Abstract
Progesterone prevents development of endometrial cancers through its receptor (PR) although the molecular mechanisms have yet to be fully characterized. In this study, we performed a global analysis of gene regulation by progesterone using human endometrial cancer cells that expressed PR endogenously or exogenously. We found progesterone strongly inhibits multiple components of the platelet derived growth factor receptor (PDGFR), Janus kinase (JAK), signal transducer and activator of transcription (STAT) pathway through PR. The PDGFR/JAK/STAT pathway signals to control numerous downstream targets including AP-1 transcription factors Fos and Jun. Treatment with inhibitors of the PDGFR/JAK/STAT pathway significantly blocked proliferation in multiple novel patient-derived organoid models of endometrial cancer, and activation of this pathway was found to be a poor prognostic signal for the survival of patients with endometrial cancer from The Cancer Genome Atlas. Our study identifies this pathway as central to the growth-limiting effects of progesterone in endometrial cancer and suggests that inhibitors of PDGFR/JAK/STAT should be considered for future therapeutic interventions.
Keywords: Progesterone receptor (PR), endometrial cancer, platelet derived growth factor receptor (PDGFR), Janus kinase (JAK), signal transducer and activator of transcription (STAT), Fos, Jun
1. Introduction
Endometrial cancer is the most common gynecologic malignancy and one of only a few cancers in the United States with an increasing incidence and mortality [1]. Remarkably, from 2008 to the present, endometrial cancer cases have increased over 30%, equating to 65,620 new cases in 2020. Moreover, in 2020, 12,590 women in the US lost their lives to this pervasive and under-studied disease [1]. Patients with early-stage disease have a reasonably good prognosis following hysterectomy; however, for advanced or recurrent cases, effective treatments are severely limited. We have proposed that the natural hormone progesterone serves as a powerful endometrial tumor suppressor [2–4]. Our objective in this study was to uncover the anti-tumor effects of progesterone at the molecular level, with the goal of using these progesterone-controlled pathways as a novel roadmap to design more effective targeted therapies for women with advanced or recurrent endometrial cancer.
Progesterone exerts differentiating effects in the endometrium through progesterone receptors A and B (PRA and PRB), which are DNA-binding transcription factors in the steroid hormone receptor family. PRA and PRB arise from alternative translation initiation sites of a single gene, PGR, with PRA being a shorter isoform due to N-terminal truncation of 164 amino acids. The two isoforms form homo- and heterodimers with different functional activities with respect to the control of gene transcription and co-localize in the cell nuclei of normal endometrium. In addition to binding directly to the progesterone response elements (PREs) in DNA, PRs are also believed to interact with other DNA-bound transcription factors such as members of the AP-1 family, including Fos and Jun, as well as other steroid receptors such as the glucocorticoid, androgen, mineralocorticoid and estrogen receptors (GR, AR, MR and ER) to further modulate gene transcription [5–7]. It is believed that both PR isoforms are required for optimal, normal endometrial differentiation [8]. PRs are also present in endometrial carcinomas; however, expression of only one isoform is common [9]. The lack of PR expression may lead to the loss of the differentiating effects of synthetic progesterone (progestins) and render hormonal therapy ineffective for advanced carcinoma [10]. It is therefore critical to assess the independent and combined transcriptional effects of the PR isoforms to fully understand how progesterone controls cell function and inhibits endometrial cancer development and progression.
The expression of PR in endometrial glands is under the control of estrogen and progesterone, where estrogen induces PR gene expression and progesterone down-regulates the expression of its own receptor [11]. Indeed, we have documented the hormone-mediated down-regulation of PR in two independent clinical trials of endometrial cancer [12, 13]. Upon exposure to progesterone, the immediate down-regulation of PR is a sign of its transcriptional activity owing to ligand-dependent destruction in the proteasome that must occur to maintain PR-dependent gene activation. On the other hand, the loss of PR in the long run is an epigenetic mechanism associated with hormone resistance [14, 15]. Thus, the lack of robust PR expression hinders the use of progestin therapy for advanced or recurrent endometrial tumors. An alternative approach would be to target signaling pathways downstream from PR to elicit endometrial differentiation in the absence of functional receptors. To identify these pathways, we examined the effects of progesterone on gene expression in cell models of PR-positive endometrial carcinoma. We demonstrate that progesterone inhibits the activity of many growth-promoting genes. These results identify new pathways worthy of exploration as targets for future therapeutics in endometrial cancer, even in PR-dysregulated tumors.
2. Materials and Methods
2.1. Ethical Approval
All studies using human tumor specimens were reviewed and approved by the University of Iowa Institutional Review Board (IRB), protocol # 201809807 and performed in accordance with the ethical standards as laid down in the 1964 Helsinki Declaration and its later amendments. Informed consent was obtained from all subjects participating in the study.
2.1. Cell Lines
Ishikawa endometrial cancer cells were purchased from ATCC. Hec50 endometrial cancer cells were provided by Dr Erlio Gurpide (New York University) [16]. An aggressive subclone, Hec50co, has been previously characterized and validated as a model for advanced endometrial cancer; these cells are negative for ER and PR [17]. Cells were cultured in DMEM (Sigma, St Louis, MO) supplemented with 10% FBS (Gemini Bio Products, Inc., Calabasas, CA) when grown prior to experiments and without FBS during hormonal treatment. Antibiotic/antimycotic solution containing 100 units/ml penicillin-G, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B (GIBCO Life Technologies, Grand Island, NY) was also added to the growth media. Cell line identity is confirmed twice per year using STR testing (BioSynthesis, Inc., Lewisville, TX).
2.2. Patient-derived organoid models
Fresh tumor tissue was collected from the patient’s debulking surgery. Samples were washed with 10 ml PBS and cut into small pieces, then collected in a 50 ml tube with 5 ml digest buffer (AdDF+++ media (Advanced DMEM-F12 with 1X Glutamax,10 mM HEPES and pen/strep) supplemented with 2 U/ml Dispase ǁ, 1 mg/ml collagenase P and 50 μg/ml DNAse I), and incubated for 0.5–1 hour at 37°C. Dissociated cells were filtered through a 40 μm cell strainer, centrifuged, washed and pelleted. Erythrocytes were removed with red blood cell lysis buffer. Cells were counted and embedded in Matrigel (Corning Life Sciences, USA) on ice and seeded as 50 μl domes in pre-warmed 24-well cell culture plates (Costar, Corning); 500 μl of Organoid Culture Media [18] was added on the top of the Matrigel to each well. Assessment of cell viability was performed as previously described [18] as follows. Organoids were collected with Organoid Harvesting Solution (Cultrex) and digested to single cells with TrypLE Express (Gibco, USA). Single cells were suspended in AdDF+++ medium with 10% Matrigel and seeded at a density of 10,000 cells/well in an ultra-low attachment 96-well U-bottom white plate (S-Bio). After 24 hours, organoids were exposed to progesterone (100 nM), cediranib (1 μM), ruxolitinib (15 μM), tofacitinib (1 μM), napabucasin (0.5 μM) or combinations for 72 hours at 37°C. At the end of the incubation, an equal volume of CellTiter-Glo 3D reagent (Promega, Fitchburg Center, WI, USA) was added to each well and incubated for 25 minutes at room temperature. Luminescence, reflective of cell viability, was measured using the Gen5 Microplate Reader (BioTek, Vermont, USA). All tests were conducted in triplicate and data normalized to untreated controls (set at 100% viability).
2.3. Adenoviral Infections
Adenoviral vectors of PRA and PRB were constructed as described previously [19]. Hec50co cells were infected the AdPRA + AdPRB for 15 h prior to treatment as previously described [19].
2.4. RNA Sequencing and Analyses
RNA quality was analyzed using the Agilent BioAnalyzer 2100 with RNA integrity number greater than eight used for the library preparation at the University of Iowa Institute of Human Genetics, Genomics Division. QC qualified RNA (500ng) was treated with DNase-1 and used to enrich for poly-A-containing transcripts using oligo-dT primers bound to beads. The enriched RNA pools were then fragmented, converted to cDNA and ligated to sequencing adaptors containing indices using the Illumina TruSeq stranded mRNA sample preparation kit (Illumina, San Diego, CA). Molar concentrations of the indexed libraries were then measured on an Agilent Model 2100 Bioanalyzer and mixed in equimolar amounts in sequencing pools. Pools were sequenced on the Illumina NovaSeq 6000 genome sequencer using 100bp paired end SBS chemistry. RNA-seq data went through rigorous quality control as outlined in our previously established quality control protocols and tools [20]. RNA-seq data alignment was performed by STAR [21] followed by gene quantification into read counts using featureCounts [22]. Differential expression analysis was carried out using DESeq2 [23]. Genes expressed at greater than 1.0 fragments per kilobase per million reads and with greater than two-fold changes in gene expression were compared using Ingenuity Pathway Analysis (Qiagen). Heatmaps of differential gene expression were generated using the average log2 fold-change relative to time-matched vehicle control in three independent biological replicates (Prism version 9.0.0 (GraphPad)).
2.5. Affymetrix Expression Microarrays
RNA was extracted using RNeasy spin columns (Qiagen Corp, Valencia, CA). The RNA quality was checked using the Agilent 2100 Bioanalyzer (Agilent Technologies, Foster City, CA) and the concentration determined with the Nanodrop (Thermo Scientific, Wilmington, DE). Functional replicate Affymetrix microarrays were performed for two independent experiments using the human HG-U133A plus 2.0 chips (Santa Clara, CA), thereby querying the expression of 54,000 transcripts. All procedures for the chip preparation and cDNA production were performed according to instructions from the manufacturer’s manual, version 701025 Rev.5. Briefly, total RNA was used to generate double-stranded cDNA with an oligo dT-primer containing the T7 RNA polymerase promoter site and the One-Cycle Target Labeling Kit. cDNA was purified via a column using the GeneChip Sample Cleanup Module, and biotinylated cRNA was synthesized by in vitro transcription using the GeneChip IVT Labeling kit. Biotin labeled cRNA was purified (GeneChip Sample Cleanup Module), and the absorbance measured at 260 nm to determine yield (Nanodrop spectrophotometer). Twenty μg of the labeled cRNA was fragmented, and its quality was assessed for purified cRNA and fragmented cRNA using the Agilent 2100 Bioanalyzer and the RNA 6000 Nano LabChip kit. The labeled fragmented cRNA was hybridized to Affymetrix GeneChip HGU133 Plus 2.0 arrays for 16 h at 45°C. Array washing and staining was performed on the Affymetrix fluidics (450) station according to the antibody amplification protocol (Fluidics script: EukGE-WS2v5). The GeneChips were scanned using the Affymetrix GeneChip Scanner 3000 (a wide-field, epifluorescent near-confocal microscope with a patented flying objective).
Initial data analysis was performed using Affymetrix Microarray Suite v 5.0 software, setting the scaling of all probe sets to a constant value of 500 for each GeneChip. Silicon Genetics’ (now Agilent Technologies) GeneSpring version 7.3 (Redwood City, CA) was used to filter data using several criteria. First, only transcripts with a fluorescent signal above the background level were retained for the subsequent 2-fold change filter. Starting with over 54,000 transcripts, this eliminated all but 5592 transcripts. Unsupervised clustering and heatmaps were constructed using the R package heatmap3 [24]. Pearson’s correlation coefficients were used as the distance between two samples. An ANOVA calculation was used to identify transcripts with a p-value less than 0.05. Heatmaps of differential gene expression for select genes were generated by plotting the log2 fold-change in response to progesterone relative to time-matched vehicle control (Prism version 9.0.0 (GraphPad)).
2.6. Quantitative Real-Time Polymerase Chain Reaction (RT-PCR)
2.6.1. Hec50 cells expressing PRA+B:
cDNA was synthesized from 1μg of total RNA and PCR reactions were carried out in 50μl reaction mixtures using 50ng of template per well. Amplification of GAPDH was used as an endogenous control to standardize the amount of RNA in each sample. The Assay on Demand ™ protocol was carried out as directed in the ABI manual (Applied Biosystems, Foster City, CA). The raw data were presented as the cycle number associated with initial amplification. The data were then normalized to an endogenous control to allow for variance in RNA template amounts added to the reverse transcription reaction. The data could then be compared to a calibrator and analyzed using the 2 −ΔΔCT method [25].
2.6.2. Ishikawa cells and patient-derived organoid models.
Ishikawa cells were treated with progesterone (100 nM) −/+ estrogen (10 nM) or vehicle control (ethanol) in serum-free media. Cells were harvested at the end of treatment and subjected to RNA purification. PDO models were treated with 100 nM progesterone or vehicle control in organoid culture media (see Supplemental Methods in ref. [18]). At the end of treatment, Matrigel was dissolved using Organoid Harvesting Solution (3700–100-01, Cultrex, R&D Systems) per the manufacturer’s instructions, and cell pellets obtained for RNA isolation. Total RNA was purified from Ishikawa cells or organoid models using the RNeasy Plus RNA purification kit (QIAGEN). Yield and purity were assessed on a Trinean DropSense 16 spectrophotometer and an Agilent Model 2100 Bioanalyzer in the Genomics Division of the Iowa Institute of Human Genetics. QC qualified total RNA (500 ng) from each vehicle only or P4 treated sample was reverse transcribed using oligo-dT primers in the SuperScript III kit following manufacturer’s recommendations (Invitrogen). The resulting cDNAs were then amplified in the presence of SYBR Green (Thermo Fisher) on an Applied Biosystems Model 7900HT platform in the Genomics Division of the Iowa Institute of Human Genetics. Locus-specific primers are shown in Table 1.
Table 1.
Primers used for quantitative PCR (qPCR).
| Locus | Primer Sequences | Amplicon | Tm(°C)* |
|---|---|---|---|
| PGR | For: ATCCTACAAACACGTCAGTGGGCA Rev: ACTGGGTTTGACTTCGTAGCCCTT |
139bp | 60.5 60.3 |
| PDGFR | For: GACTTTCGCCAAAGTGGAGGAG Rev: AGCCACCGTGAGTTCAGAACGC |
121bp | 58.0 62.0 |
| FOS | For: GCCTCTCTTACTACCACTCACC Rev: AGATGGCAGTGACCGTGGGAAT |
123bp | 56.5 60.9 |
| JUN | For: CGATCTGCACAAGATGAACCACG Rev: CTGCTGAGGTTGGTGTAAACGG |
106bp | 58.3 58.2 |
| STAT3 | For: CTTTGAGACCGAGGTGTATCACC Rev: GGTCAGCATGTTGTACCACAGG |
133bp | 57.1 58.1 |
| 18S rRNA | For: AACTTTCGATGGTAGTCGCCG Rev: CCTTGGATGTGGTAGCCGTTT |
104bp | 57.3 57.6 |
Tm is estimated in the presence of 1.5mM MgCl2 using OligoAnalyzer software available on-line at Integrated DNA Technologies (www.idtdna.com).
qPCR assays were designed using PrimerQuest software available online at Integrated DNA Technologies (www.idtdna.com). Primers were synthesized at Integrated DNA Technologies (Coralville, IA). All experiments were performed with at least three biological and three technical replicates. Raw expression values (Ct) were normalized (ΔCt) against the endogenous control 18S rRNA and then used to calculate relative expression (ΔΔCt) versus untreated cells. Fold changes in expression resulting from treatment were calculated as 2−ΔΔCt following the standard method [25, 26]. Heatmaps of fold change relative to time-matched controls were generated using Prism version 9.0.0 (GraphPad).
2.7. Immunohistochemistry
Patient tumor specimens from which organoid models were derived were subjected to immunohistochemistry for expression of PR and ER. All staining was performed by the University of Iowa Histology Research Laboratory. Formalin-fixed paraffin-embedded (FFPE) tumor blocks were sectioned to 4 μm thickness. PR (1:200 dilution, #M3568, Dako) and ER (1:50 dilution, #M7048, Dako) staining was performed on each sample as previously described [13] using a polymer-based detection system on the Biocare Medical IntelliPATH FXL system. IHC staining evaluated in a semiquantitative manner by two reviewers blinded to sample identity following the American Society of Clinical Oncology/College of American Pathologists guidelines for scoring PR and ER [27]. Per the guidelines, at least 1% of the cells in the specimen must be immunoreactive to consider the tissue positive. The intensity of tumor staining (0 to 3+) was scored using a positive control tissue as 3+, and all samples were also assessed for presence of internal positive control staining (stroma, myometrium) to ensure tissue integrity. Reviewers also scored each slide with a percent of cells staining positive (0–100), which was multiplied by the intensity score (0, 1+, 2+, or 3+) to calculate a modified H-score. The values from two independent reviewers were averaged for each specimen and biomarker. Representative images were acquired at 20X magnification.
2.8. TCGA Analysis
RNA-seq data of uterine corpus endometrial carcinoma (UCEC) (N=560) from the cancer genome atlas (TCGA) were downloaded from Genomic Data Commons. Genes of interest identified using our data were extracted from the TCGA dataset for further prognostic evaluation. Each gene of interest’s disease specific prognostic power was measured by Cox proportional hazard model in R treating gene expression as a continuous variable. Furthermore, the overall prognostic relevance of the genes of interest was evaluated using two methods. First, the overall prognostic power was measured by a meta analysis. Then, we combined genes of interest using a previously established method AESA [28] to compose a gene expression composite score (GECS) and used it for prognostic analysis.
3. Results
3.1. Gene regulation by progesterone through PR
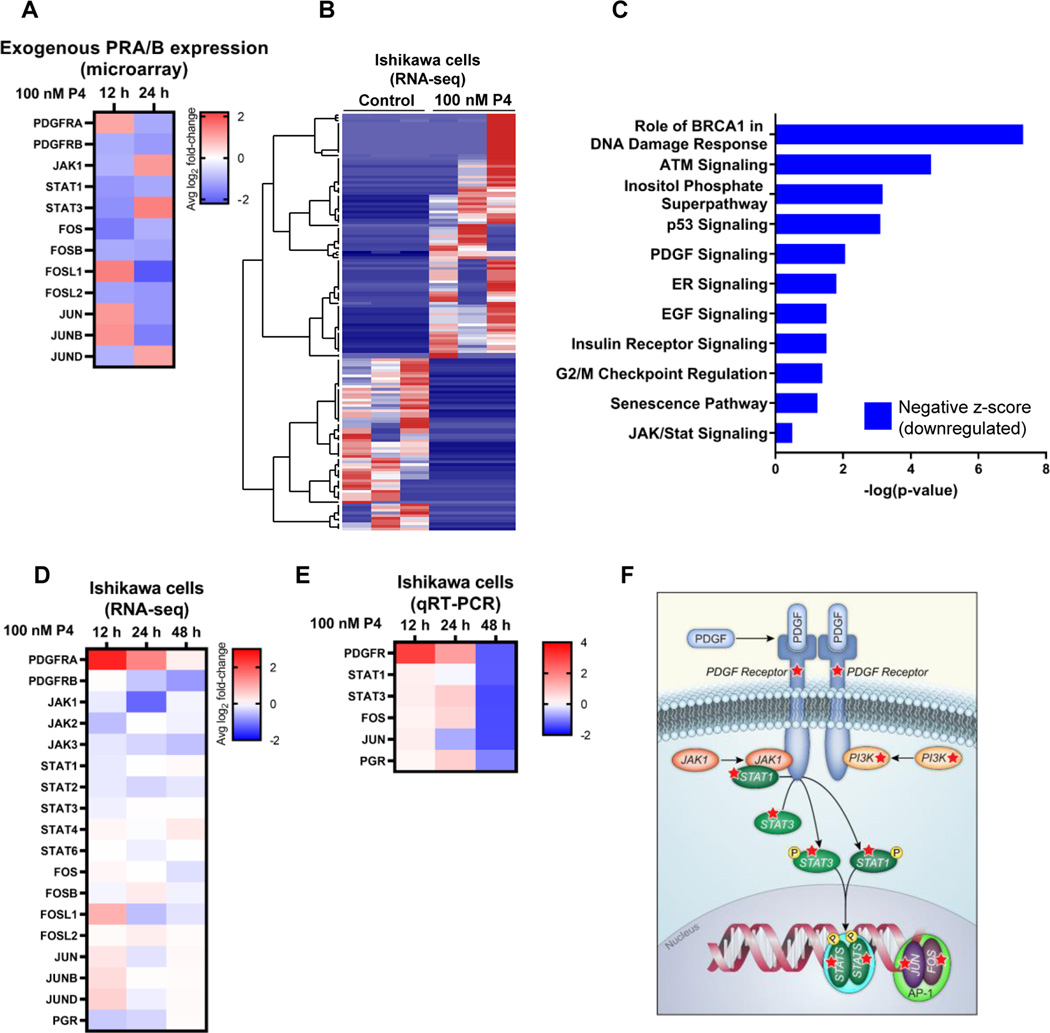
To understand the basis of progesterone’s distinct actions in endometrial cancer cells and to determine which genes this hormone regulates, we first performed studies Hec50co endometrial cancer cells engineered to transiently co-express PRA and PRB [19]. As determined by Affymetrix microarray, PR-transfected Hec50co cells responded to progesterone (P4) by down-regulating multiple STAT (signal transducer and activator of transcription) genes and the downstream AP-1 transcription factors (Fos and Jun), as well as PDGFR (platelet derived growth factor receptor) as compared to vehicle control (Figure 1A).
Figure 1. Significantly altered pathways in endometrial cancer cells response to progesterone.
(A) Heatmap of representative Affymetrix microarray data from Hec50co cells expressing exogenous PRA and PRB. Gene expression changes in response to 100 nM progesterone (P4) were compared to vehicle control. (B) Heatmap of unsupervised clustering analysis of RNA-seq data. Ishikawa cells were treated with 100 nM progesterone or vehicle control for 12 h, followed by analysis of gene expression by RNA-seq. Each column represents a biological replicate (N=3 per treatment) and each row represents a gene. (C) Ingenuity Pathway Analysis (IPA) of significantly altered pathways based on RNA-seq data from Ishikawa cells treated with P4 or vehicle control for 24 h. (D) Heatmap of representative differential gene expression from RNA-seq data from Ishikawa cells treated with either progesterone or vehicle control. (E) Heatmap of qRT-PCR of Ishikawa cells treated with P4 or vehicle control. (F) Schematic representation of the PDGFR/JAK/STAT/Fos/Jun pathway. Red stars indicate proteins in the pathway encoded by genes inhibited by progesterone. Heatmaps reflect the log2 fold-change in gene expression relative to time-matched vehicle control. Values less than 1 indicate a decrease in transcript levels in response to progesterone, whereas values greater than 1 indicate an increase. Results are the average of N=3 biological replicates.
We next asked if this pathway is also significantly altered in endometrial cancer cell lines with endogenous PR expression. Ishikawa cells express moderate levels of PR isoforms as compared to other endometrial cancer cell lines [14–16]. We performed RNA-sequencing (RNA-seq) of Ishikawa cells after treatment with progesterone (P4) for 12, 24 or 48 h and compared results to time-matched vehicle controls. Global gene expression analysis indicates a clear demarcation between control vs. progesterone-exposed cells (Figure 1B and Supplemental Figure S1). Ingenuity Pathway Analysis identified several pathways that were differentially regulated by progesterone, with the overwhelming majority being downregulated (Figure 1C). In line with the microarray results, the PDGFR and JAK/STAT signaling pathways were downregulated by P4 treatment, though progesterone required a longer time frame (48 h) in order to effect the down-regulation in cells with endogenous vs. exogenous PRs (Figure 1D). A similar pattern of effects was observed by qRT-PCR in Ishikawa cells (Figure 1E). While the magnitude and specific timecourse of the effects on genes in the PDGFR/JAK/STAT/Fos/Jun pathway differened between cells with exogenous vs. endogenous PR expression and by the method used for expression analysis, these results nevertheless indicate that the downstream transcriptional effectors of this pathway were strongly inhibited by P4. A schematic of genes in this pathway is provided in Figure 1F.
Studies were extended to a more translational model of endometrial cancer, patient-derived organoids (PDOs). To date, we have established a biorepository of 29 endometrial cancer PDO models and characterized expression of ER and PR in the primary tissue specimens from which these models were derived (Table 2).
Table 2. Expression of steroid hormone receptors PR and ER by immunohistochemistry in tumor tissue from which patient-derived organoid models were created.
Cases are organized by histologic subtype and stage/grade. Cases that were utilized for analysis of progesterone-mediated gene expression changes are underlined, whereas cases that were included in drug testing studies are italicized. N.D.: not determined; no tumor: no tumor cells were present for analysis in the stained slide.
| Modified H-Score | |||||
|---|---|---|---|---|---|
| Patient ID | Histologic subtype | Stage | Grade | PR | ER |
| ONC-6885 | Endometrioid | IA | 1 | 300 | 190 |
| ONC-7756 | Endometrioid | IA | 1 | 300 | 170 |
| ONC-6790 | Endometrioid | IA | 1 | 300 | 300 |
| ONC-6818 | Endometrioid | IA | 1 | 300 | 300 |
| ONC-6692 | Endometrioid | IA | 1 | 285 | 285 |
| ONC-8743 | Endometrioid | IA | 1 | 225 | 195 |
| ONC-8004 | Endometrioid | IA | 1 | 165 | 185 |
| ONC-7076 | Endometrioid | IA | 1 | 160 | 285 |
| ONC-6096 | Endometrioid | IA | 1 | 140 | 185 |
| ONC-8734 | Endometrioid | IA | 1 | 105 | 70 |
| ONC-6844 | Endometrioid | IA | 1 | 70 | 277.5 |
| ONC-7723 | Endometrioid | IA | 1 | 30 | 240 |
| ONC-6932 | Endometrioid | IA | 1 | 0 | 175 |
| ONC-7026 | Endometrioid | IA | 2 | 90 | 277.5 |
| ONC-6498 | Endometrioid | IA | 2 | 70 | 285 |
| ONC-6173 | Endometrioid | IA | 2 | 7.5 | 180 |
| ONC-6051 | Endometrioid | IA | 2 | 0 | 0 |
| ONC-6925 | Endometrioid | IA | 3 | 0 | 0 |
| ONC-6191 | Endometrioid | IB | 1 | 155 | 300 |
| ONC-7787 | Endometrioid | IB | 1 | 150 | 300 |
| ONC-7677 | Endometrioid | IB | 1 | 45 | 255 |
| ONC-5768 | Endometrioid | IIIC2 | 1 | 110 | 292.5 |
| ONC-7623 | Serous | IB | 45 | 70 | |
| ONC-6720 | Serous | IIIC1 | 0 | 0 | |
| ONC-7991 | Serous | IV | 45 | 217.5 | |
| ONC-6099 | Serous | IVB | 0 | 0 | |
| ONC-7757 | Clear cell | IV | 70 | 30 | |
| ONC-7367 | Mixed clear cell & endometrioid | IB | 3 | 225 | 285 |
| ONC-6742 | Mixed serous & endometrioid | IA | 285 | 300 | |
| ONC-7003 | Mixed serous & endometrioid | IA | 85 | 300 | |
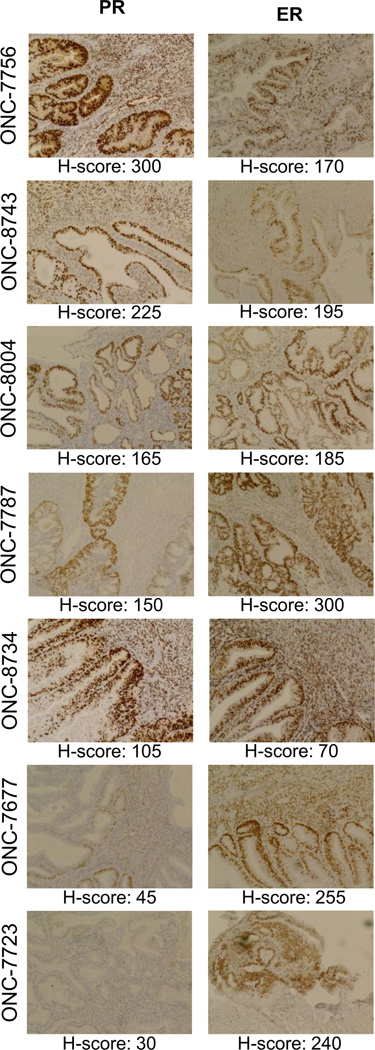
We selected PDO models of stage 1/grade 1 endometrioid endometrial cancer to determine the effects of progesterone on expression of genes in the PDGFR/JAK/STAT pathway. Expression of PR and ER in the tissue from which these models were derived is shown in Figure 2. In the majority of cases we tested, hormone receptor staining was most intense in the glandular structures and was primarily nuclear (e.g,. see ONC-8743). Whereas PR expression was variable across specimens studied as PDO models, all cases harbored relatively high ER levels (Figure 2; note the clinical standard for ER is an H-score of 75; one case, ONC-8734, is borderline high).
Figure 2: Hormone receptor expression in endometrial patient tumor specimens used to create organoid models.
Expression of progesterone receptor (PR) and estrogen receptor (ER) was determined by immunohistochemistry in patient tumor specimens from which organoid models were derived. Images are annotated with the corresponding de-identified patient ID and the average modified H-score value. Cases are ordered based on expression of PR (highest = ONC-7756; lowest = ONC-7723). All images were acquired at 20X magnification with an EVOS microscope. When possible, serial sections were stained with ER and PR. ONC-7756, 7787, 7767 and 7723 were used for gene expression studies (Fig. 3) and ONC-7756, 7767, 7723, 8004, 8734 and 8743 were used in drug response studies (Fig. 5).
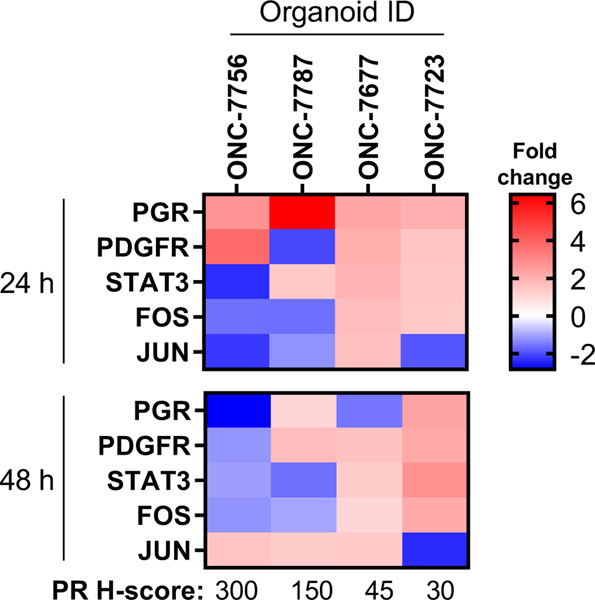
Gene expression changes were assessed in PDO models with varying levels of PR expression. Consistent with results in cell lines, we observed an overall downregulation of the genes the PDGFR/JAK/STAT/Fos/Jun pathway in response to progesterone treatment (Figure 3). The models with higher PR expression had greater downregulation in PDGFR, STAT, FOS and JUN. We also observed the anticipated increase in PR expression 24 h after P4 treatment and downregulation at 48 h in the models with PR.
Figure 3: Gene expression changes in PDO models following progesterone treatment.
Expression of the indicated genes was assessed in PDO models exposed to progesterone (P4) for 24 h or 48 h. Log2 fold-changes were calculated vs. vehicle control at the same time point and visualized as a heatmap. Data are the average of N=3 biological replicates.
3.2. Relevance of PDGFR/JAK/STAT pathway activation on outcomes in patients with endometrial cancer
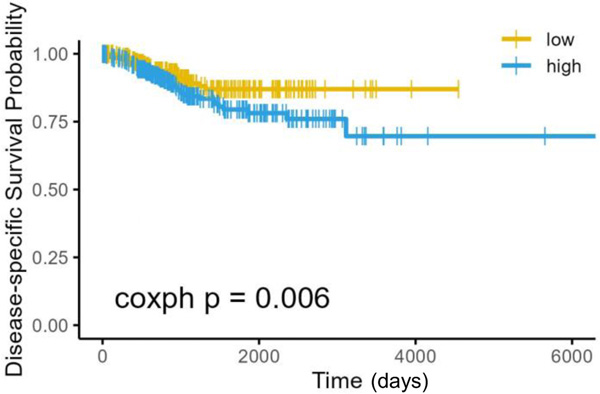
Studies were extended to understand the prognostic significance of the PDGFR/JAK/STAT/Fos/Jun pathway in endometrial cancer. The Cancer Genome Atlas (TCGA) dataset for uterine corpus endometrial cancer (UCEC) was queried for the expression of genes within the PDGFR/JAK/STAT pathway. Based on gene expression changes by microarray and RNA-seq, we selected the following ten genes for analysis: PDGFRA, STAT1, STAT2, STAT3, STAT4, STAT6, FOS, JUN, JUNB and JUND. Of these ten genes, four had significant prognostic results: PDGFRA, STAT1 and JUN were inversely associated with survival (higher expression = worse survival), whereas STAT6 was positively associated with survival (higher expression = better survival). A meta analysis of the ten genes together revealed an inverse association with survival (hazard ratio = 1.002, 95% confidence interval: 0.0006 – 1.0035). We also determined the impact of high (highest quartile) versus low (lowest quartile) expression on survival using disease-specific survival probability Kaplan-Meier curve assessment (Figure 4). Using the AESA [28] method, high expression of genes in the PDGFR/JAK/STAT/Fos/Jun pathway portended a significantly decreased survival (p = 0.006).
Figure 4. Activation of the PDGFR/JAK/STAT/Fos/Jun pathway in endometrial cancer predicts for worse survival.
Cases were separated into quartiles based on expression of PDGFRA, STAT1, STAT2, STAT3, STAT4, STAT6, FOS, JUN, JUNB and JUND. Kaplan-Meier Curve for the survival analysis was conducted on the highest and lowest quartiles using the AESA method.
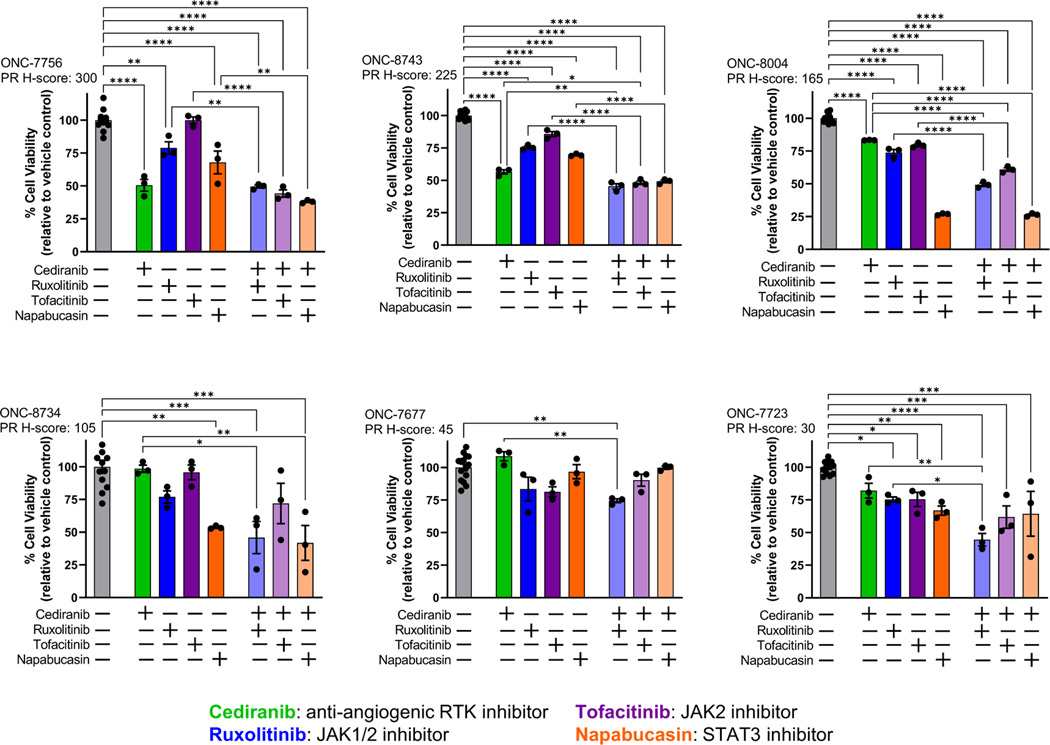
3.3. Effect of PDGFR/JAK/STAT/Fos/Jun pathway targeted inhibitors on the viability of endometrial cancer platelet derived organoids
To link progesterone activity with gene expression changes, we exposed PDOs to targeted agents that block the PDGFR/JAK/STAT/Fos/Jun pathway. The agents studied included cediranib (an anti-angiogenic inhibitor that targets multiple receptor tyrosine kinases, including PDGFR), ruxolitinib (an inhibitor of JAK1 and JAK2), tofacitinib (an inhibitor of JAK3) and napabucasin (an inhibitor of STAT3). These agents have been evaluated to a very limited degree in clinical trials of endometrial cancer (Supplemental Table S1). In general, all models displayed some sensitivity to inhibitors of the PDGFR/JAK/STAT as single agents (Figure 5). We also combined the anti-angiogenic inhibitor cediranib with the JAK or STAT targeted agents in order to evaluate the impact of dual therapy on cell viability. With the exception of one model (ONC-7677), the combinatorial regimens produced more marked reductions in cell survival as compared to single agent alone. There was only one combination for which the combination of cediranib with a JAK/STAT inhibitor was synergistic: ONC-8004, cediranib+ruxolitinib (Supplemental Figure S2). All other combinations were additive (e.g., ONC-7756, cediranib+tofacitinib) or driven predominantly by a single agent (e.g., ONC-8004, cediranib+napabucasin was similar to napabucasin alone). These data indicate that, regardless of PR expression, inhibition of the PDGFR/JAK/STAT pathway represents a potential therapeutic strategy for endometrial cancer.
Figure 5: Inhibition of the PDGFR/JAK/STAT signaling pathway is cytotoxic in patient-derived organoid models of early stage/grade endometrial cancer.
PDO models were exposed to the indicated agents for 72 hrs, followed by assessment of cell viability. Drugs concentrations are as follows: 1 μM cediranib; 20 μM ruxolitinib; 1 μM tofacitinib; 0.5 μM napabucasin. Data were normalized to control, which was set at 100% viability. Cases are ordered by PR expression (H-score). * p < 0.05; ** p < 0.01; *** p < 0001; **** p < 0.00001 by one-way ANOVA with a Tukey’s multiple comparisons test.
4. Discussion
Progesterone acts to control cell function and proliferation through PR. Gene expression profiling in response to steroid hormones through their cognate receptors has provided information on large numbers of pathways controlled by these factors. Dai et al. [5] found that progesterone inhibits endometrial cancer cell growth and invasiveness by down-regulation of cellular adhesion molecules through PR. We have also shown that progesterone inhibits inflammation by inducing anti-inflammatory cytokines and inhibiting pro-inflammatory cytokines and their receptors [29]. Indeed, given the multiple mechanisms of action of progesterone in the endometrium, it can be considered the ultimate endometrial tumor suppressor [30]. It follows that elucidating the repertoire of progesterone/PR targets will potentially shed light on other pathways with therapeutic potential in endometrial cancer and relevant even for tumors that do not express PR.
In addition to VEGF and pro-angiogenic factors, as previously reported [31], the most relevant pathway summarizing our data as assessed by Ingenuity Pathway Analysis is the PDGFR/JAK/STAT/Fos/Jun pathway, which is often constitutively up-regulated in endometrial cancer [32], and inhibited by progesterone through PR. This pathway enhances many proliferative signals in cells and is linked to tumor angiogenesis. Our analysis of TCGA dataset suggests that elevated expression of genes in the PDGFR/JAK/STAT/Fos/Jun pathway is associated with poor outcome in patients with endometrial cancer.
Like VEGF, PDGF is a key regulator of angiogenesis as well as proliferation. High intratumor microvessel density in endometrial cancer is associated with advanced clinical stage and an increased risk of recurrent disease [33–38]. A study by Slomovitz et al. showed that 33 (91%) of primary endometroid endometrial cancer tumors express PDGFR, and 8 of 11 (73%) primary uterine papillary serous tumors express PDGFR [32]. Importantly, in this study, all recurrent tumors expressed PDGFR [32]. The Gynecologic Oncology Group has also confirmed the relevance of PDGFR as a target through GOG Study 229-J wherein the tyrosine kinase inhibitor cediranib, which targets PDGFR, FGFR, and VEGFR, showed independent activity against advanced endometrial cancer [39]. Here, we have shown that progesterone inhibits PDGFR transcription as well. In breast cancer cells, PDGF ligand has been identified as a PR target gene, and exposure of smooth muscle cells, which express high levels of PDGFRs, to conditioned media from progesterone-treated PR-positive breast cancer cells results in activation of PDGFR [40]. In addition, PDGFR inhibition with imatinib blunts proliferation of PR-positive breast cancer cells [41].
The Signal Transducers and Activators of Transcription (STATs) have been implicated in cross talk between steroid hormones and growth factors. There are seven STAT genes. The activation of STATs is controlled by JAKs, which are kinases linked to the cytoplasmic tails of cytokine receptors. Once activated through phosphorylation by JAKs, STATs have dual functions as signaling molecules in the cytoplasm and as transcription factors following nuclear translocation [42]. The STAT proteins mediate diverse biological processes including cell growth, differentiation, apoptosis, transformation and survival. In endometrial cancer cells, our data consistently demonstrated that progesterone through PR inhibits the transcription of multiple pro-growth STAT genes. In contrast, ER/PR/STAT signaling is linked to estradiol and progesterone’s proliferative activity in the normal breast and possibly also in breast cancer [43]. While our data confirm down-regulation of multiple STAT transcripts by progesterone in endometrial cancer cells, treatment of T47D human breast cancer cells with the synthetic progestin R5020 results in the up-regulation of STAT3 and STAT5 proteins [44, 45]. This interesting dichotomy may relate to the potential proliferative effects of progesterone in breast cancer compared to its almost universal anti-proliferative effects in endometrial cancer.
Interestingly, signaling downstream of activated PDGFR in turn activates STAT3 [46]. STAT3 induces progression through the cell cycle and prevents apoptosis. The candidate target genes regulated by the STAT pathways, such as cyclin D1, appear to contribute to oncogenesis by inducing cell proliferation and survival [47]. It is likely that the inhibition of STAT3 contributes to the mechanisms by which progesterone through PR inhibits the cell cycle.
The most robust changes in gene expression were detected in Hec50 cells expressing artificially high levels of PR. These findings provided a clue into the signaling repertoire of progesterone and the motivation to validate studies in other models. However, a limitation of this study is the lack of endometrial cancer cell lines with endogenously high expression of PR. In Ishikawa cells, the best characterized PR-expressing cell line, we detected only modest changes in gene expression in response to progesterone. For this reason, we extended studies to the more clinically relevant model system, patient-derived organoid, and to patient datasets. Indeed, our analysis of TCGA data underscores the relevance of the PDGFR/JAK/STAT pathway as a prognostic indicator of patient outcome.
Now that signaling through the PDGFR/JAK/STAT/Fos/Jun pathway has been identified as a key proliferative pathway that likely promotes tumor progression and is inhibited by progesterone, how can this pathway be blocked therapeutically even in tumors lacking PR? Inhibitors of JAK/STAT including ruxolitinib, targeted against JAK1 and 2, and the pan-JAK inhibitor tofacitinib are now in clinical use, although they have yet to be evaluated in endometrial cancer. In addition to JAK inhibitors, a series of direct inhibitors of STAT3 are in development, including BBI-608 (napabucasin) and TTI-101, both of which have been studied in early phase clinical trials in cancer. In addition, AZD9150 is an antisense oligonucleotide against STAT3 that is in development [48]. The data provided through these experiments demonstrate that JAK/STAT inhibitors may be highly effective in blocking proliferation of early stage/grade endometrial cancer and deserve further consideration for testing.
A limitation is that the majority of studies were performed in ER-positive endometrial cancers, and future experiments in ER-negative (e.g., serous or high-grade endometrioid histologies) are necessary to understand the generalizability of our findings. Of particular interest to our group is the application of combinatorial strategies that can reactivate PR expression in advanced disease, which is commonly devoid of ER and PR expression. We have extensively studied mechanisms of PR downregulation in endometrial cancer and identified both chromatin modification through the polycomb repressor complex (PRC2) as well as DNA methylation as mechanisms of suppressed PR gene expression [14, 15, 49, 50]. Importantly, PR suppression can be overcome in vitro and in vivo with histone deacetylase inhibitors. Based on these results, we performed a non-therapeutic surgical window trial comparing medroxyprogesterone acetate (MPA) alone to MPA + a histone deacetylase inhibitor, entinostat [13]. Unfortunately, we have been unable to garner support from NCI to test the combination of MPA + a histone deacetylase inhibitor as a treatment strategy for endometrial cancer; this is due in large part to 1) the shifted emphasis on immunotherapy in endometrial cancer and competition for the same patient cohort for concurrent clinical trials; and 2) reticence from pharmaceutical companies to further pursue epigenetic modulators in solid tumors, which have shown limited efficacy as single agents. As the field makes more progress in understanding which patients are ideal candidates for immunotherapy and those that are less likely to respond, it is our hope that we will be able to renew interest in combinatorial treatment regimens using targeted agents as proposed in this study.
5. Conclusions
In summary, we have identified progesterone-controlled genes through PR to shed light on the global effects of this important tumor suppressor. We have further highlighted a proliferative pathway, PDGFR/JAK/STAT/Fos/Jun, which is inhibited by progesterone through PR. We hypothesize that JAK/STAT inhibitors may improve outcomes for patients with advanced endometrial cancer, even those who do not express PR, and hope that these preclinical data will provide the initial support for early phase clinical trials testing this concept. Use of specific inhibitors of the pathways normally down regulated by progesterone has the potential to compensate for the loss of PR in advanced endometrial cancer.
Supplementary Material
Highlights.
Progesterone inhibits PDGFR/JAK/STAT pathway in endometrial cancer preclinical models
PDGFR/JAK/STAT pathway activation predicts for worse outcomes in endometrial cancer
PDGFR/JAK/STAT inhibition is cytotoxic in organoid models of endometrial cancer
Acknowledgments:
We thank the Keck-University of New Mexico Genomics Shared Resource supported by the University of New Mexico Cancer Research and Treatment Center, the W.M. Keck foundation and the State of New Mexico, for making this study possible. We wish to express our appreciation to Marilee Morgan for her experimental assistance and expertise in the performance of the Affymetrix arrays. Additional qPCR assays and RNA sequencing were carried out in the Genomics Division of the Iowa Institute of Human Genetics (IIHG). We also acknowledge Mariah R. Leidinger and the Histology Research Laboratory at the University of Iowa for immunohistochemical staining.
Funding:
This work was supported by NIH grants R01CA99908 (KKL), K22CA263783 (KWT) and R01HG008974 (JG); Department of Defense grants through the Ovarian Cancer Research Program OC190352 (KKL) and Peer-Reviewed Cancer Research Program CA210610 (KKL and MP) and CA220729 (KWT, JG and KKL); the Cory Beach Family Fund (KKL and SD); and the Department of Obstetrics and Gynecology, the University of Iowa (SML). Studies were also carried out in the Genomics Division of the Iowa Institute of Human Genetics (IIHG) which is supported, in part, by the University of Iowa Carver College of Medicine and the Holden Comprehensive Cancer Center through the National Cancer Institute of the National Institutes of Health award P30CA086862. We also thank our benefactors, Dean and Alice Irvin and Mrs. Shirley Leslie for supporting this research. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Declarations
Conflicts of interest/Competing interests:
Financial interests: KWT is a cofounder of Immortagen, Inc. All others authors delcar they have no financial interests.
Non-financial interests: None
Code availability: N/A
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data availability:
The RNA-seq and Affymetrix array datasets generated during the current study will be deposited in GEO prior to publication. All other data generated or analyzed during this study are included in the submitted article and its supplementary information files.
References
- [1].Cancer Facts and Figures 2021, American Cancer Society, 2021. [Google Scholar]
- [2].Yang S, Thiel KW, De Geest K, Leslie KK, Endometrial cancer: reviving progesterone therapy in the molecular age, Discovery medicine, 12 (2011) 205–212. [PubMed] [Google Scholar]
- [3].Leslie KK, Thiel KW, Yang S, Endometrial cancer: potential treatment and prevention with progestin-containing intrauterine devices, Obstet Gynecol, 119 (2012) 419–420. [DOI] [PubMed] [Google Scholar]
- [4].Carlson MJ, Thiel KW, Yang S, Leslie KK, Catch it before it kills: progesterone, obesity, and the prevention of endometrial cancer, Discovery medicine, 14 (2012) 215–222. [PMC free article] [PubMed] [Google Scholar]
- [5].Dai D, Litman ES, Schonteich E, Leslie KK, Progesterone regulation of activating protein-1 transcriptional activity: a possible mechanism of progesterone inhibition of endometrial cancer cell growth, J Steroid Biochem Mol Biol, 87 (2003) 123–131. [DOI] [PubMed] [Google Scholar]
- [6].Davies S, Dai D, Pickett G, Leslie KK, Gene regulation profiles by progesterone and dexamethasone in human endometrial cancer Ishikawa H cells, Gynecol Oncol, 101 (2006) 62–70. [DOI] [PubMed] [Google Scholar]
- [7].Truong TH, Lange CA, Deciphering Steroid Receptor Crosstalk in Hormone-Driven Cancers, Endocrinology, 159 (2018) 3897–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mote PA, Balleine RL, McGowan EM, Clarke CL, Colocalization of progesterone receptors A and B by dual immunofluorescent histochemistry in human endometrium during the menstrual cycle, J Clin Endocrinol Metab, 84 (1999) 2963–2971. [DOI] [PubMed] [Google Scholar]
- [9].Leslie KK, Kumar NS, Richer J, Owen G, Takimoto G, Horwitz KB, Lange C, Differential expression of the A and B isoforms of progesterone receptor in human endometrial cancer cells. Only progesterone receptor B is induced by estrogen and associated with strong transcriptional activation, Ann N Y Acad Sci, 828 (1997) 17–26. [DOI] [PubMed] [Google Scholar]
- [10].Mote PA, Balleine RL, McGowan EM, Clarke CL, Heterogeneity of progesterone receptors A and B expression in human endometrial glands and stroma, Hum Reprod, 15 Suppl 3 (2000) 48–56. [DOI] [PubMed] [Google Scholar]
- [11].Graham JD, Clarke CL, Physiological action of progesterone in target tissues, Endocr Rev, 18 (1997) 502–519. [DOI] [PubMed] [Google Scholar]
- [12].Zaino RJ, Brady WE, Todd W, Leslie K, Fischer EG, Horowitz NS, Mannel RS, Walker JL, Ivanovic M, Duska LR, Histologic effects of medroxyprogesterone acetate on endometrioid endometrial adenocarcinoma: a Gynecologic Oncology Group study, International journal of gynecological pathology : official journal of the International Society of Gynecological Pathologists, 33 (2014) 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Duska LR, Filiaci VL, Walker JL, Holman LL, Hill EK, Moore RG, Ring KL, Pearl ML, Muller CY, Kushnir CL, Lankes HA, Samuelson MI, Carrick KS, Rajan A, Rodgers WH, Kohn EC, Piekarz R, Leslie KK, A Surgical Window Trial Evaluating Medroxyprogesterone Acetate with or without Entinostat in Patients with Endometrial Cancer and Validation of Biomarkers of Cellular Response, Clin Cancer Res, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yang S, Jia Y, Liu X, Winters C, Wang X, Zhang Y, Devor EJ, Hovey AM, Reyes HD, Xiao X, Xu Y, Dai D, Meng X, Thiel KW, Domann FE, Leslie KK, Systematic dissection of the mechanisms underlying progesterone receptor downregulation in endometrial cancer, Oncotarget, 5 (2014) 9783–9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yang S, Xiao X, Jia Y, Liu X, Zhang Y, Wang X, Winters CJ, Devor EJ, Meng X, Thiel KW, Leslie KK, Epigenetic modification restores functional PR expression in endometrial cancer cells, Curr Pharm Des, 20 (2014) 1874–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Devor EJ, Gonzalez-Bosquet J, Thiel KW, Leslie KK, Genomic characterization of five commonly used endometrial cancer cell lines, Int J Oncol, 57 (2020) 1348–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Albitar L, Pickett G, Morgan M, Davies S, Leslie KK, Models representing type I and type II human endometrial cancers: Ishikawa H and Hec50co cells, Gynecol Oncol, 106 (2007) 52–64. [DOI] [PubMed] [Google Scholar]
- [18].Bi J, Newtson AM, Zhang Y, Devor EJ, Samuelson MI, Thiel KW, Leslie KK, Successful Patient-Derived Organoid Culture of Gynecologic Cancers for Disease Modeling and Drug Sensitivity Testing, Cancers, 13 (2021) 2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dai D, Kumar NS, Wolf DM, Leslie KK, Molecular tools to reestablish progestin control of endometrial cancer cell proliferation, Am J Obstet Gynecol, 184 (2001) 790–797. [DOI] [PubMed] [Google Scholar]
- [20].Sheng Q, Vickers K, Zhao S, Wang J, Samuels DC, Koues O, Shyr Y, Guo Y, Multi-perspective quality control of Illumina RNA sequencing data analysis, Brief Funct Genomics, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner, Bioinformatics, 29 (2013) 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Liao Y, Smyth GK, Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features, Bioinformatics, 30 (2014) 923–930. [DOI] [PubMed] [Google Scholar]
- [23].Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol, 15 (2014) 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhao S, Guo Y, Sheng Q, Shyr Y, Advanced heat map and clustering analysis using heatmap3, Biomed Res Int, 2014 (2014) 986048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Livak KJ, Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method, Methods (San Diego, Calif, 25 (2001) 402–408. [DOI] [PubMed] [Google Scholar]
- [26].Schmittgen TD, Livak KJ, Analyzing real-time PCR data by the comparative C(T) method, Nat Protoc, 3 (2008) 1101–1108. [DOI] [PubMed] [Google Scholar]
- [27].Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, Hicks DG, Lester S, Love R, Mangu PB, McShane L, Miller K, Osborne CK, Paik S, Perlmutter J, Rhodes A, Sasano H, Schwartz JN, Sweep FC, Taube S, Torlakovic EE, Valenstein P, Viale G, Visscher D, Wheeler T, Williams RB, Wittliff JL, Wolff AC, American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer, Arch Pathol Lab Med, 134 (2010) 907–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ye B, Shi J, Kang H, Oyebamiji O, Hill D, Yu H, Ness S, Ye F, Ping J, He J, Edwards J, Zhao YY, Guo Y, Advancing Pan-cancer Gene Expression Survial Analysis by Inclusion of Non-coding RNA, RNA Biol, 17 (2020) 1666–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Davies S, Dai D, Wolf DM, Leslie KK, Immunomodulatory and transcriptional effects of progesterone through progesterone A and B receptors in Hec50co poorly differentiated endometrial cancer cells, J Soc Gynecol Investig, 11 (2004) 494–499. [DOI] [PubMed] [Google Scholar]
- [30].Yang S, Thiel KW, Leslie KK, Progesterone: the ultimate endometrial tumor suppressor, Trends Endocrinol Metab, 22 (2011) 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Davies S, Dai D, Pickett G, Thiel KW, Korovkina VP, Leslie KK, Effects of bevacizumab in mouse model of endometrial cancer: Defining the molecular basis for resistance, Oncol Rep, 25 (2011) 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Slomovitz BM, Broaddus RR, Schmandt R, Wu W, Oh JC, Ramondetta LM, Burke TW, Gershenson DM, Lu KH, Expression of imatinib mesylate-targeted kinases in endometrial carcinoma, Gynecol Oncol, 95 (2004) 32–36. [DOI] [PubMed] [Google Scholar]
- [33].Kirschner CV, Alanis-Amezcua JM, Martin VG, Luna N, Morgan E, Yang JJ, Yordan EL, Angiogenesis factor in endometrial carcinoma: a new prognostic indicator?, American journal of obstetrics and gynecology, 174 (1996) 1879–1882; discussion 1882–1874. [DOI] [PubMed] [Google Scholar]
- [34].Yokoyama Y, Charnock-Jones DS, Licence D, Yanaihara A, Hastings JM, Holland CM, Emoto M, Sakamoto A, Sakamoto T, Maruyama H, Sato S, Mizunuma H, Smith SK, Expression of vascular endothelial growth factor (VEGF)-D and its receptor, VEGF receptor 3, as a prognostic factor in endometrial carcinoma, Clin Cancer Res, 9 (2003) 1361–1369. [PubMed] [Google Scholar]
- [35].Holland CM, Day K, Evans A, Smith SK, Expression of the VEGF and angiopoietin genes in endometrial atypical hyperplasia and endometrial cancer, British journal of cancer, 89 (2003) 891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Giatromanolaki A, Sivridis E, Brekken R, Thorpe PE, Anastasiadis P, Gatter KC, Harris AL, Koukourakis MI, The angiogenic “vascular endothelial growth factor/flk-1(KDR) receptor” pathway in patients with endometrial carcinoma: prognostic and therapeutic implications, Cancer, 92 (2001) 2569–2577. [DOI] [PubMed] [Google Scholar]
- [37].Gornall RJ, Anthony FW, Coombes EJ, Hogston P, Woolas RP, Investigation of women with endometrial carcinoma using serum vascular endothelial growth factor (VEGF) measurement, International journal of gynecological cancer, 11 (2001) 164–166. [DOI] [PubMed] [Google Scholar]
- [38].Presta LG, Chen H, O’Connor SJ, Chisholm V, Meng YG, Krummen L, Winkler M, Ferrara N, Humanization of an anti-vascular endothelial growth factor monoclonal antibody for the therapy of solid tumors and other disorders, Cancer research, 57 (1997) 4593–4599. [PubMed] [Google Scholar]
- [39].Bender D, Sill MW, Lankes HA, Reyes HD, Darus CJ, Delmore JE, Rotmensch J, Gray HJ, Mannel RS, Schilder JM, Hunter MI, McCourt CK, Samuelson MI, Leslie KK, A phase II evaluation of cediranib in the treatment of recurrent or persistent endometrial cancer: An NRG Oncology/Gynecologic Oncology Group study, Gynecol Oncol, 138 (2015) 507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Soares R, Guerreiro S, Botelho M, Elucidating progesterone effects in breast cancer: cross talk with PDGF signaling pathway in smooth muscle cell, Journal of cellular biochemistry, 100 (2007) 174–183. [DOI] [PubMed] [Google Scholar]
- [41].Rocha A, Azevedo I, Soares R, Progesterone sensitizes breast cancer MCF7 cells to imatinib inhibitory effects, Journal of cellular biochemistry, 103 (2008) 607–614. [DOI] [PubMed] [Google Scholar]
- [42].Yu H, Jove R, The STATs of cancer--new molecular targets come of age, Nature reviews, 4 (2004) 97–105. [DOI] [PubMed] [Google Scholar]
- [43].Leehy KA, Truong TH, Mauro LJ, Lange CA, Progesterone receptors (PR) mediate STAT actions: PR and prolactin receptor signaling crosstalk in breast cancer models, J Steroid Biochem Mol Biol, 176 (2018) 88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lange CA, Richer JK, Shen T, Horwitz KB, Convergence of progesterone and epidermal growth factor signaling in breast cancer. Potentiation of mitogen-activated protein kinase pathways, J Biol Chem, 273 (1998) 31308–31316. [DOI] [PubMed] [Google Scholar]
- [45].Richer JK, Lange CA, Manning NG, Owen G, Powell R, Horwitz KB, Convergence of progesterone with growth factor and cytokine signaling in breast cancer. Progesterone receptors regulate signal transducers and activators of transcription expression and activity, J Biol Chem, 273 (1998) 31317–31326. [DOI] [PubMed] [Google Scholar]
- [46].Silvennoinen O, Schindler C, Schlessinger J, Levy DE, Ras-independent growth factor signaling by transcription factor tyrosine phosphorylation, Science, 261 (1993) 1736–1739. [DOI] [PubMed] [Google Scholar]
- [47].Benekli M, Baer MR, Baumann H, Wetzler M, Signal transducer and activator of transcription proteins in leukemias, Blood, 101 (2003) 2940–2954. [DOI] [PubMed] [Google Scholar]
- [48].Verhoeven Y, Tilborghs S, Jacobs J, De Waele J, Quatannens D, Deben C, Prenen H, Pauwels P, Trinh XB, Wouters A, Smits ELJ, Lardon F, van Dam PA, The potential and controversy of targeting STAT family members in cancer, Semin Cancer Biol, 60 (2020) 41–56. [DOI] [PubMed] [Google Scholar]
- [49].Kavlashvili T, Jia Y, Dai D, Meng X, Thiel KW, Leslie KK, Yang S, Inverse Relationship between Progesterone Receptor and Myc in Endometrial Cancer, PLoS One, 11 (2016) e0148912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Li Y, Zhou W, Li L, Li JW, Li T, Huang C, Lazaro-Camp VJ, Kavlashvili T, Zhang Y, Reyes H, Li Y, Dai D, Zhu W, Meng X, Leslie KK, Yang S, Enhancing progestin therapy via HDAC inhibitors in endometrial cancer, Am J Cancer Res, 12 (2022) 5029–5048. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq and Affymetrix array datasets generated during the current study will be deposited in GEO prior to publication. All other data generated or analyzed during this study are included in the submitted article and its supplementary information files.