Abstract
Glycoprotein E (gE) is important for full virulence potential of the alphaherpesviruses in both natural and laboratory hosts. The gE sequence of the neurovirulent bovine herpesvirus 5 (BHV-5) was determined and compared with that of the nonneurovirulent BHV-1. Alignment of the predicted amino acid sequences of BHV-1 and BHV-5 gE open reading frames showed that they had 72% identity and 77% similarity. To determine the role of gE in the differential neuropathogenesis of BHV-1 and BHV-5, we have constructed BHV-1 and BHV-5 recombinants: gE-deleted BHV-5 (BHV-5gEΔ), BHV-5 expressing BHV-1 gE (BHV-5gE1), and BHV-1 expressing BHV-5 gE (BHV-1gE5). Neurovirulence properties of these recombinant viruses were analyzed using a rabbit seizure model (S. I. Chowdhury et al., J. Comp. Pathol. 117:295–310, 1997) that distinguished wild-type BHV-1 and -5 based on their differential neuropathogenesis. Intranasal inoculation of BHV-5 gEΔ and BHV-5gE1 produced significantly reduced neurological signs that affected only 10% of the infected rabbits. The recombinant BHV-1gE5 did not invade the central nervous system (CNS). Virus isolation and immunohistochemistry data suggest that these recombinants replicate and spread significantly less efficiently in the brain than BHV-5 gE revertant or wild-type BHV-5, which produced severe neurological signs in 70 to 80% rabbits. Taken together, the results of neurological signs, brain lesions, virus isolation, and immunohistochemistry indicate that BHV-5 gE is important for efficient neural spread and neurovirulence within the CNS and could not be replaced by BHV-1 gE. However, BHV-5 gE is not required for initial viral entry into olfactory pathway.
Bovine herpesvirus 5 (BHV-5) is a neurovirulent alphaherpesvirus that causes fatal encephalitis in calves (5, 21). Nonneurovirulent BHV-1 is associated with abortions and respiratory infections (subtype 1.1) and genital infections (subtype 1.2) in cattle (57). Both BHV-1 and BHV-5 strains are neurotropic viruses and establish latency in the trigeminal ganglion (TG) following intranasal and conjunctival inoculation (1, 50). Genetically, they share 85% DNA homology; however, they differ in the ability to cause neurological disease in calves (5). In a rabbit seizure model, nonneurovirulent BHV-1.1 and neurovirulent BHV-5 infections are distinguished by their differential neuropathogenesis (14). Following intranasal inoculation, BHV-5 invades the brain via the olfactory pathway, resulting in acute neurological signs that are comparable to those seen in calves. Virus antigen and neuronal damage are located in the affected rabbit's brain within the areas connected through the olfactory pathway. These are olfactory bulb, anterior olfactory nucleus, piriform/entorhinal cortex, frontal/cingulate cortex, hippocampus/dentate gyrus, amygdala, dorsal raphe (DR), and locus coeruleus (LC) (36). These rabbits also have a few infected neurons (5 to 10/field) in the TG; however, further invasion of the virus to the pontine and spinal trigeminal nuclei (second-order neurons in the trigeminal pathway) of the pons and medulla, respectively, does not occur (36). In BHV-1-inoculated rabbits, the virus does not invade the central nervous system (CNS), and no neurological signs develop. However, BHV-1 infected rabbits also have a few (5 to 10/field) infected neurons in the TG (36).
Herpesvirus glycoproteins located in the envelope of the virus play important roles in pathogenicity by mediating entry of the virion into the host cell, maturation of virus, cell-to-cell spread of virus, and virus release (10, 19, 20, 40, 47, 49, 58). Alphaherpesviruses encode at least 10 glycoproteins found in the virion envelope (51). Two of these glycoproteins, gE and gI, have been shown to be important for virulence and spread of several herpesviruses in all animal models tested (2, 9–11, 27, 33, 34, 39, 52, 55). Although dispensable for replication in cultured cells, the genes for gE and gI are conserved in all alphaherpesviruses studied to date (3, 19, 37, 39, 48). The gE and gI proteins in alphaherpesviruses associate with each other soon after synthesis and form a noncovalent hetero-oligomeric complex (28, 29, 55, 56, 60). For some herpesviruses, the gE-gI complex functions as a receptor for the Fc domain of immunoglobulin G and consequently, may play a role in the evasion of humoral immunity (4, 6, 22, 24, 25, 29). In vitro, the gE-gI complex is involved primarily in cell-to-cell transmission, possibly by promoting cell fusion or viral release (3, 17–20, 59), and virus mutants lacking gE and/or gI characteristically display a small-plaque phenotype (3, 19, 56, 59). Three distinct functional domains of gE have been characterized in pseudorabies virus (PRV): an extracellular domain, required for gE-gI complex formation and cell-to-cell spread; a hydrophobic transmembrane domain; and a long cytoplasmic domain required for virulence, gE endocytosis and recycling, and envelope incorporation (52, 53). In vivo, the gE and gI mutants of several herpesviruses exhibit decreased virulence properties in their natural and laboratory hosts (41, 42, 49). In PRV, gE and gI null mutants have restricted neurotropism and reduced virulence phenotypes (reviewed in references 8, 23, 27). In herpes simplex virus type 1 (HSV-1), gE and gI mutants fail to spread efficiently within the nervous system and cause less neurological disease than the wild type (20). In addition, Dingwell et al. (20) proposed that gE-gI facilitates the movement of HSV across the extensive junction formed between neurons in vivo. There is also evidence that gE-gI may influence the entry of PRV into some neuronal circuits but not others (reviewed in reference 23).
As in other alphaherpesviruses, the deletion of the gE gene in BHV-1 is sufficient to reduce its virulence in calves (15, 54). In this study, we investigated the role of gE in the differential neuropathogenesis of nonneurovirulent BHV-1 and neurovirulent BHV-5. We determined the nucleotide sequence of the BHV-5 gE open reading frame (ORF) and compared it with the published coding sequence for the BHV-1 gE ORF. We generated gE-deleted BHV-5, gE-exchanged BHV-5 (carrying BHV-1 gE), and gE-exchanged BHV-1 (carrying BHV-5 gE) recombinants and analyzed their neuropathogenicity in a rabbit seizure model (14). The systemic spread of the various mutant viruses within the CNS was evaluated at different postinoculation times by immunocytochemistry, histopathology, and virus isolation. Our results indicate that BHV-5 gE is not required for entry into the CNS via the olfactory receptor neurons. However, within the CNS, BHV-5 gE is important for efficient spread, replication, and neuropathogenesis (virulence) of BHV-5 and could not be replaced by the BHV-1 gE.
MATERIALS AND METHODS
Virus strains and cell lines.
The BHV-1 Cooper (Colorado-1) strain, obtained from the American Type Culture Collection (Manassas, Va.), and BHV-5 strain TX-89 (21) were used in this study. The two viruses were propagated and titrated in Madin-Darby bovine kidney (MDBK) cells grown in Dulbecco modified Eagle's medium supplemented with heat-inactivated 10% fetal bovine serum.
Mapping, cloning, and sequencing of the BHV-5 gE gene.
The location of the BHV-5 gE gene on the virus genome and pertinent restriction sites for subcloning and the sequencing strategy are illustrated in Fig. 1. A pUC-based plasmid library containing the BamHI genomic fragments of the BHV-5 genomic DNA was developed (12). A cloned copy of the 14.6-kb BamHI-C fragment between map units 0.827 and 0.944 of BHV-5, which includes the entire unique short region of the BHV-5 genome, was digested with StuI. Then the 9.6-kb StuI/StuI fragment between map units 0.849 and 0.926 was gel purified and cloned into the SmaI site of pUC19, resulting in plasmid pKS-95-15 (Fig. 1A). To precisely map the location of the BHV-5 gE gene, clone pKS-95-15 was digested with several restriction endonucleases, separated on agarose gel, and analyzed by hybridization with a 1.8-kb AsuII subfragment containing the BHV-1 gE coding sequence (15). The DNA restriction analysis combined with the hybridization data showed that a 4.2-kb XhoI/StuI fragment contained the entire BHV-5 gE and gI genes (Fig. 1B) and a 3.2-kb NotI/StuI fragment contained the entire BHV-5 gE gene. A restriction map of the 3.2-kb NotI/StuI fragment was constructed (Fig. 1C), and subclones spanning the entire fragment were generated. Both strands of each fragment were sequenced by the method of Maxam and Gilbert (38).
FIG. 1.
BHV-5 genomic structure and schematic maps of gE recombinant plasmids. The genomic organization of BHV-5 depicted at the top consists of unique long (UL) and short (US) regions and two repeat regions (IR) and (TR). Localization of gE gene is indicated (A and B), and the region encompassing the gE gene (NotI/XhoI) is enlarged (C). A restriction site map of the NotI and XhoI fragment was generated, and various restriction endonuclease subfragments, indicated as bold lines, were cloned and sequenced. The arrow represents the BHV-5 gE ORF, and the arrowhead shows the direction of transcription. The regions of interest are shown in the schematic structures of plasmids pBHV-5gEΔβ (D), pBHV-5gE1 (E), and pBHV-1gE5 (F).
Sequence analysis and comparisons between BHV-5 gE and BHV-1 gE.
Sequence data were assembled and analyzed using the Seqaid II sequence analysis software (D. D. Rhodes and D. J. Roufa, Center for Basic Cancer Research, Kansas State University, Manhattan, Kans.). Hydropathicity analysis was performed using a 9-amino-acid (aa) window (35). The antigenicity profiles of predicted amino acids sequences were analyzed using the Seqaid II software. The predicted amino acid sequences of BHV-5 gE (this study) and BHV-1.1 and BHV-1.2 gE (37, 48) were aligned by using the GCG Gap program (Genetics Computer Group) (gap weight, 12; length weight, 4). The prediction of signal sequence was determined using the SignalP V1.1 program from the World Wide Web (43; http://www.cbs.dtu.dk/services/SignalP/output.html).
Production of anti-BHV-5 and BHV-1 gE peptide-specific polyclonal rabbit sera.
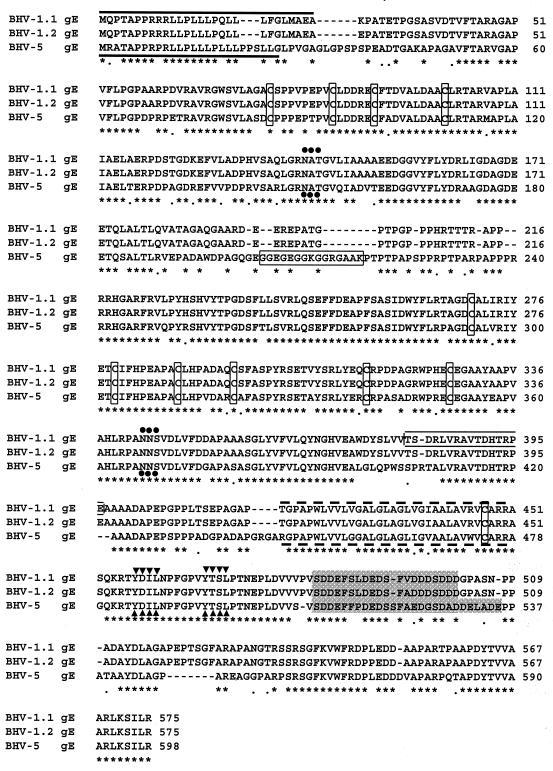
Based on the predicted regional hydropathicity and antigenicity, the predicted amino acid residues 204 to 218 ([H]-C*GGEGEGGKGGRGAAK-[OH]) from BHV-5 gE (Fig. 2) and residues 381 to 396 ([H]-TSDRLVRAVTDHTRPEC*-[OH]) from BHV-1 gE (48) were selected to make peptide antigen. To facilitate conjugation to keyhole limpet hemocyanin, an additional irrelevant cysteine was added (indicated by *) at the N terminus of peptide from BHV-5gE and at the C terminus of peptide from BHV-1 gE. The peptides were synthesized at the Biotechnology Center of Kansas State University, using the 9-fluorenylmethyloxycarbonyl chemistry on an ABI model 431A automated peptide synthesizer (Applied Biosystems, Inc., Foster City, Calif.) as described earlier (12). After conjugation with keyhole limpet hemocyanin, the peptides were used to immunize New Zealand White rabbits as described earlier (13).
FIG. 2.
Comparison of the predicted amino acid sequence of the BHV-5 gE with the predicted amino acid sequences of BHV-1.1 and BHV-1.2 gEs (37, 49). The predicted amino acid sequences of BHV-1 and BHV-5 gEs were aligned by using the GCG Gap program. Eleven conserved cysteine residues are marked by boxes, and potential N-linked glycosylation sites are indicated (•••). The presumptive signal sequences and the transmembrane anchor sequences are shown by solid and broken lines, respectively. Peptide sequences selected for raising rabbit antibodies are indicated by rectangular boxes. The YXXL motif sequences are indicated (▾▾▾), and the acidic domain is highlighted.
Construction of deletion and transfer vectors. (i) BHV-5 gE deletion/β-Gal insertion plasmid.
The BHV-5 gE gene is flanked upstream by the gI gene and downstream by the BHV-5 homologue of the HSV-1 US9 gene (Fig. 1B). To clone the BHV-5 gE coding and its flanking sequences, first the pKS95-15 DNA containing the 9.6-kb StuI/StuI fragment (Fig. 1A) was digested partially with XhoI, blunt ended by Klenow enzyme, and redigested with KpnI (plasmid site). The 4.2-kb fragment containing the XhoI/StuI fragment (Fig. 1A) was cloned into the KpnI/XhoI sites of pBC/SK (Stratagene). The resulting clone, pBHV-5gE5′3′, contained the entire gI, gE, and US9 genes (Fig. 1B). To create a deletion in the gE ORF coding region, this plasmid was digested partially with StyI and blunt ended by Klenow enzyme. The larger fragment was gel purified and ligated to a PstI linker 5′ GCTGCAGC 3′. After digestion with PstI, the fragment was gel purified again and religated. The resulting BHV-5 gE deletion clone pBHV5gEΔ5′3′ (Fig. 1B) had a deletion of 1,074 bp of BHV-5 DNA sequences (marked ⧫ in Fig. 1B) containing BHV-5 gE residues 35 to 340. To insert the β-galactosidase (β-Gal) gene, the PstI fragment (4.5 kb) of plasmid pCMVβ (Clontech, Palo Alto, Calif.) containing the bacterial β-Gal gene under the control of the human cytomegalovirus immediate-early promoter was gel purified and inserted into the previously created PstI site of plasmid pBHV5gEΔ5′3′ to yield plasmid pBHV5gEΔβ (Fig. 1D). The orientation of the β-Gal gene was verified by restriction enzyme digestion, and a clone containing the β-Gal gene in the parallel transcriptional orientations with respect to the gE was selected for further studies. In plasmid pBHV5gEΔβ, the β-Gal gene is flanked by virus-specific 1.8-kb gE upstream sequences (containing the entire gI region, the upstream gE regulatory sequence, and the amino-terminal 102 bp of gE coding region) and 1.4-kb gE downstream sequences (containing the carboxy-terminal 628 bp of gE coding region and the entire US9 gene sequence) required for recombination with the virus DNA.
(ii) BHV-5gE1 exchange/transfer vector.
To exchange the BHV-5 gE coding region with the BHV-1 gE ORF coding sequences, plasmid pBHV1gE 5′3′ containing the BHV-1 gE ORF and the 5′ and 3′ gE flanking sequences (15) was digested with AsuII and blunt ended by Klenow enzyme, and a 1.8-kb AsuII(blunt)/AsuII (blunt) fragment containing the entire BHV-1 gE ORF was gel purified and inserted in correct orientation in the PstI (blunt)/PstI (blunt) sites of the BHV-5 gE deletion plasmid pBHV5gEΔ5′3′ (described above), resulting in pBHV-5gE1 (Fig. 1E). The correct orientation of the BHV-1 gE ORF coding sequence was verified by restriction digest analysis. The resulting plasmid (pBHV5gE1) contained the entire BHV-1 gE ORF fused in frame (verified by sequencing) to the coding sequences of the first 33 aa of BHV-5 gE and flanked by BHV-5 gE upstream (1.8 kb) and downstream (1.4 kb) sequences similar to the deletion vector pBHV5gEΔ5′3′ (described above).
(iii) BHV-1gE5 exchange vector.
To replace the BHV-1 gE ORF with the BHV-5 gE ORF, the pBHV-5gE ORF DNA (Fig. 1C) was digested partially with NotI and blunt ended by Klenow enzyme. The larger fragment was gel purified and further digested with HincII. Then the 2,067-bp NotI (blunt)/HincII fragment containing the entire BHV-5 ORF coding sequence was gel purified. Plasmid pBHV-1gE5′3′, containing the BHV-1 gE and its flanking sequences, was constructed earlier and has been reported elsewhere (15). This plasmid was digested with AsuII to release the 1.8-kb BHV-1 gE ORF coding sequences. After blunt ending, the larger fragment containing the 5′ and 3′ BHV-1 gE ORF flanking sequences was gel purified. The 2,067-bp NotI (blunt)/HincII fragment containing the BHV-5 gE ORF sequences (isolated above) then was inserted at the collapsed AsuII (blunt) sites of the pBHV-1gE5′3′. The correct orientation of the BHV-5 gE ORF was verified by restriction digest analysis. The resulting plasmid pBHV-1 gE5 (Fig. 1F) contained the entire BHV-5 gE ORF flanked by 5′ and 3′ BHV-1-specific sequences. In addition, the first 13 predicted aa of the BHV-1 US9 equivalent gene were deleted.
Generation of recombinant viruses. (i) BHV-5gEΔ.
To generate the BHV-5 gE-deleted recombinant BHV-5gEΔ, linearized pBHV5gEΔβ (Fig. 1D) and full-length wild-type BHV-5 DNA were cotransfected in MDBK cells, using Lipofectamine (Gibco BRL, Life Technologies, Inc., Grand Island, N.Y.) according to the manufacturer's protocol. Recombinant viruses expressing β-Gal were plaque purified three times by screening for blue plaque under a Bluo-Gal (Gibco BRL, Life Technologies) overlay as described previously (13). The recombinant viruses were characterized further by Southern blot analysis using gE ORF coding, gE flanking, and β-Gal sequence regions as probes and confirmed by Western blot analysis with anti-BHV-5gE peptide-specific rabbit polyclonal serum.
(ii) BHV-5gE1 recombinant.
To generate the gE-exchanged recombinant BHV-5 expressing BHV-1 gE (BHV-5gE1), linearized pBHV5gE1 (Fig. 1E) and full-length BHV-5gEΔ DNA were cotransfected using the methods described above. Recombinant viruses with the BHV-1 gE gene incorporated and β-Gal gene deleted were identified as white plaques after screening with Bluo-Gal. These recombinant viruses were plaque purified and analyzed by Western blot for the expression of BHV-1 gE.
(iii) BHV-1gE5 recombinant.
To generate the gE-exchanged BHV-1 recombinant expressing the BHV-5 gE (BHV-1gE5), the linearized pBHV1gE5 DNA (Fig. 1F) and full-length BHV-1 gE-deleted recombinant BHV-1 DNA (BHV-1gEΔ3.1IBRβ) (15) were cotransfected as described above. Recombinant BHV-1 expressing BHV-5 gE was identified as white plaques after screening with Bluo-Gal. These recombinant viruses were plaque purified and characterized by Western blot using anti-BHV-5 gE peptide-specific polyclonal sera.
(iv) BHV-5 gE revertant.
To generate the BHV-5 gE revertant, plasmid pBHV-5 gE 5′3′ (Fig. 1B) was linearized and cotransfected with full-length recombinant BHV-5 gEΔ DNA as described above. The revertant virus, in which the gE deletion was rescued, was identified by white plaques after screening with Bluo-Gal. This recombinant virus was plaque purified and verified by Western blot analysis with the anti-BHV-5 gE peptide-specific polyclonal serum.
Radiolabeling of mock- and virus-infected cell proteins.
Confluent MDBK cells were infected with wild-type BHV-1 and BHV-5 or with recombinant BHV-5gEΔ, BHV-5gE1, and BHV-1gE5 at a multiplicity of infection (MOI) of 5 PFU per cell. Mock infections with virus-free media were always included in the analysis. At 4 h postinfection, the cells were deprived of serum, cysteine, and methionine for 2 h by replacing growth medium with serum-free and cysteine-methionine-free medium (Sigma, St. Louis, Mo.). At 6 h postinfection, the medium was replaced with cysteine-methionine-free medium containing 30 μCi of [35S]methionine-cysteine per ml. Infected cells then were incubated at 37°C for an additional 10 h. At the end of the labeling period, the cells were harvested by low-speed centrifugation and washed three times with Tris-buffered saline (0.15 M NaCl, 0.01 M Tris [pH 7.4]) containing proteinase inhibitors (0.1 mM phenylmethylsulfonyl fluoride and 1 μl of aprotinin per ml). The washed cells were resuspended (30% [wt/vol] suspension) in extraction buffer (0.15 M NaCl, 0.01 M Tris [pH 8.0], 0.01 M EDTA, 1.0% Nonidet P-40, 0.5% sodium deoxycholate, 1.0 mM phenylmethylsulfonyl fluoride, 2 μl of aprotinin per ml) and then incubated on ice for 1h. The suspension was centrifuged at 13,000 × g for 15 min. The resulting supernatant (cell extract) was recovered and stored at −20°C.
Immunoprecipitation.
Fifty microliters of a 10% (vol/vol) suspension of protein A-Sepharose (Sigma) in extraction buffer was mixed with 10 μl of BHV-1 or BHV-5 gE-specific polyclonal rabbit serum and incubated for overnight at 4°C with frequent mixing. After the antibody-protein A complexes were washed three times with extraction buffer, the labeled cell extracts (50 μl) were added to the complexes, and the mixture was incubated for an additional 2h at 4°C with frequent mixing. Immune complexes were collected by centrifugation and were washed four times in extraction buffer–0.1% sodium dodecyl sulfate (SDS) and once in 50 mM Tris (pH 8.0)–0.1% Nonidet P-40. The washed complexes then were resuspended in 50 μl of 2× sample buffer, heated to 100°C for 5 min, and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting (12). Labeled and precipitated proteins were visualized via autoradiography. In some cases, precipitated proteins were also identified by immunoblotting.
Virus growth curve experiment.
One-step virus growth experiments were conducted, as described earlier (13, 15), to compare the growth kinetics of BHV-5gEΔ, BHV-5gE1, and BHV-1gE5 to those of the parent viruses. A series of replicate cultures of MDBK cells were infected separately at an MOI of 5 PFU per cell. Infected cultures were harvested at successive intervals postinfection, and virus stocks were prepared for use in virus titration assays.
Animal experiments, tissue processing, and immunohistochemistry.
Four-week-old New Zealand White rabbits weighing 500 to 600 g (Myrtles Rabbitry, Thomson Station, Tenn.) were used. Rabbits were maintained in laboratory isolation cages in our vivarium throughout the experiments, with food and water freely available. All procedures were approved by the Kansas State University Animal Care and Use Committee.
(i) Virus isolation and histopathology (experiments 1 to 4).
To determine the neurovirulence properties of BHV-5gEΔ (experiment 1), BHV-5 gE revertant (experiment 2), BHV-5gE1 (experiment 3), and BHV-1gE5 (experiment 4), the rabbit seizure model described previously (14) was used. Ten rabbits were infected with gE-deleted virus, five rabbits were inoculated with BHV-1gE5 or BHV-5gE1, and four rabbits were infected with BHV-5 gE revertant. In each case, the viruses were inoculated (2 × 107 PFU/0.5 ml/nostril) in the paranasal sinuses as described earlier (14). For BHV-5gEΔ, the experiment was repeated in an additional 10 rabbits, using 108 PFU/1.0 ml/nostril. Following infection, the rabbits were observed four times a day for the appearance of neurological symptoms. The rabbits were sacrificed when they showed neurological symptoms or at 12 to 14 days postinfection (dpi). After euthanasia, swabs from the nose and olfactory mucosa (turbinate area) were collected, and virus was eluted (in 1 ml) as described earlier (36). The brain was removed and examined for gross lesions. As described earlier (14), it then was divided midsagittally; the left half of the brain and the TG were used for virus isolation, and the right half was processed for histopathological examination. All cultures without evidence of cytopathic effects were repassaged to confirm the absence of the virus.
(ii) Immunohistochemical processing (experiments 5 to 8).
To compare the neural spreads of BHV-5gEΔ (experiment 5), BHV-5gE1 (experiment 6), BHV-5 gE revertant (experiment 7), and BHV-1gE5 (experiment 8), 12 rabbits were infected intranasally with each virus as described above. The animals were euthanized 2, 4, 6, 8, 10, or 12 dpi or when they showed neurological signs. At each survival period, two rabbits were perfused transcardially by 10% buffered neutral formalin, and then the brain and TG were used for immunohistochemistry as described earlier (36).
Nucleotide sequence accession number.
The nucleotide sequence of the BHV-5 gE gene has been submitted to GenBank with accession no. AF208294.
RESULTS
Analysis of BHV-5 gE ORF and comparison of predicted amino acid sequences of BHV-5 and BHV-1 gE.
The nucleotide sequence analysis of the 2.15-kb XhoI/NotI fragment (Fig. 1C) encoding the gE gene identified only one 1,794-bp ORF (from nucleotides 155 to 1948) coding for 598 aa, large enough to encode the BHV-5 gE gene. (Fig. 2). The nucleotide composition of the BHV-5 gE ORF coding sequences showed that G+C content was 76% (41% C, 35% G), which is higher than that for the BHV-1 gE coding segment (70.4%) (48). Hydropathic analysis of the predicted protein revealed the presence of two prominent hydrophobic peaks similar to those for the BHV-1 gE, representing the signal sequence and transmembrane anchor sequence (data not shown). Amino acid residues 1 to 26 have the position, length, relative hydrophobicity, and consensus cleavage site characteristic of a signal sequence, as indicated by empirical rules for predicting signal sequences (43, 44). Similarly, amino acid residues 445 to 478 have the position, length, and relative hydrophobicity characteristic of a transmembrane anchor sequence (marked in Fig. 2).
Alignment of the predicted amino acid sequence of the BHV-5 gE gene with the corresponding sequences of BHV-1.1 and BHV-1.2 (37, 48) showed 72% identity and 77% similarity. The predicted BHV-5 gE ORF contains 598 aa (molecular mass of the corresponding unprocessed protein would be 63.3 kDa), whereas the BHV-1 gE contained 575 aa (61.1 kDa). In addition, several domains of BHV-5 gE show divergence from the predicted gE sequences for BHV-1.1 and BHV-1.2, i.e., residues P28 to A51, E191 to T220, L398 to A409, P432 to A444, and D530 to A555 (Fig. 2). The acidic domain within the BHV-5 gE cytoplasmic tail domain (S509 to E535) is slightly longer than the corresponding acidic domain of BHV-1.1 and BHV-1.2 (S482 to D502); however, the net acidic charge of the three cytoplasmic domains remained similar. The alignment also showed that the regions between V58 and V190, R241 and A397, and R445 and F513 containing the 10 cysteine residues (85, 94, 100, 110, 294, 303, 312, 320, 339, and 351); two potential N-linked glycosylation sites (residues 150, 367) (32); and the transmembrane domain (residues 445 to 478) are conserved among the three sequences (Fig. 2). Within the cytoplasmic tail, four tyrosine residues (Y484, Y494, Y542, and Y586) including the two YXXL motifs (residues 484 to 487 and 494 to 497) also are conserved among the three sequences (Fig. 2).
Identification of the BHV-5 gE protein.
The anti-BHV-5 gE peptide-specific antibody reacted specifically with a 94-kDa protein of BHV-5 and did not react to any protein of BHV-1 (Fig. 3A′). The anti-BHV-1 gE peptide-specific antibody reacted with the gE of BHV-1 (92 kDa) as well as with the 94-kDa protein of BHV-5 (Fig. 3B′). These results are consistent with the sequence alignment data: BHV-5 gE is predicted to be 2 kDa larger than BHV-1 gE, the BHV-5 gE peptide has no homology in the corresponding region of BHV-1, but the BHV-1 gE peptide has considerable homology in the corresponding region of BHV-5 (Fig. 2). Interestingly, the reactivity of the anti-BHV-1 gE peptide serum was stronger with the BHV-5 gE than with the BHV-1 gE. The 94- and 92-kDa bands were not detected by the matching preimmune serum (data not shown), and they were absent in the mock-infected MDBK cell lysates.
FIG. 3.
Immunoblotting analysis of recombinant viruses. Recombinants and the parental viruses were purified partially by ultracentrifugation through a 30% sucrose cushion as described earlier (15). SDS-PAGE and immunoblot analysis of mock-infected cell lysates and purified virion proteins was performed under reducing conditions as described earlier (12). (A) Coomassie blue-stained SDS-polyacrylamide gel containing lysates of cells infected with BHV-5, BHV-5gEΔ, BHV-5 gE revertant (BHV-5gE-R), BHV-1 gE5, BHV-5gE1, and BHV-1 and of mock-infected MDBK cells. (A′) Immunoblotting of the same proteins with BHV-5 gE-specific antipeptide rabbit polyclonal serum. (B) Coomassie blue-stained SDS-polyacrylamide gel containing lysates of cells infected with BHV-5, BHV-5gEΔ, BHV-5 gE-R, BHV-1 gE5, BHV-5gE1, BHV-1, and BHV-1 gEΔ (15) and of mock-infected MDBK cells. (B′) Immunoblotting of the same proteins with BHV-1 gE-specific antipeptide rabbit polyclonal serum.
Construction and analysis of BHV-5 and BHV-1 gE recombinants.
The DNA from BHV-5gEΔ and wild-type BHV-5 was analyzed by Southern blot hybridization for the intended deletion and insertion of a β-Gal sequence at the gE locus (data not shown). The absence of a StyI/PstI fragment sequence and the presence of a β-Gal sequence in the gE-deleted recombinant isolate but not in the parental BHV-5 demonstrated that the intended recombination in these isolates had taken place in a site-specific manner (data not shown). Consistent with this finding, the 94-kDa BHV-5 gE protein was absent in the gE-deleted BHV-5 but was detected in the parent and rescued gE revertant viruses (Fig. 3A′).
The gE-exchanged BHV-5 and BHV-1 recombinants were verified by immunoblotting with sera raised in rabbit against BHV-5 gE-specific and BHV-1 gE-specific peptides. The BHV-5 gE-specific rabbit polyclonal serum reacted specifically with BHV-5 gE expressed by BHV-5 and gE-exchanged BHV-1 (BHV-1gE5) but not with the BHV-1 gE (Fig. 3A′). As expected, based on our earlier observation, the BHV-1 gE expressed by BHV-5gE1 as well as by BHV-1 showed characteristic weaker immunoblotting reactivity (Fig. 3B′). In addition, the level of gE1 expression in the BHV-5 background appeared identical to that of authentic BHV-1 gE expression (Fig. 3B′). As described above, in the gE-exchanged BHV-5, BHV-1 gE ORF coding sequences and 18 bp BHV-1 gE upstream sequences were inserted in frame after the coding sequences of the first 34 aa of BHV-5 gE. As a consequence, the BHV-5 gE signal peptide (SP) sequence (first 26 aa) plus 8 additional aa (LPVGAGLG) of the BHV-5 gE ORF (Fig. 2) were fused in frame to the BHV-1 gE and its 18-bp upstream sequence starting at the AsuII site (6 irrelevant aa, RKGHLA, would be added). Additionally, after cleavage of the BHV-5 gE SP, the BHV-1 gE SP would still be present (only one signal sequence is predicted to be utilized). Thus, the BHV-1 gE expressed by the gE-exchanged BHV-5 would contain an additional 40 aa (8 aa BHV-5 gE immediately after the SP cleavage site, 6 irrelevant aa translated from the sequence upstream of the BHV-1 gE start site, and uncleaved 26 aa of BHV-1 signal sequence) (37, 48). As a result, the expected molecular mass of BHV-1 gE expressed by the recombinant BHV-5gE1 would be 2 to 3 kDa larger than that of the authentic BHV-1 gE. Consistent with this assumption, BHV-1 gE expressed by BHV-5gE1 was slightly larger (94 to 95 kDa) (Fig. B′).
gE-gI complex formation in the gE-exchanged BHV-5 and BHV-1.
To determine whether the exchanged gEs of BHV-5gE1 and BHV-1gE5 formed complexes with their heterologous gI counterparts, immunoprecipitation experiments were performed. Mock- and virus-infected MDBK cells were labeled with [35S]methionine and [35S]cysteine beginning at 6 h after infection. Initial results indicated that rabbit antiserum raised against BHV-5 gE peptide (aa 204 to 218) precipitated specifically gE (94 kDa) from the BHV-5-infected cell lysates under nondenaturing conditions. In addition, it coprecipitated 61.5- and 47-kDa proteins that are most likely the BHV-5 gI and proteolytic cleavage product of gI, respectively (56) (Fig. 4A). The antiserum raised against BHV-1 gE peptide (aa 381 to 396) antigen failed to precipitate BHV-1 gE and the gE-gI complex. Therefore, we used rabbit serum raised against BHV-1 gE aa 366 to 575 (56). This serum precipitated both BHV-1 gE and BHV-5 gE (92 and 94 kDa, respectively) and the BHV-1gE expressed by BHV-5gE1. Consistent with our immunoblotting results, the BHV-1 gE precipitated from BHV-5gE1-infected cell lysates (Fig. 4B) also was larger (94 to 95 kDa). In addition, it coprecipitated a 61.5-kDa protein (from the BHV-1- and BHV-5-infected cell lysates), a 45-kDa protein (BHV-1), and 47-kDa protein (BHV-5) (Fig. 4B). Immunoblotting of the same gel with rabbit serum raised against BHV-1 gI aa 86 to 380 (56) confirmed that the 61.5-kDa BHV-1 and BHV-5 proteins, coprecipitated by the BHV-1 gE-specific antibody, represent their respective gIs (Fig. 4C). Similarly, the BHV-1 gI-specific rabbit serum precipitated both the BHV-1 and BHV-5 gI (61.5 kDa) and coprecipitated 92-kDa (BHV-1) and 94-kDa (BHV-5) proteins representing their gEs (Fig. 4D). Immunoblotting of the same gel with the BHV-1 gE-specific rabbit serum confirmed that 92-kDa (BHV-1) and 94-kDa (BHV-5) proteins coprecipitated are their respective gEs. Additionally, both the gE5 (Fig. 4A) and gE1 (Fig. 4B to E) expressed by BHV-1gE5 and BHV-5gE1, respectively, formed complexes with their heterologous gI counterparts.
FIG. 4.
Immunoprecipitation, SDS-PAGE, and Western blot analyses of gE and gI complex formation in BHV-5 and BHV-1 gE recombinants. Mock- and virus-infected MDBK cells were labeled with [35S]methionine-cysteine for 18 h. Detergent extracts of infected cells were prepared and immunoprecipitated with BHV-5 gE-specific (A), BHV-1 gE-specific (B), or BHV-1 gI-specific (D) polyclonal rabbit serum. After SDS-PAGE, the precipitated proteins were analyzed by immunoblotting with BHV-1 gI-specific (C) or BHV-1 gE-specific (E) rabbit sera. Panels A, B, and D show autoradiography of labeled proteins. The positions of molecular size markers, gE, and gI are indicated. White asterisks in panels C and E indicate heavy chain of rabbit immunoglobulin G.
In vitro growth characterization of recombinant viruses in MDBK cells.
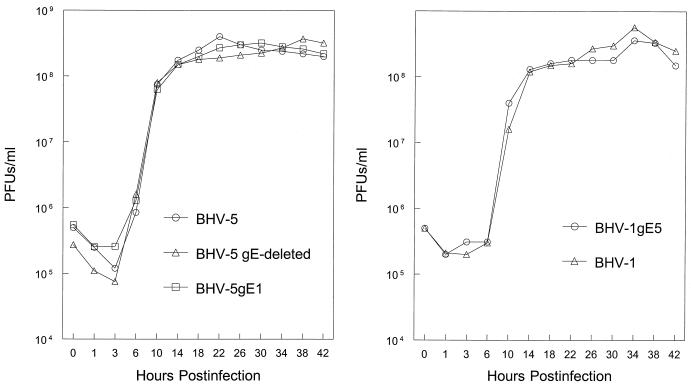
One-step growth experiments were conducted to analyze the growth kinetics of BHV-5 gEΔ, BHV-5 gE1, and BHV-1gE5 recombinants in MDBK cells in comparison to their respective parental strains BHV-5 TX-89 and BHV-1 Cooper. The virus growth curve (Fig. 5) demonstrates that the time course and yield of infectious progeny of BHV-5 gEΔ, BHV-5 gE1, and BHV-1gE5 were similar to those of their parental viruses.
FIG. 5.
One-step growth curve of recombinants and parental viruses in MDBK cells. Confluent MDBK cells were infected with viruses at an MOI of 5 PFU per cell. After 1 h of adsorption at 4°C, residual input viruses were removed. The cultures were washed three times with phosphate-buffered saline, and 5 ml of medium was added to each flask before further incubation (37°C). At indicated time intervals, replicate cultures were frozen. Virus yields were determined by titration on MDBK cells. Each data point represents the average of duplicate samples obtained from separate infections.
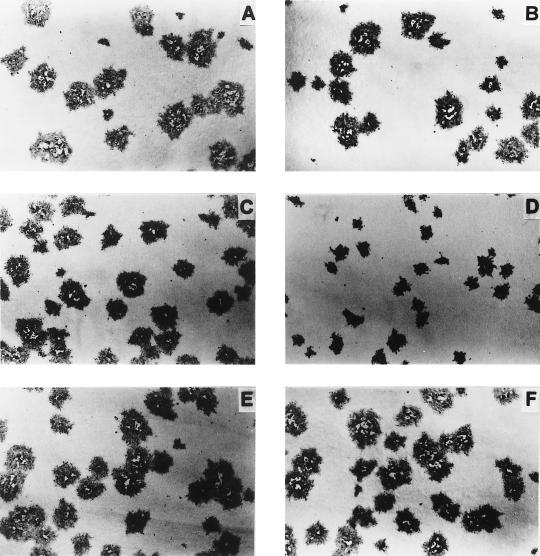
To compare the plaque size phenotypes of recombinants BHV-5 gEΔ, BHV-5 gE1, and BHV-1gE5 and those of their parental wild-type viruses, MDBK cell monolayers were infected with wild-type and mutant viruses and overlaid with medium containing 1.6% carboxymethylcellulose. The infected cells were fixed at 36 h postinfection and stained immunohistochemically with bovine anti-BHV-5 serum. The recombinant gE-deleted BHV-5 (Fig. 6D) and BHV-1 (data not shown) produced small plaques; however, replacement of the gE gene either in BHV-5 gE revertant (homologous) (Fig. 6F) or in gE-exchanged (heterologous) BHV-5 (Fig. 6E) and BHV-1 (Fig. 6C) restored the wild-type plaque phenotype (Fig. 6A and B).
FIG. 6.
Plaque morphology of recombinant viruses in MDBK cell monolayer. BHV-1 (A), BHV-5 (B), BHV-1gE5 (C), BHV-5gEΔ (D), BHV-5gE1 (E), and BHV-5gE revertant (F) were inoculated onto MDBK cell monolayers, fixed after 30 h postinfection, and immunostained with bovine anti-BHV-5 serum (12).
Pathogenicity of recombinant viruses in rabbits. (i) BHV-5gEΔ shows significantly reduced neurovirulence and neural spread.
In the first experiment, 9 of 10 rabbits infected with the BHV-5 gEΔ recombinant showed no detectable neurological signs through 14 dpi. At 10 dpi, one rabbit showed only mild neurological signs characterized by head twitching and slight trembling, which did not progress to seizures. All rabbits were euthanized at 14 dpi. Virus was isolated from the anterior and posterior cortices of the rabbit showing mild neurological signs (<100 PFU/g). The amount of virus isolated was significantly lower than that isolated from rabbits infected with the BHV-5 gE revertant (5000 to 10,000 PFU/g) (Table 1). Virus was not isolated from the brains of any other rabbit infected with BHV-5gEΔ. Histopathological changes in the brains of rabbits infected with BHV-5gEΔ were minimal (Table 1). This experiment was repeated once with a higher dose of BHV-5gEΔ and gave very similar results. As shown in Table 1, the gE-deleted virus grew less efficiently in the nasal mucosa compared to the rescued BHV-5. However, growth of both viruses was comparable in the olfactory mucosa.
TABLE 1.
Summary of clinical signs, histopathological findings, and virus isolation
| Virus | Animal no. | Neurological signsc | Histopathologye | Virus isolation
|
||||
|---|---|---|---|---|---|---|---|---|
| PFU (range)
|
Scored (no. of rabbits)
|
|||||||
| Nose | Olfactory mucosa | Olfactory bulb | Anterior cortex | Posterior cortex | ||||
| BHV-5 gE | Severe (3) | ●●● | 1,500 | 220 | ||||
| revertant | 4 | Mild (1) | ●● | (1,000–2,000) | (150–300) | ++ (3) | ++ (4) | +++ (4) |
| 300 | 150 | |||||||
| BHV-5gEΔa | 20 | Very mild (2) | ● | (250–400) | (100–250) | − | + (2) | + (2) |
| 1,400 | 350 | |||||||
| BHV-5gE1 | 5 | Mild (1) | ● | (1,000–1,800) | (250–450) | − | +b (1) | − |
| 2,000 | 1000 | |||||||
| BHV-1gE5 | 5 | None | ○ | (1,500–3,000) | (800–2,000) | − | − | − |
Data collected from two independent experiments.
Virus was recovered after repassage of the infected cells.
Numbers in parentheses indicate number of animals showing clinical signs or brain segments positive for the virus isolation.
−, no virus detected; +, 10 to 100 PFU/g of tissue; ++, 100 to 2,000 PFU/g; +++, 2,000 to 20,000 PFU/g.
○, no detectable histopathology; ●●●, severe lymphocytic leptomeningitis in the piriform and entorhinal cortices, diffuse lymphocytic infiltration in the piriform and entorhinal cortices, diffuse microgliosis throughout the cortex, perivascular cuffing in the superficial cortex, and neuronal degeneration in the cortical pyramidal layers (laminae 2 and 3); ●●, diffuse to moderate meningoencephalitis containing all of the above; ●, slight meningitis and localized encephalitis characterized by focal nodular gliosis and minimal neuronal changes.
Spread of BHV-5gEΔ in the CNS following intranasal infection was detected by immunostaining and compared with that of the wild-type BHV-5. The results are shown in Fig. 7 and 8 and summarized in the Table 2. Virus-specific antigen was detected first in the olfactory bulb at 8 dpi (Fig. 7A). At 10 dpi, a small number of neurons (25 to 50/field) in the anterior olfactory nucleus and piriform cortex also were stained for viral antigen (Fig. 7B and C). The numbers of neurons infected by the BHV-5gEΔ in all of these areas were significantly lower than the numbers in rabbits infected with the wild type. In rabbits infected with the BHV-5gEΔ, we have never observed infected neurons in the amygdala, hippocampus/dentate gyrus, frontal/cingulate cortex, DR, LC, and lateral dorsal tegmentum. In contrast, in the rabbits infected with wild-type BHV-5, immunostained neurons in the olfactory bulb was observed first at 4 to 6 dpi, and a large number of immunostained neurons were found in the anterior olfactory nucleus and piriform cortex at 8 and 9 dpi (Fig. 7). In addition, neurons in the frontal/cingulate cortices, hippocampus/dentate gyrus (Fig. 8), DR, and LC (data not shown) (36) were also infected. Taken together, the results of histopathology, virus isolation, and immunohistochemistry suggest that BHV-5gEΔ can enter the brain through the olfactory pathway; it then replicates and spreads relatively inefficiently compared to the wild-type BHV-5.
FIG. 7.
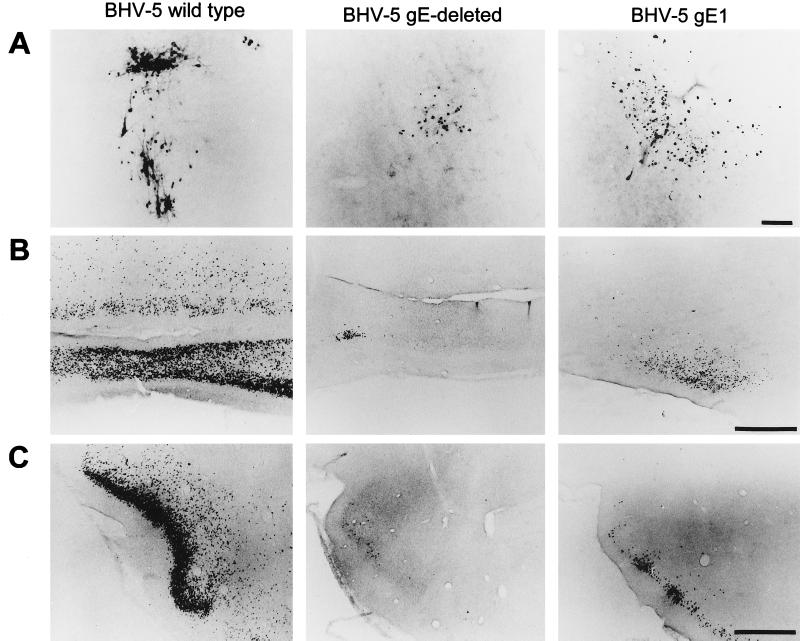
Localization of viral antigen in brain sections. Animals were inoculated intranasally with either wild-type BHV-5, BHV-5gEΔ, or BHV-5gE1 as described in Materials and Methods. The animals were euthanized on days 2, 4, 6, 8, 10, and 12 days postinfection or when they showed neurological signs, and their brains were processed for immunohistochemical analysis as described in Materials and Methods. Representative sections of the olfactory bulb (A), anterior olfactory nucleus (B), and piriform cortex (C) are pictured. In this assay, wild-type BHV-5 spread to the olfactory bulb at 4 to 6 dpi; however, labeling in the bulb was first observed at 8 and 6 dpi for the gE-deleted and gE-exchanged BHV-5, respectively. Wild-type BHV-5 spread to the anterior olfactory nucleus and piriform cortex at 8 dpi. gE-deleted and gE-exchanged BHV-5 took 10 to 12 dpi to spread to these areas. Bar in panel A, 100 μm; bar in panels B and C, 1,000 μm.
FIG. 8.
Localization of viral antigen in representative sections showing the amygdala, hippocampus/dentate gyrus, and cingulate cortex. In this assay, wild-type BHV-5 spread to amygdala (A), hippocampus/dentate gyrus (B), and cingulate cortex (C). However, BHV-5gEΔ spread only up to piriform cortex (12 dpi) and was never found in these areas. In BHV-5gE1-infected animals, a few neurons in the amygdala and several neurons in the cingulate cortex were labeled at 12 dpi, but no labeling was found in the hippocampus/dentate gyrus.
TABLE 2.
Summary of viral spread in the brain and TG after intranasal inoculation
| Virus | Presence and location of viral antigena
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OB | AON/LOT | Pir | Hippo | Amyg | CG | LC | DR | LDT | TG | |
| BHV-5 | ++ | ++++ | ++++ | ++++ | +++ | +++ | + | ++ | + | +b |
| BHV-5gEΔ | +b | + | ++b | − | − | − | − | − | − | +b |
| BHV-5gE1 | + | ++b | ++b (1) | − | +b | +b | − | − | − | +b |
| BHV-1gE5 | − | − | − | − | − | − | − | − | − | +b |
Indicated as no labeling (−) or 1 to 25 (+), 30 to 150 (++), 160 to 500 (+++), or >500 (++++) labeled neurons per field at a magnification of ×5. Abbreviations: OB, olfactory bulb; AON/LOT, anterior olfactory nucleus/lateral olfactory tract; Pir, piriform cortex; Hippo, hippocampus; Amyg, amygdala; CG, cingulate cortex; LC, locus coeruleus; DR, dorsal raphe; LDT, lateral dorsal tegmentum; TG, trigeminal ganglion.
Lower end of the range; in the case of BHV-5 gE1, one animal had more labeled neurons compared with others and is indicated by (1).
(ii) BHV-5gE1 does not show significant increases in neurovirulence and neural spread compared to its parent-deleted BHV-5 gEΔ.
One out of five rabbits infected with the BHV-5gE1 recombinant showed mild neurological signs at 9 dpi and was euthanized at 10 dpi. The remaining four rabbits did not show any detectable neurological signs through 12 dpi, when they were euthanized. The BHV-5gE1 recombinant was isolated, only after repassage, from the anterior cortex of the rabbit showing mild signs but not from the brains of any other rabbits, and the histopathological changes in the brains were mild (Table 1). Thus, the results of virus isolation and histopathology were very similar to those for BHV-5gEΔ. In the nasal and olfactory mucosa, the gE-exchanged BHV-5 also replicated with equal efficiency compared to the BHV-5 gE revertant (Table 1).
Results of spread of BHV-5gE1 in the CNS following intranasal infection in comparison to the gE-deleted and wild-type BHV-5 are shown in Fig. 7 and 8 and summarized in Table 2. At 6 dpi, immunostained neurons were detected only in the olfactory bulb. At 8 and 10 dpi, a small number of immunostained neurons were detected in the anterior olfactory nucleus (25 to 100/field) and piriform cortex (50 to 200/field). Only one animal had a few infected neurons in the cingulate cortex and amygdala (Fig. 8). This animal also showed more infected neurons in the piriform cortex (150 to 200/field) (Fig. 7C) in comparison to the other rabbits infected with BHV-5gE1 (data not shown) and BHV-5gEΔ. However, the number of infected neurons was significantly less than that resulting from infection with the wild-type BHV-5 (Fig. 7 and 8) or BHV-5 gE revertant (not shown). Taken together, the results of virus isolation and immunohistochemistry showed that the neural spread of BHV-5gE1 was slightly greater than that of BHV-5gEΔ. When the authentic BHV-5 gE was restored (BHV-5 gE revertant), the neurovirulence (Table 1) and the neural spread (data not shown) properties of the BHV-5 gE revertant also were restored to the wild-type levels. These results indicate that the BHV-1 gE could not restore neurovirulence and neural spread in BHV-5gEΔ to levels comparable to those of the wild-type or BHV-5 gE revertant.
(iii) BHV-1gE5 does not invade the CNS.
The BHV-1gE5 recombinant grew better than all the other recombinant and BHV-5 revertant viruses (Table 1) in both the nasal and olfactory epithelium, giving a yield very similar to that of the wild-type BHV-1 (data not shown) (36). However, no virus was isolated from the brains of rabbits infected with BHV-1gE5, and no infected neurons were detected by immunohistochemistry in the olfactory bulb and other olfactory pathway structures (data not shown). As with BHV-1 (36), only a few (5 to 10/field) immunostained neurons were present in the TG between 2 and 6 dpi (data not shown).
DISCUSSION
We initiated these experiments to test the role of BHV-5 gE in the differential neuropathogenesis of BHV-5 and BHV-1. The BHV-5 gE sequence was determined, and the predicted amino acids sequences of BHV-5 gE were compared with those of BHV-1 gE. Second, we constructed BHV-5gEΔ, BHV-5gE1, and BHV-1gE5 recombinants. Third, we investigated in vitro growth properties and the ability of BHV-5 and BHV-1 gEs to form complexes with their heterologous gI counterparts. Fourth, we investigated the neurovirulence and neuroinvasive properties of these recombinants and BHV-5 gE revertant viruses in the rabbit seizure model. The studies demonstrated that in vitro, gE1 and gE5 could complement each other with respect to gE-gI interaction and cell-to-cell spread. However, with respect to BHV-5 neuropathogenesis in the olfactory pathway, gE5 is important for efficient spread, replication, and virulence of BHV-5 and could not be replaced by the gE1.
To spread via the olfactory pathway to the deeper tissue of the CNS, the virus must infect the cell bodies of the first-order olfactory receptor neurons located within the olfactory epithelium and spread transynaptically to higher-order neurons (36). It is unknown whether virus replication takes place within the olfactory receptor neurons before the virus is transported to the second-order neurons in the olfactory bulb. Subsequent invasion to the third-, fourth-, and/or fifth-order neurons probably involves replication in the cell bodies and subsequent spread via the axon and synapses (36). Our data show that BHV-5gEΔ gains entry into the olfactory bulb (second-order neurons) but spreads and replicates poorly there as well as in the subsequent deeper brain tissues. In the nasal mucosa, the gE-deleted virus grew less efficiently than the BHV-5 gE revertant. In contrast, in the olfactory mucosa, BHV-5gEΔ and BHV-5 gE revertant grew equally well, yet the BHV-5gEΔ replicated and spread poorly within the CNS. Thus, in the case of rabbits infected with BHV-5gEΔ, the reduced neural invasion/spread and neurovirulence probably were due to the inability of the virus to be transported efficiently or to replicate efficiently within the CNS and not to the inability of the virus to infect the olfactory receptor neurons. The virus may need gE for efficient transport across the synapses and/or for its efficient replication in the neurons. The cumulative effect in either (or both) situations may account for the reduction of infected neurons at the second, third, or subsequent neuronal levels and may account for the labeling patterns restricted to the neuronal cell bodies. Similarly, HSV mutants unable to express gE or gI were markedly restricted in the ability to spread within the retina, produced 10-fold less virus in the retina, and spread inefficiently in the brain (20). In addition, experiments in neuronal cell culture showed that the gE-gI hetero-oligomer is required for efficient neuron-to-neuron transmission through synaptically linked neuronal pathways (20).
To determine if gE1 could replace gE5 with respect to neurovirulence and neuroinvasiveness of BHV-5, we also determined the pathogenic properties of BHV-5gE1. Our data indicated that BHV-5gE1 spread within the olfactory pathway better than the BHV-5gEΔ but notably less than the BHV-5 gE revertant or wild-type BHV-5. This result is in contrast to previous reports that BHV-1 gE and gI compensated for the virulence defect of PRV lacking its own gE and gI genes in a rodent model (30, 31). The gE1 in the BHV-5 backbone is synthesized with two signal sequences, and it appears that the gE1 signal peptide sequence was retained in the mature form of the protein. Thus, we cannot exclude the possibility that the unusual amino terminus of gE1 in the BHV-5 background diminished gE1's ability to fully complement gE5 with respect to neuropathogenesis. Additionally, in our study, we did not exchange both the gE and gI genes of BHV-5 with the gE and gI genes of BHV-1. Thus, BHV-1 gE may not be fully functional without its authentic BHV-1 gI counterpart. Our argument against this possibility is that BHV-5gE1 formed large plaques in vitro, which were similar to those of the wild-type BHV-5. In addition, we showed that gE1, in the BHV-5 backbone, formed a complex with the BHV-5 gI. Thus, we have documented that BHV-1 gE can compensate for BHV-5 gE, in vitro, with respect to the cell-to-cell spread function and gE-gI complex formation. However, our in vivo data indicate that BHV-1 gE does not fully replace BHV-5 gE.
We believe that BHV-5 gE is important for efficient neural spread and neurovirulence within the CNS. However, it is not required for initial viral entry into the olfactory pathway. This is expected since gE is not implicated in virus entry but other glycoproteins such as gB and gD may be involved (47, 51). We also believe that the explanation, in part, for the inability of gE1 to fully complement BHV-5 neuropathogenesis lies in the sequence differences between the two gEs. The ectodomain of BHV-5 gE contains a glycine-rich region (residues 204 to 218; [H]-GGEGE-GGKGGRGAAK-[OH]) showing divergence to the corresponding gE1 regions of BHV-1.1 and BHV-1.2. Because the antibody against the synthesized peptide precipitated only BHV-5 gE, it most likely constitutes a type-specific epitope. In the cytoplasmic tail, the acidic domain of gE5 (residues S509 to E535) is longer than the corresponding acidic domains of BHV-1.1 and BHV-1.2 gEs. It remains to be seen whether these and other regions showing sequence differences are functionally important with respect to gE5's role in differential neuropathogenesis of BHV-5. Several reports indicated that specific deletions and alterations of PRV gE sequences may affect the gE-mediated cell-to-cell spread, neurovirulence, and neuroinvasive properties of the virus (26, 52; P. J. Husak, M. G. Eldridge, and L. W. Enquist, Abstr. 24th Int. Herpesvirus Workshop, abstr. 6.020, 1999). The deletion of two amino acids (Val-125 and Cys-126) at the gE ectodomain resulted in a reduction of neurovirulence in PRV (26). Earlier, Tirabassi et al. (52) reported that the cytoplasmic tail of PRV gE is a virulence determinant, and the ectodomain part is involved in the neuroinvasiveness and cell-to-cell spread. Recent report (Husak et al., Abstr. 24th Int. Herpesvirus Workshop) indicated that the ectodomain and cytoplasmic tail domains of PRV gE are involved in the neurovirulence and cell-to-cell spread, respectively. Thus, the specific roles mediated by the ectodomain and cytoplasmic tail domain of the gE are still unfolding.
The cytoplasmic tail of BHV-5 gE contains two tetrapeptide YXXL tyrosine motifs. Both the YXXL motifs are conserved in all three gE sequences (BHV-5 as well as BHV-1.1 and BHV-1.2). The cytoplasmic tails of several other herpesviruses, including PRV and varicella-zoster virus, contain similar YXXL motifs and acidic domains (45, 46, 52). The YXXL motif have been implicated in tyrosine phosphorylation, mediation of antibody-induced capping and shedding of viral glycoprotein, incorporation of gE in the virion envelope, endocytosis, and cycling of gE between the trans-Golgi network and cell surface (16, 24, 45, 52, 53). Recent reports, also indicate that herpesvirus gE homologues are multifunctional and are additionally involved in the efficient maturation and release of virus from cells (7, 39). We believe that these global functions of gE are conserved among the alphaherpesviruses, which would account partially for the restricted transport and replication of the gE-deleted BHV-5 in the brain of rabbits. We assume that the slight increases in the neural spread and in the number of infected neurons in rabbits infected with BHV-5 gE1 compared with BHV-5gEΔ reflect the conserved global functions of the two gEs. This assumption is supported by the fact that in BHV-1 the second YXXL motif is important for cell-to-cell spread function (K. Bienkowska-Szewczyk, M. Rychlowski, and F. Rijsewijk, Abstr. 24th Int. Herpesvirus Workshop, abstr. 6.036, 1999). Because both YXXL motifs are conserved in BHV-1 and BHV-5, they probably are not important in determining the differential neuropathogenesis of BHV-5.
Considering the role of the cytoplasmic domain as a determinant of PRV neurovirulence (52), the differences observed between the BHV-1 and BHV-5 acidic domains could be important with respect to BHV-5 neurovirulence. In addition, other differences observed at the gE5 ectodomain also may be important themselves or may indirectly contribute to the utilization of tyrosine motifs through conformational effects. Future work with BHV-5 and BHV-1 recombinants containing gEs with hybrid cytoplasmic tail domains or recombinants containing site-specific mutations within the ectodomain and cytoplasmic tail domain would be needed to address the functional significance of the specific sequence differences.
In rabbits, BHV-1gE5 failed to invade the olfactory pathway. In PRV, the loss of US9 resulted in reduced virulence and spread in the rat eye model (A. D. Brideau and L. W. Enquist, Abstr. 24th system. Int. Herpesvirus Workshop, abstr. 10.005, 1999). Since in BHV-1gE5 the US9 gene was deleted, the failure of the virus to invade the CNS could be a consequence of the US9 deletion. We are currently pursuing additional experiments to address the role of US9 in BHV-1 and BHV-5 differential neuropathogenesis.
ACKNOWLEDGMENTS
We thank Lynn Enquist, Princeton University, for the rabbit anti-BHV-1 gE- and gI-specific antibodies and Julie Hix for the photography.
This work was supported by USDA grants 95-37204-2309 and 97-35204-4700 to S. I. Chowdhury.
Footnotes
Contribution 00-81-J, Kansas Agricultural Experiment Station.
REFERENCES
- 1.Ashbaugh S E, Thompson K E, Belknap E B, Schultheiss P C, Chowdhury S I, Collins J K. Specific detection of shedding and latency of bovine herpesvirus 1 and 5 using a nested polymerase chain reaction. J Vet Diagn Investig. 1997;9:387–394. doi: 10.1177/104063879700900408. [DOI] [PubMed] [Google Scholar]
- 2.Babic N, Klupp B, Brack A, Mettenleiter T C, Ugolini G, Flamand A. Deletion of glycoprotein gE reduces the propagation of pseudorabies virus in the nervous system of mice after intranasal inoculation. Virology. 1996;219:279–284. doi: 10.1006/viro.1996.0247. [DOI] [PubMed] [Google Scholar]
- 3.Balan P, Davis-Poynter N, Bell S, Atkinson H, Browne H, Minson T. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J Gen Virol. 1994;75:1245–1258. doi: 10.1099/0022-1317-75-6-1245. [DOI] [PubMed] [Google Scholar]
- 4.Baucke R B, Spear P G. Membrane proteins specified by herpes simplex viruses V. Identification of an Fc-binding glycoprotein. J Virol. 1979;32:779–789. doi: 10.1128/jvi.32.3.779-789.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belknap E B, Collins J K, Ayers V K, Schulteiss P C. Experimental infection of neonatal calves with neurovirulent bovine herpes virus type 1.3. Vet Pathol. 1994;31:358–365. doi: 10.1177/030098589403100309. [DOI] [PubMed] [Google Scholar]
- 6.Bell S, Cranage M, Borysiewicz L, Minson T. Induction of immunoglobulin G Fc receptors by recombinant vaccinia viruses expressing glycoproteins E and I of herpes simplex virus type 1. J Virol. 1990;64:2181–2186. doi: 10.1128/jvi.64.5.2181-2186.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brack A R, Dijkstra J M, Granzow H, Klupp B G, Mettenleiter T C. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J Virol. 1999;73:5364–5372. doi: 10.1128/jvi.73.7.5364-5372.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Card J P, Enquist L W. Neurovirulence of pseudorabies virus. Crit Rev Neurobiol. 1995;9:137–162. [PubMed] [Google Scholar]
- 9.Card J P, Rinaman L, Schawber J S, Melis R R, Whealy M E, Robbins A K, Enquist L W. Neurotropic properties of pseudorabies virus: uptake and transneuronal passage in the rat central nervous system. J Neurosci. 1990;10:1974–1994. doi: 10.1523/JNEUROSCI.10-06-01974.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Card J P, Whealy M E, Robbins A K, Enquist L W. Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat system. J Virol. 1992;66:3032–3041. doi: 10.1128/jvi.66.5.3032-3041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Card J P, Whealy M E, Robbins A K, Moore R Y, Enquist L W. Two alphaherpesvirus strains are transported differentially in the rodent visual system. Neuron. 1991;6:957–969. doi: 10.1016/0896-6273(91)90236-s. [DOI] [PubMed] [Google Scholar]
- 12.Chowdhury S I. Molecular basis of antigenic variation between the glycoprotein C of respiratory bovine herpesvirus 1 (BHV-1) and neurovirulent BHV-5. Virology. 1995;213:558–568. doi: 10.1016/s0042-6822(95)80137-5. [DOI] [PubMed] [Google Scholar]
- 13.Chowdhury S I. Construction and characterization of an attenuated bovine herpesvirus type 1 (BHV-1) recombinant virus. Vet Microbiol. 1996;52:13–23. doi: 10.1016/0378-1135(96)00043-0. [DOI] [PubMed] [Google Scholar]
- 14.Chowdhury S I, Lee B J, Mosier D, Sur J-H, Osorio F A, Kennedy G, Weiss M L. Neuropathology of bovine herpesvirus type 5 (BHV-5) meningo-encephalitis in a rabbit seizure model. J Comp Pathol. 1997;117:295–310. doi: 10.1016/s0021-9975(97)80078-3. [DOI] [PubMed] [Google Scholar]
- 15.Chowdhury S I, Ross C S D, Lee B J, Hall V, Chu H-J. Construction and characterization of a glycoprotein E gene-deleted bovine herpesvirus type 1 recombinant. Am J Vet Res. 1999;60:227–232. [PubMed] [Google Scholar]
- 16.Collawn J F, Kuhn L A, Liu L S, Tainer J A, Trowbridge I S. Transplanted LDL and mannose 6-phosphate receptor internalization signals promote high-efficiency of the transferrin receptor. EMBO J. 1991;10:3247–3253. doi: 10.1002/j.1460-2075.1991.tb04888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis-Poynter N S, Bell, Minson T, Browne H. Analysis of the contributions of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. J Virol. 1994;68:7586–7590. doi: 10.1128/jvi.68.11.7586-7590.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dingwell K S, Johnson D C. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J Virol. 1998;72:8933–8942. doi: 10.1128/jvi.72.11.8933-8942.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dingwell K S, Brunetti C R, Hendricks R L, Tang Q, Tang M, Rainbow A J, Johnson D C. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol. 1994;68:834–845. doi: 10.1128/jvi.68.2.834-845.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dingwell K S, Doering L C, Johnson D C. Glycoprotein E and I facilitate neuron-to-neuron spread of herpes simplex virus. J Virol. 1995;69:7087–7098. doi: 10.1128/jvi.69.11.7087-7098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Offay J M, Mock R E, Fulton R W. Isolation and characterization of encephalitic bovine herpesvirus type 1 isolates from cattle in North America. Am J Vet Res. 1993;54:534–539. [PubMed] [Google Scholar]
- 22.Dubin G, Socolof E, Frank I, Friedman H M. Herpes simplex virus type 1 Fc receptor protects infected cells from antibody-dependent cellular cytotoxicity. J Virol. 1990;65:7046–7050. doi: 10.1128/jvi.65.12.7046-7050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enquist L W, Husak P J, Banfield B W, Smith G A. Spread of alpha herpesviruses in the nervous system. Adv Virus Res. 1999;51:237–347. doi: 10.1016/s0065-3527(08)60787-3. [DOI] [PubMed] [Google Scholar]
- 24.Favoreel H W, Nauwynck H J, Van Oostveldt P, Mettenleiter T C, Pensaert M B. Antibody-induced and cytoskeleton-mediated redistribution and shedding of viral glycoproteins, expressed on pseudorabies virus-infected cells. J Virol. 1997;71:8254–8261. doi: 10.1128/jvi.71.11.8254-8261.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frank I, Friedman H M. A novel function of the herpes simplex virus type 1 Fc receptor: participation in bipolar bridging of antiviral immunoglobulin G. J Virol. 1989;63:4479–4488. doi: 10.1128/jvi.63.11.4479-4488.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobs L, Mulder W A M, van Oirschot J T, Gielkens A L J, Kimman T G. Deleting two amino acids in glycoprotein I pseudorabies virus decreases virulence and neurotropism for pigs, but does not affect immunogenicity. J Gen Virol. 1993;74:2201–2206. doi: 10.1099/0022-1317-74-10-2201. [DOI] [PubMed] [Google Scholar]
- 27.Jacobs L. Glycoprotein I of pseudorabies virus and homologous proteins in other alpha herpes virinae. Arch Virol. 1994;137:209–228. doi: 10.1007/BF01309470. [DOI] [PubMed] [Google Scholar]
- 28.Johnson D C, Feenstra V. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J Virol. 1987;61:2208–2216. doi: 10.1128/jvi.61.7.2208-2216.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson D C, Frame M C, Ligas M W, Cross A M, Stow N D. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J Virol. 1988;62:1347–1354. doi: 10.1128/jvi.62.4.1347-1354.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knapp A C, Enquist L W. Pseudorabies virus recombinants expressing functional virulence determinants gE and gI from bovine herpesvirus 1.1. J Virol. 1997;71:2731–2739. doi: 10.1128/jvi.71.4.2731-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knapp A C, Husak P J, Enquist L W. The gE and gI homologs from two alphaherpesviruses have conserved and divergent neuroinvasive properties. J Virol. 1997;71:5820–5827. doi: 10.1128/jvi.71.8.5820-5827.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 33.Kritas S K, Nauwynck H J, Pensaert M B. Dissemination of wild-type and gC-, gE- and gI-deleted mutants of Aujeszky's disease virus in the maxillary nerve and trigeminal ganglion of pigs after intranasal inoculation. J Gen Virol. 1995;76:2063–2066. doi: 10.1099/0022-1317-76-8-2063. [DOI] [PubMed] [Google Scholar]
- 34.Kritas S K, Pensaert M B, Mettenleiter T C. Role of envelope glycoproteins gI, gp63 and gIII in the invasion and spread of Aujeszky's disease virus in the olfactory nervous pathway of the pig. J Gen Virol. 1994;75:2319–2327. doi: 10.1099/0022-1317-75-9-2319. [DOI] [PubMed] [Google Scholar]
- 35.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 36.Lee B J, Weiss M L, Mosier D, Chowdhury S I. Spread of bovine herpesvirus type5 (BHV-5) in the rabbit brain after intranasal inoculation. J Neurovirol. 1999;5:473–483. doi: 10.3109/13550289909045376. [DOI] [PubMed] [Google Scholar]
- 37.Leung-Tack P, Audonnet J C, Rivière M. The complete DNA sequence and genetic organization of the short unique region (US) of the bovine herpes virus type 1 (ST strain) Virology. 1994;199:409–421. doi: 10.1006/viro.1994.1139. [DOI] [PubMed] [Google Scholar]
- 38.Maxam A M, Gilbert W. Chemical sequencing of DNA. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mettenleiter T C. Pseudorabies (Aujeszky's disease) virus: state of the art. Acta Vet Hung. 1994;42:153–177. [PubMed] [Google Scholar]
- 40.Mettenleiter T C, Schreurs C, Zuckermann F, Ben-Porat T. Role of pseudorabies virus glycoprotein gI in virus release from infected cells. J Virol. 1987;61:2764–2769. doi: 10.1128/jvi.61.9.2764-2769.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mettenleiter T C, Zsak L, Kaplan A S, Ben-Porat T, Lomnizci B. Role of a structural glycoprotein of pseudorabies virus in virus virulence. J Virol. 1987;61:4030–4032. doi: 10.1128/jvi.61.12.4030-4032.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulder W A, Jacobs L, Priem J, Kok G L, Wagenaar F, Kimman T G, Pol J M A. Glycoprotein gE-negative pseudorabies virus has a reduced capability to infect second- and third-order neurons of the olfactory and trigeminal routes in the porcine central nervous system. J Gen Virol. 1994;75:3095–3106. doi: 10.1099/0022-1317-75-11-3095. [DOI] [PubMed] [Google Scholar]
- 43.Neinhardt H, Schröder C H, Kaerner H C. Herpes simplex type 1 glycoprotein E is not indispensable for viral infection. J Virol. 1987;61:600–603. doi: 10.1128/jvi.61.2.600-603.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nielson H, Engelbrecht J, Brunak S, Heijne G V. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 45.Olson J K, Grose C. Endocytosis and recycling of varicella-zoster virus Fc receptor glycoprotein gE: internalization mediated by a YXXL motif in the cytoplasmic tail. J Virol. 1997;71:4042–4054. doi: 10.1128/jvi.71.5.4042-4054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olson J K, Bishop G A, Grose C. Varicella-zoster virus Fc receptor gE glycoprotein: serine/threonine and tyrosine phosphorylation of monomeric and dimeric forms. J Virol. 1997;71:110–119. doi: 10.1128/jvi.71.1.110-119.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peeters B, Pol J, Gielkens A, Moormann R. Envelope glycoprotein gp50 of pseudorabies virus is essential for virus entry but is not required for viral spread in mice. J Virol. 1993;67:170–177. doi: 10.1128/jvi.67.1.170-177.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rebordosa X, Pinol J, Peres-Pons J A, Lloberas J, Naval J, Querol E. Mapping, cloning and sequencing of a glycoprotein-coding gene from bovine herpesvirus type 1 homologous to the gE gene from HSV-1. Gene. 1994;149:203–209. doi: 10.1016/0378-1119(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 49.Rajcnai J, Herget U, Kaerner H C. Spread of herpes simplex virus (HSV) strains SC16, ANG, ANGpath and its glyC minus and glyE minus mutants in DBA-2 mice. Acta Virol. 1990;34:305–320. [PubMed] [Google Scholar]
- 50.Rock D L, Hagemoser W A, Osorio F A, Reed D E. Detection of bovine herpesvirus type 1 RNA in trigeminal ganglia of latently infected rabbits by in situ hybridization. J Gen Virol. 1986;67:2515–2520. doi: 10.1099/0022-1317-67-11-2515. [DOI] [PubMed] [Google Scholar]
- 51.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Vol. 2. New York, N.Y: Raven Press; 1996. pp. 2231–2295. [Google Scholar]
- 52.Tirabassi R S, Townley R A, Eldridge M G, Enquist L W. Characterization of pseudorabies virus mutants expressing carboxy-terminal truncations of gE: evidence for envelope incorporation, virulence, and neurotropism domains. J Virol. 1997;71:6455–6464. doi: 10.1128/jvi.71.9.6455-6464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tirabassi R S, Enquist L W. Role of envelope protein gE endocytosis in the pseudorabies virus life cycle. J Virol. 1998;72:4571–4579. doi: 10.1128/jvi.72.6.4571-4579.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Engelenburg F A C, Kaashoek J J, Rijsewijk F A M. A glycoprotein E deletion mutant of bovine herpesvirus 1 is avirulent in calves. J Gen Virol. 1994;75:2311–2318. doi: 10.1099/0022-1317-75-9-2311. [DOI] [PubMed] [Google Scholar]
- 55.Whealy M E, Card J P, Robbins A K, Dubin J R, Rhiza H J, Enquist L W. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J Virol. 1993;67:3786–3797. doi: 10.1128/jvi.67.7.3786-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitbeck J C, Knapp A C, Enquist L W, Lawrence W C, Bello L J. Synthesis, processing, and oligomerization of the bovine herpes virus 1 gE and gI membrane proteins. J Virol. 1996;70:7878–7884. doi: 10.1128/jvi.70.11.7878-7884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wyler R, Engels M, Schwyzer M. Infectious bovine rhinotracheitis/vulvovaginitis (BHV-1) In: Wittman G, editor. Herpesvirus diseases of cattle, horses and pigs. Hingham, Mass: Kluwer Academic Publishers; 1989. pp. 1–72. [Google Scholar]
- 58.Yuhasz S, Stevens J G. Glycoprotein B is a specific determinant of herpes simplex virus type 1 neuroinvasiveness. J Virol. 1993;67:5948–5954. doi: 10.1128/jvi.67.10.5948-5954.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zsak L, Zuckermann F A, Sugg N, Ben-Porat T. Glycoprotein gI of pseudorabies virus promotes cell fusion and virus spread via direct cell-to-cell transmission. J Virol. 1992;66:2316–2325. doi: 10.1128/jvi.66.4.2316-2325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zuckermann F A, Mettenleiter T C, Schreurs C, Sugg N, Ben-Porat T. Complex between glycoproteins gI and gp63 of pseudorabies virus: its effect on virus replication. J Virol. 1988;62:4622–4626. doi: 10.1128/jvi.62.12.4622-4626.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]