Abstract
Terpene aroma compounds are key quality attributes of postharvest Torreya grandis nuts, contributing to their commercial value. However, terpene biosynthesis and regulatory networks in different T. grandis cvs. are still poorly understood. Here, chief cvs. ‘Xi Fei’ and ‘Xiangya Fei’ were investigated for their differences in terpene biosynthesis and gene expression levels during postharvest ripening using headspace solid-phase microextraction (HS-SPME) coupled with gas chromatography–mass spectrometry (GC-MS) and transcriptomic datasets. A total of 28 and 22 aroma compounds were identified in ‘Xi Fei’ and ‘Xiangya Fei’, respectively. Interestingly, differences in aroma composition between the two cvs. were mostly attributed to D-limonene and α-pinene levels as key determinants in Torreya nuts’ flavor. Further, transcriptome profiling, correlation analysis, and RT-qPCR annotated two novel genes, TgTPS1 in ‘Xi Fei’ and TgTPS2 in ‘Xiangya Fei’, involved in terpene biosynthesis. In addition, six transcription factors (TFs) with comparable expression patterns to TgTPS1 and four TFs to TgTPS2 were identified via correlation analysis of a volatile and transcriptome dataset to be involved in terpene biosynthesis. Our study provides novel insight into terpene biosynthesis and its regulation at the molecular level in T. grandis nut and presents a valuable reference for metabolic engineering and aroma improvement in this less explored nut.
Keywords: Torreya grandis, GC-MS, terpene, transcriptome sequencing, gene expression
1. Introduction
Nuts (walnuts, almonds, pistachios, etc.) are rich in protein and fat and provide a balanced blend of monounsaturated and polyunsaturated fatty acids, as well as a variety of bioactive compounds that play a beneficial role in disease management such as cancer, diabetes, and hypertension [1,2]. As a result, many studies have recommended the inclusion of nuts in a healthy diet [3]. In addition, the US Food and Drug Administration (FDA) approved the first specific health claim related to nuts in 2003—that eating 1.5 ounces (42.5 g) of nuts per day can reduce the risk of heart disease. Among nuts, Torreya grandis (T. grandis) is an important specialty nut from the subtropical mountains of China, with high nutritional value, and it contains a variety of bioactive compounds such as sciadonic acid, tocopherols, and flavonoids, which have preventive effects against heart and brain disease, as well as anti-inflammatory, antiviral, and antioxidant properties [4,5,6,7].
The aroma of the fruit is a complex mixture of hundreds of different heterogeneous volatile chemicals (terpenes, esters, alcohols, aldehydes, ketones, pyrazines, acids, etc.) in highly variable concentrations (ranging from a few mg kg−1 to a few ng kg−1) [8]. The different proportions of volatile components determine the overall aroma characteristics. The aroma also has a strong influence on fruit quality, with volatile components being the decisive factor between species [9,10]. At present, headspace solid-phase microextraction (HS-SPME) coupled with gas chromatography–mass spectrometry (GC-MS) is a widely used and efficient method for aroma detection and analysis [11,12,13,14,15,16]. Aroma means ‘Xiang’ and T. grandis means ‘Xiang Fei’ in Chinese. The fact that the Chinese name for T. grandis contains the word ‘Xiang’ implies that aroma is an important sensory quality of T. grandis nuts and that their characteristic aroma contributes significantly to their overall acceptance by consumers. Our previous study revealed five major categories of aromatic components in T. grandis nuts post harvest, including terpenes, aldehydes, alcohols, alkanes, and esters using SPME GC-MS analysis. Among them, terpenes constituted the major class throughout the postharvest ripening stage, ranging from 56% to 87% [17].
The terpene biosynthesis primarily occurs through two pathways: the mevalonic acid (MVA) pathway located in the cytosol and the 2-C-methyl-D-erythritol-4-phosphate (MEP) pathway located in the plastid [18]. The MVA pathway utilizes acetyl-CoA as a substrate to generate isopentenyl pyrophosphate (IPP) [19], which is mainly involved in the biosynthesis of sesquiterpenes, carotenoids, sterols, and triterpenoids in the cytosol [20]. The MEP pathway, on the other hand, utilizes pyruvate and glyceraldehyde-3-phosphate as substrates to produce dimethylallyl pyrophosphate (DMAPP) [21], primarily participating in the biosynthesis of monoterpenes, diterpenes, gibberellins, abscisic acid, and carotenoids in the plastid [22]. The primary enzymes that catalyze the manufacture of terpenes are called terpene synthases (TPSs). Since the discovery of the first linalool synthase gene in fairy fans (Clarkia breweri) [23], many TPS genes have been cloned and investigated in Arabidopsis (Arabidopsis thaliana) [24], citrus (Citrus sinensis) [25], grape (Vitis vinifera) [26], tomato (Solanum lycopersicum) [27], etc. Terpene biosynthesis is regulated though not only by key TPSs but also by the activity of transcription factors (TFs) [28]. Many TFs involved in terpene biosynthesis have been identified, such as AP2/ERF (APETALA2/ethylene-responsive factor), MYB, WRKY, bHLH (helix-loop-helix), bZIP (basic leucine zipper), and NAC (NAM, ATAF and CUC) families [29]. For example, CitAP2.10 promotes the production of the sesquiterpene (+)-valencene via activating the CsTPS1 gene in ‘Newhall’ orange (Citrus sinensis) [30]. In peach fruit, PpbHLH1 is directly bound to the PpTPS3 promoter to regulate linalool production [31]. Furthermore, SlMYC1, SlEOT1, and SlWRKY73 have been reported to activate SlTPS5 promoter to promote linalool production in tomatoes [32].
Genotypes can also account for differences in terpene composition and content in different taxa represented by species, cv. type, etc. [33,34]. However, knowledge of the variation in terpene composition among different T. grandis cvs. during postharvest ripening is not reported in the literature. Therefore, this present study aimed to assess changes in terpene profiles from two T. grandis cvs. ‘Xi Fei’ and ‘Xiangya Fei’ during postharvest ripening as analyzed using headspace solid-phase microextraction (HS-SPME) coupled with the gas chromatography–mass spectrometry (GC-MS) technique. Furthermore, the regulatory mechanism underlying key gene expression patterns with terpene production in ‘Xi Fei’ and ‘Xiangya Fei’ was also investigated to underline the molecular mechanisms. The results presented from this study provide better insight into terpene biosynthesis and their regulatory mechanisms in T. grandis nuts.
2. Results and Discussion
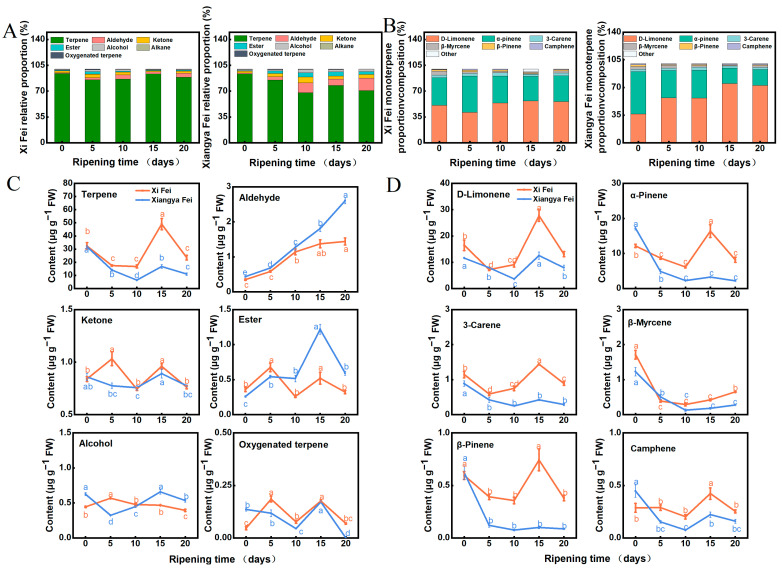
2.1. Differences in Terpene Content Account for Aroma Differences between ‘Xi Fei’ and ‘Xiangya Fei’ Nuts during Postharvest Ripening
Two T. grandis cultivars, ‘Xi Fei’ and ‘Xiangya Fei’, were placed in a postharvest ripening environment at 25 °C and 90% relative humidity (RH), and then sampled on days 0, 5, 10, 15, and 20 (Figure 1A). Volatiles were analyzed using SPME coupled with GC-MS (Figure S1). Firstly, the CIRG value of seed coats of the two T. grandis cvs. during postharvest ripening was measured. The results revealed that the changes in CIRG value and seed coat color were consistent in both varieties, with a significant overall upward trend (p < 0.05; Figure 1B), revealing that the postharvest ripening process was proceeding normally [35]. Totals of 28 and 22 volatile compounds were identified from ‘Xi Fei’ and ‘Xiangya Fei’ using GC-MS, respectively (Tables S2 and S3). The results revealed that aroma compounds were mainly classified into seven major groups, including terpenes, alcohols, aldehydes, esters, oxygenated terpenes, alkanes, and ketones, with terpenes being the most abundant, accounting for 85.19–93.90% in ‘Xi Fei’ (Figure 2A) and 67.75–93.24% in ‘Xiangya Fei’ (Figure 2B), suggesting that terpenes are the main aroma-presenting substances during the postharvest ripening process of T. grandis. We previously explored changes in the aroma composition of ‘Xi Fei’ at 20 °C and a 90% RH environment during postharvest ripening, revealing that terpenes accounted for 56–87%, and that the terpene content increased significantly on days 4 and 8 [36]. Our current results showed that the total terpene content in both cvs. significantly peaked on days 10–15 of postharvest ripening, at 49.02 µg g−1 for ‘Xi Fei’ versus 16.70 µg g−1 for ‘Xiangya Fei’, with the terpene content of ‘Xi Fei’ being 2.94 times higher than that of ‘Xiangya Fei’ (p < 0.05; Figure 2C). Meanwhile, the content of other aroma compounds was lower and not significantly different compared to the total terpenes (Figure 2C). Different temperatures and humidity levels significantly influenced aroma content during postharvest ripening, in line with our previous study [17]. Upon further analysis of the terpenes, D-limonene, α-pinene, 3-carene, β-myrcene, β-pinene, and camphene were detected in the two cvs. to account for aroma, with D-limonene and α-pinene being the most abundant (Figure 2D). Differences in the levels of D-limonene and α-pinene were also observed among cvs., with D-limonene and α-pinene in ‘Xi Fei’ accounting for 41.06–56.61% and 33.35–49.48%, with 36.31–74.94% and 19.52–53.74% in ‘Xiangya Fei’, respectively (Figure 2D). Therefore, differences in the total terpene content are likely to account for the difference in aroma between these two cvs., and as observed for cv., impact on aroma composition in apple [37], bayberry [38], pepper [39], etc. In addition, the major difference in the two cvs. was D-limonene content (Figure 2D). D-limonene is a relatively abundant monoterpene that serves as the primary terpene element of most citrus fruit essential oils, which has a broad range of medicinal, antibacterial, antioxidant, and chemotherapeutic activities [40]. Moreover, D-limonene finds extensive application in the food and beverage, cosmetic, perfume, and pharmaceutical sectors [41]. T. grandis nuts have a lot of D-limonene, which may make it easier for it to be used extensively in industrial processing. Therefore, the major differences in terpene or D-limonene content in the two cvs. may provide a theoretical basis for variety selection and quality enhancement.
Figure 1.
External changes in T. grandis ‘Xi Fei’ and ‘Xiangya Fei’ in different stages of postharvest ripening. (A) ‘Xi Fei’ and ‘Xiangya Fei’ in different stages of postharvest ripening treatment. (B) The CIRG value in the seed coat of ‘Xi Fei’ and ‘Xiangya Fei’. The different letters indicate significant differences (p < 0.05) in different stages of during postharvest ripening.
Figure 2.
Changes in volatile composition in T. grandis ‘Xi Fei’ and ‘Xiangya Fei’ in different stages during postharvest ripening. (A) Changes in the proportion of total volatile substances in ‘Xi Fei’ and ‘Xiangya Fei’. (B) Changes in the proportion of total terpenes in ‘Xi Fei’ and ‘Xiangya Fei’. (C) Changes in the content of total volatile substances in ‘Xi Fei’ and ‘Xiangya Fei’. (D) Changes in the content of total terpenes in ‘Xi Fei’ and ‘Xiangya Fei’. Bars = 1 cm. Error bars represent SE based on three biological replicates. The different letters indicate significant differences (p < 0.05) in different stages during postharvest ripening. FW, fresh weight.
2.2. Deferentially Expressed Terpenoid Biosynthetic Genes Responsible for Terpene Accumulation in ‘Xi Fei’ and ‘Xiangya Fei’ during Post-Ripening Stage
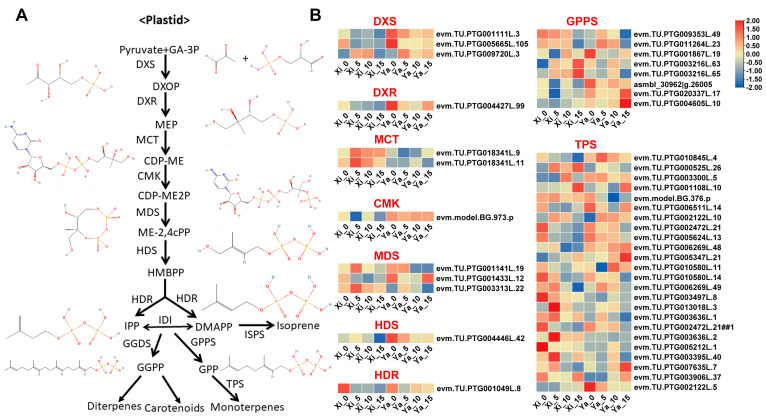
To investigate the molecular basis accounting for differences in terpene biosynthesis between these two Torreya grandis cultivars, transcriptome sequencing was performed on postharvest-treated samples of both cvs., resulting in 24 transcriptome samples. After filtering out low-quality reads, each sample yielded clean data of at least 8.74 G, with average Q20 and Q30 values of 97.25% and 92.75%, respectively, and an average GC content of 45.07% (Table S4). Additionally, principal component analysis (PCA) revealed clustering of the three biological replicates of ‘Xi Fei’ and ‘Xiangya Fei’ during the post-ripening stage, indicating that the transcriptomic data in our study were accurate for subsequent analysis (Figure S2). In this study, GC-MS analysis revealed that only for monoterpenes in T. grandis nuts did the aroma blend show differential levels among cvs. Therefore, the expression levels of 44 genes involved in monoterpene biosynthesis in the MEP pathway were analyzed (Figure 3). Several key regulatory genes in this pathway have been thoroughly studied, including DXS, which converts pyruvate and glyceraldehyde-3-phosphate to MEP and serves as a rate-limiting enzyme in the MEP pathway; DXR, the second rate-limiting enzyme in the MEP pathway that plays a crucial role in catalyzing terpenoid biosynthesis; GPPS, a short-chain isoprenyl transferase that provides the direct precursor geranyl diphosphate (GPP) for terpenoid biosynthesis [42]; and TPS, a terpene synthase that catalyzes the formation of sesquiterpenes, monoterpenes, diterpenes, and triterpene skeletons from different precursors [43].
Figure 3.
Analysis of MEP pathway genes of terpene biosynthesis in ‘Xi Fei’ and ‘Xiangya Fei’ nuts in different stages during postharvest ripening. (A) Plant terpene biosynthetic MEP pathway involved in monoterpene production [44]. Abbreviations: DXS, 1-deoxy-D-xylulose 5-phosphate synthase; DXR, 1-deoxy-D-xylulose 5-phosphate reductoisomerase; MCT, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; CMK, 4-(cytidine 50 -diphospho)-2-C-methyl-D-erythritol kinase; MDS, 2-C-methyl-D-erythritol 2,4- cyclodiphosphate synthase; HDS, (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase; HDR, (E)-4-hydroxy-3-methylbut-2-enyl diphosphate reductase; GPPS, geranyl pyrophosphate synthase; GGPPS, geranylgeranyl pyrophosphate synthase; FPPS, farnesyl pyrophosphate synthase; TPS, terpene synthase; IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate. (B) The FPKM value of MEP pathway genes in ‘Xi Fei’ and ‘Xiangya Fei’ in different stages of postharvest ripening treatment. The MEP pathway genes whose FPKM values were greater than 0 were selected based on annotation. ‘Xi’ represents ‘Xi Fei’, ‘Ya’ represents ‘Xiangya Fei’.
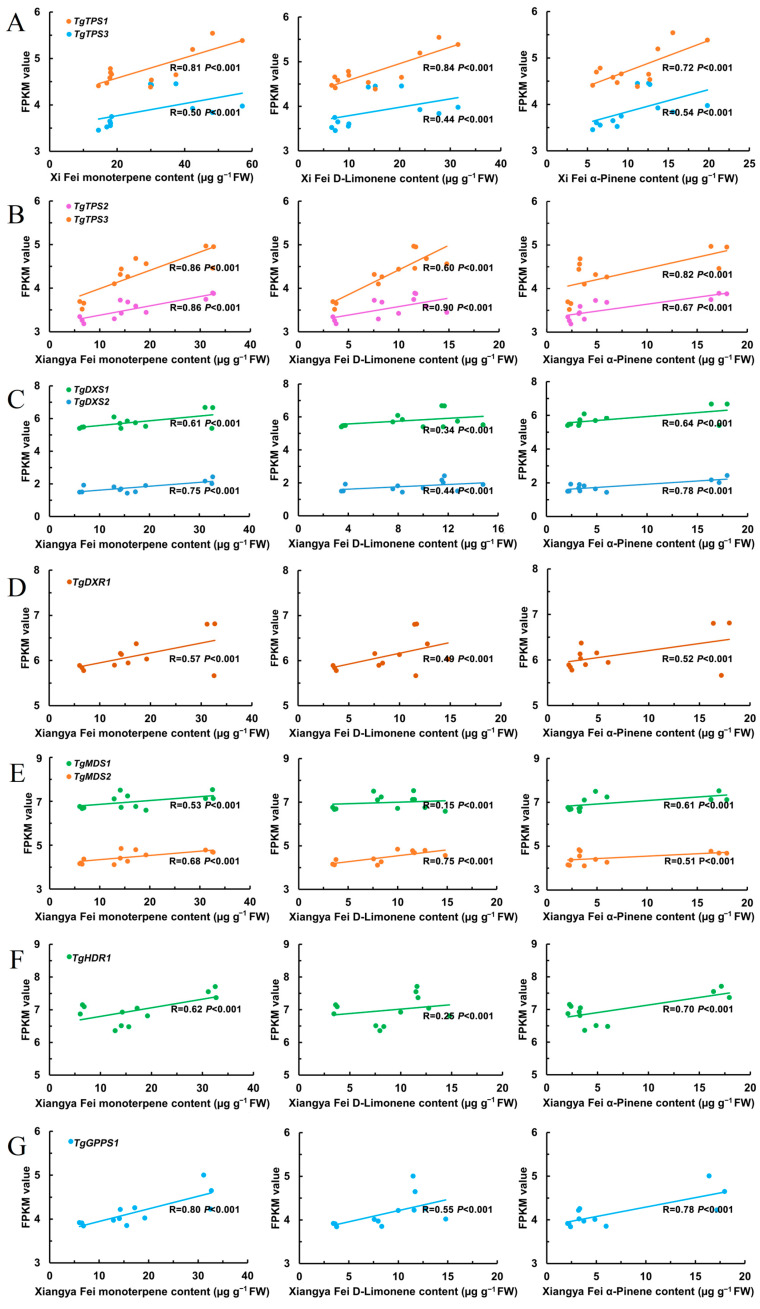
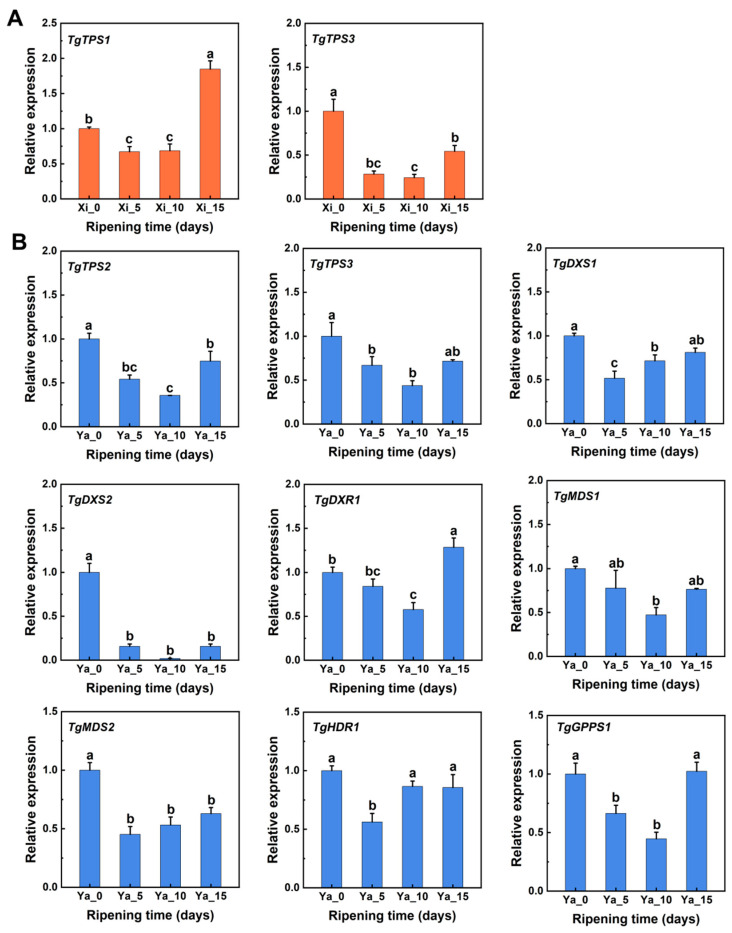
Some genes in the MEP pathway exhibited different expression patterns, and the differences in the expression levels of these genes may contribute to the total terpene differences among the two T. grandis cvs. To identify which specific genes or gene categories play a crucial role in terpene biosynthesis, a correlation analysis was conducted between the expression levels of the 44 genes in the MEP pathway and the total terpenes, D-limonene, and α-pinene in the two varieties. The results showed significant positive correlations between the FPKM value of certain genes and the content of total terpenes, viz., D-limonene, and α-pinene, including TgTPS1 (evm.TU.PTG001108L.10) and TgTPS3 (evm.TU.PTG006511L.14) in ‘Xi Fei’ (p < 0.05; Figure 4A). Among these, the FPKM value of TgTPS1 (R = 0.81) in ‘Xi Fei’ showed a stronger correlation with the total terpene content than TgTPS3 (R = 0.50) (p < 0.05; Figure 4A). In ‘Xiangya Fei’, nine MEP pathway genes were significantly correlated with the content of total terpenes, viz., D-limonene, and α-pinene, including TgTPS2 (evm.model.BG.376.p), TgTPS3 (evm.TU.PTG006511L.14) (p < 0.05; Figure 4B), TgDXS1 (evm.TU.PTG001111L.3), TgDXS2 (evm.TU.PTG005665L.105) (p < 0.05; Figure 4C), TgDXR1 (evm.TU.PTG004427L.99) (p < 0.05; Figure 4D), TgMDS1 (evm.TU.PTG001141L.19), TgMDS2 (evm.TU.PTG001433L.12) (p < 0.05; Figure 4E), TgHDR1 (evm.TU.PTG001049L.8) (p < 0.05; Figure 4F), and TgGPPS1 (evm.TU.PTG001867L.19) (p < 0.05; Figure 4G). Meanwhile, expression results for these genes were further validated using RT-qPCR experiments (Figure 5). The RT-qPCR results were consistent with the FPKM value of the transcriptome data. Since genes such as TgDXS, TgDXR, TgMDS, TgHDR, and TgGPPS provided precursors for monoterpenoid biosynthesis and did not directly affect the content of total terpenes, this study mainly focused on the effects of TPSs on terpene biosynthesis. RT-PCR analysis revealed that TgTPS1 in ‘Xi Fei’ and TgTPS2 and TgTPS3 ‘Xiangya Fei’ were their key terpene biosynthetic genes, and their expression differences accounted for differences in the total terpene content (Figure 5). Similar results were observed in two cultivars of Dendrobium officinale showing differentially expressed genes of 13 TPSs as the main factor accounting for differences in terpene composition [45].
Figure 4.
Correlation analysis of total terpene, D-limonene, and α-pinene contents and all related genes in MEP pathway in ‘Xi Fei’ and ‘Xiangya Fei’ in different stages during postharvest ripening. (A) The correlation analysis between TgTPS1 and TgTPS2 FPKM values and the total terpene, D-limonene, and α-pinene contents in ‘Xi Fei’. (B) The correlation analysis between TgTPS2 and TgTPS3 FPKM values and the total terpene, D-limonene, and α-pinene contents in ‘Xiangya Fei’. (C) The correlation analysis between TgDXS1 and TgDXS2 FPKM values and the total terpene, D-limonene, and α-pinene contents in ‘Xiangya Fei’. (D) The correlation analysis between TgDXR1 FPKM values and the total terpene, D-limonene, and α-pinene contents in ‘Xiangya Fei’. (E) The correlation analysis between TgMDS1 and TgMDS2 FPKM values and the total terpene, D-limonene, and α-pinene contents in ‘Xiangya Fei’. (F) The correlation analysis between TgHDR1 FPKM value and the total terpene, D-limonene, and α-pinene contents in ‘Xiangya Fei’. (G) The correlation analysis between TgGPPS1 FPKM value and the total terpene, D-limonene, and α-pinene contents in ‘Xiangya Fei’.
Figure 5.
Relative gene expression of all related genes in MEP pathway in ‘Xi Fei’ (A) and ‘Xiangya Fei’ (B) cvs. in different stages during postharvest ripening. The data on Day 0 were set to 1. ‘Xi’ represents ‘Xi Fei’, ‘Ya’ represents ‘Xiangya Fei’. The different letters indicate significant differences (p < 0.05) in different stages of during postharvest ripening.
2.3. Deferentially Expressed Transcription Factors Responsible for Variations in Terpenes in ‘Xi Fei’ and ‘Xiangya Fei’ during Postharvest Ripening
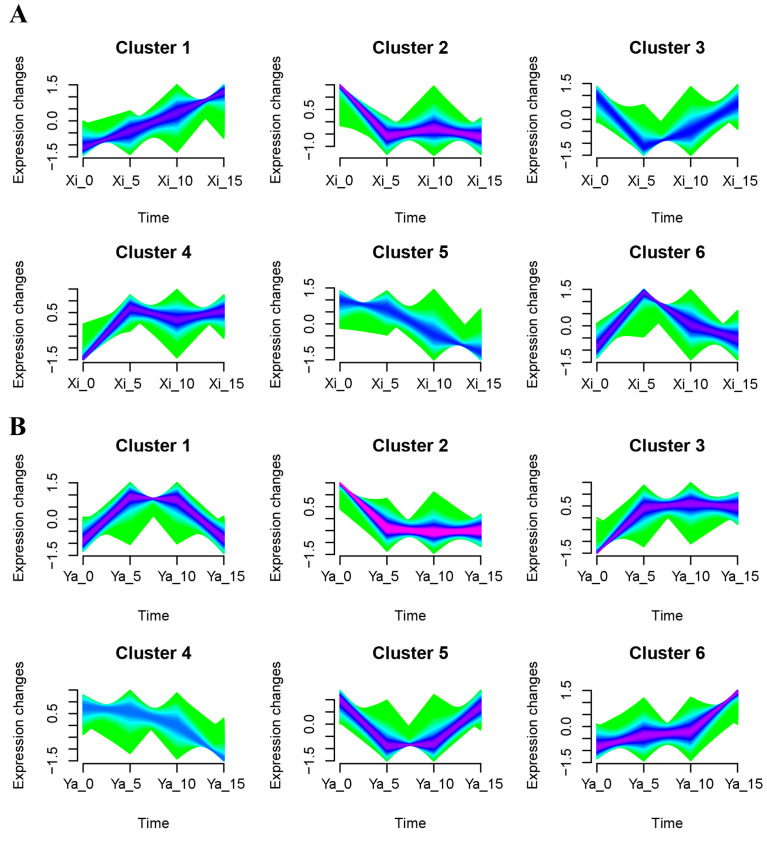
Identifying the molecular mechanism behind terpene biosynthesis is essential to improve terpene production through metabolic engineering. Transcription factors (TFs) play important roles in regulating gene expression in various plant biological processes including terpene production [46]. In this study, co-regulation cluster identification and analysis was performed based on the Mfuzz soft clustering method to reveal TFs with similar expression patterns to TgTPS1, TgTPS2, and TgTPS3. The results revealed that all genes in ‘Xi Fei’ could be classified into six clusters, with TgTPS1 in Cluster 1, implying that the TFs co-expressed with TgTPS1 could be selected from Cluster 1 (Figure 6A). In ‘Xiangya Fei’, TgDXS1, TgDXS2, TgDXR1, TgHDR1, TgGPPS1, and TgTPS2 belonged to cluster 2, TgMDS1 in cluster 4, and TgMDS2 and TgTPS3 in cluster 5 (Figure 6B). Since most of the genes that were significantly and positively correlated with terpene biosynthesis in ‘Xiangya Fei’ were located in cluster 2, TFs co-expressed with TgTPS2 in cluster 2 were further analyzed in ‘Xiangya Fei’.
Figure 6.
Mfuzz analysis of all genes from the transcriptomic datasets in ‘Xi Fei’ (A) and ‘Xiangya Fei’ (B) in different stages during postharvest ripening. ‘Xi’ represents ‘Xi Fei’, ‘Ya’ represents ‘Xiangya Fei’.
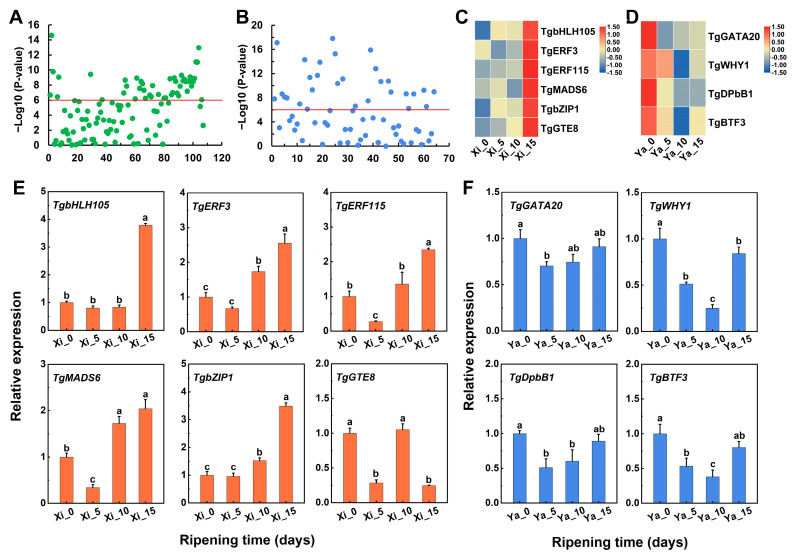
A total of 107 TFs in Cluster 1 of ‘Xi Fei’ and 64 TFs in Cluster 2 of ‘Xiangya Fei’ were analyzed for correlation values with TgTPS1 and TgTPS2, respectively (Figure 7A,B). Six TFs with −log10(p-value) > 6 and a correlation with TgTPS1 > 0.8 were selected in ‘Xi Fei’, which were TgbHLH105 (evm.TU. PTG012091L.13), TgERF3 (evm.TU.PTG002487L.23), TgERF115 (evm.TU.PTG005873L.5), TgMADS6 (evm.TU.PTG006600L.3), TgbZIP1 (evm.TU. PTG004438L.39), and TgGTE8 (evm.TU.PTG000808L.2) (Figure 7C). In addition, four TFs with −log10(p-value) > 6 and a correlation with TgTPS2 > 0.6 were selected in ‘Xiangya Fei’, namely TgGATA20 (evm.TU.PTG008279L.15), TgWHY1 (evm.TU.PTG022256L.4), TgTgDPbB1 (evm.TU.PTG005832L.37), and TgBTF3 (evm.TU.PTG001638L.23) (Figure 7D). To validate the expression patterns of TFs observed in correlation analysis, 10 annotated TFs were selected for analysis of their expression levels using RT-qPCR. The results revealed that the TFs involved in the regulation of terpene biosynthesis were different in ‘Xi Fei’ versus ‘Xiangya Fei’, and that the TFs were highly expressed in ‘Xi Fei’ on Day 15 of the post-ripening stage (Figure 7E). In contrast, TFs were highly expressed in ‘Xiangya Fei’ on Day 0 of the post-ripening stage (Figure 7F).
Figure 7.
Statistical analysis of correlations between transcription factors (TFs) and genes related to terpene biosynthesis. (A) Statistical analysis of the correlation between candidate TFs and TgTPS1 in ‘Xi Fei’. (B) Statistical analysis of the correlation between candidate TFs and TgTPS2 in ‘Xiangya Fei’. (C) FPKM value of candidate TFs in ‘Xi Fei’. Threshold level −log10(p-value) > 6, R > 0.8. (D) FPKM value of candidate TFs in ‘Xiangya Fei’. Threshold level −log10(p-value) > 6, R > 0.6. (E) RT-qPCR analysis of candidate TFs in ‘Xi Fei’. (F) RT-qPCR analysis of candidate TFs in ‘Xiangya Fei’. The different letters indicate significant differences (p < 0.05) in different stages of during postharvest ripening.
The main families of TFs involved in terpene biosynthesis in plants are bHLH [47,48], AP2/ERF [49], NAC [50], WRKY [51], MYB [52], and bZIP [53]. For example, terpene biosynthesis in Litsea cubeba was regulated by LcbHLH78 and LcTPS42, and the overexpression of LcbHLH78 significantly increased the content of α-pinene, linalool, and geraniol [54]. CitERF71 directly activates the CitTPS16 promoter and promotes the synthesis of (E)-geraniol in sweet orange fruit [55]. However, there are fewer reports on the involvement of MADS, GTE, WHY, E2F/DP, and BTF3 transcription factor families in the regulation of terpene biosynthesis. Therefore, further study shall focus on clarifying the regulatory role of these TFs on TPS of terpene biosynthesis in T. grandis nuts.
3. Material and Methods
3.1. Plant Materials
The two primary T. grandis cvs. (‘Xi Fei’ and ‘Xiangya Fei’) with a distinct morphology difference were hand-picked on 10 September 2022 at the Anji Torreya base, Zhejiang Province, China (30°38′ N, 119°41′ E), put on ice, and transported to the lab within 5 h of harvest. The nuts were cracked (ca., 525 days after flowering) and the arils was removed mechanically. After washing with purified water, nuts that were free of mechanical damage and of uniform size and color were selected for subsequent experiments. The screened nuts were mixed, divided into three groups each of 5 kg, and then placed at 25 ± 2 °C and in a 90% RH ± 2% RH environment for postharvest ripening treatment [35]. Generally, the postharvest ripening process of Torreya grandis nuts usually takes a period of 10–15 days under appropriate temperature and humidity conditions [36]. Therefore, the nuts were then sampled on days 0, 5, 10, 15, and 20. After removing the seed shells, the kernels were cut into 1 mm × 1 mm pieces, snap-frozen in liquid nitrogen, and stored at −80 °C for subsequent experiments.
3.2. Determination of the Color of the Seed Coat
The color of the seed coat was determined using the color index of red grapes (CIRG) = (180 − H)/(L* + C) [56]. Every five days during the treatment, five to six T. grandis nuts from each of the two treatments were chosen at random and shelled to see how the seed coat color changed. A CR-10 colorimeter (KONICA MINOLTA, INC., Tokyo, Japan) was used to measure four spots at 90◦ intervals at the nut equator. The results showed that L* (lightness, 1~100 value range), a* (redness, −60~60 value range), and b* (yellowness, −60~60 value range) were recorded in the appropriate ranges. The hue’s intensity and purity were then measured by converting the a* and b* values into H (hue angle) [H = arc tan (b*/a*)] and C (chroma) [C = [(a*)2 + (b*)2]0.5].
3.3. Extraction of Volatile Compounds from T. grandis Nuts
Volatiles were extracted according to our previous study [36]. Volatile compounds from T. grandis nuts were extracted using a solid-phase microextraction (SPME) autosampler fitted with a 65 μm polydimethylsiloxane and divinylbenzene (PDMS-DVB) fiber (Supeclo Co., Bellefonte, PA, USA). The fiber was conditioned in a GC injector port at 250 °C for 30 min before use. Afterward, frozen T. grandis kernel tissue (1 g) was ground into a powder in liquid nitrogen and placed into 20 mL vials containing 5 mL of saturated NaCl solution. An internal standard, 2-octanol (0.76 mg/mL, 20 µL, GC ≥ 99.5%), was purchased from Sigma-Aldrich Co. (St. Louis, MO, USA) and then added and stirred for 10 s with a vortex before sealing the vials. The samples were extracted using the PDMS-DVB fiber with constant stirring after the samples were equilibrated at 40 °C for 30 min. Subsequently, volatiles were desorbed for 5 min at 230 °C into the splitless injection port of the GC–flame ionization detector (FID). For the volatile analysis, an Agilent 6890N GC with an FID and a DB-WAX column (0.25 mm, 30 m, 0.25 μm, J & W Scientific, Folsom, CA, USA) was employed. Triplicate analyses of each sample were performed to assess for variance.
3.4. Gas Chromatography-Mass Spectrometry (GC-MS) Analysis
The GC-MS analysis was performed based on our previous study [36]. Volatile compounds were subjected to GC analysis on an Agilent 6890A GC coupled to an Agilent 5975C MS and equipped with a DB-WAX column (30 m × 0.25 mm inner diameter, 0.25 μm phase film thickness, J & W Scientific, Folsom, CA, USA). The mass spectrometer was operated in electron impact mode at a voltage of 70 eV. The flow rate of helium through the DB-WAX column was 1.0 mL min−1. The injector temperature was 250 °C and the source temperature was 230 °C. The column was initially started at 40 °C; the temperature was then increased from 40 °C to 100 °C at 3 °C min−1 and finally increased to 245 °C at a rate of 5 °C min−1. The NIST Mass Spectral Library (NIST-08 and Flavor) was used to identify volatiles detected by GC-MS by comparing electron ionization mass spectra and retention time data. Volatile levels were determined relative to the peak area of the internal standard in each sample as a reference. To assess for sensitivity of the GC-MS method, an internal standard (2-octanol) curve spanning from 1.6 to 200 ug/mL was obtained and analyzed under the same conditions used for Torreya nut analysis. The calculated limit of detection (LOD) and limit of quantification (LOQ) were 0.484 and 1.472 ug/mL, respectively. The reproducibility of the analytical method was based on injections of three different concentrations (0.4, 0.8, and 8 ug/mL) of the internal standard 2-octanol following the same method on three different days in triplicates, revealing a low coefficient of variation ranging from 0.011 to 0.082.
3.5. RNA Extraction and cDNA Library Construction and Sequencing
The CATB method [57] was used to isolate T. grandis total nut RNA, which was then subjected to electrophoresis on a 1.2% agarose gel, and the absorbance was measured at 260 nm to evaluate the total RNA purity and quality. The cDNA libraries were constructed from a total of 3 µg RNA samples, according to the standard protocol. To guarantee that the obtained library had an effective concentration greater than 2 nM, it was measured using the Qubit2.0 Fluorometer (Thermo Fisher Scientific, Carlsbad, CA, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). After the library was qualified, the different libraries were sequenced by an Illumina NovaSeq 6000 (Illumina, Inc., San Diego, CA, USA). After trimming reads containing adapters, reads containing N bases, and low-quality reads from the raw data, clean data (clean reads) were obtained. Based on the reference genome [58], the clean data were annotated accordingly.
3.6. Transcriptome Analysis
The gene expression levels were calculated as Fragments Per Kilobase per Million (FPKM). Differential expression analysis of the three conditions/groups (three biological replicates per condition) was performed using the DESeq2 R package (1.20.0). The resulting p-values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. The threshold for significant differential expression was set as padj < 0.05 and |log2(foldchange)| > 1. The Kyoto Encyclopedia of Genes and Genomes (KEGG) was used to compare the pathway enrichment of the differentially expressed genes [4].
3.7. Real-Time qPCR (RT-qPCR) Analysis
RT-qPCR analysis was performed based on our previous study [36]. For cDNA synthesis, 1000 ng of total RNA from each sample was treated with a gDNA eraser to remove genomic DNA and was then used to synthesize first-strand cDNA using the PrimeScript™ RT Master Mix (Takara, Dalian, China). RT-qPCR was performed using a CFX96 instrument with the Ssofast Eva Green Supermix Kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The primers for the RT-qPCR test were designed using NCBI/Primer-BLAST and the sequences of all primers are listed in Table S1. The qPCR reactions were performed in a total volume of 20 μL, consisting of 10 µL of SYBR PCR supermix (Bio-Rad Laboratories, Inc., Hercules, CA, USA), 6 µL of DEPC-treated H2O, 2 µL of 5-fold-diluted cDNA template, and 1 µL of each primer (10 µM). The TgActin gene was used as an internal control to normalize small differences in template amounts. The 2(−ΔΔCt) method was used to calculate the expression levels of each gene, and the data on day 0 were set as 1. Three biological replicates were used for RT-qPCR analysis.
3.8. Co-Regulation Cluster Identification and Analysis
Mfuzz is a soft-clustering method based on the fuzzy c-means algorithm, which is capable of clustering genes based on similar expression profiles, where genes with the same expression trend may be involved in the same biological process [59]. All genes in ‘Xi Fei’ and ‘Xiangya Fei’ were clustered using Mfuzz analysis based on the transcriptome data. A total of 6 clusters were classified in both T. grandis cultivars accordingly.
3.9. Statistical Analysis
The experiments were conducted using a totally random design. The data are presented as mean ± standard deviation (SD). Significant differences in volatile compounds and gene expression levels in the two T. grandis cultivars obtained in the triplicate analysis were determined by one-way ANOVA in SPSS Statistics 20.0 (SPSS Inc., Chicago, IL, USA).
4. Conclusions
In this study, volatile profiles in two T. grandis cultivars ‘Xi Fei’ and ‘Xiangya Fei’ were investigated during postharvest ripening, revealing that terpenes were the most abundant types of volatile compounds contributing to nut aroma post harvest. In addition, the terpene level in ‘Xi Fei’ was 2.94 times higher than that of ‘Xiangya Fei’. The analysis of transcriptome sequencing showed 44 genes in the MEP pathway likely to be involved in terpene biosynthesis in ‘Xi Fei’ and ‘Xiangya Fei’. Combined correlation analysis and RT-qPCR indicated that TgTPS1 in ‘Xi Fei’ and TgTPS2 in ‘Xiangya Fei’ accounted for its terpene biosynthesis as well as differences in aroma composition. Furthermore, co-regulation cluster identification and RT-qPCR validation demonstrated that the transcription factors involved in terpene biosynthesis in the two cultivars were significantly different: TgbHLH105, TgERF3, TgERF115, TgMADS6, TgbZIP1, TgGTE8 in ‘Xi Fei’ and TgGATA20, TgWHY1, TgTgDPbB1, and TgBTF3 in ‘Xiangya Fei’. Collectively, the production of terpene in T. grandis nuts during postharvest ripening appears to be the result of a combination of complex metabolic pathways, including diverse physiological processes and molecular regulation mechanisms. Our study provides valuable clues to gain better insight into terpene production in Torreya nut, and a reference for metabolic engineering and aroma improvement in the future.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25115581/s1.
Author Contributions
Z.Z. conceived the research plans and wrote the manuscript. L.G. and L.T. carried out most of the experiments and analyzed the data; T.W., J.S., Y.H. and W.Y. provided help with the experiments and data analysis. L.S. and J.W. conceived the research plans and supervised the experiments. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This work was financially supported by the National Natural Science Foundation of China (32101556 and U23A20219), the National Key Research and Development Program of China (2022YFD2200603), the Zhejiang Forestry Science and Technology Project (2022B06), and the Breeding of New Varieties of Torreya grandis Program (2021C02066-11).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Souza R.G.M., Gomes A.C., Naves M.M.V., Mota J.F. Nuts and legume seeds for cardiovascular risk reduction: Scientific evidence and mechanisms of action. Nutr. Rev. 2015;73:335–347. doi: 10.1093/nutrit/nuu008. [DOI] [PubMed] [Google Scholar]
- 2.Cardoso B.R., Duarte G.B.S., Reis B.Z., Cozzolino S.M.F. Brazil nuts: Nutritional composition, health benefits and safety aspects. Food Res. Int. 2017;100:9–18. doi: 10.1016/j.foodres.2017.08.036. [DOI] [PubMed] [Google Scholar]
- 3.De Souza R.G.M., Schincaglia R.M., Pimentel G.D., Mota J.F. Nuts and human health outcomes: A systematic review. Nutrients. 2017;9:1311. doi: 10.3390/nu9121311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suo J., Tong K., Wu J., Ding M., Chen W., Yang Y., Lou H., Hu Y., Yu W., Song L. Comparative transcriptome analysis reveals key genes in the regulation of squalene and β-sitosterol biosynthesis in Torreya grandis. Ind. Crops Prod. 2019;131:182–193. doi: 10.1016/j.indcrop.2019.01.035. [DOI] [Google Scholar]
- 5.Zhang F., Ma Z., Qiao Y., Wang Z., Chen W., Zheng S., Yu C., Song L., Lou H., Wu J. Transcriptome sequencing and metabolomics analyses provide insights into the flavonoid biosynthesis in Torreya grandis kernels. Food Chem. 2022;374:131558. doi: 10.1016/j.foodchem.2021.131558. [DOI] [PubMed] [Google Scholar]
- 6.Song L., Wen S., Ye Q., Lou H., Gao Y., Bajpai V.K., Carpena M., Prieto M.-A., Simal-Gandara J., Xiao J., et al. Advances on delta 5-unsaturated-polymethylene-interrupted fatty acids: Resources, biosynthesis, and benefits. Crit. Rev. Food Sci. Nutr. 2023;63:767–789. doi: 10.1080/10408398.2021.1953960. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z., Chen W., Tao L., Wei X., Gao L., Gao Y., Suo J., Yu W., Hu Y., Yang B., et al. Ethylene treatment promotes umami taste-active amino acids accumulation of Torreya grandis nuts post-harvest by comparative chemical and transcript analyses. Food Chem. 2023;408:135214. doi: 10.1016/j.foodchem.2022.135214. [DOI] [PubMed] [Google Scholar]
- 8.Belitz H.D., Grosch W., Schieberle P. Aroma compounds. Food Chem. 2004;5:342–408. [Google Scholar]
- 9.Solís-Solís H.M., Calderón-Santoyo M., Schorr-Galindo S., Luna-Solano G., Ragazzo-Sánchez J.A. Characterization of aroma potential of apricot varieties using different extraction techniques. Food Chem. 2007;105:829–837. doi: 10.1016/j.foodchem.2007.01.061. [DOI] [Google Scholar]
- 10.El Hadi M.A.M., Zhang F.-J., Wu F.-F., Zhou C.-H., Tao J. Advances in fruit aroma volatile research. Molecules. 2013;18:8200–8229. doi: 10.3390/molecules18078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krist S., Unterweger H., Bandion F., Buchbauer G. Volatile compound analysis of SPME headspace and extract samples from roasted italian chestnuts (Castanea sativa mill.) using GC-MS. Eur. Food Res. Technol. 2004;219:470–473. doi: 10.1007/s00217-004-0983-5. [DOI] [Google Scholar]
- 12.Sung J., Suh J.H., Chambers A.H., Crane J., Wang Y. Relationship between sensory attributes and chemical composition of different mango cultivars. J. Agric. Food Chem. 2019;67:5177–5188. doi: 10.1021/acs.jafc.9b01018. [DOI] [PubMed] [Google Scholar]
- 13.Wadood S.A., Boli G., Xiaowen Z., Raza A., Yimin W. Geographical discrimination of chinese winter wheat using volatile compound analysis by HS-SPME/GC-MS coupled with multivariate statistical analysis. J. Mass Spectrom. 2020;55:e4453. doi: 10.1002/jms.4453. [DOI] [PubMed] [Google Scholar]
- 14.Ojeda-Amador R.M., Fregapane G., Salvador M.D. Influence of cultivar and technological conditions on the volatile profile of virgin pistachio oils. Food Chem. 2020;311:125957. doi: 10.1016/j.foodchem.2019.125957. [DOI] [PubMed] [Google Scholar]
- 15.Lee Y.-G., Choi W.-S., Yang S.-O., Hwang-Bo J., Kim H.-G., Fang M., Yi T.-H., Kang S.C., Lee Y.-H., Baek N.-I. Volatile profiles of five variants of abeliophyllum distichum flowers using headspace solid-phase microextraction gas chromatography-mass spectrometry (HS-SPME-GC-MS) analysis. Plants. 2021;10:224. doi: 10.3390/plants10020224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang D., Liu Z., Chen W., Lan X., Zhan S., Sun Y., Su W., Lin C.-C., Ni L. Comparative study of the volatile fingerprints of roasted and unroasted oolong tea by sensory profiling and HS-SPME-GC-MS. Curr. Res. Food Science. 2023;6:100442. doi: 10.1016/j.crfs.2023.100442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y., Zhang Z., Hua B., Tao L., Chen W., Gao Y., Suo J., Yu W., Wu J., Song L. The interaction of temperature and relative humidity affects the main aromatic components in postharvest Torreya grandis nuts. Food Chem. 2022;368:13083. doi: 10.1016/j.foodchem.2021.130836. [DOI] [PubMed] [Google Scholar]
- 18.Lange B.M., Rujan T., Martin W., Croteau R. Isoprenoid biosynthesis: The evolution of two ancient and distinct pathways across genomes. Proc. Natl. Acad. Sci. USA. 2000;97:13172–13177. doi: 10.1073/pnas.240454797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miziorko H.M. Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch. Biochem. Biophys. 2011;505:131–143. doi: 10.1016/j.abb.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oldfield E., Lin F.-Y. Terpene biosynthesis: Modularity rules. Angew Chem. Int. 2012;51:1124–1137. doi: 10.1002/anie.201103110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohmer M. From molecular fossils of bacterial hopanoids to the formation of isoprene units: Discovery and elucidation of the methylerythritol phosphate pathway. Lipids. 2008;43:1095–1107. doi: 10.1007/s11745-008-3261-7. [DOI] [PubMed] [Google Scholar]
- 22.Abbas F., Ke Y., Yu R., Yue Y., Amanullah S., Jahangir M.M., Fan Y. Volatile terpenoids: Multiple functions, biosynthesis, modulation and manipulation by genetic engineering. Planta. 2017;246:803–816. doi: 10.1007/s00425-017-2749-x. [DOI] [PubMed] [Google Scholar]
- 23.Dudareva N., Cseke L., Blanc V.M., Pichersky E. Evolution of floral scent in clarkia: Novel patterns of s-linalool synthase gene expression in the C. breweri flower. Plant Cell. 1996;8:1137–1148. doi: 10.1105/tpc.8.7.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aubourg S., Lecharny A., Bohlmann J. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Gen. Genom. 2002;267:730–745. doi: 10.1007/s00438-002-0709-y. [DOI] [PubMed] [Google Scholar]
- 25.Dornelas M.C., Mazzafera P. A genomic approach to characterization of the citrus terpene synthase gene family. Genet. Mol. Biol. 2007;30:832–840. doi: 10.1590/S1415-47572007000500011. [DOI] [Google Scholar]
- 26.Martin D.M., Aubourg S., Schouwey M.B., Daviet L., Schalk M., Toub O., Lund S.T., Bohlmann J. Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol. 2010;10:22. doi: 10.1186/1471-2229-10-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falara V., Akhtar T.A., Nguyen T.T.H., Spyropoulou E.A., Bleeker P.M., Schauvinhold I., Matsuba Y., Bonini M.E., Schilmiller A.L., Last R.L., et al. The tomato terpene synthase gene family. Plant Physiol. 2011;157:770–789. doi: 10.1104/pp.111.179648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J., Liu X., Peng Y., Li Q., Han Y. Identification and characterization of transcription factors involved in geraniol biosynthesis in Rosa chinensis. Int. J. Mol. Sci. 2022;23:14684. doi: 10.3390/ijms232314684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y., Zhu C., Xu C., Sun J., Grierson D., Zhang B., Chen K. Integration of metabolite profiling and transcriptome analysis reveals genes related to volatile terpenoid metabolism in finger citron (C. medica var. sarcodactylis) Molecules. 2019;24:2564. doi: 10.3390/molecules24142564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen S., Yin X., Zhang B., Xie X., Jiang Q., Grierson D., Chen K. CitAP2.10 activation of the terpene synthase CsTPS1 is associated with the synthesis of (+)-valencene in ‘newhall’ orange. J. Exp. Bot. 2016;67:4105–4115. doi: 10.1093/jxb/erw189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei C., Liu H., Cao X., Zhang M., Li X., Chen K., Zhang B. Synthesis of flavour-related linalool is regulated by PpbHLH1 and associated with changes in DNA methylation during peach fruit ripening. Plant Biotechnol. J. 2021;19:2082–2096. doi: 10.1111/pbi.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spyropoulou E.A., Haring M.A., Schuurink R.C. RNA sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genom. 2014;15:402. doi: 10.1186/1471-2164-15-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao F., Liu B., Li M., Gao X., Fang Q., Liu C., Ding H., Wang L., Gao X. Identification and characterization of terpene synthase genes accounting for volatile terpene emissions in flowers of Freesia × Hybrida. J. Exp. Bot. 2018;69:4249–4265. doi: 10.1093/jxb/ery224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fei X., Qi Y., Lei Y., Wang S., Hu H., Wei A. Transcriptome and metabolome dynamics explain aroma differences between green and red prickly ash fruit. Foods. 2021;10:391. doi: 10.3390/foods10020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z., Jin H., Suo J., Yu W., Zhou M., Dai W., Song L., Hu Y., Wu J. Effect of temperature and humidity on oil quality of harvested Torreya grandis cv. merrillii nuts during the after-ripening stage. Front. Plant Sci. 2020;11:573681. doi: 10.3389/fpls.2020.573681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z., Tao L., Gao L., Gao Y., Suo J., Yu W., Hu Y., Wei C., Farag M.A., Wu J., et al. Transcription factors TgbHLH95 and TgbZIP44 cotarget terpene biosynthesis gene TgGPPS in Torreya grandis nuts. Plant Physiol. 2023;193:1161–1176. doi: 10.1093/plphys/kiad385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang S., Li D., Li S., Yang H., Zhao Z. GC-MS metabolite and transcriptome analyses reveal the differences of volatile synthesis and gene expression profiling between two apple varieties. Int. J. Mol. Sci. 2022;23:2939. doi: 10.3390/ijms23062939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng H., Chen J., Li X., Pan J., Xue S.J., Liu D., Ye X. Differentiation of the volatile profiles of chinese bayberry cultivars during storage by HS-SPME–GC/MS combined with principal component analysis. Postharvest Biol. Technol. 2015;100:59–72. doi: 10.1016/j.postharvbio.2014.09.003. [DOI] [Google Scholar]
- 39.Huang C., Sun P., Yu S., Fu G., Deng Q., Wang Z., Cheng S. Analysis of volatile aroma components and regulatory genes in different kinds and development stages of pepper fruits based on non-targeted metabolome combined with transcriptome. Int. J. Mol. Sci. 2023;24:7901. doi: 10.3390/ijms24097901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rezaeinia H., Ghorani B., Emadzadeh B., Tucker N. Electrohydrodynamic atomization of balangu (Lallemantia Royleana) seed gum for the fast-release of mentha longifolia L. essential oil: Characterization of nano-capsules and modeling the kinetics of release. Food Hydrocoll. 2019;93:374–385. doi: 10.1016/j.foodhyd.2019.02.018. [DOI] [Google Scholar]
- 41.Zhou Y., Sun S., Bei W., Zahi M.R., Yuan Q., Liang H. Preparation and antimicrobial activity of oregano essential oil pickering emulsion stabilized by cellulose nanocrystals. Int. J. Biol. Macromol. 2018;112:7–13. doi: 10.1016/j.ijbiomac.2018.01.102. [DOI] [PubMed] [Google Scholar]
- 42.Dudareva N., Negre F., Nagegowda D.A., Orlova I. Plant volatiles: Recent advances and future perspectives. Crit. Rev. Plant Sci. 2006;25:417–440. doi: 10.1080/07352680600899973. [DOI] [Google Scholar]
- 43.Degenhardt J., Köllner T.G., Gershenzon J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry. 2009;70:1621–1637. doi: 10.1016/j.phytochem.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 44.Nagegowda D.A. Plant volatile terpenoid metabolism: Biosynthetic genes, transcriptional regulation and subcellular compartmentation. FEBS Lett. 2010;584:2965–2973. doi: 10.1016/j.febslet.2010.05.045. [DOI] [PubMed] [Google Scholar]
- 45.Li N., Dong Y., Lv M., Qian L., Sun X., Liu L., Cai Y., Fan H. Combined analysis of volatile terpenoid metabolism and transcriptome reveals transcription factors related to terpene synthase in two cultivars of Dendrobium officinale flowers. Front. Genet. 2021;12:661296. doi: 10.3389/fgene.2021.661296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei J., Yang Y., Peng Y., Wang S., Zhang J., Liu X., Liu J., Wen B., Li M. Biosynthesis and the transcriptional regulation of terpenoids in tea plants (Camellia sinensis) Int. J. Mol. Sci. 2023;24:6937. doi: 10.3390/ijms24086937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu J., van Herwijnen Z.O., Dräger D.B., Sui C., Haring M.A., Schuurink R.C. SlMYC1 regulates type VI glandular trichome formation and terpene biosynthesis in tomato glandular cells. Plant Cell. 2018;30:2988–3005. doi: 10.1105/tpc.18.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Z., Li Y., Gao F., Jin W., Li S., Kimani S., Yang S., Bao T., Gao X., Wang L. MYB21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J. Exp. Bot. 2020;71:4140–4158. doi: 10.1093/jxb/eraa184. [DOI] [PubMed] [Google Scholar]
- 49.Li S., Wang H., Li F., Chen Z., Li X., Zhu L., Wang G., Yu J., Huang D., Lang Z. The maize transcription factor EREB58 mediates the jasmonate-induced production of sesquiterpene volatiles. Plant J. 2015;84:296–308. doi: 10.1111/tpj.12994. [DOI] [PubMed] [Google Scholar]
- 50.Nieuwenhuizen N.J., Chen X., Wang M.Y., Matich A.J., Perez R.L., Allan A.C., Green S.A., Atkinson R.G. Natural variation in monoterpene synthesis in kiwifruit: Transcriptional regulation of terpene synthases by NAC and ethylene-insensitive3-like transcription factors. Plant Physiol. 2015;167:1243–1258. doi: 10.1104/pp.114.254367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun W.-J., Zhan J.-Y., Zheng T.-R., Sun R., Wang T., Tang Z.-Z., Bu T.-L., Li C.-L., Wu Q., Chen H. The jasmonate-responsive transcription factor CbWRKY24 regulates terpenoid biosynthetic genes to promote saponin biosynthesis in Conyza blinii h. lév. J. Genet. 2018;97:1379–1388. doi: 10.1007/s12041-018-1026-5. [DOI] [PubMed] [Google Scholar]
- 52.Guo Y., Guo Z., Zhong J., Liang Y., Feng Y., Zhang P., Zhang Q., Sun M. Positive regulatory role of R2R3 MYBs in terpene biosynthesis in Lilium ‘siberia’. Hortic. Plant J. 2023;9:1024–1038. doi: 10.1016/j.hpj.2023.05.004. [DOI] [Google Scholar]
- 53.Michael R., Ranjan A., Kumar R.S., Pathak P.K., Trivedi P.K. Light-regulated expression of terpene synthase gene, AtTPS03, is controlled by the bZIP transcription factor, HY5, in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2020;529:437–443. doi: 10.1016/j.bbrc.2020.05.222. [DOI] [PubMed] [Google Scholar]
- 54.Yang J., Chen Y., Gao M., Wu L., Xiong S., Wang S., Gao J., Zhao Y., Wang Y. Comprehensive identification of bHLH transcription factors in Litsea cubeba reveals candidate gene involved in the monoterpene biosynthesis pathway. Front. Plant Sci. 2022;13:1081335. doi: 10.3389/fpls.2022.1081335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X., Xu Y., Shen S., Yin X., Klee H., Zhang B., Chen K., Hancock R. Transcription factor CitERF71 activates the terpene synthase gene CitTPS16 involved in the synthesis of E-geraniol in sweet orange fruit. J. Exp. Bot. 2017;68:4929–4938. doi: 10.1093/jxb/erx316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang W., Li X., Zheng J., Wang G., Sun C., Ferguson I.B., Chen K. Bioactive components and antioxidant capacity of chinese bayberry (Myrica rubra sieb. and zucc.) fruit in relation to fruit maturity and postharvest storage. Eur. Food Res. Technol. 2008;227:1091–1097. doi: 10.1007/s00217-008-0824-z. [DOI] [Google Scholar]
- 57.Chang S., Puryear J., Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993;11:113–116. doi: 10.1007/BF02670468. [DOI] [Google Scholar]
- 58.Lou H., Song L., Li X., Zi H., Chen W., Gao Y., Zheng S., Fei Z., Sun X., Wu J. The Torreya grandis genome illuminates the origin and evolution of gymnosperm-specific sciadonic acid biosynthesis. Nat. Commun. 2023;14:1315. doi: 10.1038/s41467-023-37038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar L., Futschik M.E. Mfuzz: A software package for soft clustering of microarray data. Bioinformation. 2007;2:5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.