Abstract
Hepatitis E virus (HEV) is an important etiological agent of epidemic and sporadic hepatitis, which is endemic to the Indian subcontinent and prevalent in most of the developing parts of the world. The infection is often associated with acute liver failure and high mortality, particularly in pregnant women. In order to develop methods of intervention, it is essential to understand the biology of the virus. This is particularly important as no reliable in vitro culture system is available. We have constructed a cDNA clone encompassing the complete HEV genome from independently characterized subgenomic fragments of an Indian epidemic isolate. Transfection studies were carried out with HepG2 cells using in vitro-transcribed RNA from this full-length HEV cDNA clone. The presence of negative-sense RNA, indicative of viral replication, was demonstrated in the transfected cells by strand-specific reverse transcription-PCR and slot blot hybridization. The viral proteins pORF2 and pORF3 and processed components of the pORF1 polyprotein (putative methyltransferase, helicase, and RNA-dependent RNA polymerase) were identified in the transfected cells by metabolic pulse-labeling with [35S]methionine-cysteine, followed by immunoprecipitation with respective antibodies. The expression of viral proteins in the transfected cells was also demonstrated by immunofluorescence microscopy. Viral replication was detected in the transfected cells up to 33 days posttransfection (six passages). The culture supernatant from the transfected cells was able to produce HEV infection in a rhesus monkey (Macaca mulatta) following intravenous injection, indicating the generation of viable HEV particles following transfection of cells with in vitro-synthesized genomic RNA. This transient cell culture model using in vitro-transcribed RNA should facilitate our understanding of HEV biology.
Hepatitis E virus (HEV) is established as an etiological agent of the epidemic and sporadic forms of waterborne hepatitis (6, 25). The first well-characterized HEV epidemic was reported in Delhi, India, in 1955 (35). Several other epidemics have since been described in developing countries on nearly every continent (19, 21). HEV has a positive-strand polyadenylated RNA genome ∼7.2 kb in length (27) containing three open reading frames (ORFs). Nonstructural ORF1 (5′ end) codes for a polyprotein of 1,693 amino acids (pORF1) and contain domains homologous to a viral methyltransferase, a papainlike cysteine protease, an RNA helicase, and an RNA-dependent RNA polymerase (RdRp). The second ORF (3′ end) codes for the major viral capsid protein of 660 amino acid (pORF2), while the third and smallest ORF (ORF3) codes for a 123-amino-acid-long polypeptide (pORF3) whose function is unknown (32). Apart from its coding region, the viral genome has 27- and 68-nucleotide (nt)-long noncoding regions at its 5′ and 3′ ends, respectively (32). The genome sequences of HEV have been reported from different geographical isolates and show a high degree of homology at both the nucleotide and amino acid levels (3, 4, 12, 32, 33). Expression of structural proteins pORF2 and pORF3 in prokaryotic and eukaryotic systems has been reported by different investigators (7, 11, 14, 23, 29). Earlier, we have expressed and characterized pORF2 and pORF3 in animal cells. pORF2 is an 88-kDa glycoprotein which is expressed intracellularly, as well as on the cell surface (14, 37). The ORF3 protein (pORF3) is a 13.5-kDa phosphoprotein which is phosphorylated by the cellular mitogen-activated protein kinase and associates with the cytoskeleton (36). We have also recently expressed the ORF1 polyprotein in both prokaryotic and eukaryotic systems (2).
In the absence of a reliable in vitro culture system, the replication and transcription strategy of HEV is poorly understood. The in vitro propagation and production of HEV from the primary hepatocytes of experimentally infected cynomolgus macaques has been reported (31). However, this is of limited utility due to difficulties associated with animal experimentation. Several cDNA clones of other positive-strand RNA viruses have been shown to be infectious in cell culture and experimental animal systems (9, 26). In the present study, we have analyzed the potential of a full-length, in vitro-synthesized HEV RNA from a cDNA clone of an Indian isolate of HEV. We demonstrate here the replication and expression of HEV from this RNA in cultured hepatoma cells, as well as the production of infectious virions ascertained through experimental infection of a rhesus macaque.
MATERIALS AND METHODS
Assembly of a full-length HEV cDNA clone.
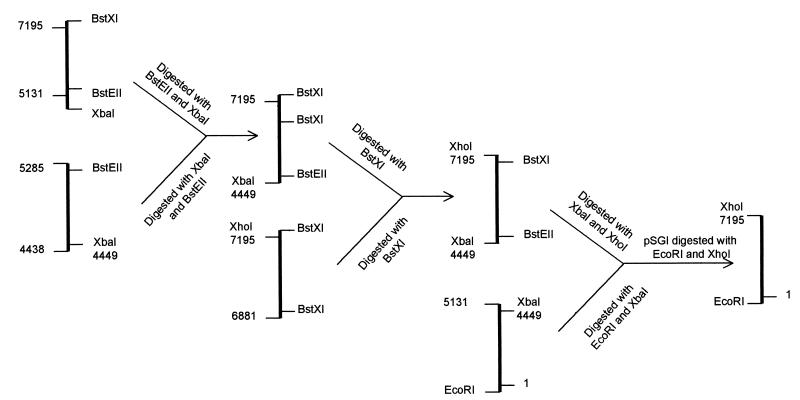
The three known ORFs (GenBank accession no. AF028091 [ORF1] and U22532 [ORF2 and ORF3]), along with the noncoding regions from an epidemic isolate of HEV from India, have been cloned by us using subgenomic PCR amplification followed by a reconstruction strategy (2, 14). We have expressed these fragments in both prokaryotic and eukaryotic systems (2, 14, 23). These subgenomic fragments were produced by PCR cloning and assembly from a single viral isolate grown in a rhesus monkey. These previously described cloned fragments were used for the reconstruction of a full-length genomic cDNA clone of HEV. Briefly, an XhoI restriction enzyme site was engineered in the primer 7195 sequence (5′GCctcgagTTTTTCAGGGAGCGCGGAACGCA3′) that had a stretch of five thymidine bases at the 3′ end to produce an ORF2-pBluescript SK(+) (Stratagene) clone. This clone was extended at the 5′ end by inserting a PCR-amplified fragment of HEV covering 4,438 to 5,285 nt using a standard cloning procedure. The HEV clone ORF1-pSGI was digested with EcoRI and XbaI to release an insert ranging from nt 1 to nt 4449, whereas the insert ranging from nt 4449 to nt 7195 was released from the nt 4438 to nt 7195 pBluescript SK(+) clone by digestion with restriction enzymes XbaI and XhoI. These inserts were cloned into a pSGI vector (13) digested with restriction enzymes EcoRI and XhoI in a three-way ligation. A schematic representation of the reconstruction is given in Fig. 1. This full-length HEV cDNA clone was completely sequenced after reconstruction and named pSGI-HEV(I).
FIG. 1.
Schematic representation of the strategy used to assemble a full-length HEV cDNA clone. The ORF2 pCR-Script SK(+) clone was extended at its 5′ end with a PCR-amplified fragment (nt 4438 to 5285) using inherent BstEII and XbaI restriction sites. An XhoI site was created at the 3′ end using another PCR-amplified fragment with the XhoI site in the primer, which replaced a fragment extending from nt 6881 to 7195. This fragment (nt 4449 to 7195) and the fragment of ORF1 (nt 1 to 4449) were used in a three-way ligation to create the complete cDNA clone of HEV [pSGI-HEV(I)] in the pSGI vector.
In vitro transcription of full-length HEV RNA.
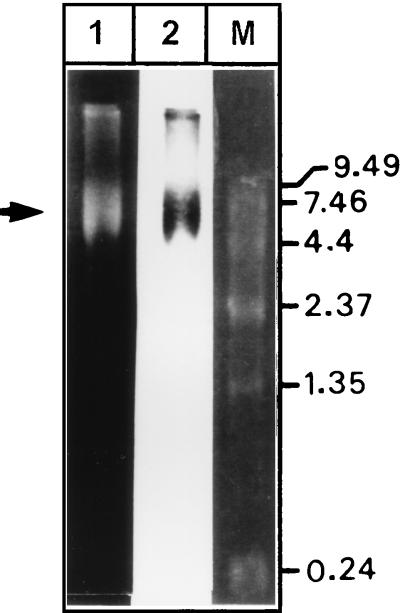
In vitro transcription of the above-described cDNA clone [pSGI-HEV(I)] was carried out using T7 RNA polymerase (Stratagene) to generate full-length HEV transcripts. Briefly, the CsCl2 gradient-purified plasmid [pSGI-HEV(I)] containing the full-length HEV cDNA was digested with XhoI to produce a linear DNA template. RNA was transcribed in vitro using 50 U of T7 RNA polymerase in a 50-μl reaction volume containing 40 mM Tris-HCl (pH 8.0), 8 mM MgCl2, 50 mM NaCl, 2 mM spermidine, 30 mM dithiothreitol, 400 μM each ribonucleoside triphosphate, and 5 μg of template DNA. The reaction mixture was incubated at 37°C for 30 min and then digested with DNase I (RQ1 RNase-free DNase; Promega) for 1 h at 37°C. The reaction mixture was extracted with phenol-chloroform and then subjected to a second round of DNase I digestion and phenol-chloroform extraction to ensure no carryover of the template DNA with the transcripts. The transcripts were ethanol precipitated at −70°C for 1 h. The RNA pellet was washed with cold 70% ethanol and dried, and its integrity was analyzed by 0.8% formaldehyde agarose gel electrophoresis, followed by ethidium bromide staining and Northern hybridization (Fig. 2).
FIG. 2.
Analysis of in vitro-transcribed full-length HEV RNA from the cDNA clone [pSG-HEV(1)] by 0.8% formaldehyde agarose gel electrophoresis. Lanes: 1, ethidium bromide-stained gel; 2, Northern hybridization using an HEV-specific probe; M, RNA molecular size markers (in kilobases; Life Technologies).
In vitro-produced RNA was resolved on a 0.8% formaldehyde denaturing agarose gel and transferred to nylon membrane (Hybond; Amersham International) in the presence of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The membrane was washed with a 10× SSC solution, air dried, and subjected to UV cross-linking in a UV cross-linker (Stratagene). The membrane was put in a prehybridization solution (6× SSC, 5× Denhardt's solution, 0.5% sodium dodecyl sulfate [SDS], 100 μg of calf thymus DNA per ml of solution) and incubated at 68°C for 6 h in a hybridization oven (Shel Lab model 1004). The hybridization was subsequently carried out in a fresh prehybridization solution containing 107 cpm of an [α-32P]dCTP-labeled probe generated from the full-length ORF2 clone of HEV (22). The probe was prepared by using a commercial random priming kit (Prime-it; Stratagene) in accordance with the manufacturer's protocol. Following hybridization for 16 h at 68°C, the membrane was washed as follows: (i) once in 2× SSC–0.1% SDS for 5 min at room temperature, (ii) twice in 0.2× SSC–0.1% SDS for 5 min at room temperature, (iii) once in 0.2× SSC–0.1% SDS for 15 min at 42°C, and (iv) once in 0.1× SSC–0.1% SDS for 15 min at 68°C.
All of the solutions were discarded as radioactive waste. Following the last wash, the membrane was wrapped in Saran Wrap and exposed to autoradiography using Kodak X-Omat AR film with Du Pont intensifying screens.
RNA transfection and metabolic pulse-labeling.
HepG2 cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum (Life Technologies). Cells at ∼50% confluency were used for transfection of HEV RNA. Twenty micrograms of in vitro-produced RNA was transfected by a liposome induction method (Lipofectamine; Life Technologies) in accordance with the manufacturer's guidelines. The plasmid vector (pSGI) served as a control for the transfection. For each 60-mm-diameter culture dish, 20 μg of the HEV RNA and 10 μl of Lipofectamine were diluted in 1.5 ml of serum-free medium. The mixture was kept at room temperature for 30 min and gently overlaid onto the monolayer. Fresh medium with 10% fetal calf serum was added after 6 h, and the cells were kept in an atmosphere of 5% CO2. After 72 h, the cells were harvested for extraction of total RNA.
Transfected cells were pulse-labeled (100 μCi/ml/60-mm-diameter plate) with [35S]methionine-cysteine (Promix; Amersham International) for 4 h at 72 h posttransfection in methionine-cysteine-deficient Dulbecco's modified Eagle's medium (Sigma). The metabolically labeled cells were harvested, and proteins were immunoprecipitated using HEV-specific polyclonal antibodies. Similar labeling experiments were also carried out at 12, 24, 36, 72, and 96 h to determine the expression kinetics of the viral RdRp. A batch of the HEV RNA-transfected cells was maintained in the culture and allowed to grow for the next 45 days (eight passages). These cells were analyzed at 3, 7, 15, 33, and 45 days posttransfection for the presence of antisense RNA replicative intermediates using strand-specific PCR.
Detection of antisense HEV RNA.
Strand-specific PCR was carried out to detect antisense and sense HEV RNAs in the transfected cells (20). Total RNA from the cells was isolated at days 3, 7, 15, 33, and 45 posttransfection by a single-step RNA isolation method (8). A serial log fold dilution of the total RNA extracted at 72 h (3 days) was carried out to determine an approximate ratio of sense and antisense strands. For sense strand detection, the HepG2 cells transfected with plasmid pSGI and serum from an HEV-infected monkey with viremia served as negative and positive controls, respectively. For antisense strand detection, RNA isolated from an HEV-infected monkey liver containing an antisense replicative intermediate served as a positive control whereas bile fluid or serum from the same viremic animal served as a negative control. For strand-specific detection, reverse transcription (RT) was carried out using either a sense or an antisense primer. Following cDNA synthesis, the RNA in the reaction mixture was degraded by digestion with RNase H (2 U) and RNase A (1 μg; Promega). Following RNase treatment, the reaction mixture was extracted once with phenol-chloroform and ethanol precipitated. The precipitated cDNA was used for PCR amplification using both sense and antisense primers.
For hybridization-based detection, total RNA (30 μg) extracted from the transfected cells was immobilized on a nylon membrane (Amersham International) using a Hybri-slot manifold (Life Technologies). RNA from cells transfected with plasmid pSGI was used as a negative control. In addition, the in vitro-transcribed, unlabeled sense and antisense HEV RNAs (2.5 μg of each) were used as positive and negative controls alternatively. Before transfer, the manifold was cleaned with 0.1 M NaOH and rinsed twice with diethyl pyrocarbonate-treated water. The membrane was cut to the size of the manifold and soaked in 20× SSC for 10 min prior to blotting. Approximately 30 μg of the total RNA isolated from the HEV RNA-transfected cells was mixed with 3 volumes of denaturing solution (5 ml of deionized formamide, 1.62 ml of 37% formaldehyde, 1 ml of MOPS buffer (0.2 M morpholinepropanesulfonic acid [MOPS], 0.5 M sodium acetate, 0.01 M EDTA [pH 8.0]) and incubated at 65°C for 15 min. The mixture was chilled on ice, diluted with 1 ml of ice-cold 20× SSC, and loaded into defined wells. Suction was continued until the samples in all of the wells were exhausted. The membrane was air dried, UV cross-linked, and incubated in prehybridization solution as described above. Sense- and antisense-specific riboprobes were prepared by transcription with T7 and SP6 polymerases (Riboprobe system-T7 and Riboprobe system-SP6; Promega) using direct and reverse-oriented clones of an HEV cDNA encompassing nt 1 to 457 in pCR-Script SK(+) (Stratagene) and pGEM-T (Promega) as vectors, respectively. The transcription reaction was carried out in the presence of [α-33P]UTP (2,500 Ci/mmol; Amersham International). Prior to in vitro transcription, the template DNA was linearized by restriction enzyme digestion at the end of the fragment. The reaction mixture was treated with DNase I, extracted with phenol-chloroform, and ethanol precipitated as described above. The hybridization and washing conditions used were similar to those described earlier. The thoroughly washed membrane was wrapped in Saran Wrap and exposed to autoradiography.
Detection of viral proteins in transfected cells.
The RNA-transfected cells were lysed in 750 μl of radioimmunoprecipitation assay buffer (10 mM Tris-HCl [pH 8.0], 140 mM NaCl, 5 mM iodoacetamide, 0.5% Triton X-100, 1% SDS, 1% sodium deoxycholate, 2 mM phenylmethylsulfonyl fluoride). The clarified lysate was incubated with 7 μl of anti-ORF2, anti-ORF3, or anti-ORF1 antibodies (putative anti-methyltransferase [anti-met], putative anti-helicase [anti-hel], or putative anti-RdRp, respectively) independently on ice for 1 h. The polyclonal antibodies were raised in rabbits against the structural proteins (pORF2 and pORF3) and components of nonstructural polyprotein pORF1 (putative methyltransferase, helicase, and RdRp regions) as described elsewhere (2, 23). The antigen-antibody complexes formed were further incubated with 100 μl of a 10% suspension of preswollen protein A-Sepharose 4B (Pharmacia, Uppsala, Sweden), and the reaction mixture was kept at 4°C with slow end-to-end shaking. After 1 h, the reaction mixture was centrifuged for 1 min at 10,000 rpm in a refrigerated microcentrifuge (Hermle). The supernatant was discarded as radioactive waste, and the beads were washed thrice with 1 ml of radioimmunoprecipitation assay buffer for 10 min at 4°C with shaking. The complex was boiled with 50 μl of 2× SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 5% 2-β-mercaptoethanol, 0.1% bromophenol blue) and analyzed by SDS–6 to 15% gradient PAGE. The gel was treated with 0.5 M sodium salicylate, washed, dried, and exposed to autoradiography as described above.
For immunofluorescence studies, transfection was carried out on cells grown on coverslips (30-mm diameter). After 72 h, the cells on the coverslips were washed twice with phosphate-buffered saline (PBS, pH 7.2) and fixed with 4% paraformaldehyde at room temperature for 30 min. Following fixation, the cells were incubated with 0.1% saponin (Sigma) on ice for 10 min, washed with PBS, and incubated with 1:100-diluted anti-pORF2, anti-pORF3, anti-met, and anti-hel antibodies for 1 h in a humid chamber at 37°C. The cells were washed with PBS and further incubated with a goat anti-rabbit fluorescein isothiocyanate conjugate (1:100 dilution; Sigma) at 37°C in the humid chamber for 45 min. After three washings with PBS, the coverslips were mounted on glass slides and observed under a confocal microscope (Bio-Rad). Similarly, immunofluorescence labeling of transfected cells with anti-RdRp antibody was carried out 24 h posttransfection. Cells transfected with the vector pSGI served as a negative control.
Experimental infection of rhesus monkeys.
Ethical clearance was obtained from the institutional primate research facility for experimentation on rhesus monkeys. The animals (M-1690, M-1761, M-1927, and M-2197) were put under quarantine, and any prior infections were ruled out. Preinoculation blood was collected aseptically. The sera were analyzed for the presence of anti-HEV immunoglobulin M (IgM), anti-HEV IgG, and HEV RNA by RT-PCR. Animals negative for these markers were used for further experiments. One rhesus monkey (M-1690) was injected intravenously with 6 ml of pooled culture supernatants from HEV RNA-transfected HepG2 cells collected at 72 h. The control monkey (M-1761) was injected only with PBS and kept under the same conditions. Two other monkeys (M-1927 and M-2197) were injected with 100 μg of in vitro-transcribed HEV RNA with animal-injectable RNase inhibitor (Promega) in the liver at multiple sites after performance of a minilaparotomy. After the inoculation, blood samples were collected aseptically twice a week and serum was stored at −70°C for further use. Alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels were monitored by using a commercial assay kit (Boehringer GmbH, Mannheim, Germany). HEV antibodies were detected by an in-house enzyme-linked immunosorbent assay system using recombinant HEV proteins (pORF2, pORF3, and pORF1) (S. K. Panda, unpublished data). The presence of HEV RNA was investigated by RT-PCR as described earlier (13).
Nucleotide sequence accession number.
The sequence of pSGI-HEV(I) was submitted to GenBank and assigned accession no. AF076239.
RESULTS
Production of HEV transcripts.
An HEV cDNA clone encompassing nucleotides 1 to 7195 with a 5-bp stretch of adenine residues at its 3′ end was reconstructed in a eukaryotic expression vector (Fig. 1). In vitro transcription of this clone generated a transcript of ∼7.2 to 7.4 kb as determined by denaturing agarose gel electrophoresis and Northern blot analysis. Most of the in vitro-transcribed material corresponded to this RNA species, as shown by a comparison of the ethidium bromide-stained and Northern blotted lanes (Fig. 2). Most of the HEV transcripts were about 7.2 kb in size, corresponding to the complete genome, when compared with standard RNA molecular weight markers (Life Technologies). The resolved RNA was blotted onto a nylon membrane, and Northern hybridization with an HEV-specific probe confirmed the full-length HEV transcripts (Fig. 2).
Detection of antisense replicative intermediate RNA.
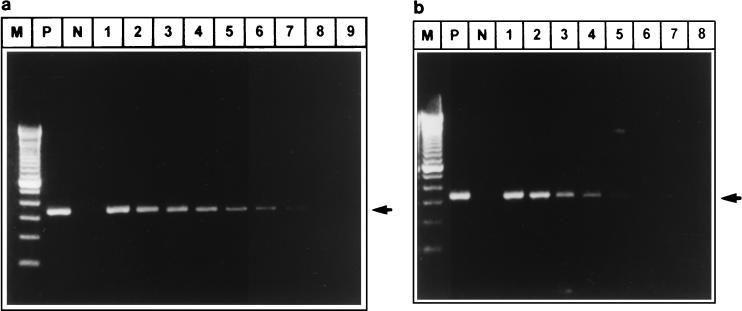
Both the sense and antisense strands of the HEV genome were detected by strand-specific RT-PCR in HEV RNA-transfected HepG2 cells. The pSGI-transfected cells remained negative for HEV sequences at all times. The HEV RNA-transfected cells were positive for the sense and antisense strands of the HEV genome at 3, 7, 15, and 33 days (data not shown). In the experiment with serial log-fold dilutions carried out at 72 h posttransfection, the negative-sense strand of the HEV genome was detected up to a dilution of 10−6 of the total HepG2 cell RNA. On the other hand, the positive-sense strand was detected up to a dilution of 10−15 (Fig. 3a and b). The positive-sense strand was more abundant than the negative-sense strand.
FIG. 3.
HEV genome amplification product resulting from strand-specific PCR carried out on total RNA extracted from HepG2 cells transfected with full-length, positive-sense, in vitro-synthesized HEV RNA. (a) Detection of the negative-sense strand of HEV RNA. Lanes: M, marker (100-bp ladder; Life technologies); P, positive control (RNA isolated from HEV-infected rhesus monkey liver); N, negative control (RNA extracted from bile fluid of a HEV-infected rhesus monkey); 1, RNA (1 μg) extracted from HepG2 cells transfected with in vitro-synthesized HEV RNA at 72 h (neat); 2 through 9, log dilutions of transfected HepG2 cell RNA from 10−1 through 10−8. (b) Detection of the positive-sense strand of HEV RNA. Lanes: M, marker (100-bp ladder; Life Technologies); P, positive control (RNA extracted from HEV-infected rhesus monkey serum); N, negative control (RNA extracted from normal control rhesus monkey serum); 1, RNA (1 μg) extracted from HepG2 cells transfected with in vitro-synthesized HEV RNA at 72 h; 2 through 8, log dilutions of total RNA from transfected HepG2 cells (2, 10−1 dilution; 3, 10−5 dilution; 4, 10−10 dilution; 5, 10−15 dilution; 6, 10−16 dilution; 7, 10−17 dilution; 8, 10−18 dilution).
To further confirm the presence of antisense RNA, a strand-specific hybridization was carried out using [α-33P]UTP-labeled riboprobes. Both sense and antisense HEV RNAs could be detected in transfected cells by this method, while control cells transfected with the pSGI vector did not show any signal on hybridization (Fig. 4). In vitro-synthesized sense and antisense HEV RNAs were used to validate the specificity of the hybridization method in detecting the strands. The presence of antisense HEV RNA was reconfirmed by hybridization.
FIG. 4.
Strand-specific slot blot hybridization for detection of positive- and negative-sense HEV RNA in total RNA extracted from HepG2 cells transfected with full-length, in vitro-transcribed HEV RNA. (A) Hybridization with [α-33P]UTP-labeled riboprobe of antisense polarity. Slots: 1, In vitro-synthesized positive-sense HEV RNA (positive control); 2, In vitro-synthesized negative-sense HEV RNA (negative control); 3, RNA isolated from HepG2 cells transfected with full-length, in vitro-transcribed HEV RNA; 4, RNA isolated from HepG2 cells transfected with the pSGI vector as a control. (B) Hybridization with [α-33P]UTP riboprobe of sense polarity: 1, In vitro-synthesized HEV RNA of negative-sense polarity (positive control); 2, In vitro-synthesized HEV RNA of positive-sense polarity (negative control); 3, RNA isolated from HepG2 cells transfected with full-length, in vitro-transcribed HEV RNA; 4, RNA isolated from HepG2 cells transfected with the pSGI vector as a control.
Detection of viral proteins in RNA-transfected cells.
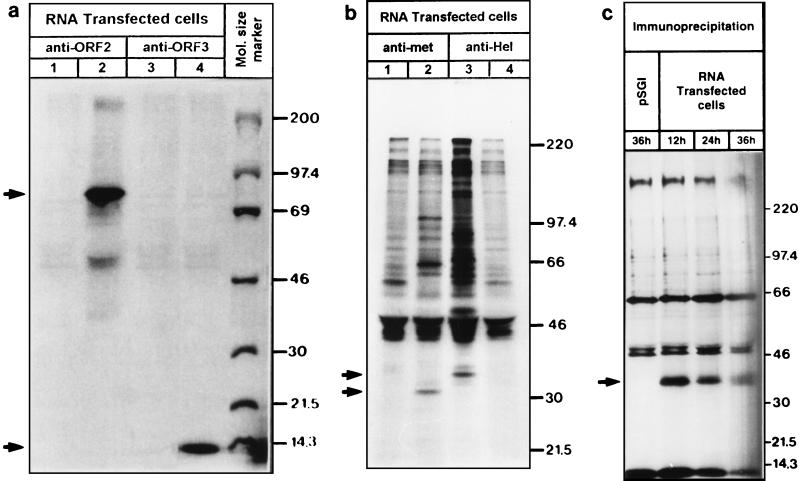
The HEV ORF2 and ORF3 proteins (pORF2 and pORF3) were detected by immunoprecipitation with the corresponding specific antibodies (Fig. 5a). pORF2 was detected as an ∼72-kDa protein by SDS–6 to 15% gradient PAGE followed by autoradiography. A protein corresponding to pORF3 (∼13.5 kDa) was also immunoprecipitated from the transfected cells (Fig. 5a). The signals corresponding to the putative methyltransferase (∼35 kDa), helicase (∼38 kDa), and RdRp (∼36 kDa) domains could be detected by immunoprecipitation (Fig. 5b and c). The putative helicase and methyltransferase were detected in samples prepared at 72 h posttransfection. However, the putative RdRp was detected only at 12, 24, and 36 h posttransfection (Fig. 5c). It was not detected in transfected cells at 72 and 96 h (data not shown). There were additional unique high-molecular-weight bands in the immunoprecipitation experiments using antibodies against the putative helicase and methyltransferase domains. These may be intermediates in the processing of the ORF1 protein (Fig. 5b). In all of the immunoprecipitation experiments, pSGI-transfected HepG2 cells were used as a control and specific polypeptide species were found to be missing from these control cells.
FIG. 5.
(a) Autoradiograph showing immunoprecipitation of structural proteins from HEV RNA-transfected HepG2 cells. The arrows indicate signals in lanes 2 and 4 that represent pORF2 (∼72 kDa) and pORF3 (13.5 kDa), respectively. pSGI-transfected cells were immunoprecipitated with anti-ORF2 and anti-ORF3 antibodies (lanes 1 and 3) to serve as controls. The immunoprecipitates were analyzed by SDS–6 to 15% gradient PAGE and visualized by fluorography. The molecular sizes of 14C-labeled markers (in kilodaltons; Amersham International) are indicated on the right. (b) Autoradiograph showing immunoprecipitation of putative domains of a nonstructural polyprotein corresponding to the methyltransferase and helicase from HEV RNA-transfected HepG2 cells. The arrows indicate signals in lanes 2 and 3 that represent the signals corresponding to the putative methyltransferase (∼35 kDa) and helicase (∼38 kDa), respectively. Mock-transfected pSGI cells were immunoprecipitated with anti-met and anti-hel antibodies (lanes 1 and 4) to serve as controls. The immunoprecipitates were analyzed by SDS–6 to 15% gradient PAGE and visualized by fluorography. Molecular size markers, in kilodaltons (Rainbow markers; Amersham International), are indicated on the right. (c) Autoradiograph showing immunoprecipitation of putative domains of a nonstructural polyprotein corresponding to RdRp from HEV RNA-transfected HepG2 cells. The immunoprecipitation was carried out using anti-RdRp antibodies at 12, 24, and 36 h posttransfection. For a control, HepG2 cells were transfected with the pSGI vector and immunoprecipitated at 36 h with the same antibody. The immunoprecipitates were analyzed by SDS–6 to 15% gradient PAGE and visualized by fluorography. Molecular size markers, in kilodaltons (Rainbow markers; Amersham International), are indicated on the right.
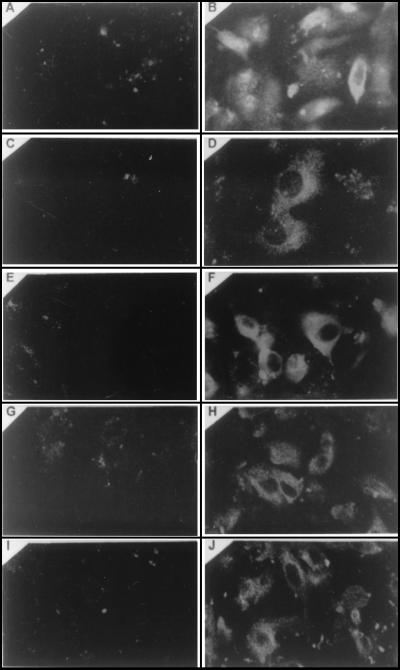
Fluorescent-antibody staining was carried out at 72 h posttransfection with anti-pORF2, anti-pORF3, anti-met, anti-hel, and anti-RdRp antibodies against the HEV proteins. In addition, staining for RdRp was performed at 24 h posttransfection because of its absence at 72 h in the immunoprecipitation experiments. Both the structural (pORF2 and pORF3) and nonstructural proteins (corresponding to the putative domains of Met, Hel, and RdRp) were demonstrated in the transfected cells following staining with the corresponding antibodies (Fig. 6). None of the antibodies showed any significant signal with control cells transfected with vector plasmid pSGI (Fig. 6).
FIG. 6.
Composite photograph showing immunofluorescent antibody staining of in vitro-synthesized HEV RNA-transfected HepG2 cells and control cells transfected with the pSGI vector. Panels B, D, F, H, and K represent the immunofluorescent staining of HEV-transfected HepG2 cells with anti-pORF2, anti-pORF3, anti-met, anti-hel, and anti-RdRp antibodies, respectively. Panels A, C, E, G, and J represent the immunostaining of control HepG2 cells with the same antibodies corresponding to the test panel. All of the immunostaining was carried out at 72 h posttransfection, except that with the anti-RdRp antibodies, which was performed 24 h posttransfection.
HEV infection of Macaca mulatta following inoculation with culture supernatant from RNA-transfected cells.
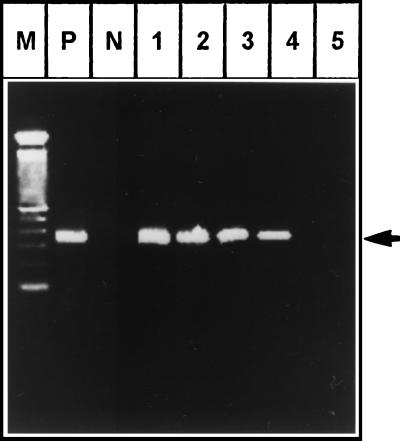
The culture supernatant from HEV RNA-transfected cells was used to produce infection in a rhesus monkey (M-1690). Following inoculation, HEV RNA was observed with the help of RT-PCR in sera collected on days 24 to 37 (Fig. 7). During this period (24 to 37 days), the AST and ALT levels increased to 1.5 to 2.5 (53 to 100 IU/liter) times normal levels. The IgM class of anti-HEV antibodies directed against the ORF1, ORF2, and ORF3 viral proteins were detected after 4 weeks and persisted for the next 14 days. The ratios of optical density between preinoculation and positive sera were in the range of 1:8 to 1:15, which is typical for HEV infection in rhesus monkeys. The animals (M-1927 and M-2197) which received in vitro-produced HEV RNA, as well as the control monkey (M-1761), remained normal, with no rise in ALT and AST levels, and no seroconversion for antibodies was observed. They also remained negative for HEV RNA in serum (viremia) throughout the follow-up period. The anti-HEV IgG antibodies were detected in the infected monkey (M-1690) 3 months after inoculation.
FIG. 7.
Agarose (2%) gel electrophoresis of RT-nested PCR products (343 bp) of the HEV genome amplified from the serum of infected rhesus monkey (M. mulatta) M-1690. P, positive control; N, control monkey M-1761; M, 100-bp DNA ladder (Life Technologies). Lanes 1 to 5 represent serum samples collected from the animal on days 24, 28, 33, 37, and 43, respectively, postinjection. Arrow, amplified fragments from HEV genome.
DISCUSSION
Infection due to HEV accounts for one-third of the sporadic acute viral hepatitis and almost all of the described epidemics on the Indian subcontinent (16, 22). It is a major health problem in tropical and subtropical parts of the world. The genome of HEV has been cloned and sequenced from several different geographical isolates (3, 12, 32, 33). However, the biology of the virus is poorly understood due to the lack of a suitable culture system. Recently, primary hepatocytes collected from HEV-infected macaques have been successfully propagated (31). However, this is time consuming and requires a suitable experimental animal facility. The alternative approach is to express the viral proteins in vitro, study their properties, and thereby define their role in the viral life cycle. Our earlier studies involved such a subgenomic expression strategy to characterize HEV structural proteins pORF2 and pORF3 (14, 23, 36, 37). Complete nonstructural ORF1 and its putative functional domains have been similarly reconstructed and expressed in prokaryotic and eukaryotic systems (2). These three ORFs, originating from a single viral isolate, were joined in the proper orientation to produce a full-length cDNA clone of HEV, which was used to generate full-length HEV RNA. Such in vitro-generated RNA was shown to be infectious in the tissue culture system. This is similar to observations with the other positive-stranded RNA viruses (9, 26, 28, 30).
Production of a replication- and transcription-competent (infectious) clone will mitigate the difficulties in the analysis of virus biology. For this purpose, the in vitro-produced full-length HEV RNA was used to study the replication of the virus via gene transfer. In this context, the HEV genome was cloned downstream of the T7 promoter and transcribed in vitro, and transcripts were characterized. The negative strand of viral RNA usually serves as the replicative intermediate in most of the positive-stranded RNA viruses. Such a species was demonstrated for HEV in transfected HepG2 cells, indicating active viral replication. The antisense strand was found to be present in a lower amount than the sense strand, as in other positive-stranded virus systems (30). While the sense strand was detected at up to a 10−15 dilution of the template RNA, the antisense strand was detected at up to a dilution of 10−6. This is possibly because the antisense pregenome tends to get converted into the sense strand faster. In addition, some of the RNA used for transfection may persist even after thorough washing and may lead to the detection of a very high level of the sense strand. In most of the positive-stranded RNA viruses, the positive and negative strands are synthesized in an unequimolar ratio; i.e., the positive strand is produced in excess of the negative strand (30). It is believed that this is due to the variation in interaction of different cellular proteins and/or RdRp involved in regulating the rate of initiation of viral RNA synthesis from a positive- or negative-sense template. The viral replication was detected in the cells for six passages (33 days). Thereafter, neither the sense nor the antisense HEV RNA could be detected (45 days).
For efficient productive infection, the viral genome has to interact with several viral, as well as cellular, proteins. These interactions determine the efficiency of replication, transcription, and translation. Therefore the in vitro-produced viral RNA transcript has to mimic the virion RNA as closely as possible. The parameters which affect infection by gene transfer are heterogeneity of the transcript population; the presence of a point mutation and the sequences at the 5′ and 3′ ends, i.e., a number of nonviral nucleotides; and the presence of a cap structure at the 5′ end and a poly(A) tail at the 3′ end. The problem of heterogeneity in the transcript population is mainly due to the poor fidelity of RNA polymerase (1, 10). As a result, it may hamper the infectivity of the transcripts (5). In our experiments, this was circumvented by use of a large quantity of RNA for transfection studies. The effect of nonviral sequences at the extreme ends of viral transcripts may also play an important role, and it has been observed that 5′ extensions generally decrease or abolish infectivity whereas 3′ extensions are tolerated to a limited extent (5). The reduced infectivity of dengue virus RNA transcripts due to the presence of nonviral sequences at the 5′ end has been reported, whereas nonviral nucleotides at the 3′ end of the dengue virus RNA transcript did not abolish infectivity (18). In other positive-stranded RNA viruses, such as poliovirus (34), hepatitis A virus (9), and Sindbis virus (28), the in vitro-produced infectious RNA from a cDNA clone had additional 5′ nonviral sequences. In these cases, the infectivity was found to be lower than that of the virion RNA. However, such a comparison was not possible in the case of HEV because it is still not possible to culture this virus. There was only one nonviral nucleotide at the 3′ end of our HEV clone following digestion of the cDNA clone with the restriction enzyme XhoI prior to in vitro transcription. The HEV transcript used in this study has 12 nonviral nucleotides at the 5′ end in addition to the complete viral genome (accession no. AF076239). These additional nucleotides in the transcript at the 5′ (12 nt) and 3′ (1 nt) ends did not abolish its competence for replication, as observed in this study. Recently, the presence of a cap structure in the HEV genome has been described (15). However, the present study demonstrated replication of HEV RNA without a cap structure. Therefore, it may be presumed that the presence of a cap structure is not obligatory for HEV genome replication.
The transfected viral genome was not only capable of replication but also expressed viral proteins in transfected cells and released infectious virus into the culture supernatant, as evaluated by experimental infection of a rhesus monkey. Nearly 20% of the cells were transfected, as observed by immunofluorescence assay. The metabolically labeled viral proteins were immunoprecipitated from transfected cells using their respective antibodies derived from both structural and putative nonstructural regions. The nonstructural polyprotein components identified from the predicted homology to the putative methyltransferase, helicase, and RdRp domains were immunoprecipitated separately from the transfected cells. The putative RdRp was detected at up to 36 h of transfection. This is possible because it is an early protein and undergoes rapid degradation. Therefore, no signal corresponding to RdRp could be detected at 72 and 96 h. However, the other proteins, which include the putative methyltransferase and helicase, were detected at 72 h posttransfection. This indicates that the protein product from the ORF1 region undergoes processing.
No processing of the ORF1 polyprotein (∼186 kDa) was observed in our earlier experiments with the expression of HEV ORF1 (2). No processed putative functional proteins could be individually identified either in an in vitro coupled translation system or in HepG2 cells transfected with the ORF1 gene. Incubation of the eukaryotic expression product of ORF1 did not reveal any degradation over 24 h at 37°C (2). However, the complete genome of HEV incorporating the same ORF1 shows processed components that could be immunoprecipitated with putative domain-specific antibodies. Therefore, it is predicted that processing of the nonstructural polyprotein occurs only in the context of the complete virus genome. The other viral proteins, either directly or indirectly through cell-dependent mechanisms, may activate proteases responsible for such processing. A putative protease domain has been identified in the ORF1 gene based on sequence comparison (17). However, this has not been characterized yet. The possibility that the viral protease needs activation cannot be ruled out. The ORF3 protein is a phosphoprotein that binds to src homology domain III. It is phosphorylated by mitogen-activated protein kinase. Therefore, this possibly can play a role in protein phosphorylation (36). Whether this protein alters the activity of any cellular or viral protease to initiate polyprotein processing needs further investigation.
Inoculation of the culture supernatant from the RNA-transfected cells was able to produce infection in one rhesus monkey. This was evidenced by a rise in serum transaminase, direct detection of the viral genome, and the appearance of IgM, and later IgG, anti-HEV antibodies in the serum of the inoculated animal. This is possible only when intact virus is released into the culture supernatant, as inoculation of in vitro-produced HEV RNA did not produce infection. This method of gene transfer is unique in the sense that it permits the recovery of an infectious agent from cells transfected with in vitro-produced RNA from an HEV cDNA clone generated by assembly of PCR-amplified subgenomic fragments. Similar assembly of PCR-amplified fragments has been described earlier (24). It has always been believed that during PCR amplification, errors in nucleotide incorporation lead to production of mutated fragments that may not be functionally active. However, our experience indicates that use of simple methods, like addition of a proofreading enzyme (Pfu DNA polymerase; Stratagene) during amplification, can avoid this problem. This model of HEV gene transfer can now be used to facilitate studies on the evolution, pathogenesis, and molecular biology of HEV and in drug development studies relevant to the understanding and control of HEV infection.
ACKNOWLEDGMENTS
This study was funded by a grant-in-aid project of the Department of Science and Technology (DST), Government of India, to S. K. Panda. I. H. Ansari is a senior research fellow of the University Grants Commission, Department of Pathology, AIIMS, New Delhi, India.
REFERENCES
- 1.Ahlquist P, Janda M. cDNA cloning and in vitro transcription of the complete brome mosaic virus genome. Mol Cell Biol. 1984;4:2876–2882. doi: 10.1128/mcb.4.12.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansari, I. H., S. K. Nanda, H. Durgapal, S. Agrawal, D. Gupta, S. Jameel, and S. K. Panda. Cloning, sequencing and expression of the hepatitis E virus non-structural open reading frame 1 (ORF1). J. Med. Virol., in press. [PubMed]
- 3.Aye T T, Uchida T, Ma X Z, Iida F, Shikata T, Zuang H, Win K M. Complete nucleotide sequence of a hepatitis E virus isolated from the Xinjiang epidemic (1986–1988) of China. Nucleic Acids Res. 1992;20:3512. doi: 10.1093/nar/20.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi S L, Purdy M A, McCaustland K A, Margolis H S, Bradley D W. The sequence from hepatitis E virus directly from a single source during an outbreak in China. Virus Res. 1993;28:233–247. doi: 10.1016/0168-1702(93)90024-h. [DOI] [PubMed] [Google Scholar]
- 5.Boyer J C, Haenni A L. Infectious transcripts and cDNA clones of RNA viruses. Virology. 1994;19:415–426. doi: 10.1006/viro.1994.1053. [DOI] [PubMed] [Google Scholar]
- 6.Bradley D W. Enterically transmitted non-A, non-B hepatitis. Br Med Bull. 1990;46:442–461. doi: 10.1093/oxfordjournals.bmb.a072409. [DOI] [PubMed] [Google Scholar]
- 7.Carl M, Issacs S N, Kaur M, He J, Tam A W, Yarbough P O, Reyes G R. Expression of hepatitis E virus putative structural proteins in recombinant vaccinia viruses. Clin Diagn Lab Immunol. 1994;1:253–256. doi: 10.1128/cdli.1.2.253-256.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Ann Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Cohen J I, Ticehurst J R, Feinstone S M, Rosenblum B, Purcell R H. Hepatitis A virus cDNA and its RNA transcripts are infectious in cell culture. J Virol. 1987;61:3035–3039. doi: 10.1128/jvi.61.10.3035-3039.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson W O, Neck D L, Knorr D A, Grantham G L. cDNA cloning of the complete genome of tobacco mosaic virus and production infectious transcripts. Proc Natl Acad Sci USA. 1986;83:1832–1836. doi: 10.1073/pnas.83.6.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He J, Tam A W, Yarbough P O, Reyes G R, Carl M. Expression and diagnostic utility of hepatitis E virus putative structural proteins expressed in insect cells. J Clin Microbiol. 1993;31:2167–2173. doi: 10.1128/jcm.31.8.2167-2173.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang C C, Nguyen D, Fernandez J, Yun K Y, Fry K E, Bradley D W, Tam A W, Reyes G R. Molecular cloning and sequencing of the Mexico isolate of hepatitis E virus (HEV) Virology. 1992;191:550–558. doi: 10.1016/0042-6822(92)90230-m. [DOI] [PubMed] [Google Scholar]
- 13.Jameel S, Durgapal H, Habibullah C M, Khuroo M S, Panda S K. Enteric non-A, non-B hepatitis: epidemics, animal transmission and hepatitis E virus detection by the polymerase chain reaction. J Med Virol. 1992;37:263–270. doi: 10.1002/jmv.1890370405. [DOI] [PubMed] [Google Scholar]
- 14.Jameel S, Zafrullah M, Ozdener M H, Panda S K. Expression in animal cells and characterization of the hepatitis E virus structural proteins. J Virol. 1996;70:207–216. doi: 10.1128/jvi.70.1.207-216.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabrane-Lazizi Y, Xiang-Jin M, Purcell R H, Emerson S U. Evidence that the genomic RNA of hepatitis E virus is capped. J Virol. 1999;73:8848–8850. doi: 10.1128/jvi.73.10.8848-8850.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khuroo M S, Deurmeyer W, Zargar S A, Ahanger M A, Shah M A. Acute sporadic non-A, non-B hepatitis in India. Am J Epidemiol. 1983;118:360–364. doi: 10.1093/oxfordjournals.aje.a113643. [DOI] [PubMed] [Google Scholar]
- 17.Koonin E V, Gorbalenya A E, Purdy M A, Rozanov M N, Reyes G R, Bradley D W. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of new group of animal and plant positive-strand RNA viruses. Proc Natl Acad Sci USA. 1992;89:8259–8263. doi: 10.1073/pnas.89.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai C J, Zhao B T, Hori H, Bray M. Infectious RNA transcribed from stably cloned full-length cDNA of dengue type 4 virus. Proc Natl Acad Sci USA. 1991;88:5139–5143. doi: 10.1073/pnas.88.12.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naik S R, Aggarwal R, Salunke P N, Mehrotra N N. A large waterborne viral hepatitis E epidemic in Kanpur, India. Bull W H O. 1992;70:597–604. [PMC free article] [PubMed] [Google Scholar]
- 20.Nanda S K, Panda S K, Durgapal H, Jameel S. Detection of negative strand of hepatitis E virus RNA in the livers of experimentally infected rhesus monkeys: evidence for viral replication. J Med Virol. 1994;42:237–240. doi: 10.1002/jmv.1890420306. [DOI] [PubMed] [Google Scholar]
- 21.Panda S K, Jameel S. Hepatitis E virus: from epidemiology to molecular biology. Viral Hepatitis Rev. 1997;3:227–251. [Google Scholar]
- 22.Panda S K, Datta R, Kaur K, Zuckerman A J, Nayak N C. Enterically transmitted non-A, non-B hepatitis: recovery of virus-like particles from an epidemic in South Delhi and transmission studies in rhesus monkeys. Hepatology. 1989;10:466–472. doi: 10.1002/hep.1840100411. [DOI] [PubMed] [Google Scholar]
- 23.Panda S K, Nanda S K, Zafrullah M, Ansari I H, Ozdener M H, Jameel S. An Indian strain of hepatitis E virus (HEV): cloning, sequencing, and expression of structural region and antibody responses in sera from individuals from an area of high-level HEV endemicity. J Clin Microbiol. 1995;33:2653–2659. doi: 10.1128/jcm.33.10.2653-2659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pugachev K V, Abernathy E S, Frey T K. Improvement of the specific infectivity of the rubella virus (RUB) infectious clone: determinants of cytopathogenicity induced by RUB map to the nonstructural proteins. J Virol. 1997;71:562–568. doi: 10.1128/jvi.71.1.562-568.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell R H, Ticehurst J R. Enterically transmitted non-A, non-B hepatitis: epidemiology and clinical characteristics. In: Zuckerman A J, editor. Viral hepatitis and liver diseases. New York, N.Y: Alan R. Liss, Inc.; 1988. pp. 131–137. [Google Scholar]
- 26.Racaniello V R, Baltimore D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science. 1981;214:916–919. doi: 10.1126/science.6272391. [DOI] [PubMed] [Google Scholar]
- 27.Reyes G R, Purdy M A, Kim J, Luk K C, Young L M, Fry K M, Bradley D W. Isolation of a cDNA from the virus responsible for enterically transmitted non-A, non-B hepatitis. Science. 1990;247:1335–1339. doi: 10.1126/science.2107574. [DOI] [PubMed] [Google Scholar]
- 28.Rice C M, Levis R, Strauss J H, Huang H V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987;61:3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson R A, Burgess W H, Emerson S U, Leibowitz R S, Sosnovtseva S A, Tsarev S, Purcell R H. Structural characterization of recombinant hepatitis E virus ORF2 proteins in baculovirus-infected insect cells. Protein Expr Purif. 1998;12:75–84. doi: 10.1006/prep.1997.0817. [DOI] [PubMed] [Google Scholar]
- 30.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tam A W, White R, Reed E, Short M, Zhang Y, Furest T R, Lanford R E. In-vitro propagation and production of hepatitis E virus from in-vivo-infected primary macaque hepatocytes. Virology. 1996;215:1–9. doi: 10.1006/viro.1996.0001. [DOI] [PubMed] [Google Scholar]
- 32.Tam A W, Smith M M, Guerra M E, Huang C C, Bradley D W, Fry K E, Reyes G R. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsarev S, Emerson S U, Reyes G R, Tsareva T S, Legters L J, Malik I A, Iqbal M, Purcell R H. Characterization of a prototype strain of hepatitis E virus. Proc Natl Acad Sci USA. 1992;89:559–563. doi: 10.1073/pnas.89.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Werf S, Bradley J, Wimmer E, Studier F W, Dunn J J. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:2330–2334. doi: 10.1073/pnas.83.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vishwanathan R. Infectious hepatitis in Delhi (1955–56). A critical study: epidemiology. Indian J Med Res. 1957;45:49–58. [Google Scholar]
- 36.Zafrullah M, Ozdener M H, Panda S K, Jameel S. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J Virol. 1997;71:9045–9053. doi: 10.1128/jvi.71.12.9045-9053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zafrullah M, Ozdener M H, Kumar R, Panda S K, Jameel S. Mutational analysis of glycosylation, membrane translocation, and cell surface expression of the hepatitis E virus ORF2 protein. J Virol. 1999;73:4074–4082. doi: 10.1128/jvi.73.5.4074-4082.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]