Abstract
Tuberculosis (TB) is the leading cause of death due to an infectious agent, with more than 1.5 million deaths attributed to TB annually worldwide. The global dissemination of drug resistance across Mycobacterium tuberculosis (Mtb) strains, the causative agent of TB, has resulted in an estimated 450,000 cases of drug-resistant (DR) TB in 2021. Dysregulated immune responses have been observed in multi-drug resistant (MDR) TB patients, but the effects of drug resistance acquisition and impact on host immunity remain obscured. This review compiles studies that span aspects of altered host-pathogen interactions and highlights research that explore how drug resistance and immunity might intersect. Understanding the immune processes differentially induced during DR TB would aid in developing rational therapeutics and vaccines for MDR TB patients.
Drug Resistance in Tuberculosis and Implications for Host-Pathogen Interactions
Antimicrobial resistance is an emerging global threat that confounds infection treatments and stifles efforts to curb disease. The World Health Organization (WHO) estimates that, in 2021 alone, there were 450,000 new tuberculosis cases (TB) resistant to the frontline antibiotic, rifampicin, worldwide[1]. The majority of the rifampicin drug resistance (RDR)-conferring mutations occur in the RNA polymerase B subunit (rpoB) of Mycobacterium tuberculosis (Mtb), causative agent of TB[2], with changes in the core transcription machinery of the bacterium associated with profound effects on bacterial physiology and metabolism[3,4]. Additionally, clinical reports note differences in the immune responses of patients infected with drug-sensitive or drug-resistant Mtb[5,6]. However, the mechanistic links between how drug resistance mutations in Mtb result in altered responses and impact clinical disease remain to be elucidated. Emerging evidence from animal models suggest that downstream effects of rpoB mutations shift host-pathogen interactions and drive differential immune responses, pathology, and outcomes[7,8]. This review covers our current understanding of the effects of RDR mutations on Mtb, compiles various observations of immune responses in multi-drug resistant (MDR) TB patients, and puts forth mechanistic insights underlying the potential impact of drug resistance on immune responses to infection.
RDR Mutations Impact Bacterial Physiology and Function
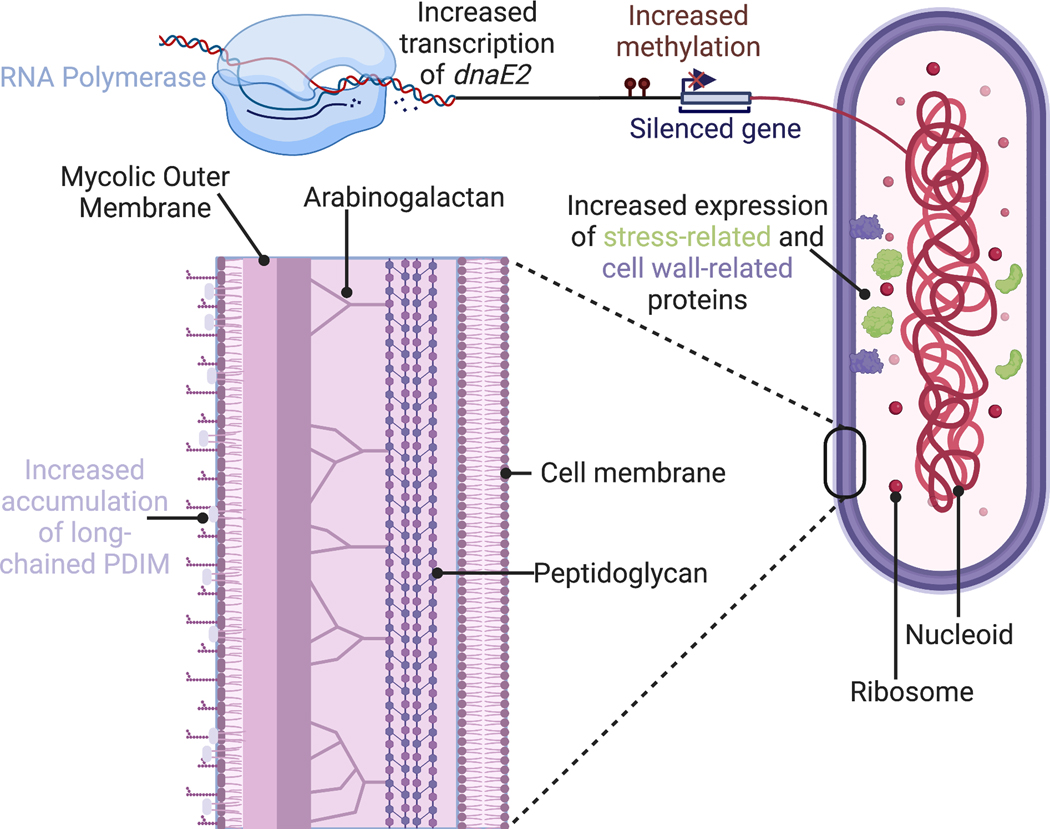
Rifampicin, or rifampin, is a frontline antibiotic used in a multidrug regimen as part of standard chemotherapy to treat TB disease, and acquisition of RDR is a prerequisite to multi-drug resistance by definition, with the WHO estimating numbers of DR TB cases without distinguishing RDR and MDR[1]. The mechanism of action for rifampicin is through binding to the core of the multi-subunit DNA-dependent RNA polymerase machinery and preventing elongation of RNA transcripts[2]. Specifically, the antibiotic binds in a pocket of rpoB and bacterial resistance to rifampicin in bacteria is mediated primarily through mutations in a section of rpoB known as the rifampicin resistance determining region (RRDR)[9]. Indeed, a single non-synonymous point mutation that has commonly been seen at amino acids D435, H445, and S450 within the RRDR, is sufficient to confer resistance to rifampicin, likely through altering the binding interaction between the antibiotic and rpoB[10]. In this section, we detail various downstream effects observed in RDR Mtb strains following the development of rpoB mutations (Figure 1).
Figure 1-. Examples of differences observed between drug sensitive and drug resistant Mtb strains.
The presence of rifampicin drug resistance mutations is associated with broad changes in bacterium physiology and function. Some changes include increased expression of DNA-damage related DNA polymerase, dnaE2[11], differences in DNA methylation patterns[12], increased expression of cell wall- and stress-related proteins[16–18], and increased accumulation of long-chain cell wall lipid phthiocerol dimycocerosate (PDIM)[3,4] This figure was created using BioRender.com
Transcription and Translation
Studies have shown broad transcriptional differences between spontaneously generated RDR Mtb mutants and their parental bacterial strains[11,12]. For example, independent RDR Mtb isolates have exhibited constitutively elevated expression of dnaE2 (an inducible low-fidelity DNA polymerase), even in stress-free environments, relative to their drug-susceptible (DS) counterparts[11]. In bacteria, dnaE2 would generally be induced in response to DNA damage to repair DNA quickly, albeit incorrectly perhaps, thus leaving behind mutations. While dnaE2 might be speculated to play a role in antibiotic stress-induced mutagenesis, sustained expression of this error-prone enzyme suggests broader and far-reaching effects of acquiring RDR. For example, an increased mutation rate, facilitated by heightened dnaE2 expression, might drive adaptation and survival of the bacterium to future stresses such as second-line antibiotics better than Mtb strains with low dnaE2 expression[13]. Indeed, another study comparing RDR Mtb strains with their parental DS strain showed hundreds of differentially methylated genes across the strains, which correlated with differences in gene expression[12]. This is relevant as acquisition of an RDR mutation leading to broad, persisting epigenetic modifications suggests that differences between susceptible and resistant bacterial strains might be reinforced beyond the initial antibiotic stress and lead to long-lasting transcriptional and translational changes between susceptible and resistant bacterial strains. Thus, acquisition of antibiotic resistance and subsequent genetic and epigenetic modifications could have far-reaching impact on cell physiology and functions, such as ability to form persister cells[14], which can influence patient outcome and/or infection clearance[15].
Differences between DS and DR strains have also been observed at the bacterial protein level. Western blot analysis comparing Mtb isolates revealed five antigenic proteins (Rv3248c, Rv0350, Rv0440, Rv0475, and Rv3588c) uniquely expressed in MDR strains[16]. Another similar study comparing a susceptible and MDR Mtb isolate found six proteins (Rv0147, Rv3597c, Rv0379, Rv3699, Rv1392, and Rv0443) through two-dimensional gel electrophoresis and mass spectrometry[17]. Broadly, these proteins can be functionally categorized to be part of the stress response pathway (Rv0350 and Rv0440), cell wall and cell processes (Rv0475 and Rv0379), metabolism and respiration (Rv3588c, Rv0147, Rv3248c, and Rv1392), potential virulence factors (Rv3597c), or conserved hypothetical proteins (Rv3699 and Rv0443)[16,17]. A possible caveat is whether the differences in the drug susceptible and MDR bacterial strains are driven directly by the drug resistance-conferring mutations or whether other unrelated mutations could drive the observed differences in protein expression. However, a study sequentially isolating Mtb from a patient that developed MDR TB also found significant overexpression of proteins involved in metabolism and respiration (Rv1437 and Rv2970c) or an Mtb virulence factor (Rv2145c) in the later resistant isolates, compared with the initial DS isolate[18]. Thus, acquiring an RDR mutation by Mtb strains unexpectedly alters the transcription and translation of various genes in the bacterium[12,16,18]. Whether these transcriptional differences could result from RDR mutations changing the interaction of the RNA polymerase with promoters and other protein factors to initiate transcription, the functionality of the RNA polymerase during transcription, or other aspects of the RNA polymerase require further investigation. Nevertheless, studies have consistently shown the upregulation of genes and increased abundance of proteins that have implications for crucial bacterial cell processes and functions such as the cell wall and metabolism, which have major ramifications for bacterial growth[19], host-pathogen interactions[20,21], and antibiotic efficacy[22].
Physiology and Metabolism
Unlike most bacteria with simple cell walls that are comprised of a layer of peptidoglycan, mycobacteria contain two additional layers[23,24]. The second layer contains arabinogalactan that is covalently attached to the peptidoglycan and then links with a layer of mycolic acids. Various types of cell wall lipids, such as sulpholipids, glycolipids, and phthiocerol dimycocerosate (PDIM), are found in the mycolic acid layer and are important virulence factors for Mtb[25]. The development of RDR in Mtb has also been connected with changes in specific cellular pathways, notably bacterial cell wall lipid production and assembly[3,4,26]. From paired clinical Mtb isolates developing drug resistance over the course of TB treatment[26], DR strains exhibited heightened expression of genes involved in the biosynthetic production of mycobacterial lipids as well as increased production of precursor molecules to PDIM. Of note, these changes were observed in Mtb strains from different lineage backgrounds, Lineages 2 (W-Beijing) and 4 (Haarlem), with differing rpoB mutations at amino acid residues H445 and S450, respectively. An unbiased mass spectrometry approach that screened nearly 10,000 cell wall lipid species from DR and DS Mtb from Lineages 2 and 4 and across three rpoB mutations, Q432E, H445Y, and S450L, found broad changes in the bacterial cell wall lipid abundance[3]. While a reduction in sulfoglycolipids was observed across all DR isolates, increases in PDIM signal were only observed in Lineage 2 DR strains. Our laboratory group has also observed that the presence of the H445Y rpoB mutation in an MDR clinical isolate and a spontaneously-generated RDR mutant was necessary and sufficient to drive increased accumulation of long-chain PDIM in the mycobacterial cell wall[4]. In this study, RDR Mtb with other rpoB mutations, such as S450L, were not found to have significant alterations to the amount or type of PDIM in their cell walls. A broader characterization of the H445D rpoB mutation in a clinical Lineage 2 isolate showed an association between the mutation and increased bacterial cell wall length, decreased cell wall thickness, and increased sensitivity to cell wall-related stress[27]. These studies highlight how RDR mutations have broader effects on Mtb and suggest roles for Mtb background lineage and mutation identity in these alterations.
A natural consequence of the changes to DR Mtb utilizing and incorporating lipids differentially in its cell wall and the upregulation of proteins involved in respiration, include other broad downstream changes to bacterial metabolic pathways. For instance, an unbiased metabolomics approach screening intracellular and cultured supernatant metabolites found significant differences between DS and MDR clinical isolates[28]. Specifically, differential metabolites were enriched for in nicotinate and nicotinamide metabolism, purine and pyrimidine metabolism, as well as arginine metabolism, which are metabolic pathways related to Mtb processes of growth[29], physiology[30], and host defense[31], respectively, suggesting that acquisition of drug resistance could have far-reaching effects on bacterial functions through changes in metabolism. Even the treatment of DS Mtb strains with sub-lethal concentrations of rifampicin resulted in significant metabolite alterations in pathways such as purine, pyrimidine, and arginine metabolism[32], which further underscores the link between rifampicin resistance and mycobacterial metabolism. Using BindingDB, a public database of small molecule and human protein interaction datasets, these metabolic changes were predicted to interact with host proteins involved in processes such as inflammation, apoptosis, and proteolysis [33]. Also, when examining methylation differences between DS and RDR Mtb[12], the nitrogen metabolism pathway was significantly enriched in RDR strains, suggesting a durable and persisting change in metabolic processes in RDR strains. Thus, we observe an indirect association between RDR mutations with mycobacterial metabolism that could shape many bacterial processes and overall host-pathogen interactions. However, how RDR mutations are linked with broad metabolic changes and the implications for downstream effects due to these metabolic differences remains to be understood.
Altered Human Immune Responses to Drug-Resistant Mtb
Immune Correlates of Protection During Mtb Infection
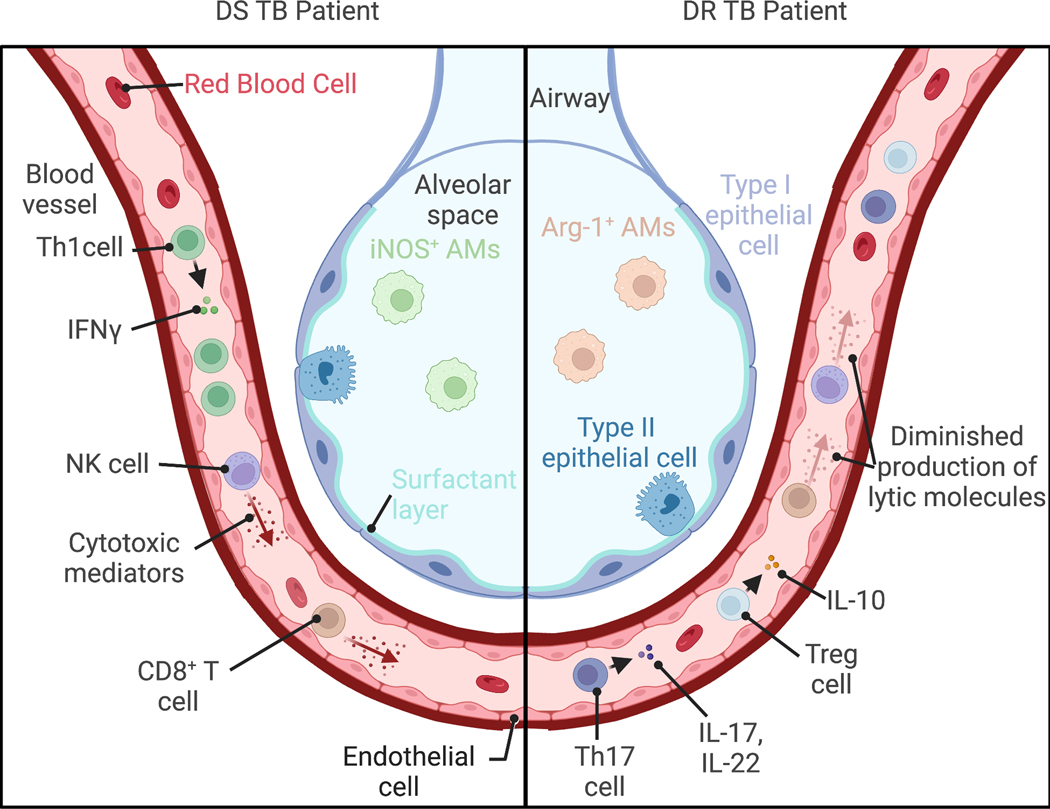
Protective immune responses to Mtb infection have been characterized via animal experimental studies[34–36] and by identifying host factors in patients that mediate susceptibility to mycobacterial disease[37]. CD4+ T cells are essential for limiting Mtb replication and controlling infection, primarily through the production of IFNγ by T helper 1 (Th1) cells[38,39]. Additionally, humans with deficiencies in IFN-γ signaling are predisposed to mycobacterial infections[40,41]. C57Bl/6 (B6) mouse models of TB disease have also shown a protective role for interleukin-17 (IL-17) and IL-22 produced by T helper 17 (Th17) cells in promoting the recruitment of Th1 cells and myeloid cells, respectively to the lungs during hypervirulent Mtb infection[42–44]. Cytotoxic CD8+ T and NK cells also play important roles during Mtb infection through IFNγ production and cytotoxic activity leading to the killing of infected cells and limiting Mtb intracellular replication as observed in B6 TB mouse models of in vivo infection and during in vitro co-culture of Mtb-infected cells with cytotoxic T lymphocyte cell lines generated from healthy donors[45–47]. From studies of human granulomas, macrophages have been determined to be vital players in TB disease and can either serve as niches for Mtb growth[48] or kill the pathogen upon activation by IFNγ through antimycobacterial mechanisms such as phago-lysosome fusion and induction of the inducible nitric oxide synthase (iNOS) that produces bactericidal reactive nitrogen species[49,50]. In this section, we detail how these various immune processes have been characterized in MDR TB patients (Figure 2).
Figure 2-. Comparison of reported immune responses between DS and DR TB patients.
Clinical studies comparing peripheral and tissue immune responses largely find divergent and diminished immune responses in drug resistant (DR) TB patients relative to drug susceptible (DS) TB counterparts. While elevated proportions of T helper 1 (Th1) CD4+ T cells producing interferon (IFN)γ are found in the peripheral blood of DS TB patients, Th17 CD4+ T cells producing IL-17 and IL-22 and T regulatory (Treg) cells producing IL-10 are elevated in DR TB patients[7,55,71]. Additionally, the CD8+ T cells and natural killer (NK) cells from peripheral blood of DR TB patients produce less cytotoxic and lytic mediators than similar cells from DS TB patients[58,60]. In the lungs greater proportions of Arginase 1+ (Arg-1+) alveolar macrophages (AMs) are found in DR TB patients, while inducible nitric oxide synthase+ (iNOS+) AMs are found in DS TB patients[62,64]. This figure was created using BioRender.com
Limited Immune Activation in DR TB Patients
Some studies have analyzed immune responses in MDR TB patients compared to DS TB patients, Mtb-experienced patients as determined through the tuberculin skin test (TST), and Mtb naïve, healthy controls [7,51,52]. MDR TB patients have been reported to exhibit lower numbers and frequencies of CD4+ T cells[7,51,52] and lower frequencies of Th1 cells[8] in peripheral blood than DS TB patients. Moreover, comparison between MDR and DS TB patients revealed decreased IFNγ stimulation of peripheral blood mononuclear cells (PBMCs) from MDR TB patients with Mtb proteins resulted in lower frequencies of CD4+ IFNγ -producing T cells (likely Th1 cells)[53] and limited production of IFNγ [54] measured in the cell supernatant, when compared with similarly stimulated PBMCs from DS TB patients. By contrast, PBMCs from MDR TB patients exhibited increased proportions of IL-17, but not IFNγ, producing CD4+ T cells than PBMCs from DS TB patients[55]. And, further stimulation of PBMCs from DS or MDR TB patients with proteins from an MDR strain expanded the CD4+ IL-17+ IFNγ– T cell population far more than comparable stimulation with proteins from a DS strain[55], suggesting that MDR Mtb strains more potently and selectively drive IL-17 production.
Additionally, increased frequencies of CD4+ IL-17+ T cells were positively correlated with high sputum Mtb in MDR TB patients[55], which is a marker for TB disease severity[56]. Conversely, increased frequencies of CD4+ IL-22+ T cells were negatively correlated numbers of bacteria in the sputum of MDR TB patients[57]. These findings imply a detrimental and protective role for IL-17 and IL-22 signaling in MDR TB patients, respectively. However, these observations run contrary to the protective role of IL-17 signaling noted above and are puzzling. Could increased IL-17 responses during MDR TB disease just be associated with increased bacterial replication or driving poorer control of MDR Mtb infection. The selective induction of IL-17 suggests modulation of immune processes, potentially through virulence factors that are uniquely upregulated by DR Mtb; these, in turn, might limit the protective impact of IL-17 signaling, although this remains conjectural and warrants robust investigation. One such mechanism could be through promoting the survival of CD4+ IL-17+ IFNγ– T cells and apoptosis of CD4+ IL-17+ IFNγ+ T cells through TGF-β signaling in MDR TB patients[6]. Nevertheless, the CD4+ T helper cell response appears to be dysregulated in MDR TB patients.
From another angle, the analysis of CD8+ T cells responses to MDR Mtb strains shows opposing results. One study found that PBMCs cultured with the MDR Mtb strain M resulted in limited expression of lytic molecules in CD8+ T cells relative to PBMCs cultured with the lab-adapted DS Mtb strain H37Rv[58]. However, another study found that after stimulation with H37Rv protein antigens, CD8+ T cells from MDR TB patients had higher cytotoxic activity than CD8+ T cells from DS TB patients and healthy controls[59]. These contrasting results might be attributed to the different stimulating agents, with the DS Mtb H37Rv antigens more potently activating CD8+ T cells than the MDR Mtb strain, although this is speculative and should be evaluated in the future. In comparison, analysis of NK cells from MDR TB patients consistently shows decreased NK cell activity relative to NK cells from matched DS TB patients[51,60,61]. Broadly, immune cells from MDR TB patients appear to show limited cytolytic activity, which might be driven by MDR Mtb strains failing to elicit these responses or, alternatively, an inability to produce cytotoxic effectors due to ‘exhaustion’ and overactivation- possibilities that remain to be tested.
From the myeloid cell perspective, studies have also documented limited monocyte and macrophage activation peripherally and at the site of infection in the lungs, which are associated with decreased control of MDR Mtb infection relative to DS Mtb infection. Staining of resected lung sections from TB patients for ‘M1-like’-polarized (iNOS+) or ‘M2-like’-polarized (Arginase-1+) CD68+ alveolar macrophages revealed a greater proportion of M2-like macrophages in DR TB lungs than in DS TB lungs, suggesting that macrophages are differentially activated in DR TB patient lungs[62]. Moreover, macrophages derived from monocytes (MDMs) isolated from MDR TB patients were unable to limit DS Mtb proliferation following ex vivo infection, unlike MDMs isolated from healthy controls[63]. Also, peripheral blood monocytes from MDR TB patients produced significantly less NO upon ex vivo infection compares with controls[64]. Further, an MDR Mtb strain, MKR, displayed increased intracellular growth, in THP-1s, a human monocyte cell line relative to H37Rv [65]. Increased MDR Mtb proliferation in vitro correlated with elevated expression of IL-36RN, a negative regulator of the IL-36 pathway. As IL-36 signaling has been implicated in host defense against Mtb[66], induction of IL-36RN suggests that monocytes are mounting a more limited response to MDR Mtb infection than to DS Mtb infection. Collectively, these studies suggest that key immune processes previously found to be essential in an effective and protective response against Mtb in DS TB patients are not robustly present in MDR TB patients.
Active Immune Suppression during MDR TB?
With ample evidence of limited immune responses in MDR TB patients, an evident question is whether MDR strains passively fail to elicit a robust immune response or actively antagonize host processes during infection. One mechanism of immune suppression characterized during Mtb infection is the induction of IL-10 production by T regulatory cells (Tregs), generally defined as CD4+ CD25+ Foxp3+ CD127–[67–69]. Accordingly, depletion of Tregs from PBMCs of active DS TB patients improved anti-microbicidal capacity of the depleted PBMCs to secondary ex vivo Mtb infection as measured through a decline in Mtb growth when compared with untreated PBMCs[70]. In the peripheral blood of MDR TB patients, the proportions of Tregs have consistently been found to be elevated above levels found in DS TB patients and healthy controls[5,53,60,71]. While some studies found a corresponding increase in IL-10 concentrations in the peripheral blood and serum of MDR TB patients as compared with DS TB patients[53,60], another study found that PBMCs isolated from MDR TB patients produced less IL-10 after stimulation with PMA and ionomycin, relative to similarly stimulated PBMCs from DS TB patients[8]. In contrast, PBMCs and monocytes from some MDR TB patients produced IL-10 to a similar extent as healthy controls in response to Mtb or its lipid antigens[7,72]. Yet, one of the upregulated proteins found in the Mtb strain that developed drug resistance (Rv2145c) has been implicated in promoting intracellular bacterial survival through IL-10 signaling[73]. Thus, while the cellular mechanisms of Treg function in MDR TB disease remain to be determined, the increased accumulation of Tregs in the peripheral blood of MDR TB patients beyond what has been observed in the blood of DS TB patients suggests that MDR Mtb strains might be actively subverting host T cell immunity through a more robust induction of this immunoregulatory cell population, warranting further investigation. Indeed, augmenting T cell responses through addition of recombinant IL-12 to in vitro PBMCs from MDR TB patients bolstered low IFNγ production[7], and addition of recombinant IL-2 to PBMCs stimulated with MDR strain M restored expression of cytolytic molecules in CD8+ T cells[58]. Similar treatment of cells stimulated with H37Rv or a different MDR strain 410 did not impact cytolytic activity. Further, in a completed phase 1 two arm-randomized controlled trial (NCT03069534) with 271 enrolled participants, addition of recombinant IL-2 to standard chemotherapy of MDR TB patients improved success rates and mycobacterial sputum clearance, the primary and secondary outcomes, compared to a matched control group with no adverse events [74]. Also, these better treatment outcomes correlated with increased proportions of Th1 cells and decreased frequencies of Th17 and Treg populations in peripheral blood of rIL-2 treated patients relative to patients in the control group. Though additional and robust randomized controlled trials are necessary to interrogate the general effectiveness of augmenting T helper cell responses in MDR TB patients, this study suggests that therapeutic intervention to boost the limited T cell responses in MDR TB patients has potential. Thus, the collected evidence suggest that a suppressive immune response is unique to some DR Mtb strains that promote DR TB pathogenesis.
Altered Immune Responses in Animal Models to Drug-Resistant Mtb
Guinea Pig Models
Studies in human MDR TB patients are limited to associations and ex vivo analysis of peripheral immune cells. Alternatively, guinea pigs recapitulate features of TB disease observed in humans, such as a well-organized granuloma with a necrotic core surrounded by myeloid and multinucleated cells and encapsulated with a lymphocyte cuff, and are used as a model to mechanistically study infection establishment and disease progression[75,76]. In one such study, guinea pigs were infected with a low dose of approximately 20 Mtb bacili by aerosolizing a liquid inoculum of either a DS or MDR strain[77]. While both MDR strains, W7642 (Lineage 2) and TN5904 (non-Lineage 2), grew to similar bacterial titers in the lungs as the DS, lab-adapted H37Rv strain (Lineage 4) at 30 days post infection (dpi), both strains declined in bacterial burden during the chronic infection period at 90 dpi. Both MDR strains also induced less pathology upon infection than the H37Rv strain as determined through parameters such as lung lesion size, necrosis, and fibrosis[77]. Another study modeled an alternate infection route by continuously exposing guinea pigs to exhaust air from MDR TB patients resulted in 75% of the animals becoming infected, as determined through a TST[78]. Unexpectedly, 22% of guinea pigs reverted to a negative TST reaction, and a third of reverting guinea pigs redeveloped a positive TST reaction. Reversion of a positive TST reaction could suggest clearance of infection without development of T memory cell responses, whereas redevelopment of a positive reaction could indicate re-infection. Even in infected guinea pigs with persisting positive TST reactions, disease severity score was relatively low after 4 months of exposure. These results indicate that MDR strains are fairly transmissible but appear to drive limited disease although these findings may be species dependent. Although more studies with closer comparative strains and a greater depth of immune profiling need to be conducted, both studies suggest that MDR strains likely induce milder disease than DS strains during infection, which coincide with clinical observations of limited or suppressed peripheral immune responses, as noted above.
Mouse Models
Mice are another widely-used model for studying TB disease through forward and reverse genetic approaches that aim to characterize the roles of specific host factors[76,79]. An early study comparing the infection responses of C57Bl/6 mice to a panel of clinically isolated Mtb strains found that RDR strains grew steadily in the lungs by 20 dpi but declined at 40 and 60 dpi[80]. However, another study found the opposite result, with MDR strains growing more slowly than the H37Rv strain in the first three weeks of infection but not after that timepoint[81]. Accordingly, comparison of clinically isolated strains from an epidemic outbreak in KwaZulu-Natal, South Africa that belong to Lineage 4 also revealed decreased MDR bacterial replication at 2 weeks post infection and reduced mortality of infected mice, relative to DS strains[82]. These seemingly discrepant results could be explained by differences in the genetic backgrounds of the RDR and MDR strains, with the drug resistance mutations impacting these DR strains differently. But, these studies collectively might suggest that the acquisition of drug resistance can result in a decrease in virulence of Mtb strains. However, a lack of characterization of immune responses in these mice limits our understanding of the host processes driving these outcomes.
We sought to understand the roles of various host immune pathways during DR and DS TB with a collection of MDR strains and a DS strain, HN878, all belonging to the W-Beijing family (Lineage 2)[4]. While single knockout mice of immune genes previously shown to be essential for protection, such as Ifngr and Nos2, exhibited increased bacterial loads across these different strain infections, Il1r1−/− mice could still control some MDR Mtb infections, notably with the W7642 strain, as evidenced by similar lung bacterial burdens in Il1r1+/+ and Il1r1−/− mice. It should be noted that the W7642 strain did not exhibit reduced virulence compared to HN878 up to 60 dpi, suggesting that acquisition of an rpoB mutation does not always impose a fitness cost on the bacteria and could be ameliorated through additional compensatory mutations. Further, in vitro infection of bone marrow derived macrophages (BMDMs) with HN878 drove high IL-1β and low type I interferon (IFN) production from BMDMs. HN878-infected BMDMs also underwent a glycolytic shift in metabolism, as measured through transcriptional changes in infected macrophages as well as lactate accumulation, relative to uninfected controls. By contrast, BMDMs infected with W7642 produced low IL-1β, high type I IFN, and did not effectively shift to glycolysis. This suggested that heightened type I IFN signaling in the MDR-infected BMDMs likely contributed to limiting this metabolic shift[83], although this remains to be further tested. Of note, whole genome sequencing of all DS and MDR strains used in the study implicated the RDR mutation rpoB-H445Y as potentially causal in driving the observed phenotypes. Indeed, spontaneous, independent DR mutants containing the rpoB-H445Y, but not other mutations such as rpoB-S450L, recapitulated the immune phenotypes observed with W7642. Treatment of HN878 Mtb-infected BMDMs with PDIM isolated from W7642 or the DR strain containing H445Y also resulted in decreased IL-1β and lactate accumulation but increased type I IFN relative to treatment with PDIM isolated from HN878. Thus, the impact on host-pathogen interactions is, in part, mediated by changes in Mtb antigens and virulence factors that arise during the acquisition of RDR mutations.
Concluding Remarks
Acquisition of RDR is known to have significant consequence for the pathogen, but emerging evidence supports that this also impacts host-pathogen interactions, which is more than meets the eye at first glance. Similar changes have been observed in other bacterial species that become resistant to rifampicin with changes to bacterial physiology[84–86] and effects on infected immune cells[87]. However, there are significant gaps in discerning the impact of RDR mutations on immunity, chiefly how point mutations in rpoB of Mtb shift host-pathogen interactions (see Outstanding questions). As noted throughout this review, many of the comparisons of DS and MDR strains extend across lineages and often times without comprehensive sequencing information. Yet, the impact of Mtb background lineage has been well documented to affect host immunity[88–91]. Also, most clinical studies referenced here highlight peripheral immune responses across DS and DR TB patients, which have yielded interesting, but limited insights. To more thoroughly understand the impact of bacterial rifampicin drug resistance on host immunity, future clinical studies should more closely match and sequence DS and DR strains to identify the specific rpoB mutations, additional compensatory mutations that might interact in an epistatic manner, as well as the strains’ lineage background[92,93], and also correlate the immune responses of DR TB patients with differences in pathology and patient outcomes. Moreover, the limited immune response to MDR TB should be better characterized through animal models to identify novel immune correlates of protection, and other immune processes important in mycobacterial host defense, such as autophagy[94,95], should be studied if they can aid in immune control of MDR Mtb infection (see Outstanding questions)[96,97]. Additionally, expanded clinical trials should be conducted to study how limited T cell immunity could be therapeutically targeted to improve MDR TB patient outcomes. Overall, a better understanding of how drug resistance mutations to a frontline antibiotic intersect with host immune responses may reveal novel biology and ideally assist in the development of more effective vaccines and host-directed therapeutics to combat the pervasive, ever-growing threat of DR TB.
Glossary
- Epistatic
Describing genetic interactions where the effects of a mutation in one gene is influenced by mutations in other genes
- Phthiocerol dimycocerosate (PDIM)
A bacterial cell wall lipid unique to Mtb that is produced intracellularly and exported to the mycobacterial outer membrane and is important for bacterial virulence
- Stress response pathway
Broad categorization of cellular responses to mediate stresses such as nutrient limitation, antimicrobial compounds, and low pH
- T helper 1 cells
CD4+ T cell subset that are important for clearance of intracellular bacterial and viral pathogens through the production of cytokines, such as IFNγ
- T helper 17 cells
CD4+ T cell subset that are important for clearance of extracellular pathogens through production of IL-17
- T regulatory cells
CD4+ T cell subset defined through Foxp3 and CD25 expression that suppress and limit immune responses
- Tuberculin skin test
A method for detecting Mtb exposure through measuring a patient’s response to intradermal injection of Mtb antigens. A positive result indicates that the patient has generated a T cell response to Mtb, likely due to infection
- Virulence factors
Molecules that contribute to a pathogen’s ability to infect, colonize, and survive in a host
REFERENCES
- 1.World Health Organization (2022) Global tuberculosis report 2022. Geneva. Licence: CC BY-NC-SA 3.0 IGO. [Google Scholar]
- 2.Goldstein BP (2014) Resistance to rifampicin: a review. J. Antibiot. (Tokyo) 67, 625–630 [DOI] [PubMed] [Google Scholar]
- 3.Lahiri N. et al. (2016) Rifampin Resistance Mutations Are Associated with Broad Chemical Remodeling of Mycobacterium tuberculosis. J. Biol. Chem. 291, 14248–14256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howard NC et al. (2018) Mycobacterium tuberculosis carrying a rifampicin drug resistance mutation reprograms macrophage metabolism through cell wall lipid changes. Nat. Microbiol. 3, 1099–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geffner L. et al. (2009) Patients with Multidrug-Resistant Tuberculosis Display Impaired Th1 Responses and Enhanced Regulatory T-Cell Levels in Response to an Outbreak of Multidrug-Resistant Mycobacterium tuberculosis M and Ra Strains. Infect. Immun. 77, 5025–5034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basile JI et al. (2017) Mycobacterium tuberculosis multi-drug-resistant strain M induces IL-17+IFNγ– CD4+ T cell expansion through an IL-23 and TGF-β-dependent mechanism in patients with MDR-TB tuberculosis. Clin. Exp. Immunol. 187, 160–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDyer JF et al. (1997) Patients with multidrug-resistant tuberculosis with low CD4+ T cell counts have impaired Th1 responses. J. Immunol. 158, 492–500 [PubMed] [Google Scholar]
- 8.Tan Q. et al. (2012) Characterization of Th1- and Th2-type immune response in human multidrug-resistant tuberculosis. Eur. J. Clin. Microbiol. Infect. Dis. 31, 1233–1242 [DOI] [PubMed] [Google Scholar]
- 9.Telenti A. et al. (1993) Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. The Lancet 341, 647–651 [DOI] [PubMed] [Google Scholar]
- 10.Alifano P. et al. (2015) Rifampicin-resistance, rpoB polymorphism and RNA polymerase genetic engineering. J. Biotechnol. 202, 60–77 [DOI] [PubMed] [Google Scholar]
- 11.Bergval IL et al. (2007) Specific mutations in the Mycobacterium tuberculosis rpoB gene are associated with increased dnaE2 expression. FEMS Microbiol. Lett. 275, 338–343 [DOI] [PubMed] [Google Scholar]
- 12.Chen L. et al. (2018) Genome-wide DNA methylation and transcriptome changes in Mycobacterium tuberculosis with rifampicin and isoniazid resistance. Int. J. Clin. Exp. Pathol. 11, 3036–3045 [PMC free article] [PubMed] [Google Scholar]
- 13.Denamur E. et al. (2005) Intermediate Mutation Frequencies Favor Evolution of Multidrug Resistance in Escherichia coli. Genetics 171, 825–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y. et al. (2021) DNA adenine methylation is involved in persister formation in E. coli. Microbiol. Res. 246, 126709 [DOI] [PubMed] [Google Scholar]
- 15.McCune RM et al. (1966) MICROBIAL PERSISTENCE. J. Exp. Med. 123, 445–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yari S. et al. (2016) Proteome-scale MDR-TB-antibody responses for identification of putative biomarkers for the diagnosis of drug-resistant Mycobacterium tuberculosis. Int. J. Mycobacteriology 5, S134–S135 [DOI] [PubMed] [Google Scholar]
- 17.Hadizadeh Tasbiti A. et al. (2021) Comparing mRNA expression and protein abundance in MDR Mycobacterium tuberculosis: Novel protein candidates, Rv0443, Rv0379 and Rv0147 as TB potential diagnostic or therapeutic targets. Biotechnol. Rep. 30, e00641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh A. et al. (2015) Comparative proteomic analysis of sequential isolates of Mycobacterium tuberculosis from a patient with pulmonary tuberculosis turning from drug sensitive to multidrug resistant. Indian J. Med. Res. 141, 27–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q. et al. (2020) PE/PPE proteins mediate nutrient transport across the outer membrane of Mycobacterium tuberculosis. Science 367, 1147–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai RPJ et al. (2021) Transcriptomic Characterization of Tuberculous Sputum Reveals a Host Warburg Effect and Microbial Cholesterol Catabolism. mBio 12, e01766–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandra P. et al. (2022) Macrophage global metabolomics identifies cholestenone as host/pathogen cometabolite present in human Mycobacterium tuberculosis infection. J. Clin. Invest. 132, e152509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koh E-I et al. (2022) Chemical–genetic interaction mapping links carbon metabolism and cell wall structure to tuberculosis drug efficacy. Proc. Natl. Acad. Sci. U. S. A. 119, e2201632119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jankute M. et al. (2015) Assembly of the Mycobacterial Cell Wall. Annu. Rev. Microbiol. 69, 405–423 [DOI] [PubMed] [Google Scholar]
- 24.Chiaradia L. et al. (2017) Dissecting the mycobacterial cell envelope and defining the composition of the native mycomembrane. Sci. Rep. 7, 12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quigley J. et al. (2017) The Cell Wall Lipid PDIM Contributes to Phagosomal Escape and Host Cell Exit of Mycobacterium tuberculosis. mBio 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bisson GP et al. (2012) Upregulation of the Phthiocerol Dimycocerosate Biosynthetic Pathway by Rifampin-Resistant, rpoB Mutant Mycobacterium tuberculosis. J. Bacteriol. 194, 6441–6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campodónico VL et al. (2018) Altered Mycobacterium tuberculosis Cell Wall Metabolism and Physiology Associated With RpoB Mutation H526D. Front. Microbiol. 9, 494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L. et al. (2023) Metabolic Profiles of Clinical Isolates of Drug-Susceptible and Multidrug-Resistant Mycobacterium tuberculosis: A Metabolomics-Based Study. Infect. Drug Resist. 16, 2667–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cloete R. et al. (2021) In silico repurposing of a Novobiocin derivative for activity against latency associated Mycobacterium tuberculosis drug target nicotinate-nucleotide adenylyl transferase (Rv2421c). PLoS ONE 16, e0259348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi K-X et al. (2019) Housecleaning of pyrimidine nucleotide pool coordinates metabolic adaptation of nongrowing Mycobacterium tuberculosis. Emerg. Microbes Infect. 8, 40–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tiwari S. et al. (2018) Arginine-deprivation–induced oxidative damage sterilizes Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 115, 9779–9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yelamanchi SD et al. (2022) Rifampicin-Mediated Metabolic Changes in Mycobacterium tuberculosis. Metabolites 12, 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilson MK et al. (2016) BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 44, D1045–D1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esaulova E. et al. (2021) The immune landscape in tuberculosis reveals populations linked to disease and latency. Cell Host Microbe 29, 165–178.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmed M. et al. (2020) Immune correlates of tuberculosis disease and risk translate across species. Sci. Transl. Med. 12, eaay0233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith CM et al. (2022) Host-pathogen genetic interactions underlie tuberculosis susceptibility in genetically diverse mice. eLife 11, e74419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casanova J-L and Abel L. (2021) Lethal infectious diseases as inborn errors of immunity: toward a synthesis of the germ and genetic theories. Annu. Rev. Pathol. 16, 23–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mogues T. et al. (2001) The Relative Importance of T Cell Subsets in Immunity and Immunopathology of Airborne Mycobacterium tuberculosis Infection in Mice. J. Exp. Med. 193, 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Green AM et al. (2013) IFN-γ from CD4 T cells is essential for host survival and enhances CD8 T cell function during Mycobacterium tuberculosis infection. J. Immunol. Baltim. Md 1950 190, 270–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosain J. et al. (2023) Human IRF1 governs macrophagic IFN-γ immunity to mycobacteria. Cell 186, 621–645.e33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kerner G. et al. (2020) Inherited human IFN-γ deficiency underlies mycobacterial disease. J. Clin. Invest. 130, 3158–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khader SA et al. (2007) IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat. Immunol. 8, 369–377 [DOI] [PubMed] [Google Scholar]
- 43.Gopal R. et al. (2014) Unexpected Role for IL-17 in Protective Immunity against Hypervirulent Mycobacterium tuberculosis HN878 Infection. PLoS Pathog. 10, e1004099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Treerat P. et al. (2017) Novel role for IL-22 in protection during chronic Mycobacterium tuberculosis HN878 infection. Mucosal Immunol. 10, 1069–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Einarsdottir T. et al. (2009) Cytotoxicity and Secretion of Gamma Interferon Are Carried Out by Distinct CD8 T Cells during Mycobacterium tuberculosis Infection. Infect. Immun. 77, 4621–4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho S. et al. (2000) Antimicrobial activity of MHC class I-restricted CD8+ T cells in human tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 97, 12210–12215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choreño Parra JA et al. (2017) Memory of Natural Killer Cells: A New Chance against Mycobacterium tuberculosis? Front. Immunol. 8, 967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peyron P. et al. (2008) Foamy Macrophages from Tuberculous Patients’ Granulomas Constitute a Nutrient-Rich Reservoir for M. tuberculosis Persistence. PLoS Pathog. 4, e1000204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mattila JT et al. (2013) Microenvironments in tuberculous granulomas are delineated by distinct populations of macrophage subsets and expression of nitric oxide synthase and arginase isoforms. J. Immunol. Baltim. Md 1950 191, 773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonecini-Almeida MG et al. (1998) Induction of In Vitro Human Macrophage Anti-Mycobacterium tuberculosis Activity: Requirement for IFN-γ and Primed Lymphocytes. J. Immunol. 160, 4490–4499 [PubMed] [Google Scholar]
- 51.Yildiz P. et al. (2001) Natural Killer Cell Activity in Multidrug-Resistant Pulmonary Tuberculosis. Respiration 68, 590–594 [DOI] [PubMed] [Google Scholar]
- 52.Kiran B. et al. (2010) Can immune parameters be used as predictors to distinguish between pulmonary multidrug-resistant and drug-sensitive tuberculosis? Arch. Med. Sci. AMS 6, 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pinheiro RO et al. (2013) Different immunosuppressive mechanisms in multi-drug-resistant tuberculosis and non-tuberculous mycobacteria patients. Clin. Exp. Immunol. 171, 210–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fortes A. et al. (2005) Detection of in vitro interferon-γ and serum tumour necrosis factor-α in multidrug-resistant tuberculosis patients. Clin. Exp. Immunol. 141, 541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Basile JI et al. (2011) Outbreaks of Mycobacterium Tuberculosis MDR Strains Induce High IL-17 T-Cell Response in Patients With MDR Tuberculosis That Is Closely Associated With High Antigen Load. J. Infect. Dis. 204, 1054–1064 [DOI] [PubMed] [Google Scholar]
- 56.Sabiiti W. et al. (2020) Tuberculosis bacillary load, an early marker of disease severity: the utility of tuberculosis Molecular Bacterial Load Assay. Thorax 75, 606–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Imperiale BR et al. (2021) Th22 response induced by Mycobacterium tuberculosis strains is closely related to severity of pulmonary lesions and bacillary load in patients with multi-drug-resistant tuberculosis. Clin. Exp. Immunol. 203, 267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geffner L. et al. (2014) Mycobacterium tuberculosis Multidrug Resistant Strain M Induces an Altered Activation of Cytotoxic CD8+ T Cells. PLOS ONE 9, e97837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sada-Ovalle I. et al. (2009) In vitro cytotoxicity of CD8+ T cells in multi-drug-resistant tuberculosis. A preliminary report. Respirology 14, 574–578 [DOI] [PubMed] [Google Scholar]
- 60.Fan R. et al. (2016) Impaired NK cells’ activity and increased numbers of CD4 + CD25+ regulatory T cells in multidrug-resistant Mycobacterium tuberculosis patients. Tuberculosis 98, 13–20 [DOI] [PubMed] [Google Scholar]
- 61.Ratcliffe LT et al. (1994) Reduced NK activity correlates with active disease in HIV- patients with multidrug-resistant pulmonary tuberculosis. Clin. Exp. Immunol. 97, 373–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cho HJ et al. (2020) Different macrophage polarization between drug-susceptible and multidrug-resistant pulmonary tuberculosis. BMC Infect. Dis. 20, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khan TA et al. (2016) Interferon-Gamma Improves Macrophages Function against M. tuberculosis in Multidrug-Resistant Tuberculosis Patients. Chemother. Res. Pract. 2016, 7295390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharma S. et al. (2004) Mycobacterium tuberculosis induces high production of nitric oxide in coordination with production of tumour necrosis factor-α in patients with fresh active tuberculosis but not in MDR tuberculosis. Immunol. Cell Biol. 82, 377–382 [DOI] [PubMed] [Google Scholar]
- 65.Prombutara P. et al. Host cell transcriptomic response to the multidrug-resistant Mycobacterium tuberculosis clonal outbreak Beijing strain reveals its pathogenic features. Virulence 13, 1810–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ahsan F. et al. (2018) IL-36/LXR axis modulates cholesterol metabolism and immune defense to Mycobacterium tuberculosis. Sci. Rep. 8, 1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.DE LA BARRERA S. et al. (2004) IL-10 down-regulates costimulatory molecules on Mycobacterium tuberculosis-pulsed macrophages and impairs the lytic activity of CD4 and CD8 CTL in tuberculosis patients. Clin. Exp. Immunol. 138, 128–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen X. et al. (2007) CD4+CD25+FoxP3+ regulatory T cells suppress Mycobacterium tuberculosis immunity in patients with active disease. Clin. Immunol. 123, 50–59 [DOI] [PubMed] [Google Scholar]
- 69.Liu W. et al. (2006) CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J. Exp. Med. 203, 1701–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stringari LL et al. (2021) Increase of CD4+CD25highFoxP3+ cells impairs in vitro human microbicidal activity against Mycobacterium tuberculosis during latent and acute pulmonary tuberculosis. PLoS Negl. Trop. Dis. 15, e0009605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li N. et al. (2015) Enrichment of regulatory T-cells in blood of patients with multidrug-resistant tuberculosis. Int. J. Tuberc. Lung Dis. Off. J. Int. Union Tuberc. Lung Dis. 19, 1230–1238 [DOI] [PubMed] [Google Scholar]
- 72.Shams Shahemabadi A. et al. (2010) IL-10 and IL-12 production in response to Mycobacterium tuberculosis total lipid antigens in multidrug resistant tuberculosis. Iran. J. Immunol. IJI 7, 57–63 [PubMed] [Google Scholar]
- 73.Park H-S et al. (2021) Mycobacterium tuberculosis Rv2145c Promotes Intracellular Survival by STAT3 and IL-10 Receptor Signaling. Front. Immunol. 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tan Q. et al. (2017) Clinical and Immunological Effects of rhIL-2 Therapy in Eastern Chinese Patients with Multidrug-resistant Tuberculosis. Sci. Rep. 7, 17854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Clark S. et al. (2015) Animal Models of Tuberculosis: Guinea Pigs. Cold Spring Harb. Perspect. Med. 5, a018572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Orme IM and Ordway DJ (2016) Mouse and Guinea Pig Models of Tuberculosis. Microbiol. Spectr. 4, 4.4.08 [DOI] [PubMed] [Google Scholar]
- 77.Palanisamy GS et al. (2009) Clinical strains of Mycobacterium tuberculosis display a wide range of virulence in guinea pigs. Tuberculosis 89, 203–209 [DOI] [PubMed] [Google Scholar]
- 78.Dharmadhikari AS et al. (2011) Natural infection of guinea pigs exposed to patients with highly drug-resistant tuberculosis. Tuberc. Edinb. Scotl. 91, 329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cooper AM (2015) Mouse Model of Tuberculosis. Cold Spring Harb. Perspect. Med. 5, a018556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ordway DJ et al. (1995) Drug-resistant strains of Mycobacterium tuberculosis exhibit a range of virulence for mice. Infect. Immun. 63, 741–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dave S. et al. (2009) Comparative growth pattern of multi drug resistance versus susceptible isolates of Mycobacterium tuberculosis in mice lungs. Indian J. Med. Res. 130, 58–62 [PubMed] [Google Scholar]
- 82.Smith KLJ et al. (2014) Reduced Virulence of an Extensively Drug-Resistant Outbreak Strain of Mycobacterium tuberculosis in a Murine Model. PLOS ONE 9, e94953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olson GS et al. (2021) Type I interferon decreases macrophage energy metabolism during mycobacterial infection. Cell Rep. 35, 109195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Patel Y. et al. Mutations in rpoB That Confer Rifampicin Resistance Can Alter Levels of Peptidoglycan Precursors and Affect β-Lactam Susceptibility. mBio 14, e03168–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morishita T. and Yura T. (1976) Altered nutritional requirements associated with mutations affecting the structures of ribonucleic acid polymerase in Lactobacillus casei. J. Bacteriol. 125, 416–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sonenshein AL et al. (1974) Isolation and Characterization of Rifampin-Resistant and Streptolydigin-Resistant Mutants of Bacillus subtilis with Altered Sporulation Properties. J. Bacteriol. 120, 253–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gao W. et al. (2013) The RpoB H481Y Rifampicin Resistance Mutation and an Active Stringent Response Reduce Virulence and Increase Resistance to Innate Immune Responses in Staphylococcus aureus. J. Infect. Dis. 207, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Manca C. et al. (2005) Hypervirulent M. tuberculosis W/Beijing Strains Upregulate Type I IFNs and Increase Expression of Negative Regulators of the Jak-Stat Pathway. J. Interferon Cytokine Res. 25, 694–701 [DOI] [PubMed] [Google Scholar]
- 89.Kremer K. et al. (2004) Definition of the Beijing/W Lineage of Mycobacterium tuberculosis on the Basis of Genetic Markers. J. Clin. Microbiol. 42, 4040–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wiens KE and Ernst JD (2016) The Mechanism for Type I Interferon Induction by Mycobacterium tuberculosis is Bacterial Strain-Dependent. PLOS Pathog. 12, e1005809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Manca C. et al. (2001) Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-α/β. Proc. Natl. Acad. Sci. U. S. A. 98, 5752–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Napier G. et al. (2020) Robust barcoding and identification of Mycobacterium tuberculosis lineages for epidemiological and clinical studies. Genome Med. 12, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shanmugam SK et al. (2020) Mycobacterium tuberculosis Lineages Associated with Mutations and Drug Resistance in Isolates from India. Microbiol. Spectr. 10, e01594–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aylan B. et al. (2023) ATG7 and ATG14 restrict cytosolic and phagosomal Mycobacterium tuberculosis replication in human macrophages. Nat. Microbiol. 8, 803–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Golovkine GR et al. (2023) Autophagy restricts Mycobacterium tuberculosis during acute infection in mice. Nat. Microbiol. 8, 819–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peláez Coyotl EA et al. (2020) Antimicrobial Peptide against Mycobacterium Tuberculosis That Activates Autophagy Is an Effective Treatment for Tuberculosis. Pharmaceutics 12, 1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Singh DK et al. (2023) Co-treatment with Clofazimine and Rapamycin eliminates drug-resistant tuberculosis by inducing polyfunctional central memory T cell responses. J. Infect. Dis. DOI: 10.1093/infdis/jiad214 [DOI] [PubMed] [Google Scholar]