Abstract
In this report, we show that the glycoprotein of vesicular stomatitis virus (VSV G) contains within its extracellular membrane-proximal stem (GS) a domain that is required for efficient VSV budding. To determine a minimal sequence in GS that provides for high-level virus assembly, we have generated a series of recombinant ΔG-VSVs which express chimeric glycoproteins having truncated stem sequences. The recombinant viruses having chimeras with 12 or more membrane-proximal residues of the G stem, and including the G protein transmembrane-cytoplasmic tail domains, produced near-wild-type levels of particles. In contrast, viruses encoding chimeras with shorter or no G-stem sequences produced ∼10- to 20-fold less. This budding domain when present in chimeric glycoproteins also promoted their incorporation into the VSV envelope. We suggest that the G-stem budding domain promotes virus release by inducing membrane curvature at sites where virus budding occurs or by recruiting condensed nucleocapsids to sites on the plasma membrane which are competent for efficient virus budding.
Vesicular stomatitis virus (VSV) is a relatively simple enveloped RNA virus from the Rhabdoviridae family that assembles at the plasma membrane of a host cell and is released from the cell by a process called budding. VSV virions consist of a helical ribonucleocapsid (RNP) core, which contains the single-stranded, nonsegmented, negative-sense RNA genome tightly encapsidated by 1,258 molecules of the nucleocapsid (N) protein (45). The viral polymerase, which consists of the phosphoprotein (P) and the large catalytic subunit (L protein), is also tightly associated with the RNP in virions. Prior to budding, the nucleocapsid core condenses upon binding to the matrix (M) protein. Initiation of virus budding occurs when the condensed core associates with the inner leaflet of the plasma membrane, presumably through M protein-dependent interactions (6, 18). During budding, the condensed core becomes enclosed within a membrane envelope that consists of host-derived lipids and approximately 1,200 molecules of the VSV spike glycoprotein (G protein) (45).
Although much progress has been made in defining domains that are important for the assembly and release of virions from the cell surface, as well as for glycoprotein incorporation into virions, relatively little is known about what drives the budding process. For example, studies examining the role of glycoprotein cytoplasmic tails (CTs) in the assembly of a variety of enveloped viruses have provided evidence that the tails are indeed important but often not essential for virus assembly or infectivity (1, 9, 14–16, 28, 30, 39, 47). In contrast, it has been shown that there is an absolute requirement for the CT of alphavirus glycoprotein in alphavirus assembly and budding (5, 19, 24, 32, 43). At the other extreme, many retroviruses (8, 11, 34, 46) and now rhabdoviruses (28, 44) have been shown not to require viral glycoproteins at all for the assembly and release of virus particles. It is likely that interactions of the late domains, found within rhabdovirus M or retroviral Gag proteins, with host factors at the plasma membrane are the primary driving force in budding and release of particles (7, 12). In the case of rhabdoviruses, typical bullet-shaped virions are produced from cells infected with recombinant viruses that either lack G protein (ΔG viruses) (28, 44) or express heterologous glycoproteins in the absence of G protein (17, 41). However, the amount of ΔG virus released is significantly less than that released from wild-type (WT)-virus-infected cells, suggesting that G protein contributes to the efficiency of virus budding.
To examine the requirements for VSV assembly and budding, we have taken a reverse genetics approach and have generated several different recombinant VSVs that encode either truncated or chimeric envelope proteins. The goal of these studies is to understand which components of the virion are essential for high-level virus budding and what factors influence the efficiency of glycoprotein incorporation into virions. In this report, we show that a relatively small domain in the membrane-proximal stem region of the G protein ectodomain contributes to efficient G protein incorporation and virus budding. We suggest that the G-stem domain contributes directly to high-level virus budding either by facilitating membrane curvature at the bud site or by selecting subdomains of the plasma membrane that are competent for virus release, perhaps by modifying the local lipid environment such that efficient virus release occurs.
MATERIALS AND METHODS
Recombinant cDNAs.
The construct GSHA (G-stem hemagglutinin epitope [HA] tagged), which encodes a truncated version of VSV G protein, was generated by PCR-mediated mutagenesis using a sense primer that overlapped the MluI site in the 5′ nontranslated region of the G protein gene of pVSVFL(+)-2 and an antisense oligonucleotide, 5′-AGGATGTTCGAAAGCGTAATCTGGTACATCATACGGATACTTGCAATTCACCCCAATG-3′, which overlapped the NspV site (underlined) at position 1280 in the G protein gene. The antisense primer linked sequences encoding a portion of the G protein signal peptide (in italics) to the HA epitope (in boldface) followed by the membrane-proximal stem region of the G protein ectodomain. The amplicon was digested with MluI and NspV and then subcloned into pBS-GMMG (42), which had been digested with the same two enzymes. A similar construct, GS (G stem), which did not contain the HA epitope, was also generated using the appropriate mutagenic primer. The sequences were confirmed by dideoxynucleotide sequencing. The GSHA or GS fragments were then moved into pVSV-FL(+)-2 (21) by replacing the G gene using the MluI and NheI sites.
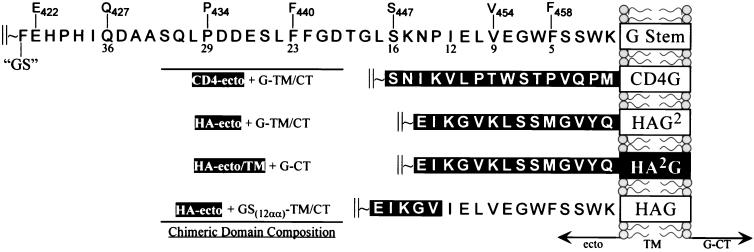
The CD4-G chimeras were constructed by PCR-mediated mutagenesis using, individually, a series of sense-strand oligonucleotides that had a common 5′ sequence (5′-ATGGCCTCGGGT…) which contained an AvaI site (underlined) followed by sequences coding for amino acids starting at Q427, P434, F440, S447, V454, and F458, respectively (complementary sequences are indicated by …), and an antisense oligonucleotide, 5′-CCAAACATGAAGCTTCTGTTGTGCATGCTTTGAGTTAC-3′, which introduced an SphI site (underlined) within the 3′ untranslated region of the GInd cDNA. In-frame chimeras were produced by ligation of a XhoI-AvaI fragment encoding the ectodomain of human CD4, with the various AvaI-SphI fragments encoding GS peptides of varying lengths (illustrated in Fig. 1). Also, similar CD4 constructs, E422 and G404-link, were produced by in-frame ligations of the CD4-encoding XhoI-AvaI fragment to either a naturally occurring NspV site or a naturally occurring KpnI site in the G-encoding sequence (at amino acid positions 422 and 404, respectively) by using either an AvaI-NspV- or an AvaI-KpnI-compatible double-stranded oligonucleotide linker that encoded amino acid residues GASKAQV or AGGGSGGGGST, respectively.
FIG. 1.
Comparison of membrane-proximal regions of the VSV GI ectodomain, and chimeras containing heterologous sequences derived from the ectodomains of human CD4 or influenza virus HA. The top schematic represents the stem region of G protein (GS), with numbers below the amino acid sequence of the ectodomain (ecto) indicating the distance in residues from the TM domain (boxes) and where residue numbering above the stem indicates their position from the N terminus of G protein. This nomenclature is used for the CD4 chimeras to indicate the position of G protein that was linked to the CD4 ectodomain. Representation of chimeric domain composition is shown at left, where domains encoding heterologous sequences are in dark boxes.
A full-length ΔG-VSV cDNA was generated by replacing the WT G protein gene in pVSVFL(+)I/NJG (21) with an oligonucleotide-derived polylinker region (5′-MluI-KpnI-XhoI-SmaI-EagI-SphI-NheI-3′) introduced between the MluI and NheI sites of pVSVFL(+)I/NJG. This plasmid (pVSVΔG-PL) was used to generate the recombinant VSV cDNAs encoding either GS, GSHA, or the CD4-G chimeras described above. A six-gene VSV cDNA, which had an additional transcription unit placed between the G and L genes, was also generated. This construct, called pVSV-ΔG-GSHA/GFP, expressed GSHA from the upstream transcription unit and green fluorescent protein (GFP) (3, 13) from the adjacent downstream transcription unit located immediately before the L gene.
Production of recombinant VSV.
Recovery and propagation of infectious ΔG viruses were similar to those described previously (44). Approximately 106 BHK-21 cells in 35-mm-diameter dishes were infected with a recombinant vaccinia virus encoding bacteriophage T7 RNA polymerase at a multiplicity of 5 for 40 min. The cells were then transfected with a DNA-liposome suspension composed of 5 μg of the appropriate pVSV-ΔG plasmid and 8, 3, 5, and 1 μg of plasmids containing the WT G, N, P, and L genes, respectively, from the Indiana serotype of VSV (VSVInd). Cationic liposomes used for the transfections were prepared as described previously (2, 35). After 4 h, the transfection mix was replaced with Dulbecco modified Eagle's medium (DMEM) containing 10% fetal bovine serum. At 48 h posttransfection, the supernatants were harvested and filtered through a 0.2-μm-pore-size filter (Millex-GS; Millipore) to remove vaccinia virus. The filtrates were applied to 106 BHK-21 cells transiently expressing G protein. For transient expression of G protein, we used pCVSVG (44). Approximately 7 × 105 BHK-21 cells were transfected with a DNA-liposome suspension composed of 10 μl of Lipofectamine (Gibco-BRL) and 2 μg of pCVSVG in 2 ml of OptiMEM (Gibco-BRL) according to the manufacturer's instructions. After 8 to 12 h, the transfection mix was replaced with DMEM containing 10% fetal bovine serum. Filtrates were added to the cells at 36 h posttransfection. Cells were examined for cytopathic effects typical of VSV infection after 24 to 72 h. The recombinant viruses were then plaque purified on cells transiently expressing G protein and subsequently passaged on G-expressing cells to produce high-titered infectious stocks (∼108 to 109 IU/ml). Titers were determined using an immunofluorescence-based assay. Cells were infected with limiting dilutions of virus supernatants and fixed with 3% paraformaldehyde at 12 to 18 h postinfection, and the number of infected cells was determined by indirect immunofluorescence using anti-M protein-specific (23H12) or N protein-specific (10G4) monoclonal antibody (23). Viral expression of the foreign or chimeric proteins was verified for the individual clones and for passaged virus stocks by indirect immunofluorescence using either a CD4-specific monoclonal antibody (Sim.2 [25] obtained from the National Institutes of Health AIDS Research and Reference Reagent Program), an HA epitope-specific monoclonal antibody (12CA5, kindly provided by Lorraine Albritton, University of Tennessee—Memphis), or an A/Japan HA-specific goat polyclonal serum (kindly provided by Robert Webster, St. Jude Children's Research Hospital).
The ΔG-GSHA virus was recovered by an alternative strategy in which a VSV minivirus (GMMG) expressing only the G and M proteins (42) was used to provide the source of complementing G protein. Approximately 106 vTF7-3 infected cells were transfected with 10 μg of pVSV-ΔG-GSHA and 3, 5, and 1 μg of plasmids encoding the WT VSVInd N, P, and L proteins, respectively. The transfection mix was removed after 3 h, and the cells were then superinfected with GMMG particles at a multiplicity of infection (MOI) of 1. Fresh medium was added directly to the cells after they adsorbed the GMMG minivirus for 1 h. Supernatants were harvested 18 h later and then filtered to remove vaccinia virus. One-half of each filtered supernatant was used to infect fresh cells. Successful recoveries were indicated when cultures showed the typical cytopathic effects of a normal VSV infection after 18 to 24 h. The titers of the supernatants from those cultures were then determined, and stocks of the cocomplementing viruses were produced by infecting fresh cells at an MOI of 0.01. Supernatants, which contained both ΔG-GSHA and GMMG, were harvested after 24 h, and the two particles were concentrated by centrifugation and then separated by banding in 20 to 45% sucrose gradients. The lower fraction, which usually contained approximately 300-fold-more ΔG-GSHA than GMMG, was recovered by side puncture, and the proportion of ΔG-GSHA to GMMG was determined by an immunofluorescence-based titering assay utilizing either an N protein-specific (for ΔG-GSHA) or a G protein-specific monoclonal antibody (I1) (23). To obtain a stock of ΔG-GSHA which was free of GMMG, the partially purified ΔG-GSHA fraction was used to infect cells expressing the G protein from the New Jersey serotype of VSV (GNJ) and then neutralizing antibody, which was specific for the Indiana serotype of VSV (VSVInd), was added to the culture medium. The amount of Indiana-specific serum used completely neutralized all virus produced by ΔG-GSHA–GMMG-coinfected cells. After three consecutive passages on cells expressing GNJ and in the presence of anti-VSVInd neutralizing antibody, the titers of ΔG-GSHA reached approximately 108 IU/ml while 0.5 ml of the supernatant contained no detectable GMMG. High-titered stocks of pure ΔG-GSHA complemented with the GInd protein were then made as described above for the CD4-G chimeric ΔG viruses.
Virus budding assay.
BHK-21 cells were infected with either WT VSV or the G-complemented ΔG viruses at an MOI of 10 for 1 h, washed three to four times with DMEM, and incubated in serum-free DMEM. At 18 h postinfection, virions were purified from the supernatants by centrifugation through a 20% sucrose cushion in an AH650 rotor at 45,000 rpm for 35 min. The pellets were suspended in equal volumes of reducing sample buffer, and one-third of each sample was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by colloidal Coomassie blue staining (Gel Code Blue; Pierce Chemical Co.). Virus yields were determined by quantifying N protein content using ImageQuant analytical software (Molecular Dynamics) from digital images of the stained gels captured using transmitted light.
Metabolic labeling of viral protein.
The CTs of WT GI protein, GSHA, HA-G, and the CD4-G chimeras were labeled with [3H]palmitate by infecting 2 × 106 BHK-21 cells with the appropriate viruses at an MOI of 10 for 1.5 h. Following infection, the cells were washed twice with DMEM, and then DMEM supplemented with 1.5% dimethyl sulfoxide and 2.5 mCi of 9,10-[3H]palmitate was added. Virions were purified from the supernatant as described above, and one-half of each of the samples was resolved on a 10 to 20% polyacrylamide gradient SDS-PAGE gel followed by fluorography.
Chemical cross-linking studies.
Samples containing approximately 250 ng of purified virions were incubated with DTSSP [dithiobis(sulfosuccinimidylpropionate)] at final concentrations ranging from 0 to 200 μM in 5 mM sodium citrate (pH 5.5) for 30 min on ice. The reactions were quenched by making the solution to 50 mM glycine. Approximately 1/10 of each sample was resolved by nonreducing SDS-PAGE and transferred to a polyvinylidene difluoride membrane. Immunoblotting was carried out using anti-G CT-specific polyclonal rabbit sera, followed by chemiluminescent detection using the ECL reagent (Pierce) according to the manufacturer's instructions.
RESULTS
Assembly phenotypes of recombinant ΔG-VSVs.
Previous studies by others using recombinant VSVs that expressed heterologous glycoproteins, either in the presence or in the absence of WT G protein, had found that some foreign glycoproteins could be efficiently incorporated into VSV particles, while others could not (17, 20, 40, 41). These results suggested that heterologous glycoprotein incorporation is not dependent on the presence of G. However, the amount of rhabdovirus particles produced in the absence of G protein was significantly less than that when G protein was expressed (28, 41). Therefore, G protein contributes to the efficiency of virus budding. To understand the basis for this phenomenon, we set out to first identify domains in G protein that contribute to high-level virus budding as seen during a WT VSV infection.
To examine the contribution of the transmembrane (TM) and CT domains in virus budding and glycoprotein incorporation, we produced several different recombinant VSVs in which the G gene was replaced by those of heterologous or chimeric proteins. To examine virus budding in the absence of a virus-encoded glycoprotein, we used a recombinant VSV described previously (44) called ΔG*, which has the G gene replaced with that encoding GFP. Figure 2 shows the results from recombinant viruses that encode either CD4 or a chimera composed of the CD4 ectodomain fused to the TM and CT of G (CD4-G). In Fig. 2A, viruses encoding either CD4-G, CD4, or GFP (no G protein) produced approximately 10- to 20-fold-fewer particles than WT VSV. These data indicate that the TM-CT of G protein is not sufficient to drive efficient virus budding, in the context of the CD4 ectodomain, nor do these domains enhance incorporation of CD4-G relative to that of CD4. Therefore, the ectodomain, or a region within the ectodomain, must contribute to high-level virus budding and efficient envelope protein incorporation as seen for WT VSV.
FIG. 2.
Budding efficiency and glycoprotein incorporation. (A) Cells infected with recombinant ΔG-VSVs were metabolically labeled with [35S]methionine, and virus released into the supernatant was purified by ultracentrifugation. The viral pellets were resuspended in identical volumes of reducing SDS sample buffer, and proteins were resolved by SDS-PAGE followed by Coomassie blue staining. Shown is a Coomassie blue-stained gel on which equivalent sample volumes were loaded (corresponding to one-third of the virus produced from 106 cells). The same gel was used for autoradiography and resulted in a protein profile indistinguishable from that shown in the figure. (B) Similar amounts of viral protein were loaded per lane as determined by 35S radioactivity. Quantitation of viral proteins was determined using a STORM phosphorimager and ImageQuant software. Virus yields were calculated from the signal intensities produced by N, P, and L proteins, either in sum or separately, and percentages were determined relative to the values obtained for WT VSV.
Characterization of the membrane-proximal G stem.
To determine if we could identify a region in the G protein ectodomain that promotes high-level virus production, we generated a virus that expressed a truncated form of G protein that has the 42 membrane-proximal residues of the G ectodomain plus the TM anchor and CT. We call this protein G stem (GS) for simplicity. The rationale for designing this construct was based on reports that membrane-anchored proteolytic cleavage products of G are transport competent and are incorporated in virions (4). These C-terminal proteolytic fragments were once thought to be essential for the budding of spikeless (or bald) ts045 virions produced at the nonpermissive temperature (29). To determine if GS was expressed on the cell surface, we constructed GSHA by replacing ∼90% of the ectodomain with a nine-residue N-terminal HA epitope tag at position F421 (Fig. 1). Cells transiently expressing GSHA showed bright surface staining when examined by indirect immunofluorescence microscopy using the HA-specific monoclonal antibody 12CA5, indicating that GSHA was transported to the plasma membrane. The GS or GSHA cDNAs were inserted into ΔG-PL to create ΔG-GS or ΔG-GSHA, respectively. A third virus, called ΔG-GSHA*, which expressed GFP from a sixth VSV transcriptional unit placed between the GSHA and L protein genes, was also produced.
In contrast to the CD4 recombinants, ΔG-GSHA-infected cells produced near-WT amounts of virus (Fig. 2A, lane 2). The results from multiple independent experiments indicated that virus yields for ΔG-GSHA ranged from 70 to 80% of WT level, whereas the yields for ΔG*, ΔG-CD4, and ΔG-CD4G ranged from approximately 5 to 10%. The amounts of CD4 or CD4-G in purified virions were virtually identical, ranging from 14 to 18% of that found for G protein when normalized to N protein and for methionine content (Fig. 2B; see also Fig. 6 legend).
FIG. 6.
Incorporation of assembly-proficient or assembly-deficient spike proteins into VSV virions. The cytoplasmic tails of the spike proteins were metabolically labeled with [3H]palmitic acid and analyzed as described in the text. Identical volumes of samples were loaded onto a 10 to 20% gradient gel, electrophoresed, and then fluorographed. The amount of spike incorporated (shown as a percentage of that for G) was determined after normalization to N protein content. The percentages of G incorporation were calculated from similar signal intensities using different exposure times of the fluorographs. The total values shown are for the processed and unprocessed forms of those proteins in which cleavage occurs; HAG(0) uncleaved and HAG(2) furin-processed C termini, or the full-length and C-terminal fragments of CD4-S447 and -V454 were combined to give an incorporation level for both species.
Envelope protein oligomerization and effects on virus assembly.
Considering that CD4 is a monomer and G protein is a trimer, we next asked if GSHA is also oligomeric, and if so, could this be the natural context by which spike proteins promote efficient assembly of VSV particles? To address whether GS has the potential to form oligomers, we conducted chemical cross-linking analysis using a membrane-impermeable reagent (DTSSP). As shown in Fig. 3, GSHA could be cross-linked into species migrating at the molecular weights expected for dimeric and trimeric forms of the protein. Similar experiments were also conducted using the membrane-permeable cross-linking reagent DSP [dithiobis(succinimidylpropionate)]. Similarly, we found that GSHA dimer and trimer species were also observed with increasing DSP concentrations (data not shown).
FIG. 3.
Chemical cross-linking analysis of the G stem. Purified virions were subjected to increasing concentrations (0, 10, 100, and 200 μM) of the membrane-impermeable cross-linking reagent DTSSP. After quenching of the reaction, the samples were resolved by SDS-PAGE under nonreducing conditions and detected by immunoblotting using anti-G CT-specific polyclonal rabbit sera. The GSHA molecule is shown to be similar to WT G protein in its ability to be chemically cross-linked into species that migrate at the approximate molecular masses expected for their dimeric and trimeric forms.
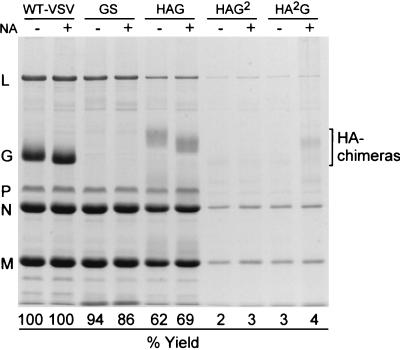
Since the G stem appears to be an oligomer, we next wanted to determine if the TM and CT domains, when presented in the context of a trimer, could drive high-level virus budding. To test this hypothesis, we constructed recombinant ΔG-VSVs that expressed influenza virus HA-G protein chimeras (Fig. 1). These chimeras had previously been shown to fold, oligomerize, and be transported to the cell surface similar to WT-HA (22, 36). The three constructs that we examined were composed of (i) the ectodomain of HA fused to the TM-CT of G (HAG2, previously referred to as HA-G-G [22]), (ii) the ectodomain-TM of HA fused to the CT of G (HA2G, previously referred to as HA-HA-G [22]), and (iii) the ectodomain of HA fused to amino acid 451 of G, which corresponds to the 12th membrane-proximal residue of GS, as well as the TM-CT of G (HAG [36]). When these viruses were grown in cells that did not express G protein, but in the presence of trypsin and neuraminidase, we found that infectious particles were released. This indicated that the HA-G chimeras were functionally incorporated into the VSV envelope. The protein profiles of the HA-G chimera-expressing ΔG-VSVs are shown in Fig. 4. Cells infected with viruses that expressed the HA chimeras containing either the TM-CT (HAG2) or only the CT of G protein (HA2G) produced less than 5% of the amount of virus released by WT VSV-infected cells. These data show that the TM-CT or only the CT domains of G, when presented under the structural constraints of an HA trimer, are not sufficient to drive high-level budding. Furthermore, these results demonstrate that the TM and/or CT does not directly confer any positive effect on virus release or glycoprotein incorporation in the absence of the G ectodomain. In contrast, when cells were infected with the ΔG-HAG virus, which has 12 TM-proximal residues of GS together with the TM-CT of G, particle production increased approximately 20- to 30-fold over that produced by the HAG2 or HA2G virus. The results indicate that as few as 12 membrane-proximal residues of GS, in the context of an HA-G chimera, are sufficient to facilitate high-level virus budding.
FIG. 4.
Virus produced by recombinant viruses expressing oligomeric spike proteins. Virions budded in the absence or presence of neuraminidase (NA) by viruses expressing chimeras consisting of either the ectodomain of HA plus 12 membrane-proximal residues of the G stem-TM-CT (HAG), the ectodomain of HA plus the G TM-CT (HAG2), or the ectodomain and TM of HA plus the G CT (HA2G) (Fig. 1) were compared to those of WT VSV and ΔG-GS. Viral proteins were analyzed by loading equivalent volumes and resolving them by SDS-PAGE. The gel was Coomassie blue stained and photographed with transmitted light using a digital camera. Virus yields were determined by quantitation of the N protein-containing bands using ImageQuant software and calculated as a percentage of WT VSV N protein content. L protein and P protein content were also calculated and found to closely agree with the results for the N protein analysis.
Effects of G-stem truncations on virus assembly and budding.
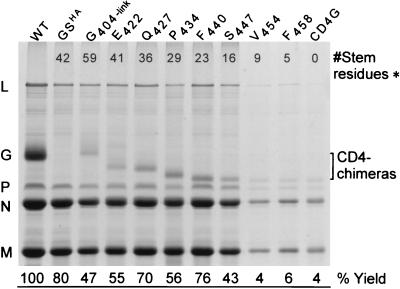
We next wanted to determine a minimal sequence within GS that was sufficient to confer high-level virus budding. To address this question, we generated a series of ΔG-VSVs that expressed chimeric glycoproteins composed of the CD4 ectodomain fused to N-terminally truncated G stems (Fig. 1). By transient transfection, these chimeras were found to be expressed on the cell surface at levels comparable to those of CD4 or CD4-G. When cells were infected with recombinant viruses expressing CD4 chimeras that possess either 9, 5, or 0 membrane-proximal GS residues (V454, F458, or CD4-G, respectively), ΔG* or basal levels of particles were produced. This corresponded to approximately 5% of the particles produced by WT VSV (Fig. 5). In contrast, the amount of virus released in the context of chimeras containing at least 16 GS residues was approximately 10- to 20-fold higher than the ΔG* levels. From multiple experiments, this corresponded to near-WT production levels (between 40 and 80%). Together with the HA-G chimera data, these results demonstrate that in the context of heterologous glycoprotein chimeras the membrane-proximal 12 to 16 residues of the G ectodomain are sufficient for high-level virus budding.
FIG. 5.
Virus budded from cells infected by recombinant ΔG-VSVs expressing CD4 chimeras containing truncated stem sequences. Virions released from cells infected with viruses expressing CD4 chimeras having different-length stem sequences were compared to those of either WT VSV or ΔG-GSHA (∗, see Fig. 1 for details). Virions were prepared as described in the text, and virus pellets were suspended in identical volumes of sample buffer. Viral proteins were analyzed as described for Fig. 4.
It seemed likely that a budding domain of an envelope protein would also facilitate its efficient incorporation into the viral envelope. As shown in Fig. 6, the GS and HAG proteins were efficiently incorporated into virions at about 80 to 90% of the level of G. However, it appeared that the assembly-proficient CD4-GS chimeric glycoproteins were not incorporated nearly as well as WT G or HAG (Fig. 4 and 5). This was unexpected, since our assumption was that efficient incorporation of a membrane-spanning protein would be associated with that protein's ability to promote high-level virus budding. Upon initial examination by Western blot analysis, using CT-specific antisera, we found that the CD4-GS chimeras were incorporated into virions as full-length glycoprotein and as C-terminal processed fragments (data not shown). Similar results for metabolically labeled proteins are shown in Fig. 6. The [3H]palmitate-labeled products of the CD4-GS molecules are heterogeneous in size as a result of cleavage near the CD4-GS junction. The full-length glycoprotein and material of approximately 12 to 16 kDa (similar in size to GS) were incorporated, indicating that the envelope contains both species (e.g., cleaved GS and full-length CD4-GS). Although small amounts of these low-molecular-weight species were also observed in WT VSV virions, they were completely absent from HAG and CD4-G virions. Thus, it appears that the processing event(s) leading to the generation of GS fragments is not obligatory for high-level virus budding or glycoprotein incorporation.
DISCUSSION
Previous reports have demonstrated that rhabdoviruses can bud in the absence of virally encoded glycoproteins (28, 44). However, the number of virus particles released from ΔG-infected cells was significantly less than that made by WT virus. For example, ΔG-rabies virus (RV) produced 30-fold-less virus (28) than did the parental WT virus. In the case of some ΔG-VSVs which encoded foreign glycoproteins, approximately 50-fold-fewer particles than from the comparable WT counterpart were released (41). Although the amount of virus released from ΔG-infected cells was significantly reduced, these initial studies provide the first direct evidence that the minimal components needed to initiate, drive, and complete the budding process are the condensed RNP core in association with the matrix (M) protein. Furthermore, these findings indicated that the mechanism of rhabdovirus budding is probably more like that of type C retroviruses, which also do not require viral glycoproteins to release particles (reviewed in reference 10). In fact, these different types of viruses share similar proline-rich domains (PPxY motif in rhabdoviral M and late domain of retroviral Gag), which have recently been implicated to be the primary driving force in particle budding (7, 12). However, it is clear that rhabdovirus budding in the absence of the WT glycoprotein is inefficient; therefore, the envelope glycoprotein contributes to virus release. Similar findings have recently been reported for baculoviruses, where efficient budding is dependent on the presence of the major envelope glycoprotein, GP64 (31).
In this report, we analyzed the budding phenotypes of several different recombinant ΔG-VSVs to identify regions of G protein that are essential for high-level assembly and budding. We found that the TM and CT domains of G are not sufficient to promote high-level virus budding even when the TM and CT are presented in the context of a trimer. Recombinant viruses expressing either CD4, CD4G, or the two HA-G chimeras (HA2G and HAG2) produced the same amount of virus as did ΔG-VSV. Although the recombinant VSVs that we used differ from those used by Schnell et al. (39), our findings corroborate their conclusions that the TM and CT domains, per se, are not the primary determinants of G protein that contribute to virus budding or to glycoprotein incorporation into virions. In this respect, VSV differs from RV in that foreign glycoprotein incorporation appears to require the RV G protein TM and CT domains (26, 27). The basis for these alternative requirements is currently not known, but what has emerged from these studies is that glycoprotein assembly into RV appears to be quite stringent. In contrast, VSV appears to be more promiscuous and can utilize a variety of different TM and CT sequences. Although there is no specific sequence requirement in the VSV CT domain, there is a strong requirement for the presence of at least a short sequence of amino acids to promote budding (39). The observation that oligomerization of the TM-CT domains of G was not sufficient to restore high-level budding of virions was somewhat unexpected, since models of virus budding propose that interactions (whether specific or nonspecific) between the CT and components of the condensed nucleocapsid ostensibly are important to initiate or to drive virus budding. Instead, we found that a C-terminal portion of G, containing a relatively small region in the membrane-proximal stem of the G protein ectodomain in conjunction with its TM and CT domains, was the primary determinant for promoting efficient virus budding.
There are now several examples where extracellular domains have been shown to be critical for glycoprotein incorporation into virions. In the case of the influenza virus M2 ion channel, a signal within the extracellular domain has been identified which mediates incorporation of M2 into influenza virus particles (33). Furthermore, M2 appears to be important for incorporation of other influenza virus envelope proteins into virions. Therefore, the ectodomain of M2 may serve in targeting the viral spikes to regions of the plasma membrane where virus particles are actively budding, rather than participating directly in the budding process. Another example where the ectodomain of a viral spike protein is critical for its incorporation into virions is the hydrophobic, tryptophan-rich region of the membrane-proximal domain of human immunodeficiency virus type 1 (HIV-1) gp41 (37). Not only is the highly conserved tryptophan-rich motif in gp41 important for viral glycoprotein incorporation into HIV virions, but also it is vital for fusogenic activity. In recent studies, we have found that the VSV GS can enhance or potentiate the membrane fusion activity of a variety of different viral fusion proteins (C. S. Robison and M. A. Whitt, 18th Annu. Meet. Am. Soc. Virol., abstr. W32-10, 1999). This suggests that, for VSV G protein, and possibly HIV gp41, the membrane-proximal domain may be the critical link between two opposing but related activities (e.g., virus budding and virus entry).
The importance of the G protein membrane-proximal domain for virus assembly and release was initially demonstrated by the phenotypes of viruses expressing membrane-anchored GS proteins. Since the GS proteins contained less than 10% of the ectodomain, it was remarkable that such a small region was able to drive virus budding to levels similar to that conferred by WT G protein. The results from both the CD4 and HA chimeric viruses further demonstrated that a subdomain of GS necessary to promote high-level budding includes at least 12 to 16 membrane-proximal residues, whereas nine or fewer residues did not support this phenotype. One mechanism that could account for these findings is that virus budding presumably requires significant modification of the lipid environment at the site of assembly (or bud site), such that membrane curvature is induced while the condensed nucleocapsid-matrix complex associates with the inner leaflet of the plasma membrane. It is conceivable that sequences in the membrane-proximal region of envelope glycoproteins have been selected to facilitate lipid reorganization either before or during the budding event. Alternatively, the stem domain may associate the glycoprotein with specific lipid types or membrane domains during transport. Since virus budding involves the concerted action of both viral and cellular components, the stem may also select for regions of the plasma membrane that are competent for virus budding. Support for this suggestion comes from immunofluorescence studies which revealed that GS-containing glycoproteins (i.e., CD4-GS and HAG), but not those lacking GS sequence (i.e., CD4, CD4-G, or HAG2), were localized in clusters or blebs on the plasma membrane (unpublished observations). These regions presumably represent sites of active virus budding, since matrix (M) protein also appeared to be concentrated at these sites. To address whether the restricted localization results from coalescence of viral components by specific interactions exclusively or from some limitation in the number or location of regions at the plasma membrane best suited for efficient virus assembly will require additional studies using confocal microscopy where high-resolution optical sections of the bud sites can be examined in greater detail. In any event, it is evident that highly efficient virus budding of VSV virions is mediated by the membrane-proximal G-stem domain through a concerted yet undefined synergy with the condensed nucleocapsid-matrix protein complex.
To extend these observations further, we examined amino acid sequence alignments of the membrane-proximal regions of several related rhabdovirus glycoproteins, as well as of some heterologous viral glycoproteins (Fig. 7). This analysis revealed some intriguing features that may be important for the assembly phenotype that we have observed. Among the vesiculoviruses, this region is highly conserved; however, significant divergence is seen when the analysis is extended to RV (a lyssavirus) and an unclassified fish rhabdovirus. Yet there are characteristics within the domains of these less-related glycoproteins that are absent from glycoproteins (such as CD4 and influenza virus HA) that result in low or basal levels of virus budding in the context of recombinant VSVs. First, there are tryptophan residues located at position 6 and at position 2 or 3 relative to the TM domain. These are reminiscent of those found in the HIV-1 gp41 discussed above. Second, there are two hydrophobic residues located at positions 9 and/or 10. The tryptophans and the hydrophobic amino acids are not present in either the A/Japan HA or CD4, both of which were not efficiently incorporated into the recombinant VSVs and did not confer high-level virus budding, unless appended to at least 12 to 16 residues of the G stem. It will be interesting to determine if the introduction of these residues into the analogous positions of the CD4 or HA stem region can confer high-level budding of VSV.
FIG. 7.
Sequence alignment of the TM-ectodomain junction of VSV G protein with analogous regions of related and unrelated type I glycoproteins. Darkly shaded boxes indicate conserved tryptophan residues within the membrane-proximal regions of the glycoprotein ectodomains which are spaced either two or three residues and/or six residues from the putative TM domain. Lightly shaded boxes indicate hydrophobic residues positioned at either the third, fourth, or seventh residue from the conserved tryptophans. A high degree of sequence homology within their membrane-proximal regions is observed for related vesiculovirus glycoproteins, whereas the glycoproteins of a fish rhabdovirus (hemorrhagic septicemia virus, an unclassified rhabdovirus), RV (a lyssavirus), ebola virus (a filovirus), and HIV-1 (a lentivirus) are similar to VSV G only in that these have a similar spacing of tryptophan and hydrophobic residues at a conserved distance from their TM domains.
We certainly cannot exclude the possibility that the VSV budding domain in GS is responsible for localization of the glycoprotein within zones of the plasma membrane where viral RNPs are actively budding. It is possible that specificity of incorporation is dictated by a protein's affinity for particular membrane lipids, which could potentially explain lipid-type selectivity that results in distinct lipid compositions of different viral envelopes (38). However, not mutually exclusively, we favor a model in which the budding domain is involved in recruitment of, or being recruited to, the condensed RNPs underlying the plasma membrane during the initial stages of virus budding. What remains to be tested is whether VSV budding occurs at different sites at the plasma membrane when the GS budding domain is present compared to when it is absent. In our most simplistic view, a protein containing a VSV budding domain, which has the capacity to synergize M protein-mediated particle budding, must therefore contribute to its own incorporation into the VSV envelope.
ACKNOWLEDGMENTS
We greatly appreciate the generous gifts of the HA-G-encoding cDNAs from Michael Roth (University of Texas Southwestern). We also thank Robert Webster (St. Jude Children's Research Hospital) for the kind gift of the HA-specific antisera and Lorraine Albritton (University of Tennessee—Memphis) for the 12CA5 hybridoma supernatant. CD4-specific antibodies were provided by the NIH AIDS Research and Reference Reagent Program. We thank Jeetendra Eswaraka and Himangi Jayakar for helpful comments on the manuscript. The technical assistance of Carolyn Matthews and Anne Timmerman is greatly appreciated. Oligonucleotides were synthesized by the Molecular Resource Center at University of Tennessee—Memphis.
This work was supported by NIH grant GM-53726 (to M.A.W.).
REFERENCES
- 1.Bilsel P, Castrucci M R, Kawaoka Y. Mutations in the cytoplasmic tail of influenza A virus neuraminidase affect incorporation into virions. J Virol. 1993;67:6762–6767. doi: 10.1128/jvi.67.11.6762-6767.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell M J. Lipofection reagents prepared by a simple ethanol injection technique. BioTechniques. 1995;18:1027–1032. [PubMed] [Google Scholar]
- 3.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 4.Chen S S, Ariel N, Huang A S. Membrane anchors of vesicular stomatitis virus: characterization and incorporation into virions. J Virol. 1988;62:2552–2556. doi: 10.1128/jvi.62.8.2552-2556.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng R H, Kuhn R J, Olson N H, Rossmann M G, Choi H K, Smith T J, Baker T S. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong L D, Rose J K. Interactions of normal and mutant vesicular stomatitis virus matrix proteins with the plasma membrane and nucleocapsids. J Virol. 1994;68:441–447. doi: 10.1128/jvi.68.1.441-447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craven R C, Harty R N, Paragas J, Palese P, Wills J W. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J Virol. 1999;73:3359–3365. doi: 10.1128/jvi.73.4.3359-3365.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delchambre M, Gheysen D, Thines D, Thiriart C, Jacobs E, Verdin E, Horth M, Burny A, Bex F. The GAG precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 1989;8:2653–2660. doi: 10.1002/j.1460-2075.1989.tb08405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia Sastre A, Palese P. Influenza virus vectors. Biologicals. 1995;23:171–178. doi: 10.1006/biol.1995.0028. [DOI] [PubMed] [Google Scholar]
- 10.Garoff H, Hewson R, Opstelten D J E. Virus maturation by budding. Microbiol Mol Biol Rev. 1998;62:1171–1190. doi: 10.1128/mmbr.62.4.1171-1190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 12.Harty R N, Paragas J, Sudol M, Palese P. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J Virol. 1999;73:2921–2929. doi: 10.1128/jvi.73.4.2921-2929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heim R, Cubitt A B, Tsien R Y. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 14.Januszeski M M, Cannon P M, Chen D, Rozenberg Y, Anderson W F. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J Virol. 1997;71:3613–3619. doi: 10.1128/jvi.71.5.3613-3619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin H, Leser G P, Lamb R A. The influenza virus hemagglutinin cytoplasmic tail is not essential for virus assembly or infectivity. EMBO J. 1994;13:5504–5515. doi: 10.1002/j.1460-2075.1994.tb06885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin H, Leser G P, Zhang J, Lamb R A. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 1997;16:1236–1247. doi: 10.1093/emboj/16.6.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson J E, Schnell M J, Buonocore L, Rose J K. Specific targeting to CD4+ cells of recombinant vesicular stomatitis viruses encoding human immunodeficiency virus envelope proteins. J Virol. 1997;71:5060–5068. doi: 10.1128/jvi.71.7.5060-5068.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Justice P A, Sun W, Li Y, Ye Z, Grigera P R, Wagner R R. Membrane vesiculation function and exocytosis of wild-type and mutant matrix proteins of vesicular stomatitis virus. J Virol. 1995;69:3156–3160. doi: 10.1128/jvi.69.5.3156-3160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kail M, Hollinshead M, Ansorge W, Pepperkok R, Frank R, Griffiths G, Vaux D. The cytoplasmic domain of alphavirus E2 glycoprotein contains a short linear recognition signal required for viral budding. EMBO J. 1991;10:2343–2351. doi: 10.1002/j.1460-2075.1991.tb07773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kretzschmar E, Buonocore L, Schnell M J, Rose J K. High-efficiency incorporation of functional influenza virus glycoproteins into recombinant vesicular stomatitis viruses. J Virol. 1997;71:5982–5989. doi: 10.1128/jvi.71.8.5982-5989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawson N D, Stillman E A, Whitt M A, Rose J K. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci USA. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazarovits J, Shia S P, Ktistakis N, Lee M S, Bird C, Roth M G. The effects of foreign transmembrane domains on the biosynthesis of the influenza virus hemagglutinin. J Biol Chem. 1990;265:4760–4767. [PubMed] [Google Scholar]
- 23.Lefrancois L, Lyles D S. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. II. Monoclonal antibodies of nonneutralizing and cross-reactive epitopes of Indiana and New Jersey serotypes. Virology. 1982;121:168–174. doi: 10.1016/0042-6822(82)90126-x. [DOI] [PubMed] [Google Scholar]
- 24.Lopez S, Yao J S, Kuhn R J, Strauss E G, Strauss J H. Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. J Virol. 1994;68:1316–1323. doi: 10.1128/jvi.68.3.1316-1323.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCallus D E, Ugen K E, Sato A I, Williams W V, Weiner D B. Construction of a recombinant bacterial human CD4 expression system producing a bioactive CD4 molecule. Viral Immunol. 1992;5:163–172. doi: 10.1089/vim.1992.5.163. [DOI] [PubMed] [Google Scholar]
- 26.Mebatsion T, Conzelmann K K. Specific infection of CD4+ target cells by recombinant rabies virus pseudotypes carrying the HIV-1 envelope spike protein. Proc Natl Acad Sci USA. 1996;93:11366–11370. doi: 10.1073/pnas.93.21.11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mebatsion T, Finke S, Weiland F, Conzelmann K K. A CXCR4/CD4 pseudotype rhabdovirus that selectively infects HIV-1 envelope protein-expressing cells. Cell. 1997;90:841–847. doi: 10.1016/s0092-8674(00)80349-9. [DOI] [PubMed] [Google Scholar]
- 28.Mebatsion T, Konig M, Conzelmann K K. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell. 1996;84:941–951. doi: 10.1016/s0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- 29.Metsikko K, Simons K. The budding mechanism of spikeless vesicular stomatitis virus particles. EMBO J. 1986;5:1913–1920. doi: 10.1002/j.1460-2075.1986.tb04444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naim H Y, Roth M G. Basis for selective incorporation of glycoproteins into the influenza virus envelope. J Virol. 1993;67:4831–4841. doi: 10.1128/jvi.67.8.4831-4841.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oomens A G, Blissard G W. Requirement for GP64 to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology. 1999;254:297–314. doi: 10.1006/viro.1998.9523. [DOI] [PubMed] [Google Scholar]
- 32.Owen K E, Kuhn R J. Alphavirus budding is dependent on the interaction between the nucleocapsid and hydrophobic amino acids on the cytoplasmic domain of the E2 envelope glycoprotein. Virology. 1997;230:187–196. doi: 10.1006/viro.1997.8480. [DOI] [PubMed] [Google Scholar]
- 33.Park E K, Castrucci M R, Portner A, Kawaoka Y. The M2 ectodomain is important for its incorporation into influenza A virions. J Virol. 1998;72:2449–2455. doi: 10.1128/jvi.72.3.2449-2455.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez L G, Davis G L, Hunter E. Mutants of the Rous sarcoma virus envelope glycoprotein that lack the transmembrane anchor and cytoplasmic domains: analysis of intracellular transport and assembly into virions. J Virol. 1987;61:2981–2988. doi: 10.1128/jvi.61.10.2981-2988.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose J K, Buonocore L, Whitt M A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 36.Roth M G, Doyle C, Sambrook J, Gething M J. Heterologous transmembrane and cytoplasmic domains direct functional chimeric influenza virus hemagglutinins into the endocytic pathway. J Cell Biol. 1986;102:1271–1283. doi: 10.1083/jcb.102.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salzwedel K, West J T, Hunter E. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J Virol. 1999;73:2469–2480. doi: 10.1128/jvi.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheiffele P, Rietveld A, Wilk T, Simons K. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J Biol Chem. 1999;274:2038–2044. doi: 10.1074/jbc.274.4.2038. [DOI] [PubMed] [Google Scholar]
- 39.Schnell M J, Buonocore L, Boritz E, Ghosh H P, Chernish R, Rose J K. Requirement for a non-specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J. 1998;17:1289–1296. doi: 10.1093/emboj/17.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnell M J, Buonocore L, Kretzschmar E, Johnson E, Rose J K. Foreign glycoproteins expressed from recombinant vesicular stomatitis viruses are incorporated efficiently into virus particles. Proc Natl Acad Sci USA. 1996;93:11359–11365. doi: 10.1073/pnas.93.21.11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnell M J, Johnson J E, Buonocore L, Rose J K. Construction of a novel virus that targets HIV-1-infected cells and controls HIV-1 infection. Cell. 1997;90:849–857. doi: 10.1016/s0092-8674(00)80350-5. [DOI] [PubMed] [Google Scholar]
- 42.Stillman E A, Rose J K, Whitt M A. Replication and amplification of novel vesicular stomatitis virus minigenomes encoding viral structural proteins. J Virol. 1995;69:2946–2953. doi: 10.1128/jvi.69.5.2946-2953.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suomalainen M, Liljestrom P, Garoff H. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J Virol. 1992;66:4737–4747. doi: 10.1128/jvi.66.8.4737-4747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takada A, Robison C, Goto H, Sanchez A, Murti K G, Whitt M A, Kawaoka Y. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas D, Newcomb W W, Brown J C, Wall J S, Hainfeld J F, Trus B L, Steven A C. Mass and molecular composition of vesicular stomatitis virus: a scanning transmission electron microscopy analysis. J Virol. 1985;54:598–607. doi: 10.1128/jvi.54.2.598-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wills J W, Craven R C, Achacoso J A. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J Virol. 1989;63:4331–4343. doi: 10.1128/jvi.63.10.4331-4343.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu X, Yuan X, McLane M F, Lee T H, Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J Virol. 1993;67:213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]