Abstract
Schizophrenia lacks a clear definition at the neuroanatomical level, capturing the sites of origin and progress of this disorder. Using a network-theory approach called epicenter mapping on cross-sectional magnetic resonance imaging from 1124 individuals with schizophrenia, we identified the most likely “source of origin” of the structural pathology. Our results suggest that the Broca’s area and adjacent frontoinsular cortex may be the epicenters of neuroanatomical pathophysiology in schizophrenia. These epicenters can predict an individual’s response to treatment for psychosis. In addition, cross-diagnostic similarities based on epicenter mapping over of 4000 individuals diagnosed with neurological, neurodevelopmental, or psychiatric disorders appear to be limited. When present, these similarities are restricted to bipolar disorder, major depressive disorder, and obsessive-compulsive disorder. We provide a comprehensive framework linking schizophrenia-specific epicenters to multiple levels of neurobiology, including cognitive processes, neurotransmitter receptors and transporters, and human brain gene expression. Epicenter mapping may be a reliable tool for identifying the potential onset sites of neural pathophysiology in schizophrenia.
Brain imaging identifies the Broca’s area and frontoinsular cortex as epicenters of neuroanatomical pathology in schizophrenia.
INTRODUCTION
The pursuit of a neuroimaging signature for schizophrenia that can accurately diagnose the disorder, predict its long-term course, and map to expected treatment response has been ongoing for years (1, 2). However, progress has been hindered by the lack of large sample sizes and high heterogeneity within the schizophrenia population, leading to inconsistent and unreliable results (3–6). For example, previous studies, including the Enhancing Neuro Imaging Genetics through Meta Analysis (ENIGMA) consortium’s efforts, have identified brain structural alterations in schizophrenia, but these alterations appear to be of modest effect size and are not specific to schizophrenia, overlapping notably with other neuropsychiatric disorders (7–10). A recent cross-disorder study reveals shared and disorder-specific neuroanatomical characteristics, such as increased volume of lateral ventricles in schizophrenia, bipolar disorder (BD), and major depressive disorder, as well as smaller hippocampus volume in schizophrenia and BD (11). Furthermore, several recent studies have indicated that structural brain alterations in schizophrenia is not uniform, as subtypes with varying degree of neuroanatomical changes, disease progression trajectories, and treatment outcomes exist within this diagnostic group (12–14). While functional magnetic resonance imaging (fMRI) has been suggested as a potential tool for discovering schizophrenia biomarkers (15), its reliability is disputed. Therefore, there is a need for robust neuroimaging methods and large datasets to identify reliable biomarkers for schizophrenia (16).
The aetiology and pathogenesis of schizophrenia is still not fully understood, but it is generally considered a network disorder or a disconnection syndrome (17, 18). Neuroanatomical abnormalities in schizophrenia, such as a thinner cortex and reduced volumes in brain structures like the hippocampus and thalamus, are present at the population level (10, 19). However, these differences do not occur simultaneously across all brain regions but rather follow a progressive trajectory (20, 21). For instance, first-episode patients tend to show subtly thinner cortical thickness mainly in the frontotemporal lobes, while chronic patients show more pronounced reductions in the parietal and occipital cortices (20, 22). Individuals with psychotic experiences, genetic high risk, or clinical high risk show progressive gray matter reductions in the temporal and frontal lobes over time (23). Moreover, these reductions persist in those who remain symptomatic or transition to psychosis (23). Longitudinal structural studies indicate a pattern of limited amelioration, indicative of reorganization that may follow connectivity patterns of large-scale functional networks (24, 25). Patients with early-onset psychosis exhibit a more pronounced progressive loss of frontal gray matter over the initial years following the onset of the illness (26). One explanation for the increasing gray matter reduction seen over time is that the disease process that begins in a specific brain region, known as the “epicenter,” then spreads “transneuronally” to other brain regions facilitated by the underlying patterns of connectivity (27, 28). This connectivity-based epicenter model has been successful in revealing the onset of disease in focal brain regions and accurately predicting the progression of neurophysiology in neurodegenerative diseases (27, 29).
Variations in connectional anatomy is now a well-known feature of human brain morphology (30). In schizophrenia, some of the interindividual variations in the patterns of neuroanatomical abnormality may occur because of either individual differences in the sites of origin per se (in which case, we may not be able to locate one or more regions as consistent epicenters) or variations in individual connectivity patterns, affecting the “transneuronal spread” despite shared anatomical origins. In this work, through robust quantitative assessment (31), we aim to contextualize the structural epicenters of schizophrenia based on gray matter morphometry (32, 33), with existing maps of human brain function (connectivity), cytoarchitecture (34), metabolism (35), neurotransmitter receptors and transporters (36), gene expression (37), and cognition-related activity (38). Our goal is to identify potential neuroimaging epicenters in schizophrenia and determine whether normative structural deviation across the epicenters can be used as a reliable magnetic resonance imaging (MRI) signature for the identification of schizophrenia and prediction of treatment response. If successful, this could ultimately help to target our enquiries on the pathophysiology of schizophrenia on a better-defined phenotype than symptom-based descriptors (27).
In pursuit of this goal, we report sites of probable origin of neuroanatomical pathology of schizophrenia, using neuroimaging-derived connectivity-based epicenter mapping, in a discovery cohort of cross-sectional MRI from 2170 individuals (1124 patients with schizophrenia) and replicated this in an independent validation cohort. Furthermore, we investigated the diagnostic specificity of neuroimaging-derived epicenters in schizophrenia by comparing normative deviation patterns with 10 other overlapping neuropsychiatric disorders. We systematically explored the associations of neuroimaging epicenters with clinical symptoms, gene expression in the human brain, neurotransmitter distributions, and expected functional activation for cognitive processes. Last, we examined the performance of neuroimaging-derived epicenters in predicting treatment response in a longitudinal follow-up sample. Our results indicate that the MRI-based epicenter mapping can be a reliable signature of the onset of pathophysiological changes specific to the diagnosis of schizophrenia and can be predictive of treatment response.
RESULTS
Experimental design
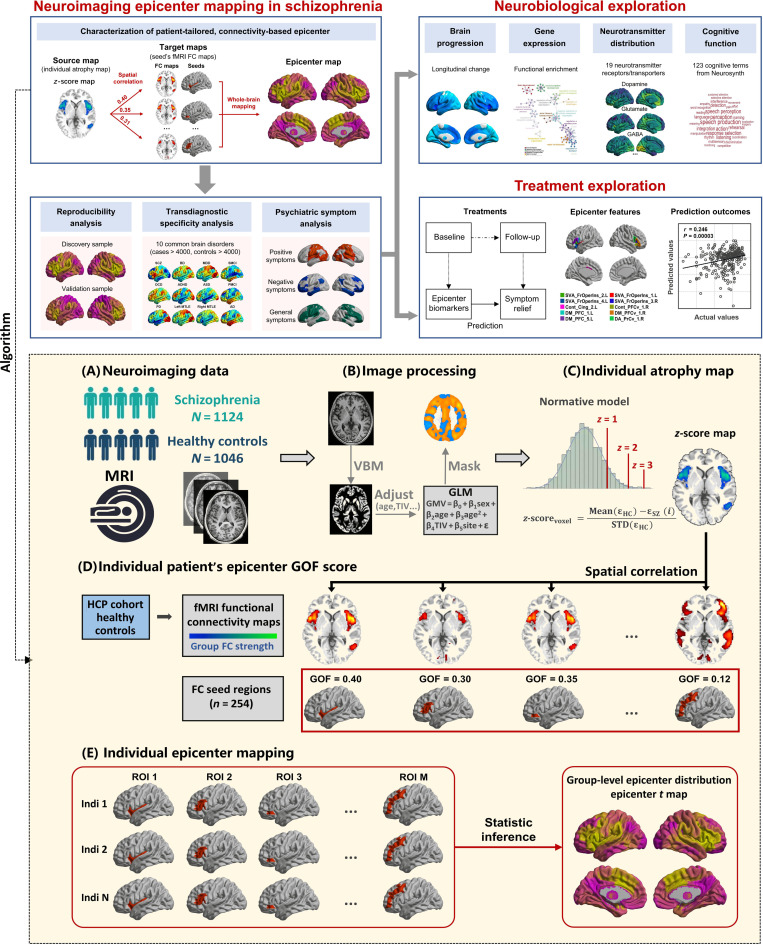
Figure 1 provides a flowchart of experimental design. First, this study detected the onset of neurophysiological pathology of schizophrenia by using a neuroimaging-derived, connectivity-based epicenter mapping algorithm based on only cross-sectional structural MRIs from 1124 patients with schizophrenia (table S1). To establish that the epicenter trait can be used as reliable biomarkers of schizophrenia, we performed exploratory analyses including that (i) examining the reproducibility of epicenter in another independent cohort; (ii) comparing the transdiagnostic specificity in more than 4000 individuals (table S2) diagnosed with schizophrenia, BD, major depression (MDD), obsessive-compulsive disorder (OCD), attention deficit hyperactivity disorder (ADHD), autism spectrum disorder (ASD), Parkinson’s disease (PD), mesial temporal lobe epilepsy (MTLE), mild cognitive impairment (MCI), or Alzheimer’s disease (AD); and (iii) mapping the specific psychiatric symptoms of schizophrenia with the degree of epicenter. Second, to offer a comprehensive cross-disciplinary perspective of function and structure of epicenters, we linked the schizophrenia-specific epicenter distribution to (i) gray matter morphometric maps including longitudinal change of schizophrenia and ENIGMA-derived thinner–cortical thickness patterns; (ii) human brain gene expression profiles; (iii) positron emission tomography (PET)–derived whole-brain neurotransmitter receptor/transporter maps; and (iv) Neurosynth-derived meta-analytic task activation maps from 123 cognitive processes. Third, we examined whether epicenter traits predicted the short-term response to antipsychotic medications.
Fig. 1. Experimental design and methodology of epicenter mapping.
The connectivity-based patient-tailored epicenter mapping approach to identify the probable origin of neuroanatomical pathology of schizophrenia is depicted here. The top panel shows the overall research approach. Neuroimaging analyses were performed to characterize the identified epicenter’s reproducibility, diagnostic specificity, and association with psychiatric symptoms. Systematic studies of the association of changes in the epicenter with signatures of brain atrophy, gene expression, the spatial distribution of neurotransmitter systems, and cognitive function were carried out. We also examined the performance of epicenter mapping in predicting treatment outcomes in a longitudinal follow-up sample. The bottom panel shows the methodology of neuroimaging-derived, patient-tailored, connectivity-based epicenter mapping. (A) Neuroimaging data included cross-sectional MRI from 2170 individuals (1124 patients with schizophrenia). (B) MRI was processed using standard voxel-based morphometry (VBM), yielding voxel-wise gray matter volume (GMV). The GMV is adjusted by regressing out the effects of gender, age, the square of age, total intracranial volume (TIV), and sites using a regression model. A gray matter mask is used to exclude non–gray matter voxels. (C) The GMV is normalized relative to control population using z-score procedure. Higher z-score represents larger deviation from the normal (i.e., more severe atrophy in this case). (D) To characterize the epicenter for a given patient’s z-score map, spatial correlation is performed between the z-score map and each seed functional connectivity (FC) map derived from an independent healthy cohort [Human Connectome Project (HCP)]. Spearman correlation coefficients are calculated to quantify epicenter GOF score. The procedure generated M GOF scores corresponding to M candidate epicenter regions. (E) One sample t test is performed on the GOF score to determine significance of a candidate epicenter. The statistical t map represented inferred epicenter degree for the whole-brain M regions at group level.
Neuroimaging epicenter distribution map in schizophrenia
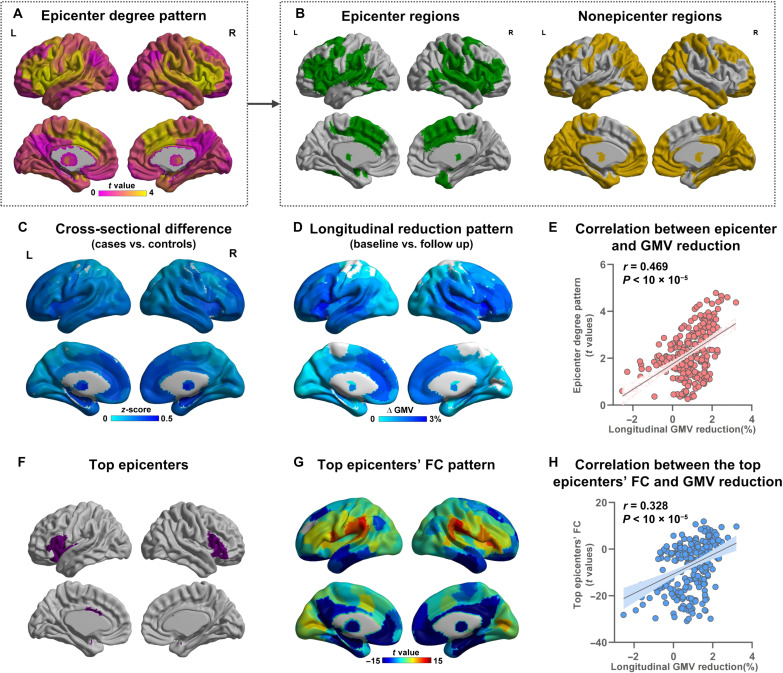
Using cross-sectional structural MRI data, we derived each patient’s epicenter distribution map (see Methods) that quantified the goodness-of-fit (GOF) score for a given region being an epicenter across whole brain. The region with a higher GOF score indicated higher probability being a candidate epicenter at the individual level. This statistical t map of epicenter provided a group-level degree of each region being a notable epicenter in 1124 individuals with schizophrenia (Fig. 2A). This epicenter t map indicated that areas involving insula, anterior cingulate, ventrolateral frontal cortex, and superior temporal cortex exhibited significantly higher degree across the whole brain, identified as notable epicenter regions after false discovery rate (FDR) correction (FDR P < 0.05) (Fig. 2B and table S3). We also characterized epicenter t map separately in schizophrenia subsamples differing in sex, illness stages, and age bins. There was no significant difference of epicenter distribution between female and male with schizophrenia after FDR correction (fig. S1). The notable epicenter regions in frontal and insular areas were replicated in a first-episode patient subgroup (fig. S2) and youth patient subgroups (fig. S3).
Fig. 2. Epicenter distribution and association with signatures of brain structure.
(A) Characterization of epicenter distribution map across the whole brain. (B) Notable epicenters (FDR P < 0.05) and nonepicenter regions. (C) GMV reduction pattern from cross-sectional MRI data in schizophrenia (i.e., case-control difference). (D) Brain change pattern from a longitudinal subsample with 12-week follow-up (i.e., the percentage change between baseline and follow-up in schizophrenia). (E) A significant spatial correlation between epicenter degree pattern and longitudinal GMV reduction pattern (r = 0.469, P < 0.00001), indicating that regions with higher epicenter degree (i.e., more likely to be the origins) have pronounced GMV reduction. (F) The top 10 regions with the highest epicenter degree were defined as the most likely epicenters. (G) Whole-brain FC pattern with the top epicenters as the seed. (H) A significant correlation between the epicenters FC pattern and longitudinal GMV reduction pattern (r = 0.328, P < 0.00001) demonstrates that brain regions with stronger connectivity with the top epicenters have more severe longitudinal change.
We further investigated the relationship between the epicenter degree pattern (i.e., epicenter t map) (Fig. 2A) and gray matter reduction pattern in schizophrenia (Fig. 2C). We found a significant spatial positive correlation between the epicenter degree pattern and cross-sectional smaller gray matter volume (GMV) patterns (i.e., case-control difference) (r = 0.223, Pspin = 0.0004). Furthermore, epicenter degree pattern was also correlated with longitudinal gray matter reduction pattern (Fig. 2D) (i.e., percentage difference between baseline and follow-up) in a longitudinal subsample (table S4) with 12-week follow-up individuals with schizophrenia (r = 0.469, Pspin < 0.00001) (Fig. 2E). Among the notable epicenter regions, the top 10 regions with the highest degree were defined as the top epicenters (Fig. 2F). We found that at 12-week follow-up, the mean percentage in GMV reduction at the top epicenters was 2.2%, which was significantly worse than epicenters (exclude top epicenters) (mean = 1.3%, t = 3.9, P = 0.00014) and nonepicenters (mean = 0.5%, t = 5.9, P < 0.00001) (fig. S4). The results indicate that regions with higher epicenter degree (i.e., more likely to be the origins) may predict pronounced GMV reduction.
Using resting-state fMRI data of healthy controls from the Human Connectome Project (HCP) cohort (n = 1089, age = 28.8 ± 3.7 years, 593 females), we derived a normative set of functional connectivity (FC) maps. The whole-brain FC pattern with the top epicenters as the seed is shown in Fig. 2G. We also observed a significant correlation between the top epicenters’ FC pattern and longitudinal GMV reduction pattern (r = 0.328, Pspin < 0.00001) (Fig. 2H), demonstrating that brain regions with stronger connections to the top epicenters had more severe regional volume reduction. These findings suggested that the top epicenters may be the origins of neuroanatomical abnormality and that pathophysiology may spread from epicenters through the inherent network patterns of the human brain.
As neuroanatomical abnormality is implicated in multiple diseases and disorders, we therefore examined whether the link of epicenter t map with neuroanatomical abnormality pattern is schizophrenia specific across different brain disorder. We separately estimated the relationship between the epicenter t map and ENIGMA-derived thinner–cortical thickness maps across eight neurological, neurodevelopmental, and psychiatric disorders (see Methods). We observed the strongest relationship between the epicenter t map and the thinner–cortical thickness pattern in the ENIGMA schizophrenia population (r = 0.519, Pspin = 0.0036), compared to other brain disorders (Extended Fig. 1). The epicenter t map did not exhibit significant association with thinner–cortical thickness maps derived from other diseases or disorders (Pspin > 0.05) despite of a correlation with BD adults (r = 0.400, Pspin = 0.0091).
Reproducibility and robustness of epicenter distribution map
We calculated the epicenter t map in two independent cohorts of schizophrenia (discovery sample, ncases = 1124; validation sample, ncases = 147) and found significant consistency between the two results (r = 0.280, Pspin < 0.001) (Extended Fig. 2). The highest epicenters still appear in the inferior frontal and adjacent frontoinsular-cingulate regions. We also verified the robustness of the epicenter distribution using a brain atlas with different parcels ranging from 68 to 600 parcels (Extended Fig. 3). These results demonstrated the reproducibility and robustness of the epicenter distribution in schizophrenia.
Epicenter mapping in neurological, neurodevelopmental, and psychiatric disorders
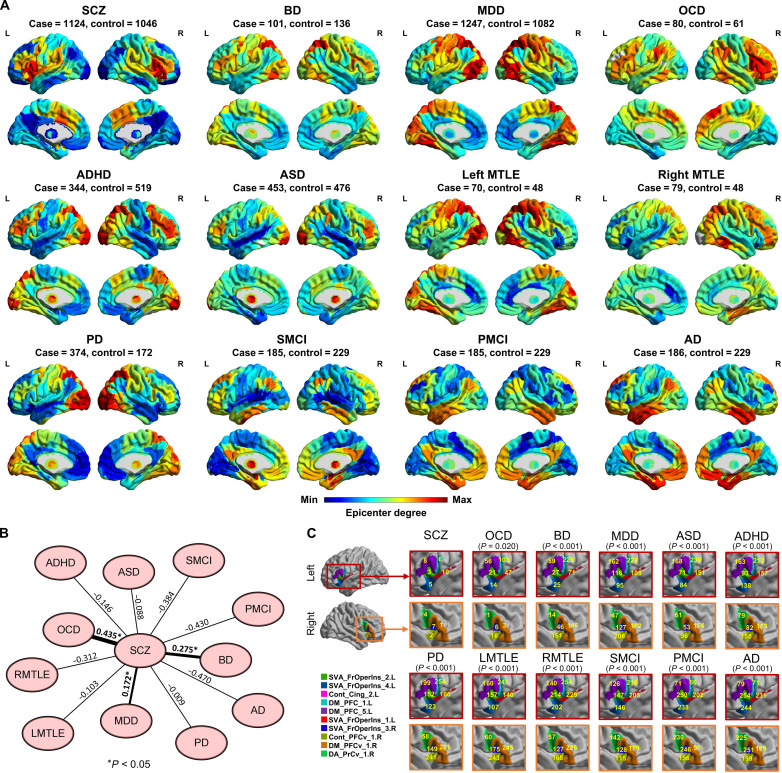
We performed epicenter mapping for individuals diagnosed with schizophrenia (ncases = 1124), BD (ncases = 101), MDD (ncases = 1247), OCD (ncases = 80), ADHD (ncases = 344), ASD (ncases = 453), PD (ncases = 374), MTLE (ncases = 149), MCI (ncases = 370), and AD (ncases = 186). Figure 3A shows the group-level epicenter t map for each disorder. Spatial correlation analysis indicated that the epicenter t map in schizophrenia spatially correlated with the epicenter t maps in BD (r = 0.276, Pspin = 0.0006), MDD (r = 0.172, Pspin = 0.024), and OCD (r = 0.435, Pspin < 0.0001) (Fig. 3B). In addition, Fig. 3C shows the ranks of insular and frontal opercular regions (that were identified as the top epicenters in schizophrenia) across whole-brain regions (n = 254) according to their epicenter degree in each disorder. The rank of all brain regions according to epicenter degree in each disorder is shown in table S5. Wilcoxon rank sum test indicated significant differences in the rank of the top epicenters (shown in Fig. 3C) between schizophrenia and any of the other brain disorders (all Ps < 0.05). These results indicated that the regions within the insular and frontal opercular cortex, which were identified as the top epicenters of schizophrenia, showed high specificity to only schizophrenia compared to BD, MDD, OCD, ADHD, ASD, PD, MTLE, MCI and AD, despite that schizophrenia also exhibited a similar global pattern in the whole-brain level with MDD, OCD, and BD. As the disease data were from different cohorts/sites, we also performed epicenter mapping for their matched healthy individuals in each disease cohort. This epicenter mapping on healthy individuals from different sites can examine whether the epicenter differences across diseases are caused by cohorts/sites. Extended Figure 4 shows a similarity pattern across healthy individual’s epicenter t maps from different cohorts/sites, suggesting that the epicenter differences across diseases are not caused by a cohort/site effect. The insular and frontal opercular regions (i.e., top epicenters in schizophrenia) were not notable epicenters in healthy controls.
Fig. 3. Schizophrenia-specific epicenter.
(A) Characterization of epicenter degree (t value) maps in additional neuroimaging datasets that included individuals diagnosed with BD, MDD, OCD, ADHD, ASD, PD, left MTLE (LMTLE), right MTLE (RMTLE), progressive MCI (PMCI), nonprogressive stable MCI (SMCI) and AD. (B) The epicenter degree map in schizophrenia was spatially correlated with the epicenter degree maps in the BD (r = 0.276, Pspin = 0.0006), MDD (r = 0.172, Pspin = 0.024), and OCD (r = 0.435, Pspin < 0.0001) by Spearman correlation test. The asterisk represents one-tailed P < 0.05. (C) Ranks of insular and frontal opercular regions (that are identified as the top epicenters in schizophrenia) across whole brain regions (n = 254) according to their epicenter degree in each disorder. P values represent significant differences in the rank of the insular and frontal opercular regions between schizophrenia and each of the other brain disorders using the Wilcoxon rank sum test.
Associations between regional GOF score and symptoms in schizophrenia
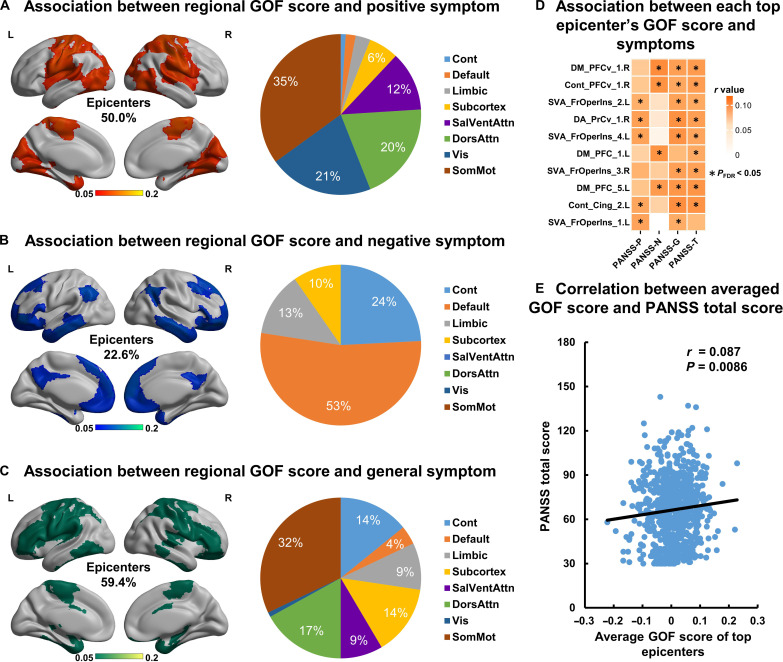
We estimated the association of different symptoms of schizophrenia (positive, negative, and psychopathological symptoms) with the GOF score for each brain region (Fig. 4, A to C). We observed that these regions (50.0% belong to epicenters) that exhibited significant association (P < 0.05, FDR correction) with positive symptoms were mainly located at the primary cortex, including the visual and sensorimotor cortex. The regions (22.6% belong to epicenters) that were associated with negative symptoms (P < 0.05, FDR correction) were mainly located at the higher-level cortices including the prefrontal, temporal, parietal, and association cortices. Within the epicenter top 10 regions, positive symptoms are mainly related to the regions belonging to the ventral attention network, while negative symptoms are mainly related to the regions belong to the default network (P < 0.05, FDR correction) (Fig. 4D). Higher GOF score of the epicenter top regions (i.e., higher probability being a candidate epicenter) correlated with higher Positive and Negative Syndrome Scale (PANSS) positive score (r = 0.071, P = 0.027), higher general score (r = 0.089, P = 0.0075), and higher PANSS total score (r = 0.087, P = 0.0086) (Fig. 4E).
Fig. 4. Associations between regional GOF score and symptoms in schizophrenia.
Brain regions whose GOF scores correlated to (A) positive symptoms, (B) negative symptoms, and (C) general symptoms are mapped to a brain template for visualization. The pie chart shows the proportion of these brain regions belonging to each resting-state network. (D) Association between the GOF score of each top epicenter and symptoms. Asterisk represents a significant correlation (P < 0.05, FDR correction). (E) Positive correlation between averaged GOF score of top epicenters and PANSS total score (r = 0.087, P = 0.0086). Cont, frontoparietal network; Default, default mode network; Limbic, limbic network; SalVentAttn, salience/ventral attention network; DorsAttn, dorsal attention network; Vis, visual network; SomMot, somatomotor network; PANSS-P, PANSS positive subscale score; PANSS-N, PANSS negative subscale score; PANSS-G, PANSS general subscale score; PANSS-T, PANSS total score.
Association between epicenter and cortical gene expression
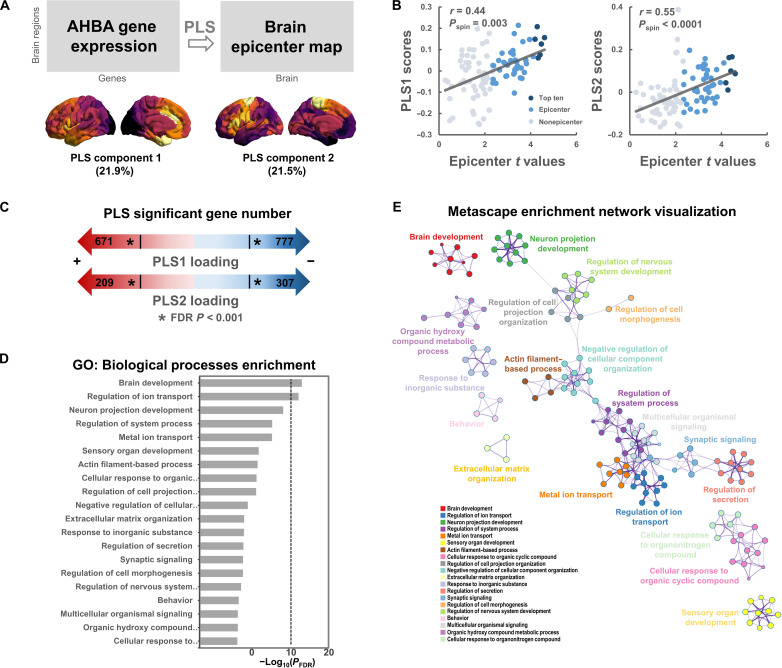
Using data from Allen Human Brain Atlas (AHBA) (37, 39), we acquired brain-wide gene expression levels in the left hemisphere. Then, Partial Least Squares (PLS) regression (40) was used to examine the spatial association between epicenter t map and gene expression maps. The first two PLS components explained 43.4% of the variance (Fig. 5A) (Pspin = 0.006), which is interpreted as regional variations in the schizophrenia-related epicenter t map that are also captured in the transcriptional architecture of human cortex. Notably, we found that the first two PLS components (PLS1 and PLS2) were spatially correlated with the epicenter t map (PLS1: r = 0.44, Pspin = 0.003; PLS2: r = 0.55, Pspin < 0.0001; Fig. 5B). We ranked the genes according to their weights (i.e., Z scores of PLS loadings) to each PLS component by bootstrapping. The list of genes with an FDR P < 0.001 (both positive Z score and negative Z score) was extracted for enrichment analysis using Metascape (41) (Fig. 5C). Enrichment analysis revealed the top 20 significant gene ontology (GO) biological processes, such as “brain development,” “regulation of ion transport,” and “neuron projection development” (Fig. 5D). To further visualize the associations between the enriched terms, a subset of enriched terms have been rendered as a network plot using Cytoscape (42) (Fig. 5E).
Fig. 5. Gene expression profiles related to epicenter distribution map.
(A) Using ENIGMA Toolbox, we obtained a matrix of transcriptional levels (100 regions × 9138 gene expression levels) in the left hemisphere from the AHBA (http://human.brain-map.org). PLS regression was used to examine the spatial association between epicenter distribution map (t values) and gene expression levels for all 9138 genes across the 100 regions within the left hemisphere. The first two PLS components explained 43.4% of the variance (Pspin = 0.006). (B) The first two PLS components (PLS1 and PLS2) are spatially correlated with the epicenter distribution map. (C) By bootstrapping, genes are ranked according to their weights (i.e., Z scores) to each PLS component. The list of genes with an FDR P < 0.001 (both positive Z score and negative Z score) was extracted for enrichment analysis. (D) Top 20 significant GO biological processes by Metascape enrichment analysis. (E) Enrichment network visualization using Cytoscape. Each node represents an enriched term, where its size is proportional to the number of input genes included in that term, and its color represents its cluster identity (i.e., nodes of the same color belong to the same cluster).
Association between epicenter distribution and brain-wide neurotransmitter distribution
Neurotransmitter alterations are implicated in the pathophysiology of schizophrenia (43, 44). To examine whether schizophrenia epicenter associated with neurotransmitter expressions, we investigated the spatial relationship between schizophrenia epicenter t map and 19 neurotransmitter receptor/transporter density maps (including dopamine, norepinephrine, serotonin, acetylcholine, glutamate, γ-aminobutyric acid (GABA), histamine, cannabinoid, and opioid), across nine different neurotransmitter systems (Extended Fig. 5A). We found that the epicenter t map in schizophrenia spatially correlated with the neurotransmitter density maps in acetylcholine [vesicular acetylcholine transporter (VAChT), r = 0.508, Pspin = 0.0010; α4β2, r = 0.371, Pspin = 0.0095], histamine [histamine H3 receptor (H3), r = 0.413, Pspin = 0.0015], opioid [mu opioid receptor (MOR), r = 0.408, Pspin = 0.0015], norepinephrine [norepinephrine transporter (NET), r = 0.385, Pspin = 0.0020] and glutamate [metabotropic glutamate receptor 5 (mGluR5), r = 0.331, Pspin = 0.0140] (Extended Fig. 5B) after the multiple comparison correction (P < 0.05, FDR correction). Details are provided in table S6.
Association between epicenter distribution and cognitive function maps
We further investigated how the spatial distribution of epicenter corresponds to expected functional activation during known cognitive processes (table S7). We used PLS analysis (40) to determine the relationship between the Neurosynth-based cognitive function matrix and schizophrenia epicenter t map (Extended Fig. 6A). The PLS first component explained 33.9% of the variance (Pspin < 0.001), indicating a spatial pattern of cognitive functions (PLS1) and epicenter t map that together captured 33.9% of the covariance between the two datasets (Extended Fig. 6B). A significant spatial correlation was observed between the PLS1 component and epicenter t map (r = 0.61, Pspin < 0.0001; Extended Fig. 6C). The cognitive terms were ranked by the Z scores of PLS loadings (a full list is provided in table S8). We found that cognitive processes with the greatest positive loading are enriched for language processes such as “speech production” and “speech perception” (Extended Fig. 6D).
Prediction of short-term treatment outcomes using epicenter mapping
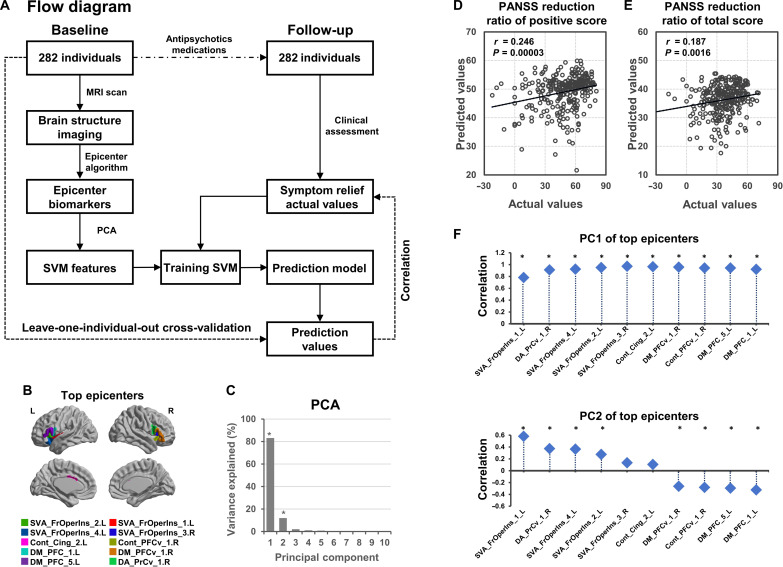
To investigate whether the baseline epicenter could predict antipsychotic medication outcomes (i.e., PANSS reduction ratio) for a given patient, we tested a model to predict each patient’s outcome (Fig. 6A) in a longitudinal follow-up sample including 282 patients with schizophrenia (table S4). First, epicenter mapping was performed for each baseline patient; the GOF scores of the top epicenters were extracted (Fig. 6B). To eliminate potential noises, principal components analysis (PCA) was used to purify the main components from the top epicenters GOF scores. Here, the first two principal components (PCs), which explained 96.9% of the variance of top epicenters’ GOFs (Fig. 6C), were used to train a support vector machine (SVM) regression model. By leave-one-individual-out cross-validation (LOOCV), we predicted the value of the PANSS reduction ratio for each patient. We observed significant correlations between predicted values and actual values for PANSS positive score (r = 0.246, P= 0.00003; Fig. 6D) and PANSS total score (r = 0.187, P= 0.0016; Fig. 6E). We also validated the significant prediction performance by K-fold cross-validation procedures (K = 5 and 10) (table S9). This suggests that the top epicenters’ GOF scores could predict the symptom changes following antipsychotic medications. Furthermore, we further investigated the relationships between the PC and each top epicenter’s GOF score using Pearson’s correlation analyses. We found that the first PC (PC1) showed highly positive correlations with all top epicenters. The second PC (PC2) showed a positive correlation with the top epicenters belonging to the ventral attention network that includes the frontoinsular cortex, as well as negative correlations with the top epicenters belonging to the default mode network (Fig. 6F). In addition, we found that training the prediction model using features from all brain regions did not improve the correlation between prediction values and actual values (Extended Fig. 7).
Fig. 6. Epicenter mapping-based prediction model predicts individual short-term symptom relief following antipsychotic medications in schizophrenia.
(A) Flow diagram of the prediction analysis to predict individual PANSS reduction score following antipsychotic medications. (B) Top epicenters (i.e., the top 10 regions with highest degree at baseline) are displayed in a brain template. The GOF scores of the top epicenters within the patient’s epicenter map at baseline were extracted as features. (C) On the basis of the PCA, the first two PCs, which explained 96.9% of the variance of the top epicenters’ GOF scores, were used for building SVM prediction model. (D) There is a significant correlation between predicted values and actual values for the PANSS positive score (r = 0.246, P = 0.00003). (E) Significant correlation between predicted values and actual values for PANSS total score (r = 0.187, P = 0.0016). (F) Pearson’s correlations between each PC and top epicenters. *Bonferroni-corrected P < 0.05.
DISCUSSION
In this work, we first detected the onset sites of neuroanatomical pathology in schizophrenia, using a patient-tailored connectivity-based epicenter mapping approach. Second, we established potential clinical applicability of epicenter mapping by verifying its specificity to schizophrenia against 10 other neurological, neurodevelopmental, or psychiatric disorders that are either differentials in diagnosis (BD and MDD), or comorbid (ADHD, ASD, and OCD) or share certain neurocognitive features (PD, MCI, and AD) with schizophrenia. Epicenter mapping also has predictive value for short-term, symptom-level efficacy of antipsychotic treatment. Third, we demonstrate a neurobiological coherence of schizophrenia epicenters by demonstrating a relationship with downstream cognitive processes (especially language), symptom domains (positive symptoms), and upstream neurotransmitter receptor/transporter distribution (multiple systems, including acetylcholine and histamine) and gene expression maps. Together, our work systematically demonstrated that epicenters are reliable neuroimaging signatures grounded on key genetic, neurochemical, neurocognitive, and psychopathological foundations of schizophrenia. An ideal use case for epicenter mapping approach will be to identify the patient-specific “origins” of pathophysiology of schizophrenia and to exploit this knowledge in providing prognostic information to patients and their families.
Previous studies using epicenter methodology have successfully identified the possible initial region of neuroanatomical abnormality in neuropsychiatric disorders (28, 45) and neurodegenerative diseases such as frontotemporal dementia (29) and AD (46). However, they were based on group-level evaluation that assumed a common epicenter for all patients, which have ignored between-patient variability. Using patient-tailored epicenter mapping, we estimate the epicenters with maximum likelihood of being the “source” sites with the most active pathophysiological process in 1024 individuals with schizophrenia. The patient-tailored approach, which allows for anatomical heterogeneity within schizophrenia (3), enables the evaluation of epicenter likelihood at individual level through comparing the patient’s unique pattern of pathophysiology to brain regional connectivity pattern. Here, we found that regions with higher likelihood being epicenters had pronounced atrophy (the term “atrophy” here represents an infranormal measures in brain structural MRI in schizophrenia compared to the healthy control population rather than a cellular process of neuron-specific changes), which was further supported by another independent cohort including patients who exhibited subtle GMV reduction (~3%) at 3-month follow-up. It is crucial to emphasize that the atrophy in brain imaging in this study cannot be solely attributed to either disease itself progression or medication effects; rather, it is likely influenced by a combination of both factors. Furthermore, this study demonstrated that regions with stronger connections to the top epicenters (the first 10 regions with largest likelihood being the sites of onset) had more severe atrophy in schizophrenia, supporting our assumption that pathophysiology may spread from epicenters through the inherent network patterns of the human brain (28). Overall, these findings argue for the epicenter mapping method being effective in identifying potential “sources of origin” and pathways of neuroanatomical spread of the neurobiological process in schizophrenia.
We determined that despite individual variations, the top epicenters of schizophrenia are located at the inferior frontal and adjacent frontoinsular-cingulate regions, with this finding replicable in another independent sample. While the role of antipsychotics in the effect of volume reduction cannot be ruled out on the basis of our design, the top epicenters with tissue reduction are more prominent in subsamples of patients with drug-naïve schizophrenia or minimal exposure to medications. These regions have been widely demonstrated to contribute to a key neuropathological role of schizophrenia (47). The inferior frontal cortex, including the core portions of language regions (i.e., Broca’s area), supports the Crow’s linguistic primacy hypothesis in schizophrenia (48). The frontoinsular-cingulate epicenter, as the key node of salience network (49), supports the model of abnormal salience processing, in line with a triple-network model of schizophrenia (50). In addition, neuroanatomical atrophy within the top epicenters have been reported to happen before the first psychotic episode in individuals who later convert to psychosis (51, 52), further supporting the notion that the frontoinsular cortex plays a role as the site of “origin.” Together, the current epicenter mapping provides evidence for the Broca’s area and adjacent frontoinsular cortex being the key sites of onset of the neuropathophysiology of schizophrenia.
Transdiagnostic pathophysiological processes assume special importance in psychiatric disorders, as successfully intervening in such processes can have a broad impact for both prevention and treatment of severe mental illnesses (53). Our study investigates transdiagnostic nature of the epicenter mapping in more than 4000 individuals diagnosed with schizophrenia, BD, MDD, OCD, ADHD, ASD, PD, MTLE, MCI, or AD. Here, we have demonstrated that the epicenter mapping successfully detects regions known to have high likelihood of being sites of onset in neurological disorders. For example, our data indicated that the top epicenters located at bilateral hippocampus for individuals diagnosed with AD or progressive MCI. For the patients with right MTLE, medial temporal regions within the right hemisphere were identified as the top epicenters. The location of epicenters was consistent with the classic lesions in neurological disorders (54–56), again confirming the validity of epicenter methodology. Applying this method to several neuropsychiatric disorders, we found that the schizophrenia-specific epicenter distribution was spatially related to other conventional psychiatric disorders OCD, BD, and MDD but not any of the neurological illnesses. The emergence of the insula and inferior frontal gyrus as epicenters was restricted only to schizophrenia but not to other disorders, including MDD, BD, and OCD. High specificity among some degree of transdiagnostic similarity suggested that while the epicenter mapping transcends categorical diagnostic criteria among severe mental illnesses, frontoinsular specificity highlights its specific utility for schizophrenia. Our longitudinal data further demonstrated that the epicenter status of the frontoinsular area at pretreatment phase could predict short-term outcomes of antipsychotic medications in schizophrenia.
In addition to clinical applicability, neurobiological interpretations of our findings are further discussed through systematically contextualizing epicenters of schizophrenia with respect to cross-disciplinary reference maps, including cognitive maps, PET-derived neurotransmitter maps, and gene expression maps. First, we found that the epicenter distribution of schizophrenia is spatially correlated with a low-dimensional representation of cognitive processes, to which the greatest positive contributions are enriched at language processes. This is in line with our observation that the language region as the possible origin of neurophysiological pathology, supporting language hypothesis in schizophrenia (57, 58). Second, we found a robust spatial concordance between the epicenter distribution of schizophrenia and gene expression profiles of cerebral cortex. These genes whose expression patterns align with epicenters in schizophrenia are significantly enriched in functional clusters with respect to neurodevelopment processes in human brain (59). As epicenters are hypothesized as onset sites that propagate pathological processes to other areas they connect to (28), it is reasonable to speculate that the neurophysiological onset of illness may coincide with atypical neurodevelopmental processes in an affected individual (60). Third, we found a prominent link between the epicenter distribution in schizophrenia and neurotransmitter distribution patterns, supporting the idea that the neurophysiological change of schizophrenia depends on the underlying chemoarchitecture (61). Collectively, our work goes beyond the traditional univariate associations and offers a comprehensive insight on the anatomy of schizophrenia from multiple scales and levels of its neurobiology. This may provide a necessary step toward uncovering the complex aetiology of schizophrenia.
The present work should be considered alongside some methodological issues. First, we used the functional connectome because fMRI can better capture long-range connections among distributed brain networks, compared to diffusion-weighted MRI (DWI). Systematic false positives are also a defect for anatomical connectomes based on current DWI tractography algorithms (62). Furthermore, we took the group average connectome from a cohort of healthy individuals (n = 1089) as the representation of intrinsic connectivity pattern, which is highly reliable across populations (63). Second, although sample size is an advantage of the current work, mixed data from sites may be affected by confounding factors, such as different cohorts, scanners, and sites. Despite the large number of cross-sectional MRI data for schizophrenia, larger longitudinal data with long-term follow-ups are needed to verify the predictive biomarkers to antipsychotic treatment. Third, some cross-sectional data were from chronic patients taking antipsychotic medications over several years. Thus, the atrophy in brain imaging, especially the generalized effects seen in nonepicenters, cannot be solely attributed to an active disease process alone, and additive or interactive effects of medications and pathophysiology are equally likely. Forth, in methodology and image processing, the parameters setting is defined by arbitrary threshold, such as spatial smooth kernel and mask definition, which requires a priori experience.
In summary, our work reveals that schizophrenia-specific epicenters are most likely located at the Broca’s area and adjacent frontoinsular cortex. The presence of epicenters highlights the existence of a reliable objective neuroimaging marker with diagnostic specificity to schizophrenia and the ability to predict response to treatments for psychosis at an individual level.
METHODS
Sample characteristics
Cross-sectional discovery sample
The discovery sample consisted of cross-sectional T1-weighted MRI scans from four hospitals including Shanghai Mental Health Centre (dataset no. 1), First Affiliated Hospital of Zhengzhou University (dataset no. 2), Taipei Veteran General Hospital (dataset no. 3), and Clinical Hospital of Chengdu Brain Science Institute in Chengdu (dataset no. 4) and from five publicly available datasets, i.e., COBRE (dataset no. 5), NMorphCH (dataset no. 6), FBIRN (dataset no. 7), NUSDAST (dataset no. 8), and DS000115 (dataset no. 9). A total of 1124 patients with schizophrenia (479 females, age = 31.1 ± 12.8 years) and 1046 healthy controls (498 females, age = 32.6 ± 12.4 years) were included after data quality control (Supplementary Materials). Patients with schizophrenia were diagnosed according to the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV). Individuals were excluded from the study if they were (i) diagnosed with schizoaffective disorder, mood disorders, or other major medical or neurologic disorders; (ii) alcohol/drug dependence; (iii) had a history of electroconvulsive therapy within 6 months; and (iv) other contraindications to MRI scanning. Symptom severity was assessed with the PANSS for patients from datasets no. 1, no. 2, no. 3, no. 4, and no. 5, with the Brief Psychiatric Rating Scale for patients from dataset no. 8, or with the Scale for the Assessment of Positive Symptoms and Scale for the Assessment of Negative Symptoms for patients from datasets no. 6, no. 8, and no. 9. Detailed information of each cohort is provided in the Supplementary Materials and table S1. Written informed consent was obtained from all participants and/or their legal guardians. The Medical Research Ethics Committees of the local hospitals approved this study [ethics number: 2017-36R (dataset no. 1), 2018-KY-88 (dataset no. 2), YM105091F (dataset no. 3), CDFH2014030501 (dataset no. 4)].
Cross-sectional validation sample
The validation sample consisted of 147 patients with schizophrenia spectrum disorders (61 female, age = 38.0 ± 14.1) and 948 healthy controls (409 female, age = 36.5 ± 14.8) from a publicly shared dataset—SRPBS Multi-disorder MRI Dataset (https://bicr-resource.atr.jp/srpbsfc/) (64). Patients with schizophrenia were diagnosed on the basis of the Structured Clinical Interview for DSM-IV Axis I Disorders-Patient Edition. Ninety individuals have illness duration (mean 14.2 ± 9.4 years) and medication information [chlorpromazine equivalents (CPZ): 581.1 ± 445.0 mg/day].
Multiple brain disorder samples
For our transdiagnostic analyses, we used MRI data from local hospitals and publicly available datasets. Included neurological, neurodevelopmental, and psychiatric disorders comprised BD (ncases = 101, ncontrols = 136), MDD (ncases = 1247, ncontrols = 1082), OCD (ncases = 80, ncontrols = 61), ADHD (ncases = 344, ncontrols = 519), ASD (ncases = 453, ncontrols = 476), PD (ncases = 374, ncontrols = 172), MTLE (ncases = 149, ncontrols = 48), and MCI (ncases = 370, ncontrols = 229), and AD (ncases = 186, ncontrols = 229). The demographic information is provided in table S2. The MDD sample came from the REST-meta-MDD consortium (http://rfmri.org/REST-meta-MDD) (65). The ADHD sample was obtained from the consortium of the International Neuroimaging Datasharing Initiative (INDI) (http://preprocessed-connectomes-project.org/adhd200/) (66). The ASD sample was acquired from the Autism Brain Imaging Data Exchange (ABIDE) initiative (http://fcon_1000.projects.nitrc.org/indi/abide/abide_II.html). The PD sample came from the Parkinson’s Progression Markers Initiative (PPMI) (www.ppmi-info.org/) (67). The MCI and AD samples were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (https://adni.loni.usc.edu/). These samples were obtained from local hospitals and have been reported in published studies for MTLE (68), OCD (69), and BD (70). We redirect the reader to the previous publications for more details on demographic information or medications. The Medical Research Ethics Committees of the local hospitals approved this study.
Longitudinal sample
A total of 282 patients with schizophrenia, from two hospitals (Shanghai Mental Health Center, ncases = 180; Peking University People’s Hospital, ncases = 102), treated with antipsychotic medication were included in the longitudinal observational analyses (table S4). All individuals met DSM-IV diagnostic criteria for schizophrenia, and no other comorbid Axis I disorders. Inclusion and exclusion criteria of individuals are provided in our previous study (22). At baseline, 260 of them were treatment-naïve first-episode schizophrenia. Following baseline MRI, patients received antipsychotic medications. Two hundred twenty-eight of 282 received monotherapy: amisulpride (n = 19), aripiprazole (n = 52), blonanserin (n = 1), clozapine (n = 2), olanzapine (n = 68), paliperidone (n = 11), paliperidone palmitate injection (n = 4), quetiapine (n = 2), risperidone (n = 67), and ziprasidone (n = 2). Fifty-four received combined therapy (two or more antipsychotic drugs). The daily dosage of drugs was converted to CPZ. The mean CPZ during medication was 375.1 ± 241.0 mg/day. The severity of symptoms was evaluated on the basis of PANSS administered by the same psychiatrist in each patient. Symptom relief indicated in PANSS total and subscale scores [reduction ratio = (baseline − follow-up)/baseline × 100% (71)] were used to measure the treatment response. The average duration of PANSS follow-up to estimate the treatment response was 10.7 weeks.
Neuroimaging acquisition and preprocessing
High spatial-resolution T1-weighted MRI acquisition protocol for each cohort has been described previously (21, 22, 72–77). Images were processed by the Computational Anatomy Toolbox (www.neuro.uni-jena.de/cat/). A fully automated procedure for standard voxel-based morphometry (VBM), including spatial registration, tissue segmentation, and bias correction of intensity nonuniformities, was conducted. The resulting modulated gray matter images were smoothed with an 8-mm full width at half maximum Gaussian kernel.
Determination of individual patient gray matter reduction z-score map
We derived voxel-wise GMV using VBM for all individuals. The GMV for each voxel was then adjusted by regressing out the effects of gender, age, the square of age, total intracranial volume (TIV), and sites using a regression model. Figure S5 shows that the site effect has been successfully removed from the adjusted GMV values by comparing site difference in healthy controls. Subsequently, a gray matter mask was used to exclude non–gray matter voxels. For everyone with schizophrenia, the adjusted GMV values were normalized relative to the control population using the z-score procedure (27), which created voxel-wise individualized maps of gray matter reduction. The z-score represents the severity of an abnormality for a voxel, in this case MRI-derived GMV. Higher z-score represents larger deviation from the normal (i.e., a smaller GMV in this case). As the goal of the current study is to detect the potential epicenter of gray matter reduction, inclusion of all voxels, regardless of whether it is “abnormal” or not, may reduce sensitivity of detection. To eliminate the bias of some voxels with subthreshold reduction, we made a spatial mask for each patient by using a threshold greater than 0.5 of GMV z-score; thus only voxels showing >0.5 z-score deviations were considered for epicenter mapping. Figure S6 shows how many voxels were included per individual for epicenter mapping. We also evaluated the consistency in using different z-score thresholds (fig. S7).
Characterization of 'disease epicenter map' in schizophrenia
To identify potential disease epicenter of neuroanatomical abnormality, we performed patient-tailored connectivity-based epicenter mapping of schizophrenia (Fig. 1). We hypothesized that an epicenter would be a region with severe GMV reduction whose intrinsic connectivity pattern most strongly resembled the patient’s GMV reduction pattern (27). First, we derived a normative set of functional connectomes from a large cohort of health control from HCP cohort (n = 1089, age = 28.8 ± 3.7 years, 593 females). We used a whole-brain parcellation composed of 200 cortical (78) and 54 subcortical regions (79) (Supplementary Materials). We also verified the robustness of the results using brain atlases with different parcels ranging from 68 to 600 parcels (78, 80–83). Using the resting-state fMRI data from the 1089 healthy individuals, each of 254 regions was used as the seed region to compute the Pearson’s correlation coefficient with other voxels, generating a seed-based intrinsic FC map. The seed-based voxel-wise FC map was transformed to FC z map by Fisher’s r-to-z transformation. One sample t test was performed on all individuals’ FC z maps to obtain group-level statistical parametric FC t map. We then ran a spatial correlation analysis between each of these 254 voxel-wise FC t maps with the gray matter reduction z-score map from each patient to detect patient-specific candidate epicenters, whose FC pattern best explained the observed pattern of GMV reduction (27). Spearman correlation coefficient was calculated and further transformed to Fisher’s z-score to quantify epicenter GOF score. The procedure generated 254 GOF scores corresponding to 254 candidate epicenter regions. The seed region with a higher GOF score represents higher probability of being a candidate epicenter. One sample t test was performed on the GOF score to determine the significance of candidate epicenters. The resulting statistical t map represented inferred epicenter degree for the 254 regions across the whole-brain at a group level. Multiple comparison correction was performed using FDR.
Associations between epicenter signature and GMV reduction pattern
To investigate the relationship between epicenter signature and GMV reduction pattern in schizophrenia, we conducted spatial association analyses in a longitudinal cohort including 148 patients with schizophrenia who had both MRI scans at baseline and 12-week follow-up. The longitudinal GMV reduction map was measured as the ratio in the change of GMV maps between the baseline and the follow-up. Spearman correlation between epicenter t map and longitudinal GMV reduction map was performed. To examine whether brain areas with stronger connections to the epicenter had more severe GMV reduction, we calculated the whole-brain FC t map with the top epicenters as the seed. Spatial correlation between the longitudinal GMV reduction map and the top epicenters’ FC map was conducted. Spatial autocorrelation-preserving permutation tests, termed “spin tests,” were used to correct for potential confounding effects of spatial autocorrelation (https://github.com/frantisekvasa/rotate_parcellation) (version 3, June 2022) (84, 85) (Supplementary Materials).
Links between epicenter signature and ENIGMA disease brain maps
To examine whether the link of epicenter signature with GMV reduction pattern in schizophrenia is specific across different brain disorders, we estimated the spin-corrected spatial correlation between the epicenter t map of schizophrenia (that is derived from our discovery sample) and ENIGMA-derived thinner–cortical thickness maps, including eight neurological, neurodevelopmental, and psychiatric disorders. ENIGMA summary statistics of thinner–cortical thickness map (i.e., effect sizes for case-control differences in cortical thickness across whole brain regions) were obtained from ENIGMA toolbox (https://github.com/MICA-MNI/ENIGMA) (version 2.0.0, July, 2022). All cortical thickness maps were collected from more than 17,000 scanned patients against almost 22,000 controls, comprising a total of eight neurological, neurodevelopmental, and psychiatric disorders including 22q11.2 deletion syndrome (22q) (86), ADHD (9), ASD (87), idiopathic generalized epilepsy, right and left temporal lobe epilepsy (88), depression (7), OCD (89), schizophrenia (19), and BD (8). FDR correction was used for multiple comparison corrections.
Epicenter characterization in multiple brain disorders
To examine whether the epicenter in schizophrenia exhibits diagnostic specificity compared with other brain disorders, we characterized the epicenter t maps from more than 4000 individuals diagnosed with either schizophrenia (ncases = 1124, ncontrols = 1046), BD (ncases = 101, ncontrols = 136), MDD (ncases = 1247, ncontrols = 1082), OCD (ncases = 80, ncontrols = 61), ADHD (ncases = 344, ncontrols = 519), ASD (ncases = 453, ncontrols = 476), PD (ncases = 374, ncontrols = 172), MTLE (ncases = 149, ncontrols = 48), MCI (ncases = 370, ncontrols = 229), and AD (ncases = 186, ncontrols = 229). The demographic information is provided in table S2. One sample t test was performed to characterize the epicenter degree map at the group level for each disease/disorder. To examine the similarity in the global pattern of epicenter t map between schizophrenia and each of the other disorders, Spearman correlation test was used to investigate the spatial correlation of epicenter t map between schizophrenia and each of the other disorders. To examine whether schizophrenia had specific epicenters compared with other disorders, we further ranked the brain regions according to their epicenter degree in each disorder. Wilcoxon rank sum test was further used to examine the significant differences in the rank of the top epicenters (the first 10 regions with the highest epicenter degree in schizophrenia, which were derived from the discovery sample) between schizophrenia and each of the other brain disorders. FDR correction was used for multiple comparison corrections.
Associations between the GOF score and specific symptoms in schizophrenia
To investigate the relationship between specific symptoms and GOF for each region in schizophrenia, we mapped specific symptoms in schizophrenia to the whole brain by regional-wise correlation analysis between the regional GOF score and PANSS positive, negative, and general subscale scores. In addition, we further explored the relationship between the mean GOF score within the top epicenters and specific symptoms in schizophrenia. FDR correction was used for multiple comparison corrections.
Correlation analysis between epicenter and gene expressions
Brain-wide gene expression profiles were acquired from the AHBA (http://human.brain-map.org). The ENIGMA Toolbox (https://enigma-toolbox.readthedocs.io/en/latest/index.html) (version 2.0.0, July 2022) (90) provides brain-wide microarray expression data collected from AHBA. Microarray expression data were first processed using abagen (91). As the AHBA dataset had only two right hemisphere data, only regions within left hemisphere (n = 100) were extracted using the same atlas (i.e., Schaefer parcellation) with the epicenter t map. In addition, genes whose similarity across donors fell below a threshold (r < 0.2) were further removed, leaving a total of 9138 genes for following analysis. PLS regression (40) was used to examine the spatial association between epicenter t map and gene expression levels for all 9138 genes across the 100 regions within the left hemisphere. Permutation testing was applied to test the null hypothesis that PLS components explained no more covariance between the epicenter t map and brain-wide gene expression than expected by chance. Spin tests were also considered for correcting spatial autocorrelation (Supplementary Materials). Z scores and corresponding P values estimated by Bootstrapping were used to rank genes according to their contribution to each significant PLS component (92). The list of genes with an FDR P < 0.001 was extracted for enrichment analysis. Gene enrichment analysis was performed using Metascape (41) (https://metascape.org/gp/index.html#/main/step1). The genes list generated from PLS analysis was input into the Metascape analysis. The threshold was set at q value = 0.01 using the Benjamini-Hochberg procedure to account for multiple testing (93). Details are provided in the Supplementary Materials.
Correlation analysis between epicenter and neurotransmitter distribution
We further investigated the association between epicenter t map and neurotransmitter distribution maps (details are provided in the Supplementary Materials). A comprehensive cortical profile of neurotransmitter receptor densities was previously obtained from PET images of more 1200 healthy individuals across multiple studies (https://github.com/netneurolab/hansen_receptors) (version 1, July, 2022). A total of 19 different neurotransmitter receptors and transporters maps, across nine different neurotransmitter systems, including dopamine, norepinephrine, serotonin, acetylcholine, glutamate, GABA, histamine, cannabinoid, and opioid, were obtained and then parcellated to 200 regions (only cortical regions are available) using the same atlas with the epicenter t map. To examine the spatial association between epicenter distribution and neurotransmitter distribution, we estimated the Spearman correlation between the epicenter t map of schizophrenia and the individual neurotransmitter receptor/transporter density map. Spin tests were used to correct the spatial autocorrelation. Multiple comparisons were corrected by FDR.
Correlation analysis between epicenter and cognitive functions
We further investigated how the spatial distribution of epicenter correspond to cognitive processes. Neurosynth (https://github.com/neurosynth/neurosynth) (version 1, July 2022), a meta-analytic tool that synthesizes results from more than 15 000 published fMRI studies, was used to derive meta-analytic task activation maps, which provide a quantitative representation of how specific brain regions are activated during multiple tasks (38) (Supplementary Materials). A total of 123 cognitive terms (table S7) were selected from Cognitive Atlas, a public ontology of cognitive science (94) including a comprehensive list of neurocognitive processes. We parcellated the activation maps into the same 254 region atlas used for epicenter t map, resulting a region × cognitive function matrix. Then, we used PLS analysis (40) and permutation testing to examine the association between cognitive function matrix and epicenter t map. Spin tests were used to correct the spatial autocorrelation. Boostrapping was used to calculate the Z scores and rank the cognitive terms according to their contribution to significant PLS component.
Individual-level prediction to short-term treatment outcomes using epicenter mapping
To investigate whether the baseline epicenters could predict treatment outcomes following short-term antipsychotic treatment for a given patient, we built a prediction model to predict each patient’s symptom relief (i.e., PANSS reduction ratio) in a longitudinal follow-up cohort including 282 patients with schizophrenia who were treated with antipsychotic medications for an average of 10.7 weeks. Figure 6A shows the flow diagram of the prediction analysis. We applied LOOCV to obtain training set and test set. In each LOOCV, one individual was used as a test set, and the remaining individuals were used as a training set. In training set, the baseline GOF scores of the top epicenters (i.e., the top 10 regions with the highest GOF derived from the discovery sample) were extracted as features. To eliminate potential noises, we performed a PCA on GOF scores to purify the main components, which were further used to train a SVM regression model to predict PANSS reduction ratio at follow-up. The test set patient’s PANSS reduction ratio was predicted based on the built SVM regression model. Last, we used the Pearson’s correlation to determine whether the predicted value is correlated with the actual value. In addition, to determine whether the GOF scores of the top epicenters would predict symptom relief better than other regions, we also used the prediction analysis using GOF scores from all brain regions. We also validate the prediction performance by using 5- and 10-fold cross-validation procedures (Supplementary Materials).
Acknowledgments
:We thank the investigators provided public access to MRI data from patients diagnosed with schizophrenia through the COBRE database funded by a Center of Biomedical Research Excellence grant 5P20RR021938/P20GM103472 from the NIH to V. Calhoun, the fBIRN data supported by grants to the Function BIRN (U24-RR021992) Testbed funded by the National Center for Research Resources at the National Institutes of Health, USA, the NMorphCH dataset funded by NIMH grant R01MH056584, and the SchizConnect funded by NIMH cooperative agreement 1 U01 MH097435.
Funding: This work was supported by the following: grant from Science and Technology Innovation 2030-Brain Science and Brain-Inspired Intelligence Project (no. 2022ZD0212800 to Y.J.); the National Natural Science Foundation of China (no. 82202242 to Y.J. and no. 82071997 to W.C.); the projects from the China Postdoctoral Science Foundation (nos. BX2021078 and 2021M700852 to Y.J.); the Shanghai Rising-Star Program (no. 21QA1408700 to W.C.) and the Shanghai Sailing Program (22YF1402800 to Y.J.) from Shanghai Science and Technology Committee; the National Key R&D Program of China (no. 2019YFA0709502 to J.F.); grant from Shanghai Municipal Science and Technology Major Project (no. 2018SHZDZX01 to J.F.), ZJ Lab, and Shanghai Center for Brain Science and Brain-Inspired Technology; grant from the 111 Project (no. B18015 to J.F.); and grants from the National Key R&D Program of China (no. 2022ZD0208500 to D.Y.) and the CAMS Innovation Fund for Medical Sciences (no. 2019-I2M-5-039 to C. Lu). L.P. acknowledges research support from the Monique H. Bourgeois Chair (McGill University) and a salary award from the Fonds de recherche du Quebec-Sante´ (FRQS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions: Conceptualization: Y.J., W.C., and J.F. Methodology: Y.J. and W.C. Investigation: C. Lu, X.C., J.Z., Y.T., T.Z., C. Li, E.Z., X.Y., W.L., D.A., D.Z., C.-C.H., S.-J.T., C.-P.L., J.C., J.W., D.Y., and ZIB Consortium. Visualization: Y.J. Supervision: J.F. Writing—original draft: Y.J. and L.P. Writing—review and editing: Y.J., L.P., W.C., and J.F.
Competing interests: L.P. reports personal fees for serving as chief editor from the Canadian Medical Association Journals; speaker/consultant fees from Janssen Canada and Otsuka Canada, SPMM Course Limited, UK, and Canadian Psychiatric Association; book royalties from Oxford University Press; and investigator-initiated educational grants from Janssen Canada, Sunovion, and Otsuka Canada. None of these potential conflicts directed the submitted work. The other authors declare that they have no competing interests.
Data and materials availability: Data of COBRE, NMorphCH, FBIRN, and NUSDAST were obtained from the SchizConnect, a publicly available website (www.schizconnect.org). The COBRE dataset was downloaded from the Center for Biomedical Research Excellence in Brain Function and Mental Illness (COBRE) (https://www.mrn.org/collaborate/data-community). The NMorphCH dataset was downloaded from http://niacal.northwestern.edu/data. The FBIRN dataset was downloaded from www.nitrc.org/projects/fbirn/. The NUSDAST dataset was downloaded from the Northwestern University Schizophrenia Data and Software Tool. The DS000115 dataset was downloaded from OpenfMRI database (www.openfmri.org/). The MDD sample came from the REST-meta-MDD consortium (http://rfmri.org/REST-meta-MDD). The ADHD sample was obtained from the consortium of the International Neuroimaging Datasharing Initiative (INDI) (http://preprocessed-connectomes-project.org/adhd200/). The ASD sample was acquired from the Autism Brain Imaging Data Exchange (ABIDE) initiative (http://fcon_1000.projects.nitrc.org/indi/abide/abide_II.html). The PD sample came from the Parkinson’s Progression Markers Initiative (PPMI) (www.ppmi-info.org/). The MCI and AD samples were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (https://adni.loni.usc.edu/). ENIGMA summary statistics of thinner–cortical thickness map were obtained from ENIGMA toolbox (https://github.com/MICA-MNI/ENIGMA) (version 2.0.0, July, 2022). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. T1-weighted images were processed using the Computational Anatomy Toolbox (www.neuro.uni-jena.de/cat/) within SPM12 (www.fil.ion.ucl.ac.uk/spm/software/spm12/). The visualization of brain mapping images was conducted using BrainNetViewer (www.nitrc.org/projects/bnv/). The code for spatial autocorrelation-preserving permutation test is available at (https://github.com/frantisekvasa/rotate_parcellation) (version 3, June 2022).
Supplementary Materials
This PDF file includes:
ZIB (Zhang-jiang International Brain Bank) Consortium Authorship List
Methods S1 to S7
Extended Figs. 1 to 7
Figs. S1 to S7
Tables S1 to S9
Legend for data S1
References
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.Falkai P., A desperate search for biomarkers in schizophrenia. What is going wrong? World Psychiatry 10, 38–39 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraguljac N. V., McDonald W. M., Widge A. S., Rodriguez C. I., Tohen M., Nemeroff C. B., Neuroimaging biomarkers in schizophrenia. Am. J. Psychiatry 178, 509–521 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brugger S. P., Howes O. D., Heterogeneity and homogeneity of regional brain structure in schizophrenia: A meta-analysis. JAMA Psychiatry 74, 1104–1111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinberg D., Lenroot R., Jacomb I., Allen K., Bruggemann J., Wells R., Balzan R., Liu D., Galletly C., Catts S. V., Weickert C. S., Weickert T. W., Cognitive subtypes of schizophrenia characterized by differential brain volumetric reductions and cognitive decline. JAMA Psychiatry 73, 1251–1259 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Alnaes D., Kaufmann T., van der Meer D., Cordova-Palomera A., Rokicki J., Moberget T., Bettella F., Agartz I., Barch D. M., Bertolino A., Brandt C. L., Cervenka S., Djurovic S., Doan N. T., Eisenacher S., Fatouros-Bergman H., Flyckt L., Di Giorgio A., Haatveit B., Jonsson E. G., Kirsch P., Lund M. J., Meyer-Lindenberg A., Pergola G., Schwarz E., Smeland O. B., Quarto T., Zink M., Andreassen O. A., Westlye L. T., C., Brain heterogeneity in schizophrenia and its association wiith polygenic risk. Psychiatry 76, 739–748 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCutcheon R. A., Pillinger T., Mizuno Y., Montgomery A., Pandian H., Vano L., Marques T. R., Howes O. D., The efficacy and heterogeneity of antipsychotic response in schizophrenia: A meta-analysis. Mol. Psychiatry 26, 1310–1320 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmaal L., Hibar D. P., Samann P. G., Hall G. B., Baune B. T., Jahanshad N., Cheung J. W., van Erp T. G. M., Bos D., Ikram M. A., Vernooij M. W., Niessen W. J., Tiemeier H., Hofman A., Wittfeld K., Grabe H. J., Janowitz D., Bulow R., Selonke M., Volzke H., Grotegerd D., Dannlowski U., Arolt V., Opel N., Heindel W., Kugel H., Hoehn D., Czisch M., Couvy-Duchesne B., Renteria M. E., Strike L. T., Wright M. J., Mills N. T., de Zubicaray G. I., McMahon K. L., Medland S. E., Martin N. G., Gillespie N. A., Goya-Maldonado R., Gruber O., Kramer B., Hatton S. N., Lagopoulos J., Hickie I. B., Frodl T., Carballedo A., Frey E. M., van Velzen L. S., Penninx B., van Tol M. J., van der Wee N. J., Davey C. G., Harrison B. J., Mwangi B., Cao B., Soares J. C., Veer I. M., Walter H., Schoepf D., Zurowski B., Konrad C., Schramm E., Normann C., Schnell K., Sacchet M. D., Gotlib I. H., MacQueen G. M., Godlewska B. R., Nickson T., McIntosh A. M., Papmeyer M., Whalley H. C., Hall J., Sussmann J. E., Li M., Walter M., Aftanas L., Brack I., Bokhan N. A., Thompson P. M., Veltman D. J., Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the ENIGMA Major Depressive Disorder Working Group. Mol. Psychiatry 22, 900–909 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hibar D. P., Westlye L. T., Doan N. T., Jahanshad N., Cheung J. W., Ching C. R. K., Versace A., Bilderbeck A. C., Uhlmann A., Mwangi B., Kramer B., Overs B., Hartberg C. B., Abe C., Dima D., Grotegerd D., Sprooten E., Boen E., Jimenez E., Howells F. M., Delvecchio G., Temmingh H., Starke J., Almeida J. R. C., Goikolea J. M., Houenou J., Beard L. M., Rauer L., Abramovic L., Bonnin M., Ponteduro M. F., Keil M., Rive M. M., Yao N., Yalin N., Najt P., Rosa P. G., Redlich R., Trost S., Hagenaars S., Fears S. C., Alonso-Lana S., van Erp T. G. M., Nickson T., Chaim-Avancini T. M., Meier T. B., Elvsashagen T., Haukvik U. K., Lee W. H., Schene A. H., Lloyd A. J., Young A. H., Nugent A., Dale A. M., Pfennig A., McIntosh A. M., Lafer B., Baune B. T., Ekman C. J., Zarate C. A., Bearden C. E., Henry C., Simhandl C., McDonald C., Bourne C., Stein D. J., Wolf D. H., Cannon D. M., Glahn D. C., Veltman D. J., Pomarol-Clotet E., Vieta E., Canales-Rodriguez E. J., Nery F. G., Duran F. L. S., Busatto G. F., Roberts G., Pearlson G. D., Goodwin G. M., Kugel H., Whalley H. C., Ruhe H. G., Soares J. C., Fullerton J. M., Rybakowski J. K., Savitz J., Chaim K. T., Fatjo-Vilas M., Soeiro-de-Souza M. G., Boks M. P., Zanetti M. V., Otaduy M. C. G., Schaufelberger M. S., Alda M., Ingvar M., Phillips M. L., Kempton M. J., Bauer M., Landen M., Lawrence N. S., van Haren N. E. M., Horn N. R., Freimer N. B., Gruber O., Schofield P. R., Mitchell P. B., Kahn R. S., Lenroot R., Machado-Vieira R., Ophoff R. A., Sarro S., Frangou S., Satterthwaite T. D., Hajek T., Dannlowski U., Malt U. F., Arolt V., Gattaz W. F., Drevets W. C., Caseras X., Agartz I., Thompson P. M., Andreassen O. A., Cortical abnormalities in bipolar disorder: An MRI analysis of 6503 individuals from the ENIGMA Bipolar Disorder Working Group. Mol. Psychiatry 23, 932–942 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoogman M., Muetzel R., Guimaraes J. P., Shumskaya E., Mennes M., Zwiers M. P., Jahanshad N., Sudre G., Wolfers T., Earl E. A., Vila J. C. S., Vives-Gilabert Y., Khadka S., Novotny S. E., Hartman C. A., Heslenfeld D. J., Schweren L. J. S., Ambrosino S., Oranje B., de Zeeuw P., Chaim-Avancini T. M., Rosa P. G. P., Zanetti M. V., Malpas C. B., Kohls G., von Polier G. G., Seitz J., Biederman J., Doyle A. E., Dale A. M., van Erp T. G. M., Epstein J. N., Jernigan T. L., Baur-Streubel R., Ziegler G. C., Zierhut K. C., Schrantee A., Hovik M. F., Lundervold A. J., Kelly C., McCarthy H., Skokauskas N., O'Gorman Tuura R. L., Calvo A., Lera-Miguel S., Nicolau R., Chantiluke K. C., Christakou A., Vance A., Cercignani M., Gabel M. C., Asherson P., Baumeister S., Brandeis D., Hohmann S., Bramati I. E., Tovar-Moll F., Fallgatter A. J., Kardatzki B., Schwarz L., Anikin A., Baranov A., Gogberashvili T., Kapilushniy D., Solovieva A., El Marroun H., White T., Karkashadze G., Namazova-Baranova L., Ethofer T., Mattos P., Banaschewski T., Coghill D., Plessen K. J., Kuntsi J., Mehta M. A., Paloyelis Y., Harrison N. A., Bellgrove M. A., Silk T. J., Cubillo A. I., Rubia K., Lazaro L., Brem S., Walitza S., Frodl T., Zentis M., Castellanos F. X., Yoncheva Y. N., Haavik J., Reneman L., Conzelmann A., Lesch K. P., Pauli P., Reif A., Tamm L., Konrad K., Weiss E. O., Busatto G. F., Louza M. R., Durston S., Hoekstra P. J., Oosterlaan J., Stevens M. C., Ramos-Quiroga J. A., Vilarroya O., Fair D. A., Nigg J. T., Thompson P. M., Buitelaar J. K., Faraone S. V., Shaw P., Tiemeier H., Bralten J., Franke B., Brain imaging of the cortex in ADHD: A coordinated analysis of large-scale clinical and population-based samples. Am. J. Psychiatry 176, 531–542 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Erp T. G., Hibar D. P., Rasmussen J. M., Glahn D. C., Pearlson G. D., Andreassen O. A., Agartz I., Westlye L. T., Haukvik U. K., Dale A. M., Melle I., Hartberg C. B., Gruber O., Kraemer B., Zilles D., Donohoe G., Kelly S., McDonald C., Morris D. W., Cannon D. M., Corvin A., Machielsen M. W., Koenders L., de Haan L., Veltman D. J., Satterthwaite T. D., Wolf D. H., Gur R. C., Gur R. E., Potkin S. G., Mathalon D. H., Mueller B. A., Preda A., Macciardi F., Ehrlich S., Walton E., Hass J., Calhoun V. D., Bockholt H. J., Sponheim S. R., Shoemaker J. M., van Haren N. E., Hulshoff Pol H. E., Ophoff R. A., Kahn R. S., Roiz-Santianez R., Crespo-Facorro B., Wang L., Alpert K. I., Jonsson E. G., Dimitrova R., Bois C., Whalley H. C., McIntosh A. M., Lawrie S. M., Hashimoto R., Thompson P. M., Turner J. A., Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol. Psychiatry 21, 547–553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okada N., Fukunaga M., Miura K., Nemoto K., Matsumoto J., Hashimoto N., Kiyota M., Morita K., Koshiyama D., Ohi K., Takahashi T., Koeda M., Yamamori H., Fujimoto M., Yasuda Y., Hasegawa N., Narita H., Yokoyama S., Mishima R., Kawashima T., Kobayashi Y., Sasabayashi D., Harada K., Yamamoto M., Hirano Y., Itahashi T., Nakataki M., Hashimoto R. I., Tha K. K., Koike S., Matsubara T., Okada G., van Erp T. G. M., Jahanshad N., Yoshimura R., Abe O., Onitsuka T., Watanabe Y., Matsuo K., Yamasue H., Okamoto Y., Suzuki M., Turner J. A., Thompson P. M., Ozaki N., Kasai K., Hashimoto R., Subcortical volumetric alterations in four major psychiatric disorders: A mega-analysis study of 5604 subjects and a volumetric data-driven approach for classification. Mol. Psychiatry 28, 5206–5216 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chand G. B., Dwyer D. B., Erus G., Sotiras A., Varol E., Srinivasan D., Doshi J., Pomponio R., Pigoni A., Dazzan P., Kahn R. S., Schnack H. G., Zanetti M. V., Meisenzahl E., Busatto G. F., Crespo-Facorro B., Pantelis C., Wood S. J., Zhuo C., Shinohara R. T., Shou H., Fan Y., Gur R. C., Gur R. E., Satterthwaite T. D., Koutsouleris N., Wolf D. H., Davatzikos C., Two distinct neuroanatomical subtypes of schizophrenia revealed using machine learning. Brain 143, 1027–1038 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang Y., Wang J., Zhou E., Palaniyappan L., Luo C., Ji G., Yang J., Wang Y., Zhang Y., Huang C.-C., Tsai S.-J., Chang X., Xie C., Zhang W., Lv J., Chen D., Shen C., Wu X., Zhang B., Kuang N., Sun Y.-J., Kang J., Zhang J., Huang H., He H., Duan M., Tang Y., Zhang T., Li C., Yu X., Si T., Yue W., Liu Z., Cui L.-B., Wang K., Cheng J., Lin C.-P., Yao D., Cheng W., Feng J.; the ZIB Consortium , Neuroimaging biomarkers define neurophysiological subtypes with distinct trajectories in schizophrenia. Nat. Mental Health 1, 186–199 (2023). [Google Scholar]
- 14.Pan Y., Pu W., Chen X., Huang X., Cai Y., Tao H., Xue Z., Mackinley M., Limongi R., Liu Z., Palaniyappan L., Morphological profiling of schizophrenia: Cluster analysis of MRI-based cortical thickness data. Schizophr. Bull. 46, 623–632 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li A., Zalesky A., Yue W., Howes O., Yan H., Liu Y., Fan L., Whitaker K. J., Xu K., Rao G., Li J., Liu S., Wang M., Sun Y., Song M., Li P., Chen J., Chen Y., Wang H., Liu W., Li Z., Yang Y., Guo H., Wan P., Lv L., Lu L., Yan J., Song Y., Wang H., Zhang H., Wu H., Ning Y., Du Y., Cheng Y., Xu J., Xu X., Zhang D., Wang X., Jiang T., Liu B., A neuroimaging biomarker for striatal dysfunction in schizophrenia. Nat. Med. 26, 558–565 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Elliott M. L., Knodt A. R., Ireland D., Morris M. L., Poulton R., Ramrakha S., Sison M. L., Moffitt T. E., Caspi A., Hariri A. R., What is the test-retest reliability of common task-functional MRI measures? New empirical evidence and a meta-analysis. Psychol. Sci. 31, 792–806 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friston K. J., Frith C. D., Schizophrenia: A disconnection syndrome? Clin. Neurosci. 3, 89–97 (1995). [PubMed] [Google Scholar]
- 18.Fornito A., Zalesky A., Pantelis C., Bullmore E. T., Schizophrenia, neuroimaging and connectomics. Neuroimage 62, 2296–2314 (2012). [DOI] [PubMed] [Google Scholar]
- 19.van Erp T. G. M., Walton E., Hibar D. P., Schmaal L., Jiang W., Glahn D. C., Pearlson G. D., Yao N., Fukunaga M., Hashimoto R., Okada N., Yamamori H., Bustillo J. R., Clark V. P., Agartz I., Mueller B. A., Cahn W., de Zwarte S. M. C., Pol H. E. H., Kahn R. S., Ophoff R. A., van Haren N. E. M., Andreassen O. A., Dale A. M., Doan N. T., Gurholt T. P., Hartberg C. B., Haukvik U. K., Jorgensen K. N., Lagerberg T. V., Melle I., Westlye L. T., Gruber O., Kraemer B., Richter A., Zilles D., Calhoun V. D., Crespo-Facorro B., Roiz-Santianez R., Tordesillas-Gutierrez D., Loughland C., Carr V. J., Catts S., Cropley V. L., Fullerton J. M., Green M. J., Henskens F. A., Jablensky A., Lenroot R. K., Mowry B. J., Michie P. T., Pantelis C., Quide Y., Schall U., Scott R. J., Cairns M. J., Seal M., Tooney P. A., Rasser P. E., Cooper G., Weickert C. S., Weickert T. W., Morris D. W., Hong E., Kochunov P., Beard L. M., Gur R. E., Gur R. C., Satterthwaite T. D., Wolf D. H., Belger A., Brown G. G., Ford J. M., Macciardi F., Mathalon D. H., O'Leary D. S., Potkin S. G., Preda A., Voyvodic J., Lim K. O., McEwen S., Yang F., Tan Y., Tan S., Wang Z., Fan F., Chen J., Xiang H., Tang S., Guo H., Wan P., Wei D., Bockholt H. J., Ehrlich S., Wolthusen R. P. F., King M. D., Shoemaker J. M., Sponheim S. R., De Haan L., Koenders L., Machielsen M. W., van Amelsvoort T., Veltman D. J., Assogna F., Banaj N., de Rossi P., Iorio M., Piras F., Spalletta G., McKenna P. J., Pomarol-Clotet E., Salvador R., Corvin A., Donohoe G., Kelly S., Whelan C. D., Dickie E. W., Rotenberg D., Voineskos A. N., Ciufolini S., Radua J., Dazzan P., Murray R., Marques T. R., Simmons A., Borgwardt S., Egloff L., Harrisberger F., Riecher-Rossler A., Smieskova R., Alpert K. I., Wang L., Jonsson E. G., Koops S., Sommer I. E. C., Bertolino A., Bonvino A., Di Giorgio A., Neilson E., Mayer A. R., Stephen J. M., Kwon J. S., Yun J. Y., Cannon D. M., McDonald C., Lebedeva I., Tomyshev A. S., Akhadov T., Kaleda V., Fatouros-Bergman H., Flyckt L., Schizophrenia P. K., Busatto G. F., Rosa P. G. P., Serpa M. H., Zanetti M. V., Hoschl C., Skoch A., Spaniel F., Tomecek D., Hagenaars S. P., McIntosh A. M., Whalley H. C., Lawrie S. M., Knochel C., Oertel-Knochel V., Stablein M., Howells F. M., Stein D. J., Temmingh H. S., Uhlmann A., Lopez-Jaramillo C., Dima D., McMahon A., Faskowitz J. I., Gutman B. A., Jahanshad N., Thompson P. M., Turner J. A., Cortical brain abnormalities in 4474 individuals with schizophrenia and 5098 control subjects via the Enhancing Neuro Imaging Genetics Through Meta Analysis (ENIGMA) Consortium. Biol. Psychiatry 84, 644–654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wannan C. M. J., Cropley V. L., Chakravarty M. M., Bousman C., Ganella E. P., Bruggemann J. M., Weickert T. W., Weickert C. S., Everall I., McGorry P., Velakoulis D., Wood S. J., Bartholomeusz C. F., Pantelis C., Zalesky A., Evidence for network-based cortical thickness reductions in schizophrenia. Am. J. Psychiatry 176, 552–563 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Jiang Y., Luo C., Li X., Duan M., He H., Chen X., Yang H., Gong J., Chang X., Woelfer M., Biswal B. B., Yao D., Progressive reduction in gray matter in patients with schizophrenia assessed with MR imaging by using causal network analysis. Radiology 287, 633–642 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Jiang Y., Wang Y., Huang H., He H., Tang Y., Su W., Xu L., Wei Y., Zhang T., Hu H., Wang J., Yao D., Wang J., Luo C., Antipsychotics effects on network-level reconfiguration of cortical morphometry in first-episode schizophrenia. Schizophr. Bull. 48, 231–240 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merritt K., Luque Laguna P., Irfan A., David A. S., Longitudinal structural MRI findings in individuals at genetic and clinical high risk for psychosis: A systematic review. Front. Psych. 12, 49 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palaniyappan L., Progressive cortical reorganisation: A framework for investigating structural changes in schizophrenia. Neurosci. Biobehav. Rev. 79, 1–13 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Chopra S., Segal A., Oldham S., Holmes A., Sabaroedin K., Orchard E. R., Francey S. M., O'Donoghue B., Cropley V., Nelson B., Graham J., Baldwin L., Tiego J., Yuen H. P., Allott K., Alvarez-Jimenez M., Harrigan S., Fulcher B. D., Aquino K., Pantelis C., Wood S. J., Bellgrove M., McGorry P. D., Fornito A., Network-based spreading of gray matter changes across different stages of psychosis. JAMA Psychiatry 80, 1246–1257 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraguas D., Diaz-Caneja C. M., Pina-Camacho L., Janssen J., Arango C., Progressive brain changes in children and adolescents with early-onset psychosis: A meta-analysis of longitudinal MRI studies. Schizophr. Res. 173, 132–139 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Brown J. A., Deng J., Neuhaus J., Sible I. J., Sias A. C., Lee S. E., Kornak J., Marx G. A., Karydas A. M., Spina S., Grinberg L. T., Coppola G., Geschwind D. H., Kramer J. H., Gorno-Tempini M. L., Miller B. L., Rosen H. J., Seeley W. W., Patient-tailored, connectivity-based forecasts of spreading brain atrophy. Neuron 104, 856–868.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hettwer M. D., Lariviere S., Park B. Y., van den Heuvel O. A., Schmaal L., Andreassen O. A., Ching C. R. K., Hoogman M., Buitelaar J., van Rooij D., Veltman D. J., Stein D. J., Franke B., van Erp T. G. M.; ENIGMA ADHD Working Group; ENIGMA Autism Working Group; ENIGMA Bipolar Disorder Working Group; ENIGMA Major Depression Working Group; ENIGMA OCD Working Group; ENIGMA Schizophrenia Working Group, Jahanshad N., Thompson P. M., Thomopoulos S. I., Bethlehem R. A. I., Bernhardt B. C., Eickhoff S. B., Valk S. L., Coordinated cortical thickness alterations across six neurodevelopmental and psychiatric disorders. Nat. Commun. 13, 6851 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shafiei G., Bazinet V., Dadar M., Manera A. L., Collins D. L., Dagher A., Borroni B., Sanchez-Valle R., Moreno F., Laforce R., Graff C., Synofzik M., Galimberti D., Rowe J. B., Masellis M., Tartaglia M. C., Finger E., Vandenberghe R., de Mendonca A., Tagliavini F., Santana I., Butler C., Gerhard A., Danek A., Levin J., Otto M., Sorbi S., Jiskoot L. C., Seelaar H., van Swieten J. C., Rohrer J. D., Misic B., Ducharme S., I. Frontotemporal Lobar Degeneration Neuroimaging, G. E. F. d., Network structure and transcriptomic vulnerability shape atrophy in frontotemporal dementia. Brain 146, 321–336 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith S., Duff E., Groves A., Nichols T. E., Jbabdi S., Westlye L. T., Tamnes C. K., Engvig A., Walhovd K. B., Fjell A. M., Johansen-Berg H., Douaud G., Structural variability in the human brain reflects fine-grained functional architecture at the population level. J. Neurosci. 39, 6136–6149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markello R. D., Hansen J. Y., Liu Z. Q., Bazinet V., Shafiei G., Suarez L. E., Blostein N., Seidlitz J., Baillet S., Satterthwaite T. D., Chakravarty M. M., Raznahan A., Misic B., Neuromaps: Structural and functional interpretation of brain maps. Nat. Methods 19, 1472–1479 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bethlehem R. A. I., Seidlitz J., White S. R., Vogel J. W., Anderson K. M., Adamson C., Adler S., Alexopoulos G. S., Anagnostou E., Areces-Gonzalez A., Astle D. E., Auyeung B., Ayub M., Bae J., Ball G., Baron-Cohen S., Beare R., Bedford S. A., Benegal V., Beyer F., Blangero J., Blesa Cabez M., Boardman J. P., Borzage M., Bosch-Bayard J. F., Bourke N., Calhoun V. D., Chakravarty M. M., Chen C., Chertavian C., Chetelat G., Chong Y. S., Cole J. H., Corvin A., Costantino M., Courchesne E., Crivello F., Cropley V. L., Crosbie J., Crossley N., Delarue M., Delorme R., Desrivieres S., Devenyi G. A., Di Biase M. A., Dolan R., Donald K. A., Donohoe G., Dunlop K., Edwards A. D., Elison J. T., Ellis C. T., Elman J. A., Eyler L., Fair D. A., Feczko E., Fletcher P. C., Fonagy P., Franz C. E., Galan-Garcia L., Gholipour A., Giedd J., Gilmore J. H., Glahn D. C., Goodyer I. M., Grant P. E., Groenewold N. A., Gunning F. M., Gur R. E., Gur R. C., Hammill C. F., Hansson O., Hedden T., Heinz A., Henson R. N., Heuer K., Hoare J., Holla B., Holmes A. J., Holt R., Huang H., Im K., Ipser J., C. R. Jack, Jr., Jackowski A. P., Jia T., Johnson K. A., Jones P. B., Jones D. T., Kahn R. S., Karlsson H., Karlsson L., Kawashima R., Kelley E. A., Kern S., Kim K. W., Kitzbichler M. G., Kremen W. S., Lalonde F., Landeau B., Lee S., Lerch J., Lewis J. D., Li J., Liao W., Liston C., Lombardo M. V., Lv J., Lynch C., Mallard T. T., Marcelis M., Markello R. D., Mathias S. R., Mazoyer B., McGuire P., Meaney M. J., Mechelli A., Medic N., Misic B., Morgan S. E., Mothersill D., Nigg J., Ong M. Q. W., Ortinau C., Ossenkoppele R., Ouyang M., Palaniyappan L., Paly L., Pan P. M., Pantelis C., Park M. M., Paus T., Pausova Z., Paz-Linares D., Pichet Binette A., Pierce K., Qian X., Qiu J., A. Qiu, A. Raznahan, Rittman T., Rodrigue A., Rollins C. K., Romero-Garcia R., Ronan L., Rosenberg M. D., Rowitch D. H., Salum G. A., Satterthwaite T. D., Schaare H. L., Schachar R. J., Schultz A. P., Schumann G., Scholl M., Sharp D., Shinohara R. T., Skoog I., Smyser C. D., Sperling R. A., Stein D. J., Stolicyn A., Suckling J., Sullivan G., Taki Y., Thyreau B., Toro R., Traut N., Tsvetanov K. A., Turk-Browne N. B., Tuulari J. J., Tzourio C., Vachon-Presseau E., Valdes-Sosa M. J., Valdes-Sosa P. A., Valk S. L., van Amelsvoort T., Vandekar S. N., Vasung L., Victoria L. W., Villeneuve S., Villringer A., Vertes P. E., Wagstyl K., Wang Y. S., Warfield S. K., Warrier V., Westman E., Westwater M. L., Whalley H. C., Witte A. V., Yang N., Yeo B., Yun H., Zalesky A., Zar H. J., Zettergren A., Zhou J. H., Ziauddeen H., Zugman A., Zuo X. N.; 3R-BRAIN; AIBL; Alzheimer’s Disease Neuroimaging Initiative; Alzheimer’s Disease Repository Without Borders Investigators; CALM Team; Cam-CAN; CCNP; COBRE; cVEDA; ENIGMA Developmental Brain Age Working Group; Developing Human Connectome Project; FinnBrain; Harvard Aging Brain Study; IMAGEN; KNE96; Mayo Clinic Study of Aging; NSPN; POND; PREVENT-AD Research Group; VETSA, Bullmore E. T., Alexander-Bloch A. F., Brain charts for the human lifespan. Nature 604, 525–533 (2022).35388223 [Google Scholar]
- 33.Ashburner J., Friston K. J., Voxel-based morphometry—The methods. Neuroimage 11, 805–821 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Scholtens L. H., de Reus M. A., de Lange S. C., Schmidt R., van den Heuvel M. P., An MRI Von Economo - Koskinas atlas. Neuroimage 170, 249–256 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Vaishnavi S. N., Vlassenko A. G., Rundle M. M., Snyder A. Z., Mintun M. A., Raichle M. E., Regional aerobic glycolysis in the human brain. Proc. Natl. Acad. Sci. U.S.A. 107, 17757–17762 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen J. Y., Shafiei G., Markello R. D., Smart K., Cox S. M. L., Norgaard M., Beliveau V., Wu Y., Gallezot J. D., Aumont E., Servaes S., Scala S. G., DuBois J. M., Wainstein G., Bezgin G., Funck T., Schmitz T. W., Spreng R. N., Galovic M., Koepp M. J., Duncan J. S., Coles J. P., Fryer T. D., Aigbirhio F. I., McGinnity C. J., Hammers A., Soucy J. P., Baillet S., Guimond S., Hietala J., Bedard M. A., Leyton M., Kobayashi E., Rosa-Neto P., Ganz M., Knudsen G. M., Palomero-Gallagher N., Shine J. M., Carson R. E., Tuominen L., Dagher A., Misic B., Mapping neurotransmitter systems to the structural and functional organization of the human neocortex. Nat. Neurosci. 25, 1569–1581 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hawrylycz M. J., Lein E. S., Guillozet-Bongaarts A. L., Shen E. H., Ng L., Miller J. A., van de Lagemaat L. N., Smith K. A., Ebbert A., Riley Z. L., Abajian C., Beckmann C. F., Bernard A., Bertagnolli D., Boe A. F., Cartagena P. M., Chakravarty M. M., Chapin M., Chong J., Dalley R. A., Daly B. D., Dang C., Datta S., Dee N., Dolbeare T. A., Faber V., Feng D., Fowler D. R., Goldy J., Gregor B. W., Haradon Z., Haynor D. R., Hohmann J. G., Horvath S., Howard R. E., Jeromin A., Jochim J. M., Kinnunen M., Lau C., Lazarz E. T., Lee C., Lemon T. A., Li L., Li Y., Morris J. A., Overly C. C., Parker P. D., Parry S. E., Reding M., Royall J. J., Schulkin J., Sequeira P. A., Slaughterbeck C. R., Smith S. C., Sodt A. J., Sunkin S. M., Swanson B. E., Vawter M. P., Williams D., Wohnoutka P., Zielke H. R., Geschwind D. H., Hof P. R., Smith S. M., Koch C., Grant S. G. N., Jones A. R., An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yarkoni T., Poldrack R. A., Nichols T. E., Van Essen D. C., Wager T. D., Large-scale automated synthesis of human functional neuroimaging data. Nat. Methods 8, 665–670 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sunkin S. M., Ng L., Lau C., Dolbeare T., Gilbert T. L., Thompson C. L., Hawrylycz M., Dang C., Allen Brain Atlas: An integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 41, D996–D1008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdi H., Partial least squares regression and projection on latent structure regression (PLS Regression). Wiley Interdiscip. Rev. Comput. Stat. 2, 97–106 (2010). [Google Scholar]
- 41.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A. H., Tanaseichuk O., Benner C., Chanda S. K., Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T., Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCutcheon R. A., Krystal J. H., Howes O. D., Dopamine and glutamate in schizophrenia: Biology, symptoms and treatment. World Psychiatry 19, 15–33 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howes O. D., Bukala B. R., Beck K., Schizophrenia: From neurochemistry to circuits, symptoms and treatments. Nat. Rev. Neurol. 20, 22–35 (2024). [DOI] [PubMed] [Google Scholar]
- 45.Shafiei G., Markello R. D., Makowski C., Talpalaru A., Kirschner M., Devenyi G. A., Guma E., Hagmann P., Cashman N. R., Lepage M., Chakravarty M. M., Dagher A., Misic B., Spatial patterning of tissue volume loss in schizophrenia reflects brain network architecture. Biol. Psychiatry 87, 727–735 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Mutlu J., Landeau B., Gaubert M., de La Sayette V., Desgranges B., Chetelat G., Distinct influence of specific versus global connectivity on the different Alzheimer's disease biomarkers. Brain 140, 3317–3328 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Fillman S. G., Weickert T. W., Lenroot R. K., Catts S. V., Bruggemann J. M., Catts V. S., Weickert C. S., Elevated peripheral cytokines characterize a subgroup of people with schizophrenia displaying poor verbal fluency and reduced Broca's area volume. Mol. Psychiatry 21, 1090–1098 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crow T. J., Is schizophrenia the price that Homo sapiens pays for language? Schizophr. Res. 28, 127–141 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Palaniyappan L., Liddle P. F., Does the salience network play a cardinal role in psychosis? An emerging hypothesis of insular dysfunction. J. Psychiatry Neurosci. 37, 17–27 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Menon V., Palaniyappan L., Supekar K., Integrative brain network and salience models of psychopathology and cognitive dysfunction in schizophrenia. Biol. Psychiatry 94, 108–120 (2023). [DOI] [PubMed] [Google Scholar]
- 51.Del Re E. C., Stone W. S., Bouix S., Seitz J., Zeng V., Guliano A., Somes N., Zhang T., Reid B., Lyall A., Lyons M., Li H., Whitfield-Gabrieli S., Keshavan M., Seidman L. J., McCarley R. W., Wang J., Tang Y., Shenton M. E., Niznikiewicz M. A., Baseline cortical thickness reductions in clinical high risk for psychosis: Brain regions associated with conversion to psychosis versus non-conversion as assessed at one-year follow-up in the Shanghai-At-Risk-for-Psychosis (SHARP) study. Schizophr. Bull. 47, 562–574 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pantelis C., Velakoulis D., McGorry P. D., Wood S. J., Suckling J., Phillips L. J., Yung A. R., Bullmore E. T., Brewer W., Soulsby B., Desmond P., McGuire P. K., Neuroanatomical abnormalities before and after onset of psychosis: A cross-sectional and longitudinal MRI comparison. Lancet 361, 281–288 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Barch D. M., What does it mean to be transdiagnostic and how would we know? Am. J. Psychiatry 177, 370–372 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Falconer M. A., Serafetinides E. A., Corsellis J. A., Etiology and pathogenesis of temporal lobe epilepsy. Arch. Neurol. 10, 233–248 (1964). [DOI] [PubMed] [Google Scholar]
- 55.Ball M. J., Fisman M., Hachinski V., Blume W., Fox A., Kral V. A., Kirshen A. J., Fox H., Merskey H., A new definition of Alzheimer's disease: A hippocampal dementia. Lancet 1, 14–16 (1985). [DOI] [PubMed] [Google Scholar]
- 56.Geddes J. W., Monaghan D. T., Cotman C. W., Lott I. T., Kim R. C., Chui H. C., Plasticity of hippocampal circuitry in Alzheimer's disease. Science 230, 1179–1181 (1985). [DOI] [PubMed] [Google Scholar]
- 57.Palaniyappan L., Homan P., Alonso-Sanchez M. F., Language network dysfunction and formal thought disorder in schizophrenia. Schizophr. Bull. 49, 486–497 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang X., Zhao W., Kang J., Xiang S., Xie C., Corona-Hernández H., Palaniyappan L., Feng J., Language abnormalities in schizophrenia: Binding core symptoms through contemporary empirical evidence. Schizophrenia 8, 1–12 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forrest M. P., Hill M. J., Kavanagh D. H., Tansey K. E., Waite A. J., Blake D. J., The psychiatric risk gene transcription dfactor 4 (TCF4) regulates neurodevelopmental pathways associated with schizophrenia, autism, and intellectual disability. Schizophr. Bull. 44, 1100–1110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rapoport J. L., Giedd J. N., Gogtay N., Neurodevelopmental model of schizophrenia: Update 2012. Mol. Psychiatry 17, 1228–1238 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tekin S., Cummings J. L., Frontal-subcortical neuronal circuits and clinical neuropsychiatry: An update. J. Psychosom. Res. 53, 647–654 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Maier-Hein K. H., Neher P. F., Houde J. C., Cote M. A., Garyfallidis E., Zhong J., Chamberland M., Yeh F. C., Lin Y. C., Ji Q., Reddick W. E., Glass J. O., Chen D. Q., Feng Y., Gao C., Wu Y., Ma J., He R., Li Q., Westin C. F., Deslauriers-Gauthier S., Gonzalez J. O. O., Paquette M., St-Jean S., Girard G., Rheault F., Sidhu J., Tax C. M. W., Guo F., Mesri H. Y., David S., Froeling M., Heemskerk A. M., Leemans A., Bore A., Pinsard B., Bedetti C., Desrosiers M., Brambati S., Doyon J., Sarica A., Vasta R., Cerasa A., Quattrone A., Yeatman J., Khan A. R., Hodges W., Alexander S., Romascano D., Barakovic M., Auria A., Esteban O., Lemkaddem A., Thiran J. P., Cetingul H. E., Odry B. L., Mailhe B., Nadar M. S., Pizzagalli F., Prasad G., Villalon-Reina J. E., Galvis J., Thompson P. M., Requejo F. S., Laguna P. L., Lacerda L. M., Barrett R., Dell'Acqua F., Catani M., Petit L., Caruyer E., Daducci A., Dyrby T. B., Holland-Letz T., Hilgetag C. C., Stieltjes B., Descoteaux M., The challenge of mapping the human connectome based on diffusion tractography. Nat. Commun. 8, 1349 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gratton C., Laumann T. O., Nielsen A. N., Greene D. J., Gordon E. M., Gilmore A. W., Nelson S. M., Coalson R. S., Snyder A. Z., Schlaggar B. L., Dosenbach N. U. F., Petersen S. E., Functional brain networks are dominated by stable group and individual factors, not cognitive or daily variation. Neuron 98, 439–452.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanaka S. C., Yamashita A., Yahata N., Itahashi T., Lisi G., Yamada T., Ichikawa N., Takamura M., Yoshihara Y., Kunimatsu A., Okada N., Hashimoto R., Okada G., Sakai Y., Morimoto J., Narumoto J., Shimada Y., Mano H., Yoshida W., Seymour B., Shimizu T., Hosomi K., Saitoh Y., Kasai K., Kato N., Takahashi H., Okamoto Y., Yamashita O., Kawato M., Imamizu H., A multi-site, multi-disorder resting-state magnetic resonance image database. Sci. Data 8, 227 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan C. G., Chen X., Li L., Castellanos F. X., Bai T. J., Bo Q. J., Cao J., Chen G. M., Chen N. X., Chen W., Cheng C., Cheng Y. Q., Cui X. L., Duan J., Fang Y. R., Gong Q. Y., Guo W. B., Hou Z. H., Hu L., Kuang L., Li F., Li K. M., Li T., Liu Y. S., Liu Z. N., Long Y. C., Luo Q. H., Meng H. Q., Peng D. H., Qiu H. T., Qiu J., Shen Y. D., Shi Y. S., Wang C. Y., Wang F., Wang K., Wang L., Wang X., Wang Y., Wu X. P., Wu X. R., Xie C. M., Xie G. R., Xie H. Y., Xie P., Xu X. F., Yang H., Yang J., Yao J. S., Yao S. Q., Yin Y. Y., Yuan Y. G., Zhang A. X., Zhang H., Zhang K. R., Zhang L., Zhang Z. J., Zhou R. B., Zhou Y. T., Zhu J. J., Zou C. J., Si T. M., Zuo X. N., Zhao J. P., Zang Y. F., Reduced default mode network functional connectivity in patients with recurrent major depressive disorder. Proc. Natl. Acad. Sci. U.S.A. 116, 9078–9083 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bellec P., Chu C., Chouinard-Decorte F., Benhajali Y., Margulies D. S., Craddock R. C., The Neuro Bureau ADHD-200 Preprocessed repository. Neuroimage 144, 275–286 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Parkinson Progression Marker Initiative , The Parkinson Progression Marker Initiative (PPMI). Prog. Neurobiol. 95, 629–635 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li X., Jiang Y., Li W., Qin Y., Li Z., Chen Y., Tong X., Xiao F., Zuo X., Gong Q., Zhou D., Yao D., An D., Luo C., Disrupted functional connectivity in white matter resting-state networks in unilateral temporal lobe epilepsy. Brain Imaging Behav. 16, 324–335 (2022). [DOI] [PubMed] [Google Scholar]
- 69.Han S., Xu Y., Guo H. R., Fang K., Wei Y., Liu L., Cheng J., Zhang Y., Cheng J., Resolving heterogeneity in obsessive-compulsive disorder through individualized differential structural covariance network analysis. Cereb. Cortex 33, 1659–1668 (2023). [DOI] [PubMed] [Google Scholar]
- 70.Huang C. C., Luo Q., Palaniyappan L., Yang A. C., Hung C. C., Chou K. H., Zac Lo C. Y., Liu M. N., Tsai S. J., Barch D. M., Feng J., Lin C. P., Robbins T. W., Transdiagnostic and illness-specific functional dysconnectivity across schizophrenia, bipolar disorder, and major depressive disorder. Biol. Psychiatry Cogn. Neurosci. Neuroimag. 5, 542–553 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Obermeier M., Mayr A., Schennach-Wolff R., Seemuller F., Moller H. J., Riedel M., Should the PANSS be rescaled? Schizophr. Bull. 36, 455–460 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu Z., Palaniyappan L., Wu X., Zhang K., Du J., Zhao Q., Xie C., Tang Y., Su W., Wei Y., Xue K., Han S., Tsai S. J., Lin C. P., Cheng J., Li C., Wang J., Sahakian B. J., Robbins T. W., Zhang J., Feng J., Resolving heterogeneity in schizophrenia through a novel systems approach to brain structure: Individualized structural covariance network analysis. Mol. Psychiatry 26, 7719–7731 (2021). [DOI] [PubMed] [Google Scholar]
- 73.Qin Y., Kang J., Jiao Z., Wang Y., Wang J., Wang H., Feng J., Jin L., Wang F., Gong X., Polygenic risk for autism spectrum disorder affects left amygdala activity and negative emotion in schizophrenia. Transl. Psychiatry 10, 322 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rolls E. T., Cheng W., Gilson M., Gong W., Deco G., Lo C. Z., Yang A. C., Tsai S. J., Liu M. E., Lin C. P., Feng J., Beyond the disconnectivity hypothesis of schizophrenia. Cereb. Cortex 30, 1213–1233 (2020). [DOI] [PubMed] [Google Scholar]
- 75.Cetin M. S., Christensen F., Abbott C. C., Stephen J. M., Mayer A. R., Canive J. M., Bustillo J. R., Pearlson G. D., Calhoun V. D., Thalamus and posterior temporal lobe show greater inter-network connectivity at rest and across sensory paradigms in schizophrenia. Neuroimage 97, 117–126 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kogan A., Alpert K., Ambite J. L., Marcus D. S., Wang L., Northwestern University schizophrenia data sharing for SchizConnect: A longitudinal dataset for large-scale integration. Neuroimage 124, 1196–1201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang L., Alpert K. I., Calhoun V. D., Cobia D. J., Keator D. B., King M. D., Kogan A., Landis D., Tallis M., Turner M. D., Potkin S. G., Turner J. A., Ambite J. L., SchizConnect: Mediating neuroimaging databases on schizophrenia and related disorders for large-scale integration. Neuroimage 124, 1155–1167 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schaefer A., Kong R., Gordon E. M., Laumann T. O., Zuo X. N., Holmes A. J., Eickhoff S. B., Yeo B. T. T., Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb. Cortex 28, 3095–3114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tian Y., Margulies D. S., Breakspear M., Zalesky A., Topographic organization of the human subcortex unveiled with functional connectivity gradients. Nat. Neurosci. 23, 1421–1432 (2020). [DOI] [PubMed] [Google Scholar]
- 80.Rolls E. T., Joliot M., Tzourio-Mazoyer N., Implementation of a new parcellation of the orbitofrontal cortex in the automated anatomical labeling atlas. Neuroimage 122, 1–5 (2015). [DOI] [PubMed] [Google Scholar]
- 81.Fan L., Li H., Zhuo J., Zhang Y., Wang J., Chen L., Yang Z., Chu C., Xie S., Laird A. R., Fox P. T., Eickhoff S. B., Yu C., Jiang T., The human Brainnetome Atlas: A new brain atlas based on connectional architecture. Cereb. Cortex 26, 3508–3526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Glasser M. F., Coalson T. S., Robinson E. C., Hacker C. D., Harwell J., Yacoub E., Ugurbil K., Andersson J., Beckmann C. F., Jenkinson M., Smith S. M., Van Essen D. C., A multi-modal parcellation of human cerebral cortex. Nature 536, 171–178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Desikan R. S., Segonne F., Fischl B., Quinn B. T., Dickerson B. C., Blacker D., Buckner R. L., Dale A. M., Maguire R. P., Hyman B. T., Albert M. S., Killiany R. J., An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31, 968–980 (2006). [DOI] [PubMed] [Google Scholar]
- 84.Alexander-Bloch A. F., Shou H., Liu S., Satterthwaite T. D., Glahn D. C., Shinohara R. T., Vandekar S. N., Raznahan A., On testing for spatial correspondence between maps of human brain structure and function. Neuroimage 178, 540–551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vasa F., Seidlitz J., Romero-Garcia R., Whitaker K. J., Rosenthal G., Vertes P. E., Shinn M., Alexander-Bloch A., Fonagy P., Dolan R. J., Jones P. B., Goodyer I. M.; NSPN consortium, Sporns O., Bullmore E. T., Adolescent tuning of association cortex in human structural brain networks. Cereb. Cortex 28, 281–294 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun D., Ching C. R. K., Lin A., Forsyth J. K., Kushan L., Vajdi A., Jalbrzikowski M., Hansen L., Villalon-Reina J. E., Qu X., Jonas R. K., van Amelsvoort T., Bakker G., Kates W. R., Antshel K. M., Fremont W., Campbell L. E., McCabe K. L., Daly E., Gudbrandsen M., Murphy C. M., Murphy D., Craig M., Vorstman J., Fiksinski A., Koops S., Ruparel K., Roalf D. R., Gur R. E., Schmitt J. E., Simon T. J., Goodrich-Hunsaker N. J., Durdle C. A., Bassett A. S., Chow E. W. C., Butcher N. J., Vila-Rodriguez F., Doherty J., Cunningham A., van den Bree M. B. M., Linden D. E. J., Moss H., Owen M. J., Murphy K. C., McDonald-McGinn D. M., Emanuel B., van Erp T. G. M., Turner J. A., Thompson P. M., Bearden C. E., Large-scale mapping of cortical alterations in 22q11.2 deletion syndrome: Convergence with idiopathic psychosis and effects of deletion size. Mol. Psychiatry 25, 1822–1834 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van Rooij D., Anagnostou E., Arango C., Auzias G., Behrmann M., Busatto G. F., Calderoni S., Daly E., Deruelle C., Di Martino A., Dinstein I., Duran F. L. S., Durston S., Ecker C., Fair D., Fedor J., Fitzgerald J., Freitag C. M., Gallagher L., Gori I., Haar S., Hoekstra L., Jahanshad N., Jalbrzikowski M., Janssen J., Lerch J., Luna B., Martinho M. M., McGrath J., Muratori F., Murphy C. M., Murphy D. G. M., O'Hearn K., Oranje B., Parellada M., Retico A., Rosa P., Rubia K., Shook D., Taylor M., Thompson P. M., Tosetti M., Wallace G. L., Zhou F., Buitelaar J. K., Cortical and subcortical brain morphometry differences between patients with autism spectrum disorder and healthy individuals across the lifespan: Results from the ENIGMA ASD Working Group. Am. J. Psychiatry 175, 359–369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Whelan C. D., Altmann A., Botia J. A., Jahanshad N., Hibar D. P., Absil J., Alhusaini S., Alvim M. K. M., Auvinen P., Bartolini E., Bergo F. P. G., Bernardes T., Blackmon K., Braga B., Caligiuri M. E., Calvo A., Carr S. J., Chen J., Chen S., Cherubini A., David P., Domin M., Foley S., Franca W., Haaker G., Isaev D., Keller S. S., Kotikalapudi R., Kowalczyk M. A., Kuzniecky R., Langner S., Lenge M., Leyden K. M., Liu M., Loi R. Q., Martin P., Mascalchi M., Morita M. E., Pariente J. C., Rodriguez-Cruces R., Rummel C., Saavalainen T., Semmelroch M. K., Severino M., Thomas R. H., Tondelli M., Tortora D., Vaudano A. E., Vivash L., von Podewils F., Wagner J., Weber B., Yao Y., Yasuda C. L., Zhang G., Bargallo N., Bender B., Bernasconi N., Bernasconi A., Bernhardt B. C., Blumcke I., Carlson C., Cavalleri G. L., Cendes F., Concha L., Delanty N., Depondt C., Devinsky O., Doherty C. P., Focke N. K., Gambardella A., Guerrini R., Hamandi K., Jackson G. D., Kalviainen R., Kochunov P., Kwan P., Labate A., McDonald C. R., Meletti S., O'Brien T. J., Ourselin S., Richardson M. P., Striano P., Thesen T., Wiest R., Zhang J., Vezzani A., Ryten M., Thompson P. M., Sisodiya S. M., Structural brain abnormalities in the common epilepsies assessed in a worldwide ENIGMA study. Brain 141, 391–408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boedhoe P. S. W., Schmaal L., Abe Y., Alonso P., Ameis S. H., Anticevic A., Arnold P. D., Batistuzzo M. C., Benedetti F., Beucke J. C., Bollettini I., Bose A., Brem S., Calvo A., Calvo R., Cheng Y., Cho K. I. K., Ciullo V., Dallaspezia S., Denys D., Feusner J. D., Fitzgerald K. D., Fouche J. P., Fridgeirsson E. A., Gruner P., Hanna G. L., Hibar D. P., Hoexter M. Q., Hu H., Huyser C., Jahanshad N., James A., Kathmann N., Kaufmann C., Koch K., Kwon J. S., Lazaro L., Lochner C., Marsh R., Martinez-Zalacain I., Mataix-Cols D., Menchon J. M., Minuzzi L., Morer A., Nakamae T., Nakao T., Narayanaswamy J. C., Nishida S., Nurmi E., O'Neill J., Piacentini J., Piras F., Piras F., Reddy Y. C. J., Reess T. J., Sakai Y., Sato J. R., Simpson H. B., Soreni N., Soriano-Mas C., Spalletta G., Stevens M. C., Szeszko P. R., Tolin D. F., van Wingen G. A., Venkatasubramanian G., Walitza S., Wang Z., Yun J. Y.; ENIGMA-OCD Working Group, Thompson P. M., Stein D. J., van den Heuvel O. A.; ENIGMA OCD Working Group , Cortical abnormalities associated with pediatric and adult obsessive-compulsive disorder: Findings from the ENIGMA Obsessive-Compulsive Disorder Working Group. Am. J. Psychiatry 175, 453–462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lariviere S., Paquola C., Park B. Y., Royer J., Wang Y., Benkarim O., Vos de Wael R., Valk S. L., Thomopoulos S. I., Kirschner M., Lewis L. B., Evans A. C., Sisodiya S. M., McDonald C. R., Thompson P. M., Bernhardt B. C., The ENIGMA Toolbox: Multiscale neural contextualization of multisite neuroimaging datasets. Nat. Methods 18, 698–700 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Arnatkeviciute A., Fulcher B. D., Fornito A., A practical guide to linking brain-wide gene expression and neuroimaging data. Neuroimage 189, 353–367 (2019). [DOI] [PubMed] [Google Scholar]
- 92.Morgan S. E., Seidlitz J., Whitaker K. J., Romero-Garcia R., Clifton N. E., Scarpazza C., van Amelsvoort T., Marcelis M., van Os J., Donohoe G., Mothersill D., Corvin A., Pocklington A., Raznahan A., McGuire P., Vertes P. E., Bullmore E. T., Cortical patterning of abnormal morphometric similarity in psychosis is associated with brain expression of schizophrenia-related genes. Proc. Natl. Acad. Sci. U.S.A. 116, 9604–9609 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hochberg Y., Benjamini Y., More powerful procedures for multiple significance testing. Stat. Med. 9, 811–818 (1990). [DOI] [PubMed] [Google Scholar]
- 94.Poldrack R. A., Kittur A., Kalar D., Miller E., Seppa C., Gil Y., Parker D. S., Sabb F. W., Bilder R. M., The cognitive atlas: Toward a knowledge foundation for cognitive neuroscience. Front. Neuroinform. 5, 17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Du J., Palaniyappan L., Liu Z., Cheng W., Gong W., Zhu M., Wang J., Zhang J., Feng J., The genetic determinants of language network dysconnectivity in drug-naïve early stage schizophrenia. NPJ Schizophr. 7, 18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Repovs G., Barch D. M., Working memory related brain network connectivity in individuals with schizophrenia and their siblings. Front. Hum. Neurosci. 6, 137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vasa F., Misic B., Null models in network neuroscience. Nat. Rev. Neurosci. 23, 493–504 (2022). [DOI] [PubMed] [Google Scholar]
- 98.Markello R. D., Misic B., Comparing spatial null models for brain maps. Neuroimage 236, 118052 (2021). [DOI] [PubMed] [Google Scholar]
- 99.J. Y. Hansen, G. Shafiei, R. D. Markello, K. Smart, S. M. Cox, Y. Wu, J.-D. Gallezot, É. Aumont, S. Servaes, S. G. Scala, Mapping neurotransmitter systems to the structural and functional organization of the human neocortex. bioRxiv 10.1101/2021.10.28.466336 [Preprint] (30 October 2021). 10.1101/2021.10.28.466336. [DOI] [PMC free article] [PubMed]
- 100.Yeo B. T., Krienen F. M., Sepulcre J., Sabuncu M. R., Lashkari D., Hollinshead M., Roffman J. L., Smoller J. W., Zollei L., Polimeni J. R., Fischl B., Liu H., Buckner R. L., The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 106, 1125–1165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ZIB (Zhang-jiang International Brain Bank) Consortium Authorship List
Methods S1 to S7
Extended Figs. 1 to 7
Figs. S1 to S7
Tables S1 to S9
Legend for data S1
References
Data S1