Abstract
A unique feature of the large hepadnavirus envelope protein (L) is its mixed transmembrane topology, resulting from partial posttranslational translocation of the pre-S domain. Using protease protection analysis, we demonstrate for duck hepatitis B virus an essential role for the small envelope protein (S) in this process, providing the first experimental evidence for an S translocation channel. Further analysis revealed that the presumed cytoplasmic loop between TM1 and TM2 in the C-terminal S domain is membrane embedded and protrudes to the particle surface. These data suggest that some L molecules form a highly folded, potentially spring-loaded topology with five membrane-spanning regions and a membrane-traversing pre-S chain.
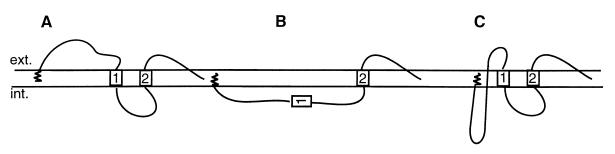
The hepadnavirus large surface protein (L) forms mixed transmembrane topologies through the partial posttranslational translocation of its N-terminal pre-S domain. This dual topology, initially recognized in hepatitis B virus (HBV) (2, 11) and confirmed for an avian hepadnavirus (6, 16), enables the pre-S domain to serve multiple, specialized functions in different cellular compartments. An external topology with a surface-exposed or translocated pre-S domain mediates binding to cell surface receptors (8, 10) (Fig. 1A), and an internal topology with the pre-S domain being cytosolically disposed serves a matrix-like function for assembly with the viral capsid (2) as well as various regulatory functions (13, 15) (Fig. 1B). A third, membrane-traversing topology, is interpreted to be an intermediate configuration in the translocation process (6) (Fig. 1C) but has no known function. Despite the fundamental importance of the mixed topologies to the replication of hepadnaviruses, the mechanism which enables either the passage of the hydrophilic pre-S domain to the fully surface-exposed ectodomain or its retention as a membrane-traversing region is still unknown but is postulated to occur through complexing of envelope subunits into a channel (14). The preservation of the intermediate topology of L in mature particles and the ability of L to release pre-S into the external conformation may, however, be an indication of a purpose in viral entry.
FIG. 1.
Models of L protein topology. (A) Model of the external topology of L with an exposed or translocated N-terminal pre-S domain; (B) model of the internal topology of L, present immediately after synthesis, with pre-S and TM1 being cytosolically disposed; (C) model of the intermediate topology, identified in mature particles, in which a small part of the C terminus of pre-S is exposed to the particle surface while the remainder is proposed to traverse the particle membrane and be located internally. The first transmembrane domain and the transmembrane anchor domain in S are indicated by boxes 1 and 2, respectively, but the third predicted but uncharacterized C-terminal transmembrane region is not shown. The N-terminal myristate is represented by the spiral. ext., exterior; int., interior.
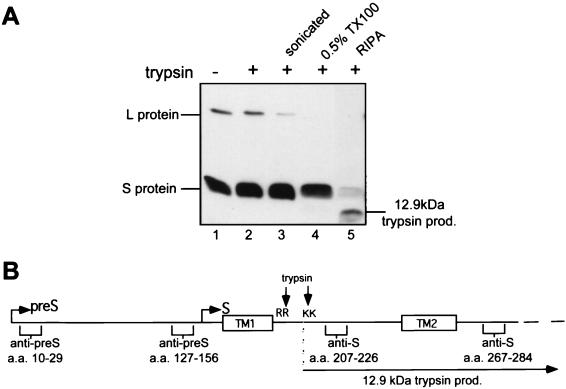
In this study, we have obtained some insight into the parameters influencing pre-S translocation, including the essential role of the S protein, comprised of the C-terminal S domain of L, by further analyzing the topologies of newly synthesized envelope proteins on microsomal membranes and in mature subviral particles (SVPs), which for duck HBV (DHBV) contain the same S/L ratio and topology of envelope proteins as virions. Previously published data have demonstrated that pre-S is protected to various extents from protease cleavage in a proportion of L molecules, consistent with a mixed L topology (1, 11, 16) but that the S domains of L and the S protein remain completely resistant to protease cleavage. These results are surprising in view of the prevailing topological model in which the loop between transmembranes 1 and 2 (TM1 and -2) is cytoplasmically disposed (Fig. 1), especially in DHBV, which, in contrast to HBV, contains arginine and lysine residues in the loop region and is thus expected to be susceptible to trypsin cleavage. As shown in Fig. 2A, the L protein becomes susceptible to trypsin digestion only upon addition of Triton X-100 (Fig. 2A, lane 4) while the S protein remains relatively resistant but is readily cleaved upon addition of radioimmunoprecipitation assay (RIPA) buffer containing the denaturants sodium dodecyl sulfate (SDS) and sodium deoxycholate, indicative of a highly complexed protein or folding of the 49-amino-acid (aa)-long loop into a protease-resistant conformation (Fig. 2A, lane 5). Sonication of the microsomes (Fig. 2A, lane 3) or addition of Triton X-100 alone (Fig. 2A, lane 4), however, rendered part of the L protein susceptible to trypsin digestion. This suggests that L may form an inverted L topology or altered topology of this region, making it resistant to digestion.
FIG. 2.
The proposed cytoplasmic loop between TM1 and TM2 is not accessible to protease cleavage. (A) Untreated (lane 1) and treated (lanes 2 to 5) microsomes from DHBV-infected primary duck hepatocytes were incubated with trypsin (25 μg/ml) for 1 h on ice, boiled with Laemmli buffer, and separated by SDS-PAGE. The envelope proteins were detected by Western blotting with anti-S domain antiserum. Lane 1, untreated microsomes; lane 2, microsomes digested with trypsin; lane 3, microsomes sonicated prior to digestion with trypsin; lane 4, microsomes solubilized with 0.5% Triton X-100 (TX100) and trypsinized; lane 5, microsomes solubilized with RIPA buffer (10 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride) and trypsinized. (B) Linear representation of L protein with its pre-S and S domains showing the epitopes of antibodies used in this study, the locations of the arginine (R) and lysine (K) residues in the loop region between the first two transmembrane domains (boxed), and the 12.9-kDa trypsin product (prod.) resulting from cleavage at these residues in the presence of detergent.
We examined this possibility by using an immunoprecipitation method enabling the differentiation of proteins with epitopes exposed to the outsides of microsomes (in a fraction bound to antibodies in the absence of detergent) from those with hidden epitopes which require solubilization or denaturation of membranes with detergents for detection. Microsomes were prepared from congenitally infected duck hepatocyte cultures (12) and immunoprecipitated with antisera to specific regions in pre-S, the cytoplasmic loop, and the lumenal loop region of the S domain (Fig. 2B, epitope map of antiserum). In a modification of the immunoprecipitation method of Haffar et al. (7), the antibody-bound microsomes were floated in a step gradient (1 ml, 66%; 2 ml, 48%; 1 ml, 10% [wt/vol] sucrose) by centrifugation at 40,000 rpm for 3 h in a SW60 rotor and recovered from the 10 to 48% interface. The microsomes were then solubilized in RIPA buffer, and the antibody-bound proteins (representing the L protein accessible to antibody prior to exposure to the detergent and denaturing agents present in the RIPA buffer) precipitated with protein G-Sepharose. The supernatant, representing proteins inaccessible prior to detergent solubilization of microsomal membranes, was recovered and further immunoprecipitated with the same antiserum. The immune complexes were resuspended in Laemmli buffer and examined by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting with antiserum derived from a different host species.
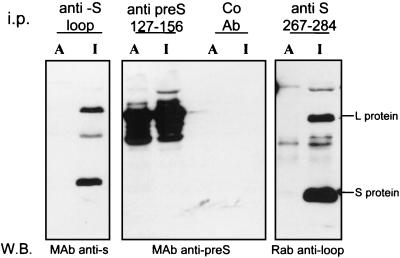
Figure 3 (left panel) shows that an antibody to the loop region between TM1 and TM2 was unable to bind this region of the envelope on microsomes and that both the L and S proteins could be immunoprecipitated with the antiloop antibody following solubilization of the microsomes with RIPA buffer. To confirm the expected orientations of pre-S and as a control for the integrity of the microsomes, we used an antibody to aa 127 to 156 of the pre-S domain and an internal (lumenal) domain of S, aa 267 to 284. The anti-pre-S antibody immunoprecipitated L protein (with and without RIPA buffer), indicative of an expected proportion of pre-S domains which are cytoplasmically exposed (center panel). The monoclonal antibody to the S domain epitope, aa 267 to 284, known to be located on the surfaces of the particles and assumed to be located within the endoplasmic reticulum lumen, resulted in immunoprecipitation of L and S only following solubilization with RIPA buffer, as expected (right panel). These results show that the topologies of the L protein conform to the known mixed inside and outside locations of the pre-S domain and the lumenal location of the region downstream of TM2 in all molecules. Furthermore, solubilisation of the above-described microsomes with Triton X-100, in the absence of denaturing agents or partial membrane inversion upon sonication, permitted antibody binding to the loop, excluding the possibility that the loop is folded in a protease-resistant conformation or complexed cytoplasmically (data not shown). Thus, in contrast with the widely accepted model for the location of the loop between TM1 and -2, this region is not accessible to the cytoplasm but appears to form a membrane-sequestered topology.
FIG. 3.
The loop region between TM1 and TM2 is not cytoplasmically exposed and accessible to specific antibody in microsomal membranes. Microsomes from DHBV-positive primary duck hepatocytes were incubated with antiserum to either the first loop region in the S domain (aa 207 to 226), the pre-S domain (aa 127 to 156), or the second loop region in the S domain (aa 267 to 284) or with a control antibody (Co Ab). The antibody-decorated microsomes were floated in a sucrose step gradient, disrupted with RIPA buffer, and immunoprecipitated (i.p.) with protein G-Sepharose. This fraction represents the proportion of envelope proteins with epitopes accessible to antibody binding (lanes A). The supernatant of this immunoprecipitation, containing detergent-solubilized membranes, was incubated with the same antibody and protein G-Sepharose. This immunoprecipitated fraction represents the envelope proteins with inaccessible or lumenal epitopes (lanes I). The immunoprecipitated envelope proteins from both fractions (A and I) were separated by SDS-PAGE, and the envelope proteins were detected by Western blotting (W.B.) with antisera from different species, as indicated below each panel. MAb, monoclonal antibody; Rab, rabbit.
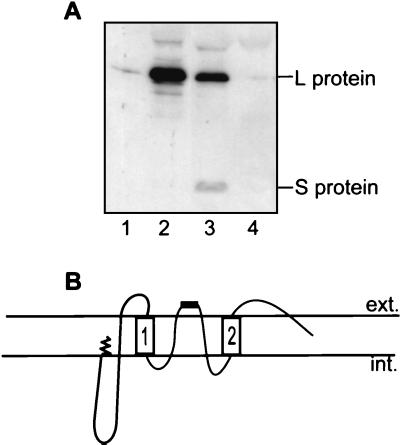
To assess the possibility that the apex of the loop even protrudes beyond the membrane and hence is accessible on the outside surface of the particle, SVPs were immunoprecipitated with the same antibody. SVPs were prepared from duck serum by sucrose gradient purification (8), and their integrity was demonstrated by the preservation of the dual topology of L and the inaccessibility of trypsin sites within the S domain following trypsin digestion. Abilities to precipitate particles with antibodies to several regions of the L protein were compared. G immunoglobulins from a specific antiserum were prebound to protein G-Sepharose and then incubated with 10 μl of SVPs and 2% heat-inactivated fetal bovine serum for 4 h at 4°C. An equal volume of 2× RIPA buffer was added, and the sample was vortexed vigorously and centrifuged in an Eppendorf benchtop centrifuge. The pellet represented protein subunits captured via binding of antibody to external or exposed epitopes on whole SVPs. Figure 4A shows that the antiloop antibody (lane 3) and, as predicted, the anti-pre-S antiserum (lane 2) were able to precipitate particles. The antiloop antibody reacted with the loop domains of both the L and S subunits, albeit very weakly to the latter. By contrast, an antibody to the end of the N terminus of pre-S was not able to precipitate particles (lane 1), consistent with a previous report (6). These data indicate that the antiloop epitope of L is exposed to the particle surface and that in the S subunits this region may be obscured.
FIG. 4.
The loop region between TM1 and TM2 protrudes through the particle surface. (A) SVPs were immunoprecipitated with antibody to aa 10 to 29 (lane 1), aa 127 to 156 (lane 2), or aa 207 to 226 (S loop) (lane 3) and with a control antiserum (lane 4). Antibody prebound to protein G-Sepharose was incubated with SVPs, which were then disrupted with RIPA buffer. The pelleted immune complex, representing protein subunits captured via binding of antibody to external or exposed epitopes on whole SVPs, was separated by SDS-PAGE and detected by Western blotting with anti-pre-S and anti-S domain (aa 267 to 284) monoclonal antibodies. (B) Revised topology of L with the loop region between TM1 and TM2 being shown as membrane embedded and with part of the loop, containing the epitope of aa 207 to 226, shown as a black bar, being exposed to the lumen and ultimately to the particle surface. ext., exterior; int., interior.
Generally, approximately 20 aa are assumed to be required to traverse a membrane. The loop region consists of 49 aa, well within the dimensions for creating two additional transmembrane regions between TM1 and TM2 and thus forming a membrane-embedded loop with the apex protruding to the particle surface as depicted by the model in Fig. 4B. Consequently, some L molecules are predicted to adopt a highly folded, protease-protected topology with five membrane-spanning regions, in which both the pre-S domain resulting from interrupted translocation and the loop region are membrane inserted (Fig. 4B). In contrast, the requirement of denaturing agents to render the S protein susceptible to trypsin (Fig. 2, lane 4) suggests that the small envelope subunits are part of a complexed, protease-resistant structure.
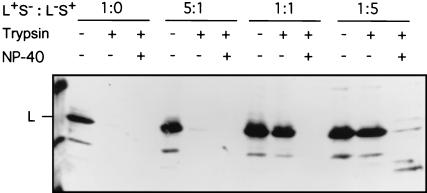
We next examined whether this apparent complexing of the S protein played a role in providing the structure to enable translocation of the pre-S domain. If such a mixed envelope protein complex was functionally significant, the presence of S would be expected to be an essential requirement for the membrane insertion or translocation of pre-S during L protein maturation. To examine this hypothesis, we assessed the protease protection of L chains that were synthesized either alone or in combination with S chains. LMH cells were transfected with a DHBV expression plasmid bearing the gene encoding full-length L protein but lacking the start site for S (plasmid L+S−), in either the absence or presence of increasing ratios of a construct carrying a stop codon in pre-S and therefore able to synthesize only S (plasmid L−S+). Microsomes were prepared and treated with trypsin, and protection of L was assessed by Western blotting. As predicted for an essential role of S, L chains were fully susceptible to trypsin digestion in the absence of S, indicative of a pre-S topology which is fully cytoplasmic (Fig. 5, second lane) (L/S ratio, 1:0). In contrast, coexpression of increasing amounts of the S protein protected L from protease attack. As a control, cells were also transfected with a construct bearing the gene encoding DHBV L with the preprolactin signal sequence at its N terminus (Sig.L), which causes all L molecules to be cotranslationally translocated (5). As predicted, Sig.L was protected, indicative of cotranslational translocation occurring in the absence of significant amounts of S expression and of microsomal integrity (data not shown).
FIG. 5.
Coexpression of S protein is required for protease protection of the L protein. Microsomal membranes were prepared from LMH cells transfected with a plasmid, L+S−, which has an altered start site for S and is defective for S production, or cotransfected with a plasmid, L−S+, containing a stop codon (G1165A) in the pre-S domain and defective in L synthesis, at L/S ratios of 5:1, 1:1, and 1:5. Each sample was divided into three aliquots, which were subjected to digestion with trypsin, with or without NP-40, or left untreated as denoted above each lane, and protease protection was analyzed by Western blotting with an anti-pre-S antiserum.
The assembly of hepadnaviruses involves a compact interaction between the surface proteins, probably through lateral interactions between their transmembrane regions in the budding membrane, a concept of envelope assembly which is becoming increasingly apparent for a number of viruses (3, 4). The major structural contribution played by S in the dense packing of the proteins in the envelope has led to the proposal that pre-S translocation may occur through a channel created from lateral interactions between the amphipathic TM regions in the S domains (6, 14). Such a channel is predicted to allow either complete pre-S translocation for the formation of external pre-S domains or retention of pre-S domains in the membrane-folded intermediate topology. The maintenance of this latter L topology throughout particle maturation and its potential to release pre-S from an apparently metastable conformation (Fig. 1C and 4B) under conditions of low pH or with chaotropic reagents (6; E. V. L. Grgacic and H. Schaller, unpublished results) suggest that this topology serves a specific purpose in the viral interaction with the host cell.
Our demonstration of an essential role of S in pre-S translocation provides the first evidence that S chains may indeed be involved in the formation of a pre-S translocation channel. Furthermore, our evidence for a membrane insertion of the loop, a region which does not exhibit the predominantly hydrophobic residues characteristic of most membrane-spanning domains, raises the possibility that this region participates in the formation of such a channel by lying within and contributing to the hydrophilic lining of a channel. In traversing the membrane twice, the 49-aa-long loop region is at the limit required for achieving this conformation, imposing a considerable constraint on the membrane-folded regions upstream of the stably anchored TM2 element (Fig. 4B). This folding in L (and possibly also in S) might thus be envisaged to create a spring-loaded, metastable structure that participates in hepadnaviral fusion. Although derived from an avian hepadnavirus, it seems reasonable to extrapolate this topological model to HBV, since the corresponding loop region is similarly resistant to protease digestion (17) and is well conserved in length and amino acid sequence between mammalian and avian hepadnaviruses. This conservation includes three cysteine residues which have been shown to be essential for HBV SVP secretion (9).
Acknowledgments
We thank J. Summers and A. Gallina for their gifts of plasmids and D. Anderson for critical reading of the manuscript.
This study was supported in part by a fellowship from the Zentrum für Molekulare Biologie, Heidelberg, Germany, the Deutsche Forschungsgemeinschaft (grant SFB 229), and the Research Fund of the Macfarlane Burnet Centre for Medical Research.
REFERENCES
- 1.Bruss V, Lu X, Thomssen R, Gerlich W H. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 1994;13:2273–2279. doi: 10.1002/j.1460-2075.1994.tb06509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruss V, Vieluf K. Functions of the internal pre-S domain of the large surface protein in hepatitis B virus particle morphogenesis. J Virol. 1995;69:6652–6657. doi: 10.1128/jvi.69.11.6652-6657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garoff H, Hewson R, Opstelten D-J E. Virus maturation by budding. Microbiol Mol Biol Rev. 1998;62:1171–1190. doi: 10.1128/mmbr.62.4.1171-1190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gavilanes F, Gonzales-Ros A, Peterson D. Structure of hepatitis B virus surface antigen: characterization of the lipid components and their association with the viral proteins. J Biol Chem. 1982;257:7770–7777. [PubMed] [Google Scholar]
- 5.Gazina E V, Lin B, Gallina A, Milanesi M, Anderson D A. Intracellular retention of duck hepatitis B virus large surface protein is independent of preS topology. Virology. 1998;242:266–278. doi: 10.1006/viro.1997.9015. [DOI] [PubMed] [Google Scholar]
- 6.Guo J-T, Pugh J C. Topology of the large surface protein of duck hepatitis B virus suggests a mechanism for membrane translocation during particle morphogenesis. J Virol. 1997;71:1107–1114. doi: 10.1128/jvi.71.2.1107-1114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haffar O K, Dowbenko D J, Berman P W. Topogenic analysis of the human immunodeficiency virus type 1 envelope glycoprotein, gp160, in microsomal membranes. J Cell Biol. 1988;107:1677–1687. doi: 10.1083/jcb.107.5.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klingmuller U, Schaller H. Hepadnavirus infection requires interaction between the viral pre-S domain and a specific hepatocellular receptor. J Virol. 1993;67:7414–7422. doi: 10.1128/jvi.67.12.7414-7422.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mangold C M T, Streeck R E. Mutational analysis of the cysteine residues in the hepatitis B virus small envelope protein. J Virol. 1993;67:4588–4597. doi: 10.1128/jvi.67.8.4588-4597.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neurath A R, Kent S B, Strick S B H, Parker K. Identification and chemical synthesis of a host cell receptor binding site on hepatitis B virus. Cell. 1986;46:429. doi: 10.1016/0092-8674(86)90663-x. [DOI] [PubMed] [Google Scholar]
- 11.Ostapchuk P, Hearing P, Ganem D. A dramatic shift in the transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO J. 1994;13:1048–1057. doi: 10.1002/j.1460-2075.1994.tb06353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prange R, Streeck R E. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 1995;14:247–256. doi: 10.1002/j.1460-2075.1995.tb06998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothman K, Schnölzer M, Radziwill G, Hildt E, Mölling K, Schaller H. Host cell-virus cross talk: phosphorylation of a hepatitis B virus envelope protein mediates intracellular signaling. J Virol. 1998;72:10138–10147. doi: 10.1128/jvi.72.12.10138-10147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stirk H J, Thornton J M, Howard C R. A topological model for hepatitis B surface antigen. Intervirology. 1992;33:148–158. doi: 10.1159/000150244. [DOI] [PubMed] [Google Scholar]
- 15.Summers J, Smith P M, Huang M, Yu M. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J Virol. 1991;65:1310–1317. doi: 10.1128/jvi.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swameye I, Schaller H. Dual topology of the large envelope protein of duck hepatitis B virus: determinants preventing pre-S translocation and glycosylation. J Virol. 1997;71:9434–9441. doi: 10.1128/jvi.71.12.9434-9441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wunderlich G, Bruss V. Characterization of early hepatitis B virus surface protein oligomers. Arch Virol. 1996;141:1191–1205. doi: 10.1007/BF01718824. [DOI] [PubMed] [Google Scholar]