Abstract
Hepatitis C virus (HCV) infection is a widespread major human health concern. Significant obstacles in the study of this virus include the absence of a reliable tissue culture system and a small-animal model. Recently, we constructed full-length HCV cDNA clones and successfully initiated HCV infection in two chimpanzees by intrahepatic injection of in vitro-transcribed RNA (A. A. Kolykhalov et al., Science 277:570–574, 1997). In order to validate potential targets for development of anti-HCV therapeutics, we constructed six mutant derivatives of this prototype infectious clone. Four clones contained point mutations ablating the activity of the NS2-3 protease, the NS3-4A serine protease, the NS3 NTPase/helicase, and the NS5B polymerase. Two additional clones contained deletions encompassing all or part of the highly conserved 98-base sequence at the 3′ terminus of the HCV genome RNA. The RNA transcript from each of the six clones was injected intrahepatically into a chimpanzee. No signs of HCV infection were detected in the 8 months following the injection. Inoculation of the same animal with nonmutant RNA transcripts resulted in productive HCV infection, as evidenced by viremia, elevated serum alanine aminotransferase, and HCV-specific seroconversion. These data suggest that these four HCV-encoded enzymatic activities and the conserved 3′ terminal RNA element are essential for productive replication in vivo.
Prior to the development of specific blood donor screening assays, hepatitis C virus (HCV) was the major cause of transfusion-associated hepatitis (see reference 25 for a review). While transfusion-associated HCV infections are rare, about 30,000 new cases of hepatitis C are estimated to occur in the United States each year. HCV is not easily cleared by the host's immunological defenses; as many as 85% of the people infected with HCV become chronically infected. Many of these persistent infections result in chronic liver disease, including cirrhosis and hepatocellular carcinoma (24). There are an estimated 170 million HCV carriers worldwide, and HCV-associated end-stage liver disease is now the leading cause of liver transplantation. In the United States alone, hepatitis C is responsible for 8,000 to 10,000 deaths annually, and without effective intervention, that number is predicted to triple in the next 10 to 20 years. There is no vaccine to prevent hepatitis C infection. Prolonged treatment of chronically HCV-infected patients with interferon or interferon plus ribavirin is the only currently approved therapy, but it results in a sustained response in fewer than 50% of the cases (37, 46).
HCV belongs to the family Flaviviridae, which comprises three genera of small enveloped positive-strand RNA viruses (see reference 47 and references therein). The 9.6-kb genome of HCV consists of a long open reading frame (ORF) flanked by 5′ and 3′ nontranslated regions (NTRs). The HCV 5′ NTR is 341 nucleotides in length and functions as an internal ribosome entry site for cap-independent translation initiation (34). The HCV polyprotein is cleaved co- and posttranslationally into at least 10 individual polypeptides (for a review, see reference 45). The structural proteins result from signal peptidase cleavages in the N terminal portion of the polyprotein. Two viral proteases mediate downstream cleavages to produce nonstructural (NS) proteins that function as components of the HCV RNA replicase. The NS2-3 protease spans the C terminal half of NS2 and the N terminal one-third of NS3 and catalyzes autocatalytic cis cleavage at the 2/3 site. The same portion of NS3 also encodes the catalytic domain of the NS3-4A serine protease that cleaves at four downstream sites. The C terminal two-thirds of NS3 is highly conserved among HCV isolates, with RNA-binding, RNA-stimulated NTPase, and RNA-unwinding activities. Although NS4B and the NS5A phosphoprotein are also likely components of the replicase, their specific roles are unknown. The C terminal polyprotein cleavage product, NS5B, is the elongation subunit of the HCV replicase possessing RNA-dependent RNA polymerase (RDRP) activity (5, 38). Following a translation stop codon, the HCV 3′ NTR consists of three subregions: (i) a 28- to 42-base sequence that varies among genotypes, (ii) an internal poly(U/UC) tract of variable length with rare A or G residues, and (iii) a highly conserved 3′ terminal 98-base sequence (33, 49, 50, 54). This recently discovered 98-base element is the most highly conserved RNA sequence in the HCV genome, but two surprising reports suggest that it is not essential for virus replication (13, 58).
The development of new and specific anti-HCV treatments is a high priority, and virus-specific functions essential for replication are the most attractive targets for drug development. In the case of HCV, it has been assumed that conserved features are essential, but this has not been experimentally testable. Assembly of functional HCV cDNA clones (31) has now allowed us to directly assess the functional importance of HCV-encoded enzymatic activities and RNA elements by site-directed mutagenesis. Here, we report the in vivo characterization of mutants defective in each of the four known HCV-encoded enzymatic activities or lacking all or part of the conserved 3′ terminal sequence.
Construction of mutant HCV full-length cDNA clones.
The infectious full-length consensus HCV cDNA clone p90/HCVFLlongpU, containing a 133-base poly(U/UC) tract and no additional 5′ terminal nucleotides (31; subsequently referred to as HCV FL), was used as the backbone for construction of six mutant clones (Fig. 1). We inactivated each of the four known HCV-encoded enzymatic activities by mutating at least two amino acid residues essential or important for function (Fig. 1A). Multiple substitutions were created to avoid the generation of same-site revertants during transcription with T7 RNA polymerase, which has a relatively high error rate (∼6 × 10−5 per nucleotide (7). In HCV FL(2-3pro−), the NS2-3 protease was inactivated by incorporating the H952A and C993A substitutions. Although it is not known if these NS2 residues participate directly in catalysis, either substitution abolishes processing at the 2/3 site (18, 22). For HCV FL(3pro−), two residues in the NS3-4A serine protease catalytic triad were changed to alanine (D1107A and S1165A). Either of these substitutions abolishes detectable processing at the downstream 3/4A, 4A/4B, 4B/5A, and 5A/5B cleavage sites (17, 22). HCV FL(hel−) contained two mutations designed to inactivate the NS3 helicase activity. Based on the presence of a DECH motif (polyprotein residues 1316 to 1319), the HCV helicase belongs to the DExH family of DEAD-box helicases (16). The first two residues of this motif are invariant, with the Asp residue binding Mg2+-ATP (44). Substitution for either of these residues disrupts NTPase and helicase activities (19); both were mutated to Ala in HCV FL(hel−). Finally, the NS5B RDRP was destroyed in HCV FL(pol−) by replacing the Gly-Asp-Asp (GDD) sequence with Ala-Ala-Gly. This polymerase motif is conserved among all plus-stranded RNA viruses (43), and mutating any of these three residues inhibits or abolishes the RDRP activity of purified HCV NS5B (26, 38).
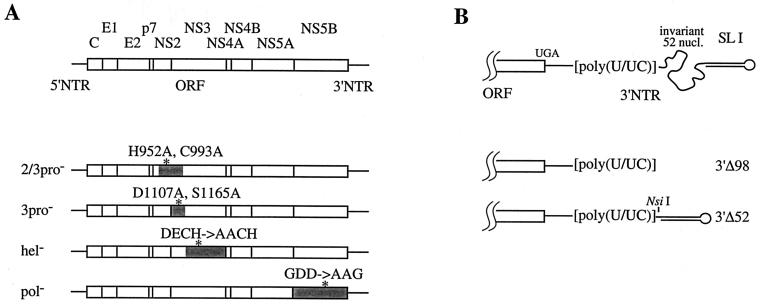
FIG. 1.
Diagram of the HCV genome and mutant constructs. All mutant derivatives were constructed on the background of HCV FL, and their structures were verified by sequence analysis. Mutants are described by the nucleotide positions and substitutions (lowercase letters) relative to HCV FL (31); sequences of the oligonucleotides used for mutagenesis, plasmid manipulations, and complete sequence files are available upon request. (A) The HCV genome organization is shown at the top with 5′ and 3′ NTRs (solid lines), and the ORF (open box) and the polyprotein cleavage products are indicated. Mutant full-length clones are shown below, highlighting the regions encoding the four enzymatic activities (shadowed), the positions of the mutations (asterisks), and the construct names (at the left). HCV FL(2-3pro−) contains the amino acid substitutions H952A (3195 to 3200; gcgtTa) and C993A (3318 to 3319; gc). HCV FL(3pro−) contains the substitutions D1107A (3660 to 3664; gcctt) and S1165A (3831 to 3836; agCgCt). HCV FL(hel−) contains the substitutions D1316A (4286 to 4289; cGca) and E1317A (4291 to 4292; ca). HCV FL(pol−) contains the substitutions G2737A (8551 to 8552; cg), D2738A (8554; c), and D2739G (8557 to 8559; gCc). (B) Organization of the 3′ portion of HCV genome RNA showing (5′ to 3′) the C-terminal part of the ORF (open box), the polyprotein translation termination codon (UGA), the variable part of the 3′ NTR (solid straight line), the poly(U/UC) tract, the highly conserved 52-base sequence (curved line), and the 3′ terminal 46-base stem-loop structure (SL I). Mutant clones are shown below with their corresponding names to the right. HCV FL(3′Δ52) is identical to HCV FL, except for an internal deletion encompassing the 5′ 52 bases of the 3′ terminal 98-base sequence. For HCV FL(3′Δ98), the entire 3′ 98-base sequence was deleted. A novel restriction site (NsiI) distinguishing HCV FL(3′Δ52) from HCV FL(3′Δ98) is indicated. nucl., nucleotides.
Two additional clones were constructed to test the functional importance of the conserved portion of the 3′ NTR (the 98-base sequence or X tail) (33, 49, 50, 54). In HCV FL(3′Δ98), the entire 3′ 98-base sequence was deleted so that runoff RNA transcripts would terminate immediately following the poly(U/UC) tract (Fig. 1B). HCV FL(3′Δ52) contained a smaller deletion encompassing the 52 invariant nucleotides between the poly(U/UC) tract and the 3′ terminal 46-base stem-loop structure (SL I; 6). Both 3′ NTR deletion constructs contained a nucleotide substitution (C519T) that was previously shown to be tolerated by HCV (31). In addition, HCV FL(3′Δ52) could be distinguished from HCV FL(3′Δ98) by an NsiI site at position 9547 that was fortuitously created by the fused sequences (Fig. 1B).
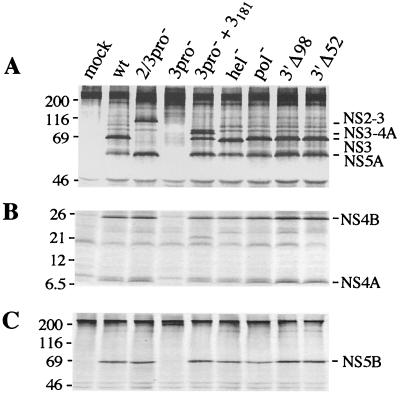
Prior to the animal experiment, translation and polyprotein processing of HCV FL and the mutant constructs were compared by transient expression in cell culture. Since the constructs contain a T7 RNA polymerase-specific promoter upstream of the HCV sequence, plasmid DNAs were transfected into BHK-21 cells previously infected with recombinant vaccinia virus vTF7-3, which expresses the T7 RNA polymerase. For all constructs, the expression level and electrophoretic mobility of the E1 glycoprotein were similar to those observed for HCV FL (data not shown). For the NS proteins, the expected processing patterns were observed (Fig. 2). Processing at the 2/3 site was abolished with HCV FL(2/3pro−), but processing at other sites was not affected, as shown by the presence of NS4A, NS4B, NS5A, and NS5B. Unprocessed E2-p7-NS2-NS3, with a molecular mass of ∼180 kDa, was also detected, presumably as a result of inefficient processing at the E2/p7 and p7/NS2 sites (35, 41, 48). For HCV FL(3pro−), processing at serine protease-dependent sites was blocked but could be restored by cotransfection of the functional protease domain (NS3181; 36). In this case, in addition to the individual NS proteins, an NS3-4A precursor was also observed as a result of inefficient trans cleavage at the 3/4A site (see, for example, reference 15). Processing of the HCV FL(hel−) polyprotein revealed patterns identical to those of HCV FL, except that NS3 migrated slightly faster than the wild-type protein as a result of the two engineered substitutions. The protein patterns of HCV FL(pol−), HCV FL(3′Δ98), and HCV FL(3′Δ52) were indistinguishable from those of HCV FL (Fig. 2).
FIG. 2.
Polyprotein processing by HCV FL and mutant derivatives. Transfection of HCV FL (wt) or mutant plasmid DNAs (indicated at the top), protein labeling, immunoprecipitation of 35S-labeled proteins with HCV-specific sera, and the analysis of the immune complexes by sodium dodecyl sulfate-polyacrylamide gel electrophoresis were conducted as described previously (32). pGEM3Zf(+) was transfected as a negative control (mock). Products were immunoprecipitated with patient serum JHF recognizing NS3, NS4A, NS4B, and NS5A (A and B) or rabbit anti-NS5B (C); separated by sodium dodecyl sulfate–12 or 9% polyacrylamide gel electrophoresis, respectively; and visualized by autoradiography. The positions of molecular weight markers are shown on the left (in kilodaltons); HCV-specific polyproteins and cleavage products are identified on the right. The ∼21-kDa species which is observed in the HCV FL(3pro−)-NS3181 cotransfection is an N terminally truncated form of the NS4B protein (32). The values on the left are molecular sizes in kilodaltons.
Infectivity of mutant HCV RNAs in vivo.
Chimpanzee 1552 (Ch1552) was used to assess the infectivity of mutant transcripts. Ch1552 had been inoculated 2 years prior to this study with RNA transcripts from 17 nonconsensus clones from an original HCV cDNA library (31). Follow-up of Ch1552 for 6 months did not reveal any evidence of productive HCV infection: serum alanine aminotransferase (ALT) remained steady at the preinoculation level, HCV RNA was not detectable by reverse transcription (RT)-PCR, no HCV-specific antibodies were registered, and no signs of hepatitis or inflammation were detected in liver biopsies. Just prior to the current study, Ch1552 was negative for HCV RNA by RT-PCR, was seronegative by anti-HCV enzyme-linked immunosorbent assay (ELISA) 3.0, and had normal ALT levels.
The six mutant plasmid DNAs were linearized following the 3′ end of the HCV cDNA and transcribed with T7 RNA polymerase. RNA transcripts were injected directly into the surgically exposed liver of Ch1552, with each transcript preparation injected into four sites. Separate injections of each RNA at different sites minimized the possibility of cotransfection of the same cell(s) with multiple RNAs capable of complementing one another, resulting in the initiation of replication and/or recombination between RNAs to generate an infectious RNA. After inoculation, Ch1552 was followed up for 32 weeks and showed no signs of productive HCV replication: ALT remained at the preinjection level: serum samples from weeks 5, 9, 15, and 18 were negative for HCV-specific antibodies, as determined by a commercial third-generation HCV ELISA; and HCV RNA was undetectable in the serum. Competitive quantitative RT-nested PCR was used to analyze pre- and postinoculation samples from weeks −9, 0, 1 to 13, 21, 25, and 32. In all of these samples, HCV RNA was undetectable (detection limit: 300 to 500 RNA molecules per ml of serum).
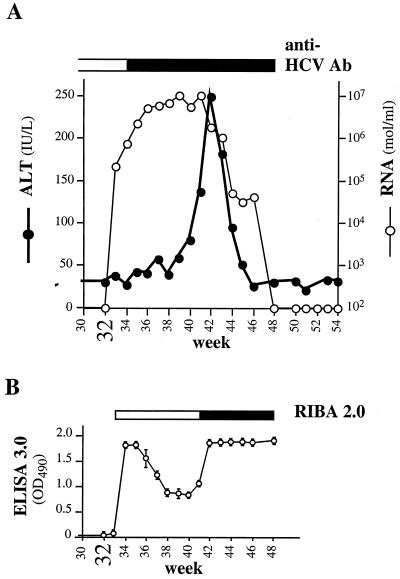
To demonstrate that Ch1552 could be productively infected by in vitro-synthesized RNA, the animal was challenged at week 32 with HCV FL RNA. To avoid another surgical procedure, intrahepatic injections were performed percutaneously, once through a biopsy needle and four times through lumbar puncture needles. After challenge, serum ALT levels indicated a typical HCV infection. Prechallenge values were observed for the first 7 weeks, followed by a sharp rise, a peak at week 42 (week 10 postchallenge), and then a return to the prechallenge level by week 45 (Fig. 3A). Circulating HCV RNA was detected 1 week after challenge, and the titers (measured using the ABI PRISM 7700 Sequence Detection System [PE Applied Biosystems, Foster City, Calif.]) gradually increased from 2 × 105 RNA molecules per ml on week 33 to ∼1 × 107 RNA molecules per ml on weeks 39 through 41. After serum ALT had peaked, HCV RNA declined to 1 × 106 to 2 × 106 on week 42, dropped sharply on week 44 (to 5 × 104 per ml), and then remained steady for 3 or 4 weeks before becoming undetectable by week 48. Even by RT-nested PCR, HCV RNA was not detected in any sample after week 48 (detection limit, 100 to 300 RNA molecules per ml of serum).
FIG. 3.
Analysis of Ch1552 samples. RNA transcription and inoculation of RNAs into the liver were performed as described previously (31). For each RNA, approximately 150 μg of RNA in phosphate-buffered saline (PBS) was injected into two separate sites, and 1 μg of an RNA-Lipofectin-PBS mixture was also injected at two separate sites. Ch1552 was challenged at week 32 with infectious HCV FL RNA transcripts using nonsurgical procedures. One hundred micrograms of RNA in 1 ml of PBS was injected into the liver percutaneously through a biopsy needle. Three additional intrahepatic injections of 100 μg of RNA in 1 ml of PBS per injection were administered with a lumbar puncture needle. A fifth lumbar puncture needle injection was performed with 3 μg of RNA mixed with 30 μl of Lipofectin and PBS in a total volume of 0.5 ml. (A) Serum ALT, HCV RNA (molecules per milliliter), and HCV-specific antibodies (Ab; as measured by HCV ELISA 3.0). (B) Detection of HCV-specific antibodies by Ortho HCV version 3.0 ELISA and by Chiron RIBA 2.0. For the RIBA 2.0, the open box indicates HCV-seronegative serum samples and the solid bar (beginning at week 41) indicates positive samples. OD490, optical density at 490 nm.
Circulating HCV-specific antibodies were analyzed using commercial third-generation HCV ELISA 3.0 (Fig. 3A and B). Antibodies were detectable as early as 2 weeks after challenge. Two response peaks were observed. After a sharp increase on week 34, ELISA-reactive antibody levels decreased during weeks 36 to 40 and then rose again starting at week 41 to week 42. The second increase in HCV-specific antibody coincided with elevated ALT. Interestingly, the presence of HCV-specific antibodies on weeks 34 through 41 was not confirmed by second-generation Chiron RIBA HCV 2.0 Strip Immunoblot Assay (RIBA 2.0). This can be explained by an early antibody response to NS5, since this antigen is present in the ELISA 3.0 but not in the RIBA 2.0. NS3-specific antibodies were detected by RIBA 2.0 beginning at week 10 postchallenge, but no other HCV-specific reactivity was detected during the course of this study.
Concluding remarks.
Although two groups have reported productive HCV replication after transfection of cell cultures with transcribed RNA (13, 58), no follow-up studies have been published, nor have these systems been used to delineate essential viral functions. In this study, we exploited the chimpanzee model, which has been extensively used for HCV studies and in particular for initiating infection by intrahepatic inoculation of RNA transcribed from functional HCV cDNA (23, 31, 55, 57). While this approach does not lend itself to mechanistic studies, we can define functions essential for replication in this stringent animal model to validate or uncover new targets for anti-HCV drug development.
We examined the four known HCV-encoded enzymes that are being actively pursued as antiviral targets. The two viral proteases mediate cleavages in the NS region that are thought to be necessary to form a functional RNA replicase. In the case of the NS3 serine protease, which is common to all members of the family Flaviviridae, previous work has demonstrated that this activity is essential for replication of the classical flaviviruses (10) and the animal pestiviruses (53). This observation can now be extended to HCV (although in no case do we know why processing of the polyprotein is important for replicase function). The NS2-3 protease, which mediates cis cleavage at the 2/3 site, is unique to HCV, although cleavage immediately upstream of the NS3 serine protease domain is also observed for both flaviviruses (see reference 47 for a review) and some pestiviruses (see reference 40 for a review). For classical flaviviruses like yellow fever virus, the functional serine protease is a heterodimer consisting of the upstream NS2B protein and NS3 (8). As in HCV, cleavage at the NS2B/3 site is autocatalytic, but for flaviviruses, the serine protease is responsible for this cleavage. Mutations which block processing at the NS2B/3 site are deleterious for yellow fever virus replication, although processing at downstream sites still occurs (9). For the pestiviruses, the situation is more complex. For noncytopathic isolates of bovine viral diarrhea virus (BVDV), the NS2-3 region remains unprocessed. In contrast, cytopathic BVDV isolates have usually undergone various RNA recombination events that lead to production of a discrete NS3 protein (40). Given the situation with noncytopathic pestiviruses, the recent report that NS2 is dispensable for BVDV replication (4) and the observations that some HCV cDNA clones fail to process at the 2/3 site (14; A. A. Kolykhalov, unpublished data), it was of interest to test the essential nature of the HCV NS2-3 protease. Our results indicate that this proteolytic activity is required for productive HCV replication, although we cannot exclude the possibility that the mutated NS2 residues are essential for some other function in the virus life cycle. The remaining two enzymatic activities, the NS3 NTPase/RNA helicase and the NS5B RDRP, are common to all members of the family and, more generally, to most positive-strand RNA viruses. The precise role(s) of such helicases in viral RNA replication is not known (29), but mutations inactivating the BVDV (E. Mendez, M. S. Collett, and C. M. Rice, unpublished data) or the HCV (this work) NS3 helicase were lethal. As expected, mutations ablating the NS5B RDRP activity were also lethal, underscoring the importance of this enzyme for HCV replication.
The HCV RNA element examined in this study was the conserved portion of the 3′ NTR. Early reports indicated that the HCV genome RNA terminated with poly(A) (20) or poly(U) (for examples, see references 21, 30, and 42). Subsequently, it was discovered that the HCV 3′ NTR is actually comprised of a short region that varies among genotypes, an internal poly(U/UC) tract, and a terminal element of 98 bases (33, 49, 50, 51, 54). The 98-base sequence consists of 52 invariant bases followed by 46 bases that form a highly stable 3′ terminal stem-loop structure (6). It has been hypothesized that this element participates in RNA replication, in particular, the initiation of minus-strand RNA synthesis. Several groups have begun to uncover host RNA-binding proteins (11, 27, 39, 52), such as polypyrimidine tract-binding protein, that may function in HCV RNA replication and translation via interaction with the 3′ NTR (28). In our study, RNAs lacking the 98-base element or those in which the 52-base invariant sequence was deleted were incapable of replication. While this report was in preparation, Yanagi et al. reported similar findings demonstrating that most HCV 3′ NTR elements were essential for productive infection in vivo, with the exception of the variable region immediately following the ORF termination codon (56). Thus, the in vivo results conflict with the two reports claiming that transfected HCV RNAs lacking this sequence can replicate in cell culture (13, 58). Further work is needed to resolve these discrepancies and determine if the RNA elements required for in vivo versus cell culture replication differ. Nonetheless, the in vivo data validate the conserved 98-base RNA element as an attractive target for antisense oligonucleotides, trans-acting ribozymes, RNA decoys, or small molecules that block critical interactions with host or viral proteins.
An interesting observation of our study was the unexpected immune response profile of Ch1552. After inoculation of Ch1552 with the seven mutant full-length RNAs, HCV replication was undetectable and no HCV-specific serological response was detectable. However, upon challenge of Ch1552 with parental infectious RNA, we observed an unusually rapid (only 2 weeks after challenge) appearance of HCV-specific antibodies. This suggested possible priming of the immune system by HCV-specific antigens before challenge, as recently reported by Beard et al. (3). Indeed, this was confirmed by analysis of peripheral blood mononuclear cells taken from the animal at week 32 prechallenge, at which time T-cell responses against core, helicase, and polymerase antigens were readily detected (T. Arichi, M. Major, H. Wedemeyer, M. Nascimbeni, S. Gagneton, A. A. Kolykhalov, J. A. Berzofsky, C. M. Rice, S. M. Finestone, and B. Rehermann, unpublished data). Priming may have occurred via direct translation of injected replication-defective mutant RNAs or possibly because one or more of the mutant RNAs was capable of low-level replication. One feature of the early antibody response postchallenge was its apparent decline in weeks 36 to 40. This could indicate that insufficient antigen is produced in the early phase of HCV infection to sustain antibody responses induced by injection and translation of input RNA. Alternatively, the apparent dip in ELISA 3.0 reactivity may reflect the sequestration of HCV-specific antibodies in immune complexes. Although further follow-up is required, Ch1552 appears to have resolved HCV infection. Whether priming of HCV-specific T-cell responses prechallenge played a role in this clinical course is difficult to determine, since a majority of naive chimpanzees are able to spontaneously resolve acute HCV infections (1, 2). Nonetheless, a detailed analysis of the humoral and T-cell responses in this animal is in progress (Arichi et al., unpublished data) and should provide further data on immune responses that correlate with resolution (12). In addition, if Ch1552 has indeed resolved the infection, then this animal will allow us to determine if the immune responses leading to resolution are sufficient to protect against challenge with a truly homologous virus isolated in the acute phase of infection after transfection with clonal infectious RNA (31).
Although our experiments were limited to a single chimpanzee, initiation of HCV infection by injection of transcribed RNA has been remarkably reproducible and this animal was productively infected by this route. Thus, our results indicate that the two HCV-encoded proteases, the NTPase/helicase, the RNA-dependent RNA polymerase, and the conserved elements of the 3′ NTR are essential for HCV replication in the chimpanzee. For mechanistic studies to determine how these enzymes and RNA elements actually function in HCV RNA replication, cell culture assay systems are sorely needed. The most straightforward approach to establishing such systems is to test the ability of transfected infectious RNAs to amplify and perhaps spread in cell culture. Toward this goal, the replication-defective mutants described in this study and validated in the chimpanzee transfection model can serve as useful negative controls to distinguish between authentic replication and persistence of transfected RNA.
Acknowledgments
We thank Scott Baginski, Keril Blight, Mara Lippa, and Tina Myers for critical reading of the manuscript.
This work was supported in part by grants from the Public Health Service to C.M.R. (CA57973 and AI40034).
REFERENCES
- 1.Bassett S E, Brasky K M, Lanford R E. Analysis of hepatitis C virus-inoculated chimpanzees reveals unexpected clinical profiles. J Virol. 1998;72:2589–2599. doi: 10.1128/jvi.72.4.2589-2599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassett S E, Thomas D L, Brasky K M, Lanford R E. Viral persistence, antibody to E1 and E2, and hypervariable region 1 sequence stability in hepatitis C virus-inoculated chimpanzees. J Virol. 1999;73:1118–1126. doi: 10.1128/jvi.73.2.1118-1126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beard M R, Abell G, Honda M, Carroll A, Gartland M, Clarke B, Suzuki K, Lanford R, Sangar D V, Lemon S M. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology. 1999;30:316–324. doi: 10.1002/hep.510300137. [DOI] [PubMed] [Google Scholar]
- 4.Behrens S-E, Grassmann C W, Thiel H-J, Meyers G, Tautz N. Characterization of an autonomous subgenomic pestivirus RNA replicon. J Virol. 1998;72:2364–2372. doi: 10.1128/jvi.72.3.2364-2372.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrens S E, Tomei L, DeFrancesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 6.Blight K J, Rice C M. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1997;71:7345–7352. doi: 10.1128/jvi.71.10.7345-7352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer J C, Bebenek K, Kunkel T A. Unequal human immunodeficiency virus type 1 reverse transcriptase error rates with RNA and DNA templates. Proc Natl Acad Sci USA. 1992;89:6919–6923. doi: 10.1073/pnas.89.15.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers T J, Grakoui A, Rice C M. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol. 1991;65:6042–6050. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers T J, Nestorowicz A, Rice C M. Mutagenesis of the yellow fever virus NS2B/3 cleavage site: determinants of cleavage site specificity and effects on polyprotein processing and viral replication. J Virol. 1995;69:1600–1605. doi: 10.1128/jvi.69.3.1600-1605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambers T J, Weir R C, Grakoui A, McCourt D W, Bazan J F, Fletterick R J, Rice C M. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci USA. 1990;87:8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung R T, Kaplan L M. Heterogeneous nuclear ribonucleoprotein I (hnRNP-I/PTB) selectively binds the conserved 3′ terminus of hepatitis C viral RNA. Biochem Biophys Res Commun. 1999;254:351–362. doi: 10.1006/bbrc.1998.9949. [DOI] [PubMed] [Google Scholar]
- 12.Cooper S, Erickson A L, Adams E J, Kansopon J, Weiner A J, Chien D Y, Houghton M, Parham P, Walker C M. Analysis of a successful immune response against hepatitis C virus. Immunity. 1999;10:439–449. doi: 10.1016/s1074-7613(00)80044-8. [DOI] [PubMed] [Google Scholar]
- 13.Dash S, Halim A-B, Tsuji H, Hiramatsu N, Gerber M A. Transfection of HepG2 cells with infectious hepatitis C virus genome. Am J Pathol. 1997;151:363–373. [PMC free article] [PubMed] [Google Scholar]
- 14.D'Souza E D, O'Sullivan E, Amphlett E M, Rowlands D J, Sangar D V, Clarke B E. Analysis of NS3-mediated processing of the hepatitis C virus non-structural region in vitro. J Gen Virol. 1994;75:3469–3476. doi: 10.1099/0022-1317-75-12-3469. [DOI] [PubMed] [Google Scholar]
- 15.Failla C, Tomei L, DeFrancesco R. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J Virol. 1994;68:3753–3760. doi: 10.1128/jvi.68.6.3753-3760.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4729. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA. 1993;90:10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross C H, Shuman S. Mutational analysis of vaccinia virus nucleoside triphosphate phosphohydrolase II, a DExH box RNA helicase. J Virol. 1995;69:4727–4736. doi: 10.1128/jvi.69.8.4727-4736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han J H, Shyamala V, Richman K H, Brauer M J, Irvine B, Urdea M S, Tekamp-Olson P, Kuo G, Choo Q-L, Houghton M. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly(A) tails at the 3′ end. Proc Natl Acad Sci USA. 1991;88:1711–1715. doi: 10.1073/pnas.88.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi N, Higashi H, Kaminaka K, Sugimoto H, Esumi M, Komatsu K, Hayashi K, Sugitani M, Suzuki K, Tadao O, Nozaki C, Mizuno K, Shikata T. Molecular cloning and heterogeneity of the human hepatitis C virus (HCV) genome. J Hepatol. 1993;17(Suppl. 3):S94–S107. doi: 10.1016/s0168-8278(05)80432-5. [DOI] [PubMed] [Google Scholar]
- 22.Hijikata M, Mizushima H, Akagi T, Mori S, Kakiuchi N, Kato N, Tanaka T, Kimura K, Shimotohno K. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol. 1993;67:4665–4675. doi: 10.1128/jvi.67.8.4665-4675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong Z, Beaudet-Miller M, Lanford R E, Guerra B, Wright-Minogue J, Skelton A, Baroudy B M, Reyes G R, Lau J Y N. Generation of transmissible hepatitis C virions from a molecular clone in chimpanzees. Virology. 1999;256:36–44. doi: 10.1006/viro.1999.9603. [DOI] [PubMed] [Google Scholar]
- 24.Hoofnagle J H. Hepatitis C: the clinical spectrum of disease. Hepatology. 1997;26:15S–20S. doi: 10.1002/hep.510260703. [DOI] [PubMed] [Google Scholar]
- 25.Houghton M. Hepatitis C viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Raven Press; 1996. pp. 1035–1058. [Google Scholar]
- 26.Ishii K, Tanaka Y, Yap C C, Aizaki H, Matsuura Y, Miyamura T. Expression of hepatitis C virus NS5B protein: characterization of its RNA polymerase activity and RNA binding. Hepatology. 1999;29:1227–1235. doi: 10.1002/hep.510290448. [DOI] [PubMed] [Google Scholar]
- 27.Ito T, Lai M M C. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J Virol. 1997;71:8698–8706. doi: 10.1128/jvi.71.11.8698-8706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito T, Tahara S M, Lai M M C. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J Virol. 1998;72:8789–8796. doi: 10.1128/jvi.72.11.8789-8796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadaré G, Haenni A-L. Virus-encoded RNA helicases. J Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolykhalov A A, Agapov E V, Blight K J, Mihalik K, Feinstone S M, Rice C M. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 32.Kolykhalov A A, Agapov E V, Rice C M. Specificity of the hepatitis C virus serine proteinase: effects of substitutions at the 3/4A, 4A/4B, 4B/5A, and 5A/5B cleavage sites on polyprotein processing. J Virol. 1994;68:7525–7533. doi: 10.1128/jvi.68.11.7525-7533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolykhalov A A, Feinstone S M, Rice C M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemon S H, Honda M. Internal ribosome entry sites within the RNA genomes of hepatitis C virus and other flaviviruses. Semin Virol. 1997;8:274–288. [Google Scholar]
- 35.Lin C, Lindenbach B D, Prágai B, McCourt D W, Rice C M. Processing of the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J Virol. 1994;68:5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin C, Prágai B, Grakoui A, Xu J, Rice C M. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J Virol. 1994;68:8147–8157. doi: 10.1128/jvi.68.12.8147-8157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindsay K L. Therapy of hepatitis C: overview. Hepatology. 1997;26:71S–77S. doi: 10.1002/hep.510260713. [DOI] [PubMed] [Google Scholar]
- 38.Lohmann V, Körner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo G. Cellular proteins bind to the poly(U) tract of the 3′ untranslated region of hepatitis C virus RNA genome. Virology. 1999;256:105–118. doi: 10.1006/viro.1999.9639. [DOI] [PubMed] [Google Scholar]
- 40.Meyers G, Thiel H-J. Molecular characterization of pestiviruses. Adv Virus Res. 1996;47:53–118. doi: 10.1016/s0065-3527(08)60734-4. [DOI] [PubMed] [Google Scholar]
- 41.Mizushima H, Hijikata H, Asabe S-I, Hirota M, Kimura K, Shimotohno K. Two hepatitis C virus glycoprotein E2 products with different C termini. J Virol. 1994;68:6215–6222. doi: 10.1128/jvi.68.10.6215-6222.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamoto H, Okada S, Sugiyama Y, Kurai K, Iizuka H, Machida A, Miyakawa Y, Mayumi M. Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: comparison with reported isolates for conserved and divergent regions. J Gen Virol. 1991;72:2697–2704. doi: 10.1099/0022-1317-72-11-2697. [DOI] [PubMed] [Google Scholar]
- 43.O'Reilly E K, Kao C C. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology. 1998;252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- 44.Pai E F, Krengel U, Petsko G A, Gody R S, Katsh W, Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-Ras p21 at 1.35A resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reed K E, Rice C M. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr Top Microbiol Immunol. 1999;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 46.Reichard O, Schvarcz R, Weiland O. Therapy of hepatitis C: alpha interferon and ribavirin. Hepatology. 1997;26:108S–111S. doi: 10.1002/hep.510260719. [DOI] [PubMed] [Google Scholar]
- 47.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 931–960. [Google Scholar]
- 48.Selby M J, Glazer E, Masiarz F, Houghton M. Complex processing and protein:protein interactions in the E2:NS2 region of HCV. Virology. 1994;204:114–122. doi: 10.1006/viro.1994.1515. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka T, Kato N, Cho M-J, Shimotohno K. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem Biophys Res Commun. 1995;215:744–749. doi: 10.1006/bbrc.1995.2526. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka T, Kato N, Cho M-J, Sugiyama K, Shimotohno K. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol. 1996;70:3307–3312. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tokita H, Okamoto H, Iizuka H, Kishimoto J, Tsuda F, Miyakawa Y, Mayumi M. The entire nucleotide sequences of three hepatitis C virus isolates in genetic groups 7–9 and comparison with those in the other eight genetic groups. J Gen Virol. 1998;79:1847–1857. doi: 10.1099/0022-1317-79-8-1847. [DOI] [PubMed] [Google Scholar]
- 52.Tsuchihara K, Tanaka T, Hijikata M, Kuge S, Toyoda H, Nomoto A, Yamamoto N, Shimotohno K. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J Virol. 1997;71:6720–6726. doi: 10.1128/jvi.71.9.6720-6726.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu J, Mendez E, Caron P R, Lin C, Murcko M A, Collett M S, Rice C M. Bovine viral diarrhea virus NS3 serine proteinase: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J Virol. 1997;71:5312–5322. doi: 10.1128/jvi.71.7.5312-5322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamada N, Tanihara K, Takada A, Yorihuzi T, Tsutsumi M, Shimomura H, Tsuji T, Date T. Genetic organization and diversity of the 3′ noncoding region of the hepatitis C virus genome. Virology. 1996;223:255–261. doi: 10.1006/viro.1996.0476. [DOI] [PubMed] [Google Scholar]
- 55.Yanagi M, Purcell R H, Emerson S U, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yanagi M, St. Claire M, Emerson S U, Purcell R H, Bukh J. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc Natl Acad Sci USA. 1999;96:2291–2295. doi: 10.1073/pnas.96.5.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yanagi M, St. Claire M, Shapiro M, Emerson S U, Purcell R H, Bukh J. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology. 1998;244:161–172. doi: 10.1006/viro.1998.9092. [DOI] [PubMed] [Google Scholar]
- 58.Yoo B J, Selby M, Choe J, Suh B S, Choi S H, Joh J S, Nuovo G J, Lee H-S, Houghton M, Han J H. Transfection of a differentiated human hepatoma cell line (Huh7) with in vitro-transcribed hepatitis C virus (HCV) RNA and establishment of a long-term culture persistently infected with HCV. J Virol. 1995;69:32–38. doi: 10.1128/jvi.69.1.32-38.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]