Abstract
The mammalian nucleotide-binding domain leucine-rich repeat containing (NLR, NBD-LRR or NOD-like receptor) family was reported over 20 years ago, although several genes later grouped in the family were already known. While it is widely known that NLRs include inflammasome receptors/sensors leading to the maturation of caspase 1, IL-1β, IL-18 and gasdermin D to drive inflammation and cell death, the other functions of NLR family members are much less known to the scientific community. Examples include CIITA, a master transcriptional activator of MHC class II genes, which is the first mammalian NBD-LRR containing protein identified and NLRC5, which regulates MHC class I genes. Other NLRs regulate key inflammatory signalling pathways or interferon responses and several NLR family members serve as check points of innate immunity by serving as negative regulators. Multiple NLRs regulate the balance of cell death, cell survival, autophagy, mitophagy and even cellular metabolism. The least mentioned is a group of NLRs that play important non-immune functions to affect the mammalian reproductive system. The focus of this review is to provide a synopsis of the NLR family, both the intensively-studied and the underappreciated members. We will focus on their function, structure and disease relevance and include issues that have received less attention in the NLR field that may serve as an impetus for future research on their conventional and nonconventional roles within and beyond the immune system.
Introduction
The initial description of the human NLR family was preceded by the identification of a related family in plants (Fig. 1C). The plant nucleotide-binding sequence leucine-rich repeat (NBS-LRR)-containing proteins are the largest group of plant disease resistance (R) proteins1. These proteins contain a nucleotide-binding domain (NBD) and LRR domain with variable C- and N-terminal domains. The majority of plant NBS-LRR proteins have either a TIR (Toll/ IL-1 receptor) domain — referred to as TNLs (TIR-NBS-LRRs) — or coiled-coil (CC) domains — referred to as CNLs (CC-NBS-LRRs) — and these domains are considered important for protein–protein interaction. NBS-LRR proteins are critical for host defense against viruses, bacteria, nematodes, fungi, oomycetes and insects2,3.
Figure 1: Nucleotide-binding and oligomerizing sensors as a universal strategy for cellular defense.
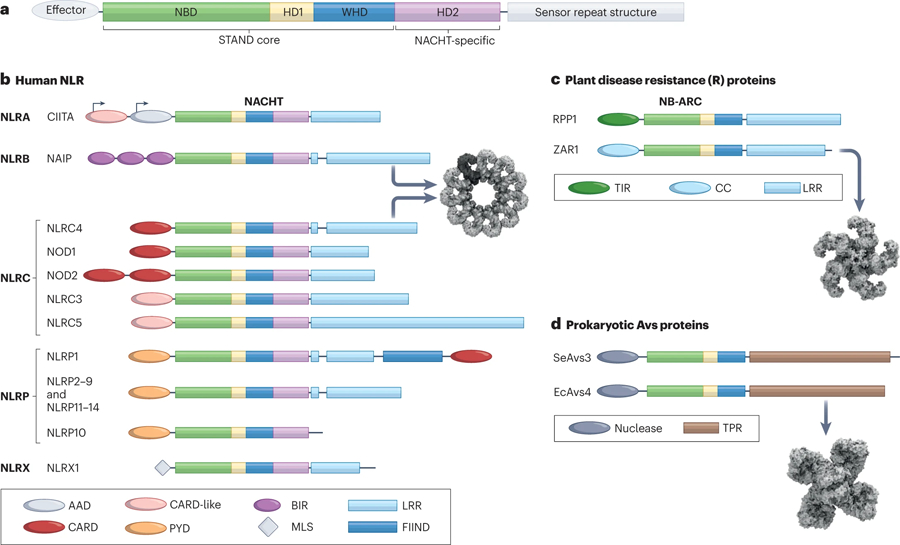
(A) The STAND ATPase module consisting of NBD, helical domain 1 (HD1), and winged helix domain (WHD) is utilized for cellular defense from prokaryotes to eukaryotes. An additional helical domain 2 (HD2) is present in many STAND proteins including a NACHT-specific domain used in the NLR family of proteins. (B) The primary structural organization of human NLRs. Here, NLRs are grouped according to subfamily which is determined by the effector domain at the N-terminus. The variable domains associated with the NACHT domain are color coded and indicated in the domain legend below. CIITA exhibits cell-type specific alternative promoter usage, and this is designated with arrows. The oligomerized inflammasome for NAIP/NLRC4 is shown (PDB:3JBL)288. (C) Two representative members of the plant disease resistance proteins with either TIR or CC domains are shown. The NB-ARC module lacks the HD2 domain. The C-terminal sensor is an LRR domain for these proteins as well. The oligomerized resistosome for ZAR1 is shown (PDB:6J5T)327. (D) Two representative members of the recently described prokaryotic Avs protein family are shown. The effector domain in these proteins is a nuclease instead of a protein-recruitment domain, and the sensor domain is composed of tetratricopeptide repeats (TPR). The oligomerized complex for EcAvs4 is shown (PDB:8DGF)27. Cryo-EM structure representations in this figure were created with BioRender.com.
In 2000, our group noted that similarities between NBS-LRR proteins, MHC class II transactivator (CIITA)4 [G] and nucleotide-binding oligomerization domain-containing protein 1 (NOD1, also known as CARD4) in terms of their nucleotide-binding domain (NBD) and LRR sequences and additionally noted the similarity in size and spacing of these domains5. Subsequently we identified a large family of 22 NBD-LRRs encoding human genes, with CIITA as the founding member6 (Fig. 1B and Fig. 2, Timeline). These genes were identified by BLAST homology searches of available genome sequences before the human genome was fully assembled. Family members have divergent N-termini, including the acidic transactivation domain of CIITA, baculovirus inhibitor of apoptosis repeat domain (BIR), pyrin domain (PYD) or caspase recruitment domain (CARD).
Figure 2: Key historical events in the NLR field.
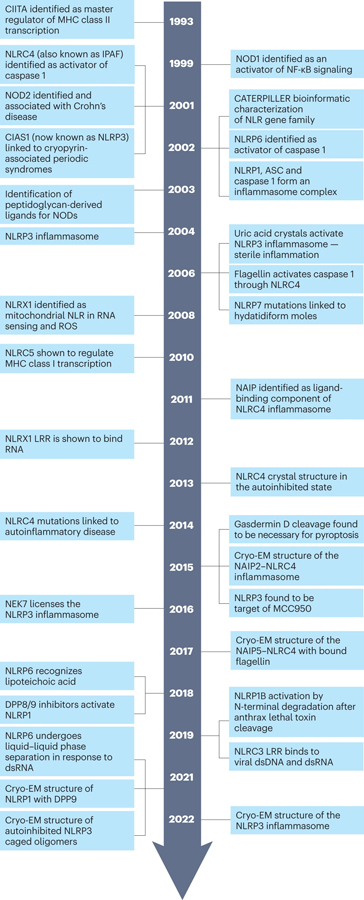
The timeline highlights some of the key discoveries and conceptual advances that have influenced the field over the last two decades. There are clearly many important contributions to the field which are not included here, and we apologize to those who have been left out due to spatial constraints.
Several other research groups had parallel findings regarding NBD-LRR family members. Human neuronal apoptosis inhibitory protein (NAIP) was originally thought to be deleted in spinal muscular atrophy7, but the deletion was later attributed to a neighboring genetic region. Nonetheless the encoded protein was noted as having BIR and NBD-LRR domain and negatively regulates apoptosis8. Koonin and Aravind used NAIP, CIITA and two other proteins to define the conserved NACHT domain [G] which includes the sequences that cover the ATP/GTPase specific P-loop, the Walker A and B motifs and five additional motifs9. Two CARD containing proteins, NOD1/CARD410,11 and NOD2/CARD1512 were found to activate NF-κB and are sensors of processed fragments of bacterial peptidoglycan13–16. Additionally, NOD2 frameshift and missense variants are genetic risk factors associated with Crohns’ disease17,18, first establishing these as pathogen recognition molecules important in inflammatory diseases. Inohara and Nunez noted similarities in the nucleotide-binding and oligomerization domain (NOD) of Apaf-1 [G], Ced-4, NOD1, NOD2 and the plant disease resistance genes and proposed that these constitute a family — the NOD family 19. Another CARD-NBD-LRR protein, IPAF (now referred to as NBD-, LRR- and CARD-containing protein 4 (NLRC4)), was discovered by Poyet et al. and found to associate with pro-caspase 1, leading to caspase 1 maturation20. Two other groups focused on the PYD-containing subgroup. Martinon, Burns and Tschopp first showed that NALP1 (now referred to as NBD-, LRR- and PYD-containing protein 1 (NLRP1)), has caspase maturation activity which they coined the term “the inflammasome” 21, and later reported a 14-member PYD-containing family22. Bertin’s group focused on the PYD-containing Apaf1-like proteins — which they named PYPAFs — and showed that PYPAF7 (now known as NLRP12) and PYPAF5 (now known as NLRP6) induced caspase 1-dependent cytokine processing activity and NF-κB-activating function when co-expressed with the ASC [G] adaptor protein23,24. The link of these proteins to human was first demonstrated by transformative human genetics study from Hoffman et al. in 2001 which identified mutations in CIAS1 (cold-induced autoinflammatory syndrome 1; now known as NLRP3) to be the cause of two rheumatological diseases, namely familial cold autoinflammatory syndrome (FCAS) and Muckle-Wells syndrome25. NLRP3 was later shown to direct the cleavage of pro-caspase 1 as well, thus representing the second inflammasome effector protein to be described26. These initial discoveries of NLR family members in the 1990s to the early 2000s kicked off over two decades of exciting progress in the field of innate immunity (Fig. 2, Timeline). In a surprising twist, a family of prokaryotic anti-viral STAND [G] (Avs) proteins was recently described that utilizes a similar oligomerizing NBD and sensor domain architecture to eukaryotic NLRs (Fig. 1D)27. Some of these Avs proteins use their sensor domains to detect the presence of phage proteins, and oligomerization brings N-terminal endonuclease domains into a complex to degrade phage DNA. Thus, this threat dependent, induced-proximity mechanism has remarkably been consistently utilized for cellular defense in all kingdoms of life.
In this Review, we will provide an overview of our current understanding of the NLR protein subgroups and discuss their biology, mechanisms of action and physiological relevance. Due to the expansiveness of this topic, we cannot credit all relevant work in the field; we apologize for this and refer the reader to other reviews for topics that have been extensively reviewed28,29. The intention here is to present a brief overview of well-known NLRs but to also discuss those members that have received less attention. NLR family members mediate a wide variety of functions, including serving as transactivators of MHC gene transcription, as inflammasome receptors and sensors, as positive and negative modulators of signalling pathways and in a variety of cell death processes. We discuss the key roles of each of these NLR functional subgroups and offer a forward-looking perspective on the field.
NLRs as transcriptional regulators
Although NLR proteins are primarily thought of as innate immune sensors, the founding member, CIITA, and the related protein NLRC5, are master transcriptional regulators of MHC class II (MHC II) and MHC class I (MHC I) genes, respectively (Fig.1, Table 1). CIITA was identified via complementation cloning using a mutant cell line devoid of MHC II expression, and mutations in CIITA were found in bare lymphocyte syndrome (BLS), an immunodeficiency caused by the lack of MHC II4. CIITA shuttles between the nucleus and cytoplasm30, and its expression pattern precisely matches that of MHC II and its accessory proteins31,32. Consistent with its role as a transcriptional co-activator, CIITA contains an N-terminal acidic transactivation domain. The NACHT domain of CIITA was shown to bind GTP although this study was not conducted with highly purified protein33. There is no clear evidence that CIITA binds DNA directly, and therefore it mediates its activity via interactions with transcription factors bound to the SXY cis-elements [G] 34 in the promoters of all MHC II genes, leading to recruitment of chromatin modifiers including histone acetyltransferases and methylases35. CIITA has interesting associations with cancer, including its absence in some cancers36,37, its transduction to active tumour immunity38, and its role as a gene fusion partner in lymphoid cancers39.
Table 1: NLR Functions and Human Disease Associations.
The table shows the 22 human NLRs with the first column indicating their principal function and the second column showing the best-characterized pathways by which they mediate their function (transcription, inflammasome, signalling pathway, etc.). Finally, the last column shows human diseases that have been linked to mutations in human NLRs. This list includes a spectrum of affected states ranging from polymorphisms that increase disease susceptibility to severe gain-of-function inflammasomopathies. We have attempted to be thorough, but we are aware that there may be some research that is not cited here.
| NLR | Function | Pathway | Diseases |
|---|---|---|---|
| CIITA | MHC-II transcriptional regulator | Regulation of MHC-II expression | Bare lymphocyte syndrome, Primary mediastinal B-cell lymphoma and Classical Hodgkin lymphoma, Multiple Sclerosis |
| NAIP | Flagellin sensing, pyroptosis, inhibition of apoptosis | TAK1-dependent JNK1 activation, inflammasome assembly | Increased susceptibility to legionella, spinal muscular atrophy |
| NOD1 | PRR for DAP | RIP2-dependent NF-κB and MAPK activation | Asthma, inflammatory bowel disease |
| NOD2 | PRR for MDP and viral ssRNA, autophagy | RIP2-dependent NF-κB and MAPK activation | Crohn’s disease, Blau syndrome, atopic eczema, atopic dermatitis, susceptibility to leprosy and tuberculosis |
| NLRC3 | Negative regulation of T cell activation and TLR response | Interaction with STING to reduce STING-TBK1 association | Prevents colorectal cancer and hepatocellular carcinoma, M. tuberculosis infection, and attenuates lymphocytic choriomeningitis virus infection and experimental autoimmune encephalomyelitis (EAE). |
| NLRC4 | PRR for flagellin and rod protein, pyroptosis, phagosome maturation | Inflammasome formation | Increased susceptibility to bacterial infection, Multiple Sclerosis, autoinflammation with infantile enterocolitis (AIFEC), NOMID and FCAS4, stroke, colitis and colitis-induced colon cancer. |
| NLRC5 | MHC-I upregulation, regulates innate immune response | MHC-I regulation, type I interferon response | hepatocellular carcinoma, liver inflammatory injury, hepatic steatosis, Liver ischemia/reperfusion injury, lymphoid cancers, CNS infection, cerebral ischemia/reperfusion injury, glioma, multiple sclerosis, and epilepsy |
| NLRP1 | PRR for MDP, anthrax lethal toxin | Inflammasome formation | Vitiligo, Alzheimer’s disease, celiac disease, Addison’s disease, type 1 diabetes, autoimmune thyroid disorders, systemic lupus erythematosus, systemic sclerosis, giant cell arteritis, congenital toxoplasmosis, rheumatoid arthritis, corneal intraepithelial dyskeratosis |
| NLRP2 | Negative regulation of NF-κB, embryonic development | Inflammasome formation | Beckwith-Wiedemann syndrome, Female infertility, hepatic steatosis |
| NLRP3 | PRR for PAMPs, DAMPs, and irritants | Inflammasome formation | Cryopyrin-associated periodic fever syndrome, gout, type 1 diabetes, celiac disease, psoriasis, Multiple Sclerosis, increased susceptibility to HIV-1 infections, Inflammatory bowel diseases, type 2 diabetes |
| NLRP4 | Negative regulation of type I interferon signaling by dsRNA, DNA, or viral infection. Reduces autophagy in response to bacterial infection | DTX4-dependent TBK1 degradation, beclin-1 dependent autophagy | Streptococcus infection, viral infection, Asthma |
| NLRP5 | Embryogenesis | Mitochondrial function, ROS | Female Infertility |
| NLRP6 | Negative regulation of NF-κB | Inflammasome formation | Colitis and colon cancer, non-alcoholic fatty liver disease, alcoholic hepatitis, hepatocarcinoma |
| NLRP7 | PRR for lipopeptide | Inflammasome formation | Recurrent hydatidiform moles, testicular seminoma, endometrial cancer, colon cancer |
| NLRP8 | Unknown | Unknown | Expressed in ovaries, testes and preimplantation embryos |
| NLRP9 | Unknown | Unknown | Expressed in ovaries, testes and preimplantation embryos |
| NLRP10 | Adaptive immunity by dendritic cells | Unknown | Increased susceptibility to bacterial infection, candida albicans, atopic dermatitis, Asthma, EAE |
| NLRP11 | Association with MAVS during viral infection, inhibits type I interferon signaling | NLRP3 Licenses NLRP11 for inflammasome activation | Cryopyrin-associated periodic syndrome (CAPS) |
| NLRP12 | Negative regulation of NF-κB | Inflammasome formation | FCAS2, Atopic dermatitis, Glioblastoma |
| NLRP13 | Unknown | Unknown | Unknown |
| NLRP14 | Spermatogenesis | Unknown | Spermatogenic failure |
| NLRX1 | ROS generation, autophagy and mitophagy | blocks STING-TBK-mediated antiviral responses, negatively regulates NF-κB pathway through TRAF6 | Increased susceptibility to chronic hepatitis B, Viral infections, Listeria monocytogenes infection, tumor suppressor in cancer, negatively regulates Group A Streptococcus (GAS) infection |
Similarly to CIITA, NLRC5 also shuttles between the nucleus and cytoplasm, and its primary function is to upregulate MHC class I and accessory protein gene expression40. NLRC5 also requires an SXY cis-acting module in MHC I promoters for transcriptional activation. In contrast to CIITA, the N-terminus of NLRC5 is composed of an atypical CARD and lacks an acidic activation domain. NLRC5 also contains a much larger C-terminal LRR region with up to 38 LRRs. In contrast to the highly restricted expression of CIITA, NLRC5 is expressed constitutively with elevated expression observed in T cells, B cells and NK cells. The role for NLRC5 in MHC I gene expression was clearly shown in the NLRC5-deficient mice generated by several groups41–43. The loss of NLRC5 has the greatest effect on MHC I expression in T cells, natural killer (NK) cells, and natural killer T (NKT) cells, whereas MHC I expression in macrophage is more modestly reduced in NLRC5-deficient mice. The role of NLRC5 in controlling MHC I expression in other cell types is modest to absent. Hence, NLRC5 differs from CIITA in that its impact is not observed in all MHC I positive cells. Similarly to CIITA, it is also a target of immune evasion in cancer44. An additional role in regulating inflammatory cytokines has been proposed for NLRC5. In one model, direct interaction between NLRC5 and IKKα/β blocks interaction with IKKγ resulting in reduced NF-κB activation45. Analysis of NLRC5-deficient mice has not consistently verified this physiological role for NLRC5, and these conflicting studies have been reviewed elsewhere46.
NLRs in inflammasomes and pyroptosis
In this section, we provide an overview of the NLR family members that form inflammasomes (Fig. 3, Table 1.). Regulated inflammasome signalling is vital for homeostasis and tissue repair, but dysregulated inflammasome signalling is central to many diseases (Fig. 3, Table 1.).
Figure 3: Inflammasome activators and related disorders.
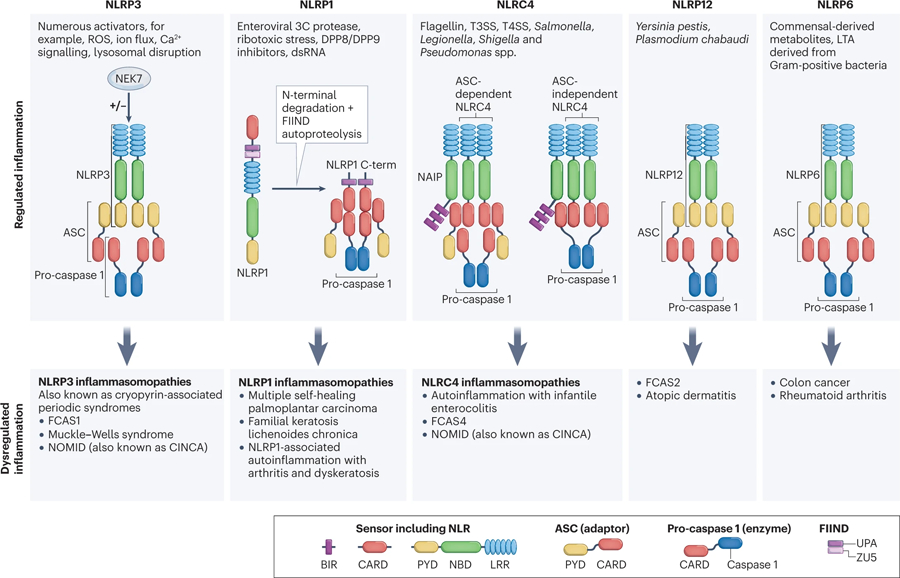
The NLRP3, NLRP1, NLRC4, NLRP12 and NLRP6 inflammasomes with their intracellular mediators involved in activation are summarised. Dysregulated or chronically activated inflammasomes may lead to inflammatory diseases. NLRP3 is a sensor for numerous PAMPs and DAMPs responding to intracellular damage induced by pathogenic or sterile insults. NLRP1 is a sensor for ribotoxic stress and dsRNA. DPP8/9 inhibitors are activators of the NLRP1 inflammasome. The current model for NLRP1 inflammasome activation involves ASC-dependent recruitment of pro-caspase 1 by the UPA-CARD C-terminal fragment of NLRP1. The NLRC4 inflammasome detects T3SS bacterial proteins via NAIPs and can assemble an inflammasome with or without ASC. NLRP12 inflammasome is assembled in response to Yersinia pestis and Plasmodium chabaudi. NLRP6 inflammasome detects commensal‐derived metabolites, LTA derived from Gram‐positive bacteria. Abbreviations used: CINCA, chronic neurologic cutaneous and articular syndrome; NLRP3, NLR family pyrin domain containing 3; NEK7, NIMA related kinase 7; LRR, leucine rich repeats; NBD, nucleotide-binding domain; PYD, pyrin domain; ASC, Apoptosis-associated speck-like protein containing a CARD; CARD, caspase recruitment domain; ROS, reactive oxygen species; FIIND, function to find domain; UPA, conserved in UNC5, the death-domain-containing protein PIDD and proteins of the ankyrin family; NLRC4, NLR family CARD domain containing 4; T3SS, type 3 secretory system, NAIP; neuronal apoptosis inhibitory protein.
NLRP3 inflammasomes.
NLRP3 is the best studied of the inflammasome sensors, and it detects pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs). Its significance in autoinflammatory diseases and its mechanisms of activation have been detailed in many excellent reviews29,47. This section is not intended to provide a comprehensive review of this topic. NLRP3 inflammasome assembly with pro-caspase 1 and ASC leads to proteolytic maturation of caspase 148. Caspase 1 in turn cleaves and activates more than 70 substrate proteins, including the pro-inflammatory cytokines IL-1β and IL-18. Cleavage of Gasdermin D (GSDMD) by caspase 1 was found to be necessary and sufficient for pyroptosis49,50. The N-terminal fragment of GSDMD forms a multimeric membrane pore to cause pyroptotic cell death51–54 and facilitate the release of IL-1β and IL-18 via a non-conventional mode of secretion. The discovery of NLRP3’s inflammasome activity helped to define its role in IL-1β release in cryopyrin-induced autoinflammatory syndromes (CAPS) [G] 25,26. CAPS include several diseases with increasing disease severity, including FCAS1, Muckle-Wells syndrome and the severely debilitating chronic infantile neurological cutaneous and articular syndrome55 (CINCA; also known as neonatal-onset multisystem inflammatory disorder (NOMID)). The NLRP3 inflammasome has also been implicated in many other diseases, ranging from autoinflammatory, metabolic and neurodegenerative to infectious diseases48.
Assembly of the fully functional NLRP3 inflammasome is initiated by two distinct steps: priming and activation. The priming step involves the recognition of PAMPs or DAMPs via receptors such as TLRs or NOD2, or the detection of TNF and IL-1β, which leads to NF-κB activation and increased cellular expression of NLRP3, caspase 1 and IL-1β56–58. In addition, post-translational modifications (PTMs) of NLRP3 including phosphorylation59 and deubiqitination60,61 promote NLRP3 activation, while ubiquitination62 and sumoylation63 suppress NLRP3 inflammasome activity. In the activation step, NLRP3 oligomerizes through homotypic interactions between NACHT domains, which serves as a scaffold to nucleate ASC through PYD–PYD interactions64,65. ASC and pro-caspase 1 combine via CARD–CARD interactions leading to the formation of prion-like filaments. Pro-caspase 1 undergoes auto-proteolytic cleavage and processing into mature caspase 1 within this complex66. Several groups identified NIMA-related kinase 7 (NEK7) as an essential component of NLRP3 inflammasome activation67–69(Fig. 3). NEK7 oligomerizes with NLRP3 to form a complex that promotes ASC speck formation and caspase 1 activation70. NEK7 interacts with NLRP3 but not with the NLRC4 and AIM2 inflammasome sensors.
Whereas the so-called ‘canonical’ activation of NLRP3 inflammasome activates caspase 1, non-canonical activation of NLRP3 inflammasome activates caspase 4 and caspase 5 in humans, and caspase 11 in mice71–73. Non-canonical NLRP3 inflammasome activation occurs in response to intracellular LPS sensing by caspase 4/11, resulting in the secretion of IL-1β and IL-18 and the induction of pyroptosis, which can lead to endotoxemia-induced death 72,74,75. Caspase 11 can be induced by TRIF and senses intracellular LPS from cytosolic gram-negative bacteria that have escaped vacuoles73. Caspase 8 can activate canonical and non-canonical NLRP3 inflammasomes in response to many pathogenic stimuli76–79. Caspase 8 initiates apoptosis in response to FASL and TNF and protects against necroptosis.
Several studies have reported that inflammasome activation can involve more than one sensor. For example, NLRP3 and NLRC4 both activate caspase 1 in response to PAMPs from Salmonella typhimurium80,81 and to DAMPs, such as lysophosphatidylcholine, which causes demyelination82. NLRP3 and AIM2 [G] inflammasomes are activated by Plasmodium parasite-derived hemozoin and DNA83 and by Aspergillus fungi84. Cytosolic DNA sensed by the cGAS–STING pathway, activates the NLRP3 and AIM2 inflammasomes via cGAMP production85. Dual-sensing mechanisms may reflect cooperative function within a cell or independent inflammasomes within a cell.
Despite extensive studies, the precise mechanism through which NLRP3 becomes activated remains unclear (see also later section on ‘Structure of NLR proteins’). However, NLRP3 can be activated by many diverse pathways, including through the chemical disruption of glycolysis86 or accumulation of cholesterol crystals87 or free fatty acids88. It is presumed that common downstream molecules associated with cellular damage are the critical activators of NLRP3. Mitochondrial DNA is a candidate for a downstream mediator that can increase NLRP3 activation89,90. RNA also has been shown to activate NLRP3, and the mitochondrial antiviral signalling protein (MAVS) has been implicated in this process91–94. Although K+ efflux is also suggested to be a common activation pathway of NLRP395, N-acetylglucosamine-induced hexokinase relocalization promotes NLRP3 inflammasome activation independently of K+ efflux in certain bacterial infections96.
The intense focus on the role of NLRP3 inflammasome activation and pyroptosis in numerous disease models has led to the development of many NLRP3 inhibitors97. While some of these inhibitors affect the conformation of NLRP3 (Oridonin, MCC950, and tranilast), others affect its function (OLT1177 and Parthenolide) or binding to signaling partners (BAY11–7082 and VI-16)98. These inhibitors show promise as future therapeutics for a range of inflammatory diseases.
NLRP1 inflammasomes.
Human NLRP121 (also known as DEFCAP, CARD7, NAC and NALP1)99–101 is a PYD- and CARD-containing NLR protein first found to induce pro-IL-1β cleavage in an in vitro, cell-free reconstitution assay when combined with caspase 1, caspase 5 and ASC. Indeed, experiments with NLRP1 led to the term ‘inflammasome’ being coined (Fig. 2). NLRP1 is highly expressed in keratinocytes and has been associated with several skin diseases102–107 (Table 1.). NLRP1 is also found in the lung and is associated with chronic obstructive pulmonary disease (COPD)108 and recurrent respiratory papillomatosis109. Finally, NLRP1 polymorphisms are associated with a novel autoinflammatory disorder, NLRP1-associated autoinflammation with arthritis and dyskeratosis (NAIAD)107.
NLRP1 exhibits significant genetic divergence in different species. Humans have one NLRP1 gene, while mice have four Nlrp1 homologue/paralogue genes110. Human and mouse NLRP1 are also divergent in their protein structure. Human NLRP1 contains an N-terminal PYD and a C-terminal CARD that flank a central NACHT, LRR and FIIND (function to find) domain, while mice have an N-terminal NR100 [G]. Although the N-termini of human and mouse NLRP1 are different, they both represent autoinhibitory domains111,112, which are removed by proteolytic cleavage of the N-termini resulting in its activation113,114. For example, the N-terminal NR100 domain of mouse NLRP1B115 and rat NLRP1116 is cleaved by B. anthrax lethal toxin (LT)117–119. In contrast, the human N-terminal PYD is resistant to LT. This cleavage exposes a new N-terminus which undergoes N-end rule-mediated degradation by ubiquitin ligase-mediated degradation, resulting in the release of the C-terminus, which is freed to interact and recruit pro-caspase 1 through its CARD, resulting in caspase 1 maturation120–122. Ubiquitin ligases and proteases that can cause NLRP1 cleavage include ubiquitin ligase UBR2122, Shigella flexneri IpaH7.8 121, and picornavirus 3C proteases123. The 3C protease cleaves human NLRP1, exposing a new N-terminus, which then undergoes N-glycine-mediated degradation, thus liberating the UPA (UNC5, PIDD and ankyrin) and CARD domains to form an inflammasome.
The FIIND domain of NLRP1 also undergoes proteolytic cleavage. The serine proteases dipeptidyl peptidase 8 (DPP8) and DPP9 interact with the FIIND domain of human NLRP1 to maintain it in an inactive state, and a DPP8/9 inhibitor, ValboroPro (VbP, or Talabostat) reverses this inhibition111,124–126. VbP causes the proteasome-dependent degradation of the N-terminus of NLRP1 and the autoproteolytic cleavage of the FIIND domain at the ZU5 (ZO-1 and UNC5) and UPA subdomains, releasing the C-terminal domain that includes CARD to recruit pro-caspase 1 113,114,127. Cryo-EM shows a ternary complex of DPP9 full-length NLRP1 and the C-terminus of NLRP1128,129. It is postulated that the full length NLRP1 inhibits activation of the C-terminal NLRP1 since an autocatalytic deficient full-length NLRP1 promotes inhibition of the C-terminal fragment. Thus, VbP weakens NLRP1-DPP9 interaction to induce inflammasome activation.
NLRP1 can also be activated by Toxoplasma parasites, bacteria, viruses and long dsRNAs130–133. Regarding the last, it was shown that human but not mouse NLRP1 binds dsRNA through its LRR domain134. NLRP1B can also be activated by energy deprivation and nutrient depletion of ATP. In contrast to other NLRs, where ATP binding is necessary for their functional activity, the ATP-binding domain in NLRP1B inhibits its function135 and this inhibition of NLRP1B by ATP also requires the FIIND region136. A recent paper sheds some light on the effect of ATP on NLRP1 activation. It shows that oxidized but not reduced thioredoxin-1 (TRX1) can interact with NLRP1 NACHT-LRR domain to restrain the activation of NLRP1, and this process is ATP-dependent process137. Furthermore, both patient-derived and ATPase-deficient mutant blocked binding to oxidized TRX1 leading to inflammasome activation. The authors suggest that under reductive stress when oxidized TRX1 is reduced, NLRP1 can be activated. Others have shown that inhibition of TRX1 activity induces the β-amyloid activated NLRP-1/caspase-1/GSDMD pyroptotic response138. Recently, another form of stress, ribosome stress, is also found to cause the hyperphosphorylation and activation of human NLRP1 by the ribotoxic stress response kinase ZAKα (also known as MAP3K20) and p38 139. Thus NLRP1 is activated by a number of cell stress inducers.
NLRC4/NAIP inflammasomes.
NLRC4 was the first NLR shown to associate with pro-caspase 1 leading to caspase 1 activation and subsequent cell death20. NLRC4 is important for caspase 1 activation after exposure to Salmonella typhimurium140 due to Salmonella flagellin141. Components of the bacterial type III secretion system (T3SS) [G] can also induce mouse NLRC4-dependent caspase 1 activation in the absence of flagellin, showing multi-component activation of the NLRC4 inflammasome142. However, NLRC4 is not the direct receptor for bacterial PAMPs, but instead, pairs with mouse NAIP5, which recognizes flagellin143, and NAIP1 and NAIP2, which respectively recognize T3SS needle144,145 and rod proteins143. Thus, the NAIP proteins recognize bacterial components and recruit NLRC4 to activate caspase 1. NAIPs contain N-terminal BIR domains, while NLRC4 displays an N-terminal CARD. Humans only have one NAIP gene and it responds to flagellin and both T3SS needle and rod protein. NLRC4 and NAIP5 also mediate pyroptosis in response to flagellin146,147, and pyroptosis induced by bacterial T3SS needle proteins is NLRC4 dependent142.
Gain-of-function mutations in human NLRC4 lead to inflammasomopathies [G] that are associated with the spontaneous formation of the NLRC4 inflammasome, production of IL-1β and IL-18 and inflammatory cell death148. Three NLRC4 inflammasomopathies have been described: autoinflammation with infantile enterocolitis (AIFEC), neonatal-onset multisystem inflammatory disease (NOMID) and FCAS4148. Endogenous short interspersed nuclear element (SINE) RNAs — which promote atrophic macular degeneration and systemic lupus erythematosus — induce NLRC4 inflammasome activation with DDX17 helicase-mediated sensing of these RNAs independent of NAIPs149. In addition, NLRC4 has been implicated in microglial cell death in models of stroke150.
NLRP6 inflammasome and inflammasome-independent functions.
NLRP6 (also known as PYPAF5 24) is expressed by immune and stromal cells, but its expression in typical lymphoid tissues is low while its expression is high in intestinal colonic myofibroblasts and epithelial cells11,151,152. Overexpression of NLRP6 leads to its assembly with ASC to form specks (that is, large protein complexes) in cells and these coordinate the activation of NF-κB and pro-caspase-1 24. It is regulated post-translationally by the deubiquitinase CYLD, which targets K63-linked ubiquitination of NLRP6 to prevent its binding to ASC153.
NLRP6 functions as an inflammasome in the dextran sodium sulfate (DSS)-induced colitis model and protects against colitis by driving IL-18 release, which promotes epithelial barrier integrity151,152,154 via the promotion of Lgf5+ stem cells and anti-microbial response155. NLRP6 also protects against colon cancer by inducing IL-18 154, resulting in reduced intestinal inflammation and reduced proliferative signals such as SMARRC1, p53, Wnt and Notch pathways all of which have been linked with intestinal tumourigenesis151. Additionally, NLRP6 enhances mucin secretion by intestinal Goblet cells through the promotion of autophagy156. NLRP6 is also reported to influence the intestinal microbiota, although this is controversial. Some have found that NLRP6 expression affects components of the microbiota such as Prevotella152, while others have failed to see this association using littermates from different animal facilities157,158.
In addition to functions in the intestine, NLRP6 is expressed in the liver and can affect disease development in this tissue. For example, expression of NLRP6 was shown to attenuate non-alcoholic fatty liver disease159, alcoholic hepatitis160, and hepatocarcinoma (HCC) in mice161. These are found to mediated by the reduced inflammatory cytokines such as TNF, reduced NF-κB and TLR4 signalling by NLRP6. By contrast, others found that C. albicans infection promotes hepatocarcinogenesis in patients may occur in an NLRP6-dependent fashion by increasing metabolites that can promote tumorigenesis 162. Thus, the impact of NLRP6 expression may be dependent on the disease and tissue context.
In the setting of infection, NLRP6 can be activated by the microbial metabolite taurine163, bacterial lipoteichoic acid164, and viral RNA through the RNA helicase Dhx15 to induce type I and type III interferon165. Furthermore, viral infection, dsRNA, TNF and type I interferon can enhance NLRP6 expression165. A seminal paper showed that during virus infection, NLRP6 is activated by binding to dsRNA and undergoes liquid–liquid phase separation (LLPS) and that a disordered poly-lysine sequence (K350–354) in the protein is important for its function166.
In addition to its role in inflammasome assembly, NLRP6 has an alternative role in suppressing inflammatory responses and signals. NLRP6-deficient mice show enhanced resistance to bacteria, accompanied by increased NF-κB and MAPK activation after TLR activation167. NLRP6 also reduces neutrophil influx and granulocytic bactericidal activity during gram-positive bacteria infection168. NLRP6 expression in inflamed periapical tissues and human periodontal ligament cells negatively regulates IL-6 and TNF by inhibiting NF-κB and ERK signaling169. Thus, NLRP6 has both inflammasome-dependent and independent functions.
In summary, a major function of multiple NLR proteins lies in inflammasome function. While the ligands for some are well defined, others are not. Furthermore, some inflammasome NLRs have alternative functions such as reducing inflammatory responses. In the next section, the focus will be on NLRs that control a variety of signalling pathways.
NLRs as regulators of diverse signalling pathways
This section focuses on NLRs that contribute to pathogen sensing and diverse immune signalling responses independently of inflammasome assembly (Fig. 4), with the exception of NLRP12 (Fig. 3) and NLRP11. NLRP12 has been reported to assemble inflammasomes in response to certain infections, but we include this NLR in this section due to its well-described role in negatively regulating inflammation. NLRP11 does not itself form an inflammasome, and is reported to regulate the NLRP3 inflammasome, as discussed below.
Figure 4: Regulatory Functions of NLRs.
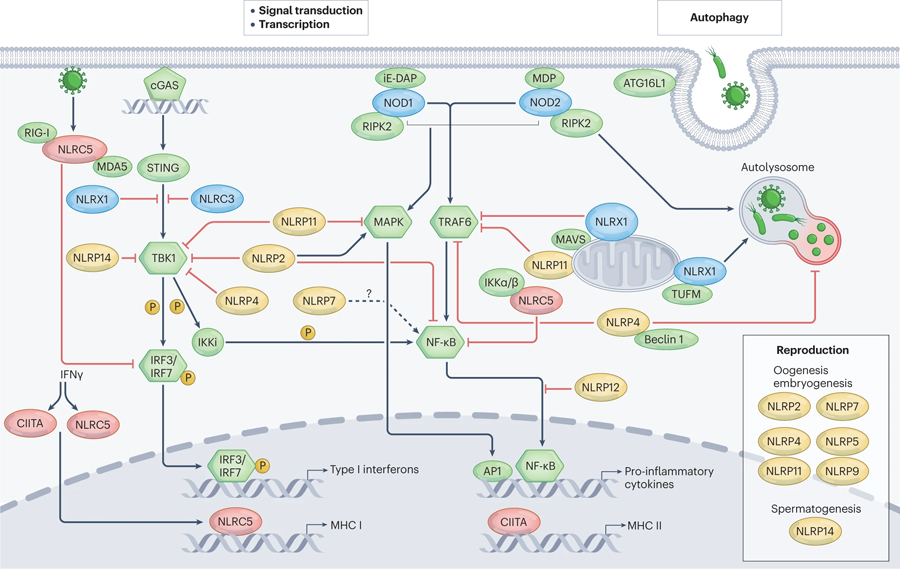
Different NLRs act as either positive or negative regulators in transcription, MAPK, NF-κB, and type I IFN signalling pathways and in autophagy. CIITA and NLRC5 induced by IFNγ acts as transactivators of MHC genes. NLRC5 also negatively regulates the NF-κB and type I IFN signalling pathways. In addition to NLRC5, several NLRs reduce either NF-κB or type I IFN signalling pathways or both. These NLRs are NLRC3, NLRX1, NLRP2, 4, 11, 12, and 14. On the other hand, NLRP7 may positively regulate NF-κB pathway, and NLRP2 promotes MAPK signalling. Likewise, NOD1 and NOD2 recognizes iE-DAP and MDP separately and interact with RIP2 to activate NF-κB and MAPK signalling pathway. Additionally, NOD2 recognizes MDP and recruits ATG16L1 to the plasma membrane to induce autophagy328. The right shows that NLRX1 interacts with TUFM to induce autophagy while NLRP4 interacts with Beclin-1 to inhibit autophagy. The right box shows NLRs that regulate development in the reproductive system. NLRP14 is the only NLRP molecule that contributes to spermatogenesis while other NLRPs including NLRP2,4,5,7,9,11 may regulate oogenesis and embryogenesis. NLR, NOD-like receptors; MAPK, mitogen-activated protein kinase; NF-κB, nuclear factor kappa B; TRAF6, tumor necrosis factor (TNF) receptor-associated factor 6; MHC, major histocompatibility complexes; STING, Stimulator of interferon genes; TBK1, TANK-binding kinase 1; IRF3, Interferon Regulatory Factor 3; iE-DAP, γ-D-glutamyl-meso-diaminopimelic acid; MDP, muramyl dipeptide; RIP2, receptor interacting protein 2; TUFM, Tu translation elongation factor of mitochondria.
Sensing of peptidoglycan fragments by NOD1 and NOD2.
NOD1 and NOD2 represent the earliest and best-characterized signalling NLRs, and they function to detect PAMPs and activate inflammatory signalling pathways. These two proteins have been exhaustively reviewed, hence only a brief synopsis is provided here170,171. In addition to a central NACHT domain and C-terminal LRR domain, the N-terminus of NOD proteins consists of a single CARD (in the case of NOD1) and two CARDs (for NOD2). NOD1 and NOD2 detect the cytosolic presence of processed fragments of peptidoglycan derived from bacterial cell walls, γ-D-glutamyl-meso-diaminopimelic acid (iE-DAP)13,14 and muramyl dipeptide (MDP)15,16, respectively. A recent study highlights the critical role of host peptidoglycan processing by showing that MDP is phosphorylated by N-acetylglucosamine kinase (NAGK), and this modification is essential for the activation of NOD2 in THP-1 cells and primary mouse macrophages172. Recognition of these PAMPs results in oligomerization, CARD-mediated recruitment of receptor-interacting serine/threonine kinase 2 (RIPK2 or RIP2) and the downstream activation of NF-κB and MAPK signalling pathways173, leading to production of inflammatory cytokines, chemokines and inflammatory cell recruitment. In addition, NOD1 can cause apoptosis in a caspase 8 and RIPK2 dependent fashion174.
NOD1 is expressed in a wide variety of cell types, including epithelial cells10, and studies in intestinal epithelial cells have illustrated key roles for NOD1 in the immune response to intestinal pathogens. By contrast, NOD2 is primarily expressed in cells of the myeloid lineage12, and specialized cells such as intestinal Paneth cells. Gain-of-function mutations in NOD2 were found to be associated with the human inflammatory disease, Blau syndrome175. Blau syndrome mutations are restricted to the NACHT domain, and the resultant amino acid changes are predicted to destabilize the autoinhibited conformation of NOD2 leading to constitutive activation176. In contrast, mutations that lead to impaired function of NOD2 represent the most significant risk factor for development of Crohn’s disease17,18. Crohn’s associated mutations are dispersed throughout both the NACHT and LRR domains potentially interfering with oligomerization, localization, or ligand sensing176. Both NOD1 and NOD2 have been shown to associate with endosomes, and this localization efficiently places these sensors in position to encounter their ligands transported by endosomal peptide transporters177,178. Membrane localization of NOD1/NOD2 is dependent on S-palmitoylation mediated by the palmitoyltransferase ZDHHC5, and mutations that disrupt palmitoylation result in impaired NF-κB signalling in response to NOD activators179.
The mitochondrial sensor NLRX1.
NLRX1 is ubiquitously expressed in mammals. It has a central NACHT and LRR domain and a unique N-terminus, which functions as a mitochondrial targeting sequence (MTS)180–182. The C-terminus of human NLRX1 was found to be required for oligomerization and it binds ssRNA, dsRNA, lipid metabolites but not DNA183–185. The precise location of NLRX1 in mitochondria may be dynamic. It is found in the mitochondrial outer membrane and thought to interact with the CARD of MAVS through its LRR domain180. Others found that it localizes to the mitochondrial matrix to regulate mitochondria reactive oxygen species (ROS) production181,182. Multiple studies have shown that NLRX1 negatively regulates MAVS-mediated activation of IRF3 and type I IFN induction in virus infection180,186,187. It also blocks STING–TBK-mediated antiviral responses in HIV-1 infection188, negatively regulates NF-κB signalling through interacting with TNF receptor-associated factor 6 (TRAF6)187,189 and has been shown to function as a tumour suppressor in cancer190,191. However, in contrast to these roles in negatively regulating IRF3- and NF-κB-mediated antiviral responses, NLRX1 has been shown to support early antiviral responses by post-transcriptionally regulating IRF1 abundance192.
NLRX1 also functions in autophagy and mitophagy186. NLRX1 was first reported to induce autophagy by interacting with the Tu elongation factor mitochondrial (TUFM)–ATG5-ATG12 pathway to promote virus-induced autophagy and to inhibit RLR-induced type I IFN production upon virus infection193. NLRX1 also interacts with Beclin-1 or E7 protein of oncogenic human papillomavirus (HPV16) to promote autophagy in cancer cells194,195. Additionally, it promotes LC3-associated phagocytosis (LAP, a non-canonical form of autophagy), and activates the MAPK pathway upon fungal infection196. Furthermore, NLRX1 acts as an LC3 interacting region (LIR)-containing mitophagy receptor to promote mitophagy during Listeria monocytogenes infection197. It also regulates mitophagy in metastatic mammary tumours198, in intestinal ischaemic–reperfusion injury199, and mediates morphine-induced immunosupression in microglia by activating an insufficient mitophagy response200. In contrast to these aforementioned studies, NLRX1 was reported to negatively regulate autophagy during Group A Streptococcus infection201. However, the findings of a majority of the studies indicate that NLRX1 may represent a therapeutic target when increased autophagy or reduced inflammation is desired. Indeed, an activator of NLRX1, NX-13, is found to attenuate inflammatory bowel disease model by decreasing multiple aspects of immune activation202.
Negative regulation of immune cell activation by NLRC3.
NLRC3 is a CARD-containing NLR and is expressed in both human and mouse immune cells6,203. Its highest expression is in T cells, and T cell activation downregulates NLRC3 expression, suggesting that it may serve as a suppressor of T cell activation203. Indeed, NLRC3 reduces K63-linked ubiquitination of TRAF6 to downregulate NF-κB signalling, expression of interferon-γ and TNF, and the mTOR pathway in CD4+ T cells in the context of viral infection, T cell-mediated autoimmunity204 and M. tuberculosis infection205. In peritoneal macrophages, NLRC3 negatively regulates TLR4- and LPS-induced NF-κB activation by interacting with TRAF6 to mediate its deubiquitination in macrophages206. Additionally, NLRC3 also attenuates innate immune cell responses to cytosolic DNA, cyclic di-GMP and DNA viruses. It does this by interacting with the DNA sensor stimulator of interferon genes (STING) and TBK1 to impede the STING–TBK1 interaction, thereby limiting interferon production and NF-κB activation207. Upon binding of viral dsDNA and dsRNA to the LRR domain, NLRC3’s ATPase activity is increased and STING–TBK1 is released185. Additionally, NLRC3 NACHT can interact with a scaffold IQGAP1 and this disrupts the NLRC3-STING association in regulating type I IFN responses208. Overall, these studies reveal the impact of NLRC3 in regulating STING–TBK-mediated type I IFN signalling and NF-κB pathways to control cellular immune responses. In human cancer, NLRC3 expression is found to be a positive prognostic factor209,210, and NLRC3 was shown to mediate protection in a mouse model of colon cancer by inhibiting the PI3K–mTOR pathway 211.
Cell-specific regulation of inflammation by NLRP12.
NLRP12 is expressed in different myeloid cell populations, with prominent expression in granulocytes and dendritic cells212. NLRP12 functions as both a negative regulator of inflammation and as an inflammasome. It attenuates canonical and non-canonical NF-κB signalling pathways by blocking IRAK1 activation213 and by increasing the degradation of the NF-κB-inducing kinase (NIK)214, respectively. Additionally, NLRP12 can interact with TRAF3, which is involved in NIK degradation. Others have shown that it associates with ASC to mediate caspase 1 activation23, leading to IL-1β and IL-18 release in bone marrow-derived macrophages (BMDMs) infected with Yersinia pestis and Plasmodium chabaudi215,216 thus serving as an inflammasome sensor.
NLRP12 activity in myeloid cells is essential for colonic homeostasis217,218. Inhibition of NF-κB and ERK signalling by NLRP12 suppresses colonic inflammation and colitis-associated colorectal cancer in mouse models. NLRP12 also mitigates colitis by regulating the gut microbiota — for instance, its expression maintains the presence of protective commensal strains of Lachnospiraceae in the gut219,220. Interestingly, colonic inflammation and dysbiosis of the microbiota in NLRP12-deficient mice can contribute to obesity via enhanced inflammation221.
NLPR12 has also been associated with cell proliferation. NLRP12-deficient microglia show increased proliferation while NLRP12-deficient tumour cells show decreased cellular proliferation in glioblastoma222. Human glioblastoma tissue shows elevated NLRP12 expression. In response to Leishmania major Nlrp12 deficient neutrophils showed increased neutrophil migration towards the chemokine CXCL1 and the neutrophil parasite Leishmania major, revealing NLRP12 as a negative regulator of directed neutrophil migration in vitro223.
Mutations in NLRP12 are associated with the systemic autoinflammatory disease FCAS2224. This is a cold-induced autosomal dominant disease characterized by noninfectious periodic fevers and inflammatory symptoms in joints, muscles, digestive organs, skin and nerves. Interestingly, the IL-1 receptor antagonist (Anakinra) [G] only partially relieves the symptoms seen in patients with FCAS2225, whereas this drug is potently protective in patients with the NLRP3-associated diseases CAPS and FMF . It remains to be explored if the clinical manifestations are reflective of the divergent functions attributed to NLRP12.
A potential anti-inflammatory role for NLRP10.
NLRP10 (also known as PYNOD) contains an N-terminal PYD in addition to a NACHT domain, but it is the only mammalian NLR family member that lacks an LRR domain226. NLRP10 is expressed in a variety of tissues, and is highly expressed in heart, skeletal muscle, brain and skin226,227. Exogenous expression studies in cell lines226 and transgenic mice227 found that overexpressed NLRP10 inhibits IL-1β processing. NLRP10 overexpressing transgenic mice were resistant to lethal doses of LPS injection, although serum levels of TNF-α were more significantly reduced when compared to IL-1β227. Attempts to examine the physiological role of NLRP10 in knockout mice were temporarily hindered by the presence of a secondary genetic mutation that was responsible for the initially described phenotype228,229. Extensively backcrossed NLRP10-deficient mice were shown to have increased inflammatory responses in response to Leishmania infection, supporting a potential anti-inflammatory role for NLRP10230. Additional NLRP10 KO lines have been created and are phenotypically normal in the absence of an inflammatory challenge231,232. However, while one line of NLRP10 KO mice showed reduced inflammation in a contact hypersensitivity model231, another group saw no difference between NLRP10-deficient and wild type mice in a similar model232. NLRP10 remains an enigmatic NLR protein, and future studies will be required to determine its physiological role.
Regulation of inflammatory signalling and autophagy by NLRP4.
NLRP4 is strongly expressed in placenta, oocytes, testis, spleen, pancreas, liver, lung, kidney, and thymus233–235. Similar to other NLRPs, NLRP4 has an N-terminal PYD, but the NLRP4 PYD contains unique features and does not interact with ASC236. Initially, NLRP4 was found to negatively regulate TNF- and IL-1β-induced NF-κB activation234. Subsequently, NLRP4 was also reported to negatively regulate type I interferon signalling in response to dsRNA, DNA or viral infection. The NACHT domain of NLRP4 interacts with the E3 ubiquitin ligase DTX4 to direct the K48-linked polyubiquitination of TBK1, resulting in its degradation and this blocks the TBK1-mediated phosphorylation and translocation of IRF3235.
Aside from its role in regulating these immune signalling pathways, NLRP4 regulates autophagy in response to bacterial infection and interacts with the autophagy regulator, Beclin-1, via its NACHT domain. Upon infection with Group A Streptococcus, NLRP4 is recruited to autophagosomes and dissociates from Beclin-1, allowing the initiation of autophagy237. Additionally, NLRP4 associates with the class C vacuolar protein-sorting complex to inhibit autophagosome maturation. NLRP4 also interacts with Rho GDP dissociation inhibitor α to regulate Rho GTPase signalling and facilitate actin-mediated antibacterial autophagy during Streptococcus infection238.
NLRP11 in the regulation of signal transduction and in NLRP3 inflammasome formation.
NLRP11 is a primate-specific protein and is highly expressed in monocytes, THP-1 (human monocyte line), B cells, B cell lymphoma lines, testis, ovary, and lung 239–242. NLRP11 is induced by type I IFN and translocates to the mitochondria to interact with MAVS by its LRR and NACHT domain upon RNA viral infection 243. Furthermore, NLRP11 binds to TRAF6 to promote its degradation in a MAVS-dependent manner, reducing type I IFN signalling and virus-induced apoptosis243. NLRP11 also inhibits TLR signalling by recruiting the ubiquitin ligase RNF19A to catalyze K48-linked ubiquitination of TRAF6 for its degradation, resulting in the suppression of NF-κB activation, MAPK signalling, and pro-inflammatory cytokine production239.
In addition to its roles in regulating these immune signalling pathways, NLRP11 has been shown to negatively regulate NLRP3 inflammasome activation in cultured human cell lines by interacting with DEAD-box protein 3 (DDX3X)244, a protein shown to activate the RLR pathway245. This interaction causes NLRP11 to inhibit DDX3X’s function in enhancing type I IFN responses and NLRP3 inflammasome activation244. In contrast, another study found that NLRP11 can support NLRP3 assembly by functioning as a scaffold. NLRP11 interacts with NLRP3 via its LRR domain and ASC via its PYD domain to promote NLRP3 inflammasome assembly and activation but does not affect the assembly of other inflammasomes246. This study also reported that NLRP11 is necessary for NLRP3 inflammasome responses driven by CAPS-linked NLRP3 mutations246. The exact reason for these differences is currently unknown. This underscores the need for a deeper investigation of these understudied NLR.
Above, we have primarily focused on the immune-associated functions of NLRs. However, NLRs also regulate other biological processes including studies to indicate their importance in the reproductive system which are discussed below.
NLRs in reproduction
There is growing evidence for a mammalian reproduction-associated NLR gene cluster which include NLRP2, NLRP4, NLRP5, NLRP7, NLRP8, NLRP9, NLRP11, NLRP13 and NLRP14. Human NLRs in this reproductive gene cluster — as well as their murine orthologues, namely Nlrp2, Nlrp4a-Nlrp4g, Nlrp5, Nlrp9a-Nlrp9c and Nlrp14, — are highly expressed in oocytes and in the ovaries241,247,248. Except for the gene encoding NLRP14, all reproduction-associated NLRP-encoding genes in the human genome are located on chromosome 19, whereas all mouse reproduction-related Nlrp genes are located on chromosome 7, with the exceptions of Nlrp4g (chr 9) and Nlrp4f (chr 13)248. In this section, we focus on our growing understanding of the functions of some of these proteins.
Regulation of nucleic acid signalling and fertilization by NLRP14.
NLRP14 is expressed in the gonads241 and several NLRP14 mutations are linked to spermatogenic failure249. Interestingly, NLRP14 was reported to function as a negative regulator of the nucleic acid-sensing (NAS) pathway in germ cells by interacting with TBK1 through its N-terminus to suppress TBK1-mediated IFNγ production for supporting fertilization250. Furthermore, NLRP14 was found to interact with MAVS and STING through its LRR domain to negatively regulate the NAS pathway, however, MAVS and STING also resulted in the degradation of NLRP14, hence revealing a feedback loop to prevent the sustained immunosuppressive function of NLRP14250. Although these studies were performed in 293T cells, another group reported that NLRP14 promotes primordial germ cell-like cell differentiation and spermatogenesis through a complex with HSPA2 and BAG2251. These studies provide physiological insights on the functions of NLRP14 and indicate its roles in both immune and gonadal regulation.
NLRP2 in inflammation, proliferation, reproduction, and genomic imprinting.
NLRP2 was one of the first NLRs found to interact with ASC to assemble an inflammasome, leading to IL-1β generation26,252. By contrast, NLRP2 also acts as an inhibitor of the NF-κB pathway and a nonfunctional allelic variant within the NLRP2 NACHT contributes to the hyperactivation of NF-κB pathway and subsequent downstream inflammatory responses252,253. Along with being an inhibitor of NF-κB, NLRP2 also regulates inflammation or cell proliferation by suppressing NF-κB activation in hepatic steatosis254, HLA-C expression in trophoblasts255 and in cancer256. Additionally, it interacts with TBK1 and negatively regulates type I IFN signalling upon viral infection257. In the reproductive system, NLRP2 is essential for early embryogenesis but not oocyte maturation in ice and humans258,259, and interacts with FAF1 (Fas-associated protein factor 1) in mouse ovaries259,260. A germline frameshift mutation in exon 6 of NLRP2 is linked to Beckwith-Wiedemann syndrome, which is a human imprinting disorder [G] 261 that is also associated with NLRP5 and NLRP7 mutations262. NLRP2 also maintains proliferation and viability in human umbilical vein endothelial cells (HUVECs) by promoting MAPK signalling263. These studies demonstrate the potential of NLRP2 in inflammasome activation but also in the dampening of inflammatory and interferon activation. The latter may contribute to the establishment of immune tolerance at the maternal–fetal interface to prevent aberrant immune activation.
NLRP5 regulates embryogenesis
MATER (Maternal Antigen That Embryos Require), a murine counterpart of NLRP5, was one of the first-discovered maternal-effect genes encoding an oocyte-specific protein. It is associated with autoimmune oophoritis and is required for embryonic development in mice264. Additionally, NLRP5 was found to localize in oocyte mitochondria and nucleoli265 and is essential for assembly of the subcortical maternal complex (SCMC), which is a multiprotein complex encoded by maternal-effect genes specifically expressed in oocytes and early embryos in mice266. NLRP5 also regulates mitochondrial biogenesis and its respiratory activity, and it may affect other cell death molecules that contributes to its function in successful preimplantation during early embryogenesis267. In human, NLRP5 is predominantly expressed by oocytes and follicular cells268,269, and has been found to represent a tissue-specific autoantigen involved in hypoparathyroidism in patients with autoimmune polyendocrine syndrome type 1 (APS-1)270. Additionally, NLRP5 variants are not only found in the Beckwith-Wiedemann syndrome, but also in multi-locus imprinting disturbance (MLID) in female patients and is associated with maternal reproductive fitness271. Its direct role in these diseases will be of interest for future investigation.
NLRP7 regulates inflammation and embryogenesis.
NLRP7 is only found in primates241. It is highly expressed in reproductive organs but also broadly expressed in cell lines and most other tissues, except for skeletal muscle, heart and brain. Expression of NLRP7 is increased in LPS- and IL-1β-stimulated PBMCs and macrophages272. NLRP7 mutations specifically affect the female (but not male) reproductive system and are associated with abnormal embryogenesis (recurrent hydatidiform moles [G] in humans)273–275. In contrast to NLRP2, which inhibits NF-κB but not IL-1β production, NLRP7 inhibits caspase 1-dependent IL-1β production272. Another report showed that NLRP7 failed to interact with ASC and activate NF-κB24. By contrast, microbial acylated lipopeptides from bacteria can activate the NLRP7 inflammasome function in macrophages leading to downstream IL-1β production but not pyroptosis 240. The precise structures of the NLRP7 PYD interacting with ASC or the LRR sensing different ligands need to be further investigated to solve these controversial findings. These results suggest that certain NLRs might be either positive or negative regulators in different signalling pathways upon different stimulations in a cell-type specific manner. Others have found that the TLR agonists LPS and Pam3CSK4 can activate NLRP7 inflammasome activity and that a deubiquitinase, STAM-binding protein, increases NLRP7 activity by preventing its trafficking to the lysosome where it is normally degraded276. Additionally, higher NLRP7 expression was reported to promote tumour cell proliferation and metastasis in human colon cancer and to promote the development of M2-like macrophages [G] by increasing NF-kB activation and CCL2 to promote tumor progression277. It will be of interest to investigate if the role of NLRP7 in regulating the inflammasome or NF-κB signaling, which has not been studied in cells of the reproductive system, can be extended to reproductive cells or disorders.
As a final point of interest, in the consensus phylogenetic cluster, the NLRP2 cluster contains NLRP7, suggesting that NLRP7 might originate from a duplication of the NLRP2/7 ancestor in primates241,248. Given that both are highly expressed in the reproductive organs and regulate inflammatory signaling, these two might have overlapping functions in regulating the physiological and pathological inflammatory processes of pregnancy.
NLRP9 in reproduction and immunity.
NLRP9 is mainly expressed in the reproductive system of human and bovine, and mice (Nlrp9a-Nlrp9c) 241,278. In mice, NLRP9 is specifically expressed in ovarian follicles during early embryonic preimplantation279,280. However Nlrp9b, but not Nlrp9a and Nlrp9c, is also uniquely expressed in intestine epithelial cells and associates with an DExH-box RNA helicase 9 (DHX9) to recognize double-strand RNA and functions as an inflammasome for viral clearance during rotavirus infection in mice281. Additionally, NLRP9 expression has been reported in lung cells, brain pericytes and endothelial cells, and microglia278. Human NLRP9 also interacts with ASC to form the inflammasome upon DHX9-mediated rotaviral RNA recognition in HEK293 T cells281. However, two groups recently have reported the crystal structure of human NLRP9 PYD (hNLRP9PYD) and showed that it forms a monomer but not oligomers in solution282,283. One paper also found that it does not nucleate ASC specks in HEK293T cells, which is in contrast to NLRP3, NLRP6 and AIM2 inflammasomes282. Additionally, hNLRP9PYD might exhibit autoinhibitory function based on: (1) charge inversions in the interfaces of hNLRP9PYD which might cause repulsive effects to prevent self- oligomerization and activation282; (2) a bent N-terminal loop of hNLRP9PYD oriented toward the interior of the helical bundle which might prevent filament formation structure and subsequent NLRP9 inflammasome assembly283. NLRP9 is also linked to several inflammatory diseases 278. One study demonstrated that the lack of NLRP9b resulted in reduced neutrophil inflammation but elevated proinflammatory cytokines, NF-κB activation, and oxidative stress in the mouse acute lung injury model284. Much remains unknown regarding NLRP9. The analysis of mice lacking this gene will help understand its role in inflammatory diseases and infections in addition to its role in reproduction system.
In the sections above we have detailed the diverse functions that are mediated by NLR family members. A better understanding of the structure of the different NLR proteins should help us to better define their biology. Indeed, impressive progress has been made in deciphering the structure of several NLR proteins described below.
Structure of NLR proteins
Apaf-1, plant R proteins, and NLRs are members of the STAND subgroup of P-loop ATPases related to the AAA+ superfamily285 [G], a large family of ATPases with diverse molecular functions and a wide assortment of accessory domains. AAA+ superfamily proteins share a common arrangement of an N-terminal αβα fold (NBD) followed by a helical bundle (HD1). Early in the field, X-ray crystallography studies for NLRs were limited to isolated domains, including the PYD, CARD, and LRR domains. A significant milestone was the determination of the crystal structure for the NACHT–LRR domain of NLRC4 in the ADP-bound, autoinhibited state286 (Fig. 5A, left panel). At the time, the structure of the NLRC4 NACHT domain was most similar to the structure determined for Apaf-1 in its inactive state287. Both structures showed that ADP binding involved contributions from the NBD, HD1, and WHD, resulting in a compact, closed conformation. For NLRC4, interdomain interactions between the first LRR and the NBD are also observed in the autoinhibited conformation. Mutations that disrupt WHD-ADP binding or the LRR–NBD interaction destabilize the closed conformation resulting in auto-activation of NLRC4 and caspase 1 processing. These two basic principles for the autoinhibited state were echoed in the crystal structure for the NACHT–LRR domains of NOD2176. Similarly to NLRC4 and Apaf-1, ADP binding involves involves interactions with the NBD, HD1, and WHD resulting in a compact, closed conformation. In addition, the first two LRR units of NOD2 interact with the HD1 and HD2 subdomains within the NACHT domain. The interaction of the LRR and NACHT domains aligns with the hypothesis that ligand binding to the LRR domain disrupts LRR–NACHT interactions and exposes a surface on the NACHT domain for oligomerization.
Figure 5: Structural Insights into NLR Activation.
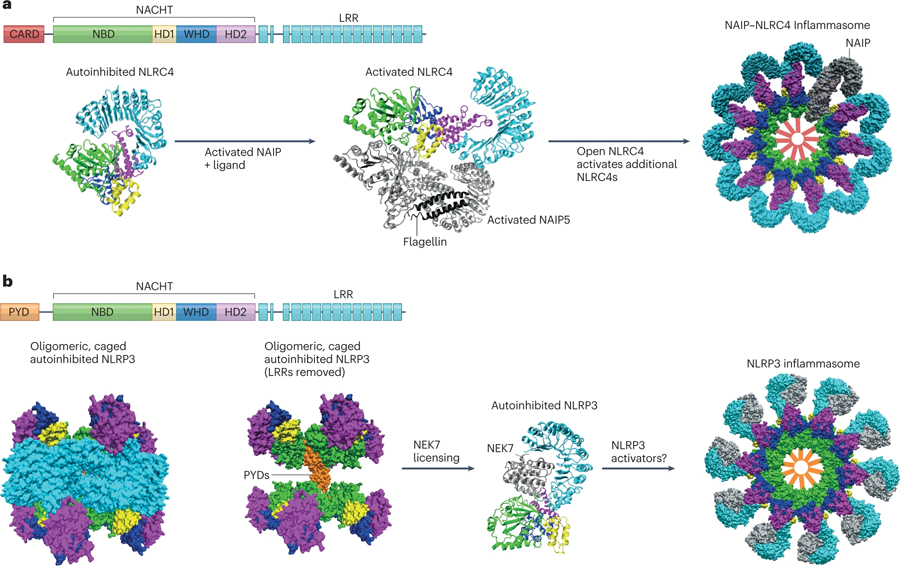
(A) Assembly of the NAIP/NLRC4 Inflammasome. The primary structure of NLRC4 is shown, and color coding for domains is replicated in the structural depictions. The crystal structure of closed, autoinhibited NLRC4 is adapted from PDB:4kxf286. Interactions between the HD2 and LRR domains stabilize the closed conformation. The cryo-EM structure of activated NLRC4 and NAIP5 is adapted from PDB:6b5b290. Activated NAIP5 (silver) with bound flagellin (black) initiates a conformational change in NLRC4 to an open, activated form which can activate downstream NLRC4 molecules. Finally, the cryo-EM structure of the assembled NAIP/NLRC4 inflammasome is adapted from PDB:3jbl288. A top view of the surface representation shows a single NAIP (gray) present in the complex with ten NLRC4 molecules. Red bars are added in the center of the complex to show the location of the NLRC4 CARDs which will recruit pro-caspase 1. (B) NLRP3 structural regulation preceding inflammasome assembly. The primary structure of NLRP3 is shown, and color coding is replicated in the structural depictions. The cryo-EM structure for autoinhibited NLRP3 alone is adapted from PDB:7pzc292. The left panel shows a surface representation of an oligomeric complex consisting of ten NLRP3 molecules. When the LRR domains are excluded from the image, the position of the PYDs within the “cage” shows how they are sequestered and prevented from initiating spurious inflammation. The cryo-EM structure of NLRP3 with bound NEK7 is adapted from PDB:6npy70. NEK7 interaction with the LRR domain of NLRP3 prevents NLRP3 from forming caged oligomers. All NLRP3 structures shown here show NLRP3 in a closed, autoinhibited conformation bound to ADP. Presumably, NEK7 licenses NLRP3 to freely interact with activating signals or ligands to form an inflammasome. Upon activation, NLRP3 assembles into an inflammasome disc with 10–11 NLRP3 molecules329. A cartoon representation is shown as the PDB file is currently unavailable. Structural representations were created using VMD330.
The advent of cryo-EM for high-resolution structural analysis of large proteins and proteins complexes allowed for breakthroughs in NLR structure determination. High-resolution structures for full-length NLRs and oligomerized complexes have now been determined. Cryo-EM structures for an inflammasome in the activated, oligomerized state were solved for NAIP2-NLRC4 inflammasomes activated by the bacterial type III secretion inner rod protein PrgJ 288,289. These studies show that PrgJ binding to NAIP2 results in a 90° rotation at a hinge site between HD1 and WHD. This conformational change exposes a surface in the NACHT of NAIP2, which can interact with an acceptor surface in NLRC4, and the activated NLRC4 can promote a similar conformational change in another NLRC4 molecule. Propagating NLRC4 activation results in the formation of a disc-like structure composed of a single NAIP with ten NLRC4 molecules (Fig. 5A, right panel). Subsequent cryo-EM studies of the NAIP5-NLRC4 complex with bound flagellin provided the first glimpse into the molecular interaction of an activating ligand with an NLR sensor. The flagellin-NAIP5 interaction shows that binding of ligand involves multiple domains including the NACHT, LRR, and BIR domains290. Whether this mode of ligand-sensor interaction extends to other NLR proteins remains to be determined.
A simplistic model for inflammasome formation involves activation of cytosolic, monomeric NLRs by a ligand leading to oligomerization and recruitment of pro-caspase 1. However, recent cryo-EM studies of NLRP3 suggest that there are additional layers of regulation beyond the presence of ligand that determine whether an inflammasome will form. Cryo-EM studies with NLRP3 alone revealed an oligomeric complex in which the pyrin domains are sequestered within a cage291–293 (Fig.5B, left panel). Isolation of pyrin domains in such a manner would prevent spurious interaction with ASC, providing protection against inappropriate inflammasome activation (Fig. 5B, middle panel). Interestingly, a previous cryo-EM structure for NLRP3 had been determined in a complex with NEK770 which is known to license NLRP3 activation (Fig. 5B, right panel). Both the caged NLRP3 and NLRP3-NEK7 structures contain NLRP3 in an autoinhibited, ADP-bound conformation. In vitro addition of NEK7 disrupts the caged oligomers, suggesting that NLRP3 inflammasome activation involves more than just the presence of activating ligand. Future studies will determine if other NLRs employ additional regulatory mechanisms.
Emerging concepts and conclusions
In the past two decades, the NLR field has gone through a revolution. However, while some NLRs are exceedingly well investigated, many are understudied. We propose some emerging concepts as well as areas that are deserving of further attention.
First, aside from well-established reports on activation and functions of NLRs in innate immunity, there are accumulated evidence suggesting NLRs function in other cell types. We have discussed the area of NLRs in the reproductive system, bolstered by fascinating genetic data supporting key roles for NLRs in this setting. Furthermore, several NLRs have roles for regulating adaptive immunity and immunometabolism in cells. NLRP3 is the first and most intensively linked to crosstalk between innate and adaptive immunity294 and metabolism29,295. Adenosine-induced NLRP11 can reduce IFN-γ and IL-17A production of human peripheral CD4+ T lymphocytes296. NLRC3 attenuates TRAF6 and modulates CD4+ T cell metabolism. NLRX1 has also been found to regulate glycolysis and oxidative phosphorylation in T cells,297,298. These studies shed light on the roles of NLRs in an array of cell types and provide a much broader perspective regarding their importance beyond innate immunity.
Second, many NLRs exhibit multiple functional roles, and this may be attributed to numerous reasons. One is that their cellular localization may be dynamic. NLRs are located in different cellular compartments and their organelle-specific, membranous or non-membranous localization may determine their ultimate functions. The mitochondrial location of NLRX1, and the nuclear localization of transcriptional regulators CIITA and NLRC5 have long been known. NLRP3 has been shown to be associated with mitochondria and the dispersed trans Golgi network299. NOD1 and NOD2 associate with the plasma membrane and endosomes177,187, and membrane localization is dependent on S-palmitoylation179 raising the possibility that post-translational modifications may influence localization of other NLRs. Another possibility is that different NLR isoforms, perhaps generated via alternative splicing or post-translational modification, may show distinct functions. One possible approach to address these different possibilities is to assess if distinct interactomes are formed by an NLR protein to conduct different functions in different cell types. Such interactomes may vary depending on all of the factors described above, including differential expression in cells with distinct protein compositions, divergent localization in organelles, varied post-translational alterations and different isoforms. Finally, it is important to re-evaluate data obtained with in vitro cell lines especially in the context of overexpressed proteins without validation in vivo, since these may result in nonphysiologic findings.
Third, several NLRs are paired to perform their function. The most prominent examples are the pairing of NAIPs and NLRC4. NLRP3 and NLRC4 are important for the simultaneous activation of inflammasomes in response to some stimuli. However, opposing functions have also been described; for example, NLRP12-dependent degradation of NOD2 results in the functional inhibition of NOD2300. This type of crosstalk is envisioned to expand the NLR regulatory network and their impact.
Finally, a common theme that emerges for NLR proteins (NLRX1, NLRC3, NLRP3, NLRP6, NLRP1) is the potential for binding to nucleic acid PAMPs and DAMPs. This flexibility would allow a single NLR to function both in host responses against pathogens as well as in host stress responses. Whether this is a shared function among many NLRs and what the divergent functional consequences and downstream signalling consequences of this may be will be of great interest to explore.
Obviously, there are numerous other areas of NLR biology that deserve further investigation, such as their therapeutic targeting and their precise roles in disease. We trust that the next decades of research on the NLR family will be just as exhilarating as the first two.
Acknowledgements
This work is supported by NIH grants R56 AI158314, R01 AI158314, AI029564, AI141333, DK094779, U19 AI067798 and R35 CA232109 to J.P.-Y.T.
Glossary
- MHC class II transactivator (CIITA)
The master transcriptional regulator of MHC class II expression.
- NACHT domain
NAIP, CIITA, HET-E (incompatibility locus protein from the fungus Podospora anserina) and TEP1 (telomerase-associated protein 1). The NACHT domain is a subgroup of the STAND class of P-loop NTPases, and is composed of four subdomains (NBD, HD1, WHD, and HD2). This domain allows for nucleotide-binding dependent conformational changes and oligomerization to influence diverse biological outcomes such as transcriptional activation, cytokine signalling, and pyroptosis.
- Apaf-1
Apoptotic protease-activating factor 1. A protein of the STAND class of P-loop ATPases that is central to initiating apoptosis upon mitochondrial cytochrome C release into the cytosol. In addition to the STAND ATPase module, Apaf-1 contains an N-terminal CARD and C-terminal WD-40 repeats. The formation of the apoptosome and activation of caspase 9 upon cytochrome C binding was a biochemical model that significantly influenced early NLR studies.
- ASC
Apoptosis-associated speck-like protein containing a CARD. Also called PYCARD and TMS1. Adaptor protein that contains a PYD and CARD allowing for inflammasome recruitment of pro-caspase 1.
- STAND
A subgroup of the AAA+ ATPase superfamily that includes both apoptotic (AP)-ATPases as well as NACHT ATPases. The model for STAND protein function involves ADP binding stabilizing a closed, inactive state, and exchange for ATP triggers a conformational change to the open, active state.
- SXY cis-elements
A regulatory module comprising four elements: S or W box, X1 box, X2 box and the Y box. When bound by their cognate transcription factors, these sites allow for assembly of the MHC enhanceosome.
- cryopyrin-induced autoinflammatory syndromes (CAPS)
Autoinflammatory diseases caused by gain-of-function mutations in NLRP3 (cryopyrin).
- AIM2
Absent in melanoma 2. An innate immune sensor that detects cytosolic dsDNA resulting in inflammasome formation. AIM2 is composed of an N-terminal PYD and a C-terminal dsDNA-binding HIN domain distinguishing it from NLR inflammasome proteins.
- NR100
N-terminal domain of rodent Nlrp1 proteins, approximately 100 amino acids. Whereas human NLRP1 possesses an N-terminal PYD, mouse Nlrp1 proteins contain this sequence of unknown function. Alphafold predicts this region to be mostly disordered.
- T3SS
Type III secretion system. A multiprotein membrane apparatus present in gram-negative bacteria used to inject proteins into host cytosol. Components of this nanomachine trigger NAIP/NLRC4 inflammasome assembly.
- Inflammasomopathies
Autoinflammatory diseases resulting from gain-of-function mutations in inflammasome-forming NLRs.
- Anakinra
A short-acting human recombinant IL-1receptor (IL-1R) antagonist that can competitively inhibit the binding of IL-1β and IL-1α to IL-1R and block IL-1 signal transduction.
- imprinting disorder
Diseases caused by genetic defects or epigenetic mutations affecting imprinted chromosomal regions or genes that are expressed in a parent-of-origin specific manner.
- hydatidiform moles
A hydatifiform mole is a rare condition in which tissue around a fertilized egg that would normally have developed into the placenta instead develops as an abnormal mass of cells.
- M2-like macrophages
M1’ and ‘M2’ are classifications historically used to define macrophages activated in vitro as pro-inflammatory (when ‘classically’ activated with IFNγ and LPS) or anti-inflammatory (when ‘alternatively’ activated with IL-4 or IL-10), respectively. However, in vivo macrophages are highly specialized, transcriptomically dynamic and extremely heterogeneous with regards to their phenotypes and functions, which are continuously shaped by their tissue microenvironment. Therefore, the M1 or M2 classification is too simplistic to explain the true nature of in vivo macrophages, although these terms are still often used to indicate whether the macrophages in question are more pro- or anti-inflammatory.
Footnotes
Competing Interests
J. Ting is a co-founder of IMMvention Therapeutix.
References
- 1.Baker B, Zambryski P, Staskawicz B & Dinesh-Kumar SP Signaling in plant-microbe interactions. Science 276, 726–733, doi: 10.1126/science.276.5313.726 (1997). [DOI] [PubMed] [Google Scholar]
- 2.McHale L, Tan X, Koehl P & Michelmore RW Plant NBS-LRR proteins: adaptable guards. Genome Biol 7, 212, doi: 10.1186/gb-2006-7-4-212 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbary A, Djian-Caporalino C, Palloix A & Castagnone-Sereno P Host genetic resistance to root-knot nematodes, Meloidogyne spp., in Solanaceae: from genes to the field. Pest Manag Sci 71, 1591–1598, doi: 10.1002/ps.4091 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Steimle V, Otten LA, Zufferey M & Mach B Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). Cell 75, 135–146 (1993). [PubMed] [Google Scholar]
- 5.Harton JA & Ting JP Class II transactivator: mastering the art of major histocompatibility complex expression. Mol Cell Biol 20, 6185–6194, doi: 10.1128/MCB.20.17.6185-6194.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harton JA, Linhoff MW, Zhang J & Ting JP Cutting edge: CATERPILLER: a large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. J Immunol 169, 4088–4093, doi: 10.4049/jimmunol.169.8.4088 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Roy N et al. The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell 80, 167–178, doi: 10.1016/0092-8674(95)90461-1 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Liston P et al. Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature 379, 349–353, doi: 10.1038/379349a0 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Koonin EV & Aravind L The NACHT family - a new group of predicted NTPases implicated in apoptosis and MHC transcription activation. Trends Biochem Sci 25, 223–224, doi: 10.1016/s0968-0004(00)01577-2 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Inohara N et al. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J Biol Chem 274, 14560–14567, doi: 10.1074/jbc.274.21.14560 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Bertin J et al. Human CARD4 protein is a novel CED-4/Apaf-1 cell death family member that activates NF-kappaB. J Biol Chem 274, 12955–12958, doi: 10.1074/jbc.274.19.12955 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Ogura Y et al. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem 276, 4812–4818, doi: 10.1074/jbc.M008072200 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Girardin SE et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 300, 1584–1587, doi: 10.1126/science.1084677 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Chamaillard M et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol 4, 702–707, doi: 10.1038/ni945 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Inohara N et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J Biol Chem 278, 5509–5512, doi: 10.1074/jbc.C200673200 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Girardin SE et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem 278, 8869–8872, doi: 10.1074/jbc.C200651200 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Hugot JP et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411, 599–603, doi: 10.1038/35079107 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Ogura Y et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411, 603–606, doi: 10.1038/35079114 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Inohara N & Nunez G The NOD: a signaling module that regulates apoptosis and host defense against pathogens. Oncogene 20, 6473–6481, doi: 10.1038/sj.onc.1204787 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Poyet JL et al. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem 276, 28309–28313, doi: 10.1074/jbc.C100250200 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Martinon F, Burns K & Tschopp J The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell 10, 417–426, doi: 10.1016/s1097-2765(02)00599-3 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Tschopp J, Martinon F & Burns K NALPs: a novel protein family involved in inflammation. Nat Rev Mol Cell Biol 4, 95–104, doi: 10.1038/nrm1019 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Wang L et al. PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-kappa B and caspase-1-dependent cytokine processing. J Biol Chem 277, 29874–29880, doi: 10.1074/jbc.M203915200 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Grenier JM et al. Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF-kappaB and caspase-1. FEBS Lett 530, 73–78, doi: 10.1016/s0014-5793(02)03416-6 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Hoffman HM, Mueller JL, Broide DH, Wanderer AA & Kolodner RD Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat Genet 29, 301–305, doi: 10.1038/ng756 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agostini L et al. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity 20, 319–325, doi: 10.1016/s1074-7613(04)00046-9 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Gao LA et al. Prokaryotic innate immunity through pattern recognition of conserved viral proteins. Science 377, eabm4096, doi: 10.1126/science.abm4096 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y & Shao F Diverse mechanisms for inflammasome sensing of cytosolic bacteria and bacterial virulence. Curr Opin Microbiol 29, 37–42, doi: 10.1016/j.mib.2015.10.003 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Sharma BR & Kanneganti TD NLRP3 inflammasome in cancer and metabolic diseases. Nat Immunol 22, 550–559, doi: 10.1038/s41590-021-00886-5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cressman DE, Chin KC, Taxman DJ & Ting JP A defect in the nuclear translocation of CIITA causes a form of type II bare lymphocyte syndrome. Immunity 10, 163–171, doi: 10.1016/s1074-7613(00)80017-5 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Steimle V, Siegrist CA, Mottet A, Lisowska-Grospierre B & Mach B Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science 265, 106–109, doi: 10.1126/science.8016643 (1994). [DOI] [PubMed] [Google Scholar]
- 32.Chang CH, Fontes JD, Peterlin M & Flavell RA Class II transactivator (CIITA) is sufficient for the inducible expression of major histocompatibility complex class II genes. J Exp Med 180, 1367–1374, doi: 10.1084/jem.180.4.1367 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harton JA, Cressman DE, Chin KC, Der CJ & Ting JP GTP binding by class II transactivator: role in nuclear import. Science 285, 1402–1405, doi: 10.1126/science.285.5432.1402 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Masternak K et al. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev 14, 1156–1166 (2000). [PMC free article] [PubMed] [Google Scholar]
- 35.Choi NM, Majumder P & Boss JM Regulation of major histocompatibility complex class II genes. Curr Opin Immunol 23, 81–87, doi: 10.1016/j.coi.2010.09.007 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Armstrong TD, Clements VK, Martin BK, Ting JP & Ostrand-Rosenberg S Major histocompatibility complex class II-transfected tumor cells present endogenous antigen and are potent inducers of tumor-specific immunity. Proc Natl Acad Sci U S A 94, 6886–6891, doi: 10.1073/pnas.94.13.6886 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yazawa T et al. Lack of class II transactivator causes severe deficiency of HLA-DR expression in small cell lung cancer. J Pathol 187, 191–199, doi: (1999). [DOI] [PubMed] [Google Scholar]
- 38.Accolla RS, Ramia E, Tedeschi A & Forlani G CIITA-Driven MHC Class II Expressing Tumor Cells as Antigen Presenting Cell Performers: Toward the Construction of an Optimal Anti-tumor Vaccine. Front Immunol 10, 1806, doi: 10.3389/fimmu.2019.01806 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steidl C et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature 471, 377–381, doi: 10.1038/nature09754 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meissner TB et al. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc Natl Acad Sci U S A 107, 13794–13799, doi: 10.1073/pnas.1008684107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robbins GR et al. Regulation of class I major histocompatibility complex (MHC) by nucleotide-binding domain, leucine-rich repeat-containing (NLR) proteins. J Biol Chem 287, 24294–24303, doi: 10.1074/jbc.M112.364604 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biswas A, Meissner TB, Kawai T & Kobayashi KS Cutting edge: impaired MHC class I expression in mice deficient for Nlrc5/class I transactivator. J Immunol 189, 516–520, doi: 10.4049/jimmunol.1200064 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staehli F et al. NLRC5 deficiency selectively impairs MHC class I- dependent lymphocyte killing by cytotoxic T cells. J Immunol 188, 3820–3828, doi: 10.4049/jimmunol.1102671 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Yoshihama S et al. NLRC5/MHC class I transactivator is a target for immune evasion in cancer. Proc Natl Acad Sci U S A 113, 5999–6004, doi: 10.1073/pnas.1602069113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui J et al. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell 141, 483–496, doi: 10.1016/j.cell.2010.03.040 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benko S, Kovacs EG, Hezel F & Kufer TA NLRC5 Functions beyond MHC I Regulation-What Do We Know So Far? Front Immunol 8, 150, doi: 10.3389/fimmu.2017.00150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Booshehri LM & Hoffman HM CAPS and NLRP3. J Clin Immunol 39, 277–286, doi: 10.1007/s10875-019-00638-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swanson KV, Deng M & Ting JP The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol 19, 477–489, doi: 10.1038/s41577-019-0165-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi J et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665, doi: 10.1038/nature15514 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Kayagaki N et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671, doi: 10.1038/nature15541 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Sborgi L et al. GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J 35, 1766–1778, doi: 10.15252/embj.201694696 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding J et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116, doi: 10.1038/nature18590 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Liu X et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158, doi: 10.1038/nature18629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aglietti RA et al. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc Natl Acad Sci U S A 113, 7858–7863, doi: 10.1073/pnas.1607769113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feldmann J et al. Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am J Hum Genet 71, 198–203, doi: 10.1086/341357 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bauernfeind FG et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol 183, 787–791, doi: 10.4049/jimmunol.0901363 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Franchi L et al. Calcium-independent phospholipase A2 beta is dispensable in inflammasome activation and its inhibition by bromoenol lactone. J Innate Immun 1, 607–617, doi: 10.1159/000227263 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xing Y et al. Cutting Edge: TRAF6 Mediates TLR/IL-1R Signaling-Induced Nontranscriptional Priming of the NLRP3 Inflammasome. J Immunol 199, 1561–1566, doi: 10.4049/jimmunol.1700175 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Song N et al. NLRP3 Phosphorylation Is an Essential Priming Event for Inflammasome Activation. Mol Cell 68, 185–197 e186, doi: 10.1016/j.molcel.2017.08.017 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Juliana C et al. Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J Biol Chem 287, 36617–36622, doi: 10.1074/jbc.M112.407130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Py BF, Kim MS, Vakifahmetoglu-Norberg H & Yuan J Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol Cell 49, 331–338, doi: 10.1016/j.molcel.2012.11.009 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Song H et al. The E3 ubiquitin ligase TRIM31 attenuates NLRP3 inflammasome activation by promoting proteasomal degradation of NLRP3. Nat Commun 7, 13727, doi: 10.1038/ncomms13727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barry R et al. SUMO-mediated regulation of NLRP3 modulates inflammasome activity. Nat Commun 9, 3001, doi: 10.1038/s41467-018-05321-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu A et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156, 1193–1206, doi: 10.1016/j.cell.2014.02.008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cai X et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 156, 1207–1222, doi: 10.1016/j.cell.2014.01.063 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boucher D et al. Caspase-1 self-cleavage is an intrinsic mechanism to terminate inflammasome activity. J Exp Med 215, 827–840, doi: 10.1084/jem.20172222 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmid-Burgk JL et al. A Genome-wide CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) Screen Identifies NEK7 as an Essential Component of NLRP3 Inflammasome Activation. J Biol Chem 291, 103–109, doi: 10.1074/jbc.C115.700492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He Y, Zeng MY, Yang D, Motro B & Nunez G NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 530, 354–357, doi: 10.1038/nature16959 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shi H et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat Immunol 17, 250–258, doi: 10.1038/ni.3333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sharif H et al. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 570, 338–343, doi: 10.1038/s41586-019-1295-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kayagaki N et al. Non-canonical inflammasome activation targets caspase-11. Nature 479, 117–121, doi: 10.1038/nature10558 (2011). [DOI] [PubMed] [Google Scholar]
- 72.Broz P et al. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature 490, 288–291, doi: 10.1038/nature11419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rathinam VA et al. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell 150, 606–619, doi: 10.1016/j.cell.2012.07.007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hagar JA, Powell DA, Aachoui Y, Ernst RK & Miao EA Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science 341, 1250–1253, doi: 10.1126/science.1240988 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shi J et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192, doi: 10.1038/nature13683 (2014). [DOI] [PubMed] [Google Scholar]
- 76.Ganesan S et al. Caspase-8 modulates dectin-1 and complement receptor 3-driven IL-1beta production in response to beta-glucans and the fungal pathogen, Candida albicans. J Immunol 193, 2519–2530, doi: 10.4049/jimmunol.1400276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gurung P et al. FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. J Immunol 192, 1835–1846, doi: 10.4049/jimmunol.1302839 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Allam R et al. Mitochondrial apoptosis is dispensable for NLRP3 inflammasome activation but non-apoptotic caspase-8 is required for inflammasome priming. EMBO Rep 15, 982–990, doi: 10.15252/embr.201438463 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gringhuis SI et al. Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1beta via a noncanonical caspase-8 inflammasome. Nat Immunol 13, 246–254, doi: 10.1038/ni.2222 (2012). [DOI] [PubMed] [Google Scholar]
- 80.Broz P et al. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J Exp Med 207, 1745–1755, doi: 10.1084/jem.20100257 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Man SM et al. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc Natl Acad Sci U S A 111, 7403–7408, doi: 10.1073/pnas.1402911111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Freeman L et al. NLR members NLRC4 and NLRP3 mediate sterile inflammasome activation in microglia and astrocytes. J Exp Med 214, 1351–1370, doi: 10.1084/jem.20150237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kalantari P et al. Dual engagement of the NLRP3 and AIM2 inflammasomes by plasmodium-derived hemozoin and DNA during malaria. Cell Rep 6, 196–210, doi: 10.1016/j.celrep.2013.12.014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Karki R et al. Concerted activation of the AIM2 and NLRP3 inflammasomes orchestrates host protection against Aspergillus infection. Cell Host Microbe 17, 357–368, doi: 10.1016/j.chom.2015.01.006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Swanson KV et al. A noncanonical function of cGAMP in inflammasome priming and activation. J Exp Med 214, 3611–3626, doi: 10.1084/jem.20171749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sanman LE et al. Disruption of glycolytic flux is a signal for inflammasome signaling and pyroptotic cell death. Elife 5, e13663, doi: 10.7554/eLife.13663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Duewell P et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 464, 1357–1361, doi: 10.1038/nature08938 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wen H et al. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat Immunol 12, 408–415, doi: 10.1038/ni.2022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou R, Yazdi AS, Menu P & Tschopp J A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225, doi: 10.1038/nature09663 (2011). [DOI] [PubMed] [Google Scholar]
- 90.Shimada K et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 36, 401–414, doi: 10.1016/j.immuni.2012.01.009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kanneganti TD et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature 440, 233–236, doi: 10.1038/nature04517 (2006). [DOI] [PubMed] [Google Scholar]
- 92.Subramanian N, Natarajan K, Clatworthy MR, Wang Z & Germain RN The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell 153, 348–361, doi: 10.1016/j.cell.2013.02.054 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Park S et al. The mitochondrial antiviral protein MAVS associates with NLRP3 and regulates its inflammasome activity. J Immunol 191, 4358–4366, doi: 10.4049/jimmunol.1301170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Franchi L et al. Cytosolic double-stranded RNA activates the NLRP3 inflammasome via MAVS-induced membrane permeabilization and K+ efflux. J Immunol 193, 4214–4222, doi: 10.4049/jimmunol.1400582 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Munoz-Planillo R et al. K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38, 1142–1153, doi: 10.1016/j.immuni.2013.05.016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wolf AJ et al. Hexokinase Is an Innate Immune Receptor for the Detection of Bacterial Peptidoglycan. Cell 166, 624–636, doi: 10.1016/j.cell.2016.05.076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Coll RC et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med 21, 248–255, doi: 10.1038/nm.3806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Coll RC, Schroder K & Pelegrin P NLRP3 and pyroptosis blockers for treating inflammatory diseases. Trends Pharmacol Sci 43, 653–668, doi: 10.1016/j.tips.2022.04.003 (2022). [DOI] [PubMed] [Google Scholar]
- 99.Bertin J & DiStefano PS The PYRIN domain: a novel motif found in apoptosis and inflammation proteins. Cell Death Differ 7, 1273–1274, doi: 10.1038/sj.cdd.4400774 (2000). [DOI] [PubMed] [Google Scholar]
- 100.Chu ZL et al. A novel enhancer of the Apaf1 apoptosome involved in cytochrome c-dependent caspase activation and apoptosis. J Biol Chem 276, 9239–9245, doi: 10.1074/jbc.M006309200 (2001). [DOI] [PubMed] [Google Scholar]
- 101.Hlaing T et al. Molecular cloning and characterization of DEFCAP-L and -S, two isoforms of a novel member of the mammalian Ced-4 family of apoptosis proteins. J Biol Chem 276, 9230–9238, doi: 10.1074/jbc.M009853200 (2001). [DOI] [PubMed] [Google Scholar]
- 102.Fenini G, Karakaya T, Hennig P, Di Filippo M & Beer HD The NLRP1 Inflammasome in Human Skin and Beyond. Int J Mol Sci 21, doi: 10.3390/ijms21134788 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jin Y et al. NALP1 in vitiligo-associated multiple autoimmune disease. N Engl J Med 356, 1216–1225, doi: 10.1056/NEJMoa061592 (2007). [DOI] [PubMed] [Google Scholar]
- 104.Levandowski CB et al. NLRP1 haplotypes associated with vitiligo and autoimmunity increase interleukin-1beta processing via the NLRP1 inflammasome. Proc Natl Acad Sci U S A 110, 2952–2956, doi: 10.1073/pnas.1222808110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ekman AK, Verma D, Fredrikson M, Bivik C & Enerback C Genetic variations of NLRP1: susceptibility in psoriasis. Br J Dermatol 171, 1517–1520, doi: 10.1111/bjd.13178 (2014). [DOI] [PubMed] [Google Scholar]
- 106.Zhong FL et al. Germline NLRP1 Mutations Cause Skin Inflammatory and Cancer Susceptibility Syndromes via Inflammasome Activation. Cell 167, 187–202 e117, doi: 10.1016/j.cell.2016.09.001 (2016). [DOI] [PubMed] [Google Scholar]
- 107.Grandemange S et al. A new autoinflammatory and autoimmune syndrome associated with NLRP1 mutations: NAIAD (NLRP1-associated autoinflammation with arthritis and dyskeratosis). Ann Rheum Dis 76, 1191–1198, doi: 10.1136/annrheumdis-2016-210021 (2017). [DOI] [PubMed] [Google Scholar]
- 108.Ozretic P et al. Association of NLRP1 Coding Polymorphism with Lung Function and Serum IL-1beta Concentration in Patients Diagnosed with Chronic Obstructive Pulmonary Disease (COPD). Genes (Basel) 10, doi: 10.3390/genes10100783 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Drutman SB et al. Homozygous NLRP1 gain-of-function mutation in siblings with a syndromic form of recurrent respiratory papillomatosis. Proc Natl Acad Sci U S A 116, 19055–19063, doi: 10.1073/pnas.1906184116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sastalla I et al. Transcriptional analysis of the three Nlrp1 paralogs in mice. BMC Genomics 14, 188, doi: 10.1186/1471-2164-14-188 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhong FL et al. Human DPP9 represses NLRP1 inflammasome and protects against autoinflammatory diseases via both peptidase activity and FIIND domain binding. J Biol Chem 293, 18864–18878, doi: 10.1074/jbc.RA118.004350 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chavarria-Smith J, Mitchell PS, Ho AM, Daugherty MD & Vance RE Functional and Evolutionary Analyses Identify Proteolysis as a General Mechanism for NLRP1 Inflammasome Activation. PLoS Pathog 12, e1006052, doi: 10.1371/journal.ppat.1006052 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Finger JN et al. Autolytic proteolysis within the function to find domain (FIIND) is required for NLRP1 inflammasome activity. J Biol Chem 287, 25030–25037, doi: 10.1074/jbc.M112.378323 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Frew BC, Joag VR & Mogridge J Proteolytic processing of Nlrp1b is required for inflammasome activity. PLoS Pathog 8, e1002659, doi: 10.1371/journal.ppat.1002659 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Boyden ED & Dietrich WF Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet 38, 240–244, doi: 10.1038/ng1724 (2006). [DOI] [PubMed] [Google Scholar]
- 116.Newman ZL et al. Susceptibility to anthrax lethal toxin-induced rat death is controlled by a single chromosome 10 locus that includes rNlrp1. PLoS Pathog 6, e1000906, doi: 10.1371/journal.ppat.1000906 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Levinsohn JL et al. Anthrax lethal factor cleavage of Nlrp1 is required for activation of the inflammasome. PLoS Pathog 8, e1002638, doi: 10.1371/journal.ppat.1002638 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hellmich KA et al. Anthrax lethal factor cleaves mouse nlrp1b in both toxin-sensitive and toxin-resistant macrophages. PLoS One 7, e49741, doi: 10.1371/journal.pone.0049741 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chavarria-Smith J & Vance RE Direct proteolytic cleavage of NLRP1B is necessary and sufficient for inflammasome activation by anthrax lethal factor. PLoS Pathog 9, e1003452, doi: 10.1371/journal.ppat.1003452 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chui AJ et al. N-terminal degradation activates the NLRP1B inflammasome. Science 364, 82–85, doi: 10.1126/science.aau1208 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sandstrom A et al. Functional degradation: A mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 364, doi: 10.1126/science.aau1330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xu H et al. The N-end rule ubiquitin ligase UBR2 mediates NLRP1B inflammasome activation by anthrax lethal toxin. EMBO J 38, e101996, doi: 10.15252/embj.2019101996 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Robinson KS et al. Enteroviral 3C protease activates the human NLRP1 inflammasome in airway epithelia. Science 370, doi: 10.1126/science.aay2002 (2020). [DOI] [PubMed] [Google Scholar]
- 124.de Vasconcelos NM et al. DPP8/DPP9 inhibition elicits canonical Nlrp1b inflammasome hallmarks in murine macrophages. Life Sci Alliance 2, doi: 10.26508/lsa.201900313 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Okondo MC et al. Inhibition of Dpp8/9 Activates the Nlrp1b Inflammasome. Cell Chem Biol 25, 262–267 e265, doi: 10.1016/j.chembiol.2017.12.013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gai K et al. DPP8/9 inhibitors are universal activators of functional NLRP1 alleles. Cell Death Dis 10, 587, doi: 10.1038/s41419-019-1817-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.D’Osualdo A et al. CARD8 and NLRP1 undergo autoproteolytic processing through a ZU5-like domain. PLoS One 6, e27396, doi: 10.1371/journal.pone.0027396 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hollingsworth LR et al. DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation. Nature 592, 778–783, doi: 10.1038/s41586-021-03350-4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huang M et al. Structural and biochemical mechanisms of NLRP1 inhibition by DPP9. Nature 592, 773–777, doi: 10.1038/s41586-021-03320-w (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Neiman-Zenevich J, Stuart S, Abdel-Nour M, Girardin SE & Mogridge J Listeria monocytogenes and Shigella flexneri Activate the NLRP1B Inflammasome. Infect Immun 85, doi: 10.1128/IAI.00338-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ewald SE, Chavarria-Smith J & Boothroyd JC NLRP1 is an inflammasome sensor for Toxoplasma gondii. Infect Immun 82, 460–468, doi: 10.1128/IAI.01170-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Okondo MC et al. DPP8 and DPP9 inhibition induces pro-caspase-1-dependent monocyte and macrophage pyroptosis. Nat Chem Biol 13, 46–53, doi: 10.1038/nchembio.2229 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gorfu G et al. Dual role for inflammasome sensors NLRP1 and NLRP3 in murine resistance to Toxoplasma gondii. mBio 5, doi: 10.1128/mBio.01117-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bauernfried S, Scherr MJ, Pichlmair A, Duderstadt KE & Hornung V Human NLRP1 is a sensor for double-stranded RNA. Science 371, doi: 10.1126/science.abd0811 (2021). [DOI] [PubMed] [Google Scholar]
- 135.Liao KC & Mogridge J Activation of the Nlrp1b inflammasome by reduction of cytosolic ATP. Infect Immun 81, 570–579, doi: 10.1128/IAI.01003-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Neiman-Zenevich J, Liao KC & Mogridge J Distinct regions of NLRP1B are required to respond to anthrax lethal toxin and metabolic inhibition. Infect Immun 82, 3697–3703, doi: 10.1128/IAI.02167-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ball DP et al. Oxidized thioredoxin-1 restrains the NLRP1 inflammasome. Sci Immunol 7, eabm7200, doi: 10.1126/sciimmunol.abm7200 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jia J, Zhang X, Xu G, Zeng X & Li L Thioredoxin-1 inhibits amyloid-beta25–35-induced activation of NLRP1/caspase-1/GSDMD pyroptotic pathway in PC12 cells. Mol Biol Rep 49, 3445–3452, doi: 10.1007/s11033-022-07177-8 (2022). [DOI] [PubMed] [Google Scholar]
- 139.Robinson KS et al. ZAKalpha-driven ribotoxic stress response activates the human NLRP1 inflammasome. Science 377, 328–335, doi: 10.1126/science.abl6324 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mariathasan S et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430, 213–218, doi: 10.1038/nature02664 (2004). [DOI] [PubMed] [Google Scholar]
- 141.Miao EA et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol 7, 569–575, doi: 10.1038/ni1344 (2006). [DOI] [PubMed] [Google Scholar]
- 142.Miao EA et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci U S A 107, 3076–3080, doi: 10.1073/pnas.0913087107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhao Y et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477, 596–600, doi: 10.1038/nature10510 (2011). [DOI] [PubMed] [Google Scholar]
- 144.Rayamajhi M, Zak DE, Chavarria-Smith J, Vance RE & Miao EA Cutting edge: Mouse NAIP1 detects the type III secretion system needle protein. J Immunol 191, 3986–3989, doi: 10.4049/jimmunol.1301549 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yang J, Zhao Y, Shi J & Shao F Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc Natl Acad Sci U S A 110, 14408–14413, doi: 10.1073/pnas.1306376110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Amer A et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem 281, 35217–35223, doi: 10.1074/jbc.M604933200 (2006). [DOI] [PubMed] [Google Scholar]
- 147.Franchi L et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol 7, 576–582, doi: 10.1038/ni1346 (2006). [DOI] [PubMed] [Google Scholar]
- 148.Romberg N, Vogel TP & Canna SW NLRC4 inflammasomopathies. Curr Opin Allergy Clin Immunol 17, 398–404, doi: 10.1097/ACI.0000000000000396 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang SB et al. DDX17 is an essential mediator of sterile NLRC4 inflammasome activation by retrotransposon RNAs. Sci Immunol 6, eabi4493, doi: 10.1126/sciimmunol.abi4493 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Poh L et al. Evidence that NLRC4 inflammasome mediates apoptotic and pyroptotic microglial death following ischemic stroke. Brain Behav Immun 75, 34–47, doi: 10.1016/j.bbi.2018.09.001 (2019). [DOI] [PubMed] [Google Scholar]
- 151.Normand S et al. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc Natl Acad Sci U S A 108, 9601–9606, doi: 10.1073/pnas.1100981108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Elinav E et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757, doi: 10.1016/j.cell.2011.04.022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mukherjee S et al. Deubiquitination of NLRP6 inflammasome by Cyld critically regulates intestinal inflammation. Nat Immunol 21, 626–635, doi: 10.1038/s41590-020-0681-x (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen GY, Liu M, Wang F, Bertin J & Nunez G A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J Immunol 186, 7187–7194, doi: 10.4049/jimmunol.1100412 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chiang HY et al. IL-22 initiates an IL-18-dependent epithelial response circuit to enforce intestinal host defence. Nat Commun 13, 874, doi: 10.1038/s41467-022-28478-3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wlodarska M et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell 156, 1045–1059, doi: 10.1016/j.cell.2014.01.026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mamantopoulos M et al. Nlrp6- and ASC-Dependent Inflammasomes Do Not Shape the Commensal Gut Microbiota Composition. Immunity 47, 339–348 e334, doi: 10.1016/j.immuni.2017.07.011 (2017). [DOI] [PubMed] [Google Scholar]
- 158.Lemire P et al. The NLR Protein NLRP6 Does Not Impact Gut Microbiota Composition. Cell Rep 21, 3653–3661, doi: 10.1016/j.celrep.2017.12.026 (2017). [DOI] [PubMed] [Google Scholar]
- 159.Henao-Mejia J et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482, 179–185, doi: 10.1038/nature10809 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ji X et al. NLRP6 exerts a protective role via NF-kB with involvement of CCL20 in a mouse model of alcoholic hepatitis. Biochem Biophys Res Commun 528, 485–492, doi: 10.1016/j.bbrc.2020.05.171 (2020). [DOI] [PubMed] [Google Scholar]
- 161.Schneider KM et al. Imbalanced gut microbiota fuels hepatocellular carcinoma development by shaping the hepatic inflammatory microenvironment. Nat Commun 13, 3964, doi: 10.1038/s41467-022-31312-5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu Z et al. Intestinal Candida albicans Promotes Hepatocarcinogenesis by Up-Regulating NLRP6. Front Microbiol 13, 812771, doi: 10.3389/fmicb.2022.812771 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Levy M et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 163, 1428–1443, doi: 10.1016/j.cell.2015.10.048 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hara H et al. The NLRP6 Inflammasome Recognizes Lipoteichoic Acid and Regulates Gram-Positive Pathogen Infection. Cell 175, 1651–1664 e1614, doi: 10.1016/j.cell.2018.09.047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang P et al. Nlrp6 regulates intestinal antiviral innate immunity. Science 350, 826–830, doi: 10.1126/science.aab3145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shen C et al. Phase separation drives RNA virus-induced activation of the NLRP6 inflammasome. Cell 184, 5759–5774 e5720, doi: 10.1016/j.cell.2021.09.032 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Anand PK et al. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature 488, 389–393, doi: 10.1038/nature11250 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ghimire L et al. NLRP6 negatively regulates pulmonary host defense in Gram-positive bacterial infection through modulating neutrophil recruitment and function. PLoS Pathog 14, e1007308, doi: 10.1371/journal.ppat.1007308 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lu WL et al. NLRP6 suppresses the inflammatory response of human periodontal ligament cells by inhibiting NF-kappaB and ERK signal pathways. Int Endod J 52, 999–1009, doi: 10.1111/iej.13091 (2019). [DOI] [PubMed] [Google Scholar]
- 170.Caruso R, Warner N, Inohara N & Nunez G NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity 41, 898–908, doi: 10.1016/j.immuni.2014.12.010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Trindade BC & Chen GY NOD1 and NOD2 in inflammatory and infectious diseases. Immunol Rev 297, 139–161, doi: 10.1111/imr.12902 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Stafford CA et al. Phosphorylation of muramyl peptides by NAGK is required for NOD2 activation. Nature 609, 590–596, doi: 10.1038/s41586-022-05125-x (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Inohara N et al. An induced proximity model for NF-kappa B activation in the Nod1/RICK and RIP signaling pathways. J Biol Chem 275, 27823–27831, doi: 10.1074/jbc.M003415200 (2000). [DOI] [PubMed] [Google Scholar]
- 174.da Silva Correia J, Miranda Y, Leonard N, Hsu J & Ulevitch RJ Regulation of Nod1-mediated signaling pathways. Cell Death Differ 14, 830–839, doi: 10.1038/sj.cdd.4402070 (2007). [DOI] [PubMed] [Google Scholar]
- 175.Miceli-Richard C et al. CARD15 mutations in Blau syndrome. Nat Genet 29, 19–20, doi: 10.1038/ng720 (2001). [DOI] [PubMed] [Google Scholar]
- 176.Maekawa S, Ohto U, Shibata T, Miyake K & Shimizu T Crystal structure of NOD2 and its implications in human disease. Nat Commun 7, 11813, doi: 10.1038/ncomms11813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Irving AT et al. The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host Microbe 15, 623–635, doi: 10.1016/j.chom.2014.04.001 (2014). [DOI] [PubMed] [Google Scholar]
- 178.Nakamura N et al. Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature 509, 240–244, doi: 10.1038/nature13133 (2014). [DOI] [PubMed] [Google Scholar]
- 179.Lu Y et al. Palmitoylation of NOD1 and NOD2 is required for bacterial sensing. Science 366, 460–467, doi: 10.1126/science.aau6391 (2019). [DOI] [PubMed] [Google Scholar]
- 180.Moore CB et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature 451, 573–577, doi: 10.1038/nature06501 (2008). [DOI] [PubMed] [Google Scholar]
- 181.Tattoli I et al. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep 9, 293–300, doi: 10.1038/sj.embor.7401161 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Arnoult D et al. An N-terminal addressing sequence targets NLRX1 to the mitochondrial matrix. J Cell Sci 122, 3161–3168, doi: 10.1242/jcs.051193 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hong M, Yoon SI & Wilson IA Structure and functional characterization of the RNA-binding element of the NLRX1 innate immune modulator. Immunity 36, 337–347, doi: 10.1016/j.immuni.2011.12.018 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lu P et al. Modeling-Enabled Characterization of Novel NLRX1 Ligands. PLoS One 10, e0145420, doi: 10.1371/journal.pone.0145420 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Li X et al. Viral DNA Binding to NLRC3, an Inhibitory Nucleic Acid Sensor, Unleashes STING, a Cyclic Dinucleotide Receptor that Activates Type I Interferon. Immunity 50, 591–599 e596, doi: 10.1016/j.immuni.2019.02.009 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fekete T, Bencze D, Biro E, Benko S & Pazmandi K Focusing on the Cell Type Specific Regulatory Actions of NLRX1. Int J Mol Sci 22, doi: 10.3390/ijms22031316 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Allen IC et al. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity 34, 854–865, doi: 10.1016/j.immuni.2011.03.026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Guo H et al. NLRX1 Sequesters STING to Negatively Regulate the Interferon Response, Thereby Facilitating the Replication of HIV-1 and DNA Viruses. Cell Host Microbe 19, 515–528, doi: 10.1016/j.chom.2016.03.001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Xia X et al. NLRX1 negatively regulates TLR-induced NF-kappaB signaling by targeting TRAF6 and IKK. Immunity 34, 843–853, doi: 10.1016/j.immuni.2011.02.022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Koblansky AA et al. The Innate Immune Receptor NLRX1 Functions as a Tumor Suppressor by Reducing Colon Tumorigenesis and Key Tumor-Promoting Signals. Cell Rep 14, 2562–2575, doi: 10.1016/j.celrep.2016.02.064 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Coutermarsh-Ott S et al. NLRX1 suppresses tumorigenesis and attenuates histiocytic sarcoma through the negative regulation of NF-kappaB signaling. Oncotarget 7, 33096–33110, doi: 10.18632/oncotarget.8861 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Feng H et al. NLRX1 promotes immediate IRF1-directed antiviral responses by limiting dsRNA-activated translational inhibition mediated by PKR. Nat Immunol 18, 1299–1309, doi: 10.1038/ni.3853 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lei Y et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity 36, 933–946, doi: 10.1016/j.immuni.2012.03.025 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Luo X et al. HPV16 drives cancer immune escape via NLRX1-mediated degradation of STING. J Clin Invest 130, 1635–1652, doi: 10.1172/JCI129497 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lei Y et al. EGFR-targeted mAb therapy modulates autophagy in head and neck squamous cell carcinoma through NLRX1-TUFM protein complex. Oncogene 35, 4698–4707, doi: 10.1038/onc.2016.11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Huang JH et al. NLRX1 Facilitates Histoplasma capsulatum-Induced LC3-Associated Phagocytosis for Cytokine Production in Macrophages. Front Immunol 9, 2761, doi: 10.3389/fimmu.2018.02761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang Y et al. Listeria hijacks host mitophagy through a novel mitophagy receptor to evade killing. Nat Immunol 20, 433–446, doi: 10.1038/s41590-019-0324-2 (2019). [DOI] [PubMed] [Google Scholar]
- 198.Singh K et al. NLRX1 regulates TNF-alpha-induced mitochondria-lysosomal crosstalk to maintain the invasive and metastatic potential of breast cancer cells. Biochim Biophys Acta Mol Basis Dis 1865, 1460–1476, doi: 10.1016/j.bbadis.2019.02.018 (2019). [DOI] [PubMed] [Google Scholar]
- 199.Li S, Zhou Y, Gu X, Zhang X & Jia Z NLRX1/FUNDC1/NIPSNAP1–2 axis regulates mitophagy and alleviates intestinal ischaemia/reperfusion injury. Cell Prolif 54, e12986, doi: 10.1111/cpr.12986 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Peng J et al. Morphine-induced microglial immunosuppression via activation of insufficient mitophagy regulated by NLRX1. J Neuroinflammation 19, 87, doi: 10.1186/s12974-022-02453-7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Aikawa C et al. NLRX1 Negatively Regulates Group A Streptococcus Invasion and Autophagy Induction by Interacting With the Beclin 1-UVRAG Complex. Front Cell Infect Microbiol 8, 403, doi: 10.3389/fcimb.2018.00403 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Leber A et al. Activation of NLRX1 by NX-13 Alleviates Inflammatory Bowel Disease through Immunometabolic Mechanisms in CD4(+) T Cells. J Immunol 203, 3407–3415, doi: 10.4049/jimmunol.1900364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Conti BJ et al. CATERPILLER 16.2 (CLR16.2), a novel NBD/LRR family member that negatively regulates T cell function. J Biol Chem 280, 18375–18385, doi: 10.1074/jbc.M413169200 (2005). [DOI] [PubMed] [Google Scholar]
- 204.Uchimura T et al. The Innate Immune Sensor NLRC3 Acts as a Rheostat that Fine-Tunes T Cell Responses in Infection and Autoimmunity. Immunity 49, 1049–1061 e1046, doi: 10.1016/j.immuni.2018.10.008 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hu S et al. NLRC3 negatively regulates CD4+ T cells and impacts protective immunity during Mycobacterium tuberculosis infection. PLoS Pathog 14, e1007266, doi: 10.1371/journal.ppat.1007266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Schneider M et al. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-kappaB. Nat Immunol 13, 823–831, doi: 10.1038/ni.2378 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhang L et al. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity 40, 329–341, doi: 10.1016/j.immuni.2014.01.010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Tocker AM et al. The Scaffolding Protein IQGAP1 Interacts with NLRC3 and Inhibits Type I IFN Production. J Immunol 199, 2896–2909, doi: 10.4049/jimmunol.1601370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wang C et al. NLRC3 High Expression Represents a Novel Predictor for Positive Overall Survival Correlated With CCL5 and CXCL9 in HCC Patients. Front Oncol 12, 815326, doi: 10.3389/fonc.2022.815326 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Liu R et al. Expression profile of innate immune receptors, NLRs and AIM2, in human colorectal cancer: correlation with cancer stages and inflammasome components. Oncotarget 6, 33456–33469, doi: 10.18632/oncotarget.5587 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Karki R et al. NLRC3 is an inhibitory sensor of PI3K-mTOR pathways in cancer. Nature 540, 583–587, doi: 10.1038/nature20597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Williams KL, Taxman DJ, Linhoff MW, Reed W & Ting JP Cutting edge: Monarch-1: a pyrin/nucleotide-binding domain/leucine-rich repeat protein that controls classical and nonclassical MHC class I genes. J Immunol 170, 5354–5358, doi: 10.4049/jimmunol.170.11.5354 (2003). [DOI] [PubMed] [Google Scholar]
- 213.Williams KL et al. The CATERPILLER protein monarch-1 is an antagonist of toll-like receptor-, tumor necrosis factor alpha-, and Mycobacterium tuberculosis-induced pro-inflammatory signals. J Biol Chem 280, 39914–39924, doi: 10.1074/jbc.M502820200 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lich JD et al. Monarch-1 suppresses non-canonical NF-kappaB activation and p52-dependent chemokine expression in monocytes. J Immunol 178, 1256–1260, doi: 10.4049/jimmunol.178.3.1256 (2007). [DOI] [PubMed] [Google Scholar]
- 215.Vladimer GI et al. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity 37, 96–107, doi: 10.1016/j.immuni.2012.07.006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ataide MA et al. Malaria-induced NLRP12/NLRP3-dependent caspase-1 activation mediates inflammation and hypersensitivity to bacterial superinfection. PLoS Pathog 10, e1003885, doi: 10.1371/journal.ppat.1003885 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Allen IC et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-kappaB signaling. Immunity 36, 742–754, doi: 10.1016/j.immuni.2012.03.012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Zaki MH et al. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell 20, 649–660, doi:S1535-6108(11)00401-6 [pii] 10.1016/j.ccr.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Chen L et al. NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat Immunol 18, 541–551, doi: 10.1038/ni.3690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sun Z et al. Gut Microbiota-Mediated NLRP12 Expression Drives the Attenuation of Dextran Sulphate Sodium-Induced Ulcerative Colitis by Qingchang Wenzhong Decoction. Evid Based Complement Alternat Med 2019, 9839474, doi: 10.1155/2019/9839474 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Truax AD et al. The Inhibitory Innate Immune Sensor NLRP12 Maintains a Threshold against Obesity by Regulating Gut Microbiota Homeostasis. Cell Host Microbe 24, 364–378 e366, doi: 10.1016/j.chom.2018.08.009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Sharma N et al. Differential Expression Profile of NLRs and AIM2 in Glioma and Implications for NLRP12 in Glioblastoma. Sci Rep 9, 8480, doi: 10.1038/s41598-019-44854-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zamoshnikova A et al. NLRP12 is a neutrophil-specific, negative regulator of in vitro cell migration but does not modulate LPS- or infection-induced NF-kappaB or ERK signalling. Immunobiology 221, 341–346, doi: 10.1016/j.imbio.2015.10.001 (2016). [DOI] [PubMed] [Google Scholar]
- 224.Jeru I et al. Mutations in NALP12 cause hereditary periodic fever syndromes. Proc Natl Acad Sci U S A 105, 1614–1619, doi: 10.1073/pnas.0708616105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Jeru I et al. Role of interleukin-1beta in NLRP12-associated autoinflammatory disorders and resistance to anti-interleukin-1 therapy. Arthritis Rheum 63, 2142–2148, doi: 10.1002/art.30378 (2011). [DOI] [PubMed] [Google Scholar]
- 226.Wang Y et al. PYNOD, a novel Apaf-1/CED4-like protein is an inhibitor of ASC and caspase-1. Int Immunol 16, 777–786, doi: 10.1093/intimm/dxh081 (2004). [DOI] [PubMed] [Google Scholar]
- 227.Imamura R et al. Anti-inflammatory activity of PYNOD and its mechanism in humans and mice. J Immunol 184, 5874–5884, doi: 10.4049/jimmunol.0900779 (2010). [DOI] [PubMed] [Google Scholar]
- 228.Eisenbarth SC et al. NLRP10 is a NOD-like receptor essential to initiate adaptive immunity by dendritic cells. Nature 484, 510–513, doi: 10.1038/nature11012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Krishnaswamy JK et al. Coincidental loss of DOCK8 function in NLRP10-deficient and C3H/HeJ mice results in defective dendritic cell migration. Proc Natl Acad Sci U S A 112, 3056–3061, doi: 10.1073/pnas.1501554112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Clay GM et al. An Anti-Inflammatory Role for NLRP10 in Murine Cutaneous Leishmaniasis. J Immunol 199, 2823–2833, doi: 10.4049/jimmunol.1500832 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Damm A, Giebeler N, Zamek J, Zigrino P & Kufer TA Epidermal NLRP10 contributes to contact hypersensitivity responses in mice. Eur J Immunol 46, 1959–1969, doi: 10.1002/eji.201646401 (2016). [DOI] [PubMed] [Google Scholar]
- 232.Nakajima S et al. Characterization of Innate and Adaptive Immune Responses in PYNOD-Deficient Mice. Immunohorizons 2, 129–141, doi: 10.4049/immunohorizons.1700074 (2018). [DOI] [PubMed] [Google Scholar]
- 233.Zhang P et al. Expression analysis of the NLRP gene family suggests a role in human preimplantation development. PLoS One 3, e2755, doi: 10.1371/journal.pone.0002755 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Fiorentino L et al. A novel PAAD-containing protein that modulates NF-kappa B induction by cytokines tumor necrosis factor-alpha and interleukin-1beta. J Biol Chem 277, 35333–35340, doi: 10.1074/jbc.M200446200 (2002). [DOI] [PubMed] [Google Scholar]
- 235.Cui J et al. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol 13, 387–395, doi: 10.1038/ni.2239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Eibl C et al. Structural and functional analysis of the NLRP4 pyrin domain. Biochemistry 51, 7330–7341, doi: 10.1021/bi3007059 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Jounai N et al. NLRP4 negatively regulates autophagic processes through an association with beclin1. J Immunol 186, 1646–1655, doi: 10.4049/jimmunol.1001654 (2011). [DOI] [PubMed] [Google Scholar]
- 238.Nozawa T, Aikawa C, Minowa-Nozawa A & Nakagawa I The intracellular microbial sensor NLRP4 directs Rho-actin signaling to facilitate Group A Streptococcus-containing autophagosome-like vacuole formation. Autophagy 13, 1841–1854, doi: 10.1080/15548627.2017.1358343 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Wu C et al. NLRP11 attenuates Toll-like receptor signalling by targeting TRAF6 for degradation via the ubiquitin ligase RNF19A. Nat Commun 8, 1977, doi: 10.1038/s41467-017-02073-3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Khare S et al. An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity 36, 464–476, doi: 10.1016/j.immuni.2012.02.001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Tian X, Pascal G & Monget P Evolution and functional divergence of NLRP genes in mammalian reproductive systems. BMC Evol Biol 9, 202, doi: 10.1186/1471-2148-9-202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ellwanger K et al. The NLR family pyrin domain-containing 11 protein contributes to the regulation of inflammatory signaling. J Biol Chem 293, 2701–2710, doi: 10.1074/jbc.RA117.000152 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Qin Y et al. NLRP11 disrupts MAVS signalosome to inhibit type I interferon signaling and virus-induced apoptosis. EMBO Rep 18, 2160–2171, doi: 10.15252/embr.201744480 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kienes I et al. DDX3X Links NLRP11 to the Regulation of Type I Interferon Responses and NLRP3 Inflammasome Activation. Front Immunol 12, 653883, doi: 10.3389/fimmu.2021.653883 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Oshiumi H, Sakai K, Matsumoto M & Seya T DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential. Eur J Immunol 40, 940–948, doi: 10.1002/eji.200940203 (2010). [DOI] [PubMed] [Google Scholar]
- 246.Gangopadhyay A et al. NLRP3 licenses NLRP11 for inflammasome activation in human macrophages. Nat Immunol 23, 892–903, doi: 10.1038/s41590-022-01220-3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Hruz T et al. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008, 420747, doi: 10.1155/2008/420747 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Van Gorp H, Kuchmiy A, Van Hauwermeiren F & Lamkanfi M NOD-like receptors interfacing the immune and reproductive systems. FEBS J 281, 4568–4582, doi: 10.1111/febs.13014 (2014). [DOI] [PubMed] [Google Scholar]
- 249.Westerveld GH et al. Mutations in the testis-specific NALP14 gene in men suffering from spermatogenic failure. Hum Reprod 21, 3178–3184, doi: 10.1093/humrep/del293 (2006). [DOI] [PubMed] [Google Scholar]
- 250.Abe T et al. Germ-Cell-Specific Inflammasome Component NLRP14 Negatively Regulates Cytosolic Nucleic Acid Sensing to Promote Fertilization. Immunity 46, 621–634, doi: 10.1016/j.immuni.2017.03.020 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Yin Y et al. A noncanonical role of NOD-like receptor NLRP14 in PGCLC differentiation and spermatogenesis. Proc Natl Acad Sci U S A 117, 22237–22248, doi: 10.1073/pnas.2005533117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Bruey JM et al. PAN1/NALP2/PYPAF2, an inducible inflammatory mediator that regulates NF-kappaB and caspase-1 activation in macrophages. J Biol Chem 279, 51897–51907, doi: 10.1074/jbc.M406741200 (2004). [DOI] [PubMed] [Google Scholar]
- 253.Fontalba A, Gutierrez O & Fernandez-Luna JL NLRP2, an inhibitor of the NF-kappaB pathway, is transcriptionally activated by NF-kappaB and exhibits a nonfunctional allelic variant. J Immunol 179, 8519–8524, doi: 10.4049/jimmunol.179.12.8519 (2007). [DOI] [PubMed] [Google Scholar]
- 254.Li C, Liu Q & Xie L Suppressing NLRP2 expression accelerates hepatic steatosis: A mechanism involving inflammation and oxidative stress. Biochem Biophys Res Commun 507, 22–29, doi: 10.1016/j.bbrc.2018.10.132 (2018). [DOI] [PubMed] [Google Scholar]
- 255.Tilburgs T et al. NLRP2 is a suppressor of NF-kB signaling and HLA-C expression in human trophoblasts. Biol Reprod 96, 831–842, doi: 10.1093/biolre/iox009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Li T et al. NLRP2 inhibits cell proliferation and migration by regulating EMT in lung adenocarcinoma cells. Cell Biol Int 46, 588–598, doi: 10.1002/cbin.11755 (2022). [DOI] [PubMed] [Google Scholar]
- 257.Yang Y et al. NLRP2 negatively regulates antiviral immunity by interacting with TBK1. Eur J Immunol 48, 1817–1825, doi: 10.1002/eji.201847589 (2018). [DOI] [PubMed] [Google Scholar]
- 258.Peng H et al. Nlrp2, a maternal effect gene required for early embryonic development in the mouse. PLoS One 7, e30344, doi: 10.1371/journal.pone.0030344 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Mu J et al. Mutations in NLRP2 and NLRP5 cause female infertility characterised by early embryonic arrest. J Med Genet 56, 471–480, doi: 10.1136/jmedgenet-2018-105936 (2019). [DOI] [PubMed] [Google Scholar]
- 260.Peng H et al. NLRP2 and FAF1 deficiency blocks early embryogenesis in the mouse. Reproduction 154, 245–251, doi: 10.1530/REP-16-0629 (2017). [DOI] [PubMed] [Google Scholar]
- 261.Meyer E et al. Germline mutation in NLRP2 (NALP2) in a familial imprinting disorder (Beckwith-Wiedemann Syndrome). PLoS Genet 5, e1000423, doi: 10.1371/journal.pgen.1000423 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Begemann M et al. Maternal variants in NLRP and other maternal effect proteins are associated with multilocus imprinting disturbance in offspring. J Med Genet 55, 497–504, doi: 10.1136/jmedgenet-2017-105190 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Zhang X, Lu X, Yu L, Gu Y & Qu F Downregulation of NLRP2 inhibits HUVEC viability by inhibiting the MAPK signaling pathway. Mol Med Rep 19, 85–92, doi: 10.3892/mmr.2018.9625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Tong ZB et al. Mater, a maternal effect gene required for early embryonic development in mice. Nat Genet 26, 267–268, doi: 10.1038/81547 (2000). [DOI] [PubMed] [Google Scholar]
- 265.Tong ZB et al. Developmental expression and subcellular localization of mouse MATER, an oocyte-specific protein essential for early development. Endocrinology 145, 1427–1434, doi: 10.1210/en.2003-1160 (2004). [DOI] [PubMed] [Google Scholar]
- 266.Li L, Baibakov B & Dean J A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell 15, 416–425, doi: 10.1016/j.devcel.2008.07.010 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Fernandes R et al. NLRP5 mediates mitochondrial function in mouse oocytes and embryos. Biol Reprod 86, 138, 131–110, doi: 10.1095/biolreprod.111.093583 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Tong ZB, Bondy CA, Zhou J & Nelson LM A human homologue of mouse Mater, a maternal effect gene essential for early embryonic development. Hum Reprod 17, 903–911, doi: 10.1093/humrep/17.4.903 (2002). [DOI] [PubMed] [Google Scholar]
- 269.Sena P et al. Human MATER localization in specific cell domains of oocytes and follicular cells. Reprod Biomed Online 18, 226–234, doi: 10.1016/s1472-6483(10)60260-x (2009). [DOI] [PubMed] [Google Scholar]
- 270.Alimohammadi M et al. Autoimmune polyendocrine syndrome type 1 and NALP5, a parathyroid autoantigen. N Engl J Med 358, 1018–1028, doi: 10.1056/NEJMoa0706487 (2008). [DOI] [PubMed] [Google Scholar]
- 271.Docherty LE et al. Mutations in NLRP5 are associated with reproductive wastage and multilocus imprinting disorders in humans. Nat Commun 6, 8086, doi: 10.1038/ncomms9086 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kinoshita T, Wang Y, Hasegawa M, Imamura R & Suda T PYPAF3, a PYRIN-containing APAF-1-like protein, is a feedback regulator of caspase-1-dependent interleukin-1beta secretion. J Biol Chem 280, 21720–21725, doi: 10.1074/jbc.M410057200 (2005). [DOI] [PubMed] [Google Scholar]
- 273.Murdoch S et al. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet 38, 300–302, doi: 10.1038/ng1740 (2006). [DOI] [PubMed] [Google Scholar]
- 274.Moglabey YB et al. Genetic mapping of a maternal locus responsible for familial hydatidiform moles. Hum Mol Genet 8, 667–671, doi: 10.1093/hmg/8.4.667 (1999). [DOI] [PubMed] [Google Scholar]
- 275.Carriere J, Dorfleutner A & Stehlik C NLRP7: From inflammasome regulation to human disease. Immunology 163, 363–376, doi: 10.1111/imm.13372 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Bednash JS et al. Targeting the deubiquitinase STAMBP inhibits NALP7 inflammasome activity. Nat Commun 8, 15203, doi: 10.1038/ncomms15203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Li B et al. NLRP7 deubiquitination by USP10 promotes tumor progression and tumor-associated macrophage polarization in colorectal cancer. J Exp Clin Cancer Res 40, 126, doi: 10.1186/s13046-021-01920-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Mullins B & Chen J NLRP9 in innate immunity and inflammation. Immunology 162, 262–267, doi: 10.1111/imm.13290 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Kanzaki S et al. Involvement of Nlrp9a/b/c in mouse preimplantation development. Reproduction 160, 181–191, doi: 10.1530/REP-19-0516 (2020). [DOI] [PubMed] [Google Scholar]
- 280.Amoushahi M et al. Maternally contributed Nlrp9b expressed in human and mouse ovarian follicles contributes to early murine preimplantation development. J Assist Reprod Genet 37, 1355–1365, doi: 10.1007/s10815-020-01767-w (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Zhu S et al. Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature 546, 667–670, doi: 10.1038/nature22967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Marleaux M, Anand K, Latz E & Geyer M Crystal structure of the human NLRP9 pyrin domain suggests a distinct mode of inflammasome assembly. FEBS Lett 594, 2383–2395, doi: 10.1002/1873-3468.13865 (2020). [DOI] [PubMed] [Google Scholar]
- 283.Ha HJ & Park HH Crystal structure of the human NLRP9 pyrin domain reveals a bent N-terminal loop that may regulate inflammasome assembly. FEBS Lett 594, 2396–2405, doi: 10.1002/1873-3468.13866 (2020). [DOI] [PubMed] [Google Scholar]
- 284.Yanling Q et al. Inhibition of NLRP9b attenuates acute lung injury through suppressing inflammation, apoptosis and oxidative stress in murine and cell models. Biochem Biophys Res Commun 503, 436–443, doi: 10.1016/j.bbrc.2018.04.079 (2018). [DOI] [PubMed] [Google Scholar]
- 285.Leipe DD, Koonin EV & Aravind L STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J Mol Biol 343, 1–28, doi: 10.1016/j.jmb.2004.08.023 (2004). [DOI] [PubMed] [Google Scholar]
- 286.Hu Z et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 341, 172–175, doi: 10.1126/science.1236381 (2013). [DOI] [PubMed] [Google Scholar]
- 287.Riedl SJ, Li W, Chao Y, Schwarzenbacher R & Shi Y Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature 434, 926–933, doi: 10.1038/nature03465 (2005). [DOI] [PubMed] [Google Scholar]
- 288.Zhang L et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 350, 404–409, doi: 10.1126/science.aac5789 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Hu Z et al. Structural and biochemical basis for induced self-propagation of NLRC4. Science 350, 399–404, doi: 10.1126/science.aac5489 (2015). [DOI] [PubMed] [Google Scholar]
- 290.Tenthorey JL et al. The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion. Science 358, 888–893, doi: 10.1126/science.aao1140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Andreeva L et al. NLRP3 cages revealed by full-length mouse NLRP3 structure control pathway activation. Cell 184, 6299–6312 e6222, doi: 10.1016/j.cell.2021.11.011 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Hochheiser IV et al. Structure of the NLRP3 decamer bound to the cytokine release inhibitor CRID3. Nature 604, 184–189, doi: 10.1038/s41586-022-04467-w (2022). [DOI] [PubMed] [Google Scholar]
- 293.Ohto U et al. Structural basis for the oligomerization-mediated regulation of NLRP3 inflammasome activation. Proc Natl Acad Sci U S A 119, e2121353119, doi: 10.1073/pnas.2121353119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Arbore G et al. T helper 1 immunity requires complement-driven NLRP3 inflammasome activity in CD4(+) T cells. Science 352, aad1210, doi: 10.1126/science.aad1210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Chou WC, Rampanelli E, Li X & Ting JP Impact of intracellular innate immune receptors on immunometabolism. Cell Mol Immunol 19, 337–351, doi: 10.1038/s41423-021-00780-y (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Ozel I, Akkaya I, Oylumlu E, Uzel G & Ciraci C Adenosine-Induced NLRP11 in B Lymphoblasts Suppresses Human CD4(+) T Helper Cell Responses. J Immunol Res 2020, 1421795, doi: 10.1155/2020/1421795 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Guo H et al. Multi-omics analyses reveal that HIV-1 alters CD4(+) T cell immunometabolism to fuel virus replication. Nat Immunol 22, 423–433, doi: 10.1038/s41590-021-00898-1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Leber A et al. NLRX1 Regulates Effector and Metabolic Functions of CD4(+) T Cells. J Immunol 198, 2260–2268, doi: 10.4049/jimmunol.1601547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Chen J & Chen ZJ PtdIns4P on dispersed trans-Golgi network mediates NLRP3 inflammasome activation. Nature 564, 71–76, doi: 10.1038/s41586-018-0761-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Normand S et al. Proteasomal degradation of NOD2 by NLRP12 in monocytes promotes bacterial tolerance and colonization by enteropathogens. Nat Commun 9, 5338, doi: 10.1038/s41467-018-07750-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Swanberg M et al. MHC2TA is associated with differential MHC molecule expression and susceptibility to rheumatoid arthritis, multiple sclerosis and myocardial infarction. Nat Genet 37, 486–494, doi: 10.1038/ng1544 (2005). [DOI] [PubMed] [Google Scholar]
- 302.Yao Q et al. A new category of autoinflammatory disease associated with NOD2 gene mutations. Arthritis Res Ther 13, R148, doi: 10.1186/ar3462 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Kabesch M et al. Association between polymorphisms in caspase recruitment domain containing protein 15 and allergy in two German populations. J Allergy Clin Immunol 111, 813–817, doi: 10.1067/mai.2003.1336 (2003). [DOI] [PubMed] [Google Scholar]
- 304.Canna SW et al. An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat Genet 46, 1140–1146, doi: 10.1038/ng.3089 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Romberg N et al. Mutation of NLRC4 causes a syndrome of enterocolitis and autoinflammation. Nat Genet 46, 1135–1139, doi: 10.1038/ng.3066 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Kitamura A, Sasaki Y, Abe T, Kano H & Yasutomo K An inherited mutation in NLRC4 causes autoinflammation in human and mice. J Exp Med 211, 2385–2396, doi: 10.1084/jem.20141091 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Steiner A et al. Recessive NLRC4-Autoinflammatory Disease Reveals an Ulcerative Colitis Locus. J Clin Immunol 42, 325–335, doi: 10.1007/s10875-021-01175-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Zupin L et al. NLRC5 polymorphism is associated with susceptibility to chronic periodontitis. Immunobiology 222, 704–708, doi: 10.1016/j.imbio.2017.01.001 (2017). [DOI] [PubMed] [Google Scholar]
- 309.Zhong J et al. NLRP3, NLRC4 and NLRC5 Gene Polymorphisms Associate with Susceptibility of Pulmonary Aspergillosis in Non-Neutropenic Patients. J Clin Med 11, doi: 10.3390/jcm11071870 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Soler VJ et al. Whole exome sequencing identifies a mutation for a novel form of corneal intraepithelial dyskeratosis. J Med Genet 50, 246–254, doi: 10.1136/jmedgenet-2012-101325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Pontillo A, Catamo E, Arosio B, Mari D & Crovella S NALP1/NLRP1 genetic variants are associated with Alzheimer disease. Alzheimer Dis Assoc Disord 26, 277–281, doi: 10.1097/WAD.0b013e318231a8ac (2012). [DOI] [PubMed] [Google Scholar]
- 312.Pontillo A, Vendramin A, Catamo E, Fabris A & Crovella S The missense variation Q705K in CIAS1/NALP3/NLRP3 gene and an NLRP1 haplotype are associated with celiac disease. Am J Gastroenterol 106, 539–544, doi: 10.1038/ajg.2010.474 (2011). [DOI] [PubMed] [Google Scholar]
- 313.Magitta NF et al. A coding polymorphism in NALP1 confers risk for autoimmune Addison’s disease and type 1 diabetes. Genes Immun 10, 120–124, doi: 10.1038/gene.2008.85 (2009). [DOI] [PubMed] [Google Scholar]
- 314.Alkhateeb A, Jarun Y & Tashtoush R Polymorphisms in NLRP1 gene and susceptibility to autoimmune thyroid disease. Autoimmunity 46, 215–221, doi: 10.3109/08916934.2013.768617 (2013). [DOI] [PubMed] [Google Scholar]
- 315.Pontillo A et al. Polimorphisms in inflammasome genes are involved in the predisposition to systemic lupus erythematosus. Autoimmunity 45, 271–278, doi: 10.3109/08916934.2011.637532 (2012). [DOI] [PubMed] [Google Scholar]
- 316.Serrano A et al. Evidence of association of the NLRP1 gene with giant cell arteritis. Ann Rheum Dis 72, 628–630, doi: 10.1136/annrheumdis-2012-202609 (2013). [DOI] [PubMed] [Google Scholar]
- 317.Witola WH et al. NALP1 influences susceptibility to human congenital toxoplasmosis, proinflammatory cytokine response, and fate of Toxoplasma gondii-infected monocytic cells. Infect Immun 79, 756–766, doi: 10.1128/IAI.00898-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Sui J et al. NLRP1 gene polymorphism influences gene transcription and is a risk factor for rheumatoid arthritis in han chinese. Arthritis Rheum 64, 647–654, doi: 10.1002/art.33370 (2012). [DOI] [PubMed] [Google Scholar]
- 319.Nakanishi H et al. NLRP3 mutation and cochlear autoinflammation cause syndromic and nonsyndromic hearing loss DFNA34 responsive to anakinra therapy. Proc Natl Acad Sci U S A 114, E7766–E7775, doi: 10.1073/pnas.1702946114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Turunen JA et al. Keratoendotheliitis Fugax Hereditaria: A Novel Cryopyrin-Associated Periodic Syndrome Caused by a Mutation in the Nucleotide-Binding Domain, Leucine-Rich Repeat Family, Pyrin Domain-Containing 3 (NLRP3) Gene. Am J Ophthalmol 188, 41–50, doi: 10.1016/j.ajo.2018.01.017 (2018). [DOI] [PubMed] [Google Scholar]
- 321.Basiorka AA et al. The NLRP3 inflammasome functions as a driver of the myelodysplastic syndrome phenotype. Blood 128, 2960–2975, doi: 10.1182/blood-2016-07-730556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Uh ST et al. Association of Genetic Variants of NLRP4 with Exacerbation of Asthma: The Effect of Smoking. DNA Cell Biol 38, 76–84, doi: 10.1089/dna.2018.4433 (2019). [DOI] [PubMed] [Google Scholar]
- 323.Lin Y & Luo Z NLRP6 facilitates the interaction between TAB2/3 and TRIM38 in rheumatoid arthritis fibroblast-like synoviocytes. FEBS Lett 591, 1141–1149, doi: 10.1002/1873-3468.12622 (2017). [DOI] [PubMed] [Google Scholar]
- 324.Tanaka N et al. Eight novel susceptibility loci and putative causal variants in atopic dermatitis. J Allergy Clin Immunol 148, 1293–1306, doi: 10.1016/j.jaci.2021.04.019 (2021). [DOI] [PubMed] [Google Scholar]
- 325.Macaluso F et al. Polymorphisms in NACHT-LRR (NLR) genes in atopic dermatitis. Exp Dermatol 16, 692–698, doi: 10.1111/j.1600-0625.2007.00589.x (2007). [DOI] [PubMed] [Google Scholar]
- 326.Zhao Q et al. Rare inborn errors associated with chronic hepatitis B virus infection. Hepatology 56, 1661–1670, doi: 10.1002/hep.25850 (2012). [DOI] [PubMed] [Google Scholar]
- 327.Wang J et al. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364, doi: 10.1126/science.aav5870 (2019). [DOI] [PubMed] [Google Scholar]
- 328.Travassos LH et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol 11, 55–62, doi: 10.1038/ni.1823 (2010). [DOI] [PubMed] [Google Scholar]
- 329.Xiao L, Magupalli VG & Wu H Cryo-EM structures of the active NLRP3 inflammasome disk. Nature, doi: 10.1038/s41586-022-05570-8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Humphrey W, Dalke A & Schulten K VMD: visual molecular dynamics. J Mol Graph 14, 33–38, 27–38, doi: 10.1016/0263-7855(96)00018-5 (1996). [DOI] [PubMed] [Google Scholar]


