Abstract
Antibodies are critical tools for research into extracellular vesicles (EVs) and other extracellular nanoparticles (ENPs), where they can be used for their identification, characterization, and isolation. However, the lack of a centralized antibody platform where researchers can share validation results thus minimizing wasted personnel time and reagents, has been a significant obstacle. Moreover, because the performance of antibodies varies among assay types and conditions, detailed information on assay variables and protocols is also of value. To facilitate sharing of results on antibodies that are relevant to EV/ENP research, the EV Antibody Database has been developed by the investigators of the Extracellular RNA Communication Consortium (ERCC). Hosted by the ExRNA Portal (https://exrna.org/resources/evabdb/), this interactive database aggregates and shares results from antibodies that have been tested by research groups in the EV/ENP field. Currently, the EV Antibody Database includes modules for antibodies tested for western Blot, EV Flow Cytometry, and EV Sandwich Assays, and holds 110 records contributed by 6 laboratories from the ERCC. Detailed information on antibody sources, assay conditions, and results is provided, including negative results. We encourage ongoing expert input and community feedback to enhance the database’s utility, making it a valuable resource for comprehensive validation data on antibodies and protocols in EV biology.
Keywords: Extracellular vesicles, Nanoparticles, Exosomes, Antibody database, Antibodies
The field of EV and ENP biology is rapidly expanding1 and the relevant nomenclature is evolving, with a growing recognition of EV and ENP heterogeneity, as defined by differential composition of a variety of molecular cargo, including protein markers.1–7 As a result, there is now a pressing need to standardize and validate antibody reagents and protocols to minimize time spent on evaluation and experimental optimization, in order to accelerate rates of discovery of EV and ENP subtypes and enable reproducible characterization across laboratories. Here, we present the EV Antibody Database, an interactive database hosted by the ExRNA Portal (exRNA.org) designed to collect and share information on curated antibodies and protocols for targets relevant to EV biology. The EV Antibody Database portal is designed to allow contributors to share procedural details for each protocol application in a self-explanatory format. Individuals wishing to contribute to the database simply have to create an account and log in and then navigate to the “Add New Antibody” tabs. Through the EV Antibody Database, we aim to streamline antibody and protocol selection, thereby expediting research efficiency and improving experimental reproducibility across the scientific community.
2. Validation of antibodies for western blot
The Western Blot analysis module of the database currently contains 62 records of antibodies raised against EV proteins contributed by multiple laboratories in the Extracellular RNA Communication Consortium (ERCC).8 Each record contains detailed information on the antibody (including the vendor and species), western blotting conditions used, and representative results. For most records, the antibody was tested using a range of western blotting conditions, including reducing and non-reducing conditions, various blocking reagents, diverse source of test materials (biofluids, cells and tissue lysates, purified EV subpopulations), and different transfer conditions (Fig. 1). Importantly, antibodies are tested using primary-source test materials, rather than cell lines engineered to overexpress the target protein or purified preparations of recombinant target proteins. Records include antibodies and conditions that failed to provide adequate signal-to-noise ratios, and those that yielded acceptable outcomes. Thus, users of the database can immediately select antibodies and conditions that produced superior results in relevant test materials, avoiding unnecessary expenditures on lower performing antibodies or the need to conduct prolonged trial-and-error optimization experiments. In addition, we provide users with a way to contact the author of a record or the database administrator regarding any difficulties with reproducing a result. Most records also link to the vendor’s website and to the Antibodypedia portal of validated antibodies (https://www.antibodypedia.com/). We anticipate that this database’s utility will expand as users contribute additional input records of antibodies against markers of vesicular or non-vesicular extracellular particles, as well as cell- and tissue-specific markers.
Fig. 1.
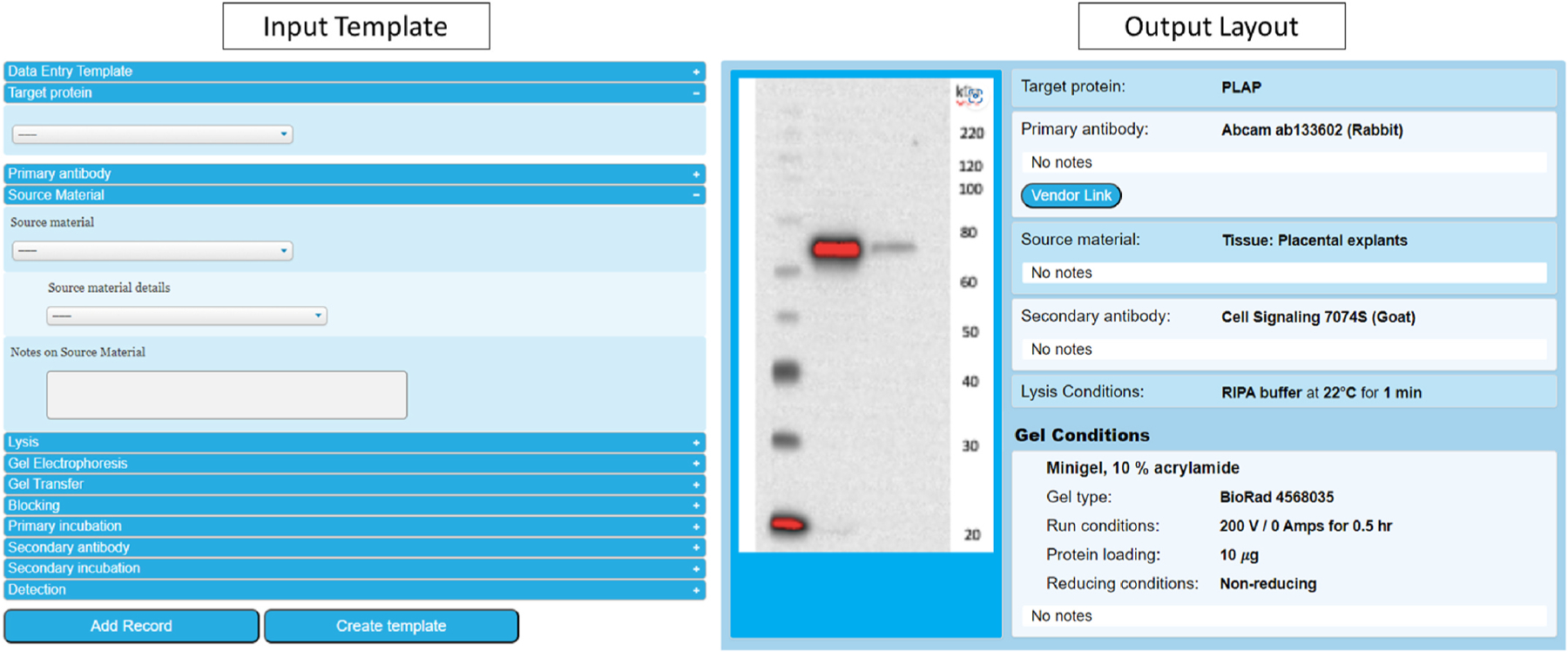
Input and output layout of the Western blot module of the EV Antibody Database. The input template contains fields for comprehensive reporting of experimental variables, a button for upload of an image of the resulting Western blot, and a field for comments. There is an option to create personalized templates, which contain pre-populated fields to save data entry time. The output page presents the experimental conditions and Western blot image for a given record in an easy-to-read format template, together with a link for the primary antibody to Antibodypedia and a button to send questions or comments to the author of the record or the database manager.
It is widely recognized that antibodies that perform well for one type of assay might not necessarily do so across other technologies. This distinction is particularly important when comparing assays run in denaturing conditions, such as most western blots, with those run on native proteins or even intact vesicles, as in the case of the various EV Flow Cytometry methodologies,9,10 intact vesicle sandwich immunoassays11,12 and many EV imaging techniques.13 Steric effects may play a far larger role in intact vesicle assays than in assays on soluble or denatured proteins, so antibodies should be selected based on downstream assay platform. Therefore, the EV Antibody Database also has modules for EV flow cytometry and sandwich assays, including both EV capture and EV detection antibodies.
3. Antibody characterization and validation for EV flow cytometry
Flow cytometry (FC) has emerged as a particularly powerful tool for the study of EVs due to its sensitivity, multi-parameter capabilities, and high throughput. Assays have been developed and validated that, when used with instruments of suitable sensitivity, can measure EV concentration, size, and molecular cargo.14–19 Antibodies are a key reagent in any type of FC, and characterization of antibody specificity, selectivity, and suitability for EV FC is critical to the generation of rigorous and reproducible results.9,10 Beyond these general properties, for any new fluorescent antibody conjugate, instrument calibration using standardized reference beads encompassing the appropriate fluorescent spectra and brightness levels, spectral calibration, generation of compensation or unmixing matrices to enable multiplexed measurements with other antibody conjugates, and titration to determine the optimal concentration for use must be performed.
The Output layout for the EV Flow Cytometry module is shown in Fig. 2. The Output display is meant to convey the basic information that a user will need to determine whether a given antibody is of potential utility for EV FC, providing graphs demonstrating the performance of the antibody on a representative sample, confirming instrument calibration, and displaying the titration curve for the specific fluorescent antibody conjugate. Information regarding the fluorescent label conjugated to the antibody, the instrument used to produce the data shown, and the positive and negative controls and calibrators used are listed. For further details on instrument configuration and the assay protocol, the user can download the associated MiFlow-Cyt EV report, which the submitter completes according to recently published International Society for Extracellular Vesicles (ISEV) guidelines10
Fig. 2.
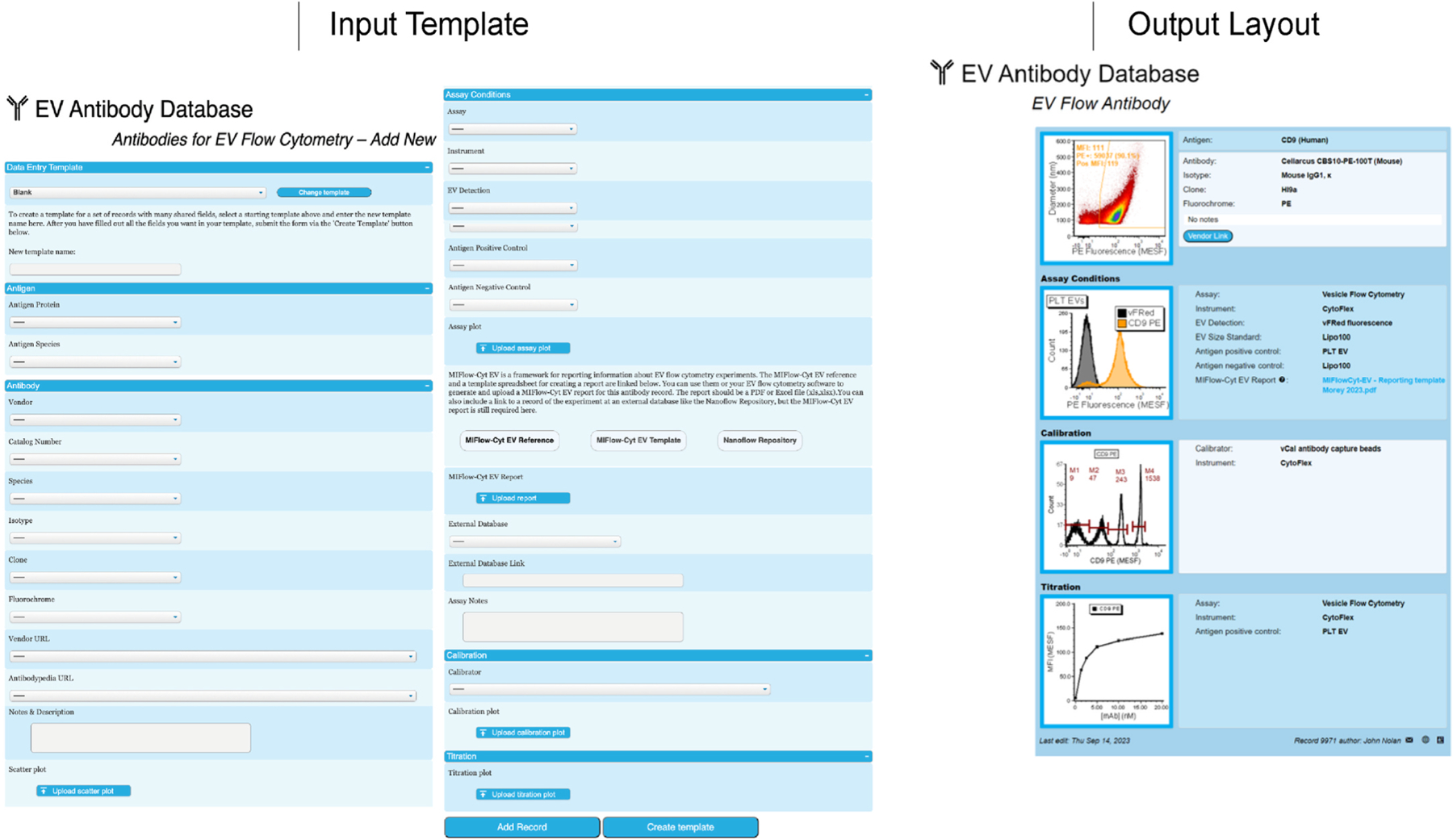
Input and Output layout of the EV Flow Cytometry module of the EV Antibody Database. The input template contains fields for comprehensive record of basic experimental information, and buttons to upload images. Data providers are also asked to upload detailed data on experimental reagents, assay conditions, calibration reagents and conditions, titration data, and instrument settings using the MIFlowCyt-EV template (Welsh et al., 2020). The output page presents the basic experimental variables and images for a given record in an easy-to-read format, as well as a button to download the MIFLowCyt-EV document.
4. Antibody characterization and validation for EV sandwich assays
Numerous commercially available and lab-derived sandwich immunoassays for detecting or quantifying EVs are available (Supplementary Table 1). EV sandwich assay formats are designed to capture intact EVs on a solid support with one or more antibodies (“capture antibodies”) followed by detection with one or more labeled antibodies (“detector antibodies”). EV sandwich assays include micro-titer plate-based single- or multiplex EV sandwich assays employing electro chemiluminescent, fluorescent, or enzyme labels, bead-based single- or multiplex EV sandwich assays with fluorescent labels, and even high-resolution microscopy-based single-EV sandwich assays. Sandwich immunoassays can provide high target specificity because they employ two specific binding interactions, through the capture and detector antibodies. In the case of EV sandwich assays, the capture and detection antibodies typically target epitopes on two distinct surface molecules on an EV; this may be two copies of the same molecule or two distinct molecules, thus allowing for many different assay configurations. Selecting suitable capture and detector antibodies for intact EV sandwich assays can be challenging and we aim to simplify this by providing a list of qualified antibodies for use as capture and/or detector and quantitative metrics of their performance in a standardized sandwich assay format.
The EV Sandwich Assay module has separate sections for capture antibodies and detector antibodies since they can be selected independently when configuring a sandwich assay. Within each section, the records, organized by target antigen, contain information on the antibody source, assay and EV-containing samples used for testing, the key performance metrics tested, as well as our assessment of whether the antibody is suitable for capture or detection. For capture antibodies, the key metric is the signal-to-background ratio, and for detector antibodies, the key metrics are the apparent dissociation constant (KD) and the signal-to-background ratio at KD.
To date, all results presented in the EV Antibody Databases for sandwich assays were produced using a commercially available ECL-based sandwich immunoassay system developed in one of our labs. This assay platform, shown schematically in Fig. 3A, was used for screening of both capture and detection antibodies targeting putative EV surface proteins. We anticipate that the results will be applicable across most sandwich immunoassay platforms. Details of how the assay system was used to compare capture and detection antibodies and the calculations involved are provided in the EV Sandwich assay module “About” tab. Fig. 3B shows the Capture Query and Output layouts for the EV Sandwich assay. The Detector Query and Output layouts for the EV Sandwich Assay module are similar in appearance and function to the Capture Query and Output, albeit with slightly different specific fields for the query and antibody performance metrics.
Fig. 3.
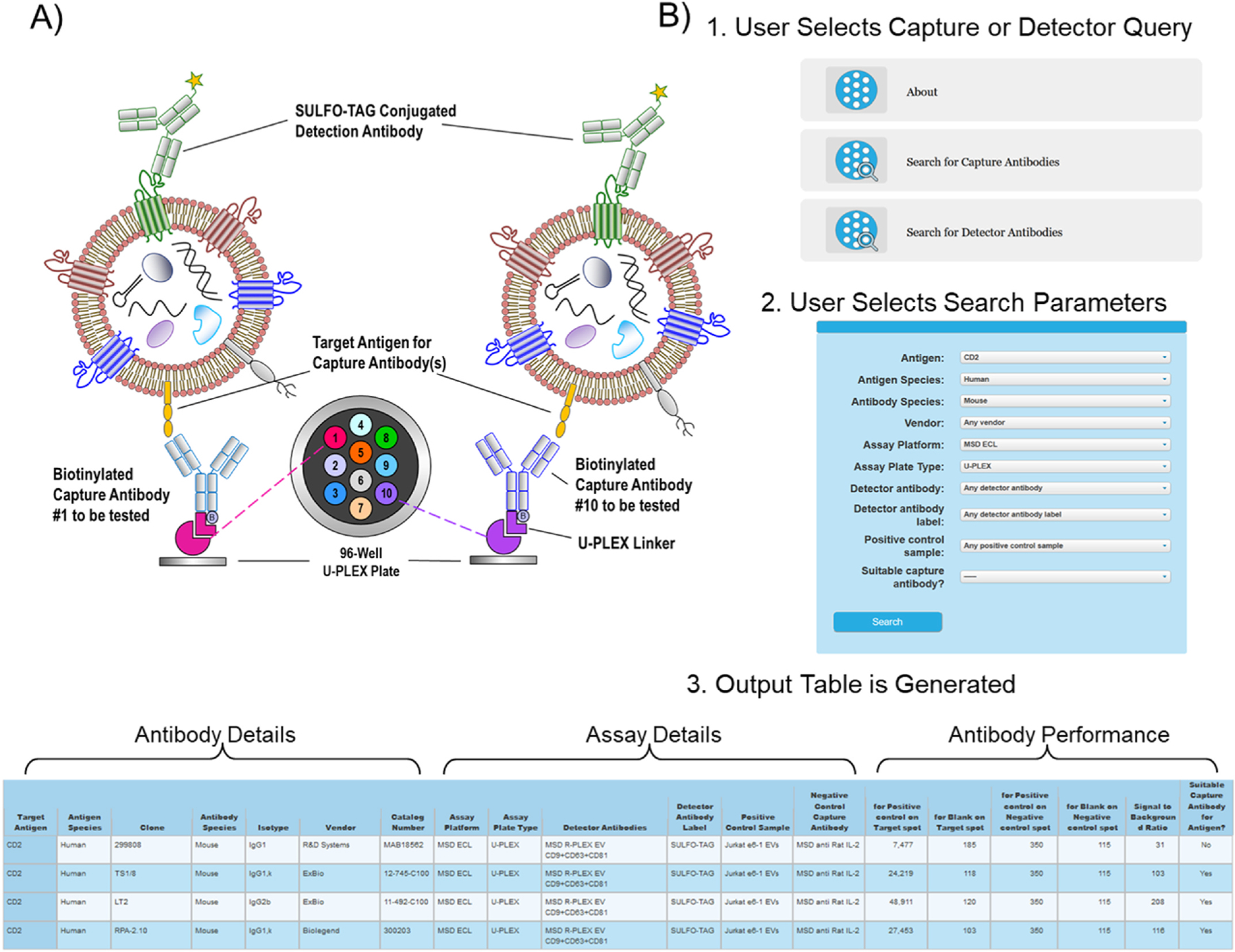
EV Sandwich Assay Module: A) Schematic of ECL assay format used to test and compare capture antibodies for intact EV sandwich assays. Assay format for comparing detectors antibodies is similar though only a single common capture antibody is used in each well and a different detection antibody is tested in each well. ©2017 Meso Scale Diagnostics, LLC. All rights reserved. B) Capture query and output layouts for the EV Sandwich Assay module. Input data are uploaded in tabular form (a template is provided). Based on the selections entered by the user on the query page, the output page presents a table that includes the antibody details, assay details and performance of capture or detector antibodies for the selected target.
In summary, the EV Antibody Database is an interactive web resource constructed to enable simple record entry and access to comprehensive validation data on curated antibodies and protocols for a variety of technologies for targets relevant to EV and ENP biology. The database currently contains data generated through projects funded by the ERCC. However, to enhance the breadth and depth of this database, groups within and outside of ERCC are encouraged to submit additional entries for consideration and integration. We will also continue to solicit records from experts in the field and incorporate contributor and user feedback in updated versions of the database. By improving the validation and comparison process for antibodies, this collective effort will not only enrich the diversity and comprehensiveness of the data, but will also ensure that the EV Antibody Database remains a useful tool for promoting rigorous high-quality, reproducible research, leading to more reliable and efficient progress in our understanding of EVs and other ENPs.
Supplementary Material
Acknowledgements
We thank the ERCC Program Staff, External Program Consultants, and Consortium investigators for their valuable input and encouragement.
Funding
This work was funded by UG3/UH3 CA241687 (AM, WC, RR, SD, PDH, JN, LCL), UG3/UH3 TR002886 (DR), U54-DA049098 (RPA), UG3/UH3 CA241685 (AC, JF), UH3 TR002878 (SD), Department of Veterans Affairs Merit grant I01BX003928 (R L R), Department of Veterans Affairs Research Career Scientist Award grant IK6BX005692 (R L R), National Institutes of Health grant UG3CA241703 (R L R), NIH R44 CA272098 and R44 GM136165 (JPN).
Footnotes
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: David Routenberg reports a relationship with Meso Scale Diagnostics LLC that includes: employment. Collin Nelson reports a relationship with Meso Scale Diagnostics LLC that includes: employment. John Nolan reports a relationship with Cellarcus Biosciences that includes: employment and equity or stocks. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.vesic.2024.100040.
References
- 1.Jeppesen DK, et al. Extracellular vesicles and nanoparticles: emerging complexities. Trends Cell Biol. 2023;33(8):667–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand S, Samuel M, Mathivanan S. Exomeres: a new member of extracellular vesicles family. Subcell Biochem. 2021;97:89–97. [DOI] [PubMed] [Google Scholar]
- 3.Bojmar L, et al. Extracellular vesicle and particle isolation from human and murine cell lines, tissues, and bodily fluids. STAR Protoc. 2021;2(1), 100225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tosar JP, Cayota A, Witwer K. Exomeres and Supermeres: monolithic or diverse? J Extracell Biol. 2022;1(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q, et al. Comprehensive isolation of extracellular vesicles and nanoparticles. Nat Protoc. 2023;18(5):1462–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Q, et al. Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nat Cell Biol. 2021;23(12):1240–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charest A Experimental and biological insights from proteomic analyses of extracellular vesicle cargos in normalcy and disease. Adv Biosyst. 2020;4(12), e2000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mateescu B, et al. Phase 2 of extracellular RNA communication consortium charts next-generation approaches for extracellular RNA research. iScience. 2022;25(8), 104653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welsh JA, et al. A compendium of single extracellular vesicle flow cytometry. J Extracell Vesicles. 2023;12(2), e12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welsh JA, et al. MIFlowCyt-EV: a framework for standardized reporting of extracellular vesicle flow cytometry experiments. J Extracell Vesicles. 2020;9(1), 1713526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y, et al. Brain tissue-derived extracellular vesicles in alzheimer’s disease display altered key protein levels including cell type-specific markers. J Alzheimers Dis. 2022;90(3):1057–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eren E, et al. Neuronal-derived EV biomarkers track cognitive decline in alzheimer’s disease. Cells. 2022;11(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saftics A, et al. Single extracellular VEsicle nanoscopy. J Extracell Vesicles. 2023;12 (7), e12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crooks ET, et al. Engineering well-expressed, V2-immunofocusing HIV-1 envelope glycoprotein membrane trimers for use in heterologous prime-boost vaccine regimens. PLoS Pathog. 2021;17(10), e1009807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh C-k, et al. S-nitrosylation of p62 inhibits autophagic flux to promote α-synuclein secretion and spread in Parkinson’s disease and lewy body dementia. J Neurosci. 2022;4(14):3011–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park DJ, et al. Serpin-loaded extracellular vesicles promote tissue repair in a mouse model of impaired wound healing. J Nanobiotechnol. 2022;20(1):474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandau US, et al. Methamphetamine use alters human plasma extracellular vesicles and their microRNA cargo: an exploratory study. J Extracell Vesicles. 2020;10(1), e12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shpigelman J, et al. Generation and application of a reporter cell line for the quantitative screen of extracellular vesicle release. Front Pharmacol. 2021;12:840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan W, et al. Cancer-cell-secreted miR-122 suppresses O-GlcNAcylation to promote skeletal muscle proteolysis. Nat Cell Biol. 2022;24(5):793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


