Abstract
Introduction
Morchella esculenta is a popular edible fungus with high economic and nutritional value. However, the rot disease caused by Lecanicillium aphanocladii, pose a serious threat to the quality and yield of M. esculenta. Biological control is one of the effective ways to control fungal diseases.
Methods and results
In this study, an effective endophytic B. subtilis A9 for the control of M. esculenta rot disease was screened, and its biocontrol mechanism was studied by transcriptome analysis. In total, 122 strains of endophytic bacteria from M. esculenta, of which the antagonistic effect of Bacillus subtilis A9 on L. aphanocladii G1 reached 72.2% in vitro tests. Biological characteristics and genomic features of B. subtilis A9 were analyzed, and key antibiotic gene clusters were detected. Scanning electron microscope (SEM) observation showed that B. subtilis A9 affected the mycelium and spores of L. aphanocladii G1. In field experiments, the biological control effect of B. subtilis A9 reached to 62.5%. Furthermore, the transcritome profiling provides evidence of B. subtilis A9 bicontrol at the molecular level. A total of 1,246 differentially expressed genes (DEGs) were identified between the treatment and control group. Gene Ontology (GO) enrichment analysis showed that a large number of DEGs were related to antioxidant activity related. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis showed that the main pathways were Nitrogen metabolism, Pentose Phosphate Pathway (PPP) and Mitogen-Activated Protein Kinases (MAPK) signal pathway. Among them, some important genes such as carbonic anhydrase CA (H6S33_007248), catalase CAT (H6S33_001409), tRNA dihydrouridine synthase DusB (H6S33_001297) and NAD(P)-binding protein NAD(P) BP (H6S33_000823) were found. Furthermore, B. subtilis A9 considerably enhanced the M. esculenta activity of Polyphenol oxidase (POD), Superoxide dismutase (SOD), Phenylal anineammonia lyase (PAL) and Catalase (CAT).
Conclusion
This study presents the innovative utilization of B. subtilis A9, for effectively controlling M. esculenta rot disease. This will lay a foundation for biological control in Morchella, which may lead to the improvement of new biocontrol agents for production.
Keywords: Morchella esculenta, fungal disease, transcriptome analysis, Lecanicillium aphanocladii, biological control
Graphical abstract
Flow chart of experimental procedures. (Created with MedPeer, www.medpeer.cn).
1. Introduction
As one of the most popular edible mushrooms, morel mushrooms are prized in many culinary applications for their rich aroma, savory taste, and crisp-tender flesh texture (Desjarden and Phillips, 1992; Du and Yang, 2021). Morel mushrooms belong to the Morchella genus of the Morchellaceae family of the Ascomycota. The prevalent cultivated species is Morchella sextelata, which has good commercial characteristics for fresh mushrooms. M. sextelata has recently been artificially cultivated in China and comprises >90% of the total area of Morchella cultivation (Deng et al., 2021; Yu et al., 2021). The fruiting bodies of morels are characterized by a unique conic head, with convex and concave folding on the surface (Tietel and Masaphy, 2018; Wu et al., 2021). M. esculenta contains all of the important nutrients, including carbohydrates, polyunsaturated fatty acids, and secondary metabolites, such as phenolic compounds. Its cultured mycelium is extensively used as a flavoring agent (Heleno et al., 2013). M. esculenta not only has a unique flavor and rich nutrition, but current scientific research has confirmed that it has antioxdant, anti-inflammatory, and immunostimulatory properties of medicinal value (Tietel and Masaphy, 2018; Sunil and Xu, 2022; Xu et al., 2022). Mushroom polysaccharides are used as anticancer drugs, including as treatments for skin and liver cancer. Morel polysaccharides also have possible uses for treatment of inflammatory diseases, and its lipopolysaccharides were shown to have potential to regulate the immune system by inhibiting the production of nitric oxide in macrophages. They are undeveloped sources of natural products and have great potential in the medicine and nutrition industries (Huang et al., 2011; Hu et al., 2012; Cai et al., 2018; Badshah et al., 2021). Therefore, the economic value of M. esculenta is very high, and market demand is constantly growing. However, the natural output of wild M. esculenta is limited, and the artificial cultivation technology of Morchella is not perfect. Thus, M. esculenta is in short supply (Liu W. et al., 2023).
Fungal diseases are important factors that seriously threaten the cultivation and growth of Morchella and are even more harmful than insect pests. Approximately 25% of the cultivation area has suffered annually from fungal diseases (He et al., 2017; Shi et al., 2022; Liu Z. et al., 2023). Currently, reported morel fungal diseases mainly include rot disease caused by Lecanicillium aphanocladii, Clonostachys rosea, and Aspergillus niger (Lv et al., 2022; Fu et al., 2023; Tu et al., 2024), white mold disease caused by Paecilomyces penicil (He et al., 2017), cobweb disease by Cladobotryum mycophilum (Liu Z. et al., 2023), and stipe spot disease by Cladosporium scabrellum (Zhang et al., 2024). Among these, L. aphanocladii is an important pathogen. Its characteristic symptom is a small white mold-like injury that first appears on the surface or top of the cap. The lesion then develops to surround the cap and gradually spreads to the stalk. Finally, white mold, which is a serious rot disease with a natural incidence of 30%, softens the fruiting body (Lv et al., 2022). This not only hinders the development of M. esculenta cultivation but also causes massive losses to farmers and agricultural companies (Yu et al., 2022). However, thus far, research on the cultivation of Morchella is mostly limited to the identification of pathogens, and methods of preventing and controlling diseases have rarely been studied or applied (Xu et al., 2022; Liu and Dong, 2023). Although the use of chemical control of disease has been studied for other edible fungi, the use of antibiotics is accompanied by increase risks of soil and water pollution, threats to food safety, damage to human health, and the development of antibiotic resistance in plant pathogens (Sánchez, 2004; Grogan, 2006; El-Baky and Amara, 2021; Gea et al., 2021; Li et al., 2023).
Thus, it is clearly necessary to develop new prevention and control strategies to eliminate or reduce the incidence of M. esculenta disease that are cost-effective and nontoxic. Biological control of plant pathogens refers to the use of beneficial microorganisms or microbial metabolites to effectively control plant diseases (Ahmed W. Q. et al., 2022; Liu Q. et al., 2022). Plant–microbe interactions play an important role in soil and plant health (Niu et al., 2020; Jia et al., 2021). The use of biocontrol microorganisms to control plant pathogens has the advantages of being green, effective, and sustainable, and it is an environmentally friendly method for the control of agricultural diseases, making it the focus of current research (Morin, 2020; Niu et al., 2020; Zhang et al., 2020). Some of the most intensively studied biological control agents are bacteria, which can use a variety of mechanisms to limit the development of plant diseases (Ahmed W. et al., 2022). Several bacterial-based products have been registered and sold as biological pesticides (Bonaterra et al., 2022). Bacillus, Pantoea, Streptomyces, Trichoderma, Clonostachys, Pseudomonas, and Burkholderia spp. have been widely studied as biocontrol agents reported, and some have been commercialized (Boro et al., 2022; Lahlali et al., 2022; Khan et al., 2023; Martinez et al., 2023). In some cases, biological control agents not only limit fungal diseases but also protect plants indirectly by triggering biochemical and molecular defense responses to a wide range of pathogens and by promoting plant growth (Kohl et al., 2019; Legein et al., 2020; Elnahal et al., 2022).
At present, bacteria have been utilized for biocontrol on many species of plants, and there have been some studies in edible fungi (Carrasco and Preston, 2020). Biocontrol bacteria are used in different hosts, and there have been some studies on biological control in edible fungi. For example, bioinoculants registered for mushrooms are reduced to the use of entomopathogenic nematodes and fungi as insecticides to fight mushroom flies, and two strains commercialized as B. subtilis QST 713 and B. amyloliquefaciens MBI 600 are registered as bioinoculants to control green mold disease in mushrooms (Carrasco and Preston, 2020). However, biocontrol of M. esculenta have not yet been reported.
Endophytic microorganisms are highly suited for biocontrol because of their stability advantage in host tissue colonization (Fontana et al., 2021). The first purpose of this study was to investigate the use of an endophytic strain of B. subtilis to alleviate rot disease. The physiological and biochemical characteristics and the whole genome of B. subtilis A9 were analyzed in order to understand the characteristics of the strain. Through the pot experiment, to explore the best way of use, and finally applied in the field to verify its biocontrol effect. Since the biocontrol mechanism of B. subtilis A9 is not clear, we further study the biocontrol mode of B. subtilis A9 through SEM observation and transcriptome analysis. Meanwhile, some evidence was provided from the point of view of transcriptome. This study has provided insight into the biocontrol mechanism of B. subtilis, and revealed its potential as a beneficial high-efficiency biocontrol agent.
2. Materials and methods
2.1. Morchella esculenta and pathogens
Morchella esculenta was sampled from a planting site in Jiaxing Zhejiang in March 2021. The whole fruiting body was collected, placed in a sterile bag, and brought back to the laboratory. The mycelium was isolated and cultured in potato dextrose agar (PDA; 200 g·L−1 potato extract, 20 g·L−1 glucose, and 15 g·L−1 agar) and grown at 28°C.
The Lecanicillium aphanocladii strains used for the present study were cultured in potato dextrose agar (PDA; 200 g·L−1 potato extract, 20 g·L−1 glucose, and 15 g·L−1 agar) and grown at 28°C (Lv et al., 2022).
2.2. Isolation and identification endophytic bacteria
Fresh M. esculenta was rinsed with sterile water for approximately 5 min. Excess water on the surface was absorbed with filter paper. Samples were washed twice with sterile water, soaked in 75% alcohol for 3 min, washed with sterile water 3 times, soaked in 3% sodium hypochlorite for 3 min, and finally washed with sterile water 4 times. The last cleaning solution was plated on medium as a control. The disinfected M. esculenta epidermis was peeled off with a sterile scalpel. To isolate the endophytic bacteria the tissue samples were ground using a mortar and pestle using an appropriate amount of disinfected sand and sterile water. After thorough grinding, the supernatant was smeared on NA and PDA medium plates and cultured in incubators at 37 and 28°C, and three repeats were set up. During culturing, the bacteria and fungi were observed every day, and novel isolates were purified using fresh solid medium. The purified endophytic bacteria, endophytic actinomycetes, and endophytic fungi of M. esculenta were transferred to NB slopes, Gaoshi No. 1 slopes, and PDA slopes, respectively, and stored in a refrigerator at 4°C (Shweta et al., 2013; Maliehe et al., 2020; Wang et al., 2020).
2.3. Antibiosis test
Antibiosis against L. aphanocladii was screened as follows. The endophytic strains were activated on NA plates, and for each one a single colony was used to inoculate NA medium and cultured at 28°C for 24 h. The L. aphanocladii G1 were activated on a PDA plate and cultured at 25°C for 7 days. The L. aphanocladii G1 and antagonistic bacteria were symmetrically spread at equal distances. A 5-mm plug of L. aphanocladii G1 was used to inoculate the center of the solid PDA medium, and the endophytic bacteria were used to inoculate the four corners of the plate center at a distance of 30 mm. Each bacterial strain was applied in a ring and cultured at 28°C for 10 days. The pathogen plate was used as the control to observe the growth of the colony on the plate. The inhibition rate Formula (1) (Liu Y. et al., 2022) of the plate confrontation test was as follows:
| (1) |
2.4. Screening and identification of antagonistic endophytes
First, the strain was morphologically identified. After the coated plate was cultured for 24 h, colony size, color, shape, edge, and transparency were observed by naked eye and recorded (Table 1).
Table 1.
List of abbreviations.
| Abbreviation | Full name |
|---|---|
| SEM | Scanning Electron Microscope |
| DEGs | Differentially Expressed Genes |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MAPK | Mitogen-Activated Protein Kinases |
| ABC | ATP-Binding Cassette |
| PPO | Polyphenol Oxidase |
| SOD | Superoxide Dismutase |
| PAL | Phenylalanine Ammonia Lyase |
| CAT | Catalase |
| BP | Biological Process |
| MF | Molecular Function |
| CC | Cellular Component |
| PPP | Pentose phosphate pathway |
| N | Nitrogen |
Bacillus subtilis A9 was then identified using molecular means. B. subtilis A9 was cultured until reaching the logarithmic growth phase. Genomic DNA was extracted using a bacterial DNA kit (Radix Scutellariae). The concentration of genomic DNA extracted from B. subtilis A9 was determined using the Epoch2 enzyme labeling instrument (BioTek, USA). The samples were then tested by Shenggong Bioengineering (Shanghai, China) for testing.
To further explore the physiological and biochemical properties indexes of B. subtilis A9, HBIG14 biochemical identification paper was used. Colonies were selected, used to inoculate a nutrient agar slope or plate, cultured at 36 ± 1°C for 24 h, and then transferred to a biochemical identification strip. If the reagent strip required a liquid test sample, a bacterial suspension method was used; if solid content was required, a zigzag line inoculation or puncture inoculation was used. After inoculation, the test strip was marked, covered, placed into the base, and cultured at 36 ± 1°C. A sealed test tube was used for anaerobic growth. After culture, the result was recorded.
2.5. Genome sequencing
After confirming this basic information, the entire B. subtilis A9 genome was sequenced. The NucleoBond® HMW DNA kit (MN NucleoBond, Germany, 740160.20) was used for high-quality genome extraction from samples. DNA concentration and purity was determined using Qubit4.0 (Thermo, Q33226) and Nanodrop (SMA4000, Taiwan, China). DNA integrity was assessed using 0.75% agarose gel electrophoresis. The gDNA was separated into two parts. One was randomly fragmented to build a library with an insertion size of 300 bp. The library was assembled on an Illumina NovaSeq 6000 platform with a paired-end 150 bp sequencing strategy. Another was performed through end-repair, 3′ adenylation, and adapter and motor protein ligations. The product was purified using Agencourt AMPure XP Beads (Beckman, A63881). Finally, fragments larger than 1 KB were screened using Single-molecule nanopore DNA sequencing on MinION Flow Cell (ONT, R9.4.1).
The raw reads were filtered and assembled using Canu with default parameters. The genomic sequences were proofread using nextpolish (v1.4.1) and Pilon (v1.18). Gene prediction and annotation were generated using Prokka (Version 1.10) and the National Center for Biotechnology Information (NCBI) nr database. The functional annotation was based on protein-coding genes using the Cluster of Orthologous Groups of Proteins (COG) (Tatusov et al., 2000) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2023) database. The Comprehensive Antibiotic Resistance Database1 was queried to predict virulence genes and antibiotic resistance genes.
The complete chromosome sequence of B. subtilis A9 was deposited in GenBank under accession number CP136257. The sequencing data were saved as FASTQ files and deposited to the National Center for Biotechnology Information (NCBI) under BioSample accession number SAMN37765015.
2.6. Scanning electron microscope (SEM) observations
To assess possible effects caused by the antagonistic bacteria on the pathogen mycelium, the mycelium was microscopically observed at the border of the growth zone in co-culture (TM4000 5 kV 11.5 mm X2.00k Mix M). Damage or hyphal changes caused by the antagonistic bacterial isolate were described by referring to the morphology of the pathogen grown in the absence of bacteria.
2.7. Pot and field assays
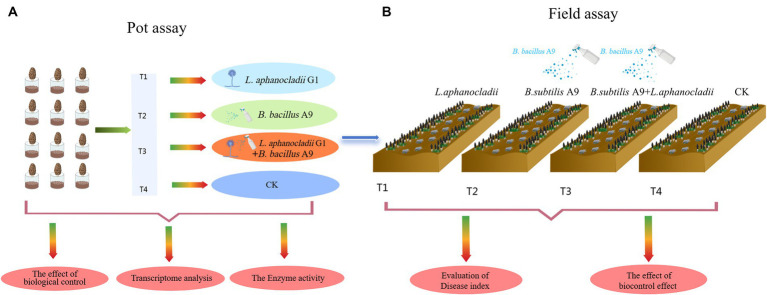
A pot assay was (Figure 1A) conducted in a growth chamber to assess the biocontrol efficacy of B. subtilis in reducing fungal load. The experiment was repeated three times and comprised four treatments (T1–T4):
Figure 1.
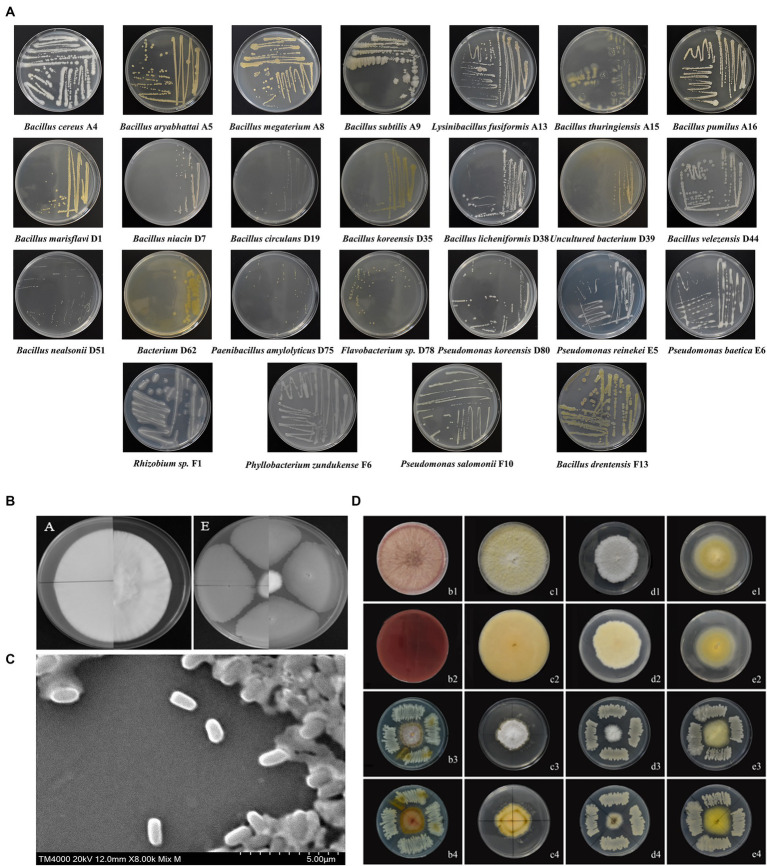
(A) Identity of endophytic bacteria. The morphological characteristics are shown, and 25 different bacterial species were identified. (B) Without inoculation of B. subtilis A9, Lecanicillium aphanocladii overgrew the PDA. In the presence of B. subtilis A9, the radial growth of L. aphanocladii was significantly inhibited, showing a clear inhibition zone between the fungal colony and the bacterial colony. (C) Observation of biocontrol bacteria B. subtilis A9 under SEM (TM4000 20 kV 12.0 mm X8.00k Mix M), a rod-shaped bacteria of 10 μ. (D) Four M. esculenta endophytes, showing stronger antagonistic activity in vitro against L. aphanocladii.
Treatment 1: M. esculenta inoculated with pathogen L. aphanocladii G1;
Treatment 2: 1 mL 2 × 109 CFU/mL B. bacillus A9 fermentation liquid applied to M. esculenta with pathogen L. aphanocladii G1 applied to M. esculenta the next day;
Treatment 3: 1 mL 2 × 109 CFU/mL B. bacillus A9 fermentation liquid applied to M. esculenta;
Treatment 4: noninoculated control.
After all experiments were performed, infected M. esculenta and soil were autoclaved before waste disposal to avoid the spread of infection.
To further study the biological control effect in the field, we also conducted field assay (Figure 1B) site at Dongzhuang Village, Liantang Town, Qingpu District, Shanghai, China. The soil of the test site was fertile, with measures for M. esculenta growth in place. Irrigation was convenient. The experiment adopted a random block distribution and was divided into four plots, the area of each block is 1 m × 20 m (Supplementary Figure S1). The following four groups were used for this experiment (T1–T4):
Treatment 1: M. esculenta inoculated with pathogen L. aphanocladii G1;
Treatment 2: 100 mL 2 × 109 CFU/mL B. bacillus A9 fermentation liquid applied to M. esculenta with pathogen L. aphanocladii G1 applied to M. esculenta the next day;
Treatment 3: 100 mL 2 × 109 CFU/mL B. bacillus A9 fermentation liquid applied to M. esculenta;
Treatment 4: noninoculated control.
After all experiments were performed, infected plants and soil were autoclaved before waste disposal to avoid the spread of infection.
Disease severity was recorded on a 0–5 scale, with 0 representing no infection and 5 denoting complete infection (Table 2). The 0–5 scale of disease severity was classified following Ma et al. (2019), Rokni and Goltapeh (2019), and Singh et al. (2021).
Table 2.
Grading standard of M. esculenta fungal disease.
| Disease level | Symptom description |
|---|---|
| 0 | No disease spots or other infection indications. |
| 1 | A few limited lesions: spots, less than 1 mm in diameter and less than 1% of total area. |
| 2 | Irregular spots dispersed: approximately 2 mm in diameter and 1–5% of total area. |
| 3 | Lesion extension: 3–4 mm in diameter, 6–20% of total area. |
| 4 | Expanded lesions: 4 mm or more in diameter, 21–50% of the total area. |
| 5 | Expanded lesions: accounting for more than 51% of the total area. |
The percentage of disease incidence was determined using the following Formula (2) (Chen et al., 2022):
| (2) |
Both the preventive and curative effects were calculated using the following Formula (3) (Chen et al., 2022):
| (3) |
2.8. Construction and sequencing of the transcriptome
Gene expression profiles were evaluated for Treatment 3 (application of only B. bacillus A9 to M. esculenta) and Treatment 4 (noninoculated control) in pot experiments. Three biological replicates for each sample were analyzed.
Total RNA was extracted from the whole M. esculenta fruiting body using Trizol Reagent (Invitrogen Life Technologies), according to the manufacturer’s protocols. Quality and integrity were determined using a NanoDrop spectrophotometer (Thermo Scientific). Three micrograms of RNA were used as input material for the RNA sample preparations. RNA libraries were constructed and size-selected using the NEBNext Ultra II RNA Library Prep Kit for Illumina (New England Biolabs Inc., Ipswich, MA, USA) and the AMPure XP system (Beckman Coulter, Beverly, CA, USA), respectively. The libraries were amplified by PCR (15 cycles) and sequenced on the NovaSeq 6,000 platform (Illumina) using Shanghai Personal Biotechnology Cp. Ltd.
Raw sequencing reads of Fastq files were filtered using Fastp (0.22.0), which masked bases with Q-scores <20 and the sequence of the removed 3′ end adapters. All subsequent analyses were high-quality analysis based on Cleandata. The reference genome and gene annotation files were downloaded from the genome website. The filtered reads were mapped to the reference genome using HISAT2 (v2.1.0). HTSeq (v0.9.1) was used to statistically compare the read count values for each gene as the original expression of the gene and to standardize the expression using FPKM. The differential expression of genes was then analyzed using DESeq (v1.38.3) with the following screening conditions: expression difference multiple |log2FoldChange| > 1 and significant p-value <0.05.
All genes were mapped to terms for the Gene Ontology (GO) database, and the number of differentially enriched genes in each term was calculated. Top GO (v2.50.0) was used to perform GO enrichment analysis on the differentially expressed genes (all DEGs/up DEGs/down DEGs), and the p-value was calculated using the hypergeometric distribution method (the standard of significant enrichment was p-value <0.05). The GO terms of significant DEGs were identified to determine their main biological functions. ClusterProfiler (v4.6.0) software was used to carry out enrichment analysis of the KEGG pathways of DEGs, focusing on significant enrichment pathways with a p-value <0.05. The Gene Set Enrichment Analysis (GSEA) (v4.1.0) tool was used for the GSEA enrichment analysis of all genes, and a GSEA enrichment analysis pathway map was drawn.
Based on the existing reference genome,2 StringTie (v2.2.1) software was used to assemble the mapped reads, and the splicing results were compared with the known transcripts to obtain annotations or transcripts of information.
2.9. Antioxidant enzymes analysis
To confirm the transcriptome results, M. esculenta samples (1.0 g) were tested. The phenylalanine ammonia-lyase (PAL), catalase (CAT), peroxidase (POD), and superoxide dismutase (SOD) activities of the parental lines were determined using assay kits as directed by the manufacturers (Suzhou Keming Biotechnology Co., Ltd.). Three biological repeats of each sample were measured.
2.10. Data treatment and statistical analysis
The diameters of the colonies were expressed as mean values ± standard deviation (SD). All assays were repeated in at least three separate experiments. All experimental data were expressed as mean ± SD. Analysis of variance (ANOVA) was completed using the Statistical Package for GraphPad Prism 8 (v8.0.1.244). One-way ANOVA followed by post hoc analysis was used to compare mean values among treatments at the 5% level of significance (p ≤ 0.05).
3. Results
3.1. Identification of 122 isolated of endophytic bacteria
A total of 122 endophytic bacteria colonies and 2 endophytic fungi colonies were isolated from M. esculenta and identified by 16SrDNA gene amplification. They are belonging to 25 bacteria and 2 fungi. The bacteria that were further studied were designated as A4, A5, A8, A9, A13, A15, A16, D1, D7, D19, D35, D38, D39, D44, D51, D62, D75, D78, D80, E5, E6, F1, F6, F10, and F13. For the endophytic bacteria, most of the colonies were orbicular, a few are irregular, most are white, and a few are yellow or light brown. The morphological characteristics of the bacteria are shown in Table 3, and details on their identification are shown in Figure 2A. The largest group among all the identified strains was Bacillus, with 15 strains, and the second largest group was Pseudomonas.
Table 3.
The morphological characteristics of endophytic bacteria.
| Strain code | Colony morphology | |||
|---|---|---|---|---|
| Form | Color | Edge condition | Surface morphology | |
| A4 | Orbicular | Milky white | Serrated | Dull, rough, opaque, waxy, flat |
| A5 | Orbicular | Yellow | Complete | Shiny, rough, opaque, easy to provoke, flat |
| A8 | Orbicular | Yellow | Complete | Shiny, rough, opaque, easy to provoke, flat |
| A9 | Irregular | Milky white | Serrated | Dull, rough, opaque, waxy, flat |
| A13 | Orbicular | Light brown | Complete | Dull, rough, opaque, waxy, flat |
| A15 | Irregular | Yellow | Complete | Shiny, smooth, opaque, sticky, convex |
| A16 | Orbicular | Milky white | Undulating | Shiny, smooth, opaque, easy to provoke, flat |
| D1 | Orbicular | Yellow | Complete | Shiny, smooth, opaque, waxy, flat |
| D7 | Orbicular | Light brown | Complete | Shiny, smooth, opaque, sticky, convex |
| D19 | Orbicular | White | Complete | Dull, smooth, translucent, easy to provoke, convex |
| D35 | Orbicular | Yellow | Complete | Dull, rough, opaque, waxy, flat |
| D38 | Irregular | Milky white | Serrated | Shiny, smooth, opaque, sticky, convex |
| D39 | Orbicular | Yellow | Complete | Shiny, smooth, opaque, easy to provoke, convex |
| D44 | Orbicular | White | Complete | Shiny, smooth, opaque, easy to provoke, convex |
| D51 | Orbicular | White | Complete | Shiny, smooth, opaque, easy to provoke, convex |
| D62 | Orbicular | Yellow | Complete | Shiny, smooth, opaque, sticky, convex |
| D75 | Orbicular | Light yellow | Complete | Shiny, smooth, opaque, easy to provoke, convex |
| D78 | Orbicular | Yellow | Complete | Shiny, smooth, opaque, easy to provoke, convex |
| D80 | Orbicular | White | Complete | Shiny, smooth, opaque, easy to provoke, convex |
| E5 | Orbicular | White | Complete | Shiny, smooth, opaque, sticky, convex |
| E6 | Orbicular | White | Complete | Shiny, smooth, opaque, sticky, convex |
| F1 | Irregular | Milky white | Complete | Shiny, smooth, translucent, sticky, convex |
| F6 | Orbicular | Milky white | Complete | Shiny, smooth, translucent, sticky, convex |
| F10 | Irregular | Light yellow | Serrated | Dull, rough, opaque, waxy, flat |
| F13 | Orbicular | Yellow | Complete | Dull, rough, opaque, waxy, flat |
122 strains of endophytic bacteria were identified as belonging to 25 species of bacteria, and the code of each specie of bacteria was listed in the table.
Figure 2.
A flow chart of the pot and field assay.
3.2. General biological characteristics and genomic features of Bacillus subtilis A9
Among the endophytic bacteria isolated from M. esculenta, nine showed stronger (IR ≥ 60%) antagonistic activity against L. aphanocladii by in vitro assays. Among the nine strains, B. subtilis A9 was most effective in suppressing the radial growth of L. aphanocladii hyphae with 72% IR (Figure 2B). B. subtilis A9 was deposited at the Chinese typical Culture Preservation Center (CCTCC), under the accession number CCTCCNO:M20221704. Additionally, as shown in Figure 2D, B. subtilis A9 also exhibited significant antagonistic activity against four other pathogenic fungi, which also cause destructive diseases of M. esculenta, indicating that B. subtilis A9 has broad-spectrum activity and great potential in the control of M. esculenta fungal diseases.
Bacillus subtilis A9 is a rod-shaped Gram-positive bacteria with a length of 10 μm. (Figure 2C), present singly or in pairs, with a central spore. On NA, B. subtilis A9 formed creamy white and surface rough colonies with irregular edges. Strain A9 was chemo-organotrophic, D-xylose, hydrolysis D-mannitol, gelatin, liquefied starch, nitrate reduction and L-arabinose were positive (Table 4).
Table 4.
Physicochemical properties of B. subtilis A9.
| Biochemical reaction | V-P test | Citrate | Propionate | D-xylose | D-xylose |
|---|---|---|---|---|---|
| Result | − | − | − | + | + |
| Biochemical reaction | HydrolysisD-mannitol | Gelatin | Liquefied starch | pH5.7 | Nitrate reduction |
| Result | + | + | + | − | + |
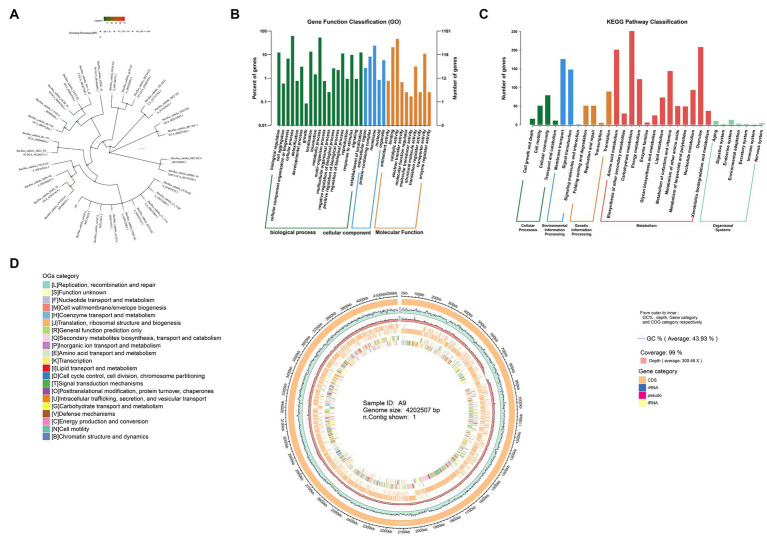
Bacillus subtilis is a widely studied biocontrol species, but thus far, no B. subtilis has been isolated from M. esculenta (NCBI data). Here, we describe the complete genome sequence and annotation. A phylogenetic tree was constructed using the FastTree program based on multiple sequence alignment and cutting using mafft software, which showed a distinct branch containing the A9 strain and Bacillus sp. (Figure 3A).
Figure 3.
Sequencing of the whole genome of B. subtilis A9. (A) Phylogenetic tree. (B) GO annotations distribute bar charts. The horizontal axis is the secondary classification of GO, and the vertical axis is the number of genes in the classification (right). Percentage indicates is the total number of annotated genes (left). Different colors represent different orthologs. (C) KEGG pathway classification bar chart. The horizontal axis is the name of the metabolic pathway involved, and the vertical axis is the number of genes annotated to that pathway. (D) Genomic circos map; displayed in the circle map are GC content, sequencing depth, gene element display, and COG function display, from outside to inside.
The total size of the genome was 4,202,507 bp with a GC content of 44%. A total of 114 RNAs, including 27 rRNAs and 87 tRNAs, were identified. The total number of genes (CDS) of the B. subtilis A9 strain was 4,075. Of these, 2,805 were assigned a COG number. The most abundant COG category was “General function prediction only” (350 proteins), followed by “Function unknown” (313 proteins), “Amino acid transport and metabolism” (279 proteins), “Transcription” (267 proteins), “Carbohydrate transport and metabolism” (237 proteins), and “Cell wall/membrane/envelope biogenesis” (194 proteins). Circos software was used to display the genome, including its GC content, sequencing depth, gene element content, and COG function display (Figure 3D). A total of 45 carbohydrate enzyme family genes were annotated. Among these, 17 glycoside hydrolase genes were found, accounting for 37.7% of the total, and 11 glycosyl transferases, accounting for 24.4%. B. subtilis A9 may act by producing glycoside hydrolase and glycosyl transferase (Supplementary Figure S2).
The predicted proteins sequences were compared with KEGG and other functional databases by BLASTX, and gene function analysis was annotated using Blast 2 GO software. Among them, there were 32,053 GO annotation genes, of which 3,785 were related to the cellular component (CC), 7,150 were related to molecular function (MF), and 21,118 were related to biological process (BP) (Figure 3B). KEGG annotated 2,458 genes, including 5 levels of signal pathways, with 164 genes related to Cellular Processes, 1,683 genes related to Metabolism, 330 genes related to Environmental Information Processing, 226 genes related to Genetic Information Processing, and 55 genes related to Organismal Systems (Figure 3C).
We then focused on genes with potential roles in antibiotic production. The secondary metabolites produced by B. subtilis A9 were predicted with the bacterial version of AntiSMASH,3 and 13 genes were detected in the genome of B. subtilis A9 (Table 5). Among the gene clusters related to secondary metabolite synthesis identified. Among them, the known antibiotic gene clusters with high similarity were clusters for production of fengycin, bacilaene, subtilin, bacillibactin, subtilosin and bacilysin, with a similarity of 100% (Supplementary Figure S3).
Table 5.
The secondary metabolites cluster by B. subtilis A9.
| Region | Type | From | To | Most similar known cluster | Similarity |
|---|---|---|---|---|---|
| Region 1 | NRPS | 360,996 | 424,430 | surfactin | 82% |
| Region 2 | lanthipeptide-class-i | 1,767,989 | 1,794,097 | – | 100% |
| Region 3 | transATPKS,PKS-like,T3PKS,NRPS | 1,816,364 | 1,921,603 | bacillaene | – |
| Region 4 | NRPS,betalactone | 1,994,208 | 2,071,170 | fengycin | 100% |
| Region 5 | terpene | 2,141,743 | 2,163,641 | – | – |
| Region 6 | T3PKS | 2,211,355 | 2,252,452 | – | – |
| Region 7 | NRPS | 3,233,422 | 3,280,558 | bacillibactin | 100% |
| Region 8 | lanthipeptide-class-i | 3,431,446 | 3,457,671 | Subtilin | 100% |
| Region 9 | CDPS | 3,586,386 | 3,607,132 | – | – |
| Region 10 | sactipeptide | 3,831,858 | 3,853,469 | subtilosin | 100% |
| Region 11 | other | 3,856,461 | 3,897,879 | bacilysin | 100% |
| Region 12 | RiPP-like | 4,086,497 | 4,099,228 | – | – |
| Region 13 | epipeptide | 4,114,416 | 4,136,114 | thailanstatin | 10% |
– indicates that there is no match to similar known cluster; − indicates that similarity <10%.
3.3. Inhibition of Bacillus subtilis A9 on mycelia and spores of Lecanicillium aphanocladii
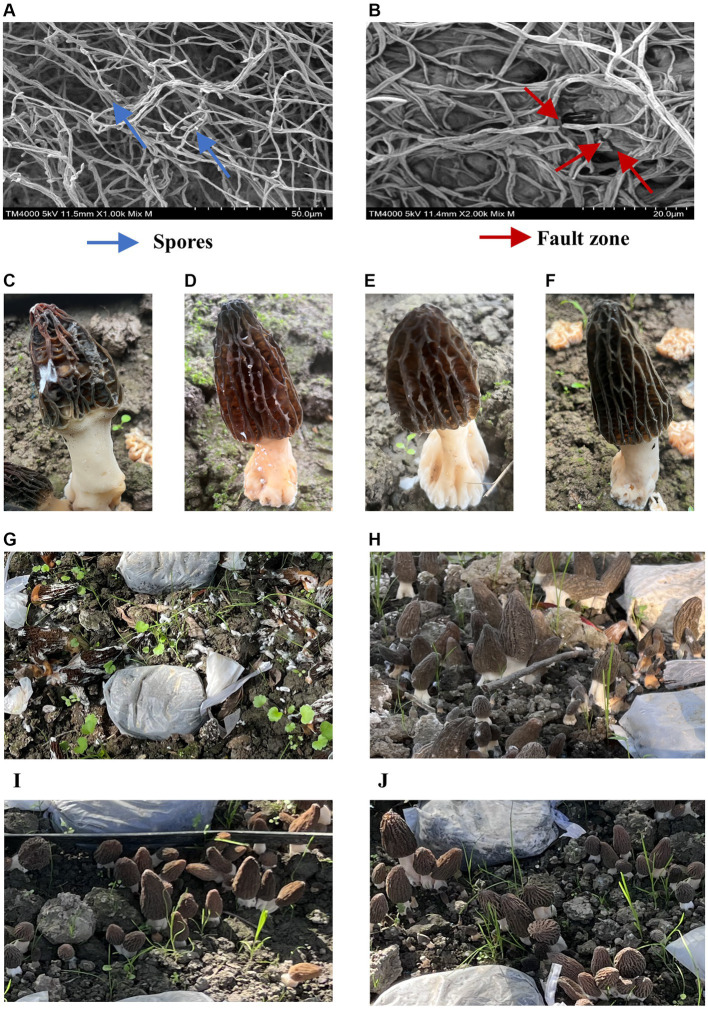
SEM observation showed that untreated L. aphanocladii G1 mycelium had uniform thickness and a smooth and complete surface (Figure 4A), while the hyphae showed branching, uneven thickness, and fractures in the presence of B. subtilis A9 (Figure 4B). In addition, the untreated hyphae was loose, while most of the treated hyphae exhibited a winding phenotype, which may have resulted from distortion of the mycelial cell wall structure. Microscopic observation revealed that the untreated mycelium produced significantly more spores than the treated group, suggesting that B. subtilis A9 can inhibit spore formation. In summary, co-cultivation of L. aphanocladii G1 with B. subtilis A9 is associated with abnormal mycelial development, compromised mycelial integrity, and decreased spore formation.
Figure 4.
Observation of B. subtilis A9 with L. aphanocladii G1 using SEM. In (A), the hyphae of the CK group were intact and uniformly distributed, and there were noticeable spores; in (B), the hyphae were clearly broken and entangled after B. subtilis A9 treatment, and the number of spores was clearly less. (C–F) Pot assay observations. (C) Morchella esculenta inoculated with L. aphanocladii G1. (D) B. subtilis A9 was administered before L. aphanocladii G1. (E) M. esculenta inoculated only with B. subtilis A9. (F) Noninoculated control. (G–J) Field assay observation. (G) M. esculenta inoculated with pathogen G1. (H) B. subtilis A9 administered before L. aphanocladii G1. (I) M. esculenta inoculated only with pathogen L. aphanocladii G1. (J) Noninoculated control.
3.4. Biocontrol of Bacillus subtilis A9 against Morchella esculenta rot disease caused by Lecanicillium aphanocladii
We then conducted a pot experiment to assess the effects of B. subtilis A9 on L. aphanocladii G1 pathogenicity of M. esculenta. After 5 days of culture, the disease symptoms of the experimental group sprayed with B. subtilis A9 were significantly lighter than those of the control group (Figures 4C,D). Moreover, spraying A9 alone had no obvious effect on M. esculenta (Figure 4E).
Pot experiments showed that the best biocontrol was achieved if the B. subtilis A9 was sprayed before inoculation with the pathogen. We then used this strategy to conduct a field experiment. The results of the field experiment (Figures 4G–J) showed that the disease incidence substantially decreased when the M. esculenta was pre-treated with the biocontrol bacteria; the disease index for the biocontrol bacteria treatment was 31.8%, compared with 84.8% when treated with the pathogen alone. The control effect of the A9 treatment was 62.5%.
3.5. Transcriptomic profiles of Morchella esculenta influenced by Bacillus subtilis A9
To study the gene expression changes in M. esculenta after spraying with B. subtilis A9, the RNA libraries of M. esculenta treated with and without B. subtilis A9 were sequenced using the Illumina Hiseq platform. A total of 39.92 Gb of clean bases were generated, and the generated clean bases reached more than 6.65 Gb per clean sample. The total number of reference genomes of M. esculenta was more than 87.71%. The proportion of clean bases was more than 93.47% (Supplementary Table S1). More than 97.57% of these readings were clean at the Q20 level, and more than 93.38% were clean at the Q30 level. The number of reads corresponding to the measured GC content of each sequence was very close to the theoretical value. These results showed that the sequencing data were of good quality and could be used for follow-up analysis.
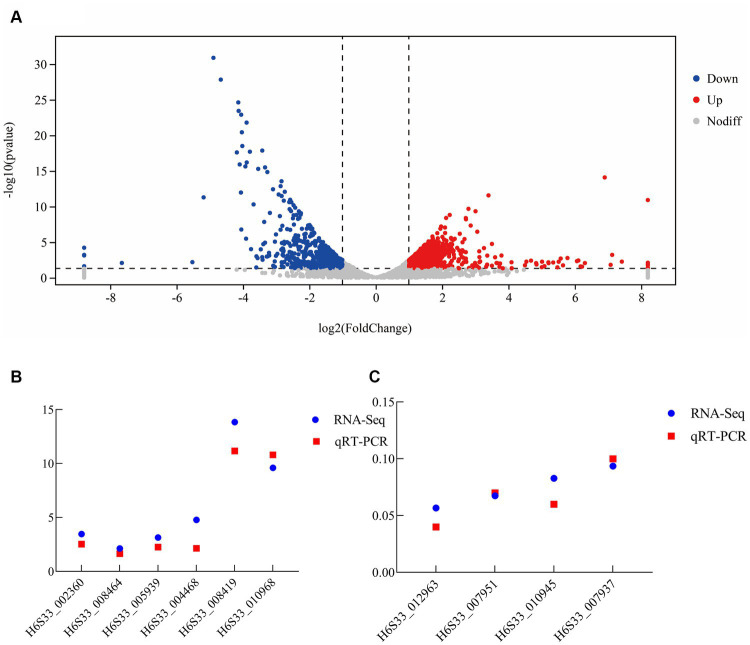
Some of the upregulated (including H6S33_004468, H6S33_002360, and H6S33_005939) (Figure 5B) and downregulated DEGs (including H6S33_007951, H6S33_010945, H6S33_012963, and H6S33_007937) (Figure 5C) were selected and validated by RT-qPCR. The results showed that the actual expression of the selected DEGs was consistent with the trend of gene expression obtained by transcriptome analysis (Supplementary Table S2).
Figure 5.
Transcriptome analysis of B. subtilis A9 on M. esculenta. (A) Differentially expressed gene analysis. (B,C) RT-qPCR verification of upregulated (B) and downregulated differentially expressed genes (C).
The main purpose of transcriptomics is to identify different genes between comparison groups to reveal their different molecular mechanisms. Thus, in the analysis results, the analysis of gene expression differences is the most important. Gene expression analysis of the two samples showed that there were 1,246 DEGs, of which 637 were upregulated and 609 were downregulated after the application of biocontrol strain B. subtilis A9 (Figure 5A). In addition, the upregulated genes showed functions, such as “oxidoreductase activity” and “response to stress,” some key genes such as peroxidase (H6S33_002360), NAD (P) H-hydrate epimerase (H6S33_008464), choline dehydrogenase (H6S33_012259) indicating that A9 may induce host Induced Systemic Resistance (ISR) by regulating the expression of genes involved in stress response and redox metabolism.
The sequencing data were saved as FASTQ files, and deposited to the National Center for Biotechnology Information (NCBI) (BioProject accessions: PRJNA1026656; BioSample accessions: SAMN37755107, SAMN37755108, SAMN37755109, SAMN37755110, SAMN37755111, SAMN37755112).
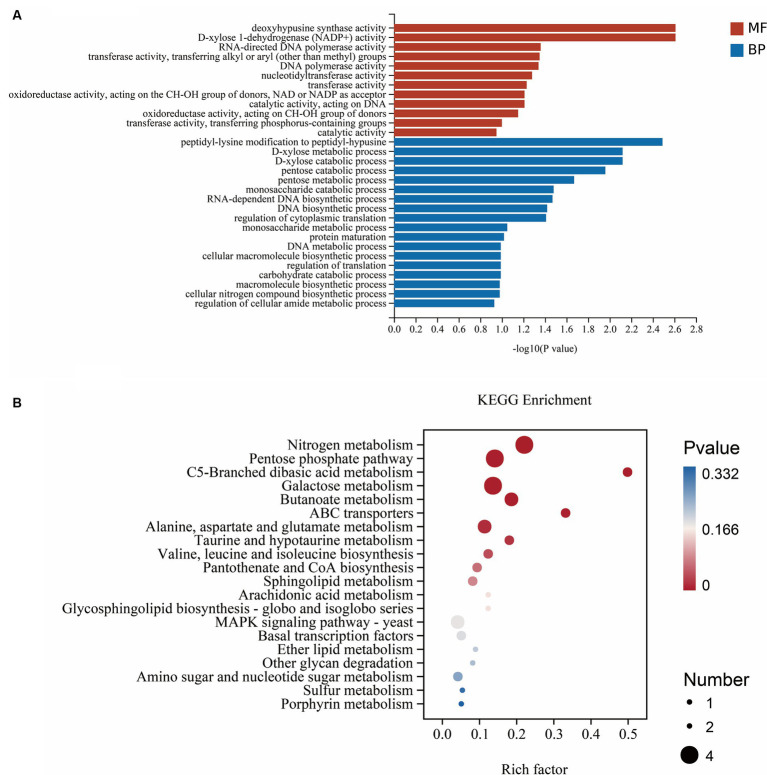
Gene Ontology (GO) was used to classify the functions of DEGs into three basic categories—BP, CC and MF. The number of significant genes in each item was calculated to determine the corresponding biological function of the active genes in the interaction between B. subtilis A9 and L. aphanocladii G1. The top 30 items enriched by GO showed that (Figure 6A) these genes were mainly enriched in two categories: BP and MF. The DEGs involved in the catabolic process and macromolecular synthesis process account for the majority of BP. The terms with the highest enrichment degree were peptidyl-lysine modification to peptidyl-hypusine, D-xylose metabolic process, and D-xylose catabolic process. The DEGs associated with MF involved mostly catalytic activity. The DEGs were significantly more abundant in deoxyhypusine synthase activity, D-xylose 1-dehydrogenase (NADP+) activity, transferase activity, oxidoreductase activity, acting on the CH-OH group of donors, and catalytic activity (Oliveira et al., 2020). In addition, genes associated with pyruvate dehydrogenase (H6S33_009813), DDE-type integrase/transposase/recombinase (H6S33_003558), Deoxyhypusine synthase (H6S33_009792) were also detected. In terms of MF, the expression level of D-xylose1-dehydrogenase (NADP+) was relatively high, and there were pathways related to enrichment of the antioxidant stress response. Thus, we speculated that antioxidant enzymes were involved in this process. GO analysis showed that B. subtilis A9 influenced M. esculenta by affecting its catalytic activity, catabolic process and macromolecular synthesis process (Dave et al., 2021).
Figure 6.
(A) GO enrichment. Abscissa is GO term, and ordinate is GO term enriched-log10 (p-value). (B) KEGG enrichment. The abscissa is the rich factor (the number of differential genes annotated to a pathway/the total number of genes annotated to the pathway), and the ordinate is the pathway. The size of the point in the map indicates the number of genes annotated in the corresponding pathway (upregulated or downregulated, related to the gene set selected at the time of analysis), and the color indicates the level of significance.
To further investigate the biological pathways mediating the effects of A9 on M. esculenta, we performed the KEGG functional annotation analysis of all DEGs identified in RNA sequencing. The top 20 enriched KEGG pathways of DEGs are analyzed and shown in Figure 6B (the lower the p-value, the greater the degree of enrichment). The results showed that the DEGs participate in three pathways: metabolism, genetic information processing and environmental information processing. DEGs were mainly enriched in nitrogen metabolism pathway, Ferredoxin-dependent glutamate synthase 1FdGOGAT (H6S33_011339) and carbonic anhydrase CA (H6S33_007248) were key genes that up regulated in this pathway. The Fold Change are 10.6 and 8.4, respectively. Followed by Pentose phosphate pathway (PPP), the irreversible oxidative section of the pathway is a major source of the reducing equivalent NADPH, for biosynthesis and maintaining the redox potential necessary to protect against oxidative stress (Xiong et al., 2009). MAPK was the signaling pathways related to disease resistance (Meng and Zhang, 2013; Dahuja et al., 2021). The genes encoding catalase CAT (H6S33_001409), tRNA dihydrouridine synthase DusB (H6S33_001297) and NAD(P)-binding protein NAD(P)BP (H6S33_000823) were induced in MAPK signaling pathway. What’s more, KEGG analysis showed that A9 influenced M. esculenta by regulating amino acid metabolism and catabolic metabolism. These results suggested that A9 considerably influenced the gene expression and metabolic pathway of M. esculenta.
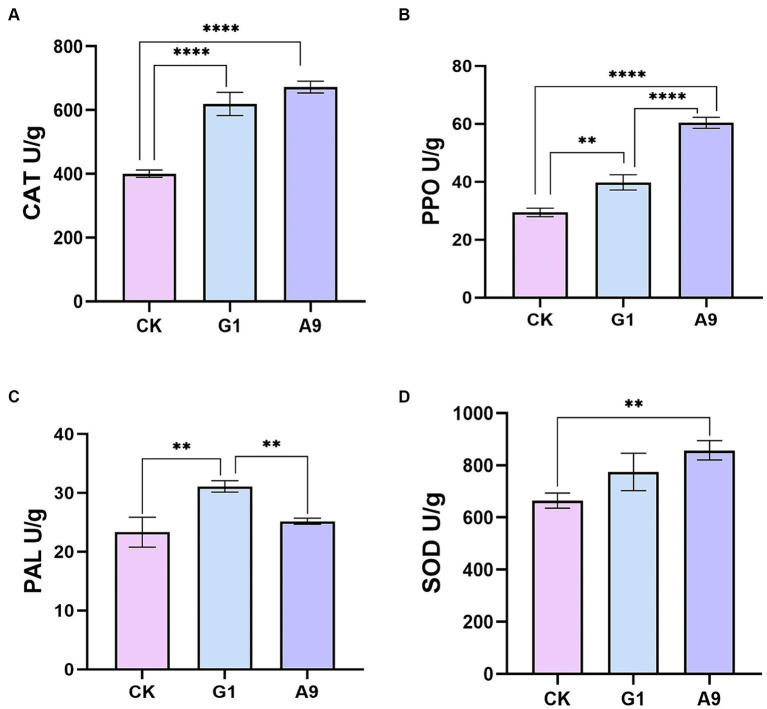
3.6. Effect of Bacillus subtilis A9 on activities of antioxidant enzymes in Morchella esculenta during biological control
Four enzymes, PPO, SOD, PAL, and CAT, were used as plant disease resistance markers to assess the effects of strain B. subtilis A9 of M. esculenta infected with pathogen L. aphanocladii G1. The activities of PPO, SOD, PAL, and CAT were higher than in M. esculenta co-inoculated with B. subtilis A9 and L. aphanocladii G1 bacteria than when treated with just the pathogen, indicating that B. subtilis A9 enhanced M. esculenta defense enzyme activities (Figure 7).
Figure 7.
Antioxidant enzyme analysis: (A) catalase (CAT), (B) polyphenol oxidase (PPO), (C) phenylalanine ammonia lyase (PAL), and (D) superoxide dismutase (SOD). All data were presented as means of three replicates ± SD, and error bars represent SD for three replicates. Means with asterisk have significant differences(∗∗p < 0.01 and ∗∗∗∗p < 0.0001).
4. Discussion
Many microorganisms have the ability to control fungal and bacterial diseases. Many of the microorganisms that have proven beneficial have been developed into products that have been commercialized. In this work, B. subtilis A9 isolated from M. esculenta was found to be effective as biocontrol bacteria. It can form spores and survive in the soil for a long time under harsh environmental conditions. Currently, several strains of Bacillus spp. are used as biocontrol agents against crop diseases (Bonaterra et al., 2022; Qiao et al., 2023; Idris et al., 2024). M. esculenta is an important edible fungus that has high economic, nutritional, and medicinal value. White mold disease in M. esculenta leads to a sharp decrease in yield. Due to the immature development of artificial cultivation technology, the decrease in the production of M. esculenta caused by rot diseases will make it even worse. Plant endophytes have been considered an excellent source of biocontrol strains and bio-inoculants in recent years due to their unique ability to thrive inside plant tissues, suppress plant pathogens, improve bacterial colonization, and promote plant growth (Chen et al., 2020). However, there are few reports of endophytes in edible fungi, and this is the first report on the endophytes of M. esculenta and their use as biocontrols.
In this study, 122 strains of endophytic bacteria were isolated from M. esculenta, of which B. subtilis A9 showed strong antagonism against L. aphanocladii G1. The greenhouse pot experiment showed that the biocontrol effect of B. subtilis A9 pre-treatment was better. This study demonstrated that the endophytic bacteria of M. esculenta are excellent as candidates for biocontrol. The use of these endophytic bacteria provides a feasible and ecologically friendly choice for the control of M. esculenta white rot disease. In our field experiment, the biocontrol effect of B. subtilis A9 was remarkable, resulting in a decrease in disease incidence of over 50%. B. subtilis has been widely studied and has been used as a biocontrol agent for several different species, and its mechanism of action is also understood to some extent. The application of B. subtilis JY-7-2L fermentation culture significantly reduced the severity of southern blight disease caused by Aconitum carmichaelii Debx by up to 30%, and the authors suggested that the possible mechanisms involved the production of hydrolytic enzymes and antimicrobial compounds (Zou et al., 2023). B. subtilis RSS-1 exerts a good inhibitory effect on Phytophthora sojae by inhibiting mycelium growth, cyst germination, and zoospore motility (Liu et al., 2019). B. subtilis E11 exhibited very strong antifungal ability, not only inhibiting of Aspergillus flavus on chili by 64% but also suppressing 81% of AFB1 at 24 h (Yuan et al., 2023). B. subtilis YPS-32 had a control efficacy of 84% against potato common scab. The mode of action of strain YPS-32 is thought to occur through with antimicrobial effects, such as surfactants and fengycin (Zhou et al., 2022). Many B. subtilis strains have been or are being registered as biological control products. Hence, B. subtilis has excellent potential for commercial biological control applications. However, the application of B. subtilis as a biocontrol bacterium in edible fungi has not been explored.
Understanding the mechanism of interaction between biocontrol bacteria and target plant pathogens is very important to strengthen the application of biocontrol bacteria in agriculture. In the past few decades, there has been a continuous understanding of the mechanism of biological control. The biocontrol mechanisms of biocontrol bacteria can now be divided into two categories. One is to inhibit the growth of pathogens directly through multiple mechanisms, such as parasitism, competition, and antibacterial activity (Fira et al., 2018; Soni and Keharia, 2021; Zhong et al., 2023). The other is to activate the expression of genes related to plant resistance to pathogens through some signal molecules, including secreted enzymes and secondary metabolites. Induce the resistance of host plants to pathogens and indirectly affect pathogens through growth promotion (Chen et al., 2022; Miftakhov et al., 2022; Yuan et al., 2022; Zhang et al., 2022). Direct and indirect biocontrol mechanisms usually cooperate to produce biocontrol effects on pathogens.
Many biocontrol bacteria can synthesize a diverse array of antibiotics, which are commonly linked by their ability to inhibit the growth of plant pathogens. Zeriouh et al. (2014) reported that the LP antibiotics bacillomycin and fengycin, synthesized by B. velezensis UMAF6614 acted directly to inhibit the growth of powdery mildew. To further determine the biological control mechanism of B. subtilis A9, we sequenced its genome. Genome-wide analysis revealed an antibiotic gene cluster, suggesting that it may exert it biocontrol effects through the production of antibiotics. Compared with the database, 13 gene clusters were discovered, and 6 of these matched known gene clusters. To the best of our knowledge, 5 of the gene clusters have not been reported. Cyclic lipopeptides from the surfactin, iturin, and fengycin families have been shown to have broad-spectrum antagonistic activities against plant pathogenic bacteria, fungi, and viruses (Fira et al., 2018; Miao et al., 2023) showed the important competitive role of bacillaene in Bacillus survival. Bacillibactin production restrained in vitro and in planta growth of the non-susceptible (to MBI600) pathogen Pseudomonas syringaepv. Tomato (Dimopoulou et al., 2021). Algburi et al. (2016) demonstrated that subtilosin prevented biofilm formation by inhibiting bacterial quorum sensing. The biological activity of these secondary metabolites is of great significance in the biological control of plant diseases. Hence, some of the clusters identified are associated with the production of antibiotics that have confirmed biocontrol effects on pathogenic bacteria, and several unknown gene clusters are interesting and worthy of further study. At the same time, we propose that the biocontrol ability of B. subtilis A9 could be enhanced by engineering the strain and to increase its production of antibiotics. To the best of our knowledge, this is the first time that B. subtilis has been isolated from M. esculenta endophytes, contributing to the improvement of the database and the further exploration of biocontrol bacteria.
We speculate that B. subtilis A9 acts directly on pathogen L. aphanocladii G1 through antibacterial metabolites. In addition, through a plate confrontation experiment in which L. aphanocladii G1 and B. subtilis A9 were co-cultured, we observed marginal hyphae under an electron microscope. The L. aphanocladii G1 hyphae treated with B. subtilis A9 showed that, compared to the control group, it produced fewer spores and featured longer hyphae that were broken and entangled. We speculate that B. subtilis A9 may cause adhesion of hyphae by destruction of the fungal cell wall, which may be its primary mode of action (Fernandes et al., 2021). In addition to acting directly on pathogens, A9 may also indirectly adjust the resistance of M. esculenta to the disease. Transcriptomic profiles shows that A9 regulates the disease resistance of M. esculenta by affecting gene expression.
Antioxidant defense systems are a prominent element in plant responses to environmental stress (Williamson and Scandalios, 1993). Increasing the activities of antioxidant enzymes and defense enzymes and up-regulating the expression of defense-related genes play an important role in inducing plant innate immunity, thus enhancing plant resistance to various types of stress (Gangireddygari et al., 2021). In this study, the peroxidase gene POD (H6S33_002360), NAD(P)H-hydrate epimerase gene (H6S33_008464) and the catalase CAT (H6S33_001409) involved in antioxidant enzyme were up-regulated. Antioxidant enzyme is an important part of the M. esculenta antioxidant system, which has the function of resisting oxidative damage (Li et al., 2016). The increased activity of catalase (CAT) and peroxidase (POD) can inhibit the accumulation of ROI (ROS intermediate oxygen intermediates) (Wagner et al., 2013), thus reducing the oxidative damage caused by oxidative stress in M. esculenta. Interestingly, the expression of CAT was increased in antioxidant enzyme experiments, and the effect of B. subtilis A9 on CAT content of M. esculenta was higher than that of L. aphanocladii G1 (Figure 7A). We speculate that A9 can induce higher levels of CAT, thereby improving the resistance of pathogenic fungi.
Nitrogen metabolism is the pathway where DEGs are most enriched. Recent results show that ROS are necessary for growth and development. Strikingly, the regulation of nitrogen (N) metabolism is closely related to ROS in response to both carbon (C) and N availability. Nitrogen (N) is one of the most important mineral nutrients required by higher plants. N metabolism can promote plant growth (Wang et al., 2014; Chaput et al., 2020). Carbonic Anhydrases CAs (H6S33_007248) is a kind of zinc-containing metalloenzymes, which plays an important role in regulating cell pH, carbon dioxide transport, electrolyte balance and cell homeostasis, and is very important for cell survival and proliferation. Carbonic anhydrase inhibitors have applied in a series of therapeutic fields (Supuran, 2018; Supuran and Capasso, 2020). Therefore, we speculate that the regulation of B. subtilis A9 on nitrogen metabolism of M. esculenta plays a key role in the process of disease resistance by regulating cell homeostasis and proliferation.
As a metabolic pathway of sugar independent of the classical EMP-TCA pathway, pentose phosphate pathway (PPP) has important biological significance, which is mainly reflected in: (1) providing reductant NADPH; for chemical reactions in organisms; (2) providing precursors for the synthesis of other chemicals in organisms; and (3) improving the disease resistance of plants (Bussell et al., 2013; Wushensky et al., 2018). DEGs mainly enriched in N metabolism, followed by PPP, so A9 was speculated that can improve the disease resistance of M. esculenta by inducing this pathway. The MAPK signal pathway is another important pathway enriched by DEGs. It regulates many important processes of plant growth and development, stress resistance, and disease resistance (Meng and Zhang, 2013; Manna et al., 2023). CAT (H6S33_001409) is an important upregulated gene enriched in this pathway. Catalases, naturally occurring oxidoreductases, are the major player of defense system against oxidative stress in all the three domains of life. Catalase activities in wild-type plants are likely to be a significant part of plant responses to changes in environmental conditions or biotic challenge (Mhamdi et al., 2010; Shaeer et al., 2021). These results explain the indirect disease resistance of B. subtilis A9 and lay a foundation for the study of its biocontrol mechanism.
Previous studies have shown that Bacillus has biocontrol effect on many plant pathogens. Bacillus strains exhibit their biocontrol capacity predominantly through inhibitory activity on the growth of plant pathogens, as well as inducing systemic resistance in plants and competing for ecological niches with plant pathogens (Fira et al., 2018; Abdelkhalek et al., 2020; Islam et al., 2022). In this study, we also revealed the action mechanism of B. subtilis A9, but there are few studies on the colonization of B. subtilis A9, so further research should be carried out. At present, there are almost no Bacillus acting on the diseases of edible fungi, we have found a strain of B. subtilis A9 which can prevent the fungal diseases of M. esculenta for the first time. The potential B. subtilis A9 is expected to be used to control the diseases of related edible fungi. The industrial production of biocontrol agents can be mainly grouped into two types: microbial pesticides and microbial fertilizers (Meng et al., 2022). Our research should develop and innovate the optimal form of B. subtilis A9 biocontrol agents. As a form of biological control, biocontrol bacteria have important value in reducing diseases and promoting growth. Meanwhile, biocontrol agents can protect the environment by reducing the use of chemical pesticides. However, there are still many problems restricting the development of biological control industry. For example, the basic research on the mechanism of microbial action is still insufficient, the role of biological pesticides is unstable, the form of biological control is single, and so on. The synthetic microbial community and developing different biocontrol agents carriers are key research directions for improving biocontrol efficiency.
5. Conclusion
In summary, we concluded that the screened endophytic B. subtilis A9 of M. esculenta significantly inhibited the occurrence of rot disease and the control effect was 62.5%. Genes related to antibiotic gene clusters were present in the genome of B. subtilis A9, which showed that A9 can produce secondary metabolites of antibiotics on pathogens. The way of B. subtilis A9 against the pathogen L. aphanocladii G1 was associated with abnormal mycelial development, compromised mycelial integrity, and decreased spore formation. Through transcriptome profiles analysis, the suggested biocontrol mechanism involved in B. subtilis A9 was to resist pathogens by regulating the activity of antioxidant enzymes and the metabolic pathways related to disease resistance of M. esculenta. Therefore, B. subtilis A9 is an ideal candidate to be developed as a biopesticide against rot disease as well as other diseases of M. esculenta.
This study provides theoretical and technical knowledge for the efficient use of microbial resources for biological control of agricultural diseases, which may lead to the improvement of new biocontrol agents for production. However, in the future, the tripartite interaction studies comprising the M. esculenta, biocontrol bacteria and the pathogens will provide new in-depth insights into the biocontrol mechanisms of B. subtilis A9. In addition, it is necessary to further explore the commercial form of B. subtilis A9 and extend the shelf-life of B. subtilis A9 based-formulations.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
XC: Data curation, Formal analysis, Investigation, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. YZ: Investigation, Methodology, Project administration, Software, Supervision, Writing – original draft, Writing – review & editing. SC: Data curation, Investigation, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. LS: Data curation, Investigation, Software, Validation, Writing – original draft, Writing – review & editing. GW: Data curation, Formal analysis, Software, Supervision, Writing – original draft, Writing – review & editing. YS: Data curation, Formal analysis, Project administration, Software, Writing – original draft, Writing – review & editing. YC: Data curation, Formal analysis, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. BL: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Writing – original draft, Writing – review & editing.
Funding Statement
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by Natural Science Foundation of Shanghai (22ZR1442500), Technical Cultivation Program Fund of Shanghai Municipal Commission of Agriculture, China (2022-02-08-00-12-F01139), National Natural Science Foundation of China (32302635), Shanghai Science and Technology Commission The belt and road project (20310750500),2024 SAAS Project on Agricultural Science and Technology Innovation Supporting Area [SAAS Application Basic Study 2024 (08)], Shanghai Academy of Agricultural Sciences 2022 (016), Shanghai Professional Technology Service Platform of Agricultural Biosafety Evaluation and Testing (23DZ2290700), Shanghai Engineering Research Center of Specialty Maize (20DZ2255300), the Science and Technology Innovation 2030 (2023ZD04062) and Grant from Shanghai Key Laboratory of Agricultural Genetics and Breeding (21DZ2271900).
Footnotes
Conflict of interest
BL was employed by Shanghai Co-Elite Agricultural Sci-Tech (Group) Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1388669/full#supplementary-material
References
- Abdelkhalek A., Behiry S. I., Al-Askar A. A. (2020). Bacillus velezensis PEA1 inhibits Fusarium oxysporum growth and induces systemic resistance to cucumber mosaic virus. Agronomy 10:1312. doi: 10.3390/agronomy10091312 [DOI] [Google Scholar]
- Ahmed W. Q., Dai Z. L., Zhang J. H., Li S. C., Ahmed A. S., Munir S., et al. (2022). Plant-microbe interaction: mining the impact of native Bacillus amyloliquefaciens WS-10 on tobacco bacterial wilt disease and rhizosphere microbial communities. Microbiol. Spectr. 10. e01471–01422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Yang J., Tan Y., Munir S., Liu Q., Zhang J., et al. (2022). Ralstonia solanacearum, a deadly pathogen: revisiting the bacterial wilt biocontrol practices in tobacco and other Solanaceae. Rhizosphere:100479: 21. doi: 10.1016/j.rhisph.2022.100479 [DOI] [Google Scholar]
- Algburi A., Zehm S., Netrebov V., Bren A. B., Chistyakov V., Chikindas M. L. (2016). Subtilosin prevents biofilm formation by inhibiting bacterial quorum sensing. Probiot. Antimicrob. Proteins 9, 81–90, [DOI] [PubMed] [Google Scholar]
- Badshah S. L., Riaz A., Muhammad A., Tel Çayan G., Çayan F., Emin Duru M., et al. (2021). Isolation, characterization, and medicinal potential of polysaccharides of Morchella esculenta. Molecules 26:1459. doi: 10.3390/molecules26051459, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaterra A., Badosa E., Daranas N., Francés J., Roselló G., Montesinos E. (2022). Bacteria as biological control agents of plant diseases. Microorganisms 10:1759. doi: 10.3390/microorganisms10091759, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boro M., Sannyasi S., Chettri D., Verma A. K. (2022). Microorganisms in biological control strategies to manage microbial plant pathogens: a review. Arch. Microbiol. 204:666. doi: 10.1007/s00203-022-03279-w, PMID: [DOI] [PubMed] [Google Scholar]
- Bussell J. D., Keech O., Fenske R., Smith S. M. (2013). Requirement for the plastidial oxidative pentose phosphate pathway for nitrate assimilation in Arabidopsis. Plant J. 75, 578–591. doi: 10.1111/tpj.12222, PMID: [DOI] [PubMed] [Google Scholar]
- Cai Z.-N., Li W., Mehmood S., Pan W.-J., Wu Q.-X., Chen Y., et al. (2018). Effect of polysaccharide FMP-1 from Morchella esculenta on melanogenesis in B16F10 cells and zebrafish. Food Funct. 9, 5007–5015. doi: 10.1039/C8FO01267A, PMID: [DOI] [PubMed] [Google Scholar]
- Carrasco J., Preston G. M. (2020). Growing edible mushrooms: a conversation between bacteria and fungi. Environ. Microbiol. 22, 858–872. doi: 10.1111/1462-2920.14765, PMID: [DOI] [PubMed] [Google Scholar]
- Chaput V., Martin A., Lejay L. (2020). Redox metabolism: the hidden player in carbon and nitrogen signaling? J. Exp. Bot. 71, 3816–3826. doi: 10.1093/jxb/eraa078, PMID: [DOI] [PubMed] [Google Scholar]
- Chen L., Wu Y. D., Chong X. Y., Xin Q. H., Wang D. X., Bian K. (2020). Seed-borne endophytic Bacillus velezensis LHSB1 mediate the biocontrol of peanut stem rot caused by Sclerotium rolfsii. J. Appl. Microbiol. 128, 803–813. doi: 10.1111/jam.14508, PMID: [DOI] [PubMed] [Google Scholar]
- Chen Q., Qiu Y., Yuan Y., Wang K., Wang H. (2022). Biocontrol activity and action mechanism of Bacillus velezensis strain SDTB038 against Fusarium crown and root rot of tomato. Front. Microbiol. 13:994716. doi: 10.3389/fmicb.2022.994716, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahuja A., Kumar R. R., Sakhare A., Watts A., Singh B., Goswami S., et al. (2021). Role of ATP-binding cassette transporters in maintaining plant homeostasis under abiotic and biotic stresses. Physiol. Plant. 171, 785–801. doi: 10.1111/ppl.13302, PMID: [DOI] [PubMed] [Google Scholar]
- Dave K., Gothalwal R., Singh M., Joshi N. (2021). Facets of rhizospheric microflora in biocontrol of phytopathogen Macrophomina phaseolina in oil crop soybean. Arch. Microbiol. 203, 405–412. doi: 10.1007/s00203-020-02046-z, PMID: [DOI] [PubMed] [Google Scholar]
- Deng K., Lan X., Fang Q., Li M., Xie G., Xie L. (2021). Untargeted metabolomics reveals alterations in the primary metabolites and potential pathways in the vegetative growth of Morchella sextelata. Front. Mol. Biosci. 8:632341. doi: 10.3389/fmolb.2021.632341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjarden D. E. (1992). [Review of Mushrooms of North America, by R. Phillips]. Mycologia. 84, 598–600. doi: 10.2307/3760330 [DOI] [Google Scholar]
- Dimopoulou A., Theologidis I., Benaki D., Koukounia M., Zervakou A., Tzima A., et al. (2021). Direct antibiotic activity of Bacillibactin broadens the biocontrol range of Bacillus amyloliquefaciens MBI600. mSphere 6:e0037621. doi: 10.1128/mSphere.00376-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X.-H., Yang Z. L. (2021). Mating systems in true morels (Morchella). Microbiol. Mol. Biol. Rev. 85:e0022020. doi: 10.1128/MMBR.00220-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Baky N. A., Amara A. (2021). Recent approaches towards control of fungal diseases in plants: an updated review. J. Fungi 7:900. doi: 10.3390/jof7110900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnahal A. S. M., El-Saadony M. T., Saad A. M., Desoky E.-S. M., El-Tahan A. M., Rady M. M., et al. (2022). The use of microbial inoculants for biological control, plant growth promotion, and sustainable agriculture: a review. Eur. J. Plant Pathol. 162, 759–792. doi: 10.1007/s10658-021-02393-7 [DOI] [Google Scholar]
- Fernandes M. F. R., Ribeiro T. G., Rouws J. R., De Barros Soares L. H., Zilli J. E. (2021). Biotechnological potential of bacteria from genera Bacillus Paraburkholderia and Pseudomonas to control seed fungal pathogens. Braz. J. Microbiol. 52, 705–714. doi: 10.1007/s42770-021-00448-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fira D., Dimkic I., Beric T., Lozo J., Stankovic S. (2018). Biological control of plant pathogens by Bacillus species. J. Biotechnol. 285, 44–55. doi: 10.1016/j.jbiotec.2018.07.044 [DOI] [PubMed] [Google Scholar]
- Fontana D. C., De Paula S., Torres A. G., De Souza V. H. M., Pascholati S. F., Schmidt D., et al. (2021). Endophytic Fungi: biological control and induced resistance to Phytopathogens and abiotic stresses. Pathogens 10:570. doi: 10.3390/pathogens10050570, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Xu X., Wu H., Li L., Wang J., Sun Y., et al. (2023). First report of Clonostachys rosea causing rot of Morchella sextelata in Anhui Province, China. Plant Dis. 107:1623. doi: 10.1094/PDIS-08-22-1794-PDN, PMID: 36210333 [DOI] [Google Scholar]
- Gangireddygari V. S. R., Chung B. N., Cho I.-S., Yoon J.-Y. (2021). Inhibitory effect of chitosan and phosphate cross-linked chitosan against cucumber mosaic virus and pepper mild mottle virus. Plant Pathol. J. 37, 632–640. doi: 10.5423/PPJ.OA.10.2021.0155, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gea F. J., Navarro M. J., Santos M., Diánez F., Carrasco J. (2021). Control of fungal diseases in mushroom crops while dealing with fungicide resistance: a review. Microorganisms 9:585. doi: 10.3390/microorganisms9030585, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan H. M. (2006). Fungicide control of mushroom cobweb disease caused by Cladobotryum strains with different benzimidazole resistance profiles. Pest Manag. Sci. 62, 153–161. doi: 10.1002/ps.1133, PMID: [DOI] [PubMed] [Google Scholar]
- Heleno S. A., Stojković D., Barros L., Glamočlija J., Soković M., Martins A., et al. (2013). A comparative study of chemical composition, antioxidant and antimicrobial properties of Morchella esculenta (L.) Pers. from Portugal and Serbia. Food Res. Int. 51, 236–243. doi: 10.1016/j.foodres.2012.12.020 [DOI] [Google Scholar]
- He X.-L., Peng W.-H., Miao R.-Y., Tang J., Chen Y., Liu L.-X., et al. (2017). White mold on cultivated morels caused by Paecilomyces penicillatus. FEMS Microbiol. Lett. 364. doi: 10.1093/femsle/fnx037, PMID: [DOI] [PubMed] [Google Scholar]
- Huang M., Zhang S., Zhang M., Ou S., Pan Z. (2011). Effects of polysaccharides from Morchella conica on nitric oxide production in lipopolysaccharide-treated macrophages. Appl. Microbiol. Biotechnol. 94, 763–771. doi: 10.1007/s00253-011-3711-7 [DOI] [PubMed] [Google Scholar]
- Hu M., Chen Y., Wang C., Cui H., Duan P., Zhai T., et al. (2012). Induction of apoptosis in HepG2 cells by polysaccharide MEP-II from the fermentation broth of Morchella esculenta. Biotechnol. Lett. 35, 1–10. doi: 10.1007/s10529-012-0917-4 [DOI] [PubMed] [Google Scholar]
- Idris A. L., Li W., Huang F., Lin F., Guan X., Huang T. (2024). Impacts of UV radiation on Bacillus biocontrol agents and their resistance mechanisms. World J. Microbiol. Biotechnol. 40:58. doi: 10.1007/s11274-023-03856-1, PMID: [DOI] [PubMed] [Google Scholar]
- Islam T., Rabbee M. F., Choi J., Baek K.-H. (2022). Biosynthesis, molecular regulation, and application of Bacilysin produced by Bacillus species. Meta 12:397. doi: 10.3390/metabo12050397, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Huang J., Qi L., Zhang X., Liu J., Guan H., et al. (2021). Bacillus subtilis strain BS06 protects soybean roots from Fusarium oxysporum infection. FEMS Microbiol. Lett. 368:fnab102. doi: 10.1093/femsle/fnab102, PMID: [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Furumichi M., Sato Y., Kawashima M., Ishiguro-Watanabe M. (2023). KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Research 51, D587–D592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., Srivastava S., Karnwal A., Malik T. (2023). Streptomyces as a promising biological control agents for plant pathogens. Front. Microbiol. 14:1285543. doi: 10.3389/fmicb.2023.1285543, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl J., Kolnaar R., Ravensberg W. J. (2019). Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front. Plant Sci. 10:845. doi: 10.3389/fpls.2019.00845, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahlali R., Ezrari S., Radouane N., Kenfaoui J., Esmaeel Q., El Hamss H., et al. (2022). Biological control of plant pathogens: a global perspective. Microorganisms 10:596. doi: 10.3390/microorganisms10030596, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legein M., Smets W., Vandenheuvel D., Eilers T., Muyshondt B., Prinsen E., et al. (2020). Modes of action of microbial biocontrol in the Phyllosphere. Front. Microbiol. 11:1619. doi: 10.3389/fmicb.2020.01619, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Limwachiranon J., Li L., Du R., Luo Z. (2016). Involvement of energy metabolism to chilling tolerance induced by hydrogen sulfide in cold-stored banana fruit. Food Chem. 208, 272–278. doi: 10.1016/j.foodchem.2016.03.113, PMID: [DOI] [PubMed] [Google Scholar]
- Liu D., Li K., Hu J., Wang W., Liu X., Gao Z. (2019). Biocontrol and action mechanism of Bacillus amyloliquefaciens and Bacillus subtilis in soybean Phytophthora blight. Int. J. Mol. Sci. 20:2908. doi: 10.3390/ijms20122908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Yang J., Ahmed W., Wan X., Wei L., Ji G. (2022). Exploiting the antibacterial mechanism of phenazine substances from Lysobacter antibioticus 13-6 against Xanthomonas oryzae pv. oryzicola. J. Microbiol. 60, 496–510. doi: 10.1007/s12275-022-1542-0, PMID: [DOI] [PubMed] [Google Scholar]
- Liu Q. Z., Dong C. H. (2023). Science and technology breakthroughs to advance artificial cultivation of true morels. Front. Microbiol. 14:1259144. doi: 10.3389/fmicb.2023.1259144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., He P., Shi X., Zhang Y., Perez-Moreno J., Yu F. (2023). Large-scale field cultivation of Morchella and relevance of basic knowledge for its steady production. J. Fungi 9:855. doi: 10.3390/jof9080855, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., He P., He P., Munir S., Ahmed A., Wu Y., et al. (2022). Potential biocontrol efficiency of Trichoderma species against oomycete pathogens. Front. Microbiol. 13:974024. doi: 10.3389/fmicb.2022.974024, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Cong Y., Sossah F. L., Lu Y., Kang J., Li Y. (2023). Characterization and genome analysis of Cladobotryum mycophilum, the causal agent of cobweb disease of Morchella sextelata in China. J. Fungi 9:411. doi: 10.3390/jof9040411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Sossah F. L., Tuo Y., Hu J., Wei Q., Li S., et al. (2023). Characterization and fungicide sensitivity of Trichoderma species causing green mold of Ganoderma sichuanense in China. Front. Microbiol. 14:1264699. doi: 10.3389/fmicb.2023.1264699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv B. B., Yu S., Chen Y. F., Yu H. L., Mo Q. (2022). First report of Lecanicillium aphanocladii causing rot of Morchella sextelata in China. Plant Dis. 106:3202. doi: 10.1094/PDIS-12-21-2656-PDN [DOI] [Google Scholar]
- Maliehe T. S., Ngidi X., Shandu J. S. E., Nxumalo C. I. (2020). Isolation of endophytic bacteria from the leaves of Anredera cordifolia CIX1 for metabolites and their biological activities. 20:300., PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Qu S., Lin J., Jia J., Baird S. M., Jiang N., et al. (2019). The complete genome of the antifungal bacterium Pseudomonas sp. strain MS82. J. Plant Dis. Protect. 126, 153–160. doi: 10.1007/s41348-019-00205-z [DOI] [Google Scholar]
- Manna M., Rengasamy B., Sinha A. K. (2023). Revisiting the role of MAPK signalling pathway in plants and its manipulation for crop improvement. Plant Cell Environ. 46, 2277–2295. doi: 10.1111/pce.14606, PMID: [DOI] [PubMed] [Google Scholar]
- Martinez Y., Ribera J., Schwarze F. W. M. R., De France K. (2023). Biotechnological development of Trichoderma-based formulations for biological control. Appl. Microbiol. Biotechnol. 107, 5595–5612. doi: 10.1007/s00253-023-12687-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J., Zhang X., Han X., Fan B. (2022). Application and development of biocontrol agents in China. Pathogens 11:1120. doi: 10.3390/pathogens11101120, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Zhang S. (2013). “MAPK cascades in plant disease resistance signaling” in Annual review of phytopathology. ed. Vanalfen N. K., vol. 51 (Palo Alto, CA: Annual Reviews; ), 245–266. [DOI] [PubMed] [Google Scholar]
- Mhamdi A., Queval G., Chaouch S., Vanderauwera S., Van Breusegem F., Noctor G. (2010). Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 61, 4197–4220. doi: 10.1093/jxb/erq282, PMID: [DOI] [PubMed] [Google Scholar]
- Miao S., Liang J., Xu Y., Yu G., Shao M. (2023). Bacillaene, sharp objects consist in the arsenal of antibiotics produced by Bacillus. J. Cell. Physiol. 239. doi: 10.1002/jcp.31228, PMID: [DOI] [PubMed] [Google Scholar]
- Miftakhov A. K., Diabankana R. G. C., Frolov M., Yusupov M. M., Validov S. Z., Afordoanyi D. M. (2022). Persistence as a constituent of a biocontrol mechanism (competition for nutrients and niches) in Pseudomonas putida PCL1760. Microorganisms 11:19. doi: 10.3390/microorganisms11010019, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin L. (2020). Progress in biological control of weeds with plant pathogens. Annu. Rev. Phytopathol. 58, 201–223. doi: 10.1146/annurev-phyto-010820-012823, PMID: [DOI] [PubMed] [Google Scholar]
- Niu B., Wang W., Yuan Z., Sederoff R. R., Sederoff H., Chiang V. L., et al. (2020). Microbial interactions within multiple-strain biological control agents impact soil-borne plant disease. Front. Microbiol. 11:585404. doi: 10.3389/fmicb.2020.585404, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira C. M., Almeida N. O., Da Rocha M. R., Rezende M. H., Carneiro R., Ulhoa C. J. (2020). Anatomical changes induced by isolates of Trichoderma spp. in soybean plants. PLoS One 15:e0242480. doi: 10.1371/journal.pone.0242480, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J., Zhang R., Liu Y., Liu Y. (2023). Evaluation of the biocontrol efficiency of Bacillus subtilis Wettable powder on pepper root rot caused by fusarium solani. Pathogens 12:225. doi: 10.3390/pathogens12020225, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokni N., Goltapeh E. M. (2019). Tolerance to dry bubble disease (Lecanicillium fungicola) in Iranian wild germplasm of button mushroom (Agaricus bisporus). Mycoscience 60, 125–131. doi: 10.1016/j.myc.2018.10.001 [DOI] [Google Scholar]
- Sánchez C. (2004). Modern aspects of mushroom culture technology. Appl. Microbiol. Biotechnol. 64, 756–762. doi: 10.1007/s00253-004-1569-7 [DOI] [PubMed] [Google Scholar]
- Shaeer A., Aslam M., Rashid N. (2021). Structural and functional analyses of a novel manganese-catalase from Bacillus subtilis R5. Int. J. Biol. Macromol. 180, 222–233. doi: 10.1016/j.ijbiomac.2021.03.074, PMID: [DOI] [PubMed] [Google Scholar]
- Shi X., Liu D., He X., Liu W., Yu F. (2022). Epidemic identification of fungal diseases in Morchella cultivation across China. J. Fungi 8:1107. doi: 10.3390/jof8101107, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shweta S., Bindu J. H., Raghu J., Suma H. K., Manjunatha B. L., Kumara P. M., et al. (2013). Isolation of endophytic bacteria producing the anti-cancer alkaloid camptothecine from Miquelia dentata Bedd. Icacinaceae 20, 913–917. doi: 10.1016/j.phymed.2013.04.004 [DOI] [PubMed] [Google Scholar]
- Singh G., Tiwari A., Gupta A., Kumar A., Hariprasad P., Sharma S. (2021). Bioformulation development via valorizing silica-rich spent mushroom substrate with Trichoderma asperellum for plant nutrient and disease management. J. Environ. Manag. 297:113278. doi: 10.1016/j.jenvman.2021.113278, PMID: [DOI] [PubMed] [Google Scholar]
- Soni R., Keharia H. (2021). Phytostimulation and biocontrol potential of gram-positive endospore-forming Bacilli. Planta 254:49. doi: 10.1007/s00425-021-03695-0, PMID: [DOI] [PubMed] [Google Scholar]
- Sunil C., Xu B. (2022). Mycochemical profile and health-promoting effects of morel mushroom Morchella esculenta (L.) – a review. Food Res. Int. 159:111571. doi: 10.1016/j.foodres.2022.111571, PMID: [DOI] [PubMed] [Google Scholar]
- Supuran C. T. (2018). Carbonic anhydrase inhibitors and their potential in a range of therapeutic areas. Expert Opin. Ther. Pat. 28, 709–712. doi: 10.1080/13543776.2018.1523897, PMID: [DOI] [PubMed] [Google Scholar]
- Supuran C. T., Capasso C. (2020). Antibacterial carbonic anhydrase inhibitors: an update on the recent literature. Expert Opin. Ther. Pat. 30, 963–982. doi: 10.1080/13543776.2020.1811853, PMID: [DOI] [PubMed] [Google Scholar]
- Tatusov R. L., Galperin M. Y., Natale D. A., Koonin E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Research 28, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietel Z., Masaphy S. (2018). True morels (Morchella)-nutritional and phytochemical composition, health benefits and flavor: a review. Crit. Rev. Food Sci. Nutr. 58, 1888–1901. doi: 10.1080/10408398.2017.1285269, PMID: [DOI] [PubMed] [Google Scholar]
- Tu S., Zhang Y., Chen X., Song L., Chen Y., Lv B. (2024). First report of Aspergillus niger causing rot of Morchella sextelata in China. Plant Dis. 108:804. doi: 10.1094/PDIS-09-23-1889-PDN [DOI] [Google Scholar]
- Wagner S., Stuttmann J., Rietz S., Guerois R., Brunstein E., Bautor J., et al. (2013). Structural basis for signaling by exclusive EDS1 Heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe 14, 619–630. doi: 10.1016/j.chom.2013.11.006 [DOI] [PubMed] [Google Scholar]
- Wang M., Shen Q. R., Xu G. H., Guo S. W. (2014). “New insight into the strategy for nitrogen metabolism in plant cells” in International review of cell and molecular biology. ed. Jeon K. W., (Burlington: Academic Press), vol. 310, 1–37. [DOI] [PubMed]
- Wang S., Ji B., Su X., Li H., Dong C., Chen S., et al. (2020). Isolation of endophytic bacteria from Rehmannia glutinosa Libosch and their potential to promote plant growth. J. Gen. Appl. Microbiol. 66, 279–288. doi: 10.2323/jgam.2019.12.001, PMID: [DOI] [PubMed] [Google Scholar]
- Williamson J. D., Scandalios J. G. (1993). Plant antioxidant gene responses to fungal pathogens. Trends Microbiol. 1:239–45. doi: 10.1016/0966-842x(93)90140-m [DOI] [PubMed] [Google Scholar]
- Wu H., Chen J., Li J., Liu Y., Park H. J., Yang L. (2021). Recent advances on bioactive ingredients of Morchella esculenta. Appl. Biochem. Biotechnol. 193, 4197–4213. doi: 10.1007/s12010-021-03670-1, PMID: [DOI] [PubMed] [Google Scholar]
- Wushensky J. A., Youngster T., Mendonca C. M., Aristilde L. (2018). Flux connections between gluconate pathway, glycolysis, and pentose–phosphate pathway during carbohydrate metabolism in Bacillus megaterium QM B1551. Front. Microbiol. 9:2789. doi: 10.3389/fmicb.2018.02789, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Defraia C., Williams D., Zhang X., Mou Z. (2009). Characterization of Arabidopsis 6-Phosphogluconolactonase T-DNA insertion mutants reveals an essential role for the oxidative section of the Plastidic pentose phosphate pathway in plant growth and development. Plant Cell Physiol. 50, 1277–1291. doi: 10.1093/pcp/pcp070, PMID: [DOI] [PubMed] [Google Scholar]
- Xu Y., Tang J., Wang Y., He X., Tan H., Yu Y., et al. (2022). Large-scale commercial cultivation of morels: current state and perspectives. Appl. Microbiol. Biotechnol. 106, 4401–4412. doi: 10.1007/s00253-022-12012-y [DOI] [PubMed] [Google Scholar]
- Yuan H., Yuan M., Shi B., Wang Z., Huang T., Qin G., et al. (2022). Biocontrol activity and action mechanism of Paenibacillus polymyxa strain Nl4 against pear Valsa canker caused by Valsa pyri. Front. Microbiol. 13:950742. doi: 10.3389/fmicb.2022.950742, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Wu Y., Jin J., Tong S., Zhang L., Cai Y. (2023). Biocontrol capabilities of Bacillus subtilis E11 against Aspergillus flavus in vitro and for dried red chili (Capsicum annuum L.). Toxins 15:308. doi: 10.3390/toxins15050308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Liu T., Liu L., Chen Y., Tang J., Peng W., et al. (2022). Application of the mushroom volatile 1-octen-3-ol to suppress a morel disease caused by Paecilomyces penicillatus. Appl. Microbiol. Biotechnol. 106, 4787–4799. doi: 10.1007/s00253-022-12038-2, PMID: [DOI] [PubMed] [Google Scholar]
- Yu Y., Tan H., Liu T., Liu L., Tang J., Peng W. (2021). Dual RNA-Seq analysis of the interaction between edible fungus Morchella sextelata and its pathogenic fungus Paecilomyces penicillatus uncovers the candidate defense and pathogenic factors. Front. Microbiol. 12:760444. doi: 10.3389/fmicb.2021.760444, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeriouh H., De Vicente A., Perez-Garcia A., Romero D. (2014). Surfactin triggers biofilm formation of Bacillus subtilis in melon phylloplane and contributes to the biocontrol activity. Environ. Microbiol. 16, 2196–2211. doi: 10.1111/1462-2920.12271, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang H., Godana E. A., Sui Y., Yang Q., Zhang X., Zhao L. (2020). Biological control as an alternative to synthetic fungicides for the management of grey and blue mould diseases of table grapes: a review. Crit. Rev. Microbiol. 46, 450–462. doi: 10.1080/1040841X.2020.1794793 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xiao J., Yang K., Wang Y., Tian Y., Liang Z. (2022). Transcriptomic and metabonomic insights into the biocontrol mechanism of Trichoderma asperellum M45a against watermelon fusarium wilt. PLoS One 17:e0272702. doi: 10.1371/journal.pone.0272702, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xu H., Ding J., Qin B., Zhou G. (2024). First report of Cladosporium scabrellum causing stipe spot disease of Morchella importuna in China. Plant Dis. 108:225. doi: 10.1094/PDIS-09-22-2152-PDN [DOI] [Google Scholar]
- Zhong J., Sui W. W., Bai X. Y., Qiu Z. L., Li X. G., Zhu J. Z. (2023). Characterization and biocontrol mechanism of Streptomyces olivoreticuli as a potential biocontrol agent against Rhizoctonia solani. Pestic. Biochem. Physiol. 197:105681. doi: 10.1016/j.pestbp.2023.105681, PMID: [DOI] [PubMed] [Google Scholar]
- Zhou Y., Li Q., Peng Z., Zhang J., Li J. (2022). Biocontrol effect of Bacillus subtilis YPS-32 on potato common scab and its complete genome sequence analysis. J. Agric. Food Chem. 70, 5339–5348. doi: 10.1021/acs.jafc.2c00274, PMID: [DOI] [PubMed] [Google Scholar]
- Zou L., Wang Q., Wu R., Zhang Y., Wu Q., Li M., et al. (2023). Biocontrol and plant growth promotion potential of endophytic Bacillus subtilis JY-7-2L on Aconitum carmichaelii Debx. Front. Microbiol. 13:1059549. doi: 10.3389/fmicb.2022.1059549, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.