Abstract
IE62, the major transcriptional activator protein encoded by varicella-zoster virus (VZV), locates to the nucleus when expressed in transfected cells. We show here that cytoplasmic forms of IE62 accumulate in transfected and VZV-infected cells as the result of the protein kinase activity associated with VZV open reading frame 66 (ORF66). Expression of the ORF66 protein kinase but not the VZV ORF47 protein kinase impaired the ability of coexpressed IE62 to transactivate promoter-reporter constructs. IE62 that was coexpressed with the ORF66 protein accumulated predominantly in the cytoplasm, whereas the normal nuclear localization of other proteins was not affected by the ORF66 protein. In cells infected with VZV, IE62 accumulated in the cytoplasm at late times of infection, whereas in cells infected with a VZV recombinant unable to express ORF66 protein (ROka66S), IE62 was completely nuclear. Point mutations introduced into the predicted serine/threonine catalytic domain and ATP binding domain of ORF66 abrogated its ability to influence IE62 nuclear localization, indicating that the protein kinase activity was required. The region of IE62 that was targeted by ORF66 was mapped to amino acids 602 to 733. IE62 peptides containing this region were specifically phosphorylated in cells coexpressing the ORF66 protein kinase and in cells infected with wild-type VZV but were not phosphorylated in cells infected with ROka66S. We conclude that the ORF66 protein kinase phosphorylates IE62 to induce its cytoplasmic accumulation, most likely by inhibiting IE62 nuclear import.
Varicella-zoster virus (VZV) is the ubiquitous human alphaherpesvirus that causes chickenpox following a primary infection and shingles upon reactivation from the latent state (reviewed in reference 1). VZV has remained a difficult virus to study because its growth in tissue culture remains highly cell associated and yields a low, unstable cell-free virus titer. However, its genomic organization is closely related to that of the better-studied herpes simplex virus type 1 (HSV-1), and this has enabled many VZV gene functions to be predicted (3, 7, 8). Evidence has also indicated that VZV gene expression is highly regulated in a fashion similar to other herpesviruses (57), where each viral gene can be categorized into one of three groups (immediate early [IE], early, and late), depending on the timing and requirements for its maximal rate of transcription (19, 20). To date, at least five VZV gene products, from open reading frame 4 (ORF4), ORF61, ORF62, ORF63, and ORF10, have been shown to influence the transcriptional events in VZV-infected cells (9, 21, 22, 43, 48).
The activation of the VZV transcriptional cascade is regulated, in part, by the product of the 1,310-amino-acid ORF62, also known as IE62. IE62 is expressed from an IE gene (14) and is a powerful transcriptional activator (4, 21, 39, 44, 52, 53). IE62 has considerable structural and functional conservation to the major HSV-1 regulatory protein ICP4, sharing many of the properties attributed to HSV-1 ICP4 and likely acting through similar mechanisms (10, 13). In VZV-infected cells, IE62 localizes to the nucleus, using a typical arginine/lysine-rich nuclear localization signal that includes amino acids 676 to 685 (2, 14, 31). Following nuclear import, IE62 binds to specific DNA sequences, although there is divergence from a consensus sequence binding motif (61, 64). An unusual property of IE62 is that high levels are found associated with the tegument of purified virions (28, 29). It has been hypothesized that virion-associated IE62 may play a role to stimulate immediate early events upon infection (28, 44).
IE62 from VZV-infected cells is a major phosphoprotein and migrates as several forms on sodium dodecyl sulfate (SDS)-polyacrylamide gels between 170 and 180 kDa (14, 29, 49). However, the correlation between specific phosphorylated forms of IE62 and its functions are largely unknown. IE62 is phosphorylated by casein kinase II in vitro (49). Amino acids 350 to 370 of IE62 contain a serine-rich tract which is also found in HSV-1 ICP4, and in ICP4 it is the predominant domain of phosphorylation by cellular protein kinases, including cyclic AMP-dependent protein kinase A and casein kinase II (65, 66). Therefore, it is likely that these kinases may also phosphorylate IE62 in the corresponding region. Phosphorylation in the alphaherpesvirus subgroup is also influenced by two virus-encoded proteins (ORF47 and ORF66 in VZV [59]) which are predicted to encode serine/threonine protein kinases (34, 58). The protein kinase from VZV ORF47 specifically phosphorylates IE62 in in vitro phosphorylation studies (49). It also drives the phosphorylation of the VZV ORF32 protein in VZV-infected cells (49). The targets of the VZV ORF66 protein kinase are not known, but the homologous protein kinases in alphaherpesviruses (sometimes referred to as the US3 protein kinases) seem to target multiple and varied proteins specific for each virus (5, 6, 38, 55, 62). Recent evidence has indicated that HSV-1 US3 protein kinase also prevents apoptosis following infection and thus may target cellular proteins (38).
The observation that the ORF47 protein kinase specifically phosphorylated the IE62 protein (49) raised the novel and intriguing possibility that VZV protein kinases might affect the functions of IE62. This prompted a further investigation of the interactions between the VZV-encoded kinases and the VZV regulatory proteins. Surprisingly, we show here that it is the ORF66 protein kinase that affects the function of the IE62 protein by promoting its accumulation in the cytoplasm of infected cells.
MATERIALS AND METHODS
Cells and virus.
VZV strain Scott (isolate 71004), a partially characterized wild-type isolate (29), was used at less than 15 passages beyond its original isolation. VZV recombinants ROka, ROka47S (deficient in expression of ORF47), and ROka66S (deficient in the expression of ORF66) have previously been described (16, 17) and were kindly provided by Jeffrey Cohen, National Institute of Allergy and Infectious Diseases, Bethesda, Md. All viruses were grown at 35°C on human foreskin fibroblasts (line 521) or on a human melanoma cell line (MeWo cells) as described previously (28, 29). Cells were maintained at 37°C in Eagle's minimal essential medium supplemented with 5% Serum Plus (Hazleton Biologics Inc., Lenexa, Kans.), 5% fetal bovine serum, and an antibiotic mixture of penicillin (100 U/ml) and streptomycin (0.1 mg/ml). Cell-free VZV was produced from VZV-infected cells by sonication, using SPGA buffer (0.2 M sucrose, 0.01 M potassium phosphate [pH 6.6], 5 mM sodium glutamate, 1% bovine serum albumin) to stabilize infectivity. Cell sonicates were filtered through 5-μm filters at 4°C prior to infection of human foreskin fibroblast monolayers grown on glass slides. Cells were harvested at times indicated in the text by fixation in acetone at −20°C for 15 min.
Antibodies.
Antibodies that recognize the product of ORF61 have been described previously (28). A novel polyclonal rabbit antibody to IE62 was generated that recognized multiple domains of the IE62 protein. The antigen used was a mixture of purified maltose binding protein-IE62 fusion proteins representing amino acids 1 to 161, 162 to 506, 506 to 1310, and 417 to 824. Protein purification and rabbit immunizations were as previously described (28). Polyclonal antibodies to HSV-1 ICP4 were a kind gift of N. DeLuca, University of Pittsburgh, Pittsburgh, Pa., and monoclonal antibodies to the simian virus 40 (SV40) T antigen were commercially purchased (Santa Cruz Biotechnology, Santa Cruz, Calif.). A mouse monoclonal antibody to ORF62 (H6) was a kind gift of A. Arvin, Stanford University, Stanford, Calif. Monoclonal antibodies which recognize a linear epitope derived from the influenza virus hemagglutinin (HA) protein (see below) were obtained from BAbCo Inc., Richmond, Calif.
DNAs and plasmid construction.
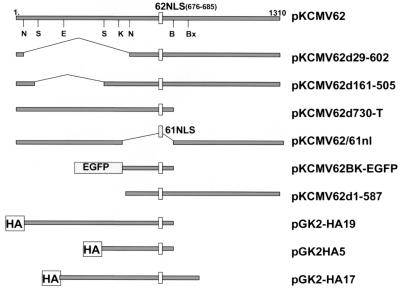
To express genes in transfected cells, complete or partial ORFs were cloned into the unique EcoRI and/or BamHI sites of plasmid pG310 (a gift from E. Mocarski, Stanford University) so as to be under the control of the complete human cytomegalovirus (CMV) IE promoter and polyadenylation signal. Some proteins were expressed in a derivative of pG310, named pGK2-HA, that contained a double-stranded oligonucleotide inserted into the EcoRI site (coding strand = 5′AATTGCCATGGGTTATCCATATGACGTCCCGGATTACGCAGTGGAATTC). This vector contained a new EcoRI site that was preceded by an initiating ATG (shown in bold) and 11 amino acids (Met-Gly-Tyr-Pro-Tyr-Asp-Val-Pro-Asp-Tyr-Ala-Val). The underlined sequence represents a linear epitope derived from the influenza virus HA that was recognized by commercially available monoclonal antibodies. Inserts were made in frame with this epitope using 8-, 10-, or 12-mer EcoRI linkers added to the ends of DNA fragments. Plasmid pKCMV62 is a derivative of the previously described pCMV62 (39, 52) but modified by deletion of the insert sequences between TthiiiI and NdeI sites (bp 104169 to 105214 with respect to the VZV sequence). This rendered a KpnI site and a BamHI site within the ORF62 coding sequence unique. Further deletions of pKCMV62 were generated by collapse of DNA sequences between rare-cutting restriction enzyme sites, followed by religation with single EcoRI linkers of either 8, 10, or 12 bases in length to maintain the reading frame. Plasmid pKCMV62d29-602 was derived by the in-frame deletion of sequences between two NotI sites present in pKCMV62 and does not express amino acids from 29 to 602. Plasmid pKCMV62d161-505 was derived by the in-frame deletion of sequences between two StuI sites and does not express amino acids 161 to 505. Plasmid pKCMV62d730-T was derived by deletion of coding sequences downstream of the unique BamHI site in ORF62 followed by insertion of DNA linkers that contained stop codons in all three reading frames (CTAGACTAGTCTAG; stop codons underlined). This plasmid expressed amino acids 1 to 734 of IE62. Plasmid pKCMV62/61nl contained a deletion of ORF62 coding sequences between the KpnI and BamHI sites (amino acids 570 to 733) followed by insertion of double-stranded complementary oligonucleotide pair (coding strand = 5′-CGCAAGGGGTGCTAAGCGCCGGTGG-3′) that encoded the ORF61 nuclear localization signal (amino acids Ala-Arg-Gly-Ala-Lys-Arg-Arg-Trp) in frame with the ORF62 coding sequence (60). Plasmid pKCMV62d1-587 expressed a portion of IE62 initiating at methionine 588, using a DNA fragment generated by PCR amplification. Primers contained additional 5′ sequences that added the appropriate EcoRI restriction site and upstream sequences from the IE62 first methionine to optimize the initiation of translation. Plasmid pGK2-HA19 contained an EcoR47III-BamHI fragment of ORF62 and expressed amino acids 7 to 734 as a fusion with the HA epitope. Plasmid pGK2-HA17 contained an EcoRV-BstXI fragment and expressed amino acids 416 to 823′ of ORF62; plasmid pGK2HA5 contained an StuI-BamHI fragment that resulted in expression of an IE62 peptide from amino acids 506 to 733 linked in frame to the HA epitope. Plasmid pKCMV62BK-EGFP was derived from pEGFP-C1 (Clontech Inc., Palo Alto, Calif.) and contained the BamHI-KpnI fragment from ORF62 (amino acids 571 to 733) expressed as a C-terminal fusion protein with the enhanced green fluorescent protein (EGFP).
Plasmid pKCMV61, which expressed complete ORF61 under the CMV IE promoter in the vector pG310, was derived by cloning a blunt-ended partial NcoI-AccI fragment of the complete ORF into pUC9 and using the flanking EcoRI and BamHI sites of that construct to insert the gene into pG310. Plasmids pKCMV47 and pKCMV66 were generated by PCR amplification of the complete ORFs from VZV strain Scott, using primers that contained additional 5′ sequences to add EcoRI and BamHI (for ORF47) or EcoRI and BglII (for ORF66) sites to facilitate cloning into the EcoRI and BamHI sites of pG310. The PCR-amplified DNAs were sequenced. pGK2-HA66 was similar to pKCMV66 except that the vector pGK2-HA was used. Plasmid P1-2 expressed HSV-1 ICP4 and was a kind gift of N. DeLuca. Plasmid pKCMV-SVTag contained the SV40 T antigen cDNA expressed in the vector pG310 and was derived by insertion of a BamHI fragment containing the entire SV40 T antigen cDNA, obtained from J. Pipas, University of Pittsburgh, Pittsburgh, Pa.
Mutations were introduced into ORF66 DNA sequences by using an established method (32) with a Mutagene kit (Bio-Rad Inc, Hercules, Calif.). For mutagenesis of ORF66, an EcoRI-to-BamHI fragment representing amino acids 1 to 336 from pKCMV66 was subcloned into the replicative forms of M13, and single-stranded DNA was prepared from a recombinant phage grown on Escherichia coli strain CJ236 dut ung. Mutagenesis was carried out according to the recommendations of the manufacturer with commercially purchased synthetic oligonucleotides. Following second-strand synthesis and transformation into E. coli DH5α F′, DNA from transforming M13 plaques was sequenced. Positive clones were used to prepare the double-stranded VZV DNA fragments containing the mutations, which were then purified and cloned to replace the wild-type sequences in the pKCMV66 or pGK2-HA66 plasmid background.
Transfections.
Transfections were carried out on MeWo cells using the Lipofectamine reagent (Gibco/Life Technologies, Inc, Gaithesburg, Md.). Briefly, trypsinized MeWo cells were seeded onto four-well chamber slides at 5 × 104/well, on 35-mm-diameter dishes seeded at 2 × 105/dish, or on 100-mm-diameter dishes at 4 × 106/dish 24 h prior to transfection. Plasmid DNA for transfection was purified by Qiagen columns (Qiagen Systems Inc., Santa Clarita, Calif.). Cells were exposed to DNA-Lipofectamine mixture at 37°C for 16 h, and then the medium was replaced with normal growth medium and further incubated for 24 h before subsequent analyses. In later experiments, Lipofectamine was used in conjunction with the Lipofectamine Plus reagent (Gibco/Life Technologies) to enhance transfection efficiency, and cells were exposed to DNA-lipid for 3 to 5 h. In all comparative and parallel transfections, the level of the CMV IE promoter was maintained constant by addition of pG310 vector.
Immunofluorescence.
Antibodies for immunofluorescence were first preabsorbed with uninfected cell sonicates at 106 cell equivalents per 100 μl of antibody. Immunofluorescence was carried out as described previously (30). Briefly, cells were fixed in acetone at −20°C for 15 min and rehydrated in phosphate-buffered saline (PBS) at room temperature. All subsequent incubations were performed in PBS containing 10% heat-inactivated goat serum at room temperature. Primary antibody incubations at a dilution of 1/100 to 1/1,000 were incubated with the cells for 1 h; following extensive washing, bound antibodies were detected with a secondary antibody of goat anti-rabbit immunoglobulin G (IgG) conjugated to either fluorescein or rhodamine or an anti-mouse IgG conjugated to rhodamine. Images were taken digitally using Bio-Rad Lasersharp software coupled with a Bio-Rad RadiancePLUS confocal microscope system.
Metabolic radiolabeling, immunoprecipitation, and immunodetection.
Procedures were modified from those described previously (49). Cells were metabolically labeled in growth media containing 1/20 the normal phosphate and [32P]orthophosphate at 500 μCi/ml. Following a 8- to 16-h labeling period, proteins were solubilized in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris HCl [pH 8.0], 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) containing a mixture of protease inhibitors (Mini-EDTA free; Boehringer Mannheim Biochemicals, Inc., Indianapolis, Ind.), 2 mM NaVO4, and 25 mM NaF. Specific proteins were precipitated using 2 to 10 μl of rabbit polyclonal antibodies to IE62 or 1 to 3 μl of HA-specific antibody in ascites fluid. Immunoprecipitated proteins were collected using a mixture of protein G-agarose and protein A-Sepharose, washed extensively in RIPA buffer, and subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE). For immunoblot analysis, procedures were similar to those described previously (28) except that bound antibodies were detected using secondary antibodies coupled to peroxidase followed by detection using SuperSignal West Dura luminescent substrate (Pierce Biochemicals, Rockford, Ill.).
Promoter-reporter constructs and reporter assay.
A construct containing the ORF4 promoter driving the expression of chloramphenicol acetyltransferase (CAT) was used to test the transactivating properties of IE62. The construct contained sequences from +36 to −190 with respect to the ORF4 transcription initiation site at +1 and was made using the vector pCATbasic (Promega Corp., Madison, Wis.). The DNA insert was prepared by PCR amplification using primers that added PstI and HindIII sites to the amplified fragment to facilitate cloning. In transfections, 1 μg of the promoter-CAT construct was transfected with 1 μg of pCMV-gal (obtained from Stratagene, Inc.), expressing β-galactosidase under the CMV IE promoter, 200 ng of pKCMV62, and a 1-μg mix of pG310 and either pKCMV66 or pKCMV47. At 24 h posttransfection, cells were washed in PBS and disrupted by repeated freeze-thaw cycles. Soluble cell extracts were assayed for CAT activity, using a 1-h incubation at 37°C with chloramphenicol and [14C]acetyl coenzyme A as the substrate, and production of [14C]acetylchloramphenicol was assayed following organic extraction with ethyl acetate and counting in a liquid scintillation counter. β-Galactosidase activity was determined using o-nitrophenylgalactopyranoside as substrate, followed by spectrophotometric assay.
RESULTS
Effect of the VZV protein kinases on IE62-mediated transactivation.
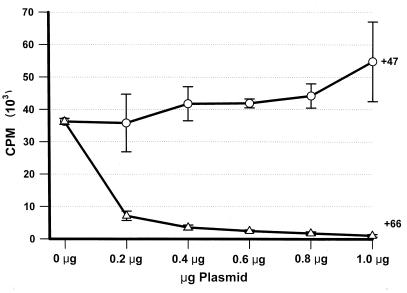
The specific phosphorylation of IE62 in vitro by the ORF47-associated protein kinase (49) raised the intriguing possibility that IE62 functions were modulated by the VZV-encoded protein kinases. To explore this possibility, we tested for the ability of the protein kinases to influence IE62-mediated transcriptional activation of an ORF4 promoter-CAT construct. All VZV ORFs were expressed from the same vector, using the constitutive human CMV IE promoter. Comparative transfections were carefully adjusted to ensure equal levels of CMV IE promoter, pCMVgal, pKCMV62, and promoter-reporter constructs. A representative experiment of the CAT activity expressed from this promoter, normalized to β-galactosidase activity, is shown in Fig. 1. The activity from transfections with pKCMV62 and without plasmids encoding protein kinases represents a 36-fold (±6.3 standard deviations) activation over the basal level of the promoter obtained from transfections without pKCMV62. From transfections containing increasing levels of pKCMV47, no significant alteration in the CAT activity was detected. However, a dramatic fall in CAT activity was observed when pKCMV66 was cotransfected with the activator and reporter constructs. Levels of CAT activity fell to 19% from transfections containing a 1:1 ratio of pKCMV66 to PKCMV62 and dropped to less than 10% from transfections containing a 2:1 ratio and higher. The effect of cotransfected pKCMV66 was specific for IE62-mediated activation and did not alter the level of constitutively expressed β-galactosidase activity expressed from the cotransfected pCMVgal. In the absence of IE62 transactivator, neither protein kinase affected the low level of basal CAT activity from the promoter (data not shown). These results indicated that the product of ORF66, and not that of ORF47, was capable of modulating functional activity of the VZV major regulatory protein. Analysis with IE62-responsive gI promoter reporter constructs yielded similar results (data not shown).
FIG. 1.
Levels of CAT activity from the ORF4 promoter in the presence of IE62 and in the presence or absence of the protein kinases from VZV ORF47 (+47) or VZV ORF66 (+66). Levels were derived from duplicate transfections on 35-mm-diameter well dishes. Transfections contained 1 μg of the ORF4 promoter CAT construct, 1 μg of pCMV-gal, 200 ng of pKCMV62, and 1 μg (total) of a mix of pG310 and either pKCMV66 or pKCMV47. The x axis indicates the amount of plasmid pKCMV47 or pKCMV66 used in the cotransfection. CAT activity at 0 μg represents a 36-fold increase over CAT activity obtained from transfections lacking pKCMV62.
Cellular localization of IE62 is influenced by the ORF66 protein.
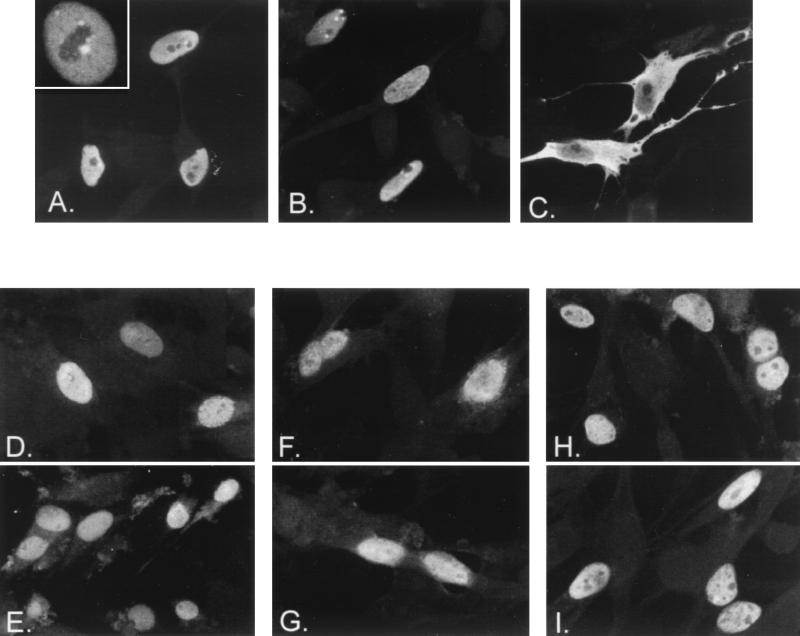
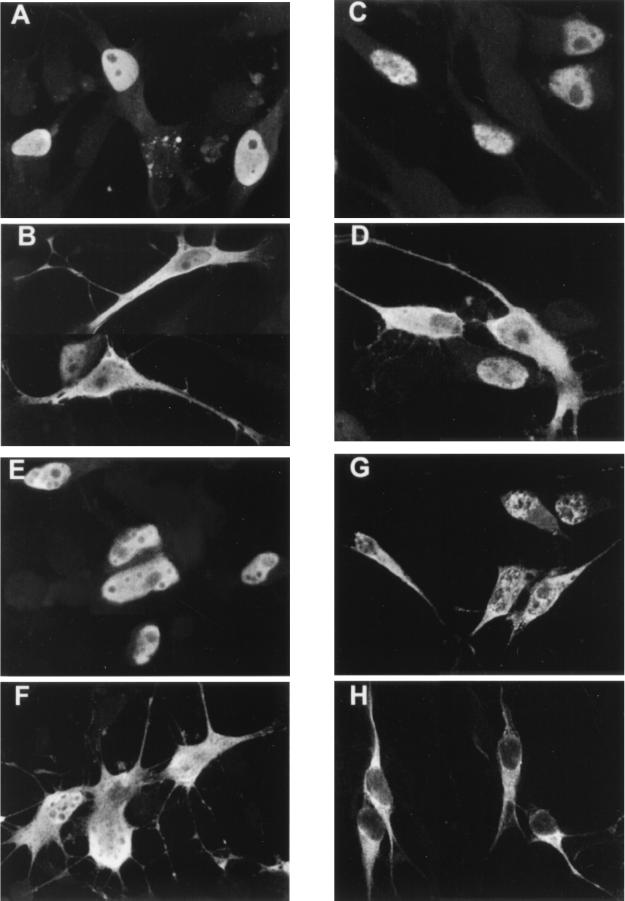
To determine the mechanism underlying the ability of the ORF66 protein to inhibit IE62-mediated transactivation, we examined the cellular location of IE62 in transfected cells following coexpression with or without the VZV protein kinases (Fig. 2A to C). IE62 protein expressed without protein kinases demonstrated an exclusively nuclear location in greater than 94% of the IE62-positive cells (Fig. 2A). It showed a diffuse staining pattern over the nucleus, with one to three subnuclear areas in which it was excluded and small, focal regions surrounding these areas where IE62 appeared more concentrated (Fig. 2A, inset). Approximately 2 to 6% of IE62-positive cells, depending on the transfection, demonstrated some additional cytoplasmic staining of IE62. Similar results were observed in transfected cells coexpressing IE62 with the ORF47 protein, indicating that the ORF47 protein kinase had little detectable influence on the cellular location of IE62 (Fig. 2B). In contrast, cells that coexpressed IE62 protein with the ORF66 protein kinase demonstrated a predominantly cytoplasmic distribution of IE62 in the majority of positive cells (Fig. 2C). Numerical estimations from three separate transfection experiments indicated that 71% of cells expressing IE62 demonstrated a predominantly cytoplasmic distribution of IE62 following a transfection that contained a 1:1 ratio of pKCMV66 to pKCMV62. Levels increased to 82% at a ratio of 3:1, respectively. These results explained the ability of ORF66 protein to inhibit IE62-mediated transactivation and implied that the ORF66 protein prevented the nuclear import of IE62.
FIG. 2.
Effect of the ORF66 protein kinase on nuclear localization of IE62 and other proteins. MeWo cells were transfected with pKCMV62, expressing IE62 (A through C), pKCMV61, expressing ORF61 (D and E), p1-2, expressing HSV-1 ICP4 (F and G), and pKCMV-SVTag, expressing SV40 T antigen (H and I). Panels show the cellular location of the protein detected by immunofluorescence using specific antibodies following cotransfection with empty vector (A, D, F, and H), threefold excess of pKCMV47 (B), or threefold excess of pKCMV66 (C, E, G, and I). The inset in panel A shows a higher magnification of a nucleus from a cell expressing IE62 protein alone. Images were captured with a 40× objective with additional magnifications of either 2× (D, F, and G) or 4× (A, inset).
We subsequently examined the nuclear localization of three other proteins in the presence of ORF66 protein to determine if ORF66 protein kinase was specific or more global for the nuclear import process (Fig. 2). VZV ORF61, a VZV transcriptional activator with homology to HSV-1 ICP0 and pseudorabies virus EPO (42, 48), was examined to determine if other VZV nucleus-localizing proteins were affected. ORF61 possesses a strong nuclear localization signal mapping to the carboxyl-terminal region of the protein (60). HSV-1 ICP4 was examined to determine if the site of action of ORF66 was conserved in the homologous protein of IE62 in a closely related alphaherpesvirus. The SV40 T antigen was examined because its nuclear import has been shown to be both positively and negatively regulated by cellular protein kinases (24). In particular, the nuclear localization of SV40 T antigen had potential relevance because threonine 680 within the heart of the IE62 nuclear localization signal is a consensus site for the cyclin-dependent kinase (CDK) p34cdc2. This kinase inhibits the nuclear localization of SV40 T antigen (23), and the possibility existed that ORF66 acted on IE62 by stimulating this cellular protein kinase. However, parallel cotransfection experiments demonstrated that all three control proteins localized exclusively to the nucleus when expressed alone or with the ORF66 protein kinase. This result strongly implied that the ORF66 protein targeted the nuclear localization process of IE62 specifically.
Distribution of ORF66 protein and IE62 in cotransfected cells.
In the cotransfection analyses just described, a minor but persistent population of IE62-positive cells continued to demonstrate exclusively nuclear forms of IE62, even following transfections with ratios of pKCMV66 to pKCMV62 as high as 5:1. Possible explanations were that this population represented a group of cells in which IE62 was immune to the effects of the ORF66 protein or, alternatively, that these cells did not express ORF66 protein. To resolve this issue, we derived a plasmid expressing ORF66 fused to an antigenic epitope at the amino terminus of the protein (plasmid pGK2-HA66). This allowed an indirect identification of the cellular location of the ORF66 protein by immunofluorescence. Previously described antibodies specific to the ORF66 protein that were provided to us (17, 59) as well as a rabbit ORF66-specific antibody made by our laboratory all failed to react efficiently with the ORF66 protein in immunofluorescence analyses. Figure 3 shows examples of cotransfected and dual-labeled cells. All cells that showed a predominantly cytoplasmic distribution of IE62 following cotransfections were found to express the HA-tagged ORF66 protein. The small fraction of IE62-positive cells retaining predominantly nuclear distribution were found not to express detectable levels of ORF66 protein. Surprisingly, the HA-tagged ORF66 protein was located mostly within the nucleus, although cytoplasmic forms were consistently observed in most cells. The cytoplasmic forms of ORF66 showed a similar distribution pattern to the cytoplasmic forms of IE62. The nuclear forms of ORF66 protein generally demonstrated two staining patterns, with either a diffuse nuclear distribution (Fig. 3B) or with concentrations of protein in three to eight subnuclear points (Fig. 3D). At this stage, we do not know the basis for the subnuclear concentration of the ORF66 protein, but the distribution patterns were found not to be dependent on the coexpression with IE62 (data not shown). These results confirmed that the ORF66 protein is directly responsible for the cytoplasmic accumulation of IE62 in transfected cells.
FIG. 3.
Cellular distribution of ORF66 protein in cotransfected cells. The images show two cells coexpressing the IE62 protein. The IE62 proteins was detected with polyclonal antibodies to IE62 and secondary anti-rabbit antibodies coupled to fluorescein (A and C), and the HA epitope-tagged ORF66 protein was detected with monoclonal antibodies to HA and with anti-mouse antibodies coupled to rhodamine (B and D). Images were collected sequentially using the lasers of the confocal microscope and a 60× objective.
IE62 distribution in VZV-infected cells.
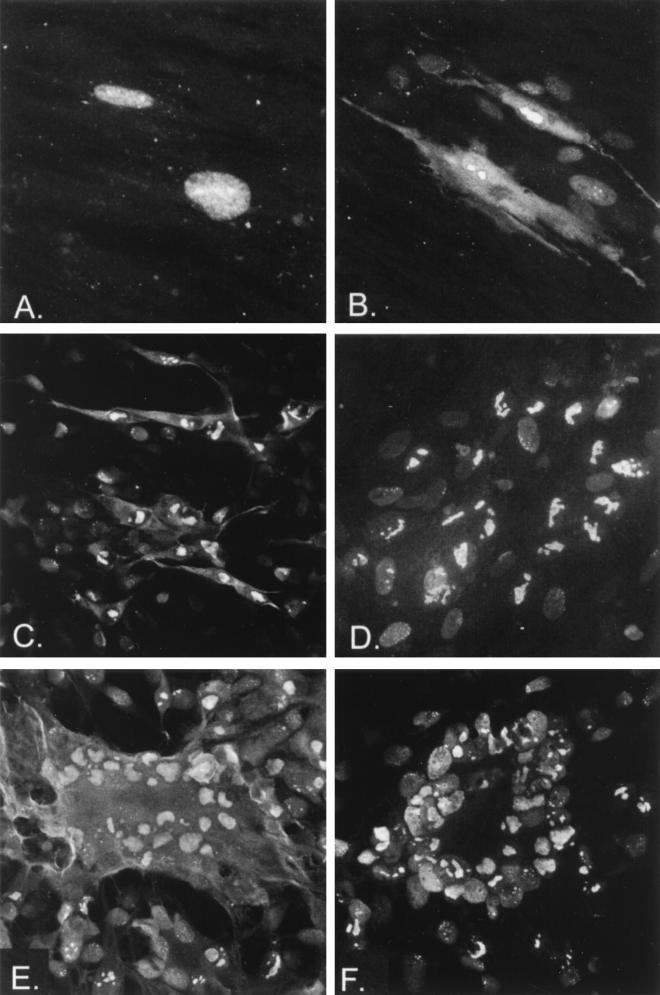
We subsequently aimed to determine if cytoplasmic forms of IE62 accumulated in VZV-infected cells. Initially, we examined a time course of infection; IE62 is encoded by an IE gene, whereas ORF66 is presumed to be an early gene (based upon transcription of the homologous HSV-1 US3 protein kinase). IE62 would be expected to show cytoplasmic forms at late times but not at very early times postinfection (p.i.). In a time course of cell-free virus-infected cells, IE62 appeared exclusively nuclear at 8 h p.i. (Fig. 4A), but cytoplasmic forms were detected at 24 h p.i. (Fig. 4B). At 24 h p.i., additional cells surrounding the primary infected cell showed a weak but predominantly nuclear staining, indicative of a second round of infection from progeny virus. These results indicated that cytoplasmic forms of IE62 accumulated during the later stages of infection.
FIG. 4.
IE62 in VZV-infected cells. Examples of the cellular distribution of IE62 are shown following infection with cell-free VZV strain Scott that was harvested at 8 (A) or 24 (B) h p.i. or in cells infected with VZV ROka47S (C), ROka (E), or ROka66S (D and F). (C and D) Cells harvested at 2 days p.i.; (E and F) cells harvested at 3 days p.i. Panels A and B were taken with a 60× objective with additional 2× magnification; panels C to F were recorded with 20× objective, with panels D, E, and F recorded with 2× magnification.
Figure 4 also shows IE62 in cells infected by ROka, ROka47S, and ROka66S, which are recombinant viruses generated from overlapping cosmids of the VZV genome. ROka47S contains a stop codon inserted into the ORF47 protein kinase gene and does not encode any protein from ORF47 (16). Likewise, ROka66S contains a stop codon engineered into the reading frame that prevents translation of any detectable protein from ORF66 (17). In ROka- and ROka47S-infected cells, IE62 demonstrated strong nuclear distribution, but it clearly showed additional cytoplasmic forms both in single infected cells (Fig. 4C) and in cells in which extensive fusion had occurred (Fig. 4E). In contrast, plaques and fused syncytia generated by infection with ROka66S demonstrated only nuclear distribution of the IE62 protein (Fig. 4D and F). Nuclear distribution was extensively marginated like that seen in ROka-infected cells. These results indicated that ORF66 mediated the cytoplasmic accumulation of IE62 in the context of VZV infection.
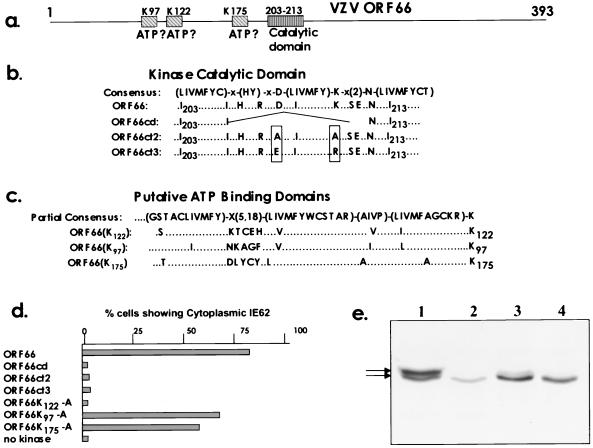
The ORF66 protein kinase domains are required for activity on IE62 nuclear localization.
Our next approach was designed to determine if the ORF66-mediated cytoplasmic accumulation of IE62 required the protein kinase activity predicted for the ORF66 protein. This prediction is based on the observation that most protein kinases are related and have conserved structural domains (15). Consensus motifs have been derived for both the catalytic and ATP binding domains (Fig. 5a). VZV ORF66 contains a match for the consensus catalytic domain of Ser/Thr protein kinases which is located from amino acids 203 to 213 (Fig. 5b). However, there is no full match in any herpesvirus protein kinase for the consensus for an ATP binding domain. As the ATP binding invariably occurs in a region amino terminal to the catalytic domain and relies on a critical lysine residue which binds ATP, we examined for key lysines with a local match to the consensus. For ORF66, lysine 122 has been predicted to be the critical ATP binding residue (36, 37). Of the seven lysines that lie N terminal to the catalytic domain, lysines 122, 175, and 97 all show close local matches to the consensus (Fig. 5c). For all three lysines, amino acids that lie further from this region diverge from the early part of the consensus sequence for an ATP binding domain (for clarity, the divergent portion of the consensus is not shown in Fig. 5c). Three ORF66 proteins were made that contained specific alterations within the predicted catalytic domain, including the deletion of seven amino acids within the consensus catalytic domain and the modification of the key aspartate and lysine residues that are predicted to be critical for catalytic activity (15). In addition, each of the putative lysines that matched the local ATP binding consensus were altered to alanines. All mutant proteins were then examined for the ability to induce the cytoplasmic accumulation of IE62 in the cotransfection assay (Fig. 5d). All three ORF66 proteins containing alterations in the predicted catalytic domain were unable to induce the cytoplasmic accumulation of IE62. Even constructs containing the very conservative alteration of aspartate 206 to glutamate and lysine 208 to arginine, which maintain the local net charge in the catalytic domain, did not express proteins capable of influencing IE62 cellular distribution. In contrast, unaltered ORF66 protein expressed from parallel transfections caused 79% of IE62-positive cells to show predominant cytoplasmic distribution of IE62. The results indicated that the amino acids in the predicted catalytic domain were critical for the ability of ORF66 to prevent IE62 nuclear localization. Regarding the mutational analysis of the predicted ATP binding domain, only alteration of lysine 122 completely abrogated influence on nuclear localization of IE62, and ORF66 proteins with mutation of either lysine 97 or lysine 175 to alanine retained the ability to cause cytoplasmic accumulation of IE62 in transfected cells. Taken together, the results strongly indicated that the protein kinase activity attributed to ORF66 was required for it to influence the cellular location of the IE62 protein.
FIG. 5.
The ORF66 protein kinase domains and effect of their alteration on IE62 nuclear accumulation. (a) Diagrammatic representation of ORF66 showing the approximate positions of the three putative ATP binding domains (indicated by K97, K122, and K175, representing the key lysine residues) and single catalytic domain (amino acids 203 to 213). (b) The full consensus for the catalytic domain of serine/threonine protein kinases is shown in single-letter amino acid code. Residues in parentheses indicate alternative residues at a specific position; X indicates any amino acid. Below the consensus is shown the single-letter amino acid sequence of the region from amino acids 203 to 213 of wild-type ORF66 that matches the consensus, with each ORF66 amino acid below its corresponding position in the consensus. Also shown are the altered amino acid sequences of three mutated ORF66 constructs created as described in the text (ORF66cd, ORF66ct2, and ORF66ct3). (c) Part of the consensus for ATP binding domain that was used to identify the putative ATP binding domains is shown, along with the local amino acid residues preceding lysines 122, 97, and 175 in wild-type ORF66. The last lysine residue (K) in each consensus is the potential active site that binds ATP. Three mutant ORF66 proteins were created in the vector pKCMV66 in which the potential active lysine residue was altered to alanine. (d) Effects of mutant ORF66 proteins on the cellular distribution of IE62, shown as percentages of IE62-positive cells demonstrating cytoplasmic distribution following cotransfection experiments with plasmids expressing wild-type or altered protein kinases. Numbers were determined from assessment of 300 IE62-positive cells from each of duplicate transfections, using digital images of fields collected at low-magnification power on the confocal microscope. (e) Immunoblot analysis of SDS-PAGE-separated proteins expressed in transfected MeWo cells from pGK2-HA-based constructs expressing the epitope-tagged forms of the wild-type ORF66 protein (lane 1), the ORF66ct2 protein (lane 2), the ORF66ct3 protein (lane 3), or the ORF66K122-A protein (lane 4). The proteins expressed were detected with commercial antibodies to the HA antigenic epitope. The lower levels of ORF66ct2 protein in this blot were the consequence of an inefficient transfection of plasmid used in this experiment. The proteins arrowed are 46 and 47 kDa in size.
The proteins expressed from selected mutant ORF66 genes that failed to mediate the cytoplasmic accumulation of IE62 were analyzed. Each ORF66 gene was expressed in the vector pGK2-HA so that the protein would be tagged with the HA epitope. When expressed in transfected cells, all proteins showed the same cellular distribution as seen for the wild-type HA-tagged protein (data not shown). SDS-PAGE and immunoblot analyses with the HA-specific antibodies indicated that the wild-type ORF66 gene expressed two close-mobility forms in transfected cells of 46 and 47kDa (Fig. 5e). In contrast, the proteins expressed from the mutant genes of ORF66ct2, ORF66ct3, and ORF66122K-A, all of which were unable to cause cytoplasmic accumulation of IE62, expressed only the faster-migrating 46-kDa form (the lower level of protein expressed by ORF66ct2 in Fig. 5e in this experiment was a result of inefficient transfection; the 47-kDa form was not detected following longer exposures). We strongly suspect that the slower-migrating 47-kDa form represented an autophosphorylated form of ORF66 and that its absence in cells expressing the mutant ORF66 proteins indicated that they were unable to autophosphorylate. This possibility has not yet been confirmed because we have been unable to obtain the ORF66 protein in a soluble form to enable it to be immunoprecipitated from transfected and VZV-infected cells.
Partial mapping of the region of IE62 that is targeted by the ORF66 protein.
The mutational analysis of the ORF66 protein kinase domains implied that ORF66 phosphorylates the IE62 protein. Previous studies based on the phosphorylation of IE62 from ROka- and ROka66S-infected cells did not indicate the specific phosphorylation of IE62 by the ORF66 protein kinase (17), and IE62 was found to be extensively phosphorylated in both ROka- and ROka66S-infected cells. We suspected that the phosphorylation of IE62 by ORF66 protein kinase was probably a minor phosphorylation event which was masked by the extensive phosphorylation of IE62 mediated by other protein kinases. Indeed, casein kinase II phosphorylates IE62 in vitro (49), and IE62 is phosphorylated when expressed in vaccinia virus-infected cells (29). Strong consensus motifs are present on IE62 which are putative targets for several cellular protein kinases (unpublished data). Therefore, we derived several IE62 constructs to map the region targeted by ORF66 protein kinase and demonstrate its specific phosphorylation of IE62. We aimed to use IE62 peptides that lacked most or all cellular kinase phosphorylation sites (Fig. 6), and this required partial mapping of the IE62 region targeted by ORF66 protein.
FIG. 6.
Diagrammatic representation of peptides and portions of IE62 that were expressed in transfected cells to identify the domain targeted by ORF66 protein kinase. Each grey line represents the portion of IE62 expressed in the corresponding plasmid designated at the right. At the top, the complete 1,310-amino-acid IE62 protein is represented with the approximate positions of restriction enzymes sites used to derive specific plasmids as described in the text (N, NotI; S, StuI; E, EcoRV; K, KpnI; B, BamHI; Bx, BstXI). 62NLS, the IE62 nuclear localization signal and its key amino acids; 61NLS, the nuclear localization signal derived from ORF61; HA, the 11-amino-acid epitope tag engineered to the amino-terminal portion of IE62 peptides; EGFP, coding sequence for EGFP.
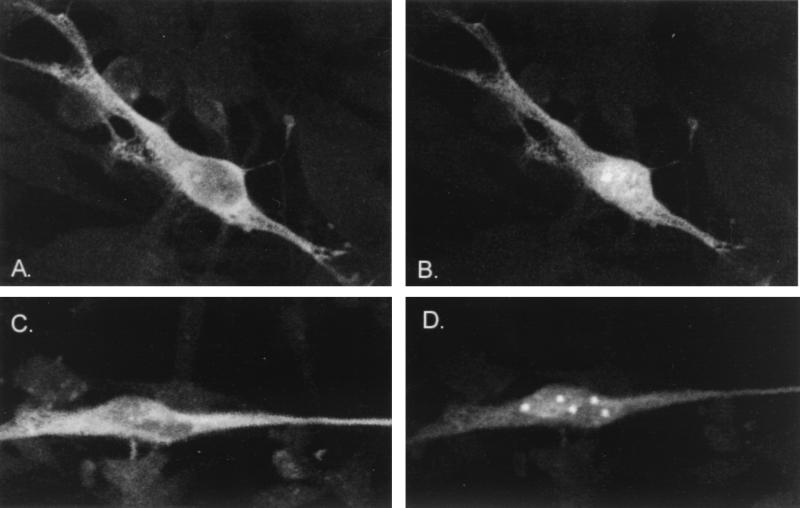
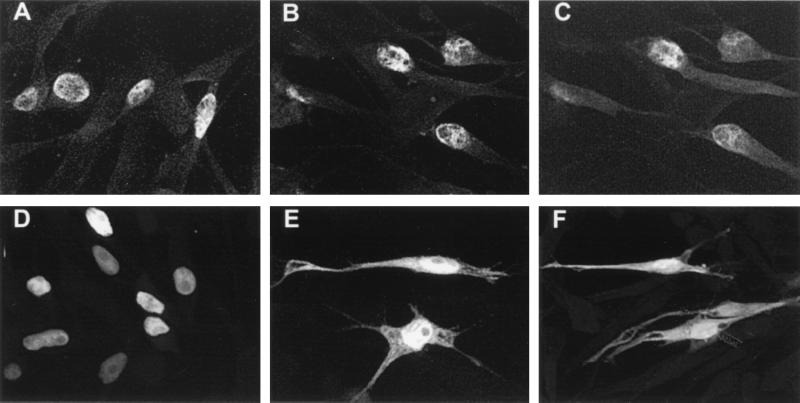
Four constructs expressing IE62 peptides with intact nuclear localization signals were expressed in transfected cells with and without the ORF66 protein kinase (Fig. 7). All contained the IE62 nuclear localization signal, which includes the arginine/lysine-rich region between amino acids 676 and 685 (31). Peptides from plasmid pKCMV62d730-T (Fig. 7A) and from pKCMV62d161-505 (Fig. 7C) both retained a strong nuclear distribution pattern similar to that observed for complete IE62 in the absence of ORF66 protein. Both peptides demonstrated predominantly cytoplasmic distribution following coexpression with ORF66 protein (in both Fig. 7B and D, a single cell containing a nuclear localizing IE62 peptide can be seen). IE62 peptides expressed from pKCMV62d29-602 in the absence of ORF66 protein also demonstrated predominantly nuclear staining, although the nuclear pattern was unusual and showed several subnuclear structures where IE62 peptide was excluded (Fig. 7E). Furthermore, while this peptide accumulated in the cytoplasm following expression with ORF66, the peptide appeared to be excluded less efficiently from the nucleus (Fig. 7F). We considered it likely that the peptide was less efficiently recognized by the ORF66 protein kinase. The fourth peptide studied, expressed from pGK2-HA5, demonstrated an inefficient nuclear localization (Fig. 7G) and showed both speckled nuclear and cytoplasmic staining in the absence of ORF66 protein. However, its coexpression with ORF66 protein resulted in an almost complete exclusion of the peptide from the nucleus (Fig. 7H). These results indicated that the nuclear localization of all four peptides retained the ability to be suppressed by the ORF66 protein. The amino acids common to all peptides included amino acids between 602 and 730, and we concluded that this domain contained the ORF66-responsive element. Further delineation of the remaining region through additional deletion analyses could not be obtained because smaller IE62 peptides demonstrated inefficient nuclear localization.
FIG. 7.
Immunofluorescent images showing nuclear localization of IE62 peptides and modulation by ORF66 protein kinase. The IE62 peptide shown in panels A and B is expressed from pKCMV62d730-T, panels C and D represent protein expressed from pKCMV62d161-505, panels E and F are from pKCMV62d29-602, and panels G and H represent the IE62 HA-tagged peptide from 506-734, expressed from plasmid pGK2HA5. Panels A, C, E, and G represent the protein expressed alone, whereas panels B, D, F, and H represent the protein expressed following transfection with a threefold excess of pKCMV66.
To confirm that the region targeted by ORF66 lay within the boundary defined by amino acids 602 to 730, two constructs expressing chimeric proteins were used. Plasmid pKCMV62/61NL expresses an IE62 protein lacking amino acids 571 to 733 but containing seven amino acids derived from the ORF66-nonresponsive nuclear localization signal of ORF61. As expected, this protein was found to be exclusively nuclear in greater than 95% of transfected cells both in the absence and in the presence of coexpressed ORF66 protein (Fig. 8A to C). Transfections were carried out using the epitope-tagged form of ORF66 to ensure that cells coexpressed both proteins (Fig. 8C). This confirmed that the predominant portion of IE62 (excluding amino acids 571 to 733) was not involved in recognition by ORF66 protein. A second chimeric protein that contained amino acids 571 to 733 of IE62 as a carboxyl-terminal fusion with EGFP was found to demonstrate exclusively nuclear fluorescence in the absence of ORF66 protein (Fig. 8D) but showed cytoplasmic distribution in addition to nuclear distribution following coexpression with ORF66 (Fig. 8E). The continued nuclear staining of the EGFP chimera in the presence of ORF66 protein was likely due to the presence of a weak nuclear localization signal in EGFP, as EGFP expressed alone from the vector showed both nuclear and cytoplasmic distribution (Fig. 8F). Taken together with the deletion mapping analyses, these studies indicated that the region of IE62 targeted by the ORF66 protein was close to the nuclear localization signal and within amino acids 602 to 733.
FIG. 8.
Immunofluorescent images showing the nuclear localization of chimeric proteins and modulation by ORF66 protein. Protein was expressed from pKCMV62/61NL (A and B) or pKCMV62-EGFP (D and E) in the absence (A and D) or presence (B and E) of a threefold excess of HA epitope-tagged ORF66 protein. (C) HA-tagged ORF66 protein in the cells shown in panel B; (F) cellular location of the EGFP expressed from the commercial vector pEGFP-C1.
The ORF66 protein phosphorylates IE62 in vivo.
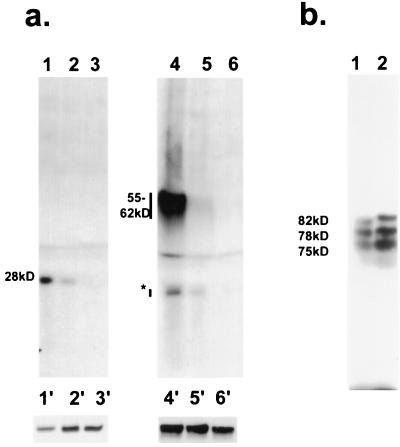
The targeting of the IE62 protein by the ORF47 protein kinase was demonstrated using in vitro phosphorylation studies with coimmunoprecipitated proteins. The in vitro demonstration of the phosphorylation of the IE62 protein by the ORF66 protein kinase has not been similarly achieved because we have not been able to solubilize the ORF66 protein from VZV-infected or from transfected cells. ORF66 was found not to be solubilized by treatment with RIPA, 10 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, pH levels from 6.5 to 8.5, high salt, and various combinations thereof (data not shown). We therefore used an alternative approach, in which we examined for specific in vivo phosphorylation of IE62 peptides in cotransfected cells. Cells were transfected with plasmids expressing IE62 peptides and with threefold excess of plasmids expressing active or mutated ORF66 proteins. Following incubation in medium containing [32P]orthophosphate, the IE62 peptides were immunoprecipitated using antibodies to IE62 (Fig. 9). The IE62 peptides expressed from pGK2-HA17 and pGK2-HA5, representing IE62 amino acids from 412 to 823 and from 506 to 734, respectively, were found to be radiolabeled following coexpression with ORF66 but were not radiolabeled following coexpression with the ORF66 protein mutated in the catalytic domain and expressed from plasmid pKCMV66ct2 (Fig. 9a). No radiolabeled proteins were precipitated when the IE62 peptides were expressed following transfections without any protein kinases (not shown). In the presence of the HA epitope-tagged form of ORF66, both IE62 peptides were also radiolabeled but appeared to be phosphorylated less efficiently (lanes 2 and 5). Analysis of whole-cell extracts from parallel and identical transfection indicated efficient expression of the IE62 peptides (lanes 1′ to 6′). These results strongly suggested that the ORF66 protein mediated phosphorylation of IE62 within the domain defined by the deletion mapping studies. In a second approach, a construct expressing IE62 sequences initiating at an internal methionine (representing IE62 from amino acids 588 to 1310) expressed three weakly phosphorylated peptides in the absence of ORF66 protein (Fig. 9b, lane 1). When this construct was cotransfected with the ORF66 protein, the three peptide species appeared to be more extensively phosphorylated, and the 78- and 82-kDa forms demonstrated a mobility shift to slower-migrating forms on SDS-PAGE (Fig. 9b, lane 2). While we do not know the origins of the multiple species, the data supported the phosphorylation of IE62 in cells coexpressing the ORF66 protein kinase. The data also suggested that a cellular protein kinase targeted IE62 in the C-terminal domain downstream of residue 823. A likely candidate cellular kinase is casein kinase II, because a very strong potential target for this kinase lies in IE62 between residues 1290 and 1295.
FIG. 9.
Phosphorylation of IE62 peptides in transfected cells by the ORF66 protein kinase. (a) Phosphorylated peptides were immunoprecipitated from cells transfected with plasmid pGK2-HA5 (lanes 1 to 3) or from pGK2-HA17 (lanes 4 to 6) following cotransfection with pKCMV66 (lanes 1 and 4), pGK2-HA66 (lanes 2 and 5), or pKCMV66ct2 (lanes 3 and 6). Proteins were immunoprecipitated with a mixture of polyclonal and monoclonal antibodies to the IE62 protein. The approximate sizes of the main phosphorylated species in kilodaltons are indicated at the left. The asterisk indicates a suspected breakdown product of the 55- to 62-kDa product. The 55- to 62-kDa species detected in lanes 4 to 6 migrate as a smear due to the close proximity of the heavy chain of IgG in the immunoprecipitates. Lanes 1′ to 6′ represent the main species of proteins detected by immunoblotting with anti-IE62 antibodies in whole-cell extracts from a parallel transfection carried out without the presence of radiolabel. (b) Phosphorylated peptides expressed from cells transfected with pKCMV62d1-587, either with (lane 2) or without (lane 1) pKCMV66 at an equimolar amount. The approximate sizes of the species detected in cells transfected without the ORF66 protein kinase are shown in kilodaltons.
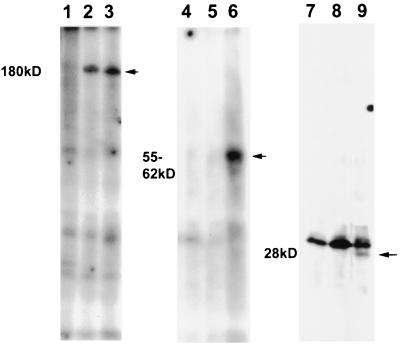
Further confirmation of the specific targeting of IE62 by the ORF66 protein was demonstrated by transfecting cells and subsequently superinfecting them with either ROka or ROka66S. Cells were transfected and then superinfected 24 h later with the recombinant VZV at a ratio of 1 infected cell to 10 transfected cells and immediately incubated in medium containing [32P]orthophosphate. At 48 h p.i., radiolabeled IE62 peptides specifically expressed from the transfected plasmids were immunoprecipitated using antibodies to the HA epitope tag. Immunoprecipitation of the complete IE62 protein using polyclonal antibodies to IE62 confirmed that IE62 protein was phosphorylated in both ROka- and ROka66S-infected cells (Fig. 10, lanes 1 to 3). When the specific peptides expressed from the transfected plasmids were immunoprecipitated, only those peptides obtained from cells that were superinfected with ROka were phosphorylated (lanes 6 and 9). These results correlated with those from the cotransfection assay and indicated that the ORF66 protein mediated a specific phosphorylation event of IE62.
FIG. 10.
Phosphorylated IE62 peptides immunoprecipitated from transfected and uninfected cells (lanes 1, 4, and 7) or from transfected cells superinfected with ROka66S (lanes 2, 5, and 8) or ROka (lanes 3, 6, and 9). Cells were initially transfected with plasmid pGK2-HA17 (lanes 3 to 6) or pGK2-HA5 (lanes 1 to 3 and 7 to 9). Superinfections were carried out using a ratio of 1 infected cell to 10 uninfected cells and were incubated for 48 h. Proteins were immunoprecipitated with either polyclonal antibodies to IE62 (lanes 1 to 3) or monoclonal antibodies to the HA epitope (lanes 4 to 9). The approximate size of the predominant radiolabeled species precipitated is indicated at the left. A nonspecific 30-kDa phosphoprotein was immunoprecipitated in all extracts transfected with pGK2HA5 (lanes 7 to 9).
DISCUSSION
The data presented here show that the ORF66 protein kinase prevents the nuclear accumulation of the major VZV regulatory protein, both in the context of transfected cells and in the context of VZV infection. This ability strongly correlates with an ORF66 protein kinase-dependent specific phosphorylation of IE62. We hypothesize that one of the functions of ORF66 may be to redirect IE62 for roles in the cytoplasm at the later stages of infection or, alternatively, to negatively regulate IE62-mediated transactivation of gene transcription at later stages of the infectious cycle. The recent demonstration that recombinant VZV lacking ORF66 is partly attenuated for growth in T cells in in vivo animal models (40) suggests that this interaction may contribute to the pathogenesis of VZV in human disease.
We have previously shown that the nuclear import of IE62, which is required for transactivation of viral transcription, relies on a lysine/arginine-rich amino acid sequence within amino acids 676 to 685 (31). This region is typical of the nuclear localization signals of other nuclear localizing proteins, and therefore IE62 most likely utilizes the cellular nuclear import pathway. The multicomponent nuclear import process initiates with the binding of a heterodimeric complex of two karyopherins (α and β) to the arginine-lysine rich region of the protein to be transported. The complex then docks to the nuclear pore complex (11, 12, 46, 51). Transportation across the nuclear pore occurs in an energy-dependent fashion after the association of a protein complex containing the GTPase protein Ran, the stimulating factor p10, and the hydrolysis of GTP to GDP (41). The unusual feature of IE62 from our studies was that IE62 nuclear accumulation was prevented by the ORF66 protein kinase, most likely as a result of phosphorylation. Nuclear localization regulated by phosphorylation has not been described for any herpesvirus protein previously, and the ability of a virus-encoded protein kinase to do this appears, so far, unique. However, similar mechanisms are used to control the function of a small group of cellular nucleus-localizing proteins involved in a variety of processes, including cell cycle regulation and the control of transcriptional activators (24, 45, 50). Nuclear localization can be both positively and negatively regulated by phosphorylation. For example, nuclear import of SV40 large T antigen, whose nuclear localization is governed by the sequence S111S112DDEATADS120QHST124PPKKKRKV, is positively regulated by casein kinase II-mediated phosphorylation of serines 111 and 112 (25, 68) or of serine 120 by the double-stranded DNA-dependent protein kinase (68). In contrast, phosphorylation of threonine 124 by the cell CDK p34cdc2 inhibits nuclear import (23). Our data indicate that IE62 belongs to this group of proteins with regulated nuclear localization signals.
The mechanism by which the ORF66 protein causes the cytoplasmic accumulation of IE62 seems highly likely to involve the phosphorylation of IE62 rather than the ORF66 protein acting through kinase-independent protein-protein interaction. IE62 was additionally phosphorylated in cells coexpressing the ORF66 protein, and the kinase domains of ORF66 were required for the influence on IE62 nuclear accumulation. While Heineman et al. (17) concluded that VZV ORF66 encoded a protein kinase activity based on differences in phosphorylation patterns in cells infected with VZV ROka and ROka66S, they did not detect differential phosphorylation of IE62 in VZV-infected and ROka66S-infected cells. Our data indicate that the ORF66-dependent phosphorylation of IE62 was probably masked in their studies by its extensive phosphorylation by cellular protein kinases. IE62 is phosphorylated in both ROka- and ROka66S-infected cells (Fig. 10) and is phosphorylated in vitro by casein kinase II. Efforts are under way in our laboratory to identify the IE62 serine or threonine residues within 602 to 730 that are targeted by ORF66. Interestingly, no serine or threonine within the IE62 sequence exactly matches the pseudorabies virus and HSV-1 US3 protein kinase consensus site RRR(R/X)(S/T)(R/Y) (35, 54). We have also not yet determined if the phosphorylation of IE62 is directly by the ORF66 protein kinase or whether there is a cellular intermediate protein kinase which is somehow activated by ORF66 that then phosphorylates IE62. Our data indicated that ORF66 did not act through enhancement of p34cdc2-mediated phosphorylation of threonine 680 of IE62, a potential site for this kinase. The question of the direct targeting of IE62 by the ORF66 protein kinase will require purification of the ORF66 protein and in vitro kinase assays with purified substrates. This may prove difficult, as our studies have indicated that the ORF66 protein is tightly associated with cell structures that make it very insoluble and difficult to obtain.
The precise mechanism by which phosphorylation causes the inefficient nuclear accumulation of IE62 has not yet been determined, but there are several possible models. The simplest model is that phosphorylation near the nuclear localization signal disrupts the charge environment and prevents binding of the karyopherins to the arginine/lysine-rich region. A second mechanism involves the phosphorylation-induced alteration of protein conformation so that the nuclear localization signal becomes masked through tertiary and/or conformational structure. For the yeast transcription SW15, phosphorylation outside the Arg/Lys-rich region of the nuclear localization signal by CDK/CDC28 results in conformational masking of the nuclear import signal (26). A similar mechanism is thought to occur for the suppression of nuclear localization of lamin B2 by protein kinase C (18). A third mechanism is demonstrated by the negative influence of the CDK p34cdc2 on SV40 T antigen, which is thought to be through the increased affinity of T antigen for a cytoplasmic retention factor. A fourth mechanism is that the cytoplasmic accumulation of IE62 is a result of the ORF66-induced activation of a nuclear export pathway for IE62. This possibility is notable because the protein kinase appeared to be distributed predominantly within the nucleus. However, experimental approaches within our laboratory have not yet indicated the movement of nuclear IE62 to the cytoplasm in the presence of ORF66 protein, in either transfected or infected cells. Therefore, we consider the most likely scenario to be that the nuclear import pathway is inhibited. Studies are in progress to examine these possibilities further.
This work establishes that both VZV protein kinases phosphorylate the major regulatory protein of VZV. This is highly novel in the alphaherpesvirus subgroup, and similar observations for the corresponding proteins of other alphaherpesviruses have not been described. HSV-1 ICP4 has forms which associate with the cytoplasm and cellular plasma membrane, but they do so in the absence of any other viral proteins (67, 69). Specific forms of ICP4 have been found in HSV-infected cells which are associated with different cellular compartments, but the identity of the protein kinases involved remains unknown (63). We have tested for the possible interaction of ICP4 and HSV US3 protein kinase and have not found evidence of similar interactions affecting nuclear localization. Published evidence rather suggests that the US3 and ORF66 protein kinases target different proteins for each alphaherpesvirus. HSV-1 US3 protein kinase affects the phosphorylation of the product of UL34, an essential viral membrane protein (55, 56). It may also target cellular proteins, as the US3 protein kinase is one factor which enables HSV-1 to overcome apoptosis (38). In contrast, the HSV-2 US3 protein kinase influences the phosphorylation of a 14- to 22-kDa product of the UL9 tegument protein, as well as the alkaline exonuclease product of UL12 (5, 6). The pseudorabies virus US3 protein kinase phosphorylates a major virion phosphoprotein of 112 kDa (70). It is, perhaps, surprising that no common target has yet been identified for the kinases in the US3 family, particularly in light of the conservation in the alphaherpesviruses. It seems likely that the various targets so far identified may reflect multiple targets which have evolved for each alphaherpesvirus kinase, and others likely remain to be identified. Our data clearly delineate an unusual functional role for the VZV member of this kinase family. We suggest that VZV has evolved this mechanism to either negatively regulate the transcriptional regulatory activities of IE62 or redirect IE62 for functional roles within the cytoplasm after the onset of early protein synthesis.
VZV ORF66 is not required for virus growth in culture (17), suggesting that the activity on IE62 nuclear localization is likely not needed for growth in culture. However, evidence has recently suggested that VZV ORF66 plays an important role in vivo, since ROka66S is attenuated for growth in T cells in the SCID-Hu mouse model (40). Thus, it seems likely that functions are probably more critical for virus growth in certain cell types. In this respect, ORF66 is similar to US3 kinases in other alphaherpesviruses, as attenuated phenotypes are also observed for HSV-2 and pseudorabies virus US3-negative viruses (27, 33, 47). It is possible that the cytoplasmic accumulation of IE62 may have an important functional significance in vivo or in a specific cell type infected in the course of a human infection.
ACKNOWLEDGMENTS
We thank Jeffrey Cohen for supplying ROka, ROka47S, and ROka66S and for reading the manuscript, and we thank David Jans for helpful discussions on nuclear localization.
This work was supported by Public Health Service grant EY 09397, CORE grant for Vision Research EY08098, The Eye & Ear Foundation, and awards from the Pittsburgh Supercomputing Center and Research to Prevent Blindness, Inc.
REFERENCES
- 1.Arvin A M. Varicella-zoster virus: overview and clinical manifestations. Semin Dermatol. 1996;15:4–7. [PubMed] [Google Scholar]
- 2.Baudoux L, Defechereux P, Schoonbroodt S, Merville M P, Rentier B, Piette J. Mutational analysis of varicella-zoster virus major immediate-early protein IE62. Nucleic Acids Res. 1995;23:1341–1349. doi: 10.1093/nar/23.8.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen I J, Straus S E. Varicella zoster virus and its replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Roizman B, Straus S E, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2525–2545. [Google Scholar]
- 4.Cohen J I, Heffel D, Seidel K. The transcriptional activation domain of varicella-zoster virus open reading frame 62 protein is not conserved with its herpes simplex virus homolog. J Virol. 1993;67:4246–4251. doi: 10.1128/jvi.67.7.4246-4251.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daikoku T, Kurachi R, Tsurumi T, Nishiyama Y. Identification of a target protein of US3 protein kinase of herpes simplex virus type 2. J Gen Virol. 1994;75:2065–2068. doi: 10.1099/0022-1317-75-8-2065. [DOI] [PubMed] [Google Scholar]
- 6.Daikoku T, Yamashita Y, Tsurumi T, Nishiyama Y. The US3 protein kinase of herpes simplex virus type 2 is associated with phosphorylation of the UL12 alkaline nuclease in vitro. Arch Virol. 1995;140:1637–1644. doi: 10.1007/BF01322537. [DOI] [PubMed] [Google Scholar]
- 7.Davison A J. Varicella-zoster virus. The Fourteenth Fleming lecture. J Gen Virol. 1991;72:475–486. doi: 10.1099/0022-1317-72-3-475. [DOI] [PubMed] [Google Scholar]
- 8.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 9.Defechereux P, Melen L, Baudoux L, Merville-Louis M P, Rentier B, Piette J. Characterization of the regulatory functions of varicella-zoster virus open reading frame 4 gene product. J Virol. 1993;67:4379–4385. doi: 10.1128/jvi.67.7.4379-4385.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Disney G H, Everett R D. A herpes simplex virus type 1 recombinant with both copies of the Vmw175 coding sequences replaced by the homologous varicella-zoster virus open reading frame. J Gen Virol. 1990;71:2681–2689. doi: 10.1099/0022-1317-71-11-2681. [DOI] [PubMed] [Google Scholar]
- 11.Enenkel C, Blobel G, Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- 12.Enenkel C, Schulke N, Blobel G. Expression in yeast of binding regions of karyopherins alpha and beta inhibits nuclear import and cell growth. Proc Natl Acad Sci USA. 1996;93:12986–12991. doi: 10.1073/pnas.93.23.12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felser J M, Kinchington P R, Inchauspe G, Straus S E, Ostrove J M. Cell lines containing varicella-zoster virus open reading frame 62 and expressing the “IE” 175 protein complement ICP4 mutants of herpes simplex virus type 1. J Virol. 1988;62:2076–2082. doi: 10.1128/jvi.62.6.2076-2082.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forghani B, Mahalingam R, Vafai A, Hurst J W, Dupuis K W. Monoclonal antibody to immediate early protein encoded by varicella-zoster virus gene 62. Virus Res. 1990;16:195–210. doi: 10.1016/0168-1702(90)90023-5. [DOI] [PubMed] [Google Scholar]
- 15.Hanks S K, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 16.Heineman T C, Cohen J I. The varicella-zoster virus (VZV) open reading frame 47 (ORF47) protein kinase is dispensable for viral replication and is not required for phosphorylation of ORF63 protein, the VZV homolog of herpes simplex virus ICP22. J Virol. 1995;69:7367–7370. doi: 10.1128/jvi.69.11.7367-7370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heineman T C, Seidel K, Cohen J I. The varicella-zoster virus ORF66 protein induces kinase activity and is dispensable for viral replication. J Virol. 1996;70:7312–7317. doi: 10.1128/jvi.70.10.7312-7317.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hennekes H, Peter M, Weber K, Nigg E A. Phosphorylation on protein kinase C sites inhibits nuclear import of lamin B2. J Cell Biol. 1993;120:1293–1304. doi: 10.1083/jcb.120.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc Natl Acad Sci USA. 1975;72:1276–1280. doi: 10.1073/pnas.72.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inchauspe G, Nagpal S, Ostrove J M. Mapping of two varicella-zoster virus-encoded genes that activate the expression of viral early and late genes. Virology. 1989;173:700–709. doi: 10.1016/0042-6822(89)90583-7. [DOI] [PubMed] [Google Scholar]
- 22.Jackers P, Defechereux P, Baudoux L, Lambert C, Massaer M, Merville-Louis M P, Rentier B, Piette J. Characterization of regulatory functions of the varicella-zoster virus gene 63-encoded protein. J Virol. 1992;66:3899–3903. doi: 10.1128/jvi.66.6.3899-3903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jans D A, Ackermann M J, Bischoff J R, Beach D H, Peters R. p34cdc2-mediated phosphorylation at T124 inhibits nuclear import of SV-40 T antigen proteins. J Cell Biol. 1991;115:1203–1212. doi: 10.1083/jcb.115.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jans D A, Hubner S. Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol Rev. 1996;76:651–685. doi: 10.1152/physrev.1996.76.3.651. [DOI] [PubMed] [Google Scholar]
- 25.Jans D A, Jans P. Negative charge at the casein kinase II site flanking the nuclear localization signal of the SV40 large T-antigen is mechanistically important for enhanced nuclear import. Oncogene. 1994;9:2961–2968. [PubMed] [Google Scholar]
- 26.Jans D A, Moll T, Nasmyth K, Jans P. Cyclin-dependent kinase site-regulated signal-dependent nuclear localization of the SW15 yeast transcription factor in mammalian cells. J Biol Chem. 1995;270:17064–17067. doi: 10.1074/jbc.270.29.17064. [DOI] [PubMed] [Google Scholar]
- 27.Kimman T G, de Wind N, De Bruin T, de Visser Y, Voermans J. Inactivation of glycoprotein gE and thymidine kinase or the US3-encoded protein kinase synergistically decreases in vivo replication of pseudorabies virus and the induction of protective immunity. Virology. 1994;205:511–518. doi: 10.1006/viro.1994.1672. [DOI] [PubMed] [Google Scholar]
- 28.Kinchington P R, Bookey D, Turse S E. The transcriptional regulatory proteins encoded by varicella-zoster virus open reading frames (ORFs) 4 and 63, but not ORF 61, are associated with purified virus particles. J Virol. 1995;69:4274–4282. doi: 10.1128/jvi.69.7.4274-4282.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinchington P R, Hougland J K, Arvin A M, Ruyechan W T, Hay J. The varicella-zoster virus immediate-early protein IE62 is a major component of virus particles. J Virol. 1992;66:359–366. doi: 10.1128/jvi.66.1.359-366.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinchington P R, Inchauspe G, Subak-Sharpe J H, Robey F, Hay J, Ruyechan W T. Identification and characterization of a varicella-zoster virus DNA-binding protein by using antisera directed against a predicted synthetic oligopeptide. J Virol. 1988;62:802–809. doi: 10.1128/jvi.62.3.802-809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinchington P R, Turse S E. Regulated nuclear localization of the varicella zoster virus major regulatory protein IE62. J Infect Dis. 1998;178:S16–S21. doi: 10.1086/514263. [DOI] [PubMed] [Google Scholar]
- 32.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 33.Kurachi R, Daikoku T, Tsurumi T, Maeno K, Nishiyama Y, Kurata T. The pathogenicity of a US3 protein kinase-deficient mutant of herpes simplex virus type 2 in mice. Arch Virol. 1993;133:259–273. doi: 10.1007/BF01313767. [DOI] [PubMed] [Google Scholar]
- 34.Leader D P. Identification of protein kinases by computer. Nature. 1988;333:308. doi: 10.1038/333308a0. [DOI] [PubMed] [Google Scholar]
- 35.Leader D P, Deana A D, Marchiori F, Purves F C, Pinna L A. Further definition of the substrate specificity of the alpha-herpesvirus protein kinase and comparison with protein kinases A and C. Biochim Biophys Acta. 1991;1091:426–431. doi: 10.1016/0167-4889(91)90210-o. [DOI] [PubMed] [Google Scholar]
- 36.Leader D P, Katan M. Viral aspects of protein phosphorylation. J Gen Virol. 1988;69:1441–1464. doi: 10.1099/0022-1317-69-7-1441. [DOI] [PubMed] [Google Scholar]
- 37.Leader D P, Purves F C. The herpesvirus protein kinase: a new departure in protein phosphorylation? Trends Biochem Sci. 1988;13:244–246. doi: 10.1016/0968-0004(88)90157-0. [DOI] [PubMed] [Google Scholar]
- 38.Leopardi R, Van Sant C, Roizman B. The herpes simplex virus 1 protein kinase US3 is required for protection from apoptosis induced by the virus. Proc Natl Acad Sci USA. 1997;94:7891–7896. doi: 10.1073/pnas.94.15.7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ling P, Kinchington P R, Sadeghi-Zadeh M, Ruyechan W T, Hay J. Transcription from varicella-zoster virus gene 67 (glycoprotein IV) J Virol. 1992;66:3690–3698. doi: 10.1128/jvi.66.6.3690-3698.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moffat J F, Zerboni L, Sommer M H, Heineman T C, Cohen J I, Kaneshima H, Arvin A M. The ORF47 and ORF66 putative protein kinases of varicella-zoster virus determine tropism for human T cells and skin in the SCID-Hu mouse. Proc Natl Acad Sci USA. 1998;95:11969–11974. doi: 10.1073/pnas.95.20.11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore M S, Blobel G. Soluble factors required for nuclear protein import. Cold Spring Harbor Symp Quant Biol. 1995;60:701–705. doi: 10.1101/sqb.1995.060.01.076. [DOI] [PubMed] [Google Scholar]
- 42.Moriuchi H, Moriuchi M, Dean H, Cheung A K, Cohen J I. Pseudorabies virus EPO is functionally homologous to varicella-zoster virus ORF61 protein and herpes simplex virus type 1 ICPO. Virology. 1995;209:281–283. doi: 10.1006/viro.1995.1256. [DOI] [PubMed] [Google Scholar]
- 43.Moriuchi H, Moriuchi M, Straus S E, Cohen J I. Varicella-zoster virus open reading frame 10 protein, the herpes simplex virus VP16 homolog, transactivates herpesvirus immediate-early gene promoters. J Virol. 1993;67:2739–2746. doi: 10.1128/jvi.67.5.2739-2746.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moriuchi M, Moriuchi H, Straus S E, Cohen J I. Varicella-zoster virus (VZV) virion-associated transactivator open reading frame 62 protein enhances the infectivity of VZV DNA. Virology. 1994;200:297–300. doi: 10.1006/viro.1994.1190. [DOI] [PubMed] [Google Scholar]
- 45.Moroianu J. Molecular mechanisms of nuclear protein transport. Crit Rev Eukaryot Gene Expr. 1997;7:61–72. doi: 10.1615/critreveukargeneexpr.v7.i1-2.40. [DOI] [PubMed] [Google Scholar]
- 46.Moroianu J, Blobel G, Radu A. The binding site of karyopherin alpha for karyopherin beta overlaps with a nuclear localization sequence. Proc Natl Acad Sci USA. 1996;93:6572–6576. doi: 10.1073/pnas.93.13.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mulder W A, Pol J M, Gruys E, Jacobs L, De Jong M C, Peeters B P, Kimman T G. Pseudorabies virus infections in pigs. Role of viral proteins in virulence, pathogenesis and transmission. Vet Res. 1997;28:1–17. [PubMed] [Google Scholar]
- 48.Nagpal S, Ostrove J M. Characterization of a potent varicella-zoster virus-encoded trans-repressor. J Virol. 1991;65:5289–5296. doi: 10.1128/jvi.65.10.5289-5296.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ng T I, Keenan L, Kinchington P R, Grose C. Phosphorylation of varicella-zoster virus open reading frame (ORF) 62 regulatory product by viral ORF 47-associated protein kinase. J Virol. 1994;68:1350–1359. doi: 10.1128/jvi.68.3.1350-1359.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 51.Ohno M, Fornerod M, Mattaj I W. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- 52.Perera L P, Mosca J D, Ruyechan W T, Hay J. Regulation of varicella-zoster virus gene expression in human T lymphocytes. J Virol. 1992;66:5298–5304. doi: 10.1128/jvi.66.9.5298-5304.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perera L P, Mosca J D, Ruyechan W T, Hayward G S, Straus S E, Hay J. A major transactivator of varicella-zoster virus, the immediate-early protein IE62, contains a potent N-terminal activation domain. J Virol. 1993;67:4474–4483. doi: 10.1128/jvi.67.8.4474-4483.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purves F C, Deana A D, Marchiori F, Leader D P, Pinna L A. The substrate specificity of the protein kinase induced in cells infected with herpesviruses: studies with synthetic substrates indicate structural requirements distinct from other protein kinases. Biochim Biophys Acta. 1986;889:208–215. doi: 10.1016/0167-4889(86)90106-0. [DOI] [PubMed] [Google Scholar]
- 55.Purves F C, Spector D, Roizman B. The herpes simplex virus 1 protein kinase encoded by the US3 gene mediates posttranslational modification of the phosphoprotein encoded by the UL34 gene. J Virol. 1991;65:5757–5764. doi: 10.1128/jvi.65.11.5757-5764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Purves F C, Spector D, Roizman B. UL34, the target of the herpes simplex virus US3 protein kinase, is a membrane protein which in its unphosphorylated state associates with novel phosphoproteins. J Virol. 1992;66:4295–4303. doi: 10.1128/jvi.66.7.4295-4303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruyechan W, Ling P, Kinchington P, Hay J. The correlation between varicella zoster virus transcription and the sequence of the viral genome. In: Wagner E K, editor. Herpesvirus transcription and its regulation. Boca Raton, Fla: CRC Press; 1991. pp. 301–318. [Google Scholar]
- 58.Smith R F, Smith T F. Identification of new protein kinase-related genes in three herpesviruses, herpes simplex virus, varicella-zoster virus, and Epstein-Barr virus. J Virol. 1989;63:450–455. doi: 10.1128/jvi.63.1.450-455.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stevenson D, Colman K L, Davison A J. Characterization of the putative protein kinases specified by varicella-zoster virus genes 47 and 66. J Gen Virol. 1994;75:317–326. doi: 10.1099/0022-1317-75-2-317. [DOI] [PubMed] [Google Scholar]
- 60.Stevenson D, Colman K L, Davison A J. Delineation of a sequence required for nuclear localization of the protein encoded by varicella-zoster virus gene 61. J Gen Virol. 1994;75:3229–3233. doi: 10.1099/0022-1317-75-11-3229. [DOI] [PubMed] [Google Scholar]
- 61.Tyler J K, Everett R D. The DNA binding domain of the varicella-zoster virus gene 62 protein interacts with multiple sequences which are similar to the binding site of the related protein of herpes simplex virus type 1. Nucleic Acids Res. 1993;21:513–522. doi: 10.1093/nar/21.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wagenaar F, Pol J M, Peeters B, Gielkens A L, de Wind N, Kimman T G. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J Gen Virol. 1995;76:1851–1859. doi: 10.1099/0022-1317-76-7-1851. [DOI] [PubMed] [Google Scholar]
- 63.Wilcox K W, Kohn A, Sklyanskaya E, Roizman B. Herpes simplex virus phosphoproteins. I. Phosphate cycles on and off some viral polypeptides and can alter their affinity for DNA. J Virol. 1980;33:167–182. doi: 10.1128/jvi.33.1.167-182.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu C L, Wilcox K W. The conserved DNA-binding domains encoded by the herpes simplex virus type 1 ICP4, pseudorabies virus IE180, and varicella-zoster virus ORF62 genes recognize similar sites in the corresponding promoters. J Virol. 1991;65:1149–1159. doi: 10.1128/jvi.65.3.1149-1159.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xia K, DeLuca N A, Knipe D M. Analysis of phosphorylation sites of herpes simplex virus type 1 ICP4. J Virol. 1996;70:1061–1071. doi: 10.1128/jvi.70.2.1061-1071.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xia K, Knipe D M, DeLuca N A. Role of protein kinase A and the serine-rich region of herpes simplex virus type 1 ICP4 in viral replication. J Virol. 1996;70:1050–1060. doi: 10.1128/jvi.70.2.1050-1060.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xia L, Courtney R J. Initial characterization of the membrane-associated form of ICP4 of herpes simplex virus type 1. J Virol. 1995;69:6548–6552. doi: 10.1128/jvi.69.10.6548-6552.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiao C Y, Hubner S, Jans D A. SV40 large tumor antigen nuclear import is regulated by the double-stranded DNA-dependent protein kinase site (serine 120) flanking the nuclear localization sequence. J Biol Chem. 1997;272:22191–22198. doi: 10.1074/jbc.272.35.22191. [DOI] [PubMed] [Google Scholar]
- 69.Yao F, Courtney R J. Association of a major transcriptional regulatory protein, ICP4, of herpes simplex virus type 1 with the plasma membrane of virus-infected cells. J Virol. 1991;65:1516–1524. doi: 10.1128/jvi.65.3.1516-1524.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang G, Stevens R, Leader D P. The protein kinase encoded in the short unique region of pseudorabies virus: description of the gene and identification of its product in virions and in infected cells. J Gen Virol. 1990;71:1757–1765. doi: 10.1099/0022-1317-71-8-1757. [DOI] [PubMed] [Google Scholar]