Abstract
The vaccinia virus H5 gene encodes a 22.3-kDa phosphoprotein that is expressed during both the early and late phases of viral gene expression. It is a major component of virosomes and has been implicated in viral transcription and, as a substrate of the B1 kinase, may participate in genome replication. To enable a genetic analysis of the role of H5 during the viral life cycle, we used clustered charge-to-alanine mutagenesis in an attempt to create a temperature-sensitive (ts) virus with a lesion in the H5 gene. Five mutant viruses were isolated, with one of them, tsH5-4, having a strong ts phenotype as assayed by plaque formation and measurements of 24-h viral yield. Surprisingly, no defects in genome replication or viral gene expression were detected at the nonpermissive temperature. By electron microscopy, we observed a profound defect in the early stages of virion morphogenesis, with arrest occurring prior to the formation of crescent membranes or immature particles. Nonfunctional, “curdled” virosomes were detected in tsH5-4 infections at the nonpermissive temperature. These structures appeared to revert to functional virosomes after a temperature shift to permissive conditions. We suggest an essential role for H5 in normal virosome formation and the initiation of virion morphogenesis. By constructing recombinant genomes containing two H5 alleles, wild type and H5-4, we determined that H5-4 exerted a dominant phenotype. tsH5-4 is the first example of a dominant ts mutant isolated and characterized in vaccinia virus.
Vaccinia virus is the prototypical member of the Poxvirus family, members of which include the human pathogens variola virus, the causative agent of smallpox, and molluscum contagiosum virus, which causes benign but long-lived skin lesions. The 192-kb double-stranded DNA genome of vaccinia virus contains approximately 200 genes, enabling the virus to replicate quite autonomously within the cytoplasm of infected cells (17, 31). The virus encodes a complex transcriptional machinery which directs the expression of three classes of temporally regulated genes: early, which encode the proteins required for replication of the viral genome, and intermediate and late, among whose products are those required for virion morphogenesis. Virion formation commences from cytoplasmic sites known as virosomes or viral factories, which contain viral DNA and viral proteins. Membrane crescents form, begin to enclose the virosome contents, and then enlarge and seal to form the visually distinguishable immature (IV) and mature (IMV) forms of the intracellular class of virions. A subset of these virions become wrapped in membranes derived from the trans-Golgi network and exit the cell. The genome also encodes a large number of effector proteins that enable the virus to evade host defenses.
Protein phosphorylation is known to play a crucial role in the regulation of many cellular processes such as initiation of DNA replication, signal transduction, and cell cycle control. Similarly, vaccinia virus regulates its life cycle by encoding two protein kinases, B1 and F10, and a protein phosphatase, H1. Their functions have been deduced, primarily, with the powerful genetic tool of conditional lethal mutants. The F10 kinase, which we have recently shown to be a dual specificity kinase that can phosphorylate Ser, Thr, and Tyr residues, is expressed late during infection and is encapsidated into virions (11a, 26, 44, 46). Temperature-sensitive (ts) mutants with lesions in the F10 gene have revealed that the kinase plays an essential role in the initial steps of virion morphogenesis. The H1 phosphatase, a dual-specificity phosphatase (20), is required for the infectivity and transcriptional competence of newly synthesized virions (28). The role of H1 in virion formation has been elucidated and its viral substrates have been identified by the construction of a recombinant virus (vindH1) in which the expression of the H1 protein is under the control of the lac operator/repressor and hence is induced by inclusion of isopropyl-β-d-thiogalactopyranoside (IPTG) in the culture medium.
The B1 Ser/Thr kinase (2, 27, 43), which is expressed during the early phase of viral gene expression, plays an essential role in viral DNA replication (35, 36). We have previously described the phenotype of two ts mutants with lesions in the B1 gene. Temperature shift experiments indicate that the B1 kinase is required throughout DNA replication, although its specific role in unknown. The S2 and Sa subunits of the 40S ribosome are cellular substrates of B1, while the only known viral substrate of the kinase is the vaccinia virus H5 protein (1, 4, 5). The H5 gene encodes a 22.3-kDa protein that is synthesized during both the early and late phases of viral gene expression (39). H5 is diffusely localized in the cytoplasm prior to DNA replication and then becomes concentrated within the virosomes (3). H5 undergoes multiple phosphorylation events, and isoforms with isolectric points of 6.8 to 5.5 have been identified (4). H5 appears to be a substrate for both viral and cellular kinases (3, 4; U. Sankar and P. Traktman, unpublished results). The protein has an extremely proline-rich N′ terminus, which has been cited as an explanation for its anomolous electrophoretic migration (35 kDa) during sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (18).
Several reports detailing possible roles for H5 have appeared in the literature. Kovacs and Moss identified H5 as a viral protein capable of stimulating late transcription two- to sixfold in vitro and designated H5 as a late gene transcription factor (VLTF-4) (25). Black et al. reported that H5 interacted both in vivo and in vitro with the viral proteins G2R and A18R, both of which play a role in the elongation and termination phases of intermediate and late viral transcription (7). Mohandas et al. reported that H5 had some affinity for both membranes and nucleic acids (30).
Because H5 is a substrate of the B1 kinase, and in light of our long-standing interest in the role of this kinase in viral replication, we chose to initiate a genetic analysis of the role of H5 in vivo. Unfortunately, no ts mutants with lesions in the H5 gene have been isolated to date, and the expression of H5 at both early and late times precludes the utility of an inducible recombinant. Therefore, we chose to use the method of “clustered charge-to-alanine” mutagenesis as an approach to generating a ts H5 mutant. In this report, we describe our success with this approach and discuss our unexpected finding that the H5 protein appears to play a critical role in viral morphogenesis.
MATERIALS AND METHODS
Materials.
Restriction endonucleases, Escherichia coli polymerase, polynucleotide kinase, the Klenow fragment of E. coli DNA polymerase, T4 DNA ligase, calf intestinal phosphatase, pancreatic RNase, and DNA molecular weight standards were purchased from New England Biolabs, Inc. (Beverly, Mass.) or Boehringer Mannheim Biochemicals (Indianapolis, Ind.) and were used as specified by the manufacturer. DNase I was purchased from Cooper Biochemical Inc. (West Chester, Pa). 32P-labeled nucleoside triphosphates and [35S]methionine were acquired from Dupont/New England Nuclear Corp. (Boston, Mass.). Cycloheximide, rifampin, and 5-bromo-2′-deoxyuridine (BrdU) were obtained from Sigma (St. Loius, Mo.). Isatin-β-thiosemicarbazone (IBT) was obtained from Pfaltz and Bauer (Waterbury, Conn.).
Cells and virus.
Monolayer cultures of BSC40 primate cells, mouse L cells, or human TK− cells were maintained in Dulbecco modified Eagle medium (DMEM) (GIBCO-BRL, Gaithersburg, Md.) containing 5% fetal calf serum (FCS). Wild-type (wt) vaccinia virus (WR strain) and vR0G8 (generously provided by B. Moss, National Institutes of Health, Bethesda, Md.) (51) were amplified in monolayer cultures of BSC40 cells or suspension cultures of L cells. Viral stocks were prepared from cytoplasmic lysates of infected cells by ultracentrifugation through 36% sucrose; titers were determined on BSC40 cells. In some cases, virions were further purified by banding on 25 to 40% sucrose gradients (28). For ts mutants, 31.5 and 39.5°C were used as the permissive and nonpermissive temperatures, respectively. Where indicated, rifampin and cycloheximide were added to final concentrations of 100 μg/ml; BrdU was added to a final concentration of 25 μg/ml.
Mutagenesis and cloning of the vaccinia virus H5 gene.
Five regions of the H5 gene were chosen for cluster charge-to-alanine mutagenesis (Fig. 1). To create each mutation, two PCRs were performed to produce overlapping products: the first with an upstream primer (UP) and a primer that engineered the mutations (U-H5-x), and the second with a downstream primer (DP) and a primer that engineered the complement of the mutations (D-H5-x). The nucleotides that encode alanine (GCN, coding strand) are underlined in the primers; the BamHI recognition sites within the UP and DP primers are shown in bold. The template for both PCRs was the HindIII H fragment of the vaccinia virus genome. A mixture of the two purified products, which overlapped by 17 bp, served as the template for a third PCR, which was performed with the UP and DP primers. This final product was gel purified, digested with BamHI, and cloned into pUC/neo, a plasmid that contains the neomycin resistance gene under the control of the vaccinia virus P7.5 strong early/late promoter (24, 28). The plasmids were sequenced to confirm the insertion of the desired mutation and the absence of any spurious changes. The primers used were as follows: D-H5-2, 5′ AAAACCGCAGCAGCCACTACTCCTCGTAA 3′; U-H5-2, 5′ GTGGCTGCTGCGGTTTTGGGCTTAGTAG 3′; D-H5-3, 5′ CCGCAGCGGCGGCAGTGGAAGAAGAAGTAGT 3′; U-H5-3, 5′ CACTGCCGCCGCTGCGGTTGATCTTTTAGTAG 3′; D-H5-4, 5′ AGTGGCAGCAGCAGTAGTTATAGAGGAATA 3′; U-H5-4, 5′ CTACTGCTGCTGCCACTTACTCCTTTTTGG 3′; D-H5-5, 5′ CGGGGCTGCTGCACCTATGGTACAAGTTGA 3′; U-H5-5, 5′ TAGGTGCAGCAGCCCCGTCGCTATCATCGA 3′; D-H5-6, 5′ TTCTGCCCTAGCGGTGGCTACCGACAATAT 3′; U-H5-6, 5′ CCACCGCTAGGGCAGAAAGATCGCTTCTAG 3′; UP (2,3,4), 5′ CGGGATCCTAGTAGATATGCTTT 3′; DP (2,3,4), 5′ CGGGATCCTTTAGTCATAGAAGTA 3′; UP2 (5,6), 5′ AAGGATCCGTAATGGCGTGGTCAA 3′; and DP2 (5,6), 5′ AAGGATCCTCTTCCCACGTTTGT 3′.
FIG. 1.
Clustered charge-to-alanine mutagenesis of the vaccinia virus H5 gene. The sequence of the vaccinia virus (strain WR) H5 gene was obtained from the public databases and confirmed by DNA sequencing. In our laboratory stock of the WR strain, the glycine residue at amino acid 95 is changed to a serine residue. Site-directed mutagenesis was performed by overlap PCR as described in Materials and Methods. For each mutant allele (H5-2, H5-3, H5-4, H5-5, and H5-6), the boxed resides were changed to alanine. The proline residues which are thought to account for the anomalous electrophoretic migration of H5 are underlined.
Transient dominant selection and isolation of mutants.
Replacement of the endogenous H5 allele with the mutant alleles was performed by the strategy of transient dominant selection (15, 24, 28). Semiconfluent 35-mm dishes of BSC40 cells were infected with wt virus at a multiplicity of infection (MOI) of 0.03 at 37°C. For each mutant, 3.5 μg of supercoiled DNA representing the appropriate pUCneo-H5-mutant plasmid was introduced into virally infected cells by calcium phosphate transfection at 3 h postinfection (p.i.), at which time the cultures were shifted to 31.5°C. To select for viruses that had incorporated the pUCneo-H5 plasmid into the viral genome, Geneticin (G418 sulfate) (GIBCO-BRL) was added to the medium at 15 h p.i. to a concentration of 3 mg/ml. After 48 h, the cells were harvested and subjected to three cycles of freezing-thawing. G418-resistant viruses were plaque purified twice, and plasmid integration was confirmed by PCR using primers complementary to sequences within the neomycin resistance gene. Selection was removed, and the virus was sequentially plaque purified in the absence of Geniticin until the loss of the neomycin resistance gene could be confirmed by PCR. DNA sequencing analysis was performed to distinguish plaques that had retained the mutant H5 allele from those that had retained the wt H5 allele. Plaques containing the mutant allele were expanded on BSC40 cells, and viral stocks were purified by centrifugation of cytoplasmic extracts through a 36% sucrose cushion.
Determination of 24-h viral yield.
Confluent 35-mm dishes of BSC40 cells were infected with wt or tsH5-4 virus at an MOI of 2 or 15 (vH5-2, vH5-3, vH5-5, and vH5-6 were also analyzed at an MOI of 2) and maintained at 31.5 or 39.5°C. Cells were harvested at 24 h p.i., collecting by centrifugation, and resuspended in 10 mM Tris (pH 9.0). The cells were disrupted by three cycles of freezing-thawing and two 15-s bursts of sonication. Viral yields were determined by titer determination on BSC40 cells at 31.5°C.
Analysis of viral DNA synthesis by dot blot hybridization.
Confluent 35-mm dishes of BSC40 cells were infected with wt or tsH5-4 virus at an MOI of 15 and incubated at 31.5 or 39.5°C. Cells were harvested at the indicated time points by scraping with a rubber policeman. The cells were collected by centrifugation, washed once with phosphate-buffered saline (PBS) (140 mM NaCl, 2 mM KCl, 10 mM Na2HPO4, 1 mM KH2PO4 [pH 7.4]), and resuspended in 300 μl of loading buffer (10× SSC [1.5 M NaCl, 0.15 M sodium citrate [pH 7.0]), 1 M ammonium acetate). They were disrupted by three cycles of freezing-thawing, and an additional 300 μl of loading buffer was added. After vortexing, 25 μl of each sample was spotted in triplicate onto a ZetaProbe blotting membrane (Bio-Rad, Richmond, Calf.) by using the Bio-Dot Microfiltration dot blot apparatus (Bio-Rad). DNA was denatured in situ with 0.5 M NaOH–1.5 M NaCl for 10 min and washed with 10× SSC twice for 5 min. The membrane was air dried, and viral DNA was detected by hybridization using a radiolabeled nick-translated probe representing the HindIII E and F fragments of the vaccinia virus genome. The blot was exposed to a phosphor screen, and data were acquired on a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.), quantitated using ImageQuant software (Molecular Dynamics), and plotted using SigmaPlot (SSPS, Chicago, Ill.).
Analysis of viral protein synthesis. (i) Metabolic labeling.
Confluent 35-mm dishes of BSC40 cells were infected with wt or tsH5-4 virus (MOI of 15) and maintained at 31.5 or 39.5°C. Individual plates of cells were rinsed with prewarmed methionine-free DMEM and incubated with prewarmed methionine-free DMEM plus 100 μCi of [35S]methionine per ml for 45 min prior to being harvested at the indicated time points (2.5, 4, 5.5, 7, and 9 h p.i.). The cells were scraped, collected by centrifugation, resuspended in PBS, and lysed by the addition of SDS sample buffer (1% SDS, 1% β-mercaptoethanol, 10% glycerol, 25 mM Tris [pH 6.8]). Samples were heated to 100°C for 3 min and analyzed by SDS-PAGE on a 12% polyacrylamide gel. After electrophoresis, the gel was dried and exposed to Kodak MR film for autoradiography.
(ii) H5 protein accumulation.
Samples prepared as above were fractionated by SDS-PAGE and transferred to nitrocellulose in 25 mM Tris–192 mM glycine–20% methanol at 4°C. The membrane was incubated with a polyclonal anti-H5 serum; an alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin was used as the secondary reagent, and the blot was developed colorimetrically using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate as specified by the manufacturer (Bio-Rad).
Genome resolution assay.
The genome resolution assay was performed as described previously (24, 29). Confluent 35-mm dishes of BSC40 cells were infected with wt or tsH5-4 virus at an MOI of 2 or vR0G8 (51) at an MOI of 15 and incubated at 31.5 or 39.5°C. An additional culture was infected with wt virus in the presence of 60 μM IBT. At 18 h p.i., cells were harvested, washed with ice-cold PBS, resuspended in 50 mM NaCl–10 mM EDTA–20 mM Tris (pH 8.0), and incubated for 10 min on ice. The samples were adjusted to 0.6% SDS and 0.7 μg of proteinase K per ml and incubated at 37°C for 6 h. The DNA was purified by organic extraction and ethanol precipitation, digested with BstEII, fractionated by electrophoresis through a 1% agarose gel cast and run in 1× TAE (40 mM Tris acetate, 1 mM EDTA), and then transferred to a ZetaProbe membrane in 20× SSC. The membrane was probed with a radiolabeled PvuII-EcoRI fragment contained within pSV9 (a generous gift from M. Merchlinsky, U.S. Department of Agriculture, Bethesda, Md.) to visualize the DNA fragments corresponding to the termini of the viral genome. The blot was washed, dried, and exposed for autoradiography.
Proteolytic processing of virion proteins.
Confluent 35-mm dishes of BSC40 cells were infected with wt or tsH5-4 virus at an MOI of 15 and incubated at 31.5 or 39.5°C. As a control, cells were infected with wt virus in the presence of 100 μg of rifampin per ml. At 8.25 h p.i., the cells were rinsed with prewarmed methionine-free DMEM and then incubated for 45 min with prewarmed methionine-free DMEM supplemented with 100 μCi of [35S]methionine per ml. The cells were harvested immediately, or the medium was aspirated and the cells were fed with prewarmed DMEM supplemented with 5% FCS and harvested at 24 h p.i. The cells were scraped and collected by centrifugation, resuspended in ice-cold PBS, and lysed by the addition of SDS sample buffer. Sample were heated to 100°C for 3 min and analyzed by SDS-PAGE on a 12% polyacrylamide gel. After electrophoresis, the gel was dried and exposed to Kodak MR film for autoradiography.
Virion banding.
Four 15-cm-diameter dishes of BSC40 cells were infected with tsH5-4 virus at an MOI of 2 and maintained at either at 31.5 or 39.5°C. At 24 h p.i., the cells were harvested and disrupted by homogenization, and virus was purified from cytoplasmic extracts by ultracentrifugation through a 36% sucrose cushion (43,000 × g for 80 min) and banding through a 25 to 40% sucrose gradient (6,000 × g for 45 min). Banded virions were removed from equivalent positions of the ultracentrifuge tube by needle aspiration, collected by sedimentation, and resuspended in 1 mM Tris (pH 9.0). The number of virion particles was measured by taking the optical density at 260 nm, assuming that 1 optical density unit is equivalent to 1.2 × 1010 virion particles per ml. Viral yields were determined by titer determination on BSC40 cells at 31.5°C.
Electron microscopy.
Confluent 60-mm dishes of BSC40 cells were infected with wt or tsH5-4 virus at an MOI of 5 and incubated at either 31.5 or 39.5°C for the time indicated. Cells were fixed in situ with 2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) for 30 min on ice, harvested, and collected by centrifugation (828 × g for 5 min). After 30 min in fresh fixative, the cells were rinsed and postfixed with 1% OsO4 for 60 min. The cells were dehydrated with a series of graded ethanols and embedded in epoxy resin. Thin sections were prepared and the grids were stained with uranyl acetate and lead citrate. The thin sections were examined with an H-600 transmission electron microscope at 75 kV.
Demonstration that the H5-4 allele is linked to the ts phenotype of tsH5-4 and causes a dominant phenotype: construction of wtVV:wtH5, wtVV:H5-4, tsH5-4:wtH5, and tsH5-4:H5-4 recombinants.
Targeted insertion and expression of a second copy of the H5 gene into the thymidine kinase (TK) locus of the vaccinia virus genome was achieved by using pJS4, a plasmid which contains strong vaccinia virus early and late promoters flanked by regions of the TK gene (9). Recombination of this plasmid into the viral genome interrupts the TK open reading frame; under selection with BrdU, only viruses which contain a disrupted TK locus will replicate. The H5 open reading frame (either the wt H5 allele or the H5-4 allele) was amplified by PCR using primers 1 (5′ GACTGGATCCTGGCGTGGTCAATTAC 3′) and 2 (5′ GCGCGAATTC TTACTTCTTACAAGTTT 3′). The purified PCR products were digested with BamHI (bold) and EcoRI (italics) and cloned downstream of one the promoters of pJS4 by ligation into the plasmid linearized by BamHI and EcoRI.
Semiconfluent 35-mm dishes of BSC40 cells were infected with either wt or tsH5-4 virus at an MOI of 0.03 at 31.5°C. For each transfection, 3.5 μg of supercoiled DNA (either pJS4/wtH5 or pJS4/H5-4) was introduced into virally infected cells by calcium phosphate transfection at 3 h p.i. After 48 h, the cells were harvested and subjected to three cycles of freezing-thawing. TK-null virus was twice plaque purified on TK− cells in the presence of BrdU, and plasmid integration was confirmed by PCR using primer 1 and a primer which annealed to the TK gene (TK-3′ primer: 5′ CGTTTGCCATACGCTCACAG 3′). Plaques containing the insertion in the TK gene were expanded on BSC40 cells. The virus was then tested for the ts phenotype as described above. To confirm the sequence of the H5 alleles within the recombinant viruses, PCR amplification of the endogenous H5 allele was performed using primers complementary to flanking sequences in the H4 gene (5′ CATTATCGCGATATCCGTTAAG 3′) and the H6 gene (primer DP2). The exogenous H5 allele was amplified using primers complementary to flanking sequences in the left and right regions of the TK locus (TK 3′ [see above] and TK-L 5′ [5′ GATTTCTCCGTGATAGG 3′]). The purified PCR products were used as templates for DNA sequencing analysis performed on an ABI Prism 310 Genetic Analyzer (Perkin-Elmer, Foster City, Calif.).
Computer analysis.
Autoradiography films and electron micrograph negatives were scanned by a SAPHIR scanner (Linotype-Hell Co., Hauppauge, N.Y.) using Adobe Photoshop software (Adobe Systems Inc., San Jose, Calif.). Tissue culture plates were scanned by an AlphaImager (Alpha Innotech, San Leandro, Calif.). Figures were labeled using Canvas software (Deneba Systems, Miami, Fla.) and were printed using a Kodak 8670 dye sublimation printer (Eastman Kodak, Rochester, N.Y.).
RESULTS
Our goal was to generate a virus with a conditionally lethal mutation in the H5 gene in order to analyze its essential functions in vivo. Clustered charge-to-alanine mutagenesis has been used successfully in vaccinia virus and other systems for this very purpose (12, 16, 21, 22, 48). The premise of this approach is the likelihood that clusters of charged residues will lie on the surface of a native protein, such that mutations in the region would be unlikely to cause a global disturbance of protein folding. Rather, the mutations might weaken protein interactions or destabilize the protein structure in a manner that would not be lethal under normal growth conditions but might become so at elevated temperatures. We decided to create several mutant alleles of the H5 gene encoding proteins with clustered-charge-to-alanine substitutions and then to isolate viral recombinants (at 31.5°C, the permissive temperature typically used for vaccinia virus ts mutants) containing these constructs in place of the endogenous H5 allele.
Isolation and characterization of vaccinia virus H5 clustered charge-to-alanine mutants: vH5-4 has a ts phenotype.
We used the technique of overlap PCR to construct alleles of the H5 gene containing the appropriate mutations. The mutant H5 alleles were then cloned into pUC/neo, a plasmid containing the neomycin resistance (Neor) gene under the control of a constitutive vaccinia virus promoter. The genomic H5 allele was replaced with the exogenous alleles by transient dominant selection (15). Supercoiled plasmids were transfected into cells infected with wt virus, and the infections were then maintained at 31.5°C in the presence of G418. G418-resistant viruses were twice plaque purified, and the recombinational insertion of the plasmid into the viral genome was confirmed by PCR using primers complementary to the Neor gene. The recombination event that inserts the Neor gene into these viruses also generates an unstable tandem duplication of the H5 locus. Consequently, when G418 selection is removed, the duplicated alleles recombine to leave a genome that has lost the inserted plasmid and retains a single allele of H5, either wt or mutant. Virus was thus subjected to plaque purification in the absence of G418 until the loss of the Neor gene could be confirmed by PCR; such viruses were then screened by DNA sequence analysis to identify those that had resolved to contain the altered H5 allele.
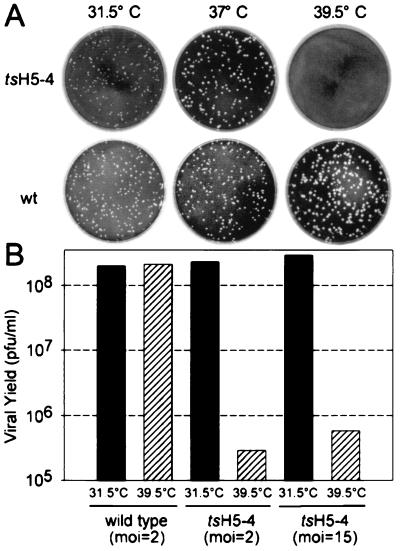
Using this method, we attempted to isolate five viral recombinants containing clustered charge-to-alanine mutations in the H5 gene: vH5-2, vH5-3, vH5-4, vH5-5, and vH5-6 (Fig. 1). In all cases, the desired recombinants could be obtained and were viable at 31.5°C. These viruses were expanded and tested for a ts phenotype with respect to plaque formation and 24-h viral yield. vH5-4, in which residues 73 to 75 of the H5 protein have been changed from glutamic acid (EEE) to alanine (AAA), was strikingly ts (Fig. 2). We have thus designated vH5-4 as tsH5-4. Plaque formation by tsH5-4 appeared to be normal at 31.5°C but was completely abrogated at 39.5°C (Fig. 2A). Surprisingly, plaque formation by tsH5-4 appeared normal at 37°C. The 24-h viral yield from cells infected with tsH5-4 at 39.5°C was decreased by more than 2 orders of magnitude relative to that obtained at 31.5°C (Fig. 2B). This dramatic temperature sensitivity in the production of infectious virus was seen at both high and low multiplicities of infection (15 and 2, respectively). The yield of tsH5-4 at 31.5°C was comparable to that obtained from wt-infected cells, suggesting that the progression of the viral life cycle of tsH5-4 is largely unperturbed at 31.5°C. A similar degree of temperature sensitivity was observed when tsH5-4 infections were performed in mouse L cells (data not shown). In all subsequent experiments with tsH5-4, 31.5°C was used as the permissive temperature and the nonpermissive temperature was rigorously maintained at 39.5°C.
FIG. 2.
tsH5-4 is defective in infectious virus production at 39.5°C. (A) Plaque assay of tsH5-4. Confluent BSC40 cells were infected with equivalent amounts of virus (either wt or tsH5-4) and maintained at the indicated temperature. After 2 days, the medium was removed and the cells were stained with crystal violet. (B) Twenty-four-hour viral yield of tsH5-4. Confluent BSC40 cells were infected with either wt or tsH5-4 virus at the indicated MOI and maintained at the indicated temperature. At 24 h p.i., the cells were harvested and disrupted and viral yield was analyzed by titer determination on BSC40 cells at 31.5°C. Each infection was performed in duplicate, and the average is shown in the bar graph.
For vH5-2, vH5-3, vH5-5, and vH5-6, no detectable ts phenotype was found by analysis of plaque formation on BSC40 cells (data not shown). Quantification of the 24-h yields of infectious virus, both in BSC40 cells and in L cells, also failed to reveal any significant temperature-sensitivity for these viruses. Therefore, further characterization of these recombinants was not performed.
tsH5-4 is not defective in viral DNA replication.
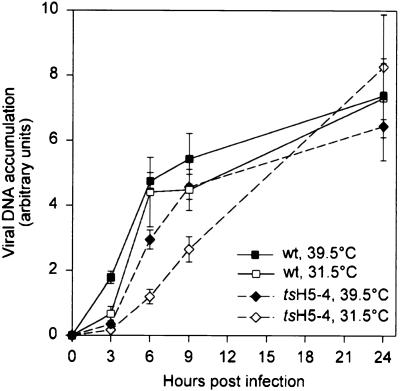
We initiated this study in part to examine if the role of the B1 kinase in viral DNA replication was somehow related to its phosphorylation of the H5 protein. Therefore, in our assessment of the stage of the viral life cycle that was impaired during nonpermissive tsH5-4 infections, we first examined viral DNA replication. Cells were infected with either wt or tsH5-4 virus at either 39.5 or 31.5°C and harvested at 3, 6, 9, or 24 h p.i. The accumulation of viral DNA was quantified by dot blot hybridization using 32P-labeled fragments of the viral genome as the probe (Fig. 3). We observed that the accumulation of viral DNA during nonpermissive tsH5-4 infections was not ts. The rate of viral DNA accumulation was somewhat lower in tsH5-4 infections than in wt infections, but this was true at both permissive and nonpermissive temperatures and did not affect the levels of DNA that accumulated by 24 h p.i.
FIG. 3.
Viral DNA replication in tsH5-4-infected cells. BSC40 cells were infected with either wt or tsH5-4 virus at an MOI of 15 and maintained at 31.5 or 39.5°C. Cells were harvested at 3, 6, 9, or 24 h p.i., washed with PBS, and resuspended in loading buffer. Uninfected cells were harvested and used for the zero time point. The cells were disrupted, and each sample was spotted onto a ZetaProbe blotting membrane. Viral DNA was detected by hybridization using a radiolabeled nick-translated probe representing the HindIII E and F fragments of the vaccinia virus genome. The blot was exposed to a phosphor screen, and data were acquired on a Storm PhosphorImager and quantified using ImageQuant software. All samples were processed in triplicate.
tsH5-4 is not defective in viral protein synthesis or H5 protein accumulation.
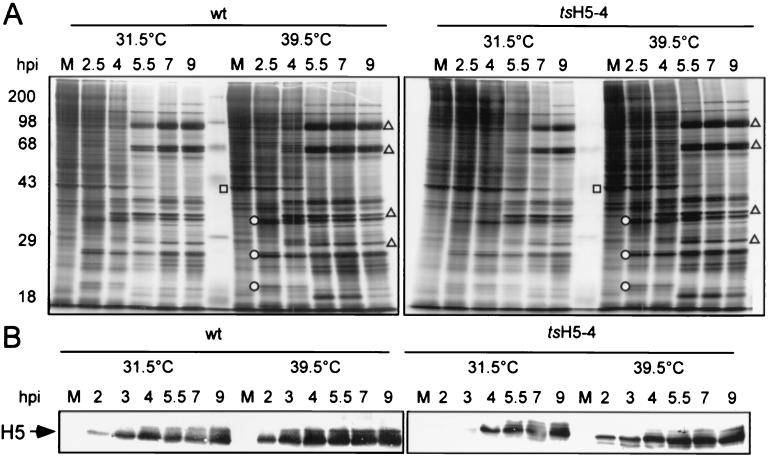
As mentioned previously, H5 has been implicated in viral transcription, as a viral late transcription factor (25) and as a component of the transcription elongation and termination machinery (7). To determine if the ts phenotype of tsH5-4 was due to altered viral gene expression, we next examined viral protein synthesis. Cells were infected with either wt or tsH5-4 virus and, at various times p.i., were metabolically labeled with [35S]methionine to enable an analysis of the temporal profile of protein synthesis. Extracts were then examined by SDS-PAGE and autoradiography. All temporally regulated classes of viral gene products were evident in tsH5-4 infections performed at the nonpermissive temperature (Fig. 4A, right panel). Early viral proteins were seen at 2.5 h p.i., and the synthesis of late viral proteins was detected at 5.5 h p.i. and continued to 9 h p.i. The expression pattern of viral proteins in tsH5-4 infections was virtually identical to that seen in wt infections (left panel). Also evident was the shutoff of host protein synthesis during viral infection. Had there been an effect on the initiation of late gene expression, we would have expected to see a decrease in the timing or level of late protein synthesis; if transcription elongation was severely affected, higher-molecular-weight proteins would have been absent (8). Clearly, if the H5 protein plays an essential role in late-gene expression, the mutations in the H5-4 allele do not compromise this activity.
FIG. 4.
Protein expression in tsH5-4-infected cells. (A) BSC40 cells were infected with wt or tsH5-4 virus at an MOI of 15 and maintained at 31.5 or 39.5°C. The cells were metabolically labeled with [35S]methionine for 45 min before being harvested at the indicated time points (2.5, 4, 5.5, 7, and 9 h p.i.). As a control, uninfected cells were labeled with [35S]methionine for 45 min and analyzed in parallel (lane M). Cells were harvested and analyzed directly by SDS-PAGE on a 12% polyacrylamide gel. After electrophoresis, the gel was dried and exposed for autoradiography. The circles indicate early viral proteins, and the triangles indicate late viral proteins. The square denotes a host protein whose production is shut off after infection. (B) H5 protein accumulation. Samples prepared as above were analyzed by SDS-PAGE, transferred to nitrocellulose, probed with a polyclonal anti-H5 antiserum, and detected using an alkaline phosphate-conjugated secondary antibody. Only the relevant portion of the blot is shown.
Frequently, the proteins encoded by ts alleles are unstable at the nonpermissive temperature. We therefore examined the production and stability of the H5-4-encoded protein. Extracts were prepared from tsH5-4 and wt-infected cells, fractionated by SDS-PAGE, and analyzed by immunoblot analysis with a rabbit polyclonal antibody raised against an H5-TrpE fusion protein (Sankar and Traktman, unpublished). In wt infections, the H5 protein appeared at 2 h p.i. and continued to accumulate throughout infection (Fig. 4B, left panel). In tsH5-4 infections, the H5-4 protein accumulated to wt levels, although the rate of accumulation was lower at 31.5°C (right panel). Most significantly, these data indicated that the H5 protein was stable at 39.5°C and thus is not thermolabile. Slower-migrating forms of H5 were seen in all cases as infection progressed, appearing at 3 to 4 h p.i. in wt infections and 4 to 5 h p.i. in the tsH5-4 infections. These forms of H5 correspond to differentially phosphorylated species; both our laboratory and others have observed that postreplicative phosphorylation events are responsible for altering the electrophoretic mobility of the H5 protein (4). The only distinguishing feature of the nonpermissive tsH5-4 infection was the appearance of a novel, faster-migrating form of H5 that was visible at all time points. The altered electrophoretic mobility of this species is likely to reflect an alteration in protein conformation, although partial proteolytic processing might also be occurring.
tsH5-4 is not defective in resolution of genomic viral DNA.
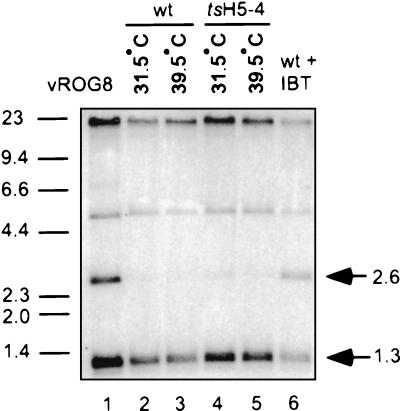
During the course of viral DNA replication, the viral genome is replicated into concatemers which are resolved into mature monomeric genomes and then encapsidated during viral morphogenesis. Resolution of the viral genome requires late viral gene expression. To assay for the resolution of the viral genome, DNA was prepared from virally infected cells and analyzed by Southern blot hybridization using a radiolabeled probe recognizing the termini of the viral genome. After restriction with BstEII, mature, monomeric genomes will release a 1.3-kb fragment while unresolved concatemers will release a 2.6-kb fragment. Unresolved concatemers were barely detectable in either wt or tsH5-4 infections performed at either 31.5 or 39.5°C (Fig. 5, lanes 2 to 5). As controls, cells were infected with either vROG8 (lane 1), a viral recombinant in which expression of the late transcription factor G8 is IPTG dependent (51), or wt virus in the presence of IBT, a drug which inhibits late viral gene expression (lane 6). Under both of these conditions, significant amounts of unresolved concatemers were detected (2.6-kb band). From these data, it can be concluded that nonpermissive tsH5-4 infections are not defective in genome resolution and that proteins required for genome resolution are synthesized normally.
FIG. 5.
Genome resolution of viral DNA. BSC40 cells were infected with wt or tsH5-4 virus at an MOI of 2 and maintained at the indicated temperature. As controls, cells were infected with vR0G8 in the absence of IPTG or infected with wt virus in the presence of 60 μM IBT throughout infection. At 18 h p.i., cells were harvested and cytoplasmic DNA was prepared and digested with BstEII. Digested DNA was resolved on a 1% TAE gel, transferred to a ZetaProbe membrane, and hybridized with a radiolabeled probe recognizing the termini of the viral genome. The arrows indicate the fragments released from resolved mature genomes (1.3 kb) and unresolved concatamer genomes (2.6 kb). The sizes of DNA standards are indicated in kilobase pairs on the left.
Proteolytic processing of late virion proteins is incomplete in nonpermissive tsH5-4 infections.
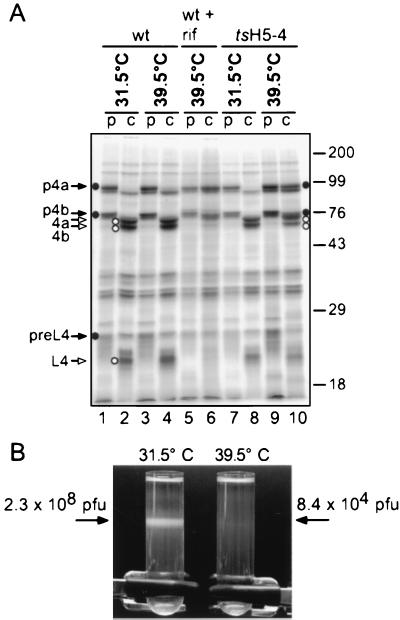
A hallmark of the formation of infectious virus is the proteolytic processing of three virion proteins from their immature forms (p4a, p4b, and preL4) to their mature forms (4a, 4b, and L4) (32, 34, 37, 52). As a first assessment of whether there might be a defect in virion morphogenesis during tsH5-4 infections, we examined the proteolytic processing of these proteins. We performed a pulse-chase experiment in which virally infected cells were metabolically labeled with [35S]methionine at 8.25 h p.i. for 45 min. Either cells were harvested immediately (pulse), or the medium was aspirated, the cells were fed with DMEM supplemented with 5% FBS, and then harvested at 24 h p.i. (chase) for analysis by SDS-PAGE (Fig. 6A). In wt infections, there was almost complete processing of p4a and p4b from their immature forms (Fig. 6A, lane 1) to their mature forms (lane 2), as well as the appearance of mature L4. In tsH5-4 infections performed at 39.5°C, approximately 50% of the p4a and p4b proteins remained unprocessed while the mature forms 4a and 4b did not accumulate to the levels seen in permissive infections (lane 10). As a comparison, we examined wt infections performed in the presence of rifampin (lanes 5 and 6), a drug that interferes with the production of the viral membrane through interactions with the D13 protein (19, 42). As can be seen in lane 6, there was little production of the mature proteins 4a, 4b, and L4 in the presence of rifampin. While the defect in proteolytic processing in nonpermissive tsH5-4 infections was not as dramatic as that seen with rifampin, these results were a first indication that tsH5-4 infections might well be compromised in some aspect of virion morphogenesis.
FIG. 6.
Virion morphogenesis during tsH5-4 infections. (A) Proteolytic processing of major virion proteins is impaired in tsH5-4 cells. BSC40 cells were infected with wt or tsH5-4 virus at an MOI of 15 and maintained at 31.5 or 39.5°C. At 8.25 h p.i., the cells were metabolically labeled with [35S]methionine for 45 min. The were harvested immediately (p [pulse]), or the medium was aspirated and the cells were fed with complete DMEM supplemented with 5% FBS and harvested at 24 h p.i. (c [chase]). Samples were analyzed directly by SDS-PAGE on a 12% polyacrylamide gel. After electrophoresis, the gel was dried and exposed for autoradiography. As a control, cells were infected with wt virus and treated with 100 μg of rifampin (rif) per ml throughout infection. Solid arrows and circles indicate the unprocessed form of virion proteins (p4a, p4b, and preL4); open arrows and circles indicate the proteolytic processed forms (4a, 4b, and L4). Protein standards are indicated on the left, with their masses shown in kilodaltons. (B) Mature virions are not produced during nonpermissive infections with tsH5-4. Four 15-cm dishes of BSC40 cells were infected with tsH5-4 virus at an MOI of 2 and maintained at 31.5 or 39.5°C for 24 h. The cells were harvested and disrupted by homogenization, and virus was purified from cytoplasmic extracts by ultracentrifugation through a 36% sucrose cushion and banding through a 25 to 40% sucrose gradient. The arrow on the left points to the band of mature virion particles produced during permissive conditions; notice the absence of virions produced during nonpermissive conditions in the tube on the right. The banded virions were removed from the 31.5°C sample by needle aspiration, and an equal volume was removed from the 39.5°C sample at the corresponding position. Virions were concentrated by centrifugation, and viral yields were obtained by titer determination on BSC40 cells at 31.5°C.
Virion production is dramatically reduced in tsH5-4 infections at the nonpermissive temperature.
To further investigate whether virion morphogenesis was defective in nonpermissive tsH5-4 infections, we attempted to purify virions from cells infected with tsH5-4 at both 31.5 and 39.5°C. Infected cells were harvested at 24 h p.i., and virion particles were purified from cytoplasmic extracts by ultracentrifugation through a 36% sucrose cushion and banding through a 25 to 40% sucrose gradient (Fig. 6B) (44). While a band of mature virions could clearly be isolated from tsH5-4-infected cells at 31.5°C (arrow, left tube), there was a striking absence of virions in the sample prepared from the 39.5°C infection (right tube). Equal volumes were removed from both tubes by needle aspiration at the position of the banded virions, and infectious virus was quantified. The amount of banded infectious virus produced at 39.5°C was 0.04% of that produced at 31.5°C. From this result, it is evident that virion production is severely abrogated in cells infected with tsH5-4 at the nonpermissive temperature.
tsH5-4 is blocked at a very early stage of morphogenesis at the nonpermissive temperature.
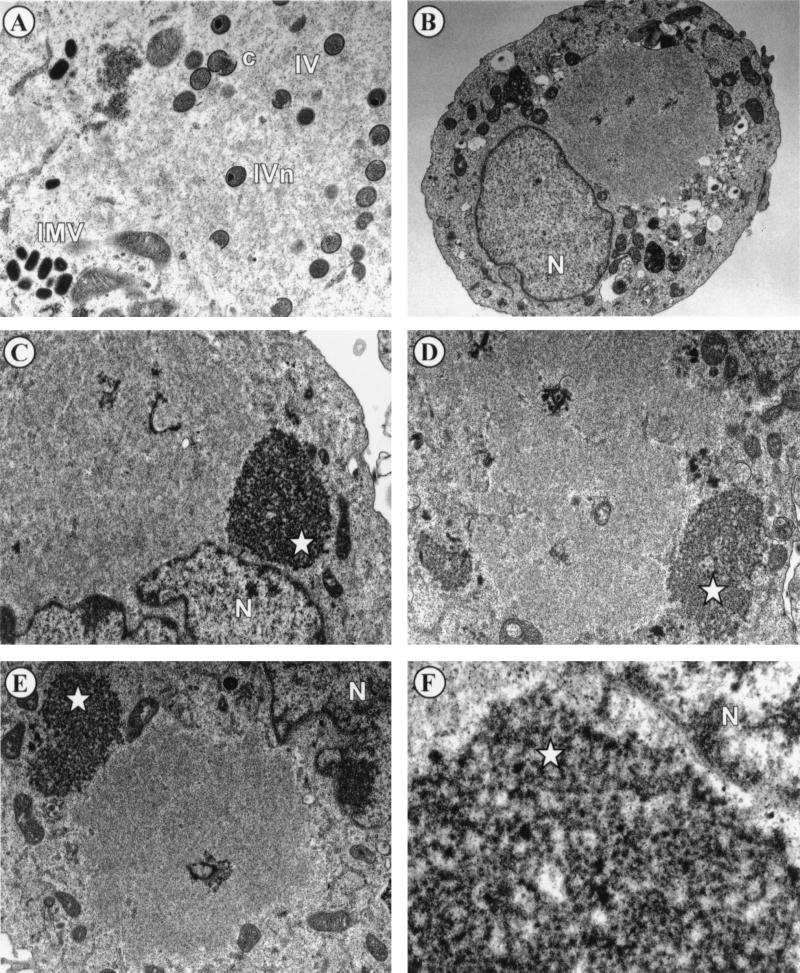
There are a number of visually distinct stages of viral morphogenesis, including the appearance of electron-dense virosomes, the emergence of membrane crescents near the virosome, and the formation of immature, immature with condensed nucleoid, and mature virions. Indeed, there are many conditionally lethal mutants of vaccinia virus that arrest at a particular state of morphogenesis during nonpermissive infections (6, 23, 24, 34, 37, 38, 44–46, 49, 50, 52, 53). To determine the stage of virion formation that is compromised during tsH5-4 infections, we analyzed virion morphogenesis by transmission electron microscopy. In cells infected with tsH5-4 at the permissive temperature and fixed at 17 h p.i., all stages of virion morphogenesis were observed (Fig. 7A), including crescents, immature virion particles, immature virion particles with nucleoids, and intracellular mature virions. Conversely, the normal hallmarks of virion morphogenesis were absent in cells infected with tsH5-4 at the nonpermissive temperature (Fig. 7B through F). There was a striking absence of crescents and immature or mature virions. The most consistent feature of these nonpermissively infected cells was a large, cleared region of the cytoplasm devoid of any cellular organelles (seen clearly in Fig. 7B). These images are evocative of the phenotype previously seen in cells infected with ts28, a virus defective in the F10 kinase (44). A second predominant feature of the cells infected with tsH5-4 at the nonpermissive temperature was the appearance of an electron-dense structure resembling a virosome but distinguished by a splotchy, “curdled” appearance (Fig. 7C through F). (We present data later in the paper to suggest why we believe these structures to be nonfunctional virosomes.) The curdled appearance of this structure is best seen at high magnification (Fig. 7F). It is clear from these data that the phenotype of tsH5-4 is due to a block at a very early stage of virion morphogenesis.
FIG. 7.
Virion morphogenesis undergoes arrest at an early stage in cells infected with tsH5-4 at the nonpermissive temperature. Confluent monolayers of BSC40 cells were infected with tsH5-4 at an MOI of 5 and maintained at 31.5°C (A) or 39.5°C (B through F) for 17 h. The cells were fixed in situ and processed for transmission electron microscopy as described in Materials and Methods. Representative fields were photographed. The stages of normal virion morphogenesis can be seen at the permissive temperature (A), including crescents (c), immature virions (IV), immature virions with nucleoid (IVn), and intracellular mature virions (IMV). In contrast, no immature or mature virion particles can be seen in cells infected with tsH5-4 at the nonpermissive temperature (B through E). An occasional viral crescent is seen (D). Large cleared areas of cytoplasm devoid of any cellular organelles are prominent (in panel B, all the cellular organelles are relegated to the periphery of the cell and a large empty region is visible above the nucleus), as are large, splotchy masses that we termed “curdled virosomes” (starred [C through E]). (F) Higher magnification of a curdled virosome. Nuclei are labeled with N. Magnifications: ×15,000 (A), ×6,000 (B), ×12,000 (C and D), ×10,000 (E), and ×50,000 (F).
The major execution point of the H5-4 allele occurs between 6 and 12 h p.i.
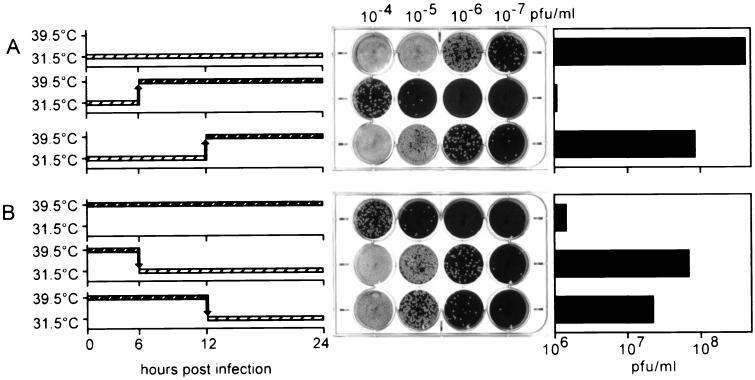
H5 is synthesized at both early and late phases of the viral life cycle, yet the H5-4 allele does not affect DNA replication or late-gene expression as one might have hypothesized. As a further examination of the tsH5-4 allele and the time at which it interferes with the viral life cycle, we performed experiments in which tsH5-4 infections were shifted from the permissive to the nonpermissive temperature (Fig. 8). When tsH5-4 infections were maintained at the permissive temperature for 6 h and then shifted to the nonpermissive temperature for the remaining 18 h (Fig. 8A, row 2), the resulting viral yield was comparable to that obtained from fully nonpermissive infections (compare Fig. 8A, row 2, with Fig. 8B, row 1), i.e., 2 orders of magnitude below that obtained from permissive infections (Fig. 8A, row 1). These data indicate that the crucial point for H5 action and/or synthesis occurs after 6 h p.i. In contrast, when tsH5-4 infections were maintained at the permissive temperature for 12 h before being shifted to the nonpermissive temperature for the remaining 12 h (Fig. 8A, row 3), the viral yield was almost 2 orders of magnitude above that seen during fully nonpermissive conditions (compare Fig. 8A, row 3, with Fig. 8B, row 1). These results suggest that the period from 6 to 12 h p.i. is crucial for the action of the H5-4 protein; this timing coincides well with the early phase of virion morphogenesis. The yield from the 12-h-shift infection remained 1 order of magnitude below that seen during fully permissive infections; these data suggest that production of infectious progeny may decline or cease after the cultures are shifted to the nonpermissive temperature.
FIG. 8.
The execution point of the H5-4 allele occurs between 6 and 12 h p.i. (A) BSC40 cells were infected with tsH5-4 virus at an MOI of 2 and incubated at 31.5°C. The temperature of the cells was either maintained for 24 h (first row) or shifted from 31.5 to 39.5°C at 6 h p.i. (second row) of 12 h p.i. (third row). At 24 h p.i., the cells were harvested and disrupted and the viral yield was analyzed by titer determination on BSC40 cells at 31.5°C (plaque assay, middle). A graphic representation of the titer is shown on the right. (B) The phenotype of tsH5-4 is reversible. BSC40 cells were infected with tsH5-4 virus at an MOI of 2 and incubated at 39.5°C. The temperature was either maintained at 39.5°C for 24 h (first row) or shifted from 39.5 to 31.5°C at 6 h p.i. (second row) or 12 h p.i. (third row). At 24 h p.i., the cells were harvested and disrupted and the viral yield was analyzed by titer determination on BSC40 cells at 31.5°C (plaque assay, middle). A graphic representation of the titer is shown on the right.
The phenotype of tsH5-4-infected cells is reversible.
During our electron microscopic examination of the cells infected nonpermissively with tsH5-4, we pondered whether the curdled virosome structures that accumulated at the nonpermissive temperature would recover if the cultures were shifted down to the permissive temperature. As a first attempt to see if the phenotype of tsH5-4 was indeed reversible, we monitored the production of infectious progeny from tsH5-4-infected cultures shifted from nonpermissive to permissive conditions at various times p.i. When the cultures were shifted down at either 6 or 12 h p.i., viral yields 1 to 2 orders of magnitude greater than those obtained from fully nonpermissive infections were obtained (Fig. 8B, compare rows 2 and 3 with row 1). These data imply that the tsH5-4 infections can recover to a significant degree even after 12 h under nonpermissive conditions.
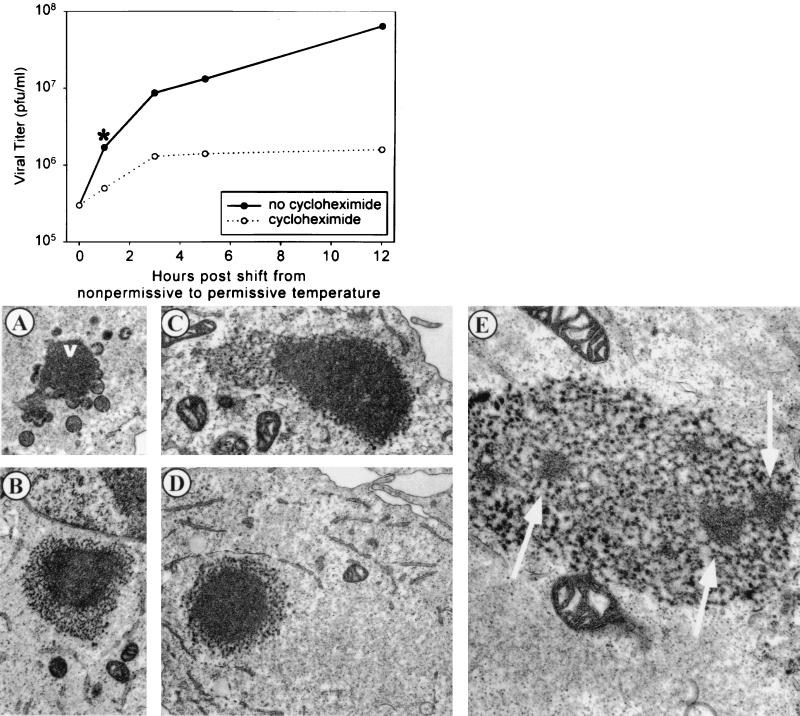
To determine whether this recovery was dependent on de novo protein synthesis, tsH5-4 infections were maintained at the nonpermissive temperature for 12 h and were then shifted to the permissive temperature in the presence or absence of the protein synthesis inhibitor cyclohexamide. Cells were harvested at 1, 3, 5, and 12 h after the temperature shift, and the viral yields were determined at 31.5°C (Fig. 9, graph). An increase in viral yield of greater than 2 orders of magnitude occurred during the 12 h postshift, when protein synthesis was permitted (solid line). In the absence of protein synthesis, an increase in viral yield was seen during the first 3 h postshift (dotted line), although the rate of production was significantly lower than during the control shift-down experiment.
FIG. 9.
Analysis of tsH5-4-infected cells after the shift from the nonpermissive to the permissive temperature. BSC40 cells were infected with tsH5-4 at an MOI of 2 and incubated at 39.5°C. The cells were either harvested at 12 h p.i. or shifted from 39.5 to 31.5°C and harvested at 13, 15, 17, or 24 h p.i. (1, 3, 5, and 12 h after the temperature shift, respectively). Where indicated, cells were treated with 100 μg cycloheximide per ml 20 min prior to the shift in temperature. The cells were disrupted, and viral yields were obtained by titer determination on BSC40 cells at 31.5°C. (A to E) Electron microscopic analysis of tsH5-4-infected cells after the shift from the nonpermissive to the permissive temperature. BSC40 cells were infected with wt (A) or tsH5-4 (B through E) virus at an MOI of 5, maintained at 39.5°C for 12 h, and then shifted to 31.5°C for 1 h (asterisk in graph). The cells were fixed in situ, harvested, and processed for transmission electron microscopy as described in Materials and Methods. Representative fields were photographed. (A) During wt infections, virosomes (v) appear as smooth electron-dense masses in the cytoplasm, with emerging crescents and immature virions. (B through E) In cells infected with tsH5-4 for 12 h at the nonpermissive temperature and shifted to the permissive temperature for 1 h, the curdled virosomes, which have a characteristic splotchy appearance, begin to form smooth, electron-dense regions from within, gradually acquiring the appearance of wt virosomes. (E) Higher magnification of a curdled virosome, with arrows pointing to emerging regions of wt density. Magnifications: ×12,000 (A through D), ×25,000 (E).
Having demonstrated that the nonpermissively infected cultures did recover after they were shifted to the permissive temperature, we used electron microscopy to investigate the fate of the curdled virosomes after the temperature shift. Was the morphogenesis arrest truly reversible, with the curdled virosomes becoming functional? Or, instead, were the curdled virosomes bypassed as dead-end structures and supplemented by new virosomes that became the sites of virion assembly? When tsH5-4 infections were shifted to the permissive temperature for 1 h after having been maintained at the nonpermissive temperature for 12 h, we indeed saw a dramatic transition occurring within the curdled virosomes (Fig. 9). Smooth, electron-dense regions appeared within the curdled masses (Fig. 9B through F); these regions were identical to the texture of wt virosomes (Fig. 9A). Figure 9E shows a curdled virosome in transition at a higher magnification: three regions with a normal, virosomal appearance are evident. By 3 h postshift, no curdled virosomes were seen; only normal virosomes with crescents and immature virions were observed (data not shown). These data suggest that the phenotype of tsH5-4 is truly reversible and that the curdled virosomes that accumulate during nonpermissive infections undergo a functional and structural change upon the temperature shift-down that permits virion morphogenesis to occur.
We also performed electron microscopy on tsH5-4 infections that were shifted from the nonpermissive to the permissive temperature in the presence of cycloheximide. We again observed the transformation of curdled virosomes to smooth, electron-dense structures (data not shown), indicating that de novo protein synthesis is not required for this transformation. However, no viral crescents or other virion intermediates were observed in these cells, suggesting that de novo protein synthesis is required for the formation and recruitment of viral membranes.
The H5-4 allele exerts a dominant phenotype.
To confirm that the phenotype of tsH5-4 was due exclusively to the targeted mutation within the H5 gene and not to superfluous mutations located elsewhere in the genome, we attempted to perform marker rescue experiments. The linearized HindIII H fragment of the wt viral genome was transfected into cells infected with tsH5-4, and cultures were then shifted to the nonpermissive temperature for 2 to 3 days. The production of temperature-insensitive virus was monitored by determination of viral yields at 39.5°C. Recombination between the viral genome and the transfected HindIII H fragment should lead to replacement of the H5-4 allele with the wt H5 allele at some frequency, and the resulting wt virus should become amplified during the incubation at 39.5°C; this approach has been used routinely to map numerous vaccinia virus ts mutations (13, 14, 33, 35, 41, 47). To our surprise, HindIII H did not rescue the growth defect of tsH5-4 (data not shown), suggesting that the lesion responsible for the ts phenotype might map elsewhere. However, the entire wt genome, either intact or HindIII restricted, was also unable to rescue the ts phenotype of tsH5-4. Control rescues of other ts mutants were always effective.
We considered two other possibilities for the failure of the rescue: either the tsH5-4 virus has a recombination defect at the nonpermissive temperature, or the H5-4 allele is in fact dominant. If the latter were true, then even if wtH5 protein were produced from recombinant genomes generated within tsH5-4-infected cells during the marker rescue experiment, the dominant H5-4 protein would block morphogenesis at the nonpermissive temperature and wt virus would not be produced. Since we had determined that the H5-4 protein was in fact stable at the nonpermissive temperature, and since we could envision a model in which the curdled virosomes might be preventing the initiation of productive virion morphogenesis even in the presence of wt H5 protein, we decided to test the hypothesis that tsH5-4 might be a dominant mutant. Our objective was to construct a panel of viruses that were diploid for the H5 gene by inserting a second allele into the nonessential TK locus. Would the H5-4 allele, when present in cis with the wtH5 allele, exert a dominant effect?
pJS4 plasmids (9) containing either the wtH5 or H5-4 alleles under the regulation of a constitutive viral promoter were transfected into cells infected with either wt or tsH5-4 virus, and the desired “diploid” recombinants were isolated at 31.5°C, as described in Materials and Methods. The genomic structure and phenotype of the generated viruses are illustrated in Fig. 10. wt virus (row 1, wtVV) formed plaques at both 31.5 and 39.5°C, as did a wtVV:wtH5 diploid (row 3). Remarkably, when the H5-4 allele was expressed from the TK locus of wt virus, it conferred a ts phenotype (row 2, virus wtVV:H5-4), allowing us to conclude that the H5-4 allele is dominant over the wt allele. The dominant phenotype was also reflected in the inability of the wtH5 gene to complement the ts phenotype of tsH5-4 when it was introduced into the TK locus (row 5, tsH5-4:wtH5). To discount the remote possibility that recombination between the two H5 genes had caused the heterozygous viruses (those that contained a wtH5 allele and an H5-4 allele) to become homozygous for the H5-4 allele, we determined the DNA sequence of both the endogenous and exogenous H5 alleles in the final recombinants. Indeed, viruses wtVV:H5-4 and tsH5-4:wtH5 retained their heterozygosity and contained both a wt allele and an H5-4 allele (data not shown). These data allowed us to conclude firmly that the H5-4 allele not only is responsible for the ts phenotype of tsH5-4 but that it also acts in a dominant fashion.
FIG. 10.
The H5-4 allele is genetically linked to the ts phenotype of tsH5-4 and acts as a dominant allele. The targeting vector pJS4 was used to insert a second copy of the H5 gene into the nonessential TK locus. Either the wt H5 gene (wtH5) or the clustered charge-to-alanine mutant gene (H5-4) was cloned into pJS4. Plasmids were transfected into cells infected with either wt vaccinia virus (wtVV) or the ts mutant (tsH5-4) at 31.5°C. Recombinant viruses with the indicated gene insertion were isolated by plaque purification as described in Materials and Methods. Viruses were then tested for a ts phenotype by plating equivalent amounts on BSC40 cells at 31.5 and 39.5°C and staining them with crystal violet after 2 days (right panels). The schematic drawings of the viral genome depict the endogenous H5 locus on the left, containing either the wtH5 or mutant H5-4 allele, and the TK locus on the right, containing either the intact TK gene or the integrated pJS4 targeting vector encoding a second copy of H5 (either wtH5 or H5-4).
DISCUSSION
We report here the first evidence that the H5 protein plays an integral role in the early events of vaccinia virus morphogenesis. We successfully used clustered charge-to-alanine mutagenesis to isolate a ts virus with a lesion in the H5 gene (tsH5-4). Altering three glutamic acid residues to three alanine residues at positions 73 to 75 allows normal viral replication at 31.5°C but decreases the yield of infectious progeny by more than 2 orders of magnitude when the virus is propagated at 39.5°C. Examination of tsH5-4-infected cells by electron microscopy shows that morphogenesis arrests prior to the formation of viral crescents or immature particles at the nonpermissive temperature. Two distinguishing hallmarks of these infections are the presence of a large, cleared area of cytoplasm devoid of cellular organelles or viral intermediates and the presence of curdled virosomes. These latter structures have a striking, splotchy texture, which is clearly abnormal; moreover, they are not accompanied by any developing membrane crescents or other intermediates in virion formation that surround normal virosomes. When nonpermissive infections are shifted down to the permissive temperature, a striking transformation of these curdled structures takes place within 1 h. Smooth, internal patches identical in texture to normal virosomes appear, and within 3 h the curdled virosomes have been fully converted to typical virosomes, which become the sites of productive virus formation. The first stage of this recovery, the transformation of the curdled virosomes to ones with a typical, smooth appearance, occurs even in the presence of cycloheximide. This finding suggests that the H5-4 protein might recover activity upon a temperature shift.
Indeed, the protein encoded by the H5-4 allele is stable during nonpermissive infections, which is atypical for ts mutants. The only sign that it is structurally abnormal is the appearance of a slightly more rapidly migrating form, which might reflect altered protein folding or partial proteolytic attack. Since the mutant protein is not absent during the nonpermissive infections, it must in some way have a loss of function. H5 has affinity both for nucleic acids and for membranes (30), and it is quite possible that one or both of these interactions is compromised during tsH5-4 infections. We propose that the H5-4 protein may localize within the curdled virosomes and fail to promote some crucial interaction at the nonpermissive temperature, thus disrupting virosomal structure and blocking the recruitment and remodeling of virion membranes. Since virosomes also fail to form during nonpermissive infections performed with ts mutants defective in the F10 kinase (44), and since the H5 protein appears to be a substrate for the F10 kinase (Sankar and Traktman, unpublished) as well as for the B1 kinase, it is possible that a defect in the phosphorylation of the H5-4 protein contributes to the phenotype we observed.
Alternatively, the H5-4 protein might fail to localize properly within the virosome. It might also sequester other viral proteins and lead to their exclusion from the virosome. We are currently initiating a comparison of the composition of normal and curdled virosomes by immunoelectron microscopy to determine if any key proteins are missing in the latter. We also plan to compare the localization of the viral A14 and A17 membrane proteins during permissive and nonpermissive infections of tsH5-4. During infections in which the expression of A14 or A17 is repressed, there is an accumulation of vesicles that are believed to be precursors to viral crescent membranes (37, 38, 45, 50). When H5 function is compromised, there is neither crescent formation nor vesicle accumulation, suggesting that the morphogenesis defect in tsH5-4 prevents membrane biogenesis.
We were unable to detect a defect in DNA replication or late gene expression during nonpermissive tsH5-4 infections. It is entirely possible that the lesion in H5-4 simply does not affect the role(s) that H5 may play in these processes. Isolation of additional alleles of the H5 gene would be informative in this regard. What the tsH5-4 allele has done, however, is to uncover a central role for H5 in virion morphogenesis that was not foreshadowed by biochemical investigations.
As described in Results, we experienced difficulty in using the marker rescue technique to conform that the ts phenotype was linked to the H5 mutation. We resolved this problem by generating recombinant viruses that were diploid for the H5 gene, carrying either the wt or H5-4 allele at the endogenous locus and either the wt or H5-4 allele within the nonessential TK locus. By examining these homozygous and heterozygous recombinants, we demonstrated that the H5-4 allele not only is responsible for the ts phenotype but exerts a dominant phenotype: a virus coexpressing wt and H5-4 protein is ts. Although it is initially surprising to consider that a loss-of-function mutant could be dominant, it is by no means unheard of if the mutant protein is stable at the nonpermissive temperature. If the loss of function blocks the progression of a multistep pathway, a normally transient intermediate can accumulate in a manner that may prevent a wt protein from being effective. Alternatively, if the loss of function acts in a dominant fashion by sequestering components of a multisubunit complex, the wt protein would not be capable of restoring the lost interactions. For example, ts mutations in the C′ terminus of the yeast Sec9 protein lead to the assembly of nonproductive SNARE complexes in vivo and can exhibit a dominant phenotype when coexpressed with wt Sec9 protein (40). Two dominant ts (DTS) mutations have also been isolated in Drosophila genes encoding beta-type subunits of the proteasome (beta6/C5 [DTS5] and beta2/Z [DTS7]) (11). The H5-4 protein appears to induce the assembly of nonproductive virosomes at 39.5°C that cannot be rescued by coexpressed wt H5 protein. The fact that the tsH5-4 allele is dominant but reversible suggests that upon shift-down to the permissive temperature, the H5-4 protein regains function and does not exert a dominant effect over newly synthesized H5.
Clustered charge-to-alanine mutagenesis has previously been successful in generating vaccinia virus ts mutants with mutations in the G2 protein and the capping enzyme (21, 22). The experience of the Condit laboratory and of investigators working with other systems suggests that approximately 30 to 40% of alleles generated by this approach will confer a ts phenotype (12, 21, 48). In our case, one of five alleles conferred a tight ts phenotype. More surprisingly, four of five alleles conferred no obvious phenotype at all and were completely viable. This would suggest that the H5 protein is elastic, i.e., capable of sustaining significant alteration without losing function in vivo. Given the multiple functions postulated for this protein, this flexibility is somewhat surprising. Although our studies have elucidated a new and intriguing role for H5 during the viral life cycle, they have also posed many new and compelling questions regarding the structure and function of H5.
ACKNOWLEDGMENTS
We thank Gang Ning for his help with the preparation of samples for electron microscopy and Usha Sankar for her help in the early stages of this project. We also thank Patrick Brennwald and Richard Condit for helpful discussions.
This work was supported by a grant to P.T. from the NIH (2 R01 GM 53601) and by a special group of donors from the Dorothy Rodbell Cohen Foundation.
REFERENCES
- 1.Banham A H, Leader D P, Smith G L. Phosphorylation of ribosomal proteins by the vaccinia virus B1R protein kinase. FEBS Lett. 1993;321:27–31. doi: 10.1016/0014-5793(93)80614-z. [DOI] [PubMed] [Google Scholar]
- 2.Banham A H, Smith G L. Vaccinia virus gene B1R encodes a 34-kDa serine/threonine protein kinase that localizes in cytoplasmic factories and is packaged into virions. Virology. 1992;191:803–812. doi: 10.1016/0042-6822(92)90256-o. [DOI] [PubMed] [Google Scholar]
- 3.Beaud G, Beaud R. Preferential virosomal location of underphosphorylated H5R protein synthesized in vaccinia virus-infected cells. J Gen Virol. 1997;78:3297–3302. doi: 10.1099/0022-1317-78-12-3297. [DOI] [PubMed] [Google Scholar]
- 4.Beaud G, Beaud R, Leader D P. Vaccinia virus gene H5R encodes a protein that is phosphorylated by the multisubstrate vaccinia virus B1R protein kinase. J Virol. 1995;69:1819–1826. doi: 10.1128/jvi.69.3.1819-1826.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaud G, Sharif A, Topa-Masse A, Leader D P. Ribosomal protein S2/Sa kinase purified from HeLa cells infected with vaccinia virus corresponds to the B1R protein kinase and phosphorylates in vitro the viral ssDNA-binding protein. J Gen Virol. 1994;75:283–293. doi: 10.1099/0022-1317-75-2-283. [DOI] [PubMed] [Google Scholar]
- 6.Betakova T, Wolffe E J, Moss B. Regulation of vaccinia virus morphogenesis: phosphorylation of the A14L and A17L membrane proteins and C-terminal truncation of the A17L protein are dependent on the F10L kinase. J Virol. 1999;3:3534–3543. doi: 10.1128/jvi.73.5.3534-3543.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black E P, Moussatche N, Condit R C. Characterization of the interactions among vaccinia virus transcription factors G2R, A18R, and H5R. Virology. 1998;245:313–322. doi: 10.1006/viro.1998.9166. [DOI] [PubMed] [Google Scholar]
- 8.Black P, Condit R C. Phenotypic characterization of mutants in vaccinia virus gene G2R, a putative transcription elongation factor. J Virol. 1996;70:47–54. doi: 10.1128/jvi.70.1.47-54.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakrabarti S, Sisler J R, Moss B. Compact, synthetic, vaccinia virus early/late promoter for protein expression. BioTechniques. 1997;23:1094–1097. doi: 10.2144/97236st07. [DOI] [PubMed] [Google Scholar]
- 10.Condit R C, Motyczka A, Spizz G. Isolation, characterization and physical mapping of temperature sensitive mutants of vaccinia virus. Virology. 1983;128:429–443. doi: 10.1016/0042-6822(83)90268-4. [DOI] [PubMed] [Google Scholar]
- 11.Covi J A, Belote J M, Mykles D L. Subunit compositions and catalytic properties of proteasomes from developmental temperature-sensitive mutants of Drosophila melanogaster. Arch Biochem Biophys. 1999;368:85–97. doi: 10.1006/abbi.1999.1294. [DOI] [PubMed] [Google Scholar]
- 11a.Derrien M, Punjabi A, Khanna M, Grubisha O, Traktman P. Tyrosine phosphorylation of A17 during vaccinia virus infection: involvement of the H1 phosphatase and the F10 kinase. J Virol. 1999;73:7287–7296. doi: 10.1128/jvi.73.9.7287-7296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diamond S E, Kirkegaard K. Clustered charged-to-alanine mutagenesis of poliovirus RNA-dependent RNA polymerase yields multiple temperature-sensitive mutants defective in RNA synthesis. J Virol. 1994;68:863–876. doi: 10.1128/jvi.68.2.863-876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ensinger M, Rovinsky M. Marker rescue of temperature sensitive mutations of vaccinia virus WR: correlation of genetic and physical maps. J Virol. 1983;48:419–428. doi: 10.1128/jvi.48.2.419-428.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans E, Traktman P. Molecular genetic analysis of a vaccinia virus gene with an essential role in DNA replication. J Virol. 1987;61:3152–3162. doi: 10.1128/jvi.61.10.3152-3162.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falkner F G, Moss B. Transient dominant selection of recombinant vaccinia viruses. J Virol. 1990;64:3108–3111. doi: 10.1128/jvi.64.6.3108-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibbs C S, Zoller M J. Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J Biol Chem. 1991;266:8923–8931. [PubMed] [Google Scholar]
- 17.Goebel S J, Johnson G P, Perkus M E, David S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 18.Gordon J, Mohandas A, Wilton S, Dales S. A prominent antigenic surface polypeptide involved in the biogenesis and function of the vaccinia virus envelope. Virology. 1991;181:671–686. doi: 10.1016/0042-6822(91)90901-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimley P M, Rosenblum E N, Mims S J, Moss B. Interruption by rifampicin of an early stage in vaccinia virus morphogenesis: accumulation of membranes which are precursors of virus envelopes. J Virol. 1970;6:519–533. doi: 10.1128/jvi.6.4.519-533.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan K, Broyles S S, Dixon J E. A tyr/ser phosphatase encoded by vaccinia virus. Nature. 1991;350:359–362. doi: 10.1038/350359a0. [DOI] [PubMed] [Google Scholar]
- 21.Hassett D E, Condit R C. Targeted construction of temperature-sensitive mutations in vaccinia virus by replacing clustered charged residues with alanine. Proc Natl Acad Sci USA. 1994;91:4554–4558. doi: 10.1073/pnas.91.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassett D E, Lewis J I, Xing X, DeLange L, Condit R C. Analysis of a temperature-sensitive vaccinia virus mutant in the viral mRNA capping enzyme isolated by clustered charge-to-alanine mutagenesis and transient dominant selection. Virology. 1997;238:391–409. doi: 10.1006/viro.1997.8820. [DOI] [PubMed] [Google Scholar]
- 23.Kane E M, Shuman S. Vaccinia virus morphogenesis is blocked by a temperature-sensitive mutation in the I7 gene that encodes a virion component. J Virol. 1993;67:2689–2698. doi: 10.1128/jvi.67.5.2689-2698.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klemperer N, Ward J, Evans E, Traktman P. The vaccinia virus I1 protein is essential for the assembly of mature virions. J Virol. 1997;71:9285–9294. doi: 10.1128/jvi.71.12.9285-9294.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovacs G R, Moss B. The vaccinia virus H5R gene encodes late gene transcription factor 4: purification, cloning, and overexpression. J Virol. 1996;70:6796–6802. doi: 10.1128/jvi.70.10.6796-6802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin S, Broyles S S. Vaccinia protein kinase 2: a second essential serine/threonine kinase encoded by vaccinia virus. Proc Natl Acad Sci USA. 1994;91:7653–7657. doi: 10.1073/pnas.91.16.7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin S, Chen W, Broyles S S. The vaccinia virus B1R gene product is a serine/threonine protein kinase. J Virol. 1992;66:2717–2723. doi: 10.1128/jvi.66.5.2717-2723.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu K, Lemon B, Traktman P. The dual specificity phosphatase encoded by vaccinia virus, VH1, is essential for viral transcription in vivo and in vitro. J Virol. 1995;69:7823–7834. doi: 10.1128/jvi.69.12.7823-7834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merchlinsky M, Moss B. Resolution of linear minimichrosomes with hairpin ends from circular plasmids containing vaccinia virus concatemer junctions. Cell. 1986;45:879–884. doi: 10.1016/0092-8674(86)90562-3. [DOI] [PubMed] [Google Scholar]
- 30.Mohandas A R, Dekaban G A, Dales S. Vaccinia virion surface polypeptide Ag35 expressed from a baculovirus vector is targeted to analogous poxvirus and insect virus components. Virology. 1994;200:207–219. doi: 10.1006/viro.1994.1179. [DOI] [PubMed] [Google Scholar]
- 31.Moss B. Poxviridae and their replication. In: Fields B N, Knipe D M, editors. Virology. New York, N.Y: Raven Press, Inc.; 1990. pp. 2079–2111. [Google Scholar]
- 32.Moss B, Rosenblum E N. Protein cleavage and poxvirus morphogenesis: tryptic peptide analysis of core precursors accumulated by blocking assembly with rifampicin. J Mol Biol. 1973;81:267–269. doi: 10.1016/0022-2836(73)90195-2. [DOI] [PubMed] [Google Scholar]
- 33.Nakano E, Panicali D, Paoletti E. Molecular genetics of vaccinia virus: demonstration of marker rescue. Proc Natl Acad Sci USA. 1982;79:1593–1596. doi: 10.1073/pnas.79.5.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ravanello M P, Hruby D E. Conditional lethal expression of the vaccinia virus L1R myristylated protein reveals a role in virion assembly. J Virol. 1994;68:6401–6410. doi: 10.1128/jvi.68.10.6401-6410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rempel R E, Anderson M K, Evans E, Traktman P. Temperature-sensitive vaccinia virus mutants identify a gene with an essential role in viral replication. J Virol. 1990;64:574–583. doi: 10.1128/jvi.64.2.574-583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rempel R E, Traktman P. Vaccinia virus B1 kinase: phenotypic analysis of temperature-sensitive mutants and enzymatic characterization of recombinant proteins. J Virol. 1992;66:4413–4426. doi: 10.1128/jvi.66.7.4413-4426.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez D, Esteban M, Rodriguez J R. Vaccinia virus A17L gene product is essential for an early step in virion morphogenesis. J Virol. 1995;69:4640–4648. doi: 10.1128/jvi.69.8.4640-4648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez J R, Risco C, Carrascosa J L, Esteban M, Rodriguez D. Vaccinia virus 15-kilodalton (A14L) protein is essential for assembly and attachment of viral crescents to virosomes. J Virol. 1998;72:1287–1296. doi: 10.1128/jvi.72.2.1287-1296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosel J, Earl P L, Weir J, Moss B. Conserved TAAATG sequence at the transcriptional and translational initiation sites of vaccinia virus late genes deduced by structural and functional analysis of the HindIII H genome fragment. J Virol. 1986;60:436–449. doi: 10.1128/jvi.60.2.436-449.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossi G, Salminen A, Rice L M, Brunger A T, Brennwald P. Analysis of a yeast SNARE complex reveals remarkable similarity to the neuronal SNARE complex and a novel function for the C terminus of the SNAP-25 homolog, Sec9. J Biol Chem. 1997;272:16610–16617. doi: 10.1074/jbc.272.26.16610. [DOI] [PubMed] [Google Scholar]
- 41.Sam C-K, Dumbell K R. Expression of poxvirus DNA in coinfected cells and marker rescue of thermosensitive mutants by subgenomic fragments of DNA. Ann Virol. 1981;132:135–150. [Google Scholar]
- 42.Tartaglia J, Piccini A, Paoletti A. Vaccinia virus rifampicin-resistance locus specified a late 63,000 Da gene product. Virology. 1986;150:45–54. doi: 10.1016/0042-6822(86)90264-3. [DOI] [PubMed] [Google Scholar]
- 43.Traktman P, Anderson M K, Rempel R E. Vaccinia virus encodes an essential gene with strong homology to protein kinases. J Biol Chem. 1989;264:21458–21461. [PubMed] [Google Scholar]
- 44.Traktman P, Caligiuri A, Jesty S A, Liu K, Sankar U. Temperature-sensitive mutants with lesions in the vaccinia virus F10 kinase undergo arrest at the earliest stage of virion morphogenesis. J Virol. 1995;69:6581–6587. doi: 10.1128/jvi.69.10.6581-6587.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Traktman, P., K. Liu, R. A. Rollins, S. A. Jesty, and B. Unger. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: construction and characterization of a tetracycline-inducible recombinant. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 46.Wang S, Shuman S. Vaccinia virus morphogenesis is blocked by temperature-sensitive mutations in the F10 gene, which encodes protein kinase 2. J Virol. 1995;69:6376–6388. doi: 10.1128/jvi.69.10.6376-6388.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weir J P, Bajszar G, Moss B. Mapping of the vaccinia virus thymidine kinase gene by marker rescue and by cell free translation of selected mRNA. Proc Natl Acad Sci USA. 1982;79:1210–1214. doi: 10.1073/pnas.79.4.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wertman K F, Drubin D G, Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams O, Wolffe E J, Weisberg A S, Merchlinsky M. Vaccinia virus WR gene A5L is required for morphogenesis of mature virions. J Virol. 1999;73:4590–4599. doi: 10.1128/jvi.73.6.4590-4599.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolffe E J, Moore D M, Peters E J, Moss B. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J Virol. 1996;70:2797–2808. doi: 10.1128/jvi.70.5.2797-2808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Keck J G, Moss B. Transcription of viral late genes is dependent on expression of the viral intermediate gene G8R in cells infected with an inducible conditional-lethal mutant vaccinia virus. J Virol. 1992;66:6470–6479. doi: 10.1128/jvi.66.11.6470-6479.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Moss B. Vaccinia virus morphogenesis is interrupted when expression of a gene encoding an 11-kilodalton phosphorylated protein is prevented by the Escherichia coli lac repressor. J Virol. 1991;65:6101–6110. doi: 10.1128/jvi.65.11.6101-6110.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Moss B. Immature viral envelope formation is interrupted at the same stage by lac operator mediated repression of the vaccinia virus D13L gene and by the drug rifampicin. Virology. 1992;187:643–653. doi: 10.1016/0042-6822(92)90467-4. [DOI] [PubMed] [Google Scholar]