Summary
The replication-dependent histone H3.1 variant, ubiquitous in multicellular eukaryotes, has been proposed to play key roles during chromatin replication due to its unique expression pattern restricted to the S phase of the cell cycle. Here, we describe recent discoveries in plants regarding molecular mechanisms and cellular pathways involving H3.1 that contribute to the maintenance of genomic and epigenomic information. First, we highlight new advances concerning the contribution of the histone chaperone CAF-1 and the TSK-H3.1 DNA repair pathway in preventing genomic instability during replication. We then summarize the evidence connecting H3.1 to specific roles required for the mitotic inheritance of epigenetic states. Finally, we discuss the recent identification of a specific interaction between H3.1 and DNA polymerase epsilon and its functional implications.
Keywords: Epigenetics, chromatin, DNA replication, histone modifications, histone variants, Arabidopsis
Introduction
In eukaryotes, DNA replication is accompanied by the faithful transmission of complex epigenetic states across the genome. The process of chromatin replication thus implicates two different but related goals—the maintenance of genetic and epigenetic information—that are both required for cellular homeostasis. As these two processes are achieved in the same temporal and spatial window during the cell cycle, they likely rely on many common factors expressed uniquely at the time of chromatin replication.
Among them, the replication-dependent histone H3.1 variant is thought to play a central role in the inheritance of genetic information and the different epigenetic states [1]. In multicellular eukaryotes, H3.1 is specifically expressed during the S phase, in contrast to replication-independent H3.3 which remains available throughout the cell cycle [2,3]. A burst of histone expression is needed during DNA synthesis to prevent nucleosome dilution on replicated DNA. However, why multicellular eukaryotes rely on a specific H3 variant during chromatin replication has remained a longstanding question in the chromatin field. The difficulty in answering this question lies with the high level of sequence similarity between H3.1 and H3.3 proteins across all organisms, with only a few amino acids distinguishing these two variants. Recent work using Arabidopsis thaliana as a model system has started to shed light on the different molecular mechanisms by which H3.1 participates in the mitotic inheritance of genetic and epigenetic information, which we summarize in this review.
Maintenance of genomic stability by the H3.1 variant
One of the challenges in uncovering specific roles for replication-dependent H3 variants is linked to the fact that these proteins, like other histones, are encoded by large gene families in most biological systems (e.g., five H3.1 genes in Arabidopsis, and 13 H3.1/H3.2 genes in humans [2,4]). This high gene copy number makes it difficult to completely inactivate H3.1 and study the resulting phenotypes. While the recent establishment of gene editing systems now allows to easily knockout or alter histone gene functions [5–10], early research on H3.1 partially bypassed the problem by inactivating the histone chaperone CAF-1. The heterotrimeric CAF-1 complex is responsible for loading H3.1 on chromatin during replication [11,12], and its inactivation results in the insertion of H3.3 instead of H3.1 in proliferative cells using a gap-filling mechanism (Figure 1a–b) [12–14]. Interestingly, CAF-1 mutants are lethal in many metazoan systems [15,16], but not in Arabidopsis [17], thus making plants a unique model system to study H3.1 via CAF-1 inactivation. Recent work taking advantage of viable CAF-1 null mutants in plants has uncovered a key role for the H3.1 chaperone in mediating genomic stability in Arabidopsis [18]. The absence of CAF-1 function leads to large tandem duplications (~50 to ~1500 kb) and deletion of rRNA gene copies (Figure 1e). Interestingly, tandem duplications in euchromatic regions led to higher transcript levels for the duplicated protein-coding genes, thus establishing one mechanism by which genomic instability can potentially affect phenotypic expression [18].
Figure 1. Interplay between H3.1 and genome stability in different genetic contexts.
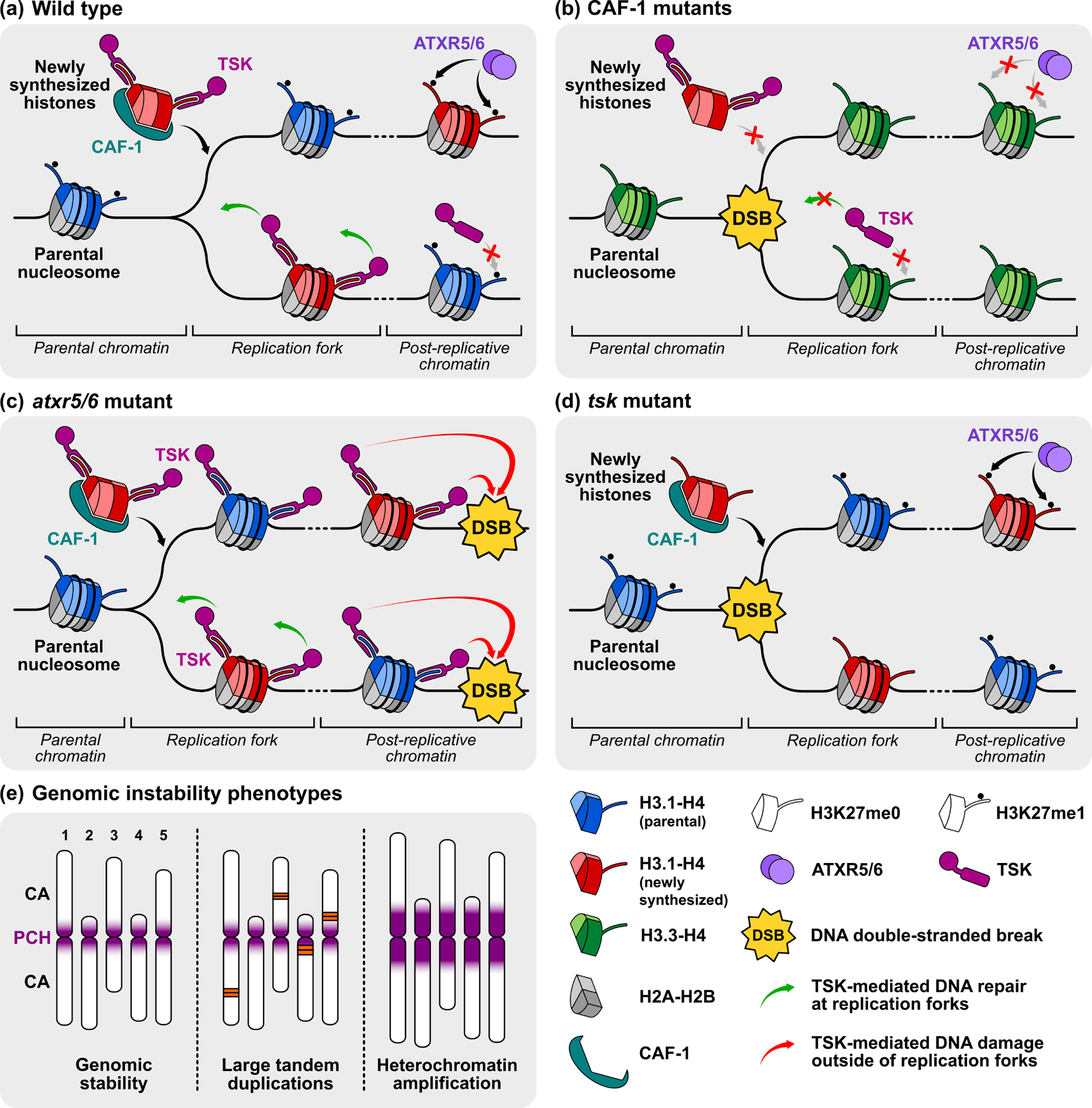
(a-d) Chromatin replication in heterochromatin is represented, where parental nucleosomes are mainly composed of the H3.1 variant, except in the CAF-1 mutant. (a) In a wild-type plant, newly synthesized H3.1-H4 tetramers are loaded by the CAF-1 complex during replication. TSK binds H3.1K27me0 and resolves broken or stalled replication forks, ensuring genomic integrity. Post-replicative maturation of chromatin involves mono-methylation of H3.1K27 by ATXR5/6, which prevents ectopic binding of TSK. (b) In CAF-1 mutants, predominance of the H3.3 variant in chromatin prevents TSK activity, resulting in DNA damage at replication forks. (c) In the atxr5/6 mutant, absence of H3.1K27me1 causes aberrant binding of TSK and genomic instability outside of replication forks. (d) In a tsk context, H3.1 is loaded and mono-methylated correctly, but TSK is not available to resolve stalled or broken replication forks. (e) Schematic depiction of known H3.1-dependent structural phenotypes in the Arabidopsis genome: genomic stability (wild-type), large tandem duplications (CAF-1 mutants), heterochromatin amplification (atxr5/6 mutants). PCH: pericentromeric heterochromatin; CA: chromosome arm.
Evidence suggests that the role of CAF-1 in the maintenance of genome stability depends on its ability to load H3.1 on chromatin during replication [19,20]. The DNA repair protein TONSOKU (TSK/BRUSHY/MGOUN3, also known as TONSOKU-LIKE/TONSL in metazoans) plays a crucial role in resolving stalled or broken replication forks [21–24]. Recently, Arabidopsis TSK was shown to use its conserved tetratricopeptide repeat domain to specifically bind H3.1 by recognizing one residue (alanine 31) that varies in H3.3 [19]. In plants, the TSK-H3.1 interaction is abrogated by mono-methylation at H3.1K27 (H3.1K27me1) [19], a mark deposited by the H3.1 mono-methyltransferases ATXR5 and ATXR6 (ATXR5/6) [25,26], with ATXR6 playing the larger role [27]. As almost all H3.1 proteins newly inserted on chromatin are rapidly mono-methylated by ATXR5/6 during replication [13], the interaction between TSK and H3.1 on chromatin is potentially confined to a short temporal and spatial window, at or near replication forks (Figure 1a). This chromatin-based mode of regulation would be hypothesized to restrict the activity of TSK to resolving stalled or broken replication forks, a seemingly critical regulatory step as depletion of H3.1K27me1 (e.g., in atxr5/6 mutants) induces genomic instability in the form of heterochromatin amplification [28], most likely due to ectopic TSK activity (Figure 1c and e) [19]. Interestingly, the reverse situation (i.e., the absence of TSK activity) may also produce genome stability defects (Figure 1d), as shown from sensitivity of tsk mutants to the genotoxic drug methyl methanesulfonate [19]. As reduced levels of TSK activity would be expected in a CAF-1 mutant due to the inability to load H3.1 variants on chromatin during replication, it is tempting to speculate that the large tandem duplications and rRNA deletions observed in the absence of CAF-1 are due to TSK-mediated resolution of impaired replication forks not being functional in this mutant background (Figure 1b and e). Overall, these studies strongly suggest a key role played by CAF-1 in recruiting the TSK-H3.1K27 DNA repair pathway to maintain genetic information during replication in plants.
H3.1 and the mitotic inheritance of epigenetic states
Aside from a role for the H3.1 variant in protecting the genome, multiple studies have hinted at functions for H3.1 in the correct transmission of epigenetic states during replication. For example, studies of CAF-1 mutants in Arabidopsis revealed various molecular and developmental phenotypes (e.g., defects in shoot and root apical meristem, and partial loss of heterochromatin silencing and organization) [17,29–32], thus arguing for epigenetic roles for H3.1 that are specific to this H3 variant. Previous work in mammalian systems has shown that some histone post-translational modifications (PTMs) are present on soluble (i.e., pre-deposition) H3.3, while soluble H3.1 proteins are mostly devoid of PTMs [33]. Thus, in the absence of CAF-1, deposition of modified H3.3 during DNA replication could affect the inheritance of epigenetic states.
The H3.1 variant was shown to be involved in the inheritance of the cell-identity marker H3K27me3 during replication [13]. The requirement for H3.1 to maintain this histone mark was proposed to depend on the mono-methyltransferase activity of ATXR5/6. In this model, H3.1K27me1 is a prerequisite for di- and tri-methylation by PRC2, which occurs during DNA replication [13]. Inheritance of epigenetic states may therefore be influenced by differential modifications on H3 variants after their deposition on chromatin, which is when ATXR5/6 are thought to be active on H3.1 based on their catalytic preference for nucleosomal substrates [34].
Other effects of H3.1 on epigenetic inheritance may be related to its interaction with TSK. The initial characterization of TSK indicated clear roles in regulating development and the maintenance of epigenetic silencing at heterochromatic loci [35,36]. More recent work has demonstrated that TSK participates in heat stress priming [37], an epigenetic phenomenon by which previous exposure to heat stress induces a molecular memory that makes plants more resistant to subsequent exposure to the same abiotic stress [38]. A possible model to explain all these results may be that, similarly to CAF-1, TSK directly participates in the deposition of the H3.1 variant during replication. Contradicting this model is the observation that morphological phenotypes between tsk and CAF-1 mutants vary significantly. An alternative function for TSK may be that by specifically binding to H3.1 (and not H3.3), it protects the N-terminal tail of H3.1 against spurious interactions during chromatin replication that may affect epigenetic inheritance. Although many pieces of the puzzle appear to be in place, more work is still needed to precisely understand the overarching mechanism by which H3.1 controls epigenetic inheritance during DNA replication.
Interplay between DNA polymerases and H3.1 during chromatin replication
The recent identification of TSK/TONSL as an H3.1 reader strongly suggests that other proteins may have a similar ability to specifically discriminate H3 variants [1]. Very recently, work in Arabidopsis revealed that the largest catalytic subunit (POL2A) of DNA polymerase epsilon (Pol ε), which catalyzes the synthesis of the leading strand, also binds specifically to H3.1 in the context of an H3.1-H4 dimer/tetramer, an activity that is conserved in POLE1, the mammalian ortholog of POL2A (Figure 2) [39]. A C-terminal CW-type zinc finger domain in POL2A recognizes H3.1 by interacting with amino acid residues A31 and S87, which both vary in H3.3 (T31 and H87). The interaction between POL2A and H3.1 was shown to be required for mediating heterochromatin condensation during meiosis in Arabidopsis [39].
Figure 2. Interplay between H3.1 and DNA polymerases during replication.
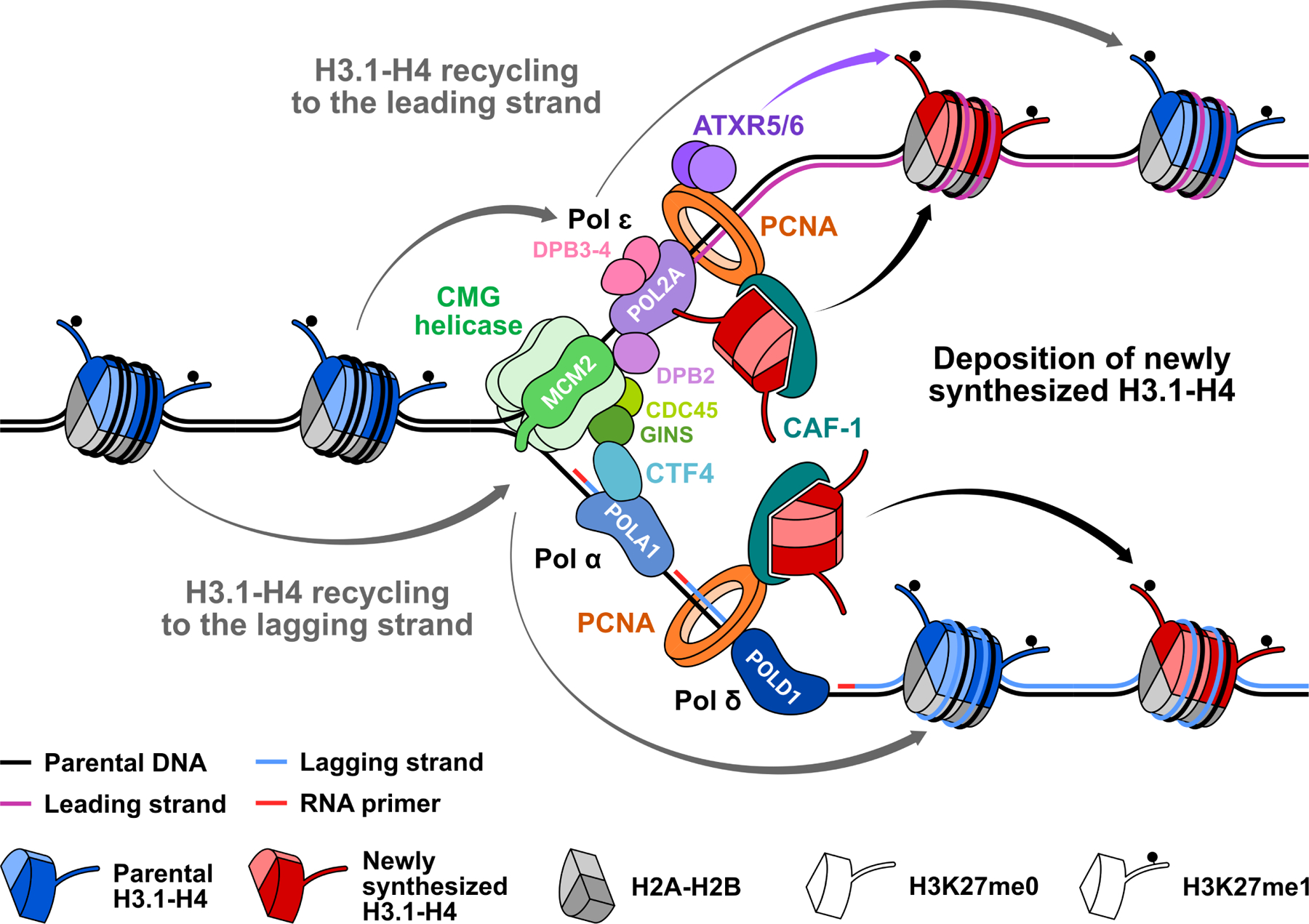
In constitutive heterochromatin, parental nucleosomes are enriched in H3.1 mono-methylated at K27. At the replication fork, the CMG helicase, which includes MCM2–7, CDC45, and GINS, unwinds parental double stranded DNA. DNA polymerase epsilon (Pol ɛ) then synthesizes the leading strand, while DNA polymerases alpha (Pol α) and delta (Pol δ) synthesize the lagging strand, with their respective catalytic subunits POL2A, POLA1, and POLD1. The CAF-1 complex interacts with the PCNA clamp to load newly synthesized H3.1-H4 tetramers on both strands of the fork, after which ATXR5/6 deposit H3K27me1. Parental H3-H4 are recycled symmetrically to the leading and lagging strands. POL2A can bind H3.1 in its unmethylated (i.e., newly synthesized) and mono-methylated (i.e., parental) forms with potential implications for epigenetic inheritance. DPB3–4 are the plant homologs of mammalian POLE3–4, two non-catalytic subunits of Pol ɛ that were shown mediate recycling of parental H3-H4 to the leading strand. The MCM2-CTF4-Pol α axis is required for recycling of parental H3-H4 to the lagging strand in mammals, and may accomplish similar functions in plants.
In the last few years, studies using new sequencing-based techniques to probe parental histone segregation during chromatin replication have uncovered specific roles for DNA polymerases and associated complexes. For example, the subunits POLE3 and POLE4 of mammalian POL ε were shown to mediate H3-H4 parental histone segregation to the leading strand [40,41], while MCM2 (a subunit of the replicative CMG helicase) and POLA1 (the catalytic subunit of DNA polymerase alpha) are involved in transferring parental H3-H4 to the lagging strand [40,42,43]. POLA1 was even shown recently to participate in the mitotic recycling of parental H2A-H2B to the lagging strand [44]. These results demonstrate a direct role for the DNA replication machinery in the inheritance of epigenetic states.
The discovery that POL2A specifically interacts with H3.1 raises interesting questions about the role of this interaction in terms of chromatin replication. Does the interaction indicate a specific role for POL2A in the insertion on chromatin of newly synthesized, unmodified H3.1 proteins over parental histones? Mono-, di-, and tri-methylation at H3K27 and H3K36 do not appear to have a major impact on the binding affinity of POL2A for H3.1 [39], arguing that modified (i.e., parental) histones may be a likely substrate for POL2A. If parental histones are inherited via POL2A, does it mean that parental H3.1-H4 dimers/tetramers are preferentially selected over parental H3.3-H4 during chromatin replication? Such a mechanism would have important implications for the mitotic inheritance of H3.3-enriched euchromatic states. A derived question would then be whether other DNA polymerase subunits or associated complexes can specifically transfer parental H3.1-H4 to the lagging strand during replication? If not, a unidirectional transfer of parental H3.1-H4 to the leading strand mediated by POL2A may be possible in some circumstances, and it could have functional ramifications for asymmetrical cell division [45]. Clearly, the discovery of the H3.1-POL2A interaction should lead to many interesting findings in the near future.
A substantial amount of work in plants has demonstrated the critical roles played by DNA polymerases in the mitotic inheritance of epigenetic states. Changes in active and repressive histone marks in hypomorphic mutants of plant DNA polymerases were shown to affect developmental genes and the maintenance of heterochromatin structure and silencing, with these effects proposed to be either direct (e.g., disruption of the physical interaction between DNA polymerase epsilon and PRC2) or indirect (e.g., loss of epigenetic information due to replication stress) [46–52]. Evidence for these roles in mediating epigenetic inheritance was obtained mainly from phenotypic characterization and genomic profiling of transcriptomic and epigenomic states in DNA polymerase mutants. It will be important to revisit these findings and apply new molecular and sequencing tools to better understand the mechanisms by which the DNA replication machinery affects chromatin replication in plant model systems.
Conclusions and perspectives
The discovery that H3.1 can specifically interact with POL2A and TSK strongly suggests that many other proteins may preferentially bind the replication-dependent H3 variant, with consequences for chromatin replication and all other DNA-localized activities (e.g., DNA repair) that must be coordinated during S phase of the cell cycle. Further advances in this field will require the identification of these H3.1-binding proteins, and their functional characterization. Similarly, other histone variants in the H1, H2A, and H2B families (a single H4 variant is present in Arabidopsis [6]) may play specific roles during replication, which could be revealed by identifying proteins that can discriminate between closely-related variants in these families. Biochemical screens involving binding and modification assays are likely to be the fastest way to identify new cellular activities that rely on distinguishing minor sequence differences between histone variants.
Plants have made major contributions to the study of mitotic inheritance of epigenetic states, principally by being great model systems for deploying genetic and epigenomic tools. It will be important to keep expanding on what plant systems can do to remain important models in this field. For example, the use and optimization of plant cell lines could provide similar advantages to mammalian cells in working out the molecular roles of proteins involved in chromatin replication, while also revealing plant-specific functional variation in these mechanisms (e.g., the plant-specific role played by H3.1F41 in mediating spatial localization of H3.1 [53]). As replication-dependent H3 variants are present in all multicellular eukaryotes, advances in this field of research will continue to rely on the use of different biological systems. This is exemplified by the recent discovery that H3.1 play a role in mediating replication timing in mammals [54,55].
Acknowledgments
We thank members of the Jacob lab for discussions and help with this review. The work was supported by grant #R35GM128661 from the National Institutes of Health to Y.J. V.J. received a postdoctoral fellowship from the Fonds de Recherche du Québec-Nature et Technologies (FRQNT) [272565].
Footnotes
Declaration of interests
Declaration of interests: none
References
- 1.Huang YC, Yuan W, Jacob Y: The Role of the TSK/TONSL-H3.1 Pathway in Maintaining Genome Stability in Multicellular Eukaryotes. Int J Mol Sci 2022, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okada T, Endo M, Singh MB, Bhalla PL: Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. The Plant journal : for cell and molecular biology 2005, 44:557–568. [DOI] [PubMed] [Google Scholar]
- 3.Wu RS, Bonner WM: Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell 1981, 27:321–330. [DOI] [PubMed] [Google Scholar]
- 4.Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ: The human and mouse replication-dependent histone genes. Genomics 2002, 80:487–498. [PubMed] [Google Scholar]
- 5.Corcoran ET, Jacob Y: Direct assessment of histone function using histone replacement. Trends Biochem Sci 2023, 48:53–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corcoran ET, LeBlanc C, Huang YC, Arias Tsang M, Sarkiss A, Hu Y, Pedmale UV, Jacob Y: Systematic histone H4 replacement in Arabidopsis thaliana reveals a role for H4R17 in regulating flowering time. Plant Cell 2022, 34:3611–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Y, Zhu Z, Meng G, Zhang R, Zhang Y: A CRISPR-Cas9 based shuffle system for endogenous histone H3 and H4 combinatorial mutagenesis. Sci Rep 2021, 11:3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sankar A, Mohammad F, Sundaramurthy AK, Wang H, Lerdrup M, Tatar T, Helin K: Histone editing elucidates the functional roles of H3K27 methylation and acetylation in mammals. Nat Genet 2022, 54:754–760. [DOI] [PubMed] [Google Scholar]; This work revealed a new gene editing strategy that can be used to create point mutations in a multiplexed manner at endogenous histone H3 genes. This was used to assess the roles of H3K27 methylation and acetylation and demonstrate that a gene replacement strategy can be successfully applied to the study histone proteins and/or residues in a mammalian cell line model.
- 9.Vasquez JJ, Wedel C, Cosentino RO, Siegel TN: Exploiting CRISPR-Cas9 technology to investigate individual histone modifications. Nucleic Acids Res 2018, 46:e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang T, Zhang Z, Dong Q, Xiong J, Zhu B: Histone H3K27 acetylation is dispensable for enhancer activity in mouse embryonic stem cells. Genome Biol 2020, 21:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benoit M, Simon L, Desset S, Duc C, Cotterell S, Poulet A, Le Goff S, Tatout C, Probst AV: Replication-coupled histone H3.1 deposition determines nucleosome composition and heterochromatin dynamics during Arabidopsis seedling development. New Phytol 2019, 221:385–398. [DOI] [PubMed] [Google Scholar]
- 12.Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, Schultz DC, Pchelintsev NA, Adams PD, Jansen LE, et al. : Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Molecular cell 2011, 44:928–941. [DOI] [PubMed] [Google Scholar]
- 13.Jiang D, Berger F: DNA replication-coupled histone modification maintains Polycomb gene silencing in plants. Science 2017, 357:1146–1149. [DOI] [PubMed] [Google Scholar]
- 14.Otero S, Desvoyes B, Peiro R, Gutierrez C: Histone H3 Dynamics Reveal Domains with Distinct Proliferation Potential in the Arabidopsis Root. Plant Cell 2016, 28:1361–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houlard M, Berlivet S, Probst AV, Quivy JP, Hery P, Almouzni G, Gerard M: CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet 2006, 2:e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song Y, He F, Xie G, Guo X, Xu Y, Chen Y, Liang X, Stagljar I, Egli D, Ma J, et al. : CAF-1 is essential for Drosophila development and involved in the maintenance of epigenetic memory. Dev Biol 2007, 311:213–222. [DOI] [PubMed] [Google Scholar]
- 17.Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T: FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell 2001, 104:131–142. [DOI] [PubMed] [Google Scholar]
- 18.Picart-Picolo A, Grob S, Picault N, Franek M, Llauro C, Halter T, Maier TR, Jobet E, Descombin J, Zhang P, et al. : Large tandem duplications affect gene expression, 3D organization, and plant-pathogen response. Genome Res 2020, 30:1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates a key role for CAF-1 in promoting genome stability via stabilization of rRNA gene copies and by preventing tandem duplications. Duplication of protein-coding genes can impact gene expression levels and plant phenotypes.
- 19.Davarinejad H, Huang YC, Mermaz B, LeBlanc C, Poulet A, Thomson G, Joly V, Munoz M, Arvanitis-Vigneault A, Valsakumar D, et al. : The histone H3.1 variant regulates TONSOKU-mediated DNA repair during replication. Science 2022, 375:1281–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work identified the DNA repair protein TSK as an H3.1 reader in plants. The interaction between TSK and H3.1 was shown to be regulated by H3.1K27me1, such that the absence of this histone mark induces ectopic TSK activity leading to genomic instability.
- 20.Huang TH, Fowler F, Chen CC, Shen ZJ, Sleckman B, Tyler JK: The Histone Chaperones ASF1 and CAF-1 Promote MMS22L-TONSL-Mediated Rad51 Loading onto ssDNA during Homologous Recombination in Human Cells. Mol Cell 2018, 69:879–892 e875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duro E, Lundin C, Ask K, Sanchez-Pulido L, MacArtney TJ, Toth R, Ponting CP, Groth A, Helleday T, Rouse J: Identification of the MMS22L-TONSL complex that promotes homologous recombination. Mol Cell 2010, 40:632–644. [DOI] [PubMed] [Google Scholar]
- 22.O’Connell BC, Adamson B, Lydeard JR, Sowa ME, Ciccia A, Bredemeyer AL, Schlabach M, Gygi SP, Elledge SJ, Harper JW: A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L-NFKBIL2 complex required for genomic stability. Mol Cell 2010, 40:645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Donnell L, Panier S, Wildenhain J, Tkach JM, Al-Hakim A, Landry MC, Escribano-Diaz C, Szilard RK, Young JT, Munro M, et al. : The MMS22L-TONSL complex mediates recovery from replication stress and homologous recombination. Mol Cell 2010, 40:619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piwko W, Olma MH, Held M, Bianco JN, Pedrioli PG, Hofmann K, Pasero P, Gerlich DW, Peter M: RNAi-based screening identifies the Mms22L-Nfkbil2 complex as a novel regulator of DNA replication in human cells. EMBO J 2010, 29:4210–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacob Y, Bergamin E, Donoghue MT, Mongeon V, LeBlanc C, Voigt P, Underwood CJ, Brunzelle JS, Michaels SD, Reinberg D, et al. : Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 2014, 343:1249–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacob Y, Feng S, LeBlanc CA, Bernatavichute YV, Stroud H, Cokus S, Johnson LM, Pellegrini M, Jacobsen SE, Michaels SD: ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nature structural & molecular biology 2009, 16:763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potok ME, Zhong Z, Picard CL, Liu Q, Do T, Jacobsen CE, Sakr O, Naranbaatar B, Thilakaratne R, Khnkoyan Z, et al. : The role of ATXR6 expression in modulating genome stability and transposable element repression in Arabidopsis. Proc Natl Acad Sci U S A 2022, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]; Previous results had suggested that the H3K27 methyltransferases ATXR5 and ATXR6 may have redundant functions in plants. This study demonstrates that they also play independent roles. In addition, it reveals that different mechanisms are involved in suppressing genomic instability in atxr5 atxr6 mutants.
- 28.Jacob Y, Stroud H, Leblanc C, Feng S, Zhuo L, Caro E, Hassel C, Gutierrez C, Michaels SD, Jacobsen SE: Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature 2010, 466:987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Exner V, Taranto P, Schonrock N, Gruissem W, Hennig L: Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development 2006, 133:4163–4172. [DOI] [PubMed] [Google Scholar]
- 30.Kirik A, Pecinka A, Wendeler E, Reiss B: The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. The Plant cell 2006, 18:2431–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramirez-Parra E, Gutierrez C: E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant physiology 2007, 144:105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schonrock N, Exner V, Probst A, Gruissem W, Hennig L: Functional genomic analysis of CAF-1 mutants in Arabidopsis thaliana. The Journal of biological chemistry 2006, 281:9560–9568. [DOI] [PubMed] [Google Scholar]
- 33.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G: PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Molecular cell 2006, 24:309–316. [DOI] [PubMed] [Google Scholar]
- 34.Bergamin E, Sarvan S, Malette J, Eram MS, Yeung S, Mongeon V, Joshi M, Brunzelle JS, Michaels SD, Blais A, et al. : Molecular basis for the methylation specificity of ATXR5 for histone H3. Nucleic Acids Res 2017, 45:6375–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guyomarc’h S, Vernoux T, Traas J, Zhou DX, Delarue M: MGOUN3, an Arabidopsis gene with TetratricoPeptide-Repeat-related motifs, regulates meristem cellular organization. J Exp Bot 2004, 55:673–684. [DOI] [PubMed] [Google Scholar]; Demonstration of another protein (i.e., POL2A) that can preferentially interact with H3.1 over H3.3. The mechanism employed by POL2A to recognize H3.1, which appears to be different from TSK, leads to regulation of chromatin organization during meiosis.
- 36.Takeda S, Tadele Z, Hofmann I, Probst AV, Angelis KJ, Kaya H, Araki T, Mengiste T, Mittelsten Scheid O, Shibahara K, et al. : BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes & development 2004, 18:782–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brzezinka K, Altmann S, Baurle I: BRUSHY1/TONSOKU/MGOUN3 is required for heat stress memory. Plant Cell Environ 2019, 42:771–781. [DOI] [PubMed] [Google Scholar]
- 38.Charng YY, Mitra S, Yu SJ: Maintenance of abiotic stress memory in plants: Lessons learned from heat acclimation. Plant Cell 2023, 35:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C, Huang J, Li Y, Zhang J, He C, Li T, Jiang D, Dong A, Ma H, Copenhaver GP, et al. : DNA polymerase epsilon binds histone H3.1-H4 and recruits MORC1 to mediate meiotic heterochromatin condensation. Proc Natl Acad Sci U S A 2022, 119:e2213540119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Z, Hua X, Serra-Cardona A, Xu X, Gan S, Zhou H, Yang WS, Chen CL, Xu RM, Zhang Z: DNA polymerase alpha interacts with H3-H4 and facilitates the transfer of parental histones to lagging strands. Sci Adv 2020, 6:eabb5820. [DOI] [PMC free article] [PubMed] [Google Scholar]; The dynamics of parental H3-H4 histones inheritance during replication have been previously investigated, but in contrast, little was known regarding the fate of H2A-H2B during the same process. This study provides evidence of recycling of parental H2A-H2B and their associated marks, an important process that serves to maintain epigenetic memory during replication.
- 41.Yu C, Gan H, Serra-Cardona A, Zhang L, Gan S, Sharma S, Johansson E, Chabes A, Xu RM, Zhang Z: A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science 2018, 361:1386–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gan H, Serra-Cardona A, Hua X, Zhou H, Labib K, Yu C, Zhang Z: The Mcm2-Ctf4-Polalpha Axis Facilitates Parental Histone H3-H4 Transfer to Lagging Strands. Mol Cell 2018, 72:140–151 e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petryk N, Dalby M, Wenger A, Stromme CB, Strandsby A, Andersson R, Groth A: MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 2018, 361:1389–1392. [DOI] [PubMed] [Google Scholar]
- 44.Flury V, Reveron-Gomez N, Alcaraz N, Stewart-Morgan KR, Wenger A, Klose RJ, Groth A: Recycling of modified H2A-H2B provides short-term memory of chromatin states. Cell 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urban JA, Ranjan R, Chen X: Asymmetric Histone Inheritance: Establishment, Recognition, and Execution. Annu Rev Genet 2022, 56:113–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bourguet P, Lopez-Gonzalez L, Gomez-Zambrano A, Pelissier T, Hesketh A, Potok ME, Pouch-Pelissier MN, Perez M, Da Ines O, Latrasse D, et al. : DNA polymerase epsilon is required for heterochromatin maintenance in Arabidopsis. Genome Biol 2020, 21:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Del Olmo I, Lopez JA, Vazquez J, Raynaud C, Pineiro M, Jarillo JA: Arabidopsis DNA polymerase ϵ recruits components of Polycomb repressor complex to mediate epigenetic gene silencing. Nucleic Acids Res 2016, 44:5597–5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.del Olmo I, Lopez-Gonzalez L, Martin-Trillo MM, Martinez-Zapater JM, Pineiro M, Jarillo JA: EARLY IN SHORT DAYS 7 (ESD7) encodes the catalytic subunit of DNA polymerase epsilon and is required for flowering repression through a mechanism involving epigenetic gene silencing. Plant J 2010, 61:623–636. [DOI] [PubMed] [Google Scholar]
- 49.Hyun Y, Yun H, Park K, Ohr H, Lee O, Kim DH, Sung S, Choi Y: The catalytic subunit of Arabidopsis DNA polymerase alpha ensures stable maintenance of histone modification. Development 2013, 140:156–166. [DOI] [PubMed] [Google Scholar]
- 50.Iglesias FM, Bruera NA, Dergan-Dylon S, Marino-Buslje C, Lorenzi H, Mateos JL, Turck F, Coupland G, Cerdan PD: The arabidopsis DNA polymerase delta has a role in the deposition of transcriptionally active epigenetic marks, development and flowering. PLoS Genet 2015, 11:e1004975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedroza-Garcia JA, De Veylder L, Raynaud C: Plant DNA Polymerases. Int J Mol Sci 2019, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J, Xie S, Cheng J, Lai J, Zhu JK, Gong Z: The Second Subunit of DNA Polymerase Delta Is Required for Genomic Stability and Epigenetic Regulation. Plant Physiol 2016, 171:1192–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu L, Chen X, Qian S, Zhong X: The plant-specific histone residue Phe41 is important for genome-wide H3.1 distribution. Nat Commun 2018, 9:630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gatto A, Forest A, Quivy JP, Almouzni G: HIRA-dependent boundaries between H3 variants shape early replication in mammals. Mol Cell 2022, 82:1909–1923 e1905. [DOI] [PubMed] [Google Scholar]
- 55.Kawamura M, Funaya S, Sugie K, Suzuki MG, Aoki F: Asymmetrical deposition and modification of histone H3 variants are essential for zygote development. Life Sci Alliance 2021, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]


