Abstract
We have investigated the secondary structure of peach latent mosaic viroid (PLMVd) in solution, and we present here the first description of the structure of a branched viroid in solution. Different PLMVd transcripts of plus polarity were produced by using the circularly permuted RNA method and the exploitation of RNA internal secondary structure to position the 5′ and 3′ termini and studied by nuclease mapping and binding shift assays using DNA and RNA oligonucleotides. We show that PLMVd folds into a complex, branched secondary structure. In general, this structure is similar to that reported previously, which was based on sequence comparison and computer modelling. The structural microheterogeneity is apparently limited to only some small domains. More importantly, this structure includes a novel pseudoknot that is conserved in all PLMVd isolates and seems to allow folding into a very compact form. This pseudoknot is also found in chrysanthemum chlorotic mottle viroid, suggesting that it is a unique feature of the viroid members of the PLMVd subgroup.
Viroids are small (∼300 nucleotides), single-stranded, circular RNAs that infect higher plants, causing significant losses in the agricultural industry (see references 7 and 13 for reviews). Viroids have been classified in two groups (groups A and B) based primarily on whether or not they possess five typical structural domains found in the group B viroids (7). Further division among the group B members depends on the sequence and length of the conserved central region. Viroids that do not possess any kind of sequence or structural similarity with the group B viroids have been classified as belonging to group A. The viroids from this group possess self-cleaving hammerhead motifs that are crucial for their replication via a rolling circle mechanism.
The group A viroids include the avocado sunblotch viroid (ASBVd), the peach latent mosaic viroid (PLMVd), and the chrysanthemum chlorotic mottle viroid (CChMVd) (10). Both PLMVd and CChMVd have been proposed to adopt branched secondary structures (Fig. 1A) instead of the rod-like ones proposed for most viroids, including ASBVd (10). The unusual conformations of PLMVd and CChMVd are supported by their insolubility in 2 M lithium chloride, whereas ASBVd and a number of non-self-cleaving viroids (i.e., the group B viroids) are soluble in this high salt solution (10). In general, secondary structures of viroids are predicted using computer software and are useful for the formulation of hypotheses on the structure-function relationships of these RNA molecules (4). Characterization of biological structures in vitro as well as in vivo is obviously more accurate for elucidating the structure-function relationship. The only secondary structure of a viroid that has been extensively studied in solution is that of the potato spindle tuber viroid (8). This group B species, which was shown to adopt a rod-like shape in solution, is responsible for most of our knowledge of the biology of viroids.
FIG. 1.
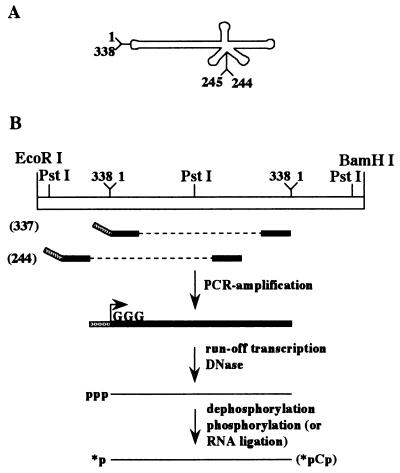
Strategy of synthesis of the PLMVd strands. (A) Schematic representation of the circular PLMVd strand of plus polarity. By convention, position 1 was arbitrarily fixed in the lefthand loop. (B) Strategy of synthesis of the linear PLMVd strands and their end labeling based on the circularly permuted RNA method. The 5′-end positions of the transcripts used in this work are indicated in parentheses. p, 5′-monophosphate; pCp, cytidine 5′-monophosphate, 3′-monophosphate; ∗, 32P.
In order to determine the secondary structure of PLMVd (the RNA species that causes peach latent mosaic disease [9]) in solution, we performed nuclease mapping and oligonucleotide binding shift assays on PLMVd synthesized by in vitro transcription. The results confirm that PLMVd folds into a complex, branched secondary structure and show that it can form a novel pseudoknot.
MATERIALS AND METHODS
Synthesis of DNA templates.
DNA templates for in vitro transcription of the three different RNA molecules used in this study were synthesized from the pPD1 vector (3). Briefly, this construct possesses two tandemly repeated PLMVd sequences (from the Armking peach cultivar [9]) cloned into the PstI restriction site of pBluescript II KS. The strategy for the production of monomeric PLMVd RNA has been reported previously (2). A first DNA template was amplified from pPD1 with Pwo DNA polymerase (Boehringer Mannheim) using sense (5′-TAATACGACTCACTATAGGGTCAAAAGTTTCGCCGC-3′) and antisense (5′-TATGAGTTTCGTCTCATTTC-3′) primers for RNA transcripts starting from position 337. A second DNA template was produced using sense (5′-TAATACGACTCACTATAGGGATTCAAACCCGGTC-3′) and antisense (5′-GGGTAGACGTCGTAATCC-3′) primers for RNA transcripts starting from position 244. The third DNA template, used for the production of the mutant harboring the sequence 212AAAA215, was prepared from pPD1 initially by using the PCR product generated with the sense primer used for transcripts starting at position 337 coupled with a mutated antisense primer (5′-TTTCTACGTTTTTACCTGGA-3′; the underlined nucleotides are the mutant ones) and, secondly, using the PCR product generated with a mutated sense primer (5′-TCCAGGTAAAAACGTAGAAA-3′) coupled with the antisense primer used for the transcripts starting at position 337. These two PCR products were mixed together and amplified using Taq DNA polymerase in order to produce a full-sized DNA template, which was then verified by dideoxynucleotide sequencing.
Runoff transcription.
In vitro transcription reaction mixtures contained unpurified PCR products, and the reactions were performed using 25 μg of purified T7 RNA polymerase (6, 14), as described previously (5). The reaction mixtures were fractionated by denaturing 5% polyacrylamide gel electrophoresis (PAGE) (ratio of acrylamide to bisacrylamide, 19:1) in buffer containing 90 mM Tris borate (pH 7.5), 7 M urea, and 2 mM EDTA. The reaction products were visualized by UV shadowing, after which the bands corresponding to the correct size (338 nucleotides) were cut out and the transcription products were eluted, passed through a G-50 Sephadex spun column (Amersham Pharmacia Biotech), and ethanol precipitated. The quantity of each product was determined by spectrophotometry at 260 nm.
5′ and 3′ end labeling of transcripts.
Transcripts (4 μg) were dephosphorylated with 0.2 U of calf intestinal alkaline phosphatase (Boehringer Mannheim) and then 5′ end labeled with 30 U of T4 polynucleotide kinase (Amersham Pharmacia Biotech) in the presence of 40 μCi of [γ-32P]ATP (3,000 mCi/mmol; Amersham Pharmacia Biotech). Transcripts (4 μg) were 3′ end labeled using 40 U of T4 RNA ligase (New England Biolabs) in the presence of 40 μCi of [32P]cytidine 5′-monophosphate, 3′-monophosphate (3,000 mCi/mmol; Amersham Pharmacia Biotech). After the labeling reaction occurred, the mixtures were separated on denaturing 5% PAGE gels and recovered as described above. The concentration of RNA was determined by Cherenkov counting.
Nuclease mapping.
Either 5′ or 3′ end labeled PLMVd transcripts (∼50,000 cpm) were resuspended in 4 μl of either 10 mM Tris-HCl (pH 8.0)–1 mM MgCl2 (low salt) or 20 mM Tris-HCl (pH 8.0)–10 mM MgCl2–100 mM NH4Cl (high salt) solution. The transcripts were then denatured by heating at 65°C for 2 min and renatured by slow cooling to room temperature. This procedure of denaturation-renaturation was performed prior to all enzymatic reactions unless indicated otherwise in the text. Either RNase T1 (0.25 U; Amersham Pharmacia Biotech), RNase T2 (6 U; Gibco BRL), RNase Phy M (0.5 U; Amersham Pharmacia Biotech), or RNase V1 (1 U; Amersham Pharmacia Biotech) was then added (in a final volume of 6 μl), and the mixtures were incubated at either 25 or 37°C for various times. For the RNase U2 reactions (0.02 U; Amersham Pharmacia Biotech), the transcripts were resuspended in a buffer containing 20 mM sodium citrate (pH 6.0). These mixtures were incubated at 37°C, and aliquots were removed at various times and quenched by the addition of 5 μl of stop solution. Ribonuclease T1 digestion was also performed in the presence of 7 M urea (at 55°C). Alkaline hydrolysis was performed to permit accurate assignment of the cleavage sites. The reaction products were fractionated on either 5, 8, or 12% denaturing PAGE gels and then either exposed to a X-ray film or fixed, dried, and exposed to a PhosphorImager screen when quantification was required.
Oligonucleotide binding shift assays.
RNA oligonucleotides were chemically synthesized using an automated oligonucleotide synthesizer (Keck Biotechnology Resource Laboratory, Yale University), deprotected, and gel purified as described previously (12). Deprotected and purified DNA oligonucleotides were purchased from Gibco BRL. Each oligonucleotide (5 pmol) was 5′ end labeled using T4 polynucleotide kinase in the presence of an excess of [γ-32P]ATP, as described above, and was purified by phenol-chloroform extraction followed by passage through two Sephadex G-50 spun columns. The quantity of labeled oligonucleotide was determined (in counts per minute) by Cherenkov counting, while the quality was verified by denaturing 20% PAGE. Dried PLMVd transcripts (500 ng) were resuspended in 8 μl of a solution containing 10 mM Tris-HCl (pH 8.0)–1 mM MgCl2 (low salt). Radiolabeled oligonucleotide (20,000 cpm, 2 μl) was added either before or after the transcripts were denatured for 2 min at 65°C and renatured by being slowly cooled to 4°C. The mixtures were then incubated at 4°C for at least 60 min prior to the addition of 2 μl of agarose dye (30% glycerol [vol/vol], 1 mM EDTA [pH 8.0], 0.25% [each] xylene cyanol and bromophenol blue) and analysis on 2.5% agarose gels. The resulting gels were fixed, dried, and exposed to X-ray films.
RESULTS
RNA synthesis strategy.
In order to probe the secondary structure of a PLMVd sequence variant isolated from the Armking peach cultivar (also named French variant [9]), a unique 32P-labeled phosphate had to be introduced at an end of the RNA strand. Initially, we tried to label PLMVd monomeric strands produced by the self-cleavage of linear concatamers. However, these RNA strands were inefficiently 32P-labeled at both the 5′ and the 3′ ends, suggesting that both extremities were located inside the molecule. Consequently, we developed a second strategy for the production of full-length PLMVd molecules that could be efficiently labeled at both ends (2). This strategy was based on the circularly permuted RNA method (11) and on the exploitation of RNA internal secondary structure to position the 5′ and 3′ termini such that they would be located in single-stranded regions which were easily accessible for efficient end labeling (Fig. 1B). In this approach, nonradioactive, full-length PLMVd transcripts possessing a 5′ triphosphate (at position 337) and a 3′ hydroxyl (at position 336) were produced and further purified. As shown in Fig. 1, positions 336 and 337 are located in the lefthand loop. The only modification from the wild-type sequence is the introduction of three guanosines at the 5′ end in place of the 337UCA1 to allow for efficient transcription. A second PLMVd-derived RNA molecule whose 5′ end is located at position 244 was produced with the appropriate oligonucleotides according to the same procedure (Fig. 1). Since positions 244 to 246 correspond to three guanosines in the wild-type sequence, no mutation was required in order to favor in vitro transcription. This transcript would confirm the results from the previous construct and more specifically would allow study of the secondary structure of the lefthand loop, including positions 336 and 337, where the other construct was opened.
Nuclease probing.
The structures of the plus polarity PLMVd strands were probed using various nucleases which cleave 3′ of different nucleotides, as follows: (i) RNase T1, which preferentially cleaves single-stranded guanosines; (ii) RNase U2, which preferentially cleaves single-stranded adenosines; (iii) RNase Phy M, which preferentially cleaves single-stranded adenosines and uracils; (iv) RNase T2, which preferentially cleaves single-stranded nucleotides regardless of base identity; and (v) RNase V1, which preferentially cleaves double-stranded nucleotides regardless of base identity. In addition, some portions of the molecules were probed with the Bacillus cereus RNase, which has a preference for single-stranded cytosines and uracils, in order to confirm the data obtained with the other enzymes. The reaction conditions for each nuclease were optimized prior to the structural probing. This optimization was required, as no carrier (i.e., tRNA) was added to the reactions to avoid any structural interference with the PLMVd strands. Regardless of whether the RNase mapping was performed at 22 or 37°C, no significant differences in the cleavage pattern were detected.
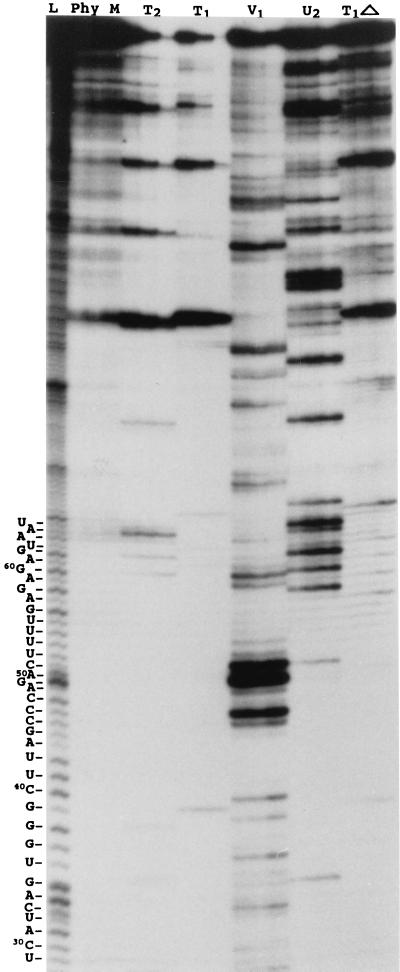
Prior to the nuclease probing, the transcripts were treated in several ways in order to favor structural homogeneity. For example, they were denatured at 65°C and then renatured by slow cooling to either room temperature or 4°C or they were partially denatured at 85°C (always in the absence of magnesium to avoid self-cleavage) and then renatured at room temperature in a two-step procedure (i.e., 2 min at 55°C followed by 2 min at room temperature) prior to addition of the magnesium. With the exception of the observation that incubation at 85°C stimulates self-cleavage of the transcripts, the various treatments all produced the same digestion pattern (data not shown). Therefore, we decided to use the simplest treatment, consisting of a 2-min denaturation at 65°C followed by slow cooling to room temperature. During the nuclease hydrolysis experiments, aliquots were removed from the reaction mixture at different times, reaction in these aliquots was stopped, and they were fractionated on 5 to 12% polyacrylamide sequencing gels. Figure 2 shows a typical autoradiogram for 5′ end-labeled PLMVd transcripts starting at position 337, under low salt conditions. Under these electrophoretic conditions the region from positions 29 to 66, which corresponds to a portion of the upper strand of the P11 stem and the P1 hairpin structure (Fig. 3), was revealed. Nucleotides 29 to 66 are primarily hydrolyzed by RNase V1, indicating that they are in a double-stranded region.
FIG. 2.
Autoradiogram of a PLMVd nuclease mapping assay under low salt conditions analyzed on a 12% PAGE gel. The nucleases used are indicated at the top. L, alkaline hydrolysis ladder; T1▵, RNase T1 reaction mixture incubated at 55°C. From left to right, the three lanes for each enzyme are for aliquots removed at 30 s, 1 min, and 2 min, respectively. The PLMVd sequence for the region from U29 to U66 is shown at the left.
FIG. 3.
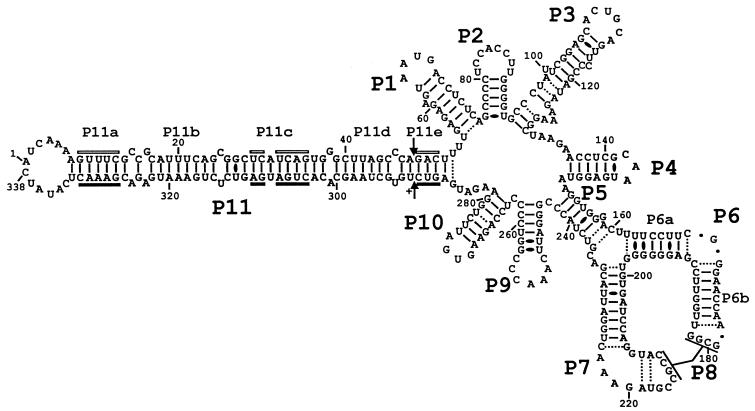
PLMVd sequence and the secondary structure proposed based on nuclease mapping under low salt conditions. The helix numbering has been arbitrarily fixed as if the circular RNA was synthesized from position 1 and follows the helices' order of appearance. GU wobble base pairs are represented with black ovals, while Watson-Crick base pairs which appeared to be unstable or for which single- and double-strand coexistence is proposed under the conditions used are illustrated with dotted lines. The base pairs of the P8 pseudoknot were not included in order to simplify the illustration.
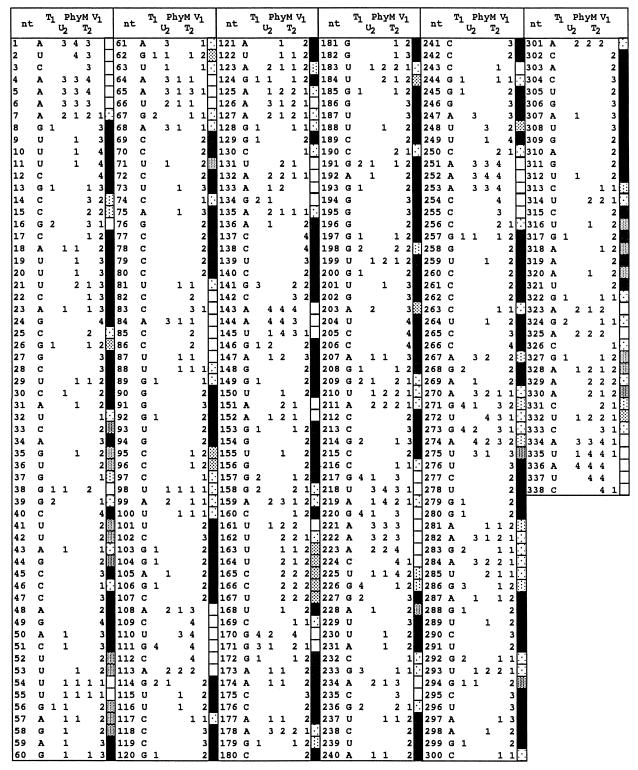
At least the first 180 nucleotides from both the 5′-end [γ-32]ATP- and 3′-end-[32P]-pCp-labeled transcripts were resolved using different migration conditions (e.g., by varying the migration times and acrylamide concentrations). The relative intensity of all radioactive bands was scored in comparison to the most intense bands found in each reaction. Scores varying from 1 to 4 (with 4 being the most intense) were attributed to each nucleotide, and then a relative intensity average at each nucleotide for each nuclease was calculated (Fig. 4). In some cases, more than eight experiments were performed to ensure the reproducibility of the results. The two RNA constructs yielded similar results, with the exception that nuclease accessibility was greater close to their respective ends. For example, the construct starting at position 337 indicated that both ends were single stranded and highly accessible, while the construct starting at position 244 confirmed the presence of the lefthand loop, although the intensity of the bands was reduced.
FIG. 4.
Compilation of band intensity observed by RNase mapping. Relative intensities (on a scale of 1 to 4, with 4 being the strongest) were determined for each pattern of RNase hydrolysis. The fill patterns of the rectangles indicate the probability of a nucleotide being in either a single- or a double-stranded region (open or closed pattern, respectively). Rectangles for bands of intermediate intensity (between closed and opened rectangles) indicate a tendency for one or the other structure, as illustrated by the dot patterns.
Description of the proposed secondary structure.
In order to facilitate the description of the secondary structure (Fig. 3), we have numbered the helices as if this circular RNA had been synthesized from position 1, which by convention is located in the lefthand loop, and numbered the stems in the order of appearance. If two helices have the potential to stack but are separated by internal loop(s), the same number was given to both stems and they are then differentiated by a letter (e.g., the P11a to P11e stems [Fig. 3]).
In general, this structure is similar to that based on sequence comparison and computer modeling (1, 3). According to this secondary structure, 86 (25.4%) and 252 (74.5%) nucleotides are located in single- and double-stranded regions, respectively, when GU wobble base pairs and Watson-Crick base pairs proven to exist in high salt conditions are considered (see below). This structure includes 11 helices (P1 to P11), with the P11 stem including four consecutive stems (P11a to P11d) that form a lefthand rod-like domain that contains the hammerhead sequences of both the plus and minus polarities. While most of the helices correspond to hairpins (i.e., stem-loop structures), the P5 stem is an internal helix which acts as a bridge closing the molecule. The proposed structure includes 17 bp for which it is not unequivocal whether the two nucleotides in question are single stranded or base paired in the presence of low salt (50 mM Tris-HCl–10 mM MgCl2). In fact, both forms may coexist. In order to verify this hypothesis and to investigate potential structural transitions caused by higher ionic strengths, nuclease probing experiments were also performed in buffer containing 50 mM Tris-HCl, 10 mM MgCl2 and 100 mM NH4Cl (raw data not shown). In general, the results were identical to those shown in Fig. 4, regardless of the difference in the processing of each enzyme. No structural transition was detected. The uncertain base pairs (Fig. 3) in the middle of the P3 stem, in the bottom of the P5 stem, in the P6a stem, and in the P10 stem appeared to be closed, eliminating the possibility of the coexistence of single- and double-stranded structures in higher salt conditions. However, the uncertain base pairs could also be due to the steric hindrance of RNases because of their large size. Thus, the proposed secondary structure appears to remain identical under both salt conditions tested.
(i) Novel P8 pseudoknot.
The proposed secondary structure for PLMVd includes a novel pseudoknot, the P8 stem. In previous reports on secondary structures (1, 3, 5, 9), nucleotides 179GCGG182 (loop P6b) and 209GUACCGCCGUAGAAA223 (loop P7) were proposed to form single-stranded regions. In this scenario, it is expected that the RNases with specificities for single-stranded nucleotides should yield hydrolysis products, while RNase V1 should not. In the structure proposed here, nucleotides 179GCGG182 (loop P6b) and 212CCGC215 (loop P7) base-pair to form a pseudoknot (Fig. 5A). We observed that RNase V1 (specific for double-stranded regions) produced several cleavage products in these regions, while both RNase T1 (specific for single-stranded G) and T2 (specific for single-stranded regions) were almost unable to hydrolyze the phosphodiester backbone of these bases. In addition, nucleotides 210UA211 and 218UA219 of the P7 loop are proposed to base-pair together. These results clearly demonstrate that the P7 loop (positions 209 to 224) appears to be highly structured, with the exception of the 220GAAA223 loop (see Fig. 4 and 5A).
FIG. 5.
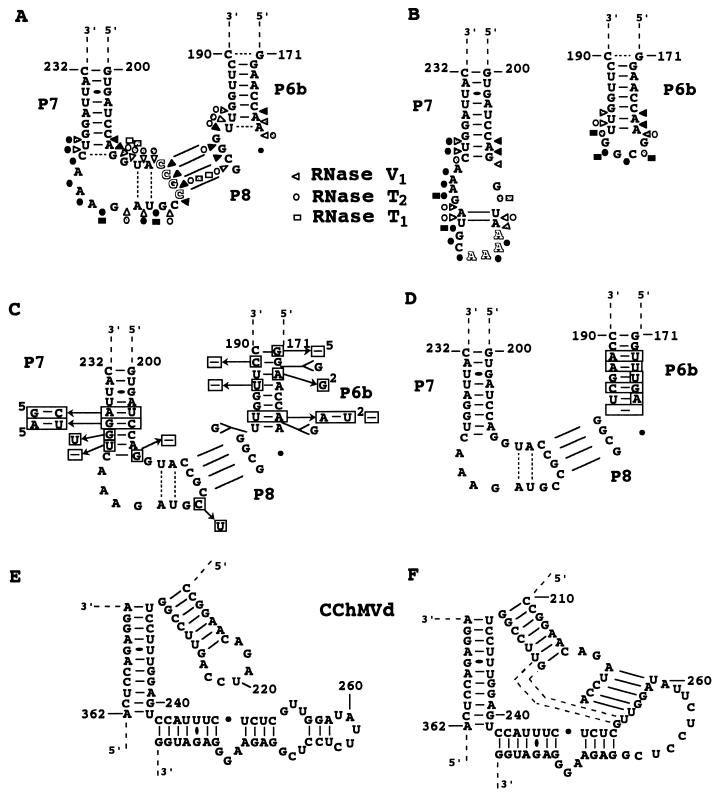
Characterization of the P8 pseudoknot and surrounding region. (A and B) Nuclease mapping of the wild-type (212CCGC215) and mutant (212AAAA215) PLMVd transcripts. The mutated bases are shadowed. Only results with RNases V1, T1, and T2 are shown. White, dotted, and black forms indicate weak, intermediate, and strong cleavage, respectively. (C and D) Nucleotide variations observed in all known natural PLMVd isolates, as of 1 August 1999 (Pelchat et al., unpublished data). (C) Sequences of 62 variants. (D) Sequence of a PLMVd isolated from Hardired cultivar (i.e., Hd6 [Pelchat et al., unpublished data] for which most of the P6b stem is different. Differences from the sequence characterized in this study are boxed. Superscript numbers in panel C indicate the number of PLMVd variants that include a mutation. (E and F) Sequences and proposed secondary structures of the corresponding region of CChMVd, with panel E showing the secondary structure as proposed previously (10) and panel F showing the secondary structure proposed here, which includes the P8 pseudoknot. Dotted lines are potential additional base pairs extending the pseudoknot.
In order to support the presence of the P8 pseudoknot, a mutant PLMVd transcript was synthesized and nuclease mapping was carried out (Fig. 5B). In contrast to the wild-type sequence, which includes the sequence 212CCGC215 in the P7 loop, the mutant possesses the sequence 212AAAA215. We chose to introduce four adenines because the hydrolysis pattern with the RNase Phy M was clear compared to those observed with other nucleases except RNase T1. With this transcript, the RNase V1 did not hydrolyze nucleotides 212AAAA215 or 179GCGG182, whereas the four adenines at positions 212 to 215 were readily hydrolyzed by RNase T2 and nucleotides 179GCGG182 were readily hydrolyzed by both RNase T2 and T1. In addition, we observed differences in the hydrolysis patterns of nucleotides 210UA211 and 218UA219, suggesting a stronger base-pairing in the mutant transcript (Fig. 5B). The latter difference is probably due to the fact that the absence of the pseudoknot reduced the constraint (i.e., stress) on the P7 loop, thereby favoring formation of the two base pairs. With the exception of the P8 pseudoknot region, the mutant gave a digestion pattern similar to that of the wild-type transcript.
The base composition of this 4-bp pseudoknot is perfectly conserved among all PLMVd variants (Fig. 5C and D) (1; M. Pelchat, D. Lévesque, J. Ouellet, S. Laurendeau, S. Lévesque, J. Lehoux, D. A. Thompson, L. J. Skrzeczkowski, and J. P. Perreault, unpublished data). This is an indirect proof of the existence of the P8 pseudoknot, although covariation would have been more conclusive. Nucleotide variations were observed on both sides of the pseudoknot, but they did not affect its length. Covariations are observed at the bottom of the P6b and P7 stems. The perfect conservation of the sequence forming the pseudoknot (i.e., four ordered GC base pairs) is probably required for formation of a helix in this stretched region of the molecule. It is also plausible that the base conservation has a biological relevance, such as binding to a host macromolecule. Surprisingly, a similar pseudoknot may be formed by CChMVd, another member of the PLMVd subgroup (Fig. 5E and F). According to the structural similarities between PLMVd and CChMVd (Pelchat et al., unpublished data), a pseudoknot may be formed between the P6 and P7 loops. The presence of this CChMVd pseudoknot prevents formation of 3 bp in the P7 stem, but it adds 5 bp, resulting in a net increase of two base pairs which serve to stabilize the structure (Fig. 5E and F).
(ii) Alternative structures.
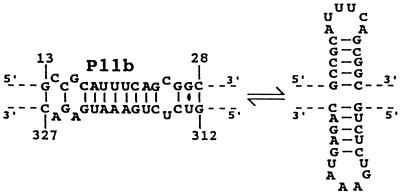
Based on sequence comparisons, it was proposed that stem P11b and the adjacent nucleotides on both sides may adopt a slightly less stable, in terms of energy, alternative structure corresponding to the hammerhead hairpin II on both strands (Fig. 6) (1). Although nuclease probing shows stem P11b to be as proposed in Fig. 3 for most RNA molecules, we did note that RNase Phy M hydrolyzed after the A and U in the 18AUUUCA23 and 316UAGAAU321 stretches, albeit at reduced levels (Fig. 4). One way to reconcile these results is to propose that the RNA strands adopt both structures but strongly prefer the most stable one, which allows for the stacking of all P11 subhelices.
FIG. 6.
Alternative structure including the hairpin II of the hammerhead motifs.
The presence of a pseudoknot formed by nucleotides of the left-handed loop and the P1 loop, specifically between the sequences 337UCAU2 and 65AUGA68, has been suggested previously (1). While the RNA construct with the 5′ end at position 337 does not allow observation of the formation of this pseudoknot, the second construct, with the 5′ end at position 245, should. The nuclease mapping results with this RNA construct suggest that both loops are located in single-stranded regions, and, therefore, the presence of this potential pseudoknot is not supported under the conditions tested (Fig. 4). However, sequence analysis of different PLMVd isolates shows that this pseudoknot may be extended 5 or 6 bp in other variants and that the stability of this motif is related to the pathogenicity of the viroid (1). Thus, it seems that this pseudoknot is not found in all PLMVd sequence variants, and consequently it may be related to an unessential function of the viroid, such as its pathogenicity (1).
Additional support from binding shift assays.
Concurrent with the nuclease mapping experiments, oligonucleotide binding shift assays were performed with wild-type PLMVd transcripts starting at position 337. Trace amounts of 5′-end-labeled DNA or RNA oligonucleotides were incubated in the presence of a large excess of PLMVd transcripts under low salt conditions, and the mixtures were fractionated on native agarose gels. The oligonucleotides were added either before or after the transcripts were denatured at 65°C and renatured by slow cooling to 4°C. Addition of the oligonucleotide prior the denaturation-renaturation should favor binding to the transcript. Among the DNA oligonucleotides tested, trace amounts (>0.1%) of PLMVd-oligonucleotide complexes were detected with D33-48 and D94-109 only when they were added prior to renaturation (Table 1). These two oligonucleotides correspond to one strand each of the P11c-e and the P3 stems and most probably hybridize to the complementary PLMVd strands. In contrast, the D39-58 oligonucleotide complementary to P11d and P11e and a portion of P1 stem produced no oligonucleotide binding, indicating that this region is tightly folded (Table 1). Oligonucleotides D153-171 and D205-224, complementary to the P5 and P6 stems and the P7 stem-loop structures, respectively, hybridized with the PLMVd transcripts. Trace amounts of D153-171 were shown to hybridize with transcripts regardless of when the oligonucleotide was added to the mixture, indicating that this region was slightly accessible to the oligonucleotide (Table 1). In contrast, D205-224 was predominantly found to hybridize when it was present during the denaturation-renaturation, whereas only trace amounts were found to hybridize when it was added after the denaturation-renaturation (Table 1). This suggests that the oligonucleotide hybridizes poorly when added to stably folded PLMVd, thereby supporting the presence of the P8 pseudoknot. The specificity of hybridization for oligonucleotides D153-171 and D205-224 was verified by RNase H hydrolysis (data not shown).
TABLE 1.
Binding shift assays
| Oligonucleotidea | Length (nt) | Polarityb | Hybridizationc
|
|
|---|---|---|---|---|
| Before | After | |||
| D33-48 | 16 | S | + | − |
| D39-58 | 20 | AS | − | − |
| D94-109 | 15 | S | + | − |
| D153-171 | 19 | AS | + | + |
| D205-224 | 20 | AS | +++ | + |
| R77-83 | 7 | AS | − | − |
| R130-135 | 6 | AS | ++ | ++ |
| R212-216 | 5 | AS | − | − |
| R212-223 | 12 | AS | ++ | + |
In the oligonucleotide designations, R and D indicate RNA and DNA natures, respectively, and the numbers correspond to positions on the PLMVd sequence (Fig. 3).
Oligonucleotide sequences are either sense (S) or antisense (AS) (complementary).
Symbols indicate proportion of shifted oligonucleotide, as follows: −, 0; +, >1%; ++, 1 to 10%; and +++, >10%. Oligonucleotides were added to the mixtures before or after the denaturation-renaturation of the transcripts.
Shorter RNA oligonucleotides were also tested in order to investigate the accessibility of short PLMVd sequences (Table 1). Oligonucleotide R77-83, which is complementary to a portion of the P2 stem-loop, constitutes a negative control and produced no complex. These results were expected, since nuclease mapping clearly indicated the presence of this stem. Oligonucleotide R130-135 hybridized efficiently to the PLMVd transcripts regardless of whether it was added before or after the denaturation-renaturation, supporting the hypothesis that the small region between stems P3 and P4 is single stranded. A short RNA oligonucleotide, R212-216 (5-mer), corresponding to the P7 loop sequence involved in formation of the P8 stem, did not bind to the PLMVd transcripts, supporting the presence of the stable pseudoknot. In contrast, oligonucleotide R212-223 (12-mer) encompassing most of the P7 loop hybridized, indicating that it has the ability to unfold the P8 pseudoknot. DNA and RNA oligonucleotide binding shift assays were also performed under different salt conditions (i.e., high salt) and different magnesium concentrations. With the exception of small differences in the binding affinity of some oligonucleotides, the results were similar regardless of the conditions used. Thus, the binding shift assays support the PLMVd secondary structure proposed with the data from the nuclease mapping experiments.
DISCUSSION
Structural homogeneity of the transcripts.
Nuclease mapping and oligonucleotide binding shift assays confirmed that PLMVd folds into a branched secondary structure in solution. The nuclease mapping data from the two transcripts (i.e., wild-type transcripts starting either at position 377 or at position 244) used here agreed perfectly except for near the termini of the RNA molecules. Clearly, both transcripts fold into a similar structure. This conclusion is supported by data from lead-induced cleavage experiments at a pH of 7.0 with both these transcripts and the 212AAAA215 mutant (J. Ouellet and J. P. Perreault, unpublished data). The patterns produced on PAGE gels by lead ion hydrolysis were similar for all three RNA transcripts, clearly suggesting that they are folded in a similar manner (data not shown). The only important difference in the cleavage patterns of these three transcripts was that the 212AAAA215 mutant's P6b and P7 loops were efficiently hydrolyzed, while those of wild-type sequence possessing the P8 pseudoknot and the base pairs 210UA211 and 218UA220 were not. The different results presented here support the hypothesis that the PLMVd transcripts characterized in this report fold into similar structures.
PLMVd structure in solution.
Complete nuclease mapping of RNA molecules longer than 100 nucleotides in length is limited to a few examples. One of these is the group B viroid potato spindle tuber viroid, which was unambiguously shown to fold into a rod-like structure in solution (8). In this report we use nuclease mapping, combined with oligonucleotide binding shift assays, to show that PLMVd adopts a complex, branched secondary structure in solution. In general, this structure is similar to that based on sequence comparison and computer modeling (1; Pelchat et al., unpublished data), having 74.5% of its nucleotides in base pairs. Structural microheterogeneity seems to be limited to small domains, including the hammerhead hairpin II and the P8 pseudoknot, of which the latter adopts primarily one structure, with only a small proportion (i.e., trace amounts) being observed to fold into alternative structures (see Results). The use of low and high salt buffers allowed us to demonstrate that some base pairs are stabilized in the latter conditions. For example, inside the P3 stem 3 bp were evident under high salt conditions, while under low salt conditions these base pairs seemed to form in only a portion of the RNA molecules.
Based on the results presented here, we believe that the PLMVd structure includes the novel P8 pseudoknot. This pseudoknot also appears to be present in CChMVd, suggesting that it is a unique feature of the viroid members of the PLMVd subgroup. The structural similarities between PLMVd and CChMVd, including the presence of this pseudoknot, may help to explain why these viroids are insoluble in 2 M lithium chloride while all other viroids proposed to adopt a rod-like structure are soluble. The branched secondary structures of PLMVd and CChMVd, in conjunction with the presence of the P8 pseudoknot, produce a compact structure that is probably the biochemical basis of this observation.
The proposed structure for PLMVd is the most stable one adopted under the conditions used. However, it must be remembered that the most stable structure is not necessarily the biologically active structure. For example, hammerhead structures are crucial for self-cleavage during the rolling circle replication of PLMVd but are not formed in the most stable PLMVd structure. Furthermore, host protein(s) may interact with PLMVd and alter its folding. Thus, a host protein may be involved in formation of the proposed pseudoknot between the P1 and P11 loops (1) and in the formation of another putative pseudoknot involving the P1 stem and the single-stranded domain between the P10 and P11 stems (Pelchat et al., unpublished data), thereby explaining why these structural motifs were not observed in this structure. Conversely, the P8 pseudoknot, whose sequence is highly conserved among all PLMVd variants, is most likely to be essential in the PLMVd life cycle.
ACKNOWLEDGMENTS
F. Bussière and J. Ouellet contributed equally to this work.
This work was sponsored by a grant from Natural Sciences and Engineering Research Council (NSERC) of Canada to J.P.P. F.B. and F.C. were the recipients of NSERC studentships. J.P.P. is a Medical Research Council (MRC) of Canada scholar.
REFERENCES
- 1.Ambros S, Hernandez C, Desvignes J C, Flores R. Genomic structure of three phenotypically different isolates of peach latent mosaic viroid: implication of the existence of constraints limiting the heterogeneity of viroid quasispecies. J Virol. 1998;72:7397–7406. doi: 10.1128/jvi.72.9.7397-7406.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaudry D, Perreault J P. An efficient strategy for the synthesis of circular RNA molecules. Nucleic Acids Res. 1995;23:3064–3066. doi: 10.1093/nar/23.15.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaudry D, Bussière F, Lareau F, Lessard C, Perreault J P. The RNA of both polarities of the peach latent mosaic viroid self-cleaves in vitro solely by single hammerhead structures. Nucleic Acids Res. 1995;23:745–752. doi: 10.1093/nar/23.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bussière F, Lafontaine D, Perreault J P. Compilation and analysis of viroid and viroid-like RNA sequences. Nucleic Acids Res. 1995;24:1793–1798. doi: 10.1093/nar/24.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bussière F, Lehoux J, Thompson D A, Skrzeczkowski L J, Perreault J P. Subcellular localization and rolling circle replication of peach latent mosaic viroid: hallmarks of group A viroids. J Virol. 1999;73:6353–6360. doi: 10.1128/jvi.73.8.6353-6360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davanoo P, Rosenberg A H, Dunn J J, Studier F W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1984;81:2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flores R, Di Serio F, Hernandez C. Viroids: the noncoding genomes. Semin Virol. 1997;8:65–73. [Google Scholar]
- 8.Gast F U, Kempe D, Spieker R L, Sänger H L. Secondary structure probing of potato spindle tuber viroid (PSTVd) and sequence comparison with other small pathogenic RNA replicaons provides evidence for central non-canonical base-pairs, large A-rich loops, and a terminal branch. J Mol Biol. 1996;262:652–670. doi: 10.1006/jmbi.1996.0543. [DOI] [PubMed] [Google Scholar]
- 9.Hernandez C, Flores R. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc Natl Acad Sci USA. 1992;89:3711–3715. doi: 10.1073/pnas.89.9.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navarro B, Flores R. Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of self-cleaving viroids with hammerhead ribozymes. Proc Natl Acad Sci USA. 1997;94:11262–11267. doi: 10.1073/pnas.94.21.11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nolan J M, Burke D H, Pace N R. Circular permuted tRNAs as specific photoaffinity probes of ribonuclease P RNA structure. Science. 1993;261:762–765. doi: 10.1126/science.7688143. [DOI] [PubMed] [Google Scholar]
- 12.Perreault J P, Altman S. Important 2′-hydroxyl groups in model substrates for M1 RNA, the catalytic subunits of RNase P from E. coli. J Mol Biol. 1992;226:399–409. doi: 10.1016/0022-2836(92)90955-j. [DOI] [PubMed] [Google Scholar]
- 13.Symons R H. Plant pathogenic RNAs and RNA catalysis. Nucleic Acids Res. 1997;25:2683–2689. doi: 10.1093/nar/25.14.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zawadski V, Gross H J. Rapid and simple purification of T7 RNA polymerase. Nucleic Acids Res. 1991;19:1948. doi: 10.1093/nar/19.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]