Abstract
Human immunodeficiency virus type 1 (HIV-1) reverse transcription is primed by the cellular tRNA3Lys molecule that binds with its 3′-terminal 18 nucleotides to the fully complementary primer-binding site (PBS) on the viral RNA genome. Besides this complementarity, annealing of the primer may be stimulated by additional base-pairing interactions between other parts of the tRNA molecule and viral sequences flanking the PBS. According to the RNA secondary structure model of the HIV-1 leader region, part of the PBS sequence is involved in base pairing to form a small stem-loop structure, termed the U5-PBS hairpin. This hairpin may be involved in the process of reverse transcription. To study the role of the U5-PBS hairpin in the viral replication cycle, we introduced mutations in the U5 region that affect the stability of this structured RNA motif. Stabilization and destabilization of the hairpin significantly inhibited virus replication. Upon prolonged culturing of the virus mutant with the stabilized hairpin, revertant viruses were obtained with additional mutations that restore the thermodynamic stability of the U5-PBS hairpin. The thermodynamic stability of the U5-PBS hairpin apparently has to stay within narrow limits for efficient HIV-1 replication. Transient transfection experiments demonstrated that transcription of the proviral genomes, translation of the viral mRNAs, and assembly of the virions with a normal RNA content is not affected by the mutations within the U5-PBS hairpin. We show that stabilization of the hairpin reduced the amount of tRNA primer that is annealed to the PBS. Destabilization of the hairpin did not affect tRNA annealing, but the viral RNA-tRNA complex was less stable. These results suggest that the U5-PBS hairpin is involved in correct placement of the tRNA primer on the viral genome. The analysis of virus mutants and revertants and the RNA structure probing experiments presented in this study are consistent with the existence of the U5-PBS hairpin as predicted in the RNA secondary structure model.
The replication cycle of human immunodeficiency virus type 1 (HIV-1) and other retroviruses is characterized by reverse transcription of the viral RNA genome into a double-stranded DNA, which subsequently becomes integrated into the host cell genome (42). This process is mediated by the virion-associated enzyme reverse transcriptase (RT), and the cellular tRNA3Lys molecule is used as a primer by HIV-1 (35). The tRNA primer binds with its 3′-terminal 18 nucleotides (nt) to a complementary sequence in the viral genome, the primer-binding site (PBS), which is located in the untranslated leader region of the viral genome (Fig. 1A). Besides the complementarity between the PBS and the 3′ end of tRNA3Lys, annealing of the primer has been proposed to be stimulated by additional base-pairing interactions between other parts of the tRNA molecule and viral sequences flanking the PBS (31).
FIG. 1.
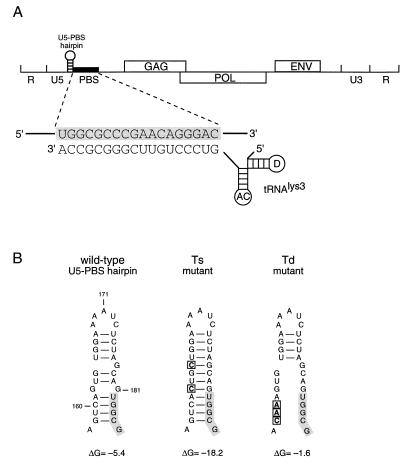
Annealing of the tRNA3Lys primer to the PBS of the HIV-1 RNA genome. (A) The tRNA3Lys primer binds with its 3′ terminus to the complementary sequence of the PBS to form an 18-bp duplex that is shown in detail (PBS sequence is marked in grey). The remainder of the tRNA cloverleaf structure is shown (AC, anticodon loop; D, D loop). Besides the base-pairing interaction with the PBS, sequences in the U5 region may interact with different parts of tRNA3Lys to stimulate primer annealing. Directly upstream of the PBS is a small hairpin structure, the U5-PBS hairpin, which is the topic of this study. (B) Shown is the wild-type U5-PBS hairpin, which was mutated to change the thermodynamic stability. In mutant Ts, the hairpin was stabilized by the introduction of an additional C nucleotide at position 165 and one nucleotide change at position 162 (G to C). In mutant Td, the hairpin is destabilized by three nucleotide substitutions at positions 158 to 160. The introduced mutations are marked by open boxes, and the PBS sequence is marked by a grey box. The thermodynamic stability of the hairpins, indicated at the bottom (ΔG in kilocalories per mole), was calculated using the Zuker algorithm (53).
Extensive secondary structure in the 5′ untranslated leader region of the HIV-1 genome has been suggested by electron microscopy, replication studies with mutant viruses, and biochemical RNase probing studies (3, 11, 17, 21, 22, 37). These results, combined with phylogenetic analyses and computer-assisted structure prediction, led to a model of the secondary RNA structure of the complete leader region of the HIV-1 genome (4). According to this model, the PBS is flanked by an upstream small stem-loop structure, the U5-PBS hairpin (Fig. 1A). This HIV-1 hairpin structure was modeled primarily based on the fact that phylogenetic analysis of different HIV and simian immunodeficiency viruses (SIV) demonstrated a conservation of the hairpin structure, despite considerable divergence in sequence (5, 7). A striking feature of the U5-PBS hairpin of different HIV and SIV isolates is that part of the PBS sequence is involved in base pairing (Fig. 1B). Several RNA secondary structures in the leader RNA have been reported to regulate important viral replication steps of HIV-1; examples are transcriptional transactivation by Tat (8, 20, 30), mRNA polyadenylation (16, 28), and dimerization of the viral RNA genome (9, 12, 38). A stem-loop structure at a similar position as the U5-PBS hairpin of HIV-1 was predicted for Rous sarcoma virus. This structure is required for efficient initiation of reverse transcription in Rous sarcoma virus (2, 13). In addition, an interaction between U5 RNA and sequences of the primer tRNA has been proposed (1) and was confirmed recently by RNA structure probing studies (37a). A detailed structure has also been proposed for the HIV-1 RNA-tRNA3Lys complex based on biochemical experiments (24, 25a). Several sequences in the U5 region upstream of the PBS were suggested to interact with different parts of the tRNA3Lys primer. According to this model, base pairing occurs between the U-rich anticodon loop of tRNA3Lys and the A-rich loop of the U5-PBS hairpin. These combined observations suggest a specific role for the U5-PBS hairpin structure in the process of reverse transcription.
To study the role of the U5-PBS hairpin in the viral replication cycle, we introduced mutations in this structured RNA motif of the HIV-1 genome. Stabilization or destabilization of the U5-PBS hairpin significantly reduced virus replication. Analysis of revertant viruses, obtained through prolonged culturing of the mutant viruses, revealed that the thermodynamic stability of the hairpin has to stay within narrow limits for efficient HIV-1 replication. Biochemical assays demonstrated the involvement of the U5-PBS hairpin in the correct placement of the tRNA3Lys primer onto the viral genome.
MATERIALS AND METHODS
DNA constructs.
A derivative of the full-length proviral HIV-1 clone pLAI was used to produce wild-type and U5-mutated viruses. This construct, pLAI-R37, was described previously (17). The 3′ long terminal repeat (3′LTR) was truncated at the SacI site within the R region, and the chloramphenicol acetyltransferase (cat) gene and simian virus 40 polyadenylation site were inserted at this position. Nucleotide numbers refer to positions on the genomic RNA transcript, with +1 being the capped G residue. For mutation of the U5-PBS hairpin, we used the construct pBlue-5′LTR (29), which contains an XbaI-ClaI fragment of HIV-1 encompassing the 5′LTR, PBS, and 5′ end of the gag gene (positions −454 to +376) cloned into pBluescript (Stratagene). The U5-PBS hairpin sequence was mutated by oligonucleotide-directed in vitro mutagenesis with a Muta-Gene phagemid in vitro mutagenesis kit (Bio-Rad). Oligonucleotides used are Ts (5′-AGACCCTTTTAGTCACTGCTGGAAAATCTCTAGC-3′) and Td (5′-CCTCAGACCCTTTTACAAAGTGTGGAAAATCTC-3′ (mutagenic positions underlined). The mutations introduced were verified by sequence analysis. Sequencing was performed with the primer AD-SD (positions +269 to +290), using a Thermo Sequenase dye terminator cycle sequencing kit (Amersham) and an Applied Biosystems 373 DNA sequencer. Subsequently, the mutated XbaI-ClaI fragments were introduced into the proviral clone pLAI-R37, which again was verified by sequence analysis. For transcription studies, the pBlue-3′LTR-luciferase reporter construct was generated by the exchange of the HindIII-BamHI fragment of pBlue-3′LTR-CAT (29), encompassing the cat gene, by the HindIII-BamHI fragment of pGL3 (Promega), encoding the luciferase gene. For construction of the pBlue-5′LTR-luciferase reporter construct, the 5′LTR-leader region of HIV-1 was PCR amplified to introduce an NcoI restriction site overlapping the gag translation start codon. The primers used are AD-R1 (positions +6 to +30) and SP6-ATG (5′-ATTTAGGTGACACTATAG CCATGGCTCTCCTTCTAGCC-3′ (start codon underlined, mutagenic positions in bold). The PCR fragment was digested with HindIII/NcoI and inserted into HindIII/NcoI-digested pBlue-3′LTR-luciferase. The control luciferase construct was generated by deletion of the XhoI/NcoI fragment, encompassing all HIV-1 sequences, filling of the recessed termini by the Klenow fragment of DNA polymerase I, and self-ligation of the vector. The expression vector pcDNA3-Tat was described previously (44).
Synthesis of RNA templates.
Plasmid pBlue-5′LTR was used as a template for PCR amplification and subsequent in vitro transcription. The 5′LTR region of HIV-1 was PCR amplified with the sense primer T7-2 (positions +1 to +20) containing the T7 RNA polymerase promoter sequence and the antisense primer AUG (positions +348 to +368). The PCR fragments were phenol extracted, precipitated, and dissolved in water. The in vitro transcription reaction was performed in 10 μl of transcription buffer (40 mM Tris [pH 7.5], 2 mM spermidine, 10 mM dithiothreitol [DTT], 12 mM MgCl2) containing 0.5 μg of DNA template, 0.06 μmol of ATP, GTP, CTP, and UTP, 10 U of T7 RNA polymerase (Boehringer), and 20 U of RNase inhibitor (Boehringer) and incubated for 4 h at 37°C. Upon DNase treatment and phenol extraction, the unincorporated free nucleotides were removed by passage through a Sephadex G-50 column. Subsequently, the RNA was ethanol precipitated and dissolved in renaturation buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl). The RNA was renatured by incubation at 85°C for 2 min, followed by slow cooling to room temperature.
RNA structure probing.
The renatured RNA (25 ng) was treated with 0.5% diethyl pyrocarbonate (DEPC) or 0.1% dimethyl sulfate (DMS) in 25 μl of 10 mM Tris (pH 7.5)–10 mM MgCl2–50 mM NaCl buffer. After incubation for 10 min at 37°C, the RNA sample was recovered by ethanol precipitation and dissolved in 5 μl of renaturation buffer. The antisense primer BB-3 (positions +216 to +245) was used to map the modified RNA positions in a primer extension reaction. This primer was end labeled with [γ-32P]ATP and T4 polynucleotide kinase (Boehringer). The labeled oligonucleotide (2 ng) was mixed with the RNA sample in a total volume of 10 μl of annealing buffer (83 mM Tris-HCl [pH 7.5], 125 mM KCl), incubated for 2 min at 85°C and for 10 min 65°C, and slowly cooled to 25°C. The primer was extended by addition of 5 μl of RT buffer (9 mM MgCl2, 30 mM DTT, 150 μg of actinomycin D per ml, 30 μM dATP, dGTP, dTTP, and dCTP) and 12.5 U of avian myeloblastosis virus (AMV) RT (Boehringer) in an incubation at 42°C for 15 min. The samples (2.5 μl) were mixed with formamide loading buffer (2.5 μl), denatured at 90°C, and analyzed on a 6% polyacrylamide–7 M urea gel.
Cells, viruses, and transfection.
SupT1 T cells were grown in RPMI 1640 medium supplemented with 10% fetal calf serum at 37°C and 5% CO2. SupT1 cells (5 × 106) were transfected with 1 and 2 μg of the HIV-1 proviral constructs by electroporation (250 V, 960 μF). After transfection, 0.5 × 106 fresh SupT1 cells were added to support viral replication. Cells were split 1 to 10 twice a week. For the selection of revertant viruses, the transfected cells were passaged up to 124 days. At the peak of virus production, 100 to 0.1 μl of the culture supernatant was used to infect fresh SupT1 cells. At each passage, cells and supernatant samples were stored at −70°C. For transcription studies, 5 × 106 SupT1 cells were transfected with 5 μg of the 5′LTR-luciferase constructs by electroporation. We added 100, 500, and 1,000 ng of pcDNA-Tat, which is within the linear range of LTR transcriptional activation. To have an equal amount of 6 μg DNA in each transfection, we added the empty pcDNA3 vector.
C33A and HeLa cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal calf serum at 37°C and 5% CO2. For the transient production of virions, C33A cells were transfected by the calcium phosphate method. Cells were grown in 20 ml of culture medium in a 75-cm2 flask to 60% confluency. Thirty micrograms of the proviral construct in 880 μl of water was mixed with 1 ml of 50 mM HEPES (pH 7.1)–250 mM NaCl–1.5 mM Na2HPO4 and 120 μl of 2 M CaCl2, incubated at room temperature for 20 min, and added to the culture medium. The culture medium was changed after 16 h. For transcription studies, HeLa cells were transfected by the DEAE-dextran method. Cells were grown in a 60- by 15-mm tissue culture dish to 60% confluency. Cells were washed two times with Tris-buffered saline (TBS) and incubated for 30 min at room temperature with the DEAE-dextran-DNA mixture, containing 1 μg of 5′LTR-luciferase construct with or without 1, 10, and 100 ng of the pcDNA3-Tat expression vector, which is within the linear range of LTR transcriptional activation, in 475 μl of TBS and 25 μl of DEAE-dextran (10 mg/ml in TBS). Finally, the cells were washed two times with TBS to remove the DEAE-dextran-DNA mixture, and culture medium was added.
Analysis of phenotypic revertants.
SupT1 cells transfected with the Ts proviral construct were pelleted by centrifugation at 4,000 rpm for 4 min and washed with phosphate-buffered saline (PBS). The cells were resuspended in 10 mM Tris-HCl (pH 8.0)–1 mM EDTA–0.5% Tween 20 and incubated with 200 μg of proteinase K per ml at 56°C for 1 h and at 95°C for 10 min to isolate total cellular DNA. The 5′LTR-leader region was PCR amplified from the total cellular DNA with the 5′ U3 region primer 5′X (positions −454 to −434) and the 3′ gag primer AD-gag (positions +442 to +463). To provide this fragment with a T7 tail for sequencing with the universal T7 dye primer, a second PCR was performed with the 5′ R region primer T7-1 (positions −54 to −34) and the 3′ primer AUG (positions +123 to +151, with six additional nucleotides at its 5′ end). These PCR products were sequenced directly with a DYEnamic Direct cycle sequencing kit (Amersham) and an Applied Biosystems 373 DNA sequencer. In addition, the 5′X/AD-gag PCR product was cloned into pBlue-5′LTR as a HindIII/NarI fragment. For analysis of individual clones, a PCR was performed with the primers T7-1 and AD-SD (positions +269 to +290), and this PCR fragment was subsequently sequenced. Finally, for insertion of the revertant sequences into the proviral plasmid pLAI-R37, XbaI/ClaI fragments of the specific clones were used to replace the corresponding wild-type sequences. Introduction of the revertant sequences into the proviral plasmid pLAI-R37 was verified by sequence analysis. Therefore, a PCR was performed with the T7-1 and AD-SD primers and the PCR fragment was sequenced.
CA-p24 and RT assay.
CA (capsid protein)-p24 levels in the culture medium were determined by enzyme-linked immunosorbent assay. RT assays were performed as described previously (49). The virus sample (10 μl) was added to 50 μl of RT buffer (60 mM Tris-HCl [pH 8.0], 1 mM EDTA, 75 mM KCl, 5 mM MgCl2, 0.1% Nonidet P-40, 4 mM DTT) supplemented with 0.25 μg of poly(A) and 8 ng of oligo(dT)18 primer and 2.5 μCi (3,000 Ci/mmol) of [α32P]dTTP. Samples (10 μl) were taken after 1, 2, and 3 h of incubation at 37°C and spotted onto DE-81 paper. The samples were dried for 5 min; the paper was subsequently washed three times in 5% Na2HPO4, washed two times in ethanol, and air dried. RT activity was quantified on a Molecular Dynamics PhosphorImager.
Luciferase assay.
Luciferase assays were performed according to Promega's luciferase assay system protocol. Two days posttransfection, HeLa cells were washed with PBS and lysed in 200 μl of reporter lysis buffer (Promega). SupT1 cells were collected by centrifugation 3 days after transfection, washed with PBS, and lysed in 200 μl of reporter lysis buffer. Luciferase activity in the samples (50 μl) was determined by addition of luciferase assay reagent (Promega) in a Berthold model LB 9501 luminometer.
Isolation of viral RNA.
Three days after transfection of C33A cells, the culture medium (20 ml) was centrifuged at 1,600 rpm for 15 min to remove cells. Subsequently the supernatant was filtered through a 0.45-μm-pore-size filter (Schleicher & Schuell), and the virions were pelleted by centrifugation at 25,000 rpm for 30 min in a Beckman SW28 rotor. Virions were resuspended in 500 μl of 10 mM Tris-HCl (pH 8.0)–100 mM NaCl–1 mM EDTA. To isolate viral RNA, the viruses were incubated for 30 min at 37°C in the presence of 100 μg of proteinase K per ml and 0.5% sodium dodecyl sulfate, followed by extraction with phenol-chloroform-isoamyl alcohol (25:24:1) and precipitation in 0.3 M sodium acetate (pH 5.2) and ethanol at −20°C. The viral RNA was pelleted by centrifugation (18,000 rpm, 20 min), washed with 70% ethanol, and dried. The pellet was dissolved in 20 μl of 10 mM Tris-HCl (pH 8.0)–1 mM EDTA and stored at −70°C.
Oligonucleotide and tRNA primer extension assays.
In the oligonucleotide and tRNA primer extension assays, viral RNA corresponding to 30 ng CA-p24 was incubated with 20 ng of oligonucleotide primer in 12 μl of 83 mM Tris-HCl (pH 7.5)–125 mM KCl at 85°C for 2 min and 65°C for 10 min, followed by cooling to room temperature in 1 h to allow annealing of the primer. The primer was extended by addition of 6 μl of RT buffer (9 mM MgCl2, 30 mM DTT, 150 μg of actinomycin D per ml, 30 μM dATP, dGTP, and dTTP, 1.5 μM dCTP), 0.5 μl of [α32P]dCTP, and either 0.5 U of HIV-1 RT (U.S. Biochemical) or 12.5 U of AMV RT (Boehringer) and incubation at 42°C for 3 min; then 10 mM (each) deoxynucleoside triphosphate (dNTP) was added, and incubation was continued for 30 min. The cDNA product was precipitated in 25 mM EDTA–0.3 M sodium acetate (pH 5.2)–70% ethanol at −20°C. For degradation of the tRNA part of the extended product in the tRNA primer extension assay, the samples were incubated with 0.5 N NaOH for 20 min at 55°C, neutralized with 0.5 M HCl, and precipitated as described above. The products were analyzed on a denaturing 6% polyacrylamide-urea sequencing gel. The antisense primers used are CN1 (positions +123 to +151) and AUG (positions +348 to +368, with six additional nucleotides at its 5′ end).
RESULTS
Design of the U5-PBS hairpin mutants.
To study the role in HIV-1 replication of the U5-PBS hairpin that is located directly upstream of the PBS, we introduced mutations in this stem-loop structure. The U5 region is encoded by the LTR that is present at both the 5′ and 3′ ends of the HIV-1 proviral genome. Mutations introduced into the U5 region of the 5′LTR will be inherited in both LTRs of the progeny. However, the presence of a wild-type 3′LTR may result in reversion of the mutant virus to the wild-type sequence by recombination with the wild-type 3′LTR sequences. For production of the mutant viruses, we therefore used a derivative of the proviral clone pLAI in which part of the 3′LTR, including the polyadenylation signal and the complete U5 region, is deleted. An SV40 polyadenylation site was placed downstream of the HIV-1 sequences to allow efficient polyadenylation of the viral transcript. Transfection of the SupT1 T-cell line with this vector results in the production of viruses with a mutant U5 region in the untranslated leader RNA. Subsequent infection of SupT1 cells by these viruses, followed by reverse transcription of the viral RNA genome, will produce proviral genomes with a complete 5′LTR and 3′LTR that both have the mutated U5 sequence.
Mutations that affect the stability of the U5-PBS hairpin were introduced in the U5 region (Fig. 1B). The mutants were carefully designed not to affect important sequence motifs, such as the attachment site for integration (positions 170 to 181) (18, 36) and the PBS sequence. All mutations were therefore introduced on the left side of the hairpin. We stabilized the hairpin in mutant Ts by generating two extra C-G base pairs. This was done by replacement of the unpaired G162 by C and by insertion of an additional C at position 165 (Fig. 1B). This results in an increase in the thermodynamic stability of the hairpin (ΔG) from −5.4 kcal/mol for the wild type to −18.2 kcal/mol for mutant Ts. In mutant Td, the hairpin was destabilized by substitution of three nucleotides at position 158 to 160. As a result, base pairing in the lower part of the stem is lost, and a relative short and instable hairpin structure is left (ΔG = −1.6 kcal/mol).
Structure probing of the wild-type and mutant U5-PBS hairpins.
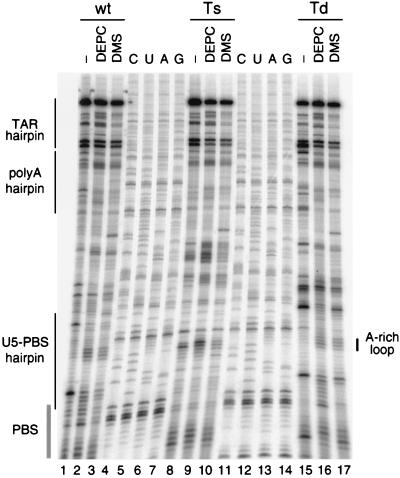
To demonstrate that the introduced mutations have no effect on folding in other parts of the leader RNA, we performed structure probing experiments. In vitro-synthesized HIV-1 leader RNA was treated with structure-specific probes, followed by primer extension analysis to localize the sites of modification. Nucleotides sensitive to DMS or DEPC are assumed not to be involved in base-pairing or base-stacking interactions. The sites of modification were identified by primer extension analysis using the DNA primer BB-3 (positions +216 to +245). No striking differences in reactivity toward the chemicals was observed between the wild-type and mutant U5-PBS hairpins (Fig. 2). The A-rich loop of the wild-type U5-PBS hairpin as well as that of the two mutants is modified by both DMS and DEPC, whereas the flanking sequences are not. The A-rich loop is even visible in the Td mutant, in which the U5-PBS hairpin was destabilized but apparently not destroyed. No major differences were observed in the upstream leader region (Fig. 2) and the region downstream of the PBS, which was also probed and analyzed with the DNA primers DIS (positions +246 to +269) and AUG (positions +348 to +368) (results not shown). Furthermore, computer modeling of a larger region of the 5′LTR leader RNA (positions +111 to +244) suggests that the introduced mutations do not affect the RNA secondary structure in this part of the genome. These combined results indicate that the mutations do not lead to an overall structural rearrangement of the HIV-1 leader.
FIG. 2.
RNA structure probing of the U5-PBS region under native conditions. In vitro-transcribed HIV-1 leader RNA of the wild-type and U5-PBS mutants was treated with a limiting amount of the single-strand-specific reagents DEPC (A specific) and DMS (A/C specific) as indicated. Modification sites were detected using primer extension analysis with the BB-3 primer. The products were analyzed on a 6% polyacrylamide–7M urea gel. For reference, the BB-3 primer was used in a DNA sequencing reaction (lanes 4 to 7 and 11 to 14). Positions of the hairpin structures in this part of the HIV-1 leader RNA and the PBS sequence are shown schematically on the left.
Replication capacity of viruses with a mutated U5-PBS hairpin.
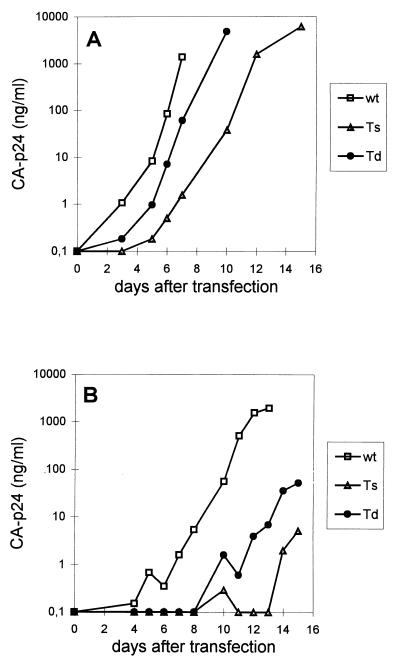
To study the replication potential of the mutant viruses, we transfected wild-type and mutant proviral genomes into SupT1 cells. These cells express the CD4-CXCR4 receptors and are fully susceptible for replication of the LAI strain. Virus production was followed by measuring CA-p24 levels in the culture medium at several days after transfection. Transfection with 2 μg of the proviral constructs showed that the replication capacity of both mutants was reduced compared with the wild-type virus (Fig. 3A). This defect is even more pronounced in transfections with 1 μg of proviral construct (Fig. 3B). No replication of mutant Ts was observed in transfections with less than 1 μg of the proviral construct. Thus, stabilization of the U5-PBS hairpin affected the replication potential of the virus more severely than destabilization of this RNA structure. These results demonstrate the importance of the U5-PBS hairpin in viral replication.
FIG. 3.
Replication of wild-type (wt) and U5-PBS hairpin mutants Ts and Td. SupT1 cells were transfected with 2 (A) or 1 (B) μg of the proviral constructs. Several days after transfection, CA-p24 production was measured in the culture medium.
Reversion of the stabilized U5-PBS hairpin mutant.
During prolonged culturing of replication-defective viruses, phenotypic revertants with an increased replication capacity can arise. The genomes of such revertant viruses should be altered in order to replicate more efficiently, and analysis of such revertant genomes may allow the identification of important RNA sequences and/or structures. This forced evolution approach can be used for most retroviruses due to their high mutation rate and has proven to be valuable in the analysis of regulatory RNA motifs (6, 15, 30). Mutant Td replicated too efficiently to allow the selection of faster-replicating revertants within a reasonable time span. We therefore focused on the evolution of the severely defective mutant Ts.
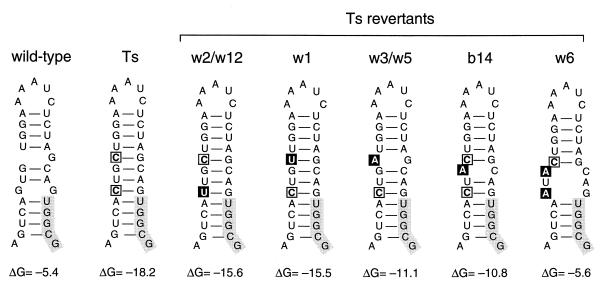
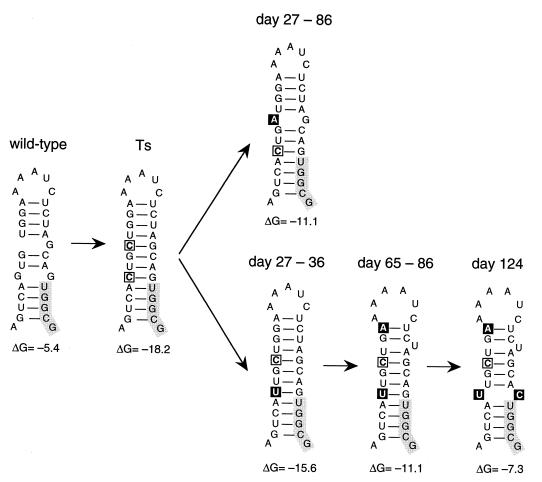
SupT1 cells transfected with the Ts proviral construct were split into several independent cultures that were maintained for 7 weeks. The replication kinetics of the viruses present in several cultures increased after a variable time. To determine the sequence of the U5-PBS hairpin of these phenotypic revertants, total cellular DNA was isolated from infected cells. The 5′LTR-leader region was PCR amplified, and we performed population-based sequencing of the DNA fragment. The predicted RNA structures for the revertant sequences are shown in Fig. 4, with the thermodynamic stability indicated below the hairpins. Remarkably, the nucleotide changes introduced in mutant Ts were frequently found to be altered in the revertant genomes, although no true wild-type reversions were observed. All acquired mutations are located on the left side of the hairpin, which is consistent with the presence of important sequence motifs on the right side. Analysis of the revertants demonstrated a variation in repair strategies, but all mutations reduced the stability of the hairpin. This is most striking in revertant w6, with two reversion-based mutations that result in a hairpin with a stability very similar to that of the wild-type U5-PBS structure. It is likely that the other revertants, all with only one nucleotide substitution within the hairpin, had not yet attained the optimal configuration and will evolve toward hairpin structures with wild-type stability over time.
FIG. 4.
Phenotypic revertants of the stabilized U5-PBS hairpin mutant Ts. Through prolonged culturing of the mutant virus Ts, several revertant viruses with nucleotide changes in the hairpin were obtained. The predicted RNA structures of these revertants and ΔG values (in kilocalories per mole) are shown. The mutations present in mutant Ts are marked by an open box, the acquired additional mutations are marked by black boxes, and the PBS is marked by a grey box. All reversion-based mutations reduce the stability of the mutant U5-PBS hairpin.
To study the evolution of the stabilized Ts hairpin in more detail, we performed an independent SupT1 transfection and monitored this culture for up to 124 days. Total cellular DNA was isolated from infected cells at several days posttransfection. Figure 5A shows the results of direct sequence analysis of the PCR-amplified U5-PBS region of the revertants. Substitutions were initially observed at the two mutated residues in mutant Ts, but two additional mutations were acquired over time. This so-called population sequencing provides information on the acquired mutations and their relative frequency in the virus quasispecies population. However, this method does not determine genetic linkages if mixed sequences are present at multiple positions (e.g., the day 27 sample). We therefore also performed clonal sequencing. To do so, the PCR fragment was cloned into pBlue-5′LTR, and multiple individual clones were sequenced for the samples obtained at day 27, 86, and 124 (Fig. 5B). In the initial phase of the evolution experiment, two revertants appeared to be present simultaneously. Both evolution routes target one of the nucleotides introduced in mutant Ts. The evolutionary pathways of Ts reversion are depicted in Fig. 6, which shows the predicted RNA structure and thermodynamic stability of the observed revertants. Alteration of C165 to A produces an A/G mismatch and destabilizes the hairpin from −18.2 to −11.1 kcal/mol. The other revertant changes C162 to U, thereby creating a weak U-G base pair that has a moderate effect on stability (ΔG = 15.6 kcal/mol). Although both genotypes are present at an approximately equimolar concentration at day 27 (Fig. 5A), the former seems to outcompete the latter, as is evident from the population sequence at days 36 and 41. However, the latter genotype reappears at day 65 due to the acquisition of another destabilizing mutation (G168 to A) that triggers a rearrangement of the upper part of the stem region and increases the loop size. This hairpin (ΔG = −11.1 kcal/mol) acquires one more substitution at day 124 that further reduces the hairpin stability (ΔG = −7.3 kcal/mol) to a value that is similar to that of the wild-type structure (ΔG = −5.4 kcal/mol).
FIG. 5.
Sequence analysis of Ts revertant genomes. (A) The sequence of the U5-PBS hairpin was determined by population sequencing several days after transfection of SupT1 cells with the Ts proviral construct. Positions of the hairpin motif and the PBS are indicated at the top. Dashes indicate nucleotides that are identical to that of the input Ts mutant. Acquired mutations are indicated in capitals if the majority of genomes carried the mutation; minor changes are in small characters. (B) The genetic linkage of the various acquired mutations was determined by sequencing of multiple individual clones for the samples taken at days 27, 86, and 124 posttransfection. The frequency of each sequence is given in parentheses. The clones tested in the replication studies are marked by asterisks.
FIG. 6.
Evolutionary pathway of the stabilized U5-PBS hairpin mutant Ts. The predicted structure for the sequences observed at several days posttransfection (Fig. 5) is shown. The period in which the intermediates were observed is indicated at the top, and the calculated thermodynamic stability (in kilocalories per mole) is indicated below the hairpins. The introduced mutations in mutant Ts are marked by an open box, the acquired mutations are marked by black boxes, and the PBS is marked by a grey box. The hairpin of mutant Ts acquires several mutations that reduce the thermodynamic stability and finally attains a stability similar to that of the wild-type U5-PBS hairpin.
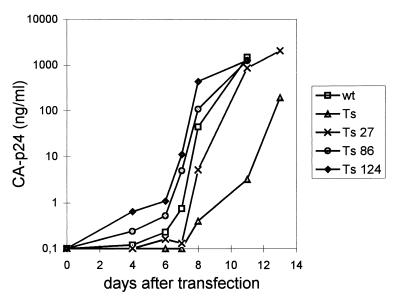
The role of the acquired U5 mutations in the phenotypic reversion of mutant Ts was demonstrated by introduction of revertant sequences observed at days 27, 86, and 124 in the wild-type proviral genome for replication studies. Consecutive intermediates in the evolutionary pathway showed gradually improved replication (Fig. 7). This finding demonstrates that replication of the mutant Ts is repaired by restoration of the hairpin stability. The combined results of the evolution studies indicate that a U5-PBS hairpin of approximately wild-type stability is optimal for virus replication.
FIG. 7.
Replication of different intermediates in the reversion of mutant Ts. Revertant genomes observed at days 27, 86, and 124 (marked by an asterisk in Fig. 5B) were introduced in the wild-type (wt) proviral genome. SupT1 cells were transfected with 2 μg of the proviral constructs. Several days after transfection, CA-p24 production was measured in the culture medium.
The U5-PBS hairpin is not involved in gene expression and virus production.
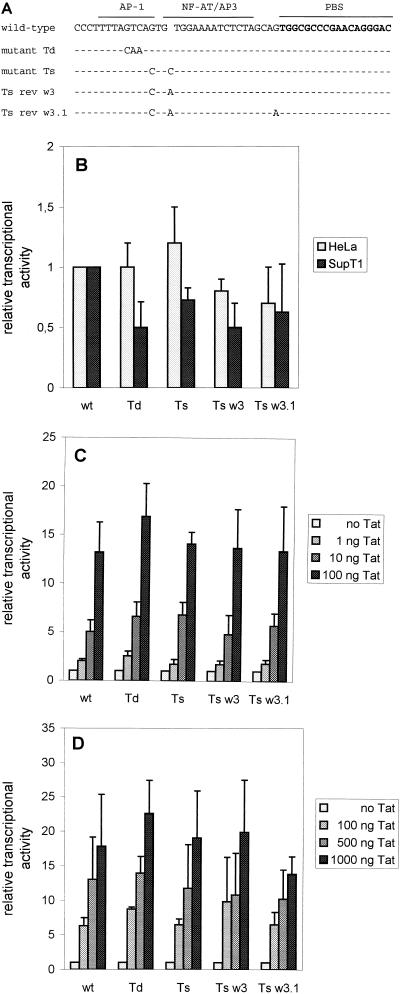
Binding sites for AP-1 and NF-AT/AP3 transcription factors were recently reported to be positioned on the proviral DNA genome directly upstream of the PBS (43). These transcription factor binding sites have been suggested to be involved in HIV-1 transcription and replication. The upstream AP-1 site is changed in mutant Td, and both binding sites are affected in mutant Ts (Fig. 8A), raising the possibility that viral transcription is affected in these mutants. Furthermore, the revertants obtained by prolonged culturing of mutant Ts possibly restore the binding of these transcription factors and thereby virus replication. To test whether the U5 mutations affect viral transcription, we transfected C33A cells (human cervix carcinoma cells not expressing CD4) with the wild-type and mutant Ts and Td proviral vectors and analyzed the level of viral gene expression and virion production. Virus production was monitored by measuring the amount of CA-p24 and virion-associated RT activity in the culture medium. Table 1 summarizes the results of two independent transfections. No significant differences were measured, as the observed differences reflect experimental variation in the electroporation protocol. The expression levels of viral proteins were also found to be similar for all constructs by Western blot analysis of total cell extracts (results not shown).
FIG. 8.
Transcriptional activity of wild-type and mutant LTR promoters. (A) The Ts and Td mutations affect transcription factor binding sites in the U5 DNA. Marked are the AP-1 and NF-AT/AP-3 transcription factor binding sites located in the U5 region immediately upstream of the PBS (43). The nucleotide changes in mutant Td, mutant Ts, and two Ts revertants (rev), w3 and w3.1 (clones obtained from revertant w3), are shown. (B) Relative basal transcriptional activity of wild-type (wt) and mutant LTRs in HeLa and SupT1 cells after transfection with 1 and 5 μg, respectively, of the different LTR-luciferase constructs. The basal transcriptional activity of the wild-type LTR was set at 1. (C and D) Relative Tat-activated transcriptional activities of wild-type and mutant LTRs in HeLa (C) and SupT1 (D) cells. HeLa cells were transfected with 1 μg of the LTR-luciferase constructs with or without 1, 10, and 100 ng of Tat expression vector. SupT1 cells were transfected with 5 μg of the LTR-luciferase constructs with or without 100, 500, and 1,000 ng of Tat expression vector. The amounts of added Tat expression vector are within the linear range of LTR transcriptional activation. The basal transcriptional activity of each individual LTR promoter was set at 1. Transfection with the control luciferase construct, containing the luciferase gene but not the LTR, and cotransfection of this construct with pcDNA3-Tat as well as transfection with the empty Tat vector (pcDNA3) revealed no luciferase activity. Cotransfection of the LTR-luciferase constructs with pcDNA3 resulted in a low basal level of LTR transcription.
TABLE 1.
Virus production upon transfection of wild-type and mutant proviral constructs into C33A cells
| Construct | Expt 1
|
Expt 2
|
||||
|---|---|---|---|---|---|---|
| CA-p24 (ng/ml) | RT activity (counts) | RT/CA-p24 | CA-p24 (ng/ml) | RT activity (counts) | RT/CA-p24 | |
| Wild type | 180 | 39,201,163 | 1.0a | 550 | 17,543,880 | 1.0a |
| Ts | 310 | 66,584,699 | 1.0 | 300 | 11,657,850 | 1.2 |
| Td | 410 | 98,476,183 | 0.9 | 380 | 11,970,842 | 1.0 |
Set at 1.
To study the transcriptional activity of the mutants Td and Ts as well as two Ts revertants in more detail, we constructed LTR-reporter vectors with the complete LTR-leader region of HIV-1 fused to the luciferase open reading frame. Transient LTR-luciferase transfection assays and cotransfections with a Tat expression vector were performed in HeLa and SupT1 cells. Transfection of episomal plasmids may not accurately reflect proviral transcription from an integrated position, but the results shown in Fig. 8B to D indicate that the wild-type, mutant, and revertant LTR constructs do not differ significantly in basal or Tat-activated transcriptional activity. These combined results suggest that the mutations in the U5 region do not affect viral gene expression (e.g., transcription and translation) and virion assembly.
The U5-PBS hairpin is not involved in packaging of the viral RNA.
The untranslated leader RNA contains important signals for packaging of the viral RNA into virion particles. To determine the RNA content of the wild-type and mutant viruses, we isolated RNA from purified virions that were produced in C33A cells. The viral RNA was measured by primer extension analysis with the CN1 oligonucleotide primer, which is complementary to the +123 to +151 region. The CA-p24 values were used to control for the amount of virions used per sample. The mutant virions contained a normal level of RT enzyme (Table 1). As summarized in Table 2, no significant differences were observed between the mutant and wild-type viruses in regard to the amount of viral RNA per virion. These results show that the U5-PBS hairpin does not contribute to the process of packaging of genomic HIV-1 RNA.
TABLE 2.
Genomic RNA content of wild-type and mutant virions as determined by primer extension
| Construct | CA-p24 (ng/ml) | Viral RNA (counts) | RNA/CA-p24 |
|---|---|---|---|
| Wild type | 1,920 | 92,238 | 100a |
| Ts | 2,000 | 92,498 | 100 |
| Td | 4,200 | 198,688 | 98 |
Set at 100%.
The U5-PBS hairpin is involved in reverse transcription.
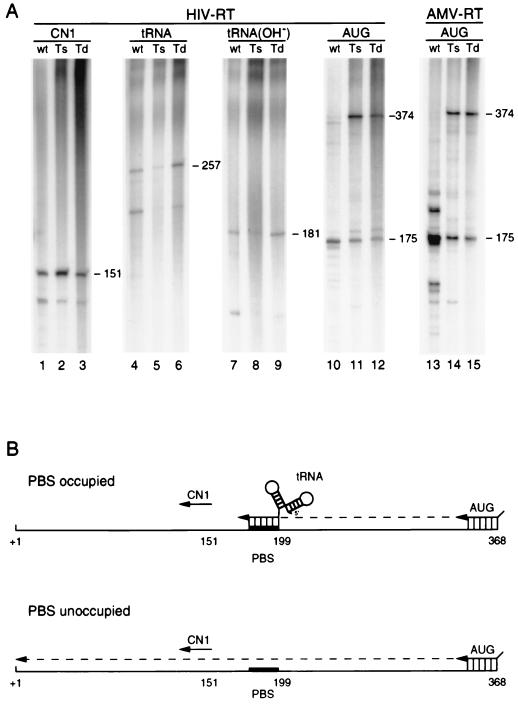
The U5-PBS structure of HIV-1 may be involved in regulation of reverse transcription, in particular because the hairpin includes part of the PBS sequence. In addition, a similar hairpin structure in avian sarcoma-leukosis virus has been found to be involved in the process of reverse transcription, and an interaction between the anticodon of the tRNA primer and the A-rich hairpin loop was proposed for HIV-1 to play a role in the process of reverse transcription. We therefore analyzed the amount of tRNA3Lys primer annealed to the PBS of the wild-type and mutant virion particles. During the isolation of viral RNA, the tRNA primer remains bound to the PBS and can be visualized by extension upon addition of HIV-1 RT enzyme and dNTPs. Extension of the tRNA primer produces a 257-nt-long tRNA-cDNA product (Fig. 1A shows a schematic; Fig. 9A, lanes 4 to 6). The identity of this product was confirmed by NaOH-mediated degradation of the tRNA part, leaving a 181-nt cDNA product (Fig. 9A, lanes 7 to 9). The extended tRNA-cDNA products were quantified and corrected for the amount of input viral RNA template as determined by CN1 primer extension (Fig. 9A, lanes 1 to 3). As summarized in Table 3, the tRNA extension efficiency of mutant Ts was reduced to 27% of the value measured for the wild-type template. The mutant Td was not affected in tRNA extension; we consistently measured a small improvement compared with the wild-type (125%).
FIG. 9.
DNA-primed and tRNA-primed reverse transcription assays with the virion-extracted RNA genome. (A) C33A cells were transfected with the wild-type (wt), Ts and Td proviral constructs. Three days posttransfection, viruses were purified and viral RNA was isolated. The amount of viral RNA was quantified by DNA primer extension with the oligonucleotide CN1 (lanes 1 to 3). Relative positions of the different primers are shown in panel B. The tRNA3Lys primer remains bound to the genomic RNA during viral RNA isolation and was extended by addition of the HIV RT enzyme and dNTPs. Extension of the tRNA primer results in a 257-nt cDNA product (lanes 4 to 6). The tRNA extension product was incubated with NaOH, resulting in the degradation of the tRNA part (76 nt), leaving a cDNA of 181 nt (lanes 7 to 9). The occupancy of the PBS with tRNA primer was determined by a primer extension assay with a primer that is positioned downstream of the PBS. The AUG primer was extended by the HIV (lanes 10 to 12) or AMV (lanes 13 to 15) RT enzyme. When the PBS is occupied by the tRNA primer, a 175-nt premature stop product is generated; in the absence of the tRNA primer, a 374-nt full-length cDNA is produced (B).
TABLE 3.
tRNA-priming efficiency and tRNA occupancy of wild-type and mutant templates
| Construct | RT activity (%)
|
AUG primer
|
||
|---|---|---|---|---|
| CN1 | tRNA | Full-length DNA | Stop product | |
| Wild type | 100a | 100a | 9.8 | 90.2 |
| Ts | 170 | 27 | 77 | 23 |
| Td | 90 | 125 | 62 | 38 |
Set at 100%.
The reduced tRNA extension efficiency of mutant Ts may be the result of less tRNA primer that is annealed to the PBS. Alternatively, normal levels of tRNA are bound, but these primers cannot be extended efficiently on the template with the stabilized U5-PBS hairpin. To discriminate between these two possibilities, the tRNA occupancy of the PBS was determined by a different assay. Viral RNA-tRNA complexes were used as a template for the extension of a primer that is positioned downstream of the PBS. The oligonucleotide primer AUG, complementary to the +348 to +368 region with six additional nucleotides at its 5′ end, was used in this experiment. Viral RNA without a tRNA primer will produce a full-length cDNA product of 374 nt (Fig. 9B). When a tRNA primer is present on the PBS, this primer will also be extended by the RT enzyme, and RNase H will subsequently degrade the RNA template strand. The AUG primer can be extended to the 3′ end of the PBS, where it encounters the annealed tRNA, which will result in a 175-nt cDNA product. Alternatively, when the tRNA is displaced by the RT enzyme, the AUG primer can be extended to the 5′ end of the PBS, producing a 193-nt cDNA product. As shown in Fig. 9A (lanes 10 to 12), extension of the downstream AUG primer produced predominantly the 175-nt stop product, indicating that the tRNA primer is not displaced by the elongating RT enzyme. A different result was reported for the avian leukosis virus, where the tRNA primer was efficiently displaced (48). Extension of the AUG primer on the wild-type template (Fig. 9, lane 10) produced predominantly the 175-nt stop product and almost no 374-nt full-length product. Quantitation of the stop and full-length cDNA products indicated that approximately 90% of the wild-type template has bound a tRNA primer (Table 3), suggesting that nearly all PBS sites of the wild-type HIV-1 RNA are occupied. In contrast, extension of the AUG primer on the mutant Ts template produced relatively less premature stop product (23%) and more full-length product (77%). This result demonstrates that the PBS of mutant Ts is only partially occupied by a tRNA. In fact, the 23% PBS occupancy measured with this assay correlates well with the 27% tRNA extension efficiency (Table 3).
Surprisingly, the PBS occupancy of mutant Td was strongly reduced (38%), even though this mutant showed no defect in the assay (125%) (Fig. 9A; Table 3). Apparently, the tRNA primer is present on the PBS and can be extended efficiently, but the tRNA is lost during the PBS occupancy assay with the downstream AUG primer. The tRNA primer is probably released during the heat denaturation step that is used to anneal the AUG primer. The results suggest that the PBS-associated tRNA primer does not optimally interact with the Td template. In fact, this means that the PBS occupancy assay is not a reliable method when RNA templates that differentially bind the tRNA primer are compared. Similar results were obtained in PBS occupancy assays performed with the RT enzyme of AMV (Fig. 9A, lanes 13 to 15). However, upon extension of the tRNA primer, we observed several additional stop products, specific for the wild-type template, at positions both upstream and downstream of the PBS. This implies a conformational difference between the viral RNA-tRNA duplex formed with the wild-type and mutant templates. Annealing of the tRNA primer onto the wild-type genome is evidently more complex and more stable than interaction with the genome of mutant Td. These results indicate that the U5-PBS hairpin is involved in the correct placement of the tRNA primer onto the viral RNA.
DISCUSSION
In this study, we demonstrate the importance of the U5-PBS hairpin structure for efficient HIV-1 replication. Both stabilization and destabilization of this RNA structure decreased the viral replication capacity. Upon prolonged culturing of the stabilized mutant Ts, several revertant viruses were obtained with an increased replication potential. All of the phenotypic revertants acquired additional mutations in the hairpin that reduce its thermodynamic stability. Thus, the mutant viruses revert by emulating the stability of the wild-type hairpin. This indicates that RNA structural effects rather than RNA or DNA sequence effects are responsible for the replication defect of these HIV-1 mutants. Apparently, the thermodynamic stability of the U5-PBS hairpin must stay within narrow limits for efficient HIV-1 replication.
Analysis of revertant viruses of the mutant Ts also revealed that the reversion-based mutations are almost exclusively present on the left side of the U5-PBS hairpin, and in particular at the nucleotide positions that were altered in the mutant Ts (first sites). This result contrasts with reversion analyses of other structured RNA motifs in which second-site reversions were observed frequently (6, 30). The nonrandom nature of U5-PBS hairpin reversion suggests that important sequence motifs are encoded by this region of the HIV-1 genome. The observation that the reversion-based mutations are predominantly present on the left side of the hairpin suggests that such motifs are encoded by the right side of the hairpin. In fact, these U5 sequences from the extreme 3′ end of the LTR, and this region is well known to contribute to proviral integration through sequence-specific interaction with the viral integrase protein (10). Recently, the importance of the 3′-terminal 12 nt (positions 170 to 181) of the U5 region has been demonstrated (18, 36, 45). Thus, the U5 motif that is critical for integration also includes part of the A-rich loop sequence, which may explain in part the importance of this sequence element for HIV-1 replication (34). In addition, the right side of the hairpin contains part of the PBS sequence (positions 182 to 185), which does not allow mutation (15, 32, 47). These two sequence motifs together constitute the right side of the U5-PBS hairpin. Thus, this part of the HIV-1 genome encodes at least three signals, of which one is recognized as part of the double-stranded DNA genome (integration motif), one as RNA sequence (PBS), and one as structured RNA motif (U5-PBS hairpin).
The fact that mainly first-site mutations were found in the revertants suggests that an important sequence motif surrounds the introduced mutations. Nevertheless, no true wild-type reversions were observed. AP-1 and NF-AT/AP3 transcription factor binding sites have been found in the U5 region (43). The mutations introduced in mutant Ts and Td affect these sites, and we therefore analyzed the transcriptional activity of these mutants in transient LTR-luciferase transfection assays. In addition, we tested whether two revertants of mutant Ts increased the LTR activity of mutant Ts. Both basal and Tat-activated transcriptional activities of the wild-type, mutant, and revertant LTRs were tested, but we measured no difference among the different promoters. Furthermore, similar levels of virus production were measured in cells transiently transfected with the proviral constructs. Thus, these transcription factor binding sites in the U5 region are either not affected by the Ts and Td mutations or not important for viral replication. Nevertheless, the nonrandom pattern of reversion suggests the presence of a sequence-specific motif in this part of the U5 region.
Biochemical assays with virion-derived RNA-tRNA complexes showed that the reduced replication potential of mutant Ts correlates with reduced tRNA priming efficiency, which is the result of decreased tRNA occupancy of the PBS. We measured nearly complete occupancy of the PBS for the wild-type template, suggesting that both copies of the dimeric HIV-1 RNA genome have an associated tRNA primer. This result differs somewhat with studies on murine leukemia virus and avian leukosis virus, for which PBS occupancies of 50 and 70% have been reported (19, 48). We found the PBS occupancy of the mutant Ts template to be reduced to 23%. Thus, inclusion of part of the PBS in an excessively stable hairpin structure inhibits the annealing of the tRNA primer. This effect apparently restricts the U5-PBS hairpin from becoming excessively stable in natural HIV and SIV isolates (5). In other words, the HIV-1 genome contains a structured RNA motif in the U5-PBS region that is at the threshold of becoming inhibitory to the process of initiation of reverse transcription. In fact, the tRNA extension efficiency could be increased to 125% of the wild-type value by opening of the U5-PBS hairpin as in mutant Td. Although speculative, this hairpin may restrict premature tRNA annealing to the viral RNA in the infected cell, but this restriction is apparently overcome in the context of the virion particle, perhaps due to viral cofactors (see below). More extended hairpin structures with greater thermodynamic stability were predicted for the HIV-2 genome and several SIV variants, but we previously stressed that these structures are characterized by having either a limited number of PBS nucleotides that are involved in base pairing or a large percentage of relatively weak G-U base pairs (7). There is recent evidence for ribozymes that terminal G-U base pairs are involved in structural rearrangements (50), which may explain the efficient tRNA annealing in HIV-2, despite the relatively stable U5-PBS structure. Obviously, there may be cofactors that facilitate tRNA annealing onto the PBS in the context of the viral particle. One such a factor is the viral nucleocapsid (NC) protein, which has been reported to facilitate the annealing of the tRNA primer to the PBS (23, 39). Apparently, the excessively stable Ts hairpin interferes with this process, and it will be of interest to study this annealing reaction in more detail in in vitro assays in the absence and presence of NC protein. Such studies are currently being performed.
A more complex defect was apparent for the Td mutant with the destabilized U5-PBS hairpin. This mutant template was at least as efficient in tRNA extension as the wild-type template, demonstrating that the PBS is occupied by tRNA. Despite efficient tRNA extension, we measured a strongly reduced PBS occupancy in tests in which reverse transcription is primed by an oligonucleotide from a position downstream of the PBS. Apparently, the tRNA primer was released during the primer-annealing step. This result indicates that the interaction between the tRNA primer and the mutant Td genome is less stable than the complex with the wild-type template, even though the two templates have identical PBSs. Moreover, several additional stop products upstream and downstream of the PBS were observed for the wild-type template during extension of the downstream primer with the AMV RT enzyme. These stops are due to tRNA annealing because the signals are not observed with the mutant Ts template. Most importantly, these stops were not observed either for the mutant Td template. These combined results indicate that a different conformation of the viral RNA-tRNA complex is reached on the wild-type template compared with mutant Td, suggesting that the U5-PBS hairpin is directly or indirectly involved in correct tRNA annealing onto the viral RNA genome. Several studies suggest that the A-rich loop of the U5-PBS hairpin interacts directly with the anticodon of tRNA3Lys (24–27, 33, 34, 40, 41, 46, 51, 52), but the interpretation of these experiments is complicated because this U5 sequence encodes multiple, overlapping replication signals. For instance, mutations in the U5-PBS region may affect the secondary structure of this part of the leader RNA (5), and the presence of overlapping integration signals (18, 36) makes it difficult to analyze the proposed interactions between the viral RNA and the tRNA primer. The phenotype of the carefully designed RNA structure mutant Td in this study does support the idea that structured RNA motifs in the U5 region contribute to functional tRNA annealing. The induction of multiple stop signals upon tRNA binding to the wild-type template suggests that a structural rearrangement occurs in the region surrounding the PBS, but the molecular nature of the additional viral RNA-tRNA interactions remains to be determined.
The mutant and revertant analysis presented in this study is consistent with the existence of the U5-PBS hairpin as depicted in Fig. 1. We previously proposed this hairpin conformation as part of a secondary RNA structure model of the complete leader region of the HIV-1 genome (4). In fact, this HIV-1 hairpin structure is not very stable but was modeled based primarily on similar structures in the HIV-2 RNA (7) and the genomes of several SIV viruses (5). Although a different conformation was recently proposed for this part of the HIV-1 genome based on RNA structure probing experiments (14), our structure probing results and in particular the functional data strongly support the existence of the U5-PBS hairpin. Furthermore, this stem-loop structure is supported by phylogenetic evidence based on the sequence of different HIV-1 subtypes (not shown). It may nevertheless be too simplistic to suggest that this part of the RNA genome has one static conformation. Several factors will bind to this region of the viral genome during discrete steps of virus replication, e.g., the tRNA primer, the NC protein, and the RNA itself during dimerization, and it is likely that the RNA conformation will change during consecutive steps of the viral replication cycle. It cannot even be excluded that this region acts as a molecular switch during replication by changing between alternative RNA conformations. We recently obtained evidence for such a conformational polymorphism of the HIV-1 leader RNA (9a).
ACKNOWLEDGMENTS
We thank Atze Das for helpful discussions, Bianca Schuijt for performing the LTR-luciferase transcription assays, and Wim van Est for photography work.
This work was supported in part by the Dutch AIDS Fund and by the Netherlands Foundation for Chemical Research with financial aid from the Netherlands Organization for Scientific Research.
REFERENCES
- 1.Aiyar A, Cobrinik D, Ge Z, Kung H J, Leis J. Interaction between retroviral U5 RNA and the TYC loop of the tRNATrp primer is required for efficient initiation of reverse transcription. J Virol. 1992;66:2464–2472. doi: 10.1128/jvi.66.4.2464-2472.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiyar A, Ge Z, Leis J. A specific orientation of RNA secondary structures is required for initiation of reverse transcription. J Virol. 1994;68:611–618. doi: 10.1128/jvi.68.2.611-618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baudin F, Marquet R, Isel C, Darlix J L, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA from defined structural domains. J Mol Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout B. The primer-binding site on the RNA genome of human and simian immunodeficiency viruses is flanked by an upstream hairpin structure. Nucleic Acids Res. 1997;25:4013–4017. doi: 10.1093/nar/25.20.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkhout B, Klaver B, Das A T. Forced evolution of a regulatory RNA helix in the HIV-1 genome. Nucleic Acids Res. 1997;25:940–947. doi: 10.1093/nar/25.5.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkhout B, Schoneveld I. Secondary structure of the HIV-2 leader RNA comprising the tRNA-primer binding site. Nucleic Acids Res. 1993;21:1171–1178. doi: 10.1093/nar/21.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkhout B, Silverman R H, Jeang K T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 9.Berkhout B, van Wamel J L B. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Berkhout, B., and J. L. B. van Wamel. The leader of the RNA genome forms a compactly folded tertiary structure. RNA, in press. [DOI] [PMC free article] [PubMed]
- 10.Brown P O. Integration. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. New York, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 161–204. [PubMed] [Google Scholar]
- 11.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cobrinik D, Aiyar A, Ge Z, Katzman M, Huang H, Leis J. Overlapping retrovirus U5 sequence elements are required for efficient integration and initiation of reverse transcription. J Virol. 1991;65:3864–3872. doi: 10.1128/jvi.65.7.3864-3872.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Damgaard C K, Dyhr-Mikkelsen H, Kjems J. Mapping the RNA binding sites for human immunodeficiency virus type-1 Gag and NC proteins within the complete HIV-1 and -2 untranslated leader regions. Nucleic Acids Res. 1998;26:3667–3676. doi: 10.1093/nar/26.16.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das A T, Klaver B, Berkhout B. Reduced replication of human immunodeficiency virus type 1 mutants that use reverse transcription primers other than the natural tRNA3Lys. J Virol. 1995;69:3090–3097. doi: 10.1128/jvi.69.5.3090-3097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das A T, Klaver B, Berkhout B. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J Virol. 1999;73:81–91. doi: 10.1128/jvi.73.1.81-91.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das A T, Klaver B, Klasens B I F, van Wamel J L B, Berkhout B. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J Virol. 1997;71:2346–2356. doi: 10.1128/jvi.71.3.2346-2356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esposito D, Craigie R. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 1998;17:5832–5843. doi: 10.1093/emboj/17.19.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu W, Ortiz-Conde B A, Gorelick R J, Hughes S H, Rein A. Placement of tRNA primer on the primer-binding site requires pol gene expression in avian but not murine retroviruses. J Virol. 1997;71:6940–6946. doi: 10.1128/jvi.71.9.6940-6946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrich D, Mavankal G, Mette-Snider A, Gaynor R B. Human immunodeficiency virus type 1 TAR element revertant viruses define RNA structures required for efficient viral gene expression and replication. J Virol. 1995;69:4906–4913. doi: 10.1128/jvi.69.8.4906-4913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison G P, Lever A M L. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoglund S, Ohagen A, Goncalves J, Panganiban A T, Gabuzda D. Ultrastructure of HIV-1 genomic RNA. Virology. 1997;233:271–279. doi: 10.1006/viro.1997.8585. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y, Khorchid A, Wang J, Parniak M A, Darlix J-L, Wainberg M A, Kleiman L. Effect of mutations in the nucleocapsid protein (NCp7) upon Pr160gag-pol and tRNALys incorporation into human immunodeficiency virus type 1. J Virol. 1997;71:4378–4384. doi: 10.1128/jvi.71.6.4378-4384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA(3Lys) (template/primer) J Mol Biol. 1995;247:236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
- 25.Isel C, Keith G, Ehresmann B, Ehresmann C, Marquet R. Mutational analysis of the tRNA3Lys/HIV-1 RNA (primer/template) complex. Nucleic Acids Res. 1998;26:1198–1204. doi: 10.1093/nar/26.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Isel C, Westhof E, Le Grice S F, Ehresmann B, Ehresmann C, Marquet R. Structural basis for the specificity of the initiation of HIV-1 reverse transcription. EMBO J. 1999;18:1038–1048. doi: 10.1093/emboj/18.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang S-M, Wakefield J K, Morrow C D. Mutations in both the U5 region and the primer-binding site influence the selection of the tRNA used for the initiation of HIV-1 reverse transcription. Virology. 1996;222:401–414. doi: 10.1006/viro.1996.0437. [DOI] [PubMed] [Google Scholar]
- 27.Kang S-M, Zhang Z, Morrow C D. Identification of a sequence within U5 required for human immunodeficiency virus type 1 to stably maintain a primer binding site complementary to tRNAMet. J Virol. 1997;71:207–217. doi: 10.1128/jvi.71.1.207-217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klasens B I F, Thiesen M, Virtanen A, Berkhout B. The ability of the HIV-1 AAUAAA signal to bind polyadenylation factors is controlled by local RNA structure. Nucleic Acids Res. 1999;27:446–454. doi: 10.1093/nar/27.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klaver B, Berkhout B. Comparison of 5′ and 3′ long terminal repeat promoter function in human immunodeficiency virus. J Virol. 1994;68:3830–3840. doi: 10.1128/jvi.68.6.3830-3840.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klaver B, Berkhout B. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 1994;13:2650–2659. doi: 10.1002/j.1460-2075.1994.tb06555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leis J, Aiyar A, Cobrinik D. Regulation of initiation of reverse transcription of retroviruses. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 33–48. [Google Scholar]
- 32.Li X, Mak J, Arts E J, Gu Z, Kleiman L, Wainberg M A, Parniak M A. Effects of alterations of primer-binding site sequences on human immunodeficiency virus type 1 replication. J Virol. 1994;68:6198–6206. doi: 10.1128/jvi.68.10.6198-6206.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Zhang Z, Wakefield J K, Kang S-M, Morrow C D. Nucleotide substitutions within U5 are critical for efficient reverse transcription of human immunodeficiency virus type 1 with a primer binding site complementary to tRNAHis. J Virol. 1997;71:6315–6322. doi: 10.1128/jvi.71.9.6315-6322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang C, Li X, Rong L, Inouye P, Quan Y, Kleiman L, Wainberg M A. The importance of the A-rich loop in human immunodeficiency virus type 1 reverse transcription and infectivity. J Virol. 1997;71:5750–5757. doi: 10.1128/jvi.71.8.5750-5757.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marquet R, Isel C, Ehresmann C, Ehresmann B. tRNAs as primer of reverse transcriptases. Biochimie. 1995;77:113–124. doi: 10.1016/0300-9084(96)88114-4. [DOI] [PubMed] [Google Scholar]
- 36.Masuda T, Kuroda M J, Harada S. Specific and independent recognition of U3 and U5 att sites by human immunodeficiency virus type 1 integrase in vivo. J Virol. 1998;72:8396–8402. doi: 10.1128/jvi.72.10.8396-8402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Morris S, Leis J. Changes in Rous sarcoma virus RNA secondary structure near the primer binding site upon tRNATrp primer annealing. J Virol. 1998;73:6307–6318. doi: 10.1128/jvi.73.8.6307-6318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paillart J-C, Berthoux L, Ottmann M, Darlix J-L, Marquet R, Ehresmann B, Ehresmann C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J Virol. 1996;70:8348–8354. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prats A C, Sarih L, Gabus C, Litvak S, Keith G, Darlix J L. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988;7:1777–1783. doi: 10.1002/j.1460-2075.1988.tb03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puglisi E V, Puglisi J D. HIV-1 A-rich RNA loop mimics the tRNA anticodon structure. Nat Med. 1998;5:1033–1036. doi: 10.1038/4141. [DOI] [PubMed] [Google Scholar]
- 41.Skripkin E, Isel C, Marquet B, Ehresmann B, Ehresmann C. Psoralen crosslinking between human immunodeficiency virus type 1 RNA and primer tRNAlys3. Nucleic Acids Res. 1996;24:509–514. doi: 10.1093/nar/24.3.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Telesnitsky A, Goff S P. Reverse transcriptase and the generation of retroviral DNA. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 121–160. [PubMed] [Google Scholar]
- 43.Van Lint C, Amella C A, Emiliani S, John M, Jie T, Verdin E. Transcription factor binding sites downstream of the human immunodeficiency virus type 1 transcription start site are important for virus infectivity. J Virol. 1997;71:6113–6127. doi: 10.1128/jvi.71.8.6113-6127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verhoef K, Koper M, Berkhout B. Determination of the minimal amount of Tat activity required for human immunodeficiency virus type 1 replication. Virology. 1997;237:228–236. doi: 10.1006/viro.1997.8786. [DOI] [PubMed] [Google Scholar]
- 45.Vicenzi E, Dimitrov D S, Engelman A, Migone T-S, Purcell D F J, Leonard J, Englund G, Martin M A. An integration-defective U5 deletion mutant of human immunodeficiency virus type 1 reverts by eliminating additional long terminal repeat sequences. J Virol. 1994;68:7879–7890. doi: 10.1128/jvi.68.12.7879-7890.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wakefield J K, Kang S-M, Morrow C D. Construction of a type 1 human immunodeficiency virus that maintains a primer binding site complementary to tRNAHis. J Virol. 1996;70:966–975. doi: 10.1128/jvi.70.2.966-975.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wakefield J K, Rhim H, Morrow C D. Minimal sequence requirements of a functional human immunodeficiency virus type 1 primer binding site. J Virol. 1994;68:1605–1614. doi: 10.1128/jvi.68.3.1605-1614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitcomb J M, Ortiz Conde B A, Hughes S H. Replication of avian leukosis viruses with mutations at the primer binding site: use of alternative tRNAs as primer. J Virol. 1995;69:6228–6238. doi: 10.1128/jvi.69.10.6228-6238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Willey R L, Smith D H, Lasky L A, Theodore T S, Earl P L, Moss B, Capon D J, Martin M A. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu M, Tinoco I. RNA folding causes secondary structure rearrangement. Proc Natl Acad Sci USA. 1999;95:11555–11560. doi: 10.1073/pnas.95.20.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Z, Kang S-M, LeBlanc A, Hajduk S L, Morrow C D. Nucleotide sequences within the U5 region of the viral RNA genome are the major determinants for an human immunodeficiency virus type 1 to maintain a primer binding site complementary to tRNAHis. Virology. 1996;226:306–317. doi: 10.1006/viro.1996.0658. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Z, Kang S-M, Li Y, Morrow C D. Genetic analysis of the U5-PBS of a novel HIV-1 reveals multiple interactions between the tRNA and RNA genome required for initiation of reverse transcription. RNA. 1998;4:394–406. [PMC free article] [PubMed] [Google Scholar]
- 53.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]