Abstract
Human pathogenic retroviruses do not have common loci of integration. However, many factors, such as chromatin structure, transcriptional activity, DNA-protein interaction, CpG methylation, and nucleotide composition of the target sequence, may influence integration site selection. These features have been investigated by in vitro integration reactions or by infection of cell lines with recombinant retroviruses. Less is known about target choice for integration in vivo. The present study was conducted in order to assess the characteristics of cellular sequences targeted for human T-cell leukemia virus type 1 (HTLV-1) integration in vivo. Sequencing integration sites from ≥200 proviruses (19 kb of sequence) isolated from 29 infected individuals revealed that HTLV-1 integration is not random at the level of the nucleotide sequence. The virus was found to integrate in A/T-rich regions with a weak consensus sequence at positions within and without of the hexameric repeat generated during integration. These features were not associated with a preference for integration near active regions or repeat elements of the host chromosomes. Most or all of the regions of the genome appear to be accessible to HTLV-1 integration. As with integration in vitro, integration specificity in vivo seems to be determined by local features rather than by the accessibility of specific regions.
In the course of retrovirus infection, a DNA copy of the viral RNA genome is synthesized, and that DNA is then permanently inserted into the genome of the host cell. The integration of the viral DNA into the host genome is a crucial step in the life cycle: it is important for the efficient expression of progeny virus, and it is responsible for the ability of these viruses to persist and cause disease (36). If most or all of the regions of the host genome are accessible to retroviral integration (14, 36, 46), the frequency of use of potential sites varies considerably. This integration specificity correlates with local DNA structural features, which govern the accessibility of specific regions (37, 50).
Indeed, integration may be favored near DNase I-hypersensitive sites (32, 33) or CpG islands (23), suggesting a preference for transcriptionally active regions (26, 37). Human immunodeficiency virus type 1 (HIV-1) integration preferentially occurs near Alu elements (41) or topoisomerase cleavage sites (19). Recently, centromeric alphoid repeats were found to be selectively absent at HIV-1 integration sites (6). A nonrandom, compartmentalized integration in GC-rich isochores has been previously described for human T-cell leukemia virus type 1 (HTLV-1) (52), bovine leukemia virus (22), hepatitis B virus (51), and Rous sarcoma virus (34), while mouse mammary tumor virus has been found to integrate into GC-poor regions (35) of the host genome. At the nucleotide level, in vitro integration reactions and analysis of the flanking sequence of cloned viruses have shown that there is a preference for integration in A/T-rich regions (15, 17, 27, 39). A consensus sequence at the direct repeat flanking HIV-1 provirus genome has been shown by sequencing integration sites derived from cells infected in vitro (6) (41, 47).
The data summarized above result from in vitro studies, and only a few flanking sequences derived from naturally infected samples have been described to date. HTLV-1 is the causative agent of adult T-cell leukemia/lymphoma (ATLL), an aggressive T-cell malignancy and of tropical spastic paraparesis/HTLV-1 associated myelopathy (TSP/HAM), a chronic progressive neuromyelopathy. A study based on the analysis of four HTLV-1 integration sites derived from uncultured ATLL cells has suggested that the regions flanking the provirus were A/T-rich with a nucleotide composition bias in the 6-bp direct repeat generated by integration (13). In a previous work based on the analysis of 24 distinct HTLV-1 integration sites derived from asymptomatic carriers, the mean A/T content of the hexameric repeat was found to be 59% (49).
An inventory of 218 distinct HTLV-1 integration sites was made from infected individuals. The virus was found to integrate in A/T-rich regions and a weakly conserved sequence was identified at in vivo integration sites. Database analysis revealed that these features were not associated with any preference for integration in transcriptionally active regions or in repeat elements of the host genome.
MATERIALS AND METHODS
Samples studied.
Samples from 29 HTLV-1-infected individuals were analyzed. These samples corresponded to 12 ATLL, 10 TSP/HAM, and 7 asymptomatic carriers. DNA was phenol-chloroform extracted and ethanol precipitated from peripheral blood mononuclear cells (PBMCs), cerebrospinal fluid (CSF), lymph nodes, or skin biopsies.
PCR.
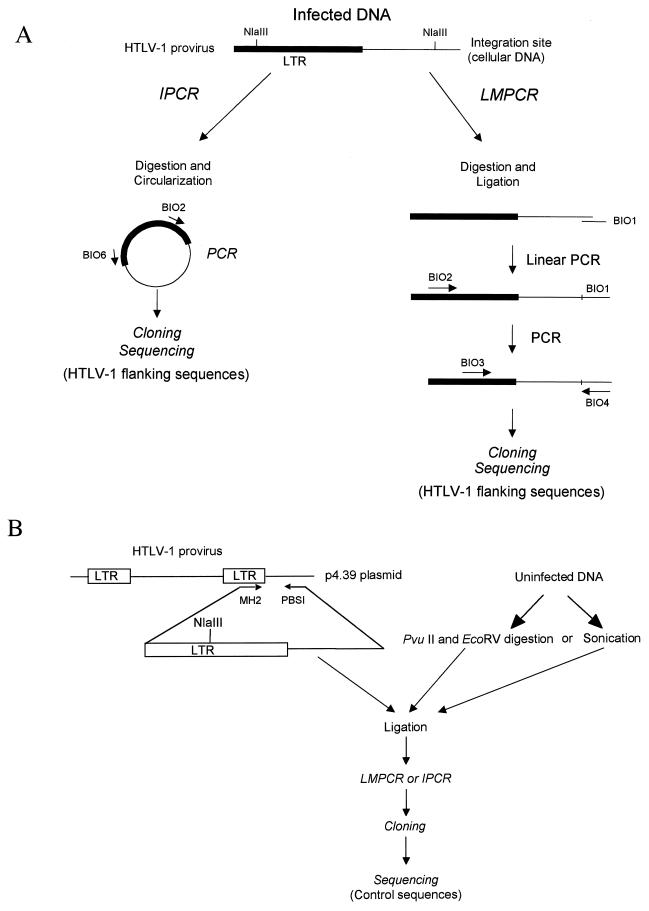
Our strategy is summarized in Fig. 1. DNA was amplified as described by ligation-mediated PCR (LMPCR) or inverse PCR (IPCR) (7–10, 49). Both of these two methods allow the amplification of the HTLV-1 3′ extremities, together with their cellular flanking sequences. For LMPCR, DNA was digested with NlaIII in 1× NlaIII buffer for 3 h at 37°C. Digestion was controlled by gel electrophoresis. DNA was phenol-chloroform extracted and ethanol precipitated. Digested DNA was ligated with BIO1 primer (49) by using T4 DNA ligase. This was followed by two phenol-chloroform extractions and precipitation. Ligated DNA was amplified for 100 cycles with the BIO2 primer alone (49). Conditions were 1× Stoffel DNA polymerase buffer, 1.5 mM MgCl2, 50 pmol BIO2, a 150 μM concentration of each deoxynucleoside triphosphate (dNTP), and 10 U of Stoffel fragment of Taq DNA polymerase (Perkin-Elmer Cetus) in a final volume of 85 μl. First, 25 μl of a 1× PCR buffer containing dNTPs and primers were first loaded into a 750-μl tube, and an Ampliwax PCR Gem 100 (Cetus) was added to each tube. After wax layer formation by incubation at 75°C 10 min and cooling at room temperature for 15 min, 60 μl of the remaining reagent and ligated products were loaded. Thermal cycling parameters were as follows: 1 cycle of 94°C for 10 min and 100 cycles of 95°C for 45 s, 60°C for 45 s, and 72°C for 2 min, followed by a final elongation step of 10 min at 72°C. Ten microliters of this linear PCR reaction was used in a classical PCR amplification with the BIO3 and BIO4 primer pair (49). Amplification conditions were as before, with 40 pmol of each primer and 2.5 U of Taq polymerase all in a final volume of 100 μl. Thermal cycling parameters were as follows: 1 cycle of 94°C for 10 min and 35 cycles of 95°C for 45 s, 58°C for 45 s, and 72°C for 1 min, followed by a final elongation step of 10 min at 72°C.
FIG. 1.
Strategy to construct HTLV-1 flanking sequences and control sequences libraries. (A) Construction of HTLV-1 integration sites library. HTLV-1-infected DNA was amplified by LMPCR or IPCR. Amplified products were cloned and sequenced as detailed in Materials and Methods. (B) A ∼1,500-bp segment spanning the 3′ extremity of the provirus was generated by PCR amplification of a HTLV-1 provirus cloned in p4.39 plasmid (10). High-molecular-weight uninfected human DNA was first sonicated or digested with PvuII and EcoRV which cut blunt ends. Sonicated or digested DNA was then ligated with the 3′ HTLV-1 purified construct as detailed in Materials and Methods. Then, LMPCR, IPCR, cloning, and sequencing were performed in the same conditions as with the infected samples.
For inverse PCR, DNA was first digested by NlaIII as in LMPCR. Digested DNA was circularized with T4 DNA ligase in 600 μl for 16 h at 14°C. This was followed by two phenol-chloroform extractions and precipitation. Circularized DNA was amplified for 40 cycles by using the BIO2 and BIO6 primer pair (10). Amplification conditions and thermal cycling parameter were as follows: 1 cycle of 95°C for 10 min and 40 cycles of 95°C for 45 s, 58°C for 45 s, and 72°C for 1 min, followed by a final elongation step of 10 min at 72°C.
The IPCR and LMPCR thresholds are 100 and 20 copies/sample, respectively (10). Since high proviral loads are invariably associated with ATLL and TSP/HAM, corresponding PBMC samples were analyzed by IPCR, while PBMC samples from asymptomatic carriers and CSF samples from TSP/HAM patients were analyzed by LMPCR. Two to four samples of noninfected DNA served as negative controls in all experiments.
Cloning and sequencing.
PCR products were phosphorylated by T4 polynucleotide kinase and ligated with SmaI-digested and dephosphorylated M13mp18 replicative-form DNA as described earlier (49). After transformation of Escherichia coli XL1 by electroporation, recombinant M13 plaques were transferred in situ to nitrocellulose filters and screened by hybridization with the HTLV-1 long terminal repeat (LTR)-specific 32P-labeled oligonucleotide BIO5 (49). Filters were first prehybridized at 42°C for 2 h in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–1× Denhardt's solution–10 μg of denatured salmon sperm DNA per ml. Hybridization was carried out in a fresh solution at 42°C, and filters were washed in 2× SSC–0.1% sodium dodecyl sulfate at 42°C. Positive plaques were picked and prepared for DNA sequencing. Single-stranded templates were sequenced by using fluorescent dideoxynucleotides. The products were resolved on an Applied Biosystems 377A DNA sequencer with 377 software.
Control experiments.
A control was used in order to compare the nucleotide composition of HTLV-1 flanking sequences with that of the uninfected DNA. As shown in Fig. 1B, a 1,494-bp segment spanning the 3′ extremity of the provirus was first generated by PCR. This was performed by using an integration site-specific primer, PBSI (10), and an LTR-specific primer, MH2 (5′-CCCGCCAATCACTCATACAACC-3′) that encompassed the 3′ extremity of the HTLV-1 provirus cloned in the plasmid p4.39 (10). Amplification conditions and thermal cycling parameters were as follows: 1 cycle of 95°C for 10 min and 35 cycles of 95°C for 1 min, 58°C for 45 s, and 72°C for 2 min, followed by a final elongation step of 10 min at 72°C. Amplified products were purified and treated with the Klenow fragment of the DNA polymerase. As shown in Fig. 1, high-molecular-weight uninfected human DNA that derived from an uninfected blood donor was blunt-end digested with PvuII and EcoRV or sonicated. These three different methods were used in order to avoid any bias in the selection of DNA sequence during fragmentation. Digested or sonicated DNA was then ligated with the purified 3′ HTLV-1 end. Then, LMPCR, IPCR, cloning, and sequencing were performed under the same conditions as with samples that were derived from infected individuals.
Comparison of integration sites with database sequences.
Sequences with integration sites longer than 25 bp were analyzed by comparison to the nonredundant human sequence (nr) database (2), the human cDNA (dbEST) database (3), and the MONTH (January 1999) database (42) by using BLASTN with Search Launcher (1), FASTA (28), and Repeat Masker (See A. F. A. Smit and P. Green, RepeatMasker, at http://ftp.genome.washington.edu/cgi-bin/RepeatMasker) (6). Default parameters were used. The 25-bp cutoff was arbitrary chosen in order to avoid nonspecific homologies that could result from the alignment of shorter sequences with those of databases. A total of 18,118 bp of human DNA corresponding to 178 integration sites that were ≥25 bp were analyzed. For the 44 control sequences of ≥25 bp, 2,991 bp were analyzed. These 21,109 bp corresponded to 96% of the 22,018 bp we have sequenced. The lengths of flanking human DNA sequences analyzed ranged from 25 to 350 bp. The lengths of control sequences analyzed ranged from 25 to 150 bp. Similarities to repeated sequences were ranked in accordance with the Smith-Waterman parameter generated by RepeatMasker or by the probability of matching by chance generated by BLASTN (1) and by FASTA (29).
RESULTS
Construction of control sequences and HTLV-1 integration sites libraries.
The goal of the study was to analyze HTLV-1 integration sites isolated by LMPCR or IPCR. Since PCR and cloning are influenced by target sequence composition and sequence size, a control was performed in order to assess the nucleotide composition of the uninfected human genome by using the same experimental procedure as that used in the analysis of HTLV-1 integration sites (see Materials and Methods and Fig. 1). This was done by sequencing flanking sequences of an HTLV-1 construct ligated in uninfected DNA. The three different methods used in the control DNA fragmentation process (PvuII digestion, EcoRV digestion, and sonication) ruled out any bias in the nucleotide composition of the control sequences. HTLV-1 flanking sequences isolated from infected individuals were subsequently compared to that of the control library.
Sixty distinct control molecular clones were sequenced. The mean sequence size was 53 bp, ranging from 12 to 150 bp (median, 37 bp). A total of 218 distinct HTLV-1 integration sites were sequenced. The mean sequence size was 84 bp (median, 69 bp; range, 6 to 365 bp). Totals of 75, 116, and 27 sequences were derived from asymptomatic carriers, TSP/HAM, and ATLL, respectively. We obtained 1 to 44 distinct integration sites (mean, 8) from each infected individual.
LMPCR products were found to be shorter than IPCR products. In addition, a weak correlation was observed between the nucleotide content and the length of flanking sequences, the longest being the more A/T-rich. Based on these results, all sequence comparisons were adjusted for size.
HTLV-1 flanking sequences are A/T-rich.
The A/T content was 51% (median, 50%; standard deviation [SD], 11%; range, 30 to 80%) after control experiments, a value significantly lower than that initially described for the human DNA (58%) (43). DNA digestion, amplification, and cloning may have accounted for the selection of such sequences. Based on these results, we performed a comparative analysis of HTLV-1 integration sites and control sequences.
For the control sequences, the percentages of A, C, G, and T nucleotides were 25, 26, 23, and 26%, respectively. The overall A/T content of HTLV-1 integration sites was 57% (median, 57%; SD, 9%; range, 33 to 79%), a value significantly higher than that of control sequences (P = 10−4, independent samples t test). The percentages of A, C, G, and T nucleotides were 29, 22, 21, and 28%, respectively. Only the A (P = 0.001) and C (P = 0.001) contents were found to be significantly different between HTLV-1 and control sequences. All of these differences remained significant when HTLV-1 and control sequences were adjusted for size. The A/T content of the hexameric repeat was 56% (median, 50%; SD, 20%; range, 17 to 100%). The percentages of A, C, G, and T nucleotides of the HTLV-1 flanking hexameric repeats were 30, 24, 20, and 26%, respectively. The T content without the hexameric repeats were 30, 24, 20, and 26%, respectively. The T content without the hexameric repeat was significantly higher than that within, i.e., 28 versus 25% (P = 0.039, paired samples t test). There was no significant difference in the distribution of the remaining nucleotides within and without the hexameric repeat. The A/T contents of flanking sequences isolated from ATLL, TSP/HAM, and carriers were 57, 59, and 55%, which were not significantly different values.
Nonhomogenous distribution of the nucleotide composition within and without the hexameric repeat.
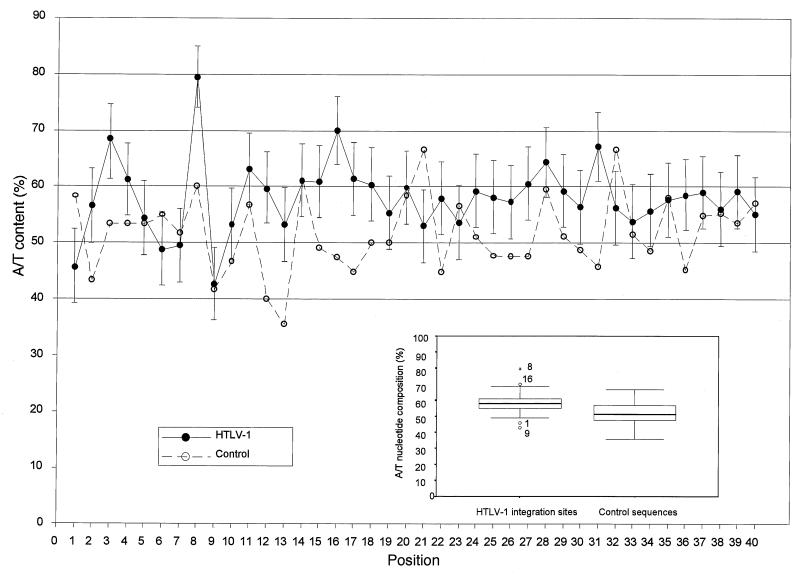
Figure 2 represents the distribution of A/T nucleotides along the 40 first positions of both the HTLV-1 integration sites and control sequences. Deletion of the 3′ LTR sequence was performed prior to sequence analysis. In addition, deletion of the first 31 bases of the p4.39 integration site, together with the 3 bases that correspond to PvuII and EcoRV restriction sites, was also performed when corresponding control sequences were studied. For the box plot analysis, points more than 1.5 times the interquartile range from the ends of the box were labeled as outliers. Points more than 3 times the interquartile range from the ends of the box were labeled as extreme values (see legend of Fig. 2).
FIG. 2.
Distribution of A/T nucleotides at each of the first 40 positions of the flanking sequences. The 218 HTLV-1 integration sites (●) and the 60 control sequences (○) were aligned, as described in the text, relative to the first base of the flanking sequences 3′ to the provirus. The standard deviation of the A/T content is shown at each position of the HTLV-1 flanking sequences. The insert shows the distribution of the AT content along the flanking sequences. The boxes represent the difference between the 75th percentile and the 25th percentile of the AT distribution (i.e., the interquartile range). Within the inset box, the median is represented by a thick horizontal line. Lines from the ends of the box extend as far as the most extreme values not considered outliers. Points more than 1.5 times the interquartile range from the ends of the box are labeled as outliers (○) or as extreme values (∗).
Figure 2 shows that the A/T content of HTLV-1 integration sites was higher than that of control sequences in 31 of 40 positions (P < 10−4). In addition, positions 8 and 16 were found to be preferentially occupied by A or T residues (79 and 70%, respectively), while positions 1 and 9 were preferentially occupied by G or C residues (54 and 57%, respectively). These four positions corresponded to outliers (positions 1, 9, and 16) or extreme values (position 8) after box plot analysis. The distribution of A/T residues was monotonous at the remaining positions. As shown in Fig. 2, no extreme value was observed along the control sequences. Finally, the box plot analysis revealed that the A/T content spread of the integration sites library was narrower than that of the control library. However, such difference in the spread of the A/T content might be explained in part by the small sample size of the control library.
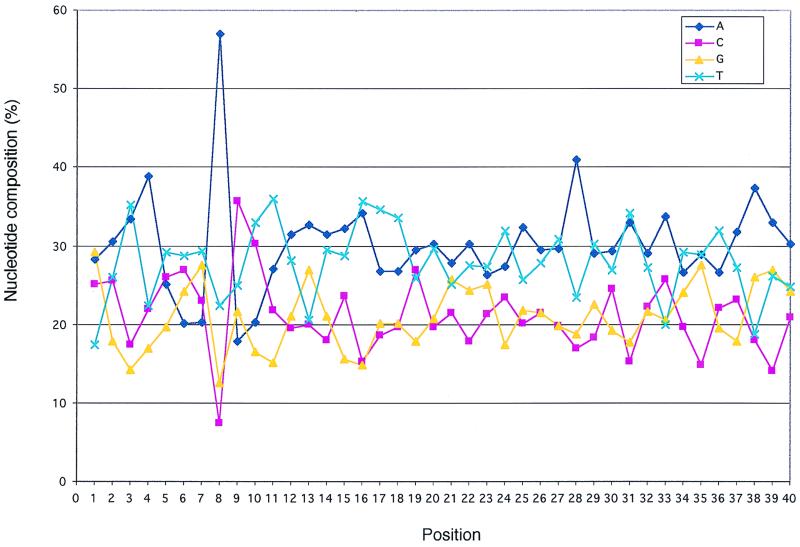
Figure 3 shows the distribution of each residue along the first 40 positions of HTLV-1 integration sites. The figure shows that the significant C/G richness observed in Fig. 2 at position 1 of HTLV-1 integration sites resulted from an excess of G residues associated with a lack of T residues. The A/T richness at position 8 resulted from a large excess of A residues, while the C/G richness at position 9 corresponded to a significant excess of C residues combined with a lack of A residues. The A/T richness at position 16 resulted from a combination of A- and T-rich sequences. In addition, Fig. 3 shows that there was an excess of A residues at positions 4 and 28. However, only the A and C contents at positions 8 and 9 and the T content at position 1 were detected as outliers after box plot analysis (not shown). No additional extreme position was noted along HTLV-1 integration sites. The percentage of HTLV-1 and control sequences harboring at least three of the four identified significant hot spots (C/G, A, C, and A/T at positions 1, 8, 9, and 16, respectively) were 34.5 and 15.3%, respectively (P < 10−4).
FIG. 3.
Nucleotide composition of the HTLV-1 flanking sequences. Sequences isolated after cloning LMPCR and IPCR products were aligned relative to the first base of the hexameric repeat sequences flanking the provirus.
An additional characteristic of the HTLV-1 library was the presence of integration sites harboring long A/T stretches. Indeed, 17% of the 194 HTLV-1 flanking sequences that were longer than 10 bp harbored A/T stretches of ≥6 bp compared to only 2% of the control sequences (P = 0.002). By contrast, the frequencies of C/G and CpG long stretches were not significantly different between the two libraries (0.5 versus 1% and 3 versus 2%, respectively).
Identification of the host sequences targeted for HTLV-1 integration in vivo.
The matches to known sequences are summarized in Table 1, while Table 2 shows the distribution of HTLV-1 3′ integration sites and control sequences with significant homologies to known nonrepetitive elements from databases. The sequences were classified as genomic noncoding nonrepetitive elements; most of these corresponded to GenBank high-throughput genomic sequences, transcription units, and repeat elements. Overall, the mean size of anonymous sequences was not significantly different than that of other sequences (100 versus 99 bp). As shown in Table 1, 84 (47%) of the 178 HTLV-1 sequences of ≥25 bp were found to match to known sequences. Of the 44 control sequences of ≥25 bp, 17 (39%) matched known sequences. For both libraries, the frequency of matches to repeat sequences was significantly higher than that for transcription units. Table 2 also shows that there was no significant difference in the distribution of matches between the HTLV-1 integration sites and the control sequences. In addition, there was no correlation between the clinical status accompanying to the DNA samples and the type of matches (not shown).
TABLE 1.
Distribution of the number and the percentage of matches to known sequences in HTLV-1 integration sites and control sequences longer than 25 bp
| Sequence type | No. of HTLV-1 integration sites (%) | No. of control sequences (%) |
|---|---|---|
| Genomic sequences | 15 (8) | 1 (2) |
| Transcription units | 10 (6) | 3 (7) |
| cDNA | 8 (5) | 3 (7) |
| Genes | 2 (1) | 0 (0) |
| Repeat elements | 59 (33) | 13 (30) |
| SINE elements | 20 (11) | 6 (14) |
| LINE elements | 20 (11) | 3 (7) |
| RLE elements | 11 (6) | 3 (7) |
| DNA transposon | 1 (1) | 1 (2) |
| DNA satellite | 3 (2) | 0 (0) |
| Simple repeat | 4 (2) | 0 (0) |
| Anonymous | 94 (53) | 27 (61) |
| Total of matched sequences | 84 (47) | 17 (39) |
TABLE 2.
Integration sites and control sequences having significant homologies to known nonrepetitive elements
| Group and sequence no. | Sample origin | Length (bp) | Hexameric repeat | AT (%) | Identified similarities
|
|
|---|---|---|---|---|---|---|
| Type | Accession no.b | |||||
| ATLL | ||||||
| 78 | Tumorousa | 200 | TTATTC | 60 | cDNA | HSU68704 |
| 84 | Nontumorous | 141 | AGCAAG | 66 | cDNA | N53238 |
| 74 | Tumourous | 99 | TCTTTC | 60 | Gene (human P protein, exon 23) | HSPPROT23 |
| 85 | Nontumorous | 28 | CCTCTC | 68 | Noncoding nonrepetitive | AL034422 |
| 91 | Tumorous | 36 | CAGCTG | 50 | Noncoding nonrepetitive | AQ223501 |
| Asymptomatic carriers | ||||||
| 10 | PBMCs | 25 | TCCGCA | 36 | cDNA | AA451666 |
| 18 | PBMCs | 32 | ACCCGC | 41 | cDNA | AA631969 |
| 27 | PBMCs | 25 | GCAACT | 60 | cDNA | AC002036 |
| 39 | PBMCs | 25 | GCAAAA | 52 | cDNA | AC004583 |
| 58 | PBMCs | 45 | TTATGT | 69 | Noncoding nonrepetitive | AC006054 |
| 23 | PBMCs | 34 | GTTATA | 71 | Noncoding nonrepetitive | AC006227 |
| 34 | PBMCs | 25 | GAGAAC | 52 | Noncoding nonrepetitive | AQ306143 |
| 37 | PBMCs | 101 | CTGTGG | 59 | Noncoding nonrepetitive | B17581 |
| 20 | PBMCs | 38 | GGTGTG | 42 | Noncoding nonrepetitive | HS232D4 |
| TSP/HAM | ||||||
| 233 | PBMCs | 26 | TAATAG | 62 | cDNA | H72803 |
| 231 | PBMCs | 69 | CTTGGT | 51 | cDNA | W57727 |
| 224 | PBMCs | 365 | GCTAGG | 61 | Gene (alpha enolase, exon 1) | HSENOAL1 |
| 220 | PBMCs | 29 | CATATG | 55 | Noncoding nonrepetitive | AC003693 |
| 216 | CSF | 25 | GCTAGG | 48 | Noncoding nonrepetitive | AC004505 |
| 183 | PBMCs | 163 | ACATTT | 59 | Noncoding nonrepetitive | AC005881 |
| 274 | PBMCs | 52 | CTGAGG | 44 | Noncoding nonrepetitive | AL021937 |
| 275 | PBMCs | 75 | TCAGTC | 55 | Noncoding nonrepetitive | AL022345 |
| 223 | PBMCs | 144 | GAGAAT | 70 | Noncoding nonrepetitive | AL031599 |
| 246 | PBMCs | 30 | TCAATC | 67 | Noncoding nonrepetitive | AL031683 |
| 204 | CSF | 41 | TAAAGT | 78 | Noncoding nonrepetitive | HUAC002307 |
| Control sequences | ||||||
| 109 | 64 | 58 | cDNA | AI001768 | ||
| 137 | 25 | 72 | cDNA | AA969105 | ||
| 151 | 30 | 43 | cDNA | AA909212 | ||
| 110 | 45 | 42 | Noncoding nonrepetitive | HS431P23 | ||
Tumorous clones correspond to malignant ATLL cellular clones.
GenBank accession number of the corresponding database sequences (2).
Centromeric alphoid repeats have been previously found to be unfavorable targets for HIV-1 integration in a cell line (6). In the present study, three HTLV-1 integration sites showed significant homologies with such satellite DNA sequences. The first clone corresponded to alpha-satellite DNA located on chromosomes 13, 14, and 21 (48). The second clone was homologous to a human middle repetitive DNA sequence (P. P. Ratnasinghe and P. R. Musich, unpublished data), while the third clone corresponded to beta-satellite DNA located on the distal and proximal short arms of the human acrocentric chromosomes (18). No satellite DNA was observed in the control library.
The A/T contents of anonymous sequences, repeat elements, genomic noncoding nonrepetitive elements, and transcription units were 59, 55, 58, and 55%, respectively. These differences were statistically significant (P = 0.0063, Kruskal-Wallis one-way analysis-of-variance). Among repeat elements, the A/T content of LINE elements was significantly higher than that of the SINE element, i.e., 60 versus 52% (P < 10−4). The A/T contents of retrovirus-like elements (RLE) and other repeat elements were 53 and 55%, respectively. There was a weak correlation between the A/T content of identified database sequences and their frequencies of match with HTLV-1 integration sites or control sequences. Indeed, the proportion of integration sites matching with the A/T-rich LINE elements was higher than that for control sequences: 11 versus 7% (Table 1). Similarly, the proportion of integration site matching with the C/G-rich LINE elements was lower than that for control sequences: 11 versus 14% (Table 1). However, these differences were not statistically significant.
DISCUSSION
In the infected cell, integrase functions as a component of the preintegration complex derived from the virus core. The main factors which may influence the choice for a chromosomal target site during infection include chromatin structure (26), target DNA sequence (15), host cell proteins, and virus-encoded proteins other than integrase (4). The present study was conducted in order to assess the selection of target sites during integration of the HTLV-1 provirus in vivo. To this end, HTLV-1 integration sites, isolated by IPCR or LMPCR amplification from naturally infected individuals, were sequenced. The nucleotide composition of the flanking sequences was analyzed, and their homologies to known sequences were identified. The nature of the HTLV-1 flanking sequences was compared to that of uninfected DNA. Since PCR and cloning are influenced by the structure and the nucleotide content of the target sequence, we designed a control experiment in order to isolate uninfected DNA sequences by using a strategy identical to that used for the analysis of HTLV-1 flanking sequences. The A/T content was found to be 51% after this control experiment, a value significantly lower than that initially described for human DNA (58%) (43). This indicates that the A/T content is underestimated by IPCR and LMPCR and emphasizes the need to perform a comparative analysis of HTLV-1 integration sites with sequences isolated by the same manner. Results from the comparative analysis show that there is a nucleotide composition bias at HTLV-1 integration sites. The virus integrates into A/T-rich regions with a nonhomogeneous distribution of residues 3′ to the provirus. These characteristics do not appear to be associated with a preference for integration in transcription units or repeat elements.
Preference for A/T-rich regions as a common feature for in vivo and in vitro integration.
HTLV-1 flanking sequences (∼19 kb) isolated from naturally infected individuals were found to be significantly more A/T-rich than control sequences. A preference for A/T-rich regions has been previously described for insertion by transposons (36) or Ty elements (28) and for integration of adenovirus (24), spleen necrosis virus (40), avian myeloblastosis virus (16, 17) and Moloney murine leukemia virus (31). By hybridization of a viral probe with compositional fractions of HTLV-1 cultured cells DNA, HTLV-1 sequences have been found to be distributed in the 46 to 61% A/T range of the host genome (52). Recently, four HTLV-1 flanking sequences (183 bp) isolated from four ATLL patients were found to have an average A/T content of 63% (13). Altogether, these data indicate that a preference for A/T-rich regions characterizes both in vitro and in vivo integration.
The target DNA structure affects HTLV-1 integration site selection.
Integration of proviral DNA into the host genome generates short direct repeats of 4 to 6 bp as a result of DNA repair to the cellular sequence flanking the integrated provirus (45). Analysis of the nucleotide sequence surrounding the integrated provirus indicates that the size of the direct repeat is characteristic of each virus. For MMLV, the middle two positions of the 4-bp direct repeat are preferentially occupied by AA, TT, or AT dinucleotides (31). Similarly, a preferential AT pairing toward the central portion of the direct repeat has been previously described in direct repeat generated by the insertion of yeast Ty elements (28) and in the target of site-specific recombination by FLP recombinase (44). For HIV, analysis of the 112 published direct repeats shows that there is a preference for a G residue at the first position, for T at the second position, for A at the third and fourth positions, and for C at the fifth position (6, 41, 47). Here, alignment of the 218 HTLV-1 clones showed a weak consensus sequence within the hexameric repeat flanking the HTLV-1 provirus. However, only the C/G content at position 1 of the hexameric repeat corresponded to an outlier as shown in Fig. 2. In fact, we have found a significant nucleotide composition bias at positions within and without the hexameric repeat. Indeed, there was a preference for C or G residues at position 1, for A residues at position 8, for C residues at position 9, and for A or T residues at position 16. These weakly conserved motifs observed in the vicinity of the provirus 3′ end may constitute a propitious binding site for the preintegration complex. Alternatively, they might correspond to binding sites for other cellular or viral factors that interact favorably with the integration machinery.
DNA bending creates favored sites for retroviral integration (4, 27). Such distortion may result from DNA-protein interaction (4). Alternatively, the target sequence itself may result in intrinsic DNA bent (27). In the present study, the presence of long runs of A/T residues was almost restricted to HTLV-1 integration sites compared to control sequences. Indeed, some of the HTLV-1 flanking sequences isolated here were found to have strong homologies with intrinsically bent A-tract DNA (21). Therefore, in addition to a preference for A/T-rich regions of the host cell chromosomes, the target choice for HTLV-1 integration appears to be influenced by the target DNA structure. In addition to an intrinsically bent target, it is possible that the consensus motifs observed along HTLV-1 integration sites may contribute to the binding of proteins that induce DNA bents.
Lack of evidence for favored HTLV-1 integration near known sequences.
Nucleotide composition of the database sequences with significant homologies with HTLV-1 integration sites reflected the nature of the corresponding matches. Indeed, transcription unit sequences harbored the lowest A/T range, which correspond to DNA regions of high gene concentration. However, there was no significant difference in the frequency and distribution of matches to known sequences between HTLV-1 and control sequences. In contrast to results from a previous analysis of HIV-1 integration in SupT1 cells (6), we found that HTLV-1 can integrate into alphoid repeats in vivo. The frequency of HTLV-1 integration in repeat element was 33%, a value similar to that previously described for HIV integration in cultured cells (6). The frequency of matches to known transcription units was not significantly different between HTLV-1 and control sequences. This suggests that transcriptionally active regions are not preferred targets for HTLV-1 integration in vivo.
HTLV-1 integration may alter gene expression in vivo.
In contrast to Ty3 elements (5, 11, 12), Ty1 elements (20), and retroviruses that induce chronic leukemia in animals (25), HTLV-1 integration does not preferentially occur near cellular sequences such as proto-oncogenes (38). However, about 6% of the HTLV-1 proviruses analyzed in the present work were found to be integrated into transcription units. This suggests that in some cells, HTLV-1 integration may alter gene expression in vivo. Given the elevated proviral load in HTLV-1-infected individuals, a substantial number of clonally expanded cells must have disrupted transcription units (6% of a large number is substantial). It is possible that some of these events could contribute further to clonal expansion.
Basis of HTLV-1 target site selection in vivo.
HTLV-1 replicates mainly via the mitosis of its host cells (49) and the number of distinct circulating clones of infected CD4 T cells reflects the number of integration events. In a previous work, the overall number of such events has been estimated in an asymptomatic carrier in whom 20 distinct clones of infected cells were evidenced (10). Although this number is higher in TSP/HAM and in ATLL (7, 9), the present collection of HTLV-1 flanking sequences appears to correspond to a representative sample of in vivo HTLV-1 integration events.
At the level of nucleotide sequence, the present study shows that HTLV-1 integration is not completely random in vivo. The differences observed between HTLV-1 integration sites and control sequences indicate that the integration machinery shows subtle preferences in vivo. Given the huge excess of potential sites, the observed subset may reflect those with a kinetic advantage in recognition, which is influenced by the sequence within and without the hexanucleotide. Present results suggest that the structure of the target DNA plays an important role for integration in vivo. However, the structural constraints evidenced here among the 218 HTLV-1 flanking sequences do not appear to be linked to a preference for integration in specific regions of host cell chromosomes. Therefore, most or all regions of the host cell genome seem to be accessible to HTLV-1 integration in vivo.
ACKNOWLEDGMENTS
This work was supported by grants from the Association pour la Recherche sur le Cancer, from the Ligue Nationale contre le Cancer (Comité Pas de Calais), and from the Fondation Contre la Leucémie. I.L. and F.M. were supported by bursaries from the Ministère de l'Enseignement Supérieur et de la Recherche.
We thank P. Wattre and collaborators, who kindly received us in their laboratories for DNA extraction, digestion, ligation, and PCR. We also thank Marie-Dominique Reynaud for assistance.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bilofsky H S, Burks C. The GenBank genetic sequence data bank. Nucleic Acids Res. 1988;16:1861–1863. doi: 10.1093/nar/16.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boguski M S, Lowe T M, Tolstoshev C M. dbEST database for “expressed sequence tags.”. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 4.Bor Y C, Bushman F D, Orgel L E. In vitro integration of human immunodeficiency virus type 1 cDNA into targets containing protein-induced bends. Proc Natl Acad Sci USA. 1995;92:10334–10338. doi: 10.1073/pnas.92.22.10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodeur G M, Sandmeyer S B, Olson M V. Consistent association between sigma elements and tRNA genes in yeast. Proc Natl Acad Sci USA. 1983;80:3292–3296. doi: 10.1073/pnas.80.11.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carteau S, Hoffmann C, Bushman F. Chromosome structure and human immunodeficiency virus type 1 cDNA integration: centromeric alphoid repeats are a disfavored target. J Virol. 1998;72:4005–4014. doi: 10.1128/jvi.72.5.4005-4014.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavrois M, Gessain A, Wain-Hobson S, Wattel E. Proliferation of HTLV-1 infected circulating cells in vivo in all asymptomatic carriers and patients with TSP/HAM. Oncogene. 1996;12:2419–2423. [PubMed] [Google Scholar]
- 8.Cavrois M, Leclercq I, Gout O, Gessain A, WainHobson S, Wattel E. Persistent oligoclonal expansion of human T-cell leukemia virus type 1 infected circulating cells in patients with Tropical spastic paraparesis/HTLV-1 associated myelopathy. Oncogene. 1998;17:77–82. doi: 10.1038/sj.onc.1201906. [DOI] [PubMed] [Google Scholar]
- 9.Cavrois M, Wain-Hobson S, Gessain A, Plumelle Y, Wattel E. Adult T-cell leukemia/lymphoma on a background of clonally expanding HTLV-1 positive cells. Blood. 1996;88:4646–4650. [PubMed] [Google Scholar]
- 10.Cavrois M, Wain-Hobson S, Wattel E. Stochastic events in the amplification of HTLV-I integration sites by linker-mediated PCR. Res Virol. 1995;146:179–184. doi: 10.1016/0923-2516(96)80578-4. [DOI] [PubMed] [Google Scholar]
- 11.Chalker D L, Sandmeyer S B. Transfer RNA genes are genomic targets for de Novo transposition of the yeast retrotransposon Ty3. Genetics. 1990;126:837–850. doi: 10.1093/genetics/126.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chalker D L, Sandmeyer S B. Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev. 1992;6:117–128. doi: 10.1101/gad.6.1.117. [DOI] [PubMed] [Google Scholar]
- 13.Chou K S, Okayama A, Su I J, Lee T H, Essex M. Preferred nucleotide sequence at the integration target site of human T-cell leukemia virus type I from patients with adult T-cell leukemia. Int J Cancer. 1996;65:20–24. doi: 10.1002/(SICI)1097-0215(19960103)65:1<20::AID-IJC4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 14.Craigie R. Hotspots and warm spots: integration specificity of retroelements. Trends Genet. 1992;8:187–190. doi: 10.1016/0168-9525(92)90223-q. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald M L, Grandgenett D P. Retroviral integration: in vitro host site selection by avian integrase. J Virol. 1994;68:4314–4321. doi: 10.1128/jvi.68.7.4314-4321.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fitzgerald M L, Vora A C, Zeh W G, Grandgenett D P. Concerted integration of viral DNA termini by purified avian myeloblastosis virus integrase. J Virol. 1992;66:6257–6263. doi: 10.1128/jvi.66.11.6257-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grandgenett D P, Inman R B, Vora A C, Fitzgerald M L. Comparison of DNA binding and integration half-site selection by avian myeloblastosis virus integrase. J Virol. 1993;67:2628–2636. doi: 10.1128/jvi.67.5.2628-2636.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greig G M, Willard H F. Beta satellite DNA: characterization and localization of two subfamilies from the distal and proximal short arms of the human acrocentric chromosomes. Genomics. 1992;12:573–580. doi: 10.1016/0888-7543(92)90450-7. [DOI] [PubMed] [Google Scholar]
- 19.Howard M T, Griffith J D. A cluster of strong topoisomerase II cleavage sites is located near an integrated human immunodeficiency virus. J Mol Biol. 1993;232:1060–1068. doi: 10.1006/jmbi.1993.1460. [DOI] [PubMed] [Google Scholar]
- 20.Ji H, Moore D P, Blomberg M A, Braiterman L T, Voytas D F, Natsoulis G, Boeke J D. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell. 1993;73:1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- 21.Kahn J D, Crothers D M. Protein-induced bending and DNA cyclization. Proc Natl Acad Sci USA. 1992;89:6343–6347. doi: 10.1073/pnas.89.14.6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kettmann R, Meunier-Rotival M, Cortadas J, Cuny G, Ghysdael J, Mammerickx M, Burny A, Bernardi G. Integration site of bovine leukemia virus DNA in the bovine genome. Arch Int Physiol Biochim. 1979;87:818–819. [PubMed] [Google Scholar]
- 23.Kitamura Y, Lee Y M, Coffin J M. Nonrandom integration of retroviral DNA in vitro: effect of CpG methylation. Proc Natl Acad Sci USA. 1992;89:5532–5536. doi: 10.1073/pnas.89.12.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knoblauch M, Schrer J, Schmitz B, Doerfler W. The structure of adenovirus type 12 DNA integration sites in the hamster cell genome. J Virol. 1996;70:3788–3796. doi: 10.1128/jvi.70.6.3788-3796.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kung H J, Boerkoel C, Carter T H. Retroviral mutagenesis of cellular oncogenes: a review with insights into the mechanisms of insertional activation. Curr Top Microbiol Immunol. 1991;171:1–25. doi: 10.1007/978-3-642-76524-7_1. [DOI] [PubMed] [Google Scholar]
- 26.Mooslehner K, Karls U, Harbers K. Retroviral integration sites in transgenic Mov mice frequently map in the vicinity of transcribed DNA regions. J Virol. 1990;64:3056–3058. doi: 10.1128/jvi.64.6.3056-3058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Müller H P, Varmus H E. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 1994;13:4704–4714. doi: 10.1002/j.1460-2075.1994.tb06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oyen T B, Gabrielsen O S. Non-random distribution of the Ty1 elements within nuclear DNA of Saccharomyces cerevisiae. FEBS Lett. 1983;161:201–206. doi: 10.1016/0014-5793(83)81007-2. [DOI] [PubMed] [Google Scholar]
- 29.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pruss D, Bushman F D, Wolffe A P. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc Natl Acad Sci USA. 1994;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pryciak P M, Varmus H E. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell. 1992;69:769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- 32.Robinson H L, Gagnon G C. Patterns of proviral insertion and deletion in avian leukosis virus-induced lymphomas. J Virol. 1986;57:28–36. doi: 10.1128/jvi.57.1.28-36.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohdewohld H, Weiher H, Reik W, Jaenisch R, Breindl M. Retrovirus integration and chromatin structure: Moloney murine leukemia proviral integration sites map near DNase I-hypersensitive sites. J Virol. 1987;61:336–343. doi: 10.1128/jvi.61.2.336-343.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rynditch A, Kadi F, Geryk J, Zoubak S, Svoboda J, Bernardi G. The isopycnic, compartmentalized integration of Rous sarcoma virus sequences. Gene. 1991;106:165–172. doi: 10.1016/0378-1119(91)90196-i. [DOI] [PubMed] [Google Scholar]
- 35.Salinas J, Zerial M, Filipski J, Crepin M, Bernardi G. Nonrandom distribution of MMTV proviral sequences in the mouse genome. Nucleic Acids Res. 1987;15:3009–3022. doi: 10.1093/nar/15.7.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandmeyer S B, Hansen L J, Chalker D L. Integration specificity of retrotransposons and retroviruses. Annu Rev Genet. 1990;24:491–518. doi: 10.1146/annurev.ge.24.120190.002423. [DOI] [PubMed] [Google Scholar]
- 37.Scherdin U, Rhodes C, Breindl M. Transcriptionally active genome regions are preferred targets for retrovirus integration. J Virol. 1990;64:907–912. doi: 10.1128/jvi.64.2.907-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seiki M, Eddy R, Shows T, Yoshida M. Nonspecific integration of the HTLV provirus genome into adult T-cell leukemia cells. Nature. 1984;309:640–642. doi: 10.1038/309640a0. [DOI] [PubMed] [Google Scholar]
- 39.Shih C C, Stoye J P, Coffin J M. Highly preferred targets for retrovirus integration. Cell. 1988;53:531–537. doi: 10.1016/0092-8674(88)90569-7. [DOI] [PubMed] [Google Scholar]
- 40.Shimotohno K, Temin H M. No apparent nucleotide sequence specificity in cellular DNA juxtaposed to retrovirus proviruses. Proc Natl Acad Sci USA. 1980;77:7357–7361. doi: 10.1073/pnas.77.12.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevens S W, Griffith J D. Sequence analysis of the human DNA flanking sites of human immunodeficiency virus type 1 integration. J Virol. 1996;70:6459–6562. doi: 10.1128/jvi.70.9.6459-6462.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoesser G, Tuli M A, Lopez R, Sterk P. The EMBL nucleotide sequence database. Nucleic Acids Res. 1999;27:18–24. doi: 10.1093/nar/27.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swartz M N, Trautner T A, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XI. Further studies on nearest neighbor base sequences in deoxyribonucleic acid. J Biol Chem. 1962;237:1961–1967. [PubMed] [Google Scholar]
- 44.Umlauf S W, Cox M M. The functional significance of DNA sequence structure in a site-specific genetic recombination reaction. EMBO J. 1988;7:1845–1852. doi: 10.1002/j.1460-2075.1988.tb03017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varmus H E. Using retroviruses as insertional mutagens to identify cellular oncogenes. Prog Clin Biol Res. 1983;119:23–35. [PubMed] [Google Scholar]
- 46.Varmus H E, Brown P O. Mobile DNA. In: Howe M M, Berg D E, editors. Retroviruses. Washington, D.C.: American Society of Microbiology; 1989. pp. 53–108. [Google Scholar]
- 47.Vincent K A, York Higgins D, Quiroga M, Brown P O. Host sequences flanking the HIV provirus. Nucleic Acids Res. 1990;18:6045–6047. doi: 10.1093/nar/18.20.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vissel B, Nagy A, Choo K H. A satellite III sequence shared by human chromosomes 13, 14, and 21 that is contiguous with alpha satellite DNA. Cytogenet Cell Genet. 1992;61:81–86. doi: 10.1159/000133374. [DOI] [PubMed] [Google Scholar]
- 49.Wattel E, Vartanian J P, Pannetier C, Wain-Hobson S. Clonal expansion of human T-cell leukemia virus type I-infected cells in asymptomatic and symptomatic carriers without malignancy. J Virol. 1995;69:2863–2868. doi: 10.1128/jvi.69.5.2863-2868.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Withers-Ward E S, Kitamura Y, Barnes J P, Coffin J M. Distribution of targets for avian retrovirus DNA integration in vivo. Genes Dev. 1994;8:1473–1787. doi: 10.1101/gad.8.12.1473. [DOI] [PubMed] [Google Scholar]
- 51.Zerial M, Salinas J, Filipski J, Bernardi G. Genomic localization of hepatitis B virus in a human hepatoma cell line. Nucleic Acids Res. 1986;14:8373–8386. doi: 10.1093/nar/14.21.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zoubak S, Richardson J H, Rynditch A, Hollsberg P, Hafler D A, Boeri E, Lever A M, Bernardi G. Regional specificity of HTLV-I proviral integration in the human genome. Gene. 1994;143:155–163. doi: 10.1016/0378-1119(94)90091-4. [DOI] [PubMed] [Google Scholar]