Abstract
A tetracycline-regulated gene expression system and a panel of novel monoclonal antibodies were used to examine the subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus (HCV) NS3-NS4A complex in inducible cell lines. The NS3 serine protease domain and the full-length NS3 protein expressed in the absence of the NS4A cofactor were diffusely distributed in the cytoplasm and nucleus. Coexpression of NS4A, however, directed NS3 to the endoplasmic reticulum (ER) or an ER-like modified compartment, as demonstrated by colocalization with 3,3′-dihexyloxacarbocyanine iodide, protein disulfide isomerase, and calnexin, as well as subcellular fractionation analyses. In addition, coexpression with NS4A dramatically increased the intracellular stability of NS3 (mean protein half-life of 26 versus 3 h) and allowed for NS4A-dependent trans-cleavage at the NS4B-NS5A junction. Deletion analyses revealed that the hydrophobic amino-terminal domain of NS4A was required for ER targeting of NS3. These results demonstrate the importance of studying HCV proteins in their biological context and define a well-characterized cell culture system for further analyses of the NS3-NS4A complex and the evaluation of novel antiviral strategies against hepatitis C.
Hepatitis C virus (HCV) is the most common etiologic agent of posttransfusion and sporadic non-A, non-B hepatitis (6, 25). The majority of HCV-infected individuals develop chronic infection which may progress to liver cirrhosis and eventually hepatocellular carcinoma (19). HCV contains a single-stranded, positive-sense RNA genome of approximately 9,600 nucleotides (nt) that encodes a polyprotein precursor of 3,010 to 3,033 amino acids (aa). The polyprotein precursor is co- and posttranslationally processed by cellular and viral proteases to yield the mature structural and nonstructural proteins (39). The structural proteins are believed to be processed by the endoplasmic reticulum (ER) signal peptidase (17, 26, 43). Cleavage at the NS2-NS3 site is mediated by a viral protease composed of NS2 and the amino-terminal one-third of NS3 (13, 18, 38). A distinct serine protease located in the amino-terminal one-third of NS3 is responsible for the downstream cleavages in the nonstructural region (1, 12, 49). The proteolytic events mediated by the NS3 serine protease have been shown to be essential for viral replication in vivo in the related yellow fever (4) and bovine viral diarrhea viruses (50), as well as very recently also in HCV (23a). This viral enzyme, therefore, has emerged as a major target in the design of novel antiviral agents against hepatitis C. The biochemical features of the NS3 serine protease have been well characterized in cell-free translation and transient cellular expression systems (39). In addition, the crystal structure of the NS3 se-rine protease (23, 31, 51) and RNA helicase (5, 22, 52) domains, the latter located in the carboxy-terminal region of NS3, have recently been elucidated. The 54-aa NS4A polypeptide functions as a cofactor for the NS3 serine protease (2, 10, 27). The central domain of NS4A has been shown to interact with NS3 and to be essential for its cofactor function (3, 29, 45, 47). The crystal structure revealed that NS4A is incorporated as an integral component into the amino-terminal β barrel of the serine protease core (23, 51).
Compared to the detailed knowledge of its biochemical features, much less is known concerning the characteristics of the NS3-NS4A complex in a cellular context. This is in part due to the lack of an efficient cell culture system for HCV and the difficulty of reliably detecting HCV proteins in naturally infected liver tissue. While previous studies indicated a cytoplasmic localization of NS3 (15, 24, 42), recent work suggested that NS3 may be a nuclear protein that, with varying efficiency among different HCV isolates and depending on the p53 status of the cell, may be retained in the cytoplasm by interaction with NS4A (36). In this context, it was speculated that nuclear NS3 may be responsible for the reported interference of NS3 with various host cell functions, such as growth control (40) and apoptosis (11). In addition, preliminary subcellular fractionation and pulse-chase analyses in transiently transfected cells suggested that expression of NS4A induced membrane association and stabilization of NS3 (28, 44, 47). However, the subcellular compartment targeted by the NS3-NS4A complex, and the requirements for membrane association have not been defined. Finally, a quantitative comparison of the intracellular stability and protein half-lives of NS3 and the NS3-NS4A complex has not been reported to date.
The aim of this study, therefore, was to examine the subcellular localization, stability, and trans-cleavage competence of the NS3-NS4A complex in vitro in tetracycline-regulated cell lines as a well-defined and highly reproducible cell culture system. In addition, a panel of novel monoclonal antibodies (MAbs) against the NS3 serine protease domain was generated to investigate the subcellular localization of this key viral enzyme in vitro and in vivo in naturally infected liver tissue from patients with chronic hepatitis C.
MATERIALS AND METHODS
Production and characterization of MAbs.
HCV genotype 1b-derived, hexahistidine-tagged HCV NS3 serine protease domain (HCV aa 1027 to 1219) was expressed in Escherichia coli and purified under denaturing conditions by nickel affinity chromatography and size exclusion chromatography by high-pressure liquid chromatography. Spleen cells from BALB/c mice immunized with recombinant protein were fused with the X63-Ag8.653 myeloma cell line (American Type Culture Collection, Rockville, Md.). Hybridomas were selected, and supernatants were screened by enzyme-linked immunosorbent assay essentially as described earlier (16). Hybridomas immunoreactive with recombinant and cellular-expressed NS3 protein were cloned twice by limiting dilution. MAb isotypes were determined with reagents from Amersham (Arlington Heights, Ill.). Epitopes were mapped by random DNase I fragment expression library screening by using the NovaTope System (Novagen, Madison, Wis.) and by competitive inhibition experiments.
MAbs 8N against HCV NS4A and 11F against NS5A were generously provided by Jan Albert Hellings and Winand Habets, Organon Teknika B.V., Boxtel, The Netherlands. MAb D3 against protein disulfide isomerase (PDI) was obtained from StressGen (Victoria, British Columbia, Canada), MAb 37 against calnexin was obtained from Dianova (Hamburg, Germany), and MAb 1D6-E1-A8 against cytochrome oxidase subunit I (COX-I) was obtained from Molecular Probes (Eugene, Oreg.). A rabbit polyclonal antiserum to Rab1 was obtained from Zymed (San Francisco, Calif.). A rabbit polyclonal antiserum to mannosidase II (Man II) (34) was kindly provided by Kelley Moremen, University of Georgia, Athens.
Establishment and characterization of inducible cell lines.
cDNA fragments comprising nt 3418 to 4022 (aa 1027 to 1227), nt 3418 to 5312 (aa 1027 to 1657), and nt 3418 to 5475 (aa 1027 to 1711) of the HCV H strain (genotype 1a) were amplified by PCR from pBRTM/HCV1-3011 (14) by using the sense primer 5′-GCACGAATTCACCATGGCGCCCATCACGGCGTACGCCCAGCAGA C-3′ (the EcoRI site is underlined; the engineered translation initiation codon is double underlined) and the reverse primers 5′-GCTGTCTAGATTAGTGGGCCACCTGGAAGCTCTGGGGCACTGC-3′, 5′-GCTGTCTAGATTACGTGACGACCTCCAGGTCGGCCGACATGC-3′, and 5′-GCTGTCTAGATTAGCACTCTTCCATCTCATCGAACTCCTGGTAG-3′ (the XbaI site is underlined, and the engineered ochre stop codon is double underlined). Amplification products were cloned into the EcoRI-XbaI sites of pUHD10-3 (36) to yield the expression constructs pUHDNS3P201, pUHDNS3, and pUHDNS3-4A, respectively. These constructs allow expression of the NS3 serine protease domain, of full-length NS3 protein, and of the NS3-NS4A complex under the transcriptional control of a tetracycline-controlled transactivator (tTA)-dependent promoter (Fig. 1A). Inducible cell lines were generated as previously described (32, 33). In brief, the constitutively tTA-expressing, U-2 OS human osteosarcoma-derived founder cell line UTA-6 (9) was cotransfected with tTA-dependent expression constructs and pBabepuro (35). G418 and puromycin double-resistant clones were isolated, screened for tightly regulated HCV gene expression, and characterized by genomic Southern, Northern, and Western blot analyses. UHCV cells, which inducibly express the complete HCV H open reading frame, have been described previously (33).
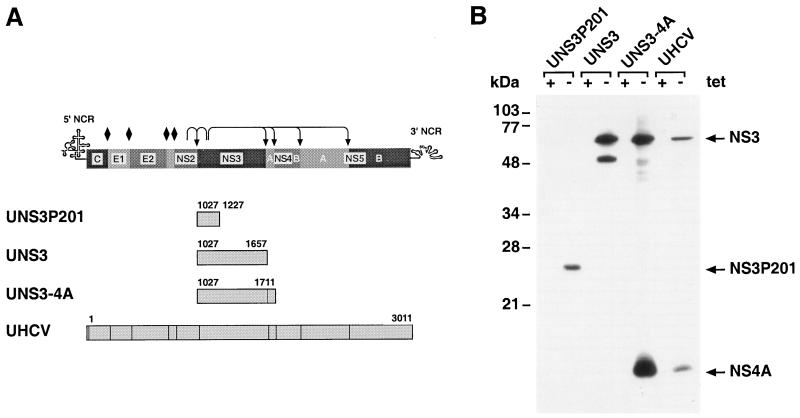
FIG. 1.
Tetracycline-regulated cell lines. (A) Expression cassettes present in UNS3P201, UNS3, UNS3-4A, and UHCV cells. The HCV amino acid positions included are indicated for each construct. The genetic organization and polyprotein processing of HCV are schematically illustrated at the top of the panel. Diamonds denote cleavages of the HCV polyprotein precursor by the ER signal peptidase, and arrows indicate cleavages by the NS2-NS3 and NS3 proteases. (B) Tightly regulated expression of NS3 protein in inducible cell lines. UNS3P201-47.10, UNS3-25, UNS3-4A-24, and UHCV-35 cells were cultured for 24 h in the presence (+ tet) or absence (− tet) of tetracycline. A total of 70 μg of protein per lane was separated by sodium dodecyl sulfate (SDS)–12% polyacrylamide gel electrophoresis (PAGE) and analyzed by immunoblot with MAbs 1H7 against NS3 and 8N against NS4A as described in Materials and Methods. Molecular mass standards are indicated on the left in kilodaltons.
Indirect immunofluorescence and confocal laser scanning microscopy.
Indirect immunofluorescence microscopy was performed as described previously (32). In brief, cells grown as monolayers on glass coverslips were fixed with 2% paraformaldehyde, permeabilized with 0.05% saponin, and incubated with primary antibodies in phosphate-buffered saline (PBS) containing 3% bovine serum albumin and 0.05% saponin. Bound primary antibody was revealed with fluorescein isothiocyanate (FITC)-conjugated goat F(ab′)2 fragment to mouse immunoglobulin G (IgG) F(ab′)2 (Cappel, Durham, N.C.) or sheep F(ab′)2 fragment to rabbit IgG (Boehringer Mannheim, Mannheim, Germany), respectively. For colocalization experiments, protein G affinity-purified MAb 1B6 was biotinylated by using the FluoReporter Biotin-XX labeling kit and revealed with Texas Red (TXR)-conjugated streptavidin (both from Molecular Probes). Coverslips were mounted in SlowFade (Molecular Probes) and examined with a Zeiss Axiovert photomicroscope equipped with an epifluorescence attachment. Confocal laser scanning microscopy was performed by using a Zeiss LSM 410 microscope, and images were processed with the Adobe Photoshop 3.0.5 program.
Immunohistochemistry.
NS3 protein was detected in liver tissue from patients with chronic hepatitis C by a two-stage indirect immunostaining procedure as previously described (41) with some modifications. Liver biopsy samples from 29 anti-HCV and HCV RNA-positive patients were included. Histological diagnosis was minimal changes in 2 patients, chronic persistent hepatitis in 9 patients, chronic active hepatitis without cirrhosis in 12 patients, and chronic active hepatitis with cirrhosis in 6 patients. Next, 5-μm cryosections were cut from snap-frozen liver tissue on siliconized coverslips, which were then air dried and stored at −80°C until use. After rehydration, sections were washed in PBS and incubated with protein G affinity-purified 2E10 MAb at a concentration of 2.5 μg/ml, followed by the addition of affinity-purified sheep IgG F(ab′)2 fragment to mouse Ig conjugated to alkaline phosphatase (Boehringer Mannheim). Unbound antibody was removed by immersion of the slides in 100 mM Tris-Cl (pH 7.4) containing 150 mM NaCl, followed by 100 mM Tris-Cl (pH 9.5) containing 100 mM NaCl and 50 mM MgCl2. Levamisole was added to the alkaline phosphatase substrate 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium (Bio-Rad, Hercules, Calif.) to block endogenous alkaline phosphatase. The specificity of NS3 staining patterns was assessed on selected positive samples before and after absorption of the MAb with recombinant NS3 protein. Recombinant hepatitis B virus-associated proteins (HBsAg and HBcAg) were included in these experiments as controls. A nonrelevant control antibody (mouse anti-human chorionic gonadotropin) and liver tissue samples from anti-HCV and HCV RNA-negative patients were used as additional negative controls.
Western blot analysis.
Western blot analysis was performed as described previously (32, 33).
Substrates for NS3 trans-cleavage assays.
cDNA fragments comprising nt 5473 to 7602 (aa 1712 to 2420) and nt 6256 to 8538 (aa 1973 to 2732) of the HCV H strain were amplified by PCR from pBRTM/HCV1-3011 by using the primer pairs sense 5′-CACGAATTCACCATGGGCTCTCAGCACTTACCGTACATCGAGCAAG-3′ (the EcoRI site is underlined, the engineered translation initiation codon is double underlined, and the engineered glycine codon for the optimal Kozak sequence is in italics) and reverse 5′-CTGTCTAGACTAGCAGCACACGACATCTTCCGTGTCGGC-3′ (the XbaI site is underlined, and the engineered amber stop codon is double underlined), as well as sense 5′-CACG GTACCACCATGGGCTCCGGCTCCTGGCTAAGGGACATCTG-3′ (the KpnI site is underlined, the engineered translation initiation codon is double underlined, and the engineered glycine codon for optimal Kozak sequence is in italics) and reverse 5′-CTGTCTAGATTAGGTGCAGTCCTGGAGCCCTGCGGCTCGACG-3′ (the XbaI site is underlined, and the engineered ochre stop codon is double underlined). Amplification products were cloned into the EcoRI-XbaI sites and KpnI-XbaI sites, respectively, of pcDNA3.1 (Invitrogen) to yield the expression constructs pCMVNS4B-5A and pCMVNS5A-5B312. These constructs allow expression of a NS4B-NS5A substrate and of a substrate comprising NS5A and the amino-terminal 312 aa of NS5B, respectively, under the control of the cytomegalovirus immediate-early promoter.
NS4A expression constructs.
cDNA fragments comprising nt 5312 to 5475 (aa 1658 to 1711) and nt 5312 to 5414 (aa 1658 to 1691) of the HCV H strain were amplified by PCR from pBRTM/HCV1-3011 by using the sense primer 5′-GCACGAATTCACCATGAGCACCTGGGTGCTCGTTGGCGGCGTCC-3′ (the EcoRI site is underlined, and the engineered translation initiation codon is double underlined) and one of the reverse primers 5′-GCTGTCTAGATTAGCACTCTTCCATCTCATCGAACTCCTGG-3′ and 5′-CTGTCTAGACTACTTCCCGGACAAGACAATCCTGCCCAC-3′ (the XbaI site is underlined, and the engineered stop codon is double underlined), respectively. A cDNA fragment comprising nt 5373 to 5475 (aa 1678 to 1711) was amplified by using the sense primer 5′ CACGAATTCACCATGGGCTGCGTGGTCATAGTGGGCAGGAT-3′ and the reverse primer 5′-GCTGTCTAGATTAGCACTCTTCCATCTCATCGAACTCCTGG-3′. Amplification products were cloned into the EcoRI-XbaI sites of pcDNA3.1 to yield the expression constructs pCMVNS4A, pCMVNS4A1-34, and pCMVNS4A21-54, respectively. These constructs allow expression of the entire NS4A protein (aa 1 to 54), the amino-terminal (aa 1 to 34), and the carboxy-terminal two-thirds (aa 21 to 54) of NS4A, respectively, under the control of the cytomegalovirus immediate-early promoter. Plasmid pCMVNS4A21-34, allowing expression of the central one-third of NS4A (aa 21 to 34), was generated by hybridization of the primer pair sense 5′-AATTCAC CATGGGCTGCGTGGTCATAGTGGGCAGGATTGTCTTGTCCGGGAA GTAGT-3′ and reverse 5′-CTAGACTACTTCCCGGACAAGACAATCCTGCCCACTATGACCACGCAGCCCATGGTG-3′ and ligation into the EcoRI-XbaI sites of pcDNA3.1.
RESULTS
NS3 MAbs and inducible cell lines.
Recombinant NS3 serine protease was expressed in E. coli and used as an antigen to generate a panel of novel MAbs. The characteristics of these antibodies, termed 1B6, 1G9, 1H7, and 2E10, are summarized in Table 1. All MAbs functioned well in indirect immunofluorescence microscopy and Western blot applications, suggesting that they recognize linear epitopes. The epitope recognized by MAb 1H7 could be mapped to HCV aa 1186 to 1219 by random DNase I fragment expression library screening. In addition, competitive inhibition experiments indicated that MAbs 1B6 and 1G9 recognized closely related epitopes in the carboxy-terminal region of the NS3 serine protease domain. MAb 2E10, on the other hand, was found to be directed against a distinct epitope in the NS3 serine protease domain.
TABLE 1.
Characteristics of MAbs specific for the NS3 serine protease domain
| MAb | Isotype | Epitope (aa)a | Detection byb:
|
||
|---|---|---|---|---|---|
| IIF | WB | IHC | |||
| 1B6 | IgG1κ | 1186–1219 | + | + | − |
| 1G9 | IgM | 1186–1219 | + | + | − |
| 1H7 | IgG1κ | 1186–1219 | + | + | − |
| 2E10 | IgG2bκ | 1027–1219 | + | + | + |
The epitope recognized by MAb 1H7 was mapped to HCV aa 1186 to 1219 by random DNase I fragment expression library screening as described in Materials and Methods. 1B6 and 1H7 competed for binding to the same or closely related epitopes. In addition, 1B6 and 1H7 showed partial competition with 1G9 in competitive inhibition assays, indicating that these three MAbs recognize closely related epitopes in the carboxy-terminal region of the NS3 serine protease domain. By contrast, MAb 2E10 did not compete with these MAbs, indicating that it recognizes a distinct epitope within the NS3 serine protease domain. The epitope values for MAbs 1B6 and 1G9 are approximate.
IIF, indirect immunofluorescence microscopy; WB, Western blot; IHC, immunohistochemistry of liver tissue sections from patients with chronic hepatitis C.
Continuous human cell lines inducibly expressing various forms of NS3 and the NS3-NS4A complex were generated by transfection of the tTA-expressing founder cell line UTA-6 with constructs allowing expression of the NS3 serine protease domain, of full-length NS3 protein, and of the NS3-NS4A complex under the transcriptional control of a tTA-dependent promoter (Fig. 1A). Screening of antibiotic double-resistant stable clones resulting from these transfections allowed the identification of several tightly regulated UNS3P201, UNS3, and UNS3-4A cell lines. These contained different numbers of HCV transgenes chromosomally integrated in a head-to-tail fashion that were inducibly expressed as a single HCV-specific transcript of the expected length, as assessed by genomic Southern and Northern blot analyses, respectively (data not illustrated). These cell lines were maintained in continuous culture for more than 12 months and for more than 50 passages without loss of their characteristics.
Data obtained with the cell lines UNS3P201-47.10, UNS3-25, and UNS3-4A-24 will be presented below. All results were confirmed in at least two additional independent clones. In addition, a previously described cell line, UHCV-35, which allows expression of NS3 in the context of the entire HCV polyprotein, was used (33). A representative Western blot of inducible cell lines cultured in the presence or absence of tetracycline is shown in Fig. 1B. A single NS3-specific product of approximately 24 kDa, corresponding to the serine protease domain, was found in UNS3P201 cells cultured in the absence of tetracycline. The 70-kDa full-length NS3 protein was detected in UNS3, UNS3-4A, and UHCV cells cultured in the absence of tetracycline. In addition, the NS4A cofactor was inducibly expressed and processed by cis-cleavage in UNS3-4A and UHCV cells.
Subcellular localization of NS3 and the NS3-NS4A complex.
The subcellular localization of the various forms of NS3 and the NS3-NS4A complex expressed in UNS3P201, UNS3, UNS3-4A, and UHCV cells was investigated by indirect immunofluorescence and confocal laser scanning microscopy. As shown in Fig. 2, the NS3 serine protease domain expressed in UNS3P201 cells was found in a diffuse cytoplasmic and nuclear staining pattern with superimposed broady distributed fine granules. Full-length NS3 protein expressed in UNS3 cells showed a diffuse cytoplasmic and nuclear localization as well. In addition, more intensively stained somewhat coarser granules were found in the perinuclear region. In about 60% of these cells, NS3 accumulated in the nucleus, with apparent sparing of the nucleoli. A minority of cells showed a diffuse exclusively cytoplasmic staining.
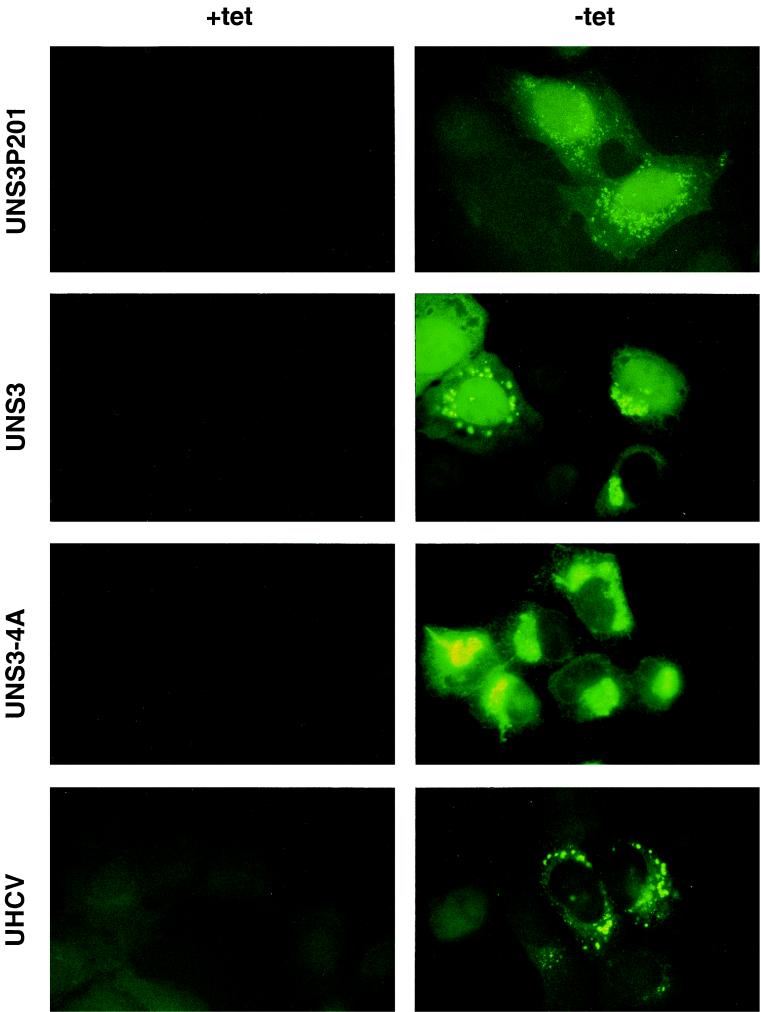
FIG. 2.
Subcellular localization of HCV NS3 protein in vitro. UNS3P201-47.10, UNS3-25, UNS3-4A-24, and UHCV-35 cells were cultured for 24 h in the presence (+tet) or absence (−tet) of tetracycline and subsequently processed for indirect immunofluorescence microscopy with MAb 1H7 as described in Materials and Methods.
In sharp contrast to the pattern observed in UNS3P201 and UNS3 cells, the NS3-NS4A complex expressed in UNS3-4A cells was found in a predominantly perinuclear reticular and granular staining pattern typical for ER proteins. No nuclear staining was detected in these cells. In addition, however, there was staining of tubular structures radiating from the perinuclear region to the periphery of the cell, a pattern typical for mitochondria. In UHCV cells, overall expression levels were lower, but the cytoplasmic reticular and granular staining pattern was similar to that observed in UNS3-4A cells.
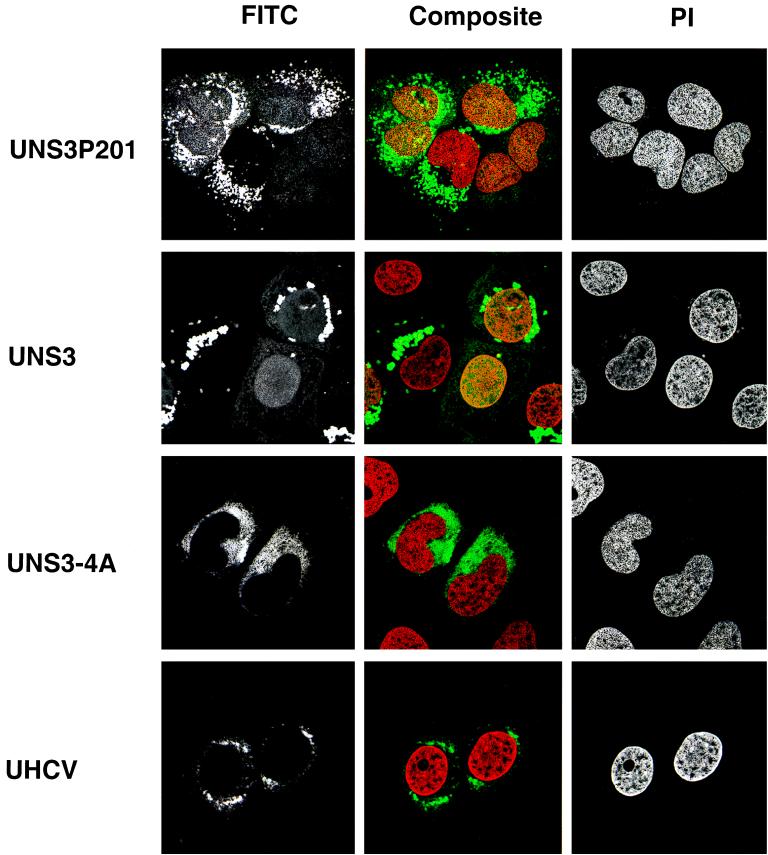
These observations were extended by confocal laser scanning microscopy. As shown in Fig. 3, sections placed through the center of the nuclei confirmed NS3 to be localized in the nucleus only in UNS3P201 and UNS3, but not in UNS3-4A and UHCV cells.
FIG. 3.
Subcellular localization of HCV NS3 protein in vitro. UNS3P201-47.10, UNS3-25, UNS3-4A-24, and UHCV-35 cells were cultured for 24 h in the absence of tetracycline and subsequently processed for confocal laser scanning microscopy with MAb 1H7 as described in Materials and Methods. Cells were counterstained with propidium iodide (PI) to visualize nuclei. Horizontal sections taken through the center of the nuclei are shown. Images recorded in green (FITC) and red (PI) channels are presented separately on the left and on the right, respectively, and composite images are shown in the middle.
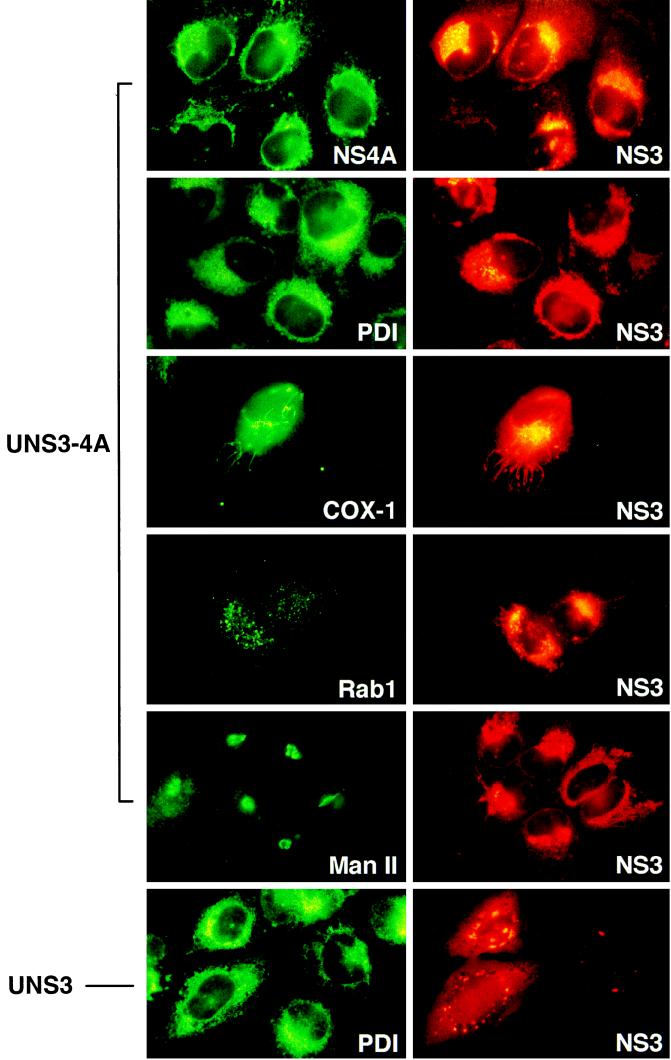
To further investigate the subcellular compartment targeted by the NS3-NS4A complex, staining with fluorescent dyes and colocalization experiments with antibodies to cellular proteins were performed. The NS3-NS4A complex expressed in UNS3-4A and UHCV cells showed a pattern very similar to that revealed by live staining of the cells with the fluorescent dye 3,3′-dihexyloxacarbocyanine iodide [DiOC6(3)], which is specific for the ER and mitochondria (48) (data not illustrated). In addition, the tubular structures observed in UNS3-4A cells corresponded to the pattern revealed by the fluorescent dye rhodamine-123, which labels mitochondria (21) (data not illustrated). These findings were examined in more detail by double-immunolabeling experiments. For this purpose, the reactivity of biotinylated MAb 1B6 against NS3 was revealed by TXR-conjugated streptavidin, and the reactivity of antibodies against cellular marker proteins was revealed by FITC-conjugated secondary antibodies, as shown in Fig. 4. As expected, NS3 and NS4A were found to colocalize in UNS3-4A cells. The NS3-NS4A complex expressed in UNS3-4A cells colocalized with PDI and calnexin (the latter is not illustrated), two markers specific for the ER, as well as with COX-I, a marker specific for mitochondria. The NS3 staining pattern observed in these cells was clearly different, however, from that revealed by antibodies directed against Rab1, a marker of the ER-to-Golgi intermediate compartment, and Man II, a marker of the Golgi apparatus. These observations indicate that the NS3-NS4A complex expressed in UNS3-4A cells is localized in the ER or an ER-like modified compartment. Analogous results were obtained in UHCV cells (data not illustrated). By contrast, the NS3 serine protease domain and full-length NS3 protein expressed in UNS3P201 and UNS3 cells, respectively, did not colocalize with DiOC6(3), rhodamine-123, PDI, calnexin, Rab1, Man II, or COX-I, as representatively shown for UNS3 cells stained with antibodies against NS3 and PDI in the bottom panel of Fig. 4.
FIG. 4.
NS3 is localized in the ER in UNS3-4A cells. UNS3-4A-24 and UNS3-25 cells were cultured for 24 h in the absence of tetracycline and subsequently processed for double immunolabeling. In brief, cells were fixed and permeabilized as described in Materials and Methods, followed by sequential incubations with (i) MAbs 8N against NS4A, 1D3 against PDI, or 1D6-E1-A8 against COX-I or with rabbit polyclonal antisera against Rab1 or Man II, as indicated; (ii) FITC-conjugated goat F(ab′) fragment to mouse IgG F(ab′)2 or sheep F(ab′)2 fragment to rabbit IgG; (iii) biotinylated MAb 1B6 against the HCV NS3 serine protease domain; and (iv) TXR-conjugated streptavidin (Molecular Probes). Slides were viewed with filter sets for TXR and FITC, respectively.
As found previously by us (32, 33) and by others (37) using the tetracycline-regulated gene expression system, there was some heterogeneity in expression levels among individual cells of a given monoclonal cell line. This inherent feature of the expression system explains the observation that not all of the cells stained with antibodies against marker proteins showed a positive staining for NS3 protein in the double-immunolabeling experiments.
The results obtained by immunofluorescence microscopy were supported by subcellular fractionation experiments which revealed NS3 protein in the cytosolic 100,000 × g supernatant (S-100 fraction) only in UNS3 but not in UNS3-4A cells (data not illustrated). In addition, NS3, NS4A, and PDI were found to peak in the same discrete fraction of sucrose gradients only in UNS3-4A cells, whereas NS3 expressed in UNS3 cells showed a broad distribution across these gradients (data not illustrated).
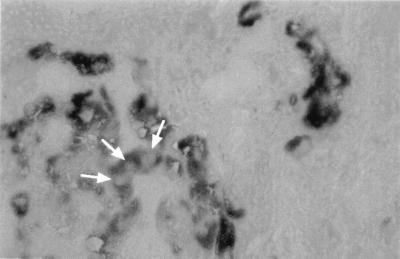
Taken together, these results indicate that the NS3 serine protease domain and full-length NS3 protein expressed in the absence of NS4A were diffusely distributed in the cytoplasm and nucleus. By contrast, coexpression of the NS4A cofactor, either in the context of the NS3-NS4A complex or in the context of the entire HCV polyprotein, directed NS3 to the ER or an ER-like modified compartment. The relevance of these observations made in cell lines in vitro was confirmed by immunohistochemical analyses of liver tissue sections from patients with chronic hepatitis C which revealed NS3 only in the cytoplasm and not in the nucleus of naturally infected hepatocytes. Using MAb 2E10, a positive signal was detected in 16 out of 29 liver biopsy specimens (55%) from patients with chronic hepatitis C. In positive samples, 30 to 80% of the hepatocytes were stained with this MAb (Fig. 5). No reactivity was found in liver tissues from HCV-negative patients. In addition, preincubation with recombinant NS3 protease domain almost completely abolished reactivity (data not illustrated).
FIG. 5.
Localization of NS3 protein in liver tissue sections from patients with chronic hepatitis C. NS3 protein was detected as diffuse and homogeneous immune reactant within the cytoplasmic compartment. Cell nuclei were consistently negative (arrows). Magnification, ×400.
Stability of NS3 and the NS3-NS4A complex.
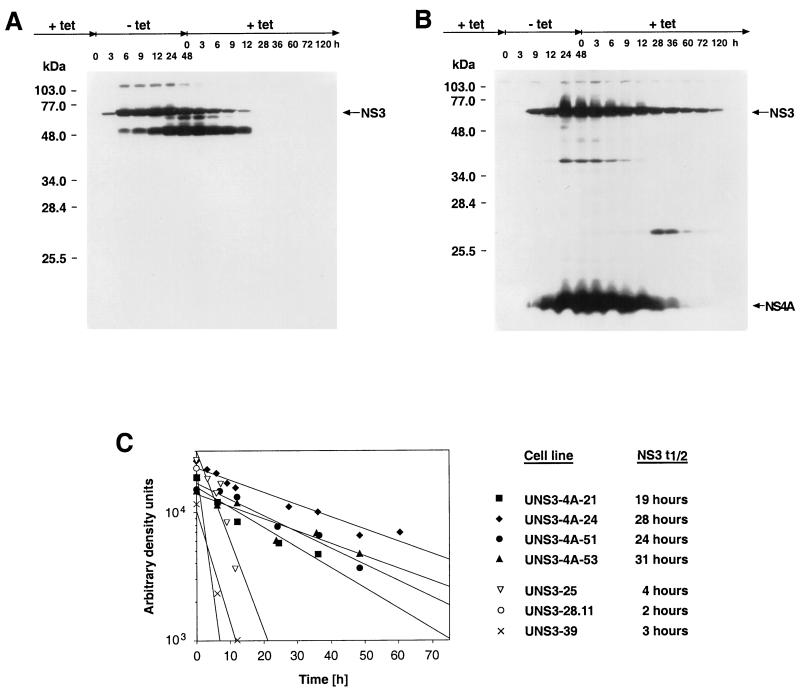
The ability to regulate protein expression in inducible cell lines allowed us to determine and compare the intracellular half-lives of NS3 and the NS3-NS4A complex. In this context, the HCV-specific mRNA transcript was found to disappear with a half-life of <1 h after addition of tetracycline to the culture medium (data not illustrated). Figure 6 shows a representative study of NS3 protein expression in UNS3 and UNS3-4A cells cultured for various times in the absence or presence of tetracycline. No HCV protein synthesis was observed in the presence of tetracycline. HCV protein expression was induced upon tetracycline withdrawal and reached a steady-state level after 24 to 48 h. When tetracycline was re-added to the culture medium, NS3 rapidly disappeared from UNS3 cells (Fig. 6A). By contrast, in UNS3-4A cells NS3 was still detectable after 120 h (Fig. 6B). By densitometry scanning, semilogarithmic plotting, and regression analyses, mean NS3 protein half-lives of 3 and 26 h were calculated for NS3 expressed in a number of independent UNS3 and UNS3-4A cell clones, respectively (Fig. 6C). Thus, coexpression of NS4A dramatically increased the stability of NS3 in this cellular context. A breakdown product of approximately 54 kDa appeared early in all UNS3 cell clones and represented roughly half of the total NS3 protein expressed at steady state (Fig. 6A). In UNS3-4A cells, two minor breakdown products of approximately 40 and 27 kDa were found, with the temporal sequence of their appearance suggesting a precursor-product relationship (Fig. 6B). Under the same experimental conditions no NS3 breakdown products were detectable in UHCV cells, suggesting a further degree of stabilization of NS3 in the context of the entire HCV polyprotein.
FIG. 6.
NS4A increases the intracellular stability of HCV NS3 protein. UNS3-25 (A) and UNS3-4A-24 (B) cells were cultured in the presence of tetracycline until time point zero, when tetracycline was withdrawn from the medium. Subsequently, cells were harvested at the times indicated on top. After 48 h tetracycline was re-added to the medium. A total of 70 μg of protein per lane was separated by SDS–12% PAGE and analyzed by immunoblot with MAbs 1H7 against NS3 and 8N against NS4A as described in Materials and Methods. Molecular mass standards are indicated on the left in kilodaltons. (C) NS3 protein half-life determinations. Four different UNS3-4A and 3 UNS3 cell clones were subjected to time course analyses as shown in panels A and B. Immunoblot signals were quantitatively examined by densitometry scanning as described in Materials and Methods. Regression equations were generated with the Cricket Graph III program (Computer Associates International, Inc., Islandia, N.Y.) and used to calculate half-lives.
trans-Cleavage competence of NS3 and the NS3-NS4A complex expressed in inducible cell lines.
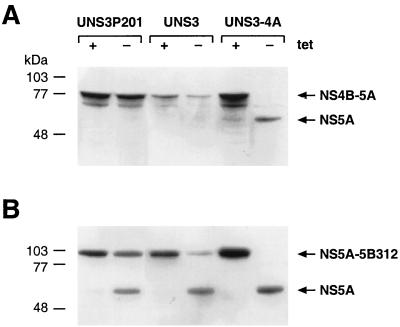
HCV NS3 protein expressed in UNS3-4A cells allowed proper cis-cleavage of NS4A, as shown in Fig. 1B. To assess the trans-cleavage competence of NS3 expressed in the different cell lines, cytomegalovirus promoter-driven expression constructs coding for NS4B-NS5A or NS5A-NS5B312 substrates of the NS3 serine protease were transiently transfected into UNS3P201, UNS3, and UNS3-4A cells cultured in the presence or absence of tetracycline. NS3 protease activity was assessed by the detection of the 56- to 58-kDa processed NS5A protein by immunoblot analysis. As shown in Fig. 7A, the NS4B-NS5A precursor was processed only in UNS3-4A but not in UNS3P201 and UNS3 cells cultured in the absence of tetracycline. This is consistent with reports from cell-free and transient cellular expression systems where cleavage at the NS4B-NS5A junction has been found to be NS4A dependent (2, 10, 27, 47). By contrast, the NS5A-5B312 fusion protein was processed in UNS3P201, UNS3, and UNS3-4A cells cultured in the absence of tetracycline, although with reduced efficiency in UNS3P201 and UNS3 compared to UNS3-4A cells (Fig. 7B). In our system, therefore, NS3 serine protease expression allowed both NS4A-dependent trans-cleavage at the NS4B-NS5A junction and NS4A-independent cleavage at the NS5A-NS5B junction.
FIG. 7.
trans-Cleavage competence of NS3 expressed in inducible cell lines. UNS3P201-47.10, UNS3-28.11, and UNS3-4A cells were cultured for 2 h in the presence or absence of tetracycline, followed by transient transfection of pCMVNS4B-5A (A) or pCMVNS5A-5B312 (B) NS3 substrate expression constructs. Cells were harvested 36 h posttransfection, and cell lysates were separated by SDS–10% PAGE, followed by immunoblotting with MAb 11H against NS5A as described in Materials and Methods. Molecular mass standards are indicated on the left in kilodaltons.
The amino-terminal two-thirds of NS4A are required for ER targeting of NS3.
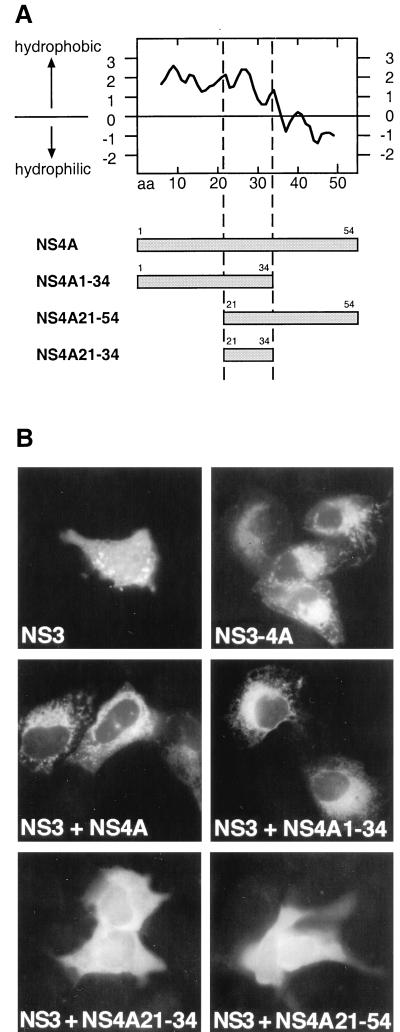
As shown in Fig. 8, the NS4A polypeptide directed NS3 to the ER not only when expressed and processed in cis in the context of a NS3-NS4A complex but also when coexpressed in trans. This observation allowed us to determine the domain of NS4A required for ER targeting of NS3 by transient cotransfection experiments. For this purpose, the expression constructs coding for the amino-terminal two-thirds (aa 1 to 34), the central one-third (aa 21 to 34), and the carboxy-terminal two-thirds (aa 21 to 54) of NS4A were cotransfected with a NS3 expression construct and the subcellular localization of NS3 was subsequently examined by immunofluorescence microscopy. The central one-third of NS4A was included in all NS4A expression constructs because this domain has been shown to interact with NS3 (3, 29, 45, 47). As shown in Fig. 8, of the different NS4A deletion constructs only the construct encoding the amino-terminal two-thirds of NS4A was able to direct NS3 to the ER. Cotransfection of NS3 with the central domain or the carboxy-terminal two-thirds resulted in a diffuse staining pattern identical to the one observed after transfection of NS3 alone. Expression of the polypeptides encoded by the different NS4A deletion constructs was verified by their ability to confer NS4A-dependent trans-cleavage competence to NS3 in transient cotransfection experiments with an NS4B-NS5A substrate (data not illustrated). Taken together, these results indicate that the amino-terminal two-thirds of NS4A are required for ER targeting of NS3.
FIG. 8.
The amino-terminal two-thirds of NS4A are required for ER targeting of NS3. (A) NS4A expression constructs. A hydrophobicity plot of NS4A as determined by the algorithm of Kyte and Doolittle is shown at the top. (B) U-2 OS cells were transiently transfected with cytomegalovirus promoter-driven expression constructs coding for NS3 or the NS3-NS4A complex or were cotransfected with NS3 and the different NS4A constructs, as indicated by the captions. Cells were subsequently processed for indirect immunofluorescence microscopy with MAb 1H7 as described in Materials and Methods.
DISCUSSION
The subcellular localization, stability, and trans-cleavage competence of the HCV NS3-NS4A complex were examined in tetracycline-regulated cell lines. Given the lack of an efficient cell culture system for HCV, these cell lines represent a highly reproducible system for studying the structural and functional properties of the NS3-NS4A complex in a cellular context. The UNS3P201, UNS3, UNS3-4A, and UHCV cell lines, which allow the tightly regulated expression of the NS3 serine protease domain, full-length NS3 protein, the NS3-NS4A complex, and NS3 in the context of the entire HCV polyprotein, respectively, were characterized in detail. These cell lines were maintained in continuous culture for more than 1 year and over 50 passages with stable characteristics and without loss of tightly regulated gene expression.
The NS3 serine protease domain and full-length NS3 protein expressed in the absence of NS4A were diffusely distributed in the cytoplasm and nucleus. By contrast, coexpression of the NS4A cofactor, either in the context of the NS3-NS4A complex or the entire HCV polyprotein, led to a cytoplasmic localization of NS3. In this case, NS3 could not be detected in the nucleus by immunofluorescence and confocal laser scanning microscopy or in a soluble cytosolic fraction by differential centrifugation analyses. Colocalization studies with fluorescent dyes and antibodies against cellular marker proteins revealed the NS3-NS4A complex to be localized in the ER. Double-immunolabeling experiments with markers for the ER-to-Golgi intermediate compartment and the Golgi apparatus showed no evidence of transport of NS3 protein beyond the ER. Interestingly, we have previously found that all investigated HCV structural and nonstructural proteins colocalized in UHCV cells (33). In addition, native HCV envelope glycoprotein complexes were found to be strictly retained in the ER of these cells (8). It is conceivable, therefore, that the ER or an ER-like modified cell compartment is the site of both membrane-associated HCV RNA replication and virus assembly.
Based on results of transient-cotransfection experiments, Muramatsu et al. recently reported that wild-type p53 mediated accumulation of NS3 in the nucleus even in the presence of NS4A (36). U-2 OS cells are wild type with respect to p53 (7). At the levels of endogenous p53 expressed in these cells, however, we were unable to confirm these findings.
The significance of the staining of mitochondria observed in UNS3-4A cells remains to be defined. This pattern was not readily apparent in UHCV cells, most likely due to the overall lower expression levels in these cells. A possible explanation for this staining pattern may be that it results from ER membranes which typically surround mitochondria in U-2 OS cells (32).
The relevance of our observations made in cell lines in vitro was confirmed by immunohistochemical analyses of liver tissue sections from patients with chronic hepatitis C. These revealed NS3 only in the cytoplasm and not in the nucleus of naturally infected hepatocytes. Interestingly, of our panel of MAbs, all of which reacted with NS3 expressed in cell culture, only MAb 2E10 was able to detect NS3 protein in liver tissue sections. The epitope within the NS3 serine protease domain recognized by this MAb, therefore, appears to be particularly accessible for this application.
In addition to determining its subcellular localization, coexpression of NS4A dramatically increased the intracellular stability of NS3. Dynamic expression studies in different UNS3 and UNS3-4A cell clones revealed protein half-lives of 3 and 26 h, respectively, for NS3 expressed alone or together with NS4A. A discrete breakdown product of approximately 54 kDa appeared early in all UNS3 cell clones and represented roughly half of the total NS3 protein expressed at steady state. This is reminiscent of an approximately 49-kDa amino-terminal fragment that has recently been reported to be derived by internal processing of NS3 (46). In UNS3-4A cells, however, two different minor breakdown products of approximately 40 and 27 kDa were found, suggesting an altered susceptibility to proteolytic cleavage as a consequence of either conformational changes or membrane association of NS3 complexed with NS4A. Finally, under the same experimental conditions no smaller size NS3 products were detectable in UHCV cells, suggesting a further degree of stabilization of NS3 in the context of the entire HCV polyprotein.
In addition to determining its subcellular localization and intracellular stability, the NS4A cofactor was found to alter the trans-cleavage competence of the NS3 serine protease expressed in tetracycline-regulated cell lines. Consistent with results obtained in cell-free and transient cellular expression systems (2, 10, 27), cleavage at the NS4B-NS5A junction was dependent and at the NS5A-NS5B junction was independent of NS4A.
Taken together, the NS4A cofactor was found to profoundly modulate important features of NS3 when expressed in a cellular context. This should be considered in studies addressing structural, functional, and immunological properties of NS3 and interactions of NS3 with host cell proteins and pathways.
NS4A directed NS3 to the ER not only when expressed and processed in cis but also when expressed in trans. This observation allowed us to map the NS4A domain responsible for ER targeting of NS3 by transient-cotransfection experiments. These experiments revealed the amino-terminal two-thirds of NS4A (aa 1 to 34) to be required for ER targeting. Consistent with data from cell-free and transient cellular expression systems (3, 29, 45, 47), however, the central domain of NS4A (aa 21 to 34) was sufficient for protease activation. The very hydrophobic amino-terminal domain of NS4A, therefore, likely represents the membrane anchor for the NS3-NS4A complex. The data shown in Fig. 7 and additional data not shown indicate that protease activation and membrane association mediated by NS4A may not be completely separated because cleavage at NS4A-dependent sites occurred more efficiently and completely when the amino-terminal two-thirds of NS4A or the entire NS4A polypeptide was present. Modulation of NS3 function and stability by NS4A may represent an important regulatory mechanism in the HCV life cycle.
NS3 is likely to interact with other HCV nonstructural proteins to form a membrane-associated replication complex (20, 30). The nonstructural proteins other than NS3 are localized in the ER also when expressed individually (D. Moradpour, T. Hügle, E. Bieck, C. M. Rice, and H. E. Blum, unpublished data). Cotransfection experiments with constructs coding for NS3 and NS2, NS4A, NS4B, NS5A, and NS5B indicated, however, that only NS4A can direct NS3 to the ER (Moradpour et al., unpublished). Current studies in our laboratories are aimed at understanding in more detail the mechanism of membrane association of NS4A.
In conclusion, using a tetracycline-regulated gene expression system to establish continuous human cell lines inducibly expressing various forms of HCV NS3 and the NS3-NS4A complex, we found that coexpression of NS4A directs NS3 to the ER or an ER-like modified compartment, dramatically increases its intracellular stability, and alters its trans-cleavage competence. By contrast, NS3 protein expressed without the NS4A cofactor showed a diffuse nuclear and cytoplasmic distribution and was very unstable. NS3 when expressed alone, therefore, has very different properties compared to the NS3-NS4A complex. These results demonstrate the importance of studying HCV proteins in their biological context. In addition, they define a well-characterized and highly reproducible cell culture system for further analyses of the NS3-NS4A complex and for the development of novel antiviral strategies against hepatitis C. In this context, these cell lines represent a unique tool to evaluate candidate protease inhibitors as antiviral agents.
ACKNOWLEDGMENTS
We gratefully acknowledge Petra Binninger and Elke Bieck for excellent technical assistance, Christoph Englert for UTA-6 cells, Jan Albert Hellings and Winand Habets for MAbs 8N and 11F, and Kelley Moremen for the antiserum against Man II.
This work was supported by grant Mo 799/1-1 from the Deutsche Forschungsgemeinschaft to D.M. and H.E.B., a grant from the Associazione Italiana per la Ricerca sul Cancro to D.S. and F.D., a grant from the German Ministry for Research and Technology to H.-G.K., and Public Health Service grants CA57973 and AI40034 to C.M.R.
REFERENCES
- 1.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J Virol. 1993;67:3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager R, Ahlborn-Laake L, Yasargil K, Mous J, Jacobsen H. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J Virol. 1994;68:5045–5055. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager R, Lohmann V, Wilkinson T, Koch J O. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J Virol. 1995;69:7519–7528. doi: 10.1128/jvi.69.12.7519-7528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambers T J, Weir R C, Grakoui A, McCourt D W, Bazan J F, Fletterick R J, Rice C M. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site specific cleavages in the viral polyprotein. Proc Natl Acad Sci USA. 1990;87:8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho H S, Ha N C, Kang L W, Chung K M, Back S H, Jang S K, Oh B H. Crystal structure of RNA helicase from genotype 1b hepatitis C virus. A feasible mechanism of unwinding duplex RNA. J Biol Chem. 1998;273:15045–15052. doi: 10.1074/jbc.273.24.15045. [DOI] [PubMed] [Google Scholar]
- 6.Choo Q-L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 7.Diller L, Kassel J, Nelson C E, Gryka M A, Litwak G, Gebhardt M, Bressac B, Ozturk M, Baker S J, Vogelstein B. p53 functions as a cell cycle control protein in osteosarcomas. Mol Cell Biol. 1990;10:5772–5781. doi: 10.1128/mcb.10.11.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duvet S, Cocquerel L, Pillez A, Cacan R, Verbert A, Moradpour D, Wychowski C, Dubuisson J. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J Biol Chem. 1998;273:32088–32095. doi: 10.1074/jbc.273.48.32088. [DOI] [PubMed] [Google Scholar]
- 9.Englert C, Hou X, Maheswaran S, Bennett P, Ngwu C, Re G G, Garvin A J, Rosner M R, Haber D A. WT1 suppresses synthesis of the epidermal growth factor receptor and induces apoptosis. EMBO J. 1995;14:4662–4675. doi: 10.1002/j.1460-2075.1995.tb00148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Failla C, Tomei L, De Francesco R. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J Virol. 1994;68:3753–3760. doi: 10.1128/jvi.68.6.3753-3760.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita T, Ishido S, Muramatsu S, Itoh M, Hotta H. Suppression of actinomycin D-induced apoptosis by the NS3 protein of hepatitis C virus. Biochem Biophys Res Commun. 1996;229:825–831. doi: 10.1006/bbrc.1996.1887. [DOI] [PubMed] [Google Scholar]
- 12.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA. 1993;90:10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grakoui A, Wychowski C, Lin C, Feinstone S M, Rice C M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada T, Kim D W, Sagawa K, Suzuki T, Takahashi K, Saito I, Matsuura Y, Miyamura T. Characterization of an established human hepatoma cell line constitutively expressing non-structural proteins of hepatitis C virus by transfection of viral cDNA. J Gen Virol. 1995;76:1215–1221. doi: 10.1099/0022-1317-76-5-1215. [DOI] [PubMed] [Google Scholar]
- 16.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 17.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci USA. 1991;88:5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hijikata M, Mizushima H, Akagi T, Mori S, Kakiuchi N, Kato N, Tanaka T, Kimura K, Shimotohno K. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol. 1993;67:4665–4675. doi: 10.1128/jvi.67.8.4665-4675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoofnagle J H. Hepatitis C: the clinical spectrum of disease. Hepatology. 1997;26(Suppl. 1):15S–20S. doi: 10.1002/hep.510260703. [DOI] [PubMed] [Google Scholar]
- 20.Ishido S, Fujita T, Hotta H. Complex formation of NS5B with NS3 and NS4A proteins of hepatitis C virus. Biochem Biophys Res Commun. 1998;244:35–40. doi: 10.1006/bbrc.1998.8202. [DOI] [PubMed] [Google Scholar]
- 21.Johnson L V, Walsh M L, Chen L B. Localization of mitochondria in living cells with rhodamine 123. Proc Natl Acad Sci USA. 1980;77:990–994. doi: 10.1073/pnas.77.2.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J L, Morgenstern K A, Griffith J P, Dwyer M D, Thomson J A, Murcko M A, Lin C, Caron P R. Hepatitis virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 23.Kim J L, Morgenstern K A, Lin C, Fox T, Dwyer M D, Landro J A, Chambers S P, Markland W, Lepre C A, O'Malley E T, Harbeson S L, Rice C M, Murcko M A, Caron P R, Thomson J A. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 23a.Kolykhalov A A, Mihalik K, Feinstone S M, Rice C M. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J Virol. 2000;74:2046–2051. doi: 10.1128/jvi.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krawczynski K, Beach M J, Bradley D W, Kuo G, Di Bisceglie A M, Houghton M, Reyes G R, Kim J P, Choo Q-L, Alter M J. Hepatitis C virus antigen in hepatocytes: immunomorphologic detection and identification. Gastroenterology. 1992;103:622–629. doi: 10.1016/0016-5085(92)90856-t. [DOI] [PubMed] [Google Scholar]
- 25.Kuo G, Choo Q-L, Alter H J, Gitnick G L, Redeker A G, Purcell R H, Miyamura T, Dienstag J L, Alter M J, Stevens C E, Tegtmeier G E, Bonino F, Colombo M, Lee W-S, Kuo C, Berger K, Shuster J R, Overby L R, Bradley D W, Houghton M. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- 26.Lin C, Lindenbach B D, Pragai B, McCourt D W, Rice C M. Processing of the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J Virol. 1994;68:5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin C, Pragai B M, Grakoui A, Xu J, Rice C M. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J Virol. 1994;68:8147–8157. doi: 10.1128/jvi.68.12.8147-8157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin C, Rice C M. The hepatitis C virus NS3 proteinase and NS4A cofactor: establishment of a cell-free trans-processing assay. Proc Natl Acad Sci USA. 1995;92:7622–7626. doi: 10.1073/pnas.92.17.7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin C, Thomson J A, Rice C M. A central region in the hepatitis C virus NS4A protein allows formation of an active NS3-NS4A serine proteinase complex in vivo and in vitro. J Virol. 1995;69:4373–4380. doi: 10.1128/jvi.69.7.4373-4380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin C, Wu J-W, Hsiao K, Su M-S. The hepatitis C virus NS4A protein: interactions with the NS4B and NS5A proteins. J Virol. 1997;71:6465–6471. doi: 10.1128/jvi.71.9.6465-6471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love R A, Parge H E, Wickersham J A, Hostomsky Z, Habuka N, Moomaw E W, Adachi T, Hostomska Z. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell. 1996;87:331–342. doi: 10.1016/s0092-8674(00)81350-1. [DOI] [PubMed] [Google Scholar]
- 32.Moradpour D, Englert C, Wakita T, Wands J R. Characterization of cell lines allowing tightly regulated expression of hepatitis C virus core protein. Virology. 1996;222:51–63. doi: 10.1006/viro.1996.0397. [DOI] [PubMed] [Google Scholar]
- 33.Moradpour D, Kary P, Rice C M, Blum H E. Continuous human cell lines inducibly expressing hepatitis C virus structural and nonstructural proteins. Hepatology. 1998;28:192–201. doi: 10.1002/hep.510280125. [DOI] [PubMed] [Google Scholar]
- 34.Moremen K W, Touster O, Robbins P W. Novel purification of the catalytic domain of Golgi alpha-mannosidase II. Characterization and comparison with the intact enzyme. J Biol Chem. 1991;266:16876–16885. [PubMed] [Google Scholar]
- 35.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muramatsu S, Ishido S, Fujita T, Itoh M, Hotta H. Nuclear localization of the NS3 protein of hepatitis C virus and factors affecting the localization. J Virol. 1997;71:4954–4961. doi: 10.1128/jvi.71.7.4954-4961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Precious B, Young D F, Bermingham A, Fearns R, Ryan M, Randall R E. Inducible expression of the P, V, and NP genes of the paramyxovirus simian virus 5 in cell lines and an examination of NP-P and NP-V interactions. J Virol. 1995;69:8001–8010. doi: 10.1128/jvi.69.12.8001-8010.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reed K E, Grakoui A, Rice C M. Hepatitis C virus-encoded NS2-3 protease: cleavage-site mutagenesis and requirements for bimolecular cleavage. J Virol. 1995;69:4127–4136. doi: 10.1128/jvi.69.7.4127-4136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reed K E, Rice C M. Molecular characterization of hepatitis C virus. Curr Stud Hematol Blood Transfus. 1998;62:1–37. doi: 10.1159/000060472. [DOI] [PubMed] [Google Scholar]
- 40.Sakamuro D, Furukawa T, Takegami T. Hepatitis C virus nonstructural protein NS3 transforms NIH 3T3 cells. J Virol. 1995;69:3893–3896. doi: 10.1128/jvi.69.6.3893-3896.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sansonno D, Cornacchiulo V, Iacobelli A R, Di Stefano R, Lospalluti M, Dammacco F. Localization of hepatitis C virus antigens in liver and skin tissues of chronic hepatitis C virus-infected patients with mixed cryoglobulinemia. Hepatology. 1995;21:305–312. [PubMed] [Google Scholar]
- 42.Sansonno D, Dammacco F. Hepatitis C virus c100 antigen in liver tissue from patients with acute and chronic infection. Hepatology. 1993;18:240–245. [PubMed] [Google Scholar]
- 43.Santolini E, Migliaccio G, La Monica N. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol. 1994;68:3631–3641. doi: 10.1128/jvi.68.6.3631-3641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Satoh S, Tanji Y, Hijikata M, Kimura K, Shimotohno K. The N-terminal region of hepatitis C virus nonstructural protein 3 (NS3) is essential for stable complex formation with NS4A. J Virol. 1995;69:4255–4260. doi: 10.1128/jvi.69.7.4255-4260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimizu Y, Yamaji K, Masuho Y, Yokota T, Inoue H, Sudo K, Satoh S, Shimotohno K. Identification of the sequence on NS4A required for enhanced cleavage of the NS5A/5B site by hepatitis C virus NS3 protease. J Virol. 1996;70:127–132. doi: 10.1128/jvi.70.1.127-132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shoji I, Suzuki T, Sato M, Aizaki H, Chiba T, Matsuura Y, Miyamura T. Internal processing of the hepatitis C virus NS3 protein. Virology. 1999;254:315–323. doi: 10.1006/viro.1998.9540. [DOI] [PubMed] [Google Scholar]
- 47.Tanji Y, Hijikata M, Satoh S, Kaneko T, Shimotohno K. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J Virol. 1995;69:1575–1581. doi: 10.1128/jvi.69.3.1575-1581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terasaki M, Song J, Wong J R, Weiss M J, Chen L B. Localization of endoplasmic reticulum in living and glutaraldehyde-fixed cells with fluorescent dyes. Cell. 1984;38:101–108. doi: 10.1016/0092-8674(84)90530-0. [DOI] [PubMed] [Google Scholar]
- 49.Tomei L, Failla C, Santolini E, De Francesco R, La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993;67:4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu J, Mendez E, Caron P R, Lin C, Murcko M A, Collett M S, Rice C M. Bovine viral diarrhea virus NS3 serine proteinase: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J Virol. 1997;71:5312–5322. doi: 10.1128/jvi.71.7.5312-5322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan Y, Li Y, Munshi S, Sardana V, Cole J L, Sardana M, Steinkuehler C, Tomei L, De Francesco R, Kuo L C, Chen Z. Complex of NS3 protease and NS4A peptide of BK strain of hepatitis C virus: a 2.2 Å resolution structure in a hexagonal crystal form. Protein Sci. 1998;7:837–847. doi: 10.1002/pro.5560070402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao N, Hesson T, Cable M, Hong Z, Kwong A D, Le H V, Weber P C. Structure of the hepatitis C virus RNA helicase domain. Nat Struct Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]