Abstract
Systemic lupus erythematosus (SLE) is a prototypic autoimmune disease characterized by antibodies to DNA (anti-DNA) and other nuclear macromolecules. Anti-DNA antibodies are markers for classification and disease activity and promote pathogenesis by forming immune complexes that deposit in the tissue or stimulate cytokine production. Studies on the antibody response to DNA have focused primarily on a conformation of DNA known as B-DNA, the classic right-handed double helix. Among other conformations of DNA, Z-DNA is a left-handed helix with a zigzag backbone; hence, the term Z-DNA. Z-DNA formation is favored by certain base sequences, with the energetically unfavorable flip from B-DNA to Z-DNA dependent on conditions. Z-DNA differs from B-DNA in its immunogenicity in animal models. Furthermore, anti-Z-DNA antibodies, but not anti-B-DNA antibodies, can be present in otherwise healthy individuals. In SLE, antibodies to Z-DNA can occur in association with antibodies to B-DNA as a cross-reactive response, rising and falling together. While formed transiently in chromosomal DNA, Z-DNA is stably present in bacterial biofilms; biofilms can provide protection against antibiotics and other challenges including elements of host defense. The high GC content of certain bacterial DNA also favors Z-DNA formation as do DNA-binding proteins of bacterial or host origin. Together, these findings suggest that sources of Z-DNA can enhance the immunogenicity of DNA and, in SLE, stimulate the production of cross-reactive antibodies that bind both B-DNA and Z-DNA. As such, DNA can act as a molecular chameleon that, when stabilized in the Z-DNA conformation, can drive autoimmunity.
Introduction
Systemic lupus erythematosus (SLE) is a prototypic autoimmune disease characterized by inflammatory disease manifestations in association with antibodies to components of the cell nucleus (antinuclear antibodies of ANAs) 1. These antibodies can bind to DNA, RNA as well as protein-nucleic acid complexes. ANA expression is so linked with SLE that a positive test for these antibodies is now required for disease classification 2 3. According to the current model for SLE, ANAs can bind to their cognate antigens to form immune complexes with two main actions: deposition in the kidney to induce nephritis and stimulation of aberrant cytokine production by interaction of the constituent DNA or RNA with internal nucleic acid sensors 4.
Among ANAs expressed in SLE, antibodies to DNA (anti-DNA) are unique since they are markers for both classification and disease activity 5–7. A large negatively charged polymer, DNA displays a repeating structure marked by the helical twist of the phosphodiester backbone (Figure 1). The main conformation of DNA is called B-DNA. B-DNA is the classic Watson-Crick right-handed double helix with two chains bound together by hydrogen bonds to form a smooth helix with a major and minor groove. Since anti-DNA antibodies from SLE patients can bind to essentially any natural double stranded DNA, the phosphodiester backbone appears to be the relevant antigenic structure.
Figure 1.
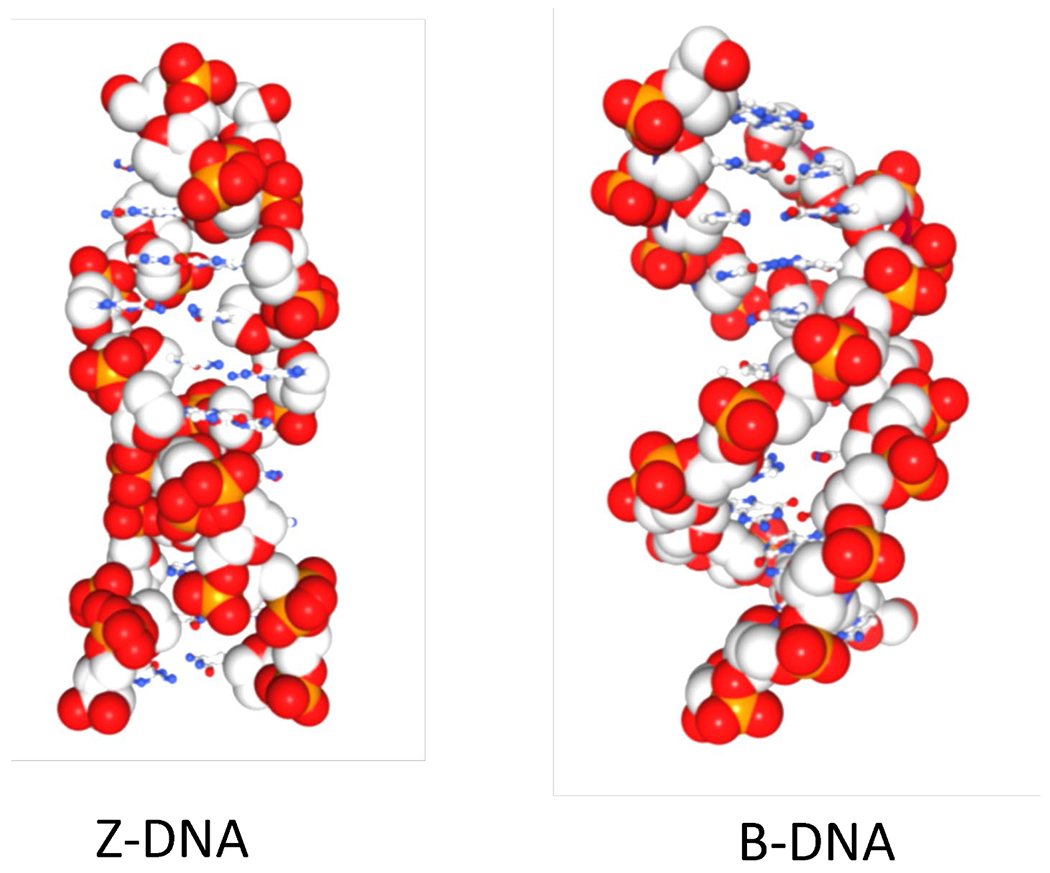
DNA chameleon colors. Left-handed Z-DNA has a zig-zag backbone resulting from the dinucleotide repeat with alternating syn- and anti-bases. In contrast, the backbone of right-handed B-DNA is smooth as all the bases have an anti-conformation. The phosphate oxygens are red, the phosphorous atom orange and the gray spheres represent deoxyribose carbons. The small blue circles are base nitrogens while the small red circles base oxygens.
Modeling anti-DNA production by immunization of animals with B-DNA has in general been unsuccessful, even with DNA bound to protein carrier such as methylated bovine serum albumin and administered in adjuvant; other carriers, however, may lead to more robust responses 8 9. The poor immunogenicity of B-DNA has been a conundrum since monoclonal anti-B-DNA from patients or mice with lupus-like disease have the hallmarks of antigen selection by negatively charged DNA (i.e., variable region somatic mutations leading to positively charged amino acids such as arginine) 10 11. The failure to establish an induced model of SLE has even suggested that a molecule other than DNA induces anti-DNA production.
As now recognized, DNA can exist in conformations other than B-DNA; in the genome, these conformations can arise from sequences called flipons 12 13. These non-B-DNA conformations, which depend on base sequence and ambient conditions, differ in the orientation and dimensions of the helix. This review will focus on the unique properties of a conformation known as Z-DNA and advance the idea that DNA can act as a molecular chameleon whose structure and immunological properties can dramatically change. A chameleon is a small lizard that can change its color depending on the surroundings. By considering DNA as a molecular chameleon, we believe that new insights into the immune response to DNA can be achieved, thereby illuminating the pathogenesis of SLE.
The structure of DNA
The double helix has been called a beautiful molecule because its structure can account so elegantly for its role in heredity. From the time that the original Watson-Crick model was first proposed, structural studies indicated that DNA can adopt conformations other than right-handed B-DNA, each just as beautiful. Indeed, the first crystal structure of double-stranded DNA showed a helical twist to the left rather than to the right 14. Instead of being smooth as in B-DNA, the backbone in this structure had a zig-zag configuration that gave rise to the name of Z-DNA (Figure 1).
The structure of Z-DNA shows a two base pair repeat, alternating between a nucleotide in the syn conformation (with its base pointing back over the sugar) and the anti conformation (with the base pointing away from the sugar as found in B-DNA) 15–17. Also, in Z-DNA, the base pairs are flipped over relative to that of B-DNA, with the flip in conformation occurring without breaks in the DNA backbone 16 18 19. The transition is dynamic and rapid, with Z-DNA formation requiring an input of energy, mostly to initiate the formation of the two junctions between B- and Z-DNA (BZj). The likelihood of a flip to Z-DNA depends on base sequence, with alternating GC sequences converting to Z-DNA most readily. Like DNA, RNA can form a Z structure that depends on sequence; Z-DNA and Z-RNA can be bound by the same proteins, including antibodies, and both are designated as Z nucleic acids.
In model studies in vitro, the flip from B-DNA to Z-DNA is influenced by a variety of factors that include the salt concentration, with 4 M NaCl or 1 M MgCl2 needed to induce the Z-DNA conformation in Z-prone polymers. Chemical modifications of DNA bases can also promote the B-DNA to Z-DNA transition under more physiological conditions by stabilizing the Z-DNA syn conformation. These modifications include bromination, methylation, and oxidation 20 21 22 23. Many biochemical and immunochemical studies to elucidate Z-DNA-dependent outcomes have used the compound brominated poly(dGdC) [Br-poly(dGdC)] as a model substrate since the polymer is locked in the Z-DNA conformation under ordinary salt conditions as shown by analysis of circular dichroism and the ratio of 260 to 295 nm UV absorbances 22.
The structure of Z-DNA and the conditions needed for its induction differ so greatly from those of B-DNA that its biological role (and even its existence inside cells) was long debated. Increasing data, however, support the relevance of Z-DNA to in vivo cell function. As discussed in recent reviews, the transition to non-B-DNA conformations such as Z-DNA can occur under physiological conditions to regulate gene transcription 15 24. Supercoiling induced by the action of RNA polymerases can promote the B to Z transition during transcription; in this setting, the flip can be modulated by methylation and other base modifications. Enzymes like topoisomerases can also affect the transition 24 25.
The antibody response to Z-DNA
Unlike B-DNA, Z-DNA is an effective immunogen and can potently induce antibodies by immunization of normal animals under conditions in which B-DNA is inactive 20 26–28. Anti-Z-DNA antibodies induced by immunization with compounds such as Br-poly(dGdC) are highly specific for Z-DNA and bind by non-ionic interactions since binding occurs in high salt conditions. At least some of antibodies to Z-DNA induced by immunization appear to recognize sequences, likely binding to base residues ordinarily buried in the major groove of B-DNA but exposed on the convex surface of Z-DNA 28.
The immunogenicity of Z-DNA as revealed in immunization models has suggested that Z-DNA may not induce canonical B cell tolerance through deletion or anergy because of its transience or low concentrations. In contrast, B-DNA is both ubiquitous and abundant and may therefore involve different mechanisms of central and peripheral B cell tolerance than Z-DNA. While tolerance to DNA is a topic of ongoing investigations, nevertheless, the differences between the antibody induction by B-DNA and Z-DNA are nevertheless striking.
In addition to immunization of animals, the expression of anti-Z-DNA occurs in SLE 29–33. Sera of patients with SLE display levels of anti-Z-DNA antibodies that are similar to those to B-DNA when measured using synthetic Z-DNA antigens [e.g., Br-poly(dGdC)]. Furthermore, antibodies to both B-DNA and Z-DNA conformations can rise and fall together, suggesting common induction or cross-reactivity (Figure 2) 29 32. Affinity adsorption experiments indicate that certain anti-DNA antibodies in SLE can bind both B-DNA and Z-DNA, while others are specific for Z-DNA 29 32. While antibodies to Z-DNA were initially described almost 40 years ago, it was not clear until recently how to interpret this discovery.
Figure 2.
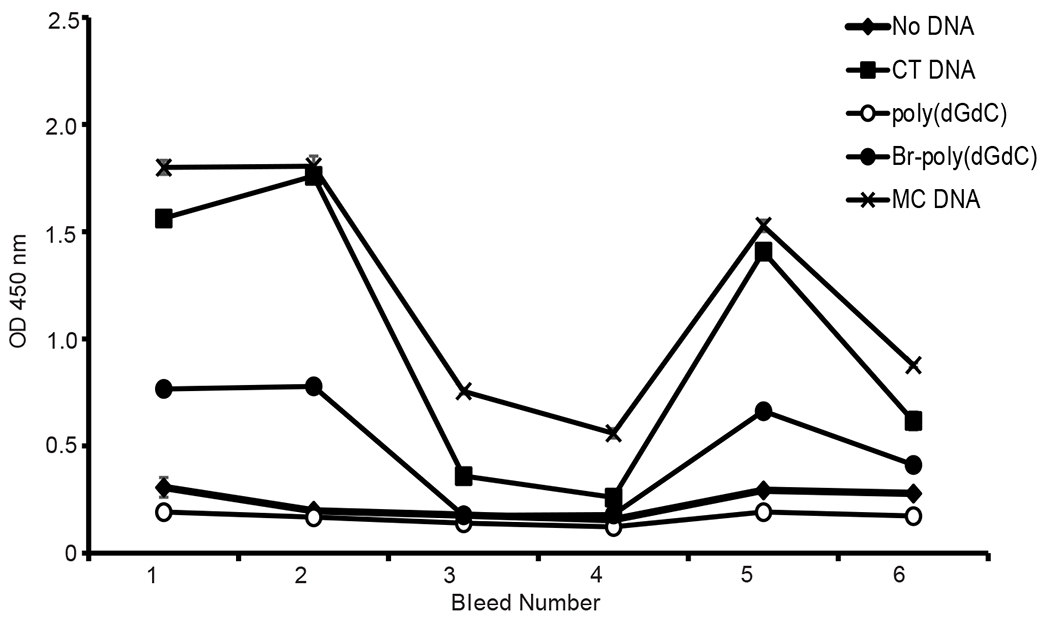
Longitudinal expression of antibodies to B-DNA and Z-DNA. The figure shows levels of IgG antibodies to different DNA antigens in longitudinal sera from a patient with SLE. The DNA antigens were from calf thymus (CT), Micrococcus luteus (MC), Br-poly(dGdC) and unbrominated poly(dGdC). Br-poly(dGdC) represents Z-DNA. CT and MC DNA both represent B-DNA although MC DNA has Z-DNA content. Antibody levels were determined by an ELISA with results reported in terms of the OD450 values of the assay. Reproduced with permission from reference 32.
Like SLE antibodies to B-DNA, antibodies to Z-DNA from patients depend on ionic interactions suggesting that the phosphodiester backbone is the main antigenic determinant 32 33. Given the differences in the structure of the phosphodiester backbone in B-DNA and Z-DNA (smooth vs zig-zag, right-handed vs left-handed), cross-reactive binding is perhaps surprising. Cross-reactivity can extend to other charged antigens such as phospholipids, however, suggesting epitope spreading 33. The presence of a broad array of anti-DNA specificities in SLE supports this mechanism in the setting of autoimmunity 34.
Unlike the antibodies to B-DNA which are specific for SLE, antibodies to Z-DNA can occur in other inflammatory diseases such as rheumatoid arthritis, inflammatory bowel disease and drug-induced lupus; in these conditions, antibodies to B-DNA are not present 31 35 36. Importantly, some studies have detected antibodies to Z-DNA in otherwise healthy humans (NHS) 32 33. Together, these findings suggest that antibodies in SLE that bind to both Z-DNA and B-DNA are indicative of a more generalized breakdown of immune tolerance. The predominance of ionic interactions in the binding of both B-DNA and Z-DNA by SLE anti-DNA could suggest a relatively “non-specific” interaction based on charge; nevertheless, specificity is possible since the anti-Z-DNA antibodies in clinical settings other than SLE do not bind to B-DNA.
In their pattern of expression and binding interactions, antibodies to Z-DNA resemble antibodies that are specific for certain bacterial DNA and viral DNA antigens 37–39. These antibodies occur in NHS as well as in patients with SLE, lack reactivity to B-DNA and are primarily the IgG2 isotype; in contrast, anti-B-DNA antibodies from patients with SLE primarily display IgG1, an isotype characteristic of T cell dependent responses 37. Because of the specificity of these antibodies for sites on DNA from only certain bacteria or viruses, these antibodies are not detected in the standard assays used to screen for anti-DNA in patients.
The role of extracellular bacterial DNA in immunity
Recent research on biofilms has dramatically revealed the chameleon-like nature of DNA in the extracellular space and provides intriguing clues about the potential origin of anti-Z-DNA antibodies in both normal and aberrant immunity.40–42. Biofilms represent communities of bacteria that are embedded in a multicomponent matrix to facilitate bacterial growth; biofilms provide nutritional support and protection from environmental influences, including antibiotics. Encounters with biofilms are almost inevitable in life since about 80% of infections involve biofilms formation; furthermore, bacteria in the microbiome grow as biofilms. Among components in the matrix, high molecular weight DNA can create extended arrays through its interaction with other macromolecules. The DNA present in biofilms can also promote immune responses through the activation of TLR9 (Toll-ike receptor 9) by the unmethylated CpG motifs common in bacterial genomes 43.
In a fascinating study on biofilm structure, Buzzo et al showed that DNA in a biofilm can undergo a B-DNA to Z-DNA transition that may explain the protective effects of the biofilm44. This transition can result from the interaction of DNA with HU (histone like protein) and IHF (integration host factor), two bacterial DNABII proteins incorporating HMG boxes that promote the B-DNA to Z-DNA transition. Both proteins bend DNA and most likely promote Z-DNA formation by stabilizing BZj 45; this stabilization overcomes the major energetic barrier to Z-DNA formation.
The data of Buzzo et al also suggest that biofilms formed by various organisms all express Z-DNA; the base composition of each genome, however, could perhaps reflect the rate at which B-DNA flips to Z-DNA. Over time, Z-DNA will accumulate in biofilms since the left-handed DNA, unlike B-DNA, is resistant to digestion by mammalian DNases 22. Indeed, a seminal study of Whitchurch et al 46 showed that DNase 1 can inhibit biofilms in vitro during its early stage of formation but cannot degrade mature films. Of note is the susceptibility of Z-DNA in biofilms to the S1 bacterial nuclease, contrasting with the resistance to mammalian DNases 22 47. Importantly, Buzzo et al were able to use a monoclonal antibody to promote the formation of biofilms in vitro by using a monoclonal antibody to bind and stabilize DNA in the Z-DNA conformation; a similar effect occurs with bacterial HU and HIS proteins 44. In contrast to the anti-Z-DNA antibodies in SLE and NHS, the, binding of the anti-Z-DNA monoclonal antibody in these experiments does not involve electrostatic interactions 32.
In addition to DNA of prokaryotic origin, eukaryotic DNA from neutrophils undergoing NETosis can also contribute to the biofilm structure. As neutrophils undergo NETosis, they release DNA which interacts with neutrophil granule proteins to form NETs or neutrophil extracellular traps 48. NETs have anti-bacterial action and can ensnare and kill bacteria (Figure 3A). Like DNA from the bacteria in the biofilm, DNA from NETs can undergo a B-DNA to Z-DNA transition under the influence of DNABII proteins. DNA in NETs may also undergo Z-DNA formation because host proteins such as HMGB1 can bend DNA to potentially stabilize BZj (Figure 3B). Interestingly, the association of HMGB1 with DNA is increased by neutrophil elastase that removes the C-terminal acidic tail 49.
Figure 3.
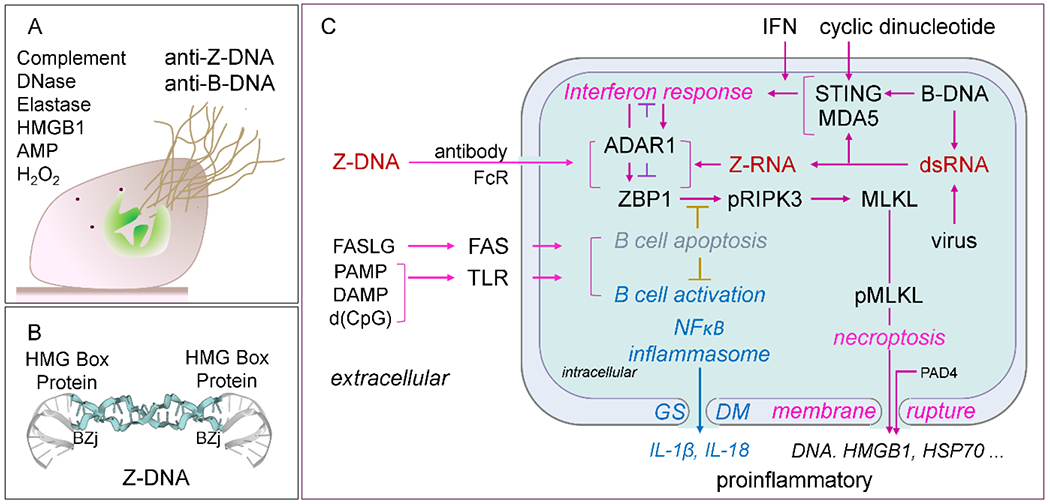
NETs, Z-DNA and Pathways. A. Extrusion of DNA from neutrophils along with other nuclear molecules leads to the formation of NETs and can be recognized by anti-B-DNA and anti-Z-DNA antibodies. The various proteases, anti-microbial proteins (AMP) and enzymes contribute to the antibacterial activity. Complement can opsonize bacterial DNA to promote clearance. B. HMG box containing proteins like HU and IHF from bacteria and HMGB1 from the host promote the formation of B-DNA/Z-DNA junctions (BZj) that stabilize Z-DNA formation by NETs. C. Different signaling pathways that regulate the proliferation of autoimmune B cells and interferon responses during inflammation. DNA can be delivered inside of cells following uptake by anti-DNA antibodies by the B cell receptor to deliver DNA into endosomes or through other pathways that deliver DNA to the cytoplasm. Once inside cells, DNA can interact with internal sensors and lead to signal transduction. These pathways complement those delivering DNA through Fc receptors (FcR) as immune complexes and through toll-like receptors to promote B cell proliferation and interferon responses. Intracellular activation of the Z-DNA binding protein 1 (ZBP1) sensor by Z-DNA or Z-RNA can activate RIPK3 (receptor interacting kinase to phosphorylate MLKL to induce pore formation that results in necroptosis, a form of inflammatory cell death, and activation of inflammasomes. GADM is gasdermin which can create pores to release cytokines or promote apoptosis. FAS and TLR signaling systems can have roles in B cell activation and apoptosis. Gasdermin (GSDM) pores can lead to release of IL-1β and IL-18 or apoptosis.
The immunogenicity of biofilm components has already received experimental support. Anti-DNA can be induced by immunization with a complex of DNA and curli, an amyloid forming protein found in biofilms 50 51. Sera of patients with SLE or otherwise healthy individuals have antibodies to DNA-curli, consistent with exposure to biofilm 52. In another setting, experimental infection of mice with Salmonella Typhimurium can induce anti-DNA autoantibody production 53. Together, these findings suggest that infection can provide a source of Z-DNA to drive antibody responses that have tell-tale serological evidence of encounters with immunogenic bacterial DNA.
While the biochemical basis of the transition to Z-DNA still requires further investigation, the results are very important for thinking about lupus. Thus, Z-DNA resists nuclease digestion by enzymes such as DNase 1 and, thus far, a mammalian enzyme specific for Z-DNA has not been described. Once Z-DNA forms in the biofilm, it can provide a persistent and high concentration of antigen to induce antibody production; in this sense, the biofilm can act as a depot to supply antigen to the system. The persistence of a local source of Z-DNA in the tissue contrasts with the fate of B-DNA in the blood which is rapidly degraded within minutes 54.
Another finding that highlights the possible immunogenic role of Z-DNA relates to the propensity of bacterial genomic DNA to display Z-DNA. The GC content of bacterial DNA varies widely, with content as low as 20% and as high as 80% 55. As shown recently, a monoclonal anti-Z-DNA antibody from a mouse and polyclonal anti-Z-DNA preparations can bind at high titers to DNA from Micrococcus luteus (MC) and Mycobacterium tuberculosis (MTb) under the ordinary conditions of an ELISA 32. The GC content of MC DNA is 73-74% and 65-66% for MTb DNA compared to 40-41% for human genomic and 44-45% for mitochondrial DNA 56 57 58. In these experiments, the anti-Z-DNA antibodies showed limited if any binding to other sources of DNA such as calf thymus DNA; in contrast, a monoclonal anti-Z-DNA bound E. coli DNA (50-51% GC) 32.
Since biofilms represent arrays of Z-DNA antigen, these structures could act as thymus-independent antigens, with the activation of naive B-cells may be driven by multiple low-affinity interactions with a Z-DNA array. Consistent with this idea, antibodies to Z-DNA in NHS are predominantly of the IgG2 isotype, rather than the IgG1 or IgG3 that are typical of T cell dependent responses 37.
The role of nucleases
Despite its origin in the cell nucleus, DNA occurs prominently in the extracellular space as a result of cell death and NETosis 48. While much of the DNA initially generated from these processes is high molecular weight, DNA in the circulation is generally of much lower molecular mass, with a mean size of 166 bases pairs 59 60. The marked reduction in size results from degradation by intracellular nucleases during phagocytosis of dead and dying cells as well as by extracellular nucleases in the tissue or blood 61. The two main extracellular DNase enzymes are DNase 1 and DNase 1 like 3 (DNase 1L3) 62. While these enzymes have structural similarity, DNase 1 prefers free or naked DNA as the substrate while DNase 1L3 targets nucleosomal DNA. DNase 1 and DNase 1L3 both play a role in clearing and degrading DNA in the extracellular space, with deficiencies of these enzymes associated with diseases with features of SLE in both humans and mice 63 64 65. While enzyme deficiencies can be genetic in origin, autoantibodies expressed by patients can inhibit enzyme activities to increase levels of extracellular DNA 66.
The role of DNases in degrading exogenous DNA in host defense is important. As shown by Lacey et al., deficiency of DNase 1 and DNase 1L3 increases the formation of biofilms during experimental infection of mice, consistent with prior studies showing the prevention of biofilm formation by DNase I in vitro 67. These results suggest that DNase 1 and DNase 1L3, in addition to targeting DNA from dead and dying cells, can target non-self-DNA, whether arising from infection, colonization, or the microbiome. With enzyme deficiency, persistence of the biofilm would favor Z-DNA formation, increasing the amount of Z-DNA incorporated.
While Z-DNA is not susceptible to nuclease digestion by DNase 1 or DNase 1L3, proteolytic digestion of proteins such as HU, IHF and HMGB1 that stabilize Z-DNA could allow the flip back to B-DNA; this digestion can occur intra- or extra-cellularly, where enzymes like DNase 1 and DNase 1L3 can mediate NET destruction. In this clearance process, the classical pathway of complement may also play a role since C1q deficiency is one cause of monogenic lupus 68 69. In other cases, antibodies binding to NETs may functionally produce C1q deficiency 70. If unresolved, the NETs stabilized by anti-Z-DNA antibodies may be long-lived because of Z-DNA’s resistance to DNase, forming a positive feedback loop for inflammation.
Interestingly, current therapy may impact on these processes. An antimalarial agents such as hydroxychloroquine can shift the conformational equilibrium of DNA to favor B-DNA and retard the development of biofilms 71. In another action, hydroxychloroquine can increase the sensitivity of B-DNA to DNase digestion 72 and thereby reduce the formation of immunostimulatory Z-DNA in the biofilm, whether of bacterial or host origin. Finally, hydroxychloroquine can inhibit PAD4 (peptidyl arginine deiminase 4) in vitro to prevent the release of DNA from neutrophils during NETosis 73.
The role of nucleic acid sensors
Incorporating bacterial Z-DNA model into a model for lupus thus requires consideration of the intricacies of sensing of the Z conformation although most of the studies on this issue have focused on the recognition of RNA from viral infection or transcription of host retroelements. Figure 3c presents an overall schema for the sensing of Z nucleic acids and the many interactions and downstream consequences. Since the issue of Z-nucleic acid sensing is beyond the scope of this article, the reader is referred to reviews which describe sensing to Z nucleic acids 74 75 76 77.
Key to signaling by Z-nucleic acids 78 74 79 are two proteins: ZBP1 (Z binding protein 1) and ADAR1 (adenosine deaminase acting on RNA 1). ZBP1 was originally discovered as a cytoplasmic DNA sensor called DAI (DNA-dependent activator of IFN-regulatory factors) with subsequent studies demonstrating Z- nucleic acids as the ligand 80. The other player in this system is ADAR1 (adenosine deaminase of RNA 1). ADAR1 has two major isoforms, p110 and p150, which catalyze the conversion of adenosine to inosine in double stranded RNA; this process is known as RNA editing. Both ADAR1 isoforms can sequester right-handed dsRNA molecules through three dsRNA binding domains; sequestration prevents the activation of other RNA sensing systems such as myeloma differentiation-associated gene 5 (MDA5) and Protein Kinase RNA specific (PKR) to limit immune responses.
ADAR1 p150 is the only isoform that can recognize Z-DNA and, like ZBP1, it can interact with Z-RNA through a Zα domain; this domain is lacking in ADAR1 p110, accounting for the difference in action. In this pathway, ADAR1 can bind to Z-RNA through its Zα domain to prevent activation of ZBP1. Interestingly, loss of function Zα domain variants of ADAR1 can produce Aicardi-Goutières syndrome, a type I interferonopathy that has some features of SLE 81. Under normal cellular conditions, Z-RNA binding by ADAR1 prevents immune responses induced by self-transcripts from retroelements that have embedded Z-forming elements 75.
Interferon can induce the expression of both ADAR1 p150 and ZBP1, with differing consequences. Whereas ADAR1 can act as a negative regulator of the interferon response, ZBP1 can produce inflammatory cell death 82. (Figure 3C). Furthermore, with activation, ZBP1 can interact with RIPK3 to cause phosphorylation of MLKL (mixed lineage domain like protein); phosphorylated MLKL can self-assemble to form membrane pores, regardless of whether ZBP1 is triggered by Z-DNA or Z-RNA. These pores allow the release of DNA from various cell compartments, with DNA from mitochondria combining with ZBP1 to activate STING to induce interferon production 83. Induction of high levels of Z-RNA from retroelements, which can occur during viral infections can amplify activation 76. NLRP3 inflammasome activation to produce IL-1 and IL-18 can also occur with the flux of ions through the MLKL pores.84 85 86. The role Z nucleic acids play in host defense may be reflected in the complex evolution of ZBP1 and ADAR1, with viruses such as smallpox and measles driving the selection of these molecules 87 88.
As these considerations suggest, sensing of Z nucleic acids can impact other systems (e.g., STING, NLRP3 inflammasome) that can exert adjuvant activity for induction of antibodies to Z-DNA. In this regard, DNA from bacteria may trigger TLR9 (via CpG motifs), cGAS-STING (presence in the cytoplasm) and ZBP1 (either through Z-forming sequences in the genome or conformational changes in the biofilm). Thus, even though sensing of Z nucleic acids frequently involves RNA, this system could significantly boost antibody responses to DNA in health and autoimmunity.
DNA as a Chameleon
To highlight a novel mechanism of host defense, we would like to apply the term chameleon to bacterial and host DNA as it transitions from B-DNA to Z-DNA in the setting of the biofilm. The term also applies to DNA from NETs which, under the influence of DNA binding proteins with an HMG domain, can form Z-DNA. These binding proteins may stabilize BZj by bending DNA to a lower the energy barrier to flip from B-DNA to Z-DNA. The formation of Z-DNA may also be promoted by the presence of oxidized bases, such as 8-oxodG (8-oxo-2’-deoxguanosine), through the release of NADPH oxidase during NETosis and by H2O2 production catalyzed by DNA 89 90 91. Furthermore, adjuvant effects could result from interferon-induced expression of Z-RNA, internalization of antibody-bound Z-DNA or the occurrence of cell death triggered by ZBP1.
The serological studies on anti-Z-DNA in humans suggest that, in normal individuals, induced antibodies obey the rules and are specific for the Z-DNA structure. In SLE, antibody cross-reactivity between B-DNA and Z-DNA may occur because of the disturbances in the B cell repertoire. In addition, disturbances in developmental checkpoints may allow retention of precursors that are polyreactive and bind broadly to DNA 92 93. In the setting of a biofilm where Z-DNA concentrations are high, T-cell independent activation of autoreactive B cells may occur, accounting for the expression of antibodies to nuclear molecules (including those from NETs) in individuals genetically prone to autoimmunity 94 95.
This model does not necessitate a role of self DNA in the induction of the anti-DNA response since foreign DNA (i.e., bacterial, or viral) has regions of both B-DNA and Z-DNA depending on the stage of the infection, the stage of biofilm or the sequence of the DNA. While not excluding a role of self or B-DNA in inducing anti-DNA autoantibody production, a model of anti-DNA based on the changing immunological properties of DNA focuses attention on the potential roles of infection, NETosis and stress in pathogenesis; this model also incorporates the role of host DNase enzymes which, while effective in clearing B-DNA, may be stymied by a Z-DNA.
Conclusion
Every analogy has its limitations but the term chameleon appears apt to describe the structural transformations that DNA can undergo. For lizards, the color change is defensive and protective, and hides the lizard from predators. In a related way, the transition from B-DNA to Z-DNA may protect the bacteria from elements of the immune system. Future studies will explore approaches to block the B-DNA to Z-DNA transition and determine the impact of anti-DNA antibodies on the course of lupus as well as the role of any ongoing infections or microbiome blooms provoking disease flares. Finally, new approaches to treating biofilms may also change the color of the immune system in both autoimmune and infectious disease.
Funding
Work completed in Dr. Pisetsky’s lab was supported by a Veterans Administration Merit Review grant (BX003772) and a National Institutes of Health grant (R01AR073935).
Footnotes
Competing and Conflicts of Interests
The authors declare no conflict of interest.
References
- 1.Tsokos GC, Lo MS, Costa Reis P, et al. New insights into the immunopathogenesis of systemic lupus erythematosus. Nature reviews Rheumatology 2016;12:716–30. doi: 10.1038/nrrheum.2016.186 [published Online First: 2016/11/23] [DOI] [PubMed] [Google Scholar]
- 2.Pisetsky DS, Lipsky PE. New insights into the role of antinuclear antibodies in systemic lupus erythematosus. Nature reviews Rheumatology 2020;16(10):565–79. doi: 10.1038/s41584-020-0480-7 [published Online First: 2020/09/05] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aringer M, et al. 2019 European League Against Rheumatism/American College of Rheumatology classification criteria for systemic lupus erythematosus. Ann Rhem Dis 2019;0:1151–59. doi: 10.1136/annrheumdis-2018-214819 [DOI] [PubMed] [Google Scholar]
- 4.Pisetsky DS. The central role of nucleic acids in the pathogenesis of systemic lupus erythematosus. F1000Research 2019;8 doi: 10.12688/f1000research.17959.1 [published Online First: 2019/04/20] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jang YJ, Stollar BD. Anti-DNA antibodies: aspects of structure and pathogenicity. Cellular and Molecular Life Sciences 2003;60(2):309–20. [published Online First: 2003/04/08] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pisetsky DS. Anti-DNA antibodies--quintessential biomarkers of SLE. Nature reviews Rheumatology 2016;12(2):102–10. doi: 10.1038/nrrheum.2015.151 [published Online First: 2015/11/20] [DOI] [PubMed] [Google Scholar]
- 7.Rekvig OP. The anti-DNA antibody: origin and impact, dogmas and controversies. Nature reviews Rheumatology 2015;11(9):530–40. doi: 10.1038/nrrheum.2015.69 [published Online First: 2015/06/03] [DOI] [PubMed] [Google Scholar]
- 8.Desai DD, Krishnan MR, Swindle JT, et al. Antigen-specific induction of antibodies against native mammalian DNA in nonautoimmune mice. Journal of immunology (Baltimore, Md : 1950) 1993;151(3):1614–26. [published Online First: 1993/08/01] [PubMed] [Google Scholar]
- 9.Desai DD, Marion TN. Induction of anti-DNA antibody with DNA-peptide complexes. International immunology 2000;12(11):1569–78. doi: 10.1093/intimm/12.11.1569 [published Online First: 2000/11/04] [DOI] [PubMed] [Google Scholar]
- 10.Radic MZ, Weigert M. Genetic and structural evidence for antigen selection of anti-DNA antibodies. Annual review of immunology 1994;12:487–520. doi: 10.1146/annurev.iy.12.040194.002415 [published Online First: 1994/01/01] [DOI] [PubMed] [Google Scholar]
- 11.Stollar BD. Why the difference between B-DNA and Z-DNA? Lupus 1997;6(3):327–8. doi: 10.1177/096120339700600327 [published Online First: 1997/01/01] [DOI] [PubMed] [Google Scholar]
- 12.Matos-Rodrigues G, Hisey JA, Nussenzweig A, et al. Detection of alternative DNA structures and its implications for human disease. Molecular cell 2023;83(20):3622–41. doi: 10.1016/j.molcel.2023.08.018 [published Online First: 2023/10/21] [DOI] [PubMed] [Google Scholar]
- 13.Herbert A. A Genetic Instruction Code Based on DNA Conformation. Trends in genetics : TIG 2019;35(12):887–90. doi: 10.1016/j.tig.2019.09.007 [published Online First: 2019/11/02] [DOI] [PubMed] [Google Scholar]
- 14.Wang AH, Quigley GJ, Kolpak FJ, et al. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 1979;282(5740):680–6. doi: 10.1038/282680a0 [published Online First: 1979/12/13] [DOI] [PubMed] [Google Scholar]
- 15.Herbert A. Z-DNA and Z-RNA in human disease. Communications biology 2019;2:7. doi: 10.1038/s42003-018-0237-x [published Online First: 2019/02/08] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rich A, Nordheim A, Wang AH. The chemistry and biology of left-handed Z-DNA. Annual review of biochemistry 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043 [published Online First: 1984/01/01] [DOI] [PubMed] [Google Scholar]
- 17.Rich A, Zhang S. Timeline: Z-DNA: the long road to biological function. Nature reviews Genetics 2003;4(7):566–72. doi: 10.1038/nrg1115 [published Online First: 2003/07/03] [DOI] [PubMed] [Google Scholar]
- 18.Ravichandran S, Subramani VK, Kim KK. Z-DNA in the genome: from structure to disease. Biophysical reviews 2019;11(3):383–87. doi: 10.1007/s12551-019-00534-1 [published Online First: 2019/05/24] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy R, Chakraborty P, Chatterjee A, et al. Comparative review on left-handed Z-DNA. Frontiers in bioscience (Landmark edition) 2021;26(5):29–35. doi: 10.52586/4922 [published Online First: 2021/05/25] [DOI] [PubMed] [Google Scholar]
- 20.Hanau LH, Santella RM, Grunberger D, et al. An immunochemical examination of acetylaminofluorene-modified poly(dG-dC) X poly(dG-dC) in the Z-conformation. The Journal of biological chemistry 1984;259(1):173–8. [published Online First: 1984/01/10] [PubMed] [Google Scholar]
- 21.Behe M, Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proceedings of the National Academy of Sciences of the United States of America 1981;78(3):1619–23. doi: 10.1073/pnas.78.3.1619 [published Online First: 1981/03/01] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Möller A, Nordheim A, Kozlowski SA, et al. Bromination stabilizes poly(dG-dC) in the Z-DNA form under low-salt conditions. Biochemistry 1984;23(1):54–62. doi: 10.1021/bi00296a009 [published Online First: 1984/01/03] [DOI] [PubMed] [Google Scholar]
- 23.Fleming AM, Zhu J, Ding Y, et al. Oxidative Modification of Guanine in a Potential Z-DNA-Forming Sequence of a Gene Promoter Impacts Gene Expression. Chemical research in toxicology 2019;32(5):899–909. doi: 10.1021/acs.chemrestox.9b00041 [published Online First: 2019/03/02] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbert A. Flipons and small RNAs accentuate the asymmetries of pervasive transcription by the reset and sequence-specific microcoding of promoter conformation. The Journal of biological chemistry 2023;299(9):105140. doi: 10.1016/j.jbc.2023.105140 [published Online First: 2023/08/07] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azorin F, Nordheim A, Rich A. Formation of Z-DNA in negatively supercoiled plasmids is sensitive to small changes in salt concentration within the physiological range. The EMBO journal 1983;2(5):649–55. doi: 10.1002/j.1460-2075.1983.tb01479.x [published Online First: 1983/01/01] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malfoy B, Leng M. Antiserum to Z-DNA. FEBS letters 1981;132(1):45–8. doi: 10.1016/0014-5793(81)80424-3 [published Online First: 1981/09/14] [DOI] [PubMed] [Google Scholar]
- 27.Edgington SM, Stollar BD. Immunogenicity of Z-DNA depends on the size of polynucleotide presented in complexes with methylated BSA. Molecular immunology 1992;29:609–17. doi: 10.1016/0161-5890(92)90197-6 [published Online First: 1992/05/01] [DOI] [PubMed] [Google Scholar]
- 28.Lafer EM, Möller A, Nordheim A, et al. Antibodies specific for left-handed Z-DNA. Proceedings of the National Academy of Sciences of the United States of America 1981;78(6):3546–50. doi: 10.1073/pnas.78.6.3546 [published Online First: 1981/06/01] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lafer EM, Valle RP, Möller A, et al. Z-DNA-specific antibodies in human systemic lupus erythematosus. The Journal of clinical investigation 1983;71(2):314–21. doi: 10.1172/jci110771 [published Online First: 1983/02/01] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas TJ, Meryhew NL, Messner RP. Enhanced binding of lupus sera to the polyamine-induced left-handed Z-DNA form of polynucleotides. Arthritis Rheum 1990;33(3):356–65. doi: 10.1002/art.1780330308 [published Online First: 1990/03/01] [DOI] [PubMed] [Google Scholar]
- 31.Sibley JT, Lee JS, Decoteau WE. Left-handed “Z” DNA antibodies in rheumatoid arthritis and systemic lupus erythematosus. The Journal of rheumatology 1984;11(5):633–7. [published Online First: 1984/10/01] [PubMed] [Google Scholar]
- 32.Spencer DM, Svenungsson E, Gunnarsson I, et al. The expression of antibodies to Z-DNA in the blood of patients with systemic lupus erythematosus: Relationship to autoantibodies to B-DNA. Clinical immunology (Orlando, Fla) 2023;255:109763. doi: 10.1016/j.clim.2023.109763 [published Online First: 2023/09/07] [DOI] [PubMed] [Google Scholar]
- 33.Krishna P, Fritzler MJ, Van de Sande JH. Interactions of anti-DNA antibodies with Z-DNA. Clinical and experimental immunology 1993;92(1):51–7. doi: 10.1111/j.1365-2249.1993.tb05947.x [published Online First: 1993/04/01] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ullal AJ, Marion TN, Pisetsky DS. The role of antigen specificity in the binding of murine monoclonal anti-DNA antibodies to microparticles from apoptotic cells. Clinical immunology (Orlando, Fla) 2014;154(2):178–87. doi: 10.1016/j.clim.2014.05.007 [published Online First: 2014/05/31] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allinquant B, Malfoy B, Schuller E, et al. Presence of Z-DNA specific antibodies in Crohn’s disease, polyradiculoneuritis and amyotrophic lateral sclerosis. Clinical and experimental immunology 1984;58(1):29–36. [published Online First: 1984/10/01] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas TJ, Seibold JR, Adams LE, et al. Hydralazine induces Z-DNA conformation in a polynucleotide and elicits anti(Z-DNA) antibodies in treated patients. The Biochemical journal 1993;294 ( Pt 2)(Pt 2):419–25. doi: 10.1042/bj2940419 [published Online First: 1993/09/01] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson CR, Gilkeson GS, Ward MM, et al. Patterns of heavy and light chain utilization in the antibody response to single-stranded bacterial DNA in normal human subjects and patients with systemic lupus erythematosus. Clinical immunology and immunopathology 1992;62(1 Pt 1):25–32. doi: 10.1016/0090-1229(92)90019-k [published Online First: 1992/01/01] [DOI] [PubMed] [Google Scholar]
- 38.Karounos DG, Grudier JP, Pisetsky DS. Spontaneous expression of antibodies to DNA of various species origin in sera of normal subjects and patients with systemic lupus erythematosus. Journal of immunology (Baltimore, Md : 1950) 1988;140(2):451–5. [published Online First: 1988/01/15] [PubMed] [Google Scholar]
- 39.Fredriksen K, Skogsholm A, Flaegstad T, et al. Antibodies to dsDNA are produced during primary BK virus infection in man, indicating that anti-dsDNA antibodies may be related to virus replication in vivo. Scandinavian journal of immunology 1993;38(4):401–6. [published Online First: 1993/10/01] [DOI] [PubMed] [Google Scholar]
- 40.Campoccia D, Montanaro L, Arciola CR. Extracellular DNA (eDNA). A Major Ubiquitous Element of the Bacterial Biofilm Architecture. International journal of molecular sciences 2021;22(16) doi: 10.3390/ijms22169100 [published Online First: 2021/08/28] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flemming HC, Wingender J, Szewzyk U, et al. Biofilms: an emergent form of bacterial life. Nature reviews Microbiology 2016;14(9):563–75. doi: 10.1038/nrmicro.2016.94 [published Online First: 2016/08/12] [DOI] [PubMed] [Google Scholar]
- 42.Panlilio H, Rice CV. The role of extracellular DNA in the formation, architecture, stability, and treatment of bacterial biofilms. Biotechnology and bioengineering 2021;118(6):2129–41. doi: 10.1002/bit.27760 [published Online First: 2021/03/23] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annual review of immunology 2002;20:709–60. doi: 10.1146/annurev.immunol.20.100301.064842 [published Online First: 2002/02/28] [DOI] [PubMed] [Google Scholar]
- 44.Buzzo JR, Devaraj A, Gloag ES, et al. Z-form extracellular DNA is a structural component of the bacterial biofilm matrix. Cell 2021;184(23):5740–58.e17. doi: 10.1016/j.cell.2021.10.010 [published Online First: 2021/11/05] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swinger KK, Lemberg KM, Zhang Y, et al. Flexible DNA bending in HU-DNA cocrystal structures. The EMBO journal 2003;22(14):3749–60. doi: 10.1093/emboj/cdg351 [published Online First: 2003/07/11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitchurch CB, Tolker-Nielsen T, Ragas PC, et al. Extracellular DNA required for bacterial biofilm formation. Science (New York, NY) 2002;295(5559):1487. doi: 10.1126/science.295.5559.1487 [published Online First: 2002/02/23] [DOI] [PubMed] [Google Scholar]
- 47.Gabriel Antonio SM, Andreas M, Celine T, et al. Extracellular G-quadruplex and Z-DNA protect biofilms from DNase I and forms a DNAzyme with peroxidase activity. bioRxiv 2023:2023.05.22.541711. doi: 10.1101/2023.05.22.541711 [DOI] [Google Scholar]
- 48.Singh J, Boettcher M, Dölling M, et al. Moonlighting chromatin: when DNA escapes nuclear control. Cell death and differentiation 2023;30(4):861–75. doi: 10.1038/s41418-023-01124-1 [published Online First: 2023/02/10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Mayorga-Flores M, Bien KG, et al. DNA-mediated proteolysis by neutrophil elastase enhances binding activities of the HMGB1 protein. The Journal of biological chemistry 2022;298(11):102577. doi: 10.1016/j.jbc.2022.102577 [published Online First: 2022/10/12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tursi SA, Lee EY, Medeiros NJ, et al. Bacterial amyloid curli acts as a carrier for DNA to elicit an autoimmune response via TLR2 and TLR9. PLoS pathogens 2017;13(4):e1006315. doi: 10.1371/journal.ppat.1006315 [published Online First: 2017/04/15] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gallo PM, Rapsinski GJ, Wilson RP, et al. Amyloid-DNA Composites of Bacterial Biofilms Stimulate Autoimmunity. Immunity 2015;42(6):1171–84. doi: 10.1016/j.immuni.2015.06.002 [published Online First: 2015/06/18] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pachucki RJ, Corradetti C, Kohler L, et al. Persistent Bacteriuria and Antibodies Recognizing Curli/eDNA Complexes From Escherichia coli Are Linked to Flares in Systemic Lupus Erythematosus. Arthritis & rheumatology (Hoboken, NJ) 2020;72(11):1872–81. doi: 10.1002/art.41400 [published Online First: 2020/08/26] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller AL, Pasternak JA, Medeiros NJ, et al. In vivo synthesis of bacterial amyloid curli contributes to joint inflammation during S Typhimurium infection. PLoS pathogens 2020;16(7):e1008591. doi: 10.1371/journal.ppat.1008591 [published Online First: 2020/07/10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emlen W, Mannik M. Effect of DNA size and strandedness on the in vivo clearance and organ localization of DNA. Clinical and experimental immunology 1984;56(1):185–92. [published Online First: 1984/04/01] [PMC free article] [PubMed] [Google Scholar]
- 55.Bohlin J, Eldholm V, Pettersson JH, et al. The nucleotide composition of microbial genomes indicates differential patterns of selection on core and accessory genomes. BMC genomics 2017;18(1):151. doi: 10.1186/s12864-017-3543-7 [published Online First: 2017/02/12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghosh A, Chaudhary SA, Apurva SR, et al. Whole-Genome Sequencing of Micrococcus luteus Strain Modasa, of Indian Origin. Genome announcements 2013;1(2):e0007613. doi: 10.1128/genomeA.00076-13 [published Online First: 2013/03/22] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garnier T, Eiglmeier K, Camus JC, et al. The complete genome sequence of Mycobacterium bovis. Proceedings of the National Academy of Sciences of the United States of America 2003;100(13):7877–82. doi: 10.1073/pnas.1130426100 [published Online First: 2003/06/06] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Piovesan A, Pelleri MC, Antonaros F, et al. On the length, weight and GC content of the human genome. BMC research notes 2019;12(1):106. doi: 10.1186/s13104-019-4137-z [published Online First: 2019/03/01] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lo YMD, Han DSC, Jiang P, et al. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science (New York, NY) 2021;372(6538) doi: 10.1126/science.aaw3616 [published Online First: 2021/04/10] [DOI] [PubMed] [Google Scholar]
- 60.Jiang P, Lo YMD. The Long and Short of Circulating Cell-Free DNA and the Ins and Outs of Molecular Diagnostics. Trends in genetics : TIG 2016;32(6):360–71. doi: 10.1016/j.tig.2016.03.009 [published Online First: 2016/05/01] [DOI] [PubMed] [Google Scholar]
- 61.Han DSC, Lo YMD. The Nexus of cfDNA and Nuclease Biology. Trends in genetics : TIG 2021;37(8):758–70. doi: 10.1016/j.tig.2021.04.005 [published Online First: 2021/05/20] [DOI] [PubMed] [Google Scholar]
- 62.Keyel PA. Dnases in health and disease. Developmental biology 2017;429(1):1–11. doi: 10.1016/j.ydbio.2017.06.028 [published Online First: 2017/07/02] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yasutomo K, Horiuchi T, Kagami S, et al. Mutation of DNASE1 in people with systemic lupus erythematosus. Nature genetics 2001;28(4):313–4. doi: 10.1038/91070 [published Online First: 2001/08/02] [DOI] [PubMed] [Google Scholar]
- 64.Al-Mayouf SM, Sunker A, Abdwani R, et al. Loss-of-function variant in DNASE1L3 causes a familial form of systemic lupus erythematosus. Nature genetics 2011;43(12):1186–8. doi: 10.1038/ng.975 [published Online First: 2011/10/25] [DOI] [PubMed] [Google Scholar]
- 65.Napirei M, Karsunky H, Zevnik B, et al. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nature genetics 2000;25(2):177–81. doi: 10.1038/76032 [published Online First: 2000/06/03] [DOI] [PubMed] [Google Scholar]
- 66.Hartl J, Serpas L, Wang Y, et al. Autoantibody-mediated impairment of DNASE1L3 activity in sporadic systemic lupus erythematosus. The Journal of experimental medicine 2021;218(5) doi: 10.1084/jem.20201138 [published Online First: 2021/03/31] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lacey KA, Serpas L, Makita S, et al. Secreted mammalian DNases protect against systemic bacterial infection by digesting biofilms. The Journal of experimental medicine 2023;220(6) doi: 10.1084/jem.20221086 [published Online First: 2023/03/18] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vinuesa CG, Shen N, Ware T. Genetics of SLE: mechanistic insights from monogenic disease and disease-associated variants. Nature reviews Nephrology 2023;19(9):558–72. doi: 10.1038/s41581-023-00732-x [published Online First: 2023/07/13] [DOI] [PubMed] [Google Scholar]
- 69.Alperin JM, Ortiz-Fernández L, Sawalha AH. Monogenic Lupus: A Developing Paradigm of Disease. Frontiers in immunology 2018;9:2496. doi: 10.3389/fimmu.2018.02496 [published Online First: 2018/11/22] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leffler J, Martin M, Gullstrand B, et al. Neutrophil extracellular traps that are not degraded in systemic lupus erythematosus activate complement exacerbating the disease. Journal of immunology (Baltimore, Md : 1950) 2012;188(7):3522–31. doi: 10.4049/jimmunol.1102404 [published Online First: 2012/02/22] [DOI] [PubMed] [Google Scholar]
- 71.Kwakye-Berko F, Meshnick S. Sequence preference of chloroquine binding to DNA and prevention of Z-DNA formation. Molecular and biochemical parasitology 1990;39(2):275–8. doi: 10.1016/0166-6851(90)90066-u [published Online First: 1990/03/01] [DOI] [PubMed] [Google Scholar]
- 72.Yang LY, Luo Q, Lu L, et al. Increased neutrophil extracellular traps promote metastasis potential of hepatocellular carcinoma via provoking tumorous inflammatory response. Journal of hematology & oncology 2020;13(1):3. doi: 10.1186/s13045-019-0836-0 [published Online First: 2020/01/08] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.D’Cruz AA, Speir M, Bliss-Moreau M, et al. The pseudokinase MLKL activates PAD4-dependent NET formation in necroptotic neutrophils. Science signaling 2018;11(546) doi: 10.1126/scisignal.aao1716 [published Online First: 2018/09/06] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.DeAntoneo C, Herbert A, Balachandran S. Z-form nucleic acid-binding protein 1 (ZBP1) as a sensor of viral and cellular Z-RNAs: walking the razor’s edge. Current opinion in immunology 2023;83:102347. doi: 10.1016/j.coi.2023.102347 [published Online First: 2023/06/05] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Herbert A. To “Z” or not to “Z”: Z-RNA, self-recognition, and the MDA5 helicase. PLoS genetics 2021;17(5):e1009513. doi: 10.1371/journal.pgen.1009513 [published Online First: 2021/05/14] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Balachandran S, Mocarski ES. Viral Z-RNA triggers ZBP1-dependent cell death. Current opinion in virology 2021;51:134–40. doi: 10.1016/j.coviro.2021.10.004 [published Online First: 2021/10/25] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maelfait J, Rehwinkel J. The Z-nucleic acid sensor ZBP1 in health and disease. The Journal of experimental medicine 2023;220(8) doi: 10.1084/jem.20221156 [published Online First: 2023/07/14] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Samuel CE. Adenosine deaminase acting on RNA (ADAR1), a suppressor of double-stranded RNA-triggered innate immune responses. The Journal of biological chemistry 2019;294(5):1710–20. doi: 10.1074/jbc.TM118.004166 [published Online First: 2019/02/03] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tang Q. Z-nucleic acids: Uncovering the functions from past to present. European journal of immunology 2022;52(11):1700–11. doi: 10.1002/eji.202249968 [published Online First: 2022/09/28] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takaoka A, Wang Z, Choi MK, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 2007;448(7152):501–5. doi: 10.1038/nature06013 [published Online First: 2007/07/10] [DOI] [PubMed] [Google Scholar]
- 81.Nakahama T, Kato Y, Shibuya T, et al. Mutations in the adenosine deaminase ADAR1 that prevent endogenous Z-RNA binding induce Aicardi-Goutieres-syndrome-like encephalopathy. Immunity 2021;54(9): 1976–88.e7. doi: 10.1016/j.immuni.2021.08.022 [published Online First: 2021/09/16] [DOI] [PubMed] [Google Scholar]
- 82.Zhang T, Yin C, Fedorov A, et al. ADAR1 masks the cancer immunotherapeutic promise of ZBP1-driven necroptosis. Nature 2022;606(7914):594–602. doi: 10.1038/s41586-022-04753-7 [published Online First: 2022/05/26] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang Y, Wu M, Cao D, et al. ZBP1-MLKL necroptotic signaling potentiates radiation-induced antitumor immunity via intratumoral STING pathway activation. Science advances 2021;7(41):eabf6290. doi: 10.1126/sciadv.abf6290 [published Online First: 2021/10/07] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hafner-Bratkovič I, Pelegrín P. Ion homeostasis and ion channels in NLRP3 inflammasome activation and regulation. Current opinion in immunology 2018;52:8–17. doi: 10.1016/j.coi.2018.03.010 [published Online First: 2018/03/21] [DOI] [PubMed] [Google Scholar]
- 85.Conos SA, Chen KW, De Nardo D, et al. Active MLKL triggers the NLRP3 inflammasome in a cell-intrinsic manner. Proceedings of the National Academy of Sciences of the United States of America 2017;114(6):E961–e69. doi: 10.1073/pnas.1613305114 [published Online First: 2017/01/18] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lawlor KE, Khan N, Mildenhall A, et al. RIPK3 promotes cell death and NLRP3 inflammasome activation in the absence of MLKL. Nature communications 2015;6:6282. doi: 10.1038/ncomms7282 [published Online First: 2015/02/19] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Águeda-Pinto A, Alves LQ, Neves F, et al. Convergent Loss of the Necroptosis Pathway in Disparate Mammalian Lineages Shapes Viruses Countermeasures. Frontiers in immunology 2021;12:747737. doi: 10.3389/fimmu.2021.747737 [published Online First: 2021/09/21] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koehler FI, Cotsmire S, Zhang T, et al. Vaccinia virus E3 prevents sensing of Z-RNA to block ZBP1-dependent necroptosis. Cell host & microbe 2021;29(8):1266–76.e5. doi: 10.1016/j.chom.2021.05.009 [published Online First: 2021/07/01] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gehrke N, Mertens C, Zillinger T, et al. Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity 2013;39(3):482–95. doi: 10.1016/j.immuni.2013.08.004 [published Online First: 2013/09/03] [DOI] [PubMed] [Google Scholar]
- 90.Wang J, Wang S, Zhong C, et al. Novel insights into a major DNA oxidative lesion: its effects on Z-DNA stabilization. Organic & biomolecular chemistry 2015;13(34):8996–9. doi: 10.1039/c5ob01340b [published Online First: 2015/08/06] [DOI] [PubMed] [Google Scholar]
- 91.Ti-Hsuan K, Nikhil R-M, Elizabeth JZ, et al. Neutrophil Extracellular Traps have DNAzyme activity that drives bactericidal potential. bioRxiv 2023:2023.10.23.563618. doi: 10.1101/2023.10.23.563618 [DOI] [Google Scholar]
- 92.Atisha-Fregoso Y, Toz B, Diamond B. Meant to B: B cells as a therapeutic target in systemic lupus erythematosus. The Journal of clinical investigation 2021;131(12) doi: 10.1172/jci149095 [published Online First: 2021/06/16] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meffre E, O’Connor KC. Impaired B-cell tolerance checkpoints promote the development of autoimmune diseases and pathogenic autoantibodies. Immunological reviews 2019;292(1):90–101. doi: 10.1111/imr.12821 [published Online First: 2019/11/14] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zuo Y, Navaz S, Tsodikov A, et al. Anti-Neutrophil Extracellular Trap Antibodies in Antiphospholipid Antibody-Positive Patients: Results From the Antiphospholipid Syndrome Alliance for Clinical Trials and International Networking Clinical Database and Repository. Arthritis & rheumatology (Hoboken, NJ) 2023;75(8):1407–14. doi: 10.1002/art.42489 [published Online First: 2023/03/03] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pisetsky DS. Antibodies to Neutrophil Extracellular Traps: Novel Markers for the Antiphospholipid Syndrome. Arthritis & rheumatology (Hoboken, NJ) 2023;75(8):1331–33. doi: 10.1002/art.42548 [published Online First: 2023/05/16] [DOI] [PubMed] [Google Scholar]


