Abstract
The murine retrovirus SL3-3 is a potent inducer of T-cell lymphomas when inoculated into susceptible newborn mice. Previously, DNAs from twenty SL3-3-induced tumors were screened by PCR for provirus integration sites. Two out of 20 tumors demonstrated clonal provirus insertion into a common region. This region has now been isolated and characterized. The region, named SL3-3 integration site 1 (Sint1), maps to the distal end of mouse chromosome 11, corresponding to human chromosome 17q25, and may be identical to a mouse mammary tumor virus integration site in a T-cell lymphoma, Pad3. Two overlapping genomic λ clones spanning about 35 kb were isolated and used as a starting point for a search for genes in the neighborhood of the virus integration sites. A genomic fragment was used as a hybridization probe to isolate a 3-kb cDNA clone, the expression of which was upregulated in one of two tumors harboring a provirus in Sint1. The cDNA clone is predicted to encode a protein which shows 97.0% identity to a human septin-like protein encoded by a gene which has been found as a fusion partner gene of MLL in an acute myeloid leukemia with a t(11;17)(q23;q25). Together these findings raise the possibility that a proto-oncogene belonging to the septin family, and located about 15 kb upstream of the provirus integration sites, is involved in murine leukemia virus-induced T-cell lymphomagenesis.
The induction of lymphomas and leukemias by the non-oncogene-bearing murine leukemia viruses (MLVs) is a complex process, comprising many steps of which the most well-defined one involves long terminal repeat (LTR) deregulation or impairment of cellular proto-oncogenes or tumor suppressor genes (29, 38). Proviral tagging of critical loci has during the last 15 years proved to be a powerful tool to identify genes associated with specific diseases. For example, c-myb, Evi1, and Evi2 are genes/common integration sites which mainly are correlated with MLV-induced myeloid leukemias (4, 23, 33, 39), while c-myc, Pim1, Mlvi1/pvt1, Evi3, and Evi5 primarily are associated with MLV-induced lymphomas (8, 13, 18, 21, 31, 37). Although there is no doubt that insertional mutagenesis plays a major role in tumor development, insertion into a common integration site for a specific tumor type has never been reported to be 100%, revealing the complexity of the process. Some proto-oncogenes (or common integration sites) may function redundantly as well as cooperate with other proto-oncogenes (17).
In addition to depending on the type of disease induced, the specific genes that will be tagged by a provirus depend on virus type. This is illustrated by the difference between SL3-3 MLV and Moloney MLV (Mo-MLV). Despite the fact that both viruses induce T-cell lymphomas with similar latency periods, the frequencies of integrations into specific sites are different. For example, while Mo-MLV and SL3-3 both exhibit insertions in the c-myc gene in 20 to 40% of the induced end-stage tumors, the frequencies of insertions observed in the Pim1 locus amount to 15 to 65% of the Mo-MLV-induced tumors but less than 1% of the SL3-3-induced tumors (1, 8, 9, 15, 24, 29, 31, 35, 36). It thus appears that even viruses of similar disease specificities may exploit different gene activation pathways.
Previously, we have by a PCR-based method isolated and determined the sequences of provirus integration sites in twenty tumors induced by SL3-3 MLV (34, 35). We found that in two independent tumors a provirus had inserted into the same region. In essence, retroviral integration is random; thus the finding of two independent insertions into a common site strongly indicates that a linked host gene plays a role in the tumorigenic process.
We report here on further analysis of the integration site region as well as on the isolation of a cDNA clone representing a gene in this region. The chromosomal mapping of the region, which was named SL3-3 integration site 1 (Sint1), to the distal end of mouse chromosome 11 suggests that Sint1 overlaps or is identical to the Pad3 locus previously identified as an integration site in an mouse mammary tumor virus (MMTV)-induced T-cell lymphoma (28). Taken together with the finding that the associated Sint1 cDNA seems to encode a protein homologous to a human septin-like protein that has been found as a fusion partner of MLL in an acute myeloid leukemia, this indicates that Sint1 cDNA may identify a novel proto-oncogene involved in mouse T-cell lymphomagenesis.
MATERIALS AND METHODS
Genomic DNA and RNA analyses.
DNA was extracted from frozen tumor tissues as previously described (14). Tumors originated from experiments described by Hallberg et al. (14). Total RNA was isolated from frozen tumor tissues or cell lines by RNA Isolator (Genosys Biotechnologies, Inc.) following the manufacturer's recommendations.
For Northern analysis 25 μg of total RNA was separated on a 1.2% agarose–formaldehyde gel, transferred to nylon membranes (Zeta-Probe; Bio-Rad) by alkaline (50 mM NaOH) blotting, and hybridized in 0.5 M Na2HPO4 (pH 7.2)–7% sodium dodecyl sulfate buffer with 32P-labeled (randomly primed DNA labeling) DNA probes. The multiple-tissue Northern and the mouse embryo Northern filters that were purchased from Clontech Laboratories, Inc., contain approximately 2 μg of poly(A)+ RNA per lane. Hybridization procedures were as recommended by the manufacturer. The following probes were used: probe A, a genomic preintegration PCR probe (35); probe B, a genomic 900-bp PstI fragment; probe C, a 700-bp EcoRI cDNA fragment; human β-actin, an internal control probe.
RNase protection assay.
The assays were performed with the RPA III (RNase protection assay) kit from Ambion Inc. In essence, the supplied instruction manual was followed; 5 μg of tumor RNA and 105 cpm of RNA probe were used. The gel-purified radiolabeled RNA probes were made by in vitro transcription from T7 promoters according to standard protocols. The Sint1 probe and the control mouse β-actin probe were transcribed from gel purified-PCR products using the following primers: Sint1 probes 5′-GCA GTA AGC TTC CCC GAA TTC AAG GGA TCC ATT TAG GTG ACA CTA TAG AAC CCT GGC TGA CAA CCC TAG AGA TGC CAT-3′ (SP6) and 5′-TAC AGA AGC TTT ACA GAA TTC CAG GGA TCC TAA TAC GAC TCA CTA TAG GCG TAA CTG TCA GCT TCA TTC GAA CCC CCT-3′ (T7); mouse β-actin probes 5′-GCA GTA AGC TTC CCC GAA TTC AAG GGA TCC ATT TAG GTG ACA CTA TAG AAC CCG CCC TAG GCA CCA GGG TGT GAT GGT-3′ (SP6) and 5′-TAC AGA AGC TTT ACA GAA TTC CAG GGA TCC TAA TAC GAC TCA CTA TAG GCG TAT CGG TGA GCA GCA CAG GGT GCT CCT-3′ (T7). The underlined parts of the primers correspond to SP6 or T7 promoters, with the boldface letter denoting the transcriptional start site, while the italicized parts correspond to the hybridizing sequences of the primers. The expected sizes of the unprotected and RNase-protected RNA probes are as follows: Sint1 probes, 378 and 325 bp, respectively; mouse β-actin probes, 263 and 210 bp, respectively. RNase-resistant products were analyzed by electrophoresis and autoradiography, and quantification of radioactive fragments was performed on the PhosphorImager SF (Molecular Dynamics, Inc.). Primers were from DNA Technology A/S, Aarhus, Denmark.
RT-PCRs.
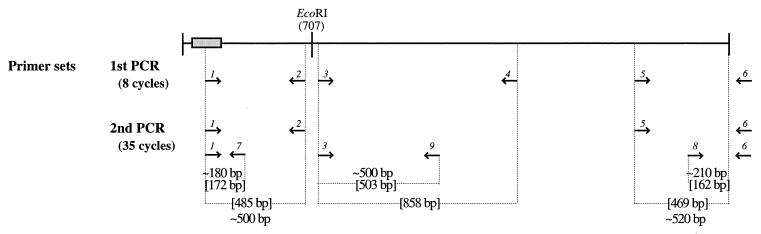
Reverse transcription-PCRs (RT-PCRs) were carried out using primer 10 (see below) for the first-strand cDNA synthesis (first-strand cDNA synthesis kit; Amersham Pharmacia Biotech) on approximately 2 to 4 μg of total RNA from tumors or spleen. PCR amplifications of the cDNAs were performed in two rounds. In the initial PCRs of eight cycles (first PCR; Fig. 1), 5 μl out of 15 μl of first-strand cDNA synthesis mixture was used as the template and the following primer sets were employed: 1 plus 2, 3 plus 4, and 5 plus 6 (Fig. 1). In the nested PCR (second PCR; Fig. 1) of 35 cycles, 2 μl out of 50 μl of the first PCR mixture was used as the template and the appropriate primer sets (1 plus 2, 1 plus 7, 3 plus 9, 5 plus 6, and 8 plus 6) were employed. For analysis, 10 μl out of 50 μl from the second PCR mixture was electrophoresed on an ethidium bromide-stained 2% agarose gel. The sequences of the primers were as follows: primer 1, 5′-TTGCAGCCAGCAGAGCCCACTTAAACT-3′; primer 2, 5′-TTCCTCCTGCAGGTACTTCTCATAT-3′; primer 3, 5′-CAGCGGATCACTGCAGACCTGCTGT-3′; primer 4, 5′-GGGCAGGCAGCTGAGGGCGCTGT-3′; primer 5, 5′-GCTAGCTTTCTGCAGCCCAGAAGT-3′; primer 6 (nested primer 10), 5′-GGGTCTAGAGCTCGAGTCACT-3′; primer 7, 5′-CTGCGCATCTGCTCCAGGATGGAGT-3′; primer 8, 5′-CTGTTCGTCTCACCAGGCCGGTCCACGT-3′; primer 9, 5′-TGGGAAGAGATGGATGGAGGCAGGT-3′; primer 10 (poly[T] primer), 5′-GGGTCTAGAGCTCGAGTCACTTTTTTTTTTTTTTTTV-3′ (V = A, G, or C). All primers were purchased from DNA Technology A/S).
FIG. 1.
Primer positions for RT-PCR analyses. The 3.0-kb Sint1 cDNA is indicated as a line with the identified exon included as a shaded box. Arrows indicate positions and numbers of primers. The expected (brackets) and observed sizes of the PCR fragments are indicated. The PCR analyses were performed in two rounds (see Materials and Methods), and the products from the second PCR were analyzed by gel electrophoresis (not shown).
Library screening.
The genomic library was made of partially MboI-digested mouse genomic (ES SV129 Dg) DNA inserted into BamHI-digested λGEM12 vectors. Hybridizations with the preintegration PCR probe A and subsequent purifications of positive clones were done by standard techniques (30). Two positive clones were obtained. The cDNA library was a mouse embryonic cDNA library in a λZAP/EcoRI vector (Stratagene). Screening with probe B and in vivo excision (according to a protocol from manufacturer) resulted in one positive clone.
Sequence analysis.
Sequencing reactions were performed with a Thermo Sequenase II dye terminator cycle sequencing kit (Amersham Pharmacia Biotech), and reaction products were analyzed on an automated DNA sequencer (373A DNA sequencer; Applied Biosystems Inc.). Nucleotide sequences were compared with sequences in the GenBank, EMBL, and EST databases by using Wisconsin package EGCG [version 8.1.0(a)] FASTA and BESTFIT programs.
Interspecific mouse backcross mapping.
Interspecific backcross progeny were generated by mating (C57BL/6J × Mus spretus) F1 females and C57BL/6J males as described previously (6). A total of 205 N2 mice were used to map the Sint1 locus (see below for details). DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, Southern blot transfer, and hybridization were performed essentially as described previously (16). All blots were prepared with Hybond-N+ nylon membrane (Amersham). The probe, a 392-bp fragment of mouse genomic DNA (probe A; Fig. 2) was labeled with [α-32P] dCTP using a randomly primed labeling kit (Stratagene); washing was done to a final stringency of 0.5× SSCP (0.06 M NaCl, 7.5 mM sodium citrate, 10 mM sodium phosphate)–0.1% sodium dodecyl sulfate at 65°C. A fragment of 4.7 kb was detected in SphI-digested C57BL/6J DNA, and a fragment of 3.8 kb was detected in SphI-digested M. spretus DNA. The presence or absence of the 3.8-kb SphI M. spretus-specific fragment in backcross mice was monitored.
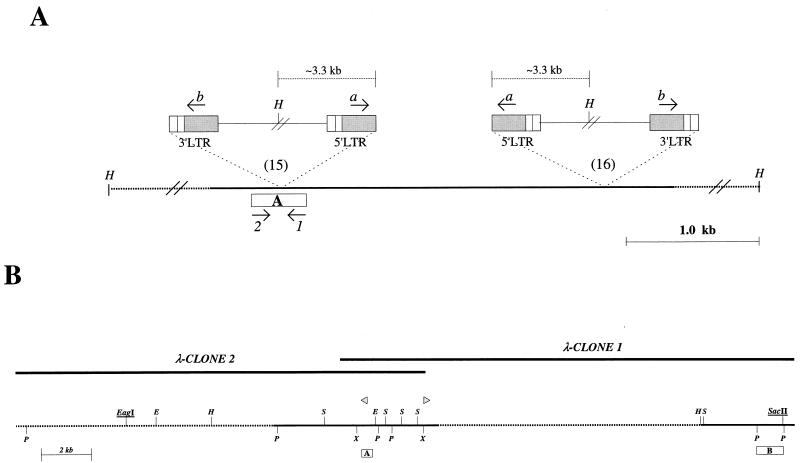
FIG. 2.
(A) Relative locations and orientations of the integrated proviruses in two independent tumors (15 and 16). Box, preintegration site PCR probe A; arrows 1 and 2, primers used to amplify probe A; arrows a and b, provirus-specific primers; H, HindIII restriction site. (B) Restriction map of the cloned Sint1 locus. Upper lines indicate the two overlapping genomic λ-phage clones, while the bottom line shows the resulting map. SacI (S), XhoI (X), PstI (P), HindIII (H), and EcoRI (E) sites are indicated. Underlined are the CpG indicator enzymes (see text). Dotted lines, regions only partly analyzed by restriction enzyme mapping and sequencing; boxes A and B, hybridization probes; triangles, provirus integration sites.
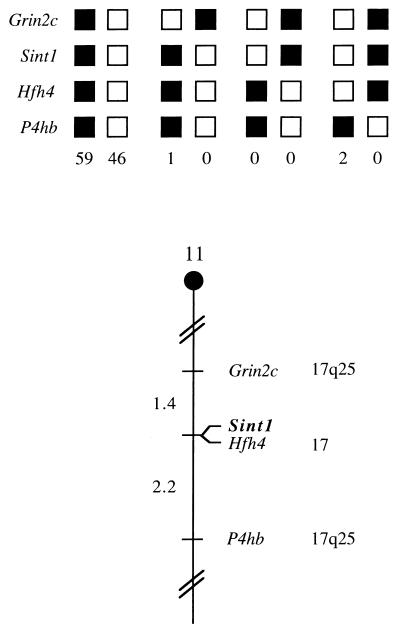
A description of the probes and restriction fragment length polymorphisms (RFLPs) for the loci linked to Sint1, including Grin2c, Hfh4, and P4hb, has been reported previously (12). Recombination distances were calculated using Map Manager, version 2.6.5. Gene order was determined by minimizing the number of recombination events required to explain the allele distribution patterns.
Nucleotide sequence accession numbers.
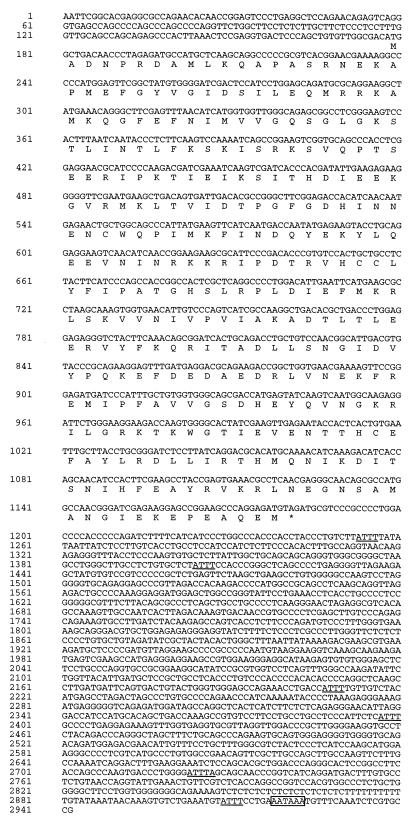
The nucleotide sequences of the Sint1 cDNA (Fig. 6) and the genomic subclone shown in Fig. 5 have been assigned EMBL data bank accession no. AJ250723 and AJ250724, respectively.
FIG. 6.
Sint1 cDNA and predicted amino acid sequences. In the 3′-UTR, the AT degradation motifs are underlined and the polyadenylation site consensus sequence is boxed.
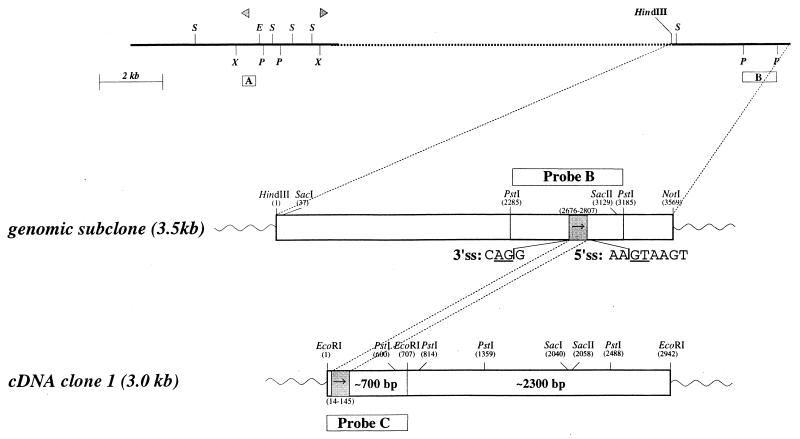
FIG. 5.
Correlation between Sint1 genomic region and Sint1 cDNA. (Top) Part of the genomic region; (middle) genomic 3.5-kb HindIII/NotI subclone; (bottom) corresponding cDNA clone. Triangles, provirus integration sites. Restriction enzyme site abbreviations are as defined for Fig. 1. Hybridization probes are shown as boxes A, B, and C. The grey box containing an arrow indicates the location and orientation of the exon in the genomic clone as well as in the cDNA clone. The 5′ and 3′ splicing signal (ss) sequences are shown with the conserved intron dinucleotides underlined.
RESULTS
Two proviruses inserted into a common region.
Previously, in order to identify proto-oncogenes involved in SL3-3 MLV-induced T-cell lymphomagenesis, we have amplified by PCR and analyzed provirus integration sites in tumor DNAs (34, 35). In brief, from one such integration site of which the flanking sequence showed no homology to any known sequences in available databases, a unique PCR probe of 392 bp was generated (probe A; Fig. 2A). This probe was hybridized to Southern blots containing HindIII-digested DNA from 20 SL3-3-induced lymphomas, and thus the clonality of the original integration was verified. Moreover, the hybridization revealed a rearranged fragment in the same region of DNA in an independent tumor (35). That this rearrangement was also due to an integrated provirus was confirmed by combinatorial PCRs using provirus-specific primers a and b together with primers 1 and 2 (primer localizations are indicated in Fig. 2A). A PCR fragment of about 3.0 kb was amplified by primer 2 and primer a. Taken together, these results demonstrated that a provirus had integrated into a common DNA region in two independent tumors, with the integration sites being separated by approximately 2.5 kb and the orientations of the integrated proviruses being opposite relative to each other (Fig. 2A). The DNA region was named SL3-3 integration site 1 (Sint1).
To obtain clones that covered the Sint1 region, a genomic λ-phage library was screened with probe A. Two overlapping clones covering about 35 kb were obtained, and a physical map of Sint1 was constructed (Fig. 2B).
Chromosomal mapping of Sint1.
The integration site sequences did not reveal any open reading frames (ORFs), and the integration site probe (probe A; Fig. 2) did not hybridize to RNA from a murine T-cell line (data not shown), suggesting that the proviruses might have inserted outside an expressed region. Accordingly, the Sint1 region could contain already-known proto-oncogenes. In order to test this, the chromosomal location of Sint1 was determined by interspecific backcross analysis using progeny derived from matings of ([C57BL/6J × M. spretus]F1 × C57BL/6J) mice. This interspecific backcross mapping panel has been typed for over 2,900 loci that are well distributed among all the autosomes as well as the X chromosome (6). C57BL/6J and M. spretus DNAs were digested with several enzymes and analyzed by Southern blot hybridization for informative RFLPs using probe A (Fig. 2). The 3.8-kb SphI M. spretus RFLP (see Materials and Methods) was used to monitor the segregation of the Sint1 locus in backcross mice. The mapping results indicated that Sint1 is located in the distal region of mouse chromosome 11 linked to Grin2c, Hfh4, and P4hh. Although 108 mice were analyzed for every marker and are shown in the segregation analysis (Fig. 3), up to 147 mice were typed for some pairs of markers. Each locus was analyzed in pairwise combinations for recombination frequencies using the additional data. The ratios of the total number of mice exhibiting recombinant chromosomes to the total number of mice analyzed for each pair of loci and the most likely gene order are as follows: centromere–Grin2c–(2 of 147)–Sint1–(0 of 128)–Hfh4–(3 of 136)–P4hb. The recombination frequencies (expressed as genetic distances in centimorgans [cM] ± the standard error) were as follows: centromere–Grin2c–(1.4 ± 1.0, cM)–(Sint1, Hfh4)–(2.2 ± 1.3 cM)–P4hb. No recombinants were detected between Sint1 and Hfh4 in 128 animals typed in common, suggesting that the two loci are within 2.3 cM of each other (upper 95% confidence limit). This same region is known to include another retroviral integration site, Pad3, identified as an MMTV integration site in DNA from a T-cell tumor (28).
FIG. 3.
Sint1 maps in the distal region of mouse chromosome 11. Sint1 was placed on mouse chromosome 11 by interspecific backcross analysis. The segregation patterns of Sint1 and flanking genes in 108 backcross animals that were typed for all loci are shown at the top. For individual pairs of loci, more than 108 animals were typed (see text). Each column represents the chromosome identified in the backcross progeny that was inherited from the (C57BL/6J × M. spretus)F1 parent. Black, boxes, presence of a C57BL/6J allele; white boxes, presence of an M. spretus allele. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. A partial chromosome 11 linkage map showing the location of Sint1 in relation to linked genes is shown at the bottom. Recombination distances between loci in centimorgans are shown to the left of the chromosome, and the positions of loci in human chromosomes, where known, are shown to the right. References for the human map positions of loci cited in this study can be obtained from the Genome Data Base, a computerized database of human linkage information maintained by the William H. Welch Medical Library of Johns Hopkins University (Baltimore, Md.).
The distal region of mouse chromosome 11 shares a region of homology with human chromosome 17q25 (summarized in Fig. 3), suggesting that the human homolog of Sint1 will map to 17q25 as well.
Identification of a Sint1-linked cDNA.
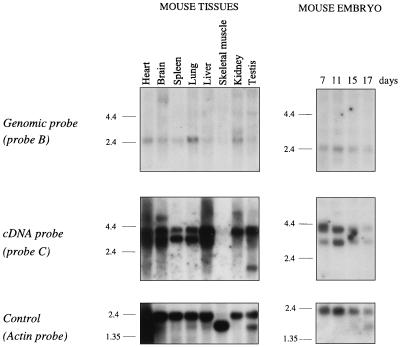
To identify possible genes localized in the Sint1 regions, the genomic λ clones were digested with the rare-cutting enzymes SacII (CCGC↓GG), EagI (C↓GGCCG), and BssHII (G↓CGCGC). A cluster of sites for these enzymes would indicate the presence of CpG islands, which are associated with the promoter regions of many genes (2, 20). The restriction endonuclease digestions of the λ clones revealed no such cluster. However, both SacII and EagI cut once in the Sint1 region as indicated in Fig. 2B. Since the SacII cutting site is located very close to the end of genomic λ clone 1, it seemed possible that this site was part of a cluster of SacII/EagI/BssHII cutting sites, and thus the 3.5-kb NotI/HindIII subclone of λ clone 1 became the starting point for a search for Sint1 genes. From this subclone, a PstI fragment (probe B; Fig. 2B) was isolated and used as a probe in Northern blot analyses with mRNA from different mouse tissues and from mouse embryos (Fig. 4). Although weakly expressed, a predominant transcript of about 2.4 kb was seen in all examined tissues except skeletal muscle. Probe B was then employed in an embryo cDNA library screen by which one cDNA clone of about 3.0 kb was obtained.
FIG. 4.
Northern blot analyses of Sint1 expression. All three panels are identical Northern blots hybridized with the probes indicated at the left. The blots contain about 2 μg of poly(A)+ RNA per lane from eight different mouse tissues (left) and from mouse embryos at different time points (right). Size markers (in kilobases) are at the left of each autoradiogram.
The sequences of the genomic 3.5-kb subclone as well as those of the cDNA clone were determined, and it was found that an exon of 132 bp with conserved 5′ and 3′ splicing signals is located in the genomic PstI fragment (Fig. 5) and that the sites of the provirus integrations are located about 15 kb 5′ of this exon. The integrity and the 3′ end of the cDNA clone were verified by RT-PCRs employing primers located both inside and outside the identified exon as shown in Fig. 1, and the obtained PCR fragments all showed the expected sizes (data not shown).
To address the apparent discrepancy between the transcript size observed in the Northern blot analysis (∼2.4 kb) and the size of the cDNA clone (∼3.0 kb), a cDNA EcoRI fragment of a 700-bp (probe C; Fig. 5) was used as a probe on the same Northern blot (Fig. 4). This time two major transcripts of about 3.0 and 4.0 kb were seen in all examined tissues except skeletal muscle. However, in some tissues such as spleen and lung, the predominant transcript was the 3.0-kb species, whereas in other tissues such as kidney and testis, only the 4.0-kb transcript seemed to be expressed. The highest expression was seen in liver, where both mRNA species were found, although the 4.0-kb transcript was expressed at much higher levels. In addition, some minor transcripts were seen, e.g., a transcript of about 5.5 kb in brain and a transcript of about 1.4 kb in testis. Likewise, Northern blot analyses of RNA from different lymphoid cell lines with probe C showed different transcript sizes with the 3.0- and 4.0-kb transcripts as the predominant ones (data not shown). Thus, it appears that the overall RNA expression pattern of this gene is complex.
Sint1 encodes a septin-like protein.
The sequence of the cDNA clone revealed a 5′-untranslated region (UTR) of at least 177 bp, an ORF of 1,002 bp, and a rather long 3′-UTR of 1,765 bp (Fig. 6). The 3′-UTR contains several copies of the ATTT(A) motif, which is thought to confer instability to mRNAs (10, 32), and a consensus poly(A) signal (AATAAA). The ORF is predicted to encode a polypeptide of 334 amino acid (aa) residues.
A comparison of the predicted amino acid sequence encoded by the Sint1 cDNA clone and sequences in the GenBank, EMBL, Swissprot, and EST databases showed a significant identity (97.0%) to a human brain cDNA-encoded protein and the human MSF (MLL septin-like fusion) protein (27) (Fig. 7). The comparison analyses in addition revealed significant homologies (>70% identity) to proteins of the septin family, a family of proteins which were originally identified by analysis of budding yeast cell division cycle mutants defective in cytokinesis (5, 22) and which were later isolated from flies, mice, and humans (19, 25, 26). The amino acid sequences of these proteins contain motifs that define the GTPase superfamily (3), although the function of nucleotide hydrolysis has not been determined. As indicated in Fig. 7, the GTP-binding motifs are also conserved in the Sint1 cDNA-encoded protein.
FIG. 7.
Comparison of the predicted amino acid sequences encoded by Sint1 cDNA and a human brain cDNA (KIAA0991; accession no., AB023208) or the human MSF cDNA (accession no., AF123052). The KIAA0991 and MSF sequences are 100% identical in the region shown. Boxed are the GTP-binding motifs. The sequences show 97.0% identity and 97.9% similarity (it should be noted, however, that the KIAA0991 and MSF sequences are 88 and 234 aa residues longer, respectively, than the Sint1 sequence). Alignment was performed with the Genetics Computer Group program BESTFIT. Vertical lines, identical residues; colons, well-conserved replacements that scored better than 0.5 in the PAM-250 matrix; dots, replacements scoring better than 0.1. The lengths of the Sint1, KIAA0991, and MSF sequences are 334, 422, and 568 aa, respectively.
Sint1 cDNA expression in tumors with proviral insertions at the Sint1 locus.
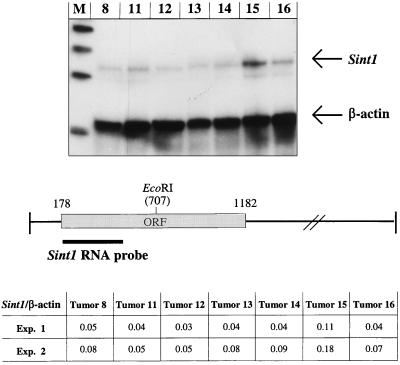
In order to examine if provirus integrations at the Sint1 locus affected the expression of the Sint1 cDNA, an RNase protection assay was performed with RNAs from seven different SL3-3-induced tumors, including the two tumors (tumor no. 15 and 16) that harbor a clonal provirus insertion at the Sint1 locus (Fig. 2). The RNA probe employed spans 325 bp of the coding region of the Sint1 cDNA (Fig. 8). When the intensities of the protected Sint1 fragments were correlated with the intensities of the protected internal control fragments (mouse β-actin), a clear upregulation (two- to threefold) of the Sint1 cDNA expression in tumor 15 was observed (Fig. 8). This indicates that a provirus integration at the Sint1 locus may have modified the expression of the linked Sint1 gene, the effect of which might have contributed to the development of the tumor. On the other hand, no significant alteration of the expression level of the Sint1 gene could be detected in tumor 16, which may suggest a more complex mode of involvement of the Sint1 gene in tumor progression.
FIG. 8.
RNase protection assay. (Top) RNase protection of RNA from seven different SL3-3 MLV-induced tumors assayed with Sint1 and mouse β-actin RNA probes. Lane M, molecular DNA size markers (200, 300, 400, and 500 bp). Arrows indicate the protected fragments, the sizes of which are 325 bp for Sint1 and 210 bp for mouse β-actin. (Bottom) Localization of the Sint1 RNA probe as well as the Sint1/β-actin values after quantification of radioactive protected fragments in two experiments (experiment 2 is the one shown at the top).
DISCUSSION
In order to identify genes involved in T-cell lymphomagenesis, we have analyzed provirus integration sites in SL3-3-induced T-cell tumors. We have identified a region of the mouse genome which has been rearranged by a retrovirus insertion in 2 of 20 tumors. Hence, this locus, which we have named Sint1, appears to define a region that is important in the development of T-cell lymphomas. This is further supported by the findings of Rajan and Dudley (28), who identified an integration site, named Pad3, in an MMTV-induced T-cell lymphoma. Pad3 cosegregates with the marker Hfh4 (HNF-3/forkhead homolog 4) as does Sint1 (Fig. 3); hence Sint1 and Pad3 may constitute the same locus.
A genomic fragment was used as a probe to isolate a linked cDNA clone of 3 kb. Enhanced levels of expression of this cDNA were detected in one of the two tumors carrying a proviral insertion in the Sint1 locus, strongly indicating that in this case overexpression of the Sint1 gene, probably due to the integrated provirus, did play a role in tumor initiation and/or progression. Regarding the other tumor harboring a provirus in the Sint1 locus, the picture is less clear, since no change of expression was detected. As the normal expression pattern of the Sint1 gene during differentiation is not known, there are several possibilities concerning how and when the Sint1 gene may be affected by the integrated provirus. For example, the distinction between regulated and deregulated expression levels may at certain stages be subtle, and a disturbance by an integrated provirus may have profound effects on differentiation and/or growth but may not necessarily have measurable effects on the expression level in the end-stage tumor.
The distance of approximately 15 kb between the integrated proviruses and the identified Sint1 exon and the fact that the two proviruses have integrated in opposite directions relative to each other indicate that the mechanism of activation is of the type referred to as enhancer insertion (29, 38). In addition, preliminary Northern blot analyses show no detectable differences in transcript sizes in RNAs from tumors harboring proviruses inserted at the Sint1 locus and from other tumors (data not shown). However, since the integrity of the Sint1 cDNA 5′ end has not been verified, the possibility still exists that the proviruses have integrated in an intron of the Sint1 gene and thereby, at least for tumor 16, which harbors a provirus inserted in the same transcriptional orientation as that of the Sint1 gene, the activation could have been 3′-LTR or 5′-LTR promotion in which a 5′ exon(s) might have been removed from the resulting transcript.
The putative Sint1 cDNA-encoded septin-like protein of 334 aa revealed a striking homology (97.0% identity; Fig. 7) to the encoded protein (568 aa) of MSF, a fusion partner gene of MLL in an acute myeloid leukemia with a t(11;17)(q23;q25) (27), further strengthening the assumption that Sint1 is a cancer-related gene. The same high degree of homology to a human brain cDNA (KIAA0991)-encoded protein (466 aa) is seen. As mentioned, the integrity of the 5′ end of the Sint1 cDNA has not been verified, and therefore we cannot exclude the possibility that the full-length Sint1 cDNA might be several hundred base pairs longer, which again might result in a longer ORF. In that case, however, the extended region would show a homology of less than 50% to the MSF (or KIAA0991) amino acid sequence. Despite this uncertainty, Sint1 and MSF seem to represent the same gene in mice and humans, respectively. First, the chromosomal locations match each other. Sint1 maps to mouse chromosome 11 in an area of human chromosome 17q25 homology (Fig. 3), which corresponds to the location of the MSF gene. Second, results of the Northern blot analyses of Sint1 and MSF in normal tissues are rather similar (Fig. 4) (27), showing two predominant transcripts of 4.0 and 3.0 kb, the 4.0 kb species being more ubiquitously expressed. In addition, a 1.7-kb transcript is seen in many human tissues, with a relatively high level of expression in heart, liver, skeletal muscle, and kidney (27). Finally, some minor transcripts can be detected in tissues from both organisms. The most prominent difference in expression pattern concerns the expression in skeletal muscle; in mice almost no Sint1 transcripts are seen; in contrast, in humans skeletal muscle does not seem to be different from other human tissues regarding MSF expression. With respect to the KIAA0991 gene, the nucleotide sequence is 100% identical to the MSF sequence in a long continuous stretch of about 2 kb, suggesting that these two sequences share several exons. Altogether, these observations make us incline to the hypothesis that Sint1 represents the mouse homolog of MSF and that MSF and KIAA0991 represent splice variants of the human gene. Due to the similarity of expression pattern, it seems reasonable to assume that the Sint1 cDNA clone characterized here thus represents one of the splice variants in mice.
The genes of the septin family are considered to be involved in cytokinesis and are characterized by a conservation of sequence motifs defining the GTPase superfamily (3, 11). Speculations about the mechanism of leukemogenesis involving members of this family might thus include disturbance of the interactions with the cytoskeletal filaments. GTP binding and hydrolysis may regulate these interactions and/or interactions with other cell cycle proteins.
In summary, we have by a PCR screening of SL3-3-induced tumors identified a common integration site, Sint1, and cloned a linked septin-like gene which seems to be affected by the integrated proviruses. The assumption of a role for the Sint1 gene in leukemogenesis is strongly supported by a recent report of a human homolog (MSF) directly involved in a human acute myeloid leukemia as a fusion partner at a translocation breakpoint (27). However, since only one tumor showed an increased level of Sint1 RNA, we must leave open the possibility of an effect of the proviral integrations on another still not identified gene(s) at the Sint1 locus.
ACKNOWLEDGMENTS
We thank Peder Lisby Nørby and Jesper Laursen for helpful technical advice and discussions. Likewise, the technical assistance of Lone Højgaard and Debra J. Gilbert is gratefully acknowledged.
This project was supported by the Danish Cancer Society, the Karen Elise Jensen Foundation, the Danish Natural Sciences and Medical Research Councils, the Danish Biotechnology Programme, DNA Technology A/S, and by the National Cancer Institute, DHHS, under contract with ABL.
REFERENCES
- 1.Amtoft H W, Sørensen A B, Bareil C, Schmidt J, Luz A, Pedersen F S. Stability of AML1 (core) site enhancer mutations in T lymphomas induced by attenuated SL3-3 murine leukemia virus mutants. J Virol. 1997;71:5080–5087. doi: 10.1128/jvi.71.7.5080-5087.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bird A P. CpG islands as gene markers in the vertebrate nucleus. Trends Genet. 1987;3:342–347. [Google Scholar]
- 3.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–126. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 4.Buchberg A M, Bedigian H G, Jenkins N A, Copeland N G. Evi-2, a common integration site involved in murine myeloid leukemogenesis. Mol Cell Biol. 1990;10:4658–4666. doi: 10.1128/mcb.10.9.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper J A, Kiehart D P. Septins may form a ubiquitous family of cytoskeletal filaments. J Cell Biol. 1996;134:1345–1348. doi: 10.1083/jcb.134.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Copeland N G, Jenkins N A. Development and applications of a molecular genetic linkage map of the mouse genome. Trends Genet. 1991;7:113–118. doi: 10.1016/0168-9525(91)90455-y. [DOI] [PubMed] [Google Scholar]
- 7.Copeland N G, Jenkins N A, Gilbert D J, Eppig J T, Maltais L J, Miller J C, Dietrich W F, Weaver A, Lincoln S E, Steen R G, Stein L D, Nadeau J H, Lander E S. A genetic linkage map of the mouse: current applications and future prospects. Science. 1993;262:57–66. doi: 10.1126/science.8211130. [DOI] [PubMed] [Google Scholar]
- 8.Corcoran L M, Adams J M, Dunn A R, Cory S. Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell. 1984;37:113–122. doi: 10.1016/0092-8674(84)90306-4. [DOI] [PubMed] [Google Scholar]
- 9.Cuypers H T, Selten G, Quint W, Zijlstra M, Maandag E R, Boelens W, van Wezenbeek P, Melief C, Berns A. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984;37:141–150. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- 10.DeMaria C T, Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 11.Dever T E, Glynias M J, Merrick W C. GTP-binding domain: three consensus sequence elements with distinct spacing. Proc Natl Acad Sci USA. 1987;84:1814–1818. doi: 10.1073/pnas.84.7.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fletcher C F, Okano H J, Gilbert D J, Yang Y, Yang C, Copeland N G, Jenkins N A, Darnell R B. Mouse chromosomal locations of nine genes encoding homologs of human paraneoplastic neurologic disorder antigens. Genomics. 1997;45:313–319. doi: 10.1006/geno.1997.4925. [DOI] [PubMed] [Google Scholar]
- 13.Graham M, Adams J M, Cory S. Murine T lymphomas with retroviral inserts in the chromosomal 15 locus for plasmacytoma variant translocations. Nature. 1985;314:740–743. doi: 10.1038/314740a0. [DOI] [PubMed] [Google Scholar]
- 14.Hallberg B, Schmidt J, Luz A, Pedersen F S, Grundström T. SL3-3 enhancer factor 1 transcriptional activators are required for tumor formation by SL3-3 murine leukemia virus. J Virol. 1991;65:4177–4181. doi: 10.1128/jvi.65.8.4177-4181.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hays E F, Bristol G, McDougall S. Mechanisms of thymic lymphomagenesis by the retrovirus SL3-3. Cancer Res. 1990;50(Suppl.):5631s–5635s. [PubMed] [Google Scholar]
- 16.Jenkins N A, Copeland N G, Taylor B A, Lee B K. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J Virol. 1982;43:26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonkers J, Berns A. Retroviral insertional mutagenesis as a strategy to identify cancer genes. Biochim Biophys Acta. 1996;1287:29–57. doi: 10.1016/0304-419x(95)00020-g. [DOI] [PubMed] [Google Scholar]
- 18.Justice M J, Morse III H C, Jenkins N A, Copeland N G. Identification of Evi-3, a novel common site of retroviral integration in mouse AKXD B-cell lymphomas. J Virol. 1994;68:1293–1300. doi: 10.1128/jvi.68.3.1293-1300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S, Tomooka Y, Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem Biophys Res Commun. 1992;185:1155–1161. doi: 10.1016/0006-291x(92)91747-e. [DOI] [PubMed] [Google Scholar]
- 20.Larsen F, Gundersen G, Prydz H. Choice of enzymes for mapping based on CpG islands in the human genome. Genet Anal Tech Appl. 1992;9:80–85. doi: 10.1016/1050-3862(92)90002-m. [DOI] [PubMed] [Google Scholar]
- 21.Liao X, Buchberg A M, Jenkins N A, Copeland N G. Evi-5, a common site of retroviral integration in AKXD T-cell lymphomas, maps near Gfi-1 on mouse chromosome 5. J Virol. 1995;69:7132–7137. doi: 10.1128/jvi.69.11.7132-7137.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longtine M S, DeMarini D J, Valencik M L, Al-Awar O S, Fares H, De Virgilio C, Pringle J R. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- 23.Morishita K, Parker D S, Mucenski M L, Jenkins N A, Copeland N G, Ihle J N. Retroviral activation of a novel gene encoding a zinc finger protein in IL-3-dependent myeloid leukemia cell lines. Cell. 1988;54:831–840. doi: 10.1016/s0092-8674(88)91175-0. [DOI] [PubMed] [Google Scholar]
- 24.Morrison H L, Soni B, Lenz J. Long terminal repeat enhancer core sequences in proviruses adjacent to c-myc in T-cell lymphomas induced by a murine retrovirus. J Virol. 1995;69:446–455. doi: 10.1128/jvi.69.1.446-455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakatsuru S, Sudo K, Nakamura Y. Molecular cloning of a novel human cDNA homologous to CDC10 in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1994;202:82–87. doi: 10.1006/bbrc.1994.1896. [DOI] [PubMed] [Google Scholar]
- 26.Neufeld T P, Rubin G M. The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell. 1994;77:371–379. doi: 10.1016/0092-8674(94)90152-x. [DOI] [PubMed] [Google Scholar]
- 27.Osaka M, Rowley J D, Zeleznik-Le N J. MSF (MLL septin-like fusion), a fusion partner gene of MLL, in a therapy-related acute myeloid leukemia with a t(11;17)(q23;q25) Proc Natl Acad Sci USA. 1999;96:6428–6433. doi: 10.1073/pnas.96.11.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajan L, Dudley J P. An MMTV integration site maps near the distal end of mouse chromosome 11. Mamm Genome. 1997;8:295–296. doi: 10.1007/s003359900420. [DOI] [PubMed] [Google Scholar]
- 29.Rosenberg N, Jolicoeur P. Retroviral pathogenesis, 475–586. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Selten G, Cuypers H T, Berns A. Proviral activation of the putative oncogene Pim-1 in MuLV induced T-cell lymphomas. EMBO J. 1985;4:1793–1798. doi: 10.1002/j.1460-2075.1985.tb03852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 33.Shen-Ong G L C, Morse III H C, Potter M, Muchinsky F. Two modes of c-myb activation in virus-induced mouse myeloid tumors. Mol Cell Biol. 1986;6:380–392. doi: 10.1128/mcb.6.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sørensen A B, Duch M, Jørgensen P, Pedersen F S. Amplification and sequence analysis of DNA flanking integrated proviruses by a simple two-step polymerase chain reaction method. J Virol. 1993;67:7118–7124. doi: 10.1128/jvi.67.12.7118-7124.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sørensen A B, Duch M, Amtoft H W, Jørgensen P, Pedersen F S. Sequence tags of provirus integration sites in DNAs of tumors induced by the murine retrovirus SL3-3. J Virol. 1996;70:4063–4070. doi: 10.1128/jvi.70.6.4063-4070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steffen D. Proviruses are adjacent to c-myc in some murine leukemia virus-induced lymphomas. Proc Natl Acad Sci USA. 1984;81:2097–2101. doi: 10.1073/pnas.81.7.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsichlis P N, Shephard B M, Bear S E. Activation of the Mlvi-1/mis1/pvt-1 locus in Moloney murine leukemia virus-induced T-cell lymphomas. Proc Natl Acad Sci USA. 1989;86:5487–5491. doi: 10.1073/pnas.86.14.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Lohuizen M, Berns A. Tumorigenesis by slow-transforming retroviruses—an update. Biochim Biophys Acta. 1990;1032:213–235. doi: 10.1016/0304-419x(90)90005-l. [DOI] [PubMed] [Google Scholar]
- 39.Wolff L. Myb-induced transformation. Crit Rev Oncog. 1996;7:245–260. doi: 10.1615/critrevoncog.v7.i3-4.60. [DOI] [PubMed] [Google Scholar]