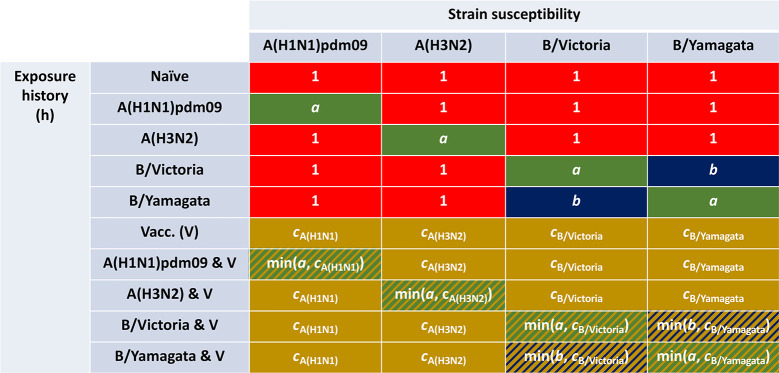
There is an error in Fig 3. The row labels for rows 4 and 5 were ordered incorrectly: the label for row 4 should read ‘B/Victoria’ and the label for row 5 should read ‘B/Yamagata’. Please see the correct Fig 3 here.
Fig 3. Infographic presenting the interaction between exposure history and susceptibility.
The interaction between exposure history h and susceptibility to strain m in the current influenza season, f(h, m), was classified into ten distinct groups: One group for the naive (uninfected and not vaccinated, row 1); one group per strain, infected but not vaccinated (rows 2–5); one group for those vaccinated and experiencing no natural infection (row 6); one group per strain for being infected and vaccinated (rows 7–10). We let a denote modified susceptibility to strain m given infection by a strain m type virus the previous influenza season (dark green shading), b modified susceptibility due to cross-reactivity between type B influenza lineages (dark blue shading), and cm the change in susceptibility to strain m given vaccination in the previous influenza season (gold shading). Unmodified susceptibilities retained a value of 1 (red shading). We enforced 0 < a,b,cm < 1.
Reference
- 1.Hill EM, Petrou S, de Lusignan S, Yonova I, Keeling MJ (2019) Seasonal influenza: Modelling approaches to capture immunity propagation. PLoS Comput Biol 15(10): e1007096. 10.1371/journal.pcbi.1007096 [DOI] [PMC free article] [PubMed] [Google Scholar]