Abstract
Introduction
Darier disease is a rare inherited disease with dominant skin manifestations including keratotic papules and plaques on sebaceous and flexural areas. Secondary infection of skin lesions is common, and Staphylococcus aureus commonly colonizes these lesions. The aim of the study was to characterize the bacterial microbiome of cutaneous Darier lesions compared to normal-looking skin and disease severity.
Methods
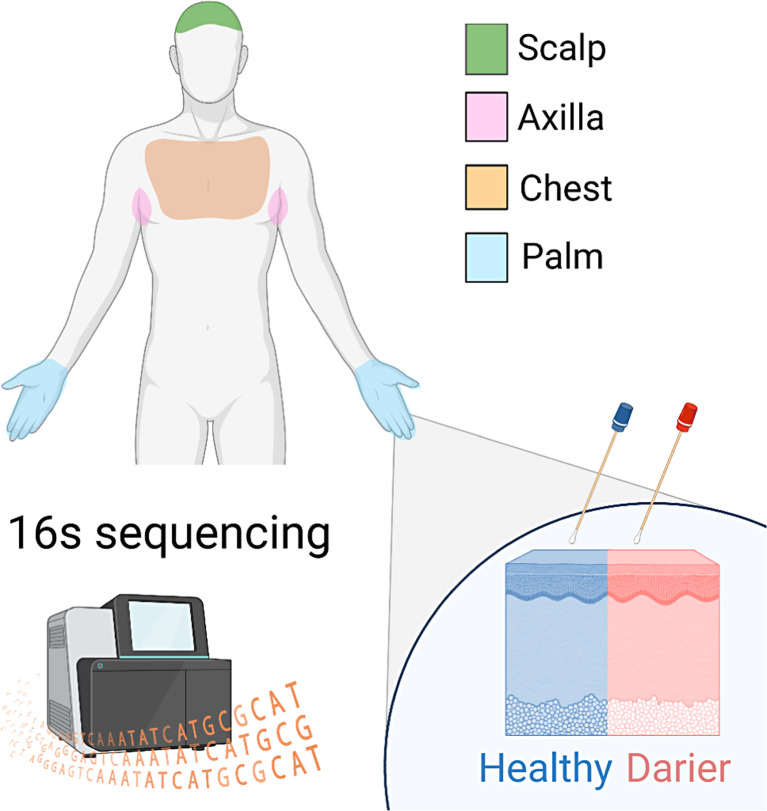
All patients with a history of Darier followed up at Emek Medical Center were invited to participate in the study. Patients that did not use antibiotics in the past month and signed informed consent had four skin sites sampled with swabs: scalp, chest, axilla, and palm. All samples were analyzed for bacterial microbiome using 16S rDNA sequencing.
Results
Two hundred and eighty microbiome samples obtained from lesional and non-lesional skin of the scalp, chest, axilla, and palm of 42 Darier patients were included in the analysis. The most abundant bacterial genera across all skin sites were Propionibacterium, Corynebacterium, Paracoccus, Micrococcus, and Anaerococcus. Scalp and chest lesions featured a distinct microbiome configuration that was mainly driven by an overabundance of Staphylococci species. Patients with more severe disease exhibited microbiome alterations in the chest, axilla, and palm compared with patients with only mild disease, driven by Peptoniphilus and Moryella genera in scalp and palmar lesions, respectively.
Conclusion
Staphylococci were significantly associated with Darier lesions and drove Darier-associated dysbiosis. Severity of the disease was associated with two other bacterial genera. Whether these associations also hold a causative role and may serve as a therapeutic target remains to be determined and requires further investigation.
Keywords: Microbiome, Darier disease, Staphylococcus, Peptoniphilus, Moryella
Introduction
The human cutaneous microbiome consists of a variety of microorganisms, including viruses, bacteria, fungi, and parasites. The four major bacterial phyla found on the skin are Actinobacteria (36–51%), Firmicutes (24–34%), Proteobacteria (11–16%), and Bacteroidetes (6–9%) [1]. However, the composition of the cutaneous microbiome varies between different body sites, with three major microenvironments: oily/sebaceous sites (e.g., forehead); moist sites (e.g., antecubital fossa); and dry sites (e.g., volar forearm) [2].
In the past few years, multiple studies have analyzed the cutaneous microbiome of patients with a variety of skin diseases, such as atopic dermatitis [3–5], seborrheic dermatitis [6], acne [7], psoriasis [8], alopecia areata [9], and mycosis fungoides [10]. These studies found the cutaneous microbiome to be altered in patients with skin diseases compared to healthy individuals [8], in lesional compared to non-lesional skin [3], and during flares of the disease compared to times of remission [4]. Interestingly, cutaneous dysbiosis has been implicated to have a pathogenic role in some disease states [11, 12].
Originally described in 1889 [13], Darier disease is a rare autosomal dominant inherited disease [14] caused by mutations in the ATP2A2 gene [15]. The disease has dominant skin manifestations, with the most common being a rash comprised of keratotic papules that may coalesce to form plaques on sebaceous areas such as the chest, back, head, and neck. Additional common manifestations are malodorous papillomatous plaques on flexural areas and wart-like papules and pits on the hands [16–18]. Darier disease may also involve the nails, oral mucosa, and other extracutaneous systems; mostly neuropsychiatric [18] but also learning disabilities [19], salivary gland obstruction [16], and ophthalmic involvement [20].
Darier disease is often associated with bacterial and viral skin infections, including reports of life-threatening bacterial infections [21–27]. Dodiuk-Gad et al. [28] found a high prevalence of S. aureus colonization in lesional skin and nares in patients with Darier disease. Moreover, an association was discovered between the severity of disease and extent of S. aureus colonization. These findings, as well the evidence of improvement of Darier lesions with antibiotic treatment [29, 30], may suggest a pathogenic role of the microbiome in this disease. However, to date, there have been no studies analyzing the cutaneous microbiome of Darier patients using next-generation sequencing methodologies. The goal of the current study was to characterize the bacterial microbiome of cutaneous Darier lesions and compare it to non-lesional skin and to the disease severity.
Materials and Methods
This cross-sectional study’s protocol was approved by the Institutional Ethics Committee.
Participants
All patients with a history of Darier disease followed up at Emek Medical Center, Israel, during 2017–2018 were invited to participate in the study. Inclusion criteria were a clinicopathological diagnosis of Darier disease and age of 18 years or older. Only patients with both a biopsy-proven diagnosis of Darier disease and typical clinical skin features were enrolled in the study. In order to apply these inclusion criteria, each participant was assessed by an experienced dermatologist (the primary investigator [PI], R.P.D-G.) prior to enrollment in the study. Exclusion criteria were the use of topical or systemic antibiotic agents in the previous month; lack of apparent Darier disease skin lesions; pregnancy; and inability to sign informed consent.
Instruments
Research Visit
All patients were contacted via a phone call and invited for a research visit, during which patients signed written, informed consent, were interviewed, had a full skin examination, and were sampled with swabs.
Data collected included date of birth, gender, age of disease onset, history of hospitalizations, previous treatments, and history of topical and systemic antibiotic treatments over the previous 3 months.
A physical examination was performed by the same dermatologist (R.P.D.-G.) in all patients to determine disease severity according to Sakuhntabhai et al. [15] (“mild” – few keratotic papules involving the torso or flexural area or limited to not more than two body sites/“moderate” – multiple keratosis papules on the torso or flexural areas involving more than two body sites or coalescing into large plaques/“severe” – papillomatous plaques merging to involve most of the torso or hypertrophic plaques with major involvement of flexural areas); disease distribution (seborrheic/flexural/segmental/acral/other [16]); and percentage of body surface area involvement.
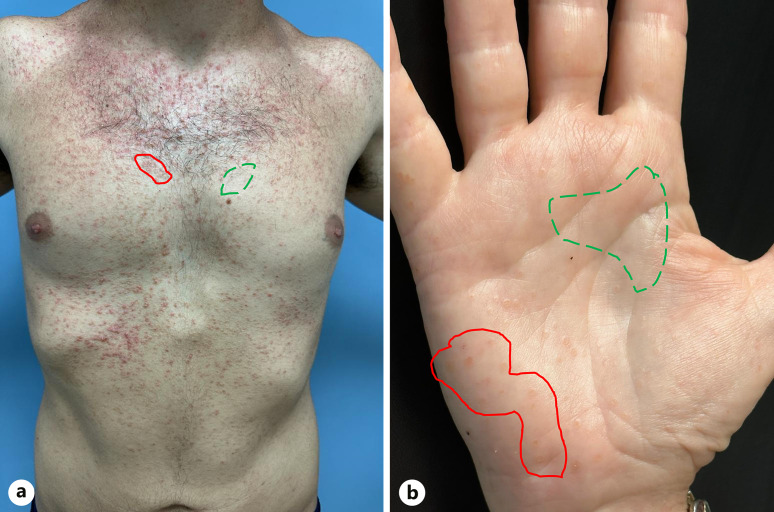
Microbiome samples were collected based on a predefined protocol from four cutaneous sites: (1) scalp, (2) chest, (3) axilla, and (4) palm. For each cutaneous site, two samples were obtained: one from a Darier lesion (when available) and one from normal-looking skin (Fig. 1). A clear method was used to differentiate between lesional and non-lesional skin for obtaining swabs from the scalp, chest, axilla, and palm. Lesions of Darier disease on the scalp, chest, and axilla included the classical lesion of Darier disease: keratotic papules with central erosion. Lesions of Darier disease on the palm included either wart-like papules on the dorsum of the hands or palmar pits – palmar papules with central depression. These lesions characterize the disease, as described in our previous publication [17]. Non-lesional skin was considered the skin without these lesions in those locations. To ensure accurate adherence to this methodology and proper diagnosis and differentiation between lesional and non-lesional skin, all skin swabs in the study were conducted by an experienced dermatologist (the primary investigator [PI], R.P.D.-G.). Lesional and non-lesional skin are demonstrated in Figure 2.
Fig. 1.
A representative scheme of the study design. We sampled the cutaneous microbiome from Darier patients using swabs in four body sites. For each Darier lesion, a control sample was collected from adjacent healthy-looking skin. All samples were sequenced by 16S rDNA sequencing.
Fig. 2.
Lesional versus non-lesional skin. An example of a classic study participant’s torso (a) and palm (b). The areas sampled as lesional skin are circled with a red straight curve and the areas sampled as non-lesional skin are circled by a green dotted curve.
All samples were obtained using a sterile rayon-tipped swab (COPAN, Brescia, Italy) soaked in sterile 0.15 m NaCl with 0.1% Tween 20 (J.T. Baker) from a 2 × 2 cm area of skin and were stored at −80°C until DNA extraction as previously described [5].
16S rDNA Sequencing
DNA was extracted from the swabs and filters using the DNeasy PowerSoil and PowerWater DNA extraction kits (QIAGEN), respectively, following the manufacturer’s instructions as previously described [5]. Metabarcoding libraries were prepared using a two-step polymerase chain reaction (PCR) protocol, in which the first PCR reaction is designed to amplify the genetic marker along with artificial overhang sequences, and the second PCR reaction is designed to attach sample-specific barcode sequences and Illumina flow cell adapters. The forward and reverse PCR primers in the first reaction were 5′-tcgtcggcagcgtcagatgtgtataagagacagCCTACGGGNGGCWGCAG-3′ and 5′-gtctcgtgggctcggagatgtgtataagagacagGACTACHVGGGTATCTAATCC-3′, respectively, including the target-specific primers for the V3–V4 region [31] with overhangs in lowercase. For the second PCR reaction, the forward and reverse primers were 5′-AATGATACGGCGACCACCGAGATCTACACtcgtcggcagcgtcagatgtgtataagagacag-3′ and 5′-CAAGCAGAAGACGGCATACGAGATXXXXXXgtctcgtgggctcgg-3′, with Illumina adapters (uppercase), overhang complementary sequences (lowercase), and sample-specific DNA barcodes (“X” sequence). The PCR reactions were carried out in triplicate, with the KAPA HiFi HotStart ReadyMix PCR Kit (KAPA Biosystems), in a volume of 25 μL, including 1 μL of DNA template and following the manufacturer’s instructions. The first PCR reaction started with a denaturation step of 3 min at 95°C, followed by 35 cycles of 20 s denaturation at 98°C, 15 s of annealing at 55°C, and 7 s of polymerization at 72°C. The reaction was finalized with another 1-min-long polymerization step. The second PCR reaction was carried out in a volume of 25 μL as well but with 10 μL of the first PCR product as DNA template. It started with a denaturation step of 3 min at 95°C, followed by 8 cycles of 20 s denaturation at 98°C, 15 s of annealing at 55°C, and 7 s of polymerization at 72°C. The second PCR reaction was also finalized with another 1-min-long polymerization step. The first and second PCR reaction products were purified using AMPure XP PCR product cleanup and size selection kit (Beckman Coulter), following the manufacturer’s instructions, and sequenced on an Illumina MiSeq to produce 250 base-pair paired-end sequence reads. The sequencing was carried out by the genomics applications laboratory at the faculty of medicine, Hebrew University.
16S rDNA Analysis
Fastq files were analyzed using QIIME 2 [32]. Using the demux emp-paired plugin and the barcode mapping file, the reads were demultiplexed and assigned to their corresponding sample. De-noising and amplicon sequencing variants tables were constructed using DADA2 plugin [33] via command dada2 denoise-paired. Samples that did not reach the established sequencing depth (500 mapped reads) were excluded from further analysis. Samples with extremely low biomass were excluded due to the possibility of contamination that could stem from the DNA extraction and sequencing process and biomass that might be present in the skin swabs. In addition, the samples were compared to negative samples to ensure there were no pronounced contaminations. Amplicon sequencing variants were assigned with taxonomic annotations by applying the feature-classifier plugin (classify-sklearn) to our representative sequences and using a naive Bayes taxonomic classifier downloaded from QIIME’s data resources page.
Statistical Analysis
PERMANOVA was used with the stratification of dependent samples (collected from the same subject) when performing the random permutations. Statistical tests were performed in Python using the SciPy package. The tests used in each analysis are indicated in the main text or figure legends.
Results
Out of 75 Darier patients followed up at Emek Medical Center, 52 patients agreed to participate in the study. After excluding 6 patients based on predetermined exclusion criteria (antibiotic treatment within the previous month, n = 2, lack of apparent Darier disease skin lesions at the time of examination, n = 4), a total of 46 patients were included in the study.
16S rDNA sequencing was performed on 317 cutaneous microbiome samples obtained from lesions and non-lesional skin on the scalp, chest, axilla, or palm of the 46 subjects with Darier disease. Thirty-five samples obtained from 10 participants were excluded from the analysis due to low quality (sequencing depth less than 500 reads), resulting in four participants having all their samples excluded. In total, 280 samples obtained from 42 patients were included in the analysis. Baseline clinical characteristics of all 42 patients and 280 samples are depicted in Table 1. Most subjects were female (57%), and the average age was 50 years. About half of patients had a mild disease.
Table 1.
Characteristics of patients (n = 42) and samples (n = 280)
| n (%) | |
|---|---|
| Age at research visit | 50 (mean, SD 13) |
| Age of disease onset | 17 (mean, SD 9) |
| Gender | |
| Male | 18 (43) |
| Female | 24 (57) |
| Disease severity | |
| Mild | 22 (52) |
| Moderate | 13 (31) |
| Severe | 7 (17) |
| Percent body surface area involvement | 12 (mean, SD 9) |
| Disease distribution* | |
| Seborrheic | 32 (76) |
| Flexural | 26 (62) |
| Segmental | 2 (5) |
| Acral | 34 (81) |
| Current topical treatments | |
| Emollients | 10 (24) |
| Steroids | 10 (24) |
| Retinoids | 0 |
| Sunscreen/sunblock | 1 (2) |
| Others | 7 (17) |
| None | 17 (40) |
| Current systemic treatment with acitretin/isotretinoin | 5 (12) |
| History of hospitalization | 6 (14) |
| Number of microbiome samples | 280 |
| Lesional samples | 132 |
| Scalp | 39 |
| Chest | 37 |
| Axilla | 18 |
| Palm | 38 |
| Non-lesional samples | 148 |
| Scalp | 38 |
| Chest | 40 |
| Axilla | 32 |
| Palm | 38 |
*An overlap of three distribution patterns was found in 43% and of two distribution patterns in 40%. Only 7 patients had a single distribution pattern.
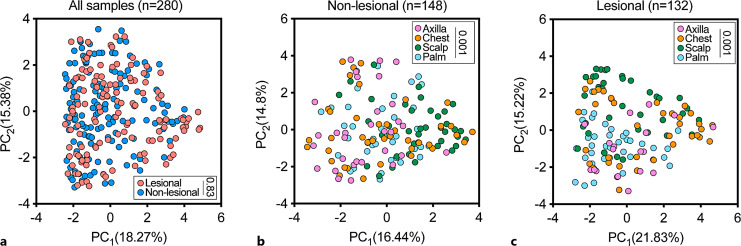
Considering all body sites together, there was no statistically significant genus-level difference between the global microbiome configuration in lesional versus non-lesional skin (Fig. 3a). Expectedly [2], a stratified analysis in both lesional (Fig. 3b) and non-lesional (Fig. 3c) skin revealed significant heterogeneity in the skin microbiome composition across body sites; hence, we next analyzed each body site independently.
Fig. 3.
Genus-level cutaneous microbiome composition of four body sites in patients with Darier. a A principal component analysis (PCA) of samples that were obtained from lesional and non-lesional skin. Inset: p value; permutational multivariate analysis of variance (PERMANOVA). b A PCA of microbiome composition in lesional skin, stratified by body site. Inset: p value; PERMNOVA. c A PCA of microbiome composition of non-lesional skin stratified by body site. Inset: p value; PERMANOVA. PC, principal component.
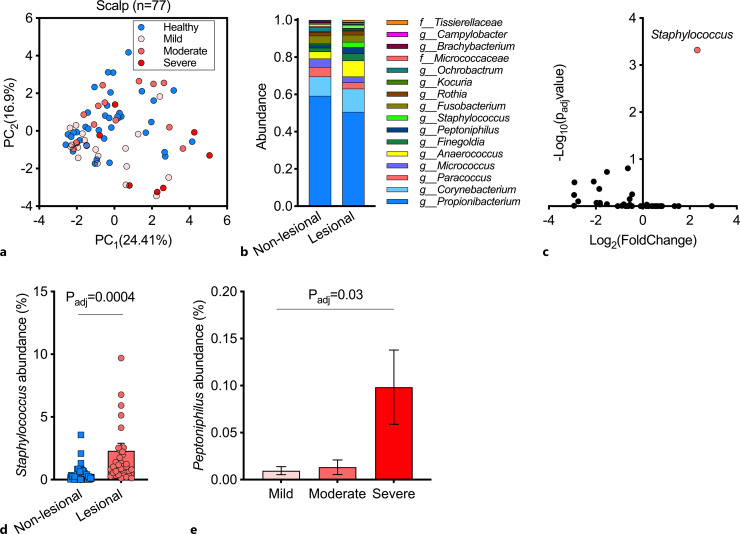
Analyzing samples obtained from the scalp revealed significant differences between lesional samples of different severity (p = 0.001, PERMANOVA); however, the difference between all lesional and non-lesional samples failed to meet the statistical threshold for significance (p = 0.06, PERMANOVA; Fig. 4a). Propionibacterium, Corynebacterium, Paracoccus, Micrococcus, and Anaerococcus constituted the most dominant genera, comprising ∼80% of the scalp microbiome in both lesional and non-lesional skin (Fig. 4b). A differential abundance analysis between lesional and non-lesional scalp samples identified members of the Staphylococci genus to drive Darier-associated microbiome alteration (Fig. 4c, d). Of note, the abundance of members of the Peptoniphilus genera significantly correlated with Darier clinical severity in scalp lesions (Fig. 4e, adjusted p = 0.03 ANOVA).
Fig. 4.
Cutaneous microbiome of the scalp in patients with Darier. a A principal component analysis (PCA) of samples that were obtained from lesional and non-lesional scalp skin. b Relative abundance of bacterial genera in samples from lesional and non-lesional scalp skin. c A differential abundance analysis of lesional and non-lesional scalp cutaneous microbiome. Two-sided Wilcoxon test, false discovery rate (FDR) correction for multiple comparisons (q < 0.05). d Distribution of staphylococcus abundance across lesional and non-lesional samples in scalp samples. Two-sided Wilcoxon test, false discovery rate (FDR) correction for multiple comparisons. e Relative abundance of Peptoniphilus as a function of disease severity (one-way analysis of variance comparing the mean abundance across all three categories, corrected by FDR for multiple comparisons of all genera). PC, principal component.
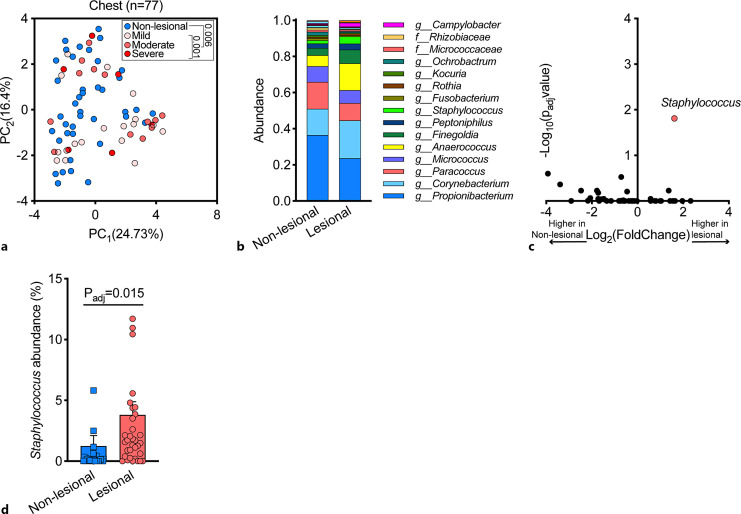
Analyzing samples obtained from the chest revealed a distinct microbiome in lesional samples compared with non-lesional samples (Fig. 5a, p = 0.006 PERMANOVA) with dependency on disease severity (Fig. 5a, p = 0.001 PERMANOVA). The most prevalent genera in the cutaneous microbiome of the chest were similar to the scalp (Fig. 5b). Also in line with our observations in scalp samples, a differential abundance analysis of lesional versus non-lesional chest skin samples identified the Staphylococcus genus as a key driver of this microbiome shift (Fig. 5c, d). However, this genus was not significantly correlated with disease severity (p = 0.48, ANOVA).
Fig. 5.
Cutaneous microbiome of the chest area in patients with Darier. a A PCA of 77 samples obtained from lesional and non-lesional skin of the chest. Inset: p value PERMANOVA. b Relative abundance of bacterial genera in samples from lesional and non-lesional chest skin. c A differential abundance analysis of lesional and non-lesional chest skin microbiome (two-sided Wilcoxon test, false discovery rate (FDR) correction for multiple comparisons (p < 0.05). d Distribution of staphylococcus abundance across lesional and non-lesional samples in chest samples. Two-sided Wilcoxon test, false discovery rate (FDR) correction for multiple comparisons. PC, principal component.
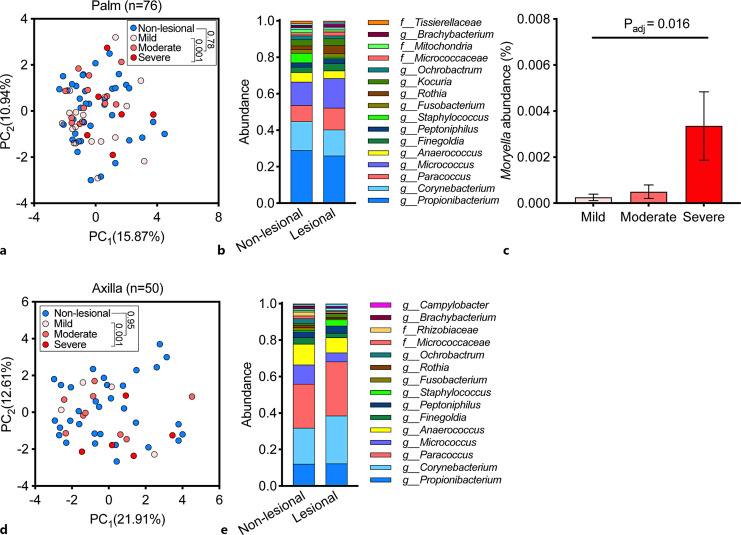
There were no significant genus-level microbiome shifts in palmar (Fig. 6a) or axillary (Fig. 6d) lesions; however, within lesional skin samples, there was a significant compositional segregation of the microbiome across disease severity (Fig. 6a, d, respectively). The most prevalent bacterial genera in the palm were similar to the scalp and chest (Fig. 6b). The Moryella genus was more abundant in lesions of higher severity and was the main driver for the microbiome compositional segregation across disease severity (Fig. 6c).
Fig. 6.
Cutaneous microbiome of the palm and axilla in patients with Darier. a A PCA of 76 samples obtained from lesional and non-lesional palmar skin. b Relative abundance of bacterial genera in samples from lesional and non-lesional palmar skin. c Relative abundance of Moryella in lesional skin across disease severity (one-way ANOVA). d A PCA of 50 samples obtained from lesional and non-lesional axillary skin. e Relative abundance of bacterial genera in samples from lesional and non-lesional axillary skin. PC, principal component.
The most dominant bacterial genera in the axilla were similar to the scalp, chest, and palm (Fig. 6e). No bacterium passed the statistical threshold for differential abundance or association with disease severity in axillary samples.
Discussion
To the best of our knowledge, this is the first study to analyze the skin bacterial microbiome in Darier patients. We analyzed the microbiome composition in Darier lesions and compared them with the microbiome of healthy-appearing adjacent skin from the same subject. This paired-sampling approach enabled us to control for the interindividual variability in skin microbiome.
We chose to sample four body sites, based both on Darier disease distribution of skin lesions and on different microbiome environments [2]. It appears that the intra-individual variability in cutaneous microbiome across body sites outweighs the variability between lesional and non-lesional skin [2], which may explain why we could not detect an overall disease-dependent microbiome configuration for all body sites combined. However, separately analyzing each body site uncovered microbiome alterations between lesional and non-lesional skin in sebaceous areas. These alterations were statistically significant in the chest. The scalp showed a similar trend; however, it did not reach the threshold for statistical significance.
We found members of the Staphylococci genus to be significantly enriched in scalp and chest Darier lesions compared to non-lesional skin. This trend was also seen in axillary lesions; however, it did not reach statistical significance, possibly owing to a smaller sample size (n = 50), a more significant interindividual variability of the microbiome in the axilla compared with other body sites, or a combination of both. Not surprisingly, this enrichment was not seen in palmar lesions, as Staphylococci preferably colonize moist areas of the skin and are less common in dry areas, such as the palms, which harbor a more diverse microbiome [34]. Our sequencing strategy limits our ability to generate species-level insights; however, one may posit that the abundance of Staphylococci represents, at least in part, the abundance of S. aureus, which was previously found to be commonly isolated from Darier patients [28].
Interestingly, within lesional samples, there was a statistically significant microbiome shift across disease severity that was recapitulated in all tested body sites. Peptoniphilus and Moryella, both members of the Clostridia class, were the key drivers of this shift in the scalp and the palm, respectively. Peptoniphilus, a gram-positive anerobic coccus, was recently found to be prevalent in diabetic foot and pressure ulcers [35]. Moreover, along with Corynebacterium, they have been suggested to have a pathogenic role in generating a biofilm in chronic wound infections [36].
Moryella is a gram-positive anerobic rod. Very few publications have previously described the isolation of this genus, specifically Moryella indoligenes, which was isolated from a buttock abscess [37]. This finding is thus novel, and its clinical significance merits further investigation.
Our data implicates Staphylococci, Peptoniphilus, and Moryella genera to be associated with Darier skin lesions and disease severity; however, the cross-sectional design of the current study limits our ability to establish their role in disease flares. Their presence in skin lesions of various types may reflect their capacity to thrive on damaged skin and adapt to inflammatory and more anerobic environment, or they may have a causative role in impairing skin integrity and hampering wound repair. Staphylococcus aureus was previously speculated to cause Darier disease exacerbation by inducing elevated cytokine levels, including IL-6, and causing haploinsufficiency by reducing intracellular ATPA2 mRNA levels [28]. As mentioned above, Peptoniphilus species are associated with chronic wound infections and biofilm creation. It therefore merits further investigation as to whether antibiotic treatment targeting this genus may result in better clinical outcomes of Darier patients, especially among patients with more severe disease with a seborrheic distribution.
This study originated from a single center and was limited by the relatively low sample size derived from the rarity of this disease and the common exposure to antibiotic agents, which precluded the participation of many patients. In addition, for the reasons described above, this study adopted a relatively permissive 1-month period of no antibiotic treatment prior to sampling.
In conclusion, in alignment with previous findings and similar to some other skin diseases such as atopic dermatitis, we found Staphylococci to be significantly associated with Darier lesions and to drive Darier-associated dysbiosis. In addition, severity of the disease was found to be associated with two other bacterial genera – Peptoniphilus and Moryella. Whether all these associations also hold a causative role remains to be determined. Future investigation is essential to explore the potential causal link between these genera and Darier disease, as they might serve as prospective therapeutic targets for patients with Darier.
Key Message
Darier lesions harbor a distinct microbiome configuration, mainly driven by an overabundance of Staphylococci species.
Statement of Ethics
This study protocol was reviewed and approved by the Institutional Review Board at Emek Medical Center, approval number 0008-17-EMC. Written informed consent was obtained from all participants.
Conflict of Interest Statement
The authors state that there is no conflict of interest.
Funding Sources
This study was supported by the Israeli Ministry of Science and Technology (Grants 3-11174, 3-16033). The funding organizations were not involved in study design, data collection, data analysis, or any other stage of the research.
Author Contributions
Ofer Reiter: conceptualization, design, analysis, writing – original draft preparation, and final approval; Avner Leshem: conceptualization, formal analysis, investigation, methodology, writing – original draft preparation, and final approval; Rivka Alexander-Shani: data acquisition, formal analysis, drafting, and final approval; Michael Brandwein: data curation, formal analysis, and writing – review and editing; Yotam Cohen: formal analysis, software, and writing – review and editing; Algit Yeshurun: data acquisition, drafting, and final approval; Michael Ziv: resources, data acquisition, drafting, and formal analysis; Eran Elinav: conceptualization, methodology, analysis, supervision, editing, and final approval; Emmilia Hodak: conceptualization, design, funding acquisition, supervision, writing – review and editing, and final approval; and Roni P. Dodiuk-Gad: conceptualization, design, data acquisition, investigation, writing – review and editing, and final approval.
Funding Statement
This study was supported by the Israeli Ministry of Science and Technology (Grants 3-11174, 3-16033). The funding organizations were not involved in study design, data collection, data analysis, or any other stage of the research.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
References
- 1. Carmona-Cruz S, Orozco-Covarrubias L, Sáez-de-Ocariz M. The human skin microbiome in selected cutaneous diseases. Front Cell Infect Microbiol. 2022;12:834135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Publ Gr. 2018;16(3):143–55. [DOI] [PubMed] [Google Scholar]
- 3. Paller AS, Kong HH, Seed P, Naik S, Scharschmidt TC, Gallo RL, et al. The microbiome in patients with atopic dermatitis. J Allergy Clin Immunol. 2019;143(1):26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22(5):850–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brandwein M, Fuks G, Israel A, Sabbah F, Hodak E, Szitenberg A, et al. Skin microbiome compositional changes in atopic dermatitis accompany dead sea climatotherapy. Photochem Photobiol. 2019;95(6):1446–53. [DOI] [PubMed] [Google Scholar]
- 6. Tanaka A, Cho O, Saito C, Saito M, Tsuboi R, Sugita T. Comprehensive pyrosequencing analysis of the bacterial microbiota of the skin of patients with seborrheic dermatitis. Microbiol Immunol. 2016;60(8):521–6. [DOI] [PubMed] [Google Scholar]
- 7. Fitz-Gibbon S, Tomida S, Chiu B-H, Nguyen L, Du C, Liu M, et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Invest Dermatol. 2013;133(9):2152–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alekseyenko AV, Perez-Perez GI, De Souza A, Strober B, Gao Z, Bihan M, et al. Community differentiation of the cutaneous microbiota in psoriasis. Microbiome. 2013;1(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pinto D, Sorbellini E, Marzani B, Rucco M, Giuliani G, Rinaldi F. Scalp bacterial shift in Alopecia areata. PLoS One. 2019;14(4):e0215206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Salava A, Deptula P, Lyyski A, Laine P, Paulin L, Väkevä L, et al. Skin microbiome in cutaneous T-cell lymphoma by 16S and whole-genome shotgun sequencing. J Invest Dermatol. 2020;140(11):2304–8.e7. [DOI] [PubMed] [Google Scholar]
- 11. Blicharz L, Rudnicka L, Samochocki Z. Staphylococcus aureus: an underestimated factor in the pathogenesis of atopic dermatitis? Postepy Dermatol Alergol. 2019;36(1):11–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Skinner RB Jr, Noah PW, Zanolli MD, Rosenberg EW. The pathogenic role of microbes in seborrheic dermatitis. Arch Dermatol. 1986;122(1):16–7. [PubMed] [Google Scholar]
- 13. Darier J. Psorospermose folliculaire vegetante. Ann Dermatol SyphiloI. 1889;(10):597–612. [Google Scholar]
- 14. Beck ALJ, Finocchio AF, White JP. Darier’s disease: a kindred with a large number of cases. Br J Dermatol. 1977;97(3):335–9. [DOI] [PubMed] [Google Scholar]
- 15. Sakuntabhai A, Ruiz-Perez V, Carter S, Jacobsen N, Burge S, Monk S, et al. Mutations in ATP2A2, encoding a Ca2+ pump, cause Darier disease. Nat Genet. 1999;21(3):271–7. [DOI] [PubMed] [Google Scholar]
- 16. Burge SM, Wilkinson JD. Darier-White disease : a review of the clinical features in 163 patients. J Am Acad Dermatol. 1992;27(1):40–50. [DOI] [PubMed] [Google Scholar]
- 17. Yeshurun A, Ziv M, Cohen-Barak E, Vered S, Rozenman D, Sah M, et al. An update on the cutaneous manifestations of darier disease. J Cutan Med Surg. 2021;25(5):498–503. [DOI] [PubMed] [Google Scholar]
- 18. Dodiuk-Gad RP, Cohen-Barak E, Khayat M, Milo H, Amariglio-Diskin L, Danial-Faran N, et al. Darier disease in Israel: combined evaluation of genetic and neuropsychiatric aspects. Br J Dermatol. 2016;174(3):562–8. [DOI] [PubMed] [Google Scholar]
- 19. Dodiuk-Gad R, Lerner M, Breznitz Z, Cohen-Barak E, Ziv M, Shani-Adir A, et al. Learning disabilities in Darier’s disease patients. J Eur Acad Dermatol Venereol. 2014;28(3):314–9. [DOI] [PubMed] [Google Scholar]
- 20. Hammad H, Adler E, Yeshurun A, Abayev L, Vered S, Briscoe D, et al. Ophthalmic assessment in patients with darier disease. Am J Ophthalmol. 2021;227:139–42. [DOI] [PubMed] [Google Scholar]
- 21. Okada E, Nagai Y, Motegi S-I, Tamura A, Ishikawa O. Fatal case of Darier’s disease with recurrent severe infections. Acta Derm Venereol. 2009;89(4):408–9. [DOI] [PubMed] [Google Scholar]
- 22. Hazen PG, Eppes RB. Eczema herpeticum caused by herpesvirus type 2. A case in a patient with Darier disease. Arch Dermatol. 1977;113(8):1085–6. [PubMed] [Google Scholar]
- 23. Toole JW, Hofstader SL, Ramsay CA. Darier’s disease and Kaposi’s varicelliform eruption. J Am Acad Dermatol. 1979;1(4):321–4. [DOI] [PubMed] [Google Scholar]
- 24. Nikkels AF, Beauthier F, Quatresooz P, Piérard GE. Fatal herpes simplex virus infection in Darier disease under corticotherapy. Eur J Dermatol. 2005;15(4):293–7. [PubMed] [Google Scholar]
- 25. Pantazi V, Potouridou I, Katsarou A, Papadogiorgaki TH, Katsambas A. Darier’s disease complicated by Kaposi’s varicelliform eruption due to herpes simplex virus. J Eur Acad Dermatol Venereol. 2000;14(3):209–11. [DOI] [PubMed] [Google Scholar]
- 26. Milavec-Puretić V, Lipozencić J, Sustić N, Pećina-Slaus N. Sepsis as an unusual event in dyskeratosis follicularis. Croat Med J. 2001;42(1):64–6. [PubMed] [Google Scholar]
- 27. Sbarouni E, Petraki M, Stavridis G, Manginas A. Infected thoracic aortic graft in a woman with Darier disease: a case report. Eur Heart J Case Rep. 2022;6(8):ytac314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dodiuk-Gad R, Cohen-Barak E, Ziv M, Shani-Adir A, Shalev S, Chazan B, et al. Bacteriological aspects of Darier’s disease. J Eur Acad Dermatol Venereol. 2013;27(11):1405–9. [DOI] [PubMed] [Google Scholar]
- 29. Sfecci A, Orion C, Darrieux L, Tisseau L, Safa G. Extensive darier disease successfully treated with doxycycline monotherapy. Case Rep Dermatol. 2015;7(3):311–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pettit C, Ulman CA, Spohn G, Kaffenberger J. A case of segmental Darier disease treated with doxycycline monotherapy. Dermatol Online J. 2018;24(3). [PubMed] [Google Scholar]
- 31. Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41(1):e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, et al. Survey of bacterial diversity in chronic wounds using Pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS One. 2008;3(10):e3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carlier J-P, K’ouas G, Han XY. Moryella indoligenes gen. nov., sp. nov., an anaerobic bacterium isolated from clinical specimens. Int J Syst Evol Microbiol. 2007;57(Pt 4):725–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.