Abstract
Satellite panicum mosaic virus (SPMV) depends on its helper virus, panicum mosaic virus (PMV), to provide trans-acting proteins for replication and movement. The 824-nucleotide (nt) genome of SPMV possesses an open reading frame encoding a 17.5-kDa capsid protein (CP), which is shown to be dispensable for SPMV replication. To localize cis-acting RNA elements required for replication and movement, a comprehensive set of SPMV cDNA deletion mutants was generated. The results showed that the 263-nt 3′ untranslated region (UTR) plus 73 nt upstream of the CP stop codon and the first 16 nt in the 5′ UTR are required for SPMV RNA amplification and/or systemic spread. A region from nt 17 to 67 within the 5′ UTR may have an accessory role in RNA accumulation, and a fragment bracketing nt 68 to 104 appears to be involved in the systemic movement of SPMV RNA in a host-dependent manner. Unexpectedly, defective RNAs (D-RNAs) accumulated de novo in millet plants coinfected with PMV and either of two SPMV mutants: SPMV-91, which is incapable of expressing the 17.5-kDa CP, and SPMV-GUG, which expresses low levels of the 17.5-kDa CP. The D-RNA derived from SPMV-91 was isolated from infected plants and used as a template to generate a cDNA clone. RNA transcripts derived from this 399-nt cDNA replicated and moved in millet plants coinoculated with PMV. The characterization of this D-RNA provided a biological confirmation that the critical RNA domains identified by the reverse genetic strategy are essential for SPMV replication and movement. The results additionally suggest that a potential “trigger” for spontaneous D-RNA accumulation may be associated with the absence or reduced accumulation of the 17.5-kDa SPMV CP. This represents the first report of a D-RNA associated with a satellite virus.
Four plant satellite viruses have been identified in association with helper viruses from natural infections, including satellite tobacco necrosis virus (STNV), satellite panicum mosaic virus (SPMV), satellite maize white line mosaic virus, and satellite tobacco mosaic virus (STMV) (4, 23). Satellite viruses are valuable tools for dissecting sequence or structural features of RNA molecules that are essential for biological and biochemical functions, such as replication, symptom induction, and spread, without perturbing the helper virus genome. These “molecular parasites” can also help us to understand the intricately coordinated interactions between helper virus-encoded proteins, the satellite virus RNAs, and the host cells (15, 23).
The SPMV genome is composed of a single-stranded, plus-sense RNA of 824 nucleotides (nt). Four open reading frames (ORFs) were originally identified, two on the viral plus-sense strand and two on the viral complementary strand (8). An ORF from nt 88 to 561 on the viral sense RNA encodes a 17.5-kDa capsid protein (CP) (8). The 5′ untranslated region (UTR) is composed of 87 nt preceding the SPMV CP start codon, and the 3′ UTR extends from nt 562 to 824. Although strong secondary structures were predicted in the 5′ and 3′ UTRs and implicated in the replication of the SPMV genome (8), cis-acting sequences essential for the replication and/or systemic movement of SPMV have not been experimentally identified.
SPMV depends on its helper virus, panicum mosaic virus (PMV), for replication and systemic spread (8, 21). PMV has the unusual characteristic of supporting two distinct types of subviral entities, SPMV and satellite RNAs, which is reflected in the naturally occurring viral complex in infected St. Augustinegrass lawns along the Gulf Coast of the United States (3). Sequence analysis revealed that the 4,326-nt single-stranded plus-sense RNA of PMV encodes six ORFs (28). The 5′-proximal p48 and p112 read-through proteins provide the core components of the PMV replicase complex, which is also proposed to be essential for the replication of SPMV and the satellite RNAs (28). A 26-kDa CP and three other smaller proteins (p15, p8, and p6.6) have been implicated in local and systemic movement of PMV (27).
RNA transcripts produced in vitro from the full-length cDNA clones of PMV and SPMV are infectious on millet host plants (9, 21, 28). In the present study, a comprehensive set of SPMV mutants was generated and analyzed for the ability to infect millet protoplasts and plants. Infectivity assays with these mutants mapped the cis-acting elements that are required for SPMV replication and movement. Furthermore, the characterization of an infectious cDNA clone derived from a defective SPMV RNA (D-RNA), which accumulated de novo in the plants coinoculated with PMV and SPMV CP-deficient mutants, also defined the cis-acting elements required by SPMV for its viability in plants.
MATERIALS AND METHODS
Host plants and RNA transcripts.
Pearl millet (Pennisetum glaucum), foxtail millet (Setaria italica cv. ‘German R’), and proso millet (Panicum miliaceum cv. ‘Sunup’ or ‘Red Turghai’) plants were used as experimental hosts and grown in the greenhouse (28 to 30°C) or in the growth chamber (28°C, 14 h of light; 24°C, 10 h of dark).
Standard molecular biology protocols were applied throughout the study (18). To supply DNA templates for the in vitro transcription reaction, cesium chloride-purified plasmids containing full-length PMV cDNA (28) were digested with EcoICR1, and the plasmids carrying SPMV or mutagenized SPMV cDNAs were digested with BglII. Plasmids containing the SPMV D-RNA cDNA were digested with PstI and treated with DNA polymerase I large fragment (Klenow). All treatments were followed by phenol-chloroform extraction and ethanol precipitation. Approximately 0.5 μg of linearized DNA was consistently used as a template for in vitro transcription reactions, as specified by the supplier of T7 RNA polymerase (Gibco Life Technologies, Gaithersburg, Md.).
Four to five millet plants at the three-leaf stage were mechanically inoculated with a mixture of ca. 6 μg of uncapped RNA transcripts and 12 μl of RNA inoculation buffer (0.05 M K2HPO4, 0.05 M glycine, 1% bentonite, 1% Celite, pH 9.0). The inoculated plants were maintained at room temperature overnight before being transferred to the greenhouse or growth chamber. Each bioassay, including monitoring of visual symptoms and detection of viral RNAs or proteins from the infected millet plants, was repeated at least three times with independently synthesized PMV and SPMV RNA transcripts. The symptoms on the inoculated plants were documented at 14 to 21 days postinoculation.
Protoplasts were isolated from pearl millet or foxtail millet seedlings 2 weeks postgermination in the growth chamber. The preparation and transfection of millet protoplasts were performed essentially as described previously (19) except that millet protoplasts were centrifuged at 210 × g instead of 70 × g in a clinical centrifuge (International Equipment Co., Needham Heights, Mass.). Approximately 5 × 106 protoplasts were transfected with ca. 6 μg of PMV RNA transcripts alone or in combination with in vitro-synthesized SPMV transcripts (ca. 6 μg each) derived from either the type strain or mutant cDNA clones through a polyethylene glycol procedure (19). The transfected protoplasts were incubated in the growth cabinet (28°C, 14 h of light; 24°C, 10 h of dark) for 40 to 45 h prior to the extraction of total RNA or protein.
Site-directed oligonucleotide mutagenesis of the SPMV cDNA.
Single-stranded DNA was generated from the SPMV cDNA phagemid using standard protocols (6). A KpnI site at nt 100 (primer 5′-GGCTCCTAAGGGTACCAGGCG-3′ [the KpnI site is underlined]) and an EcoRV site at nt 486 (primer 5′-CCAAGGGTGATATCGCCCCCAGC-3′ [the EcoRV site is underlined]) were introduced into the SPMV cDNA clone to generate a new construct, designated SPMV-KE, to allow for deletion of the CP ORF. An NcoI site (primer 5′-CCCTTTCCCATGGCAATGCC-3′ [the NcoI site is underlined]) was inserted at nt 772 for deletion analysis of the 3′ UTR. The newly introduced KpnI, EcoRV, and NcoI sites produced a combined restriction map of SPMV (Fig. 1).
FIG. 1.
Schematic diagram of SPMV constructs used to dissect SPMV RNA cis-acting elements for PMV-dependent replication and movement. The KpnI, EcoRV, and NcoI sites (in boldface) were introduced at nt 100, 486, and 772 on the SPMV cDNA clone, respectively. The combined restriction map of the SPMV cDNA is shown. The SPMV CP ORF is denoted by the hatched rectangles. The open regions with solid outlines represent a nontranslated region preceding the second in-frame AUG codon at nt 235 in the mutant SPMV-91 and also the putative out-of-frame codons fused with the N-terminal four amino acids in SPMV-ΔCP, SP2, and SP3. The solid regions indicate the retained fragments on the SPMV deletion mutants. The open regions with dashed outlines represent deletions on the SPMV cDNA. The positions of the CP start codon AUG (or GUG in the mutant SPMV-GUG) at nt 88, a second in-frame AUG at nt 235, two introduced stop codons in SPMV-91 at nt 97 and 109, and the stop codon UAA at nt 561 are indicated below the CP ORF.
A degenerate primer (5′-CCTGR[G/A]TGS[C/G]CTCCTAAGCGTTCC-3′) was used to create CP start codon context mutants (AUGC, AUGG, GUGC, and GUGG). For this study, the SPMV CP start codon context mutant GUGG was identified and designated SPMV-GUG. Mutant SPMV-91, in which two stop codons were introduced immediately downstream of the CP start codon, was generated with primer 91 (5′-GGCTCCTTAGCGTTCCAGGTGATCTAATCG-3′ [the stop codons are underlined]).
Construction of the SPMV deletion mutants.
Internal sequences from the full-length SPMV cDNA clone were deleted by excision of fragments with two selected restriction enzymes in various combinations. For example, SP1 was generated by digestion of the full-length SPMV cDNA clone with NheI and SpeI (Fig. 1). If the ends were not compatible, the linearized plasmids were subjected to Klenow treatment or mung bean nuclease digestion followed by electrophoresis and purification of DNA from the 1% agarose gel. A reverse transcription PCR (RT-PCR)-amplified product of SPMV lacking nt 572 to 811 in the 3′ UTR was generated, presumably due to internal annealing of the SPMV reverse primer (5′-AAACTGCAGGGTCCTAGGAGGGGG-3′ [the PstI site is underlined]) to positions 567 to 571. This RT-PCR product was inserted into pUC119 to make the SP13 construct. The deletions of the fragments from the full-length SPMV cDNA clones were confirmed by sequencing across the junction sites of the resultant cDNA clones (Table 1).
TABLE 1.
Features of the SPMV deletion mutants and their PMV-dependent accumulation on two species of millet plants
| Virus or deletion mutant | Restriction enzymes | Junction sequencea | Nucleotides deleted | Presenceb
|
|||
|---|---|---|---|---|---|---|---|
| Foxtail millet (S. italica)
|
Proso millet (P. miliaceum)
|
||||||
| Inoculated | Systemic | Inoculated | Systemic | ||||
| SPMV | + | + | + | + | |||
| SPMV-ΔCP | KpnI-EcoRV | TAAGGATCGC | 100–489 | + | + | + | + |
| SP1 | NheI-SpeI | GCTAGTCTCA | 17–67 | + | + | + | + |
| SP2 | BssHII-EcoRV | AACGATCGC | 307–489 | + | + | + | + |
| SP3 | KpnI-BssHII | TAAGGCCTGT | 100–311 | + | + | + | + |
| SP4 | SpeI-KpnI | ACTAGCAGGC | 68–104 | + | − | + | + |
| SP5 | SpeI-BssHII | TTGACCTG | 63–311 | + | − | + | + |
| SP6 | NheI-EcoRV | GCTAGATC | 17–489 | + | − | + | +/− |
| SP7 | SpeI-EcoRV | AACTAGATC | 68–489 | + | − | + | +/− |
| SP8 | NheI-BsrGI | GCTAGGTAC | 17–709 | − | − | − | − |
| SP9 | SpeI-BsrGI | AACTAGGTAC | 68–709 | − | − | − | − |
| SP10 | BsrGI-NcoI | TGTACCATG | 713–772 | − | − | − | − |
| SP11 | 17–67 and 713–772 | − | − | − | − | ||
| SP12 | NcoI-BglII | CCATGGATCT | 777–824 | − | − | − | − |
| SP13 | GATATCCCCC | 572–811 | − | − | − | − | |
Boldface indicates the partial restriction enzyme sequence inside the 5′-proximal portion of the junction site.
+, detectable SPMV RNA in the inoculated leaves or upper systemically infected leaves following coinfection with PMV, as determined by the RNA blot; −, SPMV RNA accumulation below the detection level 14 days postinoculation; +/−, RNA accumulation is dependent on the environmental conditions.
Isolation and cDNA cloning of SPMV D-RNA.
Total RNA was extracted from millet plants infected with PMV alone and in combination with either SPMV-91 or SPMV-GUG as previously described (20). The RNA was electrophoresed through a 1.2% agarose gel. A single SPMV-specific band, which is smaller than the genome-size SPMV RNA and designated D-RNA, was excised from the gel. The agarose block was placed on sterilized Miracloth (Calbiochem Co., La Jolla, Calif.) which was inserted into a 0.5-ml microfuge tube with a hole punctured by an 18-gauge syringe needle. The tube was then placed in a 1.5-ml microfuge tube and centrifuged at 10,000 × g for 2 min to elute the RNA from the gel matrix. The gel matrix was eluted a second time with 200 μl of 1× TE buffer (10 mM Tris, 1 mM EDTA, pH 7.5). The solutions from both elutions were combined and subjected to phenol-chloroform extraction. The isolated RNAs were then precipitated and stored at −80°C.
A previously described RT-PCR procedure (14) was adapted to amplify the D-RNA. A reverse SPMV primer (5′-AAACTGCAGGGTCCTAGGAGGGGG-3′ [the PstI site is underlined]) complementary to the 3′-terminal 811 to 824 nt was used for first-strand cDNA synthesis. A forward SPMV primer (5′-AAAGGATCCTAATACGACTCACTATAGGGTATTCCACGCTAGC-3′ [the BamHI site is underlined]) contained the first 17 nt of the 5′ end of SPMV, which was linked with an upstream T7 RNA polymerase promoter sequence (in boldface letters). Following RT-PCR of the D-RNA, the product was cloned into the SmaI site of pUC119.
RNA and protein blot analyses.
Total RNA and protein were extracted from 500 mg of inoculated or systemically infected leaves bulked from four millet plants about 14 days postinoculation or from ca. 5 × 106 protoplasts 40 to 45 h posttransfection (20). Approximately 3 μg of total plant RNAs or the entire protoplast RNA was separated in a 1.2% agarose gel and subsequently transferred to nylon membranes (Micron Separations Inc., Westborough, Mass.). PMV and SPMV RNAs were detected by hybridizing the RNA blots with [32P]dCTP-labeled PMV- or SPMV-specific probes generated by a random-priming method as previously described (28). Approximately 15 μg of total proteins were electrophoresed through a sodium dodecyl sulfate–12% polyacrylamide gel and then transferred to nitrocellulose membranes (Micron Separations Inc.). The membranes were incubated with rabbit polyclonal anti-SPMV CP antiserum or with rabbit polyclonal anti-PMV CP antiserum (21). The secondary anti-rabbit immunoglobulin G conjugated with alkaline phosphatase (Sigma, St. Louis, Mo.) or horseradish peroxidase (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.) was used at a 1:5,000 dilution. The presence of viral CP was assayed by enzymatic reactions of alkaline phosphatase or horseradish peroxidase as described previously (22).
Nucleotide sequence accession number.
The D-RNA cDNA sequence was deposited in GenBank as accession no. AF159425.
RESULTS
The SPMV CP gene is dispensable for replication and systemic infection.
The full-length SPMV cDNA clone pSPMV-1 (9) was resequenced and found to contain 824 nt instead of the previously identified 826 nt (8). The nucleotide A at position 10 in the 5′ UTR and the nucleotide C at position 766 in the 3′ UTR were absent from the SPMV cDNA clone used in this study, which is now designated the type strain.
Prior to the dissection of cis-acting elements on the SPMV type strain RNA genome, unique KpnI and EcoRV sites were introduced at nt 100 and 486, respectively, inside the CP ORF or an NcoI site was introduced at nt 772 on the 3′ UTR. The introduced KpnI site converted the CP amino acids R5 and S6 to G and T, respectively. The EcoRV site changed codons for amino acids A134 and N135 to D and I, respectively. The introduction of these sites generated a new combined restriction map of the SPMV genome (Fig. 1). The modified SPMV RNAs, with a combination of KpnI and EcoRV sites or an NcoI site, behaved essentially the same as the SPMV type strain, as determined by comparative analysis of viral titers and symptoms on millet plants coinoculated with PMV (data not shown).
To unveil the biological functions of SPMV CP, two nucleotide changes were introduced to prematurely terminate the synthesis of the CP (mutant SPMV-91) while minimally changing the RNA sequence within the CP ORF (Fig. 1). In addition, two contiguous deletion constructs, SP2 and SP3 (Fig. 1 and Table 1), were made to delete either the 3′ or the 5′ half of the SPMV CP ORF, respectively. This allowed for analysis of cis elements inside the CP ORF for replication and movement. The SP2 mutant encoded an N-terminal portion of the SPMV CP, which produced a ca. 8-kDa protein in a wheat germ in vitro translation assay, but this protein was not detectable by immunoblot analysis with protein extracts from infected plants (data not shown). SP3 was predicted to encode a putative 21-amino-acid polypeptide by the fusion of the N-terminal 4 amino acids of the CP ORF to 17 downstream, out-of-frame codons. Lastly, the major portion of the SPMV CP coding region was deleted by digestion of the SPMV cDNA clone with KpnI and EcoRV to generate SPMV-ΔCP (Fig. 1 and Table 1). This deletion forced the fusion of four N-terminal amino acids with the C-terminal amino acids of the SPMV CP ORF, but out of frame, to yield a putative 17-amino-acid polypeptide.
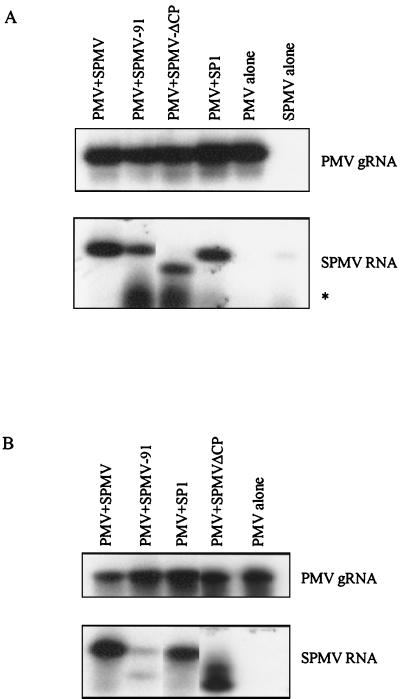
SPMV-91 was replicated by PMV in pearl millet protoplasts (Fig. 2A). Although SPMV-91 was translocated along with PMV to the upper noninoculated leaves, it accumulated less abundantly than the SPMV type strain (Fig. 2B). Removal of either the terminal half-portion of the SPMV CP in deletion mutants SP2 and SP3 or the majority of the SPMV CP ORF in SPMV-ΔCP did not impair replication or systemic movement of SPMV RNA in the millet plants coinfected with PMV (Fig. 2 and Table 1). Degraded SPMV RNAs were detected on the RNA blots from protoplasts transfected with PMV and two CP-deficient mutants, SPMV-91 and SPMV-ΔCP. This is in contrast to the RNA profiles obtained with the SPMV type strain and SP1, two constructs that express the SPMV CP (Fig. 2A). Degraded forms of PMV RNAs were not observed in parallel RNA blots (data not shown).
FIG. 2.
RNA blot analysis of SPMV replication in pearl millet protoplasts and plants. (A) Replication competency of SPMV and its derived mutants in pearl millet protoplasts coinfected with PMV. An asterisk indicates the SPMV-specific degraded RNAs. Note the residual SPMV input RNA (lane SPMV alone) in the protoplasts ca. 40 h posttransfection by SPMV alone. (B) Replication and movement competency of SPMV and its derived mutants in upper noninoculated leaves at 21 days postinoculation. The features of the mutants and the results of the RNA analyses are summarized in Fig. 1 and Table 1, respectively. gRNA, genomic RNA.
Regions of SPMV RNA that are essential for replication and systemic movement.
Since the CP gene is dispensable for the replication and systemic movement of SPMV (Fig. 2), it could be inferred that the essential elements for these functions reside in the 5′ and 3′ UTRs. To investigate this, the 5′- and 3′-proximal SPMV cDNA fragments were serially removed to evaluate their contributions to replication and movement (Fig. 1). The infectivity assays of the deletion mutants in millet plants coinoculated with PMV were performed, and the results are summarized in Table 1. Briefly, deletion of nt 17 to 67 in the mutant SP1 (Fig. 1 and Table 1) slightly reduced the accumulation of the satellite viral RNA compared to the SPMV type strain (Fig. 2). This was also reflected by the decreased amount of SPMV CP on the immunoblots in protoplasts and millet plants (data not shown). The removal of nt 68 to 104 in SP4 or nt 63 to 311 in SP5 did not affect their infectivity in the proso millet plants (Table 1). The mutants SP6 and SP7, lacking nt 17 to 489 and 68 to 489, respectively, were detectable on the upper noninoculated leaves of proso millet plants. However, the RNA accumulation was influenced by the environmental conditions in the greenhouse. The mutants SP4, SP5, SP6, and SP7 were detected on the inoculated leaves but not on the upper noninoculated leaves of foxtail millet plants (Fig. 1 and Table 1).
Deletion of nt 572 to 811 in the 3′ UTR abolished the replication of SP13 even though it retained the 3′-terminal 14 nt of the SPMV RNA. The deletion of 3′-proximal sequences from nt 713 to 772 in SP10 or 777 to 824 in SP12 also abolished the infectivity of SPMV, as these RNAs were not detected in the millet plants (Fig. 1 and Table 1). Taken together, the data suggested that deletions of any region of the 3′ UTR in the mutants SP8, SP9, SP10, SP11, SP12, and SP13 were lethal to SPMV propagation in the millet plants (Fig. 1 and Table 1).
In summary, the above data collectively provide a functional map of the SPMV RNA genome in which four domains can be tentatively assigned: (i) nt 17 to 67 have a role in regulating replication, (ii) nt 68 to 104 appear to be involved in host-dependent systemic movement, (iii) nt 100 to 489 are dispensable for SPMV infectivity, and (iv) nt 490 to 824, covering the 73 nt immediately upstream of the CP stop codon plus the entire 3′ UTR, are required for biological activity of SPMV in plants (Table 1).
The generation of an infectious cDNA clone from a defective SPMV RNA.
Aberrant RNAs that migrated faster than the genome-size SPMV RNAs were frequently detected in agarose gels (Fig. 3A). These spontaneously generated RNAs were particularly associated with serial passages of SPMV-91 or SPMV-GUG through millet plants coinoculated with PMV and were tentatively designated D-RNAs. The D-RNA derived from the SPMV-91 mutant accumulated more abundantly than those derived from SPMV-GUG (Fig. 3A). Due to double stop codons immediately downstream of the CP start codon, SPMV-91 is incapable of expressing the 17.5-kDa CP, but it does express a small amount of ca. 10-kDa protein predicted to be a carboxy-terminal portion of the CP (Fig. 1 and 3A). In contrast, SPMV-GUG expressed more ca. 10-kDa protein than SPMV-91 but very low levels of the 17.5-kDa CP compared to the SPMV type strain (Fig. 1 and 3A). The D-RNAs could represent either degenerate products of SPMV genomic RNAs or independently replicating SPMV-derived RNAs. To test these possibilities, the D-RNA from SPMV-91, which was passaged five times on proso millet plants, was isolated and subjected to RT-PCR amplification. An RT-PCR product with a T7 polymerase promoter sequence at the 5′ end was inserted into pUC119. In vitro-generated RNA transcripts derived from the SPMV-91 D-RNA cDNA template replicated efficiently and moved in millet plants when coinoculated with PMV (Fig. 3B). The results of this assay indicated that the SPMV-91 D-RNA retained the essential domains for replication and systemic movement and was viable independently of the SPMV genomic RNA.
FIG. 3.
The emergence of an SPMV D-RNA from SPMV CP-deficient mutants and the replication competency of an infectious SPMV-91 D-RNA cDNA clone. (A) The SPMV CP-specific immunoblot (top) detects different expression levels and forms of the SPMV CP from the SPMV type strain and mutants SPMV-GUG and SPMV-91 in the infected proso millet plants. The RNA blot (bottom) revealed de novo D-RNA accumulation in the infected proso millet plants at 14 days postcoinoculation with PMV plus SPMV-GUG or SPMV-91 transcripts. (B) RNA blot analysis of total RNA from upper noninoculated leaves of proso millet plants coinoculated with RNA transcripts produced from SPMV-91 D-RNA and PMV cDNA clones. RNA samples from two separate experiments were analyzed. The RNA blot was first hybridized with an SPMV-specific probe and then hybridized with a PMV-specific probe to simultaneously show the presence of the SPMV-91 D-RNA and PMV RNAs. The cDNA in parentheses indicates the D-RNAs were derived from the infectious cDNA clones. The positions of the genomic RNA (gRNA), the subgenomic RNA (sgRNA) of PMV, and the SPMV D-RNA are indicated.
The alignment of cDNA sequences from D-RNA and SPMV RNA.
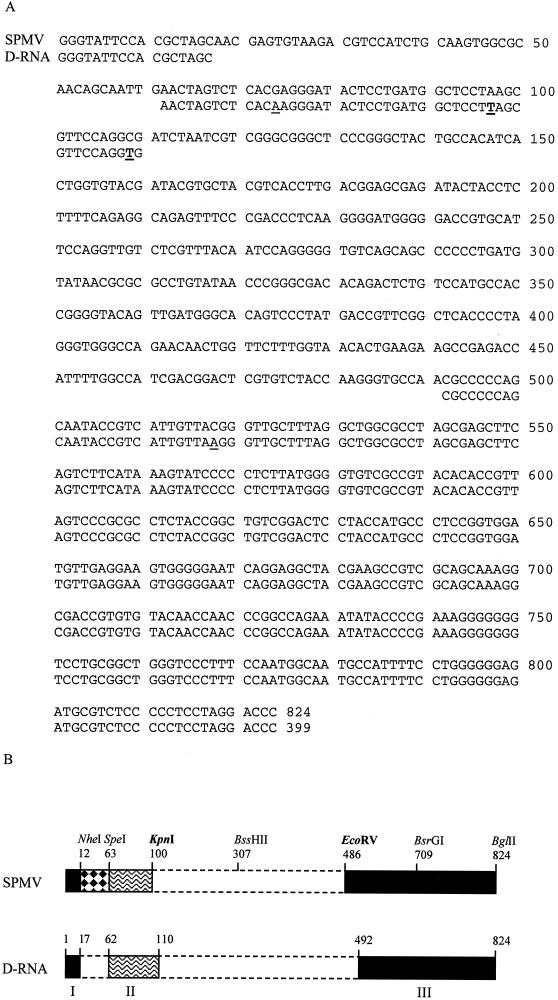
A near-perfect match was observed between cis-acting RNA domains delimited by the in vitro deletion assay, and the composition of the D-RNA spontaneously emerged from SPMV-91. The 399-nt D-RNA preserves three discrete regions from the SPMV genomic RNA, which are designated domains I, II, and III (Fig. 4B). Domain I contains the 5′-terminal 17 nt of SPMV RNA, domain II is composed of 49 nt from nt 62 to 110, and domain III consists of 333 nt from nt 491 to 824 in the 3′ end (Fig. 4A). The D-RNA was confirmed to be derived from SPMV-91 RNA by the evidence that domain II in the D-RNA cDNA contained two Ts in the exact positions where they were originally introduced in the SPMV-91 cDNA (nt 97 and 109 on the SPMV-91 genomic RNA) (Fig. 1 and 4A). More interestingly, only slight deviations were found in the two junction sites of domains I and II and domains II and III between the D-RNA and two in vitro-generated replication- or movement-competent deletion mutants, SP1 and SPMV-ΔCP. The junction site between domains I and II from SPMV-91 D-RNA cDNA is GCTAGCAACTAGT (the retained NheI and SpeI sites are underlined, and the nucleotides in domain II are shown in italics), which possesses seven more nucleotides (in boldface) than the junction site GCTAGT, created by ligation of the compatible ends of NheI and SpeI sites to form the deletion mutant SP1 (Fig. 1 and Table 1). The junction site between domains II and III of SPMV-91 D-RNA cDNA is TTAGCGTTCCAGGTGCGCCCCC (the nucleotides in italics are within domain II, and the remainder of the nucleotides are within domain III). The D-RNA has eight extra nucleotides (in boldface) compared to the junction site TAAGGATCGCCCCC in SPMV-ΔCP (Fig. 1 and Table 1) (the nucleotides inside domain II are in italics). The variations between the junction sites on SPMV-91 D-RNA and the deletion mutants, SP1 and SPMV-ΔCP, do not obviously affect the replication and movement competency of these SPMV-derived RNAs (Fig. 2 and 3).
FIG. 4.
Sequence and schematic comparisons of the infectious SPMV-91 D-RNA cDNA with the SPMV type strain cDNA. (A) Alignment and comparison of the 399-nt D-RNA cDNA (D-RNA) and the 824-nt SPMV cDNA (SPMV). The two Ts at nt 53 and 65 (shown in boldface and underlined) in the D-RNA cDNA are in the same positions where they were originally introduced in the mutant SPMV-91 cDNA by site-directed mutagenesis. The G at nt 74 and the C at nt 518 of the type strain, which were changed to A at nt 30 and 93 (underlined) in the D-RNA cDNA, are shown. (B) Schematic representation of RNA domains on the SPMV genome essential for the PMV-dependent replication and movement delineated by the reverse genetic approach and also defined by the D-RNA. D-RNA is solely composed of domains I, II, and III. Representative restriction enzyme sites with their positions on the SPMV cDNA are shown above the genome. The regions essential for replication and movement are denoted by solid rectangles. The region between domain I and II presumably having an accessory role in replication is represented by a checkered rectangle; domain II, which is depicted by wavy lines, delineates sequences which are putatively involved in host-dependent systemic movement; and nonessential regions are denoted by open rectangles with dashed outlines.
DISCUSSION
Viral RNAs normally contain critical cis-acting elements in the 5′ and 3′ UTRs which are essential for viral replication (2). Defective interfering (DI) RNAs of TBSV (30) and a piconavirus (26) represent two rare cases where the 5′ UTR or the 3′ UTR could be deleted without abolishing viability. Recent evidence has shown that the recognition of satellite virus RNAs in trans by their helper virus replicase complexes are also facilitated primarily by signals located in their 5′ or 3′ UTRs (1, 12, 16, 17). For example, the cis-acting elements necessary for the replication of STNV are embedded in three hairpin-like structures on the 5′ UTR and in two discrete regions identified as the 3′-proximal terminal sequence and the sequence immediately adjacent to the CP ORF in the 3′ UTR (1). Likewise, a replication competency assay with STMV deletion mutants revealed that the 3′ UTR sequence immediately downstream of the CP ORF was critical for STMV accumulation (16, 17). In contrast, the CP genes of both STNV and STMV were dispensable for accumulation and movement (1, 16, 17).
The crucial regions for SPMV viability on millet plants coinfected with PMV reside in three regions, (i) nt 1 to 16, (ii) nt 62 to 110 at the 5′-end of the RNA, and (iii) 73 nt upstream of the CP stop codon plus the entire 3′ UTR (nt 562 to 824) (Fig. 2, 3, and 4 and Table 1). Extensive comparisons of PMV RNA sequence with SPMV RNA did not reveal significant sequence homology except for seven identical 5′-terminal nucleotides (5′-GGGUAUU-3′) and three identical 3′-terminal nucleotides (5′-CCC-3′) (28). It can therefore be envisioned that the three identical CCC bases at the termini of PMV RNA and SPMV RNA may harbor the initial recognition signal for the PMV replicases to extend both minus-strand and plus-strand RNA. This has been demonstrated for turnip crinkle virus and its satellite RNAs (10, 24, 25). Hairpin-like structures can be formed with the nucleotides from 490 to 824 in the 3′ end of SPMV (data not shown), as predicted by the program M-fold (version 3.0; Institute for Biomedical Computing, Washington University, St. Louis, Mo. [http://mfold2.wustl.edu/∼mfold/rna/form1.cgi]), suggesting that the essential signals for replication might be embedded in the stem-loop structures. The significance of such 3′ UTR stem-loop structures for virus replication has been experimentally demonstrated for red clover necrotic mosaic virus (29) and alfalfa mosaic virus (13). Compensatory mutagenesis will be necessary to determine how these putative stem-loop structures regulate the accumulation of SPMV RNA.
The removal of the hairpin-like structures from the STNV 5′ UTR abolished its replication (1). In the adaptation of STMV to the nonoriginal helper viruses, tomato mosaic virus and green tomato atypical mosaic virus, nucleotides A and G at positions 1 and 61 in the 5′ UTR were consistently deleted (12), suggesting that the 5′ UTR plays a role in the helper virus-specific replication of STMV. However, SPMV accumulation in plants and protoplasts was reduced only slightly by the deletion of nt 17 to 67 within the 5′ UTR (Fig. 2). A further confirmation of the dispensability of this region was provided by its absence from the in vivo-generated D-RNA (Fig. 4). This short region in the 5′ UTR of SPMV may function as a cis-acting factor for RNA stability or enhance replicase binding activity. Alternatively, the domain may interact with the 3′ UTR in a long-distance manner to optimize SPMV RNA synthesis, analogous to an interaction between 5′- and 3′-terminal sequences proposed for potato virus X RNA synthesis (5). Another cis-acting RNA element, spanning nt 68 to 104 in the 5′ end, is critical for SPMV movement on foxtail millet plants (S. italica cv. ‘German R’) but is not required for systemic spread on proso millet plants (P. miliaceum cv. ‘Sunup’) (Fig. 1 and Table 1). These results suggest that this region might contain a signal for interaction with movement-associated host proteins. A similar observation was described for beet necrotic yellow vein virus in which a noncoding domain of 225 nt on the RNA3 was necessary for vascular movement of the virus (7). The exact mechanism involved in the RNA-directed host-specific transport of SPMV remains to be resolved.
The minimum RNA domains for SPMV replication and/or systemic spread in the millet plants that were mapped by the reverse genetic approach were independently confirmed by the de novo-generated D-RNA isolated from infected millet plants with SPMV-91 and PMV. A widely accepted model for the formation of DI RNAs is that viral replicase bound with its nascent RNA may dissociate from the template and reinitiate the synthesis at a new position with or without nucleotides homologous with the 3′ terminal sequences of the nascent RNA on the same template (11). Typically, DI RNAs are degenerative genomic RNAs derived directly from the parental viral genome during the replication process. The formation of a D-RNA from a satellite virus may comply with this model but differs from the typical DI RNA formation in that the satellite viral RNA replication is accomplished in trans by the helper virus replicase complex. The infectivity assay with SPMV mutants indicated that the D-RNA accumulated more abundantly in the initially infected plants that were coinoculated with SPMV-91 and PMV than in the plants coinoculated with SPMV-GUG and PMV (Fig. 3A). In contrast, D-RNAs did not spontaneously accumulate to detectable levels during the initial coinfection of the SPMV type strain and PMV on proso millet plants (Fig. 3A). Expression of the 17.5-kDa SPMV CP was abolished in the mutant SPMV-91, and the amount of the 17.5-kDa CP expressed from SPMV-GUG was less than 10% that of the SPMV type strain (Fig. 3A). Therefore, one explanation for the formation of D-RNA from two SPMV mutants could be inferred from the putative role of SPMV CP in the satellite virus life cycle. SPMV CP may act as a stabilizer to maintain the integrity of the SPMV genome through the trans-replication process by the PMV replicase complex. In other words, the PMV replicase complex may be prone to reinitiate at new positions on the SPMV genome when little or no 17.5-kDa SPMV CP is present in the infected plants. This hypothesis is indirectly supported by our observation that degraded SPMV RNAs were detected in the millet protoplasts infected with either SPMV-91 or SPMV-ΔCP, two constructs which are incapable of expressing the 17.5-kDa CP (Fig. 2A). Furthermore, SPMV D-RNAs have not been detected in St. Augustinegrass plants naturally infected with wild-type SPMV and PMV (3). However, the possibility that other mechanisms (11) may also be involved in the formation of SPMV D-RNAs cannot be ruled out.
In summary, the reverse genetic study and the characterization of an SPMV D-RNA that emerged de novo in infected millet plants conclusively demonstrate that the CP gene is not required for SPMV viability. Furthermore, the signals for replication and movement are embedded in the 5′-proximal 16 nt, nt 62 to 110 in the 5′ end, and the entire 3′ UTR plus the 73 nt upstream of the CP stop codon on SPMV RNA. The delineation of cis-acting elements in three satellite viruses, STNV (1), STMV (16, 17), and SPMV, supports the implication that the crucial signals for the initial replication cycle and systemic movement are clustered within the UTRs of the satellite viral RNA genomes. An interesting question that has not yet been addressed is why SPMV and other satellite viruses retain their respective CP genes even though they are dispensable for the essential life cycles of satellite viruses. No matter which mechanisms are involved in the formation of satellite virus D-RNAs, these unique RNAs will provide an opportunity to further dissect the features critical for virus replication and spread and to explore the mechanisms underlying the evolution of satellite viruses and satellite RNAs.
ACKNOWLEDGMENTS
We thank Massimo Turina for providing the SPMV-KE construct and helpful discussions. Critical reading of the manuscript and suggestions were generously provided by Herman Scholthof, Bénédicte Desvoyes, and Jeff Batten. We are also grateful to Gene Perry of Perry Brothers Seed (Otis, Colo.) for providing proso and foxtail millet seeds.
Funding for the research was provided by USDA-NRI Competitive Grants (96-35303-3714 and 99-35303-7974) and the Texas Agricultural Experiment Station (H-8388).
REFERENCES
- 1.Bringloe D H, Gultyaev A P, Pelpel M, Pleij C W, Coutts R H. The nucleotide sequence of satellite tobacco necrosis virus strain C and helper-assisted replication of wild-type and mutant clones of the virus. J Gen Virol. 1998;79:1539–1546. doi: 10.1099/0022-1317-79-6-1539. [DOI] [PubMed] [Google Scholar]
- 2.Buck K W. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cabrera O, Scholthof K-B G. The complex viral etiology of St. Augustine decline. Plant Dis. 1999;83:902–904. doi: 10.1094/PDIS.1999.83.10.902. [DOI] [PubMed] [Google Scholar]
- 4.Dodds J A. Satellite tobacco mosaic virus. Annu Rev Phytopathol. 1998;36:295–310. doi: 10.1146/annurev.phyto.36.1.295. [DOI] [PubMed] [Google Scholar]
- 5.Kim K-H, Hemenway C. Long distance RNA-RNA interactions and conserved sequence elements affect potato virus X plus-strand RNA accumulation. RNA. 1999;5:1–10. doi: 10.1017/s1355838299982006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 7.Lauber E, Guilley H, Tamada T, Richards K E, Jonard G. Vascular movement of beet necrotic yellow vein virus in Beta macrocarpa is probably dependent on an RNA 3 sequence domain rather than a gene product. J Gen Virol. 1998;79:385–393. doi: 10.1099/0022-1317-79-2-385. [DOI] [PubMed] [Google Scholar]
- 8.Masuta C, Zuidema D, Hunter B G, Heaton L A, Sopher D S, Jackson A O. Analysis of the genome of satellite panicum mosaic virus. Virology. 1987;159:329–338. doi: 10.1016/0042-6822(87)90471-5. [DOI] [PubMed] [Google Scholar]
- 9.Monis J, Sopher D S, Jackson A O. Biologically active cDNA clones of panicum mosaic virus satellites. Phytopathology. 1992;82:1175. [Google Scholar]
- 10.Nagy P D, Carpenter C D, Simon A E. A novel 3′ end repair mechanism in an RNA virus. Proc Natl Acad Sci USA. 1997;94:1113–1118. doi: 10.1073/pnas.94.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagy P D, Simon A E. New insights into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 12.Ngon A, Yassi M, Dodds J A. Specific sequence changes in the 5′-terminal region of the genome of satellite tobacco mosaic virus are required for adaptation to tobacco mosaic virus. J Gen Virol. 1998;79:905–913. doi: 10.1099/0022-1317-79-4-905. [DOI] [PubMed] [Google Scholar]
- 13.Olsthoorn R C L, Mertens S, Brederode F T, Bol J F. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J. 1999;18:4856–4864. doi: 10.1093/emboj/18.17.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu W P, Geske S M, Hickey C M, Moyer J M. Tomato spotted wilt Tospovirus genome reassortment and genome segment-specific adaptation. Virology. 1998;244:186–194. doi: 10.1006/viro.1998.9131. [DOI] [PubMed] [Google Scholar]
- 15.Roossinck M J, Sleat D, Palukaitis P. Satellite RNAs of plant viruses: structures and biological effects. Microbiol Rev. 1992;56:265–279. doi: 10.1128/mr.56.2.265-279.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Routh G, Dodds J A, Fitzmaurice L, Mirkov T E. Characterization of deletion and frameshift mutants of satellite tobacco mosaic virus. Virology. 1995;212:121–127. doi: 10.1006/viro.1995.1460. [DOI] [PubMed] [Google Scholar]
- 17.Routh G, Ngon M, Yassi A, Rao A L N, Mirkov E T, Dodds J A. Replication of wild-type and mutant clones of satellite tobacco mosaic virus in Nicotiana benthamiana protoplasts. J Gen Virol. 1997;78:1271–1275. doi: 10.1099/0022-1317-78-6-1271. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Scholthof H B, Morris T J, Jackson A O. The capsid protein gene of tomato bushy stunt virus is dispensable for systemic movement and can be replaced for localized expression of foreign genes. Mol Plant-Microbe Interact. 1993;6:309–322. [Google Scholar]
- 20.Scholthof H B, Scholthof K-B G, Kikkert M, Jackson A O. Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology. 1995;213:425–438. doi: 10.1006/viro.1995.0015. [DOI] [PubMed] [Google Scholar]
- 21.Scholthof K-B G. A synergism induced by satellite panicum mosaic virus. Mol Plant-Microbe Interact. 1999;12:163–166. [Google Scholar]
- 22.Scholthof K-B G, Hillman B I, Modrell B, Heaton L A, Jackson A O. Characterization and detection of sc4: a sixth gene encoded by sonchus yellow net virus. Virology. 1994;204:279–288. doi: 10.1006/viro.1994.1532. [DOI] [PubMed] [Google Scholar]
- 23.Scholthof K-B G, Jones R W, Jackson A O. Biology and structure of plant satellite viruses activated by icosahedral helper viruses. Curr Top Microbiol Immunol. 1999;239:123–143. doi: 10.1007/978-3-662-09796-0_7. [DOI] [PubMed] [Google Scholar]
- 24.Song C, Simon A E. Synthesis of novel products in vitro by an RNA-dependent RNA polymerase. J Virol. 1995;69:4020–4028. doi: 10.1128/jvi.69.7.4020-4028.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stupina V, Simon A E. Analysis in vivo of turnip crinkle virus satellite RNA C variants with mutations in the 3′-terminal minus-strand promoter. Virology. 1997;238:470–477. doi: 10.1006/viro.1997.8850. [DOI] [PubMed] [Google Scholar]
- 26.Todd S, Towner J S, Brown D M, Semler B L. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J Virol. 1997;71:8868–8874. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turina M, Desvoyes B, Scholthof K-B G. A gene cluster encoded by panicum mosaic virus is associated with virus movement. Virology. 2000;266:120–128. doi: 10.1006/viro.1999.0069. [DOI] [PubMed] [Google Scholar]
- 28.Turina M, Maruoka M, Monis J, Jackson A O, Scholthof K-B G. Nucleotide sequence and infectivity of a full-length cDNA clone of panicum mosaic virus. Virology. 1998;241:141–155. doi: 10.1006/viro.1997.8939. [DOI] [PubMed] [Google Scholar]
- 29.Turner R L, Buck K E. Mutational analysis of cis-acting sequences in the 3′- and 5′-untranslated regions of RNA2 of red clover necrotic mosaic virus. Virology. 1999;253:115–124. doi: 10.1006/viro.1998.9495. [DOI] [PubMed] [Google Scholar]
- 30.Wu B, White K A. Formation and amplification of a novel tombusvirus defective RNA which lacks the 5′ nontranslated region of the viral genome. J Virol. 1998;72:9897–9905. doi: 10.1128/jvi.72.12.9897-9905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]