Abstract
Introduction
Differences in transmissibility of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) variants of concern (VOCs) in different districts are hard to assess. To address this, our study focused on calculating the Real-time reproduction number (Rt) for these variants in different regions.
Methods
According to the criteria defined by the World Health Organization (WHO), the global landscape was categorized into six distinct regions. In each region, the predominant SARS-CoV-2 variant was first identified based on the proportion of variant sequencing analysis results. Then, using serial interval (SI) parameters, we calculated Rt for the relevant Variant of Concern (VOC) in each region. This approach enabled us to compare the Rt values of the same variant across different regions and analyze the transmissibility of each region's variant in relation to the overall situation in that region.
Results
The progression of VOC for SARS-CoV-2 shows regional variations. However, a common sequence of evolution is observed: Wild-type → Alpha → Beta → Delta → Omicron. Moreover, an increasing trend is discerned within diverse regions where the shift in Rt of distinct VOC corresponds with the overarching Rt route of SARS-CoV-2 in specific regions.
Conclusion
As the COVID-19 pandemic advances, regional epidemiological trends are aligning, likely due to similar virus mutations and shared public health strategies, suggesting opportunities for standardized global responses.
Keywords: SARS-CoV-2, Period of dominant variants, Real-time reproduction number, Six regions
1. Introduction
Since the first reported case of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2, also previously known as 2019-nCoV) in China in December 2019, the virus has rapidly spread worldwide, leading to a global pandemic [1]. As of September 12, 2022, the World Health Organization (WHO) reports over 600 million cumulative cases. A key factor in SARS-CoV-2's extensive impact is its ability to mutate into numerous variants. By September 12, 2022, 144,692 variants had been identified. WHO has categorized these variants into the following categories: Variants of Interest (VOIs), Variants under monitoring (VUMs), and Variants of Concern (VOCs) for better tracking and monitoring. The latest VOC, Omicron (B.1.1.529), was first discovered in southern Africa, and has significant impacts on public health worldwide. Omicron rapidly quickly became the dominant variant surpassing Delta within just a few months [2,3].
Previous studies have shown that each new VOC of SARS-CoV-2 tends to have a higher reproduction number (R0). The progression of R0 is as follows: Beta(1.19)/Alpha(1.22–1.51) → Gamma(1.21) → Delta(1.38) → Omicron(1.90–8.2) [[4], [5], [6]]. Most of the studies focused on comparing the transmissibility of VOCs to wild-type. The transmissibility increase was 29 % (95 % CI: 24–33 %) for Alpha, 25 % (95 % CI: 20–30 %) for Beta, 38 % (95 % CI: 29–48) for Gamma, and 97 % (95 % CI: 76–117) for Delta [7]. However, the R0 or Reff can vary significantly across different regions. For example, the Delta variant had a Reff ranging from 2.43 to 5.11 in South Africa, Denmark, China, the UK and Italy [8]. Similarly, Omciron's R0 has a mean value of 9.5, between 5.5 and 24 (median 10, interquartile range 7.25,11.88). The parameter estimates for R0 are influenced by various factors, such as the number of infections, the infection rate, and the recovery rate, all of which might be time-dependent, non-linear, and different among populations in different regions. This variability makes the timely comparison of R0 or Reff across countries challenging [9].
This study aims to address the aforementioned problem by focusing on the Rt as a metric to assess the transmissibility of VOC in six regions. By conducting such comparative analyses, the research aims to identify similarities or dissimilarities in the patterns of influence experienced by these countries when confronted with diverse dominant variant periods. The ultimate goal is to enhance the effectiveness of outbreak control strategies for handling subsequent outbreaks associated with emerging VOC in the future.
2. Methods and materials
2.1. Study design
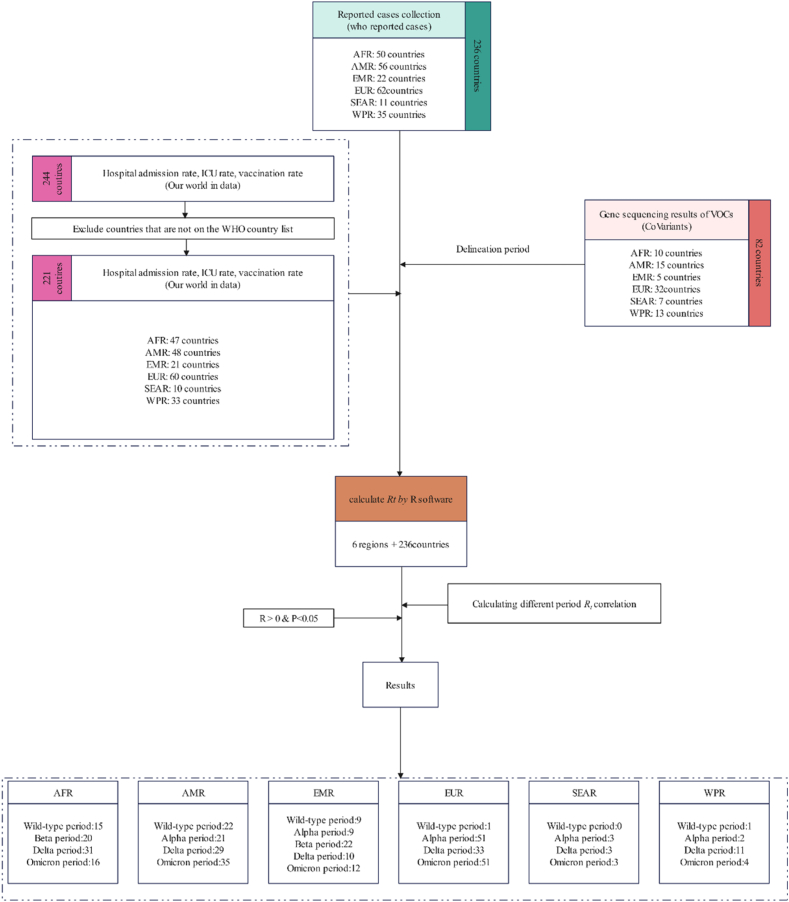
This study is based on the analysis of raw data on SARS-CoV-2 variants and reported COVID-19 cases, sourced from two publicly accessible datasets [[10], [11], [12]]. Countries with no reported COVID-19 cases or incomplete data were carefully excluded to maintain data integrity. Subsequently, a comparative assessment was conducted to evaluate the transmissibility of different VOCs in the remaining countries and regions. Additionally, the correlation between Rt in different regions and the overall regional situation was examined. The study design concept is illustrated in Fig. 1.
Fig. 1.
Schematic diagram of the study design (WPR: Western Pacific Region; SEAR: Southeast Asia Region; EMR: Eastern Mediterranean Region; EUR: European Region; AMR: American Region; AFR: African Region).
2.2. Data collection
Raw data on SARS-CoV-2 variants and reported COVID-19 cases were obtained from two publicly accessible datasets [[7], [8], [9]]. Additionally, different serial interval (SI) of VOC were obtained from previous studies (Table S1).
2.3. Period division of dominant variants
We collated the gene sequencing data of SARS-CoV-2 from the publicly available data of CoVariants [11]. Subsequently, we identified the periods during which VOC were the dominant variants, as determined by the highest percentage of variant distribution in the gene sequencing results.
2.4. Rt calculation
In this study, we measured the transmissibility of the SARS-CoV-2 VOC through Rt [[13], [14], [15]] to estimate whether the disease outbreak is growing (Rt > 1) or declining (Rt < 1), and the Rt can provide real-time information for adjusting control measures. In calculating Rt for different VOC, we used different SI with different variants (Table S1). We used 14 days as a rolling window to calculate Rt. It was estimated by EpiEstim (version 2.2.4) in the R software (version 4.1).
The method assumed that the incidence of new cases (It) on day t could be expressed as a Poisson distribution. The specific calculation steps and equations (S1) - (S5)in the online Supplementary Materials are used for the calculations.
2.5. Correlation analysis
In this study, the correlation coefficient r [16] was calculated by the following calculation method (6). In this study, the Rt values of the six regions overall were calculated, and the Rt of each country in the region was compared with the Rt of the region at the test level of 0.05 of the t-test. Countries with p < 0.05 and r>0 were considered to have the same trend with the overall trend of Rt in the region at that dominant variant period. The specific calculation steps and equations (S6) - (S7)in the online Supplementary Materials are used for the calculations.
3. Results
3.1. Timeline of different variants
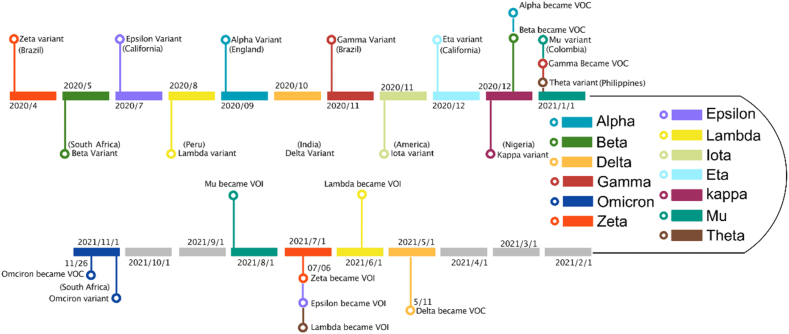
A timeline chart (Fig. 2) was constructed, encompassing the first reported occurrences of various SARS-CoV-2 variants, including VOC, VOI, and VUM. This chart comprises data from a total of 13 distinct variants. Notably, five SARS-CoV-2 variants have been officially classified as VOCs by the World Health Organization (WHO), namely Alpha, Beta, Gamma, Delta, and Omicron.
Fig. 2.
Timeline of the appearance of different variants of COVID-19.
Table S2 details the durations of VOCs in different regions. Specifically, the Delta variant had the longest duration in Southeast Asia and the shortest duration in Africa. Moreover, the Omicron variant demonstrated the most prolonged duration, lasting 294 days in Africa, while its shortest duration of 252 days was observed in America, Southeast Asia, and the Western Pacific.
Table 1 provides a comprehensive overview of the cumulative number of incidences and the Public Health and Social Measures (PHSM) stringency index values associated with the presence of different SARS-CoV-2 variants, including the wild-type, Alpha, Beta, Delta, Gamma, and Omicron, across six global regions. The data reveals that the highest PHSM stringency index was recorded in the American Region (AMR) during the wild-type phase with a value of 79.63, indicating robust control measures. In contrast, the index was notably lower during the Omicron phase across all regions, with AMR showing a significant decrease to 29.19.
Table 1.
COVID-19 global and cumulative number of incidences, cumulative number of deaths and PHSM stringency index for different variants.
| Regions | Statistical indicators | Wild-type | Alpha | Beta | Delta | Gamma | Omicron |
|---|---|---|---|---|---|---|---|
| The global | cumulative number of incidences | 894299.595 | 5555283.913 | 388183.251 | 5819618.093 | – | 23397297 |
| AFR | cumulative number of incidences | 45859.171 | – | 305080.498 | 321037.866 | – | 631959.9 |
| PHSM stringency index | 70.37 | – | 47.22 | 45.37 | – | 33.89 | |
| AMR | cumulative number of incidences | 221836.322 | 1353142.816 | – | 1427555.995 | – | 4833014.686 |
| PHSM stringency index | 79.63 | 66.2 | – | 56.48 | – | 29.19 | |
| EMR | cumulative number of incidences | 322979.958 | 274477.406 | 83102.753 | 358094.119 | – | 711963.378 |
| PHSM stringency index | 64.81 | 56.48 | 62.5 | 49.045 | – | 35.19 | |
| EUR | cumulative number of incidences | 214601.829 | 3730144.599 | – | 3106252.195 | – | 12326405.92 |
| PHSM stringency index | 55.09 | 58.8 | – | 42.59 | – | 19.11 | |
| SEAR | cumulative number of incidences | 41340.142 | 38351.268 | – | 51141.441 | – | 314722.651 |
| PHSM stringency index | 77.78 | 68.06 | – | 76.85 | – | 37.04 | |
| WPR | cumulative number of incidences | 47682.173 | 159167.824 | – | 555536.477 | – | 4579230.472 |
| PHSM stringency index | 53.24 | 51.39 | – | 55.64 | – | 40.49 |
Furthermore, the study observes that PHSM responses varied not only with the type of variant but also across different regions. For instance, the Eastern Mediterranean Region (EMR) and the European Region (EUR) displayed higher PHSM stringency indices during the wild-type phase compared to the periods dominated by later variants.
3.2. The alternation process of VOC
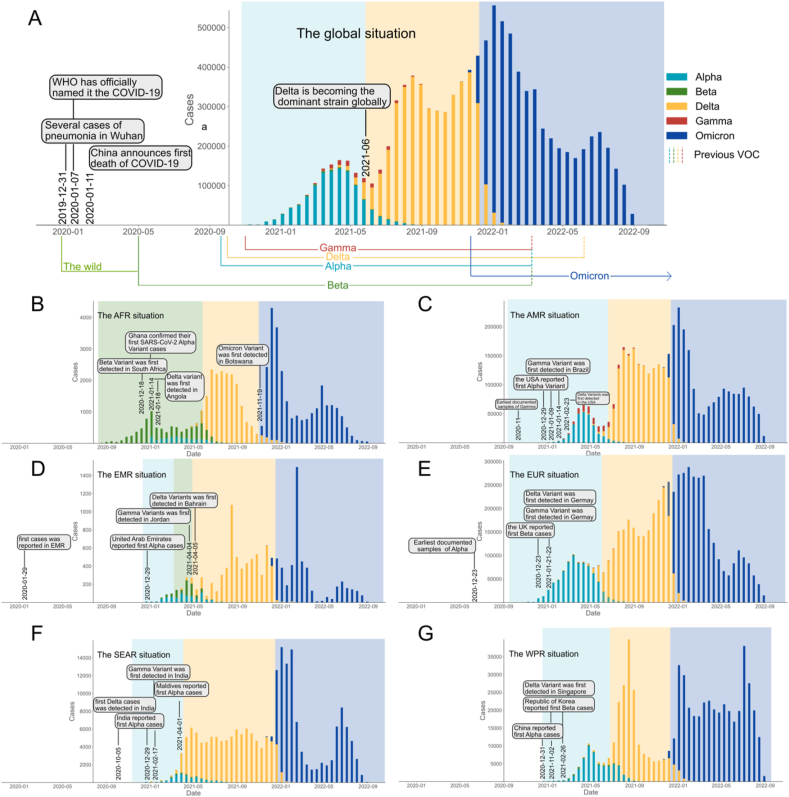
Fig. 3 (A) outlines the temporal phases of the COVID-19 pandemic, characterized by the predominance of three Variants of Concern (VOCs): Alpha, Delta, and Omicron. These periods commenced on August 17, 2020, for Alpha; June 7, 2021, for Delta; and December 20, 2021, for Omicron, with recorded cumulative infections of 151,277,111, 101,727,007, and 342,905,211, lasting 293, 195, and 292 days, respectively. The phase prior to the emergence of Alpha is identified as the wild-type virus outbreak period. The Omicron phase, still ongoing as of the study's cut-off date of September 12, 2022, has become the most extended duration of the pandemic, exceeding 292 days.
Fig. 3.
Sequencing results of SARS-CoV-2 VOCs globally and in six regions (Panel A represents the global situation, panels B–G represent AFR, AMR, EMR, EUR, SEAR, and WPR respectively.).
AFR is unique among the regions, as it experienced the Beta variant as the dominant strain for nearly 300 days [Fig. 3 (B)]. In contrast, the Alpha variant emerged as the initial dominant strain in the other five regions [Fig. 3 (C-G)], with each subsequently following a uniform transition sequence of 'Alpha→Delta→Omicron,' occurring within similar durations. Notably, EMR presents a deviation from this pattern, with a distinct 27-day period of Beta dominance interrupting the 'Alpha→Delta' succession before ultimately giving way to Omicron. Table S3 provides a detailed breakdown of these variant-dominant periods.
3.3. Comparative analysis of Rt in different regions
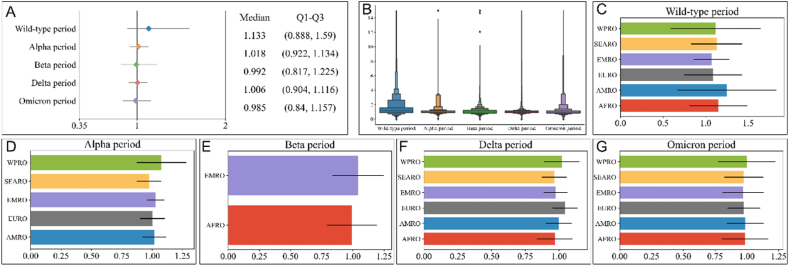
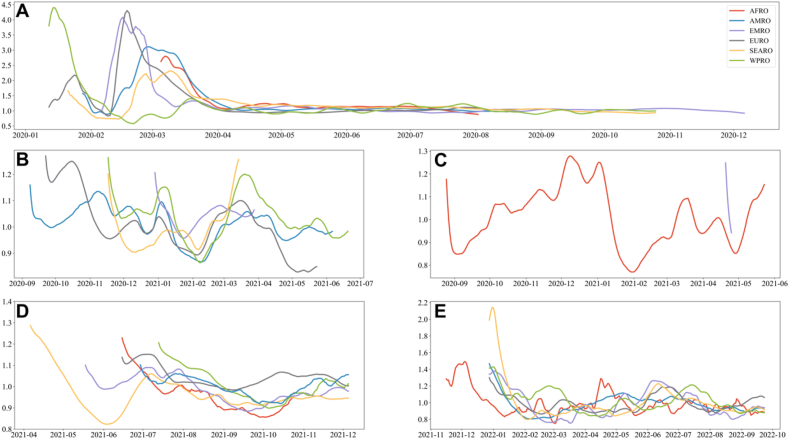
We analyzed Rt for different dominant variants during the global timeline, as depicted in Fig. 4. The period dominated by the wild-type strain registered the highest median Rt at 1.133 [Fig. 4 (A-B)], with an interquartile range (IQR) of (0.888, 1.59). In contrast, the Omicron period had a median Rt of 0.985, with an IQR of (0.840, 1.157). The specific median Rt for the different variants is presented in Table S4.
Fig. 4.
Global Rt values for different variant periods (Panel A represents the median and quartiles of Rt for 5 periods; Panel B represents box plots of all Rt values for 5 periods; Panels C–G represent the Rt values for each continent for different dominant variants).
During the wild-type period [Fig. 4 (C)], AFR demonstrated a median Rt of 1.246 with an IQR of (0.952, 2.114), while WPR exhibited the highest median Rt of 1.077 with an IQR of (0.945, 1.356) during the Alpha period [Fig. 4 (D)],. In the Beta period [Fig. 4 (E)], only two regions, AFR and EMR, were included, with EMR displaying the highest median Rt of 1.043 and an IQR of (0.890, 1.302). For the Delta period [Fig. 4 (F)], EUR had the highest median Rt of 1.045 with an IQR of (0.955, 1.142), and SEAR had the lowest median Rt of 0.965 with an IQR of (0.894, 1.078). Finally, in the Omicron period [Fig. 4 (G)], WPR exhibited the highest median Rt of 1.001 with an IQR of (0.849, 1.300), while EMR had the lowest median Rt of 0.970 with an IQR of (0.818, 1.145). Detailed variations in Rt values for the five periods across different regions are illustrated in Fig. 5 (A-E).
Fig. 5.
Rt values for different regions (Panel A: Wild-type; Panel B: Alpha; Panel C: Beta; Panel D: Delta; Panel E: Omicron).
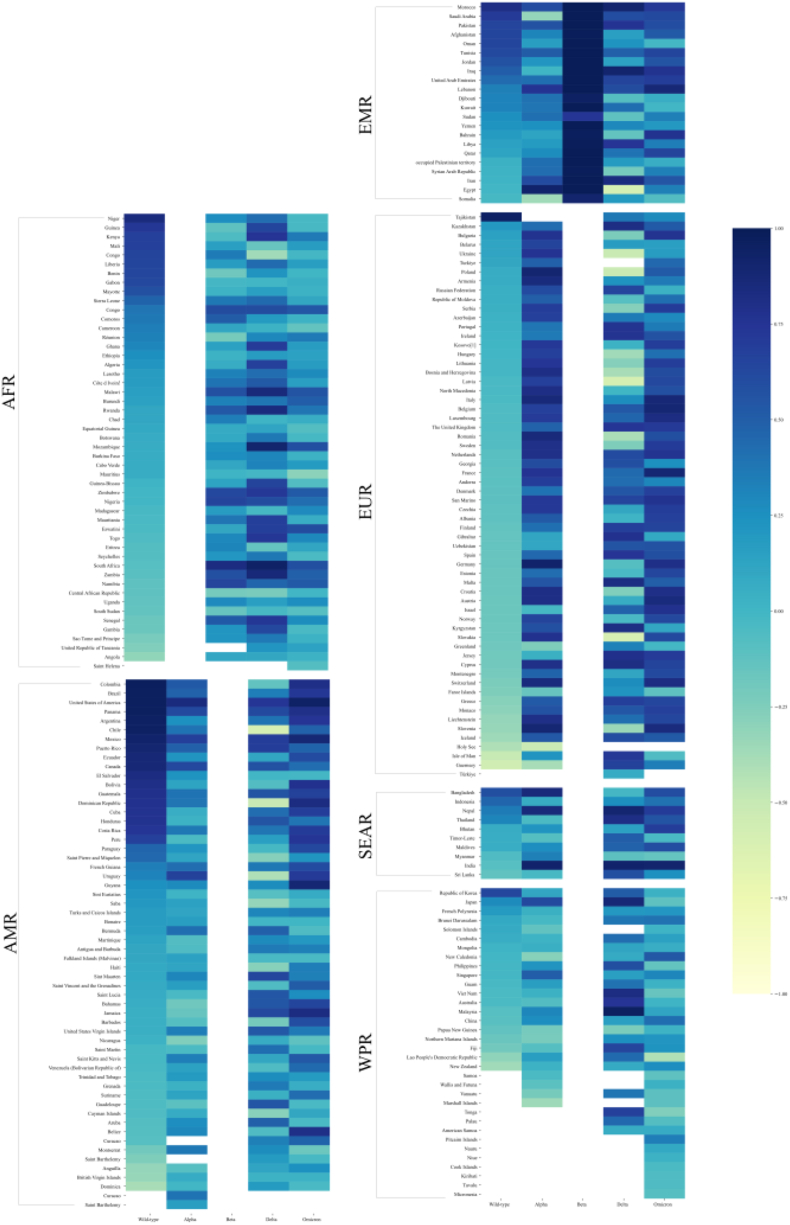
3.4. Correlation analysis of Rt
As depicted in Fig. 6 and Table S5, the trends in Rt exhibited a consistent pattern with the respective regions in different periods for various countries. Specifically, for the AFR, the Rt trends were consistent in 15, 20, 31, and 16 countries during the Wild-type, Beta, Delta, and Omicron periods, respectively. Similarly, the AMR saw similar consistency in 22, 21, 29, and 35 countries for the Wild-type, Alpha, Delta, and Omicron periods, respectively.
Fig. 6.
Consistency results of Rt values for six regions for different periods.
In the EMR, the Rt trends demonstrated consistency in 9, 9, 22, 10, and 12 countries during the Wild-type, Alpha, Beta, Delta, and Omicron periods, respectively. Furthermore, in the EUR, 1, 51, 33, and 51 countries exhibited consistent Rt trends during the Wild-type, Alpha, Delta, and Omicron periods, respectively. Notably, during the continuous evolution of the COVID-19 pandemic, the consistency of national Rt trends within the same region showed a general increasing trend and stabilization, particularly during the "Delta→Omicron" period.
4. Discussion
In this study, we segmented the global COVID-19 epidemic into distinct periods based on dominant VOC in different regions, aiming to compare and contrast disease transmissibility both within and across these regions. Our analyses sought insights into the variations and similarities in the spread and impact of different VOCs.
The progression of VOCs followed a sequence from Wild-type to Alpha, Beta, Delta, and finally Omicron. In this study, a concise systematic review of VOC was conducted, aiming to gain insights into the variations and similarities in disease spread and impact associated with different VOC across the studied regions. 1) Alpha, first detected in southeast England in September 2020, quickly spread to become the dominant variant in the UK within a few months. It then spread to at least 114 countries worldwide, owing to its significantly higher transmissibility compared to the wild-type. 2) The Beta variant dominated in Africa for a prolonged period of 294 days. Still, it did not spark a global pandemic, potentially due to prolonged viral replication influenced by HIV prevalence, strong interventions at that time, and its evolution from the wild-type. 3) Delta, with approximately 50 % higher transmissibility than Alpha in the UK and about 13 % faster spread [17], replaced Alpha and Beta to become the predominant variant in six regions. 4) The emergence of the Omicron variant, with a higher R0 value (10) [18] and a shorter incubation period [19] compared to other VOCs such as Alpha and Delta [20], has raised concerns due to its potential for rapid transmission. The short incubation period allows cases with the ability to spread to circulate within society before isolation or treatment, leading to widespread outbreaks. Vaccination [21] is considered the most effective measure to combat the SARS-CoV-2 pandemic, and many countries have initiated third and even fourth doses of vaccinations to enhance immunity. However, continuous mutation of SARS-CoV-2 and waning vaccine effectiveness have contributed to ongoing epidemics in some regions, such as the United States [22], the United Kingdom [23], and Hong Kong [24], China [25], despite relatively high vaccination rates of nearly 60 %. The emergence of the highly transmissible Omicron variant, rapidly upgraded to VOCs by the WHO only 15 days after its detection, poses further challenges due to its strong transmissibility and increased immune evasion and mutation capabilities. Many factors can influence the high transmission of Omicron. Based on the summary results of the literature, the following are the main possible reasons for this, 1) genomic sequencing data of Omicron shows that there are more than 30 mutations in the spiny mutant protein, which constantly produces novel variants during infection; 2) It N501Y mutation increases the binding affinity to ACE2 receptor [26], which is the main influencing factor for increased transmission, and Q498R binding, the binding affinity is more substantial, and the Omicron variant readily enters the host so that Omicron can be hyper-transmissible; 3) Heavy mutations in Omicron spike-in proteins are associated with increased infectivity and antibody evasion. BNT vaccinations provide more than 90 % protection against severe disease when infected with Delta, but may be significantly less effective against Omicron.
To address disparities in transmissibility influenced by region and time, we collected SI parameters of different variants. This enabled us to estimate Rt values across times and regions, balancing the effects of time and region on transmissibility. We used a consistent set of natural history parameters for different variants to analyze Rt values over time.
According to the results of the study, the overall trend of Rt values of the same variant in different regions differed. For example, the median Rt of Alpha in WPR was the highest at 1.077, while the median Rt of SEAR was 0.975, the median Rt of Delta in EUR was the highest at 1.046, while the median Rt of AFR was 0.970, the median Rt of Omicron in WPR was the highest at 1.001, while the median Rt of EMR was 0.971. During the period of VOC succession, the transition occurred from the Wild-type variant to Alpha, followed by Beta, then Delta, and finally Omicron. A gradual decline in the discrepancy of Rt values was observed, with the median difference in Rt decreasing from 0.102 to 0.076 and subsequently to 0.03. With the turnover of Variants of Concern (VOCs), the number of countries in different regions exhibiting convergence in Rt trends with the overall region is progressively increasing. This phenomenon may be attributed to the prolonged duration of the COVID-19 pandemic, spanning nearly three years, leading to a relaxation of control measures on a global scale. Consequently, new VOCs are only able to replace the previous VOC as dominant variants when they exhibit a significantly higher transmissibility. Given the high mutability of the SARS-CoV-2 virus, it is imperative for the six regions of the world to remain vigilant and acknowledge the heightened potential for another pandemic when a new VOC emerges.
The study revealed distinct regional trends in Rt values for the same SARS-CoV-2 variant. For instance, the Alpha variant exhibited a median Rt of 1.077 in the WPR, the highest among the regions, whereas in the SEAR, its median Rt was notably lower at 0.975. Similar regional disparities were observed for other variants: the Delta variant's median Rt was highest in the EUR at 1.046, compared to 0.970 in the AFR. For the Omicron variant, the highest median Rt was recorded in WPR at 1.001, while the EMR had a lower median Rt of 0.971.
During the succession of VOCs — transitioning from Wild-type to Alpha, then Beta, Delta, and finally Omicron — there was a noticeable decline in the variance of Rt values across regions. The median difference in Rt values gradually decreased from 0.102 to 0.076, eventually to 0.03. This pattern indicates an increasing convergence of Rt trends among countries within the same region, particularly through the Delta to Omicron transition. This trend may be linked to the prolonged duration of the COVID-19 pandemic, which has spanned nearly three years and led to a global relaxation of control measures. As a result, a new VOC can only supplant its predecessor as the dominant variant if it demonstrates significantly higher transmissibility. The high mutability of the SARS-CoV-2 virus necessitates continued vigilance in all six global regions, especially in recognizing the potential for new pandemics triggered by emerging VOCs.
5. Limitations
The limitations of this study encompass several aspects. First, during data collection from countries within six regions, significant variations in sample sizes among countries were observed, potentially introducing biases. However, this study employed inclusion and exclusion criteria that solely removed countries with missing data. Second, the collection of results from different variant sequencing relied on a single source, possibly leading to reporting biases. Third, there is a scarcity of similar studies for comparative analysis, resulting in a lack of comparative research findings. Considerably, more work will need to be done to gather additional genetic sequencing data and results from similar studies.
6. Conclusions
As the COVID-19 pandemic unfolds, a notable pattern is emerging across different geographic areas. Epidemiological trends, which initially varied widely from one region to another, are now showing signs of convergence. This uniformity in trends may be attributed to a variety of factors, including similar virus mutations, shared public health responses, and the global dissemination of information about the virus. The convergence of these trends suggests that regions are experiencing similar stages of the pandemic or are adopting comparable strategies in their fight against the virus. This uniformity offers an opportunity for global health organizations to develop more standardized, effective approaches to combat COVID-19 and prepare for future pandemics. Understanding and analyzing these converging patterns can lead to more coordinated and efficient global health responses.
Funding
This project was supported by National Key Research and Development Program of China (No. 2021YFC2301604), Self-supporting Program of Guangzhou Laboratory (No. SRPG22-007), Exploration and Research on a Precision Prevention and Control System for Close Contacts and Other High-Risk Individuals of COVID-19 (No. 2022SK2121) and Research on the Transmission Capacity and Prevention Effectiveness of the COVID-19 Omicron Variant Based on Mathematical Models (No. D202312058730).
Data availability statement
The data underpinning the findings of this study were sourced from open-source platforms and are publicly accessible in the CoVariants repository. For further examination and use, these data can be found at https://github.com/hodcroftlab/covariants.
Ethics declarations
Review and approval by an ethics committee was not needed for this study because it does not involve any human participants, animals, or data that could raise ethical concerns. The nature of this study is theoretical, and it strictly adheres to the general ethical guidelines for academic research. No identifiable personal or sensitive information has been used or disclosed in the course of this research. Therefore, informed consent from participants/patients was not applicable to this study.
CRediT authorship contribution statement
Hongjie Wei: Writing – review & editing, Writing – original draft, Visualization, Methodology, Formal analysis, Data curation, Conceptualization. Yunkang Zhao: Supervision, Resources. Jia Rui: Writing – review & editing, Supervision, Conceptualization. Kangguo Li: Software, Conceptualization. Buasiyamu Abudunaibi: Writing – review & editing. Zeyu Zhao: Supervision, Conceptualization. Wentao Song: Supervision, Project administration. Yao Wang: Data curation, Conceptualization. Qiuping Chen: Writing – original draft, Resources. Hong Liu: Resources. Shuo Zhang: Validation. Xiaojun Li: Writing – original draft, Validation. Kaiwei Luo: Writing – original draft, Validation. Laurent Gavotte: Supervision. Roger Frutos: Supervision. Tianmu Chen: Writing – review & editing, Supervision, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e32164.
Contributor Information
Laurent Gavotte, Email: lgavotte@gmail.com.
Roger Frutos, Email: frutossmt@gmail.com.
Tianmu Chen, Email: 13698665@qq.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.McArthur L., et al. Review of burden, clinical definitions, and management of COVID-19 cases. Am. J. Trop. Med. Hyg. 2020;103(2):625. doi: 10.4269/ajtmh.20-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Umakanthan S., et al. Origin, transmission, diagnosis and management of coronavirus disease 2019 (COVID-19) Postgrad. Med. 2020;96(1142):753–758. doi: 10.1136/postgradmedj-2020-138234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin L., et al. The disease severity and clinical outcomes of the SARS-CoV-2 variants of concern. Front. Public Health. 2021;9 doi: 10.3389/fpubh.2021.775224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manathunga S., Abeyagunawardena I., Dharmaratne S. 2022. A Comparison of Transmissibility of SARS-CoV-2 Variants of Concern. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang B., et al. Estimation of the transmission risk of the 2019-nCoV and its implication for public health interventions. J. Clin. Med. 2020;9(2):462. doi: 10.3390/jcm9020462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guerra F.M., et al. The basic reproduction number (R0) of measles: a systematic review. Lancet Infect. Dis. 2017;17(12):e420–e428. doi: 10.1016/S1473-3099(17)30307-9. [DOI] [PubMed] [Google Scholar]
- 7.Sheikhi F., et al. Estimation of the basic reproduction number of Alpha and Delta variants of COVID-19 pandemic in Iran. PLoS One. 2022;17(5) doi: 10.1371/journal.pone.0265489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du Z., et al. Reproduction number of the Omicron variant triples that of the Delta variant. Viruses. 2022;14(4):821. doi: 10.3390/v14040821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hisaka A., et al. Global comparison of changes in the number of test-positive cases and deaths by coronavirus infection (COVID-19) in the world. J. Clin. Med. 2020;9(6):1904. doi: 10.3390/jcm9061904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization . 2022 Sep 12. WHO Coronavirus (COVID-19) Dashboard: Overview.https://covid19.who.int/ [Google Scholar]
- 11.CoVariants. CoVariants: variants. 2022 Sep 01. https://covariants.org/variants
- 12.Our world in data. Coronavirus Pandemic (COVID-19): data explorer. 2022 Sep 01. https://ourworldindata.org/explorers/coronavirus-data-explorer
- 13.Cori A., et al. A new framework and software to estimate time-varying reproduction numbers during epidemics. Am. J. Epidemiol. 2013;178(9):1505–1512. doi: 10.1093/aje/kwt133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallinga J., Teunis P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am. J. Epidemiol. 2004;160(6):509–516. doi: 10.1093/aje/kwh255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reich N.G., et al. Estimating incubation period distributions with coarse data. Stat. Med. 2009;28(22):2769–2784. doi: 10.1002/sim.3659. [DOI] [PubMed] [Google Scholar]
- 16.Ezekiel M. 1930. Methods of Correlation Analysis. [Google Scholar]
- 17.Hemmer C.J., Löbermann M., Reisinger E. COVID-19: Epidemiologie und Mutationen. Radiologe. 2021;61(10):880–887. doi: 10.1007/s00117-021-00909-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferré V.M., et al. Vaccine Ab neutralization against Omicron and SARS-CoV-2 variants using neutralization and specific ELISA assays. J. Infect. 2022;84(6):834–872. doi: 10.1016/j.jinf.2022.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Del Águila-Mejía J., et al. Secondary attack rate, transmission and incubation periods, and serial interval of SARS-CoV-2 Omicron variant, Spain. Emerg. Infect. Dis. 2022;28(6):1224. doi: 10.3201/eid2806.220158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMahan K., et al. Reduced pathogenicity of the SARS-CoV-2 Omicron variant in hamsters. Med. 2022;3(4):262–268. e4. doi: 10.1016/j.medj.2022.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jena A., et al. Response to SARS-CoV-2 vaccination in immune mediated inflammatory diseases: systematic review and meta-analysis. Autoimmun. Rev. 2022;21(1) doi: 10.1016/j.autrev.2021.102927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.See I., et al. Case series of thrombosis with thrombocytopenia syndrome after COVID-19 vaccination—United States, december 2020 to August 2021. Annals of internal medicine. 2022;175(4):513–522. doi: 10.7326/M21-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen L.H., et al. Self-reported COVID-19 vaccine hesitancy and uptake among participants from different racial and ethnic groups in the United States and United Kingdom. Nat. Commun. 2022;13(1):1–9. doi: 10.1038/s41467-022-28200-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMenamin M.E., et al. Vaccine effectiveness of one, two, and three doses of BNT162b2 and CoronaVac against COVID-19 in Hong Kong: a population-based observational study. Lancet Infect Dis. 2022;22(10):1435–1443. doi: 10.1016/S1473-3099(22)00345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin C., et al. Acceptance of a third dose of COVID-19 vaccine and associated factors in China based on Health Belief Model: a national cross-sectional study. Vaccines. 2022;10(1):89. doi: 10.3390/vaccines10010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou D., et al. Evidence of escape of SARS-CoV-2 variant B. 1.351 from natural and vaccine-induced sera. Cell. 2021;184(9):2348–2361. e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underpinning the findings of this study were sourced from open-source platforms and are publicly accessible in the CoVariants repository. For further examination and use, these data can be found at https://github.com/hodcroftlab/covariants.