Abstract
The Epstein-Barr virus (EBV)-encoded replication proteins that account for the basic reactions at the replication fork are thought to be the EBV Pol holoenzyme, consisting of the BALF5 Pol catalytic and the BMRF1 Pol accessory subunits, the putative helicase-primase complex, comprising the BBLF4, BSLF1, and BBLF2/3 proteins, and the BALF2 single-stranded DNA-binding protein. Immunoprecipitation analyses using anti-BSLF1 or anti-BBLF2/3 protein-specific antibody with clarified lysates of B95-8 cells in a viral productive cycle suggested that the EBV Pol holoenzyme physically interacts with the BBLF4-BSLF1-BBLF2/3 complex to form a large complex. Although the complex was stable in 500 mM NaCl and 1% NP-40, the BALF5 protein became dissociated in the presence of 0.1% sodium dodecyl sulfate. Experiments using lysates from insect cells superinfected with combinations of recombinant baculoviruses capable of expressing each of viral replication proteins showed that not the BMRF1 Pol accessory subunit but rather the BALF5 Pol catalytic subunit directly interacts with the BBLF4-BSLF1-BBLF2/3 complex. Furthermore, double infection with pairs of recombinant viruses revealed that each component of the BBLF4-BSLF1-BBLF2/3 complex makes contact with the BALF5 Pol catalytic subunit. The interactions of the EBV DNA polymerase with the EBV putative helicase-primase complex warrant particular attention because they are thought to coordinate leading- and lagging-strand DNA synthesis at the replication fork.
Epstein-Barr virus (EBV) is a human B-lymphotropic herpesvirus which is a causative agent of infectious mononucleosis and is known to be closely associated with several human cancers, including nasopharyngeal carcinoma and Burkitt's lymphoma. The EBV genome is a linear double-stranded DNA which is 172 kbp in length (2). Like other herpesviruses, EBV has both a latent state and a lytic replication cycle. In latently infected lymphoblastoid cells, the viral genome is maintained as a circular plasmid molecule and replicated by the replication machinery of the host (1, 46). However, after induction of lytic viral replication, EBV DNA replication proteins are induced and the EBV genome is amplified 100- to 1,000-fold via the lytic-phase replication origin, oriLyt. The intermediate replication product is a large concatemeric molecule in which single genome units are arranged head to tail (15).
EBV encodes seven viral replication genes that are essential for oriLyt-dependent DNA replication (11, 12). The BZLF1 protein is an oriLyt-binding protein and also acts as the lytic transactivator (31). The BALF5 gene encodes the DNA Pol catalytic subunit (43), and the BMRF1 gene encodes the DNA Pol accessory subunit (19, 39, 40, 41). A single-stranded DNA (ssDNA)-binding protein is encoded by the BALF2 gene (42, 44). The enzymatic activities of the remaining three proteins encoded by the BBLF4, BSLF1, and BBLF2/3 genes have not been determined, but they are predicted to act as helicase, primase, and helicase-primase complex proteins, respectively, from sequence homology to the herpes simplex virus type 1 (HSV-1) UL5, UL52, and UL8 genes (12). These viral replication proteins other than BZLF1 protein conceivably work together at replication forks to synthesize leading and lagging strands of the concatemeric EBV genome.
The BALF5 Pol catalytic polypeptide is copurified with the BMRF1 Pol accessory subunit from EBV-producing lymphoblastoid cells (18, 21, 37), as demonstrated by immunoprecipitation with anti-BALF5 Pol-specific antibody (48), appearing to function as a Pol holoenzyme which exhibits both 5′-to-3′ DNA polymerase and 3′-to-5′ exonuclease activities (37). In addition, this EBV DNA Pol holoenzyme is characterized by strikingly high polymerase processivity (38, 40). Although the BALF5 Pol catalytic subunit by itself is a quasi-processive enzyme (40), complexed together with the BMRF1 Pol accessory subunit, it demonstrates stable interaction with the 3′-OH end of the primer on template DNA during polymerization and exonucleolysis (40, 41). oriLyt-dependent DNA replication would be best served by a highly processive DNA polymerase which could synthesize long DNA chains without dissociating from the template.
The BALF2 gene product has an apparent molecular weight on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of 130,000 (predicted molecular weight, 123,122) and contains a series of motifs that are conserved in a number of ssDNA-binding proteins (45). The BALF2 protein binds to ssDNA preferentially and possesses helix-destabilizing property (42, 44). The BALF2 protein greatly enhances DNA synthesis catalyzed by the EBV BALF5 Pol catalytic subunit on a primed M13 ssDNA template, suggesting functional interaction between the EBV DNA polymerase and the EBV ssDNA-binding protein (44).
As noted above, the enzymatic activities of the BBLF4, BSLF1, and BBLF2/3 proteins have yet to be demonstrated, but the BBLF4 and BSLF1 gene products share 34 and 23% sequence identities with HSV-1 UL5 and UL52 gene products and are composed of 810 and 875 amino acids (predicted relative molecular weights of 89,795 and 97,972), respectively. Although the BBLF2/3 gene product has no significant similarity with HSV-1 UL8 gene product in overall identity, it has one stretch of 55 amino acids which are similar to the UL8 protein (11). The BBLF2/3 gene product is translated from one spliced RNA derived from the two open reading frames, BBLF2 and BBLF3, and is composed of 710 amino acids (predicted relative molecular weight of 78,176). Few studies characterizing the EBV putative helicase-primase have been performed. Gao et al. (14) have provided evidence of the BSLF1-BBLF4-BBLF2/3 complexes through immunofluorescence assays with nuclear retranslocation. They expressed the three BBLF4, BSLF1, and BBLF2/3 proteins fused to the Myc epitope in Vero cells by transfecting their expression vectors. When individually transfected, Myc-BBLF2/3 showed mixed nuclear and cytoplasmic staining, Myc-BSLF1 was perinuclear, and Myc-BBLF4 localized to the cytoplasm. The concurrent presence of the all three members resulted in nuclear localization of the BBLF4, BBLF2/3, and BSLF1 proteins, suggesting the existence of a BSLF1-BBLF4-BBLF2/3 complex. Recently we have directly demonstrated and confirmed the assembly of the BBLF4, BSLF1, and BBLF2/3 proteins in the lytic phase of B95-8 cells by immunoprecipitation analyses (47). Furthermore, we designed baculovirus expression systems for the BBLF4, BSLF1, and BBLF2/3 proteins to characterize the direct protein-protein interactions among these proteins in infected insect cells and could reproduce their complex formation in recombinant baculovirus expression systems. Experiments performed with double infection of pairs of recombinant viruses revealed that each component of the BBLF4-BSLF1-BBLF2/3 complex interacts directly with the other two (47).
The economy of proteins involved in replication of the EBV genome has made it an attractive model for dissecting the protein-protein interactions that are essential for coordination of the multiple reactions that occur at a replication fork. The specific interactions that occur among the six replication proteins appear to be essential for EBV DNA replication. In this report, we document our findings for direct interactions of the EBV DNA DNA polymerase and the EBV putative helicase-primase complex.
MATERIALS AND METHODS
Cells.
B95-8 cells, a marmoset lymphoblastoid cell line immortalized with a human EBV, were grown at 37°C in a 5% CO2 atmosphere in RPMI 1640 medium (Life Technologies, Inc.) supplemented with 40 μg of kanamycin per ml and 10% fetal calf serum. Spodoptera frugiperda (Sf9) and Trichoplusia ni (High Five) insect cells were grown at 27°C in Sf-900 IISFM and Express Five SFM media (Life Technologies), respectively, supplemented with 20 μg of gentamicin per ml and 10% fetal calf serum.
Preparation of the recombinant baculoviruses.
Construction of the recombinant baculoviruses AcBALF5, AcBMRF1, AcBALF2, AcBBLF4, AcBSLF1, and AcBBLF2/3, expressing BALF5, BMRF1, BALF2, BBLF4, BSLF1, and BBLF2/3 gene products, respectively, was described previously (39, 42, 43, 47). Stocks of recombinant viruses were prepared by infecting monolayers of Sf9 cells grown to a multiplicity of infection (MOI) of 0.1 PFU/cell. After incubation for 4 days, the cells were pelleted by centrifugation at 1,500 × g for 10 min at 4°C, and the virus in the supernatant fluid was either used directly or frozen at −80°C. The final viral titers were approximately 107 to 108 PFU/ml.
Antibodies.
The seven viral replication protein-specific antibodies (purified immunoglobulin G [IgG]) were used for immunoprecipitation and Western blotting analyses. BALF5 protein-specific rabbit antibody was produced against a truncated region of the EBV BALF5 protein containing a 3′-to-5′ exonuclease domain and a nucleotide binding domain (361 amino acids) as described previously (43). BMRF1 protein-specific mouse monoclonal antibody was purchased from NEN/Dupont Inc. (29). BZLF1 protein-specific mouse monoclonal antibody was purchased from DAKO Inc. BALF2 protein-specific antibody was produced against the carboxyl-terminal two-thirds of the EBV BALF2 protein (699 amino acids) as described previously (44). BSLF1c, BBLF4, and BBLF2/3w protein-specific rabbit antibodies were produced against the BSLF1 carboxy-terminal region (504 amino acids), the BBLF4 carboxy-terminal oligopeptide (20 amino acids), and whole BBLF2/3 gene products, respectively, as described previously (43, 47).
Coupling the anti-BBLF2/3w antibody to protein A beads.
The anti-BBLF2/3w antibody was coupled to protein A beads according to standard procedures (16). Briefly, 500 μl of protein A-agarose beads (Amersham Pharmacia Biotech Inc.) was mixed with 500 μg of the anti-BBLF2/3w IgG or normal rabbit IgG (DAKO) and gently rocked for 1 h at room temperature (RT). The protein A-antibody beads were washed four times with 5 ml of 0.2 M sodium borate adjusted to pH 9.0 and resuspended in 10 ml of 0.2 M sodium borate (pH 9.0). After addition of dimethylpimelidate dihydrochloride to a final concentration of 20 mM, the beads were gently rocked for 30 min at RT and washed twice with 10 ml of ethanolamine adjusted to pH 8.0. After being suspended in 10 ml of ethanolamine and rocked for 2 h at RT, the coupled beads were washed twice with 5 ml of distilled water and 0.5 ml of 0.1 M glycine-HCl (pH 2.5). The coupled beads were then suspended in 5 ml of phosphate-buffered saline (PBS) and stored at 4°C until used.
Preparation of lysate from B95-8 cells in the virus productive cycle.
B95-8 cells were seeded at 3 × 106 cells/ml in 30 ml of culture medium with 200 ng of phorbol 12-myristate 13-acetate, 5 mM sodium n-butyrate, and 1 μM calcium ionophore A23187 for induction of the EBV lytic replication cycle. At 72 h postinduction, the cells were harvested and washed with cold PBS. Pellets were resuspended in 5 ml of hypotonic buffer (50 mM Tris-HCl [pH 7.6], 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol [DTT], 10 μg each of pepstatin A and leupeptin per ml), stored on ice for 30 min, subjected to Dounce homogenization with 30 strokes, and then centrifuged at 1,500 × g for 10 min at 4°C. Nonidet P-40 (NP-40) and sodium chloride were added to the supernatant to final concentrations of 0.2% and 150 mM, respectively, followed by centrifugation at 18,000 × g for 5 min. The clarified lysate was confirmed to contain the BALF5, BMRF1, BALF2, BSLF1, BBLF4, and BBLF2/3 proteins as judged by Western blotting analyses using each of the protein-specific antibodies, frozen in liquid nitrogen, and then stored at −80°C until used for immunoprecipitation analyses.
Preparation of cell extracts from High Five cells infected with recombinant baculoviruses.
High Five cells were seeded at 106 cells per well in six-well plates and infected or coinfected with the indicated recombinant baculovirus(es) at an MOI of 5 PFU per cell for each virus. At 48 h postinfection, the cells were harvested, washed three times with cold PBS, resuspended in 1 ml of lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride, 1 mM DTT, 10 μg each of pepstatin A and leupeptin per ml), and incubated on ice for 20 min. The resultant lysates were centrifuged at 18,000 × g for 10 min at 4°C, and the supernatants were frozen in liquid nitrogen and stored at −80°C until used for immunoprecipitation analyses.
Immunoprecipitation analyses.
Immunoprecipitation analyses were performed according to standard procedures (30). The protein extracts were mixed with indicated antibodies and gently rocked for 1 h at 4°C. After addition of protein A-agarose beads (Amersham Pharmacia Biotech), the mixtures were further incubated for 1 h at 4°C, and then the antigen-antibody-protein A bead complexes were washed six times with 1 ml of NET gel buffer (50 mM Tris-HCl [pH 7.6], 500 mM NaCl, 1% NP-40, 1 mM EDTA). Each aliquot of the immunoprecipitated beads was suspended in 70 μl of SDS sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 0.1 M DTT, 10% glycerol, 0.01% bromphenol blue) and heated at 100°C for 6 min. After centrifugation at 18,000 × g for 5 min, the supernatants were subjected to SDS-PAGE (6 or 10% polyacrylamide) and Western blotting analyses, the latter performed as described previously (13). The protein blots transferred onto nitrocellulose membranes were incubated with the indicated first antibodies for 1 h at RT, subsequently probed with peroxidase-conjugated goat anti-rabbit IgG (Zymed Laboratories, Inc.) or peroxidase-conjugated rabbit anti-mouse IgG (Biolabs Inc.) for 1 h at RT, and then visualized by using enhanced chemiluminescence (NEN Inc.).
RESULTS
Detection of physical interaction between the EBV Pol holoenzyme and BBLF4-BSLF1-BBLF2/3 complex in virus-productive-phase B95-8 cells.
We have reported that EBV BBLF4, BSLF1, and BBLF2/3 replication proteins are induced and assembled to form a tripartite complex in B95-8 cells after induction of the lytic phase of EBV DNA replication (47). To investigate whether other EBV replication proteins interact with the BBLF4-BSLF1-BBLF2/3 complex in B95-8 cells in a virus productive cycle, immunoprecipitations combined with Western blotting analyses were performed. Antibodies specific for each of the six viral replication proteins which conceivably work together at replication forks of EBV were prepared. Anti-BSLF1c and anti-BBLF2/3w rabbit IgGs were used for immunoprecipitation analyses because they could precipitate effectively their target proteins but not any of the other viral proteins alone (see Fig. 4D). As a negative control, normal rabbit IgG was used.
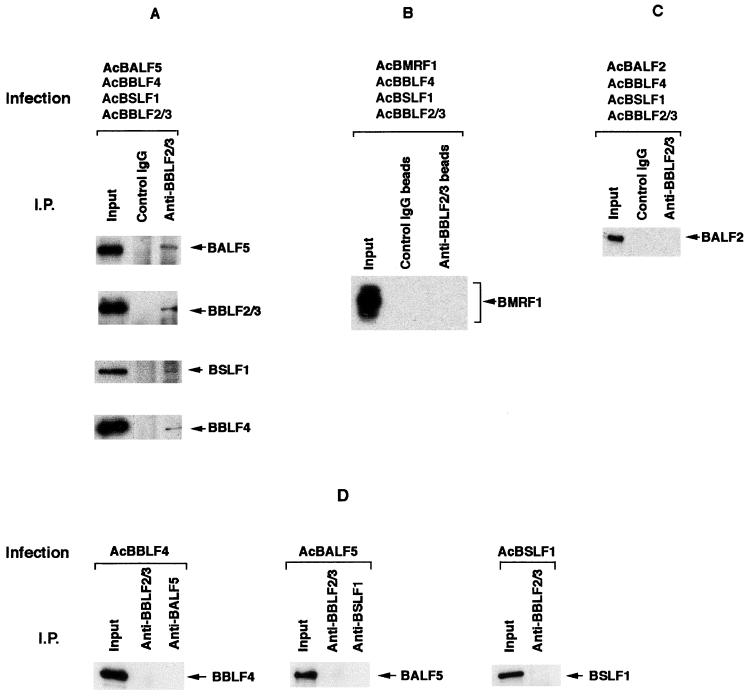
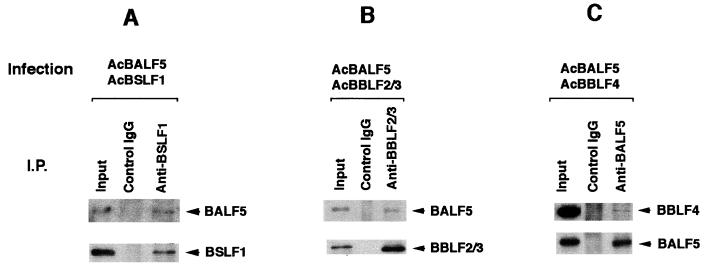
FIG. 4.
Direct interaction between the BALF5 Pol catalytic subunit and the BBLF4-BSLF1-BBLF2/3 complex in the recombinant baculovirus expression system. (A) High Five insect cells were superinfected with the recombinant baculoviruses AcBALF5, AcBBLF4, AcBSLF1, and AcBBLF2/3. Cell extracts were prepared as described in Materials and Methods and subjected to immunoprecipitation (I.P.) analyses. One microgram of the control rabbit IgG or anti-BBLF2/3w IgG was added to the lysates, and the mixtures were incubated in the presence of 150 mM NaCl and 1% NP-40. The antigen-antibody complexes were collected with 20 μg of protein A beads and washed six times with NET gel buffer containing 500 mM NaCl and 0.1% NP-40. Aliquots of the immunoprecipitated proteins (10 μl) and lysates (5 μl; Input) were resolved by SDS-PAGE and analyzed by Western blotting with the anti-BALF5, anti-BSLF1c, anti-BBLF2/3w, or anti-BBLF4 IgG. Immunoprecipitated proteins are indicated by arrowheads. (B) High Five insect cells were superinfected with the recombinant baculoviruses AcBMRF1, AcBBLF4, AcBSLF1, and AcBBLF2/3, and cell extracts were prepared and subjected to immunoprecipitation (I.P.) analyses. Five micrograms of control rabbit IgG or anti-BBLF2/3w IgG beads was added to the lysates, and the mixtures were incubated. The antigen-antibody-bead complexes were washed six times with NET gel buffer containing 500 mM NaCl and 1% NP-40. Aliquots of the immunoprecipitated proteins, (10 μl) and lysates (5 μl; Input) were resolved by SDS-PAGE and analyzed by Western blotting with the anti-BMRF1 mouse monoclonal IgG. (C) High Five cells were superinfected with the recombinant baculoviruses AcBALF2, AcBBLF4, AcBSLF1, and AcBBLF2/3, and cell extracts were prepared and subjected to immunoprecipitation analyses. One microgram of the control rabbit IgG or anti-BBLF2/3w IgG was added to the lysates, and the mixtures were incubated. The antigen-antibody complexes were collected with 20 μg of protein A beads and washed six times with NET gel buffer containing 500 mM NaCl and 1% NP-40. Aliquots of the immunoprecipitated proteins (10 μl) and lysates (5 μl; Input) were resolved by SDS-PAGE and analyzed by Western blotting with the anti-BALF2 IgG. (D) Lack of cross-reactivity of each IgG in immunoprecipitation analyses. High Five cells were infected with each of the indicated recombinant baculoviruses. The clarified lysates were prepared and subjected to immunoprecipitation analyses. One microgram of the indicated IgG was added to the lysates; the mixtures were incubated in the presence of 150 mM NaCl and 0.1% NP-40 and processed as described above. Aliquots of the immunoprecipitated proteins (10 μl) and lysates (5 μl; Input) were resolved by SDS-PAGE and analyzed by Western blotting with each of the indicated IgGs.
The EBV lytic phase of DNA replication was induced by treatment with a combination of chemical agents such as phorbol 12-myristate 13-acetate, sodium n-butyrate, and calcium ionophore A23187, as described previously (47). The cells were harvested at 72 h postinduction, and clarified lysates were prepared and confirmed to contain the six viral replication proteins (BALF5, BMRF1, BALF2, BBLF4, BSLF1, and BBLF2/3) and low amounts of BZLF1 origin-binding protein as judged by Western blotting analyses (Fig. 1; see also Fig. 3). The lysates were subjected to immunoprecipitation analysis with anti-BSLF1c or anti-BBLF2/3w IgG. The ternary antigen-antibody-protein A complexes were collected and washed extensively with NET gel buffer containing 500 mM NaCl and 1% NP-40, subjected to SDS-PAGE, and transferred onto nitrocellulose membranes. Then the protein blots were incubated with the indicated first antibodies.
FIG. 1.
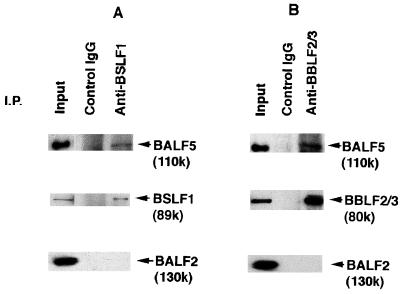
Coprecipitation of the BALF5 Pol catalytic subunit with the anti-BSLF1c or the anti-BBLF2/3w antibodies from clarified lysates of B95-8 cells in the virus productive cycle. Cells were harvested at 72 h postinduction, and 5-ml aliquots of clarified lysates were prepared as described in Materials and Methods. Then 400-μl aliquots of the lysate were subjected to immunoprecipitation (I.P.) analyses; 3-μg aliquots of the control rabbit IgG, the anti-BSLF1c IgG (A), or the anti-BBLF2/3w (B) IgG were added to the lysates, and the mixtures were gently rocked for 1 h at 4°C in the presence of 150 mM NaCl and 0.2% NP-40. The antigen-antibody complexes were collected with 20 μg of protein A beads, washed six times with NET gel buffer containing 500 mM NaCl and 1% NP-40, and suspended in 70 μl of sample buffer for SDS-PAGE. Aliquots of the immunoprecipitated proteins (10 μl) and lysates (5 μl; Input) were resolved by SDS-PAGE and analyzed by Western blotting with the anti-BALF5, anti-BSLF1c, anti-BALF2, or anti-BBLF2/3w IgG. Immunoprecipitated proteins are indicated by arrowheads.
FIG. 3.
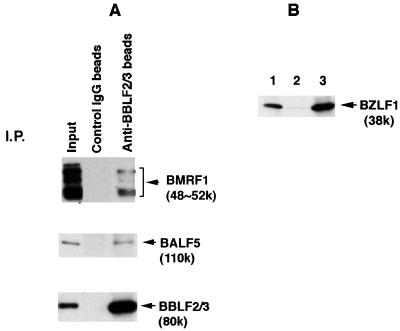
(A) Detection of the BMRF1 Pol accessory subunit with the BALF5 Pol catalytic subunit in the immunoprecipitated proteins by anti-BBLF2/3w IgG beads from lysates of B95-8 cells in the EBV productive cycle. Clarified lysates were prepared as described in Materials and Methods and subjected to immunoprecipitation (I.P.) analyses; 10 μg of control rabbit IgG or anti-BBLF2/3w IgG beads was added to the lysates, and the mixtures were incubated as described in the legend to Fig. 1. The antigen-antibody-bead complexes were washed six times with NET gel buffer containing 500 mM NaCl and 1% NP-40. Aliquots of the immunoprecipitated proteins (10 μl) and lysates (5 μl; Input) were resolved by SDS-PAGE and analyzed by Western blotting with the anti-BMRF1 monoclonal IgG, anti-BALF5 IgG, or anti-BBLF2/3w rabbit IgG. Immunoprecipitated proteins are indicated by arrowheads. (B) Fractionation of the BZLF1 protein expressed in B95-8 cells in the virus productive cycle. Cells were harvested at 72 h postinduction and homogenized as described in Materials and Methods. The whole cell lysates (5 ml) were centrifuged for fractionation into clarified lysates (5 ml) and pellets. The pellets were resuspended in 5 ml of sample buffer; 5-μl aliquots of each sample were resolved by SDS-PAGE and analyzed by Western blotting with the anti-BZLF1 protein mouse IgG. Lane 1, whole cell lysates; lane 2, clarified lysates; lane 3, pelleted fraction.
The results of immunoprecipitation analyses using the anti-BSLF1c IgG are shown in Fig. 1A. The control rabbit IgG could not precipitate any of viral replication proteins from the cell extract. In contrast, the anti-BSLF1c IgG coprecipitated the 110-kDa (110K) BALF5 Pol catalytic subunit with the corresponding 89K BSLF1 protein from the B95-8 cell extracts, although it did not precipitate the BALF5 protein directly (see Fig. 4D). The anti-BBLF2/3w IgG coimmunoprecipitated the BALF5 protein in addition to the corresponding 80K BBLF2/3 protein from the cell extract (Fig. 1B) while not precipitating the BALF5 protein directly (see Fig. 4D). Thus, we demonstrated that the BALF5 protein is associated with the BSLF1 and BBLF2/3 proteins. In contrast, neither the anti-BSLF1c nor the anti-BBLF2/3w antibody precipitated the BALF2 ssDNA-binding protein (Fig. 1). These results and our previous observation of a tripartite complex consisting of the BBLF4, BSLF1, and BBLF2/3 proteins (47) strongly suggest that the EBV BALF5 DNA catalytic subunit is associated with the BBLF4-BSLF1-BBLF2/3 complex in B95-8 cells after induction of the lytic phase of EBV DNA replication.
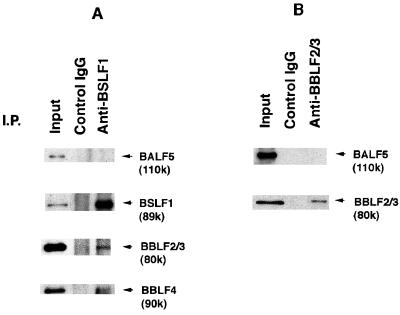
To ascertain the affinity of the BALF5 protein for the BBLF4-BSLF1-BBLF2/3 complex, the ternary antigen-antibody-protein A complexes were washed extensively under more stringent conditions with NET gel buffer containing 0.5 M NaCl, 1% NP-40, and 0.1% SDS. The anti-BSLF1c IgG could not precipitate the BALF5 Pol catalytic subunit but did coprecipitate the 80K BBLF2/3 and 90K BBLF4 proteins along with the 89K BSLF1 protein from the B95-8 cell lysate (Fig. 2A). Also, the anti-BBLF2/3w IgG did not precipitate the BALF5 protein, although the antibody could precipitate its target protein (Fig. 2B). These results indicate that the BBLF4, BSLF1, and BBLF2/3 proteins form a very tight tripartite complex. On the other hand, the association of the BALF5 protein with the BBLF4-BSLF1-BBLF2/3 complex was unstable to 0.1% SDS–500 mM NaCl–1% NP-40 but stable to 500 mM NaCl–1% NP-40.
FIG. 2.
Dissociation of the BALF5 protein from the BBLF4-BSLF1-BBLF2/3 complex. Clarified lysates from B95-8 cells in the virus productive cycle were prepared and subjected to immunoprecipitation (I.P.) analyses; 3 μg of the control rabbit IgG, the anti-BSLF1c IgG (A), or the anti-BBLF2/3w IgG (B) was added to the lysates, and the mixtures were incubated as described in the legend to Fig. 1. The antigen-antibody complexes were collected with 20 μg of protein A beads and washed six times under more stringent conditions with NET gel buffer containing 500 mM NaCl, 1% NP-40, and 0.1% SDS. Aliquots of the immunoprecipitated proteins (10 μl) and lysates (5 μl; Input) were resolved by SDS-PAGE and analyzed by Western blotting with the anti-BALF5, anti-BSLF1c, anti-BBLF2/3w, or anti-BBLF4 IgG. Immunoprecipitated proteins are indicated by arrowheads.
It is known that the BALF5 Pol catalytic subunit is functionally associated with the BMRF1 accessory subunit, acting as a Pol holoenzyme (19, 20, 39, 40, 41, 43). Furthermore, Zeng et al. (48) showed that anti-BALF5 antibody could precipitate the BMRF1 protein by immunoprecipitation analysis. To investigate whether the BBLF4-BSLF1-BBLF2/3 complex interacts with the BALF5 Pol catalytic subunit only or with the EBV Pol holoenzyme, it was determined whether the immunoprecipitated complex contains the BMRF1 Pol accessory subunit. In Western blotting analysis using the anti-BMRF1 monoclonal antibody, a thick band of the rabbit immunoglobulin heavy chain (molecular weight of about 50,000) used for immunoprecipitation analysis disturbed detection of the BMRF1 gene products (molecular weights of 48,000 to 52,000), since the peroxidase-conjugated anti-mouse IgG cross-reacted with rabbit immunoglobulin heavy chain. To avoid this problem, we prepared the BBLF2/3-specific IgG coupled to protein A-Sepharose beads (anti-BBLF2/3 IgG beads) and control rabbit IgG beads. As shown in Fig. 3, the control rabbit IgG beads could not precipitate any of the viral replication proteins from the cell extracts. Although the anti-BBLF2/3 IgG could not directly precipitate replication proteins other than its own protein (Fig. 4D), the anti-BBLF2/3 IgG beads not only precipitated the corresponding 80K BBLF2/3 protein but also coprecipitated both 48K to 52K BMRF1 and BALF5 proteins from the clarified cell extract. The mobilities of BMRF1 gene products in SDS-PAGE change dependent on the phosphorylation state (39).
Since the BZLF1 protein physically interacts with the BMRF1 protein (49) and the BBLF4-BSLF1-BBLF2/3 complex (14), it is possible that the BBLF2/3-specific IgG pulled down the BMRF1 Pol accessory protein via the BZLF1 protein. Therefore, we examined the level of the BZLF1 protein in clarified lysates of B95-8 cells with virus production. As shown by the Western blotting analyses in Fig. 3B, little of the BZLF1 protein was detected in the lysates but almost all it was fractionated into the insoluble fraction. Therefore, it is unlikely that the association of the BMRF1 protein with the BBLF4-BSLF1-BBLF2/3 complex is mediated by the BZLF1 protein. These observations strongly suggest that the EBV Pol holoenzyme is physically associated with the BBLF4-BSLF1-BBLF2/3 complex in B95-8 cells after induction of the lytic phase of EBV DNA replication.
Direct interaction between the BALF5 Pol catalytic subunit and the BBLF4-BSLF1-BBLF2/3 complex in High Five insect cells.
To determine which component of the EBV Pol holoenzyme interacts with the BBLF4-BSLF1-BBLF2/3 complex, we used recombinant baculovirus expression systems. High Five insect cells were infected alone or superinfected with the AcBALF5, AcBBLF4, AcBSLF1, and AcBBLF2/3 recombinant baculoviruses at an MOI of 5 PFU per cell for each virus and harvested 48 h postinfection. Clarified cell lysates were prepared, and immunoprecipitation analyses were performed. The anti-BBLF2/3w IgG could immunoprecipitate the corresponding BBLF2/3 protein but did not cross-react with the BALF5 Pol catalytic subunit or with the BBLF4 and BSLF1 proteins (Fig. 4D). When insect cells were superinfected with the four recombinant baculoviruses, the anti-BBLF2/3w IgG precipitated all of the BALF5, BSLF1, BBLF2/3, and BBLF4 proteins (Fig. 4A). This was not the case with the control rabbit IgG. On the other hand, immunoprecipitation analysis of the extracts from cells superinfected with the four recombinant baculoviruses AcBMRF1, AcBSLF1, AcBBLF2/3, and AcBBLF4 revealed that the anti-BBLF2/3w IgG beads could not precipitate the BMRF1 Pol accessory subunit (Fig. 4B). These observations clearly indicate that the BALF5 Pol catalytic subunit directly interacts with the BBLF4-BSLF1-BBLF2/3 complex and the BMRF1 Pol accessory subunit does not. In Fig. 3, immunoprecipitation of the BBLF2/3 protein brought down the BMRF1 protein. Thus, the BMRF1 protein appears to be associated with the BBLF4-BSLF1-BBLF2/3 complex via the BALF5 Pol catalytic subunit.
Next, we tried to show whether the interaction of the BALF2 ssDNA-binding protein and the BBLF4-BSLF1-BBLF2/3 protein complex occurs in the recombinant baculovirus expression system. As expected from experiments with B95-8 cell lysates, the anti-BBLF2/3 IgG did not precipitate the BALF2 protein from the lysates of cells superinfected with the AcBALF2, AcBBLF4, AcBSLF1, and AcBBLF2/3 recombinant viruses (Fig. 4C). Under the immunoprecipitation conditions used, the BALF2 ssDNA-binding protein is therefore unlikely to interact with the BBLF4-BSLF1-BBLF2/3 complex.
The BALF5 Pol catalytic subunit directly interacts with each component of the BBLF4-BSLF1-BBLF2/3 complex.
To determine how the BALF5 protein interacts with the BBLF4-BSLF1-BBLF2/3 complex, pairwise interactions were examined. Extracts from the insect cells doubly infected with AcBALF5 and either AcBBLF4, AcBSLF1, or AcBBLF2/3 were prepared and subjected to immunoprecipitation analyses using the anti-BSLF1c IgG, anti-BBLF2/3w IgG, or anti-BALF5 IgG (Fig. 5). The anti-BSLF1c IgG coprecipitated the BALF5 protein from the lysate of cells infected with AcBALF5 and AcBSLF1 (Fig. 5A) but did not cross-react with the BALF5 protein (Fig. 4D). Also, the anti-BBLF2/3w IgG could coprecipitate the BALF5 protein from the lysate of cells infected with AcBALF5 and AcBBLF2/3 (Fig. 5B). When the cells were doubly infected with AcBALF5 and AcBBLF4, the anti-BALF5 IgG precipitated the BBLF4 protein as well as the BALF5 protein (Fig. 5C), although the antibody did not react directly with the BBLF4 protein (Fig. 4D). Thus, we could demonstrate BALF5-BBLF4, BALF5-BSLF1, and BALF5-BBLF2/3 subassemblies in the doubly infected insect cells. These results strongly suggest that the BALF5 protein interacts directly with each component of the EBV BBLF4-BSLF1-BBLF2/3 complex in vivo.
FIG. 5.
Complex formation between the BALF5 Pol catalytic subunit and each component of the BBLF4-BSLF1-BBLF2/3 complex. High Five insect cells were doubly infected with AcBALF5 and AcBSLF1 (A), AcBBLF2/3 (B), or AcBBLF4 (C). Cell extracts were subjected to immunoprecipitation (I.P.) analyses using 1 μg of the control rabbit IgG, anti-BSLF1c IgG, anti-BBLF2/3w IgG, or anti-BALF5 IgG. The antigen-antibody complexes were collected with 20 μg of protein A beads and washed with NET gel buffer containing 500 mM NaCl and 1% NP-40. Aliquots of the immunoprecipitated proteins (10 μl) and lysates (5 μl; Input) were resolved by SDS-PAGE and analyzed by Western blotting with the anti-BALF5, anti-BSLF1c, anti-BBLF2/3, or anti-BBLF4 IgG. Immunoprecipitated proteins are indicated by arrowheads.
DISCUSSION
The multiple steps essential for DNA replication are catalyzed by a number of proteins whose enzymatic reactions must be closely coordinated. This is most apparent at the replication fork, where both leading- and lagging-strand synthesis must occur simultaneously for movement of the replication fork. Therefore, it is not surprising that replication proteins are frequently isolated as complexes or interact physically with one another, suggesting that their individual enzymatic reactions are coordinated via physical associations. The six viral proteins that account for the basic reactions at the EBV replication fork are thought to be the EBV Pol holoenzyme consisting of the BALF5 Pol catalytic subunit and the BMRF1 Pol accessory subunit, the BALF2 ssDNA-binding protein, and the EBV putative helicase-primase complex consisting of the BBLF4, BSLF1, and BBLF2/3 proteins. The specific interactions that occur among these relatively few proteins appear to be essential for EBV DNA replication. In this report we have presented evidence of a novel physical interaction between the EBV DNA Pol catalytic subunit and the EBV putative helicase-primase complex. Gao et al. have recently reported that the BALF2 protein may interact with the BBLF4-BSLF1-BBLF2/3 complex (14). Under the same immunoprecipitation conditions, however, we were not able to detect any physical interaction between the BALF2 ssDNA-binding protein and the BBLF4-BSLF1-BBLF2/3 complex, indicating that the BALF5 Pol catalytic subunit-BBLF4-BSLF1-BBLF2/3 complex interaction is considerably stronger than that between the BALF2 protein and the BBLF4-BSLF1-BBLF2/3 complex. Our findings warrant particular attention because these interactions are thought to coordinate leading- and lagging-strand DNA synthesis at the EBV replication fork.
Although the immune complexes from the clarified lysate of B95-8 cells were thoroughly washed with 500 mM NaCl and 1% NP-40, it is possible that the interaction between the EBV DNA polymerase and the BBLF4-BSLF1-BBLF2/3 complex was mediated by DNA, even if the experiments were carried out in the presence of DNase. However, in the experiments with lysates from insect cells superinfected with AcBALF5, AcBBLF4, AcBSLF1, and AcBBLF2/3, the anti-BBLF2/3w IgG precipitated all of the BALF5, BSLF1, BBLF4, and BBLF2/3 proteins (Fig. 4A). Under the washing conditions used (500 mM NaCl and 1% NP-40), the BALF5 protein by itself is unable to bind DNA and exhibits no DNA polymerase activity (41, 43). Therefore, it is reasonable to assume that the BALF5 Pol catalytic subunit directly interacts with the BBLF4-BSLF1-BBLF2/3 complex. To demonstrate the direct interaction between them, domain analyses will be needed.
It is believed that helicases translocate along ssDNA by forming oligomeric structures, either dimer, hexamer, or multiprotein complexes (23, 24). The Rep protein of Escherichia coli, also a member of superfamily I, is believed to form a dimer at the replication fork (4), whereas the helicases of T4 and T7 bacteriophages and simian virus 40 apparently form hexamers (10, 17, 28, 34). UL5 (BBLF4 analogue), UL52 (BSLF1 analogue), and UL8 (BBLF2/3 analogue) are three of the seven genes that are essential for HSV-1 DNA replication. The products of these three genes form a heterotrimeric complex with helicase and primase activities (6, 7, 9). The helicase activity presumably acts to unwind duplex DNA ahead of the progressing replication fork, thereby producing the open configuration needed for both continuous and discontinuous strand synthesis. By analogy to other primases, the primase of HSV-1 presumably initiates discontinuous DNA synthesis on the lagging strand by providing the oligonucleotide primers that are elongated by the HSV-1 DNA polymerase. Although the enzymatic activities of the EBV BBLF4-BSLF1-BBLF2/3 heterotrimeric complex have yet to be demonstrated, we assume that the BBLF4-BSLF1-BBLF2/3 complex may act as a helicase and primase like the HSV-1 UL5-UL52-UL8 complex.
Polymerase and helicase-primase complex interactions have also been observed in other replication systems. In the case of bacteriophage T7, gene 5 DNA polymerase interacts with the gene 4 helicase-primase via its carboxyl terminus (26, 27), playing an important role in its coordination of leading- and lagging-strand DNA synthesis at the replication fork (8). Also, the catalytic subunit of the HSV-1 DNA polymerase interacts with the carboxyl terminus of the UL8 protein of the HSV-1 helicase-primase heterotrimeric complex (25). Considering these observations, the interaction of the EBV DNA polymerase with the helicase-primase complex might be central to the coordination of the leading- and lagging-strand synthesis at the replication fork of EBV.
We have demonstrated that each component of the EBV BBLF4-BSLF1-BBLF2/3 complex can interact directly with the BALF5 protein. It is not clear whether a single component of the BBLF4-BSLF1-BBLF2/3 complex and the BALF5 subunit of the Pol holoenzyme are in constant contact at the replication fork or whether the interaction is dispersed over a contiguous region of the complex. If the complex rotates around the DNA axis as it translocates and unwinds double-stranded DNA, the interaction with the BALF5 protein may be changing sequentially from one component in the heterotrimer to the next to relieve torsional strain. In this case, the BALF5 protein is probably not in constant contact with a single component of the BBLF4-BSLF1-BBLF2/3 complex. If, on the other hand, the complex moves along the DNA without any relative rotation, a constant interaction between the complex and the polymerase may be maintained as a stable complex. Structural studies are required to determine where the BBLF4-BSLF1-BBLF2/3 complex is located and how it is oriented relative to the DNA polymerase at the replication fork.
The interaction of the helicase-primase and DNA polymerase is required not only for strand displacement DNA synthesis but also for priming DNA synthesis on the lagging strand of the replication fork. Little is known about DNA primase-DNA polymerase interactions involved in the transition from RNA to DNA synthesis. The bacterophage T7 gene 4A protein catalyzes the synthesis of tetraribonucleotides at specific sequences on ssDNA in a template-mediated reaction (36). These tetraribonucleotides are stabilized on the template by gene 4A protein until T7 DNA polymerase can use them as primers to initiate DNA synthesis (26). Such short oligonucleotides prime T7 DNA polymerase extremely poorly in the absence of the gene 4 protein (32, 33). The effective use of these tetraribonucleotides by T7 DNA polymerase in the presence of the gene 4 protein implies that a specific protein-protein interaction is required. In the case of bacteriophage T4, a complex of two proteins encoded by genes 41 and 61 is required to catalyze efficiently the synthesis of the pentaribonucleotide pppACN3, which primes DNA synthesis by T4 DNA polymerase on ssDNA (22). In a variety of eukaryotic systems, DNA polymerase α has been purified in a tight complex with DNA primase (3, 5, 35). Interaction between DNA primases and polymerases may in general play an important role in promoting efficient transition from RNA primer to DNA synthesis on lagging-strand DNA synthesis. Thus, the interaction of the BBLF4-BSLF1-BBLF2/3 complex and the EBV DNA polymerase at the replication fork may be an important aspect of the replication process and a possible new target for antiviral agents.
Considering other DNA replication systems, it is likely that initiation of lytic-phase EBV DNA replication involves the formation of an initiation complex at oriLyt. The first step in this process would be the binding of the BZLF1 protein to its recognition sequences within the EBV replication origin, oriLyt, to form an initial complex. The interaction of the BZLF1 protein with the BBLF4-BSLF1-BBLF2/3 complex reported by Gao et al. (14) supposes that the BZLF1 protein recruits the viral helicase-primase complex to oriLyt. The BALF2 ssDNA-binding protein appears to interact with the prepriming complex consisting of the BZLF1-BBLF4-BSLF1-BBLF2/3 complex (14). These proteins together therefore would have the potential to open up the duplex DNA in the origin region and synthesize RNA primers. The interaction between the EBV Pol holoenzyme and the BBLF4-BSLF1-BBLF2/3 complex which we have now identified may play an important role in bringing the viral polymerase into the prepriming complex to initiate DNA synthesis. It is possible, for example, that binding of the EBV DNA polymerase to the BBLF4-BSLF1-BBLF2/3 complex reduces the affinity of the BBLF4-BSLF1-BBLF2/3 complex for BZLF1, allowing the polymerase-helicase-primase complex to migrate away from oriLyt to the replication forks.
ACKNOWLEDGMENTS
We thank M. Hirata, C. Yamada, and T. Yoshida for technical assistance.
This work was supported by grants-in-aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan (11138268 to T.T.) and partly by JSPS-RFTF 97L00703.
REFERENCES
- 1.Aiyar A, Tyree C, Sugden B. The plasmid replication of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 1998;17:6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Séguin C, Tuffnell P C, Barrell B G. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 3.Banks G R, Boezi J A, Lehman I R. A high molecular weight DNA polymerase from Drosophila melanogaster embryos. J Biol Chem. 1979;254:9886–9892. [PubMed] [Google Scholar]
- 4.Chao K, Lohman T M. DNA-induced dimerization of the Escherichia coli Rep helicase. J Mol Biol. 1991;221:1165–1181. doi: 10.1016/0022-2836(91)90926-w. [DOI] [PubMed] [Google Scholar]
- 5.Conaway R C, Lehman I R. A DNA primase activity associated with DNA polymerase a from Drosophila melanogaster embryos. Proc Natl Acad Sci USA. 1982;79:2523–2527. doi: 10.1073/pnas.79.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crute J J, Mocarski E S, Lehman I R. A DNA helicase induced by herpes simplex virus type 1. Nucleic Acids Res. 1988;16:6585–6596. doi: 10.1093/nar/16.14.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crute J J, Tsurumi T, Zhu L, Weller S K, Olivo P D, Challberg M D, Mocarski E S, Lehman I R. Herpes simplex virus 1 helicase-primase: a complex of three herpes-encoded gene products. Proc Natl Acad Sci USA. 1989;86:2186–2189. doi: 10.1073/pnas.86.7.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Debyser Z, Tabor S, Richardson C C. Coordination of leading and lagging strand DNA synthesis at the replication fork of bacteriophage T7. Cell. 1994;77:157–166. doi: 10.1016/0092-8674(94)90243-7. [DOI] [PubMed] [Google Scholar]
- 9.Dodson M S, Crute J J, Bruckner R C, Lehman I R. Overexpression and assembly of the herpes simplex virus type 1 helicase-primase in insect cells. J Biol Chem. 1989;264:20853–20838. [PubMed] [Google Scholar]
- 10.Dong F, von Hippel P H. The ATP-activated hexameric helicase of bacteriophage T4 (gp41) forms a stable primosome with a single subunit of T4-coded primase (gp61) J Biol Chem. 1996;271:19625–19631. doi: 10.1074/jbc.271.32.19625. [DOI] [PubMed] [Google Scholar]
- 11.Fixman E D, Hayward G S, Hayward S D. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J Virol. 1992;66:5030–5039. doi: 10.1128/jvi.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fixman E D, Hayward G S, Hayward S D. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J Virol. 1995;69:2998–3006. doi: 10.1128/jvi.69.5.2998-3006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita M, Yamada C, Goto H, Yokoyama N, Kuzushima K, Inagaki M, Tsurumi T. Cell cycle regulation of human CDC6 Protein: intracellular localization, interaction with the human MCM complex, and CDC2 kinase-mediated hyperphosphorylation. J Biol Chem. 1999;274:25927–25932. doi: 10.1074/jbc.274.36.25927. [DOI] [PubMed] [Google Scholar]
- 14.Gao Z, Krithivas A, Finan J E, Semmes O J, Zhou S, Wang Y, Hayward S D. The Epstein-Barr virus lytic transactivator Zta interacts with the helicase-primase replication proteins. J Virol. 1998;72:8559–8567. doi: 10.1128/jvi.72.11.8559-8567.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 16.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 17.Joo W S, Kim H Y, Purviance J D, Sreekumar K R, Bullock P A. Assembly of T-antigen double hexamers on the simian virus 40 core origin requires only a subset of the available binding sites. Mol Cell Biol. 1998;18:2677–2687. doi: 10.1128/mcb.18.5.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kallin B, Sternås L, Saemundssen A K, Luka J, Jörnvall H, Eriksson B, Tao P-Z, Nilsson M T, Klein G. Purification of Epstein-Barr virus DNA polymerase from P3HR-1 cells. J Virol. 1985;54:561–568. doi: 10.1128/jvi.54.2.561-568.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiehl A, Dorsky D I. Bipartite DNA-binding region of the Epstein-Barr virus BMRF1 product essential for DNA polymerase accessory function. J Virol. 1995;69:1669–1677. doi: 10.1128/jvi.69.3.1669-1677.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiehl A, Dorsky D I. Cooperation of EBV DNA polymerase and EA-D (BMRF1) in vitro and colocalization in nuclei of infected cells. Virology. 1991;184:330–340. doi: 10.1016/0042-6822(91)90849-7. [DOI] [PubMed] [Google Scholar]
- 21.Li J-S, Zhou B-S, Dutschman G E, Grill S P, Tan R-S, Cheng Y-C. Association of Epstein-Barr virus early antigen diffuse component and virus-specified DNA polymerase activity. J Virol. 1987;61:2947–2949. doi: 10.1128/jvi.61.9.2947-2949.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C-C, Alberts B M. Pentaribonucleotides of mixed sequence are synthesized and efficiently prime de novo DNA chain starts in the T4 bacteriophage DNA replication system. Proc Natl Acad Sci USA. 1980;77:5698–5703. doi: 10.1073/pnas.77.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohman T M. Helicase-catalyzed DNA unwinding. J Biol Chem. 1993;268:2269–2272. [PubMed] [Google Scholar]
- 24.Lohman T M, Bijornson K P. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 25.Marsden H S, McLean G W, Barnard E C, Francis G J, MacEachran K, Murphy M, McVey G, Cross A, Abbotts A P, Stow N D. The catalytic subunit of the DNA polymerase of herpes simplex virus type 1 interacts specifically with the C terminus of the UL8 component of the viral helicase-primase complex. J Virol. 1997;71:6390–6397. doi: 10.1128/jvi.71.9.6390-6397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakai H, Richardson C C. Interactions of the DNA polymerase and gene 4 protein of bacteriophage T7. J Biol Chem. 1986;261:15208–15216. [PubMed] [Google Scholar]
- 27.Notarnicola S M, Mulcahy H L, Lee J, Richardson C C. The acidic carboxyl terminus of the bacteriophage T7 gene 4 helicase/primase interacts with T7 DNA polymerase. J Biol Chem. 1997;272:18425–18433. doi: 10.1074/jbc.272.29.18425. [DOI] [PubMed] [Google Scholar]
- 28.Patel S S, Hingorani M M. Oligomeric structure of bacteriophage T7 DNA primase/helicase proteins. J Biol Chem. 1993;268:10668–10675. [PubMed] [Google Scholar]
- 29.Pearson G R, Vroman B, Chase B, Sculley T, Hummel M, Kieff E. Identification of polypeptide components of the Epstein-Barr virus early antigen complex with monoclonal antibodies. J Virol. 1983;47:193–201. doi: 10.1128/jvi.47.1.193-201.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Schepers A, Pich D, Hammerschmidt W. Activation of oriLyt, the lytic origin of DNA replication of Epstein-Barr virus, by BZLF1. Virology. 1996;220:367–376. doi: 10.1006/viro.1996.0325. [DOI] [PubMed] [Google Scholar]
- 32.Scherzinger E, Lanka E, Hillenbrand G. Role of bacteriophage T7 DNA primase in the initiation of DNA strand synthesis. Nucleic Acids Res. 1977;4:4151–4163. doi: 10.1093/nar/4.12.4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scherzinger E, Lanka E, Morelli G, Seiffert D, Yuki A. Bacteriophage-T7-induced DNA-priming protein. Eur J Biochem. 1977;72:543–558. doi: 10.1111/j.1432-1033.1977.tb11278.x. [DOI] [PubMed] [Google Scholar]
- 34.Smelkova N V, Borowiec J A. Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J Virol. 1997;71:8766–8773. doi: 10.1128/jvi.71.11.8766-8773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stadlbauer F, Brueckner A, Rehfuess C, Eckerskorn C, Lottspeich F, Förster V, Tseng B Y, Nasheuer H-P. DNA replication in vitro by recombinant DNA-polymerase-a-primase. Eur J Biochem. 1994;222:781–793. doi: 10.1111/j.1432-1033.1994.tb18925.x. [DOI] [PubMed] [Google Scholar]
- 36.Tabor S, Richardson C C. Template recognition sequence for RNA primer synthesis by gene 4 protein of bacteriophage T7. Proc Natl Acad Sci USA. 1981;78:205–209. doi: 10.1073/pnas.78.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsurumi T. Characterization of 3′-to-5′ exonuclease activity associated with Epstein-Barr virus DNA polymerase. Virology. 1991;182:376–381. doi: 10.1016/0042-6822(91)90685-5. [DOI] [PubMed] [Google Scholar]
- 38.Tsurumi T. Primer terminus recognition and highly processive replication by Epstein-Barr virus DNA polymerase. Biochem J. 1991;280:703–708. doi: 10.1042/bj2800703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsurumi T. Purification and characterization of the DNA-binding activity of the Epstein-Barr virus DNA polymerase accessory protein BMRF1 gene products, as expressed in insect cells by using the baculovirus system. J Virol. 1993;67:1681–1687. doi: 10.1128/jvi.67.3.1681-1687.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsurumi T, Daikoku T, Kurachi R, Nishiyama Y. Functional interaction between Epstein-Barr virus DNA polymerase catalytic subunit and its accessory subunit in vitro. J Virol. 1993;67:7648–7653. doi: 10.1128/jvi.67.12.7648-7653.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsurumi T, Daikoku T, Nishiyama Y. Further characterization of the interaction between the Epstein-Barr virus DNA polymerase catalytic subunit and its accessory subunit with regard to the 3′-to-5′ exonucleolytic activity and stability of initiation complex at primer terminus. J Virol. 1994;68:3354–3363. doi: 10.1128/jvi.68.5.3354-3363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsurumi T, Kishore J, Yokoyama N, Fujita M, Daikoku T, Yamada H, Yamashita Y, Nishiyama Y. Overexpression, purification and helix-destabilizing properties of Epstein-Barr virus ssDNA-binding protein. J Gen Virol. 1998;79:1257–1264. doi: 10.1099/0022-1317-79-5-1257. [DOI] [PubMed] [Google Scholar]
- 43.Tsurumi T, Kobayashi A, Tamai K, Daikoku T, Kurachi R, Nishiyama Y. Functional expression and characterization of the Epstein-Barr virus DNA polymerase catalytic subunit. J Virol. 1993;67:4651–4658. doi: 10.1128/jvi.67.8.4651-4658.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsurumi T, Kobayashi A, Tamai K, Yamada H, Daikoku T, Yamashita Y, Nishiyama Y. Epstein-Barr virus single-stranded DNA-binding protein: purification, characterization, and action on DNA synthesis by the viral DNA polymerase. Virology. 1996;222:352–364. doi: 10.1006/viro.1996.0432. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Hall J D. Characterization of a major DNA-binding domain in the herpes simplex type 1 DNA-binding protein (ICP8) J Virol. 1990;64:2082–2089. doi: 10.1128/jvi.64.5.2082-2089.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yates J L, Guan N. Epstein-Barr virus-derived plasmids replication only per cell cycle and not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yokoyama N, Fujii K, Hirata M, Tamai K, Kiyono T, Kuzushima K, Nishiyama Y, Fujita M, Tsurumi T. Assembly of the Epstein-Barr virus BBLF4, BSLF1, BBLF2/3 proteins and their interactive properties. J Gen Virol. 1999;80:2879–2887. doi: 10.1099/0022-1317-80-11-2879. [DOI] [PubMed] [Google Scholar]
- 48.Zeng Y, Middeldorp J, Madjar J, Ooka T. A major DNA binding protein encoded by BALF2 open reading frame of Epstein-Barr virus (EBV) forms a complex with other EBV DNA-binding proteins: DNAase, EA-D, and DNA polymerase. Virology. 1997;239:285–295. doi: 10.1006/viro.1997.8891. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Q, Hong Y, Dorsky D, Holley-Guthrie E, Zalani S, Elshiekh N A, Kiehl A, Le T, Kenney S. Functional and physical interactions between the Epstein-Barr virus (EBV) proteins BZLF1 and BMRF1: effects on EBV transcription and lytic replication. J Virol. 1996;70:5131–5142. doi: 10.1128/jvi.70.8.5131-5142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]