Abstract
A major challenge for the next generation of human immunodeficiency virus (HIV) vaccines is the induction of potent, broad, and durable cellular immune responses. The structural protein Gag is highly conserved among the HIV type 1 (HIV-1) gene products and is believed to be an important target for the host cell-mediated immune control of the virus during natural infection. Expression of Gag proteins for vaccines has been hampered by the fact that its expression is dependent on the HIV Rev protein and the Rev-responsive element, the latter located on the env transcript. Moreover, the HIV genome employs suboptimal codon usage, which further contributes to the low expression efficiency of viral proteins. In order to achieve high-level Rev-independent expression of the Gag protein, the sequences encoding HIV-1SF2 p55Gag were modified extensively. First, the viral codons were changed to conform to the codon usage of highly expressed human genes, and second, the residual inhibitory sequences were removed. The resulting modified gag gene showed increases in p55Gag protein expression to levels that ranged from 322- to 966-fold greater than that for the native gene after transient expression of 293 cells. Additional constructs that contained the modified gag in combination with modified protease coding sequences were made, and these showed high-level Rev-independent expression of p55Gag and its cleavage products. Density gradient analysis and electron microscopy further demonstrated that the modified gag and gagprotease genes efficiently expressed particles with the density and morphology expected for HIV virus-like particles. Mice immunized with DNA plasmids containing the modified gag showed Gag-specific antibody and CD8+ cytotoxic T-lymphocyte (CTL) responses that were inducible at doses of input DNA 100-fold lower than those associated with plasmids containing the native gag gene. Most importantly, four of four rhesus monkeys that received two or three immunizations with modified gag plasmid DNA demonstrated substantial Gag-specific CTL responses. These results highlight the useful application of modified gag expression cassettes for increasing the potency of DNA and other gene delivery vaccine approaches against HIV.
The induction of long-lasting, potent humoral and cellular immune responses will be important for an effective human immunodeficiency virus (HIV) vaccine. Data from HIV-infected patients, and in particular from long-term nonprogressors, have shown that viral structural genes can elicit substantial immune responses. Gag-specific CD8+ cytotoxic T lymphocytes (CTL) have been shown to be important in controlling virus load during acute infection (4, 21) as well as during the asymptomatic stages of the infection (20, 24). Moreover, a strong Gag-specific CTL response appears to correlate inversely with the viral load of HIV-1-infected patients (7). In addition, studies of exposed but uninfected prostitutes indicate that Gag-specific CTL may be involved in protection against the establishment of a persistent HIV type 1 (HIV-1) infection (28). Combined, these studies provide convincing evidence that immune responses directed against HIV Gag proteins may be an important component of an effective HIV vaccine. The usefulness of Gag immunogens for vaccines is further indicated by the fact that the protein is relatively conserved among diverse HIV strains and subtypes, and cross-clade CTL recognition directed against Gag-specific targets has been well documented (2, 3, 11, 23).
Immunization with naked DNA or recombinant virus induces both antibody and CTL responses and has been shown to be an efficient method of eliciting protective immune responses against a broad range of pathogens in animal studies (10). However, the potency of current gene delivery methods such as naked-DNA and viral vectors must be improved to induce adequately robust responses for protection in primates (1). One means to achieve this may be through increasing the expression efficiency of encoded HIV antigens. The poor expression of the HIV structural genes in recombinant vectors is caused by a strong Rev dependency that allows efficient expression only in the presence of the viral Rev protein (25, 30). The translation efficiency and stability of gag transcripts are further decreased by the presence of a relatively high AU content and destabilizing AUUUA motifs (inhibitory sequences [INS]). In previous studies, inactivation of these INS enabled the Rev-independent expression of HIV-1 gag (29), but these modifications reduced the approximate AT content of the gag gene only from 56 to 50%. Elevated percentages of AU in human mRNAs have been shown to result in instability, increased turnover, and low expression levels (15). These findings suggest that further reductions of the AT content of the gag gene could result in improved mRNA stability and increased protein expression. In support of this, it has been shown that highly expressed human genes employ codon usage patterns different from those used by HIV genomes. For highly expressed genes, G or C is generally preferred over A or T. Furthermore, changes in the codon usage of HIV-1 env to those employed by highly expressed human codons resulted in increased Rev-independent expression (14).
In order to achieve high-level Rev-independent expression of the gag gene of HIV-1SF2, the codon usage pattern was first altered to conform to that used by highly expressed human genes (14). Further modifications were then made to remove possible residual INS motifs previously identified in the gag coding region (29). This resulted in lowering the AT content of the gag coding sequences from 56 to 32%, a level more consistent with increased mRNA stability and translation efficiency. The sequence-modified HIV-1SF2 gag gene was inserted into a high-level plasmid expression vector for in vitro transfections and DNA immunization studies with rodents and nonhuman primates. Results presented here indicate that sequence-modified gag plasmids expressed protein at dramatically higher levels and showed increased immunogenicity compared to the native gag sequence in DNA immunization experiments performed with mice and rhesus macaques. Additionally, the inclusion of modified protease-coding sequences in the modified gag resulted in high-level Rev-independent expression, processing of the Gagprotease polyprotein, and the production of virus-like particles (VLP) with the morphologies of both immature and mature HIV-1 virions.
MATERIALS AND METHODS
gag and gagprotease plasmids.
The native sequences coding for the 502 amino acids (aa) of HIV-1SF2 p55Gag (GenBank accession no. K02007) were modified to change the codon usage to that utilized by highly expressed human genes as described recently for HIV-1MN gp120 (14). In addition, regions with INS were further inactivated without altering the reading frame for the p55Gag nucleic acid sequence. The resulting modified HIV-1SF2 gag encoded a p55Gag protein with three amino acid changes (Asn377Thr, Ile403Thr, and Lys405Arg); the resulting amino acid sequence conformed to the sequences for other HIV-1 subtype B Gag proteins in the HIV sequence database (Los Alamos National Laboratory; http://hiv-web.lanl.gov/cgi-bin/hivDB3/public/wdb/ssampublic) (GenBank accession no. AF201927). To further enhance the translation efficiency of the modified gag, an optimal consensus sequence for the initiation of translation (GCCACCAUGG) was employed (22). The resulting 1,527-bp gene cassette included the SalI and EcoRI cloning sites and was constructed synthetically by the Midland Certified Reagent Company (Midland, Tex.). This modified gag sequence was cloned into the SalI and EcoRI restriction sites of the eukaryotic expression vector pCMVKm2 that employs the cytomegalovirus (CMV) immediate-early enhancer/promoter and bGH terminator (Chiron Corporation, Emeryville, Calif.) (6), resulting in the plasmid pCMVKm2.GagMod.SF2. For the comparison of expression efficiencies between the modified and the native HIV-1SF2 gag expression cassettes, three different vectors containing the native p55Gag coding sequence were used, pCMV6ap55GagPRE, pCMVKm2p55GagPRE, and pCMVLinkPREp55Gag (Chiron). The pCMVLink plasmid differs from pCMVKm2 only in its multiple cloning site. All of these use the CMV immediate-early enhancer/promoter and include the hepatitis B virus posttranscriptional regulatory element (PRE) (9, 16–18) to partially overcome the Rev dependency of gag. This was demonstrated by transfection experiments using the native HIV-1SF2 gag gene with and without PRE. The expression of p55Gag was clearly improved using PRE over that using the gag gene only (S. W. Barnett, unpublished data).
For the construction of the gagprotease expression cassettes, modifications were made in the same manner as that described for gag up to the −1 frameshift region of the pol gene. The sequence from there to the gag gene's stop codon was unaltered. The sequences from the gag stop codon to the codons for first 26 aa of the reverse transcriptase were codons either optimized with subsequent INS inactivation as described above (GP1; GenBank accession no. AF202464) or modified by INS inactivation alone (GP2; GenBank accession no. AF202465). Both versions of the gagprotease cassette were cloned into the pCMVKm2 vector as described above for the modified gag to yield the plasmids pCMVKm2.GagProtMod.SF2 (GP1) and pCMVKm2.GagProtMod.SF2 (GP2).
In vitro expression assays.
Plasmid DNA was purified using endotoxin-free columns (Qiagen, Valencia, Calif.). African green monkey kidney (COS-7; European Culture Collections Organization no. 87021302), human kidney (293; American Type Tissue Collection [ATCC; Atlanta, Ga.] no. 45504), and human rhabdomyosarcoma (RD; ATCC no. CCL-136) cells were plated 1 day prior to transfection at a density of 5 × 105 cells per 35-mm-diameter well (Corning). For the transfections, 2 μg of each plasmid DNA was mixed with the Mirus TransIT-LT1 polyamine transfection reagent (PanVera, Madison, Wis.). The green fluorescent protein (GFP) reporter gene vector pEGFP (Clontech, Palo Alto, Calif.) was used as a transfection efficiency control in co- and parallel transfections. The cells were incubated with 2 ml of medium per well (for 293 cells, Iscove's modified Dulbecco's medium, 10% fetal calf serum [FCS]; for COS-7 and RD cells, Dulbecco's modified Eagle medium, 10% FCS; Gibco, Rockville, Md.). To estimate the transfection efficiency, GFP-expressing cells were analyzed quantitatively by flow cytometry (Becton Dickinson Immunocytometry Systems, San Jose, Calif.) and directly counted under a fluorescence microscope. Supernatants were harvested 24, 48, and 60 h posttransfection and filtered through 0.45-μm-pore-size syringe filters (Pall Corp., Ann Arbor, Mich.). Cells were harvested 60 h posttransfection, washed twice in phosphate-buffered saline (PBS) and then lysed on ice in 40 μl of buffer containing 1% NP-40 (Sigma, St. Louis, Mo.) and 0.1 M Tris-HCl, pH 7.5. Cell lysates were subsequently clarified by centrifugation in an Eppendorf microcentrifuge at 4°C for 10 min to remove cellular debris. The quantitation of Gag p24 protein in cell supernatants and lysates was performed using the p24 antigen capture enzyme-linked immunosorbent assay (ELISA) (Coulter Corporation, Miami, Fla.). For immunoblot analysis, samples were electrophoresed through sodium dodecyl sulfate (SDS)–8 to 16% polyacrylamide gels (Novex, San Diego, Calif.) and then transferred onto Immobilon P membranes (Millipore, Bedford, Mass.). A prestained broad-range molecular weight marker (Bio-Rad, Hercules, Calif.) and the HIV-1 p24 protein (Chiron) were used as size standards. Membranes were then incubated with HIV-1 patient serum or mouse anti-p24 monoclonal antibody (MAb) 76C.5EG (Chiron) (31). Reactive bands were visualized using Sigma Fast 3,3′-diaminobenzidine substrate as described by the manufacturer.
Sucrose density gradients.
Supernatants from transfected 293 and COS-7 cells were collected at 24 and 48 h posttransfection, filtered through a 0.2-μm-pore-size filter, and concentrated by ultracentrifugation through a 20% (wt/wt) sucrose cushion for 2 h at 140,000 × g (24,000 rpm) using a Beckman SW28 rotor. The pellets were then suspended in PBS, loaded on a 20 to 60% sucrose gradient, and centrifuged at 285,000 × g (40,000 rpm) for 2 h in a Beckman SW41ti rotor. Each gradient was fractionated into 1-ml aliquots, and 10-μl aliquots of each fraction were electrophoresed on an SDS–8 to 16% polyacrylamide gel electrophoresis gel (Novex). In addition, 2.5 μl of the concentrated gradient preload material was also analyzed. The proteins were then transferred to Immobilon P membranes (Millipore) and probed with mouse anti-p24 MAb 76C.5EG at a dilution of 1:2,000.
Electron microscopy.
COS-7 or 293 cells (4 × 106) were transfected in 100-mm-diameter dishes (Corning), and cells were harvested at 24 or 48 h posttransfection. Cells transfected with vector DNA alone served as negative controls. After two washes with PBS the cells were fixed in 2% glutaraldehyde (Sigma), incubated for 20 min at room temperature, gently scraped from the plate, and transferred into a 15-ml polypropylene tube. The fixed cells were then stained with uranium acetate and lead citrate. Electron microscopy was carried out using a transmission electron microscope (Zeiss; 10c) at ×50,000 and ×100,000 magnifications.
Animal studies.
Female BALB/c and CB6F1 mice, 6 to 8 weeks old, were used for immunogenicity studies. For the first experiment (Fig. 5), four groups of BALB/c mice (n = 4) were immunized with either modified gag plasmid DNA (pCMVKm2.GagMod.SF2) or the native gag plasmid DNA (pCMVLink.Gag.SF2.PRE). The plasmid DNA doses for the different groups were 20, 2, 0.2, and 0.02 μg in 100 μl of sterile endotoxin-free saline (Sigma). pCMVKm2 vector DNA was used to maintain the total concentration of DNA in each dose at 20 μg/100 μl to control for effects due to the lower concentration of plasmid DNA (2-, 0.2-, and 0.02-μg doses). For experiments 2 and 3 shown in Fig. 6 and 7, the mouse strain employed was CB6F1. For experiment 3, plasmid DNA doses were further diluted to include doses as low as 0.0002 μg.
FIG. 5.
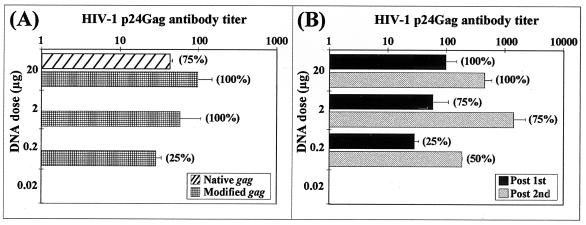
Increased immunogenicity of the modified HIV-1SF2 gag plasmid compared to that of the native gag plasmid. Groups of mice were immunized bilaterally in the tibialis anterior muscles with titrated amounts of DNA in 10-fold dilutions from 20 μg down to 0.002 μg. Sera were collected at 0 and 4 weeks and tested for HIV-1 p24-specific antibody titers by ELISA as described in Materials and Methods. (A) Comparison of humoral immune responses at different DNA doses using the native and modified gag plasmid DNA. Values represent the geometric mean antibody titers and the standard deviations of the midpoint antibody titers for each group. The values in parentheses indicate the percentages of responders (percent seroconversion) in each group. (B) Antibody responses were boosted following a second immunization with the modified gag plasmid DNA. Four weeks after the first immunization, additional groups of mice received a second immunization with the same amount of titrated plasmid DNA. Sera collected at weeks 0, 4, and 6 were analyzed by p24 antibody ELISA.
FIG. 6.
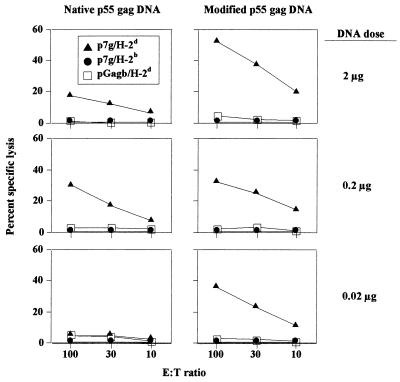
CTL responses in CB6F1 mice after a single immunization with titrated plasmid DNA. Nine weeks after immunization mice were challenged with an intraperitoneal dose of 107 PFU of rVVgag-pol. Five days later effector (E) spleen cells were tested for cytolytic activity in a 4-h 51Cr release assay using 51Cr-labeled SVBALB tumor target (T) cells (5,000 targets per well) that had been pulsed for 1 h with a 1-μg/ml concentration of the H-2Kd-binding HIV-1 Gag peptide p7g (▴). Target cells pulsed with the negative-control HIV-1 Gag peptide pgagb (□) and major histocompatibility complex-mismatched (H-2b), p7g peptide-pulsed RMA target cells (●) were employed as negative controls.
FIG. 7.
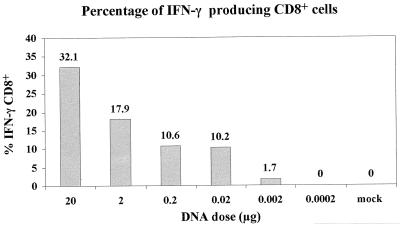
Quantification of Gag-specific, IFN-γ-producing CD8+ T lymphocytes in mice after a single immunization of titrated modified gag plasmid DNA followed by rVVgag-pol challenge. Splenic IFN-γ-positive CD8+ T lymphocytes specific for the p7g Gag peptide were enumerated by flow cytomerty as described in Materials and Methods. mock, results using spleen cells from naive mice.
For the DNA immunization study with rhesus monkeys (Macaca mulatta), four animals were immunized bilaterally in the quadriceps muscles with 1-mg doses of pCMVKm2.GagMod.SF2 plasmid DNA in saline at weeks 0, 4, and 8 and bled at weeks 0, 4, 6, 8, and 10. Animals were maintained at the Southwest Foundation for Biomedical Research (San Antonio, Tex.).
Measurements of antibody responses.
Ninety-six-well plates (Corning) were coated with 100 μl of recombinant HIV-1SF2 p24 antigen at a concentration of 2 μg per ml in 50 mM borate buffer, pH 9. Sera were diluted 1:25, followed by threefold serial dilutions in dilution buffer containing 1% casein as the blocking reagent. Pooled anti-p24 antibody-positive mouse sera served as both a positive control and an assay standard. The sera were incubated for 50 min at 37°C, washed, and incubated with a 1:22,000 dilution of goat anti-mouse immunoglobulin G (IgG)-IgM peroxidase conjugate (Pierce, Rockford, Ill.) for an additional 50 min at 37°C. After the plates were washed, the tetramethylbenzone substrate (Pierce) was added to each well, and the reaction was stopped after 30 min by the addition of 2 N H2SO4. The plates were read on an ELISA reader (312e; Bio-Tek, Winooski, Vt.) at 450 nm with a reference wavelength of 620 nm. The calculated titers are the reciprocal of the dilution of serum at a cutoff optical density of 0.4.
Recombinant vaccinia virus challenge of immunized mice.
The recombinant vaccinia virus containing the HIV-1SF2 gag and pol genes (rVVgag-pol) has been described previously (8). Nine (experiment 2, Fig. 6) and 5 (experiment 3, Fig. 7) weeks following gag DNA immunization, mice were challenged with an intraperitoneal injection of 107 PFU of rVVgag-pol. Five days later spleens were harvested and tested directly for cytolytic activity against Gag peptide-pulsed, 51Cr-labeled tumor target cells or were stimulated with Gag peptide and then stained for intracellular gamma interferon (IFN-γ), as described below. This rVVgag-pol challenge model provides a quantitative measure of CD8+ T-cell function (G. Otten, unpublished data).
CTL assays.
Spleen cells were tested for cytolytic activity in a 4-h 51Cr release assay using 51Cr-labeled SVBALB (H-2d) or RMA (H-2b) tumor target cells (5,000 targets per well) that had been pulsed for 1 h with a 1-μg/ml concentration of the H-2Kd-binding HIV-1 Gag peptide p7g (8) or the control HIV-1 Gag peptide pgagb (12, 26). After 4 h of incubation, 50 μl of culture supernatants was transferred to Lumaplates (Packard, Meriden, Conn.), dried, and counted in a Microbeta scintillation counter (Wallac, Gaithersburg, Md.). Percent specific 51Cr release was determined from the formula percent specific 51Cr release = (mean experimental release − mean spontaneous release)/(maximum release − spontaneous release) × 100%, where spontaneous release = mean counts per minute released from target cells in the absence of spleen cells and maximum release = mean counts per minute released from target cells in the presence of 0.1% Triton X-100.
Measurement of Gag-specific IFN-γ-producing CD8+ lymphocytes.
Spleens were taken 5 days post-rVVgag-pol challenge. Erythrocyte-depleted single cell suspensions were prepared by treatment with Tris-buffered NH4Cl (Sigma). Nucleated spleen cells (1 × 106 to 2 × 106) were cultured in duplicate at 37°C in the presence or absence of 10 μg of p7g peptide/ml. Monensin (Pharmingen, San Diego, Calif.) was added to block cytokine secretion. After 3 to 5 h cells were washed, incubated with anti-CD16/32 (Pharmingen) to block Fcγ receptors, and fixed in 2% (wt/vol) paraformaldehyde and stored overnight at 4°C. The following day cells were treated with 0.5% (wt/vol) saponin (Sigma) and then incubated with a phycoerythrin (Pharmingen)-conjugated mouse IFN-γ MAb in the presence of 0.1% (wt/vol) saponin. Cells were then washed free of saponin, stained with fluorescein isothiocyanate-conjugated CD8 MAb (Pharmingen), washed, and then analyzed on a FACSCalibur flow cytometer (Becton Dickinson Immunocytometry Systems). Samples were cultured and stained in duplicate.
Peptide pools.
A set of 51 Gag peptides 20 residues long, overlapping by 10 aa and spanning residues 1 to 496 of HIV-1SF2 p55Gag, was synthesized (Chiron Mimotopes, Clayton, Australia). Eight pools were made by mixing 5 to 7 overlapping peptides. Gag amino acid sequences spanned by the pools were as follows: aa 1 to 80, pool 1; aa 71 to 144, pool 2; aa 135 to 203, pool 3; aa 194 to 263, pool 4; aa 254 to 323, pool 5; aa 314 to 365, pool 6; aa 351 to 430, pool 7; aa 421 to 496, pool 8. A pool of six 20-aa overlapping peptides representing HIV-1SF2 Env served as a negative-control pool.
Purification of rhesus macaque PBMC and derivation of B-LCL.
Rhesus macaque peripheral blood mononuclear cells (PBMC) were separated from heparinized whole blood on Percoll gradients (5) and cultured at 3 × 106 to 3.5 × 106 per well in 1.5 ml in 24-well plates for 8 days in AIM-V–RPMI 1640 (50:50) culture medium (Gibco) supplemented with 10% FCS. Gag-specific cells were stimulated by the addition of either a Gag peptide pool (13.3 μg of total peptide/ml) or autologous PBMC that had been infected with rVVgag-pol and cultured in 24-well plates. Recombinant human interleukin-7 (IL-7; 15 ng/ml; R&D Systems, Minneapolis, Minn.) was added at the initiation of culture. Human recombinant IL-2 (20 IU/ml; Proleukin; Chiron) was added on days 1, 3, and 6. For the derivation of stable rhesus B-lymphoblastoid cell lines (B-LCL), PBMC were exposed to herpesvirus papio-containing culture supernatant from the S594 cell line (13, 27) in the presence of 1 μg of cyclosporine (Sigma)/ml.
Rhesus macaque CTL assay.
Autologous B-LCL were labeled overnight with Na251CrO4 (NEN, Boston, Mass.; 25 μCi per 106 B-LCL) and washed. Individual aliquots were then incubated for 1 h with 100 μg of Gag or Env peptide pool/ml. Peptide-pulsed, 51Cr-labeled B-LCL were added (2,500 per round-bottom well) to duplicate wells containing threefold serial dilutions of cultured PBMC. Unlabeled B cells (105) were added to each well to inhibit nonspecific cytolysis. After 4 h, 50 μl of culture supernatant was harvested, added to Lumaplates (Packard), and counted with a Microbeta 1450 liquid scintillation counter (Wallac). 51Cr released from lysed targets was normalized by the formula percent specific 51Cr release = 100% × (mean experimental release − mean spontaneous release)/(maximum release − spontaneous release), where spontaneous release = mean counts per minute released from target cells in the absence of spleen cells and maximum release = mean counts per minute released from target cells in the presence of 0.1% Triton X-100. Data are plotted as percent specific 51Cr release versus the culture fraction, where the culture fraction represents the fraction of the culture well (1.5 ml) added to the CTL assay microtiter plate, e.g., a culture fraction of 0.067 equals 1/15 or 0.1 ml of the initial PBMC culture. Serial threefold dilutions of the cultured PBMC were made. In separate experiments, where we have counted the cells recovered from cultures, we have determined the maximal effector cell/target cell ratios to be about 40:1 to 100:1.
RESULTS
Increased in vitro expression efficiency of sequence-modified HIV-1SF2 gag gene.
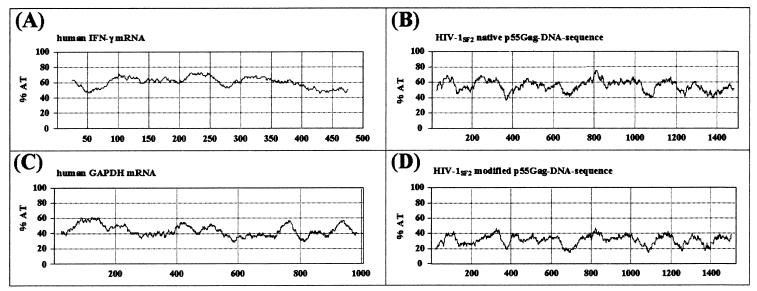
The coding sequences for the HIV-1SF2 gag gene were modified to conform to the codon usage pattern of highly expressed human genes and to eliminate residual INS motifs as described in Materials and Methods. These modifications resulted in gag coding sequences with a clear reduction in overall AT content compared to that of the native gag (Fig. 1). In fact, the percentage of A and T nucleotides was reduced from 56 to 32%, a level more consistent with increased mRNA stability and translation efficiency (14, 15). The AT content of the modified gag more closely resembled that of the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene, which encodes a relatively stable mRNA compared with the relatively unstable AU-rich human IFN-γ mRNA (Fig. 1).
FIG. 1.
Comparison of the percentages of A and T nucleotides in genes encoding relatively unstable versus stable mRNA molecules. The human IFN-γ gene and the native HIV-1SF2 gag DNA sequences both encode relatively unstable transcripts (A and B) and have an average AT content of 55 to 60%. In contrast, the stable human GAPDH gene and the modified HIV-1SF2 gag coding regions have reduced AT contents of 40 and 30%, respectively (C and D). The calculation of the AT content was done using MacVector software (Oxford Molecular Ltd.); the window size was set at 50.
The in vitro expression efficiency of the modified HIV-1SF2 gag (pCMVKm2.GagMod.SF2) was compared to that of the native SF2 gag in a construct (pCMVLink.Gag.SF2.PRE) that also contained the hepatitis B virus PRE. The pCMVLink.Gag.SF2.PRE plasmid previously had been found to express Gag at substantially higher levels than a similar plasmid containing the HIV-1SF2-derived gag gene without the PRE (S. W. Barnett, unpublished data). The expression levels for these plasmids were determined in several independent experiments after transfection of three different cell lines, RD, 293, and COS-7 (Table 1). Cell supernatants and lysates were tested at 48 and 60 h posttransfection. Gag expression levels were clearly much higher for the modified gag plasmid at all time points and in all three cell lines tested. The increased expression was most dramatic in the supernatants of the transfected human 293 cell line, where expression from the modified gag was 322- to 966-fold greater than that of the native HIV-1SF2 gag plasmid tested. The improvement in Gag expression levels in 293 cell lysates was also apparent, but less so than in the supernatants, which could be indicative of more-efficient budding of p55Gag particles in cells where expression levels are elevated. To exclude possible effects on the transfection efficiency depending on the plasmid used, flow cytometry and direct fluorescence microscope analysis were done in parallel transfections or by cotransfection using GFP plasmid DNA. On average, 70% of the cells were transfected using either method with no differences in transfection efficiency between the native and modified gag plasmids noted (data not shown).
TABLE 1.
Increased in vitro expression from modified versus native gag plasmids in supernatants and lysates from transiently transfected cells
| Experiment | Type of plasmida | Material assayedb | Cell line | Hours post-transfection | Total ng p24 (fold increase) |
|---|---|---|---|---|---|
| 1 | Nat | Sup | 293 | 48 | 3.4 |
| Mod | Sup | 293 | 48 | 1,260 (371) | |
| Nat | Sup | 293 | 60 | 3.2 | |
| Mod | Sup | 293 | 60 | 2,222 (694) | |
| 2 | Nat | Sup | 293 | 60 | 1.8 |
| Mod | Sup | 293 | 60 | 1,740 (966) | |
| 3 | Nat | Sup | 293 | 60 | 1.8 |
| Mod | Sup | 293 | 60 | 580 (322) | |
| 4 | Nat | Lys | 293 | 60 | 1.5 |
| Mod | Lys | 293 | 60 | 85 (57) | |
| 1 | Nat | Sup | RD | 48 | 5.6 |
| Mod | Sup | RD | 48 | 66 (12) | |
| Nat | Sup | RD | 60 | 7.8 | |
| Mod | Sup | RD | 60 | 70.2 (9) | |
| 2 | Nat | Lys | RD | 60 | 1.9 |
| Mod | Lys | RD | 60 | 7.8 (4) | |
| 1 | Nat | Sup | COS-7 | 48 | 0.4 |
| Mod | Sup | COS-7 | 48 | 33.4 (84) | |
| 2 | Nat | Sup | COS-7 | 48 | 0.4 |
| Mod | Sup | COS-7 | 48 | 10 (25) | |
| Nat | Lys | COS-7 | 48 | 3 | |
| Mod | Lys | COS-7 | 48 | 14 (5) |
Nat, native (pCMVLink.Gag.SF2.PRE); Mod, modified (pCMVKm2.GagMod.SF2).
Sup, supernatant; Lys, lysate.
The modified HIV-1SF2 gag gene encodes p55Gag VLP of the expected density and morphology.
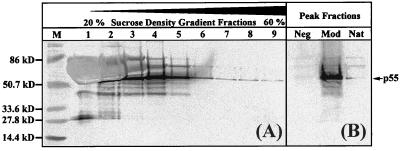
Supernatants and cell lysates from transfected 293 cells were subjected to immunoblot and density gradient sedimentation analysis. The results confirmed the previous data from the p24 capture assay with respect to the relative level of p55 expression from the modified gag plasmid. The expected p55Gag band was detected using human HIV-1 patient serum (Fig. 2) or an anti-p24 MAb for the immunostaining (data not shown). Supernatants from 293 cells transfected with the native and modified gag genes were subjected to rate zonal sedimentation to isolate p55Gag particles of the reported density (32). Gradient fractions were analyzed by p24 capture ELISA (data not shown) and Western blotting (Fig. 2A) to determine the peak fraction of each sample. Western blot analysis showed that the p55Gag band for the modified Gag expression cassette was stronger than that for the best native gag plasmid (Fig. 2B).
FIG. 2.
Increased expression in vitro of HIV-1SF2 p55Gag particles in cells transfected with the modified gag gene. 293 cells were transfected with plasmids containing either the modified or native HIV-1SF2 gag genes. Supernatants from transfected cell cultures were collected at 60 h posttransfection and centrifuged through 20 to 60% sucrose density gradients. Gradient fractions were collected, run on an SDS–8 to 16% polyacrylamide gel, and analyzed by Western blotting as described in Materials and Methods. (A) Immunoblot of fractions 1 to 9 from the sucrose density gradient from transfection supernatants of the modified gag plasmid. (B) Immunoblot comparing peak fractions collected in the density range expected for HIV-1 VLP after transfection with modified (Mod) or native (Nat) HIV-1SF2 gag plasmids. Vector alone (Neg) was used as a negative transfection control, and the prestained broad-range molecular weight marker (M) was used as the size standard.
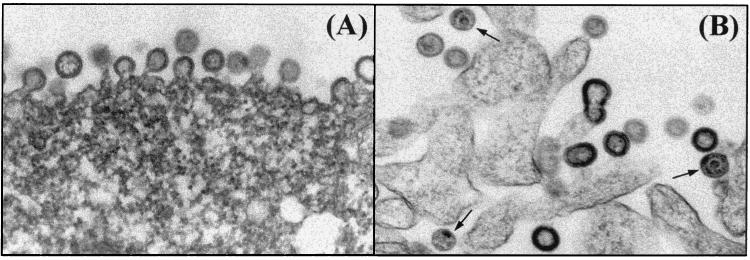
To confirm that VLP were being expressed, COS-7 cells transfected with pCMVKm2.GagMod.SF2 were harvested at 24 h posttransfection and electron microscopy was performed. As shown in Fig. 3A, budding and free immature particles could be observed. These data confirm that the sequence modifications for the gag gene did not adversely affect the p55Gag particle assembly or VLP morphology.
FIG. 3.
Modified gag and gagprotease form VLP in transiently transfected COS-7 cells. Shown are electron micrographs of immature p55Gag VLP in COS-7 cells transfected with the modified HIV-1SF2 gag (A) and mature (arrows) and immature VLP obtained using the modified HIV-1SF2 gagprotease (GP2) (B). Transfected cells were fixed at 24 (gag) or 48 h (gagprotease) posttransfection and subsequently analyzed by electron microscopy as described in Materials and Methods (magnification, ×100,000). Cells transfected with vector alone (pCMVKm2) served as the negative control (data not shown).
Construction and characterization of sequence-modified gagprotease gene cassettes.
As a first step in the design of modified HIV immunogens with increased representation of Pol-specific epitopes, two different modified gagprotease gene constructs were evaluated for expression and VLP formation. The protease coding sequences in these constructs were (i) codon optimized, with subsequent INS inactivation as described above for gag (GP1), or (ii) modified by INS inactivation alone (GP2). Like the modified gag plasmid, in the absence of Rev both versions of the modified gagprotease exhibited high-level expression of Gag proteins in supernatants and cell lysates of transiently transfected COS-7 and 293 cell lines (Table 2). In fact, the expression levels measured in lysates of 293 cells transfected with the gagprotease plasmids were higher than those seen with the modified gag alone. This result could be partially or wholly attributed to more-efficient recognition of processed Gag (mostly p24) than of unprocessed p55Gag by the Coulter p24 antigen capture assay, as has been previously described (29). This apparent increase in p24 expression in cell lysates was not observed in COS-7 cells, possibly due to lower overall expression of p55Gag in this cell line.
TABLE 2.
In vitro expression from modified gag and gagprotease plasmids in supernatants and lysates from transiently transfected cells
| Plasmida | Material assayedb | Cell line | Hours posttransfection | Total ng p24c |
|---|---|---|---|---|
| Gag | Sup | 293 | 60 | 760 |
| GagProt (1) | Sup | 293 | 60 | 380 |
| GagProt (2) | Sup | 293 | 60 | 320 |
| Gag | Lys | 293 | 60 | 78 |
| GagProt (1) | Lys | 293 | 60 | 1,250 |
| GagProt (2) | Lys | 293 | 60 | 400 |
| Gag | Sup | COS-7 | 72 | 40 |
| GagProt (1) | Sup | COS-7 | 72 | 150 |
| GagProt (2) | Sup | COS-7 | 72 | 290 |
| Gag | Lys | COS-7 | 72 | 60 |
| GagProt (1) | Lys | COS-7 | 72 | 63 |
| GagProt (2) | Lys | COS-7 | 72 | 58 |
Gag, pCMVKm2.GagMod.SF2; GagProt (1), pCMVKm2.GagProtMod.SF2 (GP1) (gagprotease with codon optimization and inactivation of INS in protease); GagProt (2), pCMVKm2.GagProtMod.SF2 (GP2) (gagprotease with only inactivation of INS in protease).
Sup, supernatant; Lys, lysate.
Representative results from three independent experiments for each cell line tested.
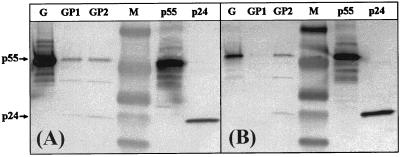
Sucrose density gradient analyses of supernatants from 293 and COS-7 cells transiently transfected with either gagprotease or gag constructs were performed, and the peak fractions were subsequently analyzed by Western blotting. The efficiency of VLP formation varied between the cell lines tested and was found to be lower for gagprotease than for the modified gag plasmid (Fig. 4). The levels of VLP formation from the two gagprotease constructs in 293 cells were similar (Fig. 4A; GP1 and GP2), but the analysis of the codon-optimized and INS-inactivated gagprotease plasmid, GP1, in COS-7 cells suggested the production of relatively small amounts of VLP (Fig. 4B). Polyproteins expressed from both of the modified versions of gagprotease were correctly processed by the encoded viral protease. Bands corresponding to unprocessed p55Gag and completely processed p24 were detectable using a MAb specific for p24 (data not shown) or HIV-1+ patient serum (Fig. 4A and B) (p17 levels were too low to be detected with the HIV+ sera used).
FIG. 4.
Expression and processing of p55Gag polyproteins in VLP using modified HIV-1 gagprotease. Supernatants from transfected cell cultures were collected at 60 h posttransfection and centrifuged through 20 to 60% sucrose density gradients. Gradient fractions were collected, and peak fractions were run on an SDS–8 to 16% polyacrylamide gel and analyzed by Western blotting using HIV-1 patient serum as described in Materials and Methods. (A) Peak fractions from 293 cells. Results for the modified gag (G) are compared to those for codon-optimized, INS-inactivated gagprotease (GP1) and for INS-inactivated-only gagprotease (GP2). (B) Immunoblot comparing peak fractions from transfected COS-7 cells using the same plasmids as those described for panel A. Purified HIV-1SF2 p24 (Chiron) and baculovirus-derived p55Gag proteins were used as additional controls. Prestained broad-range markers (Bio-Rad) were used as size standards (M).
Electron microscopic analysis of COS-7 cells transfected with the two different sequence-modified gagprotease constructs confirmed the results of the sucrose gradient analysis. COS-7 cells transfected with the codon-optimized and INS-inactivated version (GP1) showed very little VLP formation (data not shown) compared to those transfected with the nonoptimized INS-inactivated gagprotease (GP2; Fig. 3B). A possible explanation for this observation is that codon optimization of the protease coding sequences may have resulted in its overexpression relative to Gag and the prevention of the efficient budding of particles (19). The GP2 version of gagprotease, in which the INS of the protease-coding region was inactivated without codon optimization, reduced protease overexpression, and thus VLP of the mature and immature phenotypes could be detected.
Increased immunogenicity of the modified gag DNA in vivo.
To evaluate and compare the immunogenicities of the modified and the native gag plasmids, mice were immunized intramuscularly with plasmid DNA doses titrated from 20 to 0.02 μg per mouse. Serum was collected at 4 weeks postimmunization and tested in a p24Gag-specific antibody ELISA. Antibody responses to Gag could be detected in mice immunized with as little as 0.2 μg of the modified gag expression cassette, whereas the native gag cassette was able to induce an antibody response only at the 20-μg DNA dose (Fig. 5A). This represented the induction of an antibody response using the modified gag at a single DNA dose 100 fold lower than that necessary for the native gag. In parallel groups of animals, a second dose of DNA was given at 4 weeks to determine if antibody responses to the modified gag had reached maximal values at the 20-μg dose and if the lowest DNA dose of 0.02 μg could induce an antibody response after a second immunization. As shown in Fig. 5B, the Gag-specific antibody titers increased after the second immunizations for all DNA doses except for the 0.02-μg DNA dose group, which remained negative.
Measurements of the cellular immune responses following DNA immunization with the modified gag demonstrated a similar pattern. Gag-specific CTL responses were inducible at DNA amounts at least 10-fold lower than those necessary with the native gag expression cassette (Fig. 6). Gag-specific CTL were detectable after a single immunization with a dose of the modified gag plasmid DNA as low as 0.02 μg, whereas a dose of 0.2 μg of the native gag plasmid was required for the induction of detectable CTL. In a subsequent study, the modified gag plasmid DNA was further diluted (down to 0.2 ng) and used to immunize additional groups of mice. As shown in Fig. 7, Gag-specific IFN-γ-positive CD8+ T cells were scored in mice receiving as little as 2 ng of the modified gag DNA.
Induction of CTL responses in rhesus macaques immunized with the modified gag plasmid.
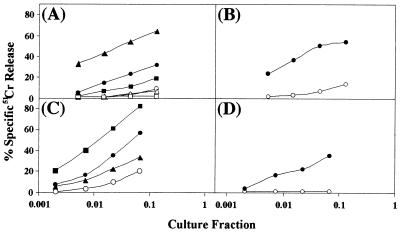
Based on the increased potency observed in mouse immunizations with the modified gag plasmid DNA, studies with nonhuman primates were initiated. Four rhesus macaques were given three intramuscular immunizations with 1-mg doses of gag plasmid at 4-week intervals. PBMC were harvested prior to immunization and at 2 weeks after the second and third immunizations. PBMC were cultured with Gag peptide pools or with rVVgag-pol-infected autologous PBMC to stimulate the expansion and differentiation of CTL and tested against Gag peptide pool-pulsed, 51Cr-labeled, autologous B-LCL targets in 4-h 51Cr release assays. No Gag-specific cytolysis in PBMC was observed prior to immunization (not shown). However, after gag DNA immunization, all four macaques showed cytolytic activity against autologous B-LCL pulsed with at least one Gag peptide pool. In addition, two of the four macaques reacted with two or three Gag peptide pools (Fig. 8). Percent specific lysis of Gag-pulsed target cells varied among animals and among pools and reached as high as 80% at the highest effector cell/target cell ratio (Fig. 8C). A Gag-specific antibody response (antibody titer, 164) was detected in one of the four animals 2 weeks after the second immunization. This animal also had an anamnestic immune response 2 weeks after the third immunization, with a fivefold increase of the antibody titer (890). A second animal had a very low titer 2 weeks after the second immunization (65), which later dropped below the detection level. These results reflect the induction of robust and relatively broad CTL responses using the modified gag plasmid following DNA immunization of nonhuman primates and warrant further study with these plasmids. This contrasts with previous results in which weak and transient CTL responses were observed in only one of four macaques given seven immunizations with 1-mg doses of the pCMVLink.Gag.SF2.PRE plasmid containing the native HIV-1SF2 gag (X. Paliard and C. Walker, unpublished data).
FIG. 8.
Cytolytic T cells from peripheral blood of four individual rhesus macaques immunized with pCMVKm2.GagMod.SF2. PBMC were isolated 2 weeks after the second immunization (A and B) or 2 weeks after the third immunization (C and D). PBMC were cultured for 8 days in the presence of pools of synthetic Gag peptides (A and B) or with rVVgag-pol-infected PBMC (C and D). PBMC cultures were harvested and serially diluted as described in Materials and Methods, and Gag-specific cytolytic activity was assayed using autologous B-LCL target cells that had been pulsed with Gag peptide pools. (A) PBMC from rhesus macaque 63 stimulated with pool 1 and assayed on targets pulsed with pool 1 (●) or pool 5 (○), stimulated with pool 4 and assayed on targets pulsed with pool 4 (▴) or pool 8 (▵), and stimulated with pool 5 and assayed on targets pulsed with pool 5 (■) or pool 1 (□); (B) PBMC from rhesus macaque 68 stimulated with pool 4 and assayed on targets pulsed with pool 4 (●) or pool 8 (○); (C) PBMC from rhesus macaque 77 stimulated with rVVgag-pol and assayed on targets pulsed with pool 1 (■), pool 5 (▴), pool 8 (●), or an Env peptide pool (○); (D) PBMC from rhesus macaque 78 stimulated with rVVgag-pol and assayed on targets pulsed with pool 2 (●) or an Env peptide pool (○).
DISCUSSION
To increase the potency of HIV-1 DNA vaccines, we modified the genes coding for HIV-1SF2 Gag and Protease to overcome Rev dependence and to increase expression levels. Changes in codon usage to that utilized by highly expressed human genes in combination with inactivation of INS regions dramatically increased Gag expression from these constructs in the absence of Rev. Expression levels from the modified gag plasmid pCMVKm2.GagMod.SF2 were increased between 322- and 966-fold in 293 cells compared with those from pCMVLink.Gag.SF2.PRE, which contained the native HIV-1SF2 gag gene. Density gradient and electron microscopy analysis demonstrated that the modified gag genes efficiently expressed particles with the density and morphology expected for HIV VLP (Fig. 2 and 3). Similarly modified gagprotease plasmids that also showed high levels of Rev-independent expression were constructed (Table 2), but the expression cassette in which the codons for protease were optimized in combination with INS inactivation showed evidence of protease overexpression and reduced formation of VLP in transfected COS-7 cells (GP1; Fig. 4B). In contrast, both immature and mature VLP were produced from gagprotease constructs in which the INS were inactivated without codon optimization (GP2; Fig. 3 and 4).
In light of the improved expression levels from the modified gag, mouse studies were conducted to evaluate immune responses to this construct when administered as a DNA vaccine. When the modified gag plasmid was employed, Gag antigen-specific IFN-γ-secreting CD8+ T cells could be measured following a single immunization with as little as 2 ng of plasmid DNA (Fig. 7). CTL responses were observed in a lysis assay after a single immunization with 20 ng of the modified gag plasmid. These results combined indicate a 10- to 100-fold improvement over the native gag plasmid, for which at least 200 ng of DNA was required for the induction of a detectable antigen-specific CTL response (Fig. 6). The improved potency of the modified gag was also reflected in the humoral responses. A single dose of 200 ng of the modified gag was sufficient to induce measurable anti-Gag antibody responses in 25% of the mice (Fig. 1A), while 100-fold more (20 μg) of the native gag plasmid was required for the detection of Gag-specific antibodies.
The improved potency of the codon-modified gag expression plasmid observed in mouse studies was confirmed with rhesus macaques. Four of four macaques had detectable Gag-specific CTL after two or three 1-mg doses of modified gag plasmid. In contrast, in a previous study, only one of four macaques given 1-mg doses of plasmid DNA encoding the wild-type HIV-1SF2 Gag showed strong CTL activity, which was not apparent until after the seventh immunization (X. Paliard and C. Walker, unpublished data). Further evidence of the potency of the modified gag plasmid was the observation that CTL from two of the four rhesus macaques reacted with three nonoverlapping Gag peptide pools, suggesting that as many as three different Gag peptides are recognized and indicating that the CTL response is polyclonal. Additional quantification and specificity studies are in progress to further characterize the T-cell responses to Gag in plasmid-immunized rhesus macaques. DNA immunization of macaques with the modified gag plasmid did not result in significant antibody responses, with only two of four animals seroconverting at low titers. In contrast, the majority of macaques in additional groups immunized with p55Gag protein seroconverted and had strong Gag-specific antibody titers (G. Otten, unpublished data). These preliminary data together with data from other investigators indicate that a prime-boost strategy, with DNA prime and protein boost, could be very promising for the induction of strong CTL and antibody responses.
These results indicate that sequence-modified high-level expression cassettes for HIV structural genes can improve the potency of plasmid-vectored HIV vaccines. Sequence-modified genes may also enhance the potency of virus-vectored vaccines and increase the production efficiency of HIV structural proteins for use in subunit vaccines.
ACKNOWLEDGMENTS
We thank Diana Atchley, Pedro Benitez, Debbie Swinarski, and Charles Vitt for their excellent help with the mouse immunization studies, Kathy Brasky and Robert Geiger from the Southwest Foundation for immunization, handling, and care of the rhesus macaques, and Benedict Yen and Ivy Hsieh at the San Francisco VA Medical Center for performing the electron microscopy.
J.M. was supported by a postdoctoral fellowship from the Ernst Schering Research Foundation (Berlin, Germany).
REFERENCES
- 1.Barnett S W, Liu M A. DNA vaccines coming of age. Annu Rep Med Chem. 1999;34:149–158. [Google Scholar]
- 2.Bertoletti A, Cham F, McAdam S, Rostron T, Rowland-Jones S, Sabally S, Corrah T, Ariyoshi K, Whittle H. Cytotoxic T cells from human immunodeficiency virus type 2-infected patients frequently cross-react with different human immunodeficiency virus type 1 clades. J Virol. 1998;72:2439–2448. doi: 10.1128/jvi.72.3.2439-2448.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betts M R, Krowka J, Santamaria C, Balsamo K, Gao F, Mulundu G, Luo C W, Gandu N N, Sheppard H, Hahn B H, Allen S, Frelinger J A. Cross-clade human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte responses in HIV-infected Zambians. J Virol. 1997;71:8908–8911. doi: 10.1128/jvi.71.11.8908-8911.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borrow P, Lewicki H, Hahn B H, Shaw G M, Oldstone M B. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Budzko D, Madden D L, London W T, Sever J L. Improved separation of rhesus monkey lymphocytes with Percoll. Lab Investig. 1985;53:586–588. [PubMed] [Google Scholar]
- 6.Chapman B S, Thayer R M, Vincent K A, Haigwood N L. Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Res. 1991;19:3979–3986. doi: 10.1093/nar/19.14.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clerici M, Shearer G. Correlates of protection in HIV infection and the progression of HIV infection to AIDS. Immunol Lett. 1996;51:69–73. doi: 10.1016/0165-2478(96)02557-6. [DOI] [PubMed] [Google Scholar]
- 8.Doe B, Walker C. HIV-1 p24 Gag-specific cytotoxic T-lymphocyte responses in mice. AIDS. 1996;10:793–794. doi: 10.1097/00002030-199606001-00015. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 9.Donello J E, Beeche A A, Smith III G J, Lucero G R, Hope T J. The hepatitis B virus posttranscriptional regulatory element is composed of two subelements. J Virol. 1996;70:4345–4351. doi: 10.1128/jvi.70.7.4345-4351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnelly J J, Ulmer J B, Shiver J W, Liu M A. DNA vaccines. Annu Rev Immunol. 1997;15:617–648. doi: 10.1146/annurev.immunol.15.1.617. [DOI] [PubMed] [Google Scholar]
- 11.Durali D, Morvan J, Letourneur F, Schmitt D, Guegan N, Dalod M, Saragosti S, Sicard D, Levy J, Gomard E. Cross-reactions between the cytotoxic T-lymphocyte responses of human immunodeficiency virus-infected African and European patients. J Virol. 1998;72:3547–3553. doi: 10.1128/jvi.72.5.3547-3553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elvin J, Potter C, Elliott T, Cerundolo V, Townsend A. A method to quantify binding of unlabeled peptides to class I MHC molecules and detect their allele specificity. J Immunol Methods. 1993;158:161–171. doi: 10.1016/0022-1759(93)90210-x. [DOI] [PubMed] [Google Scholar]
- 13.Falk L, Deinhardt F, Nonoyama M, Wolfe L G, Bergholz C. Properties of a baboon lymphotropic herpesvirus related to Epstein-Barr virus. Int J Cancer. 1976;18:798–807. doi: 10.1002/ijc.2910180611. [DOI] [PubMed] [Google Scholar]
- 14.Haas J, Park E, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 15.Hentze M. Determinants and regulation of cytoplasmic mRNA stability in eukaryotic cells. Biochim Biophys Acta. 1991;1090:281–292. doi: 10.1016/0167-4781(91)90191-n. [DOI] [PubMed] [Google Scholar]
- 16.Huang J, Liang T J. A novel hepatitis B virus (HBV) genetic element with Rev response element-like properties that is essential for expression of HBV gene products. Mol Cell Biol. 1993;13:7476–7486. doi: 10.1128/mcb.13.12.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Z M, Yen T S. Hepatitis B virus RNA element that facilitates accumulation of surface gene transcripts in the cytoplasm. J Virol. 1994;68:3193–3199. doi: 10.1128/jvi.68.5.3193-3199.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Z M, Yen T S. Role of the hepatitis B virus posttranscriptional regulatory element in export of intronless transcripts. Mol Cell Biol. 1995;15:3864–3869. doi: 10.1128/mcb.15.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karacostas V, Wolffe E J, Nagashima K, Gonda M A, Moss B. Overexpression of the HIV-1 gag-pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology. 1993;193:661–671. doi: 10.1006/viro.1993.1174. [DOI] [PubMed] [Google Scholar]
- 20.Klein M R, Vanbaalen C A, Holwerda A M, Garde S R K, Bende R J, Keet I P M, Eeftinckschattenkerk J K M, Osterhaus A D M E, Schuitemaker H, Miedema F. Kinetics of Gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J Exp Med. 1995;181:1365–1372. doi: 10.1084/jem.181.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lynch J, deSouza M, Robb M, Markowitz L, Nitayaphan S, Sapan C, Mann D, Birx D, Cox J. Cross-clade cytotoxic T cell response to human immunodeficiency virus type 1 proteins among HLA disparate North Americans and Thais. J Infect Dis. 1998;178:1040–1046. doi: 10.1086/515652. [DOI] [PubMed] [Google Scholar]
- 24.Moss P, Rowland-Jones S L, Frodsham P M, McAdam S, Giangrande P, McMichael A J, Bell J I. Persistent high frequency of human immunodeficiency virus-specific cytotoxic T cells in peripheral blood of infected donors. Proc Natl Acad Sci USA. 1995;92:5773–5777. doi: 10.1073/pnas.92.13.5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nasioulas G, Zolotukhin A S, Tabernero C, Solomin L, Cunningham C P, Pavlakis G N, Felber B K. Elements distinct from human immunodeficiency virus type 1 splice sites are responsible for the Rev dependence of env mRNA. J Virol. 1994;68:2986–2993. doi: 10.1128/jvi.68.5.2986-2993.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paliard X, Doe B, Walker C. The T cell repertoire primed by antiviral vaccination is influenced by self-tolerance. Cell Immunol. 1998;188:73–79. doi: 10.1006/cimm.1998.1338. [DOI] [PubMed] [Google Scholar]
- 27.Rabin H, Neubauer R H, Woodside N J, Cicmanec J L, Wallen W C, Lapin B A, Agrba V A, Yakoleva L A, Chuvirov G N. Virological studies of baboon (Papio hamadryas) lymphoma: isolation and characterization of foamyviruses. J Med Primatol. 1976;5:13–22. doi: 10.1159/000459903. [DOI] [PubMed] [Google Scholar]
- 28.Rowland-Jones S, Sutton J, Ariyoshi K, Dong T, Gotch F, McAdam S, Whitby D, Sabally S, Gallimore A, Corrah T, et al. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat Med. 1995;1:59–64. doi: 10.1038/nm0195-59. [DOI] [PubMed] [Google Scholar]
- 29.Schneider R, Campbell M, Nasioulas G, Felber B K, Pavlakis G N. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J Virol. 1997;71:4892–4903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwartz S, Campbell M, Nasioulas G, Harrison J, Felber B, Pavlakis G. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J Virol. 1992;66:7176–7182. doi: 10.1128/jvi.66.12.7176-7182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steimer K S, Puma J P, Power M D, Powers M A, George-Nascimento C, Stephans J C, Levy J A, Sanchez-Pescador R, Luciw P A, Barr P J. Differential antibody responses of individuals infected with AIDS-associated retroviruses surveyed using the viral core antigen p25gag expressed in bacteria. Virology. 1986;150:283–290. doi: 10.1016/0042-6822(86)90289-8. [DOI] [PubMed] [Google Scholar]
- 32.Wagner R, Fliessbach H, Wanner G, Motz M, Niedrig M, Deby G, von Brunn A, Wolf H. Studies on processing, particle formation, and immunogenicity of the HIV-1 gag gene product: a possible component of a HIV vaccine. Arch Virol. 1992;127:117–137. doi: 10.1007/BF01309579. [DOI] [PubMed] [Google Scholar]