Abstract
Background
Stomach adenocarcinoma (STAD), a frequently occurring gastrointestinal tumour, is often detected late and has a poor prognosis. Long non-coding RNAs (lncRNAs) significantly affect tumour development. Recent studies have identified disulfidptosis as a previously unexplained form of cell death. Herein, we aimed to examine the predictive value of disulfidptosis-related lncRNA models for the clinical prognosis and immunotherapy of STAD.
Methods
STAD-related transcriptomic data were obtained from The Cancer Genome Atlas (TCGA), whereas genes associated with disulfidptosis were identified from previously published papers. A risk prediction model for disulfidptosis-related lncRNAs was developed using the Cox regression and least absolute shrinkage selection algorithm methods. The accuracy of the model was confirmed using calibration curves, and the biological functions were analysed using Gene Ontology (GO) and Gene Set Enrichment Analysis (GSEA). Finally, the tumour mutation burden (TMB) and tumour immune dysfunction and exclusion (TIDE) algorithms were used to screen drugs that are sensitive to STAD.
Results
The risk prediction models were constructed using seven disulfidptosis-related lncRNAs. The validated results were consistent with the predicted ones, with significant survival differences. When combined with clinical data, the risk scores were used as independent prognostic markers. Based on the tumour mutation load, the high-risk patient group had a poorer survival rate as compared with the low-risk patient group. Further studies were conducted to understand the different groups’ inconsistent responses to immune status; subsequently, relatively sensitive drugs were identified.
Conclusions
Overall, seven markers of disulfidptosis-related lncRNAs associated with STAD were found to facilitate prognostic prediction, suggesting new ideas for immunotherapy and clinical applications.
Keywords: Disulfidptosis, stomach adenocarcinoma (STAD), long non-coding RNAs (lncRNAs), prognosis, immunotherapy
Highlight box.
Key findings
• Our study develops a risk score system based on seven disulfidptosis-related long non-coding RNAs (lncRNAs) to enhance prognosis and treatment guidance for stomach adenocarcinoma.
What is known and what is new?
• Disulfidptosis plays a critical role in tumor development.
• Our research extends this by employing disulfidptosis-related lncRNAs to accurately predict stomach adenocarcinoma outcomes.
What is the implication, and what should change now?
• This risk score system introduces a novel approach for stomach adenocarcinoma treatment, highlighting the need for further research on these lncRNAs to improve patient care.
Introduction
Stomach cancer is prevalent worldwide, with a higher incidence reported in Eastern Asia and Europe (1). Stomach adenocarcinoma (STAD), a common type of gastric cancer, is associated with age, dietary structure and Helicobacter pylori infection (2). Surgical resection is the preferred treatment for patients with early STAD, whereas a combination of chemotherapy, radiotherapy and surgical resection improves survival rates, although the prognosis remains poor, among patients with advanced STAD (3). Currently, biomarker-based methods such as tumour risk score prediction signatures are used to predict patient prognosis, and this is being gradually applied in clinical practice. Therefore, it is essential to establish prognostic markers to forecast the long-term survival of patients with STAD.
The protein solute carrier family seven member (SLC7A11) is responsible for transporting extracellular cystine into the cell and glutamate from the cell to the extracellular environment. Additionally, it plays a crucial role in regulating oxidative stress (4). Unlike healthy cells, cancer cells are highly dependent on cystine intake. SLC7A11 is highly expressed in many cancers, such as breast (5) and lung (6) cancers; this helps cancer cells escape oxidative stress and regulates their metabolism (7). Recently, a new mode of cell death known as disulfidptosis has been identified. This occurs when a large accumulation of disulphide molecules in glucose-deficient cancer cells with high SLC7A11 expression leads to abnormal disulphide bonding between actin cytoskeletal proteins, disrupting their organization and ultimately causing actin network collapse and cell death (8). This indicates that glucose transporter (GLUT) inhibitor-induced disulfidptosis may be exploited in cancer therapy.
Long non-coding RNAs (lncRNAs) control gene expression through chromatin adaptation, transcription and post-transcriptional processing (9). lncRNAs are closely linked to the development of many cancers and can be used as biomarkers or targeted therapeutics for cancers (10-12). To explore novel therapeutic targets and improve patient prognosis, it is crucial to investigate the relationship between disulfidptosis-related lncRNAs and STAD.
The impact of disulfidptosis-related lncRNAs on the existing treatment modalities for STAD warrants investigation. Herein, we used The Cancer Genome Atlas (TCGA) database to assess the biological significance of disulfidptosis-related lncRNAs and their potential for prognostic prediction in patients with STAD. We present this article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2067/rc).
Methods
Data collection and analysis
Using the TCGA database, we obtained clinical and RNA sequencing data from gastric adenocarcinoma patients. After excluding those with incomplete clinical data, our dataset included 407 tissue samples (375 STAD and 32 normal tissues). These samples were randomly split into a training set (n=204) and a test set (n=203). The clinical characteristics between the two sets exhibited no significant differences, as shown in Table 1. Ten genes associated with disulfidptosis were collected by reviewing the available literature (13). Finally, the STAD mutation data were extracted from TCGA database. The study was carried out in accordance with the Declaration of Helsinki (as revised in 2013).
Table 1. Demographics of 407 patients.
| Variables | Total cohort (n=407), n (%) | Training cohort (n=204), n (%) | Validation cohort (n=203), n (%) | P value |
|---|---|---|---|---|
| Age (years) | 0.36 | |||
| ≤65 | 183 (44.96) | 97 (47.55) | 86 (42.36) | |
| >65 | 221 (54.3) | 106 (51.96) | 115 (56.65) | |
| Unknown | 3 (0.74) | 1 (0.49) | 2 (0.99) | |
| Gender | 0.02 | |||
| Female | 144 (35.38) | 61 (29.9) | 83 (40.89) | |
| Male | 263 (64.62) | 143 (70.1) | 120 (59.11) | |
| Grade | 0.83 | |||
| G1 | 12 (2.95) | 7 (3.43) | 5 (2.46) | |
| G2 | 144 (35.38) | 73 (35.78) | 71 (34.98) | |
| G3 | 242 (59.46) | 120 (58.82) | 122 (60.1) | |
| Unknown | 9 (2.21) | 4 (1.96) | 5 (2.46) | |
| Stage | 0.34 | |||
| Stage I | 55 (13.51) | 33 (16.18) | 22 (10.84) | |
| Stage II | 122 (29.98) | 59 (28.92) | 63 (31.03) | |
| Stage III | 167 (41.03) | 78 (38.24) | 89 (43.84) | |
| Stage IV | 39 (9.58) | 21 (10.29) | 18 (8.87) | |
| Unknown | 24 (5.9) | 13 (6.37) | 11 (5.42) | |
| T stage | 0.32 | |||
| T1 | 21 (5.16) | 14 (6.86) | 7 (3.45) | |
| T2 | 86 (21.13) | 46 (22.55) | 40 (19.7) | |
| T3 | 179 (43.98) | 89 (43.63) | 90 (44.33) | |
| T4 | 113 (27.76) | 52 (25.49) | 61 (30.05) | |
| Unknown | 8 (1.97) | 3 (1.47) | 5 (2.46) | |
| N stage | 0.90 | |||
| N0 | 121 (29.73) | 64 (31.37) | 57 (28.08) | |
| N1 | 108 (26.54) | 52 (25.49) | 56 (27.59) | |
| N2 | 77 (18.92) | 38 (18.63) | 39 (19.21) | |
| N3 | 82 (20.15) | 42 (20.59) | 40 (19.7) | |
| Unknown | 19 (4.67) | 8 (3.92) | 11 (5.42) | |
| M stage | 0.54 | |||
| M0 | 362 (88.94) | 183 (89.71) | 179 (88.18) | |
| M1 | 26 (6.39) | 11 (5.39) | 15 (7.39) | |
| Unknown | 19 (4.67) | 10 (4.9) | 9 (4.43) |
Tumor staging (Stage) is based on the TNM classification, including stages I, II, III, and IV. T, N, M stages represent the tumor, lymph node, and metastasis status, respectively. Data not reported or available are marked as “unknown”. Statistical significance levels are indicated by P values.
Identification of disulfidptosis-related lncRNAs
We analyzed expression data for disulfidptosis-related genes and lncRNAs using Pearson correlations via the ‘limma’ package in R (14). Significant correlations were defined as those with |Cor| >0.4 and P<0.001. Data management was conducted with ‘dplyr’ (15), while ‘ggalluvial’ (16) and ‘ggplot2’ (17) facilitated the creation and visualization of Sankey diagrams to highlight key co-expression links.
Construction of the predictive models and analysis
First, the TCGA STAD prognostic lncRNAs were identified using univariate Cox regression analysis. The ‘glmnet’ package in R was utilized to apply the least absolute shrinkage and selection operator (LASSO) method for selecting the most predictive lncRNAs, with 10-fold cross-validation determining the optimal regularization parameter (18). A survival was assigned to each patient based on their disulfidptosis-related lncRNA expression levels and their regression coefficients [risk score = (coef LncRNA1 expr LncRNA1) + (coef LncRNA2 expr LncRNA2)… + (coef LncRNAn × expr LncRNAn)]. The risk score midpoint was used to categorise the specimens. The training set used Kaplan-Meier plots to assess the overall survival (OS) of the high- and low-risk groups (19). The ‘pROC’ package facilitated receiver operating characteristic receiver operating characteristic (ROC) curve analysis to evaluate the model’s predictive accuracy (20), while ‘pheatmap’ provided visual insights into the association between lncRNA expression and patient survival (21). Finally, the test and the full set validated the model’s accuracy.
Independent prognostic factor analysis
Univariate and multifactorial Cox regression analyses were performed using the clinical data from TCGA database to evaluate the role of the risk score as an independent predictor. The accuracy of this feature in predicting patient survival was confirmed using ROC curve analysis and concordance index (C-index) curves.
Analysis of the clinical value of the prognostic prediction models
To accurately predict the OS of the patients and provide valid prognostic information for physicians to develop appropriate treatment plans, we constructed a nomogram model based on risk categories, age and clinicopathological variables using the ‘RMS’ package. The model accuracy was evaluated by plotting calibration curves.
Principal component analysis (PCA), Gene Ontology (GO) and Gene Set Enrichment Analysis (GSEA) analysis
PCA and 3D PCA analyses were conducted using the ’scatterplot3d’ package for visualization.3D scatter plots illustrated the sample distribution across different risk scores. Differential genes between the high- and low-risk groups were identified using ‘limma’, with criteria of |log2FC| ≥1 and false discovery rate (FDR) <0.05. The ‘clusterProfiler’ package was used for differential gene analysis in the GO and GSEA enrichment studies, and P values <0.05 and FDR values <0.05 were considered statistically significant.
Immune function analysis and tumour mutation burden (TMB)
The ‘limma’ and ‘GSA’ packages were used to analyse the immune-related differences between the high- and low-risk groups, including the tumour microenvironment scores and correlations between risk scores and the tumour immune microenvironment, as demonstrated using the ‘Pheatmap’ package. The ‘Maftools’ package correlates risk scores with the TMB. The survival package compares TMB and patient survival at P<0.05.
Tumour immune dysfunction and exclusion (TIDE) and drug sensitivity
The TIDE score was used to assess the immune checkpoint blockade (ICB) response; this ICB response was used to predict the effectiveness of immunotherapeutic agents in the high- and low-risk patient populations. The therapeutic agents were screened and evaluated for drug sensitivity using P<0.001 as the filtering criterion. The analyses were based on the ‘limma’, ‘pRRophetic’ and ‘ggpubr’ software packages.
Statistical analyses
We conducted Pearson correlation analysis to investigate associations with disulfidptosis-related lncRNAs. Subsequently, we built risk models using univariate Cox regression, Lasso regression, and multivariate Cox regression. Model performance was assessed using timeROC curves and the C-index. All analyses were conducted in R version 4.3.0.
Results
Identification of disulfidptosis-related lncRNAs in STAD
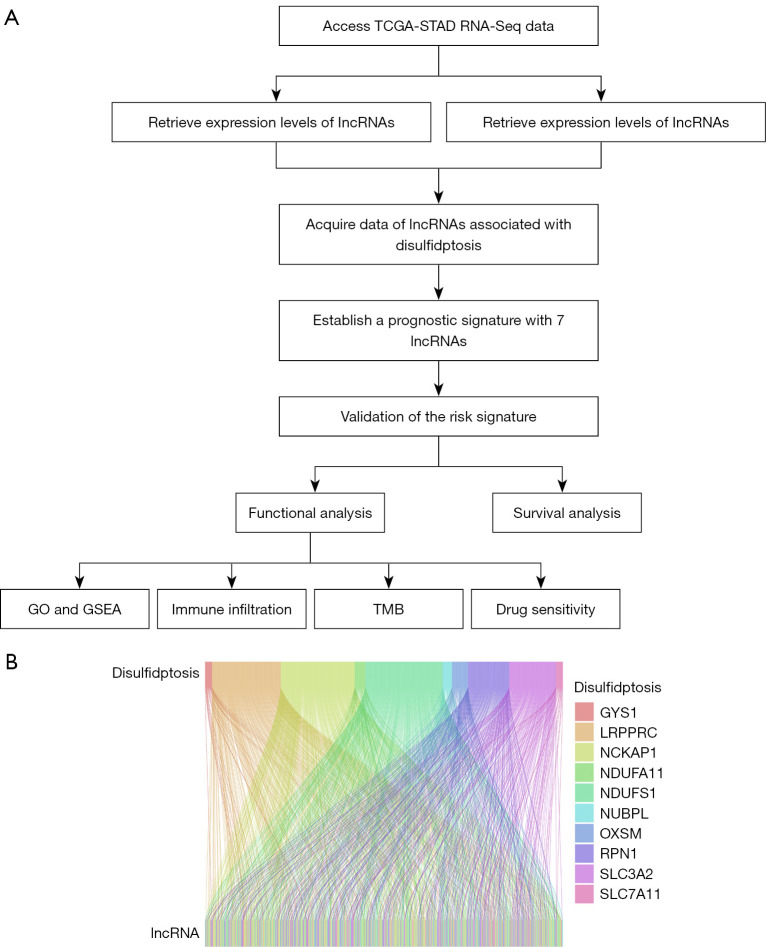
Figure 1A illustrates the schematic diagram of the research process. Based on previously published literature (13), we identified disulfidptosis genes. The transcriptome data of 407 (375 STAD and 32 normal) tissues were retrieved using TCGA database, and STAD-associated lncRNAs were extracted. A total of 694 disulfidptosis-related lncRNAs were screened using the Pearson’s correlation algorithm. Sankey diagrams were used to illustrate the relationship between the disulfidptosis genes and disulfidptosis-related lncRNAs (Figure 1B).
Figure 1.
Flow gram and Sankey diagram. (A) Analysis process of lncRNA related to disulfidptosis. (B) Sankey diagram demonstrating the co-expression of the cuproptosis gene and disulfidptosis-related lncRNAs. TCGA, The Cancer Genome Atlas; STAD, stomach adenocarcinoma; RNA-Seq, RNA sequencing; lncRNAs, long non-coding RNAs; GO, Gene Ontology; GSEA, Gene Set Enrichment Analysis; TMB, tumor mutational burden.
Risk model construction and validation
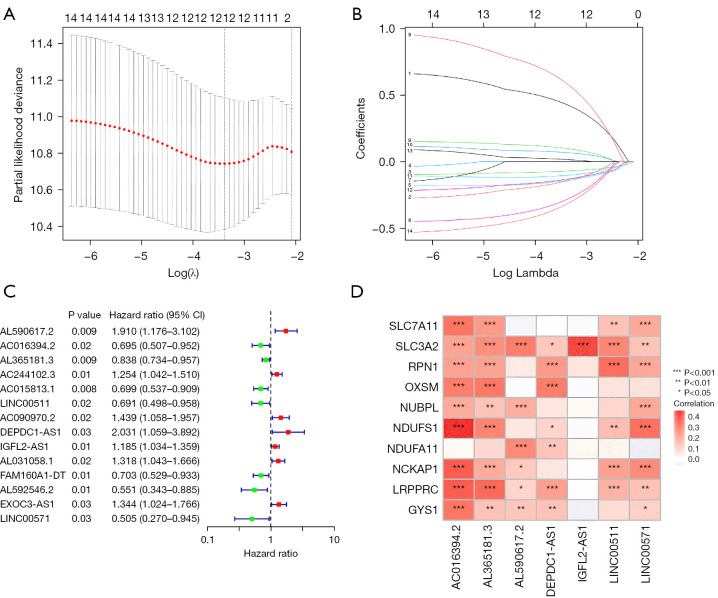
Fourteen lncRNAs were initially obtained using the uni-Cox regression approach. Subsequently, a risk score model was developed using seven prognosis-related lncRNAs identified using LASSO analysis (Figure 2A-2C). Each line in Figure 2B is color-coded to represent each lncRNA, illustrating the impact of their respective coefficients on the model’s predictive accuracy. The risk score was calculated as follows: risk score = AC016394.2 × (−0.3963) + AL365181.3 × (−0.1314) + AL590617.2 × (0.7516) + DEPDC1-AS1 × (0.8785) + IGFL2-AS1 × (0.1361) + LINC00511 × (−0.4765) + LINC00571 × (−0.7027). The correlation heat map highlighted the relationship between the disulfidptosis-related genes and the seven lncRNAs (Figure 2D).
Figure 2.
Determination of the prognostic disulfidptosis-related lncRNAs in STAD. (A) Cross-validation of LASSO. (B) LASSO coefficient distribution of the survival-related lncRNAs. The numbers on the left side of the figure correspond to different lncRNAs as follows: 1 for AC016394.2, 2 for AL365181.3, 3 for AL590617.2, 4 for DEPDC1-AS1, 5 for IGFL2-AS1, 6 for LINC00511, 7 for LINC00571, 8 for AC244102.3, 9 for AC015813.1, 10 for AC090970.2, 11 for AL031058.1, 12 for FAM160A1-DT, 13 for AL592546.2, and 14 for EXOC3-AS1. (C) Univariate Cox regression-extracted prognosis-related lncRNAs. (D) Correlation between the disulfidptosis-related genes and lncRNAs in the risk model. CI, confidence interval; lncRNA, long non-coding RNA; STAD, stomach adenocarcinoma; LASSO, least absolute shrinkage and selection operator.
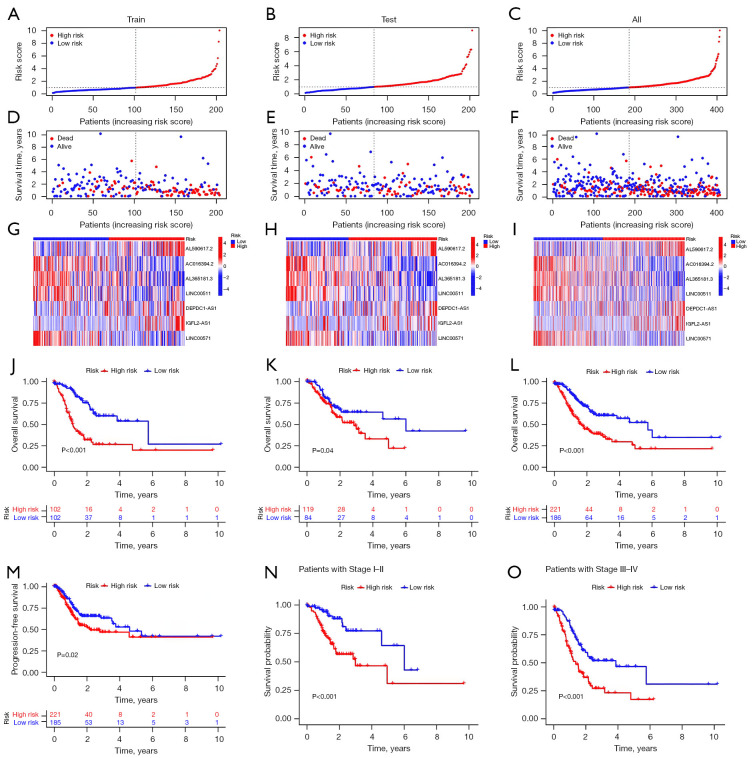
In the present study, the patients in the high-risk group exhibited significantly shorter OS and disease-free survival rates as compared to those in the low-risk group. The heat map analysis revealed a concentrated distribution of seven lncRNAs in the two risk groups. Specifically, AL590617.2, DEPDC1-AS1 and IGFL2-AS1 were predominantly expressed in the high-risk group, whereas AC016394.2, AL365181.3, LINC00511 and LINC00571 were predominantly expressed in the low-risk group (Figure 3A-3M). To investigate the correlation between the risk scores and survival outcomes in the patients with STAD, we compared the high- and low-risk groups during different periods. Our findings revealed that the survival rate was significantly lower in the high-risk group than in the low-risk group (Figure 3N,3O).
Figure 3.
Prognosis of the risk model across the various sets. (A-C) Train, test and complete set risk model demonstrations. (D-F) Relationship between the risk score and survival status for the train, test and complete set. (G-I) Heatmap of the train, test and complete lncRNA expressions. (J-L) The K-M survival graphs for the patients in the high- and low-risk groups in the train, test and complete sets. (M) PFS in the complete set. (N,O) Complete set of the K-M survival curves for the different tumour stages. LncRNA, long non-coding RNA; K-M, Kaplan-Meier; PFS, progression-free survival.
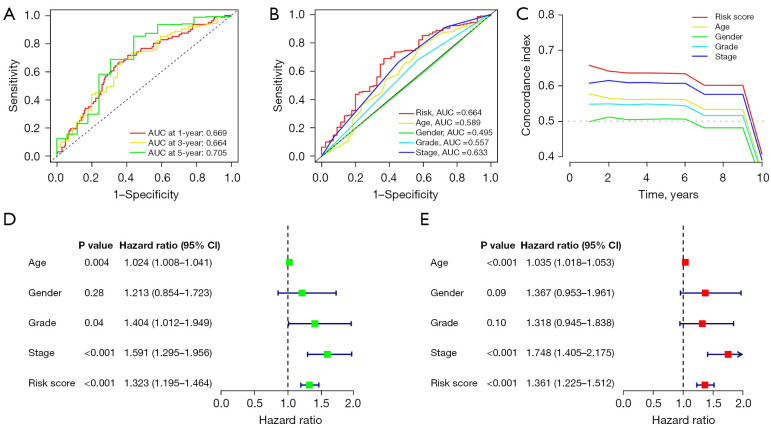
The accuracy of the predictive model risk score was evaluated using ROC curves and the C-index. The areas under the curve (AUCs) for the 1-, 3- and 5-year predictions were 0.669, 0.664 and 0.705, respectively (Figure 4A). The AUC values of the risk scores were more significant than the other clinicopathological variables. The C-index curves also demonstrated that the risk model was more accurate than the other variables (Figure 4B,4C). Furthermore, the hazard ratios (HRs) for the risk scores were 1.323 and 1.361 (P<0.001) in the single and multiple Cox regression analyses, respectively, indicating that risk characteristics are an independent prognostic factor for patients with STAD (Figure 4D,4E).
Figure 4.
Risk model assessment. (A) Complete set of ROC curves for years 1, 3 and 5. (B) ROC curves for risk scores and clinicopathological characteristics. (C) C-index curves of the risk model. (D,E) Clinicopathological and risk scores obtained using uni- and multi-Cox analyses. AUC, area under the curve; CI, confidence interval; ROC, receiver operating characteristic; C-index, concordance index; uni-Cox analysis, univariate Cox proportional hazards analysis; multi-Cox analysis, multivariate Cox proportional hazards analysis.
Nomogram construction
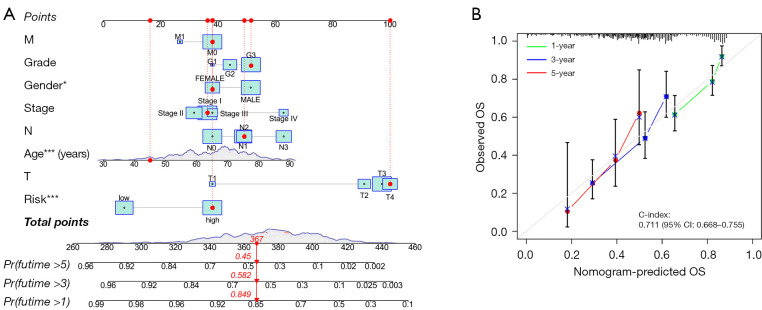
A nomogram was constructed to estimate the likelihood of OS at 1, 3 and 5 years using the risk score, age and clinicopathological factors (Figure 5A). The calibration curves demonstrated a strong correlation between the predicted and actual values (Figure 5B).
Figure 5.
Nomogram and model calibration curves. (A) OS prediction nomogram. (B) OS calibration curves at 1, 3 and 5 years. *, *** represent P<0.05, and P<0.001, respectively. Pr, probability; T, tumor; N, node; M, metastasis; OS, overall survival; CI, confidence interval; C-index, concordance index.
PCA and biological pathway analysis
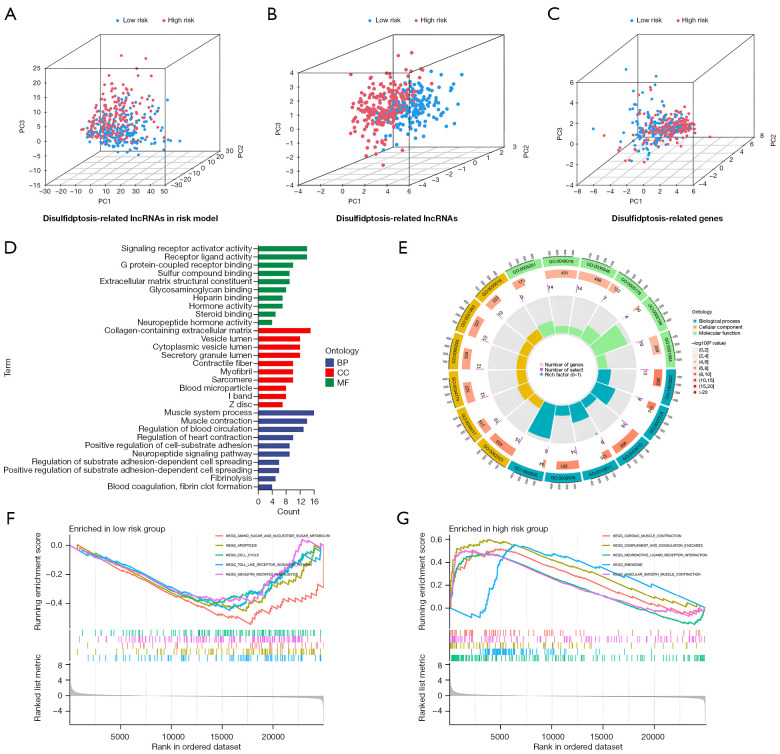
Three-dimensional scatter plots demonstrated that PCA in the high- and low-risk groups exhibited significant aggregation characteristics, indicating that these lncRNAs were persuasive for model construction (Figure 6A-6C). According to the GO analysis, the disulfidptosis-related lncRNA were enriched in muscle system processes, muscle contraction, blood circulation regulation and cardiac contraction regulation (Figure 6D,6E). The GSEA analysis showed that biological functions such as cardiac muscle traction, complement and coagulation cascades, neural activity-receptor interactions, ribosomes and vascular smooth muscle traction differed between the low- and high-risk patient groups (Figure 6F,6G).
Figure 6.
PCA and functional analysis. (A-C) PCA analysis detected sample distribution by the risk of disulfidptosis-related lncRNAs, disulfidptosis-related lncRNAs and disulfidptosis-related genes. (D,E) GO analysis in the low- and high-risk patient groups. (F,G) GSEA analysis in the low- and high-risk patient groups. PC1, 2, 3 represent the first to the third principal components of the matrix. BP, biological process; CC, cellular compartment; MF, molecular function; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PCA, principal component analysis; lncRNA, long non-coding RNA; GSEA, Gene Set Enrichment Analysis.
Immunoassay
A notable dissimilarity in the infiltration of immune cells was found between the two groups. Furthermore, significant differences were observed in the co-inhibition of the APC cells and antigen presentation mediated by MHC I (Figure 7A,7B). Moreover, high-risk patients had higher TME scores than low-risk patients (Figure 7C).
Figure 7.
Immune characteristics of the different risk groups. (A) Expression of immune cells in the different risk groups. (B) Relationship between immune-related functions and risk scores. (C) TME scores in the different risk groups. *, **, *** represent P<0.05, P<0.01, and P<0.001, respectively. APC, antigen-presenting cells; CCR, chemokine receptor; HLA, human leukocyte antigen; MHC, major histocompatibility complex; NK, natural killer; IFN, interferon; TME, tumor microenvironment.
TMB and drug sensitivity analysis
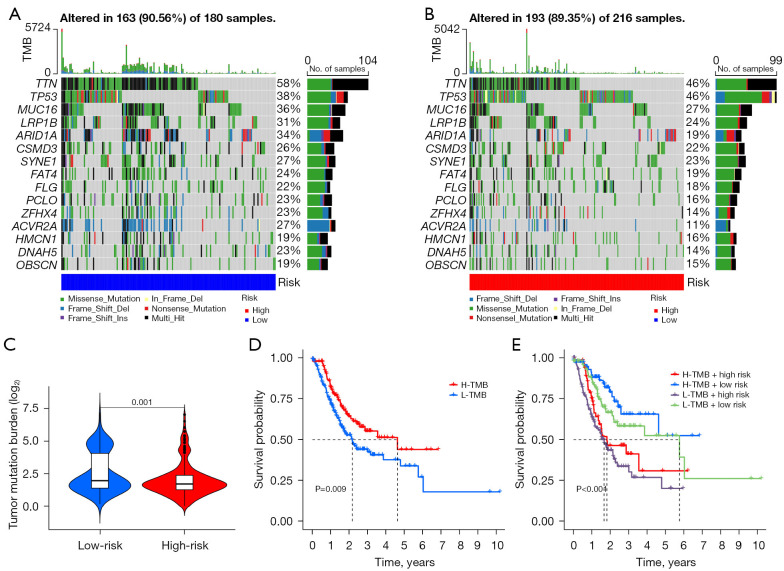
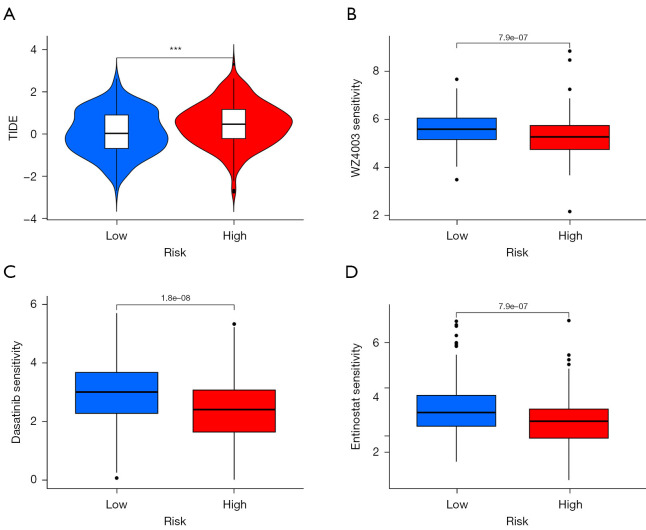
Waterfall plots were used to demonstrate somatic mutations in both the high- and low-risk groups. The frequency of mutations was found to be higher in the low-risk group than in the high-risk group (Figure 8A,8B). To determine the impact of risk categories on immunotherapy for STAD, we examined the association between the TMB and risk categories. The TMB counts were higher in the low-risk group (Figure 8C), and high TMB was associated with better OS as compared with low TMB (Figure 8D). Moreover, the low-risk group was more susceptible to immune checkpoint inhibitors than the high-risk group (Figure 8E). Using the TIDE algorithm, the response to immunotherapy was predicted in both groups, and the patients in the low-risk group were found to respond better (Figure 9A). Furthermore, analysis of drug sensitivity in high-risk STAD patients identified increased susceptibility to NUAK kinase inhibitor WZ4003, dasatinib, and entinostat (Figure 9B-9D).
Figure 8.
Association of gene mutations and TMB analysis. (A,B) Mutations in somatic cells found in the various risk groups. (C) TMB level in the different groups. (D) Different degrees of TMB survival curves. (E) K-M survival profiles with varying TMB concentrations in the different risk categories. TMB, tumor mutation burden; H, high; L, low; K-M, Kaplan-Meier.
Figure 9.
TIDE scores with survival prediction and drug sensitivity analysis. (A) TIDE scores for the different risk groups. (B-D) The drug sensitivities of WZ4003, dasatinib, and entinostat were observed. ***, P<0.001. TIDE, tumor immune dysfunction and exclusion.
Discussion
STAD, a malignant tumour, is highly prevalent worldwide and has a poor prognosis (22). Research has demonstrated that lncRNA plays a regulatory role in the progression of gastric cancer, making it a promising target for the treatment of cancers (23,24). Lin et al. identified lncRNA BC002811 as a promoter of gastric cancer metastasis through decoying SOX2 and inhibiting PTEN transcription (25). Gong et al. showed that LINC01094 promotes gastric cancer by inhibiting AZGP1, lowering PTEN, and activating AKT (26). Moreover, Wang et al. revealed that TUBA1C, targeted by lncRNA EGFR-AS1, promotes gastric cancer progression by enhancing cell proliferation, migration, and invasion (27). Although lncRNAs are important in STAD, the mechanism of its interrelationship is not fully understood.
Disulfidptosis, a new and unique programmed cell death mode, can inhibit GLUT proteins and thus induce disulfidptosis in cancer cells without affecting normal cells. In recent years, researchers have made significant progress in developing models assessing lncRNA risk in various types of cancer (e.g., constructing predictive models of lncRNAs associated with pyroptosis in STAD for clinical treatment) (28,29). However, the mechanism of disulfidptosis-related lncRNAs in STAD is still under investigation.
In this study, seven lncRNAs (AC016394.2, AL365181.3, AL590617.2, DEPDC1-AS1, IGFL2-AS1, LINC00511 and LINC00571) were chosen for building a prediction model of disulfidptosis-related lncRNAs. In previous studies, AC016394.2, AL365181.3 and LINC00571 were used as cuproptosis, ferroptosis and cellular senescence-related lncRNAs, respectively, in bioinformatic analyses for the construction of a predictive model for gastric cancer (30-32). Additionally, DEPDC1-AS1 promoted the expansion and spread of human gastric cancer cells using the human antigen R-F27R pathway (33). Research suggests that IGFL2-AS1 facilitates the advancement and spread of gastric cancer through the miR-802/ARPP19 pathway (34). LINC00511 targets miR-625-5p/STAT3 to increase gastric tumour cell growth and mobility (35). AL590617.2 was analysed for the first time in a gastric cancer study. Subsequently, the patients with STAD were randomly allocated into the training and validation cohorts in a 1:1 ratio to assess the reliability of the risk prediction model. The survival curves indicated that the low-risk group had a more favourable prognosis across the training, validation and full groups. The accuracy of the established risk prediction model was confirmed using the ROC analysis and C-index curves. Accordingly, a nomogram was developed to predict the patients with STAD. The calibration curves demonstrated agreement with the anticipated outcomes. Owing to the complexity of cancer, the molecular pathways driving gastric cancer need to be understood to accordingly develop new therapeutic targets. Therefore, functional enrichment analysis was performed, and novel signalling pathways were identified in this study, which helped identify the disulfidptosis-related genes involved in the aetiology and development of STAD.
Next, we investigated the relationship among the tumour immune microenvironment, TMB and risk score of the patients with STAD. We used the CIBERSORT and ESTIMATE techniques for immunological analysis. The results revealed that the risk score was positively correlated with CD8 cell expression. The ESTIMATE score represents the ratio of the immune and stromal components in the tumour tissue (36), and the ESTIMATE score was found to be particularly high in the high-risk group. In the context of malignancy, TMB serves as a prognostic biomarker for ICB and is associated with the effectiveness of immunotherapy (37-39). This study revealed that the low-risk group had higher TMB levels, suggesting a potential for better response to immunotherapy. Additionally, the TIDE score accurately predicted the response to ICB therapy, with a higher TIDE score indicating a weaker response to ICB (40). Thus, high-risk patients with high TIDE scores are less likely to respond to immunotherapy. Ultimately, the half-maximal inhibitory concentration (IC50) values of WZ4003, entinostat, and dasatinib were determined, showing increased effectiveness in high-risk group patients.
In summary, this study developed a prognostic model for STAD based on seven disulfidptosis-related lncRNAs. The model demonstrated outstanding performance in predicting STAD patients’ survival outcomes. Additionally, it has the potential to assess patients’ immune function, immune checkpoint marker expression, and chemotherapy drug sensitivity. This work lays a strong foundation for future anti-tumor drug development.
Our study has some limitations. First, our data were collected from TCGA database, and the results may be biased. We could have obtained different results if we combined the data from other sources. Second, we did not test the molecular transcription and expression levels, reducing the reliability of the results. In addition, this study is still in its preliminary stages, and the information collected so far may not be sufficient to justify the investigation; future studies are therefore warranted.
Conclusions
Taken together, disulfidptosis-related lncRNAs can act as specific biomarkers for predicting the progression of STAD and provide new insights into targeted therapy for the same. These findings are significant and hold promise for future research in this field.
Supplementary
The article’s supplementary files as
Acknowledgments
We thank TCGA for the availability of the data.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Footnotes
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2067/rc
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2067/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2067/coif). The authors have no conflicts of interest to declare.
References
- 1.Huang J, Lucero-Prisno DE, 3rd, Zhang L, et al. Updated epidemiology of gastrointestinal cancers in East Asia. Nat Rev Gastroenterol Hepatol 2023;20:271-87. 10.1038/s41575-022-00726-3 [DOI] [PubMed] [Google Scholar]
- 2.Salvatori S, Marafini I, Laudisi F, et al. Helicobacter pylori and Gastric Cancer: Pathogenetic Mechanisms. Int J Mol Sci 2023;24:2895. 10.3390/ijms24032895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suwaidan AA, Gordon A, Cartwright E, et al. Optimising Multimodality Treatment of Resectable Oesophago-Gastric Adenocarcinoma. Cancers (Basel) 2022;14:586. 10.3390/cancers14030586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan Y, Teng H, Hang Q, et al. SLC7A11 expression level dictates differential responses to oxidative stress in cancer cells. Nat Commun 2023;14:3673. 10.1038/s41467-023-39401-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Zhu S, Zhou H, et al. Identification of MTHFD2 as a prognostic biomarker and ferroptosis regulator in triple-negative breast cancer. Front Oncol 2023;13:1098357. 10.3389/fonc.2023.1098357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shan Q, Zhang C, Li Y, et al. SLC7A11, a potential immunotherapeutic target in lung adenocarcinoma. Sci Rep 2023;13:18302. 10.1038/s41598-023-45284-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin W, Wang C, Liu G, et al. SLC7A11/xCT in cancer: biological functions and therapeutic implications. Am J Cancer Res 2020;10:3106-26. [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H, Tang T. Pan-cancer genetic analysis of disulfidptosis-related gene set. Cancer Genet 2023;278-279:91-103. 10.1016/j.cancergen.2023.10.001 [DOI] [PubMed] [Google Scholar]
- 9.Segal D, Dostie J. The Talented LncRNAs: Meshing into Transcriptional Regulatory Networks in Cancer. Cancers (Basel) 2023;15:3433. 10.3390/cancers15133433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alahdal M, Elkord E. Non-coding RNAs in cancer immunotherapy: Predictive biomarkers and targets. Clin Transl Med 2023;13:e1425. 10.1002/ctm2.1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nemeth K, Bayraktar R, Ferracin M, et al. Non-coding RNAs in disease: from mechanisms to therapeutics. Nat Rev Genet 2024;25:211-32. 10.1038/s41576-023-00662-1 [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Fu Y, Lu Y, et al. Unravelling the complexity of lncRNAs in autophagy to improve potential cancer therapy. Biochim Biophys Acta Rev Cancer 2023;1878:188932. 10.1016/j.bbcan.2023.188932 [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Nie L, Zhang Y, et al. Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat Cell Biol 2023;25:404-14. 10.1038/s41556-023-01091-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jennifer B, Dietrich S, Don G. Introducing Data Science Techniques by Connecting Database Concepts and dplyr. J Stat Educ 2019;27:147-53. 10.1080/10691898.2019.1647768 [DOI] [Google Scholar]
- 16.Brunson JC. ggalluvial: Layered Grammar for Alluvial Plots. J Open Source Softw 2020;5:2017. 10.21105/joss.02017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gómez-Rubio V. ggplot2 - Elegant Graphics for Data Analysis (2nd Edition). J Stat Softw 2017;77:1-3. [Google Scholar]
- 18.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 2010;33:1-22. 10.18637/jss.v033.i01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan EL, Meier P. Nonparametric Estimation from Incomplete Observations. J Am Stat Assoc 1958;53:457-81. 10.1080/01621459.1958.10501452 [DOI] [Google Scholar]
- 20.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77. 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu K. Become Competent in Generating RNA-Seq Heat Maps in One Day for Novices Without Prior R Experience. Methods Mol Biol 2021;2239:269-303. 10.1007/978-1-0716-1084-8_17 [DOI] [PubMed] [Google Scholar]
- 22.Mantziari S, St Amour P, Abboretti F, et al. A Comprehensive Review of Prognostic Factors in Patients with Gastric Adenocarcinoma. Cancers (Basel) 2023;15:1628. 10.3390/cancers15051628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Usman M, Beilerli A, Sufianov A, et al. Investigations into the impact of non-coding RNA on the sensitivity of gastric cancer to radiotherapy. Front Physiol 2023;14:1149821. 10.3389/fphys.2023.1149821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Z, Mak TK, Shi Y, et al. The DNA damage repair-related lncRNAs signature predicts the prognosis and immunotherapy response in gastric cancer. Front Immunol 2023;14:1117255. 10.3389/fimmu.2023.1117255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin X, Li G, Yan X, et al. Long non-coding RNA BC002811 Promotes Gastric Cancer Metastasis by Regulating SOX2 Binding to the PTEN Promoter. Int J Biol Sci 2023;19:967-80. 10.7150/ijbs.76407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong Z, Zhang Y, Yang Y, et al. LncRNA LINC01094 Promotes Cells Proliferation and Metastasis through the PTEN/AKT Pathway by Targeting AZGP1 in Gastric Cancer. Cancers (Basel) 2023;15:1261. 10.3390/cancers15041261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Cui H, Yang X, et al. TUBA1C: a new potential target of LncRNA EGFR-AS1 promotes gastric cancer progression. BMC Cancer 2023;23:258. 10.1186/s12885-023-10707-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang M, Fang C, Ma Y. Prognosis Risk Model Based on Pyroptosis-Related lncRNAs for Gastric Cancer. Biomolecules 2023;13:469. 10.3390/biom13030469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Dai Y, Lu Y, et al. Identification and validation of a new pyroptosis-associated lncRNA signature to predict survival outcomes, immunological responses and drug sensitivity in patients with gastric cancer. Math Biosci Eng 2023;20:1856-81. 10.3934/mbe.2023085 [DOI] [PubMed] [Google Scholar]
- 30.Tu H, Zhang Q, Xue L, et al. Cuproptosis-Related lncRNA Gene Signature Establishes a Prognostic Model of Gastric Adenocarcinoma and Evaluate the Effect of Antineoplastic Drugs. Genes (Basel) 2022;13:2214. 10.3390/genes13122214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang G, Mao Z, Zhou X, et al. Construction and validation of a novel prognostic model using the cellular senescence-associated long non-coding RNA in gastric cancer: a biological analysis. J Gastrointest Oncol 2022;13:1640-55. 10.21037/jgo-22-662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang S, Zheng N, Chen X, et al. Establishment and Validation of a Ferroptosis-Related Long Non-Coding RNA Signature for Predicting the Prognosis of Stomach Adenocarcinoma. Front Genet 2022;13:818306. 10.3389/fgene.2022.818306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu W, Wang J, Xu J, et al. Long non-coding RNA DEPDC1-AS1 promotes proliferation and migration of human gastric cancer cells HGC-27 via the human antigen R-F11R pathway. J Int Med Res 2022;50:3000605221093135. 10.1177/03000605221093135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Y, Liu Y, Pu YS, et al. LncRNA IGFL2-AS1 functions as a ceRNA in regulating ARPP19 through competitive binding to miR-802 in gastric cancer. Mol Carcinog 2020;59:311-22. 10.1002/mc.23155 [DOI] [PubMed] [Google Scholar]
- 35.Cui N, Sun Q, Liu H, et al. Long non-coding RNA LINC00511 regulates the expression of microRNA-625-5p and activates signal transducers and activators of transcription 3 (STAT3) to accelerate the progression of gastric cancer. Bioengineered 2021;12:2915-27. 10.1080/21655979.2021.1940611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu W, Wang X, Le W, et al. Immune microenvironment infiltration landscape and immune-related subtypes in prostate cancer. Front Immunol 2023;13:1001297. 10.3389/fimmu.2022.1001297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fusco MJ, West HJ, Walko CM. Tumor Mutation Burden and Cancer Treatment. JAMA Oncol 2021;7:316. 10.1001/jamaoncol.2020.6371 [DOI] [PubMed] [Google Scholar]
- 38.Jang JY, Jeong SY, Kim ST. Tumor mutational burden as a potential predictive marker for the efficacy of immunotherapy in advanced gastric cancer. J Clin Oncol 2023;41:324. 10.1200/JCO.2023.41.4_suppl.324 [DOI] [Google Scholar]
- 39.Sun Q, Hong Z, Zhang C, et al. Immune checkpoint therapy for solid tumours: clinical dilemmas and future trends. Signal Transduct Target Ther 2023;8:320. 10.1038/s41392-023-01522-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keenan TE, Burke KP, Van Allen EM. Genomic correlates of response to immune checkpoint blockade. Nat Med 2019;25:389-402. 10.1038/s41591-019-0382-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as