Abstract
Introduction
Implantation failure after transferring morphologically “good‐quality” embryos in in vitro fertilization/intracytoplasmic sperm injection (IVF/ICSI) may be explained by impaired endometrial receptivity. Analyzing the endometrial transcriptome analysis may reveal the underlying processes and could help in guiding prognosis and using targeted interventions for infertility. This exploratory study investigated whether the endometrial transcriptome profile was associated with short‐term or long‐term implantation outcomes (ie success or failure).
Material and methods
Mid‐luteal phase endometrial biopsies of 107 infertile women with one full failed IVF/ICSI cycle, obtained within an endometrial scratching trial, were subjected to RNA‐sequencing and differentially expressed genes analysis with covariate adjustment (age, body mass index, luteinizing hormone [LH]‐day). Endometrial transcriptomes were compared between implantation failure and success groups in the short term (after the second fresh IVF/ICSI cycle) and long term (including all fresh and frozen cycles within 12 months). The short‐term analysis included 85/107 women (33 ongoing pregnancy vs 52 no pregnancy), excluding 22/107 women. The long‐term analysis included 46/107 women (23 ‘fertile’ group, ie infertile women with a live birth after ≤3 embryos transferred vs 23 recurrent implantation failure group, ie no live birth after ≥3 good quality embryos transferred), excluding 61/107 women not fitting these categories. As both analyses drew from the same pool of 107 samples, there was some sample overlap. Additionally, cell type enrichment scores and endometrial receptivity were analyzed, and an endometrial development pseudo‐timeline was constructed to estimate transcriptomic deviations from the optimum receptivity day (LH + 7), denoted as ΔWOI (window of implantation).
Results
There were no significantly differentially expressed genes between implantation failure and success groups in either the short‐term or long‐term analyses. Principal component analysis initially showed two clusters in the long‐term analysis, unrelated to clinical phenotype and no longer distinct following covariate adjustment. Cell type enrichment scores did not differ significantly between groups in both analyses. However, endometrial receptivity analysis demonstrated a potentially significant displacement of the WOI in the non‐pregnant group compared with the ongoing pregnant group in the short‐term analysis.
Conclusions
No distinct endometrial transcriptome profile was associated with either implantation failure or success in infertile women. However, there may be differences in the extent to which the WOI is displaced.
Keywords: assisted reproduction, endometrial receptivity, endometrium, gene expression, implantation failure, IVF/ICSI outcome
No distinct endometrial transcriptome profile associated with either implantation failure or success in infertile women in either the short term and long term was found, but there may be differences in the extent to which the window of implantation is displaced.
Abbreviations
- ART
assisted reproductive technology
- BMI
body mass index
- CPM
counts per million
- DEGs
differentially expressed genes
- ICSI
intracytoplasmic sperm injection
- IVF
in vitro fertilization
- LH
luteinizing hormone
- mRNA
messenger RNA
- OVGP1
oviductal glycoprotein 1
- PCA
principal component analysis
- PGT
preimplantation genetic testing
- pLH
pseudo‐timeline LH day
- RCT
randomized controlled trial
- RIF
recurrent implantation failure
- RIN
RNA Integrity Number
- RNA
ribonucleic acid
- RNA‐seq
RNA sequencing
- TPM
transcripts per million
- ΔLH
the difference between the pseudo‐timeline LH day and the actual LH day of the endometrial biopsy
- ΔWOI
the difference between the pseudo‐timeline LH day and the optimum endometrial receptivity day by beREADY (LH + 7)
Key message.
This exploratory study did not find a distinct endometrial transcriptome profile associated with either implantation failure or success in infertile women. However, our data may suggest differences in the extent to which the window of implantation is displaced.
1. INTRODUCTION
Approximately one in six couples is affected by infertility, 1 a condition characterized by the failure to establish a clinical pregnancy after 12 months of regular, unprotected sexual intercourse. 2 Around 20% of these couples eventually need assisted reproductive technology (ART) to address their infertility. 3 ART refers to the in vitro handling of both human oocytes and sperm and/or of embryos for the purpose of reproduction, including in vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI). 2 Although many advances have been made over the last four decades to improve IVF protocols, including improved stimulation regimens and embryo culture and selection, the success rate remains around 30% per cycle. 4 Despite the transfer of morphologically “good‐quality” embryos, implantation failure often occurs. 5 Critical for embryo implantation are both a competent blastocyst and a receptive endometrium. 6 Chromosomal abnormalities in preimplantation embryos presumably account for up to 70% of implantation failures, 7 leaving 30% that may be explained by impaired endometrial receptivity.
There are an increasing number of studies investigating molecular biomarkers for endometrial receptivity, since traditional histological dating is inaccurate in distinguishing between the endometrium of fertile and infertile women and suffers from a high interobserver variability. 8 , 9 To date, endometrial transcriptome analysis has been the most commonly applied method to investigate molecular biomarkers. 10 In addition, it may shed light on alterations in biological processes that could be responsible for the condition or provide potential therapeutic targets. 11 , 12 Thousands of potential endometrial receptivity biomarkers have already been identified in transcriptome studies, 10 and distinct gene expression profiles have been linked with either endometrial receptivity 13 , 14 , 15 , 16 , 17 , 18 or implantation failure. 19 , 20 , 21 Despite these efforts, there is poor concordance of biomarkers between studies, due to differences in experimental design and setting, sample size, selection criteria for study participants, timing of endometrial sampling, technology (eg microarray hybridization and sequencing methods), strategies for data processing and a lack of consistent standards for data presentation. 10 , 22
Nevertheless, distinct gene expression profiles, stratifying couples according to fertility prognosis, could be helpful in understanding the differences in prognosis based on underlying biology, as well as in optimizing assisted reproduction approaches, tailored to each couple's needs.
Therefore, this exploratory study describes the analysis of the endometrial transcriptome by RNA sequencing (RNA‐seq), using endometrial tissue of women who had one full failed IVF/ICSI cycle. Our objective was to investigate whether the endometrial transcriptome profile, assessed after a first full failed IVF/ICSI cycle, was associated with implantation outcome (ie success or failure) both in the short term and long term. In addition, we assessed whether endometrial receptivity profiles and cell type enrichment scored differed between these groups.
2. MATERIAL AND METHODS
2.1. Study population
Endometrial tissue, used for this exploratory substudy, was obtained from 141 infertile women participating in a previous randomized controlled trial (RCT) (ie the SCRaTCH trial) on endometrial scratching (ie biopsy), 23 who were allocated to endometrial scratching and consented to storage and future use of their tissue (see Figure 1 for the sample selection flow chart). All 141 participants underwent endometrial scratching just once during the study period. The full inclusion and exclusion criteria and study characteristics of the SCRaTCH trial have been described in detail elsewhere. 23 Briefly, women aged between 18 and 44 years, who had failed implantation after one full IVF/ICSI cycle (ie after transfer of fresh and all frozen embryos) and who were planning a new IVF/ICSI cycle, were eligible. Women undergoing oocyte donation cycles or pre‐implantation genetic testing were excluded. Women were followed‐up until 12 months after they had been randomized for endometrial scratching, or until delivery if an ongoing pregnancy was achieved within the 12‐month follow‐up period, either by spontaneous conception or resulting from ART treatments. The primary outcome of the SCRaTCH trial was live birth after a fresh embryo transfer during the second IVF/ICSI cycle (ie the first fresh cycle after randomization). Secondary outcomes included, among other outcomes, cumulative ongoing pregnancy leading to live birth within 12 months after randomization. Ongoing pregnancy was defined as a positive heartbeat on ultrasound at 10 weeks of gestational age, and live birth as the delivery of at least one live fetus after 24 weeks of gestation. 23
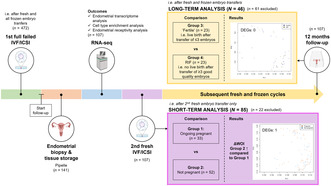
FIGURE 1.
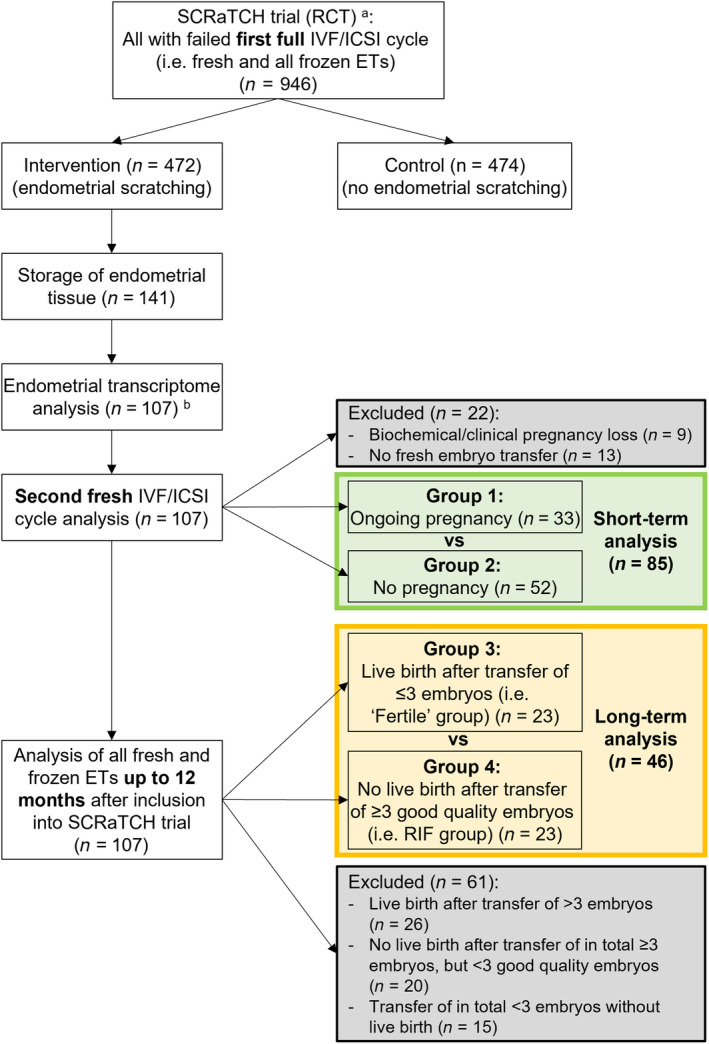
Flow chart of sample selection for the analyses of the current endometrial transcriptome study. In the short‐term analysis, we considered only the implantation outcome after the second fresh embryo transfer (ie the first fresh cycle following endometrial biopsy), whereas in the long‐term analysis, the outcome of all fresh and frozen embryo transfers within the 12‐month follow‐up period after enrollment were considered. Ongoing pregnancy was defined as a positive heartbeat on ultrasound at 10 weeks of gestational age, and no pregnancy as negative urinary or serum human chorionic gonadotropin levels 18 days after the ovum pickup. “Fertile” women were not fertile in the conventional sense but were infertile women who achieved a live birth through assisted reproductive technology after three or fewer embryos transferred. RIF was defined as no live birth after transfer of at least three good quality embryos as determined by morphology. Only the samples meeting these definitions were included in the respective analyses. ETs, embryo transfers; RCT, randomized controlled trial; RIF, recurrent implantation failure.aEndometrial biopsies (n = 141) were obtained from women undergoing endometrial scratching (biopsy) in the SCRaTCH trial, a randomized controlled trial on endometrial scratching in women with a first failed IVF/ICSI cycle (van Hoogenhuijze et al. Hum Reprod 2020;36:87–98). bOf the 141 biopsies, 107 samples had sufficient RNA quality to be sequenced and were used for further analysis.
2.2. Comparisons
As follow‐up data up to 12 months after randomization was available for all 141 participants of whom endometrial tissue was stored, we investigated whether the endometrial transcriptome was associated with implantation outcome (ie success or failure) both in the short term and long term (Figure 1). However, of 141 biopsies, 107 samples had sufficient RNA quality to be sequenced and were used for further analysis.
In the short‐term analysis, we considered only the implantation outcome after the second fresh embryo transfer (ie the first fresh embryo transfer following the endometrial biopsy), involving in total 85 women (Figure 1). Specifically, we compared the endometrial transcriptome profile of women with an ongoing pregnancy (Group 1, n = 33) with that of women with no pregnancy (Group 2, n = 52), that is, negative urinary or serum human chorionic gonadotropin (hCG) levels 18 days after the ovum pickup, after the first fresh embryo transfer in the IVF/ICSI cycle following the endometrial biopsy. All women who did not actually undergo a fresh embryo transfer during that cycle (n = 13) or had a biochemical or clinical pregnancy loss (n = 9), were excluded from this analysis. The latter were excluded as women with a pregnancy loss may have a different endometrial transcriptome profile from women who had not been able to get pregnant. 24
In the long‐term analysis, we considered the outcome of all IVF/ICSI cycles within the 12‐month follow‐up period after enrollment, involving 46 women in total (Figure 1). This includes outcomes after all fresh and frozen embryo transfer cycles. Specifically, we compared the endometrial transcriptome profile of women with implantation success after ART, hereafter referred to as “fertile” women (Group 3 n = 23), with that of women with recurrent implantation failure (RIF) (Group 4, n = 23). These samples also had been included in the short‐term analysis. Both groups are defined below. Only samples meeting these definitions were included in the long‐term analysis, resulting in the exclusion of 61 samples that did not fulfill the definition of either group (Figure 1).
We defined RIF as no live birth after transfer of at least in total three good quality embryos regardless of whether they were involved in single or double embryo transfer cycles. Embryo quality was determined by morphology. For day two, day three and day four embryos, an embryo was considered good quality when the following morphological features were present: being at a 4‐cell stage for a day two embryo, being at a ≥7‐cell stage with less than 20% fragmentation for a day three embryo and being at least at the morula stage for a day four embryo. For day five and day six embryos (ie blastocysts), the Gardner and Schoolcraft system was used. 25
We considered women ‘fertile” if they had a live birth after transfer of in total three or fewer embryos, in which the quality of the embryos was not considered, as implantation was successful. These “fertile” women were thus not fertile in the conventional sense, but were infertile women who achieved a pregnancy through ART. The criterion of fewer than three embryos was determined arbitrarily, considering the number of transferred embryos generally used in the definition of RIF. 26 , 27
2.3. Study outcomes
The primary outcome of the current substudy was the assessment of differences in the endometrial transcriptome profile between implantation success and failure groups, as defined above, by differential gene expression analysis of RNA‐seq data. Secondary outcomes included the analysis of cell type enrichment scores and endometrial receptivity gene profiles, along with the pseudo‐timeline based estimations of the deviation of the window of implantation and luteinizing hormone (LH) day, using the endometrial transcriptome data. The methods used to study these outcomes will be discussed in detail below.
2.4. Tissue sampling and storage
Endometrial tissue was obtained and stored in six hospitals in The Netherlands (Table S1). All hospitals followed the same strict protocol for endometrial biopsy and tissue storage as previously reported, 28 which is described below.
Endometrial biopsy was performed in the mid‐luteal phase of a natural cycle (5–8 days after detection of the LH surge by urinary tests) prior to the second fresh IVF/ICSI cycle, using an endometrial biopsy catheter (eg Pipelle or other similar catheters).
To limit contamination of the samples with RNases as much as possible, a sterile gown, sterile gloves and a hair cap were worn by the physician who performed the endometrial biopsy. A sterile work field was created and all instruments and materials were unpacked from their sterile casing right before the procedure. The endometrial biopsy was performed as follows: after insertion of a speculum, the cervix was cleaned with sterile water. The catheter was introduced through the cervix up to the uterine fundus. The piston of the catheter was completely drawn back to create a vacuum, and the catheter was slowly retracted within 1–2 minutes while constantly rotating 360°. After the procedure, the physician changed gloves for new sterile gloves and divided the tissue over three tissue tubes (Brooks Life Sciences, USA) using a sterile scalpel and forceps. Within 3 minutes after taking the biopsy, the tubes were snap‐frozen in liquid nitrogen and stored at −80°C as soon as possible. Tissue samples from all hospitals were transported on dry ice to the Central Biobank of the University Medical Center Utrecht for central storage.
2.5. Total RNA extraction
RNA extraction was performed by the Utrecht Sequencing Facility (USEQ) (Utrecht, The Netherlands). Tissue was thawed and homogenized using TissueLyser II (Qiagen, Germany) after adding 50 μL nuclease‐free water and stainless steel beads (Ø 5 mm, Qiagen). Each sample was incubated in 350 μL RLT Plus lysis buffer (Qiagen). Total RNA extraction, including a DNase treatment, was performed using the QIAsymphony SP and the QIAsymphony RNA Kit (Qiagen) following the manufacturer's protocol. RNA was quantified using the Qubit™ RNA BR Assay Kit (Thermo Fisher Scientific, USA) and the Qubit 4 Fluorometer (Thermo Fisher Scientific). RNA Integrity Number (RIN) was determined using the Agilent RNA 6000 Nano Kit (Agilent Technologies, USA) and the Agilent 2100 Bioanalyzer system (Agilent Technologies).
2.6. RNA sequencing
RNA‐seq was performed by USEQ. Endometrial samples with an RIN of ≥6.0 were used for library preparation with an input of 100 ng of total RNA. Libraries were prepared using the TruSeq Stranded mRNA Kit (Illumina, USA) following the manufacturer's protocol with xGen Dual Index UMI Adapters (Integrated DNA Technologies [IDT], USA) for indexing. Samples with a final library concentration of ≥1 ng/μL were further processed for sequencing. Libraries were pooled equimolarly and paired‐end (2 × 50 base pairs) sequenced on a NovaSeq 6000 S2 flow cell (Illumina).
Of 141 endometrial tissue samples, 107 samples (75.9%) had sufficient RNA quality to be sequenced. Substandard samples (n = 34) were not sequenced due to the following reasons: too low RIN (n = 24), too low RNA concentration (n = 2) or too low mRNA yield after poly(A) enrichment (n = 8).
2.7. RNA‐seq data analysis
Two independent analysts performed data analysis in parallel to check for inconsistencies.
2.7.1. Preprocessing
The dataset was handled in two ways: with and without preprocessing (trimming and quality filtering) before mapping. First, the raw reads were quality checked with FastQC tool version 0.11.9. 29 Raw fastqc files were then trimmed with TRIMMOMATIC 0.39. 30 For quality filtering, fastq_quality_filter from FASTX TOOLKIT 0.0.13 was used. 31 Both the preprocessed dataset and the dataset without preprocessing showed a high number of duplicate reads (Figure S1).
2.7.2. Alignment
Both datasets (preprocessed and not preprocessed) were mapped to human genome version 38 (GRCh38) 32 with STAR v2.7.0e. 33 The proportion of uniquely mapped reads for both datasets varied from 66% to 86%. The dataset without preprocessing had slightly more uniquely mapped reads than the preprocessed dataset. Therefore, the dataset without preprocessing was used thereafter. The datasets had a very high number of reads mapped to the mitochondrial chromosome (chrM) (Figure S2). This could explain the high number of duplicate reads in the quality control step.
A sensitivity analysis, excluding the mitochondrial transcripts, did not result in a significantly different number of differentially expressed genes (DEGs) when comparing gene expression data of the groups (data not shown). Additionally, we could not determine whether the high number of mitochondrial transcripts was due to technical issues (eg poor sample quality with a high fraction of apoptotic or lysing cells) or tissue biology. Therefore, we used the dataset with mitochondrial transcripts for our analyses.
Raw gene counts were generated with STAR during mapping with default parameters that were the same as HTSeq‐count—“union” mode. The forward‐stranded counts were used for further analysis.
2.7.3. Quality control
Mapping stats were collected with PICARD TOOLS 2.0.1. 34 To aggregate all results of our samples together, we used MULTIQC TOOL v1.9. 35
2.7.4. Normalization, differential gene expression analysis and covariates
Normalization and differential gene expression analysis were conducted using R v3.6.0 package edgeR v3.28.1. 36 , 37 , 38 , 39 To visualize the high‐dimensional transcriptome data, principal component analysis (PCA) was performed based on logarithm‐transformed counts per million (CPM) normalized counts (logCPMs). Initially, several factors, such as age, body mass index (BMI), biopsy center, smoking, LH‐timed biopsy day, infertility diagnosis, infertility type and number of previous failed embryo transfers were included in our data analysis as covariates (results not shown). However, in our final model, the covariates were narrowed down to age, BMI and LH‐timed biopsy day, as the outcomes of our analysis were comparable when limited to these factors. In addition, these three factors have previously been identified as potential confounding variables in the context of endometrial gene expression. 40 , 41 , 42 DEGs with a false discovery rate of <0.05 were considered significant.
2.8. Enrichment analysis
We intended to subject significantly DEGs to enrichment analysis to annotate these genes to functional gene sets and pathways. However, as there were fewer than two DEGs in all analyses, we did not perform enrichment analysis.
2.9. Analysis of receptivity gene profiles
Individual receptivity profiles of endometrial biopsies were analyzed with the beREADY receptivity dating model. 43 Simulating the original targeted RNA‐seq method, 68 biomarkers with four housekeeping genes were selected from the quantified whole‐genome sequencing data. 44 This filtered gene set was then further analyzed with the beREADY model. Briefly, biomarker genes were normalized using the geometric mean of the housekeeping genes. After clustering, distances to the reference receptivity groups were calculated. The two most similar receptivity profiles were found with a one‐vs‐rest exclusion method, and the input‐sample's relative probabilities for either group were output.
The beREADY receptivity test was applied to the transcriptome data of all our sequenced endometrium samples to determine the proportion of samples classified as “pre‐receptive”, “receptive” or “post‐receptive”. In addition, beREADY distinguished two transitionary groups as subgroups of the “receptive” classification, that is, “early‐receptive” and “late‐receptive”.
Due to the linear nature of the development of the endometrium as a tissue, these relative probabilities were interpreted as the transition of the endometrium from one phase to another. Based on this interpretation, we constructed an endometrial development pseudo‐timeline by finding the location of the input sample on a scale from −1 (pre‐receptive) to +1 (post‐receptive), based on endometrial gene expression data from reference samples taken on LH + 2/3, LH + 7/8 and LH + 11/12. 43 Next, we divided this pseudo‐timeline into 10 equal bins, representing LH days ranging from LH + 2 to LH + 12 of the pre‐receptive and the post‐receptive reference samples, respectively. We then estimated the location of our study samples on the pseudo‐timeline by comparing the receptivity profile of these samples to those of the reference samples. To estimate more specifically the extent of deviation from the receptive period on the pseudo‐timeline (ie where the reference samples taken on LH + 7 are located, as LH + 7 is considered the optimum endometrial receptivity day by beREADY), rather than just classifying the receptivity profile of a sample, we calculated the absolute difference between the estimated LH day of the study sample on the transcriptional pseudo‐timeline (pLH) and LH + 7 (Figure 2). This difference was termed ΔWOI and was expressed in number of days. Thus, the larger the ΔWOI, the greater the deviation from the most receptive period (LH + 7), and can express either a delay or an advancement in endometrial development. We also assessed the difference between the estimated pLH of each study sample and the actual LH day of the biopsy, by calculating the absolute difference between the two, which we termed ΔLH and which was also expressed in number of days (Figure 2). Thus, the larger the ΔLH, the greater the deviation between the actual LH day of the biopsy and the pLH estimated by the beREADY model.
FIGURE 2.
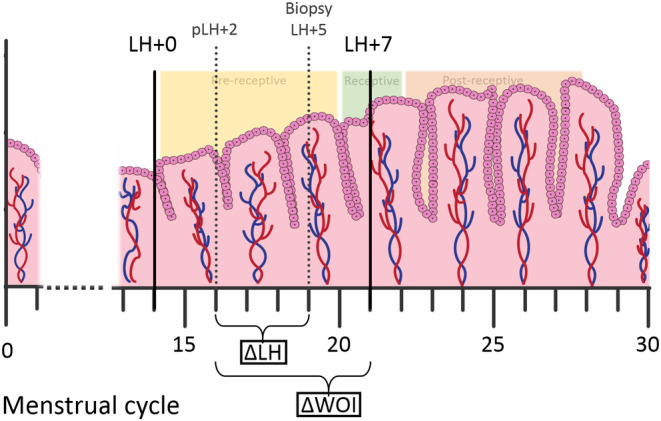
Visualization of ΔWOI and ΔLH. Based on the beREADY receptivity profiles of endometrium reference samples taken between LH + 2 and LH + 12 (Teder et al. NPJ Genom Med 2018;3:34. doi: 10.1038/s41525‐018‐0072‐5), a pseudo‐timeline was developed and divided into bins representing pseudo‐LH days from the mid‐cyclic LH‐surge (LH + 0). ΔWOI is calculated as the absolute difference in number of days between the estimated transcriptome‐based pseudo‐timeline LH day of the study sample (pLH) and LH + 7, which is considered the optimum endometrial receptivity day by beREADY. ΔLH is calculated as the absolute difference in number of days between pLH and the LH day the biopsy was taken on (Biopsy LH). In this example, the biopsy was taken on LH + 5, but the transcriptomic profile matches that of LH + 2 on the pseudo‐timeline (pLH + 2). Thus, in this case, the ΔWOI is 5 and the ΔLH is 3.
2.10. Analysis of tissue cellular heterogeneity
As our transcriptome data was generated by bulk RNA‐seq, which does not provide any information on cell type‐specific gene expression, we characterized the cellular signatures of the endometrial biopsies using xCell. xCell is a signature‐based method in which signature genes are collected from diverse pure cell‐type transcriptomes from different sources and gene expression data is converted to cell type enrichment scores. 45 We used the transcripts per million normalized gene counts as input to xCell and ran the deconvolution algorithm with additional expression data from GTEx (see GTEx 46 (diverse types of tissues) and GSE86491 and GSE119209 47 , 48 (both endometrium)) as the program performs better with heterogeneous datasets.
2.11. Statistical analyses
Continuous data of two independent groups were statistically compared using the independent t‐test in the case of normally distributed data, and the Wilcoxon rank sum test in the case of non‐normally distributed data. Categorical data were compared across groups using the Chi‐square test or Fisher's exact test when the expected frequency was <5.
For statistical comparison of the mean and median ΔLH and ΔWOI measurements between two groups, we used both a one‐sided and a two‐sided Wilcoxon rank sum test. There was no difference in our null hypothesis in mean and median ΔLH and ΔWOI between our implantation failure and implantation success groups, either in the short term or long term. Based on the results of a previous study, 43 we tested our main alternative hypothesis of a larger ΔLH and ΔWOI in the implantation failure groups compared with the implantation success groups, using the one‐sided Wilcoxon rank sum test. However, as there was a possibility that the difference in ΔLH and ΔWOI would be bidirectional (ie either smaller or larger) between the groups, we also tested the alternative hypothesis of a different ΔLH and ΔWOI, using the two‐sided Wilcoxon rank sum test.
A P‐value of <0.05 was considered to indicate a statistically significant difference. Statistical analysis was performed using SPSS Statistics v26 (IBM Corporation, USA) and R (v4.2.1).
3. RESULTS
3.1. Demographics
The baseline characteristics of the participants of the 107 sequenced endometrium samples are shown in Table S2; they did not differ from the characteristics of the participants of the initial 141 endometrium samples (Table S3).
3.2. Short‐term analysis: ongoing pregnancy (Group 1) vs no pregnancy (Group 2) after the first fresh embryo transfer in the IVF/ICSI cycle following endometrial biopsy
-
a
Clinical characteristics
The clinical baseline and cycle characteristics of the ongoing pregnancy (Group 1) and no pregnancy groups (Group 2) are shown in Table 1. A trend of a higher mean age was seen in Group 2 than in Group 1 (respectively 35.8 vs 33.9 years) but this difference was not statistically significant (P = 0.050; Table 1). None of the other characteristics differed between the groups.
-
b
Differential gene expression analysis
TABLE 1.
Baseline characteristics of ongoing pregnancy vs no pregnancy after the first fresh embryo transfer in the IVF/ICSI cycle following endometrial biopsy.
| Ongoing pregnancy (n = 33) | No pregnancy (n = 52) | P‐value | |
|---|---|---|---|
| Baseline characteristics | |||
| Female age, years | 33.9 (±4.2) | 35.8 (±4.5) | 0.050 |
| Female age groups | 0.146 | ||
| <35 years | 18 (54.5%) | 20 (38.5%) | |
| ≥35 years | 15 (45.5%) | 32 (61.5%) | |
| Female BMI, kg/m2 a | 23.1 (21.4–25.5) | 24.1 (21.7–26.8) | 0.223 |
| Duration of infertility, months | 34.0 (21.0–44.0) | 29.5 (23.0–43.0) | 0.836 |
| Female smokers a | 4 (12.1%) | 5 (9.6%) | 0.733 |
| Type of infertility of the female b | 0.069 | ||
| Primary | 20 (60.6%) | 21 (40.4%) | |
| Secondary | 13 (39.4%) | 31 (59.6%) | |
| Cause of infertility c | 0.339 | ||
| Idiopathic | 15 (45.5%) | 20 (38.5%) | |
| Male factor | 18 (54.5%) | 24 (46.2%) | |
| Tubal factor | 0 | 5 (9.6%) | |
| Endometriosis grade I/II | 0 | 1 (1.9%) | |
| Ovulatory disorder d | 0 | 1 (1.9%) | |
| Mixed diagnosis e | 0 | 1 (1.9%) | |
| No. of ETs prior to randomization per participant | 2.5 (±1.5) | 2.2 (±1.4) | 0.531 |
| Biopsy taken on LH+ days | 0.064 | ||
| 5 | 4 (15.2%) | 5 (9.6%) | |
| 6 | 14 (42.4%) | 17 (32.7%) | |
| 7 | 10 (30.3%) | 12 (23.1%) | |
| 7.5 | 1 (3.0%) | 0 | |
| 8 | 2 (6.1%) | 15 (28.8%) | |
| Missing | 2 (6.1%) | 3 (5.8%) | |
| Cycle characteristics | |||
| Treatment type | 0.259 | ||
| IVF | 13 (39.4%) | 27 (51.9%) | |
| ICSI | 20 (60.6%) | 25 (48.1%) | |
| Downregulation protocol a | 0.697 | ||
| Antagonist | 5 (15.2%) | 5 (9.6%) | |
| Agonist | 28 (84.8%) | 46 (88.5%) | |
| Starting dose FSH, IU | 150 (150–225) | 225 (150–225) | 0.062 |
| Total dose FSH, IU | 1800 (1350–2362.5) | 2025 (1500–2475) | 0.278 |
| Duration of stimulation (days) | 11 (9–12) | 10 (9–12) | 0.606 |
| No. of oocytes | 10 (5–14) | 7 (5–12) | 0.431 |
| ET performed on day | 1.000 | ||
| 2 | 3 (9.1%) | 4 (7.7%) | |
| 3 | 30 (90.9%) | 48 (92.3%) | |
| No. of embryos transferred | 1.000 | ||
| SET | 28 (84.8%) | 45 (86.5%) | |
| DET | 5 (15.2%) | 7 (13.5%) | |
| Total no. of embryos transferred | 38 | 59 | |
| Embryo quality f | 1.000 | ||
| Good | 31 (81.6%) | 47 (79.7%) | |
| Low/moderate | 7 (18.4%) | 12 (20.3%) | |
Note: Data are presented as mean (± SD), median (IQR) or number (%).
Abbreviations: BMI, body mass index; DET, double embryo transfer; ET, embryo transfer; FSH, follicle stimulating hormone; ICSI, intracytoplasmic sperm injection; IU, international units; IVF, in vitro fertilization; SET, single embryo transfer.
Data was missing for one participant in the No pregnancy group.
Primary: female has never conceived before. Secondary: female has conceived before.
No specific infertility diagnoses were excluded.
The woman in the No pregnancy group with an ovulatory disorder had a WHO group III ovulatory disorder.
The woman in the No pregnancy group with a mixed infertility diagnosis had both a male factor infertility and a WHO group II ovulatory disorder (unknown whether polycystic ovary syndrome).
Based on morphology of the embryo on the day of the embryo transfer.
The PCA did not reveal distinct separation of the groups (Figure 3A). Differential gene expression analysis resulted in only one significantly upregulated gene (Oviductal Glycoprotein 1: OVGP1) in Group 2 vs Group 1, and no significantly downregulated genes. This gene encodes a large, carbohydrate‐rich epithelial glycoprotein that is secreted from non‐ciliated oviductal epithelial cells and that associates with ovulated oocytes, blastomeres and spermatozoan acrosomal regions. 49 Adjustment of the gene expression data for age, BMI and LH‐timed biopsy day as covariates, resulted in slightly better clustering of the samples (Figure 3B). However, no distinct clustering was seen based on the clinical outcome (ie Group 1 vs Group 2). After covariate adjustment, there were no significantly DEGs in Group 2 vs Group 1, suggesting that the covariates highly influenced OVGP1 gene expression.
-
c
Receptivity gene profile
FIGURE 3.
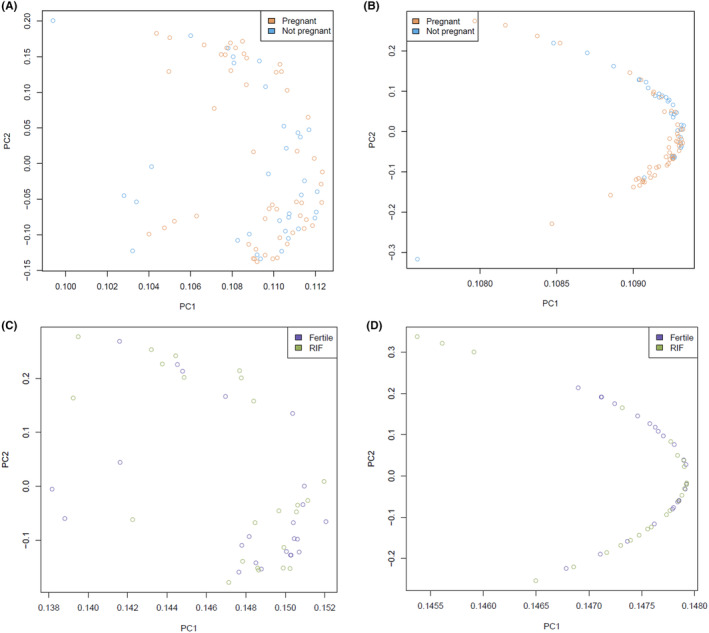
Principal component analysis (PCA) of the RNA‐sequencing data of the endometrium samples. Plots displaying the samples of the short‐term (A,B) and long‐term (C,D) analysis groups, before (A,C) and after (B,D) adjustment of the gene expression data for age, body mass index (BMI) and LH‐timed biopsy day as covariates. Two distinct clusters of samples were seen in the long‐term analysis before adjustment of the data (C), which were not explained by the clinical phenotype of the samples, whereas in the short‐term analysis no distinct clusters were seen (A). After adjustment of the data, samples clustered more in a crescent shape in both the short‐term and long‐term analyses (B,D), but no distinct clusters were present.
There were no statistically significant differences in the proportions of receptivity gene profile classifications between both groups (Table 2). Furthermore, the difference between the pseudo‐timeline LH day (pLH; based on the application of the beREADY receptivity test to the transcriptome data) and the actual LH day of the biopsy (ΔLH) or the optimal receptivity day of LH + 7 (ΔWOI) were determined for the samples of both groups. We detected a significantly larger mean and median ΔWOI in Group 2 than in Group 1 (P = 0.033; Figure S3A; Table 2) when using a one‐sided Wilcoxon rank sum test, whereas with a two‐sided test we did not find a statistically significant difference. There were no statistically significant differences in ΔLH between the two groups (Figure S3B; Table 2).
-
d
Cell type enrichment analysis
TABLE 2.
Receptivity profile analysis of samples of women with ongoing pregnancy vs no pregnancy after the first fresh embryo transfer in the IVF/ICSI cycle following endometrial biopsy.
| Ongoing pregnancy (n = 33) | No pregnancy (n = 52) | P‐value | ||
|---|---|---|---|---|
| Receptivity profile | 0.236 | |||
| Pre‐receptive | 11 (33.3%) | 15 (28.8%) | ||
| Early‐receptive | 1 (3.0%) | 8 (15.4%) | ||
| Receptive | 20 (60.6%) | 27 (51.9%) | ||
| Late‐receptive | 0 | 1 (1.9%) | ||
| Post‐receptive | 0 | 1 (1.9%) | ||
| Not classifiable | 1 (3.0%) | 0 | ||
| Median ΔWOI a | 2.0 (0–3.0) | 3.0 (1.0–4.0) | 0.082 b | 0.033 c |
| Mean ΔWOI a | 1.8 (±1.5) | 2.4 (±1.4) | ||
| ΔWOI a | 0.472 | |||
| 0 | 8 (25.8%) | 7 (14.3%) | ||
| 1 | 6 (19.4%) | 6 (12.2%) | ||
| 2 | 6 (19.4%) | 9 (18.4%) | ||
| 3 | 5 (16.1%) | 13 (26.5%) | ||
| 4 | 6 (19.4%) | 14 (28.6%) | ||
| Median ΔLH a | 2.0 (1.0–2.0) | 2.0 (1.0–3.0) | 0.143 b | 0.200 c |
| Mean ΔLH a | 1.7 (±1.1) | 2.0 (±1.0) | ||
| ΔLH a | 0.603 | |||
| 0 | 4 (12.9%) | 3 (6.1%) | ||
| 1 | 11 (35.5%) | 12 (24.5%) | ||
| 2 | 9 (29.0%) | 19 (38.8%) | ||
| 3 | 5 (16.1%) | 11 (22.4%) | ||
| 4 | 2 (6.5%) | 4 (8.2%) | ||
Note: Data are presented as mean (± SD), median (IQR) or number (%).
The difference between the pseudo‐timeline LH day and the actual LH day (ΔLH) or LH + 7 (ΔWOI). Due to missing data on actual LH day, two samples in the Ongoing pregnancy group and three samples in the No pregnancy group were excluded, leaving 31 and 49 samples, respectively.
Two‐sided Wilcoxon rank sum test.
One‐sided Wilcoxon rank sum test.
To check whether differences in cell type composition of the endometrial biopsies could have influenced the data, we estimated cell type enrichment scores based on the transcriptome data. Overall, the endometrium samples in both groups displayed the highest cell type enrichment values for smooth muscle cells (0.54–0.56), followed by epithelial cells (0.44–0.45) (Figure S4A; Table S4). There were no significant differences in the cell type enrichment scores between the two groups (Figure S4A,B; Table S4).
3.3. Long‐term analysis: fertile group (Group 3) vs recurrent implantation failure (RIF) group (Group 4)
Clinical characteristics
The clinical and cycle characteristics of the participants in the fertile group (Group 3) and the RIF group (Group 4) are shown in Table 3 and Table S5. Besides a significant difference in the number of embryo transfers per participant, there was also a statistically significant difference in the days on which the embryo transfers were performed (P < 0.001), the number of fresh and frozen‐thawed embryo transfers (P < 0.001) and the embryonic stage of transferred embryos (P = 0.005) between the groups (Table 3). A higher proportion of day three embryo transfers were performed in Group than Group 4 (respectively 85.0% vs 51.0%), and a higher proportion of day four and day five embryo transfers were performed in Group 4 than Group 3 (respectively 27.5% vs 8.3% for day four, and 19.0% vs 6.7% for day five) (Table 3). More fresh embryo transfers were performed in Group 3 than Group 4, (respectively 86.7% vs 48.4%), whereas in Group 4, more frozen‐thawed embryo transfers were performed compared with Group 3 (respectively 51.6% vs 13.3%) (Table 3). We also noted a higher proportion of cleavage stage embryos transferred in Group 3 (respectively 91.7% vs 71.2%) and a higher proportion of morulas and blastocysts transferred in Group 4 (respectively 9.2% vs 1.7% and 19.6% vs 6.7%) (Table 3).
-
b
Differential gene expression analysis
TABLE 3.
Clinical characteristics of the fertile group and recurrent implantation failure (RIF) group.
| Fertile (n = 23) | RIF (n = 23) | P‐value | |
|---|---|---|---|
| Female age, years | 34.2 (± 4.4) | 36.3 (± 3.9) | 0.095 |
| Female age groups | 0.074 | ||
| <35 years | 13 (56.5%) | 7 (30.4%) | |
| ≥35 years | 10 (43.5%) | 16 (69.6%) | |
| Female BMI, kg/m2 | 23.0 (20.7–25.9) | 23.7 (21.6–28.3) | 0.560 |
| Duration of infertility, months | 26.0 (20.0–38.0) | 29.0 (23.0–39.0) | 0.403 |
| Female smokers a | 1 (4.3%) | 1 (4.3%) | 1.000 |
| Type of infertility of the female b | 0.552 | ||
| Primary | 14 (60.9%) | 12 (52.2%) | |
| Secondary | 9 (39.1%) | 11 (47.8%) | |
| Cause of infertility c | 1.000 | ||
| Idiopathic | 9 (39.1%) | 8 (34.8%) | |
| Male factor | 13 (56.5%) | 12 (52.2%) | |
| Tubal factor | 1 (4.3%) | 2 (8.7%) | |
| Ovulatory disorder d | 0 | 1 (4.3%) | |
| No. of embryo transfers per participant | 2.4 (±0.5) | 5.7 (±2.5) | <0.001 |
| 1 | 15 (65.2%) | 0 | |
| 2 | 8 (34.8%) | 0 | |
| 3 | 0 | 5 (21.7%) | |
| 4 | 0 | 4 (17.4%) | |
| 5 | 0 | 3 (13.0%) | |
| 6 | 0 | 4 (17.4%) | |
| 7 | 0 | 1 (4.3%) | |
| 8 | 0 | 3 (13.0%) | |
| 9 | 0 | 2 (8.7%) | |
| 13 | 0 | 1 (4.3%) | |
| Total no. of embryos transferred, all participants | 60 | 153 | |
| No. of SET and DET | 0.379 | ||
| SET | 46 (86.8%) | 105 (81.4%) | |
| DET | 7 (13.2%) | 24 (18.6%) | |
| ET performed on day | <0.001 | ||
| 2 | 0 | 2 (1.3%) | |
| 3 | 51 (85.0%) | 78 (51.0%) | |
| 4 | 5 (8.3%) | 42 (27.5%) | |
| 5 | 4 (6.7%) | 29 (19.0%) | |
| 6 | 0 | 2 (1.3%) | |
| No. of fresh and frozen‐thawed ETs | <0.001 | ||
| Fresh | 52 (86.7%) | 74 (48.4%) | |
| Frozen‐thawed | 8 (13.3%) | 79 (51.6%) | |
| Embryonic stage | 0.005 | ||
| Cleavage stage embryo | 55 (91.7%) | 109 (71.2%) | |
| Morula | 1 (1.7%) | 14 (9.2%) | |
| Blastocyst | 4 (6.7%) | 30 (19.6%) | |
Note: Data are presented as mean (± SD), median (IQR) or number (%). The P‐value indicating the difference between mean number of embryo transfers per participant was determined by Independent Samples T‐test. All other P‐values were determined by Chi square test.
Abbreviations: BMI, body mass index; DET, double embryo transfer; ET, embryo transfer; SET, single embryo transfer.
Data was missing for one participant in the RIF group.
Primary: female has never conceived before. Secondary: female has conceived before.
No specific infertility diagnoses were excluded.
The woman in the RIF group with an ovulatory disorder had a WHO group III ovulatory disorder.
Differential gene expression analysis resulted in no significantly DEGs in Group 4 compared with Group 3. The PCA revealed two distinct groups that were not explained by clinical phenotype (ie RIF vs fertile), as samples of both groups appeared evenly in the two clusters (Figure 3C). After adjustment of the transcriptome data for age, BMI and LH‐timed biopsy day as covariates, the two clusters were no longer distinct, but samples of both groups remained intermingled in the PCA plot, with no significantly DEGs in Group 4 vs Group 3 (Figure 3D).
-
c
Receptivity gene profile
There were no statistically significant differences in the proportions of different receptivity gene profile classifications between the both groups (Table 4). The mean, median and proportions of ΔLH and ΔWOI were comparable between both groups (Figure S3A,B; Table 4).
-
d
Cell type enrichment analysis
TABLE 4.
Receptivity profile analysis of samples of fertile women vs women with recurrent implantation failure (RIF).
| Fertile (n = 23) | RIF (n = 23) | P‐value | ||
|---|---|---|---|---|
| Receptivity profile | 0.377 | |||
| Pre‐receptive | 11 (47.8%) | 6 (26.1%) | ||
| Early‐receptive | 2 (8.7%) | 4 (17.4%) | ||
| Receptive | 10 (43.5%) | 11 (47.8%) | ||
| Late‐receptive | 0 | 1 (4.3%) | ||
| Post‐receptive | 0 | 1 (4.3%) | ||
| Median ΔWOI a | 3.0 (1.0–4.0) | 3.0 (1.0–4.0) | 0.894 b | 0.584 c |
| Mean ΔWOI a | 2.4 (±1.5) | 2.4 (±1.6) | ||
| ΔWOI a | 1.000 | |||
| 0 | 3 (13.6%) | 4 (18.2%) | ||
| 1 | 5 (22.7%) | 4 (18.2%) | ||
| 2 | 2 (9.1%) | 1 (4.5%) | ||
| 3 | 5 (22.7%) | 5 (22.7%) | ||
| 4 | 7 (31.8%) | 8 (36.4%) | ||
| Median ΔLH a | 2.0 (1.0–3.0) | 2.0 (1.0–3.0) | 0.577 b | 0.502 c |
| Mean ΔLH a | 2.0 (±1.1) | 2.2 (±1.3) | ||
| ΔLH a | 0.767 | |||
| 0 | 2 (4.6%) | 1 (4.5%) | ||
| 1 | 5 (27.3%) | 7 (31.8%) | ||
| 2 | 9 (40.9%) | 5 (22.7%) | ||
| 3 | 4 (18.2%) | 5 (22.7%) | ||
| 4 | 2 (9.1%) | 3 (13.6%) | ||
| 5 | 0 | 1 (4.5%) | ||
Note: Data are presented as mean (± SD), median (IQR) or number (%).
The difference between the pseudo‐timeline LH day and the actual LH day (ΔLH) or LH + 7 (ΔWOI). Due to missing data on actual LH day, one sample in each group was excluded, leaving 22 samples in both groups.
Two‐sided Wilcoxon rank sum test.
One‐sided Wilcoxon rank sum test.
Cell type enrichment score analysis showed that samples of both groups had the highest enrichment values for smooth muscle cells (0.54–0.55) and epithelial cells (0.43–0.45) (Figure S4C; Table S6). There were no significant differences in the cell type enrichment scores between the samples of the two groups (Figure S4C,D; Table S6).
4. DISCUSSION
This study compared the RNA‐seq endometrial transcriptome profiles of women with at least one full failed IVF/ICSI cycle, who either had implantation success or implantation failure in the short and long term. Differential gene expression analysis did not result in any important differences in the endometrial transcriptome profile of implantation failure and success groups, either in the short or long term, indicating no association of an endometrial transcriptome profile with implantation outcome. Moreover, we did not observe a higher proportion of a receptive profile among samples of women with implantation success compared with samples of women with implantation failure, either in the short term or long term. We also determined the difference between the transcriptome data‐based pseudo‐timeline LH day (pLH) and the actual LH day of the biopsies, termed as ΔLH, as well as the difference between pLH and LH + 7, termed ΔWOI. Although we did see a significantly larger mean and median ΔWOI in the implantation failure group than in the implantation success group in the short term with a one‐sided test, there was no significant difference between both groups when using a two‐sided test. In the long term, no differences were seen in either ΔLH or ΔWOI between both groups. Lastly, cell type enrichment analysis showed that our endometrial biopsies were mostly enriched for smooth muscle cell and epithelial cell gene signatures. No differences in cell type enrichment scores were seen between the implantation failure and success groups in either the short‐term or long‐term analyses.
Previous studies comparing the endometrial transcriptome of women with (recurrent) implantation failure with those of women with implantation success reported a considerably larger amount of significantly DEGs compared with our study. 20 , 21 , 50 , 51 , 52 , 53 , 54 , 55 , 56 , 57 , 58 , 59 , 60 Although quite some heterogeneity in DEGs and enriched pathways was observed across these studies, annotations of DEGs to processes involving the immune system and cellular functions occurred in the majority of the studies.
The difference in the number of significantly DEGs between our study and previous studies can most likely be attributed to differences in study population selection criteria, as well as differences in study methodology, such as the timing of endometrial biopsies (eg natural cycle vs artificial or stimulated cycle, and differences in LH timing when performed during the mid‐luteal phase of a natural cycle), sequencing methods (eg RNA‐seq vs microarray hybridization) and data processing and analysis. For instance, there is no consensus on the definition of RIF, 26 which contributes to substantial heterogeneity across studies. Pirtea et al. reported that RIF after three euploid embryo transfers is actually rare (<5%), suggesting that ART failures mainly result from embryonic factors. 61 In our study, we assessed embryo quality based on morphologic characteristics to determine the RIF group and did not perform preimplantation genetic testing for aneuploidy (PGT‐A). Therefore, it is likely that we included women with aneuploid embryo transfers in the RIF group, which may have affected implantation chances and therefore, the selection of the study participants for the comparisons in our study. However, it is worth noting that even with PGT‐A, embryonic mosaicism can complicate the interpretation of the test results. 62 , 63 , 64
Our study showed that endometrial gene expression changed after covariate adjustment (ie age, BMI and LH‐timed biopsy day) and that samples clustered better. This finding supports that these factors can influence endometrial gene expression data, as also observed in previous studies, 40 , 41 , 42 and should be taken into consideration when attempting to identify true biological differences between implantation failure and success groups.
When considering the causes of infertility among the participants in our study, a relatively large proportion had male factor infertility (46.7%), followed by idiopathic infertility (38.3%) (Table S2). Notably, an endometrial factor as cause of the infertility is less likely in male factor infertility compared with idiopathic infertility, potentially introducing heterogeneity into our endometrial transcriptome findings. However, the proportions of male factor infertility were comparable between the implantation failure and success groups (Tables 1 and 3). Consequently, differences in the endometrial transcriptome between these groups are less likely attributable to differences in the underlying causes of infertility.
A relatively large percentage (13.1%) of our total study population were smokers (Table S2), though similar incidences were previously reported among women undergoing ART treatment. 65 Cigarette smoking has been widely associated with poor reproductive outcome. However, the effect of female smoking on ART outcome remains unclear, as it is difficult to study its precise effect due to multiple confounding factors, resulting in conflicting results from previous retrospective observational studies. 65 , 66 , 67 To our knowledge, it is unknown whether smoking affects the endometrial transcriptome. In our study, we did not observe differences in the proportions of female smokers between the comparison groups (Tables 1 and 3), therefore it is unlikely that smoking would have resulted in differences in the endometrial transcriptome profile between groups.
Furthermore, we noted a difference in the days on which the embryo transfers were performed, the number of fresh and frozen‐thawed embryo transfers and the embryonic stage of the transferred embryos between the RIF and the fertile group. We observed that more day three, fresh and cleavage stage embryos were transferred in the fertile group, whereas more day four and day five, frozen‐thawed and morula and blastocyst embryos were transferred in the RIF group (Table 3). Due to the way the fertile and RIF groups were defined in our study, there were more embryo transfers in the RIF group, which also explains the higher proportion of frozen‐thawed embryo transfers in this group. In the Netherlands, fresh embryo transfers are generally performed on day three, 68 and embryos are usually cryopreserved on day three or four, and, after thawing, transferred on day four or five. The higher proportion of fresh embryo transfers in the fertile group, and the higher proportion of frozen‐thawed embryo transfers in the RIF group explain the differences in embryo transfer days and embryonic stage of transferred embryos between the fertile and RIF group. To our knowledge, it is not recognized yet whether the stage of the embryo influences endometrial gene expression. However, we cannot rule out potential differences in the extent to which “embryonic factors” vs “endometrial factors” were involved in the fertile and the RIF group, which may have affected the endometrial transcriptome profile.
Additionally, we did not see any differences in the proportion of different endometrium receptivity profiles between samples of implantation failure and implantation success groups, either in the short term or long term. However, in the short term, we observed a significantly larger mean and median ΔWOI in the not pregnant group than in the ongoing pregnant group with one‐sided testing (P = 0.033); two‐sided testing did not result in a significant difference, albeit with a P‐value with a trend towards statistical significance (P = 0.082). This difference in statistical significance is most likely related to the small sample size of the groups in the short‐term analysis. Nevertheless, the inconclusive statistical analysis may still indicate a biologically relevant difference and may suggest that delayed endometrial development could be a potential cause of implantation failure. Previously, a displaced window of implantation (WOI), detected by endometrial transcriptome analysis, was identified as a cause of RIF. 69 A previous study that also used the beREADY model to determine receptivity profiles of endometrium samples, detected a significantly higher proportion of displaced WOI in the RIF group than in the fertile group. 43 The results of our study, however, do not indicate a displacement of the WOI in the RIF group compared with the fertile group (ie our long‐term analysis). A potential explanation for this difference could be that the endometrium samples in our study were taken prospectively. The women in our RIF group were not classified as having experienced RIF yet at the time of the biopsy and could potentially have a different endometrial gene profile than if the biopsy had been taken after RIF was confirmed. Though endometrial gene expression has been shown to remain relatively stable between natural cycles in fertile women, 70 it is unknown whether this also applies to infertile women or women with RIF. A study in which endometrial biopsies were subjected to endometrial receptivity array (ERA) testing, failed to observe a larger proportion of WOI displacement in samples of women with RIF than controls with no RIF. 71 A recent study also demonstrated no differences in the proportion of receptive or non‐receptive profiles according to ERA testing between women with and without a history of unsuccessful frozen embryo transfers. 72 However, in these studies, samples were taken in a hormone replacement therapy (HRT) cycle as opposed to in a natural cycle in our study. Although we assessed the endometrial receptivity profiles of our samples during a natural cycle, our groups (ie pregnant vs not pregnant and fertile vs RIF) were based on clinical outcomes after stimulated cycles in the short‐term analysis and a mix of stimulated and natural and/or artificial cycles in the long‐term analysis. To our knowledge, it is yet unknown whether endometrial receptivity assessment during a natural cycle similarly represents the receptivity profile during subsequent stimulated or artificial cycles. Endometrial gene expression during the mid‐secretory phase of an artificial cycle (ie on the 6th day of progesterone supplementation) has been shown to be different to that of a natural cycle (ie LH + 7). 73 Therefore, it is deemed likely that the expression of endometrial receptivity genes is altered likewise by exogenous hormones, with potentially different receptivity profiles and different clinical outcomes. In addition, endometrial gene expression in women with RIF was more similar to that in fertile controls during a natural cycle than during an artificial cycle, 73 suggesting that exogenous hormones may negatively influence endometrial gene expression towards an implantation failure profile. It could therefore be possible that our prospective natural cycle endometrial receptivity assessments were not fully representative of the participants in our study groups, who were selected based on their outcomes after stimulated, HRT and/or natural cycles. However, given all the above, evaluation of endometrial receptivity during a natural cycle presumably provides a more physiologic view on the window of implantation and its potential deviations or disruptions.
In our study, all women had a failed first full IVF/ICSI cycle, which introduced variability in the number of prior failed embryo transfers. This variation could affect subsequent pregnancy prospects and the selection of the comparison groups, which in turn could have influenced differential gene expression, representing the difference in gene expression between two different groups. However, the incorporation of the number of previous failed embryo transfers as a covariate did not yield significant alterations in the analysis outcomes (data not shown).
Additionally, all patients underwent endometrial scratching after the initial failed IVF/ICSI cycle but before the second fresh cycle to obtain tissue samples. In selecting samples for the fertile and RIF groups in our long‐term analysis, we considered the total number of successfully transferred good quality embryos after 12 months of follow‐up. Importantly, a recently published individual participant data meta‐analysis 74 confirmed that the number of prior failed embryo transfers does not impact the effectiveness of endometrial scratching. Therefore, it is reasonable to conclude that endometrial scratching did not affect the prognosis of our fertile and RIF groups, and thus did not influence the selection of patient samples for these groups.
Our endometrial biopsies showed high enrichment for smooth muscle cell and epithelial cell signatures. Although not classified as such, it seems plausible that the smooth muscle cell signature is considered part of an “endometrial stromal cell signature”. However, in xCell there is no specific signature for endometrial stromal cells. The xCell smooth muscle cell signature, along with those of chondrocytes, astrocytes and mesenchymal stem cells, has been shown to exhibit high enrichment for genes of the endometrial stromal fibroblast cluster based on single‐cell sequencing data. 75 Previous single‐cell RNA sequencing of endometrium demonstrated smooth muscle cells as a separate cell entity, but these cells expressed a similar gene profile to that of the endometrial stromal cells. 76 Finally, the estimated cellular enrichment of endometrium samples did not differ between implantation failure and implantation success groups in our study, suggesting no alterations of endometrial tissue composition in implantation failure.
Strengths of the present study are the prospective and multicenter sampling of endometrial tissue, the availability of clinical follow‐up data of the participants up to 12 months after sampling, and the large number of samples included in the RNA‐seq.
As for limitations of the study, while we included a large number of samples for RNA‐seq, our comparison groups consisted of a relatively small number of samples, which may have been insufficient to detect significant differences.
Additionally, we noticed that LH‐timed biopsy day, age and BMI affected endometrial gene expression. Our endometrial biopsies were taken between LH + 5 to LH + 8. Adhering to a stricter window (eg LH + 6 and LH + 7 only) may have resulted in less variability in gene expression profiles. However, this was not possible due to logistic constraints.
Bulk RNA‐seq was performed in this study, which provides mixed gene expression profiles of all cell types present in the endometrial tissue, rather than cell type‐specific gene expression profiles, such as is the case with single‐cell RNA‐seq. 77 Therefore, to estimate the proportions of the cell types present in the tissue, we determined cell type enrichment scores, based on gene expression data of our samples. However, as we did not perform a validation of these cell type fractions in a separate cohort, we could not adjust the transcriptome data for endometrial tissue cellular composition.
To our knowledge, this is the first study comparing the optimal receptivity LH day and actual LH day of the biopsy with the estimated LH day, based on a pseudo‐timeline constructed by applying the beREADY model on endometrial transcriptome data of reference samples taken on LH + 2/3, LH + 7/8 and LH + 11/12. This pseudo‐timeline was divided into 10 equal bins, representing the inferred transcriptome profile of each LH day. However, there could be both intra‐individual (cycle‐related) and inter‐individual variation in transcriptome data at each LH day bin; thus a bin may not represent the “true” endometrial transcriptome of a sample of an individual taken at that same LH day. This limits the use of the ΔLH only to quantifying the relative difference of the actual LH day and the LH day based on endometrial transcriptomic dating. In addition, our endometrial biopsies were taken between 5 and 8 days after the LH surge, meaning that samples taken on LH + 5/6 were compared with an ‘inferred transcriptome profile’, as compared with samples taken on LH + 7/8, which could be directly compared with the profile of reference samples taken on LH + 7/8. As for ΔWOI, the calculations do not take into account the actual LH day of the endometrial biopsy, relying only on the transcriptome profile of sample and thus its position on the pseudo‐timeline. However, we were aware that the sampling day of the biopsy does affect endometrial gene expression. In addition, the ΔWOI values depend heavily on the chosen reference profiles, which in our study were set at LH + 7, the most well‐established endometrial receptivity day by beREADY. Thus, samples taken at LH + 5 were also compared with this reference profile of LH + 7, which could already have resulted in some deviation. Taken all the above in consideration, there may be some uncertainties in both the ΔWOI and ΔLH calculations. Although valuable to provide more insight into endometrial development in respect of receptivity, the ΔWOI measurements as well as the concordance between the actual LH day and pseudo‐timeline LH‐day do require further validation.
Lastly, endometrial tissue was obtained by Pipelle biopsy. A potential limitation of this method is the inadvertent collection of endometrial tissue from the lower part of the uterus, which might not accurately represent the tissue where implantation occurs. However, in our approach, we introduced the catheter up to the fundus and conducted the biopsy from that point onwards, primarily targeting endometrial tissue within the fundus area. Additionally, as tissue was obtained within an RCT by endometrial scratching with a Pipelle catheter, we opted for this method to simplify the process of obtaining tissue and in alignment with the pragmatic nature of the study.
Our study has been unable to find a specific gene expression profile for either implantation failure or implantation success, either in the short term or long term, contrary to many previous studies. When focusing on the expression of endometrial receptivity genes only, there may a difference between both groups, which we detected only in the short term. Therefore, we cannot yet exclude a role of impaired endometrial function in implantation failure, and further research is required. In addition, multiple cell types in the endometrium are known to be involved in the process of implantation. 78 It may therefore be possible that impairment of the function of a single cell type contributes to the occurrence of implantation failure. As bulk RNA‐seq data mask gene expression levels of individual cell types present in the whole endometrial tissue, single‐cell RNA‐seq studies are required to provide more information on function per cell type and exclude a role of impairment of specific cell types in implantation failure.
Despite the multitude of omics studies in the past two decades that performed a molecular characterization of endometrial receptivity, pregnancy rates in ART have not improved. 79 Due to a lack of concordance in molecular markers between studies based on differences in study methodology (eg sampling, processing and analysis), it is difficult to compare results and to study relevant pathways in more depth.
Future omics studies with a standardized methodology, following a set of universal guidelines regarding experimental design, sample collection, preparation and analysis, data validation and data presentation, 10 are required to obtain consistencies in relevant biomarkers and pathways. To enhance reproducibility, the phenotype of our participants has been meticulously defined, furnishing all available participant data, including age, BMI, smoking status and cycle characteristics. Furthermore, a detailed description of the tissue sampling method, sample storage, sample processing and data analysis was offered, as well as a thorough examination of the study's strengths and limitations.
For transcriptome studies, single‐cell RNA‐seq studies are highly recommended, as they provide more information on cell type‐specific functions and their potential role in implantation. In addition, validation of promising biomarker sets for endometrial receptivity in RCTs will need to be undertaken. Subsequently, more studies on endometrial omics downstream of the transcriptome, such as proteomics and metabolomics, are warranted, as they are the main executors of most physiological processes. 80 , 81 Although proteins are synthesized from mRNA templates, not all mRNAs are translated to proteins, and tightly regulated post‐transcriptional to post‐translational modifications add complexity to these processes, resulting in a poor correlation between mRNA and protein expression. 82
Only with all the above can we improve our understanding of human endometrial (patho)physiology, determine whether the endometrium really plays a role in implantation failure or success, and create possibilities to improve care for infertile couples.
5. CONCLUSION
Our exploratory study did not find an endometrial transcriptome profile associated with either implantation failure or success either in the short term or long term. In the short term, a larger deviation from the WOI may potentially be an underlying cause for implantation failure; however, in the long term, our study was unable to link WOI displacement to implantation failure. Future studies with standardized methodology are required to obtain more consistencies in relevant biomarkers, which help improve our understanding of the role of the endometrium in implantation failure, and in due course potentially provide targets to improve ART outcome.
AUTHOR CONTRIBUTIONS
Frank Broekmans designed the SCRaTCH trial and obtained funding. Nienke van Hoogenhuijze, Femke Mol, Gijs Teklenburg, Jan‐Peter de Bruin, Dagmar Besselink, Linda Stevens Brentjens collected clinical data of the SCRaTCH trial participants. Nienke van Hoogenhuijze, Frank Broekmans, Femke Mol, Gijs Teklenburg, Jan‐Peter de Bruin, Dagmar Besselink, Linda Stevens Brentjens, Ron van Golde obtained endometrial biopsies. Frank Broekmans, Shari Mackens, Andres Salumets, Gaby Steba, Triin Laisk initiated this international collaborative transcriptome project. Bich Ngoc Bui, Viktorija Kukushkina, Alvin Meltsov, Catharina Olsen, Gaby Steba, Triin Laisk, Signe Altmäe, Shari Mackens, Linda Stevens Brentjens, Darina Obukhova, Masoud Zamani Esteki, Ron van Golde, Andrea Romano, Andres Salumets, Frank Broekmans actively participated in online meetings to work out ideas and discuss results of data analyses. Bich Ngoc Bui, Viktorija Kukushkina, Alvin Meltsov, Catharina Olsenanalyzed the data. Bich Ngoc Bui, Viktorija Kukushkina, Alvin Meltsovd rafted the manuscript. All authors critically appraised the pre‐final manuscript, after which Bich Ngoc Bui and Alvin Meltsov adjusted the manuscript for submission. All authors approved submission of the final version.
CONFLICT OF INTEREST STATEMENT
The SCRATCH trial and the current endometrial transcriptome substudy received funding from public sources and Merck, but none of these sources had input in the study design and analysis. In addition, several authors report having received personal fees for consultations, conference attendance and travel as follows: BB from Gedeon Richter Benelux and Guerbet; NvH from Organon; GT from Merck; GS from Guerbet; SM from Abbott Pharmaceuticals, Ferring, Abbott Pharmaceuticals, and IBSA. FB is a paid member of the advisory boards of Merck and Ferring, and also received a speakers’ fee from Besins Healthcare. All of these personal fees were unrelated to the study reported in this manuscript. The remaining authors declare no conflicts of interest relevant to this study.
FUNDING INFORMATION
The randomized controlled trial was funded by the Netherlands Organization for Health Research and Development, “ZonMw” (ZonMW project number 843002601). The current transcriptome project was funded by Merck. In addition, the work was supported by the Horizon 2020 innovation (ERIN) grant (no. EU952516) of the European Commission; the Estonian Research Council (grant PRG1076); Enterprise Estonia (grant no. EU48695); the EVA (Erfelijkheid Voortplanting & Aanleg) specialty program (grant no. KP111513) of Maastricht University Medical Center (MUMC+); the Spanish Ministry of Economy, Industry and Competitiveness (MINECO) and European Regional Development Fund (FEDER): grants RYC‐2016‐21 199 and ENDORE SAF2017‐87526‐R.
ETHICS STATEMENT
This transcriptome study is a substudy of the SCRaTCH trial, a Dutch multicenter randomized controlled trial in which endometrial tissue was obtained. The RCT was approved by the Institutional Review Board of the University Medical Center Utrecht (15‐495) on November 30, 2015 23 and registered under trial registration number NTR5342 (https://trialsearch.who.int/Trial2.aspx?TrialID=NTR5342). For this substudy, ethical approval was obtained from the Biobank Research Ethics Committee of the University Medical Center Utrecht (19‐520) on October 31, 2019. Written informed consent was obtained from all study participants.
Supporting information
Figure S1.
Figure S2.
Figure S3.
Figure S4.
Table S1.
Table S2.
Table S3.
Table S4.
Table S5.
Table S6.
ACKNOWLEDGMENTS
We thank Utrecht Sequencing Facility (USEQ) for providing sequencing service and data. USEQ is subsidized by the University Medical Center Utrecht, Hubrecht Institute, Utrecht University and The Netherlands X‐omics Initiative (NWO project 184.034.019). The graphical abstract was created with figures from BioRender.com.
Bui BN, Kukushkina V, Meltsov A, et al. The endometrial transcriptome of infertile women with and without implantation failure. Acta Obstet Gynecol Scand. 2024;103:1348‐1365. doi: 10.1111/aogs.14822
DATA AVAILABILITY STATEMENT
The data underlying this study will be shared on reasonable request to the corresponding author.
REFERENCES
- 1. Sharlip ID, Jarow JP, Belker AM, et al. Best practice policies for male infertility. Fertil Steril. 2002;77:873‐882. [DOI] [PubMed] [Google Scholar]
- 2. Zegers‐Hochschild F, Adamson GD, Dyer S, et al. The international glossary on infertility and fertility care, 2017. Hum Reprod. 2017;32:1786‐1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mansour R, Ishihara O, Adamson GD, et al. International committee for monitoring assisted reproductive technologies world report: assisted reproductive technology 2006. Hum Reprod. 2014;29:1536‐1551. [DOI] [PubMed] [Google Scholar]
- 4. Smith ADAC, Tilling K, Nelson SM, Lawlor DA. Live‐birth rate associated with repeat in vitro fertilization treatment cycles. JAMA. 2015;314:2654‐2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coughlan C. What to do when good‐quality embryos repeatedly fail to implant. Best Pract Res Clin Obstet Gynaecol. 2018;53:48‐59. [DOI] [PubMed] [Google Scholar]
- 6. Diedrich K, Fauser BCJM, Devroey P, Griesinger G. The role of the endometrium and embryo in human implantation. Hum Reprod Update. 2007;13:365‐377. [DOI] [PubMed] [Google Scholar]
- 7. Macklon NS, Stouffer RL, Giudice LC, Fauser BCJM. The science behind 25 years of ovarian stimulation for in vitro fertilization. Endocr Rev. 2006;27:170‐207. [DOI] [PubMed] [Google Scholar]
- 8. Coutifaris C, Myers ER, Guzick DS, et al. Histological dating of timed endometrial biopsy tissue is not related to fertility status. Fertil Steril. 2004;82:1264‐1272. [DOI] [PubMed] [Google Scholar]
- 9. Murray MJ, Meyer WR, Zaino RJ, et al. A critical analysis of the accuracy, reproducibility, and clinical utility of histologic endometrial dating in fertile women. Fertil Steril. 2004;81:1333‐1343. [DOI] [PubMed] [Google Scholar]
- 10. Altmäe S, Esteban FJ, Stavreus‐Evers A, et al. Guidelines for the design, analysis and interpretation of “omics” data: focus on human endometrium. Hum Reprod Update. 2014;20:12‐28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Casamassimi A, Federico A, Rienzo M, Esposito S, Ciccodicola A. Transcriptome profiling in human diseases: new advances and perspectives. Int J Mol Sci. 2017;18:1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang X, Kui L, Tang M, et al. High‐throughput transcriptome profiling in drug and biomarker discovery. Front Genet. 2020;11:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Díaz‐Gimeno P, Horcajadas JA, Martínez‐Conejero JA, et al. A genomic diagnostic tool for human endometrial receptivity based on the transcriptomic signature. Fertil Steril. 2011;95(50–60):60.e1‐60.e15. [DOI] [PubMed] [Google Scholar]
- 14. Haouzi D, Assou S, Mahmoud K, et al. Gene expression profile of human endometrial receptivity: comparison between natural and stimulated cycles for the same patients. Hum Reprod. 2009;24:1436‐1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mirkin S, Arslan M, Churikov D, et al. In search of candidate genes critically expressed in the human endometrium during the window of implantation. Hum Reprod. 2005;20:2104‐2117. [DOI] [PubMed] [Google Scholar]
- 16. Riesewijk A, Martín J, van Os R, et al. Gene expression profiling of human endometrial receptivity on days LH+2 versus LH+7 by microarray technology. Mol Hum Reprod. 2003;9:253‐264. [DOI] [PubMed] [Google Scholar]
- 17. Talbi S, Hamilton AE, Vo KC, et al. Molecular phenotyping of human endometrium distinguishes menstrual cycle phases and underlying biological processes in normo‐ovulatory women. Endocrinology. 2006;147:1097‐1121. [DOI] [PubMed] [Google Scholar]
- 18. Diaz‐Gimeno P, Sebastian‐Leon P, Sanchez‐Reyes JM, et al. Identifying and optimizing human endometrial gene expression signatures for endometrial dating. Hum Reprod. 2022;37:284‐296. [DOI] [PubMed] [Google Scholar]
- 19. Altmäe S, Martínez‐Conejero JA, Salumets A, Simón C, Horcajadas JA, Stavreus‐Evers A. Endometrial gene expression analysis at the time of embryo implantation in women with unexplained infertility. Mol Hum Reprod. 2009;16:178‐187. [DOI] [PubMed] [Google Scholar]
- 20. Koler M, Achache H, Tsafrir A, Smith Y, Revel A, Reich R. Disrupted gene pattern in patients with repeated in vitro fertilization (IVF) failure. Hum Reprod. 2009;24:2541‐2548. [DOI] [PubMed] [Google Scholar]
- 21. Koot YEM, van Hooff SR, Boomsma CM, et al. An endometrial gene expression signature accurately predicts recurrent implantation failure after IVF. Sci Rep. 2016;6:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Altmäe S, Koel M, Võsa U, et al. Meta‐signature of human endometrial receptivity: a meta‐analysis and validation study of transcriptomic biomarkers. Sci Rep. 2017;7:1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Hoogenhuijze NE, Mol F, Laven JSE, et al. Endometrial scratching in women with one failed IVF/ICSI cycle‐outcomes of a randomised controlled trial (SCRaTCH). Hum Reprod. 2021;36:87‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liaqat Ali Khan N, Nafee T, Shao T, et al. Dysregulation in multiple transcriptomic endometrial pathways is associated with recurrent implantation failure and recurrent early pregnancy loss. Int J Mol Sci. 2022;23:16051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gardner DK, Lane M, Stevens J, Schlenker T, Schoolcraft WB. Blastocyst score affects implantation and pregnancy outcome: towards a single blastocyst transfer. Fertil Steril. 2000;73:1155‐1158. [DOI] [PubMed] [Google Scholar]
- 26. Cimadomo D, Craciunas L, Vermeulen N, Vomstein K, Toth B. Definition, diagnostic and therapeutic options in recurrent implantation failure: an international survey of clinicians and embryologists. Hum Reprod. 2021;36:305‐317. [DOI] [PubMed] [Google Scholar]
- 27. Shaulov T, Sierra S, Sylvestre C. Recurrent implantation failure in IVF: a Canadian Fertility and Andrology Society clinical practice guideline. Reprod Biomed Online. 2020;41:819‐833. [DOI] [PubMed] [Google Scholar]
- 28. van Hoogenhuijze NE, Torrance HL, Mol F, et al. Endometrial scratching in women with implantation failure after a first IVF/ICSI cycle; does it lead to a higher live birth rate? The SCRaTCH study: a randomized controlled trial (NTR 5342). BMC Womens Health. 2017;17:1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data [Internet]. 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 30. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114‐2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hannon GJ. FASTX‐Toolkit [Internet]. 2010. http://hannonlab.cshl.edu/fastx_toolkit/
- 32. Schneider VA, Graves‐Lindsay T, Howe K, et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017;27:849‐864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA‐seq aligner. Bioinformatics. 2013;29:15‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Broad Institute, GitHub Repository [Internet] . Picard Toolkit . 2015. https://bradinstitute.github.io/picard/
- 35. Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047‐3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen Y, Lun ATL, Smyth GK. From reads to genes to pathways: differential expression analysis of RNA‐seq experiments using Rsubread and the edgeR quasi‐likelihood pipeline. F1000Res. 2016;5:1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139‐140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2022. https://www.r‐project.org/ [Google Scholar]
- 39. McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA‐seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288‐4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Comstock IA, Diaz‐Gimeno P, Cabanillas S, et al. Does an increased body mass index affect endometrial gene expression patterns in infertile patients? A functional genomics analysis. Fertil Steril. 2017;107:740‐748.e2. [DOI] [PubMed] [Google Scholar]
- 41. Devesa‐Peiro A, Sebastian‐Leon P, Parraga‐Leo A, Pellicer A, Diaz‐Gimeno P. Breaking the ageing paradigm in endometrium: endometrial gene expression related to cilia and ageing hallmarks in women over 35 years. Hum Reprod. 2022;37:762‐776. [DOI] [PubMed] [Google Scholar]
- 42. Devesa‐Peiro A, Sebastian‐Leon P, Pellicer A, Diaz‐Gimeno P. Guidelines for biomarker discovery in endometrium: correcting for menstrual cycle bias reveals new genes associated with uterine disorders. Mol Hum Reprod. 2021;27:1‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Meltsov A, Saare M, Teder H, et al. Targeted gene expression profiling for accurate endometrial receptivity testing. Sci Rep. 2023;13:13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Teder H, Koel M, Paluoja P, et al. TAC‐seq: targeted DNA and RNA sequencing for precise biomarker molecule counting. NPJ Genom Med. 2018;3:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. GTEx . GTEx Tissue Sampling Sites [Internet]. 2021. https://www.gtexportal.org/home/samplingSitePage
- 47. Kelleher AM, Behura SK, Burns GW, Young SL, Demayo FJ, Spencer TE. Integrative analysis of the forkhead box A2 (FOXA2) cistrome for the human endometrium. FASEB J. 2019;33:8543‐8554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sigurgeirsson B, Åmark H, Jemt A, et al. Comprehensive RNA sequencing of healthy human endometrium at two time points of the menstrual cycle. Biol Reprod. 2017;96:24‐33. [DOI] [PubMed] [Google Scholar]
- 49. O'Leary NA, Wright MW, Brister JR, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733‐D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bastu E, Demiral I, Gunel T, et al. Potential marker pathways in the endometrium that may cause recurrent implantation failure. Reprod Sci. 2019;26:879‐890. [DOI] [PubMed] [Google Scholar]
- 51. Chen MY, Liao GD, Zhou B, Kang LN, He YM, Li SW. Genome‐wide profiling of long noncoding RNA expression patterns in women with repeated implantation failure by RNA sequencing. Reprod Sci. 2019;26:18‐25. [DOI] [PubMed] [Google Scholar]
- 52. Crosby DA, Glover LE, Brennan EP, et al. Dysregulation of the interleukin‐17A pathway in endometrial tissue from women with unexplained infertility affects pregnancy outcome following assisted reproductive treatment. Hum Reprod. 2020;35:1875‐1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Guo F, Si C, Zhou M, et al. Decreased PECAM1‐mediated TGF‐beta1 expression in the mid‐secretory endometrium in women with recurrent implantation failure. Hum Reprod. 2018;33:832‐843. [DOI] [PubMed] [Google Scholar]
- 54. Lédée N, Munaut C, Aubert J, et al. Specific and extensive endometrial deregulation is present before conception in IVF/ICSI repeated implantation failures (IF) or recurrent miscarriages. J Pathol. 2011;225:554‐564. [DOI] [PubMed] [Google Scholar]
- 55. Pathare ADS, Zaveri K, Hinduja I. Downregulation of genes related to immune and inflammatory response in IVF implantation failure cases under controlled ovarian stimulation. Am J Reprod Immunol. 2017;78:1‐13. [DOI] [PubMed] [Google Scholar]
- 56. Pathare ADS, Hinduja I. Endometrial expression of cell adhesion genes in recurrent implantation failure patients in ongoing IVF cycle. Reprod Sci. 2022;29:513‐523. [DOI] [PubMed] [Google Scholar]
- 57. Shi C, Han HJ, Fan LJ, et al. Diverse endometrial mRNA signatures during the window of implantation in patients with repeated implantation failure. Hum Fertil. 2018;21:183‐194. [DOI] [PubMed] [Google Scholar]
- 58. Tapia‐Pizarro A, Figueroa P, Brito J, Marín JC, Munroe DJ, Croxatto HB. Endometrial gene expression reveals compromised progesterone signaling in women refractory to embryo implantation. Reprod Biol Endocrinol. 2014;12:1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xu H, Zhou M, Cao Y, et al. Genome‐wide analysis of long noncoding RNAs, microRNAs, and mRNAs forming a competing endogenous RNA network in repeated implantation failure. Gene. 2019;720:144056. [DOI] [PubMed] [Google Scholar]
- 60. Zhou Q, Yan G, Ding L, et al. EHD1 impairs decidualization by regulating the Wnt4/β‐catenin signaling pathway in recurrent implantation failure. EBioMedicine. 2019;50:343‐354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pirtea P, Scott RT, de Ziegler D, Ayoubi JM. Recurrent implantation failure: how common is it? Curr Opin Obstet Gynecol. 2021;33:207‐212. [DOI] [PubMed] [Google Scholar]
- 62. Munné S, Weier HUG, Grifo J, Cohen J. Chromosome mosaicism in human embryos. Biol Reprod. 1994;51:373‐379. [DOI] [PubMed] [Google Scholar]
- 63. Penzias A, Bendikson K, Butts S, et al. The use of preimplantation genetic testing for aneuploidy (PGT‐A): a committee opinion. Fertil Steril. 2018;109:429‐436. [DOI] [PubMed] [Google Scholar]
- 64. Practice Committee and Genetic Counseling Professional Group (GCPG) of the American Society for Reproductive Medicine . Clinical management of mosaic results from preimplantation genetic testing for aneuploidy (PGT‐A) of blastocysts: a committee opinion. Fertil Steril. 2020;114:246‐254. [DOI] [PubMed] [Google Scholar]
- 65. Wright KP, Trimarchi JR, Allsworth J, Keefe D. The effect of female tobacco smoking on IVF outcomes. Hum Reprod. 2006;21:2930‐2934. [DOI] [PubMed] [Google Scholar]
- 66. Cinar O, Dilbaz S, Terzioglu F, et al. Does cigarette smoking really have detrimental effects on outcomes of IVF? Eur J Obstet Gynecol Reprod Biol. 2014;174:106‐110. [DOI] [PubMed] [Google Scholar]
- 67. Freour T, Masson D, Mirallie S, et al. Active smoking compromises IVF outcome and affects ovarian reserve. Reprod Biomed Online. 2008;16:96‐102. [DOI] [PubMed] [Google Scholar]
- 68. Cornelisse S, Ramos L, Arends B, et al. Comparing the cumulative live birth rate of cleavage‐stage versus blastocyst‐stage embryo transfers between IVF cycles: a study protocol for a multicentre randomised controlled superiority trial (the ToF trial). BMJ Open. 2021;11:e042395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sebastian‐Leon P, Garrido N, Remohí J, Pellicer A, Diaz‐Gimeno P. Asynchronous and pathological windows of implantation: two causes of recurrent implantation failure. Hum Reprod. 2018;33:626‐635. [DOI] [PubMed] [Google Scholar]
- 70. Evans GE, Phillipson GTM, Sykes PH, McNoe LA, Print CG, Evans JJ. Does the endometrial gene expression of fertile women vary within and between cycles? Hum Reprod. 2018;33:452‐463. [DOI] [PubMed] [Google Scholar]
- 71. Saxtorph MH, Hallager T, Persson G, et al. Assessing endometrial receptivity after recurrent implantation failure: a prospective controlled cohort study. Reprod Biomed Online. 2020;41:998‐1006. [DOI] [PubMed] [Google Scholar]
- 72. Doyle N, Combs JC, Jahandideh S, Wilkinson V, Devine K, O'Brien JE. Live birth after transfer of a single euploid vitrified‐warmed blastocyst according to standard timing vs timing as recommended by endometrial receptivity analysis. Fertil Steril. 2022;118:314‐321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Altmäe S, Tamm‐Rosenstein K, Esteban FJ, et al. Endometrial transcriptome analysis indicates superiority of natural over artificial cycles in recurrent implantation failure patients undergoing frozen embryo transfer. Reprod Biomed Online. 2016;32:597‐613. [DOI] [PubMed] [Google Scholar]
- 74. van Hoogenhuijze NE, Lahoz Casarramona G, Lensen S, et al. Endometrial scratching in women undergoing IVF/ICSI: an individual participant data meta‐analysis. Hum Reprod Update. 2023;29:721‐740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bunis DG, Wang W, Vallvé‐Juanico J, et al. Whole‐tissue deconvolution and scRNAseq analysis identify altered endometrial cellular compositions and functionality associated with endometriosis. Front Immunol. 2022;12:788315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Garcia‐Alonso L, Handfield LF, Roberts K, et al. Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. Nat Genet. 2021;53:1698‐1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Li X, Wang CY. From bulk, single‐cell to spatial RNA sequencing. Int J Oral Sci. 2021;13:1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ashary N, Tiwari A, Modi D. Embryo implantation: war in times of love. Endocrinology. 2018;159:1188‐1198. [DOI] [PubMed] [Google Scholar]
- 79. Aplin JD, Stevens A. Use of ‘omics for endometrial timing: the cycle moves on. Hum Reprod. 2022;37:644‐650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bracewell‐Milnes T, Saso S, Abdalla H, et al. Metabolomics as a tool to identify biomarkers to predict and improve outcomes in reproductive medicine: a systematic review. Hum Reprod Update. 2017;23:723‐736. [DOI] [PubMed] [Google Scholar]
- 81. Guo X, Li TC, Chen X. The endometrial proteomic profile around the time of embryo implantation. Biol Reprod. 2021;104:11‐26. [DOI] [PubMed] [Google Scholar]
- 82. Stephens AN, Hannan NJ, Rainczuk A, et al. Post‐translational modifications and protein‐specific isoforms in endometriosis revealed by 2D DIGE. J Proteome Res. 2010;9:2438‐2449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.
Figure S2.
Figure S3.
Figure S4.
Table S1.
Table S2.
Table S3.
Table S4.
Table S5.
Table S6.
Data Availability Statement
The data underlying this study will be shared on reasonable request to the corresponding author.


