Abstract
In recent years, deep learning (DL) has been used extensively and successfully to diagnose different cancers in dermoscopic images. However, most approaches lack clinical inputs supported by dermatologists that could aid in higher accuracy and explainability. To dermatologists, the presence of telangiectasia, or narrow blood vessels that typically appear serpiginous or arborizing, is a critical indicator of basal cell carcinoma (BCC). Exploiting the feature information present in telangiectasia through a combination of DL-based techniques could create a pathway for both, improving DL results as well as aiding dermatologists in BCC diagnosis. This study demonstrates a novel “fusion” technique for BCC vs non-BCC classification using ensemble learning on a combination of (a) handcrafted features from semantically segmented telangiectasia (U-Net-based) and (b) deep learning features generated from whole lesion images (EfficientNet-B5-based). This fusion method achieves a binary classification accuracy of 97.2%, with a 1.3% improvement over the corresponding DL-only model, on a holdout test set of 395 images. An increase of 3.7% in sensitivity, 1.5% in specificity, and 1.5% in precision along with an AUC of 0.99 was also achieved. Metric improvements were demonstrated in three stages: (1) the addition of handcrafted telangiectasia features to deep learning features, (2) including areas near telangiectasia (surround areas), (3) discarding the noisy lower-importance features through feature importance. Another novel approach to feature finding with weak annotations through the examination of the surrounding areas of telangiectasia is offered in this study. The experimental results show state-of-the-art accuracy and precision in the diagnosis of BCC, compared to three benchmark techniques. Further exploration of deep learning techniques for individual dermoscopy feature detection is warranted.
Keywords: Basal cell carcinoma, Deep learning, Fusion, Telangiectasia, Transfer learning, Dermoscopy
Introduction
Basal cell carcinoma is one of the two most common types of skin cancer in the USA, with over two million new cases diagnosed yearly [1]. Dermatologists usually diagnose BCC by visual inspection. However, certain benign lesions can be confused with BCC and cause an unnecessary biopsy. Automating this diagnosis and ensuring early detection will reduce the burden on patients and healthcare professionals and produce more accurate results [2, 3].
Deep learning methodologies applied to dermoscopy images have yielded high diagnostic accuracy, now exceeding that of dermatologists [3–5]. Skin cancer diagnosis from images has advanced by implementing fusion ensembles, metadata, and some handcrafted features [6–10]. It is only pertinent to expand and explore the intersection of visually and clinically apparent features with deep learning techniques for disease detection.
Telangiectasia or thin arborizing vessels within the lesion is a crucial factor for dermatologists when looking for BCC. Their detection, either by visual inspection or any computational method, can provide pathways to make BCC diagnosis more accurate. Cheng et al. [11] investigated a local pixel color drop technique to identify vessel pixels. Kharazmi et al. [12] applied independent component analysis, k-means clustering, and shape for detecting vessels and other vascular structures. Kharazmi et al. [13] detected vessel patches by using stacked sparse autoencoders (SSAE) as their deep learning model. Maurya et al. [14] used a U-Net-based deep-learning (DL) model to perform semantic segmentation of these blood vessels. Semantically segmented precise binary masks provide means to effectively quantify the vessel feature information.
There are very few image processing or deep learning studies that utilize these vascular structures to diagnose BCC. Cheng et al. [15] used an adaptive critic design approach to discriminate vessels from competing structures, enabling BCC classification. Kharazmi et al. [16] utilize features extracted from these vascular structures to classify BCC using a random forest classifier. Kharazmi et al. [17] learned from sparse autoencoders, combined them with patient profile information and fed them to a Softmax classifier for BCC diagnosis. Recently, Serrano et.al [18] used clustering-based color features and GCLM-based texture features to train VGG16 and MLP models to extract deep learning features that they use to train another MLP model. The final MLP classifies lesions with either the presence or absence of one of seven BCC patterns, providing BCC classification. None of these studies utilizes deep learning–based vessel segmentation and classification together to achieve a BCC diagnosis. Through our fusion of telangiectasia and deep learning features, we also demonstrate how clinically observable features can be tracked better with deep learning and ultimately contribute to an improvement in diagnosis.
In this study, state-of-the-art accuracy in BCC classification was achieved by making the following unique contributions: (1) clinically inspired and explainable BCC diagnosis with deep learning–based telangiectasia mask generation as an intermediate step, (2) an ensemble learning classifier utilizing a hybrid input feature set consisting of object, shape, and color telangiectasia features integrated with deep learning features.
Materials and Methods
Image Datasets
The skin lesion images used in this study come from three datasets: the HAM10000 dataset (ISIC 2018) of Tschandl et al. [19], a publicly available skin lesion dermoscopy dataset containing over 10,000 skin images for seven diagnostic categories, the ISIC 2019 dataset [19–21], and datasets R43 from NIH studies CA153927-01 and CA101639-02A2 [22].
For training the U-Net model that generates telangiectasia masks, 127 images were selected from the HAM10000 dataset, 90 images from the ISIC 2019 dataset, and 783 images from the NIH study dataset, leading to a total of 1000 BCC images. The ISIC 2019 dataset included a few repeat images, omitted from our BCC dataset. The ground truth binary vessel masks were manually drawn by our team and verified by a dermatologist (WVS). The BCC dataset and the ground truth masks are shown in Fig. 1. For the non-BCC dataset, we selected 1000 images from the HAM10000 dataset from five benign categories: benign keratosis, nevus, actinic keratosis, dermatofibroma, and vascular lesion. The distribution for each category is shown in Fig. 2. The five categories represent the five most common benign skin conditions. Since this is a binary classification, the BCC vs non-BCC categories are balanced (1000 each). In the 1000 image non-BCC dataset, the categories maintain the same ratio as in the original HAM dataset which is illustrated in Fig. 2b. The images are 8-bit RGB of size 450 × 600 from the HAM10000 dataset and 1024 × 768 from the NIH study dataset. All the images are resized to 448 × 448 before training.
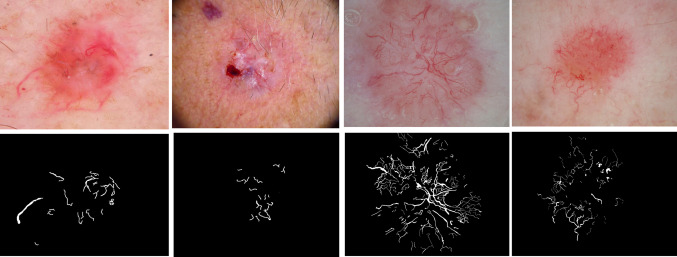
Fig. 1.
Left to right, first row: the first two images are BCC from the HAM10000 (ISIC2018) dataset; the last two images are BCC from the NIH study dataset. The second row presents telangiectasia overlays for the images in the first row
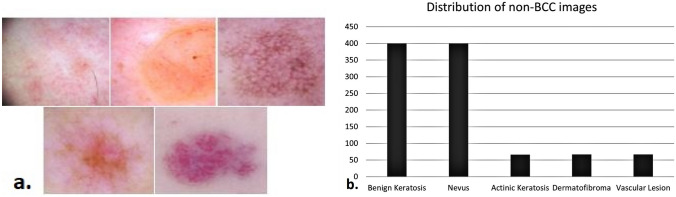
Fig 2.
a From left, first row: benign keratosis, nevus, actinic keratosis; from left second row: dermatofibroma and vascular lesion. b Number of images from each category used in the non-BCC dataset.
For both U-Net and EfficientNet-based models’ training, standard train-test splits of 80–20 were used. The training set was further split 80–20 to create a validation set. For BCC classification, the images were randomly selected, leading to subsets of 1288 for training, 324 for validation, and 395 for testing. For the U-Net model, the training set consisted of 650 images, the validation set comprised 162 images, and the test set included 195 images. These 195 BCC images combined with 200 non-BCC images make up the holdout test set for the BCC model.
Data Augmentation
For medical datasets with relatively few examples and a lack of variation, data augmentation helps create more training samples. Augmentation provides deep learning models the ability to generalize and hence provides regularization without overfitting. For both the U-Net and EfficientNet-based BCC classification models, the geometric augmentations include rotation ranging from + 30 to − 30° in reflect mode (to not distort the vessels) horizontal and vertical flip, width shift with a range of (− 0.2, + 0.2), height shift with a range of (− 0.2, + 0.2), and shear with a range (− 0.2, + 0.2). These geometric augmentations are shown in Fig. 3. The number of images in each class before and after augmentation is shown in Table 1.
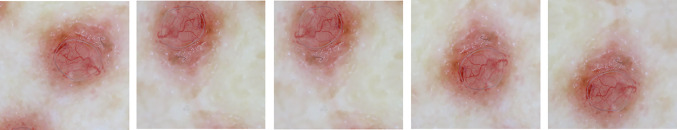
Fig. 3.
Different geometric augmentations
Table 1.
The number of images in each class before and after augmentation
| Model | Class | Before augmentation | After augmentation |
|---|---|---|---|
| U-Net | BCC | Training 650; validation162 | Training3900; validation 972 |
| EfficientNet | BCC | Training 650; validation 162 | Training 3900; validation 972 |
| EfficientNet | Non-BCC | Training 638; validation 162 | Training 3828; validation 972 |
Colors of skin lesions contain important distinguishing information and augmenting the original color of the lesions may ultimately make their classification harder. However, for telangiectasia, color augmentations can help fix the issue of vessels and red-pigmented skin being similar in color. Hence, color augmentations were utilized for training the U-Net model on the telangiectasia dataset to generate vessel masks, but not applied to the DL model responsible for calculating skin lesion features. For the U-Net model, histogram stretching to each color channel was applied, followed by contrast-limited adaptive histogram equalization, normalization, and brightness enhancement [14], as shown in Fig. 4. Due to the narrowness of the vessels, all the vessel masks are dilated with a 3 × 3 structuring element and closed with a 2 × 2 structuring element, as shown in Fig. 4. This dilation prevents the vessels in the masks from being broken when augmented. Since the goal is to identify vessels within the lesion, the vessel masks are multiplied with U-Net-generated lesion masks to yield vessels only within the lesion. For both models, the images were square-cropped and then resized to 448 × 448.
Fig. 4.
Left: color augmentations; right: vessel mask dilation
Proposed Methodology
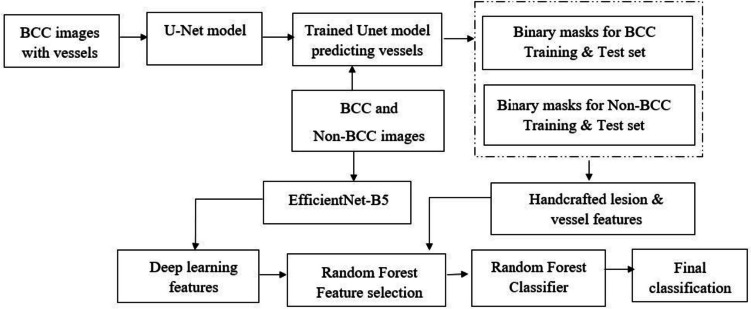
Our proposed methodology integrates clinically relevant handcrafted features of telangiectasia with high-level features extracted from a pre-trained deep learning model according to a feature importance score determined by the average Gini impurity decrease. We then utilize the higher predictive ability of ensemble learning methods to feed this hybrid feature set to a random forest classifier and create our novel fusion BCC diagnosis technique. Our method includes the following main components:
Semantically segmenting telangiectasia with a U-Net-based model to yield a binary vessel mask.
Applying image processing and statistical methods to calculate descriptive vessel features based on the objects in the vessel mask.
Extracting high-level deep learning features from fine-tuning a pre-trained EfficientNet-based model.
Calculating the feature importance score for all the features and selecting only the top-most features above a threshold.
Classifying the skin lesions into BCC or non-BCC using a random forest classifier trained on the hybrid feature set.
Our methodology is illustrated in Fig. 5.
Fig. 5.
Proposed architecture employing a fusion of deep learning and handcrafted features from vessels for BCC classification
Semantic Segmentation of Telangiectasia
In recent years, U-Net- [23] based semantic segmentation models have been the go-to for biomedical segmentation. Our vessel detection deep learning model is based on the U-Net model in [14] that generates binary vessel masks as the output. Distinguishing thin arborizing telangiectasia from sun damage telangiectasia which is sometimes present in benign skin lesions is a difficult task [11]. To add to this, since vessels occupy only 2–10% of the skin lesion image; it is quite a challenging task even for deep learning models. A combination loss function used in the U-Net model addresses this issue of severe class imbalance [14]. It is important to note that in cases of sun damage, the telangiectasia patterns are usually simpler and tend to be wider, shorter, and less defined, exhibit more variability in width, and are less densely clustered compared to those observed in BCC [11]. Hence, for training the U-Net model, only BCC images were used, since our goal is to find BCC indicating telangiectasia. For testing, masks were generated from the trained U-Net model for both the BCC and non-BCC datasets. As shown later, the U-Net model was able to find vessels in both BCC and non-BCC categories. The distinguishing properties are captured through the image processing operations explained in the next section.
Handcrafted Feature Generation from Vessel Masks
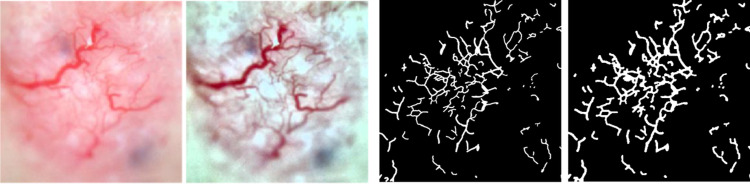
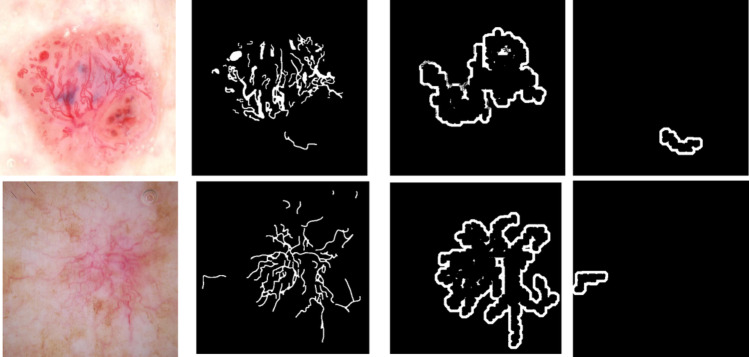
Traditional image processing techniques provide several tools to calculate telangiectasia features explicitly. Features are generated using objects in the vessel mask. From Maurya et al. [14], annotations for telangiectasia suffer from interobserver variability, fine or blurry telangiectasia, and poor contrast in images. There may be missed vessels along the boundary of annotations that need to be included. Also, regions surrounding the vessels can be helpful in differentiating basal cell carcinoma [11]. To include these probable missed features, we generate a surround mask for the vessel objects. Every object from the vessel mask is extracted and dilated with a disk structuring element of radius 12 (d1) and radius 5 (d2), resulting in two dilation variants of the object. Removing d2 from d1 gives the object surround mask, as shown in Fig. 6.
Fig. 6.
Object surround masks: Top row from left: example lesion image 1 with telangiectasia, its ground truth mask, its surround mask for object 1 (the contiguous connected vessel area), its surround mask for object 2 (the vessel on the bottom right). Bottom row from left: example lesion image 2 with telangiectasia, its ground truth mask, its surround mask for object 1 (the contiguous connected vessel area), its surround mask for object 2 (the vessel on the bottom right)
We calculate a total of 80 vessel features that include information about vessel color, geometry, shape, and statistical measure related to those features. Table 2 explains the handcrafted features we generated. Features 1 to 8 are general vessel descriptors [11]. Features 1–4 represent BCC’s narrower, longer, and more numerous vessels. Eccentricity features 5–8 are calculated to account for straighter BCC vessel structures. For the example images shown in Fig. 6, 347 and 240 vessel objects are detected. The figure shows surround masks generated for two such vessel objects. Features 5 to 8 are calculated for the surround masks and make up features 9 to 12. Features 13 to 22 include the number of objects calculated after morphologically eroding the final vessel mask with a circular structuring element of radius from 1 to 10. Features 23 to 32 include the area of objects calculated after morphologically eroding the final vessel mask with a circular structuring element of radius from 1 to 10. Features 33 to 44 are color features for vessels. Features 45 to 56 are color features in the HSV plane and features 57 to 80 are features 33 to 56 (RGB and HSV features) applied to the surround of vessel objects.
Table 2.
Different handcrafted features calculated from the U-Net binary vessel masks
| Feature number | Measure | Description | Meaning |
|---|---|---|---|
| 1 | Number of objects | Number of vessels in the final vessel mask | BCC has more vessels |
| 2 | Average object length | Average length for all vessels within a lesion | BCC vessels are longer |
| 3 | Average object width | Average width for all vessels within a lesion | BCC vessels are narrower |
| 4 | Average object area | Average area for all vessels within a lesion | BCC vessels are larger |
| 5 | Maximum eccentricity | Maximum ratio of the distance between the foci of the ellipse enclosing the vessels and its major axis length | BCC vessels are straighter |
| 6 | Minimum eccentricity | Minimum ratio of the distance between the foci of the ellipse enclosing the vessels and its major axis length | BCC vessels are straighter |
| 7 | Average eccentricity | Average eccentricity of all the vessels per vessel mask | BCC vessels are straighter |
| 8 | Standard deviation of eccentricity | Average standard deviation of eccentricity of all the vessels per vessel mask | BCC vessels are straighter and more uniform |
| 9 to 12 | Same as 5 to 8, but surround | Same as features 5 to 8 for the surrounding objects in the vessel mask | Regions around the vessel may contain distinguishing information |
| 13 to 22 | Eroded objects | Number of objects after the vessel mask is eroded with a disk structure of radius 1 to 10 | BCC objects are fewer after given number of erosions |
| 23 to 32 | Eroded objects area | Area of objects after the vessel mask is eroded with a disk structure of radius 1 to 10 | BCC object areas are smaller after given number of erosions |
| 33 to 35 | Maximum value of red, green, and blue objects | Maximum red, green, and blue value of every vessel; then averaged over total number of vessels per image | BCC vessels appear darker than lesion |
| 36 to 38 | Minimum value of red, green, and blue objects | Minimum red, green, and blue value of every vessel; then averaged over total number of vessels per image | BCC vessels appear darker than lesion |
| 39 to 41 | Average value of red, green, and blue objects | Average red, green, and blue value of every vessel; then averaged over total number of vessels per image | BCC vessels appear darker than lesion |
| 42 to 44 | Standard deviation of red, green, and blue objects | Standard deviation of red, green, and blue value of every vessel; then averaged over total number of vessels per image | BCC vessels appear darker than lesion |
| 45 to 56 | Same as 33 to 44 | Features 33 to 44 applied in HSV color space | HSV color space is more robust to lighting and shadow variations |
| 57 to 80 | Same as 33 to 56 | Features 33 to 56 applied on the surround of objects in the vessels | Regions around the vessel may contain distinguishing information |
Feature Vectors from EfficientNet
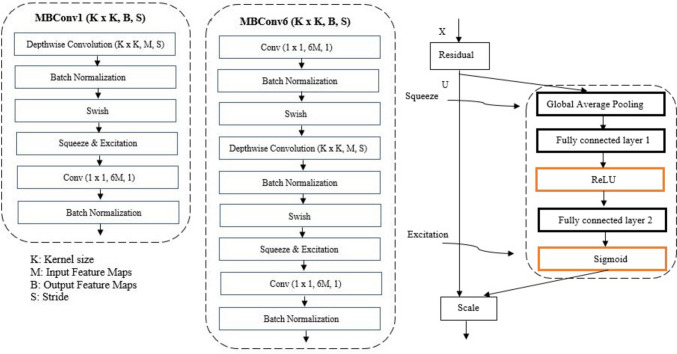
To calculate deep learning features, we use a pre-trained EfficientNet model. EfficientNets are a series of convolutional neural network models introduced by Tan et al. [24] that uniformly scale all network depth, width, or resolution dimensions by a compounding coefficient. These models achieved state-of-art top-1 accuracy on the ImageNet [25] dataset with fewer parameters. Their primary building block is a mobile inverted bottleneck called MBConv. The family of EfficientNet networks has different numbers of these MBConv blocks. The EfficientNetB5 model consists of two main blocks: MBConv1 and MBConv6. The detailed structures of these blocks are shown in Fig. 7.
Fig. 7.
Detailed structure of MBConv1, MBConv6, and squeeze and excitation block
The MBConv block or the inverted residual block improved the MobileNet [26] or residual blocks and followed a narrow- > wide- > narrow approach, which means that the connections in the MBConv blocks move from one bottleneck to another, using a residual connection. The basic MBConv block implements the following operations: 1 × 1 convolution that expands the dimensionality from the narrow channels to wider channels, a 3 × 3 or 5 × 5 channel-wise or depth-wise convolution operation to get output features, ultimately followed by another 1 × 1 convolution that downsamples the number of channels to the initial value. Since this output block and the initial block have the same dimensionality, they are added together. The primary purpose of the Squeeze operation is to extract global information from each of the channels of an image. Each block starts with a feature transformation on an image X to get features U, which are then squeezed to a single value using global average pooling [24]. This output is then fed to a fully connected layer followed by a ReLU function to add nonlinearity and reduce complexity. From here, another fully connected layer followed by a sigmoid function performs the excitation operation to get per-channel weights. The final output is achieved by rescaling these feature maps U with these activations.
Since the original EfficientNet-B5 model was built for ten-class classification, we remove the top layers to add a global average pooling layer, a dropout layer, and a final dense layer for binary classification. For our model, the initial input image size is 448 × 448 × 3.
Our model contains 14 different stages and is used first for classification, followed by feature extraction with the trained model. For the classification stage, we start with a 3 × 3 filter convolution, batch normalization, and swish activation function reducing the image dimensions in half from 448 to 224 and increasing the channels from 3 to 48. Hence, the feature map dimensions are 224 × 224 × 48. Stage 2 consists of three layers of an MBConv1 block with a 3 × 3 filter that maintains the previous stage resolution but decreases the number of channels, resulting in a feature map of size 224 × 224 × 24. Stages 3 (5 layers), 4 (5 layers), and 5 (7 layers) use three MBConv6 blocks each of kernel size 5 × 5 continuously reducing the resolution but increasing the feature map size to 28 × 28 × 128 (end of stage 5). Stages 6 (7 layers), 7 (9 layers), and 8 (3 layers) apply three more MBConv6 blocks each, with kernel sizes 3 × 3, 5 × 5, and 3 × 3, ultimately producing a feature map of size 14 × 14 × 2048. At stage 9, a 1 × 1 convolution with 2048 filters results in a feature map of size 14 × 14 × 2048. Stages 10 and 11 apply batch normalization and Softmax activation retaining the feature size as the previous layer. Stage 12 uses global average pooling to bring the resolution to 2048, followed by a dropout (stage 13) and dense layer (stage 14) leading to the final classification. The model is fine-tuned after the 200th layer, and the best model is saved. We perform feature extraction at stage 12 after the global average pooling layer, thereby resulting in a 2048-size feature vector for the training, validation, and test sets. The stages, operations, resolutions, channels, and layers are shown in Table 3.
Table 3.
Different layers of the EfficientNet-B5-based model for BCC classification
| Stage | Operator/Block | Resolution | Channels | Layers |
|---|---|---|---|---|
| 1 | Conv 3 × 3 + BN + Swish | 224 × 224 | 48 | 1 |
| 2 | MBConv1, k3 × 3 | 224 × 224 | 24 | 3 |
| 3 | MBConv6, k5 × 5 | 112 × 112 | 40 | 5 |
| 4 | MBConv6, k5 × 5 | 56 × 56 | 64 | 5 |
| 5 | MBConv6, k5 × 5 | 28 × 28 | 128 | 7 |
| 6 | MBConv6, k3 × 3 | 28 × 28 | 176 | 7 |
| 7 | MBConv6, k5 × 5 | 14 × 14 | 304 | 9 |
| 8 | MBConv6, k3 × 3 | 14 × 14 | 512 | 3 |
| 9 | Conv 1 × 1 | 14 × 14 | 2048 | 1 |
| 10 | BN | 14 × 14 | 2048 | 1 |
| 11 | Activation | 14 × 14 | 2048 | 1 |
| 12 | Global average pooling | 2048 | 1 | 1 |
| 13 | Dropout | 2048 | 1 | 1 |
| 14 | Dense | 1 | 1 | 1 |
Random Forest for Feature Importance and Final Classification
The 80 feature vectors calculated from the vessel masks and the 2048 feature vector from the EfficientNet model are ranked according to a random forest feature importance algorithm determined by the average Gini impurity decrease [27]. Once the most relevant features are selected, we employ different combinations of these fusion feature sets to a random forest classifier to yield the final classification result.
Training Details
All models were built using Keras with a TensorFlow backend in Python 3.7 and trained using a single 32 GB Nvidia V100 graphics card. The training and network parameters for the U-Net model generating the vessel masks are taken from [14]. The EfficientNet-B5 model is fine-tuned after the 200th layer. Hyperparameter information is given in Table 4.
Table 4.
Hyperparameters for the EfficientNet-based model and random forest classifier
| Model | Hyperparameter | Value |
|---|---|---|
| BCC vs non-BCC classification and DL feature generation with Efficient-Net | Epochs | 120 |
| Learning rate | 0.0001 | |
| Batch size | 20 | |
| Early stopping | Yes | |
| Patience | 5 | |
| Loss function | Binary cross-entropy | |
| Optimizer | RMSProp | |
| Dropout rate | 0.2 | |
| Random forest classifier | No. of estimators | 1000 |
| Minimum samples per split | 2 | |
| Bootstrapping | yes | |
| Maximum number of features per decision |
Experimental Results and Discussion
We present test results for the proposed architecture in the following order:
Vessel mask generation with U-Net and handcrafted features calculation.
Deep learning features generated from an Efficient-Net-based classifier.
Feature importance score results for both the features mentioned in the last two steps.
Final fusion models used for BCC vs non-BCC classification.
The classification results are evaluated on the holdout test set that consists of 195 BCC and 200 non-BCC images. The evaluation metrics used are accuracy, sensitivity, specificity, and precision (PPV) [28, 29].
Vessel Segmentation Test Results for BCC and Non-BCC Images
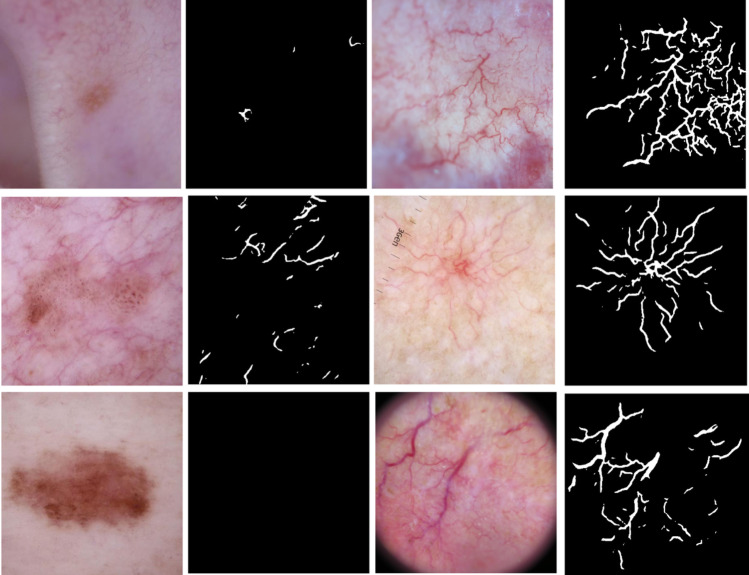
Figure 8 shows an example of vessel masks generated from the U-Net model for BCC and non-BCC images. Columns 1 and 3 show non-BCC and BCC images, respectively, whereas columns 2 and 4 show the predicted masks from the U-Net model. The second image in column 1 has some vessels outside the lesion captured in the predicted masks. Compared to the telangiectasia vessels in the BCC images, these vessels appear disconnected and thinner. Handcrafted masks can capture such distinctive properties. The 80 handcrafted features mentioned in Table 2 are then calculated from these masks and used as explained later in the “Feature Importance with Random Forest” section, where the feature importance scores for them are calculated.
Fig. 8.
From left: column 1 shows non-BCC images, column 2 shows their predicted masks, column 3 shows BCC images, and column 4 shows predicted BCC masks
Deep Learning Training Results from Fine-Tuning
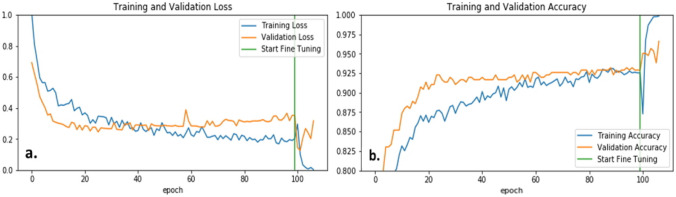
We achieved the best deep learning results from the EfficientNet-B5 baseline model. The weights after layer 200 were unfrozen, and the model was fine-tuned to our binary BCC vs. non-BCC classification. The preliminary transfer learning model converged at 100 epochs and took nine more epochs to converge after fine-tuning. Figure 9 shows the loss and accuracy curves for the training and validation sets. The validation and test set accuracies were 95.9% and 95.2%, respectively. After the model is trained, the 2048 length feature vector is extracted for the training, validation, and test sets.
Fig. 9.
Loss (a) and accuracy (b) plots before and after fine-tuning the EfficientNet-B5-based model
Feature Importance with Random Forest
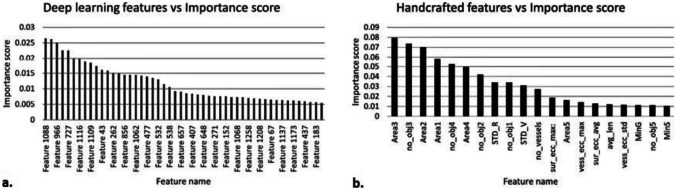
Feature importance is calculated for deep learning and handcrafted features using the random forest classifier [27]. The selection of key features results in models requiring optimal computational complexity while ensuring reduced generalization error due to noise introduced by less important features. The top 50 deep learning features with an importance score greater than 0.005, and the top 23 handcrafted features with an importance score greater than 0.01 are selected. Figure 10 shows the selected features with their importance scores. From Fig. 10b, we observe that the most important handcrafted features generally include the area of vessel objects, number of vessel objects, vessel eccentricity features, and color values of the vessel objects and the surrounding area.
Fig. 10.
Importance scores for deep learning (a) and handcrafted features (b)
Final Classification with Deep Learning and Random Forest Classifier
Table 5 shows six different fusion models that we tested. The different models are based on the following two selection criteria:
Table 5.
Performance comparison of our different fusion models
| Model | Feature set size | Accuracy | Sensitivity | Specificity | Precision |
|---|---|---|---|---|---|
| EfficientNet-B5-FT-Fusion2 | 73: 50 EfficientNet-B5-FT + 23 handcrafted | 0.972 | 0.979 | 0.965 | 0.965 |
| EfficientNet-B0-FT-Fusion2 | 73: 50 EfficientNet-B0-FT + 23 handcrafted | 0.967 | 0.979 | 0.955 | 0.960 |
| InceptionV3-FT-Fusion2 | 73: 50 InceptionV3-FT + 23 handcrafted | 0.955 | 0.965 | 0.95 | 0.95 |
| EfficientNet-B5-FT-Fusion1 | 2128: 2048 EfficientNet-B5-FT + 80 handcrafted | 0.964 | 0.967 | 0.955 | 0.95 |
| EfficientNet-B0-FT-Fusion1 | 1360: 1280 EfficientNet-B0-FT + 80 handcrafted | 0.934 | 0.945 | 0.94 | 0.933 |
| InceptionV3-FT-Fusion1 | 2128: 2048 InceptionV3-FT + 80 handcrafted | 0.93 | 0.942 | 0.934 | 0.93 |
- Pretrained model used for extracting deep learning features:
- EfficientNetB5, EfficientNetB0, and InceptionV3.
- Feature set size:
- Select deep learning and handcrafted features with the highest importance score.
-
(i)Total 73: 50 for deep learning and 23 for handcrafted.
-
(i)
- All deep learning and handcrafted features.
-
(i)EfficientNet B5: total 2128; 2048 for deep learning and 80 for handcrafted.
-
(ii)EfficientNet B0: total 1360; 1280 for deep learning and 80 for handcrafted.
-
(iii)InceptionV3: total 2128; 2048 for deep learning and 80 for handcrafted.
-
(i)
We achieve the best scores across all metrics with our Fusion 2 model that uses select critical features extracted from the fine-tuned EfficientNet-B5 model and handcrafted features extracted from vessel masks that feed a random forest classifier to yield a final classification result. We achieve an accuracy of 0.972, sensitivity of 0.979, specificity of 0.965, and precision of 0.965. We achieve an AUC of 0.995.
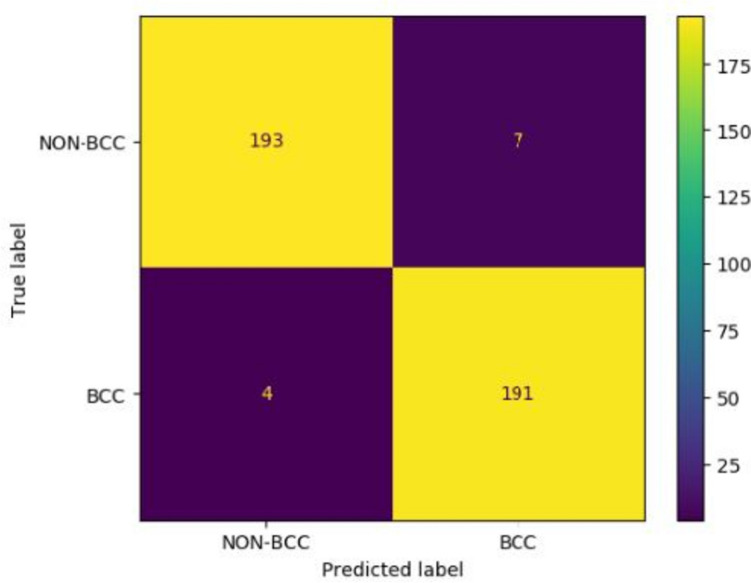
Table 6 shows the improvements in metrics as we move from the fine-tuned EfficientNet-B5 model to our fusion model. The accuracy, sensitivity, specificity, and precision improve by 1.3%, 3.7%, 1.5%, and 1.5%, respectively, suggesting the importance of adding handcrafted features. Omitting the surround features from the list of handcrafted features drops the AUC slightly, from 0.995 to 0.993. The confusion matrix is shown in Fig. 11.
Table 6.
Performance comparison with baseline deep learning model EfficientNet-B5
| Model | Feature set size | Accuracy | Sensitivity | Specificity | Precision |
|---|---|---|---|---|---|
| EfficientNet-B5-FT-Fusion2 | 73: 50 EfficientNet-B5-FT + 23 handcrafted | 0.972 | 0.979 | 0.965 | 0.965 |
| EfficientNet-B5-FT | 2048 EfficientNet-B5 | 0.959 | 0.942 | 0.950 | 0.950 |
Fig. 11.
Confusion matrix for the test set
Performance Comparison with Existing Vessel-Based BCC Detection
We compared the performance of our proposed fusion model with three other published results [16–18]. Table 7 lists the datasets, features, classifiers, and scoring metrics for the models. We achieve better accuracy and precision values with our method (bold) than the existing best values. All the methods listed except ours use some type of color and texture features; only ours semantically segments telangiectasia as an intermediate step.
Table 7.
Performance comparison with other BCC classification methods
| Manuscript | Dataset | Feature categories | Final classifier | Accuracy | Sensitivity | Precision |
|---|---|---|---|---|---|---|
| Kharazmi et al. 2017 | 659; 299 BCC and 360 non-BCC | Vascular features | Random Forest | 0.965 | 0.904 | 0.952 |
| Kharazmi et al. 2018 | 1199; 599 BCC and 600 non-BCC | Patient profile information & SAE feature learning | Softmax | 0.911 | 0.853 | 0.877 |
| Serrano et al. 2022 | 692 BCC and 671 non-BCC | Color and texture features | MLP | 0.970 | 0.993 | 0.953 |
| Proposed method | 2000; 1000 BCC and 1000 non-BCC | EfficientNet-B5 & localized vessel handcrafted color and shape features | Random Forest | 0.972 | 0.979 | 0.965 |
Discussion
A crucial clue for the clinical diagnosis of basal cell carcinoma is the presence of telangiectasia within the lesion. Classical image processing methods to detect these vessels in this study used statistical measures to quantify telangiectasia features. These measures included characteristics such as color values relative to the surrounding lesion area [11] or independent component analysis of melanin and hemoglobin components followed by thresholding and clustering [17]. From these vessel masks, different color, texture, and shape features are calculated. Our group used a deep learning–based U-Net model to detect these vessel masks with high accuracy vs. ground truth, obtaining a mean Jaccard score within the variation of human observers [14].
Recently, deep learning methods achieved superior results for detecting features such as hair [30] and globules [31]. From these deep learning–generated masks, classical features are calculated with the assumption that if the masks are more accurate, the features will be as well. Developing a single model for diagnosis without extracting individual features from whole images using pre-trained deep learning models also has been used extensively. However, it is impossible to know which features the deep learning model deems more important, contributing to its black-box nature. Our structure-based detection model partially remedies this shortcoming by detecting specific features. Moreover, this report shows improved diagnostic accuracy for BCC vs. non-BCC classification by combining deep learning and classical features with ensemble learning.
Figure 8 displays the advantages of the method presented here. Benign lesions have fewer vessels, and the total vessel area is less, as shown in columns 2 and 4 of Fig. 8. The number of vessel areas and their total number figure prominently in the most discriminatory features obtained from these masks, as shown in Fig. 10b. Eccentricity and color of objects found, all critical features of telangiectasia, are also crucial handcrafted features derived from these masks.
We achieve state-of-the-art results with this approach that are better than deep learning or traditional image processing results, indicating promise for our structure-based detection model. We also achieve clinically explainable results, opening similar pathways to solve other diagnostic challenges. Our results also confirm the superiority of ensemble learning methods for selecting a robust feature combination that improves the model’s accuracy. As seen from Table 5, all metrics improve when the features with higher importance scores are used. Another observation concerns the recent study by Serrano et al. [18]. The authors used different BCC features to annotate images with the presence or absence of features. We achieve a similar AUC but slightly better accuracy with our proposed model, using only a single automatically segmented BCC structure: telangiectasia. Due to this added local pixel information, our results achieve state-of-the-art accuracy automatically without observing and annotating every single pattern that may or may not be present. In previous work, we determined significant interobserver variability in vessel annotation [14]. However, DL can learn to detect structures with more consistency than those providing the masks for DL training. Thus, DL appears to be able to generalize from limited and inexact data sets and can detect vessel-like structures in different kinds of skin lesions, not just BCC. Figure 8 shows that our U-Net-based vessel detection model can identify these structures even in non-BCC images. Once the telangiectasia is detected, distinguishing qualities in BCC vessels (thinness, arborizing) are captured when we calculate the handcrafted features as indicated in Table 2. Using handcrafted features also helps us distinguish between vessels present outside the lesion, as they do not contribute to BCC diagnosis.
To account for the missed vessels due to blurry boundaries [14], our introduction of surround-area features also leads to an overall improvement in the AUC value. For segmentation problems, surround area detection by boundary expansion is a novel solution to feature finding that can contribute to better classification.
There are several limitations of this work. The vessel mask marking was supervised by a single dermatologist (WVS). Only one team observer (one of AM, DS, SS, or WVS) annotated each mask. The final sensitivity was less than for the study by Serrano et al.; however, the overall accuracy was higher. Even though there is a lot of intersection among the datasets of similar studies, the lack of a shared dataset with ground truths makes it harder to establish a true benchmark comparison. Hence, through this research, we share our telangiectasia masks and images openly [32].
All the comparable studies discussed, that employ deep learning models, including ours, do not include statistical significance tests. Selecting the most appropriate hypothesis testing in deep learning is challenging and is time as well as resource-consuming. The focus in deep learning has been more on predictive accuracy and model generalization rather than mean comparison among groups. However, research is progressing for the selection of appropriate statistical significance tests for model selection in machine learning. The datasets in our study as well as the ones in all comparable studies also lack diversity in skin color which limits their scope.
Conclusion and Future Work
This study proposes a telangiectasia-based fusion model approach for classifying BCC vs. non-BCC lesion images. To train our vessel identification model’s deep learning (U-Net) arm, we developed telangiectasia masks for 1000 BCC images, available at [32]. No such telangiectasia overlay database for BCC currently exists. Using the results from [14], we calculate the color and texture features from telangiectasia vessel masks and deep learning features learned from the EfficientNet-B5 model to yield a final classification result. Using a random forest model to combine features of each model provides a framework for fusion models.
Our fusion model outperforms past BCC classification models in precision and accuracy, over a larger dataset than in previous studies, one that is publicly available. Our state-of-the-art accuracy demonstrates the effectiveness of the proposed fusion techniques for this medical dataset. Our results produce more explainable results than whole-image deep learning results as we target clinically observable and relevant telangiectasia features. The current study is the only one, to the best of our knowledge, that uses semantically generated telangiectasia vessel features for BCC diagnosis.
In the future, we would like to continue this research by including more clinical features for our fusion model and employing additional statistical techniques to account for significant differences as well as ANOVA with appropriate post hoc tests. Nonetheless, as demonstrated in this study, for medical image datasets that are limited in size/quality, fusion techniques can offer a way to establish state-of-the-art diagnostic models and increase explainability as well as clinical confidence in results.
Author Contribution
Conceptualization: Akanksha Maurya, R. Joe Stanley, and William V. Stoecker; data curation: Akanksha Maurya, Gehana Patel, Daniyal Saeed, Samantha Swinfard, Colin Smith, Sadhika Jagannathan, William V. Stoecker; formal analysis: Akanksha Maurya, R. Joe Stanley, and William V. Stoecker; methodology: Akanksha Maurya, R Joe Stanley, and William V. Stoecker; resources: R. Joe Stanley; software: Akanksha Maurya, Hemanth Y. Aradhyula, Anand K. Nambisan, Norsang Lama, and Jason R. Hagerty; validation: Akanksha Maurya, R. Joe Stanley; visualization: Akanksha Maurya; writing—original draft: Akanksha Maurya; writing—review & editing: Akanksha Maurya, Norsang Lama, Anand K Nambisan, Jason R. Hagerty, R. Joe Stanley, and William V. Stoecker. All authors have read and agreed to the final version of the manuscript to be published.
Data Availability
The test set, publicly available, is referenced on the description of image data sets, 2nd page of the article. The 787 training lesions referenced have patient information and are not publicly available.
Declarations
Ethics Approval
No ethics approval is required.
Consent to Participate
NA.
Consent for Publication
All authors have read and agreed to the final version of the manuscript to be published.
Competing Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rogers HW, Weinstock MA, Feldman SR, Coldiron BM. Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the us population, 2012. JAMA Dermatol. 2015;151(10):1081–1086. doi: 10.1001/jamadermatol.2015.1187. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 3.Esteva A, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.M. A. Marchetti, N. C. F. Codella, S. W. Dusza, D. A. Gutman, B. Helba, A. Kalloo, N. Mishra, C. Carrera, M. E. Celebi, J. L. DeFazio, N. Jaimes, A. A. Marghoob, E. Quigley, A. Scope, O. Yélamos, A. C. Halpern, & International Skin Imaging Collaboration 2018 Results of the 2016 International Skin Imaging Collaboration International Symposium on Biomedical Imaging challenge: Comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images J Am Acad Dermatol 78 2 270 277. 10.1016/j.jaad.2017.08.016 [DOI] [PMC free article] [PubMed]
- 5.H. A. Haenssle, C. Fink, F. Toberer, J. Winkler, W. Stolz, T. Deinlein, R. Hofmann-Wellenhof, A. Lallas, S. Emmert, T. Buhl, M. Zutt, A. Blum, M. S. Abassi, L. Thomas, I. Tromme, P. Tschandl, A. Enk, A. Rosenberger, & Reader Study Level I and Level II Groups, “Man against machine reloaded: performance of a market-approved convolutional neural network in classifying a broad spectrum of skin lesions in comparison with 96 dermatologists working under less artificial conditions,” Annals of Oncology, vol. 31, no. 1, pp. 137–143, Jan. 2020, 10.1016/j.annonc.2019.10.013. [DOI] [PubMed]
- 6.T. Majtner, S. Yildirim-Yayilgan, and J. Y. Hardeberg, “Combining deep learning and hand-crafted features for skin lesion classification,” 2016 6th International Conference on Image Processing Theory, Tools and Applications, IPTA 2016, 2017, 10.1109/IPTA.2016.7821017.
- 7.N. Codella, J. Cai, M. Abedini, R. Garnavi, A. Halpern, and J. R. Smith, “Deep Learning, Sparse Coding, and SVM for Melanoma Recognition in Dermoscopy Images. In L. Zhou, L. Wang, Q. Wang,Y. Shi (eds) Machine Learning in Medical Imaging,” MLMI 2015, pp. 118–126, 10.1007/JRD.2017.2708299.
- 8.N. C. F. Codella et al., “Deep Learning Ensembles for Melanoma Recognition in Dermoscopy Images,” IBM J. Res. Dev., vol. 61, no. 4–5, pp. 5:1–5:15, Jul. 2017, 10.1147/978-3-319-24888-2_15.
- 9.González-Díaz I. DermaKNet: Incorporating the Knowledge of Dermatologists to Convolutional Neural Networks for Skin Lesion Diagnosis. IEEE J Biomed Health Inform. 2019;23(2):547–559. doi: 10.1109/JBHI.2018.2806962. [DOI] [PubMed] [Google Scholar]
- 10.Hagerty JR, et al. Deep Learning and Handcrafted Method Fusion: Higher Diagnostic Accuracy for Melanoma Dermoscopy Images. IEEE J Biomed Health Inform. 2019;23(4):1385–1391. doi: 10.1109/JBHI.2019.2891049. [DOI] [PubMed] [Google Scholar]
- 11.Cheng B, Erdos D, Stanley RJ, Stoecker WV, Calcara DA, Gómez DD. Automatic detection of basal cell carcinoma using telangiectasia analysis in dermoscopy skin lesion images. Skin Research and Technology. 2011;17(3):278–287. doi: 10.1111/j.1600-0846.2010.00494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kharazmi P, AlJasser MI, Lui H, Wang ZJ, Lee TK. Automated Detection and Segmentation of Vascular Structures of Skin Lesions Seen in Dermoscopy, With an Application to Basal Cell Carcinoma Classification. IEEE J Biomed Health Inform. 2017;21(6):1675–1684. doi: 10.1109/JBHI.2016.2637342. [DOI] [PubMed] [Google Scholar]
- 13.P. Kharazmi, J. Zheng, H. Lui, Z. Jane Wang, and T. K. Lee, “A Computer-Aided Decision Support System for Detection and Localization of Cutaneous Vasculature in Dermoscopy Images Via Deep Feature Learning,” Journal of Medical Systems, vol. 42, no. 2, p. 33, Jan. 2018, 10.1007/s10916-017-0885-2. [DOI] [PubMed]
- 14.Maurya A, et al. A deep learning approach to detect blood vessels in basal cell carcinoma. Skin research and technology. 2022;28(4):571–576. doi: 10.1111/srt.13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng B, Stanley RJ, Stoecker WV, Hinton K. Automatic telangiectasia analysis in dermoscopy images using adaptive critic design. Skin Research and Technology. 2012;18(4):389–396. doi: 10.1111/j.1600-0846.2011.00584.x. [DOI] [PubMed] [Google Scholar]
- 16.Kharazmi P, Kalia S, Lui H, Wang ZJ, Lee TK. A feature fusion system for basal cell carcinoma detection through data-driven feature learning and patient profile. Skin Research and Technology. 2018;24(2):256–264. doi: 10.1111/srt.12422. [DOI] [PubMed] [Google Scholar]
- 17.Serrano C, Lazo M, Serrano A, Toledo-Pastrana T, Barros-Tornay R, Acha B. Clinically Inspired Skin Lesion Classification through the Detection of Dermoscopic Criteria for Basal Cell Carcinoma. Journal of Imaging. 2022;8(7):197. doi: 10.3390/jimaging8070197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tschandl P, Rosendahl C, Kittler H. Data Descriptor: The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions Background & Summary. Nature Publishing Group. 2018 doi: 10.1038/sdata.2018.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Codella N, “Skin Lesion Analysis Toward Melanoma Detection, et al. A Challenge Hosted by the International Skin Imaging Collaboration (ISIC)”. 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) Washington, DC, USA. 2018;2018:168–172. doi: 10.1109/ISBI.2018.8363547. [DOI] [Google Scholar]
- 20.M. Combalia et al., “BCN20000: Dermoscopic Lesions in the Wild,” arXiv:1908.02288 [cs, eess], Aug. 2019, Available: https://arxiv.org/abs/1908.02288
- 21.W.V. Stoecker, Kapil Gupta, B. Shrestha, M. Wronkiewiecz, R. Chowdhury, R.J. Stanley, J. Xu, R. H Moss, M. E. Celebi, H. S. Rabinovitz, M. Oliviero, J. M. Malters, I. Kolm, “Detection of basal cell carcinoma using color and histogram measures of semitranslucent areas,” Skin Research and Technology, 2009, vol. 15, no. 3, pp.283–7. 10.1111/j.1600-0846.2009.00354.x. [DOI] [PMC free article] [PubMed]
- 22.O. Ronneberger, P. Fischer, and T. Brox, “U-Net: Convolutional Networks for Biomedical Image Segmentation.” [Online]. Available: http://lmb.informatik.uni-freiburg.de/
- 23.M. Tan and Q. Le, “Efficientnet: Rethinking model scaling for convolutional neural networks,” in International Conference on Machine Learning, 2019, pp. 6105–6114. 10.48550/arXiv.1905.11946
- 24.Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L. “Imagenet: A large-scale hierarchical image database”, in. IEEE Conference on Computer Vision and Pattern Recognition. 2009;2009:248–255. [Google Scholar]
- 25.Sandler M, Howard AG, Zhu M, Zhmoginov A, Chen L. MobileNetV2: Inverted Residuals and Linear Bottlenecks. IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2018;2018:4510–4520. [Google Scholar]
- 26.Breiman L. Random Forests. Machine Learning. 2001;45(1):5–32. doi: 10.1023/a:1010933404324. [DOI] [Google Scholar]
- 27.A.-M. Šimundić, “Measures of Diagnostic Accuracy: Basic Definitions,” EJIFCC, vol. 19, no. 4, pp. 203–11, 2009, Available: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4975285/ [PMC free article] [PubMed]
- 28.A. Baratloo, M. Hosseini, A. Negida, and G. El Ashal, “Part 1: Simple Definition and Calculation of Accuracy, Sensitivity and Specificity,” Emergency, vol. 3, no. 2, pp. 48–49, 2015, Available: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4614595/ [PMC free article] [PubMed]
- 29.Lama N, et al. ChimeraNet: U-Net for Hair Detection in Dermoscopic Skin Lesion Images. Journal of Digital Imaging. 2022 doi: 10.1007/s10278-022-00740-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nambisan AK, et al. Deep learning-based dot and globule segmentation with pixel and blob-based metrics for evaluation. Intelligent Systems with Applications. 2022;16:200126. doi: 10.1016/j.iswa.2022.200126. [DOI] [Google Scholar]
- 31.Akanksha Maurya, Ronald J Stanley, Hemanth Y Aradhyula, Norsang Lama, Anand K Nambisan, Gehana Patel, Daniyal Saaed, Samantha Swinfard, Colin Smith, Sadhika Jagannathan, Jason Hagerty, & William V Stoecker. (2023). Basal cell carcinoma diagnosis with fusion of deep learning and telangiectasia features [Data set]. Zenodo. 10.5281/zenodo.7709824 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The test set, publicly available, is referenced on the description of image data sets, 2nd page of the article. The 787 training lesions referenced have patient information and are not publicly available.