142 (2009). DOI: 10.1007/s00775-008-0431-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Doukov T, Li HY, Soltis M and Poulos TL: Single crystal structural and absorption spectral characterizations of nitric oxide synthase complexed with N-omega-hydroxy-L-arginine and diatomic ligands. Biochemistry, 48(43), 10246–10254 (2009). DOI: 10.1021/bi9009743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ilagan RP, Tejero JS, Aulak KS, Ray SS, Hemann C, Wang Z-Q, Gangoda M, Zweier JL and Stuehr DJ: Regulation of FMN subdomain interactions and function in neuronal nitric oxide synthase. Biochemistry, 48(18), 3864–3876 (2009). DOI: 10.1021/bi8021087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hollingsworth SA, Holden JK, Li H and Poulos TL: Elucidating nitric oxide synthase domain interactions by molecular dynamics. Protein Science, 25, 374–382 (2016). DOI: 10.1002/pro.2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Astashkin AV, Elmore BO, Fan W, Guillemette JG and Feng C: Pulsed EPR determination of the distance between heme iron and FMN centers in a human inducible nitric oxide synthase. J Am Chem Soc, 132(34), 12059–12067 (2010). DOI: 10.1021/ja104461p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Campbell MG, Smith BC, Potter CS, Carragher B and Marletta MA: Molecular architecture of mammalian nitric oxide synthases. Proc Natl Acad Sci USA, 111(35), E3614–E3623 (2014). DOI: 10.1073/pnas.1413763111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yokom AL, Morishima Y, Lau M, Su M, Glukhova A, Osawa Y and Southworth DR: Architecture of the nitric oxide synthase holoenzyme reveals large conformational changes and a calmodulin-driven release of the FMN domain. J Biol Chem, 289, 16855–16865 (2014). DOI: 10.1074/jbc.M114.564005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arnett DC, Persechini A, Tran Q-K, Black DJ and Johnson CK: Fluorescence quenching studies of structure and dynamics in calmodulin–eNOS complexes. FEBS Lett, 589(11), 1173–1178 (2015). DOI: 10.1016/j.febslet.2015.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haque MM, Ray SS and Stuehr DJ: Phosphorylation controls endothelial nitric-oxide synthase by regulating its conformational dynamics. J Biol Chem, 291(44), 23047–23057 (2016). DOI: 10.1074/jbc.M116.737361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Craig DH, Chapman SK and Daff S: Calmodulin activates electron transfer through neuronal nitric-oxide synthase reductase domain by releasing an NADPH-dependent conformational lock. J Biol Chem, 277(37), 33987–33994 (2002). DOI: 10.1074/jbc.M203118200 [DOI] [PubMed] [Google Scholar]
- 66.Haque MM, Kenney C, Tejero J and Stuehr DJ: A kinetic model linking protein conformational motions, interflavin electron transfer, and electron flux through a dual-flavin enzyme: Simulating the reductase activity of the endothelial and neuronal NO synthase flavoprotein domains FEBS J, 278(21), 4055–4069 (2011). DOI: 10.1111/j.1742-4658.2011.08310.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gachhui R, Presta A, Bentley DF, Abu-Soud HM, McArthur R, Brudvig G, Ghosh DK and Stuehr DJ: Characterization of the reductase domain of rat neuronal nitric oxide synthase generated in the methylotrophic yeast Pichia pastoris - Calmodulin response is complete within the reductase domain itself. J Biol Chem, 271(34), 20594–20602 (1996). DOI: 10.1074/jbc.271.34.20594 [DOI] [PubMed] [Google Scholar]
- 68.Sheta EA, McMillan K and Masters BSS: Evidence for a bidomain structure of constitutive cerebellar nitric-oxide synthase. J Biol Chem, 269(21), 15147–15153 (1994). [PubMed] [Google Scholar]
- 69.Feng CJ, Thomas C, Holliday MA, Tollin G, Salerno JC, Ghosh DK and Enemark JH: Direct measurement by laser flash photolysis of intramolecular electron transfer in a two-domain construct of murine inducible nitric oxide synthase. J Am Chem Soc, 128(11), 3808–3811 (2006). DOI: 10.1021/ja0578606 [DOI] [PubMed] [Google Scholar]
- 70.Piazza M, Taiakina V, Dieckmann T and Guillemette JG: Structural consequences of calmodulin EF hand mutations. Biochemistry, 56, 944–956 (2017). DOI: 10.1021/acs.biochem.6b01296 [DOI] [PubMed] [Google Scholar]
- 71.Piazza M, Dieckmann T and Guillemette JG: Structural studies of a complex between endothelial nitric oxide synthase and calmodulin at physiological calcium concentration. Biochemistry, 55, 5962–5971 (2016). DOI: 10.1021/acs.biochem.6b00821 [DOI] [PubMed] [Google Scholar]
- 72.Piazza M, Futrega K, Spratt DE, Dieckmann T and Guillemette JG: Structure and dynamics of calmodulin (CaM) bound to nitric oxide synthase peptides: Effects of a phosphomimetic CaM mutation. Biochemistry, 51(17), 3651–3661 (2012). DOI: 10.1021/bi300327z [DOI] [PubMed] [Google Scholar]
- 73.Volkmann N, Martásek P, Roman LJ, Xu X-P, Page C, Swift M, Hanein D and Masters BS: Holoenzyme structures of endothelial nitric oxide synthase – An allosteric role for calmodulin in pivoting the FMN domain for electron transfer. J Struct Biol, 188(1), 46–54 (2014). DOI: 10.1016/j.jsb.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He Y, Haque MM, Stuehr DJ and Lu HP: Single-molecule spectroscopy reveals how calmodulin activates NO synthase by controlling its conformational fluctuation dynamics. Proc Natl Acad Sci USA, 112(38), 11835–11840 (2015). DOI: 10.1073/pnas.1508829112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Astashkin AV, Chen L, Zhou X, Li H, Poulos TL, Liu KJ, Guillemette JG and Feng C: Pulsed electron paramagnetic resonance study of domain docking in neuronal nitric oxide synthase: The calmodulin and output state perspective. J Phys Chem A, 118(34), 6864–6872 (2014). DOI: 10.1021/jp503547w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sobolewska-Stawiarz A, Leferink NGH, Fisher K, Heyes DJ, Hay S, Rigby SEJ and Scrutton NS: Energy landscapes and catalysis in nitric-oxide synthase. J Biol Chem, 289(17), 11725–11738 (2014). DOI: 10.1074/jbc.M114.548834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hay S, Brenner S, Khara B, Quinn AM, Rigby SEJ and Scrutton NS: Nature of the energy landscape for gated electron transfer in a dynamic redox protein. J Am Chem Soc, 132(28), 9738–9745 (2010). DOI: 10.1021/ja1016206 [DOI] [PubMed] [Google Scholar]
- 78.Pudney CR, Heyes DJ, Khara B, Hay S, Rigby SEJ and Scrutton NS: Kinetic and spectroscopic probes of motions and catalysis in the cytochrome P450 reductase family of enzymes. FEBS J, 279(9), 1534–1544 (2012). DOI: 10.1111/j.1742-4658.2011.08442.x [DOI] [PubMed] [Google Scholar]
- 79.Persechini A, Tran QK, Black DJ and Gogol EP: Calmodulin-induced structural changes in endothelial nitric oxide synthase. FEBS Lett, 587(3), 297–301 (2013). DOI: 10.1016/j.febslet.2012.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jeschke G and Polyhach Y: Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys Chem Chem Phys, 9(16), 1895–910 (2007). DOI: 10.1039/b614920k [DOI] [PubMed] [Google Scholar]
- 81.Kulik LV, Dzuba SA, Grigoryev IA and Tsvetkov YD: Electron dipole-dipole interaction in ESEEM of nitroxide biradicals. Chem Phys Lett, 343(3–4), 315–324 (2001). DOI: 10.1016/S0009-2614(01)00721-7 [DOI] [Google Scholar]
- 82.Astashkin AV: Chapter ten - Mapping the structure of metalloproteins with RIDME. In: Methods in Enzymology. Ed Peter ZQ&Kurt W. Academic Press, (2015). [DOI] [PubMed] [Google Scholar]
- 83.Page CC, Moser CC, Chen XX and Dutton PL: Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature, 402(6757), 47–52 (1999). DOI: 10.1038/46972 [DOI] [PubMed] [Google Scholar]
- 84.Astashkin AV, Fan W, Elmore BO, Guillemette JG and Feng C: Pulsed ENDOR determination of relative orientation of g-frame and molecular frame of imidazole-coordinated heme center of iNOS. J Phys Chem A, 115(37), 10345–10352 (2011). DOI: 10.1021/jp204969d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ghosh DK, Ray K, Rogers AJ, Nahm NJ and Salerno JC: FMN fluorescence in inducible NOS constructs reveals a series of conformational states involved in the reductase catalytic cycle. FEBS J, 279(7), 1306–1317 (2012). DOI: 10.1111/j.1742-4658.2012.08525.x [DOI] [PubMed] [Google Scholar]
- 86.Abu-Soud HM, Feldman PL, Clark P and Stuehr DJ: Electron transfer in the nitric-oxide synthases. Characterization of L-arginine analogs that block heme iron reduction. J Biol Chem, 269(51), 32318–32326 (1994). [PubMed] [Google Scholar]
- 87.Li W, Chen L, Lu C, Elmore BO, Astashkin AV, Rousseau DL, Yeh S-R and Feng C: Regulatory role of Glu546 in flavin mononucleotide — heme electron transfer in human inducible nitric oxide synthase. Inorg Chem, 52(9), 4795–4801 (2013). DOI: 10.1021/ic3020892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li W, Fan W, Elmore BO and Feng C: Effect of solution viscosity on intraprotein electron transfer between the FMN and heme domains in inducible nitric oxide synthase. FEBS Lett, 585(16), 2622–2626 (2011). DOI: 10.1016/j.febslet.2011.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li W, Chen L, Fan W and Feng C: Comparing the temperature dependence of FMN to heme electron transfer in full length and truncated inducible nitric oxide synthase proteins. FEBS Lett, 586(2), 159–162 (2012). DOI: 10.1016/j.febslet.2011.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leys D and Scrutton NS: Electrical circuitry in biology: Emerging principles from protein structure. Curr Opin Struct Biol, 14(6), 642–647 (2004) DOI: 10.1016/j.sbi.2004.10.002 [DOI] [PubMed] [Google Scholar]
- 91.Salerno JC, Ray K, Poulos T, Li H and Ghosh DK: Calmodulin activates neuronal nitric oxide synthase by enabling transitions between conformational states. FEBS Lett, 587(1), 44–47 (2013). DOI: 10.1016/j.febslet.2012.10.039 [DOI] [PubMed] [Google Scholar]
- 92.Panda SP, Li W, Venkatakrishnan P, Chen L, Astashkin AV, Masters BSS, Feng C and Roman LJ: Differential calmodulin-modulatory and electron transfer properties of neuronal nitric oxide synthase mu compared to the alpha variant. FEBS Lett, 587(24), 3973–3978 (2013). DOI: 10.1016/j.febslet.2013.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Feng CJ, Roman LJ, Hazzard JT, Ghosh DK, Tollin G and Masters BSS: Deletion of the autoregulatory insert modulates intraprotein electron transfer in rat neuronal nitric oxide synthase. FEBS Lett, 582(18), 2768–2772 (2008). DOI: 10.1016/j.febslet.2008.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sempombe J, Elmore BO, Sun X, Dupont A, Ghosh DK, Guillemette JG, Kirk ML and Feng C: Mutations in the FMN domain modulate MCD spectra of the heme site in the oxygenase domain of inducible nitric oxide synthase. J Am Chem Soc, 131(20), 6940–6941 (2009). DOI: 10.1021/ja902141v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Panda K, Haque MM, Garcin-Hosfield ED, Durra D, Getzoff ED and Stuehr DJ: Surface charge interactions of the FMN module govern catalysis by nitric-oxide synthase. J Biol Chem, 281(48), 36819–36827 (2006). DOI: 10.1074/jbc.M606129200 [DOI] [PubMed] [Google Scholar]
- 96.Haque MM, Fadlalla M, Wang ZQ, Ray SS, Panda K and Stuehr DJ: Neutralizing a surface charge on the FMN subdomain increases the activity of neuronal nitric-oxide synthase by enhancing the oxygen reactivity of the enzyme heme-nitric oxide complex. J Biol Chem, 284(29), 19237–19247 (2009). DOI: 10.1074/jbc.M109.013144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sheng Y, Zhong L, Guo D, Lau G and Feng C: Insight into structural rearrangements and interdomain interactions related to electron transfer between flavin mononucleotide and heme in nitric oxide synthase: A molecular dynamics study. J Inorg Biochem, 153, 186–196 (2015). DOI: 10.1016/j.jinorgbio.2015.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Astashkin AV and Feng C: Solving kinetic equations for the laser flash photolysis experiment on nitric oxide synthases: Effect of conformational dynamics on the interdomain electron transfer. J Phys Chem A, 119(45), 11066–11075 (2015). DOI: 10.1021/acs.jpca.5b08414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schenter GK, Lu HP and Xie XS: Statistical analyses and theoretical models of single-molecule enzymatic dynamics. The Journal of Physical Chemistry A, 103(49), 10477–10488 (1999). DOI: 10.1021/jp992324j [DOI] [Google Scholar]
- 100.McKinney SA, Joo C and Ha T: Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J, 91(5), 1941–1951 (2006). DOI: 10.1529/biophysj.106.082487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Freeman SL, Martel A, Raven EL and Roberts GCK: Orchestrated domain movement in catalysis by cytochrome P450 reductase. Sci Rep, 7(1), 9741 (2017). DOI: 10.1038/s41598-017-09840-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Iyanagi T, Xia C and Kim J-JP: NADPH–cytochrome P450 oxidoreductase: Prototypic member of the diflavin reductase family. Arch Biochem Biophys, 528(1), 72–89 (2012). DOI: 10.1016/j.abb.2012.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Laursen T, Singha A, Rantzau N, Tutkus M, Borch J, Hedegård P, Stamou D, Møller BL and Hatzakis NS: Single molecule activity measurements of cytochrome P450 oxidoreductase reveal the existence of two discrete functional states. ACS Chem Biol, 9(3), 630–634 (2014). DOI: 10.1021/cb400708v [DOI] [PubMed] [Google Scholar]
- 104.Pommier Y and Marchand C: Interfacial inhibitors: Targeting macromolecular complexes. Nat Rev Drug Discov, 11(1), 25–36 (2012) DOI: 10.1038/nrd3404 [DOI] [PMC free article] [PubMed] [Google Scholar]


