Abstract
Six donor-recipient clusters of hepatitis C virus (HCV)-infected individuals were studied. For five clusters the period of infection of the donor could be estimated, and for all six clusters the time of infection of the recipients from the donor via blood transfusion was also precisely known. Detailed phylogenetic analyses were carried out to investigate the genomic evolution of the viral quasispecies within infected individuals in each cluster. The molecular clock analysis showed that HCV quasispecies within a patient are evolving at the same rate and that donors that have been infected for longer time tend to have a lower evolutionary rate. Phylogenetic analysis based on the split decomposition method revealed different evolutionary patterns in different donor-recipient clusters. Reactivity of antibody against the first hypervariable region (HVR1) of HCV in donor and recipient sera was evaluated and correlated to the calculated evolutionary rate. Results indicate that anti-HVR1 reactivity was related more to the overall level of humoral immune response of the host than to the HVR1 sequence itself, suggesting that the particular sequence of the HVR1 peptides is not the determinant of reactivity. Moreover, no correlation was found between the evolutionary rate or the heterogeneity of the viral quasispecies in the patients and the strength of the immune response to HVR1 epitopes. Rather, the results seem to imply that genetic drift is less dependent on immune pressure than on the rate of evolution and that the genetic drift of HCV is independent of the host immune pressure.
Hepatitis C virus (HCV) causes persistent infection in a majority of infected individuals. Among the possible mechanisms explaining persistence are the relatively poor immunogenicity of the virus, particularly of the envelope glycoproteins; the low level of viremia outside the preseroconversion period; and the considerable variability of the viral genome, leading to substantial changes in the viral epitopes over time in the same individual (22, 36). One of the main contributors to these genomic changes is the hypervariable region 1 (HVR1) located at the N terminus of the major envelope glycoprotein E2. Mutations in HVR1, which is critical for virus interaction with target cells (34, 48), produce escape mutants, a likely contributing factor to viral persistence (24, 38, 46).
The host of HCV, primate or human, seems in most cases unable to generate an effective immune response, whether humoral, as levels of antibody to the E2 protein or HVR1 are typically low or undetectable, or cellular, as no evidence of a specific T-cell response to E2 epitopes has been provided (3, 16, 17, 37). This feature was supported by data collected in vivo indicating that neither natural defenses nor passive immunotherapy was able to prevent reinfection of a chronically infected patient or animal with the same or related viruses (7, 30).
Some studies of HCV evolution over time, in the same infected individual or in different individuals infected with the same viral quasispecies, have been reported (1, 43, 46). Conflicting findings of the relative contribution of the virus itself or of the selecting pressure exercised by the host immune system were provided (18, 19, 47, 49). To further examine this question, we studied the viral population as well as the humoral immune response to HVR1 of clusters of HCV-infected blood donors and recipients of blood components from these donors. Evolutionary rates and phylogeny of donor-recipient pairs were determined and compared to the magnitude and the specificity of the anti-HVR1 response.
MATERIALS AND METHODS
Donors and patients.
Six donor-recipient clusters totaling 21 HCV-infected individuals were included in the study. Clusters were selected on the basis of blood components from each donor being the origin of HCV infection of at least two recipients. All six donors were identified as anti-HCV and HCV RNA positive between October 1991 and May 1992. They ranged in age between 34 and 45 years at the time of HCV infection diagnosis; three were males and three were females. Two had moderate elevation of alanine aminotransferase (ALT) (donors of clusters 2 and 5 [c2.d and c5.d]) with stage 3 and 0 fibrosis, respectively. The other four donors neither were tested for ALT nor had liver biopsy. Donor c1d had a level of viremia estimated to 104 genome equivalents/ml. Fifteen recipients of previous donations from these donors were identified and tested for antibody to HCV at the end of 1995 within the scope of the National Blood Service look-back program. There were seven males and eight females, ranging in age between 13 and 83 (median, 64) years. Of the 10 recipients tested for ALT, only two recipients 2 of clusters 1 and 5 [c1.r2 and c5.r2] had elevated levels (91 and 134 IU, respectively). Seven of nine patients had no clinical evidence of liver disease. Recipients c1.r3 and c1.r4 had liver histology staged at 0 and 1 for fibrosis. Estimation of HCV viremia was not part of the study protocol. The date of infection was precisely known for only two donors of clusters 4 and 6 who received a presumably infected transfusion in 1983 and 1971, respectively, and had no other risk factors. Donors of clusters 2, 3, and 5 were former intravenous drug abusers. Donors of clusters 2, 3, and 5 were active in 1970, 1972 to 1974, and 1975, respectively. The donor of cluster 1 did not have any identifiable risk factor, and his date of infection remained unknown.
All 15 recipients were diagnosed as HCV infected on the basis of seropositivity, confirmation by recombinant immunoblot assay version 3 (RIBA-3) in 13 and the presence of HCV RNA in 2. The presumably infectious transfusions were received 11.2 to 4.4 (years.months) prior to diagnosis (median, 7.5) when the studied sample was obtained. Recipients (nine males and six females) ranged in age between 13 and 82 (median, 59) years. Ten of these 15 recipients were found to be HCV RNA positive, and the HCV genome was amplified and sequenced. The other five recipients were HCV seropositive but RNA negative or, in one of them, the E1/E2 region could not be amplified. Serum samples of these five patients were used for serological analysis of HVR peptide reactivity.
HCV RNA studies.
HCV viremia was determined by reverse transcription-PCR, using nested primers in the 5′ noncoding region, as previously described (31). To determine the quasispecies diversity, the E1/E2 region was amplified and sequenced as described elsewhere (20). Nine to 15 cDNA clones were sequenced in order to identify mutations and to estimate the diversity of variants in each specimen's quasispecies. For each sample, a 258-nucleotide (nt)-long sequence encompassing the 3′ end of E1 and HVR1 (nt 1362 to 1620) (5) was obtained. The HCV genotype was determined by phylogenetic analysis using the whole sequence and reference sequences from HCV genotypes 1 to 5.
Phylogenetic analysis.
Forty-six DNA sequences from donors and recipients belonging to the different clusters were aligned with 10 HCV sequences from the EMBL database, representing HCV subtypes 1 to 5, with the Clustal software (11). The HCV sequences used as reference (with accession numbers indicated in brackets) were HCV1a [M62321], hcg2.1b [D10074], and HC.G9.1c [D14853] for subtype 1; hcj6.2a [D00944], hcj8.2b [D10988], and hcj5.2c [D10076] for subtype 2; hem265.3a [D14311] and HCV.3b [D26556] for subtype 3; ED43 [Y11604] for subtype 4a; and EUH1480.5a for subtype 5. The last sequence was kindly provided by P. Simmonds, Edinburgh, United Kingdom. In total, 56 HCV strains were aligned using a 258-nt sequence corresponding to the E1/E2 region as described above.
Phylogeny construction and evaluation were done using PHYLIP 3.572 (10). We used two different methods, neighbor joining (NJ) and maximum likelihood (ML), employing an empirical transition/transversion bias of 1.61 for both. The bias was estimated with the ML method implemented in PUZZLE (42), using the HKY model (11). The NJ tree was evaluated using 1,000 bootstrap samples (9); P values for the branches of the ML tree were also obtained.
Molecular clock analysis and evolutionary rate.
To investigate the dynamic of the viral quasispecies within each infected individual of the six donor-recipient clusters, separate alignments were obtained, each joining only the clones sequenced from the same patient. The HKY model was chosen to estimate nucleotide distance matrices and empirical transition transversion ratios for the different data sets with the program PUZZLE (42), as described above. To determine whether the viral quasispecies evolved at constant rate within a patient, we tested the molecular clock hypothesis for every data set with the likelihood ratio test (LRT), also implemented in PUZZLE. To perform the test, a phylogenetic tree is obtained, and then the branch lengths and likelihood of the tree are calculated with or without the assumption of constant evolutionary rate along the branches. If the likelihood of the tree assuming the clock (simpler model) is not significantly lower than the likelihood of the nonclock tree (more complex model) in a χ2 test with n−2 degrees of freedom, where n is the number of taxa, the molecular clock cannot be rejected (14).
Since all of the data sets showed a clock-like behavior (see Results), it was possible to calculate the evolutionary rate of HCV in most patients. In fact, for all infected individuals except c1.d, the time of infection was known with acceptable precision. Under the clock hypothesis, taxa sharing the same ancestor have the same branch length to their common ancestor. Thus for a particular data set, the evolutionary rate can be estimated by dividing one of the branch lengths connecting an HCV strain to the node representing the ancestor in common with all the other strains in the tree by the difference between time of infection and time of sampling expressed in years.
Test for positive or purifying selection.
The number of nonsynonymous and synonymous substitutions, indicated by KA and KS, respectively, was computed for each donor-recipient cluster. A KA/KS ratio significantly lower than 1 is evidence for the presence of purifying selection, whereas ratios greater than 1 indicate positive selection. Within a particular cluster, KA and KS values were estimated comparing the sequences of the different clones with another with the method of Nei and Gojobori (27) implemented in the program MEGA, and their averages were used to compute average KA/KS ratios.
Split decomposition analysis with Splitstree.
Evolutionary relationships between taxa are most often represented as phylogenetic trees, the justification being that evolution is usually a branching tree-like process. However, in certain cases, the tree-like behavior of the data in question cannot be guaranteed, in which case it can contain a number of conflicting phylogenetic signals. Thus in these situations it is appropriate to not necessarily force the data onto a tree but to allow representation of the evolutionary relationship by networks that can simultaneously display contradictory signals. To generate such networks, we used SplitsTree 2.3f (15), a program based on the split decomposition theory for analyzing metrics. Essentially, this theory allows one to canonically decompose any distance (such as the one generated from the data set presented) into the sum of split metrics plus a split prime residue. Once computed, a network represents the split metric sum (as appearing in Fig. 2) that approximates the metric in question. As this is an approximation, a fit index that indicates how well the network represents the original distance is also computed; a fit index of 100% indicates true representation, whereas lower fits imply reduction in accuracy. In general, if the distance supplied is tree-like, then SplitsTree will produce a tree; more complex networks result as the distance deviates from this ideal situation. Several options are available in SplitsTree. We will make use of the refine option, one that in certain cases allows an improvement of the resolution of the generated SplitsTrees.
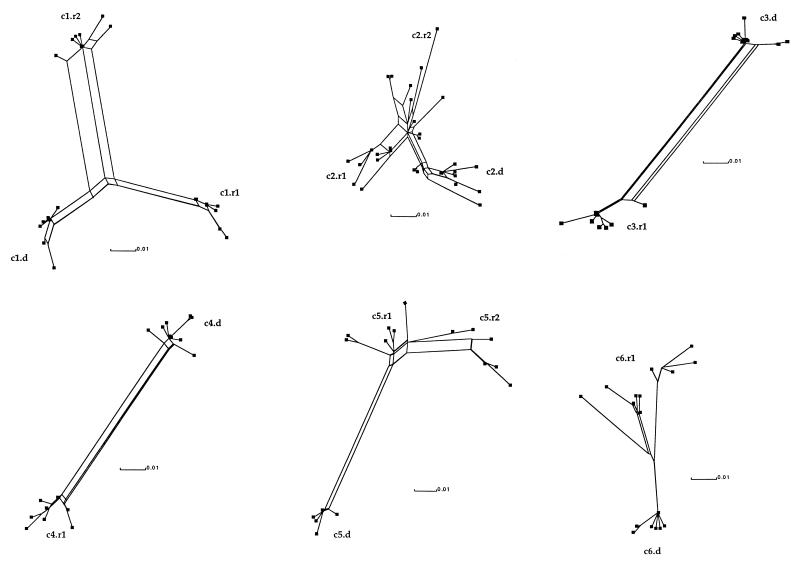
FIG. 2.
SplitsTrees obtained with the split decomposition method for the six donor-recipient clusters studied, designated as for Fig. 1. Squares indicate the different clones sequenced in each patient.
Peptide enzyme immunoassay.
Synthetic 15-mer C-terminal HVR1 (amino acids 13 to 27) peptides were prepared according to the sequences deduced from the dominant and the most divergent nucleotide sequences of each donor and recipient quasispecies. Peptides were modified by the addition of a cysteine residue at the NH2 terminus, cross-linked through this residue to activated microtiter plates (Covalab, Lyon, France), and used as antigens for the detection of anti-HVR1 in autologous sera and in sera from individuals in the same cluster (intracluster) and cross-reactivity with sera from donor and recipients from other clusters as described previously (3). Eight sera from random blood donors were also tested in each microtiter plate as negative controls to determine the assay cutoff (CO). Peptides were incubated for 30 min at room temperature, and plates were extensively washed. After addition of a blocking reagent at room temperature for another 30 min, 100 μl of 1:100-diluted serum samples were added and incubated for 60 min at 37°C. After five washings with phosphate-buffered saline containing 0.05% Tween 20 (pH 7.1), 100 μl of 1:2,000-diluted alkaline phosphatase-labeled goat anti-human rabbit immunoglobulin G (Sigma) was added, and the mixture was incubated for 60 min at 37°C. P-Nitrophenyl phosphate substrate was extemporaneously dissolved in Tris buffer; 100 μl was added to the wells, which were then incubated for 30 min at room temperature in the dark. The reaction was blocked by addition of 50 μl of 4 M sulfuric acid, and the absorbance was read at 405 nm. For each peptide tested, a CO value was calculated as the mean absorbance of eight negative control sera plus six times the standard deviation (SD). Data were expressed as the ratio between the sample absorbance and the CO value (S/CO).
RESULTS
Quasispecies and genotypes.
From the 9 to 15 clones sequenced in each patient, 5 to 12 variants were found at the nucleotide level in the E1/E2 region. The number of variants ranged between 3 and 10 when the analysis was restricted to the HVR1 region. The HVR1 region accounted for 61% of the diversity.
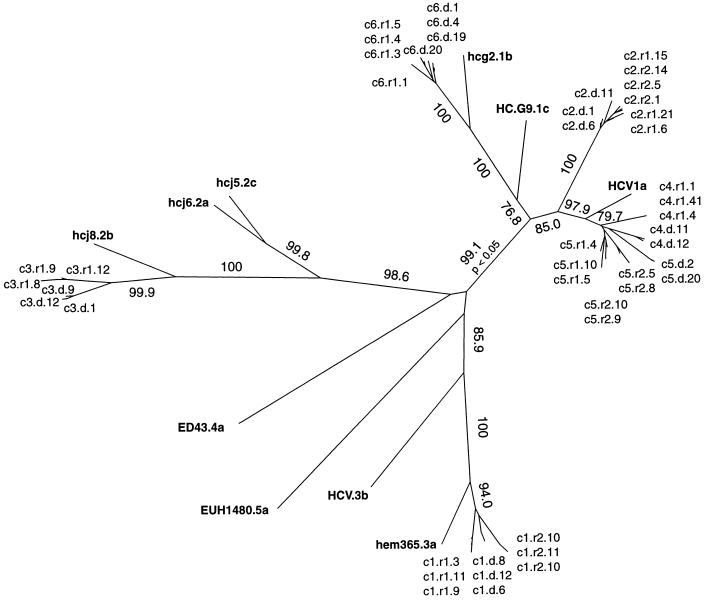
The phylogenetic analysis of the E1/E2 nucleotide sequences together with appropriate homologous reference sequences permitted genotyping. In the NJ tree shown in Fig. 1, in addition to reference sequences specific of the HCV-specific genotypes 1 to 5, the most representative variants of E1/E2 sequences from each individual in the study are indicated. The ML tree gave almost the same topology. As shown in Fig. 1, clusters 2, 4, 5, and 6 belong to genotype 1. In particular, clusters 4 and 5 are monophyletic with the HCV reference strain of the 1a subgroup (97.9% bootstrap support; P < 0.01 in the ML tree), while cluster 6 belongs to the 1b subgroup (100% bootstraps; P < 0.01). Cluster 2 is clearly separated from the other three clusters as well as from the reference sequence for genotypes 1a, 1b, and 1c with very high bootstrap support (Fig. 1). This might correspond to a separate subgroup of genotype 1 or to a divergent strain within the 1a subgroup. Clusters 1 and 3 are monophyletic with the reference sequences of genotypes 3a and 2b, respectively, with very high bootstrap support (Fig. 1). Recipient 2 of cluster 3 was found to harbor genotype 3a (data not shown), clearly different from both the donor and recipient 1 of the same cluster (2b). The history of this patient revealed that he had been transfused with an HCV-seropositive blood component 1 year prior to the index transfusion. Although no sample from the corresponding donor was available, it appeared very likely that the first infection was with an HCV strain of genotype 3a and that the index transfusion either was not infectious or did not yield superinfection with genotype 2b. This recipient was excluded from further studies.
FIG. 1.
Genotyping of six donor-recipient clusters. Unrooted NJ phylogenetic tree of the HCV 258-nt consensus E1/E2 region, including HVR1. Reference sequences of known genotypes are in boldface. Donors and recipients from the six clusters studied are indicated as follows: cluster number (c1 to c6), d for donor, r1 and r2 for recipients 1 and 2, and number of the most representative variants for each quasispecies. Bootstrap percentages of 1,000 bootstrap replicates are given along the appropriate branches. Bootstrap values <50% are not shown. All clades for which the bootstrap values are indicated were also supported by P values of <0.01 in the ML tree, except for the P < 0.05 indicated in the figure.
HCV evolutionary rates in different patients.
Several cDNA clones were sequenced and aligned for each patient to investigate the diversity of the viral quasispecies within any infected individual. Genetic distances among the different strains of a patient were computed with the HKY model (11), which provided the best-fitting model for these particular data sets (data not shown). Heterogeneity of nucleotide substitution rates across sites was tested assuming a discrete γ distribution of the rates (with eight rate categories) and estimating the α parameter of the distribution with the program PUZZLE (42). Most of the data sets gave α values greater than 50, suggesting uniform rates across sites (data not shown). Exceptions were recipient 1 in cluster 2 and the recipient in cluster 6, with α values of 0.1 and 0.28, respectively. Thus, for these two data sets, distances were estimated employing the HKY model with rate heterogeneity. Finally, the LRT for the molecular clock hypothesis was performed as described in Materials and Methods. The results of the LRT for each data set are given in Table 1. Within any infected individual, the virus accumulates mutations over time at a constant rate.
TABLE 1.
LRT for the molecular clock hypothesis of HCVa
| Cluster | Donor or recipient | No. of strains | −ln ML | −ln MLK | Δl | P value |
|---|---|---|---|---|---|---|
| 1 | d | 12 | −401.94 | −406.91 | 9.95 | 0.44 |
| r1 | 10 | −394.22 | −397.45 | 6.46 | 0.60 | |
| r2 | 11 | −426.17 | −431.93 | 11.51 | 0.24 | |
| 2 | d | 12 | −462.00 | −469.32 | 14.65 | 0.15 |
| r1 | 10 | −475.88 | −480.10 | 8.45 | 0.39 | |
| r2 | 10 | −431.01 | −436.14 | 10.26 | 0.25 | |
| 3 | d | 12 | −437.37 | −441.35 | 7.96 | 0.63 |
| r1 | 10 | −447.43 | −452.12 | 9.38 | 0.31 | |
| 4 | d | 9 | −439.42 | −443.82 | 8.81 | 0.27 |
| r1 | 8 | −430.99 | −436.11 | 10.23 | 0.12 | |
| 5 | d | 5 | −396.05 | −397.43 | 2.77 | 0.43 |
| r1 | 6 | −474.37 | −478.09 | 7.43 | 0.11 | |
| r2 | 6 | −498.87 | −502.38 | 7.03 | 0.13 | |
| 6 | d | 7 | −403.67 | −405.67 | 3.99 | 0.55 |
| r1 | 10 | −577.45 | −581.94 | 8.97 | 0.34 |
The alignments used for testing the clock hypothesis were those described in Materials and Methods. Calculations were performed by PUZZLE (42), assuming the HKY85 model with uniform substitution rate across sites, except for r1 in cluster 2 and r1 in cluster 6, where γ-distributed rates across sites were assumed (see Results). ML and MLK are the likelihoods with and without the clock, respectively. Δl is the likelihood ratio statistic; if Δl is not significant (P > 0.05 in a χ2 test with n − 2 df), the clock hypothesis cannot be rejected. Strains with identical sequences were removed from the alignment.
The estimated HCV evolutionary rate in each patient is given in Table 2. The rates range from 3.4 × 10−4 to 4.51 × 10−3 nucleotide substitutions per site per year. However, the t test shows that only in the donors of clusters 2, 5, and 6 was the evolutionary rate significantly lower than in the corresponding recipients (Table 2). Moreover, pairs of recipients infected by the same donor did not show a statistically significant difference in evolutionary rate.
TABLE 2.
HCV evolutionary rates in different clusters
| Cluster | Donor or recipient | ΔT (yr)a | Mean evolutionary rate (nt substitutions/site/year) (0.63) (10−3) | Statistical significance (P) of difference in evolutionary rateb
|
|
|---|---|---|---|---|---|
| r1 | r2 | ||||
| 1 | d | ? | ? | ? | ? |
| r1 | 5 | 1.14 (0.55) | 0.54 | ||
| r2 | 9.8 | 0.76 (0.29) | |||
| 2 | d | ≈23 | 0.65 (0.17) | 0.026 | 0.012 |
| r1 | 4.4 | 2.10 (0.63) | |||
| r2 | 5.8 | 1.60 (0.34) | |||
| 3 | d | ≈17 | 0.63 (0.23) | 0.14 | |
| r1 | 10.5 | 1.38 (0.46) | |||
| 4 | d | 9 | 0.99 (0.38) | 0.77 | |
| r1 | 8 | 1.14 (0.35) | |||
| 5 | d | ≈17 | 0.34 (0.16) | 2 × 10−12 | 1.3 × 10−5 |
| r1 | 6.6 | 2.73 (0.30) | 0.33 | ||
| r2 | 11.2 | 3.47 (0.70) | |||
| 6 | d | 21 | 0.34 (0.13) | 4 × 10−7 | |
| r1 | 8 | 4.51 (0.81) | |||
Interval from the time of sampling to the time of infection.
S standard errors are given in parentheses. Calculated by a two-tailed t test with infinite degrees of freedom. Significantly different rates (P < 0.05) are in boldface.
Analysis of selective pressure.
Table 3 indicates the average of nonsynonymous and synonymous nucleotide substitutions among the different clones sequenced in each donor-recipient cluster. KA/KS appears to be greater than 1 for clusters 1, 4, and 6 and less than 1 for clusters 2, 3, and 5. However, none of the clusters show a statistically significant difference between KA and KS values (Table 3).
TABLE 3.
Average nonsynonymous and synonymous nucleotide substitutions within different donor-recipient clusters
SplitsTree analysis.
Refined SplitsTrees obtained for each cluster are shown in Fig. 2. Unrefined SplitsTrees were almost identical. The fit indices for clusters 1, 3, 4, and 5 were good, ranging between 79 and 96%; cluster 2 had the lowest fit (68%). The pattern of donor versus recipient(s) is clear in each of these SplitsTrees except in cluster 2, where a more complex network is obtained. The donor and each of the two recipients in cluster 1, the donor and recipient in clusters 3 and 4, and the donors of clusters 5 and 6 show one or two strains at an internal node giving rise to the other strains in their respective SplitsTrees. For each of these cases, Splitstree was run separately on both the donor and recipient subclusters. In all cases except the donor subcluster of cluster 6, a tree was obtained with 100% fit.
Even though the remaining cases (donor and two recipients of cluster 2, the two recipients in cluster 5, and the recipient in cluster 6) exhibit a more complicated pattern of splits with several distinct viral lineages, it was found that the SplitsTrees for the donor and recipient subclusters were in most cases extremely tree-like; the exception was cluster 2, where one of the recipient's SplitsTrees had a net structure with 100% fit. Finally, the SplitsTrees seem to indicate differing phylogenetic relationships between the donor and recipient for each of the six clusters. In particular, in clusters 1, 2, 3, and 4, the donor subnetwork is connected to that of the recipient(s) by at least three edges, whereas in clusters 5 and 6, only two and one edges, respectively, are found. However, in most cases where several edges joined the donor and recipient, such as in clusters 1, 3, and 4, the removal of a single taxon from the data set and subsequent recomputation of the SplitsTree resulted in the disappearance of the box (data not shown). This suggests that these splits are supported by only one taxon.
Immunological reactivity with HVR1 peptides.
To examine the influence of immune pressure on the evolutionary patterns of HCV in the donor-recipient pairs, reactivities of HVR1 antibody in donor and recipient sera were tested with autologous and heterologous HVR1 peptides. Peptide sequences were derived from the dominant variant of each patient quasispecies, and when appropriate, additional peptides were derived from substantially divergent variants. As shown in Table 4, only sera from the donor of cluster 2 and recipient 1 of cluster 3 strongly reacted with the autologous peptides. Sera from six individuals did not recognize autologous peptides. In particular, no member of cluster 1 reacted with autologous peptides, and only serum from recipient 1 had low reactivity with both peptides from other cluster members. The average S/CO ± 1 SD of reactive patient sera was not significantly different between autologous, intracluster, or extracluster peptides (2.85 ± 2.77, 1.83 ± 0.80, and 2.05 ± 1.75, respectively). However, when levels of reactivity with the panel of HVR1 peptides were compared between sera, they tended to be the highest with extracluster and then intracluster peptides and lowest with autologous peptides. This analysis was in part confounded by the fact that sera from five individuals (donors from cluster 2 and 5; recipients 1 from clusters 3, 4, and 5) reacted with 14 to 18 of the 20 peptides tested. These sera were considered highly reactive against HVR1 epitopes. Sera from other patients reacted with eight or fewer peptides. As shown in Fig. 3, there was a correlation between the number of peptides recognized by each serum and the S/CO taken as a reflection of the level of antibody. This observation suggests that infected patients who develop antibody to HVR1 with high levels of reactivity to HVR1 peptides, autologous or heterologous, also have the broadest cross-reactivity.
TABLE 4.
Reactivities of 20 sera from six clusters with 20 cluster-derived HVR1 peptides
| Peptide sample | S/CO
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cluster 1
|
Cluster 2
|
Cluster 3
|
Cluster 4
|
Cluster 5
|
Cluster 6
|
|||||||||||||||
| d | r1 | r2 | d | r1a | r1b | r2 | d | r1 | r2 | d | r1 | d | r1a | r1b | r2a | r2b | d | r1a | r1b | |
| c1.d | —a | — | — | 1.0 | — | — | — | — | — | — | — | — | — | — | — | — | — | 1.0 | — | — |
| c1.r1 | 1.4 | — | 1.9 | — | — | — | — | 1.5 | — | — | — | — | 1.9 | 1.4 | — | — | 1.1 | 1.3 | — | 1.7 |
| c1.r2 | 1.3 | — | — | 1.5 | — | — | 1.2 | 1.2 | — | — | — | — | — | 2.4 | — | — | — | 1.2 | — | 1.3 |
| c1.r3 | 1.1 | — | — | 1.4 | 1.3 | — | 1.1 | 1.8 | 1.2 | — | — | — | — | — | — | 1.4 | 1.7 | 2.1 | 1.3 | 2.1 |
| c1.r4 | 1.1 | — | — | 1.2 | — | — | — | 1.7 | 1.0 | — | — | — | — | — | — | — | 1.1 | 1.4 | — | 1.4 |
| c2.d | 1.3 | 2.5 | 1.1 | 4.0 | 1.5 | 2.0 | 2.1 | 2.6 | 1.1 | 2.4 | 1.6 | 2.4 | 1.3 | — | 2.5 | — | — | 2.8 | — | 2.0 |
| c2.r1 | 1.1 | — | — | 2.4 | 1.4 | — | 1.3 | 1.8 | 1.2 | — | — | — | — | — | — | 1.2 | 1.5 | 1.9 | 1.1 | 2.0 |
| c2.r2 | 1.2 | — | — | 1.4 | 1.2 | — | — | 1.1 | 1.3 | — | — | — | − | — | — | — | — | 1.1 | — | 1.2 |
| c3.d | 1.2 | — | — | 1.6 | 1.0 | — | — | 1.2 | 1.1 | — | — | — | 1.0 | — | — | — | — | 1.3 | — | 1.1 |
| c3.r1 | 8.3 | 1.1 | 1.7 | 2.3 | 8.5 | 2.2 | 1.5 | 2.8 | 10.1 | — | 2.0 | 2.5 | 2.6 | — | 1.0 | 2.8 | 3.3 | 5.2 | 3.2 | 3.3 |
| c4.d | 1.3 | — | — | 1.2 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| c4.r1 | 1.6 | 6.9 | 1.4 | 14.6 | 1.4 | 1.4 | 2.8 | 2.9 | 1.6 | 1.7 | 1.1 | 2.2 | 1.4 | 3.7 | — | 6.5 | 2.7 | 2.0 | 2.6 | 1.9 |
| c4.r2 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | − | — | 1.0 | — | 1.1 |
| c4.r3 | 2.0 | — | — | 2.1 | 2.0 | — | — | — | 1.9 | 1.3 | — | — | 1.4 | — | 1.1 | — | — | — | — | — |
| c5.d | 1.6 | 1.0 | 4.0 | 2.9 | 3.6 | — | 3.2 | 2.1 | 1.0 | — | 3.2 | 1.3 | 1.2 | 2.8 | 1.4 | 1.5 | 2.0 | 7.3 | 1.4 | 2.4 |
| c5.r1 | 1.7 | 1.0 | 1.1 | 3.1 | 1.7 | — | 1.4 | 1.4 | 1.8 | — | — | 1.3 | 1.5 | 1.3 | 1.4 | 1.4 | 1.8 | 2.1 | 1.3 | 2.0 |
| c5.r2 | — | — | — | 1.1 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| c6.d | 1.9 | — | 2.5 | 1.7 | 3.7 | 1.3 | 1.4 | — | 1.9 | — | 1.4 | 1.3 | 1.1 | 1.9 | — | — | — | 1.7 | — | — |
| c6.r1 | 2.0 | — | — | 1.3 | 1.2 | — | — | — | 1.2 | — | — | — | 1.4 | — | — | — | 1.1 | 4.6 | — | 3.9 |
| c6.r2 | 1.7 | — | — | 1.6 | 1.6 | — | 1.1 | − | — | — | 1.0 | — | 1.2 | — | — | — | — | — | − | − |
—, negative result (S/CO < 1.0).
FIG. 3.
Correlation between the number of reactive HVR1 peptides from donors and recipients and level of reactivity by enzyme immunoassay expressed as S/CO. CO was defined as the mean of the optical density of eight negative control sera plus 6 SD. The equation of the regression line and the R2 value are indicated.
High level and frequency of reactivity were associated with peptides from donors of cluster 1 and 2 (17 and 18 of 21 sera were reactive). Five other peptides (c2.r1a, c3.d, c3.r1, c6.d, and c6.r1b) reacted with 13 of the 21 sera. These results indicate that anti-HVR1 reactivity was more related to the overall level of humoral immune response of the host than the HVR1 sequence, whether HVR1 peptides derived from autologous, intracluster, or extracluster sequences. There was no correlation between the level of reactivity and the breadth of antigen recognition of sera from HCV-infected patients tested with HVR1 peptides derived from individuals infected with unrelated HCV strains. This suggests that the particular sequence of the HVR1 peptides is not the determinant of antibody reactivity.
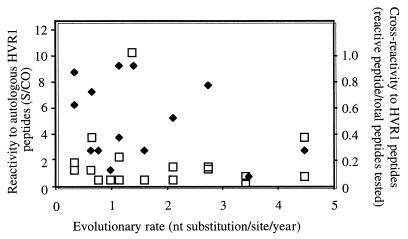
The potential correlation between evolutionary rate and anti-HVR1 immune reactivity was assessed by plotting the data presented in Table 2 with two indicators of immune response to HVR1 epitopes (Fig. 4). The first indicator was the serum reactivity with autologous HVR1 peptides expressed as S/CO; the second was the ratio between the number of extracluster HVR1 peptides recognized by each serum (S/CO ≥ 1) and the total number of peptides tested (range, 15 to 18) (Table 3). No correlation was found between the evolutionary rate and either of these immunological indicators.
FIG. 4.
Evolutionary rate and HVR1 immune response. Absence of correlation between evolutionary rate and anti-HVR1 reactivity. Open squares correspond to the reactivity of patient plasma against autologous HVR1 peptides. When two different peptides derived from a patient's quasispecies, two data points are indicated. Diamonds correspond to the number of reactive peptides of each plasma against all extracluster HVR1 peptides divided by the number of peptides tested.
DISCUSSION
HCV is an RNA virus chronically infecting humans and chimpanzees. The viral genome is submitted to a very large number of replication cycles over decades, and since transcription is not proofread, errors occur which result in the presence of a family of related genomic variants called quasispecies (23). Relatively little is known about the rate of genetic errors driving over time the drift of a given population of HCV variants (4, 28, 40, 41). The host is the second factor potentially involved in changes of the quasispecies. The immune system of the infected host develops both humoral and cellular immune responses which contribute to limiting the level of viral replication (29). Genomic viral diversity translates into different amino acid sequences and epitopes, resulting in different levels of recognition by the host immune system (7). Some epitopes are not recognized, and the corresponding variants, named escape mutants, circulate in a free form and probably benefit from a replication advantage (2, 18, 47). The others variants, more numerous, circulate as immune complexes with antibodies bound to the external glycoproteins (2, 8, 39). It has been recognized that the E1/E2 region, in particular the hypervariable N-terminal region of E2, played a major role in the escape mechanism (38, 45). However, the relative parts played by the virus's inherent ability to mutate and the ability of the host immune system to apply pressure and participate in the viral diversity by selecting particular variants have not been clearly established (3, 20, 22, 30, 43). In the work presented here, inclusion of multiple recipients of HCV from an infected blood donor ensured that two different individuals were, at a known time, chronically infected or contaminated with the same quasispecies. By studying the phylogeny of these donor-recipient pairs and the anti-HVR1 patient-specific immune response, it became possible to determine the relative contributions of the virus and the host in viral evolution.
We first established that donor-recipient pairs in each cluster were related to each other and were of the same genotype. This is clearly demonstrated in the phylogenetic tree shown in Fig. 1. The excess of clusters of genotype 1 is in accordance with the prevalence of HCV genotypes in the United Kingdom (21, 25, 44). The only exception was recipient 2 of cluster 3, whose genotype differed from the implicated donor and the other recipient in the cluster. A likely explanation for this anomaly was found in this individual's transfusion history. From previous data collected by our group and by others, it seemed that the 9 to 15 clones sequenced in each patient were adequate to provide a reliable appreciation of the quasispecies diversity (3, 20). The main feature of the data presented here is that the HCV evolutionary rate in all patients is compatible with an intrapatient molecular clock hypothesis (Table 1). The virus seems to accumulate mutations at the same rate for all variants within a particular infected individual. Since the time of the infection was known for most of the patients, it was also possible to estimate the HCV evolutionary rate in the different hosts. When these evolutionary rates were correlated with the patient level of immune response (estimated according to the number of HVR1 peptides recognized and the level of reactivity [Table 3 and Fig. 3]), no relationship was observed. It therefore appears that HCV genomic drift is mostly independent of the host level or breadth of humoral immune response to HVR1 and could be essentially driven by viral replication errors and the number of replication cycles. This conclusion is also supported by the observation that analyses of nonsynonymous and synonymous replacements within the E1/E2 region do not show the presence of strong positive or negative selection. A trend toward higher HVR1 KA/KS ratios in patients with persistent viremia has recently been reported (33). We indeed find a higher KA/KS ratio in some cases, but the difference between KA and KS is not statistically significant in any of the clusters studied, suggesting neutral evolution. This conclusion has to be viewed with caution, given the limited number of patients and the fact that we could analyze only one time point after the occurrence of HCV infection. However, the fact that results of all of the different analyses performed appear to be in agreement strengthens our confidence in the result. Viral diversity and evolutionary rates have been evaluated in agammaglobulinemic patients and found to be very low (19, 29). This finding could be taken as evidence that without immune pressure, HCV genetic drift is reduced to the viral internal clock. However, the sequences studied were taken from consensus without examining quasispecies distribution and studied at a very late stage, when patients had developed cirrhosis. In addition, the evolutionary rates found in immunocompetent controls ranged between 0.1 and 0.28 nucleotide substitutions per site per year (29), while based on the sequencing of at least 10 clones, we observed a range of 0.34 to 4.51. The data sets do not appear comparable.
The viral quasispecies were studied between 4.4 and 23 years after infection (Table 2), a relatively long period of time that does not exclude that the host immune pressure could be more visibly effective in the early part of this long-term, chronic infection. This concept is indirectly supported by the observation that antibodies to the N-terminus epitopes of HVR1 are found early in the infection, particularly when self-limiting, but not after chronic infection is established (3, 48).
It was, however, noticeable that the evolutionary rate of recipients tended to be higher than that of donors who had been infected for a considerably longer period of time (Table 2). This may reflect the relative increase of evolutionary rate during the initial phase of the infection when HCV replicates at a very high rate (doubling time of viral replication estimated at 1 day [M. Bush et al., personal communication]) in the preseroconversion period. Later during the course of the persistent infection, replication cycles are assumed to be slower and to involve fewer infected cells owing to the relative control exercised by the host (18, 30). Not surprisingly, a gross relationship was observed between evolutionary rate and viral diversity within the quasispecies. As shown in Fig. 2, both recipients in clusters 2 and 5 and recipient 1 in cluster 6, who had higher evolutionary rates (Table 1), had quasispecies composed of more divergent variants and a more network-like phylogeny of the quasispecies. Conversely, individuals with lower evolutionary rates such as donors and recipients in clusters 3 and 4 (Fig. 2) had quasispecies of limited diversity.
In this analysis, the fact that in donors who have been infected years or decades previously, several variants or lineages of variants may simultaneously replicate at approximately the same rate should also be taken into account. In contrast, in recipients, as previously shown, only a limited number of variants, presumably those circulating as free virus, preferentially infect the host and are initially submitted to intense replication cycles (2, 18, 30). The SplitsTree analysis for clusters 1, 3, and 4 showed a tree-like structure with 100% fit. Moreover, in each cluster, the splits in the recipient seem to be supported by only a single taxon (see Results). In view of the quasispecies distribution of HCV, it is possible that in the data sets we analyzed, only one variant of the donor viral strains was by chance alone the one transmitted to the recipient. The split decomposition method has already been used with some success in the study of viruses, as it does not presuppose the tree-likeness of the data analyzed (6, 32). SplitsTree seems to be a powerful tool for investigating the dynamic of HCV evolution within an infected host. It could potentially be used to make inferences about the variants transmitted during the infection of a new host and the time of infection. For example, in light of our observation and interpretation of this data set, it appears likely that the donor of cluster 1 was infected a relatively short time before infecting the first recipient.
In conclusion, it is suggested that the immune response of the host is not one of the main evolutionary forces driving the diversity of the HCV quasispecies in an infected host within the investigated E1/E2 region. The evolutionary rate and consequently the extent of the genetic drift of the quasispecies corresponded to the time of infection: the longer the time, the lower the HCV rate of evolution. This fact might have important implications for HCV pathogenesis and treatment and deserves further attention.
ACKNOWLEDGMENTS
We are grateful for the financial support from UK Department of Health grant 121-6510 to Yan Dong. The collaboration between the Division of Transfusion Medicine, Cambridge, and the Riga Institute, Leuven, was the result of the fifth European workshop on virus evolution and molecular epidemiology, 1 to 4 September 1998, Leuven, Belgium, which was sponsored by European Community grant BMH4-98-4830.
We are indebted to P. E. Hewitt, North London Blood Centre, who provided some of the samples, and to C. Llewelyn, East Anglia Blood Centre, who provided information relative to the history of donors and recipients.
REFERENCES
- 1.Abe K, Inchauspé G, Fujisawa K. Genomic characterisation and mutation rate of hepatitis C virus isolated from a patient who contracted hepatitis during an epidemic on non-A, non-B hepatitis in Japan. J Gen Virol. 1992;72:2725–2729. doi: 10.1099/0022-1317-73-10-2725. [DOI] [PubMed] [Google Scholar]
- 2.Aiyama T, Yoshioka K, Okumura A, Takayanagi M, Iwata K, Ishikawa T, Kakuma S. Sequence analysis of hypervariable region of hepatitis C virus (HCV) associated with immune complex in patients with chronic HCV infection. J Infect Dis. 1996;174:1316–1320. doi: 10.1093/infdis/174.6.1316. [DOI] [PubMed] [Google Scholar]
- 3.Allain J-P, Zhai W, Shang D, Timmers E, Alexander G J M. Hypervariable region diversity of hepatitis C virus and humoral response: comparison between patients with and without cirrhosis. J Med Virol. 1999;59:25–31. doi: 10.1002/(sici)1096-9071(199909)59:1<25::aid-jmv5>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 4.Casino C, McAllister J, Davidson F, Power J, Lawlord E, Yap P L, Simmonds P, Smith D B. Variation of hepatitis C virus following serial transmission: multiple mechanisms of diversification of the hypervariable region and evidence for convergent genome evolution. J Gen Virol. 1999;80:717–725. doi: 10.1099/0022-1317-80-3-717. [DOI] [PubMed] [Google Scholar]
- 5.Choo Q L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby R, Barr P J. Genetic organisation and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dopazo J, Dress A, von Haeseler A. Split decomposition: a technique to analyse viral evolution. Proc Natl Acad Sci USA. 1993;90:10320–10324. doi: 10.1073/pnas.90.21.10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farci P, Alter H J, Govindarajan S, Wong D C, Engle R, Lesniewski R R, Mushahwar I K, Desai S M, Miller R H, Ogata N. Lack of protective immunity against reinfection with hepatitis C infection. Science. 1992;25:135–140. doi: 10.1126/science.1279801. [DOI] [PubMed] [Google Scholar]
- 8.Farci P, Shimoda A, Wong D, Cabezon T, De Gionnis D, Strazzera A, Shimizu Y, Shapiro M, Alter H J, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci USA. 1996;93:15394–15399. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein J P. Phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 11.Hasegawa M, Kishino H, Yano T. Dating the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 12.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. CABIOS. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 13.Higgins D G, Sharp P M. Clustal: a package for performing multiple sequence alignments on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 14.Huelsenbeck J, Rannala B. Phylogenetic methods come of age: testing hypotheses in an evolutionary context. Science. 1997;276:227–232. doi: 10.1126/science.276.5310.227. [DOI] [PubMed] [Google Scholar]
- 15.Huson D H. SplitsTree: a program for analyzing and visualizing evolutionary data. Bielefeld, Germany: University of Bielefeld; 1997. [Google Scholar]
- 16.Jackson P, Petrik J, Alexander G J M, Pearson J, Allain J-P. Reactivity of synthetic peptides representing selected sections of hepatitis C virus core and envelope proteins with a panel of hepatitis C virus-seropositive human plasma. J Med Virol. 1997;51:67–79. doi: 10.1002/(sici)1096-9071(199701)51:1<67::aid-jmv11>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 17.Kato N, Sekiya H, Ootsuyama Y, Nakazawa T, Hijikata M, Ohkoshi S, Shimotohno K. Humoral immune response to hypervariable region 1 of the putative envelope glycoprotein (gp70) of hepatitis C virus. J Virol. 1993;67:3923–3930. doi: 10.1128/jvi.67.7.3923-3930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kojima M, Osuga T, Tsuda F, Tanaka T, Okamoto H. Influence of antibodies to the hypervariable region of E2/NS1 glycoprotein on the selective replication of hepatitis C virus in chimpanzees. Virology. 1994;200:665–672. doi: 10.1006/viro.1994.1582. [DOI] [PubMed] [Google Scholar]
- 19.Kumar U, Monjardino J, Thomas H C. Hypervariable region of hepatitis C virus envelope glycoprotein (E2/NS1) in an agammaglobulinemic patient. Gastroenterology. 1994;106:1072–1075. doi: 10.1016/0016-5085(94)90770-6. [DOI] [PubMed] [Google Scholar]
- 20.Lawal Z, Petrik J, Wong V S, Alexander G J M, Allain J-P. Hepatitis C virus genomic variability in untreated and immunosuppressed patients. Virology. 1997;228:107–111. doi: 10.1006/viro.1996.8359. [DOI] [PubMed] [Google Scholar]
- 21.Majid A, Holmes R, Desselberger U, Simmonds P, McKee T A. Molecular epidemiology of hepatitis C virus infection amongst intravenous drug users in rural communities. J Med Virol. 1995;46:48–51. doi: 10.1002/jmv.1890460111. [DOI] [PubMed] [Google Scholar]
- 22.Majid A, Jackson P, Lawal Z, Pearson G M J, Parker H, Alexander G J M, Allain J-P, Petrik J. Ontogeny of hepatitis C virus (HCV) hypervariable region 1 (HVR1) heterogeneity and HVR1 antibody responses over a three year period in a patient infected with HCV type 2b. J Gen Virol, 1999;80:317–325. doi: 10.1099/0022-1317-80-2-317. [DOI] [PubMed] [Google Scholar]
- 23.Martell A R, Esteban J I, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McAllister J, Casino C, Davidson F, Power J, Lawlor E, Yap P L, Simmonds P, Smith D B. Long-term evolution of the hypervariable region of hepatitis C virus in a common-source-infected cohort. J Virol. 1998;72:4893–4905. doi: 10.1128/jvi.72.6.4893-4905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McOmish F, Chan S W, Dow B C, Gillon J, Frame W D, Crawford R J, Yap P L, Follett E A C, Simmonds P. Detection of three types of hepatitis C virus in blood donors: investigation of type-specific differences in serologic reactivity and rate of alanine aminotransferase abnormalities. Transfusion. 1993;33:7–13. doi: 10.1046/j.1537-2995.1993.33193142314.x. [DOI] [PubMed] [Google Scholar]
- 26.Mondelli M U, Cerino A, Lisa A, Brambilla S, Segagni L, Cividini A, Bissolati M, Missale G, Bellati G, Meola A, Bruniercole B, Nicosia A, Galfre G, Silini E. Antibody response to hepatitis C virus hypervariable region 1: evidence for cross-reactivity and immune-mediated sequence variation. Hepatology. 1997;30:537–545. doi: 10.1002/hep.510300233. [DOI] [PubMed] [Google Scholar]
- 27.Nei M, Gojobori T. Simple method for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 28.Ni Y H, Chang M H, Chen P J, Lin H H, Hsu H Y. Evolution of hepatitis C virus quasispecies in mothers and infants infected through mother-to-infant transmission. J Hepatol. 1997;26:967–974. doi: 10.1016/s0168-8278(97)80104-3. [DOI] [PubMed] [Google Scholar]
- 29.Odeberg, J., Z. Yun, A. Sonneborg, K. Bjoro, M. Ulhen, and J. Lundenberg. Variation of hepatitis C virus hypervariable region 1 in immunocompromised patients. J. Infect. Dis. 175:938–943. [DOI] [PubMed]
- 30.Okamoto H, Mishiro S, Tokita H, Tsuda F, Miyakawa Y, Mayumi M. Superinfection of chimpanzees carrying hepatitis C virus of genotype II/1b with that of genotype III/2a or I/1a. Hepatology. 1994;20:1131–1134. [PubMed] [Google Scholar]
- 31.Petrik J, Pearson G J M, Allain J-P. High throughput PCR detection of HCV based on semiautomated multisample RNA capture. J Virol Methods. 1997;64:147–159. doi: 10.1016/s0166-0934(96)02153-2. [DOI] [PubMed] [Google Scholar]
- 32.Plikat U, Nieselt-Struwe K, Meyerhans A. Genetic drift can dominate short-term human immunodeficiency virus type 1 nef quasispecies evolution in vivo. J Virol. 1997;71:4233–4240. doi: 10.1128/jvi.71.6.4233-4240.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ray C S, Wang Y-M, Laeyendecker O, Ticehurst J R, Villano S A, Thomas D L. Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: hypervariable region 1 as a decoy. J Virol. 1999;73:2398–2346. doi: 10.1128/jvi.73.4.2938-2946.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosa D, Campagnoli S, Moretto C, Guenzi E, Cousens L, Chin M, Dong C, Weiner A J, Lau J Y N, Choo Q L, Chien D, Pileri P, Houghton M, Abrignani S. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc Natl Acad Sci USA. 1996;93:1759–1763. doi: 10.1073/pnas.93.5.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 36.Sakamoto N, Enomoto N, Kurosaki M, Marumo F, Sato C. Sequential change of the hypervariable region of the hepatitis C virus genome in acute infection. J Med Virol. 1993;42:103–108. doi: 10.1002/jmv.1890420119. [DOI] [PubMed] [Google Scholar]
- 37.Scarcelli E, Cerino A, Esposito G, Silini E, Mondelli M U, Traboni C. Occurrence of antibodies reactive with more than one variant of the putative envelope glycoprotein (gp70) hypervariable region 1 in viremic hepatitis C virus-infected patients. J Virol. 1995;60:4407–4412. doi: 10.1128/jvi.69.7.4407-4412.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimizu Y K, Hijikata M, Iwamoto A, Alter H J, Purcell R H, Yoshikura H. Neutralizing antibodies against hepatitis C virus and emergence of neutralization escape mutant viruses. J Virol. 1994;68:1494–1500. doi: 10.1128/jvi.68.3.1494-1500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimizu Y K, Igarashi H, Kiyohara T, Cabezon T, Farci P, Purcell R H, Yoshikura H. A hyperimmune serum against a synthetic peptide corresponding to the hypervariable region 1 of hepatitis C virus can prevent viral infection in cell culture. Virology. 1996;223:409–412. doi: 10.1006/viro.1996.0497. [DOI] [PubMed] [Google Scholar]
- 40.Simmonds P, Smith D B. Investigation of the pattern of diversity of hepatitis C virus in relation to times of transmission. J Viral Hepat. 1997;4:69–71. doi: 10.1111/j.1365-2893.1997.tb00163.x. [DOI] [PubMed] [Google Scholar]
- 41.Smith D B, Simmonds P. Characteristics of nucleotide substitution in the hepatitis C virus genome: constraints on sequence change in coding regions at both ends of the genome. J Mol Evol. 1997;45:238–246. doi: 10.1007/pl00006226. [DOI] [PubMed] [Google Scholar]
- 42.Strimmer K, von Haeseler A. Likelihood mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc Natl Acad Sci USA. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Doorn L J, Capriles I, Maertens G, DeLeys R, Murray K, Kos T, Schellekens H, Quint W. Sequence evolution of the hypervariable region in the putative envelope region E2/NS1 of hepatitis C virus is correlated with specific humoral immune responses. J Virol. 1995;69:773–778. doi: 10.1128/jvi.69.2.773-778.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watson J-P, Brind A M, Chapman C E, Bates C L, Gould F K, Johnson S J, Burt A D, Ferguson J, Simmonds P, Bassendine M F. Hepatitis C virus epidemiology and genotypes in the north of England. Gut. 1996;38:269–276. doi: 10.1136/gut.38.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiner A J, Geysen H M, Christopherson C, Hall J E, Mason T J, Saracco G, Bonino F, Crawford K, Marion C D, Crawford K A. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiner A J, Thaler M M, Crawford K, Ching K, Kansopon J, Chien D Y, Hall J E, Hu F, Houghton M. A unique, predominant hepatitis C virus variant found in an infant born to a mother with multiple variants. J Virol. 1993;67:4365–4368. doi: 10.1128/jvi.67.7.4365-4368.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyatt C A, Andrus L, Brotman B, Huang F, Lee D H, Prince A M. Immunity in chimpanzees chronically infected with hepatitis C virus: role of minor quasispecies in reinfection. J Virol. 1998;72:1725–1730. doi: 10.1128/jvi.72.3.1725-1730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zibert A, Schreier E, Roggendorf M. Antibodies in human sera specific to hypervariable region 1 of hepatitis C virus can block viral attachment. Virology. 1995;208:653–661. doi: 10.1006/viro.1995.1196. [DOI] [PubMed] [Google Scholar]
- 49.Zibert A, Kraas W, Meisel H, Jung G, Roggendorf M. Epitope mapping of antibodies directed against hypervariable region 1 in acute self-limiting and chronic infections due to hepatitis C virus. J Virol. 1997;71:4123–4127. doi: 10.1128/jvi.71.5.4123-4127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]