Abstract
The poly(rC) binding protein (PCBP) is a cellular protein required for poliovirus replication. PCBP specifically interacts with two domains of the poliovirus 5′ untranslated region (5′UTR), the 5′ cloverleaf structure, and the stem-loop IV of the internal ribosome entry site (IRES). Using footprinting analysis and site-directed mutagenesis, we have mapped the RNA binding site for this cellular protein within the stem-loop IV domain. A C-rich sequence in a loop at the top of this large domain is required for PCBP binding and is crucial for viral translation. PCBP binds to stem-loop IV RNA with six-times-higher affinity than to the 5′ cloverleaf structure. However, the binding of the viral protein 3CD (precursor of the viral protease 3C and the viral polymerase 3D) to the cloverleaf RNA dramatically increases the affinity of PCBP for this RNA element. The viral protein 3CD binds to the cloverleaf RNA but does not interact directly with stem-loop IV nor with other RNA elements of the viral IRES. Our results indicate that the interactions of PCBP with the poliovirus 5′UTR are modulated by the viral protein 3CD.
The highly structured 5′ untranslated region (5′UTR) of picornaviruses plays an important role in the regulation of both viral translation and RNA replication. The regulatory function of this region is mediated by its interaction with cellular and viral proteins (for a review, see references 1, 5, and 12). Using computer analysis and genetic and biochemical tests, the 5′UTR of poliovirus (the prototype member of the picornavirus family) can be divided into six well-defined RNA domains (stem-loops I to VI) (29, 30, 34).
Stem-loop I, which folds into a cloverleaf-like structure (30), is essential for viral RNA synthesis (2, 3, 17, 31, 37). This element forms a ternary RNP complex with the cellular poly(rC) binding protein (PCBP; also known as hnRNP E or α-CP) and the uncleaved precursor of the viral protease-polymerase, 3CD (15, 26). Mutations that disrupt complex formation, either within the cloverleaf RNA or within 3CD, impair viral RNA synthesis (3, 4, 31). The mechanism by which the ternary complex participates in RNA replication is poorly understood, but it was suggested that it catalyzes the initiation of positive strand RNA synthesis in trans (2). More recent evidence suggests that the ternary complex has a bifunctional role, participating in both viral translation and RNA replication (14, 33). The binding of PCBP to stem-loop B of the cloverleaf structure enhances viral translation 10-fold, while binding of 3CD decreases translation and promotes negative-strand RNA synthesis (14).
Viral translation is directed by an RNA element located within the 5′UTR (stem-loops II to VI) known as the internal ribosomal entry site (IRES). This element allows ribosomes to enter the RNA without scanning from the 5′ end (21, 27, 36). The mechanism by which the translation apparatus recognizes IRES sequences is still unknown, but it has been proposed that several canonical initiation factors, as well as other cellular proteins, participate in this process (24, 28). So far, four noncanonical factors that bind to the poliovirus IRES have been identified: the polypyrimidine tract binding protein (18), the La autoantigen (23), the cellular protein PCBP (7), and the protein encoded by the gene present upstream of N-ras, UNR (20).
PCBP was first identified as a component of the α-complex of the human α-globin mRNA, which greatly increases mRNA stability (22). More-recent evidence implicates PCBP in controlling the expression of numerous cellular and viral RNAs (for a review, see reference 25). It has been shown that PCBP specifically represses the expression of the late gene L2 in human papillomavirus (10) and enhances hepatitis A capindependent translation (16). However, the molecular mechanism by which PCBP interacts with the translation apparatus remains undefined. In poliovirus, PCBP specifically interacts with two stem-loops of the 5′UTR: the cloverleaf structure and stem-loop IV (7, 8, 15, 26). A number of observations suggest that PCBP is required for poliovirus translation. In fact, alteration of PCBP binding sites, depletion of PCBP from HeLa cell extracts, or microinjection of anti-PCBP antibodies into Xenopus oocytes inhibits poliovirus translation (8, 15).
In the present study, we further analyzed the interactions of 3CD and PCBP with the poliovirus 5′UTR and the interplay between the RNP complexes formed during the viral life cycle. Using footprinting analysis and mobility shift assays we found that PCBP forms RNP complexes with different affinities depending on the presence of the viral protein 3CD. These findings may further our understanding of the molecular mechanism by which PCBP and 3CD participate in viral translation and RNA replication.
MATERIALS AND METHODS
Footprinting analysis.
RNase treatment, primer extension, and gel electrophoresis were performed as previously described (6). Poliovirus 5′UTR RNA was synthesized in vitro with T7 polymerase, and unincorporated nucleoside triphosphates were removed by use of an RNeasy column (Qiagen). Binding reactions were performed for 10 min at room temperature with 1.5 μg of purified recombinant PCBP, 3 μg of partially purified 3CD, or 2 μg of bovine serum albumin in a final volume of 40 μl. RNase T1 or RNase T2 (Sigma) was added to the binding reaction and incubated for 15 min at room temperature. After RNase treatment, RNA was phenol extracted, ethanol precipitated, used as template for primer extension, and analyzed in 6% polyacrylamide–6 M urea sequencing gels. For the analysis of stem-loop IV, primer 1 (TCACAACTAGCGTCCCATGGCGTTAGCCATAGGTAGGCCG) was used. The recombinant proteins PCBP and 3CD were obtained as described below.
Production and purification of PCBP and 3CD.
PCBP2 was expressed in Escherichia coli as a maltose-binding protein (MBP) fusion, MBP-PCBP, as previously described (15). The fusion protein was purified by affinity chromatography with amylose resin and digested with factor X to cleave PCBP from MBP. The recombinant 3CD protein used throughout this study contained a mutation in the catalytic site of 3C (H40E). This mutation completely abolishes 3CD proteolytic activity (2). To produce recombinant 3CD, T7 expression plasmids were transformed into the E. coli BL21(DE3), which contained the T7 polymerase gene under the control of the lacUV5 promoter. An overnight culture of freshly transformed bacteria was diluted 1/10, and the culture was grown to an optical density at 550 nm of 0.5 before IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.4 mM. The induction was carried out for an additional 2 h. Cells were harvested, washed once with phosphate-buffered saline (PBS), and resuspended in lysis buffer (10 mM HEPES [pH 7.9], 20 mM KCl, 25 mM EDTA, 5 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, 1% Triton X-100). The suspension was frozen and thawed three times, sonicated for about 30 s to reduce viscosity, and centrifuged at 150,000 × g for 15 min to remove debris. Glycerol was added to 20% final concentration, and the supernatant was stored at −70°C.
RNA binding assays.
RNA binding reactions and electrophoretic mobility shift assays were performed as previously described (15). Briefly, uniformly 32P-labeled RNA probes were generated by in vitro transcription by using T7 RNA polymerase. The cloverleaf probe corresponded to the first 108 nucleotides, and the stem-loop IV probe corresponded to a fragment from nucleotides 234 to 459 of the poliovirus genome. The mutated stem-loop IV RNAs (mutants SL IV-23, SL IV-298, and SL IV-332) were generated by use of overlapping PCR, and the product DNA was directly used as a template for in vitro transcription by using T7 polymerase. Unlabeled RNAs used as competitors corresponding to stem-loop I (nucleotides 1 to 108), stem-loop II-III (nucleotides 90 to 240), stem-loop IV (nucleotides 234 to 459), stem-loop V (nucleotides 440 to 578), and stem-loop VI (nucleotides 556 to 638) were synthesized by in vitro transcription with DNA templates obtained by PCR amplification.
Dissociation constants were determined by quantifying the fraction of RNA bound (Θ) with a PhosphorImager (Molecular Dynamics). The data were fitted by using nonlinear-least-squares analysis as a function of total PCBP concentration as follows: Θ = [PCBP]/[PCBP] Kd. For PCBP–stem-loop IV RNA, which gave two band shifts, Kd values were calculated by treating bound RNA as a single species equal to the sum of both bands.
Translation in HeLa cells.
To test the translation efficiencies of wild-type and mutant Polio-Luc RNAs, 100-mm dishes containing ∼3 × 106 HeLa cells were trypsinized and transfected by standard electroporation procedures by using 20 μg of in vitro-transcribed RNA per plate. Cells were incubated at 37°C, and time point data were taken every 30 min. The cells were washed with PBS, scraped from the plates, and lysed in 200 μl of lysis buffer (Promega). Luciferase activity was measured in 10 μl of extract by using a luciferase system as recommended by the manufacturer (Promega) and quantified by using an Optocomp I luminometer.
3CD binding to biotinylated RNA.
The 5′UTR stem-loops I to VI, the IRES stem-loops II to VI, and the green fluorescent protein (GFP) mRNA were transcribed in vitro in the presence of limiting concentrations of biotin-16-UTP to incorporate two to three biotinylated nucleotides per molecule of RNA. Then, 30 μg of this RNA was incubated with 40 μl of streptoavidin beads and washed five times with PBS. The RNA present in the washes was used to estimate the efficiency of binding, which was 30 to 40% of the original amount of RNA added. Then, 4 μg of partially purified recombinant 3CD protein (2) was added together with 50 μl of the uninfected S10 HeLa fraction. The mix was incubated for 1 h on ice and, after centrifugation, the beads were washed four times with 500 μl of wash buffer (50 mM Tris-HCl, pH 7.4; 100 mM KCl; 0.2% NP-40). After the washes, the beads were resuspended in sample buffer and analyzed by Western blotting with anti-3CD antibodies.
RESULTS
Mapping the PCBP binding site within stem-loop IV of the poliovirus genome.
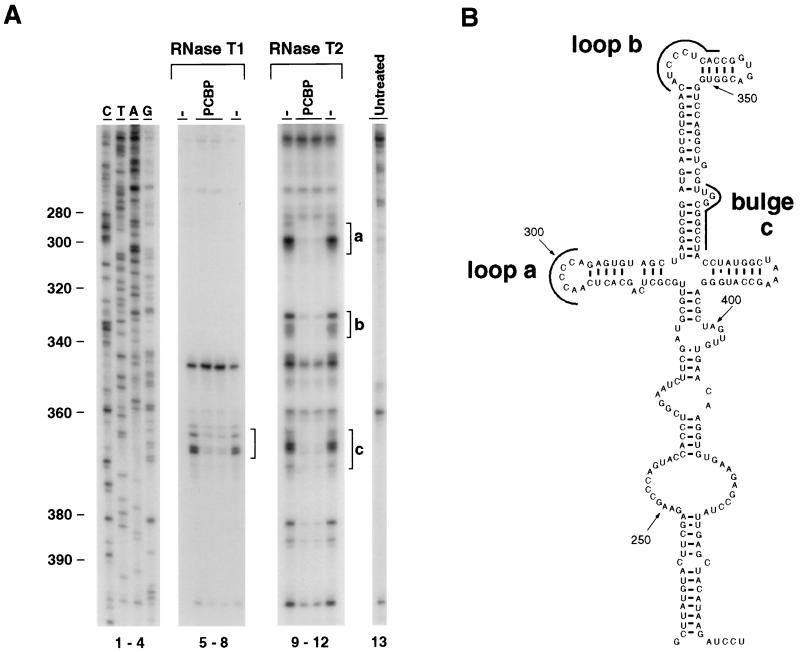
The multiple interactions of the cellular protein PCBP with the 5′UTR of the poliovirus genome are essential for viral translation and RNA replication (8, 14, 15, 26). However, many structural details and the precise mechanism by which PCBP participates in these processes remain obscure. To further define the binding site of PCBP within the poliovirus IRES, we performed RNA footprinting analysis. The complete 5′UTR of the viral genome was treated with RNase T1 or T2 in the presence or absence of recombinant PCBP. The viral RNA was then analyzed by primer extension by using primers to inspect the stem-loop IV sequence. Several RNase-hypersensitive regions were detected within the core of the IRES, corresponding to single-stranded regions of the RNA (Fig. 1A, compare lanes 5, 8, 9, and 12 with lane 13). The presence of PCBP protects nucleotides 364 to 373 from both RNase T1 and T2 digestion and nucleotides 296 to 301, 331 to 339, and 382 to 386 from RNase T2 digestion (Fig. 1A, lanes 5 to 12). According to the predicted secondary structure of stem-loop IV, the cellular protein PCBP specifically protects several regions located at the top of this large domain (Fig. 1B, loop a, loop b, and bulge c).
FIG. 1.
PCBP-RNA interactions within stem-loop IV of the poliovirus IRES. (A) Footprinting analysis reveals that PCBP protects several regions of the large stem-loop IV from RNase digestion. The viral 5′UTR was treated with RNase T1 (lanes 5 to 8) or RNase T2 (lanes 9 to 12). Lane 13 corresponds to untreated RNA. RNase was added in the presence of bovine serum albumin (−) or PCBP protein, as indicated at the top of each lane. Primer extension was performed by using a primer complementary to nucleotides 368 to 408. Lanes labeled C, T, A, and G correspond to dideoxy sequencing lanes. Brackets indicate PCBP protected regions (a, b, and c). The numbers on the left indicate the nucleotide position of the viral genome in poliovirus type 1. (B) Predicted secondary structure of stem-loop IV of the poliovirus type 1 genome. PCBP protected regions as determined by footprinting analysis in panel A are indicated by black lines (loop a, loop b, and bulge c).
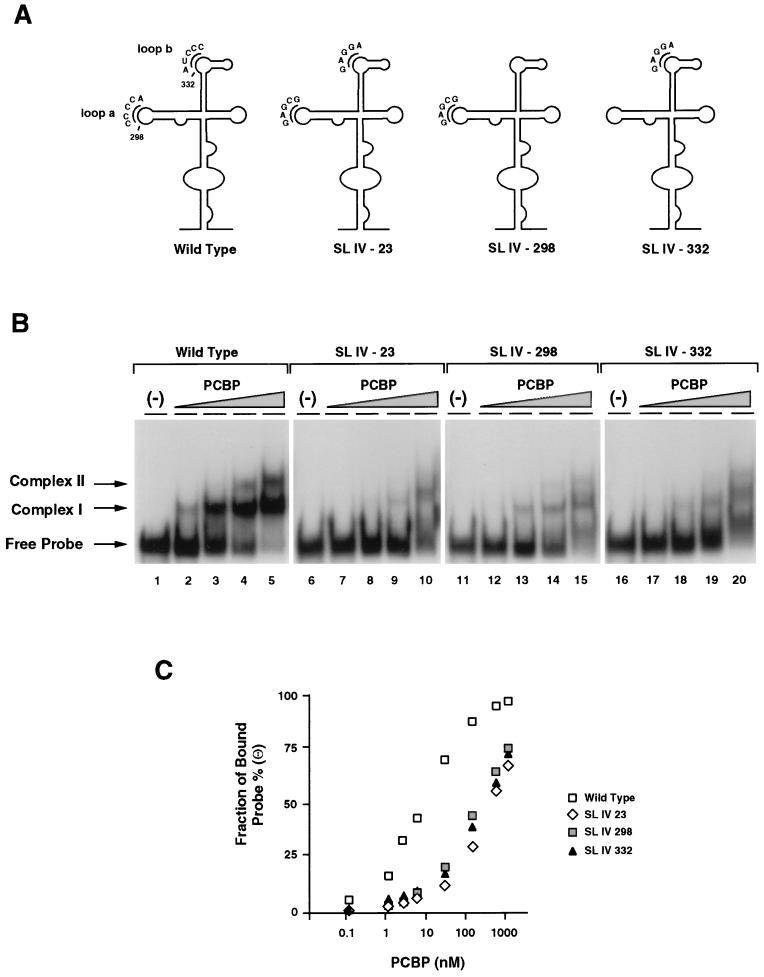
Interestingly, loop a and loop b contain single-stranded C-rich sequences which resemble the consensus RNA-binding site for PCBP found in several cellular mRNAs (19). To analyze whether these C-rich sequences are directly involved in PCBP binding to stem-loop IV, we performed site-directed mutagenesis and studied the ability of wild-type and mutated RNAs to bind PCBP in vitro. Three stem-loop IV mutants were constructed: one in which the sequence CCCCA at position 298 was replaced by GAGCG, another in which the sequence AUCCC at position 332 was replaced by GAGGA, and a third mutant combining the substitutions at positions 298 and 332 (Fig. 2A, SL IV-298, SL IV-332, and SL IV-23, respectively). The mutations introduced in loop a and loop b did not alter the predicted secondary structure observed for stem-loop IV wild type. RNA probes corresponding to wild type and the three stem-loop IV mutants were used in electrophoretic mobility shift assays. We employed PCBP purified from HeLa cell extracts because this protein binds RNA with six- to eightfold- higher affinity than the recombinant protein (data not shown). The concentration of PCBP in HeLa cells (100 nM) was estimated by comparing cell extracts with dilutions of purified recombinant PCBP by Western blotting (data not shown).
FIG. 2.
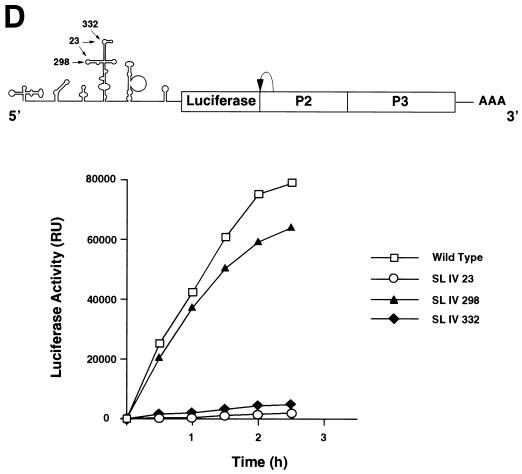
Effect of mutations within stem-loop IV on PCBP binding and viral translation. (A) Position and sequence of mutations within the predicted secondary structure of stem-loop IV. The C-rich loop a and loop b are indicated with their wild-type and mutated sequences: SL IV-23, SL IV-298, and SL IV-332. (B) RNA mobility shift analysis showing the effect of mutations within stem-loop IV probe on PCBP binding. RNA mobility shift experiments were performed with four different probes: wild-type stem-loop IV (lanes 1 to 5), SL IV-23 (lanes 6 to 10), SL IV-298 (lanes 11 to 15), and SL IV-332 (lanes 16 to 20). Each probe was incubated with buffer (−) or increasing concentrations of cellular PCBP (0.2 ng to 0.1 μg), as indicated at the top of the panel. Position of specific complexes I and II and the stem-loop IV probe (Free Probe) are indicated by arrows. (C) Binding affinity of PCBP to wild-type and mutated stem-loop IV RNAs. The fraction of radiolabeled probe bound (Θ) determined by mobility shift assays is plotted against PCBP concentration. (D) Translation efficiencies of wild type and stem-loop IV mutants in HeLa cells. At the top, a schematic representation of the poliovirus replicon carrying the luciferase reporter gene in place of the capsid proteins. The arrow indicates a recognition site for cleavage by the viral 2A protease. At the bottom, the translation of wild type and stem-loop IV mutants (SL IV-23, SL IV-298, and SL IV-332) is measured as luciferase activity and plotted as a function of the time after RNA transfections into HeLa cells.
As previously described (7, 15), PCBP interacts with the stem-loop IV RNA forming two discrete RNP complexes, complex I and complex II (Fig. 2B). None of the stem-loop IV mutants formed stable complexes with the cellular protein (Fig. 2B, lanes 6 to 20). The affinity of PCBP for the mutant SL IV-23 was about 60-fold lower than for the wild-type RNA, and the affinities for the mutants SL IV-298 and -332 were about 35- and 40-fold lower than for wild type, respectively (Fig. 2C). These results indicate that the C-rich sequences in loop a and loop b of the large stem-loop IV of the viral IRES are important determinants for binding of PCBP in vitro.
In agreement with our observations, a previous report indicated that an insertion of three nucleotides at position 325, just upstream of loop b of stem-loop IV, abolished binding of PCBP in vitro (7), presumably by altering the structure at the base of loop b. This interaction appears to be critical for replication, because a poliovirus RNA carrying the same mutation yields a nonviable virus with a major defect in translation (13, 35). Having defined more precisely the PCBP binding site, we analyzed whether the sequences of loop a and loop b are functionally important for viral translation. We introduced the three mutations described above (SL IV-298, SL IV-332, and SL IV-23) into the genome of a poliovirus subgenomic replicon in which the capsid coding region was replaced by the luciferase gene (Fig. 2D). Translation efficiencies were determined by measuring luciferase activity as a function of the time after RNA transfection into HeLa cells. Translation of the mutants SL IV-23 and SL IV-332 was reduced 100- to 1,000-fold compared with the wild-type RNA (Fig. 2D). In contrast, translation of the mutant SL IV-298 was close to wild-type levels, suggesting that not all the determinants for complex formation observed in mobility shift assays are functionally important for viral translation.
In summary, we found that a highly conserved C-rich sequence (loop b) present in stem-loop IV of the poliovirus IRES is an important determinant for PCBP binding and is essential for viral translation.
The viral protein 3CD greatly enhances the binding affinity of PCBP to the cloverleaf RNA, leading to a dissociation of PCBP from stem-loop IV.
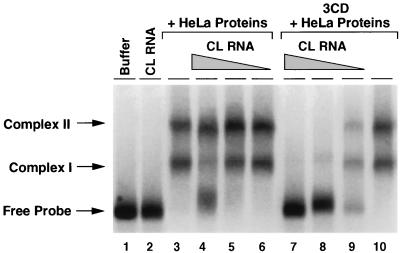
PCBP specifically interacts with two domains of the poliovirus 5′UTR: the cloverleaf and stem-loop IV. Since these two interactions appear to be important for viral translation and previous evidence suggested that PCBP dimerizes (15), we investigated whether this protein alone or together with other cellular proteins can bridge the two RNA domains. First, we determined if the cloverleaf RNA directly interacts with stem-loop IV RNA. Using gel shift analysis, we observed that an excess of unlabeled cloverleaf RNA did not modify the mobility of stem-loop IV in a native gel (Fig. 3, compare lanes 1 and 2), suggesting that these RNAs do not interact with each other in the absence of proteins. In the presence of HeLa cell proteins, two specific complexes identical to the ones observed with recombinant PCBP protein were formed (15) (Fig. 3C, lane 3). Addition of cloverleaf RNA did not alter the mobility of these complexes, indicating that even in the presence of cellular proteins the cloverleaf and stem-loop IV RNA structures do not interact with each other. Interestingly, the cloverleaf RNA was a poor competitor for the interaction of PCBP with stem-loop IV: 100-fold molar excess was required to partially compete with complex I, whereas complex II was not significantly reduced (Fig. 3, lanes 4 to 6). However, while 3CD by itself did not modify the PCBP–stem-loop IV interaction (Fig. 3, lane 10), addition of 3CD together with cloverleaf RNA effectively reduced the binding of PCBP to stem-loop IV (Fig. 3, lanes 7 to 9). In these experiments we used a bacterially expressed 3CD carrying a mutation at His 40 (H40E) within the catalytic site of the protease 3C. This mutation abrogates proteolytic activity and prevents cleavage of 3CD (2). When similar experiments were performed using mutants either in loop D of the cloverleaf or in the RNA binding domain of 3CD protein, the competition for PCBP was not observed (data not shown). These findings suggest that the binding of 3CD to the cloverleaf promotes dissociation of PCBP from stem-loop IV.
FIG. 3.
PCBP dissociates from stem-loop IV in the presence of cloverleaf RNA and 3CD. Uniformly labeled stem-loop IV (5 ng) was incubated with buffer, unlabeled cloverleaf RNA (500 ng, lane 2), S10 HeLa proteins (20 μg), and decreasing concentrations of unlabeled cloverleaf (CL) RNA (500, 50, and 5 ng) (lanes 4 to 6 and lanes 7 to 9). Binding reactions were performed in the absence (lanes 3 to 6) or in the presence (lanes 7 to 10) of 0.5 μg of recombinant 3CD. The electrophoretic mobility of the two RNP complexes formed between PCBP and stem-loop IV (complex I and complex II) and the free stem-loop IV RNA (Free Probe) is indicated on the left.
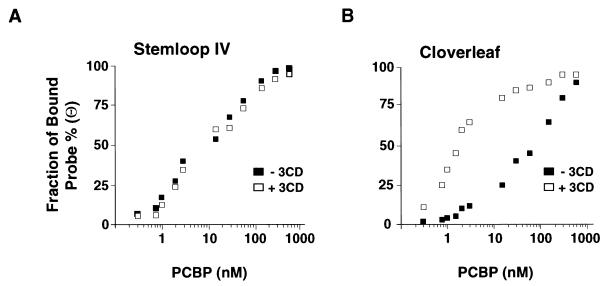
It is possible that the binding of 3CD to the RNA stabilizes the interaction of PCBP with the cloverleaf structure and, under limiting concentrations of PCBP, the cellular protein preferentially binds to the cloverleaf and not to stem-loop IV RNA. To test this hypothesis, we examined the binding affinity of PCBP either in complex with the cloverleaf or stem-loop IV in the presence or absence of the viral protein 3CD. The apparent dissociation constants for PCBP-RNA complexes were estimated from band shift titrations by using radiolabeled stem-loop IV or cloverleaf RNAs and cellular PCBP as described for Fig. 2. The estimated dissociation constant for PCBP–stem-loop IV RNA complex was ∼15 nM, and it was not significantly affected by the presence of 3CD (Fig. 4A). In contrast, the binding affinity of PCBP-cloverleaf increased 2 orders of magnitude in the presence of the viral protein, decreasing the dissociation constant from ∼95 to ∼1 nM (Fig. 4B). These results indicate that 3CD stabilizes the interaction of PCBP with the cloverleaf structure.
FIG. 4.
Effect of 3CD on the affinities of PCBP for stem-loop IV and the cloverleaf. The fraction of radiolabeled probe bound (Θ) for stem-loop IV (A) and cloverleaf (B) is plotted against PCBP concentration. The bands corresponding to free and bound probes in the mobility shift assays were quantified in a PhosphorImager. The experiment was performed in the absence (■) or in the presence (□) of 0.5 μg of recombinant 3CD protein.
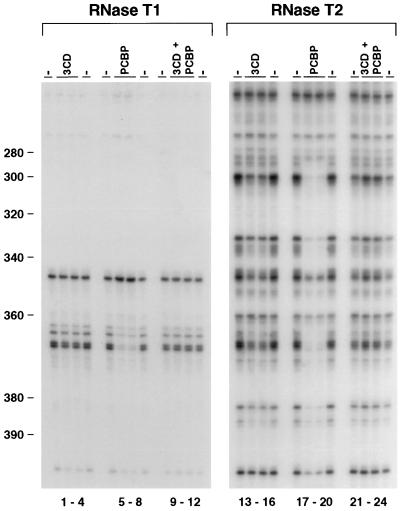
Because the mobility shift analysis in Fig. 3 was performed using the isolated RNA domains I and IV, we wanted to investigate whether 3CD was able to alter the interactions of PCBP with the viral RNA in the context of the complete 5′UTR. Thus, we employed footprinting analysis with the 750- nucleotide RNA in the presence of 3CD and/or PCBP. 3CD by itself did not alter consistently the RNase digestion pattern within the sequence of stem-loop IV (Fig. 5, lanes 1 to 4 and 13 to 16). However, when PCBP and 3CD were added together to the binding reaction, PCBP no longer protected the RNA from the attack of the RNases (Fig. 5, compare lanes 5 to 8 with 9 to 12 and lanes 17 to 20 with 21 to 24). Taken together, these results indicate that binding of 3CD to the cloverleaf RNA greatly increases the affinity of PCBP for this RNA structure and induces dissociation of PCBP from stem-loop IV RNA.
FIG. 5.
Footprinting analysis reveals that 3CD induces dissociation of PCBP from stem-loop IV. The footprinting analysis was performed as described for Fig. 1A. The viral 5′UTR was treated with RNase T1 (lanes 1 to 12) or RNase T2 (lanes 13 to 24). RNase was added in the presence of bovine serum albumin (−), 3CD, PCBP, or 3CD plus PCBP proteins, as indicated on the top of each lane. The numbers on the left indicate the nucleotide position in the poliovirus type 1 genome.
Interaction of 3CD with the poliovirus 5′UTR.
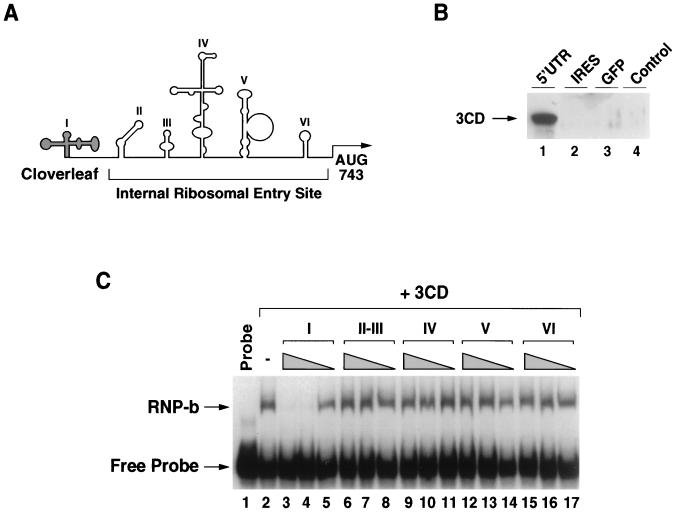
Because 3CD is able to downregulate poliovirus protein synthesis (14) and a number of stem-loops within the IRES play a crucial role in translation initiation, we investigated whether 3CD could interact with stem-loop IV RNA or with other elements of the viral IRES (Fig. 6A). We first determined whether 3CD binds to the complete 5′UTR or the IRES sequence. Immobilized RNA molecules were incubated with recombinant 3CD (see Materials and Methods) in the presence of HeLa cell extracts containing PCBP (which enhance 3CD binding) and the amount of bound 3CD was determined by Western blot analysis. The complete 5′UTR associates very efficiently with 3CD (Fig. 6C, lane 1). In contrast, the IRES (stem-loops II to VI), as well as an unrelated RNA used as a control, were unable to interact with the viral protein, suggesting that 3CD does not bind the viral IRES under our experimental conditions. To confirm this observation, we employed competition mobility shift assays. We used a radiolabeled cloverleaf as probe, bacterially expressed 3CD, and competitor RNAs corresponding to each of the predicted stem-loops I to VI of the viral 5′UTR. The complex between 3CD and the cloverleaf RNA was neither competed with nor modified by addition of stem-loops II, III, IV, V, or VI, even when used at 500-fold excess relative to the cloverleaf probe (Fig. 6C). Thus, these results further confirm that, within the viral 5′UTR, 3CD interacts only with the cloverleaf structure.
FIG. 6.
Interaction of 3CD with the 5′UTR of the poliovirus genome. (A) Schematic representation of the poliovirus 5′UTR. Predicted stem-loop I (cloverleaf) and stem-loops II to VI (IRES) are indicated. AUG at position 743 represents the authentic poliovirus initiation codon. (B) The viral protein 3CD does not interact with the poliovirus IRES. Western blot analysis with anti-3CD antibodies shows the viral protein precipitated by immobilized RNAs. The biotinylated RNA molecules used, 5′UTR, IRES, and GFP (GFP mRNA), are indicated on the top. In lane 4, no RNA was included in the reaction. The electrophoretic mobility of 3CD is indicated on the left. (C) Mobility shift competition experiments. Uniformly labeled cloverleaf RNA (1 ng, 30,000 cpm) was incubated with purified recombinant 3CD (0.5 μg) and with each of the six predicted stem-loops of the poliovirus 5′UTR as unlabeled competitors. Decreasing amounts of competitor RNAs (500, 50, and 1 ng) were used as indicated at the top of the gel with triangles. The mobility of the RNP complex corresponding to cloverleaf-3CD (RNP-b), and the free cloverleaf (Free Probe) is indicated on the left.
DISCUSSION
In this study, we have mapped the RNA binding site of PCBP within stem-loop IV of the poliovirus IRES. We found that a C-rich loop located at the top of domain IV is an important determinant for protein binding and that its sequence is essential for viral translation. Furthermore, we showed that the interactions between PCBP and the 5′UTR of the poliovirus genome are modulated by the viral protein 3CD. Since the binding of PCBP to the viral RNA is important for translation and genome replication (8, 14, 26), altering PCBP-RNA interactions could represent a mechanism for regulating these viral processes.
How can PCBP and 3CD control viral translation?
The results presented here together with previous observations indicate that the interactions of PCBP with the cloverleaf RNA and the stem-loop IV are required for IRES-mediated translation (7, 8, 14, 15, 26). How these RNP complexes assist ribosomal entry is still unknown. It has been postulated that RNA-protein interactions within the viral 5′UTR induce the appropriate spatial arrangement along the RNA to be recognized by the translation initiation machinery. We have previously reported that the specific interaction of PCBP with the cloverleaf RNA enhances viral translation 10-fold, while binding of the viral protein 3CD to the same RNA element has a negative effect on translation (14). In the present study, we inspected the interactions of PCBP and 3CD with the viral 5′UTR.
We showed that the binding of 3CD to stem-loop D of the cloverleaf RNA greatly increases the binding affinity of PCBP, changing the dissociation constant from ∼95 to ∼1 nM (Fig. 4B). The molecular basis of this effect is not yet clear. It could be caused by a favorable change in the RNA upon 3CD binding, exposing a high-affinity binding site for PCBP within the cloverleaf structure, or by direct 3CD-PCBP interaction. Regardless of the molecular basis of the complex formation, the interaction of 3CD with the PCBP-cloverleaf complex could induce structural changes that directly interfere with the ability of PCBP to enhance viral translation.
Although in the presence of 3CD the affinity of PCBP for the cloverleaf RNA increased dramatically (from Kd values of ∼95 to ∼1 nM), its affinity for stem-loop IV was not affected (∼15 nM). This suggests that, in the presence of 3CD, PCBP will bind preferentially the cloverleaf RNA. Indeed, the footprinting analysis indicates that 3CD induces dissociation of PCBP from stem-loop IV (Fig. 5). This dissociation appears to be mediated by direct competition between the cloverleaf and stem-loop IV, because an excess of PCBP restored complex formation (data not shown). We estimated that the overall cytoplasmic concentration of PCBP in HeLa cells is about 100 nM, which could be sufficient to bind both RNA targets simultaneously. If this is the case, it would be unlikely that the dissociation of PCBP from stem-loop IV induced by 3CD is the mechanism of downregulation of translation. However, because it is difficult to determine the local PCBP concentration where the viral RNA is translated, the functional relevance of the effect of 3CD within stem-loop IV warrants further investigation.
PCBP binding to picornavirus 5′UTRs.
The binding of PCBP to the cloverleaf RNA was previously mapped, and the presence of three cytosines in stem-loop B of the cloverleaf was shown to be essential for protein recognition as well as for viral viability (2, 3, 17). The interaction of PCBP with stem-loop IV was first reported by Blyn et al. (7). Here, we examined this interaction in more detail and found that two C-rich loops at position 298 (loop a) and 332 (loop b) of the large stem-loop IV are important determinants for protein binding in vitro (Fig. 2). Interestingly, the in vivo studies indicate that the sequence in loop b is crucial for poliovirus translation, while changing the sequence in loop a does not significantly alter viral translation (Fig. 2D). The behavior of this last mutant is intriguing. However, it is possible that loop a is involved in stabilizing the complex with PCBP in vitro without contributing to the in vivo binding site. In the context of the full-length poliovirus genome, the tertiary structure of the RNA or the presence of other factors could yield loop a inaccessible to PCBP. Alternatively, PCBP could make multiple contacts with the stem-loop IV RNA both in vivo and in vitro, but not all the interactions may be functionally relevant for translation. Moreover, it is possible that in vivo other RNA structures as well as cellular proteins contribute in the formation of a stable RNP complex with PCBP.
Importantly, the sequence of loop b, which is required for both binding of PCBP and viral translation (Fig. 2), is widely conserved among entero- and rhinoviruses. In addition, IRES elements of different picornaviruses such as coxsackievirus B3, human rhinovirus 14, and encephalomyocarditis virus efficiently compete with the poliovirus RNA for binding to a cellular protein later identified as PCBP, suggesting possible interactions of PCBP with those IRESs (11). More recently, it was reported that PCBP is also required for hepatitis A virus cap-independent translation, but the specific site for protein binding in this IRES remains to be defined (16). Taken together, these observations suggest that one or more binding sites for PCBP might be widely present in the 5′UTR of picornaviruses. However, more biochemical and functional studies are necessary to establish whether PCBP is indeed a general factor required for viral internal initiation of translation.
Finally, it has been shown that picornavirus IRESs not only carry signals for viral translation but also contain determinants for RNA synthesis (9, 32). Using a bicistronic construct to uncouple the synthesis of nonstructural proteins from the poliovirus IRES, it was shown that disruption of the stem-loop encompassing nucleotides 313 to 374 severely compromises viral RNA synthesis (9). Because our findings indicate that PCBP interacts specifically with part of this structure, it will be important to determine whether the interaction of PCBP with the viral IRES is also involved in RNA synthesis.
ACKNOWLEDGMENTS
We are grateful to Nina Böddeker and Debbie Silvera for useful comments on the manuscript and to Amy Corder for graphics.
This work was supported by funds provided by the Department of Microbiology and Immunology, University of California, San Francisco, and Public Health Service grant AI40085 to R.A.
REFERENCES
- 1.Andino R, Boeddeker N, Silvera D, Gamarnik A V. Intracellular determinants of picornavirus replication. Trends Microbiol. 1999;7:76–82. doi: 10.1016/s0966-842x(98)01446-2. [DOI] [PubMed] [Google Scholar]
- 2.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 4.Andino R, Rieckhof G E, Trono D, Baltimore D. Substitutions in the protease (3Cpro) gene of poliovirus can suppress a mutation in the 5′ noncoding region. J Virol. 1990;64:607–612. doi: 10.1128/jvi.64.2.607-612.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belsham G J, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black D L, Pinto A L. U5 small nuclear ribonucleoprotein: RNA structure analysis and ATP-dependent interaction with U4/U6. Mol Cell Biol. 1989;9:3350–3359. doi: 10.1128/mcb.9.8.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blyn L B, Swiderek K M, Richards O, Stahl D C, Semler B L, Ehrenfeld E. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc Natl Acad Sci USA. 1996;93:11115–11120. doi: 10.1073/pnas.93.20.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blyn L B, Towner J S, Semler B L, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borman A M, Deliat F G, Kean K M. Sequences within the poliovirus internal ribosome entry segment control viral RNA synthesis. EMBO J. 1994;13:3149–3157. doi: 10.1002/j.1460-2075.1994.tb06613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collier B, Goobar-Larsson L, Sokolowski M, Schwartz S. Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J Biol Chem. 1998;273:22648–22656. doi: 10.1074/jbc.273.35.22648. [DOI] [PubMed] [Google Scholar]
- 11.Dildine S L, Semler B L. Conservation of RNA-protein interactions among picornaviruses. J Virol. 1992;66:4364–4376. doi: 10.1128/jvi.66.7.4364-4376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrenfeld E, Semler B L. Anatomy of the poliovirus internal ribosome entry site. Curr Top Microbiol Immunol. 1995;203:65–83. doi: 10.1007/978-3-642-79663-0_3. [DOI] [PubMed] [Google Scholar]
- 13.Gamarnik A V, Andino R. Replication of poliovirus in Xenopus oocytes requires two human factors. EMBO J. 1996;15:5988–5998. [PMC free article] [PubMed] [Google Scholar]
- 14.Gamarnik A V, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamarnik A V, Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 16.Graff J, Cha J, Blyn L B, Ehrenfeld E. Interaction of poly(rC) binding protein 2 with the 5′ noncoding region of hepatitis A virus RNA and its effects on translation. J Virol. 1998;72:9668–9675. doi: 10.1128/jvi.72.12.9668-9675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris K S, Xiang W, Alexander L, Lane W S, Paul A V, Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 18.Hellen C U, Witherell G W, Schmid M, Shin S H, Pestova T V, Gil A, Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc Natl Acad Sci USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holcik M, Liebhaber S A. Four highly stable eukaryotic mRNAs assemble 3′ untranslated region RNA-protein complexes sharing cis and trans components. Proc Natl Acad Sci USA. 1997;94:2410–2414. doi: 10.1073/pnas.94.6.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunt S L, Hsuan J J, Totty N, Jackson R J. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 1999;13:437–448. doi: 10.1101/gad.13.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiledjian M, Wang X, Liebhaber S A. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J. 1995;14:4357–4364. doi: 10.1002/j.1460-2075.1995.tb00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meerovitch K, Svitkin Y V, Lee H S, Lejbkowicz F, Kenan D J, Chan E K, Agol V I, Keene J D, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer K, Petersen A, Niepmann M, Beck E. Interaction of eukaryotic initiation factor eIF-4B with a picornavirus internal translation initiation site. J Virol. 1995;69:2819–2824. doi: 10.1128/jvi.69.5.2819-2824.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostareck-Lederer A, Ostareck D H, Hentze M W. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem Sci. 1998;23:409–411. doi: 10.1016/s0968-0004(98)01301-2. [DOI] [PubMed] [Google Scholar]
- 26.Parsley T B, Towner J S, Blyn L B, Ehrenfeld E, Semler B L. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 27.Pelletier J, Kaplan G, Racaniello V R, Sonenberg N. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol Cell Biol. 1988;8:1103–1112. doi: 10.1128/mcb.8.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pestova T V, Shatsky I N, Hellen C U. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pilipenko E V, Blinov V M, Romanova L I, Sinyakov A N, Maslova S V, Agol V I. Conserved structural domains in the 5′-untranslated region of picornaviral genomes: an analysis of the segment controlling translation and neurovirulence. Virology. 1989;168:201–209. doi: 10.1016/0042-6822(89)90259-6. [DOI] [PubMed] [Google Scholar]
- 30.Rivera V M, Welsh J D, Maizel J V., Jr Comparative sequence analysis of the 5′ noncoding region of the enteroviruses and rhinoviruses. Virology. 1988;165:42–50. doi: 10.1016/0042-6822(88)90656-3. [DOI] [PubMed] [Google Scholar]
- 31.Rohll J B, Percy N, Ley R, Evans D J, Almond J W, Barclay W S. The 5′-untranslated regions of picornavirus RNAs contain independent functional domains essential for RNA replication and translation. J Virol. 1994;68:4384–4391. doi: 10.1128/jvi.68.7.4384-4391.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiroki K, Ishii T, Aoki T, Kobashi M, Ohka S, Nomoto A. A new cis-acting element for RNA replication within the 5′ noncoding region of poliovirus type 1 RNA. J Virol. 1995;69:6825–6832. doi: 10.1128/jvi.69.11.6825-6832.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simoes E A, Sarnow P. An RNA hairpin at the extreme 5′ end of the poliovirus RNA genome modulates viral translation in human cells. J Virol. 1991;65:913–921. doi: 10.1128/jvi.65.2.913-921.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skinner M A, Racaniello V R, Dunn G, Cooper J, Minor P D, Almond J W. New model for the secondary structure of the 5′ non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J Mol Biol. 1989;207:379–392. doi: 10.1016/0022-2836(89)90261-1. [DOI] [PubMed] [Google Scholar]
- 35.Trono D, Andino R, Baltimore D. An RNA sequence of hundreds of nucleotides at the 5′ end of poliovirus RNA is involved in allowing viral protein synthesis. J Virol. 1988;62:2291–2299. doi: 10.1128/jvi.62.7.2291-2299.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trono D, Pelletier J, Sonenberg N, Baltimore D. Translation in mammalian cells of a gene linked to the poliovirus 5′ noncoding region. Science. 1988;241:445–448. doi: 10.1126/science.2839901. [DOI] [PubMed] [Google Scholar]
- 37.Xiang W, Harris K S, Alexander L, Wimmer E. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J Virol. 1995;69:3658–3667. doi: 10.1128/jvi.69.6.3658-3667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]