Abstract
Mycobacterium bovis (Mb) is the causative agent of bovine tuberculosis (bTb). Genetic selection aiming to identify less susceptible animals has been proposed as a complementary measure in ongoing programs toward controlling Mb infection. However, individual animal phenotypes for bTb based on interferon-gamma (IFNɣ) and its use in bovine selective breeding programs have not been explored. In the current study, IFNɣ production was measured using a specific IFNɣ ELISA kit in bovine purified protein derivative (bPPD)-stimulated blood samples collected from Holstein cattle. DNA isolated from the peripheral blood samples collected from the animals included in the study was genotyped with the EuroG Medium Density bead Chip, and the genotypes were imputed to whole-genome sequences. A genome-wide association analysis (GWAS) revealed that the IFNɣ in response to bPPD was associated with a specific genetic profile (heritability = 0.23) and allowed the identification of 163 SNPs, 72 quantitative trait loci (QTLs), 197 candidate genes, and 8 microRNAs (miRNAs) associated with this phenotype. No negative correlations between this phenotype and other phenotypes and traits included in the Spanish breeding program were observed. Taken together, our results define a heritable and distinct immunogenetic profile associated with strong production of IFNɣ in response to Mb.
Keywords: Mycobacterium bovis, interferon-gamma, polymorphisms, breeding
1. Introduction
Animal tuberculosis is a chronic infection caused by members of the Mycobacterium tuberculosis complex (MTC). Mycobacterium bovis (Mb), an intracellular, Gram-positive pathogen, is the key pathogen responsible for bovine tuberculosis (bTb). This zoonotic disease causes important economic losses worldwide, is considered a threat to public health, and has implications for the international trade of animals [1,2].
Mb resides within phagosomes in infected host macrophages [3]. Soon after being infected, the macrophages produce interleukin 12 (IL12), which activates natural killer (NK) cells and T lymphocytes to produce interferon-gamma (IFNɣ), leading to the activation of macrophages, enhanced release of nitric oxide, and restricted bacterial multiplication [4]. However, Mb has evolved ways to evade the host defense and to replicate within infected macrophages by inhibiting phagosome–lysosome fusion. Mb suppresses the antimicrobial immune response in macrophages, facilitating intracellular survival and immune evasion through autophagy inhibition and macrophage M2 polarization [5]. In advanced stages of infection, IFNγ is involved in the pathogenesis of several immunological disorders caused by unrestrained inflammatory responses. In a vaccination–challenge study, Vordermeier et al. found that following a challenge with Mb, the IFNγ response to the early secreted antigenic target 6 kDa antigen (ESAT-6) correlated positively with lesion scores in infected animals [6]. On the other hand, previous studies have shown that IFNγ production might be regulated post-transcriptionally by micro-RNAs (miRNAs). For instance, the downregulation of miRNA-29 has been shown to upregulate IFNɣ-mediated innate and adaptative responses to Mb infection [7]. Transgenic mice in which the miRNA-29 was knocked down initiated a more potent IFNγ production in activated NK and T cells after Mb infection.
Monitoring, control, and eradication programs for bTb have been established in multiple countries. Vaccination is not used as a preventive measure in animals because of the potential interference with bTb surveillance and diagnostic tests. In Spain, all regions are subjected to a national eradication program based on animal reaction to the single intradermal tuberculin test (SITT), the slaughter of reactor animals, and post-mortem confirmation of positive animals with histopathological lesions compatible with bTb. In some countries, including Spain, IFNγ release assays (IGRAs) are being used for diagnosis in combination with the single intradermal comparative cervical test (SICCT) [8]. The sensitivity of the SITT was 63.7% (95% CI, 54.54–72.00), while the sensitivity of the IFNγ assays ranged between 60.2% and 92% [9]. Results from experimental and natural infections of cattle with Mb indicated that the IGRA can detect a cell-mediated immune response (CMI) to infection as early as two weeks post-infection, earlier than the SICCT [10,11,12,13]. In general, however, bTb diagnostic tests are unable to distinguish the infection from the disease [14]. A study by Bernitz et al. [15] observed that levels of IFNγ in unstimulated whole blood were elevated in infected buffaloes with observable pathological changes consistent with bTb in comparison with uninfected controls. Furthermore, increased IFNγ significantly correlated with increasing severity of pathological changes in the infected buffaloes, consistent with observations of associations between antigen-stimulated IFNγ and bTb pathology in cattle and badgers, demonstrating the potential of this cytokine to be used as an indicator of bTb [16,17].
The incidence of new cases of bTb in parts of Spain suggests that existing control strategies are insufficient to eradicate the disease and that additional control measures that can complement current strategies need to be explored [18,19,20]. Currently, there are no vaccines commercially available that allow differentiation between naturally infected and vaccinated individuals. Genetic selection aiming to identify and select more resistant or less susceptible animals has been proposed as an additional measure in ongoing programs toward controlling bTb [21]. The effects of this strategy are cumulative and permanent, and they are transferred to subsequent generations and might result in disease eradication.
BTb is a multifactorial disease that is the result of the interaction of genetic, environmental, and microbial factors. Previous studies quantified the genetic variation for bTb in different cattle populations and countries using a variety of trait definitions and reported heritability estimates that ranged between 0.06 and 0.18 [22,23,24,25]. More recent studies using genome-wide DNA arrays reported higher heritability estimates of 0.21–0.27 [26,27,28,29,30]. These studies demonstrated that host genetics plays an important role in the susceptibility/resistance to Mb infection and, therefore, a breeding strategy focused on increasing resistance or reducing susceptibility to Mb infection is feasible and currently used by farmers in some countries such as the UK and Ireland [21,25]. Different genetic models were investigated, and the single-step best linear unbiased prediction (BLUP) model resulted in the most accurate estimates of animal genetic merit for bTb resistance [31].
Defining the adequate phenotype is the main challenge in identifying the genetic profile of resistance or susceptibility against an infection. In the case of bTb, most previous studies combined SITT tests and postmortem data, such as records of bTb lesions and Mb bacteriological culture, to define the health status of each animal. Recently, reductionist approaches have been used to investigate a host biological subsystem, such as a key cellular function, whose performance is the phenotypical criterion for the classification of the population [32]. By performing a genome-wide association analysis (GWAS), our research group identified a total of 71 single-nucleotide polymorphisms (SNPs) associated with significant production of IFNγ in avian tuberculin-stimulated blood samples from Mycobacterium avium subsp. paratuberculosis (MAP)-infected cattle using whole-genome sequence (WGS) data [33]. In the current study, we hypothesize that animals able to induce a strong IFNɣ in response to Mb infection might also have specific host genetics. This study aimed to identify SNPs, quantitative trait loci (QTL), and candidate genes associated with susceptibility or resistance to Mb infection using IFNɣ production in response to bovine tuberculin as an indicator. The identified QTLs were compared with reported and annotated QTLs associated with health, body conformation, milk production, meat and carcass, reproduction, and length of productive life. In addition, the identified candidate genes were compared with bovine and human candidate genes previously associated with bovine and human tuberculosis, respectively. Undesirable genetic linkages between IFNɣ production and other traits included in the Spanish Holstein cattle evaluations were assessed. For this purpose, genomic estimated breeding values (gEBVs) for IFNɣ production were estimated in a larger independent population (N = 1739), and the correlations with 65 traits and phenotypes included in the Spanish evaluations of Holstein cattle were analyzed.
2. Materials and Methods
2.1. Animals and Disease Status
The animals included in this study belonged to a reference population of 986 culled Holstein cattle that were slaughtered from March 2007 to May 2010. To ensure that the animals had a mature immune system, the cows included in the reference population were older than 2 years. Sampling was systematically performed once a week at the slaughterhouse. In each visit, the first 2 to 10 animals satisfying the breed and age requirements were sampled (on average 5 animals/sampling). The cows were slaughtered in the Bilbao and Donostia municipal slaughterhouses (Basque Country, Spain) under the pertinent Basque (Basque Government Decree 454/1994), Spanish (Spanish Government Law 32/2007 and Royal decree 731/2007), and European (Council Regulation No. 1099/2009) legislation on animal welfare. The cows were not submitted to any in vivo experimentation; therefore, no specific ethical authorization was needed.
2.2. Interferon-Gamma Release Assay (IGRA)
For the IGRA, blood stimulation must be performed within the first eight hours after blood collection. Since one of the abattoirs was far from our research institute, IGRA could be performed in blood samples from only 343 of the 986 culled cows. Blood stimulation was performed as previously described [33,34]. Briefly, four 1.4 mL aliquots of lithium heparinized whole blood samples from each animal were added to four wells of a 24-well plate (Becton Dickinson, Franklin Lakes, NJ, USA). The blood samples were then stimulated with 100 µL of phosphate-buffered saline (PBS), 100 µL of avian purified protein derivative (aPPD) (0.3 µg/µL) (CZ Vaccines® SA, Porriño, Spain), 100 µL of bovine purified protein derivative (bPPD) (0.3 µg/µL) (CZ Vaccines® SA, Porriño, Spain), and 100 µL of lectin (1 μg/mL) as a positive control. BPPD is derived from Mb, strain AN-5. After incubating for 16 to 24 h at 37 °C in a 5% CO2 incubator, the plasma was separated by centrifugation at 500× g for 10 min at room temperature (RT) and then was frozen at −20 °C until testing. Subsequently, IFNɣ levels were measured in triplicate from the plasma samples using a specific IFNɣ ELISA test according to the manufacturer’s instructions (BovigamTM, Prionics, Schlieren, Switzerland). IFNɣ levels were expressed as the OD of the aPPD or bPPD-stimulated plasmas minus the OD of the PBS-stimulated samples. In the current study, 117 cows from the 343 animals with an IGRA record were selected because they had an OD (bPPD-PBS) higher than the OD (aPPD-PBS) and the OD (bPPD) was higher than the OD (PBS) as well. This selection avoids false positive reactions with MAP infection and negative OD values, respectively. According to the interpretation criteria of the kit, 76 of the 117 cows included in the study were bTb-positive because the OD of the bPPD-PBS was >0.05 and the OD (bPPD-PBS) was higher than the OD (aPPD-PBS).
2.3. Genotyping and Imputation to Whole-Genome Sequence (WGS)
Peripheral blood (PB) samples were collected into 10 mL Vacutainer EDTA tubes (Becton Dickinson, Franklin Lakes, NJ, USA) at the time of slaughter. Total DNA was extracted from the PB samples using the QIAmp DNA Blood Mini Kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). Purified DNA was then quantified by spectrophotometry and genotyped using the EuroG Medium-Density Bead Chip (Illumina) at the molecular genetic laboratory service of the Spanish Federation of Holstein Cattle (CONAFE) using the InfiniumTM iScan software for allele assignation (Illumina, San Diego, CA, USA). The individual genotypes were imputed to WGS as previously described [35]. Briefly, genotypes were phased using Eagle 2.4 [36] and imputed with minimac4 [37] to the Bovine High-Density Bead Chip (581,712 SNPs) using a reference panel of 1278 Holstein bulls from Run7.0 of the 1000 Bull Genomes project. Imputation to the WGS level (ARS-UCD1.2) was then undertaken using the same phasing and imputation procedure and a reference population of 2333 Bos taurus from Run7.0 of the 1000 Bull Genomes project [38]. Finally, the following filters were applied: call rate > 0.80, minimum allele frequency (MAF) > 0.01, and imputation score (r2) > 0.7. The final number of SNPs per animal was 11,122,500.
2.4. GWAS Analysis, Variance Components, and h2 Estimation
The IFNɣ levels in response to the bPPD, OD (bPPD-PBS), was the quantitative phenotype in the GWAS analysis. The variance components and h2 explained by all the SNPs were calculated using the genome-wide complex trait analysis (GCTA) software 1.93.2 [39], according to the following formula:
where represents the variance explained by all the SNPs and the residual variance. These WGS data and the OD of these bPPD-PBS data were analyzed using the mixed linear model association analysis of the GCTA 1.93.2 software, expressed as y = a + bx + g + e. In the model, y is the phenotype, a is the mean term, b is the allele effect, x is the genotype of the SNP coded as 0, 1, or 2 depending on the copies of the minor allele, g is the polygenic effect as a random effect (assumed to be distributed as N~(0,), and e is the residual effect (also assumed to be distributed as N~(0,)). Age was included as a covariate in the analysis. To account for multiple testing, a 5% genome-wide false discovery rate (FDR) was used. A threshold of p-value ≤ 5 × 10−7 was used as suggested by the Wellcome Trust Case Control Consortium [40]. The inflation factor (λ) and quantile–quantile plots were used to compare the observed distributions of the −log (p-values) to the expected distribution under the no-association model. λ values close to 1 suggest appropriate adjustment for the potential substructure, and λ > 1.2 suggests population stratification. The SNP effects (b-values) were also calculated using the GCTA 1.93.2 software. If the sign of the b-value is positive, it implies that there is a positive relationship between the variables SNP and IFNɣ levels.
2.5. GWAS Data Post-Processing
SNPs identified in the GWAS analysis were filtered based on the p-values and clumping. Briefly, clumping is a process that first selects the most significant SNP and clumps (removes) other SNPs in a particular window that are in linkage disequilibrium with the selected SNP. The clumping was performed using the software PLINK1.9 [41] with a window of 500 Kbp and a linkage disequilibrium-based correlation index (r2) of 0.9.
2.6. SNPs, Quantitative Trait Loci (QTLs), QTLs Enrichment Analysis, Candidate Genes Identification, and Protein-to-Protein Interaction Networks
After clumping, the localization of the significant SNPs (FDR ≤ 0.05, p-value ≤ 5 × 10−7), QTLs, and candidate genes was performed using the ARS-UCD1.3 reference genome as previously described [33]. The genomic localization of the identified SNPs in the ARS-UCD1.3 reference genome was determined using the Ensembl Variant Effect Predictor (VEP) [42].
The R package Genomic functional Annotation in Livestock for positional candidate Loci (GALLO) [43] was used to identify in the cattle QTL database release 52 [44] annotated QTLs within an interval of 500 Kbp of the identified SNPs. Overlapping QTLs were merged to create a single QTL. To determine which of the annotated QTLs were overrepresented, a QTL enrichment analysis was performed using GALLO.
Candidate genes within the QTLs were identified using Ensembl [42]. Candidate genes were compared with bovine and human candidate genes that were previously associated with bovine and human tuberculosis [45]. The function of the candidate genes was searched in GeneCards [46] by searching their gene symbol. To investigate the potential innate immune function of the identified candidate genes further, we searched the Innate DB database [47]. Protein-to-protein interaction networks were analyzed using String v2.0., setting the minimum required interaction score to 0.7.
2.7. Genomic Estimated Breeding Values (gEBVs) for IFNγ Production in Response to Bovine Tuberculin and Correlations with Other Bovine Phenotypes and Traits
gEBVs for IFNγ production were calculated for each animal in the study population based on the effect of each SNP with evidence of association with the IFNγ production (PFDR ≤ 0.05) using the genomic best linear unbiased prediction (gBLUP) model of GCTA v1.93.2. [39,48]. Subsequently, gEBVs for IFNγ production were predicted in a larger population of 1739 Friesian cattle. Correlations between the gEBVs for IFNγ levels and 65 phenotypes and traits included in the evaluations of Spanish Holstein cattle were calculated in the larger population (N = 1739) with the Spearman’s rank correlation (ρ) implemented in R v4.1.2.
3. Results
3.1. Assessment of IFNɣ Production in Response to Bovine Tuberculin Stimulation
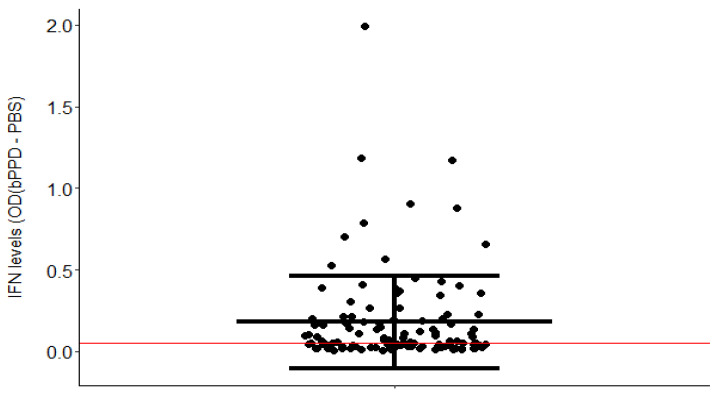
Only 117 of the 343 cows with an IGRA record were included in the study because they had an OD (bPPD-PBS) higher than the OD (aPPD-PBS), and the OD (bPPD) was higher than the OD (PBS). IFNɣ levels after blood stimulation with bPPD (OD (bPPD-PBS)) took values between 0.01 and 1.99, with most of the animals showing OD values between 0.01 and 0.5. The mean OD (bPPD-PBS) of the population was 0.17.
Ten animals had IFNɣ levels after blood stimulation with bPPD > 0.5. According to the interpretation criteria of the kit, 76 of the 117 cows included in the study had a positive IGRA result in response to bPPD. Figure 1 shows the mean of the IFNɣ production after blood stimulation with bPPD.
Figure 1.
IFNγ production. Error plot of the levels of IFNɣ in stimulated blood samples from the 117 cows included in the study. The central line represents the mean of the group, and the whiskers represent the standard error. According to the interpretation criteria of the kit, 76 of the 117 cows included in the study were bTb-positive because the OD (bPPD-PBS) was >0.05 and the OD (bPPD-PBS) > OD (aPPD-PBS).
3.2. Heritability (h2) Estimate, Variance Components, and GWAS Results
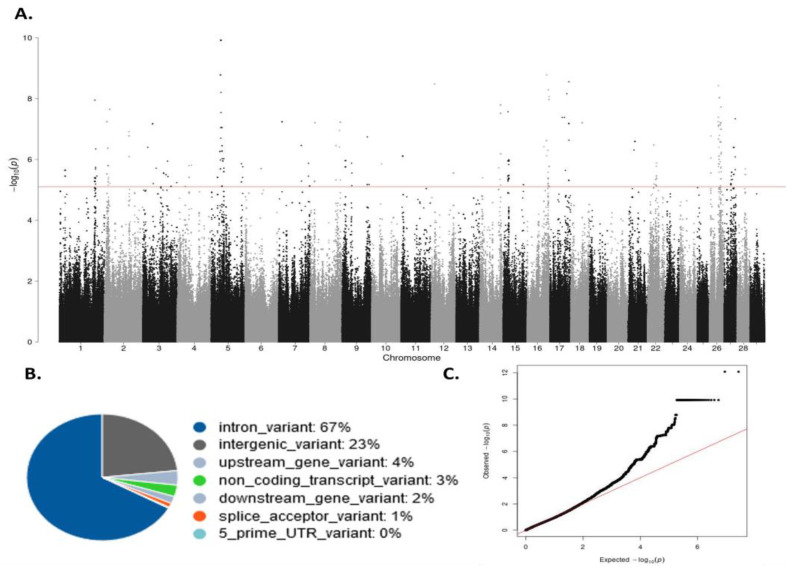
The associations between genome-wide imputed SNPs and IFNɣ levels after blood stimulation with bPPD (OD (bPPD-PBS)) (N = 117) were analyzed using GCTA1.93.2. The heritability and variance components of the IFNɣ levels in the study population were estimated as 0.23 (σG = 0.01823, σe = 0.06146). The Manhattan plot shows –log10 (p-values) of the association test between IFNɣ levels and each SNP is represented in Figure 2A.
Figure 2.
Results of the GWAS analysis. (A) Manhattan plot of the –log10 of the p-values of the association test between each SNP and the IFNɣ levels. Each dot represents one SNP. Chromosome localization of the SNPs is indicated on the x-axis. The horizontal red line is drawn at –log10 (5 × 10−7); (B) Genomic distribution of the 163 SNPs surpassing the threshold (p-value ≤ 5 × 10−7) according to the Ensembl Variant Effect Predictor (VEP); (C) Quantile–quantile plot comparing the observed distribution of –log (p-values) to the expected values under the null hypothesis.
After clumping, the total number of SNPs that surpassed the significance criteria (PFDR ≤ 0.05, p-value ≤ 5 × 10−7) was 163. The 163 significant SNPs were located on 20 different Bos taurus chromosomes. As seen in Figure 2B, most of the 163 SNPs were in intronic regions (67%), while the remaining identified SNPs were in intergenic regions (23%) or were upstream (4%) non-coding transcript variants (3%), or downstream (2%), variants. The quantile–quantile plot comparing the observed distribution of −log (p-values) to the expected p-values under the null hypothesis is shown in Figure 2C. The median inflation factor was 1.007588, which indicates the absence of population stratification.
3.3. SNPs, QTLs, and Candidate Genes Associated with IFNγ Production in Response to Bovine Tuberculin Stimulation
After clumping, a total of 163 SNPs, 72 QTLs, 197 candidate genes and 8 miRNAs (bta-mir-2285cf, bta-mir-2351, bta-mir-12005-2, bta-mir-12000, bta-mir-6121, bta-mir-6524, bta-mir-4680, and bta-mir-2399), were associated with production of IFNγ after stimulation with bPPD. p-values, QTLs positions, candidate genes, and miRNAs within each QTL are presented in Table 1. Pathway analysis failed to reveal significantly enriched biological processes and metabolic pathways when bovine-specific pathway data were considered.
Table 1.
QTLs, candidate genes, and miRNAs associated with high levels of IFNɣ in response to bovine tuberculin.
| BTA 1 | QTL Start (bp) |
QTL End (bp) |
Peak p-Value |
Genes in QTL 2 | No. SNPs in QTL |
|---|---|---|---|---|---|
| 1 | 124,889,999 | 124,889,999 | 4.05 × 10−6 | ENSBTAG00000059771, DIPK2A, ENSBTAG00000064298, ENSBTAG00000060304, SLC9A9 | 1 |
| 1 | 125,545,373 | 125,555,920 | 5.26 × 10−6 | 2 | |
| 1 | 127,196,414 | 127,784,694 | 2.06 × 10−6 | GRK7, RNF7, RASA2, U6, bta-mir-2285cf, ZBTB38, PXYLP1, ENSBTAG00000050470, ENSBTAG00000059581, SPSB4 | 5 |
| 1 | 20,524,991 | 20,585,846 | 2.29 × 10−6 | 4 | |
| 2 | 13,171,064 | 13,171,064 | 1.57 × 10−6 | 1 | |
| 2 | 14,086,412 | 14,102,103 | 1.64 × 10−6 | ENSBTAG00000053080, ENSBTAG00000058959, PDE1A | 2 |
| 2 | 15,276,423 | 15,276,423 | 3.92 × 10−6 | ENSBTAG00000064762 | 1 |
| 2 | 19,826,914 | 19,958,921 | 2.24 × 10−8 | ENSBTAG00000069400, ENSBTAG00000069332, ENSBTAG00000064902 | 2 |
| 2 | 87,618,721 | 87,987,549 | 1.21 × 10−7 | SATB2 | 2 |
| 2 | 9,485,693 | 9,772,662 | 5.72 × 10−8 | FAM171B, ZSWIM2, ITGAV, bta-mir-2351 | 5 |
| 3 | 47,301,950 | 47,301,950 | 1.94 × 10−6 | 1 | |
| 3 | 63,633,887 | 63,633,887 | 7.87 × 10−6 | 1 | |
| 3 | 73,139,474 | 73,139,474 | 2.82 × 10−6 | NEGR1 | 1 |
| 3 | 81,693,793 | 81,693,793 | 3.16 × 10−6 | ROR1 | 1 |
| 3 | 87,101,478 | 87,101,478 | 1.14 × 10−6 | ENSBTAG00000066077 | 1 |
| 3 | 88,629,927 | 88,629,927 | 3.65 × 10−6 | DAB1 | 1 |
| 4 | 28,439,718 | 28,439,718 | 7.73 × 10−6 | POLR1F, TMEM196 | 1 |
| 5 | 106,699,607 | 106,710,516 | 5.50 × 10−6 | TSPAN9, ENSBTAG00000060879 | 2 |
| 5 | 22,620,852 | 22,963,465 | 3.86 × 10−6 | PLEKHG7, U6, EEA1, ENSBTAG00000065664, ENSBTAG00000068135 | 5 |
| 5 | 31,376,781 | 31,376,781 | 5.49 × 10−7 | OR8S27, OR8S25, OR8S10 | 1 |
| 5 | 32,917,758 | 33,096,745 | 1.66 × 10−9 | ENSBTAG00000059572, ENSBTAG00000038027, ENSBTAG00000060945, ENSBTAG00000067834, PCED1B | 2 |
| 5 | 33,790,732 | 34,674,103 | 1.21 × 10−10 | ENSBTAG00000062330, ENSBTAG00000062600, ENSBTAG00000058122, SLC38A2, ENSBTAG00000067121, SLC38A1, SCAF11, ARID2, ENSBTAG00000058264, ENSBTAG00000069144, ENSBTAG00000067082 | 6 |
| 5 | 37,435,087 | 38,572,334 | 8.97 × 10−8 | ENSBTAG00000067640, SNORA62, bta-mir-12005-2, PRICKLE1, ENSBTAG00000060964, PPHLN1, ZCRB1, YAF2, GXYLT1 | 8 |
| 5 | 44,552,213 | 44,780,908 | 9.47 × 10−7 | U6, LYZ, CPSF6, ENSBTAG00000066263, ENSBTAG00000069891, ENSBTAG00000057583, ENSBTAG00000064106, ENSBTAG00000002741, ENSBTAG00000044636, ENSBTAG00000064663, ENSBTAG00000069326 | 3 |
| 6 | 56,297,100 | 56,297,100 | 2.00 × 10−6 | 1 | |
| 7 | 109,402,049 | 109,402,049 | 7.49 × 10−6 | 1 | |
| 7 | 12,351,770 | 12,389,993 | 5.77 × 10−8 | CACNA1A, bta-mir-12000, ENSBTAG00000060372, ENSBTAG00000062006, IER2, STX10 | 2 |
| 7 | 80,281,173 | 80,281,173 | 3.50 × 10−7 | TENM2 | 1 |
| 8 | 106,878,958 | 107,205,388 | 5.95 × 10−8 | TLR4, ENSBTAG00000068729, ENSBTAG00000061668 | 3 |
| 8 | 16,064,815 | 16,064,815 | 5.10 × 10−6 | LINGO2, ENSBTAG00000066439 | 1 |
| 8 | 16,591,629 | 16,755,960 | 5.32 × 10−6 | ENSBTAG00000067505, ENSBTAG00000060335, IFNK | 3 |
| 8 | 92,008,532 | 92,008,532 | 4.67 × 10−6 | 1 | |
| 9 | 11,599,785 | 11,938,116 | 1.09 × 10−6 | RIMS1 | 2 |
| 9 | 33,772,705 | 33,772,705 | 1.33 × 10−6 | RFX6 | 1 |
| 9 | 34,608,918 | 34,906,969 | 2.80 × 10−6 | NT5DC1, SYNE1 | 2 |
| 9 | 89,314,702 | 89,389,719 | 6.70 × 10−6 | SYNE1, ENSBTAG00000052173 | 2 |
| 9 | 95,898,153 | 95,898,153 | 6.69 × 10−6 | ENSBTAG00000052316 | 1 |
| 11 | 6,223,550 | 6,223,550 | 7.77 × 10−7 | CNOT11, ENSBTAG00000067224, SNORD89, ENSBTAG00000064562, RNF149, CREG2 | 1 |
| 12 | 11,410,693 | 11,410,693 | 3.30 × 10−9 | KBTBD7, MTRF1 | 1 |
| 12 | 78,341,192 | 78,341,192 | 2.79 × 10−6 | FGF14 | 1 |
| 14 | 10,240,609 | 10,240,609 | 4.03 × 10−6 | ASAP1 | 1 |
| 14 | 74,566,478 | 74,566,478 | 1.61 × 10−8 | MMP16 | 1 |
| 15 | 19,232,962 | 19,232,962 | 4.12 × 10−6 | 1 | |
| 15 | 20,351,354 | 20,406,919 | 1.04 × 10−6 | RDX, ENSBTAG00000068159, ENSBTAG00000066977 | 2 |
| 15 | 72,080,092 | 72,080,539 | 6.78 × 10−6 | 2 | |
| 16 | 59,681,912 | 59,681,912 | 3.82 × 10−7 | 1 | |
| 16 | 68,538,475 | 68,542,979 | 2.40 × 10−7 | KCNK2 | 2 |
| 16 | 69,071,657 | 69,071,657 | 1.66 × 10−9 | PTPN14 | 1 |
| 16 | 73,402,526 | 73,402,526 | 5.00 × 10−7 | SYT14, ENSBTAG00000065259, ENSBTAG00000064536, UTP25 | 1 |
| 16 | 74,037,392 | 74,222,831 | 2.63 × 10−6 | 3 | |
| 16 | 74,743,716 | 75,220,658 | 5.16 × 10−9 | ENSBTAG00000042659, PLXNA2, bta-mir-6121 | 5 |
| 16 | 77,902,129 | 77,902,129 | 1.70 × 10−6 | ENSBTAG00000059177 | 1 |
| 17 | 50,628,634 | 50,628,634 | 4.15 × 10−8 | TMEM132B | 1 |
| 17 | 59,585,737 | 59,585,737 | 6.92 × 10−9 | U2 | 1 |
| 17 | 66,464,708 | 66,600,043 | 2.81 × 10−9 | 5 | |
| 21 | 21,803,104 | 21,803,104 | 4.91 × 10−7 | MAN2A2, FES, FURIN, ENSBTAG00000057133, BLM | 1 |
| 21 | 25,085,953 | 25,085,953 | 2.56 × 10−7 | BTBD1, ENSBTAG00000066584, ENSBTAG00000063590 | 1 |
| 22 | 27,941,466 | 28,433,940 | 4.60 × 10−6 | ENSBTAG00000069336, ENSBTAG00000069185, PDZRN3 | 3 |
| 22 | 31,251,423 | 31,507,701 | 1.30 × 10−6 | ENSBTAG00000064803, ENSBTAG00000061892, MDFIC2 | 4 |
| 26 | 31,413,153 | 31,413,153 | 5.68 × 10−8 | RBM20, ENSBTAG00000061228, ENSBTAG00000061228, ENSBTAG00000059480, PDCD4, bta-mir-6524, bta-mir-4680 | 1 |
| 26 | 31,943,002 | 32,482,052 | 8.36 × 10−13 | ENSBTAG00000059653, ENSBTAG00000062595, ENSBTAG00000066814, ENSBTAG00000064546, ENSBTAG00000059196, ENSBTAG00000061789 | 8 |
| 26 | 33,800,182 | 34,212,369 | 7.72 × 10−7 | HABP2, NRAP, CASP7, PLEKHS1 | 4 |
| 26 | 35,012,876 | 35,056,491 | 9.30 × 10−9 | ABLIM1 | 3 |
| 26 | 37,967,031 | 38,185,671 | 6.19 × 10−8 | 5 | |
| 26 | 39,549,032 | 39,664,208 | 6.19 × 10−8 | ENSBTAG00000063944, ENSBTAG00000056024, RGS10, TIAL1 | 3 |
| 26 | 40,347,155 | 41,671,467 | 1.90 × 10−8 | 9 | |
| 27 | 23,351,726 | 23,817,855 | 6.29 × 10−6 | ENSBTAG00000060821, ENSBTAG00000068762, C27H8orf48, DLC1 | 2 |
| 27 | 25,170,223 | 25,252,376 | 3.51 × 10−6 | ENSBTAG00000060632, PPP1R3B, U6 | 2 |
| 27 | 26,556,329 | 26,560,206 | 2.63 × 10−6 | RBPMS, ENSBTAG00000053684, bta-mir-2399 | 2 |
| 27 | 29,107,302 | 29,107,302 | 4.53 × 10−6 | ENSBTAG00000057376, FUT10, ENSBTAG00000054272 | 1 |
| 27 | 34,187,524 | 34,187,953 | 4.04 × 10−7 | HTRA4, TM2D2, ENSBTAG00000068068, ADAM9 | 2 |
| 28 | 29,347,932 | 29,347,932 | 2.05 × 10−6 | MRPS16, CFAP70, ANXA7, ENSBTAG00000054455 | 1 |
1 Chromosome QTL location, 2 Candidate genes within the identified QTL.
The identified QTLs were distributed along the Bos taurus genome; chromosome 26 harbors the highest number of SNPs (N = 33). The QTL that harbored the SNP with the strongest association was on chromosome 26 (p-value = 8.36 × 10−13) and a b-value of 1.186. The b-values of all the identified SNPs were positive, which suggests that all the variants were associated with a significant IFNɣ production in response to Mb.
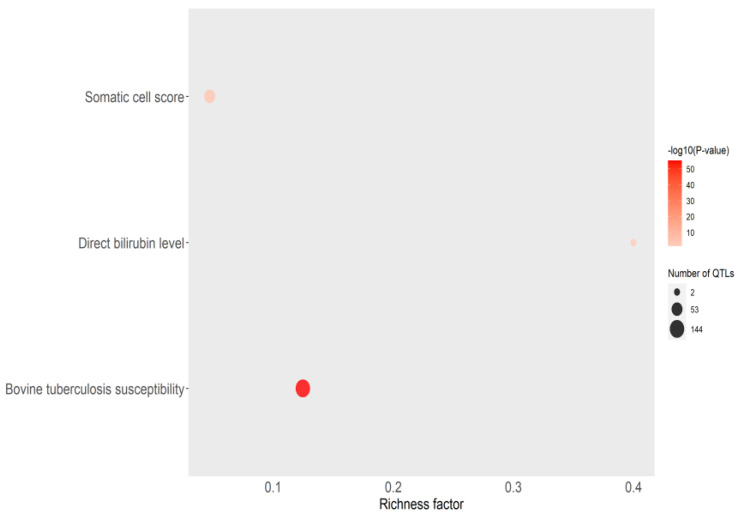
Overlapping between the QTLs identified in the current study and QTLs previously associated with exterior, meat and carcass, production, reproduction, and health traits was observed. More specifically, overlapping between some of the identified QTLs and QTLs associated with 30 health traits was observed. The identified QTLs overlapped with 159 QTLs previously associated with bovine tuberculosis [29,49,50], 158 QTLs associated with somatic cell score [51,52,53,54,55,56,57,58,59], 105 QTLs associated with bovine respiratory disease susceptibility [60,61,62], and 14 QTLs associated with bovine paratuberculosis susceptibility [63,64,65,66,67,68]. QTLs enrichment analysis revealed the enrichment of 33 QTLs associated with exterior, meat and carcass, production, reproduction, and health (Supplementary Table S1). Of these, significant enrichment of QTLs previously associated with three health traits was observed, including bovine tuberculosis susceptibility (P-adjusted = 5.39 × 10−56), somatic cell score (P-adjusted = 4.43 × 10−6), and direct bilirubin level (P-adjusted = 0.04). The health traits identified in the QTL enrichment analysis are presented in Figure 3.
Figure 3.
Bubble plot displaying the QTL enrichment results for health traits. The darker the red shade in the circles, the more significant the enrichment. The area of the circles is proportional to the number of QTLs. The x-axis shows a richness factor obtained by the ratio of the number of QTLs annotated and the total number of each QTL in the reference database.
We searched the animal genome database and discovered that 13% of the 91 identified candidate genes with a recognized gene name matched with candidate genes previously associated with bovine diseases such as bovine respiratory disease, mastitis, and bovine paratuberculosis. However, we did not find any matches with candidate genes associated with bovine tuberculosis. Some of the identified candidate genes, such as G Protein-Coupled Receptor Kinase 7 (GRK7), ADAM metallopeptidase domain 9 (ADAM9), furin, paired basic amino acid cleaving enzyme (FURIN), and Integrin Subunit Alpha V (ITGAV) were previously associated with bovine respiratory disease susceptibility [62]. Both FURIN and ITGAV mediate tumor growth factor β1 (TGFβ1) activation. Several candidate genes identified in our study were previously associated with somatic cell counts such as Receptor tyrosine kinase-like orphan receptor 1 (ROR1), DAB adaptor protein 1 (DAB1), PC-esterase domain containing 1B (PCED1B), Glucoside xylosyltransferase 1 (GXYLT1), Solute carrier family 38 member 1 (SLC38A1), and Toll-like receptor 4 (TLR4) [54,58].
Some of the identified candidate genes located on BTA11 (6,223,550–6,223,550 bp), such as CCR4-NOT Transcription Complex, Subunit 11 (CNOT11), RING-Type E3 Ubiquitin Transferase RNF149 (RNF149), and Cellular Repressor Of E1A Stimulated Genes 2 (CREG2), were previously associated with the presence of PTB-associated multifocal lesions [69]. The RFN149 negatively regulates Mitogen-Activated Protein Kinase (MAPK) cascade, and genetic variants in the vicinity of this gene were previously associated with the humoral immune response to MAP infection in dairy cattle [65]. CNOT11 is linked to various cellular processes, including bulk mRNA degradation, miRNA-mediated repression, translational repression during translational initiation, and general transcription regulation. Candidate genes identified on BTA3 (63,633,887–63,633,887 bp) such as Hyaluronan Binding Protein 2 (HABP2), Nebulin Related Anchoring Protein (NRAP), Caspase 7 (CASP7), and Pleckstrin Homology Domain Containing S1 (PLEKHS1) were previously associated with the presence of PTB-associated diffuse lesions [69]. HABP2 is involved in coagulation and fibrinolysis systems by activating coagulation factor VII and may function as a tumor suppressor, negatively regulating cell proliferation and cell migration. Mutations in this gene have been previously associated with nonmedullary thyroid cancer in humans [70] and susceptibility to venous thromboembolism due to thrombin defects [71]. CASP7 is a thiol protease involved in different programmed cell death processes, such as apoptosis, pyroptosis, or granzyme-mediated programmed cell death, by proteolytically cleaving target proteins and acts as a key regulator of the inflammatory response in response to bacterial infection by catalyzing the cleavage and activation of the sphingomyelin phosphodiesterase (SMPD1) in the extracellular milieu, thereby promoting membrane repair [72,73]. CASP7 also acts as an inhibitor of type I interferon production during pathogen-induced apoptosis by mediating cleavage of the antiviral proteins CGAS, IRF3, and MAVS, thereby preventing cytokine overproduction. Mutations in CASP7 might counteract these effects, resulting in cytokine overproduction and aberrant inflammation.
Some of the identified candidate genes matched with genes in the animal genome database previously associated with bovine paratuberculosis susceptibility, such as Toll-like receptor 4 (TLR4) [66,67] and the Periphilin 1 (PPHLN1), respectively. The PPHLN1 contributes to epidermal integrity and barrier formation and was previously associated with MAP resistance in Holstein cattle [63]. Matrix Metallopeptidase 16 (MMP16) located on BTA14 was previously associated with positive ELISA, PCR, and bacteriological culture results for MAP infection detection [35], and it was also identified in the current study. MMP16 is a protein of the matrix metalloproteinase family involved in the breakdown of extracellular matrix components such as collagen type III and fibronectin in normal physiological processes. In the lung, several MMPs contribute to tissue homeostasis, such as MMP-7, -16, -19, -21, -24, -25, and -28 [74]. Allelic variants affecting MMPs might cause improper ECM remodeling and disease progression in pathological circumstances.
Some of the identified candidate genes were previously associated with several bovine traits, highlighting their importance not just in health but also in milk production, fertility, body conformation, and length of productive life. For instance, TLR4 was identified in our study and was previously found to be associated with 18 bovine traits, some of them health traits including basophil and lymphocyte number, somatic cell counts, and MAP infection susceptibility [66,67]. TLR4 plays a fundamental role in pathogen recognition and activation of innate immunity, and mutations in this receptor might result in inadequate antigen recognition and processing, leading to immune tolerance.
By searching the human GWAs catalog, we found that PC-esterase domain containing 1BP (PCED1B), a hydrolase involved in macrophage apoptosis and autophagy, and ArfGAP With SH3 Domain, Ankyrin Repeat And PH Domain 1 (ASAP1) were the only candidate genes identified in our study that were previously associated with human tuberculosis [75]. PCED1B was also previously associated with bovine respiratory disease susceptibility [62]. Susceptibility to human tuberculosis was associated with variants in the ASAP1 gene encoding a regulator of dendritic cell migration [76]. Most of the 91 candidate genes with a recognized gene symbol were included in the InnateDB database and had a role in signaling pathways involved in the bovine immune response against microbial infections, including TLR4 and programmed cell death protein 4 (PDCD4), among others.
3.4. Protein-to-Protein Interaction Analysis
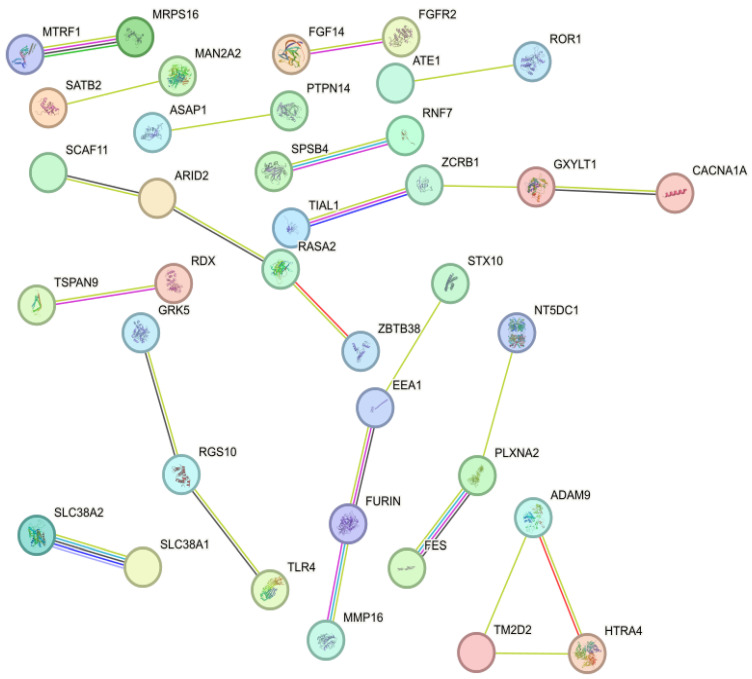
The 197 identified candidates were analyzed for protein-to-protein interactions. The analysis revealed five networks: GRK5-RGS10-TLR4, STX10-EEA1-FURIN-MMP16, TIAL1-ZCRB1-GXYLT1-CACNA1A, TM2D2-ADAM9-HTRA4, and SCAF11-ARID2-RASA2-ZBTB38 (Figure 4).
Figure 4.
Protein-to-protein network analysis using the identified candidate genes. Individual nodes represent proteins with relationships represented by edges. The green lines represent gene neighborhood, red lines represent gene fusions, blue lines represent gene co-occurrence, black lines represent co-expression, pale blue lines represent homology, and purple lines represent experimentally determined interaction. The candidate proteins with no associations to other proteins were hidden.
3.5. gEBVs for IFNγ Production and Correlations with Other Bovine Traits
gEBVs for IFNγ production were calculated for each animal in the study population and then predicted in a larger population of 1739 Friesian cattle. Correlations between the gEBVs for IFNγ levels in this larger population and 65 phenotypes and traits included in the evaluations of Spanish Holstein cattle were calculated with Spearman’s correlation test. For the phenotypes and traits with a significant correlation (p ≤ 0.05) with IFNγ production, the absolute value of Spearman’s correlation index was lower than 0.12.
4. Discussion
The use of cellular immunity traits in genetic linkage studies in Holstein cattle is scarce. Recently, a strong effect of host genetics on IFNγ production in response to the avian tuberculin was observed [33]. In the current study, we measured the IFNγ production in response to the bPPD in 343 Holstein cows in two steps consisting of (i) incubating whole blood samples from the selected animals with bPPD and (ii) detecting the presence of IFNγ released by sensitized lymphocytes in the whole blood sample to indicate a CMI to the specific antigen. The number of samples (N = 117) used in this study could be considered small in comparison with traditional GWAS analysis. The main limitation that arises from having a small sample size is the decrease in statistical power. However, the assessment of IFNɣ levels in stimulated blood is a functional and controlled trait that allowed us to identify 163 SNPs significantly associated with high IFNɣ production. Similar reductionist phenotypes, such as the assessment of the macrophages’ performance by measuring MAP load within MDMs, used only 61 samples [77].
Previous studies quantified the genetic variation for bTb in different cattle populations and countries using a variety of trait definitions and reported heritability estimates that ranged between 0.06 and 0.18 [22,23,24,25]. In the current study, we demonstrated that IFNγ production in response to Mb is, at least to some extent, dependent on host genetics (h2 = 0.23). Putative QTLs associated with Mb infection in Holstein cattle have been reported on the Bos taurus chromosome 1 (BTA1) [28], BTA2 and BTA13 [22], BTA6 [78,79], BTA22 [80], and BTA23 [28,49]. In most of these studies, case cows had a positive CITT, histopathological lesions, and a positive bacteriological culture result, and all other cattle present in the herd were considered control cows. In the current study, none of the identified genomic regions associated with IFNγ production were located on BTA23. Differences between studies may be due to differences in the phenotypes used, population structure, methodologies used across studies, and the large polygenic inheritance of the trait. In addition to the individual QTL differences between studies, our QTLs enrichment analysis revealed a total of three health traits with QTLs overlapping the QTLs identified in our study, including bovine tuberculosis susceptibility as the most significant trait. This finding suggests that the capacity to produce a strong IFNγ in response to Mb is an indicator of bTb susceptibility in cattle. Previous studies demonstrated that strong IFNγ production correlated with bTb pathology in cattle and badgers [14].
We identified 163 SNPs, 72 QTLs, 197 candidate genes, and 8 miRNAs associated with high IFNγ production in response to Mb infection. The 163 significant SNPs were located on 20 different Bos taurus chromosomes. This suggests that IFNɣ production is a polygenic trait depending on many SNPs located on different chromosomes. The identified QTLs overlapped with a total of 159 QTLs in chromosomes 3, 5, 16, 17, 22, and 27 that were previously associated with bTb susceptibility [29,49,50]. Our study identified candidate genes that might result in an inadequate recognition of Mb antigens, reduced autophagy, inflammasome activation, uncontrolled extracellular matrix degradation, and reduced immune cell migration. The RAS p21 protein activator 2 (RASA2) gene identified in our study encodes a protein that has been previously demonstrated to serve as a tumor suppressor in melanoma, and therefore, mutations affecting RASA2 might be associated with uncontrolled inflammation [81]. In the protein network analysis, we found a network (SCAF11-ARID2-RASA2-ZBTB38) centered in RASA2, together with the chromatin remodeling factor ARID2, also mutated in various cancer types [82].
Another candidate gene identified in our study was the TLR4 receptor. In the protein network analysis, we found a network (GRK5-RGS10-TLR4) containing the TLR4 gene. G Protein-Coupled Receptor Kinase 5 (GRK5) regulates the motility of polymorphonuclear leukocytes, and TLR4 recognizes pathogen-associated molecular patterns (PAMPs) that are expressed in mycobacteria and mediate the production of cytokines necessary for the development of effective immunity. Polymorphisms in the TLR4 gene have been shown to affect MAP recognition and have been associated with increased susceptibility of cattle to paratuberculosis [83]. A recent study has shown that knocking out the TLR4 gene in bovine MAC-T cells enhances inflammation in response to MAP infection [84]. TLR4 mutant C3H/HeJ mice can control Mb BCG infection as well as C3H/HeOUJ control mice, with efficient macrophage recruitment and activation, but they have arrested body weight and develop chronic exacerbated inflammation at later stages of infection [85]. Under a similar bacterial burden, inflammation was exacerbated and persisted longer in TLR4-deficient mice, suggesting that a “switch off” signal for inflammation was missing in the absence of a functional TLR4. Our study is the first to identify RASA2, GRK5, and TLR4 as candidate genes associated with strong IFNγ production in response to Mb infection. Mutations affecting these genes might cause improper Mb antigen recognition and uncontrolled inflammation.
Interestingly, the PCED1B, a hydrolase involved in apoptosis and autophagy of infected macrophages, was identified in our study, and it was also associated with human Tb susceptibility [75]. PCED1B plays a crucial role in apoptosis and autophagy, which are important mechanisms of innate immunity against Mtb [86]. Mutations in PCED1B might result in impaired autophagy and increased inflammasome activation in both human and bovine Tb. The ASAP1 gene was identified in our study and previously associated with human Tb [76]. Susceptibility to human tuberculosis is associated with variants in the ASAP1 gene, which encodes a regulator of dendritic cell (DC) migration. SNPs in ASAP1 were significantly associated with Tb in the Russian population [78], the Han Chinese population [87], and the Xinjiang Muslim population [88]. Impaired migration of mycobacteria-infected DCs, caused by the genetically determined excessive reduction of ASAP1 expression, may contribute to human and bovine tuberculosis. This may be one of the mechanisms that lead to the slow migration of DCs to lymph nodes and the delay of the adaptive immune response during the early stages of tuberculosis infection [76].
A total of eight miRNAs (bta-mir-2285cf, bta-mir-2351, bta-mir-12005-2, bta-mir-12000, bta-mir-6121, bta-mir-6524, bta-mir-4680, and bta-mir-2399) were associated with high production of IFNγ after stimulation with bPPD. MiRNAs are important regulators of innate and adaptative immune responses. More specifically, several miRNAs have been found to regulate T-cell functions or the innate function of macrophages, dendritic cells, and NK cells [89,90]. For example, bta-miR-4680 is expressed in bovine alveolar macrophages [91], suggesting its potential implication in infectious respiratory diseases. On the other hand, the identified bta-miR-2285cf is part of the bta-miR-2285 family, with 40 members spanning the entire bovine genome, which is expressed in response to Gram-positive bacteria infection [92] and in macrophages infected with Streptococcus agalactiae [93]. bta-mir-2399 is part of the repertoire of bovine miRNAs and miRNAs-like small regulatory RNAs expressed upon viral infections [94]. Future functional studies are required to confirm the association of these miRNAs, IFNγ production, and susceptibility to Mb infection.
Despite the vast research about the immune response mechanisms of human tuberculosis caused by Mtb, the knowledge of bovine tuberculosis’s immunology, particularly regarding the innate immune response, remains scarce. Our study advances the understanding of the role of bovine IFNγ in mycobacterial infections. It was interesting to find that none of the SNPs and QTLs identified in the current study overlapped with QTLs associated with strong production of IFNγ in blood samples stimulated with aPPD [33]. These findings show that different genetic variations are associated with enhanced IFNγ production in response to MAP or Mb [95]. Early after MAP infection, a strong IFNγ production correlates with resistance [33]. In contrast, strong IFNγ production in response to bovine tuberculin correlates with bTb susceptibility in more advanced stages of Mb infection. An IGRA to distinguish between Mb infection and bTb disease has not yet been developed [96].
Our results open the possibility of ranking Holstein cows based on predicted IFNɣ production, which would allow producers to select cattle less susceptible to bTb, ultimately reducing the prevalence of the disease, preventing economic losses, and increasing the length of cattle’s productive life. Genetic selection of less susceptible cattle would be particularly useful in low and middle-income countries where test-and-slaughter-based control programs are unfeasible. Importantly, the correlations between IFNɣ production and other animal traits were close to zero, suggesting that selective breeding to reduce animal susceptibility to bTb would not compromise improvements in other traits [25].
5. Conclusions
We report a new phenotype, the production of IFNɣ in response to bovine tuberculin, for evaluating susceptibility to bTb in dairy cows. The identified SNPs, QTLs, and candidate genes revealed an association between high IFNɣ levels after stimulation of blood samples with bPPD and host genetic susceptibility to bTb. QTLs enrichment analysis revealed a total of three health traits, with QTLs overlapping the QTL identified in this study, including bovine tuberculosis susceptibility, which is the most significant trait. To our knowledge, this is the first study revealing a genetic association between bovine IFNγ production and candidate genes involved in various crucial biological processes, including the recognition of bacterial antigens, apoptosis, autophagy, extracellular matrix remodeling, and the migration of immune cells. Furthermore, our results have important implications regarding the use of genetic evaluations for IFNγ production complementing bTb control.
Acknowledgments
Technical and human support provided by the DIPC Supercomputing Center is gratefully acknowledged. We gratefully acknowledge the 1000 Bull Genomes Consortium for providing accessibility to the WGS data that was used in this study. We are grateful to Kyle P. Hearn for the careful editing of the manuscript.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25116165/s1.
Author Contributions
Conceptualization, M.A.-H.; methodology, M.A.-H., G.B.-B., M.C., P.V., J.M.G., R.A.J., J.A.J., O.G.-R. and A.F.; formal analysis, G.B.-B., M.C., R.A.J., O.G.-R., A.F. and M.A.-H.; resources, R.A.J., JG, P.V., M.C., J.A.J., O.G.-R., A.F.; writing—original draft preparation, G.B.-B.; supervision, O.G.-R., A.F. and M.A.-H.; visualization, G.B.-B. and M.A.-H.; writing—review and editing, M.A.-H.; project administration, M.A.-H.; funding acquisition, M.A.-H. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Ethical review and approval were waived for this study because the cattle included in this study were not submitted to any in vivo experimentation.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary material. Further inquiries can be directed to the corresponding author. Sequence data used in this study for the imputation to WGS are owned by the 1000 Bull Genomes Project Consortium. Individual genotype data used in this study are managed by a third party, the Spanish Friesian Cattle National Federation (CONAFE). Request for individual genotype data can be made to CONAFE, Ctra. De Andalucia, km. 23,600–28,340 Valdemoro, Madrid, Spain; email: conafe@conafe.com; phone: +34-(91)-8952412; Fax: 918-951-471; website: www.conafe.com. CONAFE is a member of the Eurogenomics Cooperative U.A.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
This research was funded by MCIN/AEI/10.13039/501100011033 and ERDF “A way of making Europe”, grants RTI2018-094192-R-C21 and PID2021-122197OR-C21 to MAH. GB-B has been awarded a fellowship (PRE2019-090562) from MCIN/AEI/10.13039/501100011033 and ESF “Investing in your future”.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Smith R.L., Tauer L.W., Sanderson M.W., Gröhn Y.T. Minimum Cost to Control Bovine Tuberculosis in Cow-Calf Herds. Prev. Vet. Med. 2014;115:18–28. doi: 10.1016/j.prevetmed.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olea-Popelka F., Muwonge A., Perera A., Dean A.S., Mumford E., Erlacher-Vindel E., Forcella S., Silk B.J., Ditiu L., El Idrissi A., et al. Zoonotic Tuberculosis in Human Beings Caused by Mycobacterium bovis—A Call for Action. Lancet Infect. Dis. 2017;17:e21–e25. doi: 10.1016/S1473-3099(16)30139-6. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong J.A., Hart P.D. Response of Cultured Macrophages to Mycobacterium tuberculosis, with Observations on Fusion of Lysosomes with Phagosomes. J. Exp. Med. 1971;134:713–740. doi: 10.1084/jem.134.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buddle B.M., Wedlock D.N., Denis M., Skinner M.A. Identification of Immune Response Correlates for Protection against Bovine Tuberculosis. Vet. Immunol. Immunopathol. 2005;108:45–51. doi: 10.1016/j.vetimm.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Gutierrez M.G., Master S.S., Singh S.B., Taylor G.A., Colombo M.I., Deretic V. Autophagy Is a Defense Mechanism Inhibiting BCG and Mycobacterium tuberculosis Survival in Infected Macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 6.Vordermeier H.M., Chambers M.A., Cockle P.J., Whelan A.O., Simmons J., Hewinson R.G. Correlation of ESAT-6-Specific Gamma Interferon Production with Pathology in Cattle Following Mycobacterium bovis BCG Vaccination against Experimental Bovine Tuberculosis. Infect. Immun. 2002;70:3026–3032. doi: 10.1128/IAI.70.6.3026-3032.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma F., Xu S., Liu X., Zhang Q., Xu X., Liu M., Hua M., Li N., Yao H., Cao X. The MicroRNA MiR-29 Controls Innate and Adaptive Immune Responses to Intracellular Bacterial Infection by Targeting Interferon-γ. Nat. Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 8.Llamazares O.R.G., Martín C.B.G., Nistal D.A., de la Puente Redondo V.A., Rodríguez L.D., Ferri E.F.R. Field Evaluation of the Single Intradermal Cervical Tuberculin Test and the Interferon-γ Assay for Detection and Eradication of Bovine Tuberculosis in Spain. Vet. Microbiol. 1999;70:55–66. doi: 10.1016/S0378-1135(99)00121-2. [DOI] [PubMed] [Google Scholar]
- 9.Casal C., Infantes J.A., Risalde M.A., Díez-Guerrier A., Domínguez M., Moreno I., Romero B., de Juan L., Sáez J.L., Juste R., et al. Antibody Detection Tests Improve the Sensitivity of Tuberculosis Diagnosis in Cattle. Res. Vet. Sci. 2017;112:214–221. doi: 10.1016/j.rvsc.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Buddle B.M., McCarthy A.R., Heslop J., Aldwell F.E., Nolan A., Jackson R., Pfeiffer D.U. Evaluation of Three Serological Assays for the Diagnosis of Mycobacterium bovis Infection in Brushtail Possums. N. Z. Vet. J. 1995;43:91–95. doi: 10.1080/00480169.1995.35860. [DOI] [PubMed] [Google Scholar]
- 11.Pollock J.M., Welsh M.D., McNair J. Immune Responses in Bovine Tuberculosis: Towards New Strategies for the Diagnosis and Control of Disease. Vet. Immunol. Immunopathol. 2005;108:37–43. doi: 10.1016/j.vetimm.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Waters W.R., Palmer M.V., Thacker T.C., Davis W.C., Sreevatsan S., Coussens P., Meade K.G., Hope J.C., Estes D.M. Tuberculosis Immunity: Opportunities from Studies with Cattle. Clin. Dev. Immunol. 2011;2011:768542. doi: 10.1155/2011/768542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clegg T.A., Doyle M., Ryan E., More S.J., Gormley E. Characteristics of Mycobacterium bovis Infected Herds Tested with the Interferon-Gamma Assay. Prev. Vet. Med. 2019;168:52–59. doi: 10.1016/j.prevetmed.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Smith K., Kleynhans L., Warren R.M., Goosen W.J., Miller M.A. Cell-Mediated Immunological Biomarkers and Their Diagnostic Application in Livestock and Wildlife Infected with Mycobacterium bovis. Front. Immunol. 2021;12:639605. doi: 10.3389/fimmu.2021.639605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernitz N., Kerr T.J., Goosen W.J., Higgitt R.L., de Waal C., Clarke C., Cooper D.V., Warren R.M., van Helden P.D., Parsons S.D.C., et al. Impact of Mycobacterium bovis-Induced Pathology on Interpretation of QuantiFERON®-TB Gold Assay Results in African Buffaloes (Syncerus Caffer) Vet. Immunol. Immunopathol. 2019;217:109923. doi: 10.1016/j.vetimm.2019.109923. [DOI] [PubMed] [Google Scholar]
- 16.Vordermeier H.M., Whelan A., Cockle P.J., Farrant L., Palmer N., Hewinson R.G. Use of Synthetic Peptides Derived from the Antigens ESAT-6 and CFP-10 for Differential Diagnosis of Bovine Tuberculosis in Cattle. Clin. Diagn. Lab. Immunol. 2001;8:571–578. doi: 10.1128/CDLI.8.3.571-578.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomlinson A.J., Chambers M.A., Mcdonald R.A., Delahay R.J. Association of Quantitative Interferon-γ Responses with the Progression of Naturally Acquired Mycobacterium bovis Infection in Wild European Badgers (Meles Meles) Immunology. 2015;144:263–270. doi: 10.1111/imm.12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Álvarez J., Perez A., Marqués S., Bezos J., Grau A., de la Cruz M.L., Romero B., Saez J.L., del Rosario Esquivel M., del Carmen Martínez M., et al. Risk Factors Associated with Negative In-Vivo diagnostic Results in Bovine Tuberculosis-Infected Cattle in Spain. BMC Vet. Res. 2014;10:14. doi: 10.1186/1746-6148-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allepuz A., Casal J., Napp S., Saez M., Alba A., Vilar M., Domingo M., González M.A., Duran-Ferrer M., Vicente J., et al. Analysis of the Spatial Variation of Bovine Tuberculosis Disease Risk in Spain (2006–2009) Prev. Vet. Med. 2011;100:44–52. doi: 10.1016/j.prevetmed.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 20.Humblet M.-F., Boschiroli M.L., Saegerman C. Classification of Worldwide Bovine Tuberculosis Risk Factors in Cattle: A Stratified Approach. Vet. Res. 2009;40:50. doi: 10.1051/vetres/2009033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banos G. Selective Breeding Can Contribute to Bovine Tuberculosis Control and Eradication. Ir. Vet. J. 2023;76:19. doi: 10.1186/s13620-023-00250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bermingham M.L., More S.J., Good M., Cromie A.R., Higgins I.M., Brotherstone S., Berry D.P. Genetics of Tuberculosis in Irish Holstein-Friesian Dairy Herds. J. Dairy Sci. 2009;92:3447–3456. doi: 10.3168/jds.2008-1848. [DOI] [PubMed] [Google Scholar]
- 23.Brotherstone S., White I.M.S., Coffey M., Downs S.H., Mitchell A.P., Clifton-Hadley R.S., More S.J., Good M., Woolliams J.A. Evidence of Genetic Resistance of Cattle to Infection with Mycobacterium bovis. J. Dairy Sci. 2010;93:1234–1242. doi: 10.3168/jds.2009-2609. [DOI] [PubMed] [Google Scholar]
- 24.Richardson I.W., Bradley D.G., Higgins I.M., More S.J., Jennifer M., Berry D.P. Variance Components for Susceptibility to Mycobacterium bovis Infection in Dairy and Beef Cattle. Genet. Sel. Evol. 2014;46:77. doi: 10.1186/s12711-014-0077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banos G., Winters M., Mrode R., Mitchell A.P., Bishop S.C., Woolliams J.A., Coffey M.P. Genetic Evaluation for Bovine Tuberculosis Resistance in Dairy Cattle. J. Dairy Sci. 2017;100:1272–1281. doi: 10.3168/jds.2016-11897. [DOI] [PubMed] [Google Scholar]
- 26.Bermingham M.L., Bishop S.C., Woolliams J.A., Pong-Wong R., Allen A.R., McBride S.H., Ryder J.J., Wright D.M., Skuce R.A., McDowell S.W., et al. Genome-Wide Association Study Identifies Novel Loci Associated with Resistance to Bovine Tuberculosis. Hered. Edinb. 2014;112:543–551. doi: 10.1038/hdy.2013.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsairidou S., Woolliams J.A., Allen A.R., Skuce R.A., McBride S.H., Wright D.M., Bermingham M.L., Pong-Wong R., Matika O., McDowell S.W.J., et al. Genomic Prediction for Tuberculosis Resistance in Dairy Cattle. PLoS ONE. 2014;9:0096728. doi: 10.1371/journal.pone.0096728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raphaka K., Matika O., Sánchez-Molano E., Mrode R., Coffey M.P., Riggio V., Glass E.J., Woolliams J.A., Bishop S.C., Banos G. Genomic Regions Underlying Susceptibility to Bovine Tuberculosis in Holstein-Friesian Cattle. BMC Genet. 2017;18:27. doi: 10.1186/s12863-017-0493-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ring S.C., Purfield D.C., Good M., Breslin P., Ryan E., Blom A., Evans R.D., Doherty M.L., Bradley D.G., Berry D.P. Variance Components for Bovine Tuberculosis Infection and Multi-Breed Genome-Wide Association Analysis Using Imputed Whole Genome Sequence Data. PLoS ONE. 2019;14:0212067. doi: 10.1371/journal.pone.0212067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Callaby R., Kelly R., Mazeri S., Egbe F., Benedictus L., Clark E., Doeschl-Wilson A., Bronsvoort B., Salavati M., Muwonge A. Genetic Diversity of Cameroon Cattle and a Putative Genomic Map for Resistance to Bovine Tuberculosis. Front. Genet. 2020;11:550215. doi: 10.3389/fgene.2020.550215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mrode R., Ojango J.M.K., Okeyo A.M., Mwacharo J.M. Genomic Selection and Use of Molecular Tools in Breeding Programs for Indigenous and Crossbred Cattle in Developing Countries: Current Status and Future Prospects. Front. Genet. 2019;9:694. doi: 10.3389/fgene.2018.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ko D.C., Shukla K.P., Fong C., Wasnick M., Brittnacher M.J., Wurfel M.M., Holden T.D., O’Keefe G.E., Van Yserloo B., Akey J.M., et al. A Genome-Wide In Vitro Bacterial-Infection Screen Reveals Human Variation in the Host Response Associated with Inflammatory Disease. Am. J. Hum. Genet. 2009;85:214–227. doi: 10.1016/j.ajhg.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Badia-Bringué G., Canive M., Vázquez P., Garrido J.M., Fernández A., Juste R.A., Jiménez J.A., González-Recio O., Alonso-Hearn M. Association between High Interferon-Gamma Production in Avian Tuberculin-Stimulated Blood from Mycobacterium avium subsp. Paratuberculosis-Infected Cattle and Candidate Genes Implicated in Necroptosis. Microorganisms. 2023;11:1817. doi: 10.3390/microorganisms11071817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vázquez P., Ruiz-Larrañaga O., Garrido J.M., Iriondo M., Manzano C., Agirre M., Estonba A., Juste R.A. Genetic Association Analysis of Paratuberculosis Forms in Holstein-Friesian Cattle. Vet. Med. Int. 2014;2014:321327. doi: 10.1155/2014/321327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Canive M., González-Recio O., Fernández A., Vázquez P., Badia-Bringué G., Lavín J.L., Garrido J.M., Juste R.A., Alonso-Hearn M. Identification of Loci Associated with Susceptibility to Mycobacterium avium subsp. Paratuberculosis Infection in Holstein Cattle Using Combinations of Diagnostic Tests and Imputed Whole-Genome Sequence Data. PLoS ONE. 2021;16:e0256091. doi: 10.1371/journal.pone.0256091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loh P.-R., Palamara P.F., Price A.L. Fast and Accurate Long-Range Phasing in a UK Biobank Cohort. Nat. Genet. 2016;48:811–816. doi: 10.1038/ng.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M., et al. Next-Generation Genotype Imputation Service and Methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayes B.J., Daetwyler H.D. 1000 Bull Genomes Project to Map Simple and Complex Genetic Traits in Cattle: Applications and Outcomes. Annu. Rev. Anim. Biosci. 2019;7:89–102. doi: 10.1146/annurev-animal-020518-115024. [DOI] [PubMed] [Google Scholar]
- 39.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: A Tool for Genome-Wide Complex Trait Analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burton P.R., Clayton D.G., Cardon L.R., Craddock N., Deloukas P., Duncanson A., Kwiatkowski D.P., McCarthy M.I., Ouwehand W.H., Samani N.J., et al. Genome-Wide Association Study of 14,000 Cases of Seven Common Diseases and 3000 Shared Controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., De Bakker P.I.W., Daly M.J., et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flicek P., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., et al. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fonseca P.A.S., Suárez-Vega A., Marras G., Cánovas Á. GALLO: An R Package for Genomic Annotation and Integration of Multiple Data Sources in Livestock for Positional Candidate Loci. Gigascience. 2020;9:giaa149. doi: 10.1093/gigascience/giaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu Z.-L., Park C.A., Reecy J.M. Bringing the Animal QTLdb and CorrDB into the Future: Meeting New Challenges and Providing Updated Services. Nucleic Acids Res. 2022;50:D956–D961. doi: 10.1093/nar/gkab1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.GWAS Catalog. [(accessed on 18 March 2024)]. Available online: https://www.ebi.ac.uk/gwas/
- 46.GeneCards—Human Genes|Gene Database|Gene Search. [(accessed on 18 March 2024)]. Available online: https://www.genecards.org/
- 47.InnateDB: Systems Biology of the Innate Immune Response. [(accessed on 18 March 2024)]. Available online: https://www.innatedb.com.
- 48.VanRaden P.M., Sullivan P.G. International Genomic Evaluation Methods for Dairy Cattle. Genet. Sel. Evol. 2010;42:7. doi: 10.1186/1297-9686-42-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richardson I.W., Berry D.P., Wiencko H.L., Higgins I.M., More S.J., McClure J., Lynn D.J., Bradley D.G. A Genome-Wide Association Study for Genetic Susceptibility to Mycobacterium bovis Infection in Dairy Cattle Identifies a Susceptibility QTL on Chromosome 23. Genet. Sel. Evol. 2016;48:19. doi: 10.1186/s12711-016-0197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.González-Ruiz S., Strillacci M.G., Durán-Aguilar M., Cantó-Alarcón G.J., Herrera-Rodríguez S.E., Bagnato A., Guzmán L.F., Milián-Suazo F., Román-Ponce S.I. Genome-Wide Association Study in Mexican Holstein Cattle Reveals Novel Quantitative Trait Loci Regions and Confirms Mapped Loci for Resistance to Bovine Tuberculosis. Animals. 2019;9:636. doi: 10.3390/ani9090636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ilie D.E., Mizeranschi A.E., Mihali C.V., Neamț R.I., Goilean G.V., Georgescu O.I., Zaharie D., Carabaș M., Huțu I. Genome-Wide Association Studies for Milk Somatic Cell Score in Romanian Dairy Cattle. Genes. 2021;12:1495. doi: 10.3390/genes12101495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moretti R., Soglia D., Chessa S., Sartore S., Finocchiaro R., Rasero R., Sacchi P. Identification of SNPs Associated with Somatic Cell Score in Candidate Genes in Italian Holstein Friesian Bulls. Animals. 2021;11:366. doi: 10.3390/ani11020366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miles A.M., Huson H.J. Time- and Population-Dependent Genetic Patterns Underlie Bovine Milk Somatic Cell Count. J. Dairy Sci. 2020;103:8292–8304. doi: 10.3168/jds.2020-18322. [DOI] [PubMed] [Google Scholar]
- 54.Jiang J., Ma L., Prakapenka D., VanRaden P.M., Cole J.B., Da Y. A Large-Scale Genome-Wide Association Study in U.S. Holstein Cattle. Front. Genet. 2019;10:412. doi: 10.3389/fgene.2019.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X., Ma P., Liu J., Zhang Q., Zhang Y., Ding X., Jiang L., Wang Y., Zhang Y., Sun D., et al. Genome-Wide Association Study in Chinese Holstein Cows Reveal Two Candidate Genes for Somatic Cell Score as an Indicator for Mastitis Susceptibility. BMC Genet. 2015;16:111. doi: 10.1186/s12863-015-0263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X.P., Luoreng Z.M., Gao S.X., Guo D.S., Li J.Y., Gao X., Xu S.Z., Li F., Chen G., Wang J.R. Haplotype Analysis of TLR4 Gene and Its Effects on Milk Somatic Cell Score in Chinese Commercial Cattle. Mol. Biol. Rep. 2014;41:2345–2351. doi: 10.1007/s11033-014-3088-7. [DOI] [PubMed] [Google Scholar]
- 57.Strillacci M.G., Frigo E., Schiavini F., Samoré A.B., Canavesi F., Vevey M., Cozzi M.C., Soller M., Lipkin E., Bagnato A. Genome-Wide Association Study for Somatic Cell Score in Valdostana Red Pied Cattle Breed Using Pooled DNA. BMC Genet. 2014;15:106. doi: 10.1186/s12863-014-0106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cole J.B., Wiggans G.R., Ma L., Sonstegard T.S., Lawlor T.J., Crooker B.A., Van Tassell C.P., Yang J., Wang S., Matukumalli L.K., et al. Genome-Wide Association Analysis of Thirty One Production, Health, Reproduction and Body Conformation Traits in Contemporary U.S. Holstein Cows. BMC Genom. 2011;12:408. doi: 10.1186/1471-2164-12-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kolbehdari D., Wang Z., Grant J.R., Murdoch B., Prasad A., Xiu Z., Marques E., Stothard P., Moore S.S. A Whole Genome Scan to Map QTL for Milk Production Traits and Somatic Cell Score in Canadian Holstein Bulls. J. Anim. Breed. Genet. 2009;126:216–227. doi: 10.1111/j.1439-0388.2008.00793.x. [DOI] [PubMed] [Google Scholar]
- 60.Kiser J.N., Neibergs H.L. Identifying Loci Associated with Bovine Corona Virus Infection and Bovine Respiratory Disease in Dairy and Feedlot Cattle. Front. Vet. Sci. 2021;8:679074. doi: 10.3389/fvets.2021.679074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keele J.W., Kuehn L.A., McDaneld T.G., Tait R.G., Jones S.A., Smith T.P.L., Shackelford S.D., King D.A., Wheeler T.L., Lindholm-Perry A.K., et al. Genomewide Association Study of Lung Lesions in Cattle Using Sample Pooling. J. Anim. Sci. 2015;93:956. doi: 10.2527/jas.2014-8492. [DOI] [PubMed] [Google Scholar]
- 62.Neupane M., Kiser J.N., Neibergs H.L. Gene Set Enrichment Analysis of SNP Data in Dairy and Beef Cattle with Bovine Respiratory Disease. Anim. Genet. 2018;49:527–538. doi: 10.1111/age.12718. [DOI] [PubMed] [Google Scholar]
- 63.Pant S.D., Schenkel F.S., Verschoor C.P., You Q., Kelton D.F., Moore S.S., Karrow N.A. A Principal Component Regression Based Genome Wide Analysis Approach Reveals the Presence of a Novel QTL on BTA7 for MAP Resistance in Holstein Cattle. Genomics. 2010;95:176–182. doi: 10.1016/j.ygeno.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 64.Kirkpatrick B.W., Shi X., Shook G.E., Collins M.T. Whole-Genome Association Analysis of Susceptibility to Paratuberculosis in Holstein Cattle. Anim. Genet. 2011;42:149–160. doi: 10.1111/j.1365-2052.2010.02097.x. [DOI] [PubMed] [Google Scholar]
- 65.McGovern S.P., Purfield D.C., Ring S.C., Carthy T.R., Graham D.A., Berry D.P. Candidate Genes Associated with the Heritable Humoral Response to Mycobacterium avium ssp. Paratuberculosis in Dairy Cows Have Factors in Common with Gastrointestinal Diseases in Humans. J. Dairy Sci. 2019;102:4249–4263. doi: 10.3168/jds.2018-15906. [DOI] [PubMed] [Google Scholar]
- 66.Kumar S., Kumar S., Singh R.V., Chauhan A., Kumar A., Sulabh S., Bharati J., Singh S.V. Genetic Association of Polymorphisms in Bovine TLR2 and TLR4 Genes with Mycobacterium avium Subspecies Paratuberculosis Infection in Indian Cattle Population. Vet. Res. Commun. 2019;43:105–114. doi: 10.1007/s11259-019-09750-2. [DOI] [PubMed] [Google Scholar]
- 67.Gopi B., Singh R.V., Kumar S., Kumar S., Chauhan A., Kumar A., Singh S.V. Single-Nucleotide Polymorphisms in CLEC7A, CD209 and TLR4 Gene and Their Association with Susceptibility to Paratuberculosis in Indian Cattle. J. Genet. 2020;99:14. doi: 10.1007/s12041-019-1172-4. [DOI] [PubMed] [Google Scholar]
- 68.Kirkpatrick B.W., Cooke M.E., Frie M., Sporer K.R.B., Lett B., Wells S.J., Coussens P.M. Genome-Wide Association Analysis for Susceptibility to Infection by Mycobacterium avium ssp. Paratuberculosis in US Holsteins. J. Dairy Sci. 2022;105:4301–4313. doi: 10.3168/jds.2021-21276. [DOI] [PubMed] [Google Scholar]
- 69.Canive M., Badia-Bringué G., Vázquez P., González-Recio O., Fernández A., Garrido J., Juste R.A., Alonso-Hearn M. Identification of loci associated with pathological outcomes in Holstein cattle infected with Mycobacterium avium subsp. paratuberculosis using whole-genome sequence data. Sci. Rep. 2021;11:20177. doi: 10.1038/s41598-021-99672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wasmuth H.E., Tag C.G., Van de Leur E., Hellerbrand C., Mueller T., Berg T., Puhl G., Neuhaus P., Samuel D., Trautwein C., et al. The Marburg I Variant (G534E) of the Factor VII-Activating Protease Determines Liver Fibrosis in Hepatitis C Infection by Reduced Proteolysis of Platelet-Derived Growth Factor BB. Hepatology. 2009;49:775–780. doi: 10.1002/hep.22707. [DOI] [PubMed] [Google Scholar]
- 71.Hoppe B., Tolou F., Radtke H., Kiesewetter H., Dörner T., Salama A. Marburg I Polymorphism of Factor VII–Activating Protease Is Associated with Idiopathic Venous Thromboembolism. Blood. 2005;105:1549–1551. doi: 10.1182/blood-2004-08-3328. [DOI] [PubMed] [Google Scholar]
- 72.Edelmann B., Bertsch U., Tchikov V., Winoto-Morbach S., Perrotta C., Jakob M., Adam-Klages S., Kabelitz D., Schütze S. Caspase-8 and Caspase-7 Sequentially Mediate Proteolytic Activation of Acid Sphingomyelinase in TNF-R1 Receptosomes. EMBO J. 2011;30:379–394. doi: 10.1038/emboj.2010.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garcia-Lozano J.R., Torres B., Fernandez O., Orozco G., Alvarez-Marquez A., Garcia A., Gonzalez-Gay M.A., Garcia A., Nunez-Roldan A., Martin J., et al. Caspase 7 Influences Susceptibility to Rheumatoid Arthritis. Rheumatology. 2007;46:1243–1247. doi: 10.1093/rheumatology/kem096. [DOI] [PubMed] [Google Scholar]
- 74.Rohlwink U., Walker N., Ordonez A., Li Y., Tucker E., Elkington P., Wilkinson R., Wilkinson K. Matrix Metalloproteinases in Pulmonary and Central Nervous System Tuberculosis—A Review. Int. J. Mol. Sci. 2019;20:1350. doi: 10.3390/ijms20061350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gelemanović A., Ćatipović Ardalić T., Pribisalić A., Hayward C., Kolčić I., Polašek O. Genome-Wide Meta-Analysis Identifies Multiple Novel Rare Variants to Predict Common Human Infectious Diseases Risk. Int. J. Mol. Sci. 2023;24:7006. doi: 10.3390/ijms24087006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Curtis J., Luo Y., Zenner H.L., Cuchet-Lourenço D., Wu C., Lo K., Maes M., Alisaac A., Stebbings E., Liu J.Z., et al. Susceptibility to Tuberculosis Is Associated with Variants in the ASAP1 Gene Encoding a Regulator of Dendritic Cell Migration. Nat. Genet. 2015;47:523–527. doi: 10.1038/ng.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Badia-Bringué G., Canive M., Alonso-Hearn M. Control of Mycobacterium avium subsp. Paratuberculosis Load within Infected Bovine Monocyte-Derived Macrophages Is Associated with Host Genetics. Front. Immunol. 2023;14:1042638. doi: 10.3389/fimmu.2023.1042638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kassahun Y., Mattiangeli V., Ameni G., Hailu E., Aseffa A., Young D.B., Hewinson R.G., Vordermeier H.M., Bradley D.G. Admixture Mapping of Tuberculosis and Pigmentation-Related Traits in an African-European Hybrid Cattle Population. Front. Genet. 2015;6:210. doi: 10.3389/fgene.2015.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsairidou S., Allen A., Banos G., Coffey M., Anacleto O., Byrne A.W., Skuce R.A., Glass E.J., Woolliams J.A., Doeschl-Wilson A.B. Can We Breed Cattle for Lower Bovine TB Infectivity? Front. Vet. Sci. 2018;5:310. doi: 10.3389/fvets.2018.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Finlay E.K., Berry D.P., Wickham B., Gormley E.P., Bradley D.G. A Genome Wide Association Scan of Bovine Tuberculosis Susceptibility in Holstein-Friesian Dairy Cattle. PLoS ONE. 2012;7:30545. doi: 10.1371/journal.pone.0030545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arafeh R., Qutob N., Emmanuel R., Keren-Paz A., Madore J., Elkahloun A., Wilmott J.S., Gartner J.J., Di Pizio A., Winograd-Katz S., et al. Recurrent Inactivating RASA2 Mutations in Melanoma. Nat. Genet. 2015;47:1408–1410. doi: 10.1038/ng.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chaisaingmongkol J., Budhu A., Dang H., Rabibhadana S., Pupacdi B., Kwon S.M., Forgues M., Pomyen Y., Bhudhisawasdi V., Lertprasertsuke N., et al. Common Molecular Subtypes Among Asian Hepatocellular Carcinoma and Cholangiocarcinoma. Cancer Cell. 2017;32:57–70.e3. doi: 10.1016/j.ccell.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ruiz-Larrañaga O., Manzano C., Iriondo M., Garrido J.M., Molina E., Vazquez P., Juste R.A., Estonba A. Genetic Variation of Toll-like Receptor Genes and Infection by Mycobacterium avium ssp. Paratuberculosis in Holstein-Friesian Cattle. J. Dairy Sci. 2011;94:3635–3641. doi: 10.3168/jds.2010-3788. [DOI] [PubMed] [Google Scholar]
- 84.Shandilya U.K., Wu X., McAllister C., Mutharia L., Karrow N.A. Role of Toll-Like Receptor 4 in Mycobacterium avium subsp. Paratuberculosis Infection of Bovine Mammary Epithelial (MAC-T) Cells In Vitro. Microbiol. Spectr. 2023;11:e04393-22. doi: 10.1128/spectrum.04393-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fremond C.M.C., Nicolle D.M.M., Torres D.S., Quesniaux V.F.J. Control of Mycobacterium bovis BCG Infection with Increased Inflammation in TLR4-Deficient Mice. Microbes. Infect. 2003;5:1070–1081. doi: 10.1016/j.micinf.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 86.Li M., Cui J., Niu W., Huang J., Feng T., Sun B., Yao H. Long Non-Coding PCED1B-AS1 Regulates Macrophage Apoptosis and Autophagy by Sponging MiR-155 in Active Tuberculosis. Biochem. Biophys. Res. Commun. 2019;509:803–809. doi: 10.1016/j.bbrc.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 87.Chen C., Zhao Q., Shao Y., Li Y., Song H., Li G., Zhu L., Lu W., Xu B. A Common Variant of ASAP1 Is Associated with Tuberculosis Susceptibility in the Han Chinese Population. Dis. Markers. 2019;2019:7945429. doi: 10.1155/2019/7945429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang X., Ma A., Han X., Litifu A., Xue F. ASAP1 Gene Polymorphisms Are Associated with Susceptibility to Tuberculosis in a Chinese Xinjiang Muslim Population. Exp. Ther. Med. 2018;15:3392–3398. doi: 10.3892/etm.2018.5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taganov K.D., Boldin M.P., Chang K.-J., Baltimore D. NF-ΚB-Dependent Induction of MicroRNA MiR-146, an Inhibitor Targeted to Signaling Proteins of Innate Immune Responses. Proc. Natl. Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bezman N.A., Cedars E., Steiner D.F., Blelloch R., Hesslein D.G.T., Lanier L.L. Distinct Requirements of MicroRNAs in NK Cell Activation, Survival, and Function. J. Immunol. 2010;185:3835–3846. doi: 10.4049/jimmunol.1000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vegh P., Foroushani A.B.K., Magee D.A., McCabe M.S., Browne J.A., Nalpas N.C., Conlon K.M., Gordon S.V., Bradley D.G., MacHugh D.E., et al. Profiling MicroRNA Expression in Bovine Alveolar Macrophages Using RNA-Seq. Vet. Immunol. Immunopathol. 2013;155:238–244. doi: 10.1016/j.vetimm.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 92.Lawless N., Foroushani A.B.K., McCabe M.S., O’Farrelly C., Lynn D.J. Next Generation Sequencing Reveals the Expression of a Unique MiRNA Profile in Response to a Gram-Positive Bacterial Infection. PLoS ONE. 2013;8:e57543. doi: 10.1371/journal.pone.0057543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lewandowska-Sabat A.M., Hansen S.F., Solberg T.R., Østerås O., Heringstad B., Boysen P., Olsaker I. MicroRNA Expression Profiles of Bovine Monocyte-Derived Macrophages Infected in Vitro with Two Strains of Streptococcus Agalactiae. BMC Genom. 2018;19:241. doi: 10.1186/s12864-018-4591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Glazov E.A., Kongsuwan K., Assavalapsakul W., Horwood P.F., Mitter N., Mahony T.J. Repertoire of Bovine MiRNA and MiRNA-Like Small Regulatory RNAs Expressed upon Viral Infection. PLoS ONE. 2009;4:e6349. doi: 10.1371/journal.pone.0006349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stabel J.R., Waters W.R., Bannantine J.P., Palmer M.V. Comparative Cellular Immune Responses in Calves after Infection with Mycobacterium avium subsp. Paratuberculosis, M. Avium subsp. Avium, M. Kansasii and M. Bovis. Vet. Immunol. Immunopathol. 2021;237:110268. doi: 10.1016/j.vetimm.2021.110268. [DOI] [PubMed] [Google Scholar]
- 96.Xin T., Gao X., Yang H., Li P., Liang Q., Hou S., Sui X., Guo X., Yuan W., Zhu H., et al. Limitations of Using IL-17A and IFN-γ-Induced Protein 10 to Detect Bovine Tuberculosis. Front. Vet. Sci. 2018;5:28. doi: 10.3389/fvets.2018.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary material. Further inquiries can be directed to the corresponding author. Sequence data used in this study for the imputation to WGS are owned by the 1000 Bull Genomes Project Consortium. Individual genotype data used in this study are managed by a third party, the Spanish Friesian Cattle National Federation (CONAFE). Request for individual genotype data can be made to CONAFE, Ctra. De Andalucia, km. 23,600–28,340 Valdemoro, Madrid, Spain; email: conafe@conafe.com; phone: +34-(91)-8952412; Fax: 918-951-471; website: www.conafe.com. CONAFE is a member of the Eurogenomics Cooperative U.A.