Abstract
A HeLa cell clone (2A7d) that inducibly expresses the gene for poliovirus protease 2A (2Apro) under the control of tetracycline has been obtained. Synthesis of 2Apro induces severe morphological changes in 2A7d cells. One day after tetracycline removal, cells round up and a few hours later die. Poliovirus 2Apro cleaves both forms of initiation factor eIF4G, causing extensive inhibition of capped-mRNA translation a few hours after protease induction. Methoxysuccinyl-Ala-Ala-Pro-Val-chloromethylketone, a selective inhibitor of 2Apro, prevents both eIF4G cleavage and inhibition of translation but not cellular death. Expression of 2Apro still allows both the replication of poliovirus and the translation of mRNAs containing a picornavirus leader sequence, while vaccinia virus replication is drastically inhibited. Translation of transfected capped mRNA is blocked in 2A7d-On cells, while luciferase synthesis from a mRNA bearing a picornavirus internal ribosome entry site (IRES) sequence is enhanced by the presence of 2Apro. Moreover, synthesis of 2Apro in 2A7d cells complements the translational defect of a poliovirus 2Apro-defective variant. These results show that poliovirus 2Apro expression mimics some phenotypical characteristics of poliovirus-infected cells, such as cell rounding, inhibition of protein synthesis and enhancement of IRES-driven translation. This cell line constitutes a useful tool to further analyze 2Apro functions, to complement poliovirus 2Apro mutants, and to test antiviral compounds.
Infection with most cytolytic animal viruses is characterized by a marked inhibition of host transcription and translation (5, 58). This inhibition is particularly evident when HeLa cells are infected with poliovirus (9, 16). The selective blockade of macromolecular synthesis in virus-infected cells is accompanied by massive poliovirus RNA replication and almost exclusive synthesis of viral proteins. Poliovirus RNA translation starts at a single AUG initiation codon and continues through a long open reading frame that encodes a huge polyprotein precursor (67). This precursor is not found as such in the infected cells, since it is proteolytically processed while still attached to ribosomes as a growing peptide (32, 51). Two virus-encoded proteases, 2Apro and 3Cpro (or its precursor 3CDpro), are responsible for all but one of these proteolytic cleavages (32, 59). These two viral proteases have been implicated in the inhibition of host translation (2Apro) and transcription (3Cpro) (12, 13, 16, 17, 70). Most of the proteolytic reactions that generate the mature viral proteins are accomplished by 3Cpro (or 3CDpro). 2Apro catalyzes only two cleavages on the polyprotein, one between the capsid protein precursor (P1) and itself and another on 3CD to generate 3C′ and 3D′, two products of unknown function (35). 2Apro also bisects other cellular proteins, including initiation factor eIF4G, leading to inactivation of the eIF4F* complex for cap-dependent translation (16, 17).
The eIF4F* complex is composed of three polypeptides: eIF4E, eIF4A, and eIF4G (9, 41). eIF4E is the cap-binding subunit; eIF4A exhibits RNA helicase activity and is thought to unwind the secondary structure present at the 5′ leader sequences of mRNAs, while eIF4G serves as a scaffold to bring together eIF4E, eIF4A, and eIF3, attaching the mRNA to the small ribosomal subunit (9, 41, 56). Recently, a new homologue of eIF4G (termed eIF4GII) has been identified (23). These two forms of eIF4G seem to function identically. Not all picornaviruses cleave eIF4G; enteroviruses, rhinoviruses, and aphthoviruses encode proteases that cleave eIF4G internally to generate two fragments (16, 17), while cardioviruses do not cleave eIF4G but may activate the suppressor of cap-dependent translation, 4E-BP1, by dephosphorylation (21). The bisection of eIF4G by poliovirus 2Apro separates the two functional domains of this factor: the N-terminal domain of eIF4G, which interacts with eIF4E, and the C-terminal domain, which binds eIF4A and eIF3 (34, 72). Although, eIF4GI cleavage was linked initially to the shutoff of host translation in poliovirus-infected cells, several findings suggested a lack of correlation between eIF4GI cleavage and inhibition of host translation (10, 29, 47, 52). Recent findings suggest a correlation between the kinetics of eIF4GII bisection and the inhibition of cellular protein synthesis after poliovirus and rhinovirus infection (24, 61).
Picornavirus RNAs, in contrast to cellular mRNAs, do not contain a 5′ cap structure (9). Their translation is mediated by ribosome binding to an internal ribosome entry site (IRES) that is present within the 5′-untranslated region (9). This mode of cap-independent translation does not require initiation factor eIF4E. Only the C-terminal fragment of eIF4G that interacts with eIF4A and eIF3 is necessary to support IRES-dependent translation (9, 16, 34, 72). Moreover, the synthesis of proteins from mRNAs containing the poliovirus leader sequence is stimulated after cleavage of eIF4G by 2Apro (25, 72). However, the presence of cleaved eIF4G is not sufficient for this enhancement to take place since the presence of an active 2Apro is necessary to stimulate IRES-driven translation (54, 66). The initiation of protein synthesis on some cellular mRNAs shows similarities to picornavirus mRNA translation (40). This is the case for mRNAs that encode some heat shock proteins and that, despite the presence of typical cap structures at the 5′ ends, are still translated after shutoff of cellular translation in poliovirus-infected cells (45). This is due to the presence of a leader sequence that does not require the eIF4F* complex and the typical cap recognition step (28, 53).
In addition to eIF4G, a number of cellular proteins were found to be proteolysed during poliovirus infection (63). Thus, it has been recently reported that poliovirus 2Apro cleaves transcription factor TFIID in cell-free systems (69). However, this cleavage does not affect in vitro transcription. There may be other unidentified cellular substrates for 2Apro. In addition to being implicated in the proteolysis of viral precursors and host proteins and stimulation of translation of mRNAs bearing the picornavirus IRES, 2Apro has been implicated in poliovirus RNA replication, which occurs by mechanisms that are still poorly understood but that could be independent of its protease activity (38, 43, 71). In conclusion, the multifunctionality of poliovirus 2Apro examplifies how picornaviruses have evolved to maximize their genetic information in a short RNA genome.
The potential cellular toxicity of the individual expression of 2Apro has been analyzed in different cellular systems, including bacteria where this protease was nontoxic (4, 48), yeast (7, 8, 31), and mammalian cells. A variety of expression systems have been examined in cultured cells. The synthesis of poliovirus 2Apro activated by human immunodeficiency virus tat blocks the expression of a reporter gene in HeLa cells (60). Transient expression of 2Apro in COS cells has a major impact on transcription compared to translation of the reporter gene analyzed (14). Attempts to express 2Apro from recombinant vaccinia viruses have been hampered by 2Apro toxicity (30, 62). Transient expression of poliovirus 2Apro has been achieved by infection with recombinant vaccinia virus that expresses the T7 RNA polymerase (2, 3, 19). Poliovirus 2Apro strongly inhibits vaccinia virus gene expression at the translational level (19). Microinjection of mRNA encoding coxsackievirus 2Apro in Xenopus oocytes leads to eIF4G cleavage and to strong inhibition of translation of luciferase mRNA, while ongoing cellular translation is much less affected (15). Similar results were found using a different approach to deliver hybrid proteins containing poliovirus 2Apro directly into cells. Efficient eIF4G cleavage occurs upon entry of these hybrid proteins promoted by adenovirus particles (47), but ongoing cellular translation was less inhibited than the expression of a reporter gene (46). The results obtained with these systems show that 2Apro certainly affects the expression of a reporter gene but that its effect may depend on factors that vary with the expression system tested.
In order to further assay the effects of poliovirus 2Apro on both gene expression and ongoing cellular translation, we decided to use a tetracycline (tet)-dependent expression system (22). The Tet-Off system used to express poliovirus 2Apro avoids the presence of additional viral proteins, which characterizes most eukaryotic expression systems thus far analyzed. To this end, stably transformed HeLa cell clones that express poliovirus 2Apro under the control of an inducible, tightly regulated promoter were obtained. Thus, a biochemical and morphological analysis of the consequences of poliovirus 2Apro expression in human cells has been conducted.
MATERIALS AND METHODS
Cells and viruses.
A HeLa-derived cell line, designated HeLa Tet-Off, that expresses the chimeric tet-responsive transcriptional activator (tTA) (22) was purchased from Clontech. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% newborn calf serum and 100 μg of G418 (Geneticin; GIBCO BRL, Gaithersburg, Md.)/ml.
Infection of HeLa cells with wild-type (wt) or 2Apro mutant polioviruses was carried out as described previously (66). Poliovirus type 1 (Mahoney strain) and vaccinia virus were grown in HeLa cells with DMEM supplemented with 2% calf serum. Encephalomyocarditis virus (EMCV) was grown in L929 cells in the same medium. The recombinant vaccinia virus bearing the T7 RNA polymerase (VT7) (20) was kindly provided by B. Moss (National Institutes of Health, Bethesda, Md.). Infection of HeLa cells with vaccinia virus and VT7 has been previously described (1, 3).
General recombinant DNA protocols.
Construction and purification of vectors were carried out by standard procedures (57). pTRE.2A was generated by ligation of the EcoRI/BamHI fragment of pGBT.2A (65) containing the poliovirus 2Apro sequence with pTRE (Clontech) digested with the same enzymes. Plasmids pTM1.2C, pT7.2C, pAR.2C, pKS.act (encoding human β-actin), pKS.luc, and pKS.L1.luc have been described previously (1, 33, 66).
Generation of stably transformed HeLa cells bearing the poliovirus 2Apro gene. Induction of 2Apro synthesis.
A 60-mm-diameter plate seeded with 106 HeLa Tet-Off cells was transfected with 5 μg of pTRE.2A encoding the poliovirus 2Apro under the control of the tTA-responsive promoter and mixed with 0.25 μg of plasmid pTK-Hyg containing the hygromycin resistance gene and 10 μg of Lipofectin. After 8 h of incubation, plasmids and Lipofectin were removed and the cells were grown in DMEM supplemented with 10% newborn calf serum for 24 h before the initiation of selection with hygromycin B. Plasmid pTRE lacking an insert was used to generate mock-transformed cell lines. Positive clones were selected with 200 μg of hygromycin B/ml and 2 μg of tet/ml. The concentration of tet was reduced to 0.02 μg/ml to maintain repression of the selected clones during the experiments. Hygromycin-resistant clones were screened for the cleavage of eIF4G 24 h after induction of 2Apro in the absence of tet. eIF4G cleavage was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting. These analyses test for both protease expression and activity.
For individual experiments, subconfluent HeLa cell cultures were detached by treatment with trypsin-EDTA and 105 cells were plated in 24-well dishes. After 12 to 24 h cells were washed five times with DMEM over a period of 1 h in order to remove tet and to induce 2Apro expression. Finally, cells were placed in DMEM supplemented with 2% fetal calf serum without tet (induced) or with 0.02 μg of tet/ml (uninduced).
Transient expression and lipofection of mRNA.
HeLa cells plated in 24-well dishes were induced at the times indicated in each experiment before infection with VT7 (multiplicity of infection [MOI] of 5). After 45 min of virus adsorption, a mixture of DNA (0.5 μg/well) and Lipofectin (2 μg/well) was added to cells in DMEM as described by the manufacturer (GIBCO BRL). Cells were harvested at the times indicated in the figure legends for each experiment. RNA transfections were carried out as already described (66).
Protein labeling and Western blotting.
Cells were labeled with 25 μCi/ml of [35S]Pro-Mix (>1,000 Ci/mmol; Amersham) added to methionine-free medium. The radiolabeled cell monolayers were dissolved in sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 0.1 M dithiothreitol, 17% glycerol, 0.024% bromophenol blue [indicator]) and loaded onto SDS–15% PAGE gels. Immunoblotting was carried out as described previously (6). The anti-2C (55), -2A and -3C (8), -eIF4GI (19), and -eIF4GII (23) sera were obtained and used as described previously. The anti-eIF4FG-2 antiserum was a generous gift from N. Sonenberg (McGill University).
RNA extraction and Northern blotting.
RNA was extracted from 106 cells as described previously (18). mRNA isolation was carried out using a QuickPrep Micro mRNA purification kit purchased from Pharmacia Biotech. 32P-labeled probes were obtained using the Rediprime DNA labeling system (Amersham) employing a restriction fragment bearing poliovirus 2Apro. Northern blot analysis was performed by standard procedures (57).
RESULTS
Isolation of HeLa cell clones expressing poliovirus 2Apro under the control of tet.
Gossen and Bujard (22) developed a transcriptional regulation system for mammalian cells based on the bacterial tet resistance operon. A chimeric protein composed of the tet repressor fused to the activation domain of the herpesvirus transcriptional activator, VP16, served as the tTA. The presence of tet prevents the binding of the tTA to a synthetic promoter, while tet removal induces transcription of the gene by the binding of tTA to the promoter. Different cell lines stably transfected with tTA are now available. We used the cell line HeLa Tet-Off (Clontech). Initially, these cells were transiently transfected with pTRE.2A, a plasmid bearing the poliovirus 2Apro gene downstream from the synthetic tet response element, in order to test the regulatability of 2Apro expression in HeLa Tet-Off cells. Cleavage of eIF4G was detected 46 h after transfection in the absence of tet, while eIF4G remained intact in the presence of the antibiotic (results not shown). To obtain stably transformed cell lines expressing 2Apro, HeLa Tet-Off cells were cotransfected with pTRE.2A and pTK-Hyg bearing the selectable marker for hygromycin B, as detailed in Materials and Methods. After selection and cell cloning, 26 hygromycin B-resistant cell clones were screened for conditional eIF4G cleavage. Three of them, 2A5, 2A7, and 2A24, were clearly positive in this analysis. These clones displayed highly regulated expression of poliovirus 2Apro, but all three clones showed some basal level of eIF4G cleavage in the presence of tet. The 2A7 clone was further subcloned by limit dilution. The different clones thus obtained exhibited a phenotype similar to the parental one. The clone designated 2A7d was used in all subsequent experiments. In the following, the terms 2A7d-On and 2A7d-Off refer, respectively, to HeLa 2A7d cells that either were or were not induced to express poliovirus 2Apro. Cell clones transfected with vectors pTRE and pTK-Hyg were also isolated by following the same protocol in order to use them as controls.
A number of alterations in cellular morphology were apparent upon induction of 2Apro synthesis in all positive clones. Phase-contrast microscopy showed that most 2A7d-Off cells growing in the presence of tet had a normal phenotype; only a few cells appeared rounded and were nonadherent (Fig. 1A, left). The number of HeLa cells which were rounded and refringent rapidly increased from 28 h after tet depletion (Fig. 1A, middle). Thus, 46 h after induction, less than 1% of 2A7d-On cells remained attached to the plate (Fig. 1A, right). The few remaining cells that grew in the absence of tet seem to have lost the capacity to express poliovirus 2Apro (results not shown). The rounded and nonadherent cells remained viable for several hours before they became permeable to trypan blue (results not shown). Immunofluorescence microscopy of doubly stained cells with antitubulin and anti-2Apro antibodies showed that 2Apro expression provoked the collapse of the microtubule network, which occurs in poliovirus-infected cells (36), and almost 100% of the cells synthesized 2Apro (Fig. 1B).
FIG. 1.
tet regulates poliovirus 2Apro expression in a stable HeLa cell clone: morphological alteration. (A) Phase-contrast microscopy of HeLa 2A7d cells at different times after induction of 2Apro. OFF, uninduced 2A7d cells; ON28, 2A7d-On cells 28 h postinduction; ON46, 2A7d-On cells 46 h postinduction. (B) Immunofluorescence microscopy of 2Apro-expressing cells. 2A7d cells were fixed and doubly stained with anti-2A antibodies (α-2A; rabbit polyclonal antiserum) and with a mouse monoclonal antibody against tubulin (α-tub) after 25 h in the presence (off) or absence (on) of tet. Immunofluorescence microscopy of HeLa cells was carried out as described previously (1). (C) Northern blot of mRNAs isolated at 24 h postinduction from 2A7d cells that express (on) or do not express (off) poliovirus 2Apro hybridized with a 2A 32P-labeled probe. As a loading control, the same samples were hybridized to a human β-actin mRNA probe. (D) HeLa 2A7d cells grown in 24-well dishes were treated with the indicated concentrations of tet. tet was present (+) or removed (−) from the medium by extensive washing to induce 2Apro expression as described in Materials and Methods. Cells were harvested at 24 h postinduction, extracts were analyzed by SDS-PAGE, and the gel was immunoblotted with anti-eIF4GI antiserum. CP, cleavage products. Extracts from Hela cells infected with poliovirus (p) or not infected (m) were used as controls for eIF4G cleavage.
Expression of poliovirus 2Apro mRNA synthesis, as well as 2Apro activity in clone 2A7d, took place and was strongly regulated (Fig. 1C and D). A major band of mRNA that specifically hybridized with a 2Apro probe appeared upon induction (Fig. 1C). As a loading control the same samples were hybridized with an actin mRNA probe (Fig. 1C). A concentration of 2 μg of tet/ml was employed to select clones, but induction of 2Apro was much more rapid when cells were grown in the presence of 0.02 μg of tet/ml prior to removal. This treatment did not increase the basal expression level of 2Apro, as judged by eIF4G cleavage (Fig. 1D).
Effects of poliovirus 2Apro on cellular protein synthesis.
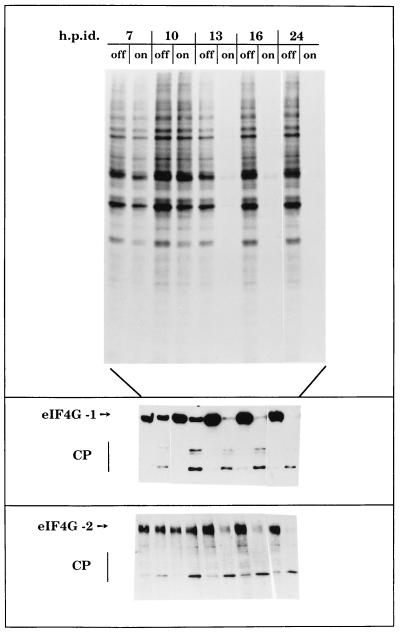
The action of poliovirus 2Apro on translation was examined in parallel with eIF4G cleavage in 2A7d cells. Inhibition of protein synthesis was consistently observed between 10 and 13 h after induction of 2Apro (Fig. 2). This inhibition correlated with eIF4G cleavage, and almost no intact eIF4G was detectable at the times when translation inhibition was already apparent (Fig. 2). The functional characterization of an eIF4G homologue designated eIF4GII has been recently described (23). Both forms of eIF4G were extensively cleaved in 2A7d-On cells, with similar kinetics (Fig. 2). The decrease of eIF4G fragments observed at late times of induction probably reflects the activity of endogenous cellular proteases. Despite the strong blockade of protein synthesis and complete cleavage of eIF4GI and eIF4GII observed, an appreciable amount of protein synthesis remained, even 24 h after 2Apro induction (estimated by densitometric analysis as 5 to 10% of control levels).
FIG. 2.
Kinetics of protein synthesis and eIF4G cleavage at different times after induction of poliovirus 2Apro in HeLa 2A7d cells. (Top) Kinetics of protein synthesis. HeLa 2A7d cells grown in 24-well dishes were extensively washed to remove tet to induce 2Apro expression. Cell monolayers were then labeled with 25 μCi of [35S]methionine/ml for 1 h at the indicated times (hours) postinduction (h.p.id.). Cell extracts were analyzed by SDS–15% PAGE. Aliquots of the same samples were also analyzed by immunoblotting. The blots were incubated with anti-eIF4GI antiserum (eIF4GI) (middle) or with an antiserum against the C-terminal domain of eIF4GII (eIF4GII; bottom). CP, cleavage products.
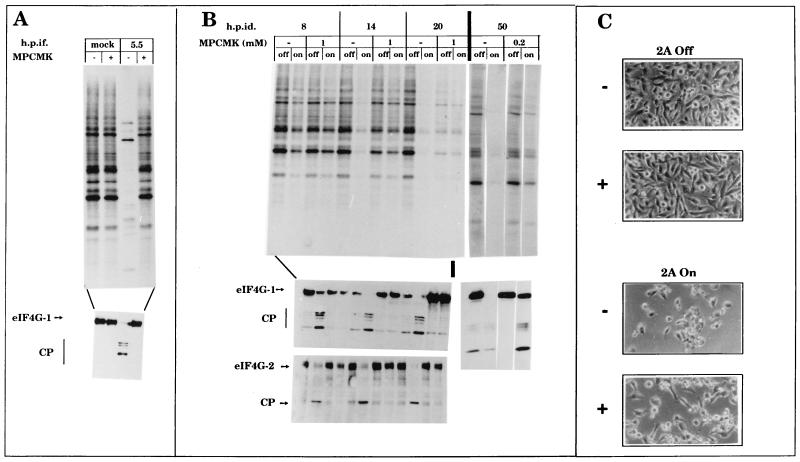
The elastase-specific inhibitor methoxysuccinyl-Ala-Ala-Pro-Val-chloromethylketone (MPCMK) potently blocks poliovirus 2Apro both in vivo and in cell-free systems, reducing the production of infectious virus particles (42). Figure 3A shows that a concentration of 1 mM MPCMK abrogates the inhibition of host translation and inhibits both the synthesis of viral proteins and eIF4G cleavage in poliovirus-infected HeLa cells. MPCMK prevented cleavage of both forms of eIF4G in 2A7d-On cells even after 20 h of induction (Fig. 3B). However, after prolonged treatment, MPCMK was toxic for cells, since neither 2A7d-Off nor 2A7d-On cells synthesized proteins 24 h after induction in the presence of 1 mM MPCMK. Lowering the concentration of MPCMK to 0.2 mM partially prevented both eIF4GI cleavage and inhibition of translation even at 50 h of induction (Fig. 3B). Therefore, there was a good correlation between inhibition of translation, eIF4G cleavage, and 2Apro activity. However, it was not proven whether MPCMK prevents the inhibition of translation by directly blocking cleavage of eIF4G or by inhibiting the effects of 2Apro on other cellular substrates. Surprisingly, MPCMK used at nontoxic concentrations (0.2 mM) delayed by a few hours, but did not prevent, the death of 2A7d-On cells (Fig. 3C), despite the fact that eIF4G remained partially intact (Fig. 3B, bottom). Therefore, it is possible to separate eIF4G cleavage from cell death induced by poliovirus 2Apro.
FIG. 3.
Effect of MPCMK on the shutoff of protein synthesis and eIF4G cleavage in poliovirus-infected cells and in 2A7d-On cells. (A) HeLa 2A7d-Off cells grown in 24-well dishes were infected with poliovirus at a MOI of 10 PFU/cell and treated with 1 mM MPCMK after virus adsorption (+) or not treated (−). At 5.5 h postinfection cells were labeled with 25 μCi of [35S]methionine/ml for 1 h and extracts were analyzed by SDS–15% PAGE (top). Aliquots of these samples were also assayed by immunoblotting with anti-eIF4GI antiserum (bottom). CP, cleavage products. (B) Poliovirus 2Apro was induced in 2A7d cells grown in 24-well dishes by tet removal. MPCMK was added (+) or not added (−) to the culture medium from the beginning of 2Apro induction. At the indicated times (hours) postinduction (h.p.id.), cells were labeled with 25 μCi of [35S]methionine/ml for 1 h. Cell extracts were analyzed by SDS–15% PAGE (top). Aliquots were also assayed by immunoblotting using anti-eIF4GI antiserum (middle) or anti-eIF4GII antiserum (bottom). (C) Phase-contrast microscopy at 50 h postinduction of 2A7d-Off and 2A7d-On cells treated (+) or not treated (−) with 0.2 mM MPCMK during the time of induction.
2Apro allows picornavirus translation in 2A7d-infected cells.
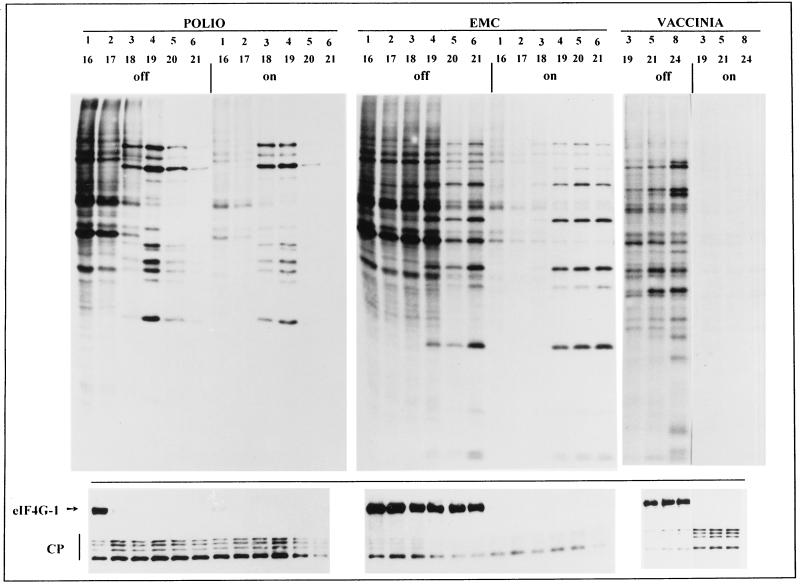
Once it was found that inhibition of host translation takes place in 2A7d-On cells, it was of interest to analyze whether 2Apro facilitates IRES-driven translation; if so the presence of this protease from the beginning of the infection would be beneficial for picornavirus gene expression. To test this possibility, two picornaviruses were chosen, poliovirus and EMCV. These viruses differ in the type of IRES present in the 5′ leader sequence (9, 26). Additionally, poliovirus cleaves eIF4G during infection, while EMCV does not (44). Moreover, cardiovirus EMCV 2A protein has no sequence homology with the enterovirus and rhinovirus 2Apro, lacking a protease consensus motif and detectable proteolytic activity (37). 2A7d HeLa cells were infected with poliovirus or EMCV 14 h after induction of 2Apro and pulse-labeled with [35S]methionine at different times postinfection, and labeled proteins were analyzed by SDS-PAGE (Fig. 4). The levels of viral protein synthesis in 2A7d-On and 2A7d-Off cells were similar, even when virus infection started 24 h after induction (results not shown). For poliovirus at the MOI used, mature viral proteins were already detectable 3 h postinfection in 2A7d-On cells and no viral proteins were made after 5 h postinfection, whereas the peak of poliovirus protein synthesis occurred between 4 and 6 h postinfection in 2A7d-Off cells (Fig. 4). For EMCV, cellular translation was inhibited earlier, such that viral proteins were exclusively synthesized in 2A7d-On cells (Fig. 4). Therefore, picornavirus proteins were synthesized in 2Apro-expressing cells. By contrast, vaccinia virus replication was powerfully inhibited in 2A7d-On cells, in good agreement with the result showing that poliovirus 2Apro interferes with vaccinia virus translation (19). Although picornavirus translation in 2A7d cells was allowed, there was no stimulation of the total amount of viral protein synthesized, resulting in levels of virus production similar to those in 2A7d-Off cells. In addition, these results show that 2A7d-On cells synthesize picornavirus proteins efficiently at a time when cellular translation is completely blocked and hence that the metabolic energy, ribosomes, and other components involved in translation are not limiting.
FIG. 4.
Kinetics of protein synthesis in 2A7d-Off and 2A7d-On cells infected with poliovirus, EMCV, or vaccinia virus. HeLa 2A7d cells grown in 24-well dishes were induced (on) or not induced (off) to synthesize poliovirus 2Apro by tet removal. After 15 h, cells were infected with poliovirus (10 PFU/cell), with EMCV (8 PFU/cell), or with vaccinia virus (5 PFU/cell). Sixteen hours postinduction coincides with 1 h postinfection. Protein synthesis was estimated by pulse-labeling the cell monolayers with 25 μCi of [35S]Met-Cys/ml as described in Materials and Methods. The same extracts were analyzed in parallel by immunoblotting using anti-eIF4GI antiserum as indicated for Fig. 1 (bottom). CP, eIF4GI cleavage products.
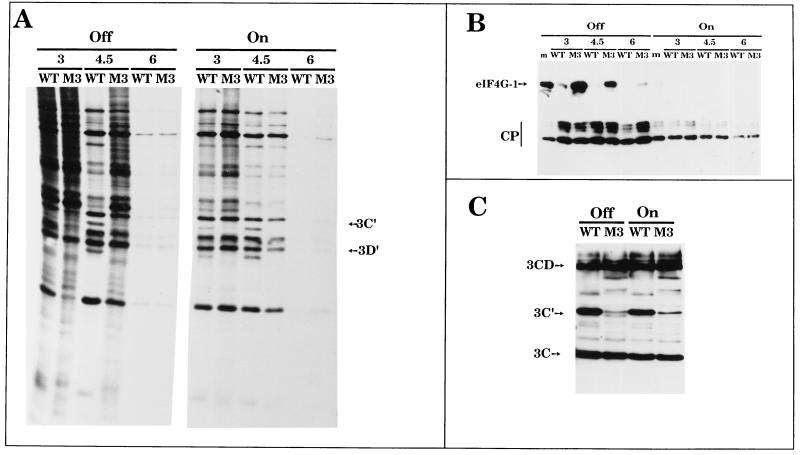
It was of interest to determine whether 2Apro expression in 2A7d-On cells could complement a poliovirus 2A-deficient mutant. M3 (2Apro; S66F) is a poliovirus variant previously isolated and characterized in our laboratory (8, 64). The point mutation S66F in 2Apro attenuates virus mRNA translation, delays the shutoff of host protein synthesis, and greatly reduces virus growth and the cytopathic effect (8). The mutant shows no transactivation of an IRES-containing mRNA and defective cleavage of 3CD by 2Apro to yield 3C′ and 3D′. Efficient cleavage of eIF4G at late times postinfection occurs in M3-infected HeLa cells, although the kinetics of this cleavage are slower than those of wt poliovirus infection (8). M3 protein synthesis was enhanced in 2A7d-On cells at different times of infection, recovering the phenotypic translation of wt poliovirus (Fig. 5A). In addition, M3 was able to induce the cytopathic effect in 2A7d-On cells (results not shown). Cleavage of eIF4GI (Fig. 5B) and eIF4GII (results not shown) correlated with the inhibition of cellular translation. However, close inspection of the pattern of viral proteins showed that 3C′ and 3D′ were still absent in M3-infected 2A7d-On cells, indicating that 2Apro expressed in trans in these cells cannot complement this defect or that the mutated M3 2Apro is transdominant in this reaction (Fig. 5C). Alternatively the levels of 2Apro in 2A7d-On cells may not be sufficient to cleave 3CD.
FIG. 5.
Complementation of a poliovirus 2Apro variant (M3) in 2A7d-On cells. HeLa 2A7d cells grown in 24-well dishes were induced to synthesize poliovirus 2Apro (on) or not induced (off). After 13.5 h of induction, 2A7d cells were infected with wt virus or with poliovirus mutant M3 (10 PFU/cell). At the indicated times postinfection (hours), cells were labeled for 1 h with [35S]Met-Cys and samples were analyzed by SDS-PAGE (A) or by immunoblotting using the corresponding specific antisera for eIF4GI (B) and poliovirus 3C (C) as described in Materials and Methods. m, mock-infected cells. The poliovirus obtained by transfection of HeLa cells with pT7XLD was used (8). The positions of 3C′, 3D′, 3CD, 3C, and eIF4GI and its cleavage products (CP) are indicated.
To determine if 2Apro could rescue the growth defects of M3, a plaque-forming assay was carried out. Surprisingly, despite the rescue of viral translation by 2Apro expression, the production of mutant virus could not be fully restored. The titers obtained with M3 were considerably lower than those of wt poliovirus, both in 2A7d-Off and 2A7d-On cells (results not shown). The plaques obtained with M3 maintained their characteristic small-size phenotype, even in 2A7d-On cells. This finding raises the possibility that 2A7d-On cells complement the defects that M3 2Apro has on translation but cannot complement one or more effects of M3 2Apro on other steps of virus replication, such as viral RNA synthesis (38, 43, 71). The fact that the levels of 3C′ (and presumably 3D′) are lower in M3-infected 2A7d-On cells further points to a function of those proteins during the poliovirus life cycle (66).
Translation of mRNAs bearing an IRES leader sequence in 2A7d cells.
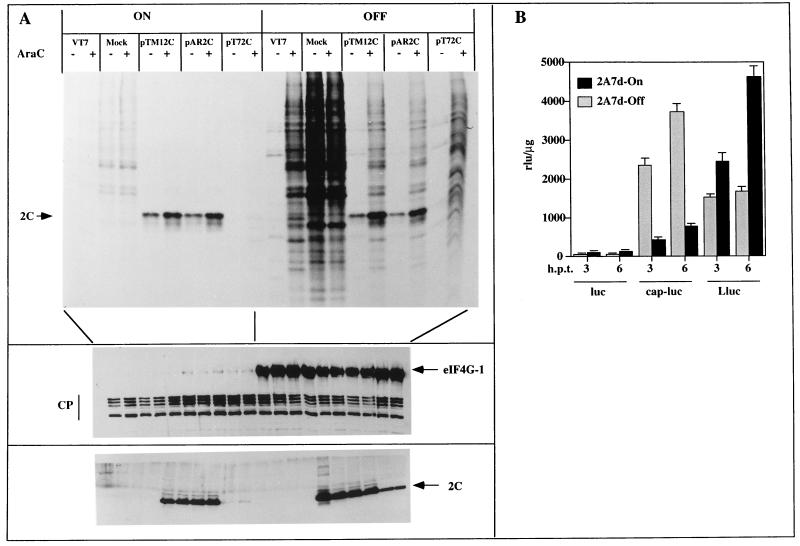
Poliovirus 2Apro causes two opposite effects on gene expression: one is the blockade of cellular gene expression at both the transcriptional and translational levels, and the other is the stimulation of translation of poliovirus and other picornavirus mRNAs (9). To test the effects of eIF4G cleavage and 2Apro on the translation of mRNAs bearing a picornavirus leader sequence, 2A7d-On and 2A7d-Off cells were transfected with plasmids pTM1.2C, pAR.2C, and pT7.2C (1, 33). All these plasmids carry the poliovirus 2C gene under a T7 promoter but are preceded by the EMCV 5′-untranslated region (UTR) (pTM1.2C), by the poliovirus 5′-UTR (pAR.2C), or by no further regulatory sequences (pT7.2C). Expression of 2C was triggered by infection with a recombinant vaccinia virus bearing the T7 RNA polymerase, just before transfection of these plasmids (20). A protein band corresponding to poliovirus 2C was apparent when both 2A7d-Off and 2A7d-On cells were pulse-labeled at 16 h postinfection and transfected with pTM1.2C or pAR.2C, but 2C synthesis was not observed with pT7.2C (Fig. 6A, top). Immunoblot analysis with anti-2C antibody shows that the amounts of 2C protein accumulated in 2A7d-On and 2A7d-Off cells were similar when the cells were transfected with pTM1.2C and pAR.2C but that the amount was very much reduced in 2A7d-On cells transfected with pT7.2C (Fig. 6A, bottom). Poliovirus 2Apro blocked host and vaccinia virus translation when eIF4G was cleaved in 2A7d-On cells, while the synthesis of poliovirus 2C was not affected when a picornavirus IRES sequence preceded the 2C gene. Similar results were found when vaccinia virus replication was inhibited with araC, although in this case the levels of poliovirus 2C synthesis achieved were higher, both in 2A7d-On and in 2A7d-Off cells (Fig. 6A). The inhibitory effect of 2Apro on vaccinia virus and consequently on T7 RNA polymerase synthesis may compensate for the potential stimulation of IRES-driven translation.
FIG. 6.
Effect of eIF4G cleavage on the translation of mRNAs bearing different leader sequences. (A) After 8 h of induction of 2A expression, HeLa 2A7d cells were infected with recombinant vaccinia virus VT7 (5 PFU/cell). Cells were transfected after 45 min of VT7 adsorption with plasmid pTM1.2C, pAR.2C, or pT7.2C. AraC (40 μg/ml) was present (+) or not present (−) during transfection. At 16 h postinfection cells were labeled with [35S]Met-Cys for 1 h as described in Materials and Methods. Proteins were analyzed by SDS-PAGE (top) or by immunoblotting. Blots were incubated with anti-eIF4GI antiserum (middle) or with poliovirus anti-2C antiserum (bottom). Positions of poliovirus 2C and eIF4GI and its cleavage products (CP) are indicated. (B) 2A7d cells grown in 35-mm-diameter dishes were induced to express 2Apro for 15 h, and then cells were transfected with 1-μg portions of different mRNAs encoding luciferase under different leader sequences: uncapped (luc), capped (cap-luc), or uncapped luciferase mRNA bearing the 5′ leader sequence of poliovirus (Lluc). After 3 and 6 h posttransfection, cells were harvested and analyzed for luciferase activity as described previously (66). rlu, relative luciferase units.
To avoid the possible inhibitory effect on T7-RNA polymerase by 2Apro, we analyzed the effect of this protease on the translation of different mRNAs that were directly transfected into 2A7d cells. These mRNAs encode luciferase as the reporter gene product. Some are uncapped (luc), while others bear a cap structure at the 5′ end (capped luc [cap-luc]) or the poliovirus IRES leader luc (Lluc) sequence. These mRNAs were introduced by Lipofectin transfection 14 h postinduction, when eIF4G had already been cleaved, and cells were harvested 3 h later to assay for luciferase activity. Figure 6B shows that poliovirus 2Apro caused about a threefold increase in luciferase activity when the IRES Lluc mRNAs were analyzed, while a sixfold inhibition was found with cap-luc mRNAs (Fig. 6B). Translation of uncapped luc mRNAs (luc) was very poor, both in 2A7d-Off and 2A7d-On cells. These results indicate that enhancement of IRES-driven translation and inhibition of cap-dependent translation occur in 2A7d-On cells in the absence of any other poliovirus- or vaccinia virus-encoded protein.
DISCUSSION
eIF4G cleavage and inhibition of cellular translation; is there a cause-and-effect relationship?
eIF4G is a key factor in the regulation of the initiation of translation in eukaryotes (27). This polypeptide orchestrates the interaction of different macromolecules to promote the binding of the mRNA to the small ribosomal subunit to form the 48S initiation complex (56). Few experimental approaches have been directed to analyzing the consequences of eIF4G cleavage by poliovirus 2Apro for ongoing cellular translation in cultured cells.
Our present findings indicate that inducible expression of poliovirus 2Apro in HeLa cells leads to efficient cleavage of both forms of eIF4G and to inhibition of cellular translation. Previous findings have illustrated that a high level of poliovirus 2Apro synthesis induces inhibition of ongoing cellular translation but that there are also conditions where eIF4G cleavage can be separated from this inhibition. When poliovirus 2Apro is synthesized at high levels using the VT7 system, an inhibition of host protein synthesis similar to that observed in 2A7d-On cells occurs (2, 3, 19). However, the synthesis of vaccinia virus proteins makes it difficult to evaluate the mechanism of this inhibition because vaccinia virus itself induces a shutoff of cellular translation. Moreover, vaccinia virus complements 2Apro function by a still-unknown mechanism (50). Injection of mRNAs encoding coxsackievirus 2Apro in Xenopus oocytes leads to efficient cleavage of eIF4G and blocks the translation of exogenous globin mRNA, while ongoing cellular translation is much less affected (15). This finding agrees well with a different approach based on the internalization of hybrid proteins bearing poliovirus 2Apro into HeLa cells (46, 47). No intact eIF4G is detected several hours after treatment with hybrid toxins, but ongoing translation takes place at substantial levels. In contrast, translation of a newly made luc mRNA was drastically blocked. These systems provided evidence that the inhibition of ongoing protein synthesis could be separated from eIF4G cleavage. The hypothesis that eIF4G participates in the very first initiation event and may not be required for further reinitiation rounds helps to explain most of the results obtained, both with cell-free systems and intact cells (49). The possibility that the inhibition of translation by 2Apro in 2A7d cells is the consequence of cleavage of eIF4G and other cellular substrates is still open. Cleavage of these putative cellular substrates would not occur with hybrid toxins bearing poliovirus 2Apro or upon injection of coxsackievirus 2Apro mRNAs into Xenopus oocytes. However, our present results show that poliovirus 2Apro alone potently blocks ongoing cellular translation and that this inhibition correlates with eIF4G degradation.
Although hybrid proteins containing poliovirus 2Apro provoke an extensive cleavage of eIF4G, 2Apro is present inside cells at low levels in a form that differs from that of genuine 2Apro. This system has been useful to test the repercussions of eIF4G cleavage on gene expression but not to analyze the consequences of genuine poliovirus 2Apro synthesis for cellular metabolism and morphology. Two important differences between the present results and those obtained previously should be noted: (i) ongoing protein synthesis was inhibited more than 90% in 2A7d-On cells, while in cells permeabilized to hybrid proteins about 50% of cellular translation continued for up to 24 h (47); (ii) 1 day after tet removal, 2A7d-On cells begin to round up and lose adherence, while cells permeabilized to poliovirus 2Apro do not die; instead, they recover their normal phenotype. The low level of 2Apro inside cells may be sufficient to induce eIF4G cleavage (11, 68) but not to cleave other putative cellular substrates that could affect cellular metabolism and morphology. Moreover, the presence of fusion proteins ligated to 2Apro could interfere with its intracellular localization and/or functioning.
The presence of two forms of eIF4G and their different kinetics of cleavage during poliovirus infection (24) may help to explain previous findings that eIF4GI cleavage was dissociated from the inhibition of cellular translation (10, 29, 47, 52). However, it is evident that the strong inhibition of translation and morphological alterations observed in 2A7d-On cells imply that 2Apro participates in events other than eIF4G cleavage. In fact, these phenomena can be separated using the 2Apro inhibitor MPCMK. Under these conditions, cleavage of both forms of eIF4G is prevented by MPCMK but cell death still occurs in 2A7d-On cells. It is probable that there are additional substrates for this protease and/or that 2Apro possesses other functions in addition to its proteolytic activity that are involved in cell killing. The 2A7d cell clone could be very useful for identifying such substrates or activities; for example, the analysis of additional 2Apro inhibitors could distinguish between different activities present in this protease.
Poliovirus 2Apro has a dual role in gene expression.
Poliovirus 2Apro has two opposite effects on gene expression. This protease depresses the expression of cellular genes but, on the other hand, augments the synthesis of picornavirus proteins (9). Inhibition of cellular translation may benefit virus replication in different ways, for instance, by precluding competition with host mRNAs for the translation machinery or by preventing the establishment of an interferon response. Both effects of 2Apro on gene expression may be achieved by proteolytic degradation of eIF4G. The data shown in this work indicate a correlation between eIF4G degradation and both inhibition of cellular protein synthesis and stimulation of picornavirus translation. If additional events are necessary for both processes to take place in the infected cells, these events could be mediated by 2Apro alone.
The poliovirus IRES sequence is efficiently recognized by the HeLa translation machinery, even in the absence of 2Apro. A number of cellular proteins have been implicated in this recognition (9, 16). Therefore, 2Apro is not an absolute requirement for IRES-driven translation, but the protease facilitates the functioning of several picornavirus IRESs with different secondary structures (54). Several hypotheses can be put forward to account for 2Apro-induced transactivation of picornavirus IRES. One is that this protease binds directly to the IRES stimulating its translation. Genetic evidence for a direct interaction between 2Apro and the poliovirus IRES has been provided (39). Another possibility is that IRES stimulation is a consequence of 2Apro proteolytic activity, either by inactivation of an inhibitor of IRES function or by the generation upon proteolysis of an activator of IRES activity or both (8, 54). The C-terminal fragment of eIF4G is a potential candidate for participating in this function. The activation of the picornavirus IRES by 2Apro seems to be independent of the loss of competition from capped mRNAs. IRES activity was not stimulated when cap-dependent protein synthesis was blocked by the translational repressor 4E-BP2 in the absence of any viral protease (54). Furthermore, genetic evidence obtained using poliovirus 2Apro mutants has shown that IRES-dependent translation is not stimulated by cleaved eIF4G per se but rather requires the presence of active 2Apro (8, 66). Moreover, stimulation of IRES activity was not achieved by the coexpression of the C-terminal fragment of eIF4G (54). Therefore, in addition to eIF4G cleavage, other 2Apro activities are necessary for the transactivation of an IRES sequence. Thus, it is possible that 2Apro transactivates IRES by direct binding to this element as previously suggested (39, 66) or by inducing the cleavage of another cellular protein. If so, cleavage of this unknown substrate also occurs in 2A7d-On cells.
Usefulness of the HeLa cell clone that inducibly expresses poliovirus 2Apro.
The HeLa 2A7d cell clone described in this work provides a good model system for studying the different functions of poliovirus 2Apro. As documented in this work, the clone can be used to analyze the molecular details that lead to inhibition of host translation, cell death, and the transactivation of mRNAs bearing an IRES sequence. It may also be used to rescue defective poliovirus 2Apro variants. It is even possible that this HeLa cell line may complement other picornavirus mutants. In addition, 2A7d cells can be used to investigate the action of known 2Apro inhibitors in vivo or to screen new compounds with antiprotease activity. This system has the advantage that simply removing tet from the culture medium induces cell death in a few hours. Potential antiviral compounds could be easily identified as inhibitors of this cytotoxicity. This assay may constitute an easy and rapid screening method for new antiviral agents targeted not only to poliovirus 2Apro but also to any toxic virus protein of therapeutic interest.
ACKNOWLEDGMENTS
The expert technical assistance of M. A. Sanz is acknowledged. We thank N. Sonenberg for generously providing eIF4GII antiserum.
A.B. is the holder of a CSIC postdoctoral fellowship. DGICYT project number PB94-0148 and an institutional grant to the CBM by Fundación Ramón Areces are acknowledged for financial support.
REFERENCES
- 1.Aldabe R, Carrasco L. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem Biophys Res Commun. 1995;206:64–76. doi: 10.1006/bbrc.1995.1010. [DOI] [PubMed] [Google Scholar]
- 2.Aldabe R, Feduchi E, Novoa I, Carrasco L. Expression of poliovirus 2Apro in mammalian cells: effects on translation. FEBS Lett. 1995;377:1–5. doi: 10.1016/0014-5793(95)01269-9. [DOI] [PubMed] [Google Scholar]
- 3.Aldabe R, Feduchi E, Novoa I, Carrasco L. Efficient cleavage of p220 by poliovirus 2Apro expression in mammalian cells: effects on vaccinia virus. Biochem Biophys Res Commun. 1995;215:928–936. doi: 10.1006/bbrc.1995.2553. [DOI] [PubMed] [Google Scholar]
- 4.Alvey J C, Wyckoff E E, Yu S F, Lloyd R, Ehrenfeld E. cis and trans cleavage activities of poliovirus 2A protease expressed in Escherichia coli. J Virol. 1991;65:6077–6083. doi: 10.1128/jvi.65.11.6077-6083.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aranda M, Maule A. Virus-induced host gene shutoff in animals and plants. Virology. 1998;243:261–267. doi: 10.1006/viro.1998.9032. [DOI] [PubMed] [Google Scholar]
- 6.Barco A, Carrasco L. A human virus protein, poliovirus protein 2BC, induces membrane proliferation and blocks the exocytic pathway in the yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3349–3364. doi: 10.1002/j.1460-2075.1995.tb07341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barco A, Carrasco L. Poliovirus 2Apro expression inhibits growth of yeast cells. FEBS Lett. 1995;371:4–8. doi: 10.1016/0014-5793(95)00841-v. [DOI] [PubMed] [Google Scholar]
- 8.Barco A, Ventoso I, Carrasco L. The yeast Saccharomyces cerevisiae as a genetic system for obtaining variants of poliovirus protease 2A. J Biol Chem. 1997;272:12683–12691. doi: 10.1074/jbc.272.19.12683. [DOI] [PubMed] [Google Scholar]
- 9.Belsham G J, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonneau A M, Sonenberg N. Proteolysis of the p220 component of the cap-binding protein complex is not sufficient for complete inhibition of host cell protein synthesis after poliovirus infection. J Virol. 1987;61:986–991. doi: 10.1128/jvi.61.4.986-991.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bovee M L, Marissen W E, Zamora M, Lloyd R E. The predominant eIF-4G-specific cleavage activity in poliovirus-infected HeLa cells is distinct from 2A protease. Virology. 1998;245:229–240. doi: 10.1006/viro.1998.9171. [DOI] [PubMed] [Google Scholar]
- 12.Clark M, Hämemerle T, Wimmer E, Dasgupta A. Poliovirus proteinase 3C converts an active form of transcription factor IIIC to an inactive form: a mechanism for inhibition of host cell polymerase III transcription by poliovirus. EMBO J. 1991;10:2941–2947. doi: 10.1002/j.1460-2075.1991.tb07844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark M E, Lieberman P M, Berk A J, Dasgupta A. Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Mol Cell Biol. 1993;13:1232–1237. doi: 10.1128/mcb.13.2.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies M V, Pelletier J, Meerovitch K, Sonenberg N, Kaufman R J. The effect of poliovirus proteinase 2Apro on cellular metabolism. J Biol Chem. 1991;266:14714–14720. [PubMed] [Google Scholar]
- 15.Donnelly M L L, Gani D, Flint M, Monaghan S, Ryan M D. The cleavage activities of aphthovirus and cardiovirus 2A proteins. J Gen Virol. 1997;78:13–21. doi: 10.1099/0022-1317-78-1-13. [DOI] [PubMed] [Google Scholar]
- 16.Ehrenfeld E. Initiation of translation by picornavirus RNAs. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 549–573. [Google Scholar]
- 17.Etchison D, Milburn S C, Edery I, Sonenberg N, Hershey J W. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- 18.Favaloro J, Treisman R, Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65:718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- 19.Feduchi E, Aldabe R, Novoa I, Carrasco L. Effects of poliovirus 2Apro on vaccinia virus gene expression. Eur J Biochem. 1995;234:849–854. doi: 10.1111/j.1432-1033.1995.849_a.x. [DOI] [PubMed] [Google Scholar]
- 20.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gingras A C, Svitkin Y, Belsham G J, Pause A, Sonenberg N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc Natl Acad Sci USA. 1996;93:5578–5583. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gradi A, Svitkin Y V, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hambidge S J, Sarnow P. Translational enhancement of the poliovirus 5′ noncoding region mediated by virus-encoded polypeptide 2A. Proc Natl Acad Sci USA. 1992;89:10272–10276. doi: 10.1073/pnas.89.21.10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hellen C U T, Wimmer E. Translation of encephalomyocarditis virus RNA by internal ribosomal entry. Curr Top Microbiol Immunol. 1995;203:31–63. doi: 10.1007/978-3-642-79663-0_2. [DOI] [PubMed] [Google Scholar]
- 27.Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 28.Iizuka N, Chen C, Yang Q, Johannes G, Sarnow P. Cap-independent translation and internal initiation of translation in eukaryotic cellular mRNA molecules. Curr Top Microbiol Immunol. 1995;203:155–177. doi: 10.1007/978-3-642-79663-0_8. [DOI] [PubMed] [Google Scholar]
- 29.Irurzun A, Sanchez-Palomino S, Novoa I, Carrasco L. Monensin and nigericin prevent the inhibition of host translation by poliovirus, without affecting p220 cleavage. J Virol. 1995;69:7453–7460. doi: 10.1128/jvi.69.12.7453-7460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jewell J E, Ball L A, Rueckert R. Limited expression of poliovirus by vaccinia virus recombinants due to inhibition of the vector by proteinase 2A. J Virol. 1990;64:1388–1393. doi: 10.1128/jvi.64.3.1388-1393.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klump H, Auer H, Liebig H D, Kuechler E, Skern T. Proteolytically active 2A proteinase of human rhinovirus 2 is toxic for Saccharomyces cerevisiae but does not cleave the homologues of eIF4G in vivo or in vitro. Virology. 1996;220:109–118. doi: 10.1006/viro.1996.0291. [DOI] [PubMed] [Google Scholar]
- 32.Krausslich H G, Wimmer E. Viral proteinases. Annu Rev Biochem. 1988;57:701–754. doi: 10.1146/annurev.bi.57.070188.003413. [DOI] [PubMed] [Google Scholar]
- 33.Lama J, Guinea R, Martinez-Abarca F, Carrasco L. Cloning and inducible synthesis of poliovirus nonstructural proteins. Gene. 1992;117:185–192. doi: 10.1016/0378-1119(92)90728-8. [DOI] [PubMed] [Google Scholar]
- 34.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases—implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 35.Lee C K, Wimmer E. Proteolytic processing of poliovirus polyprotein: elimination of 2Apro-mediated, alternative cleavage of polypeptide 3CD by in vitro mutagenesis. Virology. 1988;166:405–414. doi: 10.1016/0042-6822(88)90511-9. [DOI] [PubMed] [Google Scholar]
- 36.Lenk R, Penman S. The cytoskeletal framework and poliovirus metabolism. Cell. 1979;16:289–301. doi: 10.1016/0092-8674(79)90006-0. [DOI] [PubMed] [Google Scholar]
- 37.Lloyd R E, Grubman M J, Ehrenfeld E. Relationship of p220 cleavage during picornavirus infection to 2A proteinase sequencing. J Virol. 1988;62:4216–4223. doi: 10.1128/jvi.62.11.4216-4223.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu H H, Li X Y, Cuconati A, Wimmer E. Analysis of picornavirus 2Apro proteins: separation of proteinase from translation and replication functions. J Virol. 1995;69:7445–7452. doi: 10.1128/jvi.69.12.7445-7452.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macadam A J, Ferguson G, Fleming T, Stone D M, Almond J W, Minor P D. Role for poliovirus protease 2A in cap independent translation. EMBO J. 1994;13:924–927. doi: 10.1002/j.1460-2075.1994.tb06336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macejak D, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 41.Merrick W C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 31–69. [Google Scholar]
- 42.Molla A, Hellen C U T, Wimmer E. Inhibition of proteolytic activity of poliovirus and rhinovirus 2A proteinases by elastase-specific inhibitors. J Virol. 1993;67:4688–4695. doi: 10.1128/jvi.67.8.4688-4695.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molla A, Paul A V, Schmid M, Jang S K, Wimmer E. Studies on dicistronic polioviruses implicate viral proteinase 2Apro in RNA replication. Virology. 1993;196:739–747. doi: 10.1006/viro.1993.1531. [DOI] [PubMed] [Google Scholar]
- 44.Mosenkis J, Daniels-McQueen S, Janovec S, Duncan R, Hershey J W, Grifo J A, Merrick W C, Thach R E. Shutoff of host translation by encephalomyocarditis virus infection does not involve cleavage of the eucaryotic initiation factor 4F polypeptide that accompanies poliovirus infection. J Virol. 1985;54:643–645. doi: 10.1128/jvi.54.2.643-645.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muñoz A, Alonso M A, Carrasco L. Synthesis of heat-shock proteins in HeLa cells: inhibition by virus infection. Virology. 1984;137:150–159. doi: 10.1016/0042-6822(84)90018-7. [DOI] [PubMed] [Google Scholar]
- 46.Novoa I, Carrasco L. Cleavage of eukaryotic translation initiation factor 4G by exogenously added hybrid proteins containing poliovirus 2Apro in HeLa cells: effects on gene expression. Mol Cell Biol. 1999;19:2445–2454. doi: 10.1128/mcb.19.4.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novoa I, Cotten M, Carrasco L. Hybrid proteins between Pseudomonas aeruginosa exotoxin A and poliovirus 2Apro cleave p220 in HeLa cells. J Virol. 1996;70:3319–3324. doi: 10.1128/jvi.70.5.3319-3324.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Novoa I, Feduchi E, Carrasco L. Hybrid proteins between Pseudomonas exotoxin A and poliovirus 2Apro. FEBS Lett. 1994;355:45–48. doi: 10.1016/0014-5793(94)01157-5. [DOI] [PubMed] [Google Scholar]
- 49.Novoa I, Martínez-Abarca F, Fortes P, Ortín J, Carrasco L. Cleavage of p220 by purified poliovirus 2Apro in cell-free systems. Effects on translation of capped and uncapped mRNAs. Biochemistry. 1997;36:7802–7809. doi: 10.1021/bi9631172. [DOI] [PubMed] [Google Scholar]
- 50.Pal-Ghosh R, Morrow C D. A poliovirus minireplicon containing an inactive 2A proteinase is expressed in vaccinia virus-infected cells. J Virol. 1993;67:4621–4629. doi: 10.1128/jvi.67.8.4621-4629.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palmenberg A. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- 52.Perez L, Carrasco L. Lack of direct correlation between p220 cleavage and the shut-off of host translation after poliovirus infection. Virology. 1992;189:178–186. doi: 10.1016/0042-6822(92)90693-j. [DOI] [PubMed] [Google Scholar]
- 53.Rhoads R E, Lamphear B J. Cap-independent translation of heat shock messenger RNAs. Curr Top Microbiol Immunol. 1995;203:131–153. doi: 10.1007/978-3-642-79663-0_7. [DOI] [PubMed] [Google Scholar]
- 54.Roberts L A, Seamons R A, Belsham G J. Recognition of picornavirus internal ribosome entry sites within cells; influence of cellular and viral proteins. RNA. 1998;4:520–529. doi: 10.1017/s1355838298971989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodríguez P L, Carrasco L. Gliotoxin: inhibitor of poliovirus RNA synthesis that blocks the viral RNA polymerase 3Dpol. J Virol. 1992;66:1971–1976. doi: 10.1128/jvi.66.4.1971-1976.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook J, Fritsch E R, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 58.Schneider R J, Shenk T. Impact of virus infection on host cell protein synthesis. Annu Rev Biochem. 1987;56:317–332. doi: 10.1146/annurev.bi.56.070187.001533. [DOI] [PubMed] [Google Scholar]
- 59.Skern T, Liebig H-D. Picornains 2A and 3C. Methods Enzymol. 1994;244:583–595. doi: 10.1016/0076-6879(94)44042-5. [DOI] [PubMed] [Google Scholar]
- 60.Sun X H, Baltimore D. Human immunodeficiency virus tat-activated expression of poliovirus protein 2A inhibits mRNA translation. Proc Natl Acad Sci USA. 1989;86:2143–2146. doi: 10.1073/pnas.86.7.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Svintkin Y V, Gradi A, Imataka H, Morino S, Sonenberg N. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J Virol. 1999;73:3467–3472. doi: 10.1128/jvi.73.4.3467-3472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turner P C, Young D C, Flanegan J B, Moyer R W. Interference with vaccinia virus growth caused by insertion of the coding sequence for poliovirus protease 2A. Virology. 1989;173:509–521. doi: 10.1016/0042-6822(89)90563-1. [DOI] [PubMed] [Google Scholar]
- 63.Urzáinqui A, Carrasco L. Degradation of cellular proteins during poliovirus infection: studies by two-dimensional gel electrophoresis. J Virol. 1989;63:4729–4735. doi: 10.1128/jvi.63.11.4729-4735.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ventoso I, Barco A, Carrasco L. Mutational analysis of poliovirus 2Apro. Distinct inhibitory functions of 2Apro on translation and transcription. J Biol Chem. 1998;273:27960–27967. doi: 10.1074/jbc.273.43.27960. [DOI] [PubMed] [Google Scholar]
- 65.Ventoso I, Barco A, Carrasco L. Genetic selection of poliovirus 2Apro-binding peptides. J Virol. 1999;73:814–818. doi: 10.1128/jvi.73.1.814-818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ventoso I, Carrasco L. A poliovirus 2Apro mutant unable to cleave 3CD shows inefficient viral protein synthesis and transactivation defects. J Virol. 1995;69:6280–6288. doi: 10.1128/jvi.69.10.6280-6288.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wimmer E, Hellen C U T, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 68.Wyckoff E E, Lloyd R E, Ehrenfeld E. Relationship of eukaryotic initiation factor 3 to poliovirus-induced p220 cleavage activity. J Virol. 1992;66:2943–2951. doi: 10.1128/jvi.66.5.2943-2951.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yalamanchili P, Banerjee R, Dasgupta A. Poliovirus-encoded protease 2Apro cleaves the TATA-binding protein but does not inhibit host cell RNA polymerase II transcription in vitro. J Virol. 1997;71:6881–6886. doi: 10.1128/jvi.71.9.6881-6886.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yalamanchili P, Harris K, Wimmer E, Dasgupta A. Inhibition of basal transcription by poliovirus: a virus-encoded protease (3Cpro) inhibits formation of TBP-TATA box complex in vitro. J Virol. 1996;70:2922–2929. doi: 10.1128/jvi.70.5.2922-2929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu S F, Benton P, Bovee M, Sessions J, Lloyd R E. Defective RNA replication by poliovirus mutants deficient in 2A protease cleavage activity. J Virol. 1995;69:247–252. doi: 10.1128/jvi.69.1.247-252.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ziegler E, Borman A M, Deliat F G, Liebig H D, Jugovic D, Kean K M, Skern T, Kuechler E. Picornavirus 2A proteinase-mediated stimulation of internal initiation of translation is dependent on enzymatic activity and the cleavage products of cellular proteins. Virology. 1995;213:549–557. doi: 10.1016/s0042-6822(95)90001-2. [DOI] [PubMed] [Google Scholar]