Abstract
The polyprotein of infectious pancreatic necrosis virus (IPNV), a birnavirus, is processed by the viral protease VP4 (also named NS) to generate three polypeptides: pVP2, VP4, and VP3. Site-directed mutagenesis at 42 positions of the IPNV VP4 protein was performed to determine the active site and the important residues for the protease activity. Two residues (serine 633 and lysine 674) were critical for cleavage activity at both the pVP2-VP4 and the VP4-VP3 junctions. Wild-type activity at the pVP2-VP4 junction and a partial block (with an alteration of the cleavage specificity) at the VP4-VP3 junction were observed when replacement occurred at histidines 547 and 679. A similar observation was made when aspartic acid 693 was replaced by leucine, but wild-type activity and specificity were found when substituted by glutamine or asparagine. Sequence comparison between IPNV and two birnavirus (infectious bursal disease virus and Drosophila X virus) VP4s revealed that serine 633 and lysine 674 are conserved in these viruses, in contrast to histidines 547 and 679. The importance of serine 633 and lysine 674 is reminiscent of the protease active site of bacterial leader peptidases and their mitochondrial homologs and of the bacterial LexA-like proteases. Self-cleavage sites of IPNV VP4 were determined at the pVP2-VP4 and VP4-VP3 junctions by N-terminal sequencing and mutagenesis. Two alternative cleavage sites were also identified in the carboxyl domain of pVP2 by cumulative mutagenesis. The results suggest that VP4 cleaves the (Ser/Thr)-X-Ala↓(Ser/Ala)-Gly motif, a target sequence with similarities to bacterial leader peptidases and herpesvirus protease cleavage sites.
The proteolytic processing of viral precursor proteins is a crucial step in the life cycle of a majority of viruses that infect eucaryotic cells, and virus-encoded proteases are generally associated with these events. Each viral family may use different strategies for the generation of the cleavage products, which does occur in time and in a specific cellular environment for virus assembly (4). Most of the virus-encoded proteases are related to the chymotrypsin-like cellular proteases with a distinctive double β-barrel fold, but only a few, like the Alphavirus capsid protease and the Flaviviridae NS3 proteases, contain the His-Asp-Ser catalytic triad found in the cellular world (14, 31). A large number of viral proteases structurally related (or not) to chymotrypsin-like proteases replace the serine to cysteine as the catalytic nucleophile, as for picornavirus 3C protease or adenovirus protease (1, 9). In addition to this substitution, a number of viral enzymes (also found in adenovirus and some picornaviruses) utilize glutamic acid, instead of the catalytic aspartic acid (9, 22). The human cytomegalovirus (HCMV) protease consists of a single β-barrel structure and presents a catalytic triad His-His-Ser (26). Retroviruses, such as human immunodeficiency virus, have an active aspartic protease which is only generated when two identical amino acid domains, each of them bearing a catalytic aspartic acid, join to create a homodimer (35).
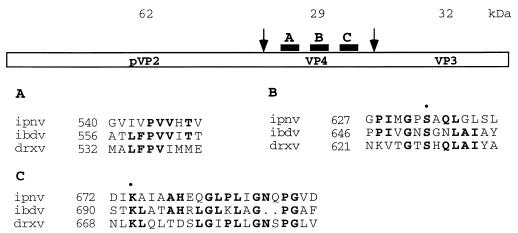
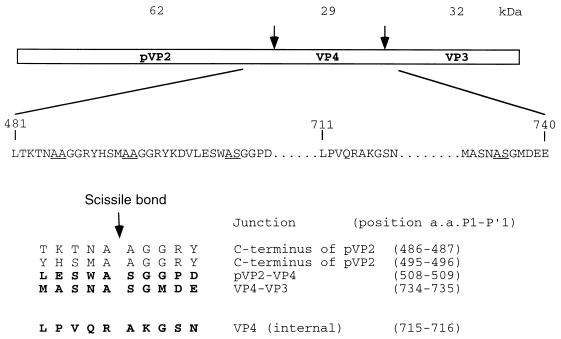
Infectious pancreatic necrosis virus (IPNV) is a pathogen that causes an acute, contagious disease of young salmonid fishes (10). IPNV is a member of the birnaviruses, a family of double-stranded RNA viruses with two genomic segments of 2.8 (segment B encoding the RNA polymerase) and 3.1 (segment A) kilobases (11). Translation of the segment A yields a polyprotein and a small protein, VP5. The polyprotein, whose protein order is NH2-pVP2-VP4-VP3-COOH, is cotranslationally processed and cleaved at the pVP2-VP4 and the VP4-VP3 junctions by the VP4 (also named NS)-associated protease activity (20, 21). The pVP2-to-VP2 conversion involves the cleavage (or cleavages) of pVP2 near the carboxy end as has been shown for infectious bursal disease virus (IBDV) (3), a birnavirus which causes a highly contagious disease of chickens (6). This processing is slow and has been proposed for IPNV to be most likely due to host cell proteases rather than the proteolytic action of the VP4 (11). The peptide bonds cleaved in birnavirus polyprotein are ill defined, but dibasic residue motifs have been proposed to be cleaved in IPNV and IBDV (11). Low sequence homology (<20%) is observed between birnavirus VP4 proteases (Fig. 1).
FIG. 1.
Patterns of conserved amino acids in birnavirus VP4s. Arrows indicate approximate positions of the cleavage sites. Three regions, A to C, containing the most conserved patterns of amino acids, are shown. The comparison includes the deduced amino acid sequences of VP4 of IPNV strain SP (accession number U56907), IBDV (accession number P15480), and DSX (accession number U60650). Identical residues are in boldface when present at least in two sequences. Numbers refer to the position of the first amino acid of each domain in the respective VP4s. Residues proposed to be part of the catalytic site of VP4s (•) are indicated.
In this study, we constructed IPNV VP4 mutants by site-directed mutagenesis and analyzed the processing of the mutated polyprotein in vitro. We identified two VP4 residues (serine 633 and lysine 674), which have properties for active-site residues. The behavior of the polyprotein altered at these positions suggests that VP4 may use a serine-lysine mechanism similar to those proposed for signal peptidases from Escherichia coli and Bacillus subtilis, bacterial LexA-like and Tsp proteases, the class A β-lactamases and, in eucaryotes, the two subunits of the mitochondrial inner-membrane protease (characterized in the yeast Saccharomyces cerevisiae) (23, 24, 30, 32, 33). The exact cleavage sites at the pVP2-VP4 and at the VP4-VP3 junctions were determined and are characterized by the Ser/Thr-X-Ala-Ser/Ala sequence, a motif which shares some similarities with bacterial signal peptidase and herpes simplex virus cleavage sites.
MATERIALS AND METHODS
Construction of IPNA expression vector SK-ΔIPNA.
The pBluescript II SK(−) phagemid (Stratagene) was cut with SspI and HincII and self-ligated to delete the T7 promoter (plasmid SKΔ). The complete IPNA segment of the IPN Sp strain (French isolate 31-75) (13) was cloned by reverse transcription (RT)-PCR by using standard techniques and procedures (27) by using the oligonucleotides 5′-CGTCGGATCCTAATACGACTCACTATAGGAAAGAGAGTTTCAACGTTAGTGGT and 5′-TGCAGCATCGATGGGGCCCCCTGGGGGGCCGGGGTT and cloned into SKΔ by using restriction sites BamHI and ClaI to generate the plasmid SKΔIPNA. Thus, the 5′ terminus of the IPNA segment (GGAAAGAGA…) was downstream from the T7 promoter. The entire segment was sequenced and compared to the published segment A sequences. The sequence encoding the polyprotein was found to be 98% identical to the published IPNA Sp strain (U56907), resulting in nine amino acid differences (V52I, V152A, K192R, R212S, N234S, Q249R, D252N, K255R, and M883T).
Site-directed mutagenesis of SK-ΔIPNA.
The mutations were introduced by using the Pfu DNA polymerase with the QuikChange Site-Directed Mutagenesis Kit (Stratagene) as described by the manufacturer. A novel restriction site was introduced for some mutations (Table 1) and sequence analyses (5′-GGGGTCCTCTTTTGACCACTCGTA) were carried out to confirm amino acid changes and the integrity of reading frames by using the following primers: 5′-GAAAATTCTCCCGAGCCCTCAAG, 5′-CCACAGGGACGATGACTCCTTTTG, 5′-GAACGACATCGAGGACGGAGTTCC, 5′-TACCATCCTTGGAACTCCGTCCTC, and 5′-GGGGTCCTCTTTTGACCACTCGTA.
TABLE 1.
Amino acid substitutions introduced into the IPNV VP2-VP4-VP3 polyprotein
| Construct | Wild-type sequencea | Mutated sequenceb | Novel site |
|---|---|---|---|
| ipnA AA486–7QL | AACGCAGCAGGC | AACCAGCTGGGC | PvuII |
| ipnA AA(486–7,495–6)QL,AS508–9QL | AACGCAGCAGGC, | AACCAGCTGGGC | PvuII |
| ATGGCCGCAGGA, | ATGCAGCTGGGA | PvuII | |
| TGGGCAAGCGGA | TGGCAGCTGGGA | PvuII | |
| ipnA AA495–6QL | ATGGCCGCAGGA | ATGCAGCTGGGA | PvuII |
| ipnA AA495–6QL,AS508–9QL | ATGGCCGCAGGA, | ATGCAGCTGGGA | PvuII |
| TGGGCAAGCGGA | TGGCAGCTGGGA | PvuII | |
| ipnA AS508–9QL | TGGGCAAGCGGA | TGGCAGCTGGGA | PvuII |
| ipnA A508G | TGGGCAAGC | TGGGGATCC | BamHI |
| ipnA S509T | GCAAGCGGA | GCAACCGGT | AgeI |
| ipnA G510T | AGCGGAGGG | AGTACTGGG | ScaI |
| ipnA E525A | CTGGAGTCC | gCTAGCGTCC | NheI |
| ipnA EE530–1AA | TACGAGGAAGTC | TACGCTGCAGTC | PstI |
| ipnA E533A | GTCGAGCTT | GTCGCGCTT | |
| ipnA V543A | ATCGTCCCT | ATCGCACCGgt | AgeI |
| ipnA P544A | GTCCCTGTG | GTCGCTGTG | |
| ipnA V545A | CCTGTGGTG | CCTGCTGTAca | BsrGI |
| ipnA H547S | GTGCACACA | GTGAGTACT | ScaI |
| ipnA H547S,H679L | GTGCACACA, GCCCACGAA | GTGAGTACT, GCCCTCGAG | ScaI, XhoI |
| ipnA E570G | CCCGAGCTT | CCCGGGCTT | XmaI |
| ipnA D573Q | CTAGATGCC | CTGCAGGCC | PstI |
| ipnA D585I | AACGACACC | AATATTACC | SspI |
| ipnA E594S | GGAGAGGAC | GGATCCGAC | BamHI |
| ipnA D595L | GAGGACATA | GAGCTCATA | SacI |
| ipnA D601S | GGAGACAAC | GGATCCAAC | BamHI |
| ipnA G631R | ATGGGTCCC | ATGAGGCCT | StuI |
| ipnA S633G | CCCTCTGCT | CCCGGGGCT | XmaI |
| ipnA S633A | CCCTCTGCT | CCCGCTGCT | |
| ipnA S633V | CCCTCTGCT | CCCGTTGCT | |
| ipnA S633C | CCCTCTGCT | CCCTGCGCA | FspI |
| ipnA S633T | CCCTCTGCT | CCCACTGCT | |
| ipnA L636A | CAACTAGGA | CAAGCAGGA | |
| ipnA L638A | GGACTGTCC | GGAGCGTCC | |
| ipnA S639Q | CTGTCCCTG | CTGCAGCTG | PstI |
| ipnA D644I | AACGACATC | AATATTATC | SspI |
| ipnA DD660–1GS | GCCGACGACGAG | GCCGGATCCGAG | BamHI |
| ipnA D672N | GTAGACATC | GTAAATATT | SspI |
| ipnA I673A | GACATCAAA | GACGCCAAA | |
| ipnA K674A | ATCAAAGCC | ATCGCAGCC | |
| ipnA K674D | ATCAAAGCC | ATCGATGCC | Bsp106i |
| ipnA K674R | ATCAAAGCC | ATCCGAGCC | |
| ipnA K674Q | ATCAAAGCC | ATCCAAGCC | |
| ipnA K674H | ATCAAAGCC | ATCCACGCC | |
| ipnA A675D | AAAGCCATC | AAAGATATC | EcoRV |
| ipnA I676A | GCCATCGCA | GCCGCCGCA | |
| ipnA I676S | GCCATCGCA | GCCAGCGCT | Eco47III |
| ipnA A677D | ATCGCAGCC | ATCGATGCC | Bsp106i |
| ipnA A678S | GCAGCCCAC | GCTAGCCAC | NheI |
| ipnA H679L | GCCCACGAA | GCCCTCGAG | XhoI |
| ipnA E680M | CACGAACAA | CATATGCAA | NdeI |
| ipnA G682L | CAAGGGCTG | CAATTGCTG | MunI |
| ipnA L683A | GGGCTGCCA | GGTGCTCCA | |
| ipnA L683S | GGGCTGCCA | GGATCCCCA | BamHI |
| ipnA L683I | GGGCTGCCA | GGGATCCCA | |
| ipnA P684Q | CTGCCACTC | CTGCAGCTC | PstI |
| ipnA L685A | CCACTCATC | CCAGCGATCg | MfeI |
| ipnA L685R | CCACTCATC | CCTAGGATC | AvrII |
| ipnA L685I | CCACTCATC | CCAATCATC | |
| ipnA I686A | CTCATCGGC | gCTAGCCGGC | NheI |
| ipnA I686Q | CTCATCGGC | CTGCAGGGC | PstI |
| ipnA G687A | ATCGGCAAC | ATCGCGAAC | NruI |
| ipnA G687N | ATCGGCAAC | ATTAATAAC | AseI |
| ipnA Q689I | AACCAACCA | AATATTCCA | SspI |
| ipnA D693L | GTGGACGAG | GTGCTCGAG | XhoI |
| ipnA D693E | GTGGACGAG | GTGGAAGAG | EarI |
| ipnA D693N | GTGGACGAG | GTGAACGAG | |
| ipnA AA702-3QL | CTGGCCGCGCAC | CTGCAGCTGCAC | PvuII |
| ipnA H704S | GCGCACCTG | GCTAGCCTG | NheI |
| ipnA ASNA731-2-4EFNS | ATGGCATCAAATGCATCC | ATGGAATTCAATTCATCC | EcoRI |
| ipnA AS734-5LE | AATGCATCCGGG | AATCTCGAGGGG | XhoI |
The codon(s) for the wild-type residues is shown in boldface.
The nucleotide substitutions are shown in boldface; novel restriction sites are underlined.
Construction of vectors expressing truncated IPNA segments.
A number of SK-ΔIPNA-derived constructs encoding putative cleavage sites were used to generate a set of T7 expression plasmids from which N- and C-terminally truncated forms of the polyprotein could be expressed in E. coli or in vitro. To obtain a reading frame carrying the putative pVP2-VP4 and VP4-VP3 cleavage sites (construct VP4-VP3), SK-ΔIPNA was digested with NcoI (nucleotide 1596) and SalI (present in the polylinker) and the insert was cloned NcoI-XhoI in a modified pET-22b (Novagen), in which the pelB leader sequence was deleted and the NcoI restriction site was located on the initiation codon. A series of VP4-VP3 Δ0 to Δ5 constructs were generated by using PCR with the Pfu DNA polymerase and adequate oligonucleotides for cloning into the BamHI restriction site of pET-28b (Novagen). Thus, six histidines and a T7 epitope were present at the N terminus of the open reading frame (ORF) and six histidines were present at the C terminus of VP3, resulting in the addition of 33 and 21 residues at their N and C termini, respectively. In the VP4-VP3 Δ1 AS734-5LE construct, the VP4-VP3 Δ1 construct was subjected to mutagenesis as described above to convert the Ala-Ser dipeptide at positions 734 and 735 to the dipeptide Leu-Glu. Generation of the VP4-His Δ1 construct was carried out by PCR by using adequate oligonucleotides for cloning into the BamHI-XhoI restriction sites of pET-28b. A silent mutation was introduced in the oligonucleotide in 5′ position to destroy the XhoI site in nucleotide position 1628.
In vitro expression and protein labeling.
In vitro, T7-derived expression was carried out using the TNT Quick Coupled Transcription/Translation System (Promega) as described by the manufacturer, except that reactions were performed in a final volume of 11 μl. The DNA template (1 μg) was incubated 1.5 h at 30°C. After incubation, aliquots of 2 to 3 μl were submitted to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (15), and gels were dried and exposed for autoradiography.
Expression of 5′-truncated IPNA polyprotein in E. coli, purification, and N-terminal sequence determination of generated polypeptides.
Overnight liquid cultures (250 ml) of BL21(DE3) carrying the constructs ipnVP4-VP3Δ1, ipnVP4-VP3Δ1AS734-5LE, and ipnVP4-HisΔ1 were half-diluted in L medium with kanamycin (50 μg/ml) and induced by 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at room temperature. Cells were collected by centrifugation at 5,000 × g for 5 min, washed in 50 ml of 50 mM Tris-HCl (pH 8)–2 mM EDTA and resuspended in 15 ml of 50 mM Tris-HCl (pH 8)–60 mM NaCl with a cocktail of protease inhibitors without EDTA (Boehringer Mannheim). Lysozyme was added to a concentration of 800 μg/ml, and the mixture was placed in an ice bath for 30 min. Then, 50 μl of benzonase (Boehringer) and 42 μl of 1 M MgCl2 were added for 10 min. The lysates were centrifugated at 13,000 rpm for 1 h at 4°C. The supernatants were diluted in 20 mM Tris-HCl (pH 8)–5 mM imidazole–0.5 M NaCl. The pellet was resuspended in the same buffer complemented with 6 M urea and solubilized overnight at 4°C under mild agitation. This material was submitted to centrifugation (13,000 × g, 30 min) for clarification. Aliquots of primary and second supernatants were submitted to SDS-PAGE. For the constructs VP4-VP3 Δ1 and VP4-VP3 Δ1 AS734-5LE, the VP3 polypeptides were found solubilized in the second supernatant, and they were processed for Ni2+ affinity chromatography under denaturing conditions as described by Novagen. The VP3 polypeptides were found eluted by the 200 mM imidazole buffer. Approximately 50 pmol was processed for automated N-terminal Edman sequencing by using a Perkin-Elmer/Applied Biosystems Procise 494A sequencer with reagents and methods of the manufacturer. The VP4 polypeptide was found in the primary supernatant of the construct VP4-HisΔ1. Supernatants (10 μl) were loaded onto a polyacrylamide gel for separation and transferred to a ProBlott membrane (Applied Biosystems) for N-terminal sequencing. The band was visualized on the membrane by Coomassie blue coloration.
High-performance liquid chromatography coupled with micro-IS-MS.
The reversed-phase high-performance liquid chromatography (RPLC) was online coupled with an ion-spray mass spectrometer (IS-MS) (Sciex API100; Perkin-Elmer). RPLC was run using a Perkin-Elmer device (Applied Biosystems 140D pump and 785 UV detector with U-shaped fused silica tubing of 7-mm pathlength) on a 0.5-by-150-mm column packed with 5-μm (300 Å) C18 silica in 4 mM CH3COONH4–0.1% HCOOH with a CH3CN linear gradient (4.5% for 5 min, from 4.5 to 9% in 15 min, from 9 to 51% in 100 min at a flow rate of 5 μl/min). After being monitored for absorbance at 215 nm, the flow was split between the micro-IS source (0.2 μl/min) and the microblotter (Perkin-Elmer/Applied Biosystems 173A) for polyvinylidene difluoride membrane blotting. IS-MS experiments were controlled with the Sample Control 1.3 software by using a positive mode from 400 to 2,100 amu with 0.2 amu steps and a 0.4-ms dwell time. The micro-IS voltage was 5,000 V, and the orifice plate voltage was 40 V. MS data were analyzed with Perkin-Elmer Sciex Bio-Multi-View 1.2 software. The average molar masses were calculated from sequences by using the Perkin-Elmer Sciex Peptide Map 2.2 software.
RESULTS
Candidates for functionally important residues of the VP4 protease.
A site-directed mutagenesis approach was used to map VP4 residues that are functionally important for protease activity. Positions for mutagenesis were chosen on the basis of several criteria. First, residues must be conserved in the different sequences of IPNV strains available in sequence databases. Second, each of the (conserved) histidines, the aspartic acids, the seven glutamic acids, and the two serine residues were mutated because these residues are often important components of known proteolytic enzymes (such as the chymotrypsin-like proteases or aspartyl proteases, enzymes encoded by a large number of viruses). Third, glycine 631 and leucines 636 and 638 were mutated because these residues fall in a small region that shows homology with other birnaviruses (Fig. 1, region B) and with an active-site region of chymotrypsin (MGPSA in VP4; MGDSG in chymotrypsin). Finally, alignments between the VP4 of IPNV, IBDV, and Drosophila X virus (DXV) shown in Fig. 1 were used as a guide, since these proteins share about 40% identity in the three regions shown but have little homology in other regions. Altogether, 42 positions were selected, full-length VP2, VP3, and VP4 ORF proteins carrying mutations were constructed (Table 1) and expressed in a rabbit reticulocyte lysate system, and their processing was analyzed by SDS-PAGE.
Activity of VP4 variants.
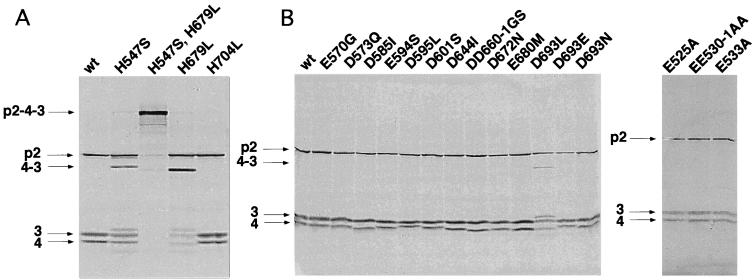
Processing of the wild-type polyprotein yielded the expected cleavage products pVP2 (62 kDa), VP3 (32 kDa), and VP4 (29 kDa) (Fig. 2A) as observed previously (21). In a first set of experiments, all conserved histidine and aspartic acid residues of VP4 were replaced by different residues. As shown in Fig. 2A, H547S and H679L mutants showed an efficient activity in the cleavage of the pVP2-VP4 junction but had a strongly reduced activity of approximately 50 and 20% of the wild-type activity for the VP4-VP3 junction, respectively. Two additional bands of 34 and 27 kDa were visualized with these mutants. Immunoprecipitation with the anti-VP3 4F4 monoclonal antibody (kindly provided by J. Dominguez) showed that the 34-kDa band was a longer VP3, suggesting that cleavage specificity was altered in these mutants with a subsidiary cleavage site present in the carboxyl part of VP4 (not shown). A double mutant, H547S H679L, was constructed. These changes resulted in a nearly complete loss of cleavage activity at both VP4 junctions (Fig. 2A). In contrast, the H704S mutant showed a wild-type activity. The D693L mutant showed band patterns similar to those of H547S and H679L mutants, with a cleavage at the VP2-VP4 junction and a partial cleavage at the VP4-VP3 junction, whereas the seven mutants D573Q, D585I, D595L, D601S, D644I, DD660-1GS, and D672N had a wild-type activity (Fig. 2B). Two additional substitutions (with glutamic acid and asparagine) were carried out on position D693. The two mutants exhibited a wild-type activity (Fig. 2B). To assess the possible role of the glutamic acids at positions 525, 530, 531, 533, 570, 594, and 680, these residues were also replaced by other residues. As shown in Fig. 2B, all of these mutants have an activity similar to that of the wild type. These first data indicated that H547, H679, and D693 residues are important for both cleavage activity and specificity between VP4 and VP3 but are not essential for the cleavage between pVP2 and VP4.
FIG. 2.
Mutagenesis of the conserved histidine, aspartic acid, and glutamic acid residues of the IPNV VP4. The autoradiographs show the results obtained with SKΔIPNA wild type (wt) and a set of mutant SKΔIPNA-derived constructs encoding proteins with a single (or double) amino acid substitution(s). (A) Substitution of histidine residues. (B) Substitution of aspartic acid and glutamic acid residues. The constructs were expressed with the rabbit reticulocyte expression system (Promega), and expression products were analyzed by SDS-PAGE. p2, pVP2; 3, VP3; 4, VP4; 4-3, VP4-VP3; p2-4-3, uncleaved polyprotein precursor. Note that single mutations can strongly affect the behavior of the VP4 in SDS-PAGE.
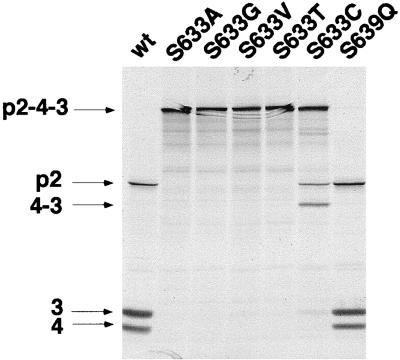
To investigate the possible role of serine 633 and 639 (see Fig. 1, B region) in protease activity, these residues were replaced by alanine and glutamine residues, respectively (Fig. 3). While replacing serine 639 had no effect on protease activity, a change of serine 633 completely abolished its function. Serine 633 was also replaced by more related residues, such as cysteine and threonine. Interestingly, the S633T mutant was not active, whereas the S633C mutant conserved approximately 20% activity for cleaving the VP2-VP4 junction, but none was detectable for the VP4-VP3 junction. Moreover, pulse-chase experiments with the S633C mutant showed that 60% of the polyprotein was cleaved at the VP2-VP4 junction and only 10% at the VP4-VP3 junction after 3.5 h of chase (not shown). No cleavage product was observed with the S633T mutant after the chase. Thus, serine 633 is critical for VP4 to function as a protease.
FIG. 3.
Mutagenesis of the conserved serine residues in the B region of IPNV VP4. A number of substitutions were introduced at the position of serines 633 and 639. Expression was carried out as described for Fig. 2.
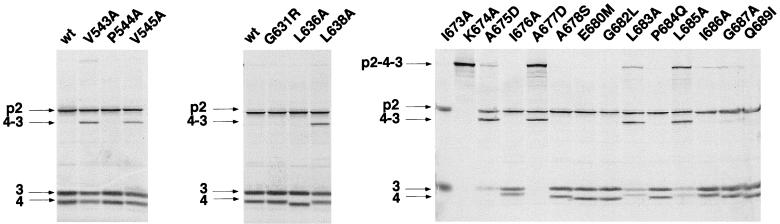
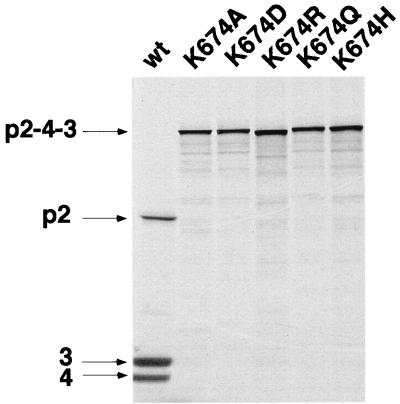
To evaluate the importance of the residues of IPNV VP4 which are in regions showing homologies with other birnaviruses, additional mutants were engineered. Valines 543 and 545 and proline 544 (region A) could be replaced by alanine without a loss of activity for the pVP2-VP4 junction, but a reduced activity was observed for two mutants at the VP4-VP3 junction (Fig. 4, left panel). In the B region which contains the serine 633, three additional residues were mutated. A change of glycine 631 to arginine and leucines 636 and 638 to alanine resulted in an efficient activity in the cleavage of the VP2-VP4 junction and in a reduced activity at the VP4-VP3 junction only for the leucine 638 substitution (Fig. 4, middle panel). In the C region, 14 positions were mutated (Fig. 4, right panel). The replacement of lysine 674 by alanine completely abolished processing between pVP2 and VP4 and between VP4 and VP3. The six mutants A675D, A677D, L683A, L685A, I686A, and G687A cleaved the pVP2-VP4 and VP4-VP3 junctions at an efficiency of approximately 40 to 80% of the wild-type efficiency. No detectable cleavage activity at the VP4-VP3 junction with mutant A677D was observed. It is of interest that the VP4 band, which was expected to be present in the same ratio as the VP3 band, was absent in the running gel in mutants I673A and A675D. This was attributed to an instability of the generated VP4 mutant. The six mutants I676A, A678S, E680M, G682L, P684Q, and Q689I exhibited wild-type activity. Additional further conservative substitutions were carried out to evaluate the importance of lysine 674. Processing at the pVP2-VP4 and VP4-VP3 junctions was completely abolished by replacement with aspartic acid, glutamine, histidine, and arginine again (Fig. 5). The results demonstrate that VP4 protease activity is highly sensitive to subtle replacements at the position of lysine 674, as previously reported for serine 633.
FIG. 4.
Mutagenesis of conserved residues in the regions A, B, and C of VP4. The autoradiographs show the results obtained with SKΔIPNA wild type (wt) and a set of mutant SKΔIPNA-derived constructs encoding proteins with a single amino acid substitution. Region A, left panel; region B, middle panel; region C, right panel. Expression was carried out as described for Fig. 2.
FIG. 5.
Mutagenesis of the lysine 674 of IPNV VP4. A number of substitutions were introduced at the position of lysine 674. Expression was carried out as described for Fig. 2.
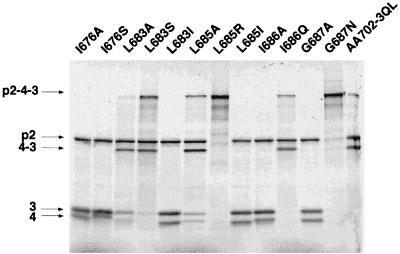
Additional mutants were engineered to gain information on the importance of hydrophobic residues of the C region in the protease activity (Fig. 6). The four mutants L683S, L685R, I686Q, and G687N cleaved the two sites mentioned above with reduced efficiency compared to their alanine mutant homologs. In contrast, the I676 position appeared to be more permissive. These results showed the relative importance of several residues of the C region for the VP4 protease activity.
FIG. 6.
Mutagenesis of hydrophobic residues in the region C of the IPNV VP4. Expression was carried out as described for Fig. 2.
Identification and mutagenesis of the cleavage site(s) at the VP4-VP3 junction.
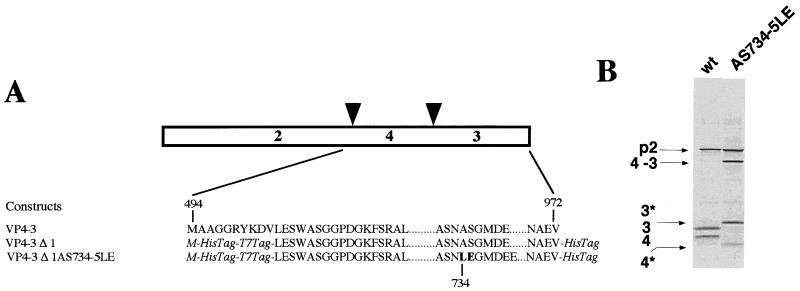
Considering that VP4 protease was shown to be functional in E. coli (20), we used this expression system to map the VP4 cleavage sites. We engineered two constructs encoding NH2-truncated forms of the polyprotein: VP4-VP3 with the methionine 494 as initiation methionine and VP4-VP3 Δ1 with a six-His tag located at both termini of the polyprotein (Fig. 7A). The latter construct VP3 product was expected to be easily purified by nickel affinity chromatography and further analyzed by N-terminal sequencing. Preliminary results indicated that (i) cleavage was efficient at the VP4-VP3 junction with the two constructs expressed in E. coli or in reticulocyte lysates and (ii) the VP3-His polypeptides immunoprecipitated with an anti-VP3 antibody comigrated, suggesting that cleavage occurred at the same position in both expression systems (not shown). E. coli-expressed VP3-His protein was purified, and its first six amino acids were determined through N-terminal sequencing: NH2-Ser-Gly-Met-Asp-Glu-Glu. The sequence is identical to the IPN sequence starting at serine residue 735. We further tested the effect of the double substitution AS734-5LE on the polyprotein processing in the reticulocyte expression system (Fig. 8B). As expected, this double mutation resulted in the generation of the uncleaved VP4-VP3 polypeptide, but two additional polypeptides of 34 and 27 kDa were also identified. Thus, in this mutant, the cleavage site generating VP3 (32 kDa) and VP4 (29 kDa) was not functional, but part of the VP4-VP3 polypeptide was cleaved at another site.
FIG. 7.
Mapping of the cleavage site at the VP4-VP3 junction. (A) Scheme of the set of three 5′-truncated expression products. Numbers refer to the amino acid position on the full-length ORF polyprotein, and additional sequences are indicated in italics. The HisTag-T7Tag (NH2 extremity) and the HisTag (COOH extremity), which derive from pET-28 vector, are made of 33 residues and 21 residues, respectively. Mutated residues are indicated in boldface. (B) Expression in reticulocyte lysates of the polyprotein mutant carrying a double substitution at the P1-P′1 position of the cleavage site between the VP4 and VP3 proteins. 3*, 34-kDa band; 4*, 27-kDa band.
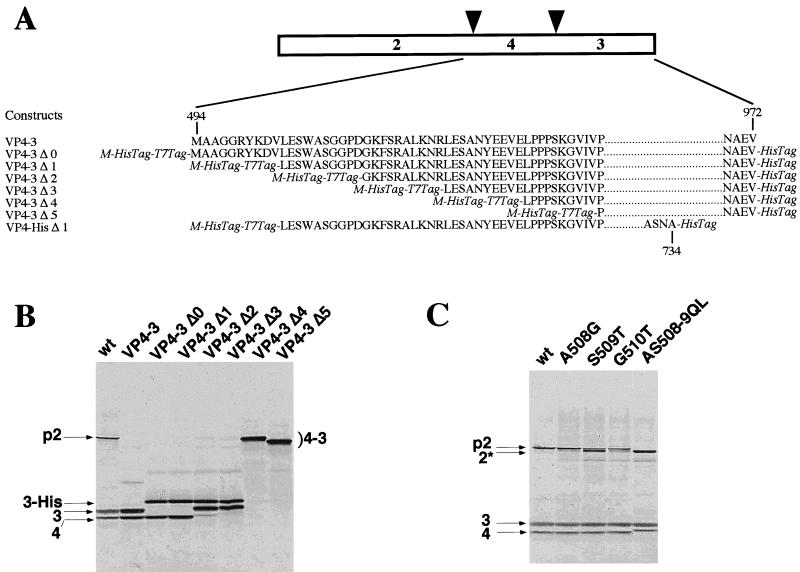
FIG. 8.
Mapping of the cleavage site at the pVP2-VP4 junction. (A) Schematic representation of the set of 5′-truncated expression products. Numbers refer to the amino acid position on the full-length ORF polyprotein, and additional sequences indicated in italics are as described in Fig. 8. (B) Expression was carried out as described for Fig. 2. (C) Mutagenesis of the cleavage site. Expression in reticulocyte lysates of polyprotein mutants carrying double (at the P1-P′1 position) or single (at the P1, P′1, and P′2 positions) substitutions of the cleavage site between the pVP2 and VP4 proteins. 2*, short pVP2.
Mapping of this subsidiary site was accomplished using a construct derived from the VP4-VP3 Δ1 and carrying the double substitution AS734-5LE (Fig. 8A). The VP4-VP3 Δ1 AS734-5LE polypeptides expressed in E. coli and in reticulocyte lysates were immunoprecipitated with an anti-VP3 antibody for analysis by SDS-PAGE. The resulting VP3s comigrated, suggesting that the subsidiary cleavage site was functional in both expression systems (not shown). The VP3 form expressed in E. coli was purified by nickel affinity chromatography and its first eight amino acids were determined through N-terminal sequencing: NH2-X-Lys-Gly-Ser-Asn-Lys-Arg-Ile. These data suggest that cleavage occurs between the Arg-Ala dipeptide 715-716 or the Ala-Lys dipeptide 716-717. Deformylation and N-terminal sequencing of this VP3 form were carried out to determine if the lysine identified in position 2 was (or not) the N-terminal residue posttranslationally modified. The lysine was again identified at position 2. The VP3 form was submitted to MS electrospray, and a molecular mass of 31,471.5 Da was determined, a finding compatible with the presence of an alanine as the first N-terminal residue. It was concluded that cleavage at the VP4-VP3 junction occurs between alanine 734 and serine 735, but a subsidiary cleavage site for VP4 exists between arginine 715 and alanine 716.
Identification and mutagenesis of the cleavage site at the pVP2-VP4 junction.
The sizes of the VP4s generated by the proteolytic processing of either the VP4-VP3 construct or the wild-type polyprotein were first compared (Fig. 8). Samples were run side by side in the same gel (Fig. 8B). As a result, the VP4s comigrated, suggesting that VP4 expressed from VP4-VP3 contained the natural N terminus of VP4. A series of NH2-truncated forms of the wild-type polyprotein with an increment of 10 residues and six-His tagged at both termini were constructed (Fig. 8A). The VP4 expressed from VP4-VP3 Δ0 and VP4-VP3 Δ1 comigrated with VP4, suggesting that the N terminus of VP4 was present in these constructs (Fig. 8B). In contrast, the VP4 expressed from VP4-VP3 Δ2 and Δ3 migrated more slowly than the natural VP4, suggesting that the cleavage site at the pVP2-VP4 junction was not present in these constructs. Thus, the domain located between valine 503 and glycine 514 contains determinants associated with (or close to) the cleavage site between pVP2 and VP4. These results also suggest that the residues which are deleted in constructs Δ4 and Δ5 are necessary for the cleavage at the VP3-VP4 junction.
Taking advantage of the fact that the cleavage site was identified at the VP4-VP3 junction, a construct derived from VP4-VP3 Δ1 with a six-His tag sequence fused in frame at the alanine 734 was constructed to allow purification and N-terminal sequencing of the VP4 (Fig. 8A). The protein was expressed in E. coli, directly subjected to SDS-PAGE, and blotted onto a membrane for N-terminal sequencing. Its first seven amino acids were identified as: NH2-Ser-Gly-Gly-Pro-Asp-Gly-Lys, thus showing, in accordance with experiments carried out in reticulocyte lysates, that cleavage between pVP2 and VP4 occurred between residues 508 and 509. (It should be noted that efforts to purify this form of tagged VP4 by nickel affinity chromatography failed, probably because of an efficient cleavage activity on the subsidiary cleavage site at positions 715 and 716.) The effect of the double substitution AS5089QL, as well as point mutations (A508G, S509T, and G510T), on the polyprotein processing was tested by using the reticulocyte expression system. Substitutions were expected to interfere strongly with processing between pVP2 and VP4. Interestingly, none of these mutations resulted in the generation of uncleaved pVP2-VP4 polypeptide; however, all of them modulated the cleavage at this junction (Fig. 8C). Specifically, the double mutation resulted in the generation of a shorter pVP2 and a longer VP4, indicating that the previously described pVP2-VP4 cleavage was blocked and an alternative cleavage site was used in the C terminus of pVP2. The three point substitutions at positions 508, 509, and 510 resulted in a less dramatic effect on pVP2-VP4 cleavage. The presence of pVP2 was detected on these mutants, but the generation of a shorter pVP2 was also clearly visualized, especially with the conservative S509T substitution which generated approximately 60% of shorter pVP2. These experiments revealed that the cleavage site between pVP2 and VP4 is located between alanine 508 and serine 509 and that an alternative cleavage site(s) is present in the carboxyl domain of pVP2.
Mutagenesis of potential additional cleavage site(s) in the carboxyl domain of pVP2.
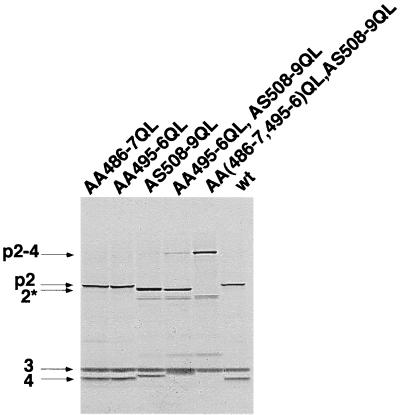
Figure 9 shows a sequence comparison between the pVP2-VP4 and the VP4-VP3 cleavage site regions. The Ala-Ser dipeptide was found as the conserved scissible peptidyl group bound in both cleavage sites. A serine at the P3 position and a glycine at the P′2 position also appeared to be conserved within the two cleavage sites. Two other putative cleavage sites corresponding to the (Ser/Thr)-X-Ala-Ala-Gly motif were also found in the COOH terminus of pVP2. To determine whether these sites are possible targets for VP4, the effect of Ala-Ala to Gln-Leu substitutions at positions 486 and 487 and positions 495 and 496 was analyzed. Substitutions present simultaneously on the same mutant in positions 486 and 487, 495 and 496, and 508 and 509 indeed completely inhibited the generation of pVP2 and shorter pVP2 forms, suggesting that all potential cleavage sites at the pVP2-VP4 junction domain are blocked in this construct (Fig. 10). The two cumulated substitutions at positions 495 and 496 and positions 508 and 509 generated a shorter pVP2 and a larger VP4 than when the residues at positions 508 and 509 were only substituted, suggesting that the Ala-Ala dipeptide at positions 495 and 496 is cleaved in the mutant AS508-9QL and that the Ala-Ala dipeptide 486-487 is efficiently used in the mutant with the cumulated substitutions AA495-6QL and AS508-9QL. These data indicate that, in addition to the cleavage site at the pVP2-VP4 junction identified between residues 508 and 509, two subsidiary cleavage sites at positions 495 and 496 and positions 486 and 487 in the C-terminal part of pVP2 were found as potential targets of the VP4 protease.
FIG. 9.
Cleavage sites (and putative cleavage sites) for the IPNV VP4 in the polyprotein. (Top) Schematic representation of the IPNA polyprotein. The amino acids of the VP2-VP4 and VP4-VP3 domains are indicated. The P1-P′1 cleavage site positions are underlined. (Bottom) The sequences relative to cleavage sites identified by N-terminal sequencing are in boldface. The sequences in regular typeface were identified by sequence homology and probed by mutagenesis.
FIG. 10.
Mutagenesis of potential cleavage sites for the VP4 protease in the COOH part of pVP2. Expression in reticulocyte lysates of the polyprotein mutants carrying double substitution(s) at the P1-P′1 position of the potential cleavage sites (positions 486 and 487 and positions 495 and 496). p2-4, VP2-VP4; 2∗, short pVP2.
DISCUSSION
The initial objective of this study was to identify amino acid residues that play an essential role in the catalytic activity of IPNV VP4 protease and to characterize its substrate cleavage sites. The relative importance of residues belonging to conserved domains in the birnavirus VP4 homologs was also addressed.
Active-site mutations.
To identify critical residues for the VP4 protease function, we first postulated that VP4 is either a serine or an aspartyl protease, and we mutated conserved residues possibly associated with the catalytic site. Thus, histidines, aspartic acids, glutamic acids, and serines found in the glycine-X-serine signature of serine hydrolases were substituted in the VP4 domain. Only changes at the serine 633 completely eliminated polyprotein processing at both pVP2-VP4 and VP4-VP3 junctions (Table 2). Replacing serine 633 by a cysteine resulted in a partially active protease, while replacing it by a threonine, alanine, or glycine completely blocked protease activity. The sulfhydryl group of the cysteine is expected to react chemically in a way similar to the serine hydroxyl group and is known to act as a nucleophile in cysteine proteases. These findings make serine 633 the strongest candidate active-site nucleophile so far identified and lead us to postulate that the IPNV VP4 is a member of the serine proteinase superfamily. The possibility that the VP4 belongs to aspartic or metallo superfamilies is unlikely. First, unlike aspartic proteases, VP4 was shown to be insensitive to Pepstatin-A and H-261 (11). In addition, none of the mutations of the nine conserved aspartic acids leads to an inactive protease. Second, unlike metalloproteases, VP4 activity was not inhibited by the metal chelator, EDTA (11). Moreover, none of the substitutions of a conserved histidine completely eliminated protease activity.
TABLE 2.
Cleavage efficiency of VP4 mutants at the pVP2-VP4 and the VP4-VP3 junctions
| Construct | Cleavage efficiencya at:
|
|
|---|---|---|
| pVP2-VP4 | VP4-VP3 | |
| ipnA | ++ | ++ |
| ipnA E525A | ++ | ++ |
| ipnA EE530–1AA | ++ | ++ |
| ipnA E533A | ++ | ++ |
| ipnA V543A | ++ | + |
| ipnA P544A | ++ | ++ |
| ipnA V545A | ++ | + |
| ipnA H547S | ++ | +* |
| ipnA H547S,H679L | − | − |
| ipnA E570G | ++ | ++ |
| ipnA D573Q | ++ | ++ |
| ipnA D585I | ++ | ++ |
| ipnA E594S | ++ | ++ |
| ipnA D595L | ++ | ++ |
| ipnA D601S | ++ | ++ |
| ipnA G631R | ++ | ++ |
| ipnA S633G | − | − |
| ipnA S633A | − | − |
| ipnA S633V | − | − |
| ipnA S633C | +/− | − |
| ipnA S633T | − | − |
| ipnA L636A | ++ | ++ |
| ipnA L638A | ++ | + |
| ipnA S639Q | ++ | ++ |
| ipnA D644I | ++ | ++ |
| ipnA DD660–1GS | ++ | ++ |
| ipnA D672N | ++ | ++ |
| ipnA I673A | ++ | ++** |
| ipnA K674A | − | − |
| ipnA K674D | − | − |
| ipnA K674R | − | − |
| ipnA K674Q | − | − |
| ipnA K674H | − | − |
| ipnA A675D | ++ | +/−** |
| ipnA I676A | ++ | ++ |
| ipnA I676S | ++ | ++ |
| ipnA I676S | ++ | ++ |
| ipnA A677D | + | − |
| ipnA A678S | ++ | ++ |
| ipnA H679L | ++ | +/−* |
| ipnA E680M | ++ | ++ |
| ipnA G682L | ++ | ++ |
| ipnA L683A | + | +/− |
| ipnA L683S | + | − |
| ipnA L683I | ++ | ++ |
| ipnA P684Q | ++ | ++ |
| ipnA L685A | + | +/− |
| ipnA L685R | − | − |
| ipnA L685I | ++ | ++ |
| ipnA I686A | ++ | ++ |
| ipnA I686Q | + | − |
| ipnA G687A | ++ | ++ |
| ipnA G687N | − | − |
| ipnA Q689I | ++ | ++ |
| ipnA D693L | ++ | +/−* |
| ipnA D693E | ++ | ++ |
| ipnA D693N | ++ | ++ |
| ipnA AA702–3QL | + | − |
| ipnA H704S | ++ | ++ |
++, 100% cleavage; +, 80% to 20% wild-type cleavage efficiency; +/−, <20% wild-type cleavage efficiency, −, no cleavage; ∗, altered cleavage specificity; ∗∗, VP4 instability.
The catalytic mechanism of the cellular serine proteases involves an active-site triad consisting of a serine, a histidine, and an aspartic acid. In a large number of viral serine proteases, substitutions were found in the catalytic triad (4, 12). Here, our results ruled out the possibility that IPNV VP4 contains the classical catalytic triad found since mutations of histidines and aspartic acids never completely blocked polyprotein processing. Our inability to identify an aspartic or a glutamic acid residue as a member of the active-site triad is reminiscent of what was observed in the serine protease of HCMV and herpesvirus. The crystal structure of these serine proteases revealed that they possess a histidine-histidine-serine catalytic triad (26). Mutagenesis and chemical modification studies carried out on members of this family of proteases showed a strict requirement of the serine (position 132 for HCMV) and histidine 63 but failed to identify the histidine 157 as the third member of the triad (8, 18, 29, 34). By analogy with the structure of the catalytic site of HCMV protease, it could be proposed that the birnavirus VP4 may be constituted by serine 633 and one of the two histidines (position 679 and 547) as the additional critical catalytic residue. Our inability to identify a histidine substitution that completely blocks polyprotein processing raises a concern regarding the validity of this hypothesis. In addition, multiple alignment of the IPNV, IBDV, and DXV VP4s did not reveal a conserved histidine.
Our data indicate that the IPNV VP4 might be a new type of viral serine protease. In some serine proteases and hydrolases (mainly found in procaryotic organisms), a lysine residue replaced the histidine base (5, 17, 23, 24, 25, 30, 33). Multiple alignment of the birnavirus VP4s shows that the lysine 674 (which is found in all strains of IPNV) is absolutely conserved in all strains of IBDV (lysine 692) and DXV (lysine 670) (Fig. 1), whereas the percentage of strictly conserved residues between the three proteases is about 5%. Substitutions of the lysine 674 by alanine, aspartic acid, glutamine, histidine, or arginine resulted in a complete block of the protease activity at both pVP2-VP4 and VP4-VP3 junctions. These results are in accordance with the observations made on LexA and its mutants K156A, K156H, and K156R (16, 28) and on B. subtilis signal peptidase with its inactive mutants K83A, K83H, and K83R (33). In addition, the activity of VP4 was not affected by usual inhibitors of serine proteases (reference 11 and unpublished results), a feature that VP4 shares with bacterial signal peptidases. These observations led us to propose that the IPNV VP4 may use a serine-lysine catalytic dyad: the hydroxyl group of the side chain of serine 633 would act as the nucleophile that attacks the carbonyl carbon of the scissible peptide bond and the lysine 674 ɛ-amino group would serve to activate the hydroxyl group of serine 633.
Cleavage sites.
Two IPNV VP4 cleavage sites, located at the pVP2-VP4 and VP4-VP3 junctions, were identified by N-terminal sequence analysis of cleavage products produced in E. coli and probed by site-directed mutagenesis. They are characterized by the Ser-X-Ala↓Ser-Gly motif. Two other additional cleavage sites in the carboxyl part of pVP2 (P1 and P′1 positions 486 and 487 and positions 495 and 496) were first identified by sequence comparison (Fig. 10). The P1 and the P′2 residues appeared to be conserved as an alanine and a glycine, respectively. The P3 serine residue can substitute to a threonine (cleavage site positions 486 and 487), whereas the P′1 serine residue was notably substituted to an alanine. The cleavage between pVP2 and VP4 was abolished only when the Ala-Ser 508-509 and the two Ala-Ala 495-496 and 486-487 pairs were mutated together. These observations do not prove definitively the existence (and the functionality) of these two additional cleavage sites. However, these data correlate well with the fact that pVP2 is slowly processed at its carboxy end (3) and strongly suggest that conversion of pVP2 to VP2 is associated with these VP4 substrate cleavage sites. But these data also raise a question. Why does expression of the polyprotein in in vitro translation experiments (11; this study) or in a baculovirus-derived system (19) only produce pVP2 without further processing of pVP2 to VP2, as observed in IPNV-infected cells? A possible explanation may be that the pVP2 product, which results from the self-cleavage of the polyprotein, is an intramolecular process, in contrast to the pVP2-to-VP2 conversion that involves a bimolecular reaction. Thus, the local concentration of pVP2 and VP4 may dramatically influence the rate of conversion to VP2, and it is likely that, in infected cells, the concentration of viral proteins is higher in the capsid assembly site than with recombinant expression systems. It cannot be excluded that host cell proteins may also contribute to the VP2 conversion. It was also remarkable that, compared to the pVP2-VP4 cleavage, processing of the VP4-VP3 junction was much more sensitive to substitutions in the VP4 (positions H547 and H679 for instance). This possibly indicates that the two cleavage sites have different structural properties and/or the association of the protease with as-yet-unidentified cofactors.
Interestingly, the IPNV VP4 cleavage site motif defined as (Ser/Thr)-X-Ala↓(Ser/Ala)-Gly shares similarities with both consensus cleavage sites of HCMV, with herpesvirus, and with bacterial leader peptidases. For these viruses, cleavage occurs between alanine and serine in the consensus sequence V/L-X-A↓S (34). For bacterial signal peptidases, cleavage sites are characterized by the Ala-X-Ala motif with a near essential alanine at P1 position, whereas the P3 alanine is less critical (2, 23). In addition, the identification of the IPNV VP4 cleavage site motif with the one of DXV as Ala-X-Ser↓Ala (7) allows the prediction of the motif Ala-X-Ala-Ala-Ser present at the pVP2-VP4 and VP4-VP3 junction domains of IBDV polyprotein as a possible target of the VP4 protease of IBDV.
To conclude, no sequence similarities were identified between the birnavirus proteases, herpesvirus proteases, and procaryotic leader peptidases. In contrast, sequence homologies were visualized between IPNV, IBDV, and DSX VP4s, suggesting that structural constraints exist for the folding of these proteases, which probably define a novel group in the serine proteases. The long-term objective is to solve the structure by X-ray crystallography to characterize their protein folding for comparison with other serine peptidases.
ACKNOWLEDGMENTS
We thank Javier Dominguez for the kind gift of the 4F4 anti-VP3 IPNV monoclonal antibody, Patrice Vende and Cynthia Jaeger for help in the sequencing work, Michel Bremont and Jean-François Eleouet for helpful discussions, and Scott Kramer for revising the English.
REFERENCES
- 1.Allaire M, Chernaia M M, Malcolm B A, James M N G. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases. Nature. 1994;369:72–76. doi: 10.1038/369072a0. [DOI] [PubMed] [Google Scholar]
- 2.Allsop A E, Ashby M, Brooks G, Bruton G, Coulton S, Edwards P D, Elsmere S A, Hatton I K, Kaura A C, McLean S D, Pearson M J, Pearson N D, Perry C R, Smale T, Southgate R. Inhibition of protein export in bacteria: the signalling of a new role for β-lactams. In: Bentley P H, O'Hanlon P J, editors. Anti-infectives: recent advances in chemistry and structure-activity relationships—1997. Cambridge, England: The Royal Society of Chemistry; 1997. pp. 61–72. [Google Scholar]
- 3.Azad A A, Jagadish M N, Brown M A, Hudson P J. Deletion mapping and expression in Escherichia coli of the large genomic segment of a birnavirus. Virology. 1987;161:145–152. doi: 10.1016/0042-6822(87)90180-2. [DOI] [PubMed] [Google Scholar]
- 4.Babé L A, Craik C S. Viral proteases: evolution of diverse structural motifs to optimize function. Cell. 1997;91:427–430. doi: 10.1016/s0092-8674(00)80426-2. [DOI] [PubMed] [Google Scholar]
- 5.Barrett A J, Rawlings N D. Families and clans of serine peptidases. Arch Biochem Biophys. 1995;318:247–250. doi: 10.1006/abbi.1995.1227. [DOI] [PubMed] [Google Scholar]
- 6.Brown F. The classification and nomenclature of viruses: summary of results of meetings of the International Committee on Taxonomy of Viruses in Sendai, September 1984. Intervirology. 1986;25:140–143. doi: 10.1159/000150091. [DOI] [PubMed] [Google Scholar]
- 7.Chung H K, Kordyban S, Cameron L, Dobos P. Sequence analysis of the bicistronic Drosophila X virus genome segment A and its encoded polypeptides. Virology. 1996;225:359–368. doi: 10.1006/viro.1996.0610. [DOI] [PubMed] [Google Scholar]
- 8.Cox G A, Wakulchik M, Sassmannshausen L M, Gibson W, Villarreal E C. Human cytomegalovirus proteinase: candidate glutamic acid identified as third member of putative active-site triad. J Virol. 1995;69:4524–4528. doi: 10.1128/jvi.69.7.4524-4528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding J, McGrath W J, Sweet R M, Mangel W F. Crystal structure of the human adenovirus proteinase with its amino acid cofactor. EMBO J. 1996;15:1778–1783. [PMC free article] [PubMed] [Google Scholar]
- 10.Dobos P, Hill B J, Hallett R, Kells D T, Becht H, Teninges D. Biophysical and biochemical characterization of five animal viruses with bisegmented double-stranded RNA genomes. Virology. 1979;32:593–605. doi: 10.1128/jvi.32.2.593-605.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobos P. The molecular biology of infectious pancreatic necrosis virus (IPNV) Annu Rev Fish Dis. 1995;5:25–54. [Google Scholar]
- 12.Dodson G, Wlodawer A. Catalytic triads and their relatives. Trends Biochem Sci. 1998;23:347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- 13.Dorson M, Castric J, Torchy C. Infectious pancreatic necrosis virus of salmonids: biological and antigenic features of a pathogenic strain and of a nonpathogenic variant selected in RTG-2 cells. J Fish Dis. 1978;1:309–320. [Google Scholar]
- 14.Kim J L, Morgenstern K A, Lin C, Fox T, Dwyer M D, Landro J A, Chambers S P, Markland W, Lepre C A, O'Malley E T, Harbeson S L, Rice C M, Murcko M A, Caron P R, Thomson J A. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Lin L-L, Little J W. Autodigestion and RecA-dependent cleavage of Ind− mutant LexA proteins. J Mol Biol. 1989;210:439–452. doi: 10.1016/0022-2836(89)90121-6. [DOI] [PubMed] [Google Scholar]
- 17.Little J W. LexA cleavage and other self-processing reactions. J Bacteriol. 1993;175:4943–4950. doi: 10.1128/jb.175.16.4943-4950.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu F, Roizman B. Differentiation of multiple domains in the herpes simplex virus 1 protease encoded by the UL26 gene. Proc Natl Acad Sci USA. 1992;89:2076–2080. doi: 10.1073/pnas.89.6.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magyar G, Dobos P. Expression of infectious pancreatic necrosis virus polyprotein and VP1 in insect cells and the detection of the polyprotein in purified virus. Virology. 1994;198:437–445. doi: 10.1006/viro.1994.1055. [DOI] [PubMed] [Google Scholar]
- 20.Manning D S, Leong J C. Expression in Escherichia coli of the large genomic segment of infectious pancreatic necrosis virus. Virology. 1990;179:16–25. doi: 10.1016/0042-6822(90)90268-v. [DOI] [PubMed] [Google Scholar]
- 21.Manning D S, Mason C L, Leong J C. Cell-free translational analysis of the processing of infectious pancreatic necrosis virus polyprotein. Virology. 1990;179:9–15. doi: 10.1016/0042-6822(90)90267-u. [DOI] [PubMed] [Google Scholar]
- 22.Matthews D A, Smith W W, Ferre R A, Condon B, Budahazi G, McElroy H E, Gribskov C L, Worland S. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell. 1994;77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 23.Nunnari J, Fox T D, Walter P. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science. 1993;262:1997–2004. doi: 10.1126/science.8266095. [DOI] [PubMed] [Google Scholar]
- 24.Paetzel M, Dalbey R E, Strynadka N C J. Crystal structure of a bacterial signal peptidase in complex with a β-lactam inhibitor. Nature. 1998;396:186–190. doi: 10.1038/24196. [DOI] [PubMed] [Google Scholar]
- 25.Peat T S, Frank E G, McDonald J P, Levine A S, Woodgate R, Hendrickson W A. Structure of the UmuD′ protein and its regulation in response to DNA damage. Nature. 1996;380:727–730. doi: 10.1038/380727a0. [DOI] [PubMed] [Google Scholar]
- 26.Qiu X, Culp J S, Dilella A G, Hellmig B, Hoog S S, Janson C A, Smith W W, Abdel-Meguid S. Unique fold and active site in cytomegalovirus protease. Nature. 1996;383:275–279. doi: 10.1038/383275a0. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Slilaty S N, Little J W. Lysine-156 and serine-119 are required for LexA repressor cleavage: a possible mechanism. Proc Natl Acad Sci USA. 1987;84:3987–3991. doi: 10.1073/pnas.84.12.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevens J T, Mapelli C, Tsao J, Hail M, O'Boyle II D, Weinheimer S P, Diianni C L. In vitro proteolytic activity and active-site identification of the human cytomegalovirus protease. Eur J Biochem. 1994;226:361–367. doi: 10.1111/j.1432-1033.1994.tb20060.x. [DOI] [PubMed] [Google Scholar]
- 30.Strynadka N C J, Adachi H, Jensen S E, Johns K, Sielecki A, Betzel C, Sutoh K, James M N G. Molecular structure of the acyl-enzyme intermediate in β-lactam hydrolysis at 1.7 Å resolution. Nature. 1992;359:700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- 31.Tong L, Wengler G, Rossmann M G. Refined structure of Sindbis virus core protein and comparison with other chymotrypsin-like serine proteinase structures. J Mol Biol. 1993;230:228–247. doi: 10.1006/jmbi.1993.1139. [DOI] [PubMed] [Google Scholar]
- 32.Tschantz W R, Sung M, Delgado-Partin V M, Dalbey R E. A serine and a lysine residue implicated in the catalytic mechanism of the Escherichia coli leader peptidase. J Biol Chem. 1993;268:27349–27354. [PubMed] [Google Scholar]
- 33.van Dijl J M, de Jong A, Venema G, Bron S. Identification of the potential active site of signal peptidase SipS of Bacillus subtilis. J Biol Chem. 1995;270:3611–3618. doi: 10.1074/jbc.270.8.3611. [DOI] [PubMed] [Google Scholar]
- 34.Welch A R, McNally L M, Hall M R, Gibson W. Herpesvirus proteinase: site-directed mutagenesis used to study maturational, release, and inactivation cleavage sites of precursor and to identify a possible catalytic site serine and histidine. J Virol. 1993;67:7360–7372. doi: 10.1128/jvi.67.12.7360-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wlodawer A, Miller M, Jaskolski M, Sathyanarayana B K, Baldwin E, Weber I T, Selk L M, Clawson L, Schneider J, Kent S B H. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science. 1989;245:619–621. doi: 10.1126/science.2548279. [DOI] [PubMed] [Google Scholar]