Abstract
We have previously shown that the herpes simplex virus tegument protein VP22 localizes predominantly to the cytoplasm of expressing cells. We have also shown that VP22 has the unusual property of intercellular spread, which involves the movement of VP22 from the cytoplasm of these expressing cells into the nuclei of nonexpressing cells. Thus, VP22 can localize in two distinct subcellular patterns. By utilizing time-lapse confocal microscopy of live cells expressing a green fluorescent protein-tagged protein, we now report in detail the intracellular trafficking properties of VP22 in expressing cells, as opposed to the intercellular trafficking of VP22 between expressing and nonexpressing cells. Our results show that during interphase VP22 appears to be targeted exclusively to the cytoplasm of the expressing cell. However, at the early stages of mitosis VP22 translocates from the cytoplasm to the nucleus, where it immediately binds to the condensing cellular chromatin and remains bound there through all stages of mitosis and chromatin decondensation into the G1 stage of the next cycle. Hence, in VP22-expressing cells the subcellular localization of the protein is regulated by the cell cycle such that initially cytoplasmic protein becomes nuclear during cell division, resulting in a gradual increase over time in the number of nuclear VP22-expressing cells. Importantly, we demonstrate that this process is a feature not only of VP22 expressed in isolation but also of VP22 expressed during virus infection. Thus, VP22 utilizes an unusual pathway for nuclear targeting in cells expressing the protein which differs from the nuclear targeting pathway used during intercellular trafficking.
Herpesviruses have a well-defined replication phase within the nucleus, where they are known to exploit many of the cellular processes performed there. Upon virus entry into the host cell, the viral DNA genome is directed into the nucleus by an as-yet-undefined mechanism and is subsequently transcribed and replicated by a combination of host cell machinery and virus gene products (1, 17, 22). At later stages in the replication cycle, assembly of the herpesvirus particle is initiated within the nucleus as the newly replicated virus DNA genome is packaged into assembling capsids (39, 41). As a consequence, herpesviruses must target several classes of their gene products, including transcription factors, DNA replication factors, scaffold proteins, and capsid proteins, to the nucleus. A number of virus proteins, such as the immediate-early proteins ICP0 (13, 30) and ICP27 (19, 28), the DNA replication protein encoded by gene UL9 (27), and the capsid protein VP19C (40), have been shown to contain classical nuclear localization signals (NLSs), which are defined in the primary amino acid sequence of these proteins (15, 32). Such NLS-containing proteins are translocated from the cytoplasm into the nucleus through the nuclear pores, a process mediated by cellular proteins typified by the heterodimeric complex of importin α and β proteins (15, 32). Thus, transient expression of these proteins in isolation from other virus products is sufficient to allow their localization to the nucleus.
However, in many cases protein localization observed by transient expression of individual virus genes does not correlate with the subcellular targeting of the same proteins during virus infection, and there are several examples of virus proteins which lack recognizable NLSs but which are nonetheless directed to the nucleus during virus infection. Several proteins have been shown to piggyback into the nucleus via an interaction with an NLS-containing partner either of viral origin, as is the case with the capsid proteins VP5 (31, 40) and VP23 (40), or of cellular origin, as has been suggested for the transactivator of immediate-early gene expression VP16, which appears to be directed into the nucleus by the cellular protein HCF (25). Thus, herpesviruses may employ a range of nuclear targeting mechanisms to ensure the correct cellular compartmentalization of their gene products.
The herpes simplex virus (HSV) structural protein VP22 is a major component of the virion tegument (18, 23, 42), that is, the virus compartment located between the capsid and the envelope (4). The role of VP22, which is encoded by gene UL49 (11), is unclear, but it does not contain a recognizable NLS, thereby suggesting that VP22 would not be targeted to the nucleus by the classical pathway during virus infection. We have previously studied the subcellular localization of VP22 using immunofluorescence of transiently transfected cells and have shown that, consistent with the lack of an NLS, transiently expressed VP22 localizes primarily in the cytoplasm of expressing cells (5, 6). However, our studies of transient transfection also revealed the presence of VP22 in the nuclei of a subpopulation of cells (5), but we have demonstrated that the majority of cells containing such nuclear VP22 have obtained it by an unusual process which we have termed intercellular spread (5). In this situation, VP22 is taken up into nonexpressing cells either from neighboring cells or from cell culture medium and is transported rapidly and efficiently to the nucleus (5). Thus, while transiently expressed VP22 clearly exhibits differential subcellular localization, this appears to be determined by whether or not the cell is expressing VP22 (in which case it is cytoplasmic) or has taken up the protein (in which case it is nuclear). Furthermore, we have also recently addressed the issue of VP22 targeting during infection by constructing an HSV type 1 (HSV-1) recombinant virus expressing green fluorescent protein (GFP)-tagged VP22 in place of wild-type VP22 (8). Time-lapse analyses of cells infected with this virus showed that, in cells infected at high multiplicity, GFP-22 was detected in a predominantly cytoplasmic location throughout the virus life cycle (8). Therefore, our results from virus infection suggest that, at high multiplicity at least, VP22 localizes primarily in the cytoplasm. However, the issue of VP22 compartmentalization is complicated by a number of other reports which suggest that VP22 may, in certain circumstances, localize within the nucleus during infection. For example, several biochemical studies have indicated that VP22 may be present in the nuclear fraction of infected cells (23, 35), while some infected-cell immunofluorescence studies have led to the proposal that VP22 may be targeted to the nucleus at certain stages of the HSV-1 replication cycle (29, 37). Nevertheless, these latter results are somewhat ambiguous, with one report suggesting that VP22 is nuclear at early times in infection and cytoplasmic later (29), while another recent study suggested that VP22 was cytoplasmic at early times and moved to the nucleus at later times during infection (37).
These previous observations on the complexity of VP22 localization, and its potential for different subcellular patterns, led us to use GFP-tagged VP22 expressed either by transient transfection or by virus infection to investigate further the trafficking and compartmentalization of VP22 in cells actively expressing the protein, rather than cells which have taken up the protein by intercellular spread. In this report, we demonstrate that GFP-22 expressed in isolation localizes in several different patterns ranging from exclusively cytoplasmic to exclusively nuclear. Using time-lapse analysis of GFP-22-expressing cells, we have further shown that these patterns represent VP22 localization at different stages of the cell cycle. Thus, the initial expression of GFP-22 in an interphase cell results in the cytoplasmic concentration of the protein, with no GFP-22 fluorescence detectable in the nucleus. However, upon entry into mitosis, the previously cytoplasmic GFP-22 becomes tightly associated with mitotic chromatin and remains bound there throughout cell division and subsequent nuclear membrane reformation. Moreover, once located in the nucleus, GFP-22 appears to be retained there, resulting in an exclusively nuclear pattern of GFP fluorescence throughout the G1 stage of the cell cycle. Thus, GFP-22 translocation from the cytoplasm to the nucleus of cells expressing the protein is regulated entirely by the process of mitosis. Furthermore, we have demonstrated the same pathway of M-phase nuclear translocation and retention using the GFP-22-expressing virus. Moreover, multiplicity of infection appears to influence this pathway, as it is observed principally in cells infected at low multiplicity, when the timing of the virus replication cycle may allow a greater proportion of infected cells to traverse cell division. Thus, we demonstrate an alternative and unusual mechanism for nuclear entry by a normally cytoplasmic virus protein and show that such translocation is an independent feature of VP22 which does not require the presence of additional virus proteins. The function of such differential cell-cycle-dependent localization of a herpesvirus tegument protein remains to be determined.
MATERIALS AND METHODS
Plasmids.
The eukaryotic expression vector pGE155 was constructed by inserting the BglII fragment from plasmid pGE109 (11), encoding the VP22 open reading frame, into the BglII site of pEGFPC1 (Clontech). This vector expresses a GFP-VP22 fusion protein under the control of the cytomegalovirus immediate-early promoter.
Cells, transfections, and virus infections.
COS-1 and Vero cells were maintained in Dulbecco's modified minimal essential medium containing 10% newborn calf serum. For transfection, COS-1 cells were plated at a density of 105 cells per well of a two-well coverglass chamber (LabTek) and transfected 24 h later with 100 ng of the plasmid DNA. Transfections were carried out by the calcium phosphate precipitation technique modified with BES [N,N-bis(2-hydroxyl)-2-aminoethanesulfonic acid]-buffered saline in place of HEPES-buffered saline. Transfected cells were analyzed by live-cell microscopy at various times up to 48 h after transfection.
Virus infections were carried out using the HSV-1 recombinant 166v, described previously (8), which expresses GFP-22 in place of parental VP22. Vero cells were plated at a density of 5 × 105 cells per well of a two-well coverglass chamber (LabTek) and infected 20 h later with 166v at a multiplicity of 0.001. Infected cell cultures were maintained in Dulbecco's modified minimal essential medium containing 2% newborn calf serum and analyzed by live-cell microscopy at various times up to 40 h after infection.
Live-cell microscopy and time-lapse analysis.
All live-cell microscopy of GFP-22-expressing cells was carried out using a Zeiss LSM 410 inverted confocal microscope, with resulting images processed using Adobe Photoshop software. Cells for short-term live analysis were examined directly in the two-well coverglass chambers in which they were grown. Cells for long-term time-lapse analysis were plated onto 42-mm-diameter coverslips contained in 60-mm-diameter dishes, at the appropriate density. Prior to analysis, the coverslip was transferred to a Bachhoffer POC chamber (obtained from Carl Zeiss) in open cultivation mode. This chamber was placed on a Saur heated frame (obtained from Carl Zeiss) seated on the microscope and covered with a Perspex lid through which a constant supply of 5% CO2 was fed. XYZT software from Zeiss was used to collect a Z series of images for each time point in the time series, which were then merged to produce an individual Z image for each time point. Animation of the time series was carried out using NIH Image software, and each series was saved as a Quicktime video. All time-lapse animations can be found elsewhere (http://www.mcri.ac.uk/VirusAssembly/timelapse.html).
RESULTS
Localization of transiently expressed GFP-22 in live cells.
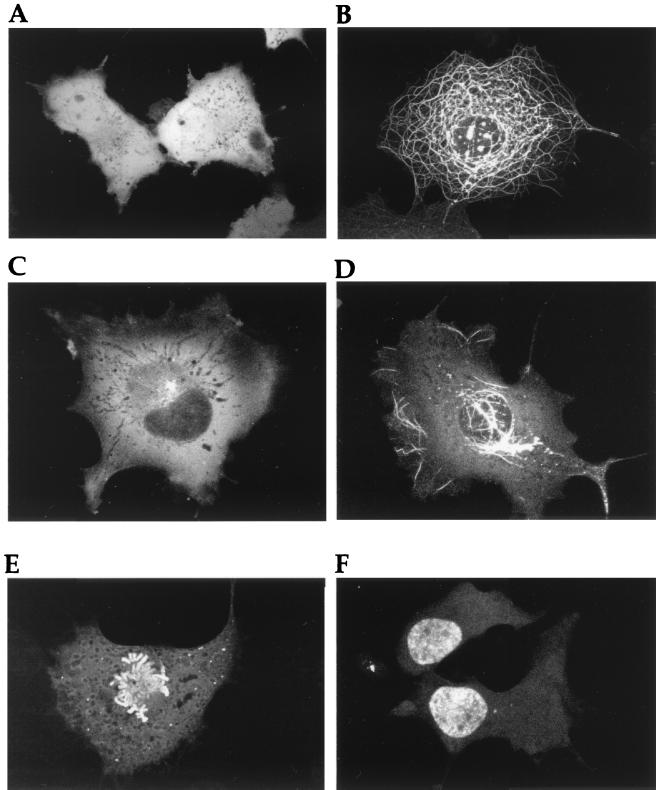
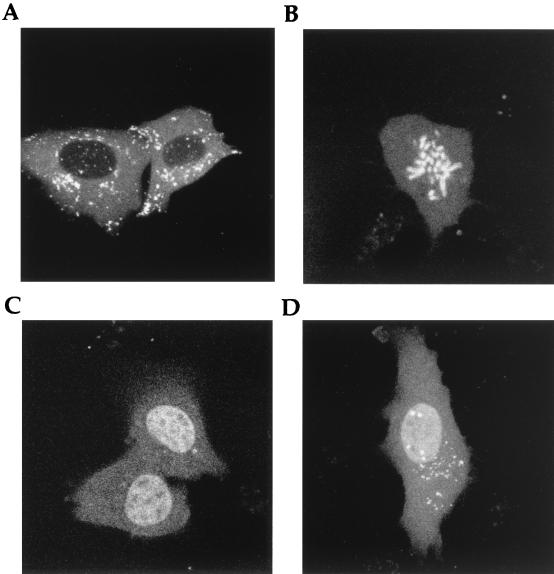
To initially characterize GFP-VP22 localization in expressing cells, COS-1 cells were transiently transfected with an expression vector for the GFP-22 fusion protein, and expression was allowed to continue for up to 48 h after transfection. At various times after transfection, the cells expressing the GFP-22 fusion protein were examined live and variations in the fluorescence patterns were compared to those observed for unfused GFP. At all times, the localization pattern of unfused GFP was the same, with the protein localizing in a diffuse cytoplasmic and nuclear pattern (Fig. 1A). By contrast, at least five GFP-VP22 patterns were identified which were markedly different from that observed for unfused GFP (Fig. 1B to F). At 24 to 36 h posttransfection, the predominant pattern of VP22 localization was that of thick cytoplasmic filaments (Fig. 1B) similar to those previously observed by immunofluorescence of fixed cells expressing VP22 (5, 6) and which we have shown to represent the reorganization and bundling of microtubules by high levels of expressed VP22 (6). However, at earlier times posttransfection, a large number of cells contained GFP-22 either in a diffuse cytoplasmic location (Fig. 1C), or in a diffuse cytoplasmic pattern on which was superimposed a small number of filaments (Fig. 1D). The likely explanation for the early diffuse patterns of GFP-22 localization is that they are the precursors of the extensive filamentous pattern shown in Fig. 1B. Strikingly, in all these cases GFP-22 was detectable only within the cytoplasm, with no detectable fluorescence in the nucleus, suggesting that, unlike GFP, the GFP-22 protein was retained within the cytoplasm of the cell (Fig. 1; compare panel A with panels B, C, and D).
FIG. 1.
Subcellular localization of GFP-22 expressed by transient transfection. COS-1 cells grown in a two-well coverslip chamber were transfected with either plasmid pEGFPC1 (A) or plasmid pGE155 (B to F), expressing GFP or GFP-22, respectively. The cells were examined live by confocal microscopy at 28 (A and B), 16 (C and D), or 44 (E and F) h after transfection.
However, at later times of up to 48 h posttransfection two very different patterns of localization were observed in a small population of GFP-22-expressing cells (Fig. 1E and F). The first of these clearly depicts cells in the process of mitosis, with GFP-22 specifically bound to condensed chromosomes (Fig. 1E), and while the example shown here is a cell in an early stage of mitosis, i.e., prometaphase, cells at all stages of mitosis were easily identified within the population of transfected cells. The second pattern consisted of GFP-22 localized specifically in the nucleus, with little or no fluorescence detectable in the cytoplasm (Fig. 1F). In these cases, the fluorescent nuclei always occurred in doublets.
Microtubule bundling by VP22 results in a block to cell division.
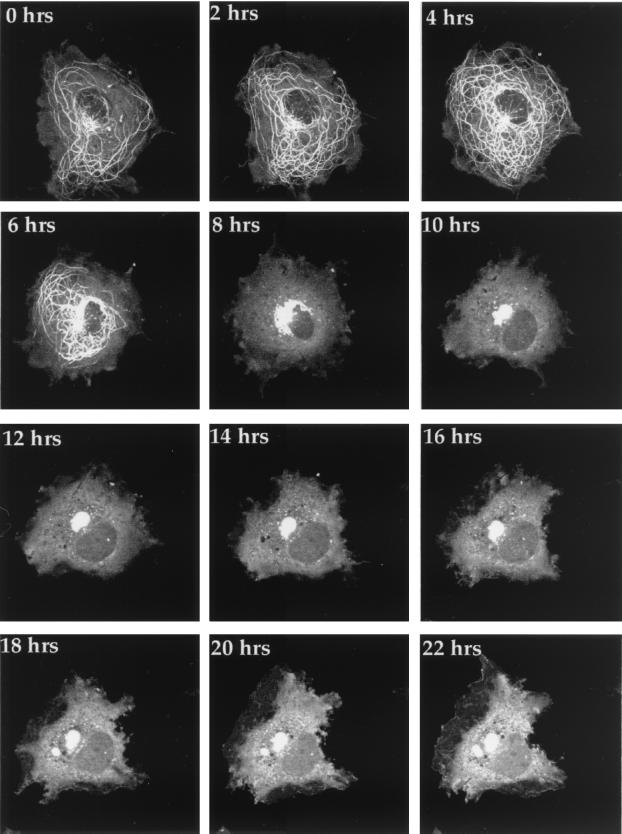
The diverse range of GFP-22 localization patterns identified during transfection, together with the binding of GFP-22 to mitotic chromatin, led us to speculate that GFP-22 compartmentalization may in some way be related to the cell cycle. Thus, to investigate the ultimate fate of cells expressing GFP-22, we initiated time-lapse analyses of COS-1 cells containing GFP-22 in its various locations. Cells grown on a coverslip were transfected with the expression vector for GFP-22, and 20 h later, the coverslip was transferred to a microscope chamber on a heated stage and allowed to equilibrate for 30 min. A suitable cell containing, in the first instance, filamentous GFP-22 (similar to Fig. 1B) was selected for further observation, and images were collected every 10 min over a period of 24 h. The resulting time-lapse analysis was processed to produce both an animation (found as Fig. 2 at http://www.mcri.ac.uk/VirusAssembly/timelapse.html) and a gallery of 2-hourly time points (Fig. 2). Between 2 and 4 h later, an increase in the concentration of bundles around the nucleus was observed (Fig. 2, 2 hrs and 4 hrs) while 6 h from the start of the analysis, the microtubule bundles began to contract into the perinuclear region (Fig. 2, 6 hrs), eventually becoming localized in a large mass at the side of the nucleus (Fig. 2, e.g., 10 hrs). The pattern of VP22 in this cell did not alter over a further period of 12 h, during which time a normal cell would have been expected to progress through mitosis. Taken together with a number of other time-lapse analyses which we have conducted (data not shown), we believe that this animation demonstrates that microtubule bundling by GFP-22 inhibits cell division.
FIG. 2.
The fate of VP22-induced microtubule bundles in transiently transfected cells. COS-1 cells grown on 42-mm-diameter coverslips were transfected with the expression vector for GFP-22 and transferred to a heated chamber 20 h later. A single cell containing bundled microtubules was selected (0 hrs), and images were collected every 10 min for a period of 24 h. Two-hourly images are shown in the gallery, and the corresponding animation can be found elsewhere (http://www.mcri.ac.uk/VirusAssembly/timelapse.html).
Soluble GFP-22 translocates from the cytoplasm to the nucleus during mitosis.
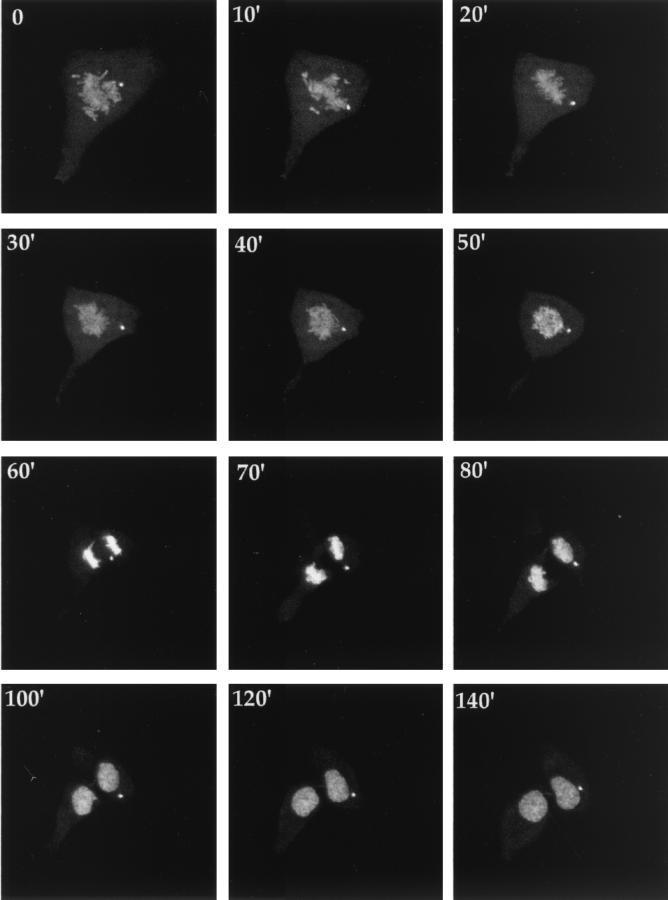
As the above data indicate that cells containing VP22-induced microtubule bundles were unable to progress through mitosis, we next addressed the possible sources of GFP-22 responsible for the protein binding to mitotic chromatin. In having observed a large number of cells expressing GFP-22, it was clear that we never found individual nuclei containing the fluorescent protein, only nuclei in pairs, suggesting that GFP-22 may localize in its nuclear pattern only after cell division. Thus, to investigate events immediately prior to the onset of mitosis, we wished to carry out a time-lapse analysis on GFP-22-expressing cells as they traversed the G2/M boundary of the cell cycle. COS-1 cells were transfected with the GFP-22 expression vector as described above and transferred to the heated chamber, and cells were examined for the characteristic appearance of rounding up, indicating the start of M phase. A single cell, which at the beginning of the time series contained GFP-22 in a diffuse cytoplasmic localization, similar to that shown in Fig. 1C, was chosen for further analysis. Images were then collected every 2 min for a total of 80 min, and the resulting time series has been presented as an animation (found at http://www.mcri.ac.uk/VirusAssembly/timelapse.html) and as a gallery of selected images (Fig. 3). Over the first 10 min of the time series, the pattern of diffuse cytoplasmic GFP-22 was unaltered (Fig. 3, 0′ and 10′). However, by 14 min a large amount of fluorescence had appeared within the nucleus, suggesting that at least some of the GFP-22 had translocated from the cytoplasm to the nucleus (Fig. 3, 14′). Two minutes later, the mitotic chromatin had condensed and was entirely decorated with bound GFP-22 (Fig. 3, 16′). Mitosis then progressed normally (see also Fig. 4), with GFP-22 remaining bound to the individual chromosomes throughout metaphase (Fig. 3, 40′ and 48′) and anaphase (Fig. 3, 64′). These results suggest then that diffuse cytoplasmic GFP-22 (Fig. 1C) progresses into chromatin-bound GFP-22 (Fig. 1F) at the start of mitosis.
FIG. 3.
Translocation of GFP-22 from cytoplasm to nucleus during cell division. COS-1 cells grown on 42-mm-diameter coverslips were transfected with the expression vector for GFP-22 and transferred to a heated chamber 24 h later. A single cell containing diffuse cytoplasmic GFP-22, which had the appearance of a cell entering mitosis, was selected (0′), and images were collected every 2 min for a total time of 80 min. Representative images are shown in the gallery, and the corresponding animation can be found elsewhere (http://www.mcri.ac.uk/VirusAssembly/timelapse.html).
FIG. 4.
Chromatin-associated GFP-22 is retained in the nucleus after cell division. COS-1 cells were treated as described in the legend to Fig. 3, and a cell in the early stages of mitosis was selected for further analysis (0′). Images of this cell were collected every minute for 150 min, with 10-min intervals presented in the gallery. The corresponding animation can be found elsewhere (http://www.mcri.ac.uk/VirusAssembly/timelapse.html).
To determine the fate of such mitotic cells with GFP-22 bound to condensed chromatin, we carried out time-lapse analysis of a mitotic cell beginning at an early stage of mitosis, in which GFP-22 was already outlining the condensed chromosomes. Images were collected every minute for a total of 160 min, and a time series was produced from the resulting images, which is presented as both an animation (found as Fig. 4 at http://www.mcri.ac.uk/VirusAssembly/timelapse.html) and a gallery of representative static images (Fig. 4). At the first time point (Fig. 4, 0′), the GFP-22-expressing cell was already in the early stages of mitosis, with the fluorescent chromosomes localizing in a prometaphase pattern. Over the next 30 min, the chromosomes became more organized into a metaphase pattern, until by 60 min after the start of the time lapse, the two sets of sister chromatids had pulled apart through anaphase (Fig. 4, 60′). Over the next 40 min, the now fluorescently decorated chromosomes decondensed, the nuclear membrane appeared to reform on the two daughter nuclei, and the cell underwent cytokinesis, at which time two daughter cells became apparent (Fig. 4, 100′). The time lapse was carried out for a further 60 min, during which time the GFP-22 fluorescence remained in the nucleus and showed no sign of returning to the cytoplasm. These results demonstrate that the double-nucleus pattern of GFP-22 localization (Fig. 1F) is the consequence of cell division and that the mitotic pattern of GFP-22 (Fig. 1E) progresses into the nuclear pattern of GFP-22 (Fig. 1F). Therefore, the process of mitosis alters the compartmentalization of GFP-22 from a cytoplasmic to a nuclear location.
Heterogeneous localization of GFP-22 in HSV-1-infected cells.
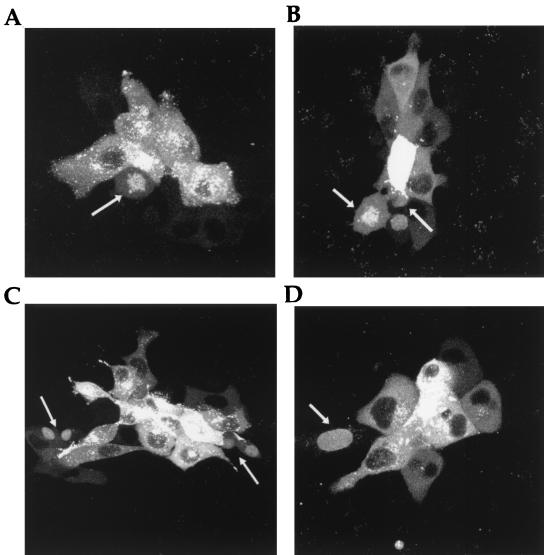
Our previous studies of cells infected with HSV-1 expressing GFP-22 had suggested that during virus infection VP22 was exclusively cytoplasmic (8). However, most of these results were obtained from high-multiplicity infections in which the virus replication cycle would be relatively rapid, reducing the possibility of infected cells progressing through cell division. We therefore reasoned that, to identify infected cells either in mitosis or postdivision, infection would have to be carried out at a low multiplicity to ensure that cells were infected with single infectious particles. Thus, Vero cells were infected with the GFP-22-expressing virus (166v) at a multiplicity of 0.001, and infection was allowed to progress for 36 h. The cells were then examined for foci of infection which were the consequence of virus spreading from individual infected cells into neighboring cells, thereby capturing infected cells over a range of stages of the replication cycle. As observed before in high-multiplicity infections (8), a large number of these infected cells contained GFP-22 in an exclusively cytoplasmic location. However, we consistently observed smaller numbers of cells on the edges of these centers of infection which contained GFP-22 in either its mitotic localization (Fig. 5A and B, arrowed) or its double nuclear location (Fig. 5B and C, arrowed). Furthermore, in addition to these patterns we also frequently observed GFP-22 in a single nuclear localization (Fig. 5D, arrowed), a pattern which we had never observed during transient expression of the protein.
FIG. 5.
Subcellular localization of GFP-22 in HSV-1 plaques. Vero cells grown in two-coverslip chambers were infected with HSV-1 expressing GFP-22, 166v, at a multiplicity of 0.001. Thirty-six hours later, the cells were examined live by confocal microscopy and fluorescent foci of infection were analyzed. Four representative foci are shown (A to D). Arrows indicate infected cells either in mitosis or containing nuclear GFP-22.
These results indicated that infection with the GFP-22-expressing virus at low multiplicity enabled us to detect GFP-22 in a pattern other than its predominant cytoplasmic pattern. To investigate this in more detail, a similar experiment was conducted whereby Vero cells were infected at a multiplicity of 0.001, but in this case the cells were examined at 16 h postinfection to analyze the range and ratio of infected-cell GFP-22 patterns. At this early stage, we were able to detect GFP-22 in four different patterns, namely, cytoplasmic (Fig. 6A), mitotic (Fig. 6B), double nuclear (Fig. 6C), and single nuclear (Fig. 6D). Moreover, the percentage of each of these patterns, with 44% being cytoplasmic, 2% being mitotic, and 54% being nuclear, seemed to approximately reflect the number of cells which should have progressed through cell division over a 16-h period.
FIG. 6.
Heterogeneity in GFP-22 subcellular localization at early times in a low-multiplicity HSV-1 infection. Vero cells grown in two-coverslip chambers were infected with HSV-1 expressing GFP-22 at a multiplicity of 0.01. Sixteen hours later, the cells were examined live by confocal microscopy and fluorescent cells were analyzed for variations in GFP-22 localization. GFP-22 was found localized exclusively in the cytoplasm (A), bound to mitotic chromatin (B), and in pairs of nuclei (C) or in single nuclei (D).
GFP-22 translocation at M phase occurs in HSV-1-infected cells.
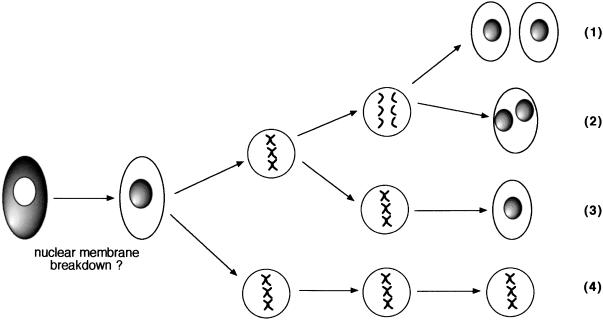
To confirm that the progression of mitotic cell-bound GFP-22 in infection was the same as that observed in the transiently transfected cells, we carried out time-lapse analyses on a range of infected mitotic cells, the results of which are summarized in Fig. 7. While a number of these animations demonstrated that infected GFP-22 behaved in a manner similar to that of transiently expressed GFP-22 (Fig. 7, pathway 1), we also found a range of alternative fates for these mitotic cells, which all seemed to be the result of aberrant mitoses. In particular, we observed that the single nuclear pattern shown in Fig. 6D was also a result of mitosis, but in this case the condensed chromatin did not separate into two populations but decondensed within a single nucleus (Fig. 7, pathway 3). In addition, there were a number of cells which progressed with a normal mitosis but failed to undergo cytokinesis, resulting in cells containing two fluorescent nuclei (Fig. 7, pathway 2). Last, we have evidence that some infected mitotic cells underwent an extremely prolonged mitosis (up to 16 h) before dividing as normal (Fig. 7, pathway 4). Thus, while the underlying process of GFP-22 translocation from the cytoplasm to the nucleus is a feature of both transiently expressing and infected cells, the end result of infected cells in mitosis is complicated by these cells exhibiting a range of cell division defects.
FIG. 7.
Schematic diagram illustrating the four pathways of mitosis identified in cells infected with the GFP-22-expressing virus. Vero cells grown on 42-mm-diameter coverslips were infected with the GFP-22-expressing virus at a multiplicity of 0.01. Sixteen hours later, the cells were transferred to the heated stage, and cells in the early stages of mitosis were identified by their content of GFP-22-decorated condensed chromatin. Time-lapse analysis was carried out on a range of these mitotic cells for periods up to 16 h, and four different outcomes were identified: (1) mitosis proceeds as normal; (2) mitosis proceeds as normal with the exception that cytokinesis fails; (3) condensed chromatin decondenses at metaphase without separation of chromosomes, resulting in a single nucleus; and (4) cells remain in mitosis for prolonged periods and then continue with any of the above three pathways.
DISCUSSION
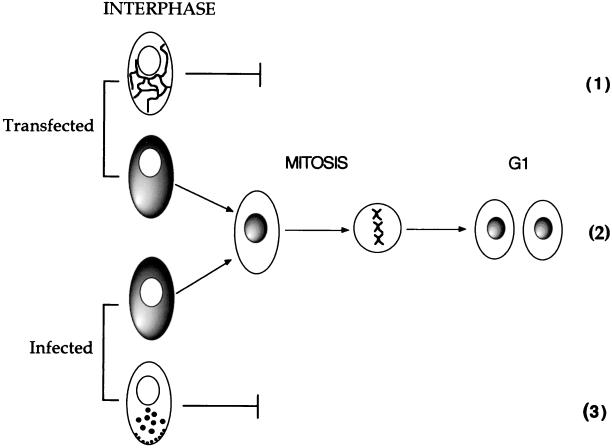
In this paper, we demonstrate that the herpesvirus tegument protein VP22 exhibits cell-cycle-dependent compartmentalization. We have exploited GFP technology to track the fate of individual GFP-22-expressing cells and to elucidate the VP22 trafficking pathway through the cell cycle. This trafficking begins with the protein localizing exclusively to the cytoplasm of the expressing cell during interphase. In a subpopulation of transiently expressing cells, this cytoplasmic VP22 binds cellular microtubules, resulting in their bundling and stabilization. In this situation, mitosis appears to be inhibited by the increased stabilization of the microtubule network (Fig. 8, pathway 1), a feature which is common to the overexpression of many other microtubule-stabilizing proteins (2, 21, 26, 34, 38). However, in another population of VP22-expressing cells, VP22 remains diffuse in the cytoplasm and upon entry into mitosis immediately associates with the condensed cellular chromatin where it remains throughout M phase and into G1 of the next cell cycle. Cytoplasm-to-nucleus translocation of VP22 is therefore a consequence of cell division (described in Fig. 8, pathway 2). The result of such differential compartmentalization is that at any time in a population of nonsynchronized expressing cells, VP22 can be found in a range of patterns, including exclusively cytoplasmic, exclusively nuclear, or both nuclear and cytoplasmic. Moreover, the chromatin interaction of VP22 at mitosis ensures that VP22 is transferred into the two daughter cells, providing an efficient mechanism for equal protein distribution through cell division. Thus, in transiently transfected cells expressing VP22 there are two potential outcomes dictating VP22 localization—either the cells proceed through mitosis, in which case VP22 translocates from the cytoplasm to the nucleus, or the cells are blocked for mitosis by microtubule stabilization and VP22 remains in the cytoplasm associated with the microtubule network (summarized in Fig. 8). While we do not yet understand the mechanism which determines whether VP22 binds microtubules or whether it remains diffuse in the cytoplasm, we would speculate that it is due to either the overall concentration of VP22 in the cytoplasm or the timing of VP22 expression and accumulation with respect to the cell cycle. In a similar manner, our studies on the intracellular trafficking of GFP-22 in HSV-1-infected cells have shown that there are also two potential outcomes of virus infection dictating the localization of VP22 (Fig. 8), which are likely to be determined by the cell cycle stage of the host cell at the time of infection. While the mitosis-dependent cytoplasm-to-nuclear translocation of VP22 also occurs during infection and is primarily observed in cells infected at low multiplicity (Fig. 8, pathway 2), the more frequent outcome involves the completion of the entire virus replication pathway before the cells can enter mitosis, resulting in the restriction of VP22 localization to the cytoplasm of the infected cell (Fig. 8, pathway 3).
FIG. 8.
Model for the compartmentalization of VP22 through the cell cycle. In both transfected and infected cells, we propose that VP22 localizes primarily to the cytoplasm of interphase-expressing cells. If such a cell enters mitosis, VP22 translocates from the cytoplasm to the nucleus and remains there through chromatin decondensation and nuclear envelope reformation (2). However, in both transfected and infected cells mitosis may be inhibited either by VP22-induced microtubule bundling (1) or by later stages in virus replication (3).
In this paper, we have addressed the issue of VP22 intracellular trafficking in cells which are expressing the protein, rather than in cells into which the protein has spread (5). We have previously shown that the efficiency of GFP-22 intercellular movement is lower than that of unfused VP22 (5), to the extent that GFP-22 spread cannot be detected in live cells expressing the fused protein and often requires antibody enhancement for detection (7). Thus, our live-cell analysis and time-lapse animations detect only the initial GFP-22-expressing cell and not the surrounding cells which have taken up the protein. However, taken together with our previous observations on intercellular trafficking, the results presented here suggest that VP22 uses two quite distinct pathways to enter the nucleus—either the mitosis-dependent pathway of the expressing cell or the mitosis-independent pathway of the nonexpressing cell, confirming that VP22 exhibits a complex range of cellular localization and trafficking pathways.
The finding of this unusual intracellular trafficking property of VP22 in cells expressing the protein provides an explanation for at least some of the variations in VP22 localization observed in previous reports (29, 37). In particular, a recent report by Pomeranz and Blaho (37) used immunofluorescence to show VP22 in the nuclei of cells located toward the exterior of virus plaques, in the same manner as we observed in our live cells infected with the GFP-22-expressing virus (demonstrated in Fig. 5). Moreover, as the cell fixation techniques used for immunofluorescence are known to cause a loss of a large percentage of mitotic cells which are only weakly attached to the substrate, the mitotic patterns of VP22 could easily have been missed in this previous study. However, it is clear that the mechanism of VP22 translocation to the nucleus during cell division could not be responsible for the apparent accumulation of the protein in the nuclei of all infected cells, as described previously for either early times in infection (29) or late times in infection (37). While there is evidently a discrepancy between these two previous reports concerning the timing and extent of VP22 localization to the nucleus, our own model based on the live-cell studies described here, suggesting that VP22 is almost exclusively cytoplasmic until cell division, differs from both scenarios. Thus, there may still be several issues concerning VP22 localization during virus infection which need to be addressed.
The vast majority of proteins which are targeted to the nucleus do so by the use of classical NLSs, consisting of clusters of basic residues which associate with cellular proteins to translocate them through the nuclear pores (15, 32). Some cellular proteins, such as basal transcription factors, appear to be targeted to the nucleus constitutively, while others are targeted only when their NLSs are somehow activated, for example, by phosphorylation (14, 16, 43). By contrast, the cell-cycle-regulated nuclear translocation of VP22 does not seem to involve the classical nuclear import pathway, as there are no consensus NLS sequences within the VP22 open reading frame. However, VP22 compartmentalization and translocation are strikingly similar to those of the cellular protein cyclin B1. During interphase, cyclin B1 is observed exclusively in the cytoplasm, where it appears to localize to microtubules (20, 36). However, at the start of M phase and prior to nuclear envelope breakdown, cyclin B1 is phosphorylated at its amino terminus, resulting in the creation of a nuclear import signal followed by the rapid translocation of the protein from the cytoplasm to the nucleus (16). Once there, cyclin B1 is observed in close association with condensed chromatin and remains bound to the dividing chromatin until anaphase, when the protein is destroyed. While we have previously identified two phosphorylation sites within the VP22 open reading frame which are substrates for cellular kinases (9, 10), we have not yet investigated the cell cycle characteristics of these sites. Furthermore, we do not yet know if VP22 moves into the nucleus before or after nuclear envelope breakdown. While the role of VP22 nuclear translocation and subsequent chromatin binding during virus infection is as yet unclear, it is interesting to speculate that it may be functioning in a similar manner to that of cyclin B1 and participating in the regulation of mitosis in infected cells. It is also noteworthy that while the chromatin labeling by GFP-22 in infected cells revealed a range of aberrant mitoses (Fig. 7), these effects were not caused by VP22 but as shown previously are likely to be due to binding of the immediate-early protein IE110 to chromosome centromeres (12).
Although VP22 is a major structural component of the tegument, it is unlikely that the nuclear localization of VP22 described here is required for the protein to be incorporated into the virus particle, as we have previously demonstrated GFP-22 trafficking during infection at high multiplicity in which GFP fluorescence was detected exclusively in the cytoplasm (8). VP22 translocation during mitosis may therefore be indicative of a function separate from its role during virus assembly and in addition to its properties of both microtubule stabilization (6) and intercellular transport (5), which we have previously described. Thus, as is the case for the tegument protein VP16, which performs roles as both a transactivator protein (3, 24, 33) and an essential structural protein (18, 44), it would seem that VP22 contributes multiple functions to the virus replication cycle.
ACKNOWLEDGMENT
This work was funded by Marie Curie Cancer Care.
REFERENCES
- 1.Boehmer P E, Lehman I R. Herpes simplex virus DNA replication. Annu Rev Biochem. 1997;66:347–384. doi: 10.1146/annurev.biochem.66.1.347. [DOI] [PubMed] [Google Scholar]
- 2.Burgin K E, Ludin B, Feralli J, Matus A. Bundling of microtubules in transfected cells does not involve an autonomous dimerization site on the MAP2 molecule. Mol Biol Cell. 1994;5:511–517. doi: 10.1091/mbc.5.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell M E, Palfreyman J W, Preston C M. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J Mol Biol. 1984;180:1–19. doi: 10.1016/0022-2836(84)90427-3. [DOI] [PubMed] [Google Scholar]
- 4.Dargin D. The structure and assembly of herpes viruses. In: Harris J R, Horne R W, editors. Electronmicroscopy of proteins. 5. Virus structure. London, United Kingdom: Academic Press; 1986. pp. 359–437. [Google Scholar]
- 5.Elliott G, O'Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 6.Elliott G, O'Hare P. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J Virol. 1998;72:6448–6455. doi: 10.1128/jvi.72.8.6448-6455.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott G, O'Hare P. Intercellular trafficking of VP22-GFP fusion proteins. Gene Ther. 1999;6:149–151. doi: 10.1038/sj.gt.3300850. [DOI] [PubMed] [Google Scholar]
- 8.Elliott G, O'Hare P. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J Virol. 1999;73:4110–4119. doi: 10.1128/jvi.73.5.4110-4119.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott G, O'Reilly D, O'Hare P. Phosphorylation of the herpes simplex virus type 1 tegument protein VP22. Virology. 1996;226:140–145. doi: 10.1006/viro.1996.0638. [DOI] [PubMed] [Google Scholar]
- 10.Elliott G, O'Reilly D, O'Hare P. Identification of phosphorylation sites within the herpes simplex virus tegument protein VP22. J Virol. 1999;73:6203–6206. doi: 10.1128/jvi.73.7.6203-6206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott G D, Meredith D M. The herpes simplex virus type 1 tegument protein VP22 is encoded by gene UL49. J Gen Virol. 1992;73:723–726. doi: 10.1099/0022-1317-73-3-723. [DOI] [PubMed] [Google Scholar]
- 12.Everett R, Earnshaw W C, Findlay J, Lomonte P. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 1999;18:1526–1538. doi: 10.1093/emboj/18.6.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett R D. Analysis of functional domains of herpes simplex virus type 1 immediate-early polypeptide Vmw110. J Mol Biol. 1988;202:87–96. doi: 10.1016/0022-2836(88)90521-9. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez F A, Seth A, Raden D L, Bowman D S, Fay F S, Davis R J. Serum-induced translocation of mitogen-activated protein kinase to the cell surface ruffling membrane and the nucleus. J Cell Biol. 1993;122:1089–1101. doi: 10.1083/jcb.122.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 16.Hagting A, Jackman M, Simpson K, Pines J. Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation-dependent nuclear import signal. Curr Biol. 1999;9:680–689. doi: 10.1016/s0960-9822(99)80308-x. [DOI] [PubMed] [Google Scholar]
- 17.Hayward G S. Immediate-early gene regulation in herpes simplex virus. Semin Virol. 1993;4:15–23. [Google Scholar]
- 18.Heine J W, Honess R W, Cassai E, Roizman B. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J Virol. 1974;14:640–651. doi: 10.1128/jvi.14.3.640-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hibbard M K, Sandri-Goldin R M. Arginine-rich regions succeeding the nuclear localization region of the herpes simplex virus type 1 regulatory protein ICP27 are required for efficient nuclear localization and late gene expression. J Virol. 1995;69:4656–4667. doi: 10.1128/jvi.69.8.4656-4667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackman M, Firth M, Pines J. Human cyclins B1 and B2 are localised to strikingly different structures: B1 to microtubules, B2 primarily to the Golgi apparatus. EMBO J. 1995;14:1646–1654. doi: 10.1002/j.1460-2075.1995.tb07153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanai Y, Chen J, Hirokawa N. Microtubule bundling by tau proteins in vivo: analysis of functional domains. EMBO J. 1992;11:3953–3961. doi: 10.1002/j.1460-2075.1992.tb05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knipe D M. The role of viral and cellular nuclear proteins in herpes simplex virus replication. Adv Virus Res. 1989;37:85–123. doi: 10.1016/s0065-3527(08)60833-7. [DOI] [PubMed] [Google Scholar]
- 23.Knopf K W, Kaerner H C. Virus-specific basic phosphoproteins associated with herpes simplex virus type 1 (HSV-1) particles and the chromatin of HSV-1 infected cells. J Gen Virol. 1980;46:405–414. doi: 10.1099/0022-1317-46-2-405. [DOI] [PubMed] [Google Scholar]
- 24.Kristie T M, LeBowitz J H, Sharp P A. The octamer-binding proteins form multi-protein–DNA complexes with the HSV alpha TIF regulatory protein. EMBO J. 1989;8:4229–4238. doi: 10.1002/j.1460-2075.1989.tb08608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaBoissiere S, Hughs T, O'Hare P. HCF-dependent nuclear import of VP16. EMBO J. 1999;18:480–489. doi: 10.1093/emboj/18.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee G, Brandt R. Microtubule-bundling studies revisited: is there a role for MAPs. Trends Cell Biol. 1992;2:286–289. doi: 10.1016/0962-8924(92)90106-w. [DOI] [PubMed] [Google Scholar]
- 27.Malik A K, Shao L, Shanley J D, Weller S K. Intracellular localisation of the herpes simplex virus type-1 origin binding protein, UL9. Virology. 1996;224:380–389. doi: 10.1006/viro.1996.0545. [DOI] [PubMed] [Google Scholar]
- 28.Mears W E, Lam V, Rice S A. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J Virol. 1995;69:935–947. doi: 10.1128/jvi.69.2.935-947.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison E E, Stevenson A J, Wang Y-F, Meredith D M. Differences in the intracellular localisation and fate of herpes simplex virus tegument proteins early in the infection of Vero cells. J Gen Virol. 1998;79:2515–2528. doi: 10.1099/0022-1317-79-10-2517. [DOI] [PubMed] [Google Scholar]
- 30.Mullen M-A, Ciufo D M, Hayward G S. Mapping of intracellular localization domains and evidence for colocalization interactions between the IE110 and IE175 nuclear transactivator proteins of herpes simplex virus. J Virol. 1994;68:3250–3266. doi: 10.1128/jvi.68.5.3250-3266.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholson P, Addison C, Cross A M, Kennard J, Preston V G, Rixon F J. Localisation of the herpes simplex virus type 1 major capsid protein VP5 to the cell nucleus requires the abundant scaffolding protein VP22a. J Gen Virol. 1994;75:1091–1099. doi: 10.1099/0022-1317-75-5-1091. [DOI] [PubMed] [Google Scholar]
- 32.Nigg E A. Nucleocytoplasmic transport: mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 33.O'Hare P, Goding C R, Haigh A. Direct combinatorial interaction between a herpes simplex virus regulatory protein and a cellular octamer-binding factor mediates specific induction of virus immediate-early gene expression. EMBO J. 1988;7:4231–4238. doi: 10.1002/j.1460-2075.1988.tb03320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson K, McIntosh J, Olmsted J. Analysis of MAP4 function in living cells using green fluorescent protein (GFP) chimeras. J Cell Biol. 1995;130:639–650. doi: 10.1083/jcb.130.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinard M F, Simard R, Bibor-Hardy V. DNA-binding proteins of herpes simplex virus type 1-infected BHK cell nuclear matrices. J Gen Virol. 1987;68:727–735. doi: 10.1099/0022-1317-68-3-727. [DOI] [PubMed] [Google Scholar]
- 36.Pines J, Hunter T. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pomeranz L E, Blaho J A. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J Virol. 1999;73:6769–6781. doi: 10.1128/jvi.73.8.6769-6781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preuss U, Biernat J, Mandelkow E-M, Mandelkow E. The ‘jaws’ model of tau-microtubule interaction examined in CHO cells. J Cell Sci. 1997;110:789–800. doi: 10.1242/jcs.110.6.789. [DOI] [PubMed] [Google Scholar]
- 39.Rixon F. Structure and assembly of herpesviruses. Semin Virol. 1993;4:135–144. [Google Scholar]
- 40.Rixon F J, Addison C, McGregor A, MacNab S J, Nicholson P, Preston V G, Tatman J D. Multiple interactions control the intracellular localisation of the herpes simplex virus type 1 capsid proteins. J Gen Virol. 1996;77:2251–2260. doi: 10.1099/0022-1317-77-9-2251. [DOI] [PubMed] [Google Scholar]
- 41.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, editors. Fundamental virology. New York, N.Y: Raven Press; 1991. pp. 849–895. [Google Scholar]
- 42.Spear P G, Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J Virol. 1972;9:143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tagawa T, Kuroki T, Vogt P K, Chida K. The cell cycle-dependent nuclear import of v-Jun is regulated by phosphorylation of a serine adjacent to the nuclear localisation signal. J Cell Biol. 1995;130:255–263. doi: 10.1083/jcb.130.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinheimer S P, Boyd B A, Durham S K, Resnick J L, O'Boyle D D. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J Virol. 1992;66:258–269. doi: 10.1128/jvi.66.1.258-269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]