Abstract
The Rep78 and Rep68 proteins of adeno-associated virus (AAV) type 2 are involved in DNA replication, regulation of gene expression, and targeting site-specific integration. They bind to a specific Rep recognition sequence (RRS) found in both the viral inverted terminal repeats and the AAVS1 integration locus on human chromosome 19. Previous in vitro studies implied that an N-terminal segment of Rep is involved in DNA recognition, while additional domains might stabilize binding and mediate multimerization. In order to define the minimal requirements for Rep to recognize its target site in the human genome, we developed one-hybrid assays in which DNA-protein interactions are detected in vivo. Chimeric proteins consisting of the N terminus of Rep fused to different oligomerization motifs and a transcriptional activation domain were analyzed for oligomerization, DNA binding, and activation of reporter gene expression. Expression of reporter genes was driven from RRS motifs cloned upstream of minimal promoters and examined in mammalian cells from transfected plasmids and in Saccharomyces cerevisiae from a reporter cassette integrated into the yeast genome. Our results show for the first time that chimeric proteins containing the amino-terminal 244 residues of Rep are able to target the RRS in vitro and in vivo when incorporated into artificial multimers. These studies suggest that chimeric proteins may be used to harness the unique targeting feature of AAV for gene therapy applications.
Adeno-associated virus (AAV) type 2 is a nonpathogenic human parvovirus that relies upon a helper virus for efficient replication (5). Under conditions that are not permissive for replication, AAV infection results in integration of the viral genome into the host chromosome (6, 30, 44). A unique characteristic of AAV integration is that in human cell lines it can be targeted in about 70% of cases to a specific site on chromosome 19q13.3-qter (24, 25, 45). The preintegration locus, termed AAVS1, has been cloned and sequenced (23). Site-specific integration by AAV into the preintegration locus requires cis-acting sequences within the viral origin of replication and AAVS1 as well as the DNA-binding activity of the Rep proteins (31, 60). It would be particularly attractive to harness this unique targeting feature for gene transfer vehicles, since this would decrease the chances of insertional mutagenesis associated with random integration.
The AAV genome is a single-stranded linear DNA molecule with inverted terminal repeats (ITRs) that fold into hairpin structures and serve as the origins for DNA replication (5). The ITR is involved in regulation of gene expression, initiation of DNA replication, packaging of the viral genome, site-specific integration, and rescue from the integrated state. The genome contains two open reading frames (ORFs) encoding nonstructural (Rep) and structural (Cap) proteins. Expression is regulated by three viral promoters, p5, p19, and p40. The rep gene encodes four overlapping multifunctional Rep proteins, named according to their apparent molecular mass (in kilodaltons). Rep78 and Rep68 are translated from unspliced and spliced transcripts that initiate from the p5 promoter. Rep52 and Rep40 are translated from unspliced and spliced transcripts initiating from the p19 promoter. The large Rep proteins, Rep78 and Rep68, have been shown to stimulate replication in vitro (36, 54) and in vivo (51). Their activities include DNA binding (19), as well as site-specific endonuclease (18), helicase (18, 61), and ATPase (61) activities. Rep78 and Rep68 have been shown to regulate transcription from the AAV p5, p19, and p40 promoters in vivo (26, 27, 32, 39, 52). The large Rep proteins also repress and activate transcription from heterologous promoters and inhibit cellular transformation, viral replication, and cell growth (13–16, 22, 28, 58, 63, 66, 67).
Binding of the Rep proteins to DNA substrates is a key step in replication, gene regulation, and targeting site-specific integration. Electrophoretic mobility shift assays (EMSAs) have shown that the Rep78 and -68 proteins bind to a specific Rep recognition sequence (RRS) in the viral ITRs that consists of GCTC repeating motifs (2, 4, 7, 19, 31, 34, 38). We have identified a similar RRS within the AAVS1 integration locus and have shown that the large Rep proteins can form a bridge between the viral ITR and the binding site in AAVS1, a reaction proposed to promote targeted integration (60). Rep78 and Rep68 proteins also bind to an RRS in the viral p5 promoter to autoregulate rep expression (27).
The Rep proteins are composed of functional domains that are partly distinct but may show some interdependence for full Rep activities (33, 38, 59, 68). The DNA-binding function has been suggested to be bipartite, with the first 241 amino acids determining binding specificity, together with stabilizing interactions imparted by amino acids 242 to 476 (33, 38, 59, 69). This is consistent with the observation that the shorter Rep52 and Rep40 proteins lacking this region do not bind DNA (20, 38, 60). Other evidence implied that Rep78 and Rep68 bind to DNA as oligomers and that the domain required for maximal self-association comprises elements within the central region of Rep78 (38, 48, 59). Direct Rep-Rep protein interactions have been shown in vivo by a mammalian two-hybrid system (48) and in vitro by coimmunoprecipitation, far-Western, and chemical cross-linking assays (17, 48). Protein-protein interaction regions within the Rep proteins include two coiled-coil repeats (amino acids 164 to 182 and 441 to 483), the region around the nucleoside triphosphate-binding motif (amino acids 332 to 346), and a predicted alpha-helical structure (amino acids 371 to 393) (9, 48). The characteristic pattern of multiple bands observed in the gel mobility shift assay may also reflect different oligomeric states of the Rep proteins (19, 37, 38). Moreover, we previously showed that truncated Rep proteins when mixed with full-length Rep68 could form hetero-oligomers on the AAV hairpin ITR substrate (59). Recently, cross-linking experiments have suggested that Rep78 forms hexameric oligomers in the presence of AAV ori sequences (48).
All studies analyzing DNA binding by the AAV Rep proteins so far have utilized gel mobility shift assays to study interactions in vitro. Because DNA binding was examined in the context of the whole Rep protein, the results are difficult to interpret in light of the interdependence of DNA recognition and other Rep functions. We therefore sought to develop a DNA binding assay to define the minimal requirements for DNA recognition in vivo. Our assay follows the principle of the one-hybrid system, in which DNA-protein interactions are detected by a simple phenotypic readout. Based on previous studies of DNA recognition by Rep proteins, we fused the Rep N terminus to a strong transcriptional activation domain. We also incorporated variants of two different oligomerization motifs (from yeast GCN4 and human p53 proteins) to allow multimerization of the chimeric proteins. Reporters were designed that would respond to binding of the chimeric Rep proteins by cloning RRS motifs upstream of minimal promoters. In cultured mammalian cells, reporter gene expression from cotransfected plasmid DNA was analyzed, while in the yeast Saccharomyces cerevisiae, an integrated RRS reporter cassette was used. Our results show for the first time that the N-terminal 244 residues of Rep78 and the RRS are sufficient for interaction in vitro and in vivo in an oligomerization state-dependent manner. The availability of yeast RRS reporter strains will enable further in vivo studies to examine Rep functions and identify cellular proteins interacting with Rep. Since DNA recognition by Rep is the crucial first step in targeted integration, additional studies of Rep-DNA interactions will allow this unique feature of AAV to be harnessed for gene therapy applications.
MATERIALS AND METHODS
Plasmids for mammalian cells.
The plasmids encoding the chimeric Rep fusion proteins were designed to allow easy swapping of functional domains by use of unique restriction sites flanking the different domains. The sequence from the 5′ end is as follows: HindIII–EcoRI–Rep DNA-binding domain–NheI–oligomerization domain–NotI–transcriptional activation domain–HpaI–nuclear localization domain–ApaI–Myc/His tag–PmeI.
In a first cloning step, the sequence encompassing the amino-terminal 244 residues of Rep78 were tagged with a Myc epitope. The rep gene was PCR amplified with pAAV2 as a template, digested with EcoRI-ApaI, and subcloned into an EcoRI-ApaI backbone fragment of pcDNA3.1/Myc-His (Invitrogen). To generate pcDNA.Rep.NLS, two antiparallel oligonucleotides encoding a unique HpaI site and the simian virus 40 (SV40) large T nuclear localization signal were inserted into the ApaI site of pcDNA.Rep.Myc. The sequences coding for the GCN4-based oligomerization domains, the wild-type leucine zipper (LZ) domain or an engineered leucine zipper (TZ) domain that assembles as a four-stranded coiled coil (56), were PCR amplified with pGEMhp53LZ335Q and pGEMhp53TZ334NR (both kindly provided by Thanos Halazonetis) as templates. In both cases, the left-hand primers additionally encoded an NheI site and the right-hand primers coded for a unique NotI site. The PCR products were inserted directly into HpaI-linearized pcDNA.Rep.NLS, thereby restoring the HpaI site. The resulting plasmids were named pcDNA.Rep.LZ and pcDNA.Rep.TZ, respectively. In a next step, the sequence coding for the transcriptional activation domain of VP16 (AD) was inserted into pcDNA.Rep.LZ and pcDNA.Rep.TZ. A NotI-HpaI-digested PCR amplification product coding for residues 147 to 226 of GAL4-VP16 was subcloned into the respective NotI-HpaI backbone fragments to give rise to pcDNA.Rep.LZ.AD and pcDNA.Rep.TZ.AD. Plasmid pcDNA.Rep.AD was constructed by deleting the NheI-NotI fragment of pcDNA.Rep.LZ.AD, followed by a Klenow fill-in reaction and religation. The N- or C-terminal deletion mutants were prepared by PCR of the Rep DNA-binding domain by using primers leading to amplification of the nucleotide sequences encoding Rep78 residues 13 to 244, 1 to 220, 1 to 200, and 1 to 180, respectively. The internal deletion mutants were constructed by overlap extension PCR with internal primers to link Rep78 codon 61 to 88 or 113 to 123, respectively. The resulting fragments were digested with EcoRI and XbaI and inserted into an EcoRI-NheI backbone fragment of pcDNA.Rep.TZ.AD.
To generate the chimeric Rep constructs harboring the p53 oligomerization domains, a region encompassing residues 315 to 363 of human p53 was amplified by PCR. In addition to the wild-type sequence, we amplified mutant variants L348A/L350A (49), L344A (55), and I322A (kindly provided by Jayne M. Stommel). The PCR products were digested with SpeI and NotI and subcloned into a NheI-NotI backbone fragment of pcDNA.Rep.TZ.AD. The resulting plasmids were named pcDNA.Rep.TD.AD, pcDNA.Rep.CD.AD, pcDNA.Rep.DD.AD, and pcDNA.Rep.MD.AD, respectively.
To generate the reporter plasmid pRRS.tk.Luc, a set of two antiparallel oligonucleotides containing one copy of the RRS, as contained in the viral ITR, were inserted into the polylinker of plasmid tk-Luc (kindly provided by Ron Evans): 5′-AGCTTCAGTGAGCGAGCGAGCGCGCAGG and 5′-TCGACCTGCGCGCTCGCTCGCTCACTGA (RRS is underlined).
All plasmids were sequenced to confirm the expected structures.
Yeast expression plasmids.
A HindIII-PmeI fragment of pcDNA.Rep.TZ.AD containing the entire Rep.TZ.AD ORF was subcloned into a HindIII backbone fragment of the yeast expression plasmid pGAD424 (Clontech) to give rise to pG.Rep.TZ.AD. Plasmids pG.Rep.AD, pG.Rep.LZ.AD, and pG.Rep.TZ were obtained by subcloning an EcoRI-HpaI fragment of the respective pcDNA construct into the EcoRI-HpaI backbone fragment of pG.Rep.TZ.AD. Yeast expression plasmids for Rep68 and Rep78 were generated as follows. BglII-XbaI fragments of pcDNA.Rep68 or pcDNA.Rep78, which encompass the entire ORF of either Rep68 or Rep78, were digested with NcoI. The resulting NcoI-XbaI fragment was subcloned into an NcoI-NheI backbone fragment of pG.Rep.TZ.AD. In a second step, an ApaI-BssHII fragment of the obtained plasmids was deleted to remove the remaining 303 bp of the Rep.TZ.AD ORF. The backbone was blunted by a Klenow fill-in reaction and religated to give rise to pG.Rep68 and pG.Rep78. For high-level expression of the chimeric proteins, the ORFs of the plasmids presented above were subcloned under control of the full-length yeast ADH1 promoter. An AatII-HindIII fragment of pGAD GH (Clontech) containing the full-length promoter was subcloned into the pG expression plasmid series to replace a corresponding fragment containing a truncated version of the ADH1 promoter, giving rise to pADH.Rep68, pADH.Rep78, pADH.Rep.AD, pADH.Rep.LZ.AD, pADH.Rep.TZ.AD, and pADH.Rep.TZ. Plasmid pADH.Keratin was kindly provided by Jeanette Ducut.
Cell culture and immunoblotting of nuclear extracts.
293 (human embryonic kidney) and HeLa (human cervical carcinoma) cells were obtained from American Type Culture Collection and grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Subconfluent monolayers of 293 cells in 150-mm-diameter plates were transfected with 40 μg of plasmid DNA encoding chimeric Rep fusion proteins by calcium phosphate precipitation. Cells were harvested 48 h posttransfection in phosphate-buffered saline (PBS), and nuclear extracts were prepared as described elsewhere (1). Equivalent amounts of proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes (Millipore). Membranes were blocked with 1% bovine serum albumin–5% skim milk powder in TBS-T (10 mM Tris [pH 8.0], 150 mM NaCl, 0.05% Tween 20) overnight at room temperature and then incubated with a Myc-specific antibody (1:5,000 dilution; Invitrogen) for 1 h at room temperature in TBS-T supplemented with 1% bovine serum albumin. Proteins were visualized after incubation with a horseradish peroxidase-conjugated secondary antibody (1:3,000 dilution; Jackson Laboratories) for 1 h at room temperature by enhanced chemiluminescence (NEN).
In vitro translation and protein cross-linking.
The chimeric Rep fusion proteins were in vitro translated in the presence of Tran-35S label (ICN) by using the T7 polymerase-based TNT Coupled Reticulocyte Lysate System (Promega) according to the manufacturer's instructions. One microliter of in vitro-synthesized protein was diluted with 9 μl of PBS in the absence or presence of 0.05% formaldehyde. The reaction was allowed to proceed for 30 min at 37°C before an equal volume of 2× quencher dye (800 mM glycine, 6% SDS, 6% β-mercaptoethanol, 20% glycerol, 0.01% bromophenol blue) was added. The extent of cross-linking was analyzed by 4 to 15% polyacrylamide gradient SDS-PAGE followed by autoradiography.
EMSAs.
The EMSA was basically performed as described previously (59). Briefly, 2 μl of primed rabbit reticulocyte lysate was mixed with 5,000 cpm of a 32P-labeled DNA substrate in binding buffer and incubated for 15 min at 30°C. The DNA probe contains an RRS corresponding to the RRS in human chromosome 19 or a mutated RRS (mRRS) as described previously (60). The core sequences of the probes are as follows: RRS, 5′-GC(GCTC)3GCTGGG-3′; and mRRS, 5′-GC(CCTC)3CCTGGG-3′. For supershifts, 1 μl of a diluted antibody solution was added to the binding reaction: the anti-Myc antibody (Invitrogen) as a 1:5 dilution in water and the anti-Rep antiserum (41) as a 1:30 dilution in water or undiluted. In competition experiments, 5-, 25-, or 125-fold molar excess of unlabeled DNA substrate was added to the binding reaction mixture.
Reporter assays.
HeLa cells in 35-mm-diameter wells were transfected with the indicated plasmids by calcium phosphate precipitation in duplicate. Total DNA concentrations were maintained at 4 μg per well for all experiments. A typical experiment included 1 μg of the reporter, 2 μg of a plasmid encoding the Rep fusion proteins, or empty pcDNA vector DNA. To normalize for transfection efficiency between individual experiments, 1 μg of pCMVβ was included. Cells were harvested 32 h after transfection in Reporter lysis buffer (Promega). Luciferase and β-galactosidase activity were measured in a luminometer (Berthold) by using Luciferase assay substrate (Promega) or GalctoLight (Tropix) according to the manufacturers' instructions.
Yeast strains and assays.
All yeast manipulations were basically performed as described in the One-Hybrid System User Manual or the Yeast Protocols Handbook (Clontech, Palo Alto, Calif.). Briefly, reporter plasmid pRRS3.LacZ was generated by inserting two sets of two antiparallel oligonucleotides containing one or two copies of the RRS, respectively, into the polylinker of pLacZi (Clontech): 5′-AGCTTCAGTGAGCGAGCGAGCGCGCAGG, 5′-TCGACCTGCGCGCTCGCTCGCTCACTGA, 5′-TCGAAGTGAGCGAGCGAGCGCGCAGGTGAGCGAGCGAGCGCGCAGC, and 5′-TCGAGCTGC-GCGCTCGCTCGCTCACCTGCGCGCTCGCTCGCTCACT (RRS is underlined). The resulting plasmid was linearized with NcoI and used to transform yeast strain YM4271 (Clontech). The transformation mixture was plated on SD/−Ura plates to select for colonies with an integrated reporter gene. After 3 days, large colonies were picked and patched on SD/−Ura plates to determine lacZ background expression by a colony-lift filter assay. Clones producing small amounts of β-galactosidase were identified and used as reporter strain YM.RRS3.LacZ.
For the reporter assay, YM.RRS3.LacZ was transformed with 0.5 μg of the Rep effector plasmids by the LiAc transformation procedure. Double-transformed clones were selected by plating the transformation mix on SD/−Ura, −Leu agar plates. After 3 days, large colonies were picked and patched on SD/−Ura, −Leu plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and BU salts (26 mM Na2HPO4, 25 mM NaH2PO4) to perform an in vivo plate assay. Plates were incubated for 2 days at 30°C and assessed for the development of blue cells. For quantitative analysis of lacZ expression, double-transformed colonies were used to inoculate an SD/−Ura, −Leu liquid culture. Cells were harvested in mid-log phase, and the optical density at 600 nm (OD600) was read. To lyse the cells, repeated freeze-thaw cycles in Z-buffer were performed. Cell lysates were mixed with Galacton Star reaction mixture (Tropix), and β-galactosidase activity was determined in a luminometer. To normalize for cell number, the activity was calculated as relative light units (RLU) per OD600 of cell culture.
RESULTS
Characterization of chimeric Rep fusion proteins.
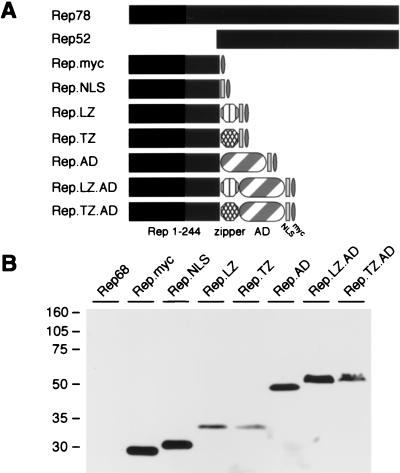
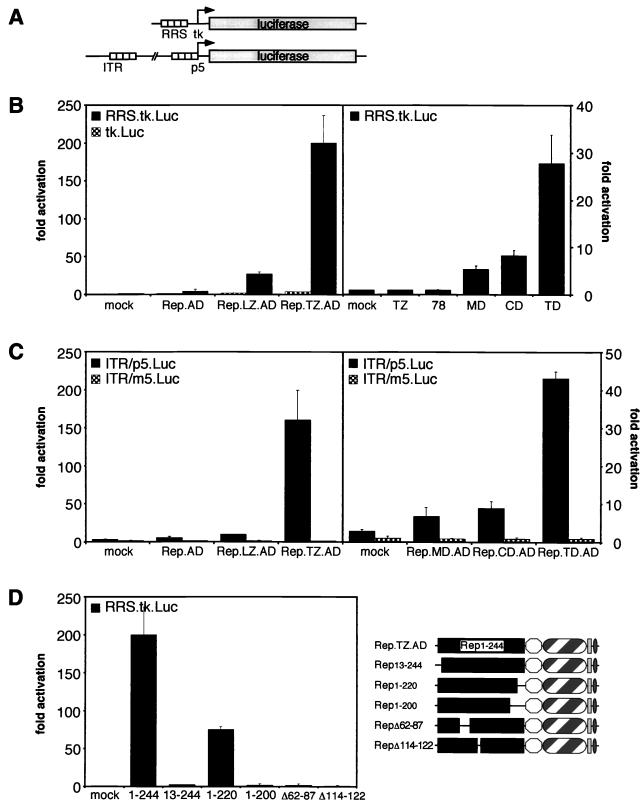
In order to study the requirements for DNA recognition by the large AAV Rep proteins, we designed chimeric Rep proteins to be used in domain-swap experiments (Fig. 1A). Since it was expected that the major DNA-binding activity resides within the N terminus of Rep78, residues 1 to 244 were cloned into pcDNA.Myc. To direct the chimeric proteins to the nucleus, we incorporated the nuclear localization signal (NLS) from the SV40 large T antigen. Our previous studies had suggested that oligomerization was important for DNA binding. We therefore included a dimerization domain of the yeast transcription factor GCN4 or a mutant GCN4 zipper that is predicted to assemble into a parallel tetramer (12, 56). To generate proteins that would activate transcription upon Rep binding to the RRS, we added the activation domain of VP16 to some constructs.
FIG. 1.
Rep proteins and chimeric proteins generated to study DNA binding. (A) Schematic of wild-type and chimeric Rep proteins. The amino-terminal 244 residues of Rep78 were tagged with a Myc epitope, joined to the SV40 large T NLS, and fused to the LZ or a modified zipper (TZ) of GCN4. The transcriptional activation domain of VP16 (AD) was included in some fusion proteins. Drawings are not to scale. (B) Western blot analysis of chimeric Rep proteins. Nuclear extracts were made from 293 cells transfected with plasmids expressing the indicated fusion proteins. Proteins were separated on a 10% polyacrylamide gel by SDS-PAGE, and chimeric proteins were detected with an anti-Myc antibody. The positions of molecular size markers are indicated on the left.
Plasmids for the chimeric Rep proteins were transfected into 293 cells. Immunoblotting of nuclear extracts with a Myc-specific antibody detected expression of the tagged proteins in lysates from transfected cells (Fig. 1B). All constructs generated proteins of the expected size. The immunoblot also indicated equivalent steady-state levels of the chimeric proteins containing the VP16 activation domain. Indirect immunofluorescence confirmed that the chimeric Rep proteins with the NLS were located predominantly (>90%) in the nucleus (data not shown).
Oligomeric status of Rep fusion proteins.
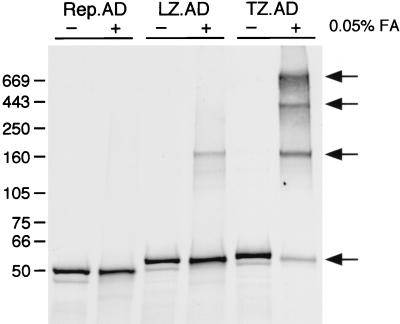
To examine the oligomeric state of the chimeric Rep proteins, we employed chemical cross-linking of in vitro-synthesized, 35S-labeled proteins in 0.05% formaldehyde. The products were assessed under denaturing conditions on a 4 to 15% gradient polyacrylamide gel and detected by autoradiography (Fig. 2). In all cases, the amounts of labeled protein synthesized were similar. The Rep.AD protein was unaffected by the cross-linker, confirming that it exists as a monomer. For Rep.LZ.AD, which contains the wild-type GCN4 leucine zipper, a slower-migrating band was detected in the presence of the cross-linker. It is difficult to determine the oligomeric state of this complex from gel electrophoresis alone. In addition, the formaldehyde cross-linking reagent may alter the mobility of proteins in the gel. However, based on the predicted molecular weight of approximately 45,000 Da, the migration of this band suggests that Rep.LZ.AD might form a tetramer. Rep.TZ.AD, which contains the modified leucine zipper, showed a band of similar size to Rep.LZ.AD in the presence of the cross-linker and additionally yielded slower-migrating bands presumably representing high-order multimers. In general, Rep.TZ.AD was more efficient at the formation of multimers than Rep.LZ.AD.
FIG. 2.
Oligomerization of chimeric Rep proteins. In vitro-synthesized, [35S]methionine-labeled proteins were incubated at 37°C in the absence (−) or presence (+) of 0.05% formaldehyde (FA) diluted in PBS. After 30 min, the reaction was stopped, and cross-linked proteins were separated on a 4 to 15% polyacrylamide gradient gel by SDS-PAGE. The gel was stained with Coomassie brilliant blue to visualize the molecular size markers, dried, and exposed to X-ray film. The positions of the molecular size markers (in kilodaltons) are marked on the left, and the different oligomeric states are indicated by arrows. LZ.AD, Rep.LZ.AD; TZ.AD, Rep.TZ.AD.
In summary, these results show that in the absence of an oligomerization motif, the chimeric Rep.AD protein is present as a monomer. Incorporation of leucine zipper sequences was able to shift the oligomeric status of the chimeric Rep proteins from a monomeric form towards higher-order multimers.
Oligomerization promotes binding of the Rep N terminus to the RRS.
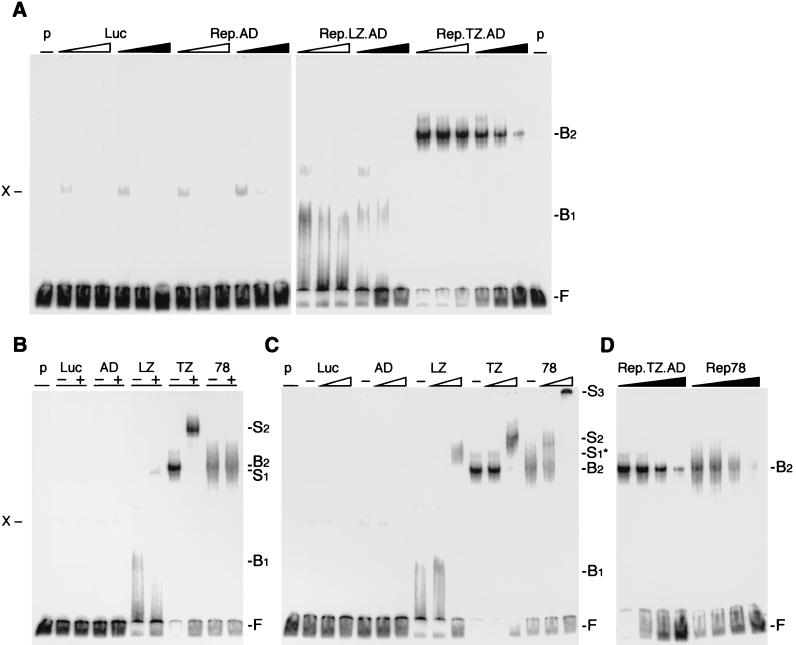
In order to assess the effect of oligomerization on DNA binding by the N terminus of Rep, we first utilized the well-established EMSA. Rep fusion proteins were synthesized in vitro and assayed for their ability to bind a 32P-labeled, RRS-containing DNA substrate (Fig. 3). The specificity of binding was confirmed by competition experiments with increasing amounts (5-, 25-, and 125-fold molar excess) of unlabeled DNA substrate (Fig. 3A) containing either the wild-type RRS or a mutant RRS (mRRS) as competitor. No distinguishable specific band was identified with the monomeric Rep.AD or a control reaction. A weak band (X) was detected in all reactions, which disappeared with increasing amounts of nonspecific competitor DNA. The Rep.LZ.AD and Rep.TZ.AD proteins were both capable of binding to the RRS, as detected by a mobility shift. Addition of the Rep.LZ.AD protein produced a faint smear (B1), whereas Rep.TZ.AD generated a more significant shift (B2). The slower migration of the Rep.TZ.AD-containing complex (B2), compared to the Rep.LZ.AD-induced shift (B1), could represent the higher oligomeric status of Rep.TZ.AD. Binding to the RRS could be competed by unlabeled fragments containing the RRS, but not by similar fragments with a mutation in the RRS (Fig. 3A), thus establishing that binding of the chimeric Rep proteins was sequence specific for the RRS.
FIG. 3.
EMSAs of Rep fusion proteins. Chimeric Rep proteins were synthesized in vitro in a rabbit reticulocyte lysate and incubated with the 3′-end-labeled RRS probe. In vitro-translated luciferase (Luc) served as a negative control for the lysate, and nonspecific bands are indicated (X). The positions of free (F), bound (B), and supershifted (S) DNA substrate are indicated on the right. (See text for an explanation of the bound and supershifted complexes.) (A) The chimeric Rep proteins bind to the RRS in a specific manner. Increasing molar ratios (5×, 25×, and 125×) of unlabeled DNA fragments containing the RRS (black triangles) or a mutant RRS (open triangles) were added to the reaction as competitors. (B and C) The DNA-bound, chimeric Rep proteins are supershifted with specific antibodies to the Myc tag (B) or Rep (C). The absence (−) or presence (+) of the antibody is indicated on top. Triangles indicate increasing amounts (1/30 and 1 μl) of the polyclonal anti-Rep antibody. (D) Rep.TZ.AD binds to the RRS with similar affinity as Rep78. Increasing molar ratios of unlabeled RRS probe (0, 5×, 25×, 125×) were added as competitor to the reaction. p, probe; Luc, luciferase; AD, Rep.AD; LZ, Rep.LZ.AD; TZ, Rep.TZ.AD; 78, Rep78.
To verify that the chimeric Rep proteins were actually components of the shifted bands, an antibody specific to the Myc epitope was added to the binding reaction (Fig. 3B). A novel slower-migrating band was detected for both Rep.LZ.AD (S1) and Rep.TZ.AD (S2). In a control reaction, in vitro-translated Rep78 was not supershifted by the Myc antibody, confirming that the antibody was specific for the Myc tag present in the chimeric fusion proteins. The same experiment was repeated with a Rep-specific rabbit polyclonal antiserum (Fig. 3C). Supershifted bands were detected for all complexes, including Rep.LZ.AD (S1*), Rep.TZ.AD (S2) and Rep78 (S2 and S3). Addition of the Rep-specific antibody induced a more pronounced shift of the DNA-Rep.LZ.AD complex (S1*), compared to the Myc antibody. It also increased the overall amount of shifted probe for Rep.LZ.AD, which could be explained by pseudo-oligomerization of the N-terminal Rep domain and increased binding to the RRS. In contrast to Rep78, more of the Rep-specific antibody was required to supershift the Rep.LZ.AD- and Rep.TZ.AD-containing complexes. This is probably due to the fact that the polyclonal Rep antibody recognized more epitopes for the full-length Rep protein, compared to just the N terminus in the chimeric proteins. Larger amounts of the Rep antibody caused aggregation of the DNA-Rep78-antibody complex (S3).
To compare the binding affinity of Rep.TZ.AD with that of Rep78, competition experiments with increasing amounts (0-, 5-, 25-, and 125-fold molar excess) of unlabeled RRS probe were performed (Fig. 3D). The experiment showed that the chimeric Rep.TZ.AD protein bound to the RRS with an affinity similar to or greater than that of the wild-type Rep78 protein (B2).
These results clearly demonstrated that the monomeric Rep.AD was incapable of binding to the RRS, but that oligomerization of the fusion proteins could confer DNA-binding ability to the N terminus of the Rep protein. The Rep.TZ.AD protein produced bands with slower migration than Rep.LZ.AD, supporting the hypothesis that the multiple banding pattern observed with the wild-type Rep proteins might reflect different oligomeric states of Rep bound to the substrate.
Characterization of chimeric Rep proteins fused to p53 oligomerization motifs.
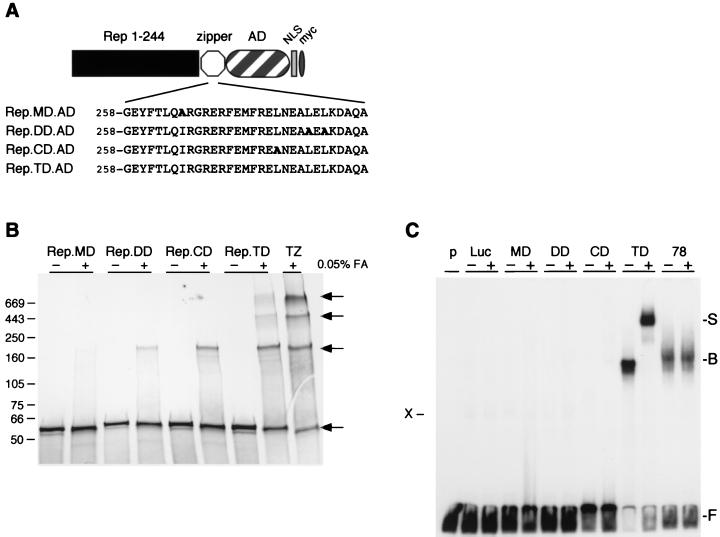
In order to verify the results we obtained for the chimeric Rep proteins fused to the GCN4 oligomerization domains, we designed a new series of chimeric proteins containing either the wild-type tetramerization domain of the human p53 protein or mutant variants thereof (Fig. 4A). The p53-based oligomerization motifs were predicted to mediate assembly into tetramers (TD), dimers (CD or DD), or monomers (MD), dependent on the position of the alanine exchange within the zipper domain (49, 55).
FIG. 4.
Chimeric Rep proteins containing p53 oligomerization domains. (A) Schematic of chimeric Rep-p53 proteins. The chimeric proteins consist of the amino-terminal 244 residues of Rep78 fused to mutant or wild-type p53 oligomerization motifs (MD, DD, CD, and TD), the VP16 transcriptional activation domain (AD), the SV40 large T NLS, and a Myc tag. The amino acid sequences of the respective oligomerization domains are indicated below. Residues in boldface indicate changes to the p53 wild-type sequence. The drawings are not to scale. (B) Oligomerization of chimeric Rep-p53 proteins. In vitro-synthesized, 35S-labeled proteins were incubated at 37°C in the absence (−) or presence (+) of 0.05% formaldehyde (FA) diluted in PBS as described in the legend to Fig. 2. The positions of the molecular size markers (in kilodaltons) are marked on the left, and the different oligomeric states are indicated on the right. (C) EMSAs of Rep-p53 fusion proteins. Chimeric proteins were synthesized in vitro and incubated with the 3′-end-labeled RRS probe as described in the legend to Fig. 3. DNA-bound proteins were supershifted with a specific antibody to the Myc tag. The absence (−) or presence (+) of the antibody is indicated on top. The positions of free (F), bound (B), and supershifted (S) DNA substrate are indicated on the right; the position of a nonspecific band is indicated on the left (X). p, probe; Luc, luciferase; MD, Rep.MD.AD; DD, Rep.DD.AD; CD, Rep.CD.AD; TD, Rep.TD.AD; 78, Rep78; TZ, Rep.TZ.AD.
To analyze their oligomeric state, the chimeric proteins were synthesized in vitro and analyzed by cross-linking (Fig. 4B). The Rep.MD.AD protein was unaffected by the cross-linker, confirming that the introduced mutation prevented multimerization of the protein. For Rep.DD.AD and Rep.CD.AD, which contain modified oligomerization domains predicted to form dimers, slower-migrating bands were detected in the presence of the cross-linker. As seen with Rep.LZ.AD, the migration of these bands suggests that Rep.DD.AD and Rep.CD.AD might form tetramers, although this has not been rigorously addressed. Cross-linking of Rep.TD.AD, which contains the p53 wild-type oligomerization motif, yielded a band of similar size, as well as additional slower-migrating bands. Compared to the Rep.GCN4 fusion proteins, the multimerization domains from p53 were less efficient at the formation of higher-order oligomers. The absence of detectable dimeric forms of chimeric Rep fusion proteins suggests that the coiled coil identified in the N terminus of Rep (8) may mediate further multimerization of the chimeric proteins once two amino-terminal Rep entities are in close proximity. Alternatively, multimeric but not monomeric forms of the Rep fusion proteins may interact with cellular reticulocyte proteins.
The effect of oligomerization on DNA binding was assayed by the EMSA. In vitro-synthesized Rep-p53 fusion proteins were incubated with a 32P-labeled, RRS-containing DNA substrate in the absence (−) or presence (+) of the anti-Myc antibody (Fig. 4C). No distinguishable band was seen for Rep.MD.AD, Rep.DD.AD, Rep.CD.AD, or a control reaction. Since the Rep.CD.AD and Rep.DD.AD proteins formed cross-linked products, it is possible that they bind weakly to DNA, but that complex formation was too weak to be detected in the in vitro EMSA. In contrast, Rep.TD.AD bound to the RRS efficiently, as detected by the mobility shift (B). In the presence of the antibody, a novel slower-migrating band (S) was detected. In a control reaction, in vitro-translated Rep78 was not supershifted by the Myc antibody.
A one-hybrid assay to study Rep binding to the RRS in cultured mammalian cells.
Having shown that chimeric Rep proteins with oligomerization motifs could multimerize and bind to the RRS in vitro, we developed an in vivo assay for Rep binding (Fig. 5). In this assay, we used activation of luciferase expression as a read-out for binding of the chimeric Rep proteins to the RRS. Two reporter constructs were used (Fig. 5A). For the first reporter, the RRS was cloned upstream of a minimal thymidine kinase promoter driving expression of the luciferase gene. In the second reporter, nucleotides 1 to 320 of the AAV genome, comprising the viral ITR and the p5 promoter, were fused to the luciferase gene (8). HeLa cells were cotransfected with the RRS.tk.Luc reporter and expression plasmids encoding the chimeric Rep proteins (Fig. 5B). The monomeric Rep.AD fusion protein gave very little activation of the reporter gene, consistent with the results showing that it cannot oligomerize and as a consequence cannot bind to the RRS in vitro (Fig. 5B, left panel). Rep.LZ.AD and Rep.TZ.AD activated the reporter in a dose-dependent fashion (data not shown). At the highest DNA concentration used, expression of Rep.LZ.AD led to an approximately 30-fold activation (Fig. 5B). The strongest activation was by the Rep.TZ.AD protein, which activated reporter gene expression about 200-fold. Transactivation was specific for the RRS, because the parental tk.Luc control reporter was unaffected by expression of any Rep fusion protein. Chimeric proteins lacking the VP16 activation domain showed no activity, as did the wild-type Rep78 protein (Fig. 5B, right panel). In the same experiment, the activities of the Rep-p53 fusion proteins were tested (Fig. 5B, right panel). Similarly, the chimeric protein harboring the tetramerization domain (Rep.TD.AD) induced the highest luciferase activity (28-fold), whereas Rep.CD.AD activated luciferase expression about 8-fold and Rep.MD.AD activated it about 6-fold. Although we were unable to detect DNA binding by the Rep.CD.AD protein in the in vitro EMSA, a low level of activation was observed in the in vivo assay. This either suggests that the in vivo assay is a more sensitive readout of DNA binding or that cellular proteins stabilize the interaction.
FIG. 5.
One-hybrid assay for Rep binding to the RRS in cultured cells. (A) Schematic overview of the reporter constructs. Reporter plasmid RRS.tk.Luc contains the luciferase gene downstream of an RRS motif and a minimal thymidine kinase promoter. The reporter ITR/p5.Luc contains nucleotides 1 to 320 of the AAV genome fused to the luciferase gene. (B) Transactivation of the RRS reporter by chimeric Rep proteins in HeLa cells. Plasmids encoding the chimeric effector proteins were cotransfected with either the RRS.tk.Luc reporter (black columns) or with the parental control plasmid lacking the RRS (checked columns). After 30 h, cells were harvested, and luciferase activity was determined in a luminometer. Luciferase activity is indicated as relative activity compared to that of cells cotransfected with a mock effector plasmid. (C) Transactivation of the ITR/p5 reporter by chimeric Rep proteins. Plasmids encoding the chimeric effector proteins were cotransfected with either the ITR/p5.Luc reporter (black columns) or with a control reporter containing mutations in the RRS of the p5 promoter (checked columns). Assays were performed as for panel B. (D) Transactivation of the RRS reporter by Rep.TZ.AD proteins with deletions in the Rep DNA-binding domain. Plasmids encoding truncated Rep.TZ.AD effector proteins (on the right) were cotransfected with the RRS.tk.Luc reporter into HeLa cells. Assays were performed as for panel B. In all cases, individual experiments were repeated at least twice in duplicate. Columns and error bars reflect the average value and the standard deviation of a representative experiment performed in duplicate. All values were normalized for transfection efficiency by evaluating β-galactosidase activity from a cotransfected LacZ expression plasmid.
The chimeric Rep-VP16 proteins were also able to activate transcription from a reporter containing the autologous AAV ITR/p5 promoter. Luciferase expression is regulated by an RRS element in the ITR, acting as an enhancer element, and a second RRS upstream of the transcriptional start site of the p5 promoter (Fig. 5A). Cotransfection of the Rep.AD expression plasmid together with the ITR/p5.Luc reporter did not activate luciferase expression (Fig. 5C, left panel). As with the RRS.tk.Luc reporter, expression of Rep.LZ.AD activated luciferase expression moderately (10-fold), and Rep.TZ.AD led to the strongest activation (160-fold). In a similar way, the chimeric Rep proteins fused to p53 oligomerization domains activated reporter gene expression from the ITR/p5 promoter (Fig. 5C, right panel). Rep.TD.AD, which contains the wild-type tetramerization domain, induced the highest luciferase activity (44-fold). The activity of Rep.DD.AD, the second construct that harbors a dimerization domain, was in both experiments comparable to that of Rep.CD.AD (data not shown). The somewhat lower levels of activation observed for the Rep-p53 fusion proteins, compared to those for the Rep-GCN4 chimeric proteins, could reflect their less efficient oligomerization, as determined in the cross-linking experiments (Fig. 2 and 3C). No activation was detected when a reporter plasmid carrying mutations in the RRS of the p5 promoter (ITR/m5.Luc) was used (27). Despite the presence of an RRS in the ITR of this construct, the reporter was not activated. This is probably due to repression of p5 promoter activity by the cellular YY1 protein (47).
In order to define a minimal region of Rep required for DNA binding, chimeric Rep.TZ.AD proteins were generated that contain internal deletions or truncations of the Rep DNA-binding domain. The proteins were in vitro translated and assayed for their ability to oligomerize. As determined by cross-linking experiments, all deletion mutants were able to multimerize like the parental Rep.TZ.AD protein (data not shown). Their ability to activate the RRS.tk.Luc reporter was examined in HeLa cells (Fig. 5D). A 12-residue deletion from the N terminus abolished the ability to drive luciferase expression. This is consistent with in vitro EMSA experiments in which deletions of the N terminus prevented DNA binding (59). A Rep.TZ.AD deletion mutant comprising residues 1 to 220 of the Rep protein was sufficient to drive luciferase expression, although the activity dropped by more than 50% compared to that of the parental protein. Further deletions from the C terminus of the Rep DNA-binding domain abolished the transactivating activity. All proteins were expressed at equivalent levels as determined by immunoblotting (data not shown).
Based on a report by Yang and Trempe (69) on the analysis of ITR binding by mutant Rep proteins, we generated Rep.TZ.AD mutants with internal deletions. Although the deletion mutants were reported to bind to ITR sequences in vitro in the context of the wild-type Rep78 protein (69), the Rep.TZ.AD internal deletion mutants were not able to activate luciferase expression in our in vivo system. The discrepancy between the reported in vitro data and our in vivo results cannot be attributed to differences between the two experimental systems, since our results clearly demonstrate that the in vivo reporter assay accurately reflects the preceding in vitro data. Binding of full-length Rep78 to an additional sequence within the ITR other than the RRS (34, 43) might compensate for decreased binding affinity of the N-terminal Rep DNA-binding segment. The reduction in DNA-binding affinity caused by the internal deletions, however, could not be compensated for on the linear RRS present in our reporters.
In summary, the reporter assays with HeLa cells show that multimerization of the N terminus of Rep is a prerequisite for activation of the reporter gene. Only chimeric proteins with the potential to form high-order multimers were able to bind to the RRS and efficiently activate transcription. The Rep sequences involved in mediating DNA binding seem to encompass the first 240 amino-terminal residues, because any further deletion affected its ability to activate reporter gene expression from RRS motifs. The results from this in vivo assay closely reflect conclusions drawn from the in vitro assays.
A yeast one-hybrid assay to study DNA binding by the Rep protein in vivo.
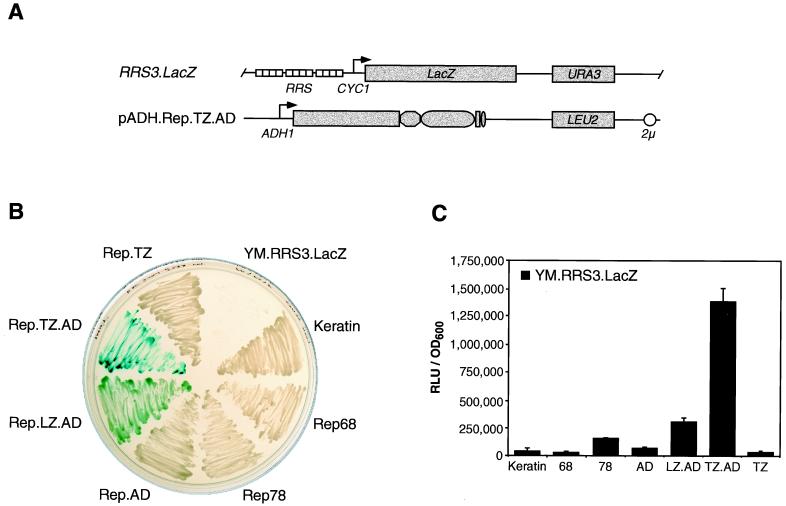
The yeast S. cerevisiae provides a powerful tool with which to examine protein-DNA interactions in a model organism. We therefore adapted a one-hybrid assay for Rep binding in yeast. The reporter construct pRRS3.LacZ contains three tandem copies of the RRS cloned upstream of the minimal CYC1 yeast promoter, followed by the lacZ reporter gene (Fig. 6A). To generate the reporter strain YM.RRS3.LacZ, plasmid pRRS3.LacZ was linearized within the nutritional marker gene URA3 and used to transform the yeast strain YM4271. Integration into the mutated ura3 locus confers a Ura+ phenotype on transformants, allowing selection in uracil-deficient medium. The resulting RRS reporter strain was then transformed with the Rep effector plasmids, which were generated by subcloning the reading frames of the chimeric Rep proteins into yeast expression vectors under control of the ADH1 promoter. The effector plasmids contain the nutritional selection marker LEU2 and a 2μ origin of replication (Fig. 6A). Activation of the reporter gene was assessed both qualitatively by an in vivo plate assay (Fig. 6B) and quantitatively by liquid culture assays (Fig. 6C).
FIG. 6.
In vivo assay for Rep binding to the RRS in S. cerevisiae. (A) Schematic overview of yeast constructs. The reporter strain YM.RRS3.LacZ contains an integrated lacZ gene in the URA3 locus. β-Galactosidase expression is driven from a minimal promoter of the yeast CYC1 gene and three upstream tandem copies of the RRS. The effector proteins are expressed from the yeast ADH1 promoter. (pADH.Rep.TZ.AD is shown as an example.) The nutritional selection markers URA3 and LEU2 and the 2μ origin of replication are indicated. The drawings are not to scale. (B) Qualitative β-galactosidase assay. The reporter strain YM.RRS3.LacZ was transformed with effector plasmids encoding the chimeric Rep proteins, wild-type Rep proteins, or a control plasmid expressing keratin. Transformants were selected by streaking cells on agar plates containing minimum selection medium lacking uracil and leucine. After 3 days, colonies were patched on agar plates containing X-Gal in the selection medium, and the development of blue cells was recorded 2 days later. The parental reporter strain was plated as a control. (C) Quantitative β-galactosidase assay. Liquid cultures of transformants were harvested in mid-log phase. β-Galactosidase activity was determined with Galacton-Star as a substrate. Individual experiments were repeated twice in duplicate. Columns and error bars reflect the average value and the standard deviation of a representative experiment performed in duplicate. All values were normalized for cell number by recording the OD600.
For a qualitative β-galactosidase assay, transformed cells were plated and selected on minimum medium lacking both uracil and leucine. After 3 days, colonies were patched on agar plates containing X-Gal in the selection medium and grown for another 2 days. Expression of β-galactosidase in the presence of X-Gal leads to development of blue-stained yeast cells. Figure 6B shows that the Rep.TZ.AD-transformed reporter strain expressed high levels of β-galactosidase, leading to a dark blue staining of the cells. Rep.LZ.AD-expressing reporter cells also turned blue, but to a lesser extent. All other transformants remained white, indicating that low levels or no β-galactosidase was expressed. As expected, the untransformed reporter strain did not grow. The experiment was also performed with expression plasmids containing a truncated version of the ADH1 promoter (pG series [data not shown]). In contrast to expression from pADH plasmids, protein levels expressed from the pG plasmids were not detectable by Western blot analysis (data not shown). Nevertheless, the small amount of chimeric protein produced in pG.Rep.TZ.AD-transformed cells was sufficient to stain the cells blue after 3 days of incubation at 30°C.
For quantitation of β-galactosidase expression, liquid cultures of transformants were grown and harvested in mid-log phase. Yeast cells were lysed, and β-galactosidase activity was determined in a luminometer with a chemiluminescent substrate (Fig. 6C). In this assay, the pADH.Rep.TZ.AD-transformed YM.RRS3.LacZ reporter strain showed the highest levels of β-galactosidase expression. Compared to the control transformants, cells transformed with pADH.Keratin or pADH.Rep.TZ, the measured β-galactosidase activity was 35-fold higher. β-Galactosidase activity in the reporter strain transformed with pADH.Rep68 and pADH.Rep.AD was in the background range, whereas Rep78- and Rep.LZ.AD-expressing cells revealed moderate LacZ expression (four- and ninefold above the background level, respectively). The liquid culture assay is more sensitive than the in vivo plate assay and thus detected the weak activation by Rep78, which was not observed in the plate assay. An intrinsic transcriptional transactivation activity of Rep78 in yeast was reported previously by fusing Rep78 to the GAL4 DNA-binding domain (10, 57).
In summary, the yeast experiments demonstrate for the first time the feasibility of target site selection by a chimeric Rep protein in vivo. A small amount of a chimeric protein was able to target integrated RRS motifs and activate transcription upon binding. The experiments confirm our previous data by showing a dependency of reporter gene activation on the oligomeric state of the chimeric activator protein.
DISCUSSION
DNA binding by the Rep proteins of AAV is an essential step in replication and targeted integration. Previous analyses suggested that the major DNA-binding activity is contained within the N terminus, but that multimerization is likely to be important for all Rep functions. To study DNA binding, we designed chimeric proteins with the N terminus of Rep linked to sequences that promote oligomerization. Variants of two unrelated multimerization sequences from heterologous proteins of yeast or human origin were incorporated into Rep fusion proteins and shown to promote oligomerization. We used in vitro and in vivo assays to establish a correlation between oligomeric status and specific binding to the RRS sequence.
In the EMSA, the monomeric forms of the Rep fusion protein could not cause a mobility shift of the RRS probe. Fusion proteins containing dimerization sequences gave only weak DNA-protein interactions, whereas those containing sequences predicted to induce tetramers demonstrated strong DNA-binding activity. Binding by the Rep N terminus in our fusion proteins was specific, as demonstrated by antibody-induced supershifts and competition experiments with unlabeled DNA substrates. The chimeric fusion proteins were also tested for DNA binding in cultured HeLa cells and in yeast cells by assays in which binding to the RRS led to activation of a reporter gene. This is the first time that DNA recognition by the Rep DNA-binding domain has been demonstrated in vivo. The in vivo assays faithfully reflected results from the in vitro binding experiments. The degree of activation from reporter genes directly correlated with the ability to form higher-order oligomers and shift the RRS probe in the EMSA. The in vitro and cell-based assays provide the first demonstration that the N terminus of Rep contains the necessary elements to bind DNA in a sequence-specific manner through recognition of the RRS and that binding is reliant on multimerization of the protein.
Identification of protein domains and amino acid residues responsible for individual activities of Rep will advance the understanding of how these proteins carry out their many functions. In vitro studies using insertions, deletions, and specific mutations have identified key residues involved in DNA recognition and other functions of Rep (9, 33, 53, 68), but the reliance on multimerization for many Rep functions has complicated interpretation of these mutants. We have been able to separate DNA binding from a requirement for Rep multimerization sequences, enabling comprehensive analysis of DNA binding requirements at the amino acid level. The assays that we have developed provide tools to analyze Rep binding to its natural RRS target in vivo and will facilitate screening of mutants. The presence of sequences similar to the RRS in the promoter or promoter-proximal regions of a number of cellular genes (62, 63) raises the possibility that a cellular protein recognizes the same sequence as the viral Rep protein. Using the yeast RRS reporter strains, screens can be performed to address this issue. The use of novel chimeric Rep proteins, with regions swapped for domains from heterologous proteins, may also be useful in the analysis of other Rep functions, such as nicking and oligomerization.
There is increasing interest in recombinant AAV as a potential gene delivery vector for human gene therapy (11, 35, 64). AAV vectors have normally had all viral genes deleted and consist of the ITRs flanking the foreign gene of interest. Recombinant AAV vectors are still capable of integration but do not target the AAVS1 locus at the high frequency observed for wild-type virus (21, 40, 42, 46, 65). However, when supplied in trans, Rep can retarget integration into AAVS1 (3, 29, 50). The mechanism for site-specific integration by AAV remains unclear, but a model has been proposed involving a Rep-mediated complex between AAV and the target site (31, 60). It would be very attractive to harness the targeting ability of AAV into a gene delivery system, because this would decrease the hazards of insertional mutagenesis associated with random integration. Since wild-type Rep has been associated with cytostatic effects, its application is not desirable, but the use of chimeric Rep proteins might bypass these shortcomings. The demonstrated target site selection of such proteins in our yeast in vivo assay suggests that the use of chimeric Rep proteins for targeted insertion of therapeutic genes might be feasible.
ACKNOWLEDGMENTS
We thank Nicolas Genoud, who was involved in an early phase of this project; Luz Beatriz Gilbert for technical assistance; Christopher J. Larson for advice on cross-linking; Brian H. Spain, Sook Shin, Jeanette Ducut, and Susan Forsburg for advice on yeast work; Ron Evans for the tk.Luc reporter plasmid; Thanos Halazonetis, Jayne Stommel, and Geoffrey Wahl for p53 plasmids; John Colicelli for yeast plasmids; and Joanne Chory for use of the luminometer. We also thank Mirta Grifman, Travis Stracker, Rick Bushman, and Roland Owens for helpful discussions and comments on the manuscript.
This work was supported by consecutive fellowships from the Swiss National Science Foundation and the Swiss Foundation for Medical Biological Grants (T.C.), by a grant from the NIH (M.D.W.), and by gifts from Odette Wurzburger and the Oracle Corporate Giving Program (M.D.W.).
REFERENCES
- 1.Andrews N C, Faller D V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashktorab H, Srivastava A. Identification of nuclear proteins that specifically interact with adeno-associated virus type 2 inverted terminal repeat hairpin DNA. J Virol. 1989;63:3034–3039. doi: 10.1128/jvi.63.7.3034-3039.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balagué C, Kalla M, Zhang W-W. Adeno-associated virus Rep78 protein and terminal repeats enhance integration of DNA sequences into the cellular genome. J Virol. 1997;71:3299–3306. doi: 10.1128/jvi.71.4.3299-3306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batchu R B, Kotin R M, Hermonat P L. The regulatory Rep protein of adeno-associated virus binds to sequences within the c-H-ras promoter. Cancer Lett. 1994;86:23–31. doi: 10.1016/0304-3835(94)90176-7. [DOI] [PubMed] [Google Scholar]
- 5.Berns K I. Parvoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2173–2197. [Google Scholar]
- 6.Cheung A K M, Hoggan M D, Hauswirth W W, Berns K I. Integration of the adeno-associated virus genome into cellular DNA in latently infected human Detroit 6 cells. J Virol. 1980;33:739–748. doi: 10.1128/jvi.33.2.739-748.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiorini J A, Weitzman M D, Owens R A, Urcelay E, Safer B, Kotin R M. Biologically active Rep proteins of adeno-associated virus type 2 produced as fusion proteins in Escherichia coli. J Virol. 1994;68:797–804. doi: 10.1128/jvi.68.2.797-804.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costello E, Saudan P, Winocour E, Pizer L, Beard P. High mobility group chromosomal protein 1 binds to the adeno-associated virus replication protein (Rep) and promotes Rep-mediated site-specific cleavage of DNA, ATPase activity and transcriptional repression. EMBO J. 1997;16:5943–5954. doi: 10.1093/emboj/16.19.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis M D, Wonderling R S, Walker S L, Owens R A. Analysis of the effects of charge cluster mutations in adeno-associated virus Rep68 protein in vitro. J Virol. 1999;73:2084–2093. doi: 10.1128/jvi.73.3.2084-2093.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Pasquale G, Stacey S N. Adeno-associated virus Rep78 protein interacts with protein kinase A and its homolog PRKX and inhibits CREB-dependent transcriptional activation. J Virol. 1998;72:7916–7925. doi: 10.1128/jvi.72.10.7916-7925.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flotte T R, Carter B J. Adeno-associated virus vectors for gene therapy. Gene Ther. 1995;2:357–362. [PubMed] [Google Scholar]
- 12.Harbury P B, Zhang T, Kim P S, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 13.Heilbronn R, Bürkle A, Stephan S, zur Hausen H. The adeno-associated virus rep gene suppresses herpes simplex virus-induced DNA amplification. J Virol. 1990;64:3012–3018. doi: 10.1128/jvi.64.6.3012-3018.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermonat P L. The adeno-associated virus rep78 gene inhibits cellular transformation induced by bovine papillomavirus. Virology. 1989;172:253–261. doi: 10.1016/0042-6822(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 15.Hermonat P L. Inhibition of bovine papillomavirus plasmid DNA replication by adeno-associated virus. Virology. 1992;189:329–333. doi: 10.1016/0042-6822(92)90710-7. [DOI] [PubMed] [Google Scholar]
- 16.Hermonat P L. Inhibition of H-ras expression by the adeno-associated virus Rep78 transformation suppressor gene product. Cancer Res. 1991;51:3373–3377. [PubMed] [Google Scholar]
- 17.Hermonat P L, Batchu R B. The adeno-associated virus Rep78 major regulatory protein forms multimeric complexes and the domain for this activity is contained within the carboxy-half of the molecule. FEBS Lett. 1997;401:180–184. doi: 10.1016/s0014-5793(96)01469-x. [DOI] [PubMed] [Google Scholar]
- 18.Im D-S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 19.Im D-S, Muzyczka N. Factors that bind to adeno-associated virus terminal repeats. J Virol. 1989;63:3095–3104. doi: 10.1128/jvi.63.7.3095-3104.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Im D S, Muzyczka N. Partial purification of adeno-associated virus Rep78, Rep52, and Rep40 and their biochemical characterization. J Virol. 1992;66:1119–1128. doi: 10.1128/jvi.66.2.1119-1128.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kearns W G, Afione S A, Fulmer S B, Pang M G, Erikson D, Egan M, Landrum M J, Flotte T R, Cutting G R. Recombinant adeno-associated virus (AAV-CFTR) vectors do not integrate in a site-specific fashion in an immortalized epithelial cell line. Gene Ther. 1996;3:748–755. [PubMed] [Google Scholar]
- 22.Khleif S N, Myers T, Carter B J, Trempe J P. Inhibition of cellular transformation by the adeno-associated virus rep gene. Virology. 1991;181:738–741. doi: 10.1016/0042-6822(91)90909-u. [DOI] [PubMed] [Google Scholar]
- 23.Kotin R M, Linden R M, Berns K I. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 1992;11:5071–5078. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotin R M, Menninger J C, Ward D C, Berns K I. Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13-qter. Genomics. 1991;10:831–834. doi: 10.1016/0888-7543(91)90470-y. [DOI] [PubMed] [Google Scholar]
- 25.Kotin R M, Siniscalco M, Samulski R J, Zhu X, Hunter L, Laughlin C A, McLaughlin S, Muzyczka N, Rocchi M, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kyöstiö S R M, Owens R A, Weitzman M D, Antoni B A, Chejanovsky N, Carter B J. Analysis of adeno-associated virus (AAV) wild-type and mutant Rep proteins for their abilities to negatively regulate AAV p5 and p19 mRNA levels. J Virol. 1994;68:2947–2957. doi: 10.1128/jvi.68.5.2947-2957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kyöstiö S R M, Wonderling R S, Owens R A. Negative regulation of the adeno-associated virus (AAV) P5 promoter involves both the P5 Rep binding site and the consensus ATP-binding motif of the AAV Rep68 protein. J Virol. 1995;69:6787–6796. doi: 10.1128/jvi.69.11.6787-6796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labow M A, Graf L H, Jr, Berns K I. Adeno-associated virus gene expression inhibits cellular transformation by heterologous genes. Mol Cell Biol. 1987;7:1320–1325. doi: 10.1128/mcb.7.4.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamartina S, Roscilli G, Rinaudo D, Delmastro P, Toniatti C. Lipofection of purified adeno-associated virus Rep68 protein: toward a chromosome-targeting nonviral particle. J Virol. 1998;72:7653–7658. doi: 10.1128/jvi.72.9.7653-7658.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laughlin C A, Cardellichio C B, Coon H C. Latent infection of KB cells with adeno-associated virus type 2. J Virol. 1986;60:515–524. doi: 10.1128/jvi.60.2.515-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linden R M, Winocour E, Berns K I. The recombination signals for adeno-associated virus site-specific integration. Proc Natl Acad Sci USA. 1996;93:7966–7972. doi: 10.1073/pnas.93.15.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarty D M, Christensen M, Muzyczka N. Sequences required for coordinate induction of adeno-associated virus p19 and p40 promoters by Rep protein. J Virol. 1991;65:2936–2945. doi: 10.1128/jvi.65.6.2936-2945.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarty D M, Ni T-H, Muzyczka N. Analysis of mutations in adeno-associated virus Rep protein in vivo and in vitro. J Virol. 1992;66:4050–4057. doi: 10.1128/jvi.66.7.4050-4057.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCarty D M, Ryan J H, Zolotukhin S, Zhou X, Muzyczka N. Interaction of the adeno-associated virus Rep protein with a sequence within the A palindrome of the viral terminal repeat. J Virol. 1994;68:4998–5006. doi: 10.1128/jvi.68.8.4998-5006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 36.Ni T-H, Zhou X, McCarty D M, Zolotukhin I, Muzyczka N. In vitro replication of adeno-associated virus DNA. J Virol. 1994;68:1128–1138. doi: 10.1128/jvi.68.2.1128-1138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owens R A, Trempe J P, Chejanovsky N, Carter B J. Adeno-associated virus Rep proteins produced in insect and mammalian expression systems: wild-type and dominant-negative mutant proteins bind to the viral replication origin. Virology. 1991;184:14–22. doi: 10.1016/0042-6822(91)90817-u. [DOI] [PubMed] [Google Scholar]
- 38.Owens R A, Weitzman M D, Kyöstiö S R M, Carter B J. Identification of a DNA-binding domain in the amino terminus of adeno-associated virus Rep proteins. J Virol. 1993;67:997–1005. doi: 10.1128/jvi.67.2.997-1005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pereira D J, McCarty D M, Muzyczka N. The adeno-associated virus (AAV) Rep protein acts as both a repressor and an activator to regulate AAV transcription during a productive infection. J Virol. 1997;71:1079–1088. doi: 10.1128/jvi.71.2.1079-1088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ponnazhagan S, Erikson D, Kearns W G, Zhou S Z, Nahreini P, Wang X S, Srivastava A. Lack of site-specific integration of the recombinant adeno-associated virus 2 genomes in human cells. Hum Gene Ther. 1997;8:275–284. doi: 10.1089/hum.1997.8.3-275. [DOI] [PubMed] [Google Scholar]
- 41.Prasad K M R, Zhou C, Trempe J P. Characterization of the Rep78/Adeno-associated virus complex. Virology. 1997;229:183–192. doi: 10.1006/viro.1996.8431. [DOI] [PubMed] [Google Scholar]
- 42.Rutledge E A, Russell D W. Adeno-associated virus vector integration junctions. J Virol. 1997;71:8429–8436. doi: 10.1128/jvi.71.11.8429-8436.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryan J H, Zolotukhin S, Muzyczka N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J Virol. 1996;70:1542–1553. doi: 10.1128/jvi.70.3.1542-1553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samulski R J. Adeno-associated virus: integration at a specific chromosomal locus. Curr Opin Biotechnol. 1993;3:74–80. doi: 10.1016/s0959-437x(05)80344-2. [DOI] [PubMed] [Google Scholar]
- 45.Samulski R J, Zhu X, Xiao X, Brook J D, Housman D E, Epstein N, Hunter L A. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shelling A N, Smith M G. Targeted integration of transfected and infected adeno-associated virus vectors containing the neomycin resistance gene. Gene Ther. 1994;1:165–169. [PubMed] [Google Scholar]
- 47.Shi Y, Seto E, Chang L S, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 48.Smith R H, Spano A J, Kotin R M. The Rep78 gene product of adeno-associated virus (AAV) self-associates to form a hexameric complex in the presence of AAV ori sequences. J Virol. 1997;71:4461–4471. doi: 10.1128/jvi.71.6.4461-4471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stommel J M, Marchenko N D, Jimenez G S, Moll U M, Hope T J, Wahl G M. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Surosky R T, Urabe M, Godwin S G, McQuiston S A, Kurtzman G J, Ozawa K, Natsoulis G. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J Virol. 1997;71:7951–7959. doi: 10.1128/jvi.71.10.7951-7959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tratschin J-D, Miller I L, Carter B J. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J Virol. 1984;51:611–619. doi: 10.1128/jvi.51.3.611-619.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tratschin J D, Tal J, Carter B J. Negative and positive regulation in trans of gene expression from adeno-associated virus vectors in mammalian cells by a viral rep gene product. Mol Cell Biol. 1986;6:2884–2894. doi: 10.1128/mcb.6.8.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Urabe M, Hasumi Y, Kume A, Surosky R T, Kurtzman G J, Tobita K, Ozawa K. Charged-to-alanine scanning mutagenesis of the N-terminal half of adeno-associated virus type 2 Rep78 protein. J Virol. 1999;73:2682–2693. doi: 10.1128/jvi.73.4.2682-2693.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ward P, Urcelay E, Kotin R, Safer B, Berns K I. Adeno-associated virus DNA replication in vitro: activation by a maltose binding protein/Rep 68 fusion protein. J Virol. 1994;68:6029–6037. doi: 10.1128/jvi.68.9.6029-6037.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waterman J L, Shenk J L, Halazonetis T D. The dihedral symmetry of the p53 tetramerization domain mandates a conformational switch upon DNA binding. EMBO J. 1995;14:512–519. doi: 10.1002/j.1460-2075.1995.tb07027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waterman M J F, Waterman J L F, Halazonetis T D. An engineered four-stranded coiled coil substitutes for the tetramerization domain of wild-type p53 and alleviates transdominant inhibition by tumor-derived p53 mutants. Cancer Res. 1996;56:158–163. [PubMed] [Google Scholar]
- 57.Weger S, Wendland M, Kleinschmidt J A, Heilbronn R. The adeno-associated virus type 2 regulatory proteins Rep78 and Rep68 interact with the transcriptional coactivator PC4. J Virol. 1999;73:260–269. doi: 10.1128/jvi.73.1.260-269.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weitzman M D, Fisher K J, Wilson J M. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J Virol. 1996;70:1845–1854. doi: 10.1128/jvi.70.3.1845-1854.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weitzman M D, Kyöstiö S R M, Carter B J, Owens R A. Interaction of wild-type and mutant adeno-associated virus (AAV) Rep proteins on AAV hairpin DNA. J Virol. 1996;70:2440–2448. doi: 10.1128/jvi.70.4.2440-2448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weitzman M D, Kyöstiö S R M, Kotin R M, Owens R A. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wonderling R S, Kyöstiö S R M, Owens R A. A maltose-binding protein/adeno-associated virus Rep68 fusion protein has DNA-RNA helicase and ATPase activities. J Virol. 1995;69:3542–3548. doi: 10.1128/jvi.69.6.3542-3548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wonderling R S, Owens R A. Binding sites for adeno-associated virus Rep proteins within the human genome. J Virol. 1997;71:2528–2534. doi: 10.1128/jvi.71.3.2528-2534.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wonderling R S, Owens R A. The Rep68 protein of adeno-associated virus type 2 stimulates expression of the platelet-derived growth factor B c-sis proto-oncogene. J Virol. 1996;70:4783–4786. doi: 10.1128/jvi.70.7.4783-4786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiao X, Li J, McCown T J, Samulski R J. Gene transfer by adeno-associated virus vectors into the central nervous system. Exp Neurol. 1997;144:113–124. doi: 10.1006/exnr.1996.6396. [DOI] [PubMed] [Google Scholar]
- 65.Yang C C, Xiao X, Zhu X, Ansardi D C, Epstein N D, Frey M R, Matera A G, Samulski R J. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J Virol. 1997;71:9231–9247. doi: 10.1128/jvi.71.12.9231-9247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Q, Chen F, Ross J, Trempe J P. Inhibition of cellular and SV40 DNA replication by the adeno-associated virus Rep proteins. Virology. 1995;207:246–250. doi: 10.1006/viro.1995.1072. [DOI] [PubMed] [Google Scholar]
- 67.Yang Q, Chen F, Trempe J P. Characterization of cell lines that inducibly express the adeno-associated virus Rep proteins. J Virol. 1994;68:4847–4856. doi: 10.1128/jvi.68.8.4847-4856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang Q, Kadam A, Trempe J P. Mutational analysis of the adeno-associated virus rep gene. J Virol. 1992;66:6058–6069. doi: 10.1128/jvi.66.10.6058-6069.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang Q, Trempe J P. Analysis of the terminal repeat binding abilities of mutant adeno-associated virus replication proteins. J Virol. 1993;67:4442–4447. doi: 10.1128/jvi.67.7.4442-4447.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]