Abstract
Individuals with Alcohol Use Disorder (AUD) typically have comorbid chronic health conditions, including anxiety and depression disorders, increased sleep disruption, and poor nutrition status, along with gut microbial dysbiosis. To better understand the effects of gut dysbiosis previously shown in individuals with AUD, gut microbiome and metabolome were investigated between three cohorts. Two groups of individuals with AUD included treatment-seeking newly abstinent for at least six weeks (AB: N = 10) and non-treatment-seeking currently drinking (CD: N = 9) individuals. The third group was age, gender, and BMI-matched healthy controls (HC: N = 12). Deep phenotyping during two weeks of outpatient National Institutes of Health Clinical Center visits was performed, including clinical, psychological, medical, metabolic, dietary, and experimental assessments. Alpha and beta diversity and differential microbial taxa and metabolite abundance of the gut microbiome were examined across the three groups. Metabolites derived from the lipid super-pathway were identified to be more abundant in the AB group compared to CD and HC groups. The AB individuals appeared to be most clinically different from CD and HC individuals with respect to their gut microbiome and metabolome. These findings highlight the potential long-term effects of chronic alcohol use in individuals with AUD, even during short-term abstinence.
Introduction
AUD is defined by the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5), as “a problematic pattern of alcohol use leading to clinically significant impairment or distress” [1], and both binge and heavy drinking are associated with AUD. The National Institute on Alcohol Abuse and Alcoholism (NIAAA) defines binge drinking as the consumption of ≥ 4 or 5 drinks respectively for women or men in about 2 hours, which brings the blood alcohol concentration to meet or ≥ 0.08% [2]. Furthermore, the NIAAA defines heavy drinking as the consumption of ≥ 8 or 15 drinks/week for women or men, respectively [3].
Gut microbiome and metabolome alterations have been linked to the pathophysiology of several mental health conditions, such as psychotic, depressive, and anxiety disorders [4], and, most recently, to socio-emotional impairment and alcohol craving in the context of binge drinking and AUD [5]. Chronic heavy drinking can affect all systems of the body, including the gut microbiome [6]; in fact, heavy alcohol intake has been shown to influence gut dysbiosis, gut mucosal damage, and increased gut permeability [7–10]. The latter paves the way to endotoxemia, which is the result of the translocation from the intestinal lumen to the blood circulation of microbial products, mainly lipopolysaccharide, peptidoglycans, lipoteichoic acid, and flagellin [11]. When these toxins reach the liver via the portal vein, they can induce inflammation and contribute to hepatocyte death and subsequent fibrotic response [12, 13]. Gut microbiome is affected not only by chronic heavy drinking, but also by many common diseases and prescription medications [14].
Several studies have focused on determining gut dysbiosis and metabolic dysfunction in animal models of alcohol intake and individuals with AUD. Studies in rodent models of alcohol intake found decreased gut microbial alpha- and beta-diversity (diversity within and between microbial communities, respectively), as well as differential relative abundance of phyla, such as Firmicutes and Bacteroidetes [15, 16] (the Firmicutes/Bacteroidetes ratio is typically elevated in alcohol drinking animals) [17]. Preliminary work from our group on the gut microbiome of growth hormone secretagogue receptor (GHSR) knockout vs wild-type rats after alcohol binge-like exposure showed a decrease in overall gut microbial diversity and changes in the proportion of Bacteroidales order following alcohol binge-drinking [18]. A minority of rodent studies analyzed the gut metabolome, revealing alcohol-related changes in metabolites belonging to glycerophospholipids, carboxylic acids, and fatty acyls [17, 19–21], classified as amino acids, bile acids, fatty acids, peptides, and phenols in the subclass level [21–23].
Other non-human primate studies investigated the relationship between alcohol use and the gut microbiome and metabolome [22, 23]. Two studies in Rhesus macaques showed an increased in inflammatory bacterial taxa, such as Firmicutes [22], and metabolomic changes primarily associated with differences in glycolysis [23]. A recent study from our group in olive baboons [24], modeling both binge and heavy drinking patterns [25, 26], found changes in the gut microbiome and metabolome, primarily loss of gut microbial alpha- and beta-diversity and enhanced energy metabolism. These changes occurred after long-term (~12 years), but not relatively short-term (~3 years) excessive drinking and were only partially affected by acute abstinence from alcohol.
Preliminary human studies point to an association between gut dysbiosis and chronic heavy drinking in individuals with AUD, typically investigated in inpatient settings [7–9]. There is evidence that alcohol misuse increases the abundance of Firmicutes and reduces Bacteroidetes [15, 27]. Fluctuations in gut luminal content of amino acids, bile acids, lipids, neurotransmitters, and markers of oxidative stress, likely reflecting alterations in the gut bacteria, have also been observed [28–30]. Individuals with greater gut dysbiosis show intestinal hyperpermeability and inflammatory response [7, 8, 10] and are also more likely to have liver damage [27]. Yet, gut dysbiosis is observed only in a subset of individuals with chronic heavy drinking [7, 8] and abstinence does not always lead to reversal of dysbiosis [7–10]. A study in individuals with AUD during detoxification found an association between gut dysbiosis, intestinal hyperpermeability, and high depression, anxiety, and alcohol craving [8], with the latter three being important predictors of relapse [31]. A study in recently abstinent inpatients with AUD found that very heavy drinkers (≥10 drinks/day) presented greater changes in the gut microbiome following abstinence vs less heavy drinkers (<10 drinks/day), yet these changes were not correlated with shifts in depression or anxiety [32].
In summary, existing human literature points to a complex relationship between AUD and gut microbial composition and function. A number of gaps exist, including the failure to account for lifestyle and environmental factors known to affect gut bacteria contributing to intra- and inter-individual heterogeneity; in addition to retrospective measures of alcohol consumption which are subject to recall bias and strictly controlled inpatient settings, failure to properly account for these factors may deter researchers from making conclusive findings on whether gut dysbiosis and metabolic alterations are due to alcohol abstinence vs factors intrinsic to inpatient care (e.g., diet, controlled environment) or other factors [14, 33]. The aim of this case-control clinical study was to analyze and compare the gut microbiome and metabolome among three groups of individuals with AUD in an outpatient setting: individuals with AUD that were both abstinent (AB) for ≥6 weeks, and non-treatment-seeking who continued drinking (CD), and healthy controls (HC) not drinking or drinking in moderation. Consistent with our previous microbiome/metabolome study in a baboon model of binge drinking [24], we hypothesized loss of gut microbial alpha- and beta-diversity and enhanced energy metabolism in the CD group, compared to the other groups. Furthermore, deep phenotyping was performed, including clinical, psychological, medical, metabolic, and dietary assessments such that other lifestyle factors could be considered to help to explain any microbiome/metabolome differences found between groups.
Materials and methods
Study participants
Recruitment for the current study occurred from August 28th 2017 to February 25th 2020 where it was terminated earlier than originally planned due to the COVID-19 pandemic. Study inclusion and exclusion criteria are listed in the Supplementary Methods. All participants provided written informed consent and were compensated for their time and participation. The study was approved by the NIH Institutional Review Board Approval Institution (Protocol Number: 17-DA-0093).
The three groups in the study are described as: (1) abstinent individuals (AB, N = 10), who met DSM-5 criteria for AUD [1] and recently completed at least four weeks of inpatient treatment at the NIH Clinical Center under the NIH/NIAAA treatment protocol 14-AA-0181, followed by at least two weeks of abstinence in “real life” environment upon discharge; these individuals had to continue to remain abstinent for the entire duration of the study; (2) currently drinking individuals (CD, N = 9), who met DSM-5 criteria for AUD, but were not actively seeking treatment for AUD at the time of study participation and who satisfied NIAAA criteria for heavy drinking [3] from at least four weeks prior to screening up to the day of enrollment; and healthy controls (HC, N = 12), who had no current or past diagnosis of AUD and with limiting alcohol intake to ≤ 1 or 2 drinks/day for women or men, respectively [34]. HCs were enrolled to match sex, race, mean age (±5 years), and mean BMI (±20%) of the AUD groups (ABs and CDs). In this study, drinks corresponded to U.S. standard drinking unit, containing 14 g of alcohol [35]. For a full description of the study design, see Supplementary Methods.
Alcohol-related clinical assessment and psychological measures
Alcohol-related clinical characteristics and psychopathological features were assessed at screening and/or at study visits using structured clinical interviews and self-report questionnaires (for a detailed description see Supplementary Methods). Diagnosis of AUD and of other possible comorbid psychiatric disorders was made at screening according to the DSM-5 criteria [1] via the Structured Clinical Interview for DSM-5 (SCID-5) [36].
AUD severity was assessed at screening using the number of endorsed SCID-5 AUD criteria, the number of heavy drinking years per the Lifetime Drinking History (LDH) [37], and the total scores of the Alcohol Use Disorders Identification Test (AUDIT) [38] and the Alcohol Dependence Scale (ADS) [39]. Alcohol intake was measured via the Timeline FollowBack (TLFB) [40]: retrospectively to reflect the 90 days prior to inpatient admission for the AB group and prior to study start for the CD and HC groups. Alcohol intake was assessed at each study visit for the groups. Alcohol related variables utilized in this study included the average number of drinks per day and the number of heavy drinking days. Other alcohol related measures such as craving and withdrawal were collected using the Penn Alcohol Craving Scale (PACS) [41] and the Clinical Institute Withdrawal Assessment of Alcohol Scale, Revised (CIWA-Ar) [42]. Quality of sleep and presence of depressive and anxiety symptoms were evaluated at screening via the PSQI [43] and an abbreviated version of the Comprehensive Psychopathological Rating Scale (CPRS) [44], which encompasses the Montgomery-Åsberg Depression Rating Scale (MADRS) [45] and the Brief Scale for Anxiety (BSA) [46]. Transient mood states were assessed during the study timeframe, specifically at the first and last study visits, using the POMS anxiety and POMS depression and both were averaged between the first and last visits [47].
Other clinical measures
Liver transient elastography
Transient elastography was performed using FibroScan® (Echosens, France) at the first study visit. FibroScan® is a non-invasive diagnostic ultrasound-based device used to assess liver stiffness (hardness), a measure of fibrosis, and liver fatty changes, a measure of steatosis, which are present in a variety of liver diseases [48] (see detailed description in Supplementary Methods). The degree of fibrosis and steatosis were quantified using a liver stiffness measurement (LSM, in kilopascals [kPa]) and a controlled attenuation parameter (CAP, in decibels/meter [dB/m]), respectively [49].
Medical comorbidity and medication data collection
Self-reported pre-existing medical conditions and concomitant medications were collected as part of the participants’ medical history and physical examination at the first study visit; they were then categorized as shown in S1 Table and based on the classification by Jackson et al. [14].The latter used the gut microbiome profiles of the >2,700 members of the deeply phenotyped Twins UK cohort to carry out gut microbiome association analyses with 38 common diseases (e.g., hypercholesterolemia, hypertension, type 2 diabetes, respiratory allergies, anxiety, osteoarthritis) and 51 medications (e.g., statins, proton pump inhibitors, vitamin D3, iron, calcium) that have been linked to gut dysbiosis; their findings show significant associations with gut microbial diversity measures and select taxa. Since pre-existing medical conditions and concomitant medications were reported by many participants in this study, we looked them up and cross-checked them with those listed by Jackson et al. to determine those linked with gut microbiota alterations (20% FDR).
Dietary intake assessment
Dietary intake was assessed using food records throughout study participation. At each study visit, food records were collected and reviewed by trained nutrition staff. Dietary intake reflected foods and beverages consumed prior to each study visit, starting from day after the previous visit. Gathering of dietary information corresponded to the in-clinic and at-home fecal sample collection. For further details, please see Supplemental Methods.
Gastrointestinal permeability testing
Gastrointestinal permeability, which is an index of gut mucosal integrity, was assessed using four nondigestible sugars that are excreted and measurable in the urine, with the urine sugar ratios providing a measure of permeability [50] (see full description in Supplementary Methods). After an overnight (≥12 hours) fast and baseline urine collection, participants drank a 100 mL solution containing sucrose (10 g/dL), lactulose (5 g/dL), mannitol (1 g/dL), and sucralose (1 g/L), and urine was collected for 5 hours. Participants were not allowed to eat or drink during these 5 hours, except for 1 L of water provided.
Fecal sample collection and processing
A total of 209 fecal samples were obtained from the 31 study participants which included depositions while participants were at home and also during the study visits while at the NIH Clinical Center. Fecal sample handling, processing, and analysis for microbiome and metabolome characterization have been described previously [24]. For full details of fecal sample collection and processing, see Supplementary Methods.
Gut microbiome extraction and sequencing
Gut microbiome analysis consisted of fecal DNA extraction and 16S rRNA gene sequencing, followed by data processing as described in Supplementary Methods.
Gut metabolomic profiling
Detailed methods for gut metabolite profiling are described in the Supplementary Methods. Following removal of metabolites with 50% or more missing values, the values were median centered, log10 transformed, and mean scaled prior to statistical analyses. Missing values, if any, were replaced by actual limits of detection (equal to 1/5 of the minimum positive value of each variable) [51]. The resulting metabolomics dataset comprised 1032 metabolites, all of which had known chemical identities.
Statistics
Study population
We compared the study participants’ demographic, alcohol-related, psychological, and other clinical characteristics, which were non-normally distributed (all p’s < .05 at the Shapiro-Wilk test), using, for categorical variables, Chi-squared (χ2) test for comparisons among groups (AB vs. CD vs. HC) and between groups (AB vs. CD, AB vs. HC, CD vs. HC). For continuous variables, we used a Kruskal-Wallis (H) test for comparisons among groups and a Wilcoxon rank sum test for comparisons between two groups. Categorical variables were summarized using the total N sample size and the percent of the total sample, continuous variables as means and standard deviations. The JMPv16 statistical software (SAS Headquarters, Cary, NC, USA) and Prism 9.4.1 for Mac OS X (GraphPad Software, Boston, MA, USA) were used for data analysis and visualization.
Repeated sampling
Repeated measures for each participant’s fecal samples and dietary intake records over the six study visits were averaged to control for intra-individual variability. Repeated measures ANOVA was run to ensure that there were no statistically significant differences among study visits within participants (all p-values≥.05). Gut microbiome and dietary intake data presented are based on this averaging unless otherwise specified.
Participant characteristics
We compared the study participants’ demographic, alcohol-related, psychological, and other clinical characteristics, which were non-normally distributed (all p’s < .05 at the Shapiro-Wilk test), using, for categorical variables, Chi-squared (χ2) test for comparisons among groups and between groups (AB vs. CD, AB vs. HC, CD vs. HC). For continuous variables, a Kruskal-Wallis (H) test for comparisons among groups and a Wilcoxon rank sum test for comparisons between two groups was used. Categorical variables were summarized using the total N sample size and the percent of the total sample, continuous variables as means and standard deviations. A p < .05 (two-tailed) was considered statistically significant.
Microbiome data
Filtering and normalization, computation of microbial measures, and data visualization were conducted using MicrobiomeAnalyst v. 2.0 [52–54], JMPv16, and Prism. Data filtering parameters: minimum count = 4, 20% prevalence in samples, percentage to remove based on IQR = 10%, sample size = 10,000. The data were then normalized, reducing the ASVs from 986 to 203. Alpha diversity metrics included observed ASVs, Chao1, Shannon, and Simpson. A PCoA of Bray-Curtis distances (Bray-Curtis dissimilarity) was used to measure beta-diversity with permutational ANOVA (PERMANOVA) test for differential analyses among groups.
Three tests were used to analyze the 71 genera (from the 203 ASVs): linear discriminant analysis (LDA) effect size [55, 56], microbiome multivariable associations with linear models 2 (MaAsLin2) [57], and heat tree analysis (Wilcoxon rank sum test) [56]. Each test was used to conduct pairwise comparisons (AB vs. HC, AB vs. CD, CD vs. HC). LEfSe is an algorithm that first employs a Kruskal-Wallis (H) test to identify features with significant differential abundance with respect to the class of interest; then, it performs a set of pairwise tests among subclasses, using Wilcoxon rank sum test to assess the contribution of differences between groups; as a last step, it uses LDA to estimate the effect size of each differentially abundant feature and to rank the relevance of the different biological aspects [55]. A size-effect threshold of 2.0 for the logarithmic LDA score was applied for discriminative microbial features between groups. MaAsLin2 is a statistical framework using general linear models to find associations between microbial features and experimental metadata. A linear model is fit to each microbial feature that includes the primary metadata, covariate, and blocking factor variables [57]. MaAslin2 was run using age and BMI adjustment in the linear model, and covariate plots of the resulting significant genera were created. Heat tree analysis [58] displays statistics associated with taxa (relative abundances) in a tree format. It leverages the hierarchical nature of the data to depict taxonomic differences quantitatively and statistically (Wilcoxon rank sum test) between microbial communities.
Genera found to be statistically significant at any of the three tests are reported. All testing procedures selected genera at p < .05. Statistically significant genera were assessed for multiple comparisons and none passed the adjustment, thus the genera p-values reported are unadjusted.
Metabolomic data
A total of 1032 identified metabolites were used to investigate metabolomic profiles among and between groups using both a Kruskal-Wallis [7] [H] test for and post-hoc Wilcoxon rank sum test for pairwise comparisons. Any metabolite with missing values in ≥ 50% of the samples was excluded from the analysis. Data were normalized by median centering, and mean scaling and were further transformed using a logarithm base 10 transformation. The final dataset submitted for analysis after filtering was 946 metabolites. Multiple comparisons were controlled for using a Benjamini-Hochberg (FDR) correction [59] and those passing a corrected p < .05 (KW test) were considered significant. A multivariate unsupervised principal component analysis (PCA) and supervised analyses (partial least squared discriminant analysis [PLS-DA]) were carried out using MetaboAnalyst v. 5.0 [60]. Variable importance in projection (VIP) measures were extracted in PLS-DA and calculated as a weighted sum of the squared correlations between the PLS-DA components and the original variable.
Gut microbiome and metabolome data correlation
To integrate the microbiome and the metabolome datasets, a correlation analysis between differential gut taxa and differential metabolites was performed using Spearman’s correlation. Relative taxa abundances and normalized Log10 transformed metabolites were used in the Spearman correlation analysis. A significant association was reported if the association was found to have an absolute Spearman ρ ≥0.50 and p < .05 (unadjusted). In addition, a Spearman’s correlation analysis was conducted between all taxa/metabolites within each group in R (Hmisc package v. 5.1–0). Only bacterial genera and fecal metabolites that had more than 50% non-zero values across participants were included in the analysis. Multiple comparisons were corrected using Bonferroni’s correction [61]. A microbiome-metabolite correlation was included in the results if Bonferroni corrected p < .05 and an absolute Spearman’s ρ of ≥0.85 was found. Significant microbiome-metabolite correlations within each group are presented using circos plots (R package circlize v. 0.4.16) in the Supplemental Material.
Study approval
The study was approved by the NIH Institutional Review Board. All participants provided informed consent and were compensated for their time and participation.
Results
Study population demographic and laboratory characteristics
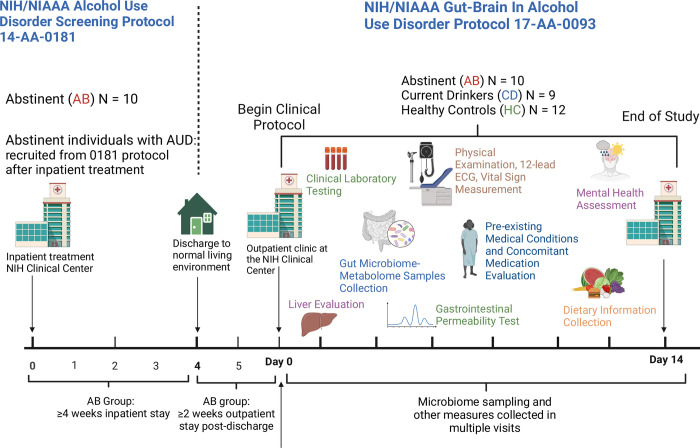
Enrollment and data collection for this study included six outpatient study visits (over 14-days ranging between 6–20 days in total) to the NIH Clinical Center (Fig 1).
Fig 1. Schematic representation of the study design.
Abstinent individuals with AUD (AB) were enrolled in the study after ≥4 weeks of inpatient treatment (NIH/NIAAA treatment protocol 14-AA-0181), followed by ≥2 weeks of “real life” (living their normal life). Non-treatment-seeking, currently drinking individuals with AUD (CD) and matched healthy controls (HC) were also enrolled. Fecal samples from the study participants were collected and processed for gut microbiome and metabolome analysis. Physical examination, 12-lead ECG, vital sign measurements, and laboratory tests were performed. Information on physical and mental health (including information on medical conditions and medications) and dietary intake was gathered and analyzed. Transient liver elastography and gastrointestinal permeability testing were carried out.
Main demographic and laboratory characteristics of the groups are presented in Table 1. Overall, no significant differences in sex, race, ethnicity, age, and BMI were observed among groups. As expected, ABs and CDs had a higher proportion of smokers than HCs, who were all non-smokers (p = .007). Average liver function tests during the study were within normal range and did not differ among groups. Of note, we found higher values of AST (AB: 55.8±49.9, CD:21.8±6.00, HC:22.3±7.03, p = .023) and GGT (AB:134±175, CD:30.8±12.3, HC:22.0±12.8, p = .0032) in the AB group collected under 14-AA-0181 screening protocol, i.e., before the inpatient treatment at the NIH Clinical Center. The inflammatory biomarker c-reactive protein (CRP) and other parameters, including blood cell counts, coagulation factors, lipids, thyroid hormones, and urinalysis were all within normal range and did not differ among groups. At the FibroScan®, no participant had an exclusionary LSM score ≥17.6 kPa. FibroScan® data showed no difference among groups in LSM and CAP.
Table 1. Main characteristics of study population.
| Variable | Abstinent (AB, N = 10) | Current Drinkers (CD, N = 9) | Healthy Controls (HC, N = 12) | Test Statistic | P-value | |
|---|---|---|---|---|---|---|
| Demographic | ||||||
| Sex, N (%) | Female | 2 (20.0) | 2 (22.2) | 4 (33.3) | χ2 = 0.6 | .744 |
| Male | 8 (80.0) | 7 (78.8) | 8 (66.7) | |||
| Race, N (%) | White/Caucasian | 6 (60.0) | 7 (77.8) | 7 (58.3) | χ2 = 3.1 | .546 |
| Black/African American | 3 (3.0) | 2 (22.2) | 5 (41.7) | |||
| Asian | 1 (1.0) | 0 | 0 | |||
| Ethnicity, N (%) | Hispanic/Latino | 0 | 0 | 1 (8.4) | χ2 = 1.6 | .441 |
| Non-Hispanic/Latino | 10 (100.0) | 9 (100.0) | 11 (91.6) | |||
| Age, years, mean (SD) | 45.9 (11.4) | 45.0 (12.6) | 48.8 (12.0) | H = 0.5 | .766 | |
| BMI, kg/m2, mean (SD) | 26.7 (5.1) | 25.7 (4.0) | 28.8 (4.9) | H = 2.4 | .299 | |
| Smoking status, N (%) | Smoker | 6 (60) | 4 (44.4) | 0 | χ2 = 9.8 | .007 |
| Non-Smoker | 4 (40) | 5 (55.5) | 12 (100.0) | |||
| Laboratory mean (SD) | ||||||
| ALT (U/L) | 22.7 (13) | 22.9 (11.4) | 22.7 (10.7) | H = 0.1 | .973 | |
| AST (U/L) | 24.9 (9.6) | 22.3 (6.3) | 22.1 (5.4) | H = 0.3 | .861 | |
| ALP (U/L) | 83.2 (46.9) | 64.6 (12.9) | 70.2 (28.1) | H = 0.4 | .801 | |
| GG (U/L) | 47.6 (72.3) | 36.6 (18.1) | 22 (13.4) | H = 3.8 | .149 | |
| Total bilirubin (mg/dL) | 0.6 (0.3) | 0.5 (0.2) | 0.5 (0.3) | H = 0.4 | .824 | |
| CRP (mg/dL) | 1.4 (0.8) | 2.8 (1.6) | 2.1 (2.8) | H = 0.8 | .654 | |
| Alcohol-related mean (SD) | ||||||
| Number of DSM-5 AUD criteria | 9 (2) | 5.5. | 0 | H = 26.8 | < .001 | |
| AUDIT | 28 (7.7) | 16 (5.6) | 2 (1.6) | H = 24.7 | < .001 | |
| ADS | 22 (9.2) | 10 (6.1) | 0 | H = 25.2 | < .001 | |
| PACS | 7 (5) | 17.5 | 0.5 | H = 20.4 | < .001 | |
| LDH heavy drinking years | 16.4 (9.4) | 12.1 (13.8) | 0 | H = 15.2 | < .001 | |
| CIWA-Ar | 0.9 (1.1) | 0.33 (0.7) | 0 (0.0) | H = 2.3 | .294 | |
| 90-day TLFB average drinks/daya | 13.6 (8.0) | 3.7 (1.2) | 0.3 (0.3) | H = 25.7 | < .001 | |
| 90-day TLFB heavy drinking daysa | 72.6 (23.6) | 66.0 (20.8) | 13.6 (14.3) | H = 20.4 | < .001 | |
| In-study average drinks/dayb | 0 (0) | 3.6 (0.7) | 1.4 (0.6) | H = 9.3 | .007 | |
| Psychological mean (SD) | ||||||
| PSQI | 12 (3.5) | 5 (4.0) | 2.5 (4.0) | H = 16.2 | < .001 | |
| MADRSc | 15 (7.8) | 0.38 (1.1) | 1.3 (1.7) | H = 19.1 | < .001 | |
| BSAc | 11 (6.6) | 0.11 (0.3) | 1.2 (1.7) | H = 21.1 | < .001 | |
| POMS depressiond | 5.3 (6.6) | 3.9 (4.5) | 1.2 (1.7) | H = 7.5 | .023 | |
| POMS anxietyd | 4.3 (3.0) | 3.4 (2.6) | 2.4 (1.8) | H = 2.5 | .292 | |
| FibroScan® mean (SD) | ||||||
| LSMe | 4.4 (1.5) | 4.5 (1.2) | 4.0 (0.9) | H = 0.9 | .624 | |
| CAPe | 229.5 (79.7) | 220.1 (80.4) | 244.3 (69.7) | H = 0.2 | .916 | |
| Medical conditions and medications mean (SD) | ||||||
| Number of conditions | 4.0 (1.8) | 1.3 (1.2) | 1.6 (1.3) | H = 11.1 | .004 | |
| Number of medications | 2.2 (1.6) | 0.7 (1.4) | 0.7 (1.1) | H = 7.2 | .028 | |
Significant p-values (Benjamini-Hochberg corrected) in bold. Abbreviations: ADS = Alcohol Dependence Scale; AUDIT = Alcohol Use Disorders Identification Test; BMI = body mass index; BSA = Brief Scale for Anxiety; CAP = Controlled Attenuation Parameter; CIWA-Ar = Clinical Institute Withdrawal Assessment of Alcohol Scale, Revised; LDH = Lifetime Drinking History; LSM = Liver Stiffness Measurement; MADRS = Montgomery-Åsberg Depression Rating Scale; PACS = Penn Alcohol Craving Scale; POMS = Profile of Mood States; PSQI = Pittsburgh Sleep Quality Index; SD = Standard Deviation; TLFB = Timeline FollowBack.
aPrior 90-day TLFB reflects 90 days prior to inpatient admission for ABs and 90 days prior to study start for CDs and HCs.
bIn-study TLFB reflects days of study participation for the groups.
cMADRS and BSA were administered at screening.
dPOMS depression and POMS anxiety scores are mean scores of the first and last study visits.
eLSM and CAP scores correspond to the median values of 10 valid measurements. Counts and percentages (%) are shown for categorical variables; means/standard deviations (SDs) are shown for continuous variables. Chi-squared (χ2) test was used for categorical variables and Kruskal-Wallis (H) test was used for continuous variables. Normal range of alanine transferase (ALT) is 23–69 U/L, of aspartate transferase (AST) is 19–62 U/L, of alkaline phosphatase (ALP) is 0–555 U/L, of gamma-glutamyl transferase (GGT) is 5–40 U/L of total bilirubin is 0.1–1.2 mg/dL of c-reactive protein (CRP) is 0.3–1.0 mg/dL.
Alcohol-related clinical and psychological characteristics
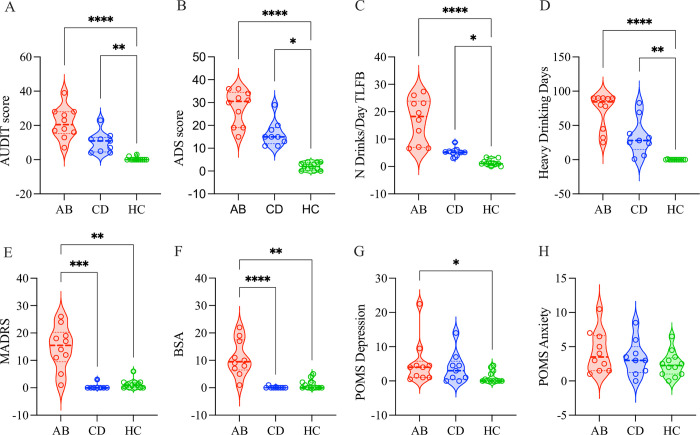
Average AUDIT scores were different among groups (p < .001) and between groups (AB vs. HC, p < .001; CD vs. HC, p = .012) (Fig 2A). ADS scores were different among (p < .001) and between groups (AB vs. HC, p = .001; CD vs. HC, p < .009) (Fig 2B). Per prior 90-day TLFB, ABs and CDs reported more average drinks per day than HCs (p < .001 and p = .019, respectively, Fig 2C), as well as more heavy drinking days than HCs (p < .001 and p < .007, respectively, Fig 2D). Average drinks per day and heavy drinking days between ABs and CDs were not significant (p = .074 and p = .285, respectively), however in both cases, the averages were higher for ABs. Depression (MADRS), anxiety (BSA) and sleep quality (PSQI) were evaluated across all individuals at screening, and all were different among groups (p’s < .001) (Fig 2E and 2F); ABs had higher depression than CDs and HCs (p < .001 and p = .003, respectively) and higher anxiety than CDs and HCs (p < .001 and p < .001, respectively). Interestingly, CDs and HCs did not differ in depression and anxiety. POMS depression scores were different among groups (p < .023) and the only significant difference was between ABs and HCs (p < .001) (Fig 2G). POMS anxiety scores were higher but not significant among groups (Fig 2H).
Fig 2. AUD severity and mental health status.
(A) AUDIT scores were significantly different among groups (p < .001) and between groups (AB vs. HC, p < .001; CD vs. HC, p = .012). (B) ADS scores were significantly different among (p < .001) and between groups (AB vs. HC, p = .001; CD vs. HC, p < .009). (C) Prior 90-day TLFB average drinks per day were significantly different among (p < .001) and between groups (AB vs. HC, p < .001; CD vs. HC, p = .019). (D) Prior 90-day TLFB heavy drinking days were significantly different among (p < .001) and between groups (AB vs. HC, p < .001; CD vs. HC p = .007). (E) MADRS scores were significantly different among (p < .001) and between groups (AB vs. HC, p < .001; AB vs. CD p = .003). (F) BSA scores were significantly different among (p < .001) and between groups (AB vs. HC, p < .001; AB vs. CD, p < .001). (G) POMS depression scores were significantly different among (p < .023) and between groups (AB vs. HC, p < .001). (H) POMS anxiety scores did not differ significantly among and between groups.
Gut microbiome across groups
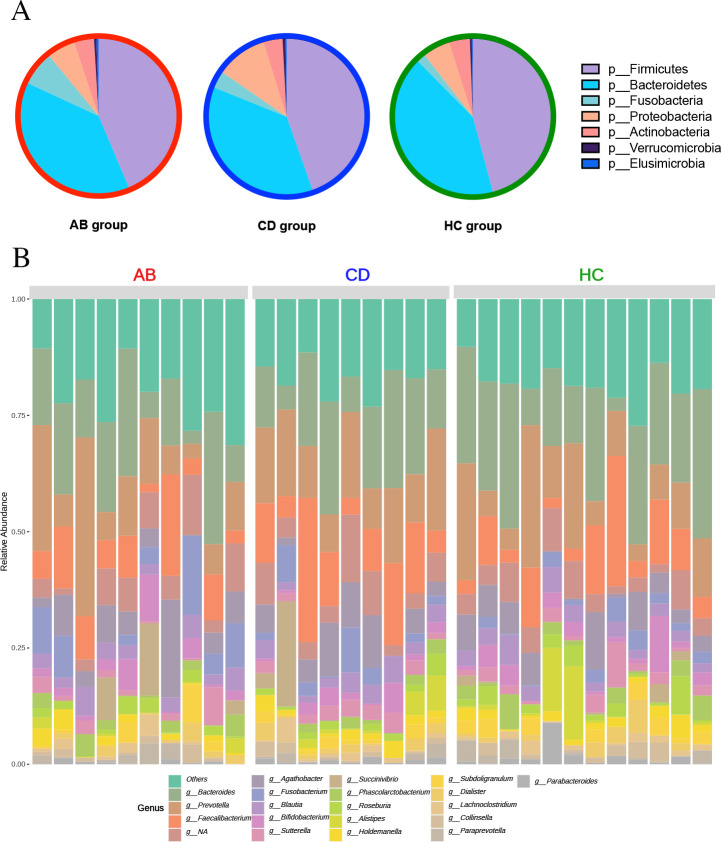
The overall distribution of the phyla across the groups showed that Firmicutes was most abundant with Bacteroides being second most dominant. Actinobacteria represented 6.19%, 2.56% and 3.34% in ABs, CDs, and HCs, respectively, and Proteobacteria represented 5.74%, 10.77%, and 8.14%, respectively (Fig 3A). At the genus level, Bacteroides was the most abundant genus in all groups, with a relative abundance of 21.30% in ABs, 17.87% in CDs, and 23.61% in HCs. Faecalibacterium and Agathobacter were second and third most dominant genera. (Fig 3B).
Fig 3. Gut microbiome of study participants.
(A) Phylum-pie chart across entire study population. Legend shows top seven most abundant phyla. (B) Relative abundance bar chart at genus level across all participants. Legend shows the top 20 most abundant genera.
Gut microbial alpha- and beta-diversity
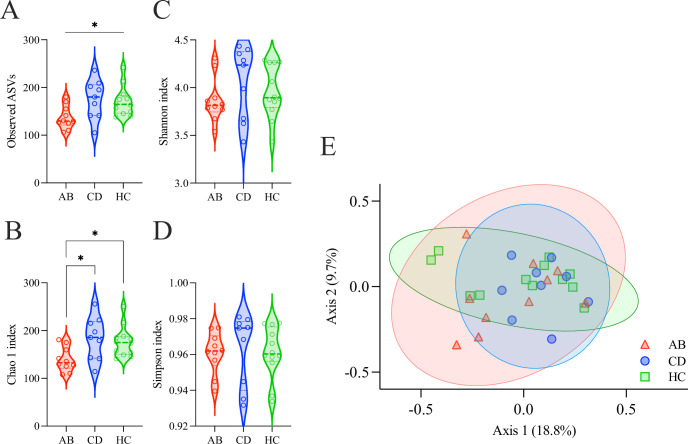
In total, 986 ASVs (618,267 reads) were identified across the 31 study participants (19,944 average reads/participant). Alpha diversity measures Chao1, Shannon, Simpson, and observed ASVs indexes were calculated on the full set of 986 ASVs. Average alpha diversity measures showed only species richness (Observed ASVs and Chao1) exhibiting significant differences among groups (Fig 4A–4D and Table 2).
Fig 4. Alpha- and beta-diversity of the gut microbiome.
(A) Alpha-diversity, calculated with Observed ASVs richness, differed significantly among ABs, CDs, and HCs (p = .026). (B) Alpha-diversity, calculated with Chao1, differed significantly among groups (p = .012). (C) Shannon diversity index did not differ significantly among groups (p = .376). (D) Simpson diversity index did not differ significantly among groups (p = .626). (E) Principal coordinates analysis (PCoA) plot comparing gut microbial beta-diversity (Bray-Curtis dissimilarity) among ABs, CDs and HCs, exhibited no separation among groups.
Table 2. Alpha diversity measures.
| Alpha-Diversity Indexes | Abstinent (AB, N = 10) | Current Drinkers (CD, N = 9) | Healthy Controls (HC, N = 12) | Kruskal-Wallis | P-value |
|---|---|---|---|---|---|
| Observed ASVs | 137 (24.32) | 175 (40.81) | 171 (30.95) | H = 7.263 | .026 |
| Chao1 | 139 (25.22) | 184 (44.88) | 177 (31.90) | H = 8.835 | .012 |
| Shannon | 3.87 (0.24) | 4.05 (0.38) | 3.96 (0.28) | H = 1.954 | .376 |
| Simpson | 0.96 (0.01) | 0.963 (0.02) | 0.961 (0.01) | H = 0.936 | .626 |
Means and standard deviations (SDs) are shown for each alpha-diversity index. Kruskal-Wallis (H) test was used to compare the median of each index among the three study groups, i.e., abstinent (AB) current drinkers (CD), and healthy controls (HC). p < .05 shown in bold. Abbreviations: ASV = Amplicon Sequence Variant; SD = Standard Deviation.
There were no significant beta diversity differences observed among groups using Principal Coordinate of Analysis (PCoA) (Fig 4E, F-value = 0.911; R-squared = 0.061; p = .619). Alpha diversity measures (Observed, Chao1, Simpson, and Shannon) were correlated with alcohol severity scores (AUDIT and ADS) within each group and no significant associations were observed.
Group wise genus-level differential abundance
The 986 ASVs were filtered, removing low abundance and low variance features, resulting in 203 ASVs that classified to 71 genera. Differential abundance was assessed between groups, i.e., AB vs. CD, AB vs. HC, and CD vs. HC using three testing procedures: LEfSe, MaAsLin2, and heat tree analysis (Wilcoxon rank sum test).
Twelve taxa were found to be significantly different between groups in at least one test (no adjustment). Taxa relative abundances and fold changes of the pairwise comparisons are shown in Table 3.
Table 3. Log2 fold change of 12 genera found to be significantly different between groups.
| Relative Abundance (%) Mean (SD) |
Log2 Fold Change | |||||
|---|---|---|---|---|---|---|
| Taxa | AB | CD | HC | AB vs.CD | AB vs. HC | CD vs. HC |
| Akkermansia | 0.936 | 1.084 | 1.297 | -3.8 ⱡ↓ | -2.19↓ | -1.61↓ |
| Candidatus Stoquefichus | 0.169 | 0.001 | 0.049 | 3.02 * ↑ | 1.34 ↑ | 1.69↑ |
| Dorea | 0.802 | 2.150 | 0.897 | -1.35¥↓ | 0.265↑ | -1.61¥↓ |
| Eisenbergiella | 0.147 | 0.012 | 0.027 | 1.79 * ↑ | 1.27↑ | 0.515↑ |
| Fusicatenibacter | 1.194 | 1.971 | 2.499 | -3.77¥↓ | -3.92 * ↓ | 0.152↑ |
| Lachnospira | 0.560 | 0.719 | 1.658 | -1.44↓ | -1.95 * ↓ | 0.51 ⱡ↑ |
| Lachnospiraceae_FCS020_group | 0.003 | 0.016 | 0.059 | -0.47↓ | -1.27↓ | 0.798↑ |
| Lachnospiraceae_UCG_001 | 0.000 | 0.065 | 0.093 | -0.613↓ | -1.32 * ↓ | 0.704↑ |
| Roseburia | 0.841 | 1.386 | 3.961 | -1.66↓ | -2.29 * ↓ | 0.632↑ |
| Ruminococcaceae_UCG_014 | 0.100 | 0.362 | 1.198 | -2.09↓ | -2.48¥↓ | 0.39↑ |
| Streptococcus | 0.614 | 0.140 | 0.129 | 2.44 * ↑ | 2.63 * ↑ | -0.193↓ |
| Victivallis | 0.008 | 0.199 | 0.108 | -4.39↓ | -1.66↓ | -2.73 ↓ |
*p < .05 on all 3 tests
ⱡ p < .05 only on 2 tests
¥ p < .05 only on 1 test. ↑ and ↓ indicates minuend direction of change.
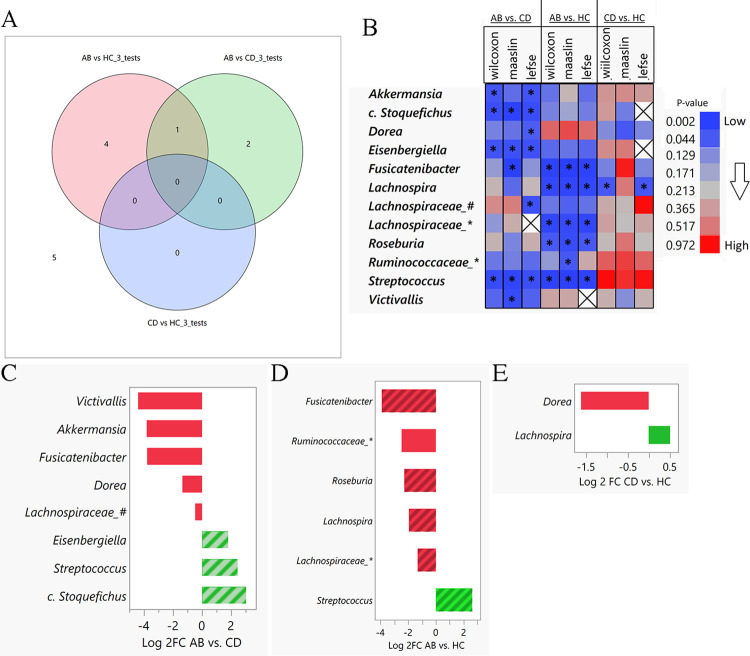
Seven taxa (in bold in Table 3) were found to be significantly different (p < .05) in at least one of the two group comparisons by all three tests. The overlap between the three pairwise comparisons of the seven taxa was assessed using a Venn Diagram analysis (Fig 5A). In addition, a color map showing p-values for all tests and group comparisons is shown in Fig 5B.
Fig 5. Differentially abundant taxa among groups.
(A) Venn Diagram overlap comparing significant genera by all three tests (Wilcoxon, MaAsLin2, LEfSe) across each comparison. (B) Heat map of p-values for 12 significant genera (y-axis) in any of three testing procedures and group wise comparisons (x-axis). (* indicates p < .05 for comparison and test; X indicates that the genus was not assessed using LEfSe test procedure due to filtering). The color gradient shows the exact p-value to which the color corresponds to. Bar plot of (C) AB vs. CD, (D) AB vs. HC and (E) CD vs. HC Log2 fold change (x-axis) of genera (y-axis) found to be statistically different in comparison of interest. Shaded boxes indicate significant genus in all three test procedures. Neither taxon was significant by all three tests for CD vs. HC comparison. (Abbreviations: c. Stoquefichus = Candidatus_Stoquefichus; Ruminococcaceae_* = Ruminococcaceae_UCG_014; Lachnospiraceae_# = Lachnospiraceae_FCS020_group; Lachnospiraceae_* = Lachnospiraceae_UCG_001).
Eight genera were found to be different in AB vs. CD groups by any test. Three of those genera were significantly different and more abundant in AB in all tests: Streptococcus, Eisenbergiella, and candidatus Stoquefichus (genus candidate) (Fig 5C). Five genera were found to be different in ABs vs. HCs in all tests: Streptococcus, Fusicatenibacter, Lachnospira, Lachnospiraceae_UCG_001, and Roseburia. (Fig 5D). In CD vs. HC group comparison, two genera were found significant in at least one test (Fig 5E).
Results from each individual testing procedure are presented in S1–S3 Figs. Briefly, eight taxa were found to be significantly different using the Wilcoxon Sign Rank Sum test procedure (p<0.05) (S1A Fig and S2 Table) and the Venn Diagram overlap analysis is shown in S1B Fig. Heat trees are shown in S1C–S1E Fig (S3C–S3E Table). Results for LEfSe showed 10 genera to be statistically significantly different among any of the three comparisons p < .05. Each LDA effect score is plotted in S2A–S2C Fig for each group wise comparison. When considering group comparisons made using MaAsLin2, 10 significant taxa were found, and covariate plots were created to show the results from each test before and after covariate correction (S3 Fig).
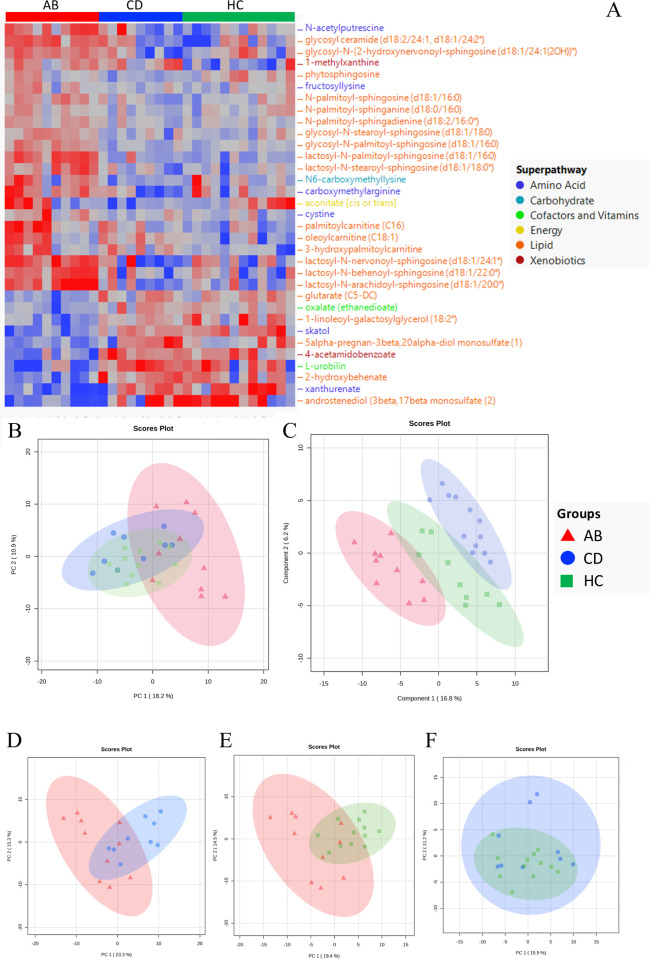
Gut untargeted metabolomics
Untargeted metabolomic analysis identified 1032 metabolites. After excluding features with 50% or more missing values, a final set of 946 metabolites was analyzed and 33 differentially abundant metabolites were found among groups (Fig 6A and S4 Table). The top 30% differentially most abundant metabolites are shown in S4A Fig. Most of them (21/33) were lipids: Five fatty acid metabolites, five lactosylceramides, four hexosylceramides, and four ceramides, together with two steroids and one metabolite ascribed to sphingolipids synthesis. Different amino acids across groups include two biochemicals involved in tryptophan metabolism, one ascribed to lysine metabolism, one to cysteine metabolism, one to arginine metabolism, and one to polyamine metabolism. Most metabolites were more abundant in the AB individuals. Using PCA, AB group clustered separately from CD and HC groups (Fig 6B). With PLS-DA (Fig 6C), while observing a relative separation among groups, we found that the AB group showed the highest and lowest metabolite values (S4B Fig). Most metabolite differences were due to the AB group, with 28/33 (85%) metabolites significantly different in both the AB vs. CD and AB vs. HC comparisons. Pairwise comparisons (Fig 6D–6F and S4 Table) showed that amino acids belonging to tryptophan metabolism, including skatol and xanthurenate, were significantly less abundant in AB group compared to CD (p = .002 and p = 004, respectively) and HC (p < .001 and p < .001) groups. Biochemicals involved in lysine, cysteine, and arginine metabolism were more abundant in AB group than CD and HC groups, namely fructosyllysine (ABs vs. CDs, p < .001; ABs vs. HCs, p = .001), cystine (AB vs. CD, p = .002; AB vs. HC, p < .001), and carboxymethylarginine (ABs vs. CDs, p < .001; ABs vs. HCs p = .006). Differently, the two cofactors oxalate and l-urobilin, along with the xenobiotic 4-acetamidobenzoate, were less abundant among ABs than in CD and HC. Lastly, a xenobiotic belonging to xanthine metabolism (1-methylxanthine) and one carbohydrate (N6-carboxymethyllysine) were more abundant among AB individuals in both comparisons. Five metabolites exhibited significant differences in CDs vs. HCs comparison.
Fig 6. Metabolome differences among groups.
(A) Hierarchical clustering of the 33 significantly different metabolites colored by super pathway among groups (p < .05, FDR corrected, Kruskal-Wallis test). Unsupervised (B) and supervised (C) ordination methods show separation between AB group vs. CD and HC groups (PCA), and AB, CD, and HC groups (PLSDA) respectively. The quality of the PLS-DA model was assessed by its ability to predict variance of the data with four components (Q2 = 0.575). Pairwise comparisons were observed by PCA in AB vs. CD (D), AB vs. HC (E), and CD vs. HC (F) comparisons. (AB: red, CD: blue, HC: green).
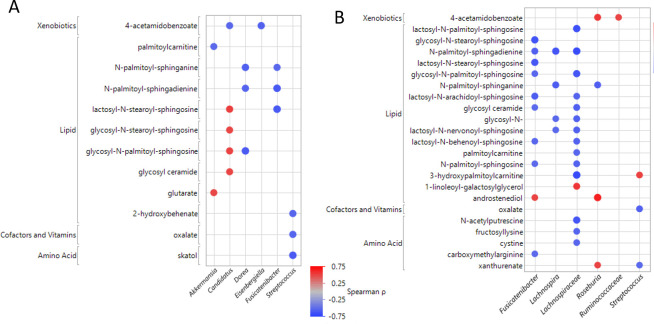
Gut microbiome and metabolome associations
Differential features identified by fecal microbiome and metabolome analyses were correlated from pairwise comparisons (AB vs. CD, AB vs. HC, and CD vs. HC). When considering only those eight gut taxa found to be different between AB vs. CD groups from any test previously described and the 30 metabolites from this comparison, 17 significant correlations were identified. Six taxa were found to be significantly correlated with 12 metabolites (Fig 7A). Streptococcus negatively correlated with three metabolites from the vitamin/cofactors and amino acids while many associations were with metabolites derived from the lipid super pathway. There were two taxa showing positive associations with lipids: Akkermansia and candidatus Stoquefichus.
Fig 7. Associations between differential gut microbes and metabolites.
(A) Differential AB vs. CD metabolites compared to microbes using Spearman correlation. (B) Differential AB vs. HC metabolites compared to microbes using Spearman correlation. Blue dot indicates negative correlation, red dot indicated positive correlation.
Likewise, in the AB vs. HC comparison, six taxa were correlated to 31 metabolites. This analysis revealed 37 significant associations (Fig 7B). The Lachnospiraceae_UCG_001 genus showed associations with 15 metabolites and was negatively correlated with many lipid-related metabolites and some metabolites related to amino acid and vitamin/cofactor pathways. Fusicatenibacter genus was associated with ten metabolites from the lipid super pathway. There were seven microbiome/metabolite pairs that were positively associated and three of those were with the Roseburia genus. We didn’t any significant correlation when investigating CDs vs. HCs. In addition, we performed a multivariate analysis comparing all gut taxa to all gut metabolites in order to understand the relationships within each group (S3 Table). All by all Spearman correlations were computed within each group and a total of 149 significant correlation pairs were found within ABs (S5A Fig), 258 were found within the CD group (S5B Fig), and 33 within the HC group (S5C Fig).
Other ancillary collected measures
No significant differences were observed in dietary intake across groups in total calorie intake (kcal) nor in percent calories from carbohydrate, protein, or fat (S5 Table). As anticipated, calories from alcohol were different among groups (p‘s < .001) with the CD individuals consuming the most.
The number of self-reported pre-existing medical conditions and concomitant medications was significantly different among the three groups (p = .004 and p = .028, respectively). Overall, ABs reported significantly more medical conditions and medications than CDs and HCs. AB individuals reported a total of 40 medical conditions, with a 4±1.8 per subject, while CD group reported 12 conditions (1.3±1.2 per subject) and HC group 19 conditions (1.6±1.3 per subject), adding up to 71 conditions reported (Table 1 and S6A Fig). These medical conditions were cross-checked with the list of diseases reported by Jackson et al., 37/71 (52.1%) conditions reported by our sample were found to be in common with the list; 21/40 (52.5%) conditions reported by ABs, 4/12 (33.3%) by CDs, and 12/19 (63.1%) by HCs overlapped with those listed by Jackson et al. (S6B Fig). ABs reported taking 22 medications (2.2±1.6 per participant), while CDs reported 6 (0.7±1.4 per participant) and HCs 8 medications (0.7±1.1 per participant), adding up to 36 medications taken by all (Table 1 and S7A Fig). When these medications were cross-checked with the list of medications by Jackson et al., 17/36 (47.2%) medications taken by our sample were found to be in common (11/22 (50%) by ABs, 3/6 (50%) by CDs, and 3/8 (37.5%) by HCs) overlapped (S7B Fig). A total of 37 diseases and 17 medications reported by the study participants across the AB, CD, and HC groups matched those found by Jackson et al. to have a significant association with a gut microbiome marker.
The gastrointestinal permeability assays showed inconsistencies in the participants’ fasting status of 16 participants; therefore, no conclusions were made using these data. However, sugar excretion curves over the sampling times (sucrose: S8A Fig, sucralose: S8B Fig and lactulose/mannitol ratio: S8C Fig) were plotted for the 15 participants who did not have sugar detected at the baseline samples.
Discussion
This study reports the results of a case-control clinical investigation in an outpatient setting aimed at comparing the gut microbiome and metabolome among three groups of individuals: individuals with AUD who were treatment-seeking but newly abstinent (ABs), non-treatment-seeking individuals who were currently drinking (CDs), and healthy controls (HCs). All samples and measures were collected over approximately two weeks at the National Institutes of Health Clinical Center. In addition to fecal sample collection, other measures including addiction- and psychopathology-related phenotyping was carried out. The overall findings show that the healthy controls and the current drinkers did not differ much in the gut microbiome and metabolome features but that the abstinent individuals differed the most. Furthermore, the clinical phenotypic characterization showed that AB individuals had greater sleep disruption, poorer mental health status, and a higher number of medical comorbidities and concomitant medications. The greater differences observed in the AB group could be attributed to some biological processes going on in early abstinence after cessation of chronic alcohol use along with more severe AUD and comorbidities, a known characteristic of treatment-seeking (compared to nontreatment-seeking) individuals with AUD. Finally, while this experimental case-control study brings light to these facts, it is not possible to disentangle these two confounds, which could be contributing to the difference in AB individuals. Our findings are in contrast with other studies showing that individuals with AUD who abstain from alcohol may present with partial recovery of the gut microbiome [23, 32]. However, there are not enough studies nor enough studies with large sample sizes to conclude on the reversibility, or irreversibility, of alcohol-induced changes to the gut microbiota. Of note, other studies did not replicate these findings and found little or no changes in the gut microbiome after abstinence [7, 8, 10]. It bears mentioning that inpatient studies allow for a well-controlled, experimentally rigorous setting, but they are unable to control for other confounders such as changes in diet, general lifestyle habits and changes due to acute alcohol withdrawal. Additionally, inpatient studies allow for a well-controlled experimentally rigorous setting, but they are unable to control for other confounders such as changes in diet and general lifestyle habits and changes due to acute alcohol withdrawal. This current study provided an exemplary opportunity to capture gut microbiome samples in an outpatient setting allowing the participants to continue their normal daily lifestyle choices.
Gut microbiome differences
The gut microbial diversity of observed ASVs and Chao1 indexes was significantly lower in the AB group compared to CD and HC groups; however, other diversity measures considering taxa relative abundance (i.e., Simpson and Shannon indexes) were not statistically different. In a study on the effects of moderate levels of alcohol consumption on the gut microbiome in both rats and humans, alcohol-treated rats had significantly lower species richness compared. Still, a human study looking at alcohol consumers did not display a similar alteration [62]. Firmicutes and Bacteroidetes phyla are dominant in human fecal samples, and previous animal and human studies point to increased Firmicutes following chronic heavy alcohol use [15–17, 27]. Taxonomic differences between groups were explored using a three-test procedure, finding seven taxa that passed all tests at a p < .05 for any comparison. Specifically, ABs presented a depletion of the beneficial genera Akkermansia, Lachnospira, Roseburia, Fusicatenibacter, and Lachnospiraceae_UCG_001 and an increase in Eisenbergiella, Streptococcus, and candidatus Stoquefichus. These genera belong to the Lachnospiraceae family which have been implicated as part of the core of the gut microbiota, colonizing the gut lumen from birth. All members of this family are anaerobic, fermentative, and chemoorganotrophic, and some display strong hydrolyzing activities, for example through the activity of pectin methyl-esterase, α- and β-galactosidase, α- and β-glucosidase, and α-amylase [63]. Lachnospiraceae family plays a crucial role in the metabolism of carbohydrates, being able to utilize diet-derived polysaccharides, including starch, inulin, and arabinoxylan, to produce butyrate and other short-chain fatty acids (SCFAs). SCAFs have known, profound, beneficial effects on human health [64, 65]. In a previous study, Eisenbergiella has been shown to decrease after chronic alcohol drinking [66], which is discordant with what was found in this study, being more abundant in the AB group compared to CD and HC groups. Similarly, regarding the members of the Lachnospiraceae family, Fusicatenibacter, shown to be depleted in ABs compared to both CDs and HCs in this study, is an anaerobic, fermentative genus that supplies nutrients and energy to the host. Among the ABs, genera Streptococcus and candidatus Stoquefichus genera were more represented. Members of the Streptococcus genus are generally considered harmful and are common pathogens responsible for bacterial infections in individuals with AUD, especially if complicated by ALD [67]. Candidatus Stoquefichus, a-yet uncultured genus, has been found to be increased in the gut microbiota of a mouse model of obesity [68].
Gut metabolomic differences
Fecal metabolic profiling of the study participants mirrored the results of the gut microbiome analysis. Both univariate and multivariate analyses showed the ABs’ fecal metabolome to be different from CDs and HCs. The 33 metabolites found to be significantly different among groups were primarily from the lipid and amino acid pathways. AB individuals had a lower abundance of tryptophan metabolites, such as skatol and xanthurenate. Tryptophan is an essential amino acid for protein synthesis that has emerged as a key player in the gut-brain axis. It is the only precursor for serotonin, which acts as a neurotransmitter in both the central and the enteric nervous systems [69, 70]. Additionally, tryptophan and its metabolites have a key role in maintaining gut-brain homeostasis; dysregulation of tryptophan metabolites plays a central role in the pathogenesis of many neuropsychiatric disorders, including AUD [71]. Gut bacteria influence tryptophan metabolism directly and indirectly, with corresponding changes in behavior and cognition. AB group, compared to CD and HC groups, had a higher abundance of metabolites of the lipid pathway, including lactosyl-N-palmitoyl-sphingosine, glycosyl-N-palmitoyl-sphingosine, and lactosyl-N-arachidoyl-sphingosine. Ingested triacylglycerides can be sequentially broken down to diacylglycerols (DAG), and then monoacylglycerols (MAG) [9] by the activity of secreted pancreatic lipases. The free fatty acids (FFAs) generated from each step, as well as the MAGs, can then be taken up by the epithelial cells of the intestinal lumen and repackaged into chylomicrons for distribution throughout the body (where at the tissue level they are broken down again to free fatty acids) [72]. As such, differences in DAGs, MAGs, and FFAs may indicate a difference in the action of lipases (bile secretion) and/or uptake of lipids. Lipids show an increase in various liver diseases, including alcohol-associated steatosis [73]. Given the known role of tryptophan and lipids in depression and other mental health disorders, we suggest that the differences in the gut metabolome observed in this study may be more reflective of the mental health comorbidities present among the AB group.
Associations of the gut microbiome and metabolome
A correlation analysis to investigate the functional associations between differential gut taxa and metabolites was conducted. In the pairwise comparisons we found that Streptococcus genus, differentially more abundant among the ABs and found in higher abundance in individuals with AUD in other studies [10, 74, 75], negatively correlated with two tryptophan metabolism biochemicals, skatol and xanthurenate (ABs vs. CDs and AB vs. HCs comparisons, respectively). Conversely, Roseburia genus (less abundant in ABs when compared to HCs) and known SCFAs producer [76], was positively associated with xanthurenate metabolite. It has been showed that tryptophan metabolism pathways in the gut are differentially affected in diseases but remains interconnected and that these pathways are controlled by the gut microbiota [77]. The role played by Streptococcus and Roseburia genera still needs to be elucidated. Candidatus Stoquefichus (found in higher abundance among the ABs) exhibit positive correlations with all the hexosylceramides (more abundant in AB group compared to CD and HC groups). These metabolites are intermediates in sphingolipid metabolism that are involved in cell proliferation and differentiation, cell senescence, and apoptosis. Alteration of sphingolipid pathways contributed to alcohol-associated liver diseases [78, 79]. The differentially abundant ceramides were negatively associated with known SCAFs producers [80] Fusicatenibacter genus (AB vs. HC and AB vs. CD comparisons), Dorea (AB vs. CD comparison), Roseburia, Lachnospira and Lachnospiraceae UCG 001 genera (AB vs. HC comparison). While this study does not allow to infer potential causal roles of these functional associations, future studies should investigate whether these gut microbiome-metabolome relationships may play a role in alcohol drinking and AUD.
Clinical characteristics and differences across groups
There were no differences observed in demographic characteristics, blood marker liver tests, or in the FibroScan® evaluation in this study. Noteworthy, the liver markers AST and GGT were higher in the AB group at screening but not during the study (after the inpatient treatment period), showing how even short periods of abstinence may improve liver tests in people with AUD. When other clinical data was investigated, the AB individuals showed more severe traits related to AUD and psychological and mental health comorbidities than CDs and HCs. In fact, AB individuals had greater severity of AUD and significantly higher alcohol intake (pre-abstinence). Additionally, the AB group presented higher levels of depression and anxiety and poorer quality of sleep. These findings may partly explain the greater disruption of the gut microbiome and metabolome in AB individuals compared to CDs and HCs. The fact that the AB group (which were treatment-seekers) had clinically more severe characteristics, conditions, and medical and mental health comorbidities than CDs (non-treatment seekers) is consistent with previous studies examining treatment seeking and non-treatment seeking individuals with AUD [81–84]. Other potential explanations for the different gut microbiome and metabolome found among AB individuals may be attributed to the mental health differences observed. Previous studies have shown perturbations of the gut microbiome in multiple psychiatric disorders [4, 85, 86] as well as sleep disturbances [87]. A recent review and meta-analysis of 34 studies on the gut microbiome of individuals affected by various psychiatric disorders revealed a significant decrease in microbial community richness compared with healthy controls. It also showed no significant differences in diversity Shannon and Simpson indexes, in line with our findings. A transdiagnostic pattern of gut microbiota signatures was found: Depleted levels of Faecalibacterium and Coprococcus genera and enriched levels of Eggerthella genus were shared between major depressive disorder and anxiety, suggesting that these disorders are characterized by a reduction of anti-inflammatory, SCFA-producing bacteria, while pro-inflammatory ones are enriched.
Consistent with the conclusions above, AB group reported being affected by significantly more medical conditions and therefore taking significantly more medications than the other groups. Many of these conditions and medications overlapped with those that were found in the Jackson et al.’s study [14] to be associated with dysbiosis. These findings imply that gut microbiome and metabolome differences observed in AB individuals compared to CDs and HCs may therefore be due to a higher prevalence of diseases and a higher intake of medications in the AB group. This limits the ability to translate gut microbiome and metabolome findings from animal models to individuals with AUD, unless these variables are controlled for which was difficult to implement in this study due to the small sample size of the groups included.
Dietary intake patterns across groups
Dietary intake showed no differences among groups during the two weeks of dietary intake assessment, other than in total alcohol intake, as expected. Some large population-wide studies have demonstrated that non-alcohol energy intake increases or remains unchanged with light and moderate drinking but decreases with heavy alcohol consumption to compensate for calories from alcohol [88]. Additionally, previous research has linked higher alcohol consumption with lower carbohydrate intake [89]. However, in this study, CD individuals consumed 14.4% of their energy from alcohol, whereas prior literature reports that the upper third of drinking Americans have an average of 20% of energy intake from alcohol [90]; this discrepancy could be due to CD group not having severe AUD than other studies. In addition to this, CDs may have underreported alcohol intake and other dietary intake variables. Furthermore, few studies have explored diet quality in abstinent and currently drinking individuals with AUD and more research needs to be conducted in this area to be able to better understand these findings.
This study presents a comprehensive gut microbiome and metabolome analysis, associated with a deep phenotyping of clinical and experimental measures of the three cohorts. The statistical strength of the study was limited due to the small groups’ sample size, and the lack of a pre-abstinence sampling timepoint preventing any longitudinal analyses. Additionally, the HC group was slightly overweight and reported some medication and pre-existing conditions which could have further confounded the comparisons. Additional long-term longitudinal outpatient studies are needed for better understanding of how prolonged abstinence (and lack of) may affect the gut microbial community.
Conclusions
In conclusion, the present case-control study provides novel data on the gut microbiome and metabolome, along with a deep phenotyping characterization, in people with AUD from both newly abstinent treatment-seeking, non-treatment-seeking current drinkers, and matched controls. These individuals were investigated in their “real world” life across multiple outpatient visits, an important strength that gives more translational value to the findings described. Of note, however, our hypothesis was not proven. That is, we hypothesized gut microbiome/metabolome similarities between healthy controls and newly abstinent individuals with AUD, while the CD group was going to be different than the other two groups. By contrast, it was the AB group to separate itself, from a gut microbiome/metabolome standpoint, from the other two groups. The deep clinical phenotyping we conducted in this study allowed us to describe how the AB group had a more severe AUD history and more medical and psychiatric comorbidities. Therefore, all results considered, we speculate that the microbiome-metabolome differences here observed reflect mental health-related transdiagnostic traits and their sequelae (including more comorbidities and related medications) rather than an AUD-specific phenotype.
Supporting information
(XLSX)
(DOCX)
(DOCX)
Pairwise comparison at the genus level between each group using Wilcoxon Test. (A) Dot plot of 9 genera (y-axis) found to be significant (p < .05, unadjusted) in pairwise comparison (x-axis). (B) Venn Diagram showing genera found to be significant that overlap between 3 pairwise tests. (C-E) The heat tree analysis leverages the hierarchical structure of taxonomic classifications to quantitatively (using the mean abundance) and statistically (using the non-parametric Wilcoxon Rank Sum test with p < .05) depict taxonomic differences between microbial communities. Red boxes show the end of the leaf (genus) in the heat tree for each plot. Taxa colored in red are more abundant in the AB (C, D) and CD (E) groups, taxa colored in blue are more abundant in the HC (C, E) and CD (D) groups. AB: Abstinent, CD: current drinkers, HC: healthy controls.
(TIF)
LEfSe LDA bar plots at the genus level of fecal microbial samples displaying the pairwise LDA scores between the three groups; abstinent group (AB), continuous alcohol drinking group (CD), and the healthy control group (HC). The bars represent the effect size (LDA) for a particular genus in a certain group. The length of the bar represents a log10 transformed LDA score. The heat maps show the LDA effect size from blue (negative) to red (positive). Shaded bars with an ‘*’ are genera that were found to be significant in the pairwise test (p < .05). (A) represents the pairwise comparison of AB to CD; (B) represents the pairwise comparison of AB to HC; and (C) represents the pairwise comparison of CD to HC. AB: Abstinent, CD: current drinkers, HC: healthy controls.
(TIF)
Results of covariates adjustment (y axis) and without adjustment (x axis). P values for the comparisons between (A) AB and CD, (B) AB and HC, and (C) CD and HC groups did not improve with covariate adjustment (BMI and age), resulting in no significantly different taxa. AB: Abstinent, CD: current drinkers, HC: healthy controls.
(TIF)
Variable importance in prediction (VIP) scores extracted by PLS-DA of metabolites and heatmap of the average scaled expression values of the indicated VIP metabolites in all samples. The top 30 metabolites driving differences between group are represented (lowest VIP score = 2.261). Metabolites in AB group samples show the lowest (in most cases) or the highest abundance compared to HC and CD groups. AB: Abstinent, CD: current drinkers, HC: healthy controls.
(TIF)
The correlations were computed within each group and a total of 149 significant correlation pairs were found within the ABs (A), 258 were found within the CDs (B), and 33 within the HCs (C). Most of the taxa within the AB group found to significantly correlate with metabolites, were represented in the CDs as well. Specifically, across the three comparisons, a high number of correlations were found in members from Lachnospiraceae and Oscillospiraceae families (Bacillota phylum): 62% in the CD group, 47% in the AB group, and 66% in the HC group. Streptococcus genus was found to be significantly more abundant in the AB group when compared to CD and HC groups. In the CD group Streptococcus genus was positively correlated with glyerolphosphoethanolamine (r = 0.950, p < .001), glycerophosphoserine (r = 0.917, p < .001), mannose (r = 0.917, p < .001), deoxycarnitine (r = 0.867, p = .002), and N,1, acetylspermidine (r = 0.850, p = .004). Conversely, in the same group we found negative correlations of Streptococcus with heptenedioate (C7:1-DC) (r = -0.865, p = .003), 2’-O-methylcytidine (r = -0.865, p = .003), and hexadecanedioate, C16 (r = -0.850, p = .004). In the AB group, Streptococcus genus correlated negatively with 7-ketocholesterol (r = -0.952, p < .001). Lachnospira genus, significantly less abundant in the AB group, was positively correlated with 3-hydroxyoctanoate (r = 0.931, p < .001) and with 1-palmitoyl-2-oleoyl-GPE (16:0/18:1) (r = 0.855, p = .002) in the AB group, while in the CD group correlated negatively with N-formylanthranilic acid (r = -0.900, p < .001), 2,hydroxybehenate (r = -0.900, p < .001), N-acetylglycine (r = -0.883, p = .002), urate (r = -0.867, p = .002), and pseudouridine (r = -0.850, p = .004). Lachnospira genus was positively correlated with indolepropionate (r = 0.867, p = .002) among the CDs. Roseburia genus, significantly less abundant in the AB group, correlated positively in the AB group with N,N,dimethylalanine (r = 0.867, p = .001), 12-ketolithocholate (r = 0.903, p < .001), and 3-dehydrodeoxycholate (r = .939, p < .001). The same genus was negatively associated with 3-(3-hydroxyphenyl) propionate (r = -0.883, p = .002), p-Cresol sulfate (r = -0.867, p = .002), and succinate (r = -0.850, p = .004) in the CD group. Fusicatenibacter genus, significantly less abundant in the AB group, negatively correlated with dibutyl sulfosuccinate (r = -0.865, p = .001) in the AB group. In the CD group this genus correlated negatively with Deoxycytidine monophosphate (dCMP) (r = -0.865, p = .003) and positively with methionine sulfoxide (r = 0.883, p = .002). Of note, we didn’t find any taxon/metabolite correlation passing the r± 0.85 cutoff in any group of candidatus Stoqueficus and Eisenbergiella (more abundant taxa among the ABs), as well as Lachnospiraceae_UCG_001 family, taxon depleted among ABs (Table 3).
(TIF)
A) Reported participant preexisting conditions use over entire patient population. Blue dots indicate condition of AB participants, red dots indicate condition of CD participants and green dots indicate condition of HC participants. B) Preexisting conditions that overlap with Jackson et al. (2018) when selecting at a 20% FDR in S5 Table (Jackson et al., 2018). (AB: Abstinent, CD: current drinkers, HC: healthy controls).
(TIF)
A) Reported participant medication use over entire patient population. Blue dots indicate medication taken by AB participants, red dots indicate medication taken by CD participants and green dot indicates medication taken by HC participants. B) Medications that overlap with Jackson et al. (2018) when selecting at a 20% FDR in S4 Table (AB: Abstinent, CD: current drinkers, HC: healthy controls).
(TIF)
Detection of sucrose (A), Sucralose (B) and Lactulose/Mannitol ratio (C) by ultra-performance liquid chromatography mass spectrometry (UPLC-MS) analysis in suspected non-fasters participants. L/M ratio corresponded to the fractional excretion (FE) of lactulose and mannitol = (urine concentration from MS x total urine volume excreted)/sugar input). L/M ratio was calculated as FE lactulose/FE mannitol. Data are presented as mean values +/- SD. AB: Abstinent, CD: current drinkers, HC: healthy controls. Baseline = 40 minutes before starting the experiment, sampling times during the experiment: 1st ≈ 90 min, 2nd ≈ 100 min, 3rd ≈ 180 min, 4th ≈ 240 min, 5th ≈ 280 min AB: Abstinent, CD: current drinkers, HC: healthy controls.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
The authors would like to thank the clinical research staff involved in patient care, data collection and support at the NIAAA DICBR, especially in the joint NIAAA/NIDA Clinical Psychoneuroendocrinology and Neuropsychopharmacology Section (notably Navan Shah and Brittney Browning). The authors would also like to thank the nursing and clinical staff at the NIH Clinical Center, especially Tom Lionetti and Dave Spero, and Shavonne Pocock (NINR) and Dr. Paule Joseph (NIAAA/NINR). The authors would also like to thank the Genomics Resource Center at the Institute for Genome Sciences (University of Maryland School of Medicine) for the 16 S rRNA amplicon sequencing. Finally, the authors would like to express their gratitude to all the participants who made this study possible.
Data Availability
All raw microbiome sequence files related to this study can be found at NCBI SRA under project ID PRJNA966972 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA966972).
Funding Statement
Funding: This study was supported by (A) NIH Intramural Research Program funding ZIA-DA000635 (Clinical Psychoneuroendocrinology and Neuropsychopharmacology Section; PI: Lorenzo Leggio), jointly supported by the NIDA IRP and the NIAAA Division of Intramural Clinical and Biological Research (DICBR), and (B) Peter G. Dodge Foundation (PGDF) funding (Exploring Gut-Brain and Brain-Gut Interactions in Alcohol Use Disorder via Microbiota Investigations: A Pilot Study; PI: Lorenzo Leggio). The authors are also grateful to the Center on Compulsive Behaviors funding, NIH IRP (Fellowship to Daria Piacentino) and to the University of Maryland School of Medicine Dean’s Endowed Professorship (Claire M. Fraser). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Association AP. Diagnostic and statistical manual of mental disorders: DSM-5. 2013:xliv, 947-xliv,.
- 2.National Institute of Alcohol Abuse and Alcoholism. NIAAA Council Approves Definition of Binge Drinking2004; (3):[3 p.].
- 3.National Institute of Alcohol Abuse and Alcoholism. Drinking Levels Defined [updated 2020. Available from: https://www.niaaa.nih.gov/alcohol-health/overview-alcohol-consumption/moderate-binge-drinking.
- 4.Nikolova VL, Smith MRB, Hall LJ, Cleare AJ, Stone JM, Young AH. Perturbations in Gut Microbiota Composition in Psychiatric Disorders: A Review and Meta-analysis. JAMA Psychiatry. 2021;78(12):1343–54. doi: 10.1001/jamapsychiatry.2021.2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carbia C, Bastiaanssen TFS, Iannone LF, García-Cabrerizo R, Boscaini S, Berding K, et al. The Microbiome-Gut-Brain axis regulates social cognition & craving in young binge drinkers. EBioMedicine. 2023;89:104442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pohl KA-O, Moodley P, Dhanda AD. Alcohol’s Impact on the Gut and Liver. LID—10.3390/nu13093170 [doi] LID—3170. (2072–6643 (Electronic)).
- 7.Mutlu EA, Gillevet PM, Rangwala H, Sikaroodi M, Naqvi A, Engen PA, et al. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol. 2012;302(9):G966–78. doi: 10.1152/ajpgi.00380.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leclercq S, Matamoros S, Cani PD, Neyrinck AM, Jamar F, Stärkel P, et al. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci U S A. 2014;111(42):E4485–93. doi: 10.1073/pnas.1415174111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubinkina VB, Tyakht AV, Odintsova VY, Yarygin KS, Kovarsky BA, Pavlenko AV, et al. Links of gut microbiota composition with alcohol dependence syndrome and alcoholic liver disease. Microbiome. 2017;5(1):141. doi: 10.1186/s40168-017-0359-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maccioni L, Gao B, Leclercq S, Pirlot B, Horsmans Y, De Timary P, et al. Intestinal permeability, microbial translocation, changes in duodenal and fecal microbiota, and their associations with alcoholic liver disease progression in humans. Gut Microbes. 2020;12(1):1782157. doi: 10.1080/19490976.2020.1782157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meroni M, Longo M, Dongiovanni P. Alcohol or Gut Microbiota: Who Is the Guilty? Int J Mol Sci. 2019;20(18). doi: 10.3390/ijms20184568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fairfield B, Schnabl B. Gut dysbiosis as a driver in alcohol-induced liver injury. JHEP Rep. 2021;3(2):100220. doi: 10.1016/j.jhepr.2020.100220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dukić M, Radonjić T, Jovanović I, Zdravković M, Todorović Z, Kraišnik N, et al. Alcohol, Inflammation, and Microbiota in Alcoholic Liver Disease. Int J Mol Sci. 2023;24(4). doi: 10.3390/ijms24043735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson MA, Verdi S, Maxan ME, Shin CM, Zierer J, Bowyer RCE, et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat Commun. 2018;9(1):2655. doi: 10.1038/s41467-018-05184-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chivero ET, Sil S, Kumar M, Buch S. Substance use, microbiome and psychiatric disorders. Pharmacol Biochem Behav. 2022;219:173432. doi: 10.1016/j.pbb.2022.173432 [DOI] [PubMed] [Google Scholar]
- 16.Hofford RS, Kiraly DD. Clinical and preclinical evidence for gut microbiome mechanisms in substance use disorder. Biol Psychiatry. 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Li L, Bian C, Bai M, Yu H, Gao H, et al. Alterations and correlations of gut microbiota, fecal, and serum metabolome characteristics in a rat model of alcohol use disorder. Front Microbiol. 2022;13:1068825. doi: 10.3389/fmicb.2022.1068825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zallar LJ, Beurmann S, Tunstall BJ, Fraser CM, Koob GF, Vendruscolo LF, et al. Ghrelin receptor deletion reduces binge-like alcohol drinking in rats. J Neuroendocrinol. 2019;31(7):e12663. doi: 10.1111/jne.12663 [DOI] [PubMed] [Google Scholar]
- 19.Xie G, Zhong W, Li H, Li Q, Qiu Y, Zheng X, et al. Alteration of bile acid metabolism in the rat induced by chronic ethanol consumption. Faseb j. 2013;27(9):3583–93. doi: 10.1096/fj.13-231860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen P, Torralba M, Tan J, Embree M, Zengler K, Stärkel P, et al. Supplementation of saturated long-chain fatty acids maintains intestinal eubiosis and reduces ethanol-induced liver injury in mice. Gastroenterology. 2015;148(1):203–14.e16. doi: 10.1053/j.gastro.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G, Liu Q, Guo L, Zeng H, Ding C, Zhang W, et al. Gut Microbiota and Relevant Metabolites Analysis in Alcohol Dependent Mice. Front Microbiol. 2018;9:1874. doi: 10.3389/fmicb.2018.01874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barr T, Sureshchandra S, Ruegger P, Zhang J, Ma W, Borneman J, et al. Concurrent gut transcriptome and microbiota profiling following chronic ethanol consumption in nonhuman primates. Gut Microbes. 2018;9(4):338–56. doi: 10.1080/19490976.2018.1441663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Yasuda K, Gilmore RA, Westmoreland SV, Platt DM, Miller GM, et al. Alcohol-induced changes in the gut microbiome and metabolome of rhesus macaques. Psychopharmacology (Berl). 2019;236(5):1531–44. doi: 10.1007/s00213-019-05217-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piacentino D, Grant-Beurmann S, Vizioli C, Li X, Moore CF, Ruiz-Rodado V, et al. Gut microbiome and metabolome in a non-human primate model of chronic excessive alcohol drinking. Transl Psychiatry. 2021;11(1):609. doi: 10.1038/s41398-021-01728-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weerts EM, Goodwin AK, Kaminski BJ, Hienz RD. Environmental cues, alcohol seeking, and consumption in baboons: effects of response requirement and duration of alcohol abstinence. Alcohol Clin Exp Res. 2006;30(12):2026–36. doi: 10.1111/j.1530-0277.2006.00249.x [DOI] [PubMed] [Google Scholar]
- 26.Holtyn AF, Kaminski BJ, Wand GS, Weerts EM. Differences in extinction of cue-maintained conditioned responses associated with self-administration: alcohol versus a nonalcoholic reinforcer. Alcohol Clin Exp Res. 2014;38(10):2639–46. doi: 10.1111/acer.12537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Litwinowicz K, Choroszy M, Waszczuk E. Changes in the composition of the human intestinal microbiome in alcohol use disorder: a systematic review. Am J Drug Alcohol Abuse. 2020;46(1):4–12. doi: 10.1080/00952990.2019.1669629 [DOI] [PubMed] [Google Scholar]
- 28.Kakiyama G, Hylemon PB, Zhou H, Pandak WM, Heuman DM, Kang DJ, et al. Colonic inflammation and secondary bile acids in alcoholic cirrhosis. Am J Physiol Gastrointest Liver Physiol. 2014;306(11):G929–37. doi: 10.1152/ajpgi.00315.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Couch RD, Dailey A, Zaidi F, Navarro K, Forsyth CB, Mutlu E, et al. Alcohol induced alterations to the human fecal VOC metabolome. PLoS One. 2015;10(3):e0119362. doi: 10.1371/journal.pone.0119362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Llopis M, Cassard AM, Wrzosek L, Boschat L, Bruneau A, Ferrere G, et al. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut. 2016;65(5):830–9. doi: 10.1136/gutjnl-2015-310585 [DOI] [PubMed] [Google Scholar]
- 31.Sinha R. New findings on biological factors predicting addiction relapse vulnerability. Curr Psychiatry Rep. 2011;13(5):398–405. doi: 10.1007/s11920-011-0224-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ames NJ, Barb JJ, Schuebel K, Mudra S, Meeks BK, Tuason RTS, et al. Longitudinal gut microbiome changes in alcohol use disorder are influenced by abstinence and drinking quantity. Gut Microbes. 2020;11(6):1608–31. doi: 10.1080/19490976.2020.1758010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cresci GA. The gut microbiome: a new frontier for alcohol investigation. Alcohol Clin Exp Res. 2015;39(6):947–9. doi: 10.1111/acer.12732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.US Department of Health and Human Services and U.S Department of Agriculture. Dietary Guidline for Americans 2020–2025 2020 [updated December 2020. 9:[Available from: https://www.dietaryguidelines.gov/sites/default/files/2020-12/Dietary_Guidelines_for_Americans_2020-2025.pdf.
- 35.National Institute of Alcohol Abuse and Alcoholism. Rethinking drinking: What’s a standard drink? [
- 36.Michael First JW, Rhonda Karg, Robert Spitzer, American Psychiatric Association. Structured Clinical Interview for DSM-5—Research Version (SCID-5 for DSM-5, Research Version; SCID-5-RV). Arlington, VA2015.
- 37.Lemmens PH, Volovics L, Haan YD. Measurement of lifetime exposure to alcohol: data quality of a self-administered questionnaire and impact on risk assessment. Contemporary Drug Problems. 1997;24(3):581–600. [Google Scholar]
- 38.Babor TF, Higgins-Biddle JC, Saunders JB, Monteiro MG, Organization WH. AUDIT: the alcohol use disorders identification test: guidelines for use in primary health care. World Health Organization; 1989. [Google Scholar]
- 39.Skinner HA, Allen BA. Alcohol dependence syndrome: Measurement and validation. Journal of Abnormal Psychology. 1982;91(3):199–209. doi: 10.1037//0021-843x.91.3.199 [DOI] [PubMed] [Google Scholar]
- 40.Sobell LC, Sobell MB. Timeline follow-back: A technique for assessing self-reported alcohol consumption. Measuring alcohol consumption: Psychosocial and biochemical methods. Totowa, NJ, US: Humana Press/Springer Nature; 1992. p. 41–72. [Google Scholar]
- 41.Flannery B, Volpicelli J, Pettinati H. Psychometric properties of the Penn alcohol craving scale. Alcoholism: Clinical and Experimental Research. 1999;23(8):1289–95. [PubMed] [Google Scholar]
- 42.Sullivan JT, Sykora K, Schneiderman J, Naranjo CA, Sellers EM. Assessment of alcohol withdrawal: the revised clinical institute withdrawal assessment for alcohol scale (CIWA-Ar). Br J Addict. 1989;84(11):1353–7. doi: 10.1111/j.1360-0443.1989.tb00737.x [DOI] [PubMed] [Google Scholar]
- 43.Buysse DJ, Reynolds CF 3rd, Monk TH, Berman SR, Kupfer DJ. The Pittsburgh Sleep Quality Index: a new instrument for psychiatric practice and research. Psychiatry Res. 1989;28(2):193–213. doi: 10.1016/0165-1781(89)90047-4 [DOI] [PubMed] [Google Scholar]
- 44.Asberg M, Montgomery SA, Perris C, Schalling D, Sedvall G. A comprehensive psychopathological rating scale. Acta Psychiatr Scand Suppl. 1978(271):5–27. doi: 10.1111/j.1600-0447.1978.tb02357.x [DOI] [PubMed] [Google Scholar]
- 45.Montgomery SA, Asberg M. A new depression scale designed to be sensitive to change. Br J Psychiatry. 1979;134:382–9. doi: 10.1192/bjp.134.4.382 [DOI] [PubMed] [Google Scholar]
- 46.Tyrer P, Owen RT, Cicchetti DV. The brief scale for anxiety: a subdivision of the comprehensive psychopathological rating scale. J Neurol Neurosurg Psychiatry. 1984;47(9):970–5. doi: 10.1136/jnnp.47.9.970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lorr M, McNair D, Droppleman L. Manual: profile of mood states. San Diego, CA: Educational and Industrial Testing Service. 1971. [Google Scholar]
- 48.Afdhal NH. Fibroscan (transient elastography) for the measurement of liver fibrosis. Gastroenterol Hepatol (N Y). 2012;8(9):605–7. [PMC free article] [PubMed] [Google Scholar]
- 49.Oeda S, Tanaka K, Oshima A, Matsumoto Y, Sueoka E, Takahashi H. Diagnostic Accuracy of FibroScan and Factors Affecting Measurements. Diagnostics (Basel). 2020;10(11). doi: 10.3390/diagnostics10110940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Del Valle-Pinero AY, Van Deventer HE, Fourie NH, Martino AC, Patel NS, Remaley AT, et al. Gastrointestinal permeability in patients with irritable bowel syndrome assessed using a four probe permeability solution. Clin Chim Acta. 2013;418:97–101. doi: 10.1016/j.cca.2012.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Do KT, Wahl S, Raffler J, Molnos S, Laimighofer M, Adamski J, et al. Characterization of missing values in untargeted MS-based metabolomics data and evaluation of missing data handling strategies. Metabolomics [Internet]. 2018. 2018/09//; 14(10):[128 p.]. Available from: http://europepmc.org/abstract/MED/30830398 doi: 10.1007/s11306-018-1420-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu Y, Zhou G, Ewald J, Pang Z, Shiri T, Xia J. MicrobiomeAnalyst 2.0: comprehensive statistical, functional and integrative analysis of microbiome data. Nucleic Acids Research. 2023;51(W1):W310–W8. doi: 10.1093/nar/gkad407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chong J, Liu P, Zhou G, Xia J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc. 2020;15(3):799–821. doi: 10.1038/s41596-019-0264-1 [DOI] [PubMed] [Google Scholar]
- 54.Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia J. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Research. 2017;45:W180—W8. doi: 10.1093/nar/gkx295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–8. doi: 10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mallick H, Rahnavard A, McIver LJ, Ma S, Zhang Y, Nguyen LH, et al. Multivariable association discovery in population-scale meta-omics studies. PLOS Computational Biology. 2021;17(11):e1009442. doi: 10.1371/journal.pcbi.1009442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foster ZSL, Sharpton TJ, Grünwald NJ. Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLOS Computational Biology. 2017;13(2):e1005404. doi: 10.1371/journal.pcbi.1005404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal statistical society: series B (Methodological). 1995;57(1):289–300. [Google Scholar]
- 60.Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49(W1):W388–W96. doi: 10.1093/nar/gkab382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Curtin F, Schulz P. Multiple correlations and bonferroni’s correction. Biological Psychiatry. 1998;44(8):775–7. doi: 10.1016/s0006-3223(98)00043-2 [DOI] [PubMed] [Google Scholar]
- 62.Kosnicki KL, Penprase JC, Cintora P, Torres PJ, Harris GL, Brasser SM, et al. Effects of moderate, voluntary ethanol consumption on the rat and human gut microbiome. Addict Biol. 2019;24(4):617–30. doi: 10.1111/adb.12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stackebrandt E. The Family Lachnospiraceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes: Firmicutes and Tenericutes. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. p. 197–201. [Google Scholar]
- 64.Biddle A, Stewart L, Blanchard J, Leschine S. Untangling the Genetic Basis of Fibrolytic Specialization by Lachnospiraceae and Ruminococcaceae in Diverse Gut Communities. Diversity. 2013;5(3):627–40. [Google Scholar]
- 65.Devillard E, McIntosh FM, Duncan SH, Wallace RJ. Metabolism of linoleic acid by human gut bacteria: different routes for biosynthesis of conjugated linoleic acid. J Bacteriol. 2007;189(6):2566–70. doi: 10.1128/JB.01359-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Plieskatt JL, Deenonpoe R, Mulvenna JP, Krause L, Sripa B, Bethony JM, et al. Infection with the carcinogenic liver fluke Opisthorchis viverrini modifies intestinal and biliary microbiome. Faseb j. 2013;27(11):4572–84. doi: 10.1096/fj.13-232751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Riordan SM, Williams R. The intestinal flora and bacterial infection in cirrhosis. J Hepatol. 2006;45(5):744–57. doi: 10.1016/j.jhep.2006.08.001 [DOI] [PubMed] [Google Scholar]
- 68.Guo K, Figueroa-Romero C, Noureldein M, Hinder LM, Sakowski SA, Rumora AE, et al. Gut microbiota in a mouse model of obesity and peripheral neuropathy associated with plasma and nerve lipidomics and nerve transcriptomics. Microbiome. 2023;11(1):52. doi: 10.1186/s40168-022-01436-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O’Mahony SM, Clarke G, Borre YE, Dinan TG, Cryan JF. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav Brain Res. 2015;277:32–48. doi: 10.1016/j.bbr.2014.07.027 [DOI] [PubMed] [Google Scholar]
- 70.Roth W, Zadeh K, Vekariya R, Ge Y, Mohamadzadeh M. Tryptophan Metabolism and Gut-Brain Homeostasis. Int J Mol Sci. 2021;22(6). doi: 10.3390/ijms22062973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fernstrom JD, Fernstrom MH. Tyrosine, phenylalanine, and catecholamine synthesis and function in the brain. J Nutr. 2007;137(6 Suppl 1):1539S–47S; discussion 48S. doi: 10.1093/jn/137.6.1539S [DOI] [PubMed] [Google Scholar]
- 72.Schoeler M, Caesar R. Dietary lipids, gut microbiota and lipid metabolism. Rev Endocr Metab Disord. 2019;20(4):461–72. doi: 10.1007/s11154-019-09512-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Faquih TO, van Klinken JB, Li-Gao R, Noordam R, van Heemst D, Boone S, et al. Hepatic triglyceride content is intricately associated with numerous metabolites and biochemical pathways. Liver Int. 2023;43(7):1458–72. doi: 10.1111/liv.15575 [DOI] [PubMed] [Google Scholar]
- 74.Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, et al. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54(2):562–72. doi: 10.1002/hep.24423 [DOI] [PubMed] [Google Scholar]
- 75.Chen Y, Guo J, Qian G, Fang D, Shi D, Guo L, et al. Gut dysbiosis in acute-on-chronic liver failure and its predictive value for mortality. J Gastroenterol Hepatol. 2015;30(9):1429–37. doi: 10.1111/jgh.12932 [DOI] [PubMed] [Google Scholar]
- 76.Tamanai-Shacoori Z, Smida I, Bousarghin L, Loreal O, Meuric V, Fong SB, et al. Roseburia spp.: a marker of health? Future Microbiol. 2017;12:157–70. doi: 10.2217/fmb-2016-0130 [DOI] [PubMed] [Google Scholar]
- 77.Agus A, Planchais J, Sokol H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe. 2018;23(6):716–24. doi: 10.1016/j.chom.2018.05.003 [DOI] [PubMed] [Google Scholar]
- 78.Barron KA, Jeffries KA, Krupenko NI. Sphingolipids and the link between alcohol and cancer. Chem Biol Interact. 2020;322:109058. doi: 10.1016/j.cbi.2020.109058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nikolova-Karakashian M. Alcoholic and non-alcoholic fatty liver disease: Focus on ceramide. Adv Biol Regul. 2018;70:40–50. doi: 10.1016/j.jbior.2018.11.004 [DOI] [PubMed] [Google Scholar]
- 80.Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19(1):29–41. doi: 10.1111/1462-2920.13589 [DOI] [PubMed] [Google Scholar]
- 81.Rohn MC, Lee MR, Kleuter SB, Schwandt ML, Falk DE, Leggio L. Differences Between Treatment-Seeking and Nontreatment-Seeking Alcohol-Dependent Research Participants: An Exploratory Analysis. Alcohol Clin Exp Res. 2017;41(2):414–20. doi: 10.1111/acer.13304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ray LA, Bujarski S, Yardley MM, Roche DJO, Hartwell EE. Differences between treatment-seeking and non-treatment-seeking participants in medication studies for alcoholism: do they matter? The American journal of drug and alcohol abuse. 2017;43(6):703–10. doi: 10.1080/00952990.2017.1312423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee MR, Sankar V, Hammer A, Kennedy WG, Barb JJ, McQueen PG, et al. Using Machine Learning to Classify Individuals With Alcohol Use Disorder Based on Treatment Seeking Status. EClinicalMedicine. 2019;12:70–8. doi: 10.1016/j.eclinm.2019.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haass-Koffler CL, Piacentino D, Li X, Long VM, Lee MR, Swift RM, et al. Differences in Sociodemographic and Alcohol-Related Clinical Characteristics Between Treatment Seekers and Nontreatment Seekers and Their Role in Predicting Outcomes in the COMBINE Study for Alcohol Use Disorder. Alcohol Clin Exp Res. 2020;44(10):2097–108. doi: 10.1111/acer.14428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Andrioaie IM, Duhaniuc A, Nastase EV, Iancu LS, Luncă C, Trofin F, et al. The Role of the Gut Microbiome in Psychiatric Disorders. Microorganisms. 2022;10(12). doi: 10.3390/microorganisms10122436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Borkent J, Ioannou M, Laman JD, Haarman BCM, Sommer IEC. Role of the gut microbiome in three major psychiatric disorders. Psychol Med. 2022;52(7):1222–42. doi: 10.1017/S0033291722000897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barb JJ, Brooks AT, Kazmi N, Yang L, Chakravorty S, Wallen GR. A lower sleep regularity index (SRI) is associated with relapse in individuals with alcohol use disorder following inpatient treatment. Sci Rep. 2022;12(1):21583. doi: 10.1038/s41598-022-26019-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lieber CS. Relationships between nutrition, alcohol use, and liver disease. Alcohol Res Health. 2003;27(3):220–31. [PMC free article] [PubMed] [Google Scholar]
- 89.Cummings JR, Gearhardt AN, Ray LA, Choi AK, Tomiyama AJ. Experimental and observational studies on alcohol use and dietary intake: a systematic review. Obes Rev. 2020;21(2):e12950. doi: 10.1111/obr.12950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nielsen SJ, Kit BK, Fakhouri T, Ogden CL. Calories consumed from alcoholic beverages by U.S. adults, 2007–2010. NCHS Data Brief. 2012(110):1–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(DOCX)
(DOCX)
Pairwise comparison at the genus level between each group using Wilcoxon Test. (A) Dot plot of 9 genera (y-axis) found to be significant (p < .05, unadjusted) in pairwise comparison (x-axis). (B) Venn Diagram showing genera found to be significant that overlap between 3 pairwise tests. (C-E) The heat tree analysis leverages the hierarchical structure of taxonomic classifications to quantitatively (using the mean abundance) and statistically (using the non-parametric Wilcoxon Rank Sum test with p < .05) depict taxonomic differences between microbial communities. Red boxes show the end of the leaf (genus) in the heat tree for each plot. Taxa colored in red are more abundant in the AB (C, D) and CD (E) groups, taxa colored in blue are more abundant in the HC (C, E) and CD (D) groups. AB: Abstinent, CD: current drinkers, HC: healthy controls.
(TIF)
LEfSe LDA bar plots at the genus level of fecal microbial samples displaying the pairwise LDA scores between the three groups; abstinent group (AB), continuous alcohol drinking group (CD), and the healthy control group (HC). The bars represent the effect size (LDA) for a particular genus in a certain group. The length of the bar represents a log10 transformed LDA score. The heat maps show the LDA effect size from blue (negative) to red (positive). Shaded bars with an ‘*’ are genera that were found to be significant in the pairwise test (p < .05). (A) represents the pairwise comparison of AB to CD; (B) represents the pairwise comparison of AB to HC; and (C) represents the pairwise comparison of CD to HC. AB: Abstinent, CD: current drinkers, HC: healthy controls.
(TIF)
Results of covariates adjustment (y axis) and without adjustment (x axis). P values for the comparisons between (A) AB and CD, (B) AB and HC, and (C) CD and HC groups did not improve with covariate adjustment (BMI and age), resulting in no significantly different taxa. AB: Abstinent, CD: current drinkers, HC: healthy controls.
(TIF)
Variable importance in prediction (VIP) scores extracted by PLS-DA of metabolites and heatmap of the average scaled expression values of the indicated VIP metabolites in all samples. The top 30 metabolites driving differences between group are represented (lowest VIP score = 2.261). Metabolites in AB group samples show the lowest (in most cases) or the highest abundance compared to HC and CD groups. AB: Abstinent, CD: current drinkers, HC: healthy controls.
(TIF)
The correlations were computed within each group and a total of 149 significant correlation pairs were found within the ABs (A), 258 were found within the CDs (B), and 33 within the HCs (C). Most of the taxa within the AB group found to significantly correlate with metabolites, were represented in the CDs as well. Specifically, across the three comparisons, a high number of correlations were found in members from Lachnospiraceae and Oscillospiraceae families (Bacillota phylum): 62% in the CD group, 47% in the AB group, and 66% in the HC group. Streptococcus genus was found to be significantly more abundant in the AB group when compared to CD and HC groups. In the CD group Streptococcus genus was positively correlated with glyerolphosphoethanolamine (r = 0.950, p < .001), glycerophosphoserine (r = 0.917, p < .001), mannose (r = 0.917, p < .001), deoxycarnitine (r = 0.867, p = .002), and N,1, acetylspermidine (r = 0.850, p = .004). Conversely, in the same group we found negative correlations of Streptococcus with heptenedioate (C7:1-DC) (r = -0.865, p = .003), 2’-O-methylcytidine (r = -0.865, p = .003), and hexadecanedioate, C16 (r = -0.850, p = .004). In the AB group, Streptococcus genus correlated negatively with 7-ketocholesterol (r = -0.952, p < .001). Lachnospira genus, significantly less abundant in the AB group, was positively correlated with 3-hydroxyoctanoate (r = 0.931, p < .001) and with 1-palmitoyl-2-oleoyl-GPE (16:0/18:1) (r = 0.855, p = .002) in the AB group, while in the CD group correlated negatively with N-formylanthranilic acid (r = -0.900, p < .001), 2,hydroxybehenate (r = -0.900, p < .001), N-acetylglycine (r = -0.883, p = .002), urate (r = -0.867, p = .002), and pseudouridine (r = -0.850, p = .004). Lachnospira genus was positively correlated with indolepropionate (r = 0.867, p = .002) among the CDs. Roseburia genus, significantly less abundant in the AB group, correlated positively in the AB group with N,N,dimethylalanine (r = 0.867, p = .001), 12-ketolithocholate (r = 0.903, p < .001), and 3-dehydrodeoxycholate (r = .939, p < .001). The same genus was negatively associated with 3-(3-hydroxyphenyl) propionate (r = -0.883, p = .002), p-Cresol sulfate (r = -0.867, p = .002), and succinate (r = -0.850, p = .004) in the CD group. Fusicatenibacter genus, significantly less abundant in the AB group, negatively correlated with dibutyl sulfosuccinate (r = -0.865, p = .001) in the AB group. In the CD group this genus correlated negatively with Deoxycytidine monophosphate (dCMP) (r = -0.865, p = .003) and positively with methionine sulfoxide (r = 0.883, p = .002). Of note, we didn’t find any taxon/metabolite correlation passing the r± 0.85 cutoff in any group of candidatus Stoqueficus and Eisenbergiella (more abundant taxa among the ABs), as well as Lachnospiraceae_UCG_001 family, taxon depleted among ABs (Table 3).
(TIF)
A) Reported participant preexisting conditions use over entire patient population. Blue dots indicate condition of AB participants, red dots indicate condition of CD participants and green dots indicate condition of HC participants. B) Preexisting conditions that overlap with Jackson et al. (2018) when selecting at a 20% FDR in S5 Table (Jackson et al., 2018). (AB: Abstinent, CD: current drinkers, HC: healthy controls).
(TIF)
A) Reported participant medication use over entire patient population. Blue dots indicate medication taken by AB participants, red dots indicate medication taken by CD participants and green dot indicates medication taken by HC participants. B) Medications that overlap with Jackson et al. (2018) when selecting at a 20% FDR in S4 Table (AB: Abstinent, CD: current drinkers, HC: healthy controls).
(TIF)
Detection of sucrose (A), Sucralose (B) and Lactulose/Mannitol ratio (C) by ultra-performance liquid chromatography mass spectrometry (UPLC-MS) analysis in suspected non-fasters participants. L/M ratio corresponded to the fractional excretion (FE) of lactulose and mannitol = (urine concentration from MS x total urine volume excreted)/sugar input). L/M ratio was calculated as FE lactulose/FE mannitol. Data are presented as mean values +/- SD. AB: Abstinent, CD: current drinkers, HC: healthy controls. Baseline = 40 minutes before starting the experiment, sampling times during the experiment: 1st ≈ 90 min, 2nd ≈ 100 min, 3rd ≈ 180 min, 4th ≈ 240 min, 5th ≈ 280 min AB: Abstinent, CD: current drinkers, HC: healthy controls.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All raw microbiome sequence files related to this study can be found at NCBI SRA under project ID PRJNA966972 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA966972).