Abstract
Type B leukemogenic virus (TBLV) induces rapidly appearing T-cell leukemias. TBLV insertions near the c-myc gene were detectable in 2 of 30 tumors tested, whereas 80% of the tumors showed c-myc overexpression. TBLV insertions on chromosome 15 (including a newly identified locus, Pad7) may cause c-myc overexpression by cis-acting effects at a distance.
Mouse mammary tumor virus (MMTV) is a latently oncogenic B-type retrovirus that primarily causes murine mammary cancers but also induces T-cell lymphomas at a lower frequency (6, 7, 29). Like other slowly oncogenic retroviruses, MMTV exerts its tumorigenic effects by the activation of cellular proto-oncogenes in the vicinity of integrated proviruses. Using the “proviral tagging” approach, nine common sites of integration, namely, Wnt1, Fgf3, Notch4, Wnt3, Cyp19, Fgf4, Fgf8, Int6, and Wnt10b, have been identified in MMTV-induced mammary tumors (22, 24, 25, 35). Like mammary tumors, MMTV-induced T-cell tumors have acquired new copies of MMTV proviruses in chromosomal DNA in addition to endogenous MMTVs found in the germ line (7, 29). These acquired proviruses have a large deletion (350 to 500 bp) of negative regulatory elements (14) in the long terminal repeat (LTR) U3 region compared to mammotropic MMTVs, and often these deletions are accompanied by duplications of the sequences flanking the deletion (5, 23, 28, 38).
Type B leukemogenic virus (TBLV) is a replication-competent thymotropic retrovirus whose genome is >98% identical to that of milk-borne MMTV (3, 6). Unlike MMTV, which induces mammary tumors after a long latent period (6 to 9 months), TBLV induces a high incidence of T-cell lymphomas in mice after a very short latent period (42 to 55 days) (4). Like changes in other MMTVs that induce lymphomas, the alterations in the U3 region of the TBLV LTR include a deletion of 440 nucleotides and a 122-nucleotide substitution, consisting of sequences flanking the region (5). When linked to a c-myc transgene, the TBLV LTR has been shown to induce CD4+ CD8+ murine T-cell lymphomas, similar to those produced by TBLV injection (31). These results suggest that overexpression of c-myc RNA from the TBLV LTR is sufficient to initiate T-cell disease in transgenic mice. Additional experiments indicate that infection of TBLV LTR-myc-transgenic mice with MuLV accelerates leukemogenesis. Thus, activation of c-myc leads to initiation of lymphomas, but other genes are required for progression (10).
Currently, only one “common” region of proviral integration (Tblvi1) has been identified in 20% of 55 primary TBLV-induced tumors; this site maps to the mouse X chromosome and activates one or more hitherto-unidentified genes (30). To identify additional common integration sites, 30 TBLV-induced primary tumors were generated by intrathymic inoculation of newborn mice with concentrated virus as described previously (27). Tumors arose after 2 to 3 months in approximately 40 to 50% of the mice, within the thymus, spleen, and axial or mesenteric lymph nodes and often in more than one of these tissues. Southern blot analysis of tumor cells revealed T-cell receptor β-chain rearrangements in the T8, T16, and T17 tumors compared to the germ line configuration of the locus (27), consistent with the T-cell phenotype of the lymphomas (data not shown).
The c-myc locus is rearranged in TBLV-induced primary tumors.
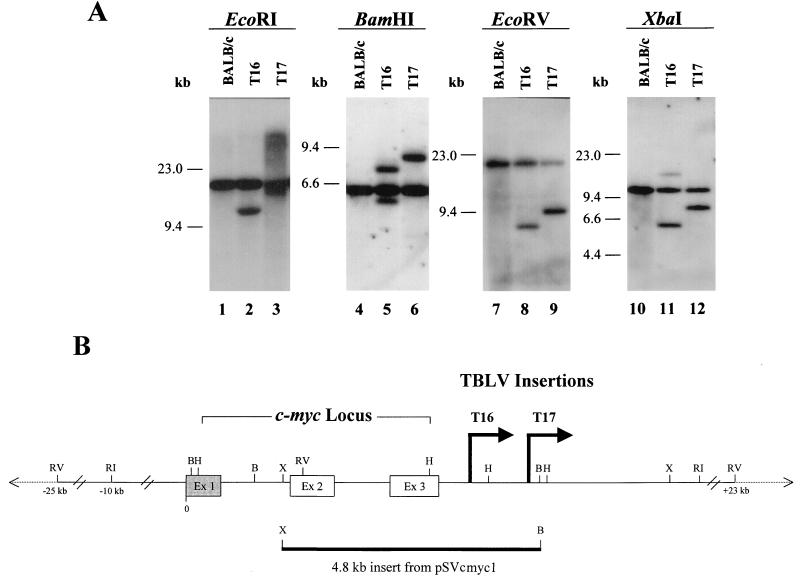
Proviral insertions near the c-myc proto-oncogene are relatively common among lymphoid tumors induced by retroviral infection. Therefore, independently derived TBLV-induced tumors were screened for virally induced rearrangements near c-myc. Of the 30 tumors screened, 2 (7%), namely, T16 and T17, showed novel bands hybridizing to the pSVcmyc1 probe (19) by Southern blotting (Fig. 1A). PCR and Southern blotting analysis indicated that the proviral integrations in the T16 and T17 tumors are ca. 0.5 and 1.9 kb, respectively, downstream and in the same transcriptional orientation as the c-myc gene (Fig. 1B).
FIG. 1.
Analysis of proviral insertions near the c-myc locus in TBLV-induced T-cell tumors. (A) Southern blot analysis of TBLV-induced tumors. Tumors were induced by intrathymic inoculation of concentrated TBLV from the 485-10 cell line (6) into BALB/c or C57BL/6 mice. Tumor DNAs (15 μg) were digested with EcoRI, BamHI, EcoRV, or XbaI prior to Southern blotting and hybridization with the pSVcmyc1 probe (19); this probe contains genomic DNA spanning c-myc exons 2 and 3. DNA extractions, Southern blotting, and hybridizations were performed as described previously (27). Genomic tumor cell DNA derived from different organs within the same animal showed identical patterns of additional TBLV integrations, indicating a common origin (data not shown). No rearrangements were detected in DNA from other TBLV-induced tumors spanning approximately 48 kb of the c-myc locus (25 kb upstream and 23 kb downstream of the locus) (data not shown). The positions of molecular size markers are shown. (B) Diagram of TBLV insertions in the c-myc locus. The c-myc gene is transcribed (5′ to 3′) from left to right, in the same transcriptional orientation as the proviruses in the T16 and T17 tumors. The location of TBLV insertions was confirmed using one primer from the c-myc gene [c-myc 695(+) 5′ GGA CTG TAA GCT TCA GCC ATA 3′] and another from the TBLV LTR [TBLV LTR 546(−) 5′ TTG GGA ACC GCA CCT GTT CTT 3′]. PCR was performed using the Expand long-template PCR system (Boehringer Mannheim). Sequencing was performed as described previously (36). The positions of some restriction enzyme sites are shown: RV, EcoRV; B, BamHI; X, XbaI; H, HindIII; RI, EcoRI. The two coding exons are shown as white boxes, and the 5′ noncoding exon is shown by a shaded box. The location of the pSVcmyc1 probe, used for Southern hybridizations, is shown.
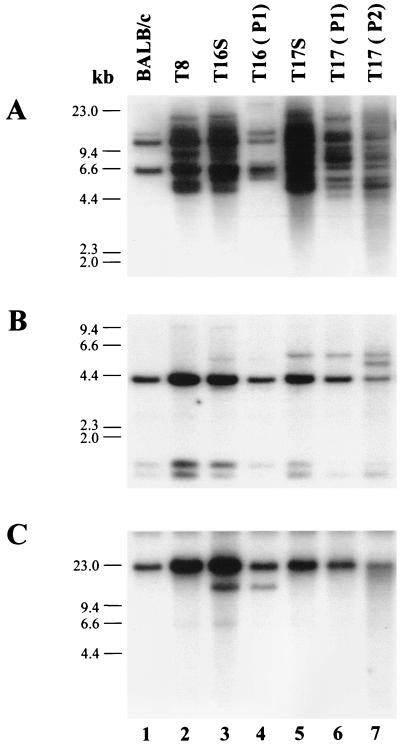
To determine if the tumors induced by TBLV were clonal, DNA was extracted from the T16 and T17 tumors after intraperitoneal passage in weanling BALB/c mice. As detected by Southern blotting, both the T16 and T17 tumors showed a change in the proviral integration pattern during passage, and the T16 integration pattern simplified during passage (Fig. 2A). These results suggested that the original tumor was a heterogeneous population, although a dominant cell type could be detected as assessed by T-cell receptor β-chain rearrangements (see above).
FIG. 2.
Southern blotting of DNA extracted from the T16 and T17 tumors after in vivo passage in mice. (A) Altered TBLV integration pattern after tumor passage. DNA (10 μg) isolated from normal BALB/c mouse liver or tumor DNAs was digested with HindIII and hybridized to an MMTV envelope probe (8). Primary tumor DNA was used in lanes 2, 3, and 5. DNA isolated after one passage in animals (P1) was used in lanes 4 and 6, whereas DNA isolated after two passages in animals (P2) was used in lane 7. The positions of molecular size markers (HindIII-digested lambda DNA) are shown. (B) TBLV integrations near c-myc are selected during in vivo passage in animals. Tumor DNA (10 μg) from primary or passaged tumors was digested with HindIII, subjected to Southern blotting, and hybridized to the pSVcmyc1 probe. (C) TBLV integrations near c-myc are selected during in vivo passage in animals. Tumor DNAs were digested with EcoRI and subjected to Southern blotting and hybridization as for panel B.
To determine if tumor cell growth selects for TBLV integrations near c-myc, Southern blotting was used to monitor viral integration sites after passage of the T16 and T17 tumors in vivo (Fig. 2B and C). As expected, data obtained after one or two tumor passages in mice showed retention of the TBLV provirus near c-myc in the primary tumor. In addition, we observed a second TBLV integration close to c-myc after two passages of the T17 tumor in mice (Fig. 2B, lane 7). Thus, it appears that TBLV insertions near c-myc are selected during tumor passage in vivo.
Activation of c-myc expression in TBLV-induced tumors.
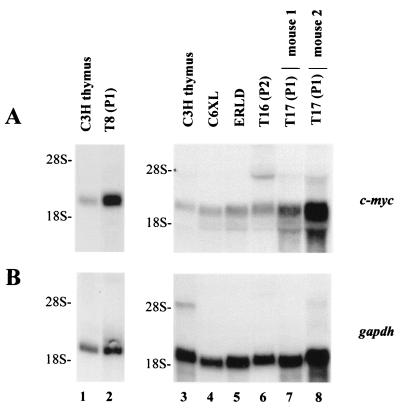
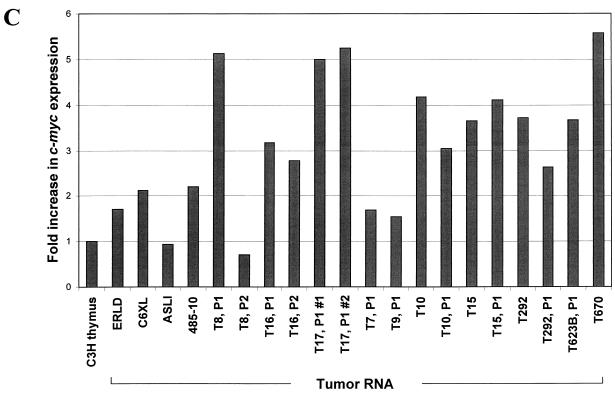
To determine if c-myc was overexpressed in TBLV-induced tumors, we performed Northern analysis on total RNA extracted from a number of TBLV-induced tumors. The T16 and T17 tumors showed a three- to fivefold overexpression of c-myc RNA relative to normal thymus RNA after normalization for RNA loading on the gel transfers by hybridization to a probe for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (9) (Fig. 3A and B). No novel c-myc transcripts were detected. With the exception of the T7 and T9 tumors, similar results were obtained with other TBLV-induced tumors (2- to 5.6-fold c-myc overexpression). Three T-cell lymphomas that were not induced by TBLV (ERLD, C6XL, and ASL1) had c-myc levels that were 0.9 to 2.1 times the level in normal C3H mouse thymus (Fig. 3C). Thus, the level of c-myc expression was elevated in most TBLV-induced tumors tested, although many tumors had no TBLV integration near c-myc detectable by Southern blotting (data not shown).
FIG. 3.
Elevated c-myc expression in TBLV-induced T-cell lymphomas. Total RNA (20 μg) was analyzed using 1.0% agarose gels containing 2.2 M formaldehyde as described previously (27). The positions of the 18S and 28S rRNAs are shown. RNA was extracted as described previously (37). C3H mouse normal thymus RNA was used in lanes 1 and 3, whereas RNA extracted after one passage of the T8 tumor was used in lane 2. RNAs extracted from TBLV-induced tumors after one or two intraperitoneal passages (P1 or P2) in mice are shown in lanes 6 to 8. RNAs from the T17 tumor were obtained from two separately injected mice. Lanes 1 and 2 were derived from a different gel than lanes 3 to 8. (A) Northern blot hybridized to the pSVcmyc1 probe (19). (B) Northern blot hybridized to a GAPDH probe. The blots used for panel A were stripped prior to hybridization with the GAPDH probe. (C) Levels of c-myc RNA in TBLV-induced T-cell lymphomas. Total RNAs from normal thymus or T-cell tumors were analyzed on Northern blots and hybridized to c-myc and GAPDH probes. Levels of c-myc RNA after normalization to GAPDH levels relative to normal C3H mouse thymus RNA (assigned a value of 1) are shown. In some cases, intact RNA was not available from primary tumors. The T-cell lines ERLD, C6XL, and ASL-1 (2, 7, 27) were derived from spontaneous or X-ray-induced tumors appearing in animals that were not infected with TBLV. RNA from passage 1 of the T17 tumor was prepared from two different animals.
Cloning and chromosomal mapping of the Pad7 locus.
Because of the low number of TBLV-induced tumors with detectable c-myc integration, we attempted to identify additional common TBLV integration sites. After screening of a partial lambda phage library from the T8 tumor (containing three TBLV insertions), we obtained an integrated TBLV provirus and cellular flanking sequences. The locus identified by this probe was designated Pad7. Southern analysis on other tumor DNAs using three different restriction enzymes suggested that the Pad7 locus is not a common integration site in TBLV-induced tumors.
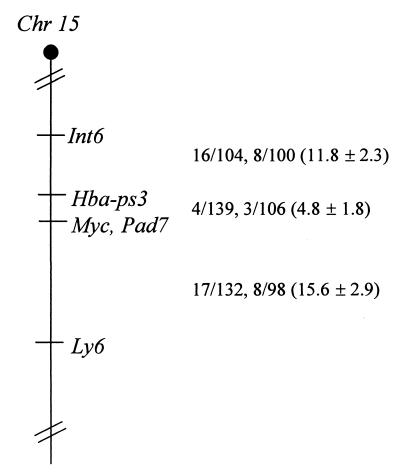
To determine the chromosomal map position of the Pad7 locus and its possible colocalization with known cellular oncogenes, we tested the progeny of two sets of multilocus crosses: (NFS/N or C58/J × Mus musculus musculus) × M. m. musculus (18) and (NFS/N × Mus spretus) × M. spretus or C58/J (1) by Southern blotting and hybridization with the Pad7 probe. No recombination was observed between Myc and Pad7 in 198 mice, indicating that, at the upper limit of the 95% confidence level, Myc and Pad7 are within 1.5 centimorgans of each other (Fig. 4).
FIG. 4.
Genetic map location of Pad7 on mouse chromosome 15. The centromere is toward the top. To obtain the Pad7 probe, BglII-digested DNA (enriched for 6- to 10-kb fragments) was ligated to the vector arms of bacteriophage EMBL3 and packaged using Gigapack II packaging extract (Stratagene). The primary library was screened with a radiolabeled MMTV LTR probe (8). The Pad7 probe was subcloned from the 3′ flanking DNA of a lambda clone containing an acquired TBLV provirus. The Pad7 locus was mapped using Southern analysis of chromosomal DNA from the progeny of two different backcross panels as described previously (32). Recombination fractions for adjacent loci are shown to the right of the map. The first fraction given is from the M. m. musculus cross, whereas the second fraction is from the M. spretus cross. Recombination distances (± standard errors) are shown. For DNAs from the M. m. musculus cross, we detected a 6.0-kb band for M. m. musculus and a 5.5-kb band for NFS/N or C58/J mice with the enzyme ApaI and the Pad7 probe. For DNAs from the M. spretus cross, we detected a 6.7-kb band for M. spretus and a 19.3-kb band for NFS/N or C58/J mice with ScaI digestion and the Pad7 probe. Progeny of these crosses have been typed for over 1,200 markers distributed over the 19 autosomes and the X chromosome. A 1.4-kb cDNA probe for Pvt1 and a 560-bp exon 1 probe for Myc were provided by K. Huppi (National Cancer Institute, Bethesda, Md.) for mapping experiments. Recombination distance was determined according to the method of Green (11), and the loci were ordered by minimizing the number of recombinants using the program LOCUS (C. E. Buckler, National Institute of Allergy and Infectious Diseases, Bethesda, Md.).
DNAs digested with 13 rare-cutting enzymes were analyzed by pulsed-field gel electrophoresis (15) to identify fragments containing Pad7, Myc, and Pvt1. The Pvt1 locus also is a common integration site for murine leukemia viruses on mouse chromosome 15 and is located approximately 270 kb from c-myc (17, 34). No physical linkage could be established between Pad7 and either Myc or Pvt1 (data not shown).
As in leukemias induced by other retroviruses, c-myc appears to be activated two- to sixfold by an enhancer insertion mechanism in most TBLV-induced T-cell lymphomas, and this conclusion is supported by our failure to detect new c-myc transcripts that are initiated within the TBLV LTR. It has been proposed that murine leukemia virus proviruses integrated in the Pvt1 or Mlvi4 locus activate c-myc expression by long-range (up to 300 kb) cis effects (20, 21, 33). Similar long-range effects on c-myc expression also have been observed in mouse or human tumors carrying chromosomal translocations that juxtapose the c-myc gene to other cellular gene enhancers (12, 13, 16, 26, 39). Thus, one explanation for c-myc overexpression in the T8 tumor (origin of the Pad7 locus) is that the TBLV proviral enhancer exerts its effect over a very long distance (>300 kb). Long-range effects of TBLV integrations could be due to proviral insertions that affect a locus control region; such regions may coordinate control of multiple genes, including those near Pad7.
Acknowledgments
We acknowledge members of the Dudley lab and Susan Ross for helpful comments on the manuscript.
This work was supported by grant CA34780 from the National Institutes of Health (J.P.D.) and NIAID (C.A.K.).
REFERENCES
- 1.Adamson M C, Silver J, Kozak C A. The mouse homolog of the gibbon ape leukemia virus receptor: genetic mapping and a possible receptor function in rodents. Virology. 1991;183:778–781. doi: 10.1016/0042-6822(91)91010-e. [DOI] [PubMed] [Google Scholar]
- 2.Allison J P, McIntyre B W, Bloch D. Tumor-specific antigen of murine T-lymphoma defined with monoclonal antibody. J Immunol. 1982;129:2293–2300. [PubMed] [Google Scholar]
- 3.Ball J K, Arthur L O, Dekaban G A. The involvement of a type-B retrovirus in the induction of thymic lymphomas. Virology. 1985;140:159–172. doi: 10.1016/0042-6822(85)90455-6. [DOI] [PubMed] [Google Scholar]
- 4.Ball J K, Dekaban G A. Characterization of early molecular biological events associated with thymic lymphoma induction following infection with a thymotropic type-B retrovirus. Virology. 1987;161:357–365. doi: 10.1016/0042-6822(87)90128-0. [DOI] [PubMed] [Google Scholar]
- 5.Ball J K, Diggelmann H, Dekaban G A, Grossi G F, Semmler R, Waight P A, Fletcher R F. Alterations in the U3 region of the long terminal repeat of an infectious thymotropic type B retrovirus. J Virol. 1988;62:2985–2993. doi: 10.1128/jvi.62.8.2985-2993.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dekaban G A, Ball J K. Integration of type B retroviral DNA in virus-induced primary murine thymic lymphomas. J Virol. 1984;52:784–792. doi: 10.1128/jvi.52.3.784-792.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dudley J, Risser R. Amplification and novel locations of endogenous mouse mammary tumor virus genomes in mouse T-cell lymphomas. J Virol. 1984;49:92–101. doi: 10.1128/jvi.49.1.92-101.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dudley J P, Varmus H E. Purification and translation of murine mammary tumor virus mRNA's. J Virol. 1981;39:207–218. doi: 10.1128/jvi.39.1.207-218.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fort P, Marty L, Piechaczyk M, el Sabrouty S, Dani C, Jeanteur P, Blanchard J M. Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 1985;13:1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Girard L, Hanna Z, Beaulieu N, Hoemann C D, Simard C, Kozak C A, Jolicoeur P. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTV/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 1996;10:1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- 11.Green E L. Genetics and probability in animal breeding experiments. New York, N.Y: MacMillan Publishing Co., Inc.; 1981. [Google Scholar]
- 12.Haluska F G, Tsujimoto Y, Croce C M. The t(8;14) breakpoint of the EW 36 undifferentiated lymphoma cell line lies 5′ of MYC in a region prone to involvement in endemic Burkitt's lymphomas. Nucleic Acids Res. 1988;16:2077–2085. doi: 10.1093/nar/16.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henglein B, Synovzik H, Groitl P, Bornkamm G W, Hartl P, Lipp M. Three breakpoints of variant t(2;8) translocations in Burkitt's lymphoma cells fall within a region 140 kilobases distal from c-myc. Mol Cell Biol. 1989;9:2105–2113. doi: 10.1128/mcb.9.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu C L, Fabritius C, Dudley J. Mouse mammary tumor virus proviruses in T-cell lymphomas lack a negative regulatory element in the long terminal repeat. J Virol. 1988;62:4644–4652. doi: 10.1128/jvi.62.12.4644-4652.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang X, Villeneuve L, Turmel C, Kozak C A, Jolicoeur P. The Myb and Ahi-1 genes are physically very closely linked on mouse chromosome 10. Mamm Genome. 1994;5:142–148. doi: 10.1007/BF00352344. [DOI] [PubMed] [Google Scholar]
- 16.Joos S, Haluska F G, Falk M H, Henglein B, Hameister H, Croce C M, Bornkamm G W. Mapping chromosomal breakpoints of Burkitt's t(8;14) translocations far upstream of c-myc. Cancer Res. 1992;52:6547–6552. [PubMed] [Google Scholar]
- 17.Koehne C F, Lazo P A, Alves K, Lee J S, Tsichlis P N, O'Donnell P V. The Mlvi-1 locus involved in the induction of rat T-cell lymphomas and the pvt-1/Mis-1 locus are identical. J Virol. 1989;63:2366–2369. doi: 10.1128/jvi.63.5.2366-2369.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak C A, Peyser M, Krall M, Mariano T M, Kumar C S, Pestka S, Mock B A. Molecular genetic markers spanning mouse chromosome 10. Genomics. 1990;8:519–524. doi: 10.1016/0888-7543(90)90039-w. [DOI] [PubMed] [Google Scholar]
- 19.Land H, Parada L F, Weinberg R A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 20.Lazo P A, Lee J S, Tsichlis P N. Long-distance activation of the myc protooncogene by provirus insertion in Mlvi-1 or Mlvi-4 in rat T-cell lymphomas. Proc Natl Acad Sci USA. 1990;87:170–173. doi: 10.1073/pnas.87.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazo P A, Tsichlis P N. Biology and pathogenesis of retroviruses. Semin Oncol. 1990;17:269–294. [PubMed] [Google Scholar]
- 22.Lee F S, Lane T F, Kuo A, Shackleford G M, Leder P. Insertional mutagenesis identifies a member of the Wnt gene family as a candidate oncogene in the mammary epithelium of int-2/Fgf-3 transgenic mice. Proc Natl Acad Sci USA. 1995;92:2268–2272. doi: 10.1073/pnas.92.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee W T, Prakash O, Klein D, Sarkar N H. Structural alterations in the long terminal repeat of an acquired mouse mammary tumor virus provirus in a T-cell leukemia of DBA/2 mice. Virology. 1987;159:39–48. doi: 10.1016/0042-6822(87)90345-x. [DOI] [PubMed] [Google Scholar]
- 24.MacArthur C A, Shankar D B, Shackleford G M. Fgf-8, activated by proviral insertion, cooperates with the Wnt-1 transgene in murine mammary tumorigenesis. J Virol. 1995;69:2501–2507. doi: 10.1128/jvi.69.4.2501-2507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchetti A, Buttitta F, Miyazaki S, Gallahan D, Smith G H, Callahan R. Int-6, a highly conserved, widely expressed gene, is mutated by mouse mammary tumor virus in mammary preneoplasia. J Virol. 1995;69:1932–1938. doi: 10.1128/jvi.69.3.1932-1938.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mengle-Gaw L, Rabbitts T H. A human chromosome 8 region with abnormalities in B cell, HTLV-I+ T cell and c-myc amplified tumours. EMBO J. 1987;6:1959–1965. doi: 10.1002/j.1460-2075.1987.tb02458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyers S, Gottlieb P D, Dudley J P. Lymphomas with acquired mouse mammary tumor virus proviruses resemble distinct prethymic and intrathymic phenotypes defined in vivo. J Immunol. 1989;142:3342–3350. [PubMed] [Google Scholar]
- 28.Michalides R, Wagenaar E. Site-specific rearrangements in the long terminal repeat of extra mouse mammary tumor proviruses in murine T-cell leukemias. Virology. 1986;154:76–84. doi: 10.1016/0042-6822(86)90431-9. [DOI] [PubMed] [Google Scholar]
- 29.Michalides R, Wagenaar E, Hilkens J, Hilgers J, Groner B, Hynes N E. Acquisition of proviral DNA of mouse mammary tumor virus in thymic leukemia cells from GR mice. J Virol. 1982;43:819–829. doi: 10.1128/jvi.43.3.819-829.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mueller R E, Baggio L, Kozak C A, Ball J K. A common integration locus in type B retrovirus-induced thymic lymphomas. Virology. 1992;191:628–637. doi: 10.1016/0042-6822(92)90238-k. [DOI] [PubMed] [Google Scholar]
- 31.Paquette Y, Doyon L, Laperrière A, Hanna Z, Ball J, Sekaly R P, Jolicoeur P. A viral long terminal repeat expressed in CD4+ CD8+ precursors is downregulated in mature peripheral CD4− CD8+ or CD4+ CD8− T cells. Mol Cell Biol. 1992;12:3522–3530. doi: 10.1128/mcb.12.8.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajan L, Kozak C A, Dudley J P. Chromosomal localization of acquired MMTV proviral integration sites in T-cell lymphomas. Mamm Genome. 1998;9:84–85. doi: 10.1007/s003359900687. [DOI] [PubMed] [Google Scholar]
- 33.Tsichlis P N, Lee J S, Bear S E, Lazo P A, Patriotis C, Gustafson E, Shinton S, Jenkins N A, Copeland N G, Huebner K, Croce C, Levan G, Hanson C. Activation of multiple genes by provirus integration in the Mlvi-4 locus in T-cell lymphomas induced by Moloney murine leukemia virus. J Virol. 1990;64:2236–2244. doi: 10.1128/jvi.64.5.2236-2244.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsichlis P N, Shepherd B M, Bear S E. Activation of the Mlvi-1/mis1/pvt-1 locus in Moloney murine leukemia virus-induced T-cell lymphomas. Proc Natl Acad Sci USA. 1989;86:5487–5491. doi: 10.1073/pnas.86.14.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Leeuwen F, Nusse R. Oncogene activation and oncogene cooperation in MMTV-induced mouse mammary cancer. Semin Cancer Biol. 1995;6:127–133. doi: 10.1006/scbi.1995.0018. [DOI] [PubMed] [Google Scholar]
- 36.Wrona T J, Lozano M, Binhazim A A, Dudley J P. Mutational and functional analysis of the C-terminal region of the C3H mouse mammary tumor virus superantigen. J Virol. 1998;72:4746–4755. doi: 10.1128/jvi.72.6.4746-4755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu L, Wrona T J, Dudley J P. Exogenous mouse mammary tumor virus (MMTV) infection induces endogenous MMTV sag expression. Virology. 1996;215:113–123. doi: 10.1006/viro.1996.0014. [DOI] [PubMed] [Google Scholar]
- 38.Yanagawa S, Murakami A, Tanaka H. Extra mouse mammary tumor proviruses in DBA/2 mouse lymphomas acquire a selective advantage in lymphocytes by alteration in the U3 region of the long terminal repeat. J Virol. 1990;64:2474–2483. doi: 10.1128/jvi.64.6.2474-2483.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeidler R, Joos S, Delecluse H J, Klobeck G, Vuillaume M, Lenoir G M, Bornkamm G W, Lipp M. Breakpoints of Burkitt's lymphoma t(8;22) translocations map within a distance of 300 kb downstream of MYC. Genes Chromosomes Cancer. 1994;9:282–287. doi: 10.1002/gcc.2870090408. [DOI] [PubMed] [Google Scholar]