Abstract
Efficient infection with adenovirus (Ad) vectors based on serotype 5 (Ad5) requires the presence of coxsackievirus-adenovirus receptors (CAR) and αv integrins on cells. The paucity of these cellular receptors is thought to be a limiting factor for Ad gene transfer into hematopoietic stem cells. In a systematic approach, we screened different Ad serotypes for interaction with noncycling human CD34+ cells and K562 cells on the level of virus attachment, internalization, and replication. From these studies, serotype 35 emerged as the variant with the highest tropism for CD34+ cells. A chimeric vector (Ad5GFP/F35) was generated which contained the short-shafted Ad35 fiber incorporated into an Ad5 capsid. This substitution was sufficient to transplant all infection properties from Ad35 to the chimeric vector. The retargeted, chimeric vector attached to a receptor different from CAR and entered cells by an αv integrin-independent pathway. In transduction studies, Ad5GFP/F35 expressed green fluorescent protein (GFP) in 54% of CD34+ cells. In comparison, the standard Ad5GFP vector conferred GFP expression to only 25% of CD34+ cells. Importantly, Ad5GFP transduction, but not Ad5GFP/F35, was restricted to a specific subset of CD34+ cells expressing αv integrins. The actual transduction efficiency was even higher than 50% because Ad5GFP/F35 viral genomes were found in GFP-negative CD34+ cell fractions, indicating that the cytomegalovirus promoter used for transgene expression was not active in all transduced cells. The chimeric vector allowed for gene transfer into a broader spectrum of CD34+ cells, including subsets with potential stem cell capacity. Fifty-five percent of CD34+ c-Kit+ cells expressed GFP after infection with Ad5GFP/F35, whereas only 13% of CD34+ c-Kit+ cells were GFP positive after infection with Ad5GFP. These findings represent the basis for studies aimed toward stable gene transfer into hematopoietic stem cells.
Human hematopoietic stem cells (HSCs) represent an important target for gene therapy. However, CD34+-enriched human bone marrow cells can be only poorly transduced by the most commonly used viral vectors. HSCs are believed to be in a quiescent state and, when induced to divide, tend to lose their stem cell capacity (4, 7, 35). Another limiting factor for viral gene transfer into HSCs is the scarcity of corresponding cellular receptors for virus binding and/or internalization (14, 53).
Key features which make recombinant adenoviruses (Ad) an attractive vehicle for gene transfer into hematopoietic cells include the ability to easily prepare high-titer stocks of purified virus, the remarkable efficiency of each step in the Ad cell/nucleus entry process leading to high-level gene expression, and the ability to transport its double-stranded DNA genome into the nucleus, allowing for transduction of nondividing cells. However, transduction with first-generation, E1/E3-deleted recombinant Ad vectors is associated with toxicity and immune responses against Ad proteins expressed in transduced cells, limiting the duration of transgene expression. Other important disadvantages include the episomal status of Ad DNA in transduced cells and the restricted tropism of recombinant Ad vectors which are based on the well-characterized, nontumorigenic serotype 5 (Ad5) (24).
Attachment to the cell surface of Ad5 is mediated by its fiber protein (11; for a review, see reference 64). The fiber molecule is a homotrimer forming 12 vertices per virion. The distal, C-terminal domain of the trimeric fiber molecule terminates in a knob, which binds with high affinity (Ka = 109 to 1010 M−1 per site) to a specific primary receptor identified recently as the coxsackievirus B-adenovirus receptor (CAR) (3). After binding, Arg-Gly-Asp (RGD) motifs in the penton base interact with cellular integrins of the αvβ3 and αvβ5 types which function as secondary Ad5 receptors (77). This interaction triggers cellular internalization whereby the virion resides within the endosome. The endosomal membrane is lysed in a process mediated by the penton base, releasing the contents of the endosome to the cytoplasm. During these processes, the virion is gradually uncoated and the Ad DNA is transported into the nucleus.
The efficiency of Ad5 infection depends on CAR and integrin density (21, 76). Binding of virions occurs with increasing cooperativity, which permits the virion to bind to several receptors simultaneously. It is known that Ad5-based vectors can infect cells that lack CAR and/or αv integrin expression when virus is applied at very high multiplicities of infection (MOIs). Furthermore, fiberless particles demonstrate infectivity (36). In both cases, low-affinity interactions, e.g., between penton base or hexon with cell surface proteins or receptors, may be utilized as alternative cell entry strategies. Importantly, infection with high MOIs is associated with cytotoxicity and immunogenicity in vivo and is therefore not practical for gene therapy approaches.
Due to the lack of corresponding primary and/or secondary receptors, Ad5 gene transfer is inefficient to a number of tissues, such as endothelia (70, 78), smooth muscle (78), differentiated airway epithelia (82), brain tissue (10), and peripheral blood cells (73). There are controversial reports with regards to the ability of Ad5-based vectors to transduce hematopoietic cells. A number of investigators noted that bone marrow cells are refractory to Ad5 vector infection and used this property to purge cancer cells from bone marrow (9, 12, 27, 30, 80). On the other hand, it has been shown that Ad5 vectors are capable of infecting CD34+ cells (5, 6, 16, 19, 46, 50, 74). Drawing an overall conclusion on Ad5 vector transduction of CD34+ cells is complicated by the fact that culture conditions, proliferative status of CD34+ cells, transgene promoter, and viral doses significantly differed between studies. In some of these studies, only MOIs of 500 to 1,000 PFU/cell (6, 50) allowed for transduction; in other studies, these same MOIs were associated with severe cytotoxicity (5, 19, 46). Furthermore, pretreatment of CD34+ cells with cytokines prior to infection (5, 74) may have changed the spectrum of potential viral receptors (2, 22, 28).
Since the cell types that can be infected with Ad5 vectors are restricted by the presence of CAR and αv integrins, attempts were made to broaden the tropism of Ad vectors. These approaches included complexing Ad with lipids or polycations (6, 56), using bispecific antibodies directed against Ad fiber and an internalizing cellular receptor (75, 78), and engineering peptide ligands into the fiber (31, 45, 76), hexon (13), or penton (79). Most of these approaches did not abrogate CAR tropism. The most commonly used method to retarget Ad tropism is by swapping part or all of the fiber from one serotype to another (10, 32, 70). There are at least 49 known Ad serotypes, which have been classified into six subgroups (A to F), based in part on amino acid sequence homology, hemagglutination pattern, and oncogenic potential. The various serotypes of human Ad show differences in tissue tropism and cause different pathologies mostly localized to the gastrointestinal tract, respiratory tract, genitourinary tract, or conjunctiva and cornea (25). Since the nucleotide and amino acid sequences of fibers differ significantly among different Ad serotypes, it has been suggested that they can recognize different receptors (1). This implies that fiber substitution would be sufficient to change virus tropism and has been convincingly shown for Ad5-Ad3 chimeras (70), Ad5-Ad7 chimeras (20), and Ad5-Ad17 fiber chimeras (10). In these studies, the Ad5 fiber or fiber knob was genetically substituted by heterologous sequences derived from the other serotype.
Previously, we have demonstrated that inverted repeat sequences inserted into first-generation Ad vector genomes mediate precise genomic rearrangements resulting in vector genomes devoid of all viral genes that are efficiently packaged into functional Ad capsids (68). As a specific application of these replication derivatives, we developed an integrating ΔAd.AAV hybrid vector devoid of all viral genes (40). The ΔAd.AAV genome contains two adeno-associated virus (AAV) inverted terminal repeats—elements that stimulated transgene integration into cellular DNA with a frequency comparable to that of recombinant AAV. So far, these vectors were based on Ad5-derived capsids, which may limit their application for gene transfer into HSCs. Our long-term goal is to adapt integrating, gutless ΔAd.AAV hybrid vectors for stable gene transfer into HSCs by modifying their tropism. On the way to reach this goal, the present study tested the ability of selected Ad serotypes to interact with and transduce human CD34+ cells. Based on these data, we generated a chimeric, first-generation Ad vector containing heterologous fiber molecules, which allowed for efficient transduction of human CD34+ cells with potential stem cell capacity.
MATERIALS AND METHODS
Cells and viruses.
HeLa (human cervix carcinoma; ATCC [American Type Culture Collection] CCL-2.2), CHO (Chinese hamster ovary; ATCC CCL-61), K562 (human erythroleukemia; ATCC 45506), HEp-2 (human larynx carcinoma, ATCC CCL-23), and 293 (human embryonic kidney; Microbix, Toronto, Ontario, Canada) cells were maintained in Dulbecco modified Eagle medium (DMEM)–10% fetal calf serum (FCS)–2 mM glutamine–penicillin–streptomycin. Culture medium for CHO cells was supplemented with 200 μM asparagine and 200 μM proline. Human CD34+-enriched bone marrow cells were purified from peripheral blood after mobilization using MiniMACS VS+ separation columns (Miltenyi Biotec, Auburn, Calif.) according to the manufacturer's instructions. Aliquots were stored in liquid nitrogen. Sixteen hours before the experiment, cells were recovered from the frozen stock and incubated in Iscove modified Dulbecco medium supplemented with 20% FCS, 10−4 M β-mercaptoethanol, 100 μg of DNase I per ml, 2 mM glutamine, 10 U of interleukin-3, and 50 ng of stem cell factor (SCF) or 2 ng of thrombopoietin per ml. The purity of CD34+ preparations was verified by flow cytometry and was consistently greater than 90%.
Ad3 (VR-3), Ad4 (VR1081), Ad5 (VR-5), Ad9 (VR1086), Ad35 (VR-716), and Ad41 (VR-930) were purchased from the ATCC. VR-716 was purchased from the ATCC labeled as serotype 34; however, it was found to be serotype 35 upon sequencing of the fiber region. For amplification, the corresponding Ads were infected onto HeLa, 293, or HEp-2 cells under conditions that prevented cross-contamination. Virus was banded in CsCl gradients, dialyzed, and stored in aliquots as described elsewhere (41). Plaque titering was performed as follows: confluent 293 cells plated in six-well plates were incubated for 24 h with virus in a total volume of 1 ml; 2 weeks after infection, plaques were counted on cultures overlaid with 1% agarose–minimal essential medium–10% FCS.
Labeling of Ads with methyl-3H-thymidine.
Serotypes were labeled with [methyl-3H]thymidine as described in detail elsewhere (59). Briefly, 5 × 107 HeLa or 293 cells were grown in 175-cm2 flasks with 15 ml of DMEM–10% FCS and infected with wild-type Ad at an MOI of 50 or higher. Twelve hours postinfection, 1 mCi of [methyl-3H]thymidine (Amersham, Arlington Heights, Ill.) was added to the medium, and cells were further incubated at 37°C until complete cytopathic effect was observed. Then cells were harvested, pelleted, washed once with cold phosphate-buffered saline (PBS), and resuspended in 5 ml of PBS. Virus was released from the cells by four freeze-thaw cycles. Cell debris was removed by centrifugation, and viral material was subjected to ultracentrifugation in CsCl gradients and subsequent dialysis as previously described (41). Virus purification and dialysis removed unincorporated radioactivity. Wild-type Ad particle concentrations were determined spectrophotometrically by measuring the optical density at 260 nm (OD260), using the extinction coefficient for wild-type Ad5, ɛ260 = 9.09 × 10−13 OD ml cm virion−1 (42). The virion-specific radioactivity was measured by a liquid scintillation counter and was always in the range of 10−5 to 10−4 cpm per virion. For selected variants, the fiber gene was PCR amplified and sequenced to ensure identity and the absence of cross-contamination.
Electron microscopy (EM) studies.
CsCl-banded Ad stocks were thawed and diluted with 0.5% glutaraldehyde. Grids were prepared as described earlier (47). After staining with 2% methylamine tungstate (Nanoprobes, Stony Brook, N.Y.), the carbon-coated grids were evaluated and photomicrographed with a Philips 410 electron microscope, operated at 80 kV (final magnification, ×85,000). For each particular Ad serotype, the number of morphologically deficient viral particles per 100 was counted in five random fields.
Attachment and internalization assays.
The studies were performed based on a protocol published elsewhere (77). In preliminary experiments, we found that labeled Ad5 virions reached equilibrium in attachment to HeLa cells after 45 min at 4°C with an MOI of 400 PFU per cell. For attachment studies, 3.5 × 105 cells were incubated for 1 h on ice with equal amounts of 3H-labeled Ad OD particles equivalent to an MOI of 400 PFU/cell for Ad5 in 100 μl of ice-cold adhesion buffer (DMEM supplemented with 2 mM MgCl2, 1% bovine serum albumin, and 20 mM HEPES). Next, the cells were pelleted by centrifugation for 4 min at 1,000 × g and washed two times with 0.5 ml of ice-cold PBS. After the last wash, the cells were pelleted at 1,500 × g, the supernatant was removed, and the cell-associated radioactivity was determined by a scintillation counter. The number of viral particles bound per cell was calculated using the virion-specific radioactivity and the number of cells. To determine the fraction of internalized 3H-labeled Ad particles, cells were incubated on ice for 1 h with the corresponding virus, washed with PBS as described above, resuspended in 100 μl of adhesion buffer, and then incubated at 37°C for 30 min. Following this incubation, cells were diluted threefold with cold 0.05% trypsin–0.5 mM EDTA solution and incubated at 37°C for an additional 5 to 10 min. This treatment removed 99% of attached radioactivity. Finally, the cells were pelleted at 1,500 × g for 5 min, the supernatant was removed, and the protease-resistant counts per minute were measured. This protocol minimizes the possibility that the internalization data were affected by receptor recycling (58). Nonspecific binding of Ad particles to cells on ice was determined in the presence of 100-fold excess of unlabeled virus. This value routinely represented less than 0.1% of viral load.
Flow cytometry.
Adherent cells (CHO and HeLa) grown in non-tissue culture-treated 10-cm-diameter dishes (Falcon, Franklin Lakes, N.J.) were detached by treatment with 1 mM EDTA and washed three times with wash buffer (WB), consisting of PBS supplemented with 1% FCS. Cells grown in suspension (K562 and CD34+) were washed three times with WB. After washing, cells were resuspended in WB at 2 × 106 cells/ml; 2 × 105 cells were incubated in WB for 1 h at 37°C with monoclonal antibodies (MAbs) specific for αv integrins (L230 [ATCC] or HB-8448 [59]; 1/30 final dilution]), CAR (RmcB [3, 26] [1/400 final dilution]), or bromodeoxyuridine (BrdU; Amersham; 1/100 final dilution). Subsequently, cells were washed with WB and incubated with fluorescein isothiocyanate (FITC)-labeled horse anti-mouse immunoglobulin G (IgG) antibodies (Vector Laboratories, Burlingame, Calif.; 1/100, final dilution) or phycoerythrin (PE)-labeled goat anti-mouse IgG antibodies (Calbiochem, La Jolla, Calif.; 1:100 dilution) for 30 min at 4°C. After incubation with secondary antibodies, cells were washed two times with WB and 104 cells per sample were analyzed in duplicate by flow cytometry.
For the analysis of CD34 and c-Kit expression on transduced CD34+ cells and for fluorescence-activated cell sorting (FACS), purified human CD34+ cells were incubated with a PE-conjugated anti-CD34 MAb (Becton Dickinson Immunocytochemistry Systems, San Jose, Calif.) or with PE-labeled anti-CD117 (c-Kit) MAb 95C3 (Immunotech, Beckman Coulter, Marseille, France) according to the manufacturer's protocol, followed by flow cytometry analysis. All analyses and sortings were performed on a FACStar Plus flow cytometer (Becton Dickinson, Franklin Lakes, N.J.) equipped with 488-nm argon and 633-nm HeNe lasers. For analysis of c-Kit expression and FACS purification of CD34+ c-Kit+ cells, SCF was not added to the medium during culturing of CD34+ cells.
Quantitative replication assay.
CD34+ or K562 cells (105) were infected in 100 μl of growth medium with different MOIs of Ad5, Ad9, or Ad35 which had been amplified in 293 cells expressing the XhoI DNA methyltransferase isoschizomer PaeR7 (51). After 2 h of incubation at 37°C, the cells were centrifuged at 1,000 × g for 5 min, the virus-containing medium was removed, and the cells were resuspended in 100 μl of fresh medium and then incubated at 37°C until harvesting. At 16 h postinfection for K562 cells or 36 h postinfection for CD34+ cells, 5 μg of pBlueScript (Stratagene, La Jolla, Calif.) plasmid DNA was added as a carrier which could also be used as a loading control. Genomic DNA was extracted as described previously (41). One-fourth of the purified cellular DNA (equivalent to 2.5 × 104 cells) was digested with HindIII, XhoI, or both HindIII and XhoI at 37°C overnight and subsequently separated in a 1% agarose gel followed by Southern blot with chimeric Ad5/9 or Ad5/35 DNA probes. The chimeric probes, containing sequences of Ad5 and Ad9 (Ad5/9) or Ad5 and Ad35 (Ad5/35), were generated by a two-step PCR amplification using Pfu-Turbo DNA polymerase (Stratagene) and viral DNA from purified particles as a template. The following primers were used for PCR (Ad5 sequences and nucleotide numbers are underlined): Ad5F1 (nucleotides [nt] 32775 to 32805), 5′-GCC CAA GAA TAA AGA ATC GTT TGT GTT ATG-3′; Ad5R1 (nt 33651-33621), 5′-AGC TGG TCT AGA ATG GTG GTG GAT GGC GCC A-3′; chimeric Ad5/9F (nt 31150 to 31177; nt 181 to 208), 5′-AAT GGG TTT CAA GAG AGT CCC CCT GGA GTC CTG TCA CTC AAA CTA GCT GAC CCA-3′; chimeric Ad5/9R (nt 32805 to 32775; nt 1149 to 1113), 5′-CAT AAC ACA AAC GAT TCT TTA TTC TTG GGC TTC ATT CTT GGG CGA TAT AGG AAA AGG-3′; chimeric Ad5/35F (nt 31150 to 31177; nt 132 to 159), 5′-AAT GGG TTT CAA GAG AGT CCCCCT GGA GTT CTT ACT TTA AAA TGT TTA ACC CCA-3′; chimeric Ad5/35R (nt 32805 to 32775; nt 991 to 958), 5′-CAT AAC ACA AAC GAT TCT TTA TTC TTG GGC ATT TTA GTT GTC GTC TTC TGT AAT GTA AG-3′. Nucleotide numbers are given according to the sequences obtained from the National Center for Biotechnology Information GenBank (accession no. M73260/M29978 for Ad5, X74659 for Ad9, and U10272 for Ad35). After the first amplification, the 968-bp-long Ad9, an 859-bp-long Ad35 DNA fragment corresponding to the fiber genes, and an 876-bp-long Ad5 fragment corresponding to the Ad5 E4 region (located immediately downstream of Ad5 fiber gene) were purified by agarose gel electrophoresis. To generate chimeric DNA probes, amplified Ad5 DNA was mixed with Ad9 or Ad35 fragments obtained during the first step of PCR and subjected to a second PCR amplification using Ad5/9F or Ad5/35F primers and the Ad5R1 primer. The resulting Ad5/9 or Ad5/35 chimeric DNA fragments (see Fig. 4C) were purified, and their concentrations were measured spectrophotometrically. Corresponding chimeric DNA fragments were loaded as concentration standards on agarose gels or labeled with [32P]dCTP and used as probes for Southern analysis. The number of viral genomes per DNA sample was calculated after quantitative phosphorimager analysis. In preliminary experiments, no preferential hybridization of chimeric DNA probes to DNA of any particular viral serotype was detected.
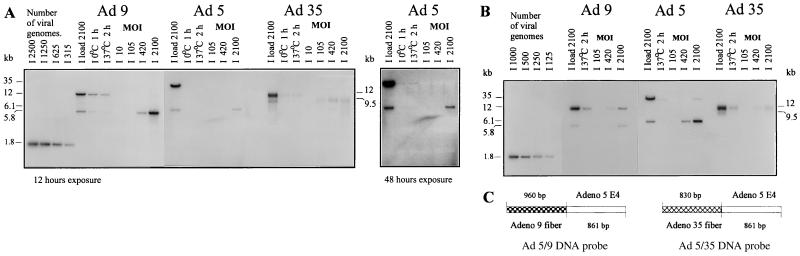
FIG. 4.
Analysis of viral replication in K562 and CD34+ cells by Southern blot analysis of methylated viral DNA. Replication studies were performed with 105 K562 (A) or CD34+ (B) cells infected with methylated Ad5, Ad9, or Ad35. The lanes labeled “load” represent DNA that was extracted from the medium-cell mixture immediately after adding the indicated viral dose to cells. The intensities of bands corresponding to methylated and unmethylated viral DNA indicate that ∼85% of the input virus was methylated. To quantify adsorption and internalization, DNA analysis was performed after prior incubation of virus with cells at 0°C (adsorption) or 37°C (internalization). For dose-dependent replication studies, the indicated viral dose (expressed as the number of genomes per cell) was added, and cellular genomic DNA together with viral DNA was extracted 16 or 36 h postinfection for K562 and CD34+ cells, respectively. Identical amounts of sample DNA were analyzed by Southern blotting. For quantification purposes, Ad9 replication was analyzed together with Ad5, using an Ad5/9 chimeric probe that hybridizes with DNA of both serotypes (C). The analysis of Ad5 versus Ad35 replication was performed with the corresponding Ad5/35 chimeric probe. Since separate hybridizations with both Ad5/35 and Ad5/9 probes gave identical signal intensities for Ad5 DNA, only one panel is shown for Ad5 replication in test cells. To produce distinguishable fragments specific for the methylated or nonmethylated status of viral genomes, Ad5 DNA was digested with XhoI, while Ad9 and Ad35 DNA was digested with XhoI and HindIII. The bands specific for methylated (not replicated) viral DNA were ∼12 kb for Ad9, 35 kb for Ad5, and ∼12 kb for Ad35. The fragments specific for nonmethylated DNA were 5.8 kb for Ad9, 6.1 kb for Ad5, and 9.5 kb for Ad35. Chimeric Ad5/9 and Ad5/35 DNA fragments (1.8 kb) were used as quantification standards and applied onto gels together with digested viral or cellular DNA (shown on the left). Hybridized blots were subjected to quantitative phosphorimager analysis or exposed to X-ray films for 12 h. Lanes representing Ad5 replication data for K562 cells were also exposed for 48 h (A, left panel).
Construction of chimeric Ad vectors.
For transduction studies, we constructed two Ad vectors, Ad5GFP and Ad5GFP/F35, containing a chimeric Ad5/35 fiber gene. Both Ad vectors contained a 2.3-kb, cytomegalovirus (CMV) promoter-driven enhanced green fluorescent protein (EGFP) gene (derived from pEGFP-1 [Clontech, Palo Alto, Calif.]) inserted into the E3 region of Ad5. The EGFP expression cassette was cloned between Ad5 positions 25191 to 28191 and 30818 to 32507 into a shuttle plasmid, which contained the E3 deletion described for pBHG10 (Microbix). The resulting plasmid was named pAdGFP. For the chimeric vector, the Ad5 fiber gene in pAdGFP was substituted by an Ad5/35 chimeric fiber gene generated by the two-step PCR protocol outlined above. In the first PCR step, three DNA fragments corresponding to (i) the Ad5 fiber 5′ nontranslated region and the first 132 bp of the fiber tail domain (nt 30798 to 31178), (ii) the Ad35 shaft and knob domains (nt 132 to 991), and (iii) the Ad5 E4 region including the Ad5 fiber polyadenylation signal (nt 32775 to 33651) were amplified by Pfu-Turbo DNA polymerase. The following primers were used: for the Ad5 tail, Ad5F-2 (nt 30798 to 30825; 5′-CGC GAT ATC GAT TGG ATC CAT TAA CTA-3′) and Ad5R-2 (nt 31178 to 31153; 5′-CAG GGG GAC TCT CTT GAA ACC CAT T-3′); for the Ad35 shaft and knob, primers Ad5/35F and Ad5/35R (see above); for the Ad5E4 and polyA, primers Ad5F-1 and Ad5R-1 (see above). After 10 PCR cycles, the products were purified by agarose gel electrophoresis, combined, and then subjected to a second PCR with primers Ad5F-2 and Ad5R-1. The resulting 2,115-bp-long chimeric fiber gene contained the Ad5 tail and the Ad35 shaft and knob domains. This product was used as a substitute for the SalI/XbaI Ad5 fiber gene containing fragment in pAdGFP. The resulting plasmid was named pAdGFP/F35. To generate full-length E1/E3 vector genomes, pAdGFP and pAdGFP/F35 were inserted in pAdHM4 (49) by recombination in Escherichia coli (8). To do this, the RecA+ E. coli strain BJ5183 was cotransformed with pAdHM4 linearized by SrfI mixed with the XbaI fragments containing the GFP genes, the Ad5 or Ad5/35 fiber genes, and the Ad5 homology regions. The resulting recombinants were analyzed by restriction analysis. Correct recombinants were amplified in E. coli HB101 and purified by double CsCl gradient banding. The plasmids were named pAd5GFP and pAd5GFP/F35. The correct structure of the Ad5/35 chimeric fiber gene was confirmed by endonuclease digestion and sequencing part of pAd5GFP/F35. To produce the corresponding viruses, pAd5GFP and pAd5GFP/F35 were digested with PacI to release the viral genomes and transfected onto 293 cells as described elsewhere (41). Plaques developed 7 to 10 days posttransfection in overlaid cultures. Recombinant viruses were propagated in 293 cells and purified by standard methods described elsewhere (41).
Hemagglutination assay.
Twenty-five-microliter aliquots of serial dilutions of Ad5, Ad35, or chimeric Ad5GFP/F35 virions in McIlvaine-NaCl buffer (0.1 M citric acid–0.2 M Na2HPO4 [pH 7.2], diluted 1:50 with 0.87% NaCl) were loaded onto 96-well plates. To each dilution, 25 μl of a 1% suspension of monkey erythrocytes (in McIlvaine-NaCl buffer) was added. The sedimentation pattern was determined after incubation for 1 h at 37°C. All tests were performed in quadruplicate in at least two independent experiments.
Southern blotting.
Extraction of genomic DNA, labeling of DNA fragments, and hybridization were performed as described earlier (41).
RESULTS
CAR and αv integrin expression on test cells.
It is generally accepted that CD34+ cells possess bone marrow-repopulating activity. Therefore, we used human CD34+ cells as the target for our studies to identify Ad serotypes with HSC tropism and constructing new viral vectors. Studies were performed on mobilized, CD34-positive peripheral blood cells from one donor under conditions which are known to retain CD34+ cells in a quiescent stage (37, 57). More than 90% of purified cells were CD34 positive by flow cytometry. Furthermore, we included into our Ad tropism studies the cell line K562, which is considered to be an adequate model for studying gene transfer into human hematopoietic cells (44). HeLa cells, which are readily infectible by Ad5, and CHO cells, which are refractory to Ad5 infection (3), were used as positive and negative control cell lines, respectively.
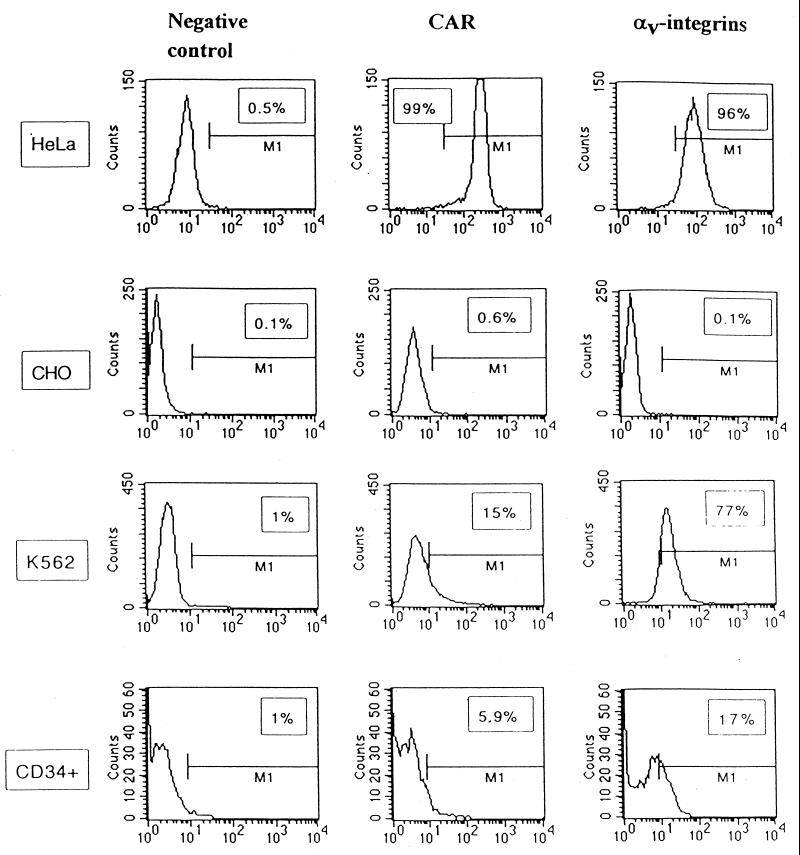
For Ad5, both binding to the primary receptor and binding to αvβ3 and αvβ5 integrins are important for high-efficiency infection of target cells. The expression of CAR and αv integrins on test cells was analyzed by flow cytometry using MAbs against CAR (RmcB [3, 26]) and αv integrins (L230 [59]) (Fig. 1). As expected, nearly all HeLa cells expressed high levels of CAR and αv integrins, whereas CHO cells lacked significant CAR and αv integrin expression. Fifteen and 77% of K562 cells expressed CAR and αv integrins, respectively. Only ∼6% of the CD34+ cells used in our studies expressed CAR, and 17% were positive for αv integrins. Notably, the preparation of CD34+ cells represents a mixture of different cell types. The absent or low expression of primary and secondary Ad5 receptors on noncycling human CD34+ cells is in agreement with previous reports (27, 50, 73).
FIG. 1.
Expression of CAR and αv integrins on test cells. For flow cytometry analysis, HeLa, CHO, K562, and CD34+ cells were incubated with an anti-CAR (RmcB; 1:400 dilution) or anti-αv integrin (L230; 1:30 dilution) MAb. As a negative control, cells were incubated with an irrelevant mouse MAb (anti-BrdU; 1:100 dilution). The binding of primary antibodies was developed with anti-mouse IgG-FITC conjugates (1:100 dilution). Data shown represent the average results of quadruplicate analyses performed on 104 cells.
Serotype screening.
It is thought that different Ad serotypes bind to different cellular receptor proteins and use different entry mechanisms (15, 43). A set of human Ad serotypes was obtained from the ATCC to be tested for tropism to CD34+ cells. These included serotypes 3, 4, 5, 9, 35, and 41 representing different subtypes (Table 1). We hypothesized that these serotypes would use different cellular attachment and internalization strategies due to differing lengths of fiber shafts (11, 60), the presence or absence of RGD motifs within the penton base, and differing tissue tropism. The relatively little characterized Ad35 was selected because it was found in immunocompromised hosts, particularly in bone marrow recipients (17, 18, 65). The latter observations prompted us to speculate that bone marrow cells may be among the natural reservoirs for Ad35.
TABLE 1.
Properties of selected serotypesa
| Serotype | Subgroup | Hemagglutinationbc | β repeats in fiber shaftd | RGD motifs in pentone | Tissue tropismb |
|---|---|---|---|---|---|
| 3 | B1 | +++ (ME) | 6 | + | Respiratory |
| 4 | E | 12 | + | Eye, respiratory | |
| 5 | C | + (RE) | 22 | + | Respiratory |
| 9 | D | +++ (RE) | 8 | + | Eye |
| 35 | B2 | +++ (ME) | 7 | ? | Genitourinary |
| 41 | F | 22/12 | − | Gastrointestinal |
Members of subgroup A were not analyzed due to their high tumorigenicity. The length of the fiber shaft seems to be specific for the virus subgroup and may be a determining factor for cell entry and/or tissue tropism (59, 60). The rod-like shaft of variable length contains repeats of a 15-amino-acid-long β sheet with the number of repeats ranging from 6 (e.g., for Ad3 or Ad7) to 22 (for Ad2 and Ad5). The protruding RGD motifs in the penton base are critical for interaction with αv integrins and virus internalization.
According to reference 25.
ME, hemagglutination of monkey erythrocytes; RE, hemagglutination of rat erythrocytes.
According to reference 11. Ad41 contains a short-shafted and long-shafted fiber.
According to reference 60. The sequence for the Ad35 penton and information about the presence of RGD motifs penton base were not available.
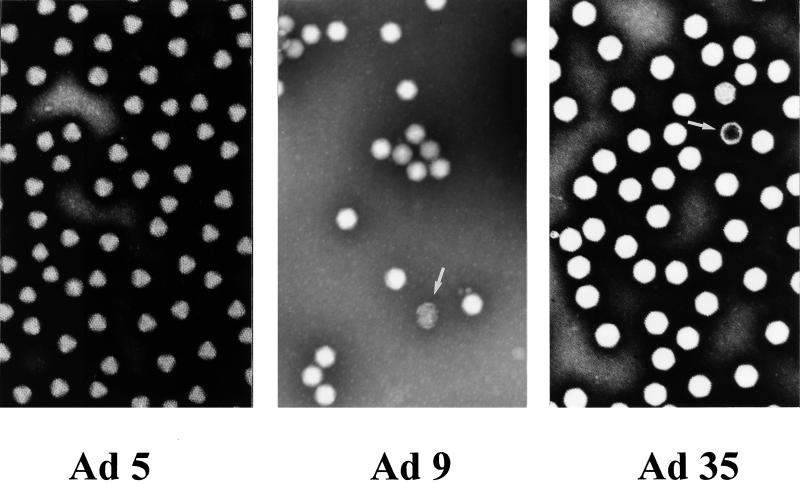
Little is known about the stability of particles from serotypes other than Ad5. Since the intactness of viral particles was crucial for comparative interaction studies, virions from the serotypes specified above were analyzed by EM. EM studies of negative-contrast-stained Ad suspensions demonstrated that the percentage of defective particles (loss of icosahedral shape or luminal staining) did not exceed 5%, indicating that serotype preparations had comparable qualities. Representative EM photographs are shown for Ad5, Ad9, and Ad35 (Fig. 2).
FIG. 2.
EM analysis of Ad particles. Purified particles from Ad5, Ad9, and Ad35 were negative contrast stained and analyzed at an original magnification of ×85,000. Defective particles are highlighted by arrows.
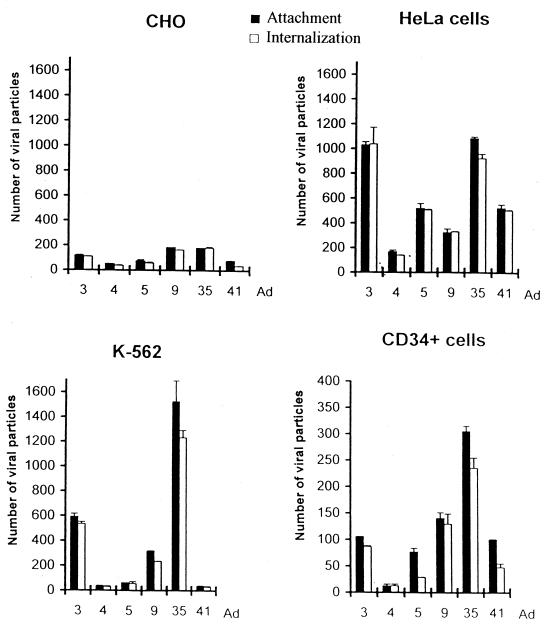
To analyze attachment of Ad particles to target cells and subsequent internalization, the selected serotypes were metabolically labeled with [3H]thymidine, which is incorporated into viral DNA during replication. Adsorption and internalization can be experimentally dissociated by taking advantage of the observation that at low temperature (0 to 4°C) only virus cell attachment occurs, whereas internalization requires incubation at higher temperatures. The number of particles adsorbed or internalized per cell was calculated using the virion-specific radioactivity and used to quantify interaction of Ad serotypes 3, 4, 5, 9, 35, and 41 with CD34+, K562, HeLa, and CHO cells (Fig. 3). The serotypes varied significantly in the ability to attach to and to be internalized by the different cell lines. For Ad5, the degree of attachment to the cell lines tested correlated with the level of CAR expression. In CHO cells, which were previously shown to be refractory to Ad5 infection, the level of attachment and internalization was about 50 to 70 viral particles per cell. This number was hereafter assumed negative in terms of susceptibility of a given cell type for Ad5. Interaction of the other serotypes with CHO cells was not significantly higher, indicating that corresponding receptors were absent on CHO cells. All serotypes tested interacted with HeLa cells, Ad3 and Ad35 being the most efficient variants. The presence of distinct Ad3 and Ad5 receptors on HeLa cells was demonstrated previously (69). Ad4, Ad5, and Ad41 did not bind to K562 cells. In contrast, Ad9 as well as the members of subgroup B, Ad3 and Ad35, efficiently interacted with K562 cells, Ad35 having the highest number of adsorbed and internalized particles. About 25 times more Ad5 than Ad35 particles were attached, and three-fourths of these were internalized by K562 cells. Viral interactions with CD34+ cells were generally weaker. Among the serotypes tested, only Ad9 and Ad35 were significantly internalized by noncycling CD34+ cells. Internalization of Ad9 and that of Ad35 were, respectively, four and eight times more efficient than for Ad5 particles. The number of Ad35 virions internalized by CD34+ cells was almost half of that seen for Ad5 in HeLa cells, which can be readily infected with Ad5-based vectors.
FIG. 3.
Analysis of attachment and internalization of different serotypes to CHO, HeLa, K562, and CD34+ cells. Equal amounts of [3H]thymidine-labeled virions of Ad serotypes 3, 4, 5, 9, 35, and 41 (measured by OD260 and equivalent to an MOI of 400 PFU per cell for Ad5) were incubated for 1 h on ice as described in Materials and Methods. Cells were then washed, and the number of labeled virions bound per cell was determined. For internalization studies, viruses were first allowed to attach to cells for 1 h on ice, and then unbound viral particles were washed out. Cells were then incubated at 37°C for 30 min, treated with trypsin-EDTA, and washed to remove uninternalized viral particles. The data were obtained from two to four independent experiments performed in triplicate. Note the different scales on the y axes for CD34+ cells.
In summary, of all serotypes tested, Ad9, Ad3, and Ad35 demonstrated the most efficient attachment to and internalization with K562 and CD34+ cells. Based on adsorption/internalization data, Ad9 and Ad35 as representatives for subgroups D and B were selected for further tropism studies.
Replication of selected serotypes in K562 and CD34+ cells.
Adsorption/internalization studies do not ultimately prove viral transduction, a process often defined as gene transfer that allows for viral or heterologous gene expression in host cells. Intracellular trafficking, including endosomal lysis, transport to the nucleus, and nuclear import of the viral genome, depends on structural capsid proteins and thus varies between different serotypes (15, 48). We hypothesized that analysis of viral gene expression would be a means to verify successful nuclear import of viral genomes and that this would be a good criterion for selection of serotypes able to efficiently infect our target cells. To do this, we studied Ad DNA replication in infected cells; viral DNA synthesis can occur only after de novo expression of Ad early genes. We used a site-specific methylation strategy to monitor viral DNA replication within infected cells (51). Methylated Ad serotypes were produced by the addition of a methyl group onto the N6 position of the adenine base of XhoI sites, CTCGAG, during propagation of the viruses in 293 cells expressing the XhoI isoschizomer PaeR7 methyltransferase (33). Loss of methylation through viral replication restores XhoI cleavage and can be detected by Southern blots of XhoI-digested genomic DNA from infected cells.
Ad replication studies were performed in K562 and CD34+ cells with Ad9 and Ad35, in comparison to Ad5. For replication studies, the infectious titer (in PFU per milliliter) and genome titer (in genomes per milliliter) were determined (by plaque assay on 293 cells and by quantitative Southern blot, respectively) for methylated and unmethylated Ad5, Ad9, and Ad35 (Table 2). The ratio of PFU to genome titer was comparable for methylated and unmethylated virus, demonstrating that DNA methylation had not altered transduction properties (data not shown). About 85% of the Ad5, Ad9, and Ad35 used for infection was methylated, as calculated based on the intensity of fragments specific for methylated and nonmethylated viral DNA present in the viral load (Fig. 4). The numbers of genomes detected after adsorption (1 h, 0°C) or internalization (2 h, 37°C) correlated well with studies shown in Fig. 3. Ad9 and Ad35 interacted more efficiently than Ad5 with K562 and CD34+ cells. Dose-dependent replication studies in K562 and CD34+ cells were performed with the same genome numbers of Ad5, Ad9, and Ad35 (Fig. 4). The replication rate was measured based on the ratio of methylated to demethylated viral DNA after infection with different MOIs (2,100, 420, and 105 genomes per cell). In K562 cells, efficient replication (100% conversion from methylated to unmethylated DNA) was detected for Ad9 at MOIs of ≥420 and for Ad35 at MOIs of ≥105. The signal from internalized Ad5 genomes was relatively weak, reflecting the low level of Ad5 interaction with K562 cells. This complicated the analysis of Ad5 replication. Clearly detectable replication of Ad5 was observed only at MOIs of ≥2,100 genomes per cell. Taken together, these findings demonstrated that Ad35 transduced K562 cells with the highest efficiency. In CD34+ cells, the replication rates were 100% for Ad5 and 31% for Ad9 after infection with an MOI of 420. Although methylated Ad35 viral DNA was present in CD34+ cells, viral replication was undetectable for Ad35. In summary, while viral replication studies in K562 cells confirmed data obtained for Ad5, Ad9, and Ad35 adsorption and internalization, there was a discrepancy between earlier results and the poor replication of Ad9 and, particularly, Ad35 in CD34+ cells. As outlined later, replication analysis in heterogeneous cell populations, like CD34+ cells, may not allow for definitive conclusions on tropism of a particular serotype.
TABLE 2.
Virus titers and genome-to-PFU ratios for wild-type Ad5, Ad9, and Ad35a
| Methylated virus | PFU/ml | Viral genomes/ml | Ratio, genomes/PFU |
|---|---|---|---|
| Ad5 | 1.25 × 1011 | 7.5 × 1011 | 6 |
| Ad9 | 4.1 × 106 | 1.5 × 1010 | 3,600 |
| Ad35 | 2.1 × 1010 | 1.2 × 1011 | 5.6 |
a The genome titer in preparations of purified methylated Ad was measured by quantitative Southern blotting as described earlier (41). The plaque titer was determined on 293 cells under conditions outlined in Materials and Methods. The different genome-to-PFU ratios confirm the different transduction properties of the given serotypes for 293 cells.
Taking all the screening data together, Ad9 and Ad35 emerged as the variants with the strongest tropism for K562 and CD34+ cells. It is thought that Ad9 can bind to CAR; however, it preferentially uses αv integrins for cell entry (59). This entry strategy may not be optimal for efficient infection of CD34+ cells, as only less than 17% of them express αv integrins (Fig. 1). Therefore, we decided to concentrate on Ad35 as a source for heterologous fiber to be used for construction of a chimeric vector based on an Ad5 backbone.
Construction and characterization of chimeric fiber.
Previously, it was shown that exchanging the fiber knob was sufficient to alter the tropism of chimeric Ad vectors (10, 31, 70). As outlined above, the length of the fiber shaft may critically determine the entry strategy of a particular serotype. Therefore, we decided to replace not only the Ad5 fiber knob but also the shaft. The chimeric Ad5/35 fiber contained the Ad5 tail (amino acids 1 to 44) necessary for interaction with the Ad5 penton base linked to 279 amino acids from Ad35 including the shaft with seven β sheets and the knob (Fig. 5A). The endogenous Ad5 fiber poly(A) signal was used to terminate transcription of the chimeric fiber gene. The combination of the Ad5 capsid including the RGD motif containing penton base with a short-shafted fiber could be risky because the natural distance between the fiber knob and the RGD motifs was disturbed. Nevertheless, the Ad5 fiber was replaced by the chimeric fiber sequences based on an E1/E3-deleted Ad vector. This vector carried a CMV promoter-GFP reporter gene cassette inserted into the E3 region. The corresponding chimeric virus (Ad5GFP/F35) was produced in 293 cells at a titer of >2 × 1012 genomes per ml. For comparison, an E1/E3-deleted Ad vector containing the original Ad5 fiber gene and the GFP expression cassette was generated (Ad5GFP). The titer and the ratio of physical to infectious particles were similar between Ad5GFP and Ad5GFP/F35, indicating that the fiber modification did not significantly alter the stability and/or growth properties of the chimeric vector (data not shown). The correctness of the fiber modification was confirmed by restriction analysis of the Ad5GFP/F35 viral genome followed by Southern blot hybridization (Fig. 5B), direct sequencing of the fiber-coding region, and a functional test for hemagglutination of monkey erythrocytes. The agglutination of erythrocytes is fiber knob mediated; it is known that Ad5 does not agglutinate monkey erythrocytes, whereas Ad35 efficiently does (55). In hemagglutination tests, Ad5GFP/F35 agglutinated monkey erythrocytes with the same efficiency as Ad35 at dilutions of up to 1:512. In contrast, no hemagglutination was observed with equivalent Ad5 dilutions. This clearly confirmed the functional activity of the chimeric Ad5/35 fiber incorporated into Ad5 capsid.
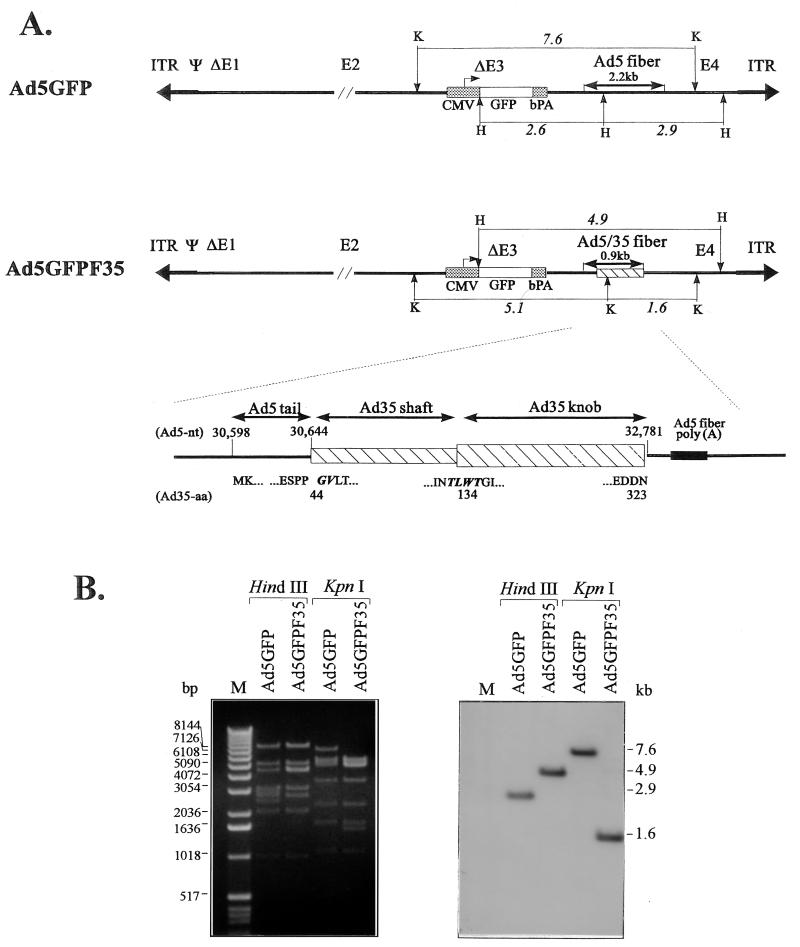
FIG. 5.
Structure of Ad5GFP and chimeric Ad5GFP/F35 vectors. (A) Schematic diagram of the original E1/E3-deleted Ad5-based vector with GFP expression cassette inserted into the E3 region (Ad5GFP) and the chimeric vector Ad5GFP/F35 containing the Ad5/35 fiber gene. The 2.2-kb Ad5 fiber gene was replaced by a 0.9-kb chimeric fiber gene encoding the short shaft and knob of Ad35 by a technique that involved PCR cloning and recombination in E. coli. KpnI (K) and HindIII (H) sites localized within or around the fiber genes are indicated. The lower panel shows the detailed structure of the chimeric fiber region. The Ad5 fiber tail (amino acids [aa] 1 to 44) were joined in frame to the Ad35 fiber shaft starting from its first two amino acids (GV), which are conserved among many serotypes. A conserved stretch of amino acids, TLWT, marks the boundary between the last β sheet of Ad35 shaft and the globular knob. The Ad35 fiber chain termination codon is followed by the Ad5 fiber polyadenylation signal. The region of Ad5GFP/F35 encoding for chimeric fiber was completely sequenced with Ad5-specific primers (see Materials and Methods). ITR, inverted terminal repeat; bPA, bovine growth hormone polyadenylation signal. (B) Restriction analysis of viral genomes. Viral DNA was isolated from purified Ad5GFP and Ad5GFP/F35 particles as described elsewhere (41). One microgram of DNA was digested with HindIII or KpnI and separated in ethidium bromide-stained agarose gels (left) which were subsequently blotted and analyzed by Southern blot with an Ad5 E4-specific probe (nt 32775 to 33651) (right). Specific patterns designating the correct structure for both viral vectors were detected. The HindIII fragments specific for Ad5GFP and Ad5GFP/F35 were 2.9 and 4.9 kb, respectively. The KpnI fragment that confirmed the correct Ad5GFP/F35 structure was 1.6 kb, compared to a 7.6-kb Ad5GFP fragment. M, 1-kb ladder (Gibco-BRL, Grand Island, N.Y.).
Competition studies.
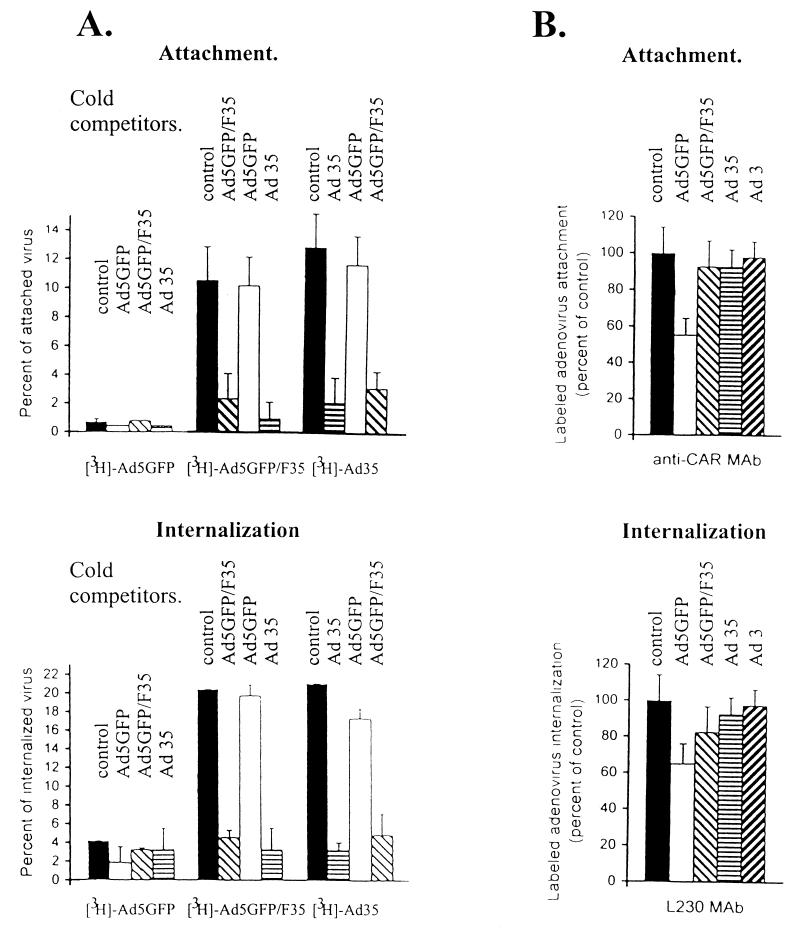
Cross-competition studies between Ad5, Ad35, and Ad5GFP/F35 for binding and internalization were performed to investigate in more detail the pathways used by the chimeric vector to infect target cells. Wild-type Ad35 and the chimeric vector Ad5GFP/F35 could recognize the same primary receptor, as they competed with each other for attachment to K562 cells (Fig. 6A, upper panel). This primary receptor is different from that used by Ad5, since neither Ad5 viral particles nor anti-CAR MAbs (Fig. 6B, upper panel) were able to abrogate Ad35 or Ad5GFP/F35 binding. In competition studies for internalization, Ad35 and Ad5GFP/F35 competed with each other with equal efficiency. Ad5 and anti-αv integrin MAb L230 (Fig. 6, lower panels) did not inhibit internalization of Ad35 or the chimeric virus. To consolidate these data, K562 cells were infected with Ad5GFP and Ad5GFP/F35 after prior incubation of cells with anti-CAR or anti-αv integrin MAbs followed by analysis of GFP-expressing cells. The transduction data mirror the results obtained in adsorption/internalization studies (data not shown). In summary, this demonstrated that Ad35 and Ad5GFP/F35 use a CAR- and αv integrin-independent pathway for infection of K562 cells; the structural elements which account for these specific properties are located within the Ad35 fiber and can be transplanted into Ad5 by fiber substitution.
FIG. 6.
Cross-competition for attachment and internalization of labeled Ad5GFP, Ad35, and chimeric Ad5GFP/F35 virions with unlabeled viruses and with anti-CAR or anti-αv integrin MAb. (A) For attachment studies, 105 K562 cells were preincubated with a 100-fold excess of unlabeled competitor virus (cold competitors) at 4°C for 1 h; then equal amounts of [3H]Ad5GFP, [3H]Ad5GFP/F35, or [3H]Ad35, at a dose equivalent to an MOI of 100 PFU per cell determined for Ad5GFP, were added to cells, which were incubated at 4°C for 1 h. Cells were then washed with ice-cold PBS and pelleted, and the percentage of attached virus (cell-associated counts per minute) was determined. For analysis of cross-competition for internalization, cells were preincubated with a 100-fold excess of competitor virus at 37°C for 30 min before labeled virus was added. After an additional incubation at 37°C for 30 min, cells were treated with trypsin-EDTA for 5 min at 37°C, washed with ice-cold PBS, and pelleted, and the percentage of internalized virus was determined. For controls, cells were incubated with labeled viruses without any competitors. Preliminary experiments had shown that the conditions chosen for competition studies allowed for saturation in attachment or internalization on K562 cells for all unlabeled competitors. As a control, 3H-labeled viruses were incubated with cells without any competitor. (B) K562 cells (105) were preincubated for 1 h at 4°C with an anti-CAR (RmcB; diluted 1:100) or anti-αv integrin (L230; diluted 1:30) MAb and then incubated with labeled viruses according to the protocols for attachment or for internalization as described above. For each particular serotype, the percentage of attached or internalized virus was compared to the control settings, where cells were preincubated under the same conditions with a 1:100 dilution of an irrelevant antibody (anti-BrdU MAb) before addition of the labeled virus. Note that the specific competitors but not the corresponding controls significantly inhibited Ad5 internalization to a degree that is in agreement with published data (59). n ≥ 4.
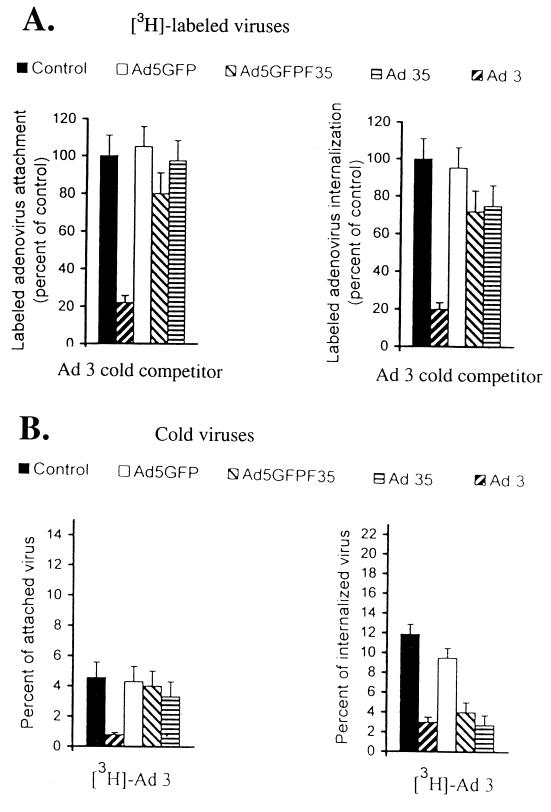
We demonstrated earlier that Ad3 can efficiently interact with K562 cells (Fig. 3). Although Ad3 and Ad35 belong to the same subgroup (B), the homology between amino acid sequences of their fibers is only about 60%. Therefore, we decided to test whether Ad3 could compete with Ad35 and Ad5GFP/F35 for attachment and internalization (Fig. 7). These studies demonstrated that Ad35 binding was not inhibited by Ad3, indicating the use of different receptors. Interestingly, Ad3 slightly inhibited attachment of Ad5GFP/F35 (Fig. 7A, left panel). We speculate that in addition to binding to the receptor common for the Ad35 and Ad5GFP/F35 fiber, the chimeric capsid (e.g., the Ad5 penton RGD motifs) also interacts with a second cellular receptor that overlaps with elements involved in Ad3 binding. In cross-competition for internalization, preincubation of cells at 37°C with Ad35 and with chimeric virus significantly decreased internalization of 3H-labeled Ad3 (Fig. B, right panel). In the reverse experiment, Ad3 as competitor decreased the level of internalization by 30% for both Ad35 and the chimeric virus (Fig. 7A, right panel). As expected, Ad5 and Ad3 did not compete for adsorption or internalization. As shown before (Fig. 6B), anti-CAR and anti-αv integrin antibodies did not block Ad3 interaction with K562 cells. In summary, we concluded that Ad35 and Ad5GFP/F35 bind to a receptor(s) different from that of Ad3. However, Ad35, Ad5GFP/F35, and Ad3 appear to use common structural elements for internalization, which are different from αv integrins.
FIG. 7.
Cross-competition for attachment and internalization of Ad5GFP, Ad35, and Ad5GFP/F35 with Ad3. (A) K562 cells (105) were preincubated with a 100-fold excess of unlabeled Ad3 according to attachment or internalization protocols described for Fig. 6. Equal amounts of [3H]Ad5GFP, [3H]Ad5GFP/F35, [3H]Ad35, or [3H]Ad3 were added to cells at a dose equivalent to an MOI of 100 PFU per cell for Ad5GFP. Control settings represent attachment or internalization of [3H]Ad3 without competitor. (B) 3H-labeled Ad3 was incubated with a 100-fold excess of cold (unlabeled) virus (Ad5GFP, Ad5GFP/F35, Ad35, or Ad3). In control settings, cells were incubated with labeled viruses without any competitors. n = 4.
Infection studies with chimeric virus.
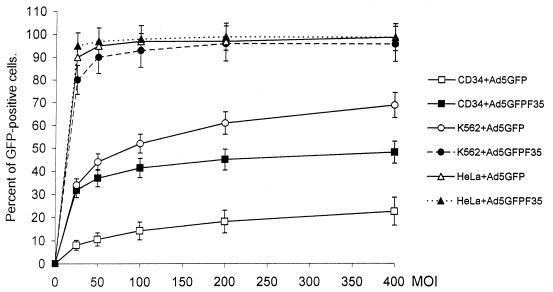
So far, we have established that Ad5GFP/F35 infected K562 cells by a CAR- and αv integrin-independent pathway. We hypothesized that this property may allow for efficient transduction of noncycling CD34+ cells, which express scarcely CAR and αv integrins. To test this, the transduction properties of Ad5GFP and Ad5GFP/F35 vectors were analyzed on CD34+, K562, and HeLa cells. Figure 8 shows the percentage of transduced, GFP-expressing cells depending on the MOI used for infection. Nearly 100% of HeLa cells were transduced with Ad5GFP and Ad5GFP/F35 at MOIs of ≥25. More than 95% of the K562 cells were transduced with Ad5GFP/F35 at MOIs of ≥100, whereas the transduction rate was significantly lower with Ad5, in which case it increased, with the MOI reaching a plateau at ∼70% GFP-positive cells after infection with an MOI of 400. Transduction of CD34+ cells was about threefold more efficient with Ad5GFP/F35 than with Ad5GFP at all MOIs analyzed. Interestingly, at higher MOIs, the transduction rate did not rise proportionally with the viral dose and soon reached a plateau, indicating that in both cases only specific subsets of CD34+ cells were permissive to infection.
FIG. 8.
Transduction of CD34+, K562, and HeLa cells with Ad5GFP and chimeric Ad5GFP/F35 vectors. Cells (105) were infected with different MOIs (PFU per cell) of viruses in 100 μl of medium for 6 h at 37°C. Virus-containing medium was then removed, and the cells were resuspended in fresh medium followed by incubation for 18 h at 37°C. The percentage of GFP-expressing cells was determined by flow cytometry. n = 3.
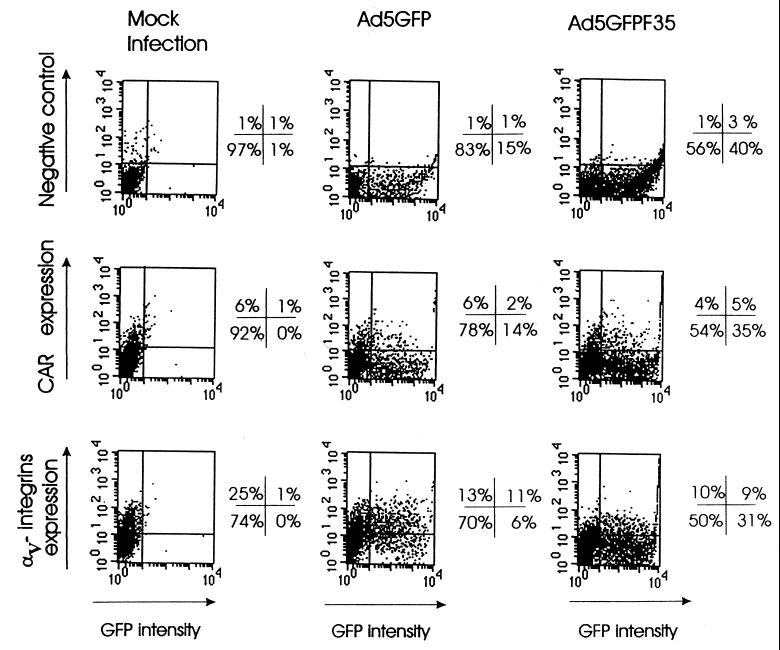
To characterize in more detail these specific, permissive subsets, additional transduction studies were performed. First, the percentage of GFP-expressing cells was determined in CD34+ fractions that were stained for αv integrins or CARs (Fig. 9). The low number of CAR-positive CD34+ cells complicated accurate colabeling studies; however, it appears that there was no correlation between CAR expression and the proportion of transduced cells among CD34+ cells infected with Ad5GFP or Ad5GFP/F35. Interestingly, for Ad5GFP, 65% of all GFP-expressing cells were positive for αv integrins, whereas less than 22% of GFP-positive cells infected with the chimeric virus stained positive for αv integrin expression. While only 17% of the whole CD34+ population expressed GFP after Ad5GFP infection, the percentage of GFP-expressing cells in the CD34+ αv integrin-positive fraction was 50%. This indicates that Ad5GFP vector-mediated GFP expression was preferentially localized to αv integrin-positive CD34+ subsets, whereas after infection with the Ad5GFP/F35 vector, GFP was expressed in a broader spectrum of CD34+ cells, most of them being αv integrin negative. It remains to be determined whether there is a correlation between αv integrin expression and the differentiation status of CD34+ cells.
FIG. 9.
Distribution of GFP-positive cells in subpopulations of human CD34+ cells expressing CAR or αv integrins. CD34+ cells (105) were infected with Ad5GFP or Ad5GFP/F35 at an MOI of 200 PFU/cell as described for Fig. 8. Twenty-four hours after infection, cells were incubated with anti-CAR (1:100 final dilution) or anti-αv integrin (1:30 final dilution) primary MAb for 1 h at 37°C. Binding of primary antibodies was developed with anti-mouse IgG-PE secondary MAbs (1:100 final dilution) at 4°C for 30 min. For each variant, 104 cells were analyzed by flow cytometry. The mock infection variants represent cells incubated with virus dilution buffer only. The quadrant borders were set based on the background signals obtained with both the GFP- and PE-matched negative controls. The percentages of stained cells found in each quadrant are indicated. The data shown are representative of three independent experiments.
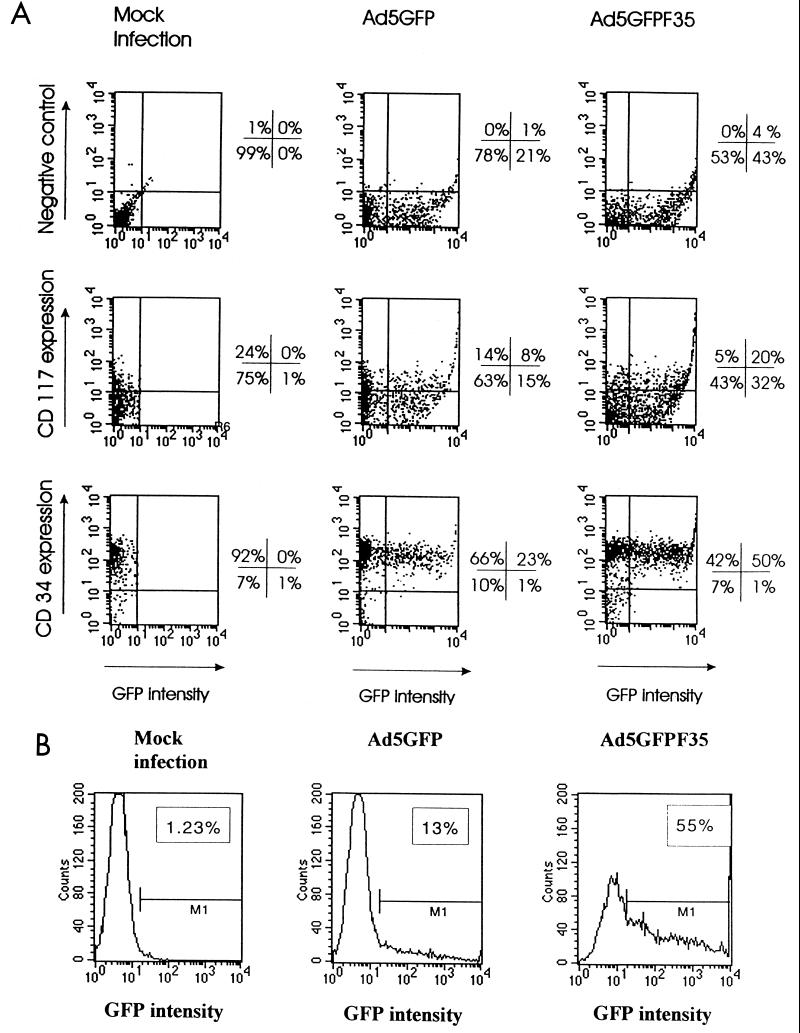
Next, transduced cells were simultaneously analyzed for GFP as well as for CD34 and CD117 markers. As mentioned before, only about 90% of all cells used in our analysis were positive for CD34 at the time of infection, hence the multiparameter analysis for CD34 and GFP. A population of CD34+ cells is extraordinarily heterogeneous in morphology and stem cell capacity. The subpopulation of CD34+ and CD117+ cells resembles very primitive hematopoietic cells (29, 67). Figure 10 summarizes the analyses of GFP expression in correlation with these specific stem cell markers. A total of 92%, 89 (66 + 23)%, or 92 (42 + 50)% of all analyzed cells were CD34 positive in the mock-infected, Ad5GFP-infected, or Ad5/35GFP-infected samples, respectively (Fig. 10A, lower panel). From these CD34-positive cells, 54% (50 out of 92) were also positive for GFP after infection with the Ad5GFP/F35 chimeric vector, whereas only 25% (23 out of 89) of all CD34-positive cells expressed GFP after infection with the Ad5 vector. Analogously, the percentage of GFP-expressing cells among all CD117-positive cells was calculated. From all CD117-positive cells, 80% (20 out of 25) were also positive for GFP after infection with the Ad5GFP/F35 chimeric vector, whereas only 36% (8 out of 22) of all CD117-positive cells expressed GFP after infection with the Ad5 vector (Fig. 10A, middle panel). In an additional experiment, CD34+ cells were sorted for CD117 expression prior to infection with Ad5GFP or Ad5GFP/F35; 24 h postinfection, GFP expression was analyzed in this specific fraction (Fig. 10B). This analysis revealed that the chimeric vectors transduced fourfold more CD34+ CD117+ cells than the Ad5GFP vector.
FIG. 10.
Distribution of GFP-positive cells in a subpopulation of human CD34+ cells expressing CD34 and CD117 (c-Kit). (A) Colocalization of GFP expression with CD34 or CD117. CD34+ cells were infected with Ad5GFP or Ad5GFP/F35 at an MOI of 200 PFU/cell under the conditions described for Fig. 8. Twenty-four hours after infection, cells were incubated with anti-CD34 PE-conjugated MAbs (final dilution, 1:2) or with anti-CD117 PE-conjugated MAbs (final dilution, 1:5) for 30 min on ice, and 104 cells per variant were subjected to two-color flow cytometry analysis. For negative control staining, no antibodies were added to the cells before analysis. The mock infection variants represent cells incubated with virus dilution buffer only. The quadrant borders were set based on the background signals obtained with both the GFP- and PE-matched negative controls. The percentages of stained cells found in each quadrant are indicated. The experiment was performed two times in triplicate, and typically obtained results are shown. The standard error of the mean was less than 10% of the statistical average. (B) Transduction of CD34+ CD117+ cells with Ad5GFP and chimeric Ad5GFP/F35 virus vectors. CD34+ cells, cultured overnight before staining in media without SCF, were incubated with PE-labeled anti-CD117 MAb for 30 min on ice. The fraction of CD117-positive cells was sorted by FACS. More than 97% of sorted cells were positive for CD117. CD117+ CD34+ cells (105) were infected with Ad5GFP or Ad5GFP/F35 at an MOI of 200 PFU/cell as for Fig. 8. Twenty-four hours postinfection, the percentage of GFP-positive cells was determined by flow cytometry. For mock infection, CD117+ CD34+ cells were incubated with virus dilution buffer only. The infections were done in triplicate, and the average percentage of GFP-expressing cells is indicated on the corresponding histogram. The standard error of the mean was less than 10% of the statistical average.
In conclusion, these results demonstrated that the chimeric Ad5GFP/F35 vector was clearly superior to the Ad5GFP vector in targeting and transduction of CD34+ cells. Furthermore, the data suggest that the spectrum of CD34+ cell subsets permissive for Ad infection was significantly different for the chimeric vector than for the Ad5 vector.
Analysis of viral genomes within CD34+ cells infected with the Ad5 and chimeric vectors.
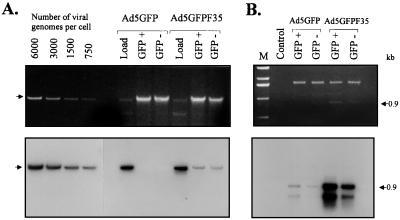
So far, the transduction rate of CD34+ cells was measured based on GFP expression after infection with Ad5GFP and Ad5GFP/F35. Considering the extraordinary heterogeneity of CD34+ cells in morphological and functional parameters, GFP may not be expressed in all cell types that were efficiently infected; reasons for this include that the CMV promoter is not active in all cell types or that the regulation of transgene expression differs between subsets on a posttranscriptional or posttranslational level. To test this, we quantified the number of intracellular (transduced) viral genomes within GFP-positive and GFP-negative fractions of CD34+ cells infected with Ad5GFP and Ad5GFP/F35. To do this, 24 h after infection, CD34+ cells were sorted for GFP-positive and GFP-negative fractions, which were subsequently used to isolate genomic DNA together with transduced viral DNA. The number of viral genomes was determined by quantitative Southern blot as described for Fig. 4. Per GFP-positive CD34+ cell, about 270 copies of the Ad5GFP/F35 viral genome were detected. Interestingly, a remarkable 200 copies of the Ad5GFP/F35 viral genome were found per GFP-negative CD34+ cell (Fig. 11A). This demonstrated that not all infected cells expressed GFP and implies that the actual transduction rate was higher than 54% (GFP-positive cells). We speculated that the CMV promoter was not active in all transduced CD34+ subsets. No Ad5GFP vector-specific signal was detected within infected CD34+ (GFP-positive or -negative) fractions by Southern blotting which had a detection limit of 14 viral genomes per cell. From this, we can conclude that the vector DNA concentration per transduced cell was at least 20 times higher for Ad5GFP/F35 than for Ad5GFP.
FIG. 11.
Southern analysis of viral genomes in GFP-positive and GFP-negative fractions of CD34+ cells infected with the Ad5GFP and chimeric Ad5GFP/F35 vectors. CD34+ cells were infected with viruses at an MOI of 100 as described for Fig. 8. Twenty-four hours postinfection, cells were sorted by FACS for GFP-positive and GFP-negative fractions; 105 cells from each fraction were used to isolate genomic DNA together with viral DNA. Before cell lysis, a rigorous treatment with trypsin and DNase followed by washing was performed to exclude that genomic DNA samples were contaminated by extracellular viral DNA. (A) The upper panel shows the ethidium bromide-stained 1% agarose gel before blotting, demonstrating that similar amounts of genomic DNA were loaded. This amount corresponded to DNA isolated from ∼25,000 GFP+ or GFP− cells. The lane labeled “Load” represents viral DNA purified from Ad5GFP or Ad5GFP/F35 virions mixed with pBluescript plasmid DNA (Stratagene) as a carrier and applied on a gel at the amount that was actually used to infect 25,000 cells. As a concentration standard, a serial dilution of Ad5GFP genomes was loaded on the gel (left). For Southern analysis (lower panel), an 8-kb-long HindIII fragment corresponding to the E2 region of Ad5 was used as a labeled probe. Hybridized filters were subjected to phosphorimager analysis and then exposed to Kodak X-Omat film for 48 h at −70°C. The cellular/viral genomic DNA is indicated by an arrow. (B) To detect Ad5GFP genomes in transduced cells, PCR amplification followed by Southern blot hybridization was performed on the same samples as used for quantitative Southern blot hybridization in panel A. DNA purified from ∼2,500 cells was subjected to PCR (95°C for 1 min, 53°C for 1 min, 72°C for 1 min; 20 cycles with primers Ad5-F1 and Ad5-R1). One-fifth of the PCR product was subjected to agarose gel electrophoresis (upper panel). A 0.9-kb-long DNA fragment specific to the E4 region of Ad5 was detected for transduced Ad5GFP/F35 genomes. DNA then was blotted onto Nybond-N+ membrane, and Southern blot hybridization (lower panel) with an Ad5 E4-specific DNA probe was performed. In addition to the 0.9-kb DNA fragment, the PCR primers generated a smaller 0.5-kb-long fragment that also hybridized with the E4 region probe.
Ad5GFP DNA was detectable in DNA samples from infected CD34+ cells by Southern blotting only after prior PCR amplification with vector-specific primers (Fig. 11B). This indicates that the replication deficient Ad5 vector is present but at a very low copy number, which may be limited by intracellular genome stability. Using the PCR-Southern detection method, Ad5 vector DNA was also detected in GFP-negative cells, supporting our hypothesis that the CMV promoter may not have been the optimal choice for transduction studies. It is notable that studies by others on viral genome analyses after infection of CD34+ cells with Ad5 vectors were performed only after prior PCR amplification (46, 50).
DISCUSSION
Our long-term goal is the development of viral vectors for stable gene transfer into human HSCs. Towards this end, we tested the interaction of selected Ad serotypes with CD34+ cells. As a result of this screening, we constructed a first-generation, Ad5-based vector whose fiber was substituted with the fiber derived from Ad35. We demonstrated that this capsid modification allowed for efficient viral transduction of potential HSCs by the corresponding chimeric Ad vectors.
All tropism and transduction studies were performed with noncycling CD34+ cells, which are thought to include HSCs. The quiescent stage of CD34+ cells purified from mobilized blood is important because induction of cell proliferation is associated with a loss of the ability to reconstitute hematopoiesis and with changes in the spectrum of cellular receptors (2, 22, 28).
Although it was reported earlier that fiber knobs derived from Ad serotypes 2, 9, 4, and 41L can bind to CAR in slot blot assays (60), it is not clear whether this binding occurs with an affinity that is physiologically relevant and whether this would confer virus infection. Furthermore, as shown for the Ad5 interaction between the penton and integrins, a secondary receptor is required to induce virus internalization. We demonstrated that different serotypes interacted differently with the K562 or CD34+ target cells. Ad5, Ad4, and Ad41 were not able to efficiently attach to and be internalized by K562 and CD34+ cells. Although Ad4 belongs to a separate subgroup (E), it is thought that Ad4 represents a natural hybrid between subgroup B and C viruses with a fiber related to Ad5 (23). Therefore, it was not surprising that Ad4 has binding properties similar to those of Ad5. The subgroup F serotype Ad41 has been shown to contain distinct fibers, a long-shafted and a short-shafted fiber allowing for different cell entry pathways (71). The Ad41 penton base does not contain RGD motifs, suggesting that this virus may use αv integrin-independent pathways for cell entry. However, these features did not improve interaction with CD34+ cells. Ad9, Ad3, and Ad35 did interact with CD34+ cells more efficiently than Ad5. Out of all the serotypes tested, Ad35 demonstrated the most efficient attachment and internalization with K562 and CD34+ cells. Although the short-shafted Ad9 can bind to CAR, it preferentially uses αv integrins for cell entry (59). Therefore, the low level of αv integrin expression on certain subsets of CD34+ cells may account for the observed susceptibility to Ad9.
Viral replication studies in K562 cells confirmed the data obtained for Ad5, Ad9, and Ad35 adsorption and internalization. However, there was a discrepancy between the interaction data and the replication data for CD34+ cells, where Ad9 replicated only poorly and no replication was seen for Ad35. Ad DNA replication is initiated only upon the production of a critical threshold of early viral proteins, which in turn is directly dependent on the number of viral genomes present in the nuclei of infected cells. Therefore, the outcome of replication studies may be affected by the rate of nuclear import of viral genomes, by the activity of viral promoters, and/or by the intracellular stability of viral DNA or RNA. These parameters may vary, on one hand, between different subsets of CD34+ and/or, on the other hand, between different Ad serotypes. This implies that viral replication analyses performed with different Ad serotypes in CD34+ cells may not predict the actual transduction properties of chimeric vectors based on Ad5 backbones.
Recently, an Ad serotype screening strategy was used to identify variants with tropism for primary fetal rat central nervous system cortex cells or human umbilical vein endothelial cells. The optimal serotype (Ad17) was selected based on immunohistochemistry for hexon production 48 h after infection (10). However, this approach is problematic because, at least in our hands, antibodies developed against Ad5 hexon did not efficiently cross-react with all other serotypes. Also, hexon is expressed only after onset of replication. As outlined above, the kinetics of intracellular trafficking, viral gene expression, and replication significantly vary between serotypes (15, 48).
In addition to being the most efficient serotype in terms of interaction with CD34+ cells, Ad35 is also interesting because it interacts with receptors different from those with which Ad5 interacts. Ad35 and Ad5GFP/F35 attachment was not inhibited by Ad5 or anti-CAR antibodies, suggesting that Ad35 binding was CAR independent. Furthermore, our data indicate that infection with Ad35 and the chimeric Ad5GFP/F35 vector does not involve αv integrins. First, Ad5 did not compete with Ad35 and Ad5GFP/F35 in internalization or infection studies, indicating that αvβ3/5 integrins may not be involved in viral entry. Second, function-blocking antibodies against αv integrins did not compete with Ad35 and Ad5GFP/F35 for internalization into K562 cells, whereas these antibodies did inhibit Ad5 internalization. Third, in contrast to Ad5-based vectors, GFP expression after infection with Ad5GFP/F35 was not restricted to αv integrin-expressing CD34+ cells. In this context, the presence or absence of RGD motifs within Ad35 penton base remains to be determined by sequencing the corresponding genome region. Cross-competition assays demonstrated that Ad35 and Ad5GFP/F35 bind to a receptor that is different from the Ad3 receptor. Although Ad3 and Ad35 belong to the same subgroup, they have been divided into two DNA homology clusters, B1 and B2; the amino acids composing their fibers are only 60% homologous. Furthermore, the target tissues for the two viruses are different; Ad3 can cause acute respiratory infections, whereas Ad35 is associated with kidney infections (25). Therefore, it was not surprising to find that Ad3 and Ad35 recognize different receptors.
In conclusion, Ad35 and the chimeric vector enter the cells by a CAR- and αv integrin-independent pathway. We speculate that Ad35 and the chimeric vector binds primarily to its fiber receptor. This interaction may be sufficient to trigger internalization. On the other hand, Ad35 internalization may involve cellular proteins which appear to overlap with those for Ad3 internalization and may represent β2 integrins, which protrude more from the cell surface than αv integrins (27).
Fiber substitution was sufficient to swap cell tropism from Ad5 to Ad35. The Ad5GFP/F35 capsid chimera contained the short-shafted Ad35 fiber incorporated into an Ad5 capsid, instead of the naturally occurring long-shafted Ad5 fiber. The length of fiber shaft and the precise spatial arrangement of knob and RGD motifs appear to be critical for the viral entry strategies. So far, most of the chimeric viruses have been generated by substituting only the Ad5 knob while maintaining the long Ad5 fiber shaft (10, 32, 69, 70). The exception was an Ad5/7 chimeric virus (20), where the whole Ad5 fiber was substituted by the short-shafted Ad7 fiber. However, similar to the parental Ad5, the Ad5/7 chimera still required αv integrins for infection. Importantly, the Ad5/35 capsid chimera allows for efficient infection of K652 and CD34+ cells, suggesting that the protruding RGD motives in the Ad5 penton base do not affect the interaction with the primary Ad35 receptor. We hypothesize that despite the presence of RGD motifs within the Ad5 penton, the chimeric virus uses cell entry pathways determined primarily by the receptor specificity of the short-shafted heterologous fiber.
Our data suggest that infection with Ad5-based vectors is restricted to a specific subset of CD34+ cells. The percentage of GFP-expressing cells after Ad5GFP infection of CD34+ cells reached a plateau at MOIs higher than 100, indicating that only a limited fraction of CD34+ cells was permissive to Ad5. Also, strong replication of wild-type Ad5 in infected CD34+ cells may be the result of preferential transduction of a specific subpopulation of CD34+, resulting in an expression of early viral genes at a level sufficient to initiate viral replication. The presence of a specific subpopulation of CD34+ cells permissive to Ad5 vector infection was suggested by others (6, 50). In the present report, we further characterized this subpopulation and demonstrated that Ad5-based vectors preferentially infected αv integrin-positive CD34+ cells. Integrins (including αv) are thought to be important for homing and trafficking of transplanted hematopoietic cells; however, little is known about the correlation between αv integrin expression and the differentiation status of hematopoietic cells (54, 61). Importantly, infection with the chimeric Ad5GFP/F35 vector was not restricted to the αv-positive CD34+ subpopulation.
Among CD34+ cells, the subpopulation of CD34+ and CD117+ cells resembles very primitive hematopoietic cells (29, 67). The receptor for stem cell factor, CD117 (c-Kit), belongs to a tyrosine kinase family. It was previously shown that c-Kit+ CD34+ cord blood cells contain a high fraction (16%) of hematopoietic progenitors (52) and that early in ontogeny CD34+ CD117+ cells have long-term repopulating activity (62). In our studies, the chimeric vector expressed GFP in 54% CD34+ cells and 80% of CD34+ c-Kit+ cells. The actual viral transduction rate could be even higher because transduced Ad5GFP/F35 vector DNA was also found in GFP-negative fractions of infected cells. This indicates that the CMV promoter used to drive GFP expression in our vectors was not active in all transduced cells. We selected the CMV promoter for transgene expression based on published data demonstrating that phosphoglycerate kinase and CMV promoters allowed for efficient transgene expression in CD34+ cells, whereas the human T-cell leukemia virus type 1 and Rous sarcoma virus promoters were almost inactive (6, 7). On the other hand, studies by Watanabe et al. (74) suggest that the CMV promoter is not active or rapidly silenced in certain CD34+ subsets. Our data underscore this observation. Considering retroviral transduction studies, the retroviral murine leukemia virus promoter may have been a better candidate for transduction studies in hematopoietic cells (5).
After having demonstrated that the Ad5GFP/F35 vector efficiently transduced cells carrying stem cell-specific markers, the next logical step would be to perform colony assays with presorted GFP-positive and -negative cells. However, this assay is complicated by the fact that infection with first-generation Ad vectors is cytotoxic and affects the formation and growth of progenitor colonies in semi-solid cultures (46, 74). This side effect is caused by the expression of Ad proteins within transduced cells (41, 63, 81). Some of these proteins (e.g., E4-orf4, pTP, and E3-11.6k) have proapoptotic activity or induce cell cycle arrest (34, 39, 66, 72). We plan to perform colony assays or, preferably, repopulation assays in SCID-NOD mice with gutless vectors (68) or integrating ΔAd.AAV vectors devoid of all viral genes (40) generated based on Ad5GFP/F35 chimeric capsids. Alternatively, gutless, retargeted vectors could be used to transiently express a retroviral receptor on CD34+ cells to increase their susceptibility to infection with retroviral vectors based on an approach that we have published earlier (38).
Our finding that Ad5GFP/F35 can efficiently transduce hematopoietic cells with potential stem cell capacity represents an important step toward stable gene transfer into HSCs and gene therapy of blood disorders. Furthermore, the virological aspects of our study contribute to a better understanding of Ad-cell interactions.
ACKNOWLEDGMENTS
We thank Denise Farrer and Zong-Yi Li for excellent technical assistance. We are grateful to Shelly Heimfeld (Fred Hutchinson Cancer Research Center, Seattle, Wash.) for generously providing human CD34+ cells. We acknowledge Jeffrey Bergelson (University of Pennsylvania School of Medicine) for providing the antibodies against CAR. We thank Cheryl Carlson and Dirk Steinwaerder for critical discussions.
This work was supported by NIH grants R01 CA80192 (A.L.), R21 DK55590 (A.L.), and P01 HL53750 (G.S., A.L.).
REFERENCES
- 1.Bailey A, Mautner V. Phylogenetic relationships among adenovirus serotypes. Virology. 1994;205:438–452. doi: 10.1006/viro.1994.1664. [DOI] [PubMed] [Google Scholar]
- 2.Becker P S, Nilsson S K, Li Z, Berrios V M, Dooner M S, Cooper C L, Hsieh C C, Quesenberry P J. Adhesion receptor expression by hematopoietic cell lines and murine progenitors: modulation by cytokines and cell cycle status. Exp Hematol. 1999;27:533–541. doi: 10.1016/s0301-472x(98)00037-x. [DOI] [PubMed] [Google Scholar]
- 3.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 4.Bodine D M, Moritz T, Donahue R E, Luskey B D, Kessler S W, Martin D I K, Orkin S H, Nienhuis A W, Williams D A. Long-term in vivo expansion of a murine adenosine deaminase gene in rhesus monkey hematopoietic cells of multiple lineages after retroviral mediated gene transfer into CD34+ bone marrow cells. Blood. 1993;82:1975–1980. [PubMed] [Google Scholar]
- 5.Bregni M, Shammah S, Malaffo F, Di Nicola M, Milanesi M. Adenovirus vectors for gene transduction into mobilized blood CD34+ cells. Gene Ther. 1998;5:465–472. doi: 10.1038/sj.gt.3300620. [DOI] [PubMed] [Google Scholar]
- 6.Byk T, Haddada H, Vainchenker W, Louache F. Lipofectamine and related cationic lipids strongly improve adenoviral infection efficiency of primitive human hematopoietic cells. Hum Gene Ther. 1998;9:2493–2502. doi: 10.1089/hum.1998.9.17-2493. [DOI] [PubMed] [Google Scholar]
- 7.Case S S, Price M A, Jordan C T, Yu X J, Wang L, Bauer G, Haas D L, Xu D, Stripecke R, Naldini L, Kohn D B, Crooks G M. Stable transduction of quiescent CD34+CD38− human hematopoietic cells by HIV-1-based lentiviral vectors. Proc Natl Acad Sci USA. 1999;96:2988–2993. doi: 10.1073/pnas.96.6.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Pulsipher M, Chen D, Sieff C, Elias A, Fine H A, Kufe D W. Selective transgene expression for detection and elimination of contaminating carcinoma cells in hematopoietic stem cell sources. J Clin Investig. 1996;98:2539–2548. doi: 10.1172/JCI119072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chillon M, Bosch A, Zabner J, Law L, Armentano D, Welsh M, Davidson B L. Group D adenoviruses infect primary central nervous system cells more efficiently than those from group C. J Virol. 1999;73:2537–2540. doi: 10.1128/jvi.73.3.2537-2540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chroboczek J, Ruigrok R W H, Cusack S. Adenovirus fiber. In: Doerfler P B W, editor. The molecular repertoire of adenoviruses. Vol. 1. Berlin, Germany: Springer-Verlag; 1995. pp. 163–200. [DOI] [PubMed] [Google Scholar]
- 12.Clarke M F, Apel I J, Benedict M A, Eipers P G, Sumantran V, Gonzalez-Garcia M, Doedens M, Fukunaga N, Davidson B, Dick J E. A recombinant bcl-x adenovirus selectively induces apoptosis in cancer cells but not in normal bone marrow cells. Proc Natl Acad Sci USA. 1995;92:11024–11028. doi: 10.1073/pnas.92.24.11024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crompton J, Toogood C I, Wallis N, Hay R T. Expression of a foreign epitope on the surface of the adenovirus hexon. J Gen Virol. 1994;75:133–139. doi: 10.1099/0022-1317-75-1-133. [DOI] [PubMed] [Google Scholar]
- 14.Crooks G M, Kohn D B. Growth factors increase amphotropic retrovirus binding to human CD34+ bone marrow progenitor cells. Blood. 1993;82:3290–3297. [PubMed] [Google Scholar]
- 15.Defer C, Belin M-T, Caillet-Boudin M-C, Boulanger P. Human adenovirus-host cell interactions: comparative study with members of subgroups B and C. J Virol. 1990;64:3661–3673. doi: 10.1128/jvi.64.8.3661-3673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldman E, Ahmed T, Luton J D, Farley T, Tani K, Freud M, Asano S, Abraham N G. Adenovirus mediated alpha-interferon gene transfer into CD34+ cells and CML mononuclear cells. Stem Cells. 1997;15:386–395. doi: 10.1002/stem.150386. [DOI] [PubMed] [Google Scholar]
- 17.Flomenberg P, Babbitt J, Drobyski W, Ash R C, Camitta B, Horowitz M M, Bunin N, Casper J T. Increasing incidence of adenovirus disease in bone marrow transplant recipients. J Infect Dis. 1994;169:775–781. doi: 10.1093/infdis/169.4.775. [DOI] [PubMed] [Google Scholar]
- 18.Flomenberg P R, Chen M, Munk G, Horwitz M S. Molecular epidemiology of adenovirus type 35 infections in immunocompromised hosts. J Infect Dis. 1987;155:1127–1134. doi: 10.1093/infdis/155.6.1127. [DOI] [PubMed] [Google Scholar]
- 19.Frey B M, Hackett N R, Bergelson J M, Finberg R, Crystal R G, Moore M A S, Rafii S. High-efficiency gene transfer into ex vivo expanded human hematopoietic progenitors and precursor cells by adenovirus vectors. Blood. 1998;91:2781–2792. [PubMed] [Google Scholar]
- 20.Gall J, Kass-Eisler A, Leinwand L, Falck-Pedersen E. Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralizing epitopes. J Virol. 1996;70:2116–2123. doi: 10.1128/jvi.70.4.2116-2123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldman M, Su Q, Wilson J M. Gradient of RGD-dependent entry of adenoviral vector in nasal and intrapulmonary epithelia: implication for gene therapy of cystic fibrosis. Gene Ther. 1996;3:811–818. [PubMed] [Google Scholar]
- 22.Gonzalez R, Verecque R, Wickham T J, Vanrumbeke M, Kovesdi I, Bauters F, Fenaux P, Quesnel B. Increased gene transfer in acute myeloid leukemic cells by an adenovirus vector containing a modified fiber protein. Gene Ther. 1999;6:314–320. doi: 10.1038/sj.gt.3300836. [DOI] [PubMed] [Google Scholar]
- 23.Gruber W C, Russell D J, Tibbetts C. Fiber gene and genomic origin of human adenovirus type 4. Virology. 1993;196:603–611. doi: 10.1006/viro.1993.1516. [DOI] [PubMed] [Google Scholar]
- 24.Hitt M M, Addison C J, Graham F J. Human adenoviral vectors for gene transfer into mammalian cells. Adv Pharmacol. 1997;40:137–205. doi: 10.1016/s1054-3589(08)60140-4. [DOI] [PubMed] [Google Scholar]
- 25.Horwitz M S. Adenoviruses. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers Inc.; 1996. pp. 2149–2171. [Google Scholar]
- 26.Hsu K-H, Lonberg-Holm K, Alstein B, Crowell R L. A monoclonal antibody specific for the cellular receptor for the group B coxsackieviruses. J Virol. 1988;62:1647–1652. doi: 10.1128/jvi.62.5.1647-1652.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang S, Kamata Y, Takada Y, Ruggeri Z M, Nemerow G R. Adenovirus interaction with distinct integrins mediates separate events in cell entry and gene delivery to hematopoietic cells. J Virol. 1996;70:4502–4508. doi: 10.1128/jvi.70.7.4502-4508.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang S, Endo R I, Nemerow G R. Upregulation of integrins αv-β3 and αv-β5 on human monocytes and T lymphocytes facilitates adenovirus-mediated gene delivery. J Virol. 1995;69:2257–2263. doi: 10.1128/jvi.69.4.2257-2263.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikuta K, Weissman I L. Evidence that hematopoietic stem cells express mouse c-kit but do not depend on steel factor for their generation. Proc Natl Acad Sci USA. 1992;89:1502–1506. doi: 10.1073/pnas.89.4.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim M H, Wright M, Deshane J, Accavitti M A, Tilden A, Saleh M, Vaughan W P, Carabasi M H, Rogers M D, Hockett R D, Grizzle W E, Curiel D T. A novel gene therapy strategy for elimination of prostate carcinoma cells from human bone marrow. Hum Gene Ther. 1997;8:157–170. doi: 10.1089/hum.1997.8.2-157. [DOI] [PubMed] [Google Scholar]
- 31.Krasnykh V, Dmitriev I, Mikheeva G, Miller C R, Belousova N, Curiel D T. Characterization of an adenovirus vector containing a heterologous peptide epitope in the HI loop of the fiber knob. J Virol. 1998;72:1844–1852. doi: 10.1128/jvi.72.3.1844-1852.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krasnykh V N, Mikheeva G V, Douglas J T, Curiel D T. Generation of recombinant adenovirus vectors with modified fibers for altering viral tropism. J Virol. 1996;70:6839–6846. doi: 10.1128/jvi.70.10.6839-6846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwoh T J, Kwoh D Y, McCue A W, Davis G R, Patrick D, Gingeras T R. Introduction and expression of the bacterial PaeR7 methylase gene in mammalian cells. Proc Natl Acad Sci USA. 1986;83:7713–7717. doi: 10.1073/pnas.83.20.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langer S J, Schaak J. 293 cell lines that inducibly express high level adenovirus type 5 precursor terminal protein. Virology. 1996;221:172–179. doi: 10.1006/viro.1996.0363. [DOI] [PubMed] [Google Scholar]
- 35.Larochelle A, Vormoor J, Hananber H, Wang J C Y, Bhatia M, Lapidot T, Moritz T, Murdoch B, Xiao X L, Kato I, Williams D A, Dick J E. Identification of primitive human hematopoietic cells capable of repopulating NOD/SCID mouse bone marrow: implication for gene therapy. Nat Med. 1996;2:1329–1337. doi: 10.1038/nm1296-1329. [DOI] [PubMed] [Google Scholar]
- 36.Legrand V, Spehner D, Schlesinger Y, Settelen N, Pavirani A, Mehtali M. Fiberless recombinant adenoviruses: virus maturation and infectivity in the absence of fiber. J Virol. 1999;73:907–919. doi: 10.1128/jvi.73.2.907-919.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leitner A, Strobl F, Fischmeister M, Kurz K, Romanakis O A, Haas D, Pintz P, Buchinger S, Bauer H, Gadner H, Fritsch G. Lack of DNA synthesis among CD34+ cells in cord blood and in cytokine mobilized blood. Br J Haematol. 1996;92:255–262. doi: 10.1046/j.1365-2141.1996.d01-1473.x. [DOI] [PubMed] [Google Scholar]
- 38.Lieber A, Vrancken Peeters M-J, Kay M A. Adenovirus-mediated transfer of the amphotropic retrovirus receptor cDNA increases retroviral transduction in cultured cells. Hum Gene Ther. 1995;6:5–11. doi: 10.1089/hum.1995.6.1-5. [DOI] [PubMed] [Google Scholar]
- 39.Lieber A, He C-Y, Meuse L, Himeda C, Wilson C, Kay M A. Inhibition of NF-κB activation in combination with Bcl-2 expression allows for persistence of first-generation adenovirus vectors in mouse livers. J Virol. 1998;72:9267–9277. doi: 10.1128/jvi.72.11.9267-9277.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lieber A, Steinwaerder D, Carlson C A, Li Z Y, Kay M A. Integrating adenovirus–adeno-associated virus hybrid vectors devoid of all viral genes. J Virol. 1999;73:9314–9324. doi: 10.1128/jvi.73.11.9314-9324.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lieber A, He C-Y, Kirillova I, Kay M A. Recombinant adenoviruses with large deletions generated by Cre-mediated excision exhibit different biological properties compared with first-generation vectors in vitro and in vivo. J Virol. 1996;70:8944–8960. doi: 10.1128/jvi.70.12.8944-8960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maizel J V, White D, Scharff M D. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7a, and 12. Virology. 1968;36:115–125. doi: 10.1016/0042-6822(68)90121-9. [DOI] [PubMed] [Google Scholar]
- 43.Mathias P, Wickham T, Moore M, Nemerow G. Multiple adenovirus serotypes use α-v integrins for infection. J Virol. 1994;68:6811–6814. doi: 10.1128/jvi.68.10.6811-6814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGuckin C P, Liu W M, Gordon-Smith E C, Uhr M R. A novel approach to investigating the erythroid lineage, using both receptor analysis and haemaglobin detection. Br J Haematol. 1996;95:457–460. doi: 10.1046/j.1365-2141.1996.d01-1942.x. [DOI] [PubMed] [Google Scholar]
- 45.Michael S I, Hong J S, Curiel D T, Engler J A. Addition of a short peptide ligand to the adenovirus fiber protein. Gene Ther. 1995;2:660–668. [PubMed] [Google Scholar]
- 46.Mitani K, Graham F L, Caskey C T. Transduction of human bone marrow by adenoviral vectors. Hum Gene Ther. 1994;5:941–948. doi: 10.1089/hum.1994.5.8-941. [DOI] [PubMed] [Google Scholar]
- 47.Mittereder N, March K L, Trapnell B C. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J Virol. 1996;70:7498–7509. doi: 10.1128/jvi.70.11.7498-7509.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyazawa N, Leopold P L, Hackett N R, Ferris B, Worgall S, Falck-Pedersen E, Crystal R. Fiber swap between adenovirus subgroups B and C alters intracellular trafficking of adenovirus gene transfer vectors. J Virol. 1999;73:6056–6065. doi: 10.1128/jvi.73.7.6056-6065.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizuguchi H, Kay M A. Efficient construction of recombinant adenovirus vectors by an improved in vitro ligation method. Hum Gene Ther. 1998;9:2577–2583. doi: 10.1089/hum.1998.9.17-2577. [DOI] [PubMed] [Google Scholar]
- 50.Neering S J, Hardy S F, Minamoto D, Spratt S K, Jordan C T. Transduction of primitive human hematopoietic cells with recombinant adenovirus vectors. Blood. 1996;88:1147–1155. [PubMed] [Google Scholar]
- 51.Nelson J, Kay M A. Persistence of recombinant adenovirus in vivo is not dependent on vector replication. J Virol. 1997;71:8902–8907. doi: 10.1128/jvi.71.11.8902-8907.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neu S, Geiselhart A, Kuci S, Baur F, Niethammer D, Handgretinger R. Isolation and phenotypic characterization of CD117 positive cells. Leuk Res. 1996;20:960–971. doi: 10.1016/s0145-2126(96)00077-x. [DOI] [PubMed] [Google Scholar]
- 53.Orlic D, Girard L J, Anderson S M, Do B K Q, Seidel N E, Jordan C T, Bodine D M. Transduction efficiency of cell lines and hematopoietic stem cells correlates with retrovirus receptor mRNA levels. Stem Cells. 1997;15:23–28. doi: 10.1002/stem.5530150805. [DOI] [PubMed] [Google Scholar]
- 54.Papayannopoulou T, Craddock C. Homing and trafficking of hematopoietic progenitor cells. Acta Haematol. 1997;97:97–104. doi: 10.1159/000203665. [DOI] [PubMed] [Google Scholar]
- 55.Pring Akerblom P, Heim A, Trijssenaar F J. Molecular characterization of hemagglutination domains on the fibers of subgenus D adenoviruses. J Virol. 1998;72:2297–2304. doi: 10.1128/jvi.72.3.2297-2304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qui C, De Young M B, Finn A, Dichek D A. Cationic liposomes enhance adenovirus entry via a pathway independent of the fiber receptor and αv integrins. Hum Gene Ther. 1998;9:507–520. doi: 10.1089/hum.1998.9.4-507. [DOI] [PubMed] [Google Scholar]
- 57.Roberts A W, Metcalf D. Noncycling state of peripheral blood progenitors cells mobilized by granulocyte colony-stimulating factor and other cytokines. Blood. 1995;86:1600–1605. [PubMed] [Google Scholar]
- 58.Rodriguez E, Everitt E. Adenovirus cellular receptor site recirculation of HeLa cells upon receptor mediated endocytolysis is not low pH dependent. Arch Virol. 1999;144:787–795. doi: 10.1007/s007050050544. [DOI] [PubMed] [Google Scholar]
- 59.Roelvink P W, Kovesdi J, Wickham T J. Comparative analysis of adenovirus fiber-cell interaction: adenovirus type 2 (Ad2) and Ad9 utilize the same fiber receptor but use different binding strategies for attachment. J Virol. 1996;70:7614–7621. doi: 10.1128/jvi.70.11.7614-7621.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roelvink P W, Lizonova A, Lee J G, Li Y, Bergelson J M, Finberg R W, Brough D E, Kovesdi I, Wickham T J. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J Virol. 1998;72:7909–7915. doi: 10.1128/jvi.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roy V, Verfaillie C M. Expression and function of cell adhesion molecules on fetal liver, cord blood, and bone marrow hematopoietic progenitors; implications for anatomical localization and developmental stage specific regulation of hematopoiesis. Exp Hematol. 1999;27:302–312. doi: 10.1016/s0301-472x(98)00031-9. [DOI] [PubMed] [Google Scholar]
- 62.Sanchez M J, Holmes A, Miles C, Dzierzak E. Characterization of the first definitive hematopoietic stem cells in the AGM and liver of mouse embryo. Immunity. 1996;5:513–525. doi: 10.1016/s1074-7613(00)80267-8. [DOI] [PubMed] [Google Scholar]
- 63.Schiedner G, Morral N, Parks R J, Wu Y, Koopmans S C, Langston C, Graham F L, Beaudet A L, Kochanek S. Genomic DNA transfer with a high-capacity adenovirus vector results in improved in vivo gene expression and decreased toxicity. Nat Genet. 1998;18:180–183. doi: 10.1038/ng0298-180. [DOI] [PubMed] [Google Scholar]
- 64.Shenk T. Adenoviridae. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publisher; 1996. pp. 2111–2148. [Google Scholar]
- 65.Shields A F, Hackman R C, Fife K H, Corey L, Meyers J D. Adenovirus infections in patients undergoing bone-marrow transplantations. N Eng J Med. 1985;312:529–533. doi: 10.1056/NEJM198502283120901. [DOI] [PubMed] [Google Scholar]
- 66.Shtrichman R, Kleinberger T. Adenovirus type 5 E4 open reading frame 4 protein induces apoptosis in transformed cells. J Virol. 1998;72:2975–2983. doi: 10.1128/jvi.72.4.2975-2982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simmons P J, Aylett G W, Niuatta S, To J B, Juttner C A, Ashman J K. c-kit is expressed by primitive human hematopoietic cells that give rise to colony-forming cells in stroma-dependent or cytokine-supplemented culture. Exp Hematol. 1994;22:157–165. [PubMed] [Google Scholar]
- 68.Steinwaerder D S, Carlson C A, Lieber A. Generation of adenoviral vectors devoid of all viral genes by recombination between inverted repeats. J Virol. 1999;73:9303–9313. doi: 10.1128/jvi.73.11.9303-9313.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stevenson S C, Rollence M, White B, Wever L, McClelland A. Human adenovirus serotypes 3 and 5 bind to two different cellular receptors via the fiber head domain. J Virol. 1995;69:2850–2857. doi: 10.1128/jvi.69.5.2850-2857.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stevenson S C, Rollence M, Marshall-Neff J, McClelland A. Selective targeting of human cells by a chimeric adenovirus vector containing a modified fiber protein. J Virol. 1997;71:4782–4790. doi: 10.1128/jvi.71.6.4782-4790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tiemessen C T, Kidd A H. The subgroup F adenoviruses. J Gen Virol. 1995;76:481–497. doi: 10.1099/0022-1317-76-3-481. [DOI] [PubMed] [Google Scholar]
- 72.Tollefson A E, Scaria A, Hermiston T W, Ryerse J S, Wold L J, Wold W S M. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J Virol. 1996;70:2296–2306. doi: 10.1128/jvi.70.4.2296-2306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tomko R P, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Watanabe T, Kuszynski C, Ino K, Heiman D G, Shephard H M, Yasul Y, Maneval D C, Talmadge J E. Gene transfer into human bone marrow hematopoietic cells mediated by adenovirus vectors. Blood. 1996;87:5032–5039. [PubMed] [Google Scholar]
- 75.Watkins S J, Mesyanzhinov V V, Kurochkina L P, Hawkins R E. The antibody approach to viral targeting-specific and enhanced adenoviral gene delivery. Gene Ther. 1997;4:1004–1012. doi: 10.1038/sj.gt.3300511. [DOI] [PubMed] [Google Scholar]
- 76.Wickham T, Roelvink P, Brough D, Kovesdi I. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat Biotechnol. 1996;14:1570–1573. doi: 10.1038/nbt1196-1570. [DOI] [PubMed] [Google Scholar]
- 77.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Integrins alpha-v-beta3 and alpha-v-beta5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 78.Wickham T J, Segal D M, Roelvink P W, Carrion M E, Lizonova G M, Kovesdi I. Targeted adenovirus gene transfer to endothelial and smooth muscle cells by using bispecific antibodies. J Virol. 1996;70:6831–6838. doi: 10.1128/jvi.70.10.6831-6838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wickham T J, Carrion M E, Kovesdi I. Targeting of adenovirus penton base to new receptors through replacement of its RGD motif with other receptor-specific peptide motifs. Gene Ther. 1995;2:750–756. [PubMed] [Google Scholar]
- 80.Wroblewski J M, Lay L T, Vant Zant G, Phillips G, Seth P, Curiel D, Meeker T C. Selective elimination of contaminating malignant cells from hematopoietic stem cell autografts using recombinant adenovirus. Cancer Gene Ther. 1996;3:257–264. [PubMed] [Google Scholar]
- 81.Yang Y, Nunes F A, Berencsi K, Furth E E, Gonczol E, Wilson J M. Cellular immunity to viral antigens limits E1-deleted adenoviruses for gene therapy. Proc Natl Acad Sci USA. 1994;91:4407–4411. doi: 10.1073/pnas.91.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zabner J, Puga A, Freimuth P, Welsh M J. Lack of high affinity fiber receptor activity explains the resistance of ciliated airway epithelia to adenovirus infection. J Clin Investig. 1997;100:1144–1149. doi: 10.1172/JCI119625. [DOI] [PMC free article] [PubMed] [Google Scholar]